Abstract
Contributions of glial cells to neuroenergetics have been the focus of extensive debate. Here we provide the first positron emission tomography (PET) evidence that activation of the astrocytic glutamate transport via GLT-1 triggers widespread but graded glucose uptake in the rodent brain. Our results highlight the need for a reevaluation of the interpretation of [18F]FDG PET data, whereby astrocytes would be recognized to contribute significantly to the [18F]FDG signal.
Keywords: [18F]FDG, ceftriaxone, GLT-1, glutamate, glucose, PET
Glucose constitutes the major source of energy in the brain with its utilization correlating with neuronal activity1. This key concept provides the basis for the interpretation of activity-dependent accumulation of the radiofluorinated glucose analog 2-deoxy-2-[18F]fluoro-D-glucose ([18F]FDG)2 visualized using positron emission tomography (PET). For over 30 years, brain [18F]FDG PET uptake has been viewed as a proxy of neuronal activity3. Despite widespread utilization in both clinical settings and basic research, the identity of the cell type(s) contributing to the [18F]FDG PET signal, as well as the mechanisms regulating its variations, remain highly controversial4, 5.
Over the past decades, astrocytes have been implicated in several dynamic mechanisms involving synaptic transmission and plasticity, including a crucial role in terms of glucose consumption6. Indeed, strong evidence supports the concept that glutamate recycling in astrocytes activates aerobic glycolysis, with neurons being partially fueled by lactate derived from astrocytes7, 8. More specifically, astroglial glutamate transport – via GLT-1 or GLAST – has been shown to act as a trigger, signaling for glucose uptake by astrocytes9. In fact, it has been shown that GLT-1 or GLAST knockout mice have reduced [14C]-2-deoxyglucose uptake in the somatosensory cortex after whisker stimulation10, 11. Based on these observations, it seems likely that [18F]FDG PET signal may largely reflect glucose consumption in astrocytes rather than in neurons12, although no PET evidence exists so far to support this claim. To further test this hypothesis, we conducted a microPET study using [18F]FDG to assess whether ceftriaxone (CEF) – a known stimulator of astrocytic glutamate transport via GLT-113, 14 – was capable of modulating cerebral [18F]FDG consumption in awake adult rats (Fig. 1a).
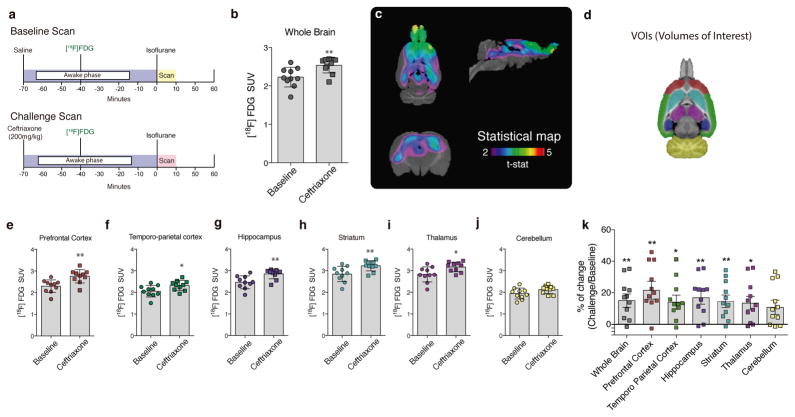
Figure 1. Astrocytic glutamate transport activation via GLT-1 triggers cerebral [18F]FDG uptake.
(a) Study design: rats received a tail vein injection of saline or CEF (200 mg/kg) 70 minutes before the scan. Thirty minutes later they received an [18F]FDG tail vein injection and were allowed to freely move in their cages (awake) during a 40 minutes uptake phase. After that period, a static scan of 10 minutes was acquired. (b) Whole brain [18F]FDG uptake. (c) Statistical parametric images (t-statistical map (CEF challenge > baseline) overlaid on a histological template. (d) Brain mask showing volume of interest (VOIs) overlaid on a histological template. Regional [18F]FDG uptake in the prefrontal (e) and temporo-parietal cortices (f), hippocampus (g), striatum (h), thalamus (i) and cerebellum (j). Regional percentage of change in VOIs (challenge > baseline, k). n = 10 rats per group. *p < 0.05, and **p < 0.01. Two tailed paired t-test. Data are presented as mean values ± s.d. and individual scatter plots. Note that each region has a correspondent scatter plot color, which follows regions defined in the VOIs mask; prefrontal cortex (red), temporo-parietal cortex (green), hippocampus (dark-purple), striatum (light-green), thalamus (light-purple) and cerebellum (yellow).
Using cultured adult cortical astrocytes, we demonstrate that acute CEF exposure enhances glutamate and glucose uptake, which is in accordance with the previously described mechanism of glutamate-induced aerobic glycolysis10, without altering GLT-1 expression. Additionally, we demonstrated that the blockade of GLT-1 transport by dihydrokainic acid (DHK) abolished CEF’s effect on glucose uptake (Supplementary Fig. 1). However, the precise molecular mechanism by which CEF acutely exerts its effect on glutamate transport via GLT-1 in astrocytes remains to be determined.
Our in vivo data demonstrate that CEF induces a global increase in [18F]FDG standard uptake value (SUV) (t(9) = 3.309, p = 0.0091, Fig. 1b) without affecting spontaneous locomotion in the open field test (Supplementary Fig. 2). A voxel-wise t-statistical analysis showed a marked increase in several brain regions encompassing cortical and subcortical areas, with a peak effect in the prefrontal cortex (peak t(9) = 5.24, p = 0.0005, Fig. 1c). Analysis of regional mRNA expression data from the Allen Brain Atlas (http://mouse.brain-map.org/gene/show/20273) suggested that regions exhibiting high GLT-1 expression were more susceptible to the CEF challenge, showing more pronounced increases in [18F]FDG uptake in our study15 (Supplementary Fig.3 and 4). To support this claim, we delineated six large volumes of interest (VOIs; Fig. 1d) to estimate regional [18F]FDG uptake. Regional VOI analysis showed increased [18F]FDG SUV in the frontal cortex (t(9) = 3.349, p = 0.0085), temporo-parietal cortex (t(9) = 2.874, p = 0.0184), hippocampus (t(9) = 3.918, p = 0.0035), striatum (t(9) = 3.420, p = 0.0076), and thalamus (t(9) = 2.975, p = 0.0156), regions known for high GLT-1 expression. In contrast, CEF did not significantly alter [18F]FDG uptake in the cerebellum (t(9) = 2.083, p = 0.070), a region of low GLT-1 expression15 (Fig. 1e–j). For percentage of change, see (Fig. 1k).
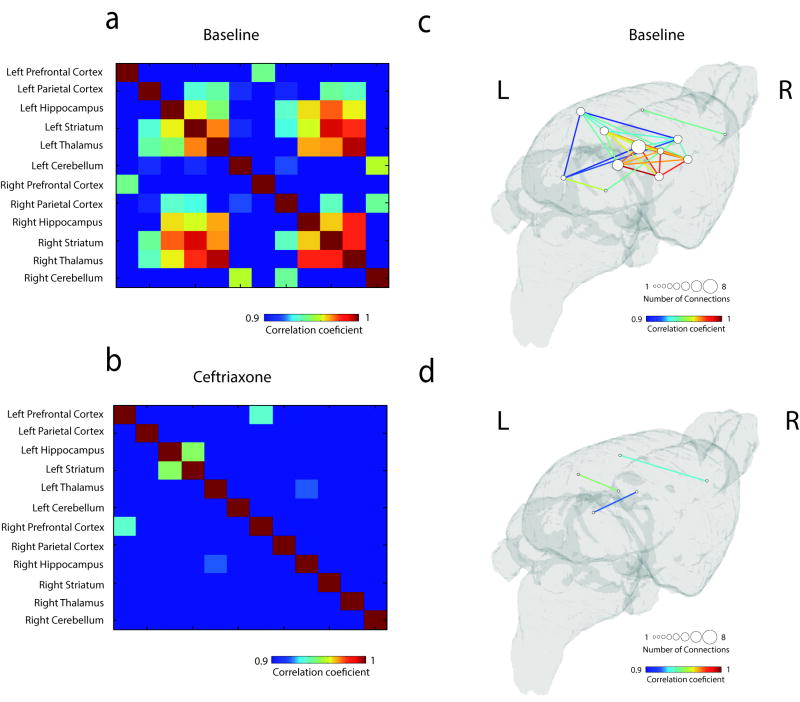
Currently, it is well established that cerebral [18F]FDG uptake is proportional to brain energy consumption and, consequently, synaptic activity16, 17. This coupling between brain activity and metabolism allows derivation of metabolic networks through inter-subject [18F]FDG analysis and consequent identification of patterns of brain metabolism18. In fact, through metabolic interregional analysis across the previously delineated VOIs, we found that acute activation of astrocytic glutamate transport via GLT-1 altered a wide range of connections within the metabolic network. Our findings may indicate that astrocytic activation per se is capable of reshaping brain metabolic architecture (p < 0.05, Bonferroni corrected, Fig. 2a–d). However, we cannot rule out a regionally dependent CEF effect due to the heterogeneous expression of cerebral GLT-1. For false discovery rate (FDR)-corrected results, see (Supplementary Fig. 5).
Figure 2. Astrocytic glutamate transport activation via GLT-1 disrupts region-toregion metabolic synchronicity.
Cross-correlation matrices: inter-subject cross-correlation maps displaying region-to-region associations in the baseline (a) and CEF challenge (b) conditions. Metabolic networks: 3D brain surfaces displaying large-scale metabolic cross-correlation maps in the baseline (c) and in the CEF challenge (d) conditions. n = 10 rats per group. Data presented as correlation values with Bonferonni corrected threshold, p < 0.05.
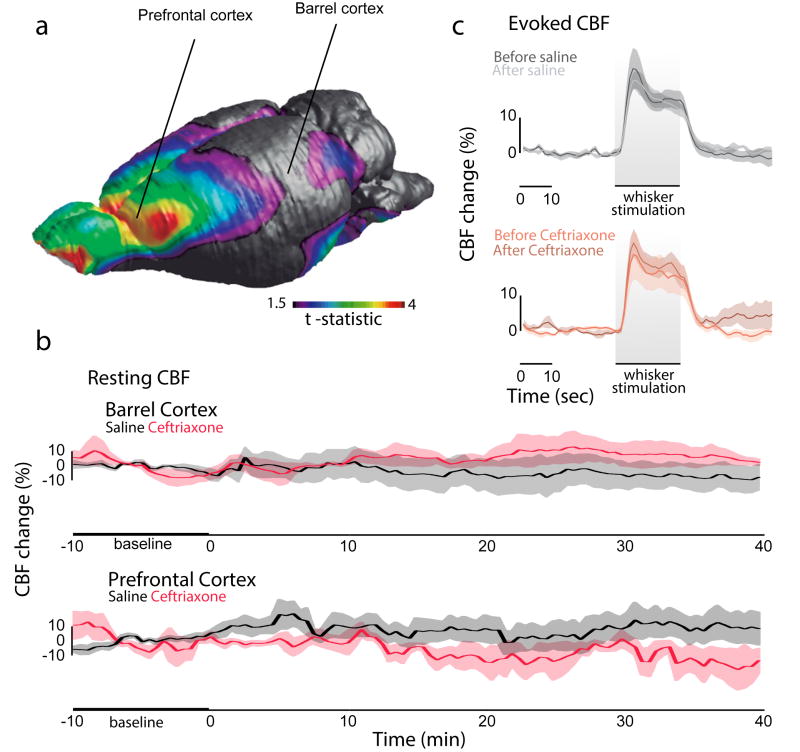
Based on the coupling between neuronal activity and cerebral blood flow (CBF)19, we then decided to investigate the coupling between [18F]FDG uptake and CBF response in the barrel and prefrontal cortices (Fig. 3a). The CEF challenge did not alter resting CBF in the barrel (F(1.888, 5.665) = 1.648, p = 0.2714; Fig. 3b) and prefrontal cortices (F(1.645, 6.579) = 0.3977, p = 0.6495, Fig. 3b). The evoked CBF after whisker stimulation did not differ between groups (interaction, F(237, 1264) = 0.4291, p > 0.999; whiskers stimulation effect: F(79, 1264) = 60.10, p < 0.001; group effect: F(3,16) = 0.612, p = 0.6170; Fig. 3c). Our findings reveal an uncoupling between cerebral glucose uptake and blood flow responses following astroglial glutamate transport activation via GLT-1, which supports the concept that the increased glucose utilization under CEF challenge is driven by astrocytic glutamate transport.
Figure 3. Astrocytic glutamate transport activation via GLT-1 uncouples [18F]FDG uptake and cerebral blood flow response.
(a) 3D brain surface displaying highest effects of CEF challenge and probe locations. (b) Temporal resting CBF flow analysis during baseline and after injection of saline or CEF in the barrel (repeated measure one-way ANOVA, p = 0.2714) and prefrontal cortices (repeated measure one-way ANOVA, p = 0.6495). (c) Evoked CBF analysis in the barrel cortex (repeated measure two-way ANOVA, interaction: p > 0.999; whiskers stimulation effect: p < 0.001; group effect: p = 0.6170). n= 5 per group for evoked CBF; n = 5 per group for resting CBF in the PFC; and n = 4 per group for resting CBF in the barrel cortex. Data are presented as mean values ± s.e.m (shadows).
Our results provide the first in vivo PET evidence for astrocytic glutamate transport via GLT-1 triggering glucose uptake. In fact, we show that [18F]FDG PET signal is substantially affected by the activation of astroglial glutamate transport. Furthermore, combining our in vivo data with several previous studies (for review see8), argues for the view that glucose utilization by astrocytes determines part of the [18F]FDG PET signal. The notion that [18F]FDG PET signal also reflects astrocyte activity can change how we decode brain metabolic images made with [18F]FDG PET. For example, in the context of Alzheimer’s disease, [18F]FDG hypometabolism is utilized as a biomarker of synaptic dysfunction of neuronal nature20. Our data indicate that [18F]FDG PET signal can be driven by astrocytes, urgently calling for a reevaluation in the way we interpret imaging of brain disorders using [18F]FDG PET.
Online Methods
Animals
Animals were kept in a room with controlled temperature (21°C) under a 12-h light/dark cycle (lights on 7 am) and had access to food and water ad libitum. All procedures were carried out according to the guide to the care and use of experimental animals (Ed2) of the Canadian Council on Animal Care. Animal experiments were performed according to the institutional guidelines of the Gwangju Institute of Science and Technology and the Brazilian guidelines of animal experimentation number 11794/08, which are compliant with McGill University guidelines for experimental research (Montreal, QC, Canada).
Imaging Procedures
Ten male two-month old Sprague-Dawley rats (body weight: 245–302 g) were scanned for baseline and one day after challenge. The scanning was made between 10:30 am – 1:30 pm. Animals received an intravenous bolus injection (0.2 mL) of [18F]FDG (mean ± s.d.: 8.14 ± 0.555 MBq) into the tail vein. Rats were maintained awake for a 40 minutes uptake period, followed by a 10 minutes static acquisition under isoflurane anesthesia. A CT transmission scan was acquired to correct for attenuation. PET measurements were performed on an Inveon microPET/CT scanner (Siemens Medical Solution, Knoxville, TN). Rats lay in prone position with the head immobilized by both a body holder and the nose cone of the anesthesia system (2% isoflurane at 0.5 L/min oxygen flow). The brain was positioned in the center of the field of view. The body temperature was maintained at 36.5 ± 1°C, and vital signs, including respiration, heart rate, and body temperature, were monitored throughout scanning procedures (BioVet; m2m Imaging Corp, Newark, NJ, USA). Images were reconstructed by fully-3D ordered subset expectation maximization (3D-OSEM) algorithm, normalized and corrected for scatter, dead time and decay.
Challenge experimental design
Rats were scanned twice under different pretreatment conditions. Firstly, animals were anesthetized (2% isoflurane at 0.5 L/min oxygen flow) and positioned in the scanner. For the baseline scan, they received a tail vein injection of saline 30 minutes before the [18F]FDG injection. For the challenge scan, they received a tail vein injection of CEF 200 mg/kg (Rocephin®, Basel, Switzerland) Roche, 30 minutes before the [18F]FDG injection (Fig. 1a).
Image analysis
Imaging analysis was conducted using minc-tools (http://www.bic.mni.mcgill.ca/ServicesSoftware/HomePage). MicroPET images were manually co-registered to a standard rat histological template. Standardized uptake value (SUV) was calculated using the following equation: SUV = (radioactivity)/(dose injected/body weight). Regional SUV was calculated based on manually delimited volumes of interest (VOIs), defined on the rat template for the following brain regions: frontal cortex (162.66 mm3), parietal cortex (380.36 mm3), hippocampus (63.25 mm3), striatum (88.72 mm3), thalamus (62.43 mm3), and cerebellum (155.64 mm3). For metabolic network analyses, [18F]FDG data were normalized by dose injected and body mass, to account for differences in total tracer tissue concentration. Matrices were constructed using cross-correlation coefficient symmetric analyses (VOIs above-stated, left and right hemispheres, 12×12 matrices). For further details about microPET imaging analyses see ref. 21. Data normalization by a reference region was not recommended in our analyses due to the lack of a reference region, since GLT-1 is widely present in the whole brain22.
In vivo cerebral blood flow (CBF) measurements
Two-month old Sprague-Dawley male rats (body weight: 280–320g, 4–5 per group) were anesthetized with urethane (1g/kg, ip) on a heating blanket to keep body temperature at 37°C. The right femoral vein was cannulated for drug injection, while a heparin-catheter was placed in the artery for blood pressure (MAP), heart rate, and blood gas measurements (pH, pO2 and pCO2). Rats were then placed in a stereotaxic frame and bones over the left prefrontal (AP: + 4.7 mm; lateral: + 1 mm from Bregma) and barrel (AP 2.5 to 3 mm, ML 6 to 7 mm from Bregma) cortices were thinned to translucency for positioning of two Laser-Doppler probes (Transonic Systems). Resting CBF was continuously recorded for 10 minutes prior to CEF injection (baseline) and for 40 minutes following CEF injection. The CBF values from laser Doppler flowmetry were averaged every second and expressed as percent change from the mean of 10 minutes.
Whisker stimulation in vivo
Right whiskers were stimulated for 20 seconds (repeated every minute) using an electrical brush (for 20 seconds, ~10 Hz) at baseline, and 40 minutes after drug injection. The evoked CBF response, 1 second averages were computed for 1 minute 20 seconds (30 seconds before, 20 seconds stimulation, 30 seconds after stimulation).
Open field
Thirty, two-month old Sprague Dawley rats were submitted to the open field test under two different pretreatment conditions. Animals were briefly anesthetized (2% isoflurane at 0.5 L/min oxygen flow) and received a received a single tail vein injection of saline or CEF 200 mg/kg 70 minutes before the open field test. The apparatus for this test was a grey acrylic box (50 cm × 50 cm × 50 cm). Experiments were conducted in a sound-attenuated room under low-intensity light (12 lx) during the light cycle. Rats were placed in the center of the box and spontaneous locomotion parameters were recorded with a video camera for 10 min. All analyses were performed using Any-maze, a computer-operated tracking system (Any-maze, Stoelting, Woods Dale, IL).
Adult astrocyte cell culture preparation and maintenance
Three-month old male Wistar rats were euthanized by decapitation, had their cerebral cortices aseptically dissected and meninges removed. The tissue was digested using trypsin at 37 °C and papain, as previously described 23. After mechanical dissociation and centrifugation, the cells were plated in 6- or 24-well plates with DMEM/F12 medium (Life technologies [11330-032], Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, Life technologies [12657-029], Carlsbad, CA) changed regularly; during the 1st week, the medium was replaced once every 2 days and from the 2nd week on, once every four days. From the 2nd week on, astrocytes received medium supplemented with 20% FBS until they reached confluence (at approximately the 3rd week). No dibutyryl cAMP was added to the culture medium in order to observe the naive response of the cells. Expression of specific neuronal (anti-NeuN, EDM Millipore [MAB377], Billerica, MA) and microglial (Anti-Cd11b/c, Invitrogen [MR6200], Carlsbad, CA) proteins was evaluated in order to determine the purity of the astrocyte cultures, which was found to be around 95 %. Experiments involving adult culture of astrocytes were replicated at least 3 times.
Immunocytochemistry
Immunocytochemistry was performed as previously described23. Briefly, cell cultures were fixed with 4% paraformaldehyde for 20 min and permeabilized with 0.1% Triton X-100 in PBS for 5 min at room temperature. After blocking overnight with 4% albumin, the cells were incubated overnight with anti-GFAP (1:400; Dako [Z0334], Carpienteria, CA), anti-GLT-1 (1:1000; Alpha diagnostic [GLT11-A], San Antonio, TX), anti-vimentin (1:1000; Sigma Aldrich [V6630], St. Louis, MO) or anti-NeuN (1:50; EDM Millipore [MAB377], Billerica, MA) at 4°C, followed by PBS washes and incubation with a specific secondary antibody (Jackson ImmunoResearch, West Grove, PA) conjugated with Alexa Fluor® 488 (green staining, [111-545-003]) or 594 (red staining, [315-585-003]) for 1 h at room temperature. For all the immunostaining-negative controls, reactions were performed omitting the primary antibody. No reactivity was observed when the primary antibody was excluded. Cell nuclei were stained with 0.2 mg/mL of 4′,6′-diamidino-2-phenylindole (DAPI; EDM Millipore [268298], Billerica, MA). The cells were visualized with a Nikon inverted microscope and the images were transferred to a computer with a digital camera (Sound Vision Inc.).
Western Blot Analyses
Cells were solubilized in lysis solution containing 4 % SDS, 2 mM EDTA, 50 mM Tris– HCl (pH 6.8). Protein content was measured, the samples were standardized in sample buffer (62.5 mM Tris–HCl (pH 6.8), 4 % (v/v) glycerol, 0.002 % (w/v) bromophenol blue) and boiled at 95 °C for 5 min. Samples were separated by SDS/PAGE (10 mg protein per sample), and transferred to nitrocellulose membranes, as previously described23. Adequate loading of each sample was confirmed using Ponceau S staining. Membranes were incubated overnight (4 °C) with anti-GLT-1 (1:1000, Alpha diagnostic [GLT11-A], San Antonio, TX) and anti-GAPDH (1:1000, EMD Millipore [G9545], Billerica, MA). The membranes were then washed and incubated with a peroxidase-conjugated anti-rabbit (GE healthcare [NA934V], Little Chalfont, UK) immunoglobulin at a dilution of 1:4000 for 2 h. Chemiluminescence signals were detected with an Image Quant LAS4000 system (GE Healthcare) using ECL kit. Full-length gels and membranes with molecular weight standards were shown on Supplementary Figure 6.
[3H]D-Aspartate uptake
After the cells reached confluence, D-Aspartate uptake was performed as previously described24 with some modifications. Briefly, cells were rinsed once with HBSS and then the medium (C5793, Sigma Aldrich, St. Louis MO) was replaced with fresh DMEM/F12 1% FBS in presence or absence of 0.1 μM CEF for 70 min at 37 °C. The medium was then replaced by DMEM/F12 1% FBS containing 1mCi/mL D-[2,3-3H] Aspartate (Perkin Elmer [NET581001MC], Boston, MA) in presence or absence of 0.1 μM CEF. The incubation was stopped after 5 min by removal of the medium and rinsing the cells three times with HBSS. The cells were then lysed in a solution containing 0.5 M NaOH. Incorporated radioactivity was measured in a scintillation counter. Unspecific uptake was carried out at 4 °C and subtracted from total uptake.
2-Deoxy-D-[2,63H]glucose ([3H]2DG) uptake
Basal and CEF-stimulated deoxyglucose uptake were assessed as previously described with some modifications9. Briefly, cells in 6- or 24- well plates were rinsed once with HBSS and then the medium was replaced with fresh DMEM/F12 1% FBS in presence or absence of CEF 0.1 uM for 70 min at 37 °C. Adult astrocytes were then incubated with DMEM/F12 1%FBS containing 1 mCi/ml [3H]2DG (GE healthcare [TRK672], Little Chalfont, UK) for 20 min at 37 °C. After incubation, the cells were rinsed three times with HBSS and lysed overnight with NaOH 0.5 M. The incorporated radioactivity was measured in a scintillation counter. Cytochalasin B 10 mM (Sigma Aldrich [C2743], Saint Louis, MO) was used as a specific glucose transporter inhibitor. Deoxyglucose uptake was determined by subtracting the uptake in the presence of cytochalasin B from the total uptake. For GLT-1 pharmacological blockade, DHK (ABCAM [ab120066], Cambrigde, UK) 100 μM was added to the medium 6 minutes before [3H]2DG uptake.
High-performance liquid chromatography (HPLC) procedure
Free amino acids concentration in the culture medium was determined by high-performance liquid chromatography (HPLC) as previously described25. Medium cell-free supernatant aliquots were used to quantify glutamate. The analysis was performed using a reverse phase column (Supelcosil LC-18, 250 mm × 4.6 mm × 5 μm, Supelco) in a Shimadzu Instruments liquid chromatograph (50 μL loop valve injection, 40 uL injection volume) and fluorescent detection after pre-column derivatization with 100,5 μL OPA (5.4 mg OPA in 1 mL 0.2 M sodium borate pH 9.5) plus 25,5 μL 4% mercaptoethanol. The mobile phase flowed at a rate of 1.4 mL/min and column temperature was 24 °C. Buffer composition is A: 0.04 mol/L sodium dihydrogen phosphate monohydrate buffer, pH 5.5, containing 80% of methanol; B: 0.01 mol/L sodium dihydrogen phosphate monohydrate buffer, pH 5.5, containing 20% of methanol. The gradient profile was modified according to the content of buffer B in the mobile phase: 100% at 0.10 min, 90% at 15 min, 48% at 10 min,100% at 60 min. Absorbance was read at 360 nm and 455 nm, excitation and emission respectively, in a Shimadzu fluorescence detector. Samples of 20 μL were used and concentration was expressed in μM (as mean ± s.e.m). Glutamate was identified by its retention time and was quantitatively determined by using chromatographic peak area. A standard amino acid mixture was used for HPLC quality control and calibration.
Statistical analysis
Data distribution was first tested for normality using Kolmogorov-Smirnov test. Sample size was evaluated using G*power 3.1 for Mac (effect size dz = 1.77, noncentrality parameter δ = 4.68, Critical t = 2.44 and actual power = 0.97) based on a previous publication26. Regional brain binding differences between baseline and challenge were estimated across the six VOIs using two-tailed paired Student t-test. Comparisons between baseline and challenge were conducted at the voxel level using t-statistic analysis (Statistical Parametric Mapping, Rminc, https://github.com/mcvaneede/RMINC). All analyses were conducted using minctools, which is available online at https://github.com/BIC-MNI/minc-tools. T-statistical maps were corrected for multiple comparisons using random field theory27. Resting CBF was analyzed by repeated measure two-way ANOVA, and evoked CBF with repeated measure one-way ANOVA. Data acquisitions were not performed blind to the conditions of the experiments. However, data analyses (imaging, biochemistry and behavior) were conducted in blind conditions. Animals were randomly assigned for all experiments, however no formal randomization protocol was utilized in this study. Open field task and In Vitro studies were analyzed by two-tailed unpaired Student-t test. [3H]2DG uptake data was analyzed by one-way ANOVA. SPSS 18.0 and MATLAB were used for all analysis. Differences were considered statistically significant at p < 0.05.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
This work was supported by a grant from iMSE from GIST and by National Research Foundation of Korea (NRF:2013R1A2A2A01067890, HK), the Canadian Institutes of Health Research (CIHR) [MOP-11-51-31, PRN] and [MOP-142417, EH], the Alan Tiffin Foundation, the Alzheimer’s Association [NIRG-08-92090, PRN], the Fonds de la recherche en santé du Québec (Chercheur boursier), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), the Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS), the INCT for Excitotoxicity and Neuroprotection and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). Work in the laboratory of LP is financially supported by the department of Physiology, University of Lausanne. We would like to thank Prof. Diogo Onofre Souza, Dr. Luis Valmor Cruz Portela, Dr. Fernanda Urruth Fontella and Ms. Andréia Silva da Rocha from the Department of Biochemistry/UFRGS for the fruitful discussion and for helping in the conduction of biochemical assays.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Author contributions
All authors participated on the conceptualization, design and interpretation of the experiments. E.Z., M.P., A.L., H.K. and P.R. were responsible for conducting imaging acquisitions and analysis. E.Z., D.S., and L.P. were responsible for conducting in vitro studies in cell cultures. C.L. and E.H. were responsible for conducting Laser-Doppler acquisitions and analysis. All authors critically revised and approved the final version of the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Sokoloff L. Developmental neuroscience. 1993;15:194–206. doi: 10.1159/000111335. [DOI] [PubMed] [Google Scholar]
- 2.Mazzilotta JC, Phelps ME, Miller J, Kuhl DE. Neurology. 1981;31:503–516. doi: 10.1212/wnl.31.5.503. [DOI] [PubMed] [Google Scholar]
- 3.Mosconi L, et al. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2008;49:390–398. doi: 10.2967/jnumed.107.045385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nehlig A, Coles JA. Glia. 2007;55:1238–1250. doi: 10.1002/glia.20376. [DOI] [PubMed] [Google Scholar]
- 5.Figley CR, Stroman PW. The European journal of neuroscience. 2011;33:577–588. doi: 10.1111/j.1460-9568.2010.07584.x. [DOI] [PubMed] [Google Scholar]
- 6.Volterra A, Meldolesi J. Nature reviews. Neuroscience. 2005;6:626–640. doi: 10.1038/nrn1722. [DOI] [PubMed] [Google Scholar]
- 7.Pellerin L, Magistretti PJ. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2003;23:1282–1286. doi: 10.1097/01.WCB.0000096064.12129.3D. [DOI] [PubMed] [Google Scholar]
- 8.Pellerin L, Magistretti PJ. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2012;32:1152–1166. doi: 10.1038/jcbfm.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pellerin L, Magistretti PJ. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voutsinos-Porche B, et al. Cerebral cortex. 2003;13:1110–1121. doi: 10.1093/cercor/13.10.1110. [DOI] [PubMed] [Google Scholar]
- 11.Voutsinos-Porche B, et al. Neuron. 2003;37:275–286. doi: 10.1016/s0896-6273(02)01170-4. [DOI] [PubMed] [Google Scholar]
- 12.Magistretti PJ, Pellerin L. Molecular psychiatry. 1996;1:445–452. [PubMed] [Google Scholar]
- 13.Rothstein JD, et al. Nature. 2005;433:73–77. doi: 10.1038/nature03180. [DOI] [PubMed] [Google Scholar]
- 14.Zimmer ER, et al. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2015 [Google Scholar]
- 15.Lehre KP, Levy LM, Ottersen OP, Storm-Mathisen J, Danbolt NC. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1995;15:1835–1853. doi: 10.1523/JNEUROSCI.15-03-01835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chugani HT, Phelps ME, Mazziotta JC. Annals of neurology. 1987;22:487–497. doi: 10.1002/ana.410220408. [DOI] [PubMed] [Google Scholar]
- 17.Phelps ME, Mazziotta JC. Science. 1985;228:799–809. doi: 10.1126/science.2860723. [DOI] [PubMed] [Google Scholar]
- 18.Choi H, et al. NeuroImage. 2014;99:226–236. doi: 10.1016/j.neuroimage.2014.05.039. [DOI] [PubMed] [Google Scholar]
- 19.Lauritzen M. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2001;21:1367–1383. doi: 10.1097/00004647-200112000-00001. [DOI] [PubMed] [Google Scholar]
- 20.Jack CR, Jr, et al. The Lancet. Neurology. 2013;12:207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmer ER, Parent MJ, Cuello AC, Gauthier S, Rosa-Neto P. MicroPET imaging and transgenic models: a blueprint for Alzheimer's disease clinical research. Trends Neurosci. 2014 Nov;37(11):629–41. doi: 10.1016/j.tins.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Furuta A, Rothstein JD, Martin LJ. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1997;17:8363–8375. doi: 10.1523/JNEUROSCI.17-21-08363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Souza DG, Bellaver B, Souza DO, Quincozes-Santos A. PloS one. 2013;8:e60282. doi: 10.1371/journal.pone.0060282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debernardi R, Magistretti PJ, Pellerin L. Brain research. 1999;850:39–46. doi: 10.1016/s0006-8993(99)02022-3. [DOI] [PubMed] [Google Scholar]
- 25.Souza DG, et al. Neurochemical research. 2016;41:1578–1586. doi: 10.1007/s11064-016-1871-7. [DOI] [PubMed] [Google Scholar]
- 26.Zimmer ER, et al. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2015;35:1169–1174. doi: 10.1038/jcbfm.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Worsley KJ, et al. Human brain mapping. 1996;4:58–73. doi: 10.1002/(SICI)1097-0193(1996)4:1<58::AID-HBM4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.