Abstract
Objective
The molecular basis of endothelial cell (EC)–specific gene expression is poorly understood. Roundabout 4 (Robo4) is expressed exclusively in ECs. We previously reported that the 3-kb 5′-flanking region of the human Robo4 gene contains information for lineage-specific expression in the ECs. Our studies implicated a critical role for GA-binding protein and specificity protein 1 (SP1) in mediating overall expression levels. However, these transcription factors are also expressed in non-ECs. In this study, we tested the hypothesis that epigenetic mechanisms contribute to EC-specific Robo4 gene expression.
Methods and Results
Bisulfite sequencing analysis indicated that the proximal promoter of Robo4 is methylated in non-ECs but not in ECs. Treatment with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine increased Robo4 gene expression in non-ECs but not in ECs. Proximal promoter methylation significantly decreased the promoter activity in ECs. Electrophoretic mobility shift assays showed that DNA methylation of the proximal promoter inhibited SP1 binding to the −42 SP1 site. In DNase hypersensitivity assays, chromatin condensation of the Robo4 promoter was observed in some but not all nonexpressing cell types. In Hprt (hypoxanthine phosphoribosyltransferase)-targeted mice, a 0.3-kb proximal promoter directed cell-type–specific expression in the endothelium. Bisulfite sequencing analysis using embryonic stem cell–derived mesodermal cells and ECs indicated that the EC-specific methylation pattern of the promoter is determined by demethylation during differentiation and that binding of GA-binding protein and SP1 to the proximal promoter is not essential for demethylation.
Conclusions
The EC-specific DNA methylation pattern of the Robo4 proximal promoter is determined during cell differentiation and contributes to regulation of EC-specific Robo4 gene expression.
Keywords: DNA methylation, endothelial cells, epigenomics
Roundabout 4 (Robo4) is a transmembrane receptor that belongs to the Robo family of neural cell adhesion molecules. Robo4 has been shown to play a role in endothelial cell (EC) migration, proliferation, angiogenesis, and stabilization of the vasculature.1–5 Robo4 is expressed specifically in ECs6,7 in the developing embryo,1 placenta,6 normal adult tissues,1,8 and tumors.6,8 Recently, shear stress was reported to suppress Robo4 gene expression.9
We previously reported that a 3-kb fragment of the 5′-flanking region of the human Robo4 gene contains information for EC-specific expression in vivo.10 We demonstrated that the Robo4 promoter is activated by the transcription factors GA-binding protein (GABP) and SP1 through an E26 transformation-specific (ETS)-binding site at −119 and 2 SP1-binding sites at −42 and −153, respectively.10,11 Although GABP and SP1 are essential for Robo4 promoter activation, both factors are known to be expressed ubiquitously. In keeping with these data, previous studies exploring the transcriptional regulation of other EC-specific genes, including von Willebrand factor, PECAM-1 (platelet endothelial cell adhesion molecule; CD31), vascular endothelial growth factor receptors 1 and -2, Tie2, and E-selectin, have consistently implicated a role for noncell-type–specific transcription factors, such as SP1, ETS family proteins, nuclear factor-κB, and GATA proteins.12 Although some transcription factors are enriched in ECs (eg, Vezf1 [vascular endothelial zinc finger], HoxA9 [homeobox], GATA2, and KLF2 [Krüppel-like factor]), none has been shown to be restricted to the endothelium.
Collectively, these findings argue against the existence of an endothelial-specific master regulator that mediates differentiation and expression of multiple differentiation markers, as MyoD (myogenic differentiation) does in skeletal myocytes.13 One possibility is that EC-specific gene expression is mediated by the cooperative activity of multiple noncell-type–specific transcription factors. Alternatively, unique post-translational modifications or alternative spliced transcripts may play a role. Finally, there is increasing evidence that epigenetic mechanisms contribute to cell-type–specific expression of genes in the endothelium.14,15
Epigenetic control of gene transcription involves DNA methylation, histone modification, and chromatin remodeling. DNA methylation refers to the addition of a methyl group to the 5′-position of cytosine to create 5-methyl-cytosine. DNA methylation at cytosine residues occurs almost exclusively in the context of the CpG sequence in the vertebrate genome. DNA methylation is a repressive mark that is associated with transcriptional silencing.16 It has been implicated in many cellular processes, including X chromosome inactivation, genomic imprinting, embryonic development, lineage specification, and cancer pathogenesis.15 Two general mechanisms have been proposed for CpG methylation–mediated gene suppression. First, DNA methylation may inhibit the binding of transcription factors to CpG dinucleotide–containing cis-regulatory elements. Second, DNA methylation results in the recruitment of methyl CpG–binding proteins. These proteins recruit large chromatin-modifying complexes and promote chromatin condensation that represses transcription by reducing DNA accessibility.
In this study, we hypothesized that EC-specific Robo4 gene expression is regulated by epigenetic mechanisms. We compared the DNA methylation pattern of the Robo4 promoter between EC and non-EC cell types and found that the proximal promoter region is methylated only in non-ECs. This cell-type–specific methylation inhibited the binding of SP1 to the −42 SP1 site and suppressed Robo4 gene expression. Finally, using transgenic mice and an ES cell differentiation system, we demonstrated that the 0.3-kb proximal promoter, including 11 CpG sites, contained the information for EC-specific gene expression and that the EC-specific methylation pattern of the promoter is determined during cell differentiation. Collectively, these data support a novel model of EC-specific gene expression that involves DNA methylation–mediated inhibition of SP1 activity in non-ECs.
Results
Methylation Pattern of the Robo4 Promoter in ECs and Non-ECs
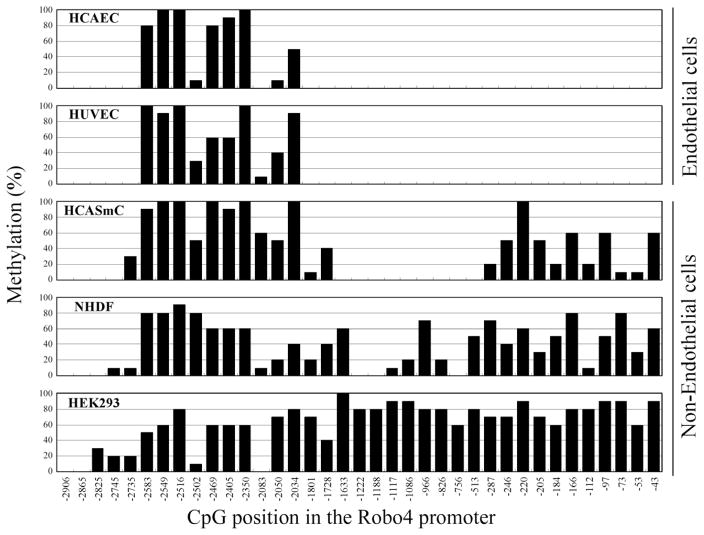
The upstream promoter (−3000 to +1) of the human Robo4 gene contains a total of 37 CpG sites and no typical CpG islands (Figure I in the online-only Data Supplement). To investigate the methylation pattern of the endogenous Robo4 promoter in ECs (human coronary artery ECs [HCAEC] and human umbilical vein ECs) and non-ECs (human coronary artery smooth muscle cells [HCASmC], normal human dermal fibroblasts, and human embryonic kidney [HEK] 293), the methylation status of 37 CpG sites in the 3-kb promoter was analyzed by bisulfite sequencing (Figure 1; Figure II in the online-only Data Supplement). In ECs, CpG methylation was restricted to an upstream region between −2583 and −2034. In contrast, non-ECs demonstrated heavy methylation not only in the upstream region but also at the proximal promoter (11 CpG sites between −287 and −43). Analysis of reduced representation bisulfite sequencing from the ENCODE (Encyclopedia of DNA Elements) consortium revealed similar differences in Robo4 promoter methylation across a wide range of ECs and non-ECs (Figure III in the online-only Data Supplement). Thus, the 300-bp upstream promoter of Robo4 is differentially methylated in expressing and nonexpressing cell types.
Figure 1.
Bisulfite sequencing of the human roundabout 4 (Robo4) promoter. Summary of the 3-kb human Robo4 promoter bisulfite sequencing results for 10 cloned polymerase chain reaction products prepared with bisulfite-treated genomic DNA from 2 endothelial cell types (human coronary artery endothelial cells [HCAEC] and human umbilical vein endothelial cells [HUVEC]) and 3 nonendothelial cell types (human coronary artery smooth muscle cells [HCASmC], normal human dermal fibroblasts [NHDF], and human embryonic kidney [HEK] 293). Each graph indicates the CpG position in the promoter and the percentage of methylated CpG.
Effects of DNA Methylation on Robo4 Gene Expression in ECs and Non-ECs
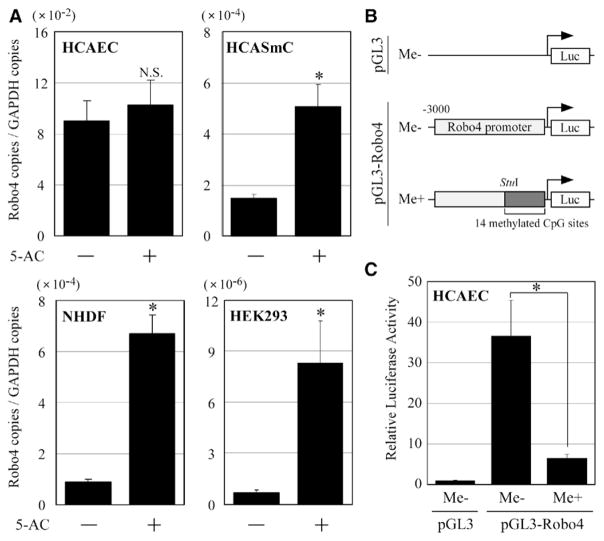
We next asked whether cell-type–specific methylation of the Robo4 promoter contributes to EC-specific expression of Robo4. To investigate the effects of promoter methylation on Robo4 gene expression, ECs (HCAEC) and non-ECs (HCASmC, normal human dermal fibroblasts, and HEK293) were treated with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine, and expression levels of the Robo4 mRNA were measured by real-time reverse transcription polymerase chain reaction. Treatment with 5-aza-2′-deoxycytidine did not affect Robo4 mRNA expression in HCAEC but significantly increased mRNA levels in HCASmC, normal human dermal fibroblasts, and HEK293 cells (by 4-fold, 7-fold, and 30-fold, respectively; Figure 2A). These results suggest that DNA methylation of the Robo4 promoter serves to silence expression in non-ECs.
Figure 2.
Effects of DNA methylation on roundabout 4 (Robo4) gene expression in endothelial cells (ECs) and non-ECs. A, ECs and non-ECs were treated with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-AC). The expression levels of Robo4 and GAPDH (as an internal control) were measured using real-time polymerase chain reaction with cDNAs prepared from human coronary artery endothelial cells (HCAEC), human coronary artery smooth muscle cells (HCASmC), normal human dermal fibroblasts (NHDF), and human embryonic kidney (HEK) 293 treated with or without 10 μmol/L 5-AC for 4 days. Copy numbers were calculated from standard curves prepared by measuring known amounts of plasmids encoding Robo4 or GAPDH. The Robo4 expression level was normalized to that of GAPDH. The data are represented as mean±SE. (n=3, Student t test, *P<0.05). B, Schematic representation of a nonmethylated control luciferase vector (pGL-3-Me−) and the Robo4 promoter–luciferase constructs with or without methylation of 14 CpG sites (pGL3-Robo4-Me+ or -Me−, respectively). The CpG sites in the proximal promoter (−928 [StuI site] to +10) were methylated by methyltransferase SssI. C, pGL3-Robo4-Me−, Me+, or pGL3-Me− was transfected into HCAEC. Luciferase activity was measured 48 hours after the transfection. The data are represented as mean±SE (n=5, Student t test, *P<0.05).
To determine the effect of CpG methylation on promoter activity, we performed transient transfection assays using Robo4 promoter–luciferase plasmids in which the proximal promoter (CpG sites from −826 to −43) was either unmethylated or methylated in vitro using Sss1 methylase (Figure 2B). As shown in Figure 2B, methylation of the proximal promoter resulted in a significant reduction of Robo4 promoter activity in HCAEC. Thus, DNA methylation of the proximal promoter suppresses Robo4 gene expression.
Effect of DNA Methylation on GABP and SP1 Binding to the Robo4 Promoter
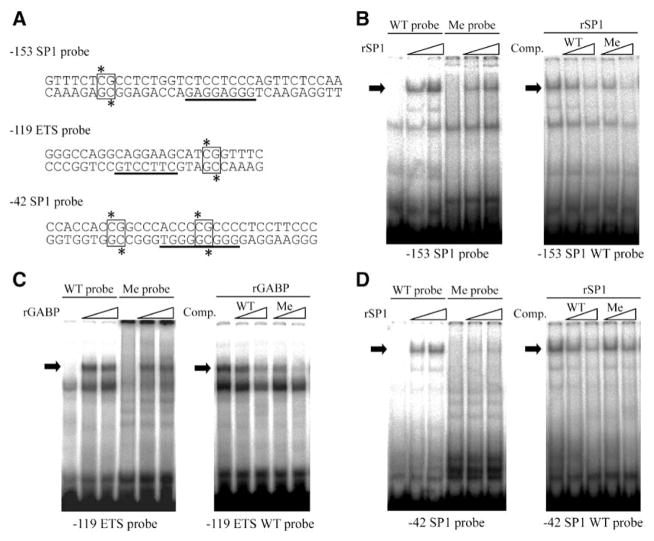
One mechanism underlying the silencing effect of DNA methylation on gene expression is direct inhibition of transcription factor binding. We have previously shown that GABP and SP1 bind to an ETS site at −119 and SP1 sites at −42 and −153. All 3 DNA elements are flanked by CpG sites (Figure 3A). In addition, the −42 SP1 site has a CpG dinucleotide internal to the consensus motif. To investigate whether DNA methylation affects the binding of GABP and SP1 to their cis-elements, elecrophoretic mobility shift assay was performed using oligonucleotide probes in which these CpG sites were methylated or unmethylated (Figure 3A). Methylation of the −153 SP1 probe did not affect SP1 binding to the −153 SP1 site (Figure 3B, left). Furthermore, in competition assays, cold unmethylated and methylated probes were effective in inhibiting binding of SP1 to the wild-type (WT) probe (Figure 3B, right). Similarly, methylation of the −119 ETS probe had no effect on GABP binding (Figure 3C). In contrast, methylation of the −42 SP1 probe significantly inhibited SP1 binding, as evidenced by a loss of binding to the radiolabeled methylated probe (Figure 3D, left) and lack of competition of SP1 binding to a WT unmethylated probe (Figure 3D, right). These results suggest that DNA methylation suppresses Robo4 gene expression in non-ECs by inhibiting SP1 binding to the −42 SP1 site.
Figure 3.
Effect of DNA methylation on the binding of SP1 and GA-binding protein (GABP) to the promoter. A, Oligonucleotides used for the probes. Underlined and boxed sequences indicate the transcription factor binding sites and CpG sites, respectively. To prepare the methylated probes, cytosines of the CpG sites in both strands were methylated. B, Left, Results of elecrophoretic mobility shift assay performed with a 32P-labeled nonmethylated (wild-type [WT]) or methylated (Me) −153 SP1 probe using recombinant SP1 (rSP1) protein. Right, Results of a competition assay performed with the −153 SP1 WT probe and competitors (a cold WT or Me probe). C and D, The same experiments as shown in B were performed with the −119 ETS and −42 SP1 probes using recombinant GABP (rGABP) and SP1 protein, respectively.
Chromatin Condensation of the Robo4 Promoter in ECs and Non-ECs
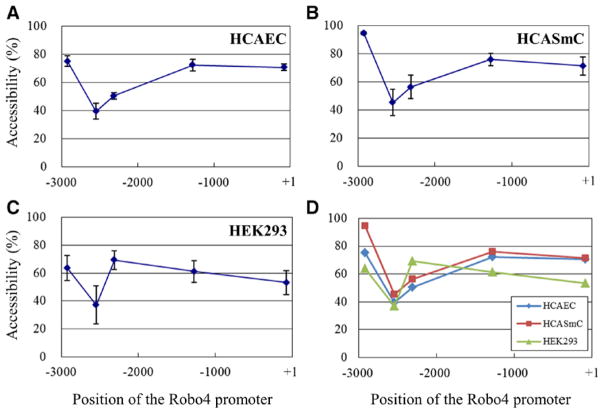
A second mechanism underlying the silencing effect of DNA methylation on gene expression is indirect inhibition of transcription factor binding through chromatin condensation. To investigate whether chromatin condensation plays a role in inhibiting Robo4 gene expression in non-ECs, we performed DNase hypersensitivity assays using micrococcal nuclease digestion of genomic DNA from HCAEC, HCASmC, and HEK293 cells (Figure 4). The chromatin was highly condensed around −2.5 kb in both ECs and non-ECs, which correlates with dense methylation in this region (Figure 1). In the proximal promoter region, chromatin condensation was low in ECs. Surprisingly, a similar pattern was observed in HCASmC. In contrast, chromatin condensation was higher in HEK293 cells. These data argue against a consistent role for chromatin condensation in silencing Robo4 gene expression in non-ECs.
Figure 4.
Chromatin condensation of the roundabout 4 (Robo4) promoter in endothelial cells (ECs) and non-ECs. A, DNase hypersensitivity assays were performed with micrococcal nuclease and chromatin isolated from human coronary artery endothelial cells (HCAEC). The digested genomic fragments were purified and measured by real-time polymerase chain reaction. Accessibility was calculated by comparing the copy numbers between digested and undigested samples. B and C, The same assays were performed with human coronary artery smooth muscle cells (HCASmC; B) and human embryonic kidney (HEK) 293 cells (C). D, Summarized data of A, B, and C.
EC-Specific Gene Expression of a 0.3-kb Proximal Promoter Fragment of Robo4 In Vivo
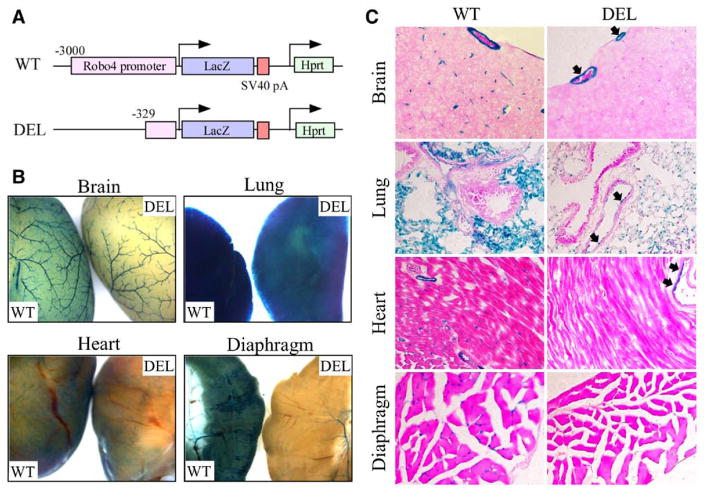
Our data suggest that the immediate 0.3-kb upstream promoter of Robo4 constitutes a differentially methylated region in ECs and non-ECs. To determine whether this region, which contains 11 CpG sites, is sufficient for mediating cell-type specificity in vivo, we generated transgenic mice carrying the 0.3-kb Robo4 promoter coupled to the LacZ gene (DEL [deletant]; Figure 5A) and compared LacZ expression with mice carrying the 3-kb full-length promoter (WT; Figure 5A). X-gal staining of whole-mount organs showed that LacZ expression in the brain, lung, heart, and diaphragm was significantly lower in DEL mice compared with WT mice (Figure 5B). X-Gal staining of tissue sections showed a similar reduction in LacZ expression in the microvasculature of all 4 organs, whereas LacZ expression was still observed in ECs of larger blood vessels in the brain, lung, and diaphragm (Figure 5C). These data indicate that the 0.3-kb proximal promoter is sufficient for mediating EC-specific gene expression in vivo, although its promoter activity is weaker than that of the full-length promoter.
Figure 5.
LacZ expression in transgenic mice containing 0.3-kb roundabout 4 (Robo4) proximal promoter. A, Schematic of the wild-type (WT) and deletant (DEL) hypoxanthine phosphoribosyltransferase (Hprt)-targeted alleles. The 3-kb or 0.3-kb human Robo4 promoter, LacZ cDNA, and SV40 polyadenylation signal upstream of the Hprt gene are indicated. B and C, Organs (brain, lung, heart, and diaphragm) were harvested from WT and DEL mice and processed for whole mount and section staining with X-Gal. Arrows indicate the LacZ-expressing endothelial cells in blood vessels of DEL mice.
Differentiation Stage–Specific Methylation of the Robo4 Promoter During Differentiation of Hprt-Targeted ES Cells
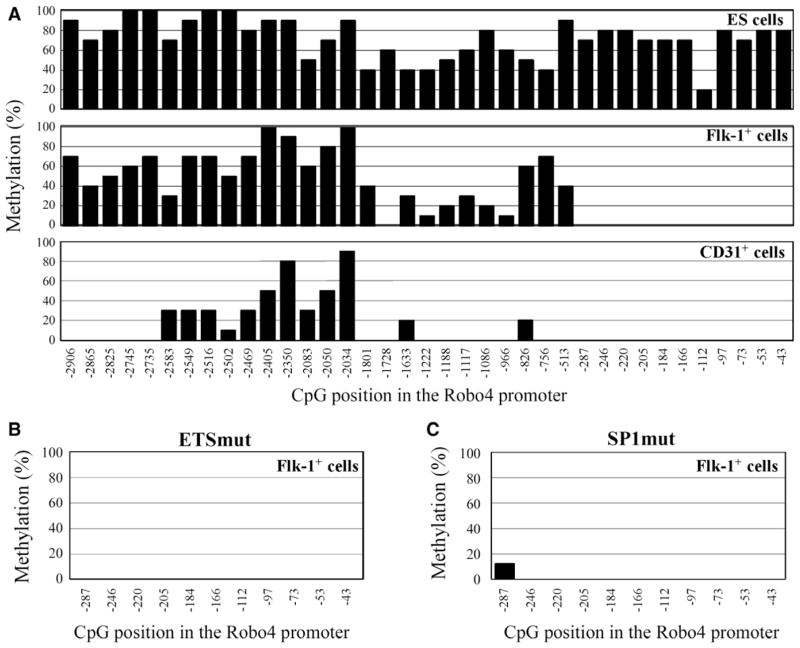
To investigate the methylation pattern of the Robo4 promoter during cell differentiation, Hprt-targeted ES cells carrying 3-kb Robo4 promoter-LacZ were differentiated into mesodermal (Flk-1+ [fetal liver kinase]) cells and endothelial (CD31+) cells. The methylation patterns of the promoter in undifferentiated ES cells, Flk-1+ cells, and CD31+ cells were analyzed by bisulfite sequencing (Figure 6; Figure IV in the online-only Data Supplement). In undifferentiated ES cells, a high degree of methylation was observed throughout the promoter. DNA methylation progressively decreased during cell differentiation. Importantly, the 11 CpG sites in the proximal promoter were completely unmethylated in the Flk-1+ and CD31+ cells (Figure 6; Figure IV in the online-only Data Supplement). These findings suggest that the EC-specific methylation pattern of the Robo4 promoter is determined by demethylation during cell differentiation.
Figure 6.
Differentiation stage–specific DNA methylation of the roundabout 4 (Robo4) promoter in targeted ES cells. A, Targeted ES cells containing the wild-type Robo4 promoter were differentiated, and Flk-1+ and CD31+ cells were separated by magnetic activated cell sorting. The methylation pattern of the targeted Robo4 promoter in undifferentiated and differentiated cells was analyzed by bisulfite sequencing. B and C, Targeted embryonic stem (ES) cells containing the Robo4 promoters with either the −119 ETS mutation (B) or the −42 and −153 SP1 double mutation (C) were differentiated into fetal liver kinase (Flk)-1+ cells. The methylation patterns of the targeted Robo4 promoters were analyzed by bisulfite sequencing.
To investigate the mechanism involved in demethylation of the proximal promoter during cell differentiation from ES cells into Flk-1+ cells, we analyzed whether GABP and SP1 binding to the promoter contributes to demethylation. Two ES cell lines containing the 3-kb Robo4 promoter with mutations either at the −119 ETS site or at the 2 SP1 sites (−153 and −142 SP1 sites) were differentiated, and methylation patterns of the proximal promoter in Flk-1+ cells were analyzed. In both mutant promoters, the proximal region showed a normal hypomethylated pattern similar to that of the WT promoter (Figure 6B and 6C). These results indicate that binding of GABP and SP1 to the proximal promoter is not essential for promoter demethylation.
Robo4 Expression and DNA Methylation in ECs Exposed to Shear Stress
Our data raise the interesting question of whether DNA methylation plays a physiological role in mediating temporal changes in Robo4 expression in ECs. A previous study by Bicknell et al showed that laminar shear stress suppresses Robo4 gene expression.9 We asked whether suppression was regulated through the methylation of the proximal promoter. Human umbilical vein ECs were grown under static conditions or exposed to laminar shear stress (20 dyn/cm2) for 24 hours and harvested for RNA and genomic DNA. In real-time reverse transcription polymerase chain reaction assays, laminar flow resulted in a significant reduction of Robo4 and an induction of KLF2 mRNA levels (Figure VA in the online-only Data Supplement). However, shear stress did not induce proximal promoter methylation in ECs (Figure VB in the online-only Data Supplement). These findings suggest that DNA methylation is not involved in mediating shear stress–dependent repression of Robo4.
Discussion
Robo4 is selectively expressed in ECs. Thus, an understanding of its transcriptional control may provide insights into mechanisms of EC-specific gene expression. We have previously shown that GABP and SP1 positively regulate Robo4 promoter activity. However, these transcription factors are also expressed in other cell types. Thus, other mechanisms must be involved in mediating cell-type–specific expression of Robo4. Recent studies from the Marsden laboratory have demonstrated an important role for DNA methylation in mediating EC-specific expression of several genes, including those encoding endothelial nitric oxide synthase, vascular endothelial cadherin, CD31, von Willebrand factor, and intercellular adhesion molecule-2.14,17 The results of the present study support a similar role for DNA methylation in governing cell-type–specific expression of Robo4.
The methylation pattern of the 5′-flanking region of Robo4 differed between ECs and non-ECs. This effect was most pronounced in the immediate 0.3-kb proximal promoter, in which all 11 CpG sites were heavily methylated in non-ECs but completely unmethylated in ECs. Studies in which the Robo4 promoter was either methylated or demethylated demonstrated a functional role for DNA methylation in repressing Robo4 expression in non-ECs. In Hprt-targeted mice, the 0.3-kb proximal promoter (containing the 11 CpG sites) region retained information for EC-specific gene expression. Taken together, these findings strongly suggest that differential DNA methylation in ECs and non-ECs contributes to cell-type–specific expression of Robo4. Two general mechanisms have been proposed for CpG methylation–mediated gene suppression. First, methylated CpG sites with or without methyl CpG–binding proteins prevent the binding of transcription factors, such as AP (activating protein)-2, HIF (hypoxia inducible factor)-1, and c-Myc (v-myc myelocytomatosis viral oncogene homolog), to their binding motifs in the promoters.18–20 Second, methyl CpG–binding proteins recruit large chromatin-modifying complexes that reduce DNA accessibility by changing chromatin structure.
We first investigated whether DNA methylation interferes with binding of transcription factors to the Robo4 promoter. In elecrophoretic mobility shift assay, DNA methylation had no effect on GABP binding to the −119 ETS or SP1 binding to the −153 SP1 site. In contrast, DNA methylation inhibited binding of SP1 to the −42 SP1 site. Because −42 SP1, but not to the −153 SP1, site contains an internal CpG site, methylation-dependent inhibition of transcription factor binding seems to be more effective when the SP1 and CpG sites overlap. In any event, our data suggest that DNA methylation at the −42 SP1 site plays a role in repressing Robo4 expression in non-ECs.
We next investigated whether DNA methylation of the promoter induces chromatin condensation, which is associated with inhibition of transcription factor binding to the promoter. Our DNase hypersensitivity assays indicated that chromatin is strongly condensed at −2.5 kb, where highly methylated CpG sites were observed in both ECs and non-ECs. In contrast, chromatin condensation of the proximal promoter region, while low in ECs, was variable in non-ECs. Thus, chromatin condensation is not a universal requirement for Robo4 gene suppression in non-ECs.
We have shown that the proximal promoter of Robo4 is demethylated during ES cell differentiation into Flk-1+ cells. We repeated these assays using ES cells targeted with a Robo4 promoter containing mutated GABP- or SP1-binding sites. The WT and mutant promoters demonstrated comparable differentiation-dependent demethylation. Thus, binding of GABP and SP1 to the proximal promoter does not seem to mediate demethylation in differentiating ECs. Additional studies are required to determine the underlying mechanism.
It is interesting to speculate that DNA methylation is dynamically regulated in ECs and contributes to physiological modulation of Robo4 expression. A previous study demonstrated that Robo4 expression is inhibited by shear stress. We confirmed these findings but were unable to demonstrate flow-mediated changes in Robo4 promoter methylation. Thus, other mechanisms are likely to be responsible for Robo4 gene repression under shear stress conditions. Perhaps this is not surprising because the methylation status of DNA is a relatively stable mark and may change over longer time scales than those used in the present experiments.
A comparison of LacZ expression in Hprt-targeted mice carrying the 3-kb and 0.3-kb upstream promoter regions of Robo4 indicates that although the shorter DNA fragment is sufficient for EC-specific expression, DNA sequences between 0.3 and 3 kb are necessary for full activity in the microvasculature. We identified enhancer elements at −2.5 kb (REn1 [Robo4 enhancer element]) and −2.9 kb (REn2) that are essential for maximal promoter activation.10 Because REn1 is located in the region where DNA methylation and chromatin condensation are observed, REn1 may contribute to activation of the Robo4 promoter through epigenetic control.
In summary, our study supports a model in which EC-specific expression of Robo4 is mediated, at least in part, by DNA methylation–dependent inhibition of SP1 binding to −42 SP1 in non-ECs. Furthermore, the data suggest the existence of an EC-specific regulator that induces demethylation of Robo4 (and perhaps, by extension, other EC-specific genes) during cell differentiation.
MATERIALS AND METHODS
Cell culture
Human coronary artery endothelial cells (HCAEC), human umbilical vein endothelial cells (HUVEC), human coronary artery smooth muscle cells (HCASmC), and human dermal fibroblasts (NHDF) were purchased from Lonza (Basel, Switzerland). Primary ECs, HCASmC, and NHDF were cultured in EGM-2-MV, SmGM-2, and FGM-2 media, respectively. Human embryonic kidney cells (HEK293 cells) were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100 μg/ml streptomycin. All cells were cultured at 37ºC under 5% CO2.
Bisulfite sequencing
Genomic DNA was extracted using the ISOGEN reagent (Nippon Gene, Tokyo, Japan). The resulting DNA (3 μg) was treated with a bisulfite reagent using the MethylEasy Xceed Rapid DNA Bisulfite Modification Kit (Human Genetic Signatures, Sydney, Australia). Robo4 promoter fragments were amplified by PCR using the bisulfite-treated DNA and region-specific primers (sequences are shown in Table SI). The resulting fragments were cloned into the pCR2.1 vector using a TOPO TA cloning kit (Invitrogen), and the resulting plasmids were transfected into DH5α cells. Plasmids were prepared from 10 randomly picked colonies, and DNA sequences were analyzed. In all of the clones, 100% C to T conversions at non-CpG sites were observed, indicating efficient sodium bisulfite reactions.
Electrophoretic mobility shift assay (EMSA)
Recombinant SP1 and GABP (GABPα, β, and γ subunits) were prepared using the TNT Quick coupled transcription/translation system (Promega, Madison, WI) and 1 μg of expression vectors. EMSA was performed as described previously.1 Briefly, 32P-labeled oligonucleotide probes spanning GABP or SP1 binding sites with or without cytosine methylation were mixed with 2 4 μl recombinant SP1 or GABP for 40 min at 4ºC. The resulting protein-DNA complexes were analyzed by gel electrophoresis using 4% native polyacrylamide gel and 0.5× TBE buffer at 120 V. Oligonucleotide sequences for the probes are shown in Table SI.
Treatment of cells with 5-aza-2′-deoxycytidine (5-AC) and real-time RT-PCR
HCAEC, HCASmC, NHDF, and HEK293 cells were treated in cell culture media containing 10 μM 5-AC (Sigma-Aldrich, St. Louis, MO) for 4 days. The media were replaced with fresh media containing 5-AC 2 days after beginning the culture. Total RNA was prepared using the RNeasy Mini Kit (Qiagen, Hilden, Germany). To generate cDNA, 0.5 or 1 μg of total RNA was reverse transcribed with Superscript III reverse transcriptase (Invitrogen). Real-time PCR measurements were performed using the cDNA, primers (shown in Table S1), and QuantiTect SYBR Green PCR Kit (Qiagen). Copy numbers were calculated from the standard curve prepared by measuring known amounts of plasmids including target sequences. The expression level of the Robo4 was normalized against GAPDH. Data were collected from at least 3 independent experiments.
Preparation of plasmid and targeting vectors
Preparation of the Robo4 promoter-reporter construct, pGL3-Robo4 and expression vectors for SP1 and GABP has been previously described.1 To generate the Hprt-targeting vector, pGL3-Robo4 was digested with BamHII and NheI. The fragment containing the 0.3-kb promoter was purified and cloned into the pSDK-lacZ vector containing lacZ cDNA to generate pRobo4-Del5-lacZ. pRobo4-Del5-lacZ was then digested with PmeI and NotI. The resulting transgenic cassette was purified and cloned into the Hprt-targeting vector, pMP8II.
Transient transfection assay using methylated plasmid
The Robo4 promoter-reporter construct contains many CpG sites in the promoter, the luciferase reporter, and in the vector backbone. To assess the role of Robo4 proximal promoter-specific methylation in the absence of both reporter and vector backbone methylation, a non-methylated Robo4-promoter-reporter construct (NM-pGL3-Robo4) was prepared by amplifying pGL3-Robo4 in Dam and Dcm methylase deficient E. coli strain SCS110 (Agilent Technologies, Santa Clara, CA). To prepare the methylated pGL3-Robo4 (M-pGL3-Robo4) in which the CpG sites in the proximal promoter were methylated, NM-pGL3-Robo4 was digested with StuI and XhoI. The resulting short promoter fragment was methylated by SssI (New England Biolabs, Ipswitch, MA) and cloned into the StuI-XhoI site of NM-pGL3-Robo4 to generate M-pGL3-Robo4. The NM-pGL3-Robo4, M-pGL3-Robo4, and pGL3 as a control were transfected into HCAEC, and promoter activities were evaluated as described previously.1 Data was collected from 5 independent assays.
Generation and analysis of Hprt-targeted mice
Generation of Hprt-targeted mice containing the wild type Robo4 promoter-lacZ was described previously. 1 To generate ES cells containing the 0.3-kb truncated Robo4 promoter (Del)-lacZ, the targeting vector was linearized by digesting with SalI, and electroporated into Hprt-deficient BK4 ES cells. Homologous recombinants were selected in ES cell culture medium containing HAT (Sigma-Aldrich). The targeted ES cell clones were used for generating Robo4 promoter-lacZ chimeric mice. Chimeric males were bred to C57BL/6 females to obtain agouti offspring. Mouse lines were generated from two independent ES clones. Organs from the generated mice were fixed with the PBS containing 2% formaldehyde and 0.2% glutaraldehyde, and stained with the PBS containing 0.02% NP-40, 0.01% SDS, 2 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 1% X-gal at 37 ºC for 20 h.
DNase hypersensitivity assay
DNase hypersensitivity assays were performed as described previously.2 HCAEC, HCASmC, and HEK293 cells were lysed on ice in Lysis buffer (10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 0.5% Nonidet P-40, 150 μM spermine, 500 μM spermidine) and centrifuged. The pelleted nuclei were washed with Wash buffer (10 mM Tris-HCl (pH 7.4), 15 mM NaCl, 60 mM KCl, 150 μM spermine, 150 μM spermidine) and resuspended in Digestion buffer (wash buffer containing 1 mM CaCl2). The resulting suspension was digested with or without 7 units of micrococcal nuclease (TAKARA, Shiga, Japan) for 5 min at 25°C. The reaction was stopped by adding Stop buffer (20 mM EDTA, 2 mM EGTA, 1% SDS) followed by incubation on ice for 10 min. Proteins in the resulting samples were digested by proteinase K. The DNA was extracted by phenol-chloroform extraction and ethanol precipitation, and resuspended in TE (pH 8.0). Real-time PCR analyses were performed in triplicate with the resulting DNA samples and region-specific primers (sequences are shown in Table SI). Copy numbers of each promoter fragment was calculated from the standard curve prepared by measuring serial dilutions of pGL3-Robo4. The percentage of accessibility was calculated by comparing the copy numbers of promoter fragments in the digested and undigested DNA samples.
In vitro ES cell differentiation
Generation of Hprt locus-targeted ES cells (Robo4 promoter-lacZ and Robo4(ETSmut)-lacZ) were described previously.1, 3 The ES cells containing the Robo4 promoter with SP1 double mutation were generated using the plasmid pGL3-SP1(1,2)mut1 and the same method used for Robo4(ETSmut)-lacZ as previously described.3 To prepare Flk-1+ or ECs, these targeted ES cells were seeded onto OP9 cells and cultured for 5 days in αMEM supplemented with 20% FBS, 0.1 mM non-essential amino acids, 2 mM L-glutamine, 100 IU/ml penicillin, and 100 μg/ml streptomycin. Flk-1+ cells were purified from the differentiated cells by MACS using an anti-mouse Flk1 antibody (BD Pharmingen, San Diego, CA). The resulting Flk1+ cells were seeded on collagen IV-coated plates (Becton Dickinson, Franklin Lakes, NJ) and cultured for 3 days in αMEM supplemented with 50 ng/ml human VEGF165 (R&D systems, Minneapolis, MN), 0.5 mM 8-bromo cAMP (Nacalai Tesque, Kyoto, Japan), 10% FBS, and 50 μM 2-mercaptoethanol. CD31+ ECs were purified by MACS using an anti-mouse CD31 antibody (BD Pharmingen). Undifferentiated ES cells, Flk-1+ cells, and CD31+ cells were used for the bisulfite sequencing analysis,
Supplementary Material
Significance.
Cell-type–specific transcription factors or tissue-specific combinations of noncell-type–specific transcription factors are thought to regulate cell-type–specific gene expression. Although the regulation of various endothelial cell (EC)–specific genes has been studied, the transcription factors and their combinations that regulate EC-specific gene expression have not been fully identified. To identify the transcription factors that regulate EC-specific gene expression, we previously investigated the regulation of the Robo4 gene and identified SP1 and GA-binding protein as essential regulators for Robo4 promoter activation. However, because these factors are known to be expressed in other tissues, we could not explain the mechanism that induces Robo4 gene expression only in ECs. In this study, we hypothesized that cell-type–specific gene expression was regulated by epigenetics, as well as transcription factors, and succeeded in demonstrating the importance of DNA methylation for EC-specific Robo4 gene expression.
Acknowledgments
We thank Dr Naoki Mochizuki for his technical support and helpful suggestions. We also thank Dr Sarah Bronson for gifting BK4 cells.
Sources of Funding
This work was supported by MEXT KAKENHI, JSPS KAKENHI, a Health and Labor Sciences Research Grant from the Ministry of Health, Labor, and Welfare of Japan, Takeda Science Foundation, the Uehara Memorial Foundation, Senri Life Science Foundation, Suzuken Memorial Foundation, and Daiichi-Sankyo Foundation of Life Science.
Nonstandard Abbreviations and Acronyms
- EC
endothelial cell
- GABP
GA-binding protein
- HCAEC
human coronary artery endothelial cells
- HCASmC
human coronary artery smooth muscle cells
- HEK
human embryonic kidney
- NHDF
normal human dermal fibroblasts
- Robo4
roundabout4
Footnotes
Disclosures
None.
References
- 1.Huminiecki L, Gorn M, Suchting S, Poulsom R, Bicknell R. Magic roundabout is a new member of the roundabout receptor family that is endothelial specific and expressed at sites of active angiogenesis. Genomics. 2002;79:547–552. doi: 10.1006/geno.2002.6745. [DOI] [PubMed] [Google Scholar]
- 2.Huminiecki L, Bicknell R. In silico cloning of novel endothelial-specific genes. Genome Res. 2000;10:1796–1806. doi: 10.1101/gr.150700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park KW, Morrison CM, Sorensen LK, Jones CA, Rao Y, Chien CB, Wu JY, Urness LD, Li DY. Robo4 is a vascular-specific receptor that inhibits endothelial migration. Dev Biol. 2003;261:251–267. doi: 10.1016/s0012-1606(03)00258-6. [DOI] [PubMed] [Google Scholar]
- 4.Seth P, Lin Y, Hanai J, Shivalingappa V, Duyao MP, Sukhatme VP. Magic roundabout, a tumor endothelial marker: expression and signaling. Biochem Biophys Res Commun. 2005;332:533–541. doi: 10.1016/j.bbrc.2005.03.250. [DOI] [PubMed] [Google Scholar]
- 5.Wang B, Xiao Y, Ding BB, Zhang N, Yuan Xb, Gui L, Qian KX, Duan S, Chen Z, Rao Y, Geng JG. Induction of tumor angiogenesis by Slit-Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell. 2003;4:19–29. doi: 10.1016/s1535-6108(03)00164-8. [DOI] [PubMed] [Google Scholar]
- 6.Suchting S, Heal P, Tahtis K, Stewart LM, Bicknell R. Soluble Robo4 receptor inhibits in vivo angiogenesis and endothelial cell migration. FASEB J. 2005;19:121–123. doi: 10.1096/fj.04-1991fje. [DOI] [PubMed] [Google Scholar]
- 7.Bedell VM, Yeo SY, Park KW, Chung J, Seth P, Shivalingappa V, Zhao J, Obara T, Sukhatme VP, Drummond IA, Li DY, Ramchandran R. round-about4 is essential for angiogenesis in vivo. Proc Natl Acad Sci U S A. 2005;102:6373–6378. doi: 10.1073/pnas.0408318102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones CA, London NR, Chen H, et al. Robo4 stabilizes the vascular network by inhibiting pathologic angiogenesis and endothelial hyperpermeability. Nat Med. 2008;14:448–453. doi: 10.1038/nm1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mura M, Swain RK, Zhuang X, et al. Identification and angiogenic role of the novel tumor endothelial marker CLEC14A. Oncogene. 2012;31:293–305. doi: 10.1038/onc.2011.233. [DOI] [PubMed] [Google Scholar]
- 10.Okada Y, Yano K, Jin E, Funahashi N, Kitayama M, Doi T, Spokes K, Beeler DL, Shih SC, Okada H, Danilov TA, Maynard E, Minami T, Oettgen P, Aird WC. A three-kilobase fragment of the human Robo4 promoter directs cell type-specific expression in endothelium. Circ Res. 2007;100:1712–1722. doi: 10.1161/01.RES.0000269779.10644.dc. [DOI] [PubMed] [Google Scholar]
- 11.Okada Y, Jin E, Nikolova-Krstevski V, Yano K, Liu J, Beeler D, Spokes K, Kitayama M, Funahashi N, Doi T, Janes L, Minami T, Oettgen P, Aird WC. A GABP-binding element in the Robo4 promoter is necessary for endothelial expression in vivo. Blood. 2008;112:2336–2339. doi: 10.1182/blood-2008-01-135079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minami T, Aird WC. Endothelial cell gene regulation. Trends Cardiovasc Med. 2005;15:174–184. doi: 10.1016/j.tcm.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirodkar AV, St Bernard R, Gavryushova A, Kop A, Knight BJ, Yan MS, Man HS, Sud M, Hebbel RP, Oettgen P, Aird WC, Marsden PA. A mechanistic role for DNA methylation in endothelial cell (EC)-enriched gene expression: relationship with DNA replication timing. Blood. 2013;121:3531–3540. doi: 10.1182/blood-2013-01-479170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan MS, Matouk CC, Marsden PA. Epigenetics of the vascular endothelium. J Appl Physiol (1985) 2010;109:916–926. doi: 10.1152/japplphysiol.00131.2010. [DOI] [PubMed] [Google Scholar]
- 16.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 17.Chan Y, Fish JE, D’Abreo C, Lin S, Robb GB, Teichert AM, Karantzoulis-Fegaras F, Keightley A, Steer BM, Marsden PA. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J Biol Chem. 2004;279:35087–35100. doi: 10.1074/jbc.M405063200. [DOI] [PubMed] [Google Scholar]
- 18.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wenger RH, Kvietikova I, Rolfs A, Camenisch G, Gassmann M. Oxygen-regulated erythropoietin gene expression is dependent on a CpG methylation-free hypoxia-inducible factor-1 DNA-binding site. Eur J Biochem. 1998;253:771–777. doi: 10.1046/j.1432-1327.1998.2530771.x. [DOI] [PubMed] [Google Scholar]
- 20.Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
REFERENCES
- 1.Okada Y, Yano K, Jin E, Funahashi N, Kitayama M, Doi T, Spokes K, Beeler DL, Shih SC, Okada H, Danilov TA, Maynard E, Minami T, Oettgen P, Aird WC. A three-kilobase fragment of the human robo4 promoter directs cell type-specific expression in endothelium. Circ Res. 2007;100:1712–1722. doi: 10.1161/01.RES.0000269779.10644.dc. [DOI] [PubMed] [Google Scholar]
- 2.Fish JE, Yan MS, Matouk CC, St Bernard R, Ho JJ, Gavryushova A, Srivastava D, Marsden PA. Hypoxic repression of endothelial nitric-oxide synthase transcription is coupled with eviction of promoter histones. J Biol Chem. 2010;285:810–826. doi: 10.1074/jbc.M109.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okada Y, Jin E, Nikolova-Krstevski V, Yano K, Liu J, Beeler D, Spokes K, Kitayama M, Funahashi N, Doi T, Janes L, Minami T, Oettgen P, Aird WC. A gabp-binding element in the robo4 promoter is necessary for endothelial expression in vivo. Blood. 2008;112:2336–2339. doi: 10.1182/blood-2008-01-135079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.