Abstract
Bacterial fruit blotch (BFB), which is caused by Acidovorax citrulli, is a serious threat to watermelon growers around the world. The present study was conducted to screen effective rhizobacterial isolates against 35 different A. citrulli isolates and determine their efficacy on BFB and growth parameters of watermelon. Two rhizobacterial isolates viz. Paenibacillus polymyxa (SN-22), Sinomonas atrocyanea (NSB-27) showed high inhibitory activity in the preliminary screening and were further evaluated for their effect on BFB and growth parameters of three different watermelon varieties under greenhouse conditions. The greenhouse experiment result revealed that SN-22 and NSB-27 significantly reduced BFB and had significant stimulatory effect on total chlorophyll content, plant height, total fresh weight and total dry weight compared to uninoculated plants across the tested three watermelon varieties. Analysis of the 16S ribosomal RNA (rRNA) sequences revealed that strains SN-22 belong to P. polymyxa and NSB-27 to S. atrocyanea with the bootstrap value of 99% and 98%, respectively. The isolates SN-22 and NSB-27 were tested for antagonistic and PGP traits. The result showed that the tested isolates produced siderophore, hydrolytic enzymes (protease and cellulose), chitinase, starch hydrolytic enzymes and they showed phosphate as well as zinc solubilizing capacity. This is the first report of P. polymyxa (SN-22) and S. atrocyanea (NSB-27) as biocontrol-plant growth promoting rhizobacteria on watermelon.
Keywords: Acidovorax citrulli, biocontrol, plant growth promoting rhizobacteria, rhizosphere, watermelon
Watermelon is one of the important cash crops in South Korea, the 9th largest watermelon producing country in the world (FAO, 2011). In recent years, it was found that bacterial diseases were on rise due to abnormal rainfall and high fluctuating temperature, and constitutes serious constraints in watermelon crops. Bacterial fruit blotch (BFB) caused significant loss in production of watermelon during 2011–2012 in Korea (Noh et al., 2014).
BFB, which is caused by Acidovorax citrulli, has been becoming a serious threat to farmers as it spreads through contaminated seeds and caused a significant economic losses in cucurbits worldwide (Bahar et al., 2009). The symptoms of BFB include greasy-appearing, water-soaked dark, olive-green stain, or blotch that develops on the upper surface of fruit which makes the fruit unmarketable and cause severe economic loss (Latin and Hopkins, 1995).
Recently, several studies reported the efficacy of antagonistic rhizobacteria against bacterial diseases in plant (Mesanza et al., 2016; Tolba and Soliman, 2013). Fessehaie and Walcott (2005) reported that use of Acidovorax avenae subsp. avenae Manns AAA99-2 suppressed BFB and reduced transmission by the seeds. Simmilarly, the application of Pseudomonas fluorescens Migula A506 suppressed the growth of the pathogen and reduced infestation in the seeds of fruits produced (Fessehaie and Walcott, 2005).
In this connection, there is an increase interest of developing environmentally friendly plant diseases controlling alternatives. Hence, the present study was conducted to screen effective rhizobacterial isolates against A. avenae isolates and determine the efficacy of rhizobacterial isolates on BFB and growth parameters of watermelon.
Materials and Methods
Isolation and selection of antagonistic bacteria
Twelve antagonistic bacterial isolates viz., NSB-1, NSB-4, NSB-10, NSB-12, NSB-17, NSB-18, NSB-27, NSB-36, NSB-37, NSB-40, NSB-41, and NSB-43 were isolated from soil samples collected from different locations of Nigeria in 2013 (Table 1). In addition, nine antagonistic bacterial isolates (SN-2, SN-9, SN-12, SN-15, SN-17, SN-18, SN-21, SN-22, and SN-23) from used rockwool, five (KFB09, KFB24, KFB25, KFB59, and KSB01) from greenhouse soil and four (KBM, KBM1, KBM2, and KBM3) from soil of pepper fields in Korea were isolated (Table 1). All the soil samples were taken at a depth of 15–20 cm after removing approximately 3 cm of the top soil surface. Soil samples were dried for 4 days and large roots and stones were removed and then sieved through an autoclave-sterilized brass sieve of a 2 mm aperture size. Bacterial isolates from the respective soil samples were serially diluted and identified based on their morphological and microscopic characteristics. Six vertical cores of rockwool per slab were taken with cork borer. These cores were pooled and three composite samples were formed per each five slab. Ten gram of rockwool was then thoroughly homogenized in 100 ml of sterile distilled water and then bacteria were isolated through serial dilution technique. A total of 30 antagonistic bacterial isolates were selected based on their potent antagonistic activity against eight fungal plant pathogens (Fusarium oxysporum, Fusarium solani, Colletotrichum gloesporoides, Colletotrichum coccodes, Sclerotinia minor, Rhizoctonia solani, Sclerotinia sclerotiorum, and Colletotrichum dematium) (data not shown). The selected candidates were grown on tryptic soy agar (TSA) plates and antagonistic test was performed with paper disc method. The experiment was arranged in completely randomized design (CRD) with three replications. The plates were incubated at 25°C for a week and the presence of inhibition zone was measured.
Table 1.
List of antagonistic bacteria identified
| Isolation code | Collection site | Name of bacteria | Gram reaction | GenBank accession no. |
|---|---|---|---|---|
| SN-2 | Rockwool | Pseudomonas fluorescens | − | KR010169 |
| SN-9 | Rockwool | Bacillus amyloliquefaciens | + | KR010170 |
| SN-12 | Rockwool | Bacillus cereus | + | KR010171 |
| SN-15 | Rockwool | B. amyloliquefaciens | + | KR010172 |
| SN-17 | Rockwool | Bacillus thuringiensis | + | KR010173 |
| SN-18 | Rockwool | P. fluorescens | − | KR010174 |
| SN-21 | Rockwool | P. fluorescens | − | KR010175 |
| SN-22 | Rockwool | Paenibacillus polymyxa | + | KR010176 |
| SN-23 | Rockwool | B. amyloliquefaciens | + | KR010177 |
| NSB1 | Crop field soil | B. amyloliquefaciens | + | KR010178 |
| NSB4 | Crop field soil | Bacillus subtilis | + | KR010179 |
| NSB10 | Crop field soil | Bacillus pumilus | + | KR010180 |
| NSB12 | Crop field soil | Stenotrophomonas maltophilia | − | KR010181 |
| NSB17 | Crop field soil | B. cereus | + | KR010182 |
| NSB18 | Crop field soil | Microbacterium paraoxydans | + | KR010183 |
| NSB27 | Crop field soil | Sinomonas atrocyanea | + | KR010184 |
| NSB36 | Crop field soil | Terrabacter terrae | + | KR010185 |
| NSB37 | Crop field soil | B. cereus | − | KR010186 |
| NSB40 | Crop field soil | Rhodococcus canchipurensis | + | KR010187 |
| NSB41 | Crop field soil | B. cereus | + | KR010188 |
| NSB43 | Crop field soil | Microbacterium oxydans | + | KR010189 |
| KFB09 | Greenhouse soil | P. polymyxa | + | KJ195693 |
| KFB24 | Greenhouse soil | Alcaligenes faecalis | − | KJ195694 |
| KFB25 | Greenhouse soil | B. pumilus | + | KJ195695 |
| KFB59 | Greenhouse soil | Bacillus megaterium | + | KJ195696 |
| KSB01 | Greenhouse soil | B. amyloliquefaciens | + | KJ195697 |
| KBM | Pepper field | Staphylococcus aureus | + | KM488320 |
| KBM1 | Pepper field | Achromobacter sp. | − | KM488321 |
| KBM2 | Pepper field | B. amyloliquefaciens | + | KM488321 |
| KBM3 | Pepper field | B. subtilis | + | KM488322 |
Pathogen inoculum preparation
In this study, thirty five different strains of A. citrulli were used as indicated in Table 2. All A. citrulli strains, which were collected from the BFB infected cucurbit field in 2011–2014, were obtained from National Institute of Horticultural and Herbal Science, Korea and Rural Development Administration (RDA)-GenBank. Pathogen inoculum was prepared using tryptic soy broth (TSB; tryptone 17.0 g, soytone 3.0 g, glucose 2.5 g, sodium chloride 5.0 g, di-potassium phosphate using chemical formula 2.5 g, in 1 l of water, maintaining pH 7.3 ± 0.2 at 25°C) medium.
Table 2.
Details of the Acidovorax citrulli strains used in vitro in the selection of antagonistic rhizobacteria
| Isolate no. | Host | Location of isolation | Symptom |
|---|---|---|---|
| 11-073 | Water melon | Yeongam | Leaf rot |
| 11-147 | Water melon | Gochang | Leaf spot |
| 11-162 | Water melon | Buyeo | Seedling |
| 11-163 | Water melon | Buyeo | Seedling |
| 11-164 | Water melon | Nonsan | Seedling |
| 11-165 | Water melon | Nonsan | Seedling |
| 11-171 | Cucumber | Anseong | Leaf spot |
| 11-201 | Water melon | Jinju | Leaf spot |
| 11-246 | Melon | Gokseong | Leaf spot |
| 11-247 | Melon | Gokseong | Leaf spot |
| 11-248 | Melon | Gokseong | Leaf spot |
| 11-251 | Water melon | Suwon | Leaf spot |
| 11-259 | Water melon | Hamyang | Leaf spot |
| 12-089 | Water melon | Buyeo | Fruit spot |
| 12-090 | Water melon | Buyeo | Fruit rot |
| 12-091 | Water melon | Buyeo | Stem blight |
| 12-158 | Water melon | Nonsan | Leaf spot |
| 12-170 | Cucumber | Nonsan | Leaf spot |
| 13-034 | Water melon | Anseong | Leaf spot |
| 13-211 | Water melon | Yeongam | Bacterial leaf spot |
| 13-217 | Cucumber | Buyeo | Bacterial leaf spot |
| 13-255 | Cucumber | Yeongam | Bacterial leaf rot |
| 14-044 | Cucumber | Anseong | Leaf spot |
| 14-194 | Melon | Gokseong | Cotyledon scab |
| 14-201 | Melon | Gwangju | Leaf spot |
| 14-202 | Melon | Gokseong | Leaf spot |
| KACC17000 | Water melon | Buyeo | Fruit blotch |
| KACC 17001 | Water melon | Nonsan | Fruit blotch |
| KACC 17002 | Cucumber | Anseong | Fruit blotch |
| KACC 17005 | Water melon | Suwon | Fruit blotch |
| KACC 17909 | Water melon | Andong | Fruit blotch |
| KACC 17910 | Water melon | Andong | Fruit blotch |
| KACC 17911 | Water melon | Andong | Fruit blotch |
| KACC 17912 | Water melon | Andong | Fruit blotch |
| KACC 17913 | Pumpkin seed | Busan | Fruit blotch |
Identification of antagonistic bacterial strains
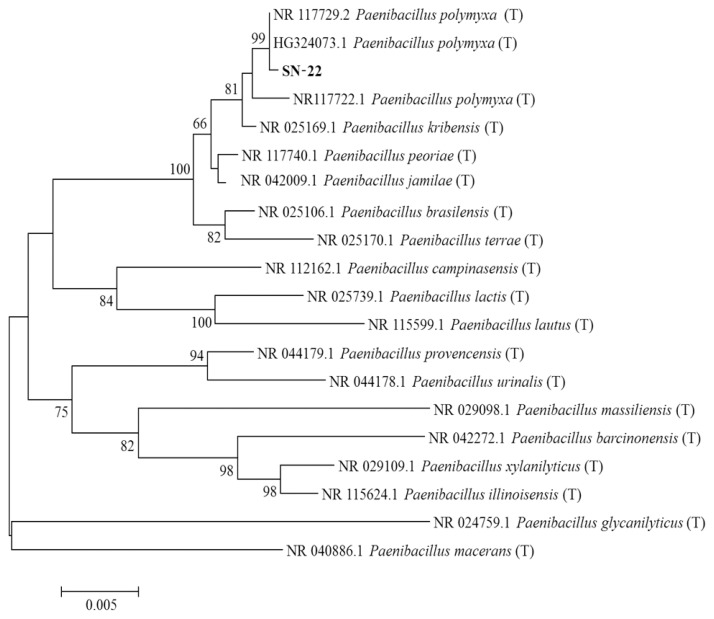
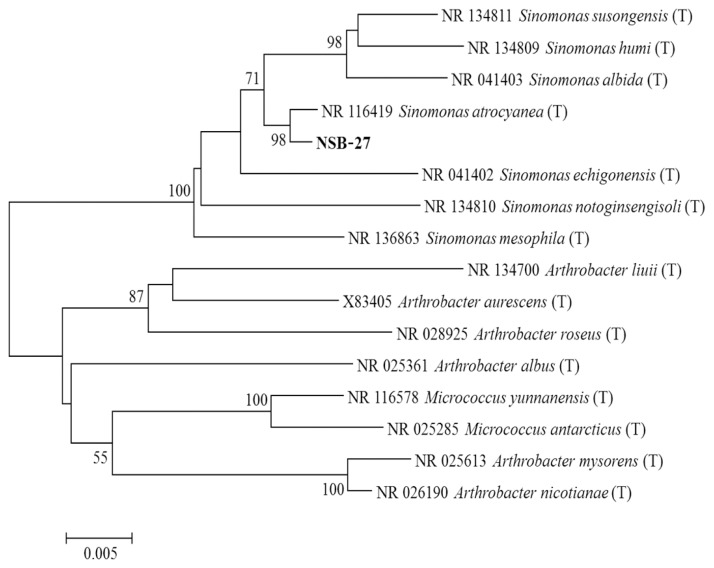
Antagonistic bacterial isolates were identified by using 16S ribosomal RNA (rRNA) sequence analysis. DNA extraction was carried out the by harvesting bacterial cells from 10 ml overnight culture. Pellets were lysed in 1 ml lysis buffer (25% sucrose, 20 mM EDTA, 50 mM Tris-HCl, and 5 mg/ml of lysozyme). The 16S rRNA was amplified using PCR with the universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-CGGTTACCTTGTTACGACTT-3′). The PCR was performed in a Thermos cycler using a 35 amplification cycles at 94°C (45 s), 55°C (60 s), and 72°C (60 s) with final extension for 7 min at 72°C. The PCR products of approximately 1,400 bp were sequenced by using universal primers 518F (5′-CCAGCAGCCGCGGTAATACG-3′) and 800R (5′-TACCAGGGTATCTAATCC-3′) and the sequencing was performed by using big dye terminator cycle sequencing kit v.3.1 (Applied BioSystems, Foster City, CA, USA). Sequencing products were resolved on Applied Biosystems model 3730XL automated DNA sequencing system (Applied BioSystems) at Macrogen Inc. (Seoul, Korea). Antagonistic bacterial isolates were identified by comparing the sequence similarities using the National Center for Biotechnology Information (NCBI) BLAST program (http://www.ncbi.nlm.nih.gov/Blast/). Phylogenetic relationships were analyzed using molecular evolutionary genetics analysis version 6.0. (MEGA6) software (Tamura et al., 2013). The neighbor-joining tree was constructed using the Kimura 2-parameter substitution model (Kimura, 1980). Bootstrap analysis was performed with 1,000 replications to determine the support for each clade (Fig. 1).
Fig. 1.
Phylogenetic relationship of 16S ribosomal RNA (rRNA) sequence between selected isolate of Paenibacillus polymyxa (SN-22) obtained from soil samples in Korea. The tree was constructed by MEGA6 program by neighbor-joining method with bootstrap values. The mark (T) indicates the type strain. Numerical values (> 500) on branches are the bootstrap values as percentage of bootstrap replication from a 1,000 replicate analysis. The bar indicates the number of substitutions per site.
Biochemical characterization of antagonistic rhizobacterial isolates
Catalase test
In this method, 48-hour-old bacterial culture on a clean tube was mixed using a sterile tooth pick and 2–3 drops of 3% hydrogen peroxide (H2O2) was added. The effervescence indicates catalase activity.
Chitinase activity
The bacterial cultures were cultured on minimal agar plates which was amended with 0.3% colloidal chitin and incubated at 30°C for 7 days. Development of halo zone around the colony after addition of iodine confirmed the chitinase positive (Roberts and Selitrennikoff, 1988).
Starch hydrolysis
Starch agar plates (peptone 5 g, beef extract 3 g, agar 15 g, distilled water 1,000 ml) were made and inoculated with pure bacterial culture and incubated at 25–29°C for 48 h. Iodine (3%) was poured into the plates after incubation. Blue black color formation indicated the positive test (Cappuccino and Sherman, 2006).
Protein hydrolysis
Skim milk agar plates (skim milk 100 g, peptone 5 g, agar 15 g, distilled water 100 ml) were prepared and inoculated with pure bacterial culture. The inoculated plated were incubated at 28°C for 48 h and plates were observed for clear zone around the wells (Cappuccino and Sherman, 2006).
Cellulose production
Cellulose producing ability of the selected antagonistic isolates was determined according to the procedures followed by (Cappuccino and Sherman, 2006). For cellulose production, 1% cellulose in basal medium (NaNO3 1 g, K2HPO4 1 g, MgSO4·7H2O 0.5 g, yeast extract 0.5 g, glucose 1 g, distilled water 100 ml, agar 15 g) were prepared. Ten milliliters of the bacterial cell suspension was inoculated into the wells and incubated for 5 days at 28°C. After incubation, grams iodine (3%) was poured in the cellulose agar media. Dark blue zone indicated the cellulose production.
Siderophore production
Chrome azurol S (CAS) agar plates were prepared and inoculated with the 10 μl of exponentially growing test bacterial culture (0.5 OD at 620 nm) and incubated for 7 days at 28°C. Yellow-orange halo zone formation confirmed the positive test for siderophore production (Schwyn and Neilands, 1987).
Phosphate solubilization
Phosphate solubilization activity of the selected rhizobacterial isolates was screened by means of Pikovskaya (PVK) agar (glucose 10 g, ammonium sulphate 1 g, potassium chloride 0.2 g, dipotassium hydrogen phosphate 0.2 g, magnesium sulphate 0.1 g, yeast extract 0.2 g, distilled water 100 ml). Clear halo zone formation indicates the phosphate solubilization (Pikovskaya, 1948). Phosphate solubilization efficiency (SE) of the isolates was calculated and determined using the following formula (Ramesh et al., 2014).
Zinc solubilization
The potential of the selected isolates for their zinc solubilization ability was performed on the modified Pikovskaya medium (Pikovskaya, 1948). The selected antagonistic bacterial isolates inoculated into modified PVK medium containing 0.1% insoluble zinc compounds (ZNO, ZnCO3, and ZnS). Zinc SE of the isolates was calculated and determined using the following formula (Ramesh et al., 2014).
Greenhouse tests
Bacterial isolates, namely Paenibacillus polymyxa (SN-22) and Sinomonas atrocyanea (NSB-27) with potent antibacterial activity in the preliminary screening were further evaluated in green house for their in vivo effect on different varieties of watermelon. The varieties used for the experiment were Coolnara, Supercool, and Fomina. Seeds were surface sterilized for 20 min in 20% sodium hypochlorite and were air dried on the sieve for 24 h at room temperature. Seeds were then sown in a sterile plastic trays. After two weeks, seedlings were transferred to a plastic pot containing commercial soil (Baroker; Seoul Bio Co., Ltd., Eumseong, Korea). Inoculation of selected bacterial isolates was carried out according to methods described by Shi et al. (2012). A week after transplanting, the pot soil was inoculated with A. citrulli suspensions at the concentration of 107 cfu g−1 soil. Five days after inoculation, the pot soil was amended with SN-22 and NSB-27 at the concentration of 107 cfu g−1 soil. Pathogen inoculated pots treated with sterile distilled water (untreated control) and Hinosan served as negative and positive control, respectively. Plants were grown twelve to fourteen days until water soaked lesions were visible. The experiment was laid out in a CRD in five replications.
Disease assessment
Disease was recorded according to the standard classification evaluation system (0–5 scale). Disease severity was recorded using the following 0 to 5 scale: 0, no incidence; 1, lesions limited to lower 1/5 of leaf area; 2, lesions present on lower 1/3 of leaf area; 3, lesions present on more than 1/3 of leaf area; 4, lesions present on more than 2/3 of leaf area; and 5, severe infections on all leaves. Disease index (DI) and the control effect (CE) were also determined using the following formulas Shi et al. (2012):
Plant growth parameters
Plant height, total fresh weight and total dry weight were examined 15 days after transplanting. Dry weight was taken after drying at 65°C for 48 h.
Statistical data analysis
The data were subjected to analysis of variance (ANOVA) using SAS software version 9.2 (SAS Institute, 2002). Means were separated using least significant difference (LSD) at (P ≤ 0.05).
Results and Discussion
A total of 30 bacterial isolates, which were collected from soil of Nigeria and Korea, were tested for antagonistic activity against A. citrulli. Among the tested 30 bacterial isolates, 15 belonged to different species of Bacillus, 2 belonged to Pseudomonas spp. and 13 belonged to other different genera (Table 1). According to staining procedure, majority of the bacterial isolates were identified as gram positive bacteria and some bacterial isolates viz. P. fluorescens, Stenotrophomonas maltophilia, Bacillus cereus, Alcaligenes faecalis, and Achromobacter sp. were identified as gram negative bacteria (Table 1).
Molecular characterization of the most promising strains viz., SN-22 and NSB-27 is indicated in Fig. 2 and Fig. 3, respectively. The sequences were compared with reference internal transcribed spacer (ITS) sequences of GenBank at NCBI using the basic local alignment search tool. The nucleotide sequences of SN-22 and NSB-27 is stored in GenBank and assigned their own GenBank accession number of KR010176 and KR010184, respectively. Analysis of the 16S rRNA sequences revealed that strains SN-22 belong to P. polymyxa and NSB-27 to S. atrocyanea with the bootstrap value of 99% and 98%, respectively (Fig. 1, 2).
Fig. 2.
Phylogenetic relationship of 16S ribosomal RNA (rRNA) sequence between selected isolate of Sinomonas atrocyanea (NSB-27) obtained from crop soil samples in Korea. The tree was constructed by MEGA6 program by neighbor-joining method with bootstrap values. The mark (T) indicates the type strain. Numerical values (> 500) on branches are the bootstrap values as percentage of bootstrap replication from a 1,000 replicate analysis. The bar indicates the number of substitutions per site.
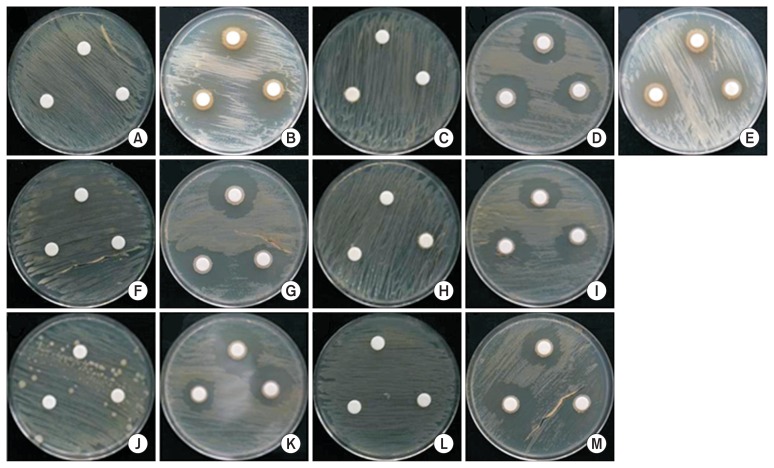
Fig. 3.
Multi paper disc culture of Acidovorax citrulli with bacterial isolates. (A) 11-201 control, (B) NBS-27; (C) 11-247 control, (D) SN-22, (E) NBS-27; (F) 11-251 control, (G) SN-22; (H) 12-090 control, (I) SN-22; (J) 13-211 control, (K) SN-22; (L) 13-217 control, (M) SN-22.
The antagonistic activity of the rhizosphere bacterial isolates against A. citrulli was assayed using dual culture method. The result revealed that out of the 30 rhizosphere isolates tested, two isolates viz., SN-22 and NSB-27 showed by far the highest antagonistic effect compared to the remaining tested isolates, which showed no or only minor inhibition activity against the different strains of the test pathogen (Table 3).
Table 3.
Effect of different antagonistic bacterial isolates on the growth of different strains of bacterial fruit blotch species
| Antagonistic bacterial isolates | Isolate no. | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||||
| 11-073 | 11-147 | 11-162 | 11-163 | 11-164 | 11-165 | 11-171 | 11-201 | 11-246 | 11-247 | 11-248 | 11-251 | 11-259 | 12-089 | 12-090 | 12-091 | 12-158 | 12-170 | |
| SN-2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| SN-9 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ++ | + | + | + |
| SN-12 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| SN-15 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| SN-17 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| SN-18 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| SN-21 | + | + | + | + | + | + | + | + | ++ | ++ | ++ | +++ | + | ++ | ++ | + | ++ | ++ |
| SN-22 | +++ | + | + | +++ | + | + | +++ | +++ | +++ | +++++ | +++ | +++++ | +++ | ++ | ++++ | + | + | + |
| SN-23 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-1 | ++ | + | + | ++ | + | + | ++ | + | + | ++ | + | + | + | + | + | + | + | + |
| NSB-4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-10 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-12 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-17 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-18 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-27 | ++ | + | +++ | +++ | + | + | +++ | ++++ | ++ | ++++ | + | +++ | + | ++ | +++ | + | ++ | ++ |
| NSB-36 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-37 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-40 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-41 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-43 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KFB09 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KFB24 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KFB25 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KFB59 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KSB01 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KBM | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KBM1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KBM2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KBM3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Antagonistic bacterial isolates | Isolate no. | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||
| 13-034 | 13-211 | 13-217 | 13-255 | 14-044 | 14-194 | 14-201 | 14-202 | 17000 | 17001 | 17002 | 17005 | 17909 | 17910 | 17911 | 17912 | 17913 | |
| SN-2 | + | + | + | + | + | + | + | + | ++ | + | ++ | + | + | + | ++ | + | + |
| SN-9 | + | ++ | + | + | + | + | + | + | +++ | + | ++ | + | + | + | + | + | + |
| SN-12 | + | + | + | + | + | + | + | + | ++ | + | ++ | + | + | + | + | + | + |
| SN-15 | + | + | + | + | + | + | + | + | ++ | + | + | + | + | + | + | + | + |
| SN-17 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| SN-18 | + | + | + | + | + | + | + | + | ++ | + | + | + | + | + | + | + | + |
| SN-21 | + | +++ | ++ | + | ++ | ++ | + | + | +++ | +++ | +++ | +++ | + | + | ++ | + | + |
| SN-22 | ++ | +++++ | +++++ | ++ | ++ | + | + | + | +++ | +++ | +++ | +++ | + | + | +++ | + | + |
| SN-23 | + | + | + | + | + | + | + | + | + | ++ | + | + | + | + | ++ | + | + |
| NSB-1 | + | ++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-10 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-12 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-17 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-18 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-27 | ++ | +++ | ++ | ++ | ++ | ++ | + | + | + | + | + | + | + | + | ++ | +++ | + |
| NSB-36 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-37 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-40 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-41 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NSB-43 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KFB09 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KFB24 | ++ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KFB25 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KFB59 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KSB01 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KBM | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KBM1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KBM2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| KBM3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
Zone of inhibition in dual culture assay: +, 0–2 mm; ++, 2–5 mm; +++, 5–7 mm; ++++, 7–9; +++++, > 9 mm.
The present study has demonstrated the antagonistic potential of rhizosphere bacterial isolates that corresponded with previous research works (Dawwam et al., 2013; Mu’minah et al., 2015) who reported that showed the in vitro suppresion of plant pathogenic bacterial by rhizosphere bacterial isolates.
In addition, in this study it has been recorded that some of the rhizobacterial isolates showed little inhibitory activity to some of the pathogen isolates and no activity to the remaining pathogen isolates (Table 3). This is in agreement the previous finding (Ryan et al., 2004) that the success of biocontrol varies among pathogen isolates.
This suggests that the mode of action exerted and/or the type of antibacterial produced by the bacterial isolates may vary, and that the A. citrulli isolates are taxonomically different from each other. It has been reported that bacterial diseases can be controlled using antagonistic microbes (Hammami et al., 2012; Ryan et al., 2004).
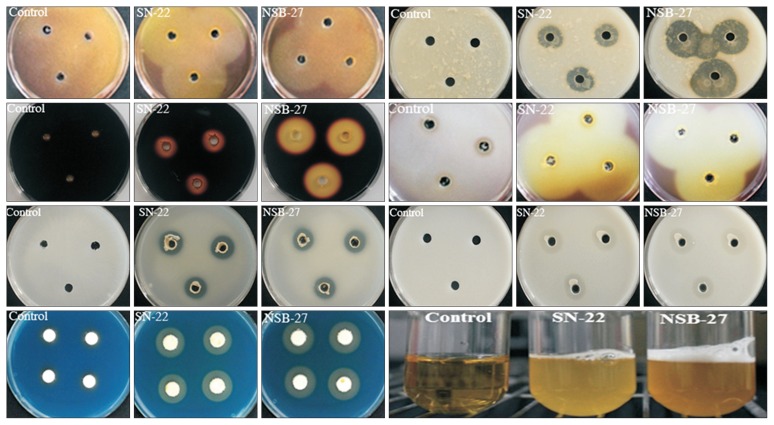
Two isolates, namely P. polymyxa (SN-22), S. atrocyanea (NSB-27), which showed high antagonistic potential in the preliminary screening, were assessed for their biochemical characteristics. The biochemical characterization result showed that those selected isolates were positive for starch and protein hydrolysis and were able to produce cellulose and siderophore. The isolates were catalase and chitinase positive. The isolates have shown positive result for phosphate and zinc solubilisation (Table 4, Fig. 4). The present result is in agreement with previous reports (He et al., 2007; Raza and Shen, 2010) that P. polymyxa is positive for catalase, siderophore production, starch hydrolysis and phosphate solubilisation. The antagonistic efficacy of the tested isolates may be due to due to the production of antimicrobial substances such as chitinolytic enzymes, cellulase and siderophore and this result is in agreement with the previous findings (Chung et al., 2008; Singh et al., 2008).
Table 4.
Production of various antibacterial compounds by selected rhizobacterial isolates against Acidovorax citrulli
| Biochemical characterization | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Catalase test | Cellulose test | Chitinase test | Starch hydrolysis | Protein hydrolysis | Siderophore production (mm)* | Phosphate solubilization (mm)* | Zinc solubilization (mm)* | |
| SN-22 | ++ | +++ | ++ | ++ | ++ | 0.8 | 0.9 | 1.1 |
| NSB-27 | +++ | +++ | +++ | +++ | +++ | 0.9 | 0.9 | 1.2 |
+++, high; ++, medium; +, low.
mm, clear halo zone production in millimeter.
Fig. 4.
Biochemical characterization activity of selected rhizobacterial isolates from left to right, chitinase production, protein hydrolysis, starch hydrolysis, cellulose production, phosphate solubilization, zinc solubilization, siderophore production, and catalase test, respectively.
The two potential antagonistic rhizobacterial isolates viz., SN-22 and NSB-27 from in vitro antagonistic test were further evaluated for their BFB suppression activity under greenhouse condition. Green house result showed that the tested rhizobacterial isolates were significantly (P ≤ 0.05) more effective in controlling BFB severity than negative and positive controls on the tested three watermelon varieties (Table 5). Among the treatments used, NSB-27 showed the lowest BFB DI with no statistical difference with SN-22 (Table 5). The highest DI across all varieties was recorded from untreated control (Fig. 5). Reports by Jiang et al. (2015) also showed that bacteria strains have the potential to control BFB of watermelon. However, the effect of the present study isolates, P. polymyxa and S. atrocyanea, against BFB of watermelon have not yet been reported. Paenibacillus originally belonged to Bacillus. In 1994, Ash et al. (1994) established Paenibacillus and appointed P. polymyxa as the model species for this genus. There have been many studies on the antagonistic potentiality of P. polymyxa (Beatty and Jensen, 2002; Karpunina et al., 2003; Tong et al., 2004). In connection with this, P. polymyxa G-14 has been reported as a broad spectrum antagonistic bacterial isolate against the bacterial disease of muskmelon (Shi et al., 2012). The other potential antagonistic isolate NSB27 (S. atrocyanea) is a member of the family Micrococcaceae in the phylum Actonobacteria was first proposed by Zhou et al. (2012). Raj et al. (2014) reported that S. atrocyanea induced systemic resistance in plants against a broad-spectrum of root and foliar pathogens.
Table 5.
Effect of antagonistic strains on bacterial fruit blotch and growth parameters of watermelon seedlings
| Varieties | Treatment | DI | CE | Chl | PHT (cm) | FWT (gm) | DWT (gm) |
|---|---|---|---|---|---|---|---|
| Supercool | NSB-27 | 44.34b | 45.33 | 30.70a | 60.16a | 22.69b | 3.38b |
| SN-22 | 42.94c | 47.05 | 30.73a | 63.61a | 25.43a | 4.14a | |
| Hinosan | 55.32b | 31.78 | 25.00b | 52.92b | 12.99c | 2.38c | |
| Untreated control | 81.10a | - | 19.54c | 45.42c | 10.23d | 1.20d | |
| CV (%) | 4.07 | 5.32 | 6.32 | 4.82 | 5.13 | ||
| LSD | 2.12 | 3.11 | 3.93 | 1.31 | 0.61 | ||
| Coolnara | NSB-27 | 47.61 | 41.75 | 30.26a | 60.74a | 22.61d | 3.76a |
| SN-22 | 43.01c | 47.08 | 30.75a | 62.79a | 25.02c | 3.86a | |
| Hinosan | 55.20b | 32.10 | 24.61b | 52.31b | 13.12c | 2.48b | |
| Untreated control | 81.30a | - | 19.35c | 42.68c | 10.25b | 1.76c | |
| CV (%) | 2.67 | 3.56 | 4.31 | 6.12 | 7.32 | ||
| LSD | 2.11 | 2.59 | 10.15 | 1.98 | 0.53 | ||
| Fomina | NSB-27 | 47.08c | 43.89 | 34.79a | 56.08a | 21.54a | 3.56a |
| SN-22 | 49.70c | 40.76 | 29.14b | 60.36a | 23.03a | 3.64a | |
| Hinosan | 61.39b | 26.83 | 22.60c | 48.59b | 11.92b | 2.52b | |
| Untreated control | 83.90a | - | 17.05d | 43.74c | 8.60c | 1.52c | |
| CV (%) | 5.86 | 6.53 | 6.53 | 9.68 | 11.25 | ||
| LSD | 4.75 | 2.23 | 4.78 | 2.11 | 0.43 |
Growth parameters assessed 15 days after transplanting: DI, disease index; CE, control effect; Chl, chlorophyll content (squad units); PHT, plant height; FWT, fresh weight; DWT, dry weight.
CV, coefficient of variation; LSD, least significant difference.
Means followed by the same letter(s) are not significantly different (P ≤ 0.05).
Fig. 5.
Disease incidence on Supercool variety of Supercool (A–D), Coolnara (E–H), and Fomina (I–L) varieties of watermelon after treatment. Disease was recorded according to the standard classification evaluation system (0–5 scale). 0, no incidence; 1, lesions limited to lower 1/5 of leaf area; 2, lesions present on lower 1/3 of leaf area; 3, lesions present on more than 1/3 of leaf area; 4, lesions present on more than 2/3 of leaf area; and 5, severe infections on all leaves.
The effect of plant growth promoting rhizobacteria (PGPR) strains viz., NSB-27 and SN-22 on the growth of three watermelon varieties was determined under greenhouse conditions (Fig. 6). The result showed that the highest chlorophyll content, plant height, total fresh weight and total dry weight on all watermelon varieties were obtained from application of the tested rhizobacteria strains (Table 5). In agreement with the present result, Ryu et al. (2005) reported that P. polymyxa have been used to enhance the growth of various crops. Similarly, Kokalis-Burelle et al. (2003) discussed the positive effects of PGPR treatments on shoot weight, shoot length and stem diameter of muskmelon and watermelon transplant. S. atrocyanea has also been reported as a promising plant growth promoting bacteria (Raj et al., 2014). The authors stated that S. atrocyanea showed various plant growth promoting mechanisms like solubilization of inorganic phosphates, increased iron nutrition through iron chelating siderophores.
Fig. 6.
Bacterial fruit blotch disease suppression and growth promotion in Supercool (A), Coolnara (B), and Fomina (C) varieties of watermelon by antagonistic bacterial isolates. Seeds were surface sterilized for 20 min in 20% sodium hypochlorite solution and were air dried on the sieve for 24 h at room temperature. Seeds were then sown in a sterile plastic trays. Plants were grown twelve to fourteen days until water soaked lesions were visible. The experiment was laid out in a completely randomized design in five replications.
In recent years, it was found that bacterial diseases were on rise due to abnormal rainfall and high fluctuating temperature, and constitutes serious constraints in watermelon production (Noh et al., 2014). The degree of infection varied with the cultivars, and use of resistant cultivars was an effective means of preventing the occurrence of disease (Hopkins et al., 1993). Use of biological control agents could be a feasible option for managing the disease (Fessehaie and Walcott, 2005). In connection with this, rhizobacteria are used not only for controlling plant diseases but they are also useful for sustainable agricultural development through by protecting the environment (Sivasakhti et al., 2014). Ahemad and Kibret (2014) discussed that plant-beneficial rhizobacteria may decrease the global dependence on hazardous agricultural chemicals which destabilize the agro-ecosystems.
In conclusion, the present study manifested that among tested 30 rhizosphere bacterial isolates, two bacterial isolates, namely P. polymyxa (SN-22) from rockwool in Korea and S. atrocyanea (NSB-27) from Nigerian soil reduced BFB of watermelon. In addition, SN-22 and NSB-27 showed high plant growth promoting activities. The isolates SN-22 and NSB-27 had positive result for catalase test, production, chitinase activity, starch hydrolysis, protein hydrolysis, cellulose production, siderophore production, ammonia production, phosphate solubilization, zinc solubilization. Further studies are warranted on ameliorating the efficiency of SN-22 and NSB-27 condition though various formulations and application techniques under different environmental condition. This is the first report of P. polymyxa and S. atrocyanea as biocontrol-PGPR on watermelon.
Acknowledgments
Authors are thankful to GSP (Golden Seed project) Vegetable Seed Center (Project No. 213002042CGS00), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA), and Korea Forest Service (KFS) for their financial support.
References
- Ahemad M, Kibret M. Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J King Saud Univ Sci. 2014;26:1–20. doi: 10.1016/j.jksus.2013.05.001. [DOI] [Google Scholar]
- Ash C, Priest FG, Collins MD. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek. 1994;64:253–260. doi: 10.1007/BF00873085. [DOI] [PubMed] [Google Scholar]
- Bahar O, Kritzman G, Burdman S. Bacterial fruit blotch of melon: screens for disease tolerance and role of seed transmission in pathogenicity. Eur J Plant Pathol. 2009;123:71–83. doi: 10.1007/s10658-008-9345-7. [DOI] [Google Scholar]
- Beatty PH, Jensen SE. Paenibacillus polymyxa produces fusaricidin-type antifungal antibiotics active against Leptosphaeria maculans, the causative agent of blackleg disease of canola. Can J Microbiol. 2002;48:159–169. doi: 10.1139/w02-002. [DOI] [PubMed] [Google Scholar]
- Cappuccino JG, Sherman N. Microbiology: a laboratory manual. 6th ed. Pearson Education; San Francisco, CA, USA: 2006. [Google Scholar]
- Chung S, Kong H, Buyer JS, Lakshman DK, Lydon J, Kim SD, Roberts DP. Isolation and partial characterization of Bacillus subtilis ME488 for suppression of soilborne pathogens of cucumber and pepper. Appl Microbiol Biotechnol. 2008;80:115–123. doi: 10.1007/s00253-008-1520-4. [DOI] [PubMed] [Google Scholar]
- Dawwam GE, Elbeltagy A, Emara HM, Abbas IH, Hassan MM. Beneficial effect of plant growth promoting bacteria isolated from the roots of potato plant. Ann Agric Sci. 2013;58:195–201. [Google Scholar]
- [FAO] Food and Agriculture Organization. FAOSTAT; 2011 onwards. [24 August 2016]. URL http://www.fao.org/faostat/ [Google Scholar]
- Fessehaie A, Walcott RR. Biological control to protect watermelon blossoms and seed from infection by Acidovorax avenae subsp. citrulli. Phytopathology. 2005;95:413–419. doi: 10.1094/PHYTO-95-0413. [DOI] [PubMed] [Google Scholar]
- Hammami I, Jaouadi B, Bacha AB, Rebai A, Bejar S, Nesme X, Rhouma A. Bacillus subtilis bacteriocin Bac 14B with a broad inhibitory spectrum: purification, amino acid sequence analysis, and physicochemical characterization. Biotechnol Bioprocess Eng. 2012;17:41–49. doi: 10.1007/s12257-010-0401-8. [DOI] [Google Scholar]
- He Z, Kisla D, Zhang L, Yuan C, Green-Church KB, Yousef AE. Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. Appl Environ Microbiol. 2007;73:168–178. doi: 10.1128/AEM.02023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins DL, Thompson CM, Elmstrom GW. Resistance of watermelon seedlings and fruit blotch bacterium. HortScience. 1993;28:122–123. [Google Scholar]
- Jiang CH, Wu F, Yu ZY, Xie P, Ke HJ, Li HW, Yu YY, Guo JH. Study on screening and antagonistic mechanisms of Bacillus amyloliquefaciens 54 against bacterial fruit blotch (BFB) caused by Acidovorax avenae subsp. citrulli. Microbiol Res. 2015;170:95–104. doi: 10.1016/j.micres.2014.08.009. [DOI] [PubMed] [Google Scholar]
- Karpunina LV, Mel’nikova UY, Konnova SA. Biological role of lectins from the nitrogen-fixing Paenibacillus polymyxa strain 1460 during bacterial-plant-root interactions. Curr Microbiol. 2003;47:376–378. doi: 10.1007/s00284-002-3987-z. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kokalis-Burelle N, Vavrina CS, Reddy MS, Kloepper JW. Amendment of muskmelon and watermelon transplant media with plant growth promoting rhizobacteria: effects on transplant quality, disease, and nematode resistance. HortTechnology. 2003;13:476–482. [Google Scholar]
- Latin RX, Hopkins DL. Bacterial fruit blotch of watermelon: the hypothetical exam question becomes reality. Plant Dis. 1995;79:761–765. doi: 10.1094/PD-79-0761. [DOI] [Google Scholar]
- Mesanza N, Iturritxa E, Patten CL. Native rhizobacteria as biocontrol agents of Heterobasidion annosum s.s. and Armillaria mellea infection of Pinus radiata. Biol Control. 2016;101:8–16. doi: 10.1016/j.biocontrol.2016.06.003. [DOI] [Google Scholar]
- Mu’minah Baharuddin Subair H, Fahruddin Isolation and screening bacterial exopolysaccharide (EPS) from potato rhizosphere in highland and the potential as a producer Indole Acetic Acid (IAA) Procedia Food Sci. 2015;3:74–81. doi: 10.1016/j.profoo.2015.01.007. [DOI] [Google Scholar]
- Noh J, Kim JH, Lim JH, Kim TB, Seong MH, Jung GT, Kim JM, Cheong SS, Oh NK, Lee WH. Occurrence of diseases and case of clinical diagnosis on watermelon in South Korea, 2008–2012. Res Plant Dis. 2014;20:8–14. doi: 10.5423/RPD.2014.20.1.008. [DOI] [Google Scholar]
- Pikovskaya RI. Mobilization of phosphorus in soil in connection with vital activity of some microbes species. Microbiology. 1948;17:362–370. [Google Scholar]
- Raj DPRS, Linda R, Babyson RS. Molecular characterization of phosphate solubilizing bacteria (PSB) and plant growth promoting rhizobacteria (PGPR) from pristine soil. Int J Innov Sci Eng Technol. 2014;1:317–324. [Google Scholar]
- Ramesh A, Sharma SK, Sharma MP, Yadav N, Joshi OP. Inoculation of zinc solubilizing Bacillus aryabhattai strains for improved growth, mobilization and biofortification of zinc in soybean and wheat cultivated in Vertisils of central India. Appl Soil Ecol. 2014;73:87–96. doi: 10.1016/j.apsoil.2013.08.009. [DOI] [Google Scholar]
- Raza W, Shen Q. Growth, Fe3+ reductase activity, and siderophore production by Paenibacillus polymyxa SQR-21 under differential iron conditions. Curr Microbiol. 2010;61:390–395. doi: 10.1007/s00284-010-9624-3. [DOI] [PubMed] [Google Scholar]
- Roberts WK, Selitrennikoff CP. Plant and bacterial chitinases differ in antifungal activity. J Gen Microbiol. 1988;134:169–176. [Google Scholar]
- Ryan AD, Kinkel LL, Schottel JL. Effect of pathogen isolate, potato cultivar, and antagonist strain on potato scab severity and biological control. Biocontrol Sci Technol. 2004;14:301–311. doi: 10.1080/09583150410001665187. [DOI] [Google Scholar]
- Ryu CM, Kim JW, Choi O, Park SY, Park SH, Park CS. Nature of a root-associated Paenibacillus polymyxa from field grown winter barley in Korea. J Microbiol Biotechnol. 2005;15:984–991. [Google Scholar]
- SAS Institute. SAS/STAT® 9.2 user’s guide for personal computers. Version 9.2 edition. SAS Institute; Cary, NC, USA: 2002. [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Shi Y, Yang L, Wang X, Gao Y, Liu W, Lou K. Biocontrol of bacterial spot diseases of muskmelon using Paenibacillus polymyxa G-14. Afr J Biotechnol. 2012;11:16845–16851. [Google Scholar]
- Singh N, Pandey P, Dubey RC, Maheshwari DK. Biological control of root rot fungus Macrophomina phaseolina and growth enhancement of Pinus roxburghii (Sarg.) by rhizosphere competent Bacillus subtilis BN1. World J Microbiol Biotechnol. 2008;24:1669–1679. doi: 10.1007/s11274-008-9680-z. [DOI] [Google Scholar]
- Sivasakhti S, Usharani G, Saranraj P. Biocontrol potentiality of plant growth promoting bacteria (PGPR)--Pseudomonas fluorescence and Bacillus subtilis: a review. Afr J Agric Res. 2014;9:1265–1277. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolba IH, Soliman MA. Efficacy of native antagonistic bacterial isolates in biological control of crown gall disease in Egypt. Ann Agric Sci. 2013;58:43–49. [Google Scholar]
- Tong Y, Guo G, Xu J, Ji Z, Chen X. Induced resistance to gray mold in tomato plant by antagonistic bacteria. Chin J Biol Control. 2004;20:187–189. [Google Scholar]
- Zhou Y, Chen X, Zhang Y, Wang W, Xu J. Description of Sinomonas soli sp. nov., reclassification of Arthrobacter echigonensis and Arthrobacter albidus (Ding et al. 2009) as Sinomonas echigonensis comb. nov. and Sinomonas albida comb. nov., respectively, and emended description of the genus Sinomonas. Int J Syst Evol Microbiol. 2012;62:764–769. doi: 10.1099/ijs.0.030361-0. [DOI] [PubMed] [Google Scholar]