Abstract
Endothelial cells are highly sensitive to changes in the extracellular milieu. Sepsis results in activation of inflammatory and coagulation pathways. We hypothesized that sepsis-associated mediators may alter the response capacity (so-called “set point”) of endothelial cells. Human umbilical vein endothelial cells (HUVEC) were preincubated in the presence or absence of tumor necrosis factor (TNF)-α, lipopolysaccharide (LPS), hypoxia, hyperthermia, and/or high glucose; treated with or without thrombin for 4 h; and then processed for RNase protection assays of selected activation markers. Priming with TNF-α and LPS significantly inhibited thrombin-mediated induction of vascular cell adhesion molecule-1, intercellular adhesion molecule-1, tissue factor, and E-selectin, but not platelet-derived growth factor-A or CD44. In electrophoretic mobility shift assays, thrombin-treated HUVEC demonstrated inducible binding of p65 NF-κB, an effect that was significantly blunted by pretreatment of cells with TNF-α and LPS. Consistent with these results, TNF-α and LPS attenuated the effect of thrombin on IκB phosphorylation, total cytoplasmic IκB, and nuclear translocation of p65 NF-κB. The inhibitory effect of TNF-α on thrombin signaling persisted for up to 24 h following removal of the cytokine. Taken together, these data suggest that inflammatory mediators prime endothelial cells to modulate subsequent thrombin response.
Keywords: endothelium, NF-κB, sepsis, thrombin, HUVEC
The endothelium participates in many homeostatic functions, including the maintenance of blood fluidity, the regulation of leukocyte adhesion and transmigration, the production of inflammatory mediators, and the control of permeability (reviewed in ref 1).
Sepsis, which is defined as systemic inflammatory response to infection, involves the activation of both inflammatory and coagulation pathways. The host response is complex and involves a nonlinear array of cells (monocytes/macrophages, neutrophils, platelets, and endothelial cells) and soluble mediators (cytokines, chemokines, compliment, growth factors; changes in temperature, glucose, pH, and oxygen). Increasing evidence points to the endothelium as both victim and perpetrator of the sepsis cascade (reviewed in ref 2). For example, previous studies have demonstrated structural changes and apoptosis of endothelial cells in sepsis. Moreover, a common thread in successful treatments of severe sepsis is their capacity to attenuate endothelial cell dysfunction (3). Whether or not the endothelium represents a valuable therapeutic target will depend on an improved understanding of this organ in health and disease.
Many studies in endothelial cell biology focus on the effect of one or another extracellular mediator on unperturbed monolayers of cultured cells. As one example, thrombin has been shown to disrupt barrier function, alter cell shape, activate myriad signaling pathways and transcriptional networks, induce leukocyte adhesion, and shift endothelial-derived hemostatic balance (reviewed in ref 4). However, the extent to which these results may be extrapolated in vivo is not clear. In the context of the intact organism, the endothelium is exposed to many biomechanical and biochemical factors that vary in both space and time. In patients with severe sepsis, the endothelial microenvironment undergoes significant changes, including increased circulating levels of cytokines and chemokines, activation of serine proteases, hyperglycemia, hyperthermia, acidosis, and/or hypoxia.
Given the remarkable capacity of the endothelium to integrate extracellular signals, we hypothesized that a change in microenvironment—as occurs in sepsis—is likely to alter the cell’s subsequent capacity to respond to one or another agonist. To test this hypothesis, we exposed primary human endothelial cells to a variety of preconditioning regimens commonly associated with sepsis (including hyperthermia, hypoxia, high glucose, tumor necrosis factor [TNF]-α, and lipopolysaccharide [LPS]), and then incubated the cells with thrombin. Using gene transcription as our read out, we demonstrate that the net signal input is indeed an important determinant of the cell’s subsequent response.
MATERIALS AND METHODS
Cell culture
Human umbilical vein endothelial cells (HUVEC) and human coronary artery endothelial cells (HCAEC) were purchased from Clonetics (Hopkinton, MA) and grown in endothelial growth medium-2-MV (EGM-2-MV; Clonetics). After reaching confluence, cells were cultivated in serum-starved medium (EBM-2 containing 0.5% FBS) for 6 h, and exposed to one or more of the following conditions for 14 h: 1) 4.5 g/L (25 mM) glucose, 2) 39.5°C using temperaturecontrolled incubator, 3) 1% O2 using an airtight Plexiglass chamber with an atmosphere of 5% CO2/95% N2 at 37°C, 4) 10 µg/ml LPS (Sigma, St. Louis, MO), and/or 5) 20 ng/ml TNF-α (Invitrogen, Carlsbad, CA). Following preconditioning, the cells were incubated in fresh serum-starved medium in the presence or absence of 1.5 U/ml thrombin (Calbiochem, La Jolla, CA) for the times specified. In “washout” experiments, preconditioning medium (containing TNF-α) was replaced with fresh medium for 24 h before thrombin treatment. All experiments were carried out with cells at passage 6 or less.
RNase protection assays
Reverse transcriptase-polymerase chain reaction (RT-PCR) fragments were generated using SuperScript First-Strand Synthesis system (Invitrogen) from human genomic DNA solution (Roche, Basel, Switzerland) and subcloned into the pCRII vector (Invitrogen). After verification of the cDNA inserts, plasmids were amplified and purified using Qiagen plasmid Midi Kit (Qiagen, Valencia, CA). The plasmids were linearized and used as in vitro transcription templates. The cDNA plasmid template for β-actin was purchased from Ambion (Austin, TX). Following preconditioning and thrombin stimulation, cells were harvested for RNA using TRIzol reagent (Invitrogen). Total RNA was extracted according to the manufacturer’s instructions. α- [32P] UTP-labeled riboprobes were synthesized from the cDNA templates, using either T3 or T7 RNA polymerase (Ambion) and subsequently gel purified. RNase protection assays were carried out using the RPA III kit (Ambion) as described previously (5).
Quantitative real-time PCR
Total RNA was prepared using the RNeasy RNA extraction kit with DNase I treatment following the manufacturer’s instructions (Qiagen). To generate cDNA, total RNA (100 ng) from each of triplicate samples was mixed and converted into cDNA using random primers and SuperscriptIII reverse transcriptase (Invitrogen). All cDNA samples were aliquoted and stored at −80°C. Primers were designed using the Primer Express oligo design software (Applied BioSystems, Foster City, CA) and synthesized by Genemed Synthesis (South San Francisco, CA). All primer sets were subjected to rigorous database searches to identify potential conflicting transcript matches to pseudogenes or homologous domains within related genes. Amplicons generated from the primer set were analyzed for melting point temperatures using the first derivative primer melting curve software supplied by Applied BioSystems. The sequences of all real-time PCR primers used in this paper are listed in Table 1. The SYBR Green I assay and the ABI Prism 7700 Sequence Detection System were used for detecting real-time PCR products from the reverse-transcribed cDNA samples, as described previously (6). β-Actin, which exhibits a constant expression level across all of the HUVEC samples in comparison with 18S rRNA, was used as the normalizer. PCR reactions for each sample were performed in duplicate, and copy numbers were measured as described previously (6). The level of target gene expression was normalized against the β-actin expression in each sample and the data presented as mRNA copies per 106 β-actin copies.
Table 1.
Real-time PCR primer sequences
| Gene Name | Real-time PCR Primer (5' to 3') | |
|---|---|---|
| Forward | Reverse | |
| CD44 | CGGAGAGGCCAGCAAGTC | ACTGGTCTGGAGTTTCTGACGAC |
| COX-2 | CCTGGCGCTCAGCCATAC | CCACACTCATACATACACCTCGGT |
| ICAM-1 | CTCCAATGTGCCAGGCTTG | CAGTGGGAAAGTGCCATCCT |
| PDGF-A | CACAGCATCCGGGACCTC | AGGCTGGTGTCCAAAGAATCC |
| E-Selectin | TTCCGGGAAAGATCAACATGA | CATTGAGCGTCCATCCTTCA |
| Tissue Factor | CAACAGACACAGAGTGTGACCTCA | AGGAGAAGACCCGTGCCAA |
| u-PA | GGAAAACCTCATCCTACACAAGGA | CGGATCTTCAGCAAGGCAAT |
| VCAM-1 | TTCCCTAGAGATCCAGAAATCGAG | CTTGCAGCTTACAGTGACAGAGC |
| β-Actin | CCAGGATCAAGGTCGGAAAG | GCATCGGACTTGCAGACCA |
Oligonucleotide microarray analysis of gene expression
The transcriptional profile of control or thrombin-treated HUVEC preconditioned with or without LPS or TNF-α was characterized by oligonucleotide microarray analysis of HUVEC by using the human U133A Affymetrix GeneChip, according to previously described protocols for total RNA extraction and purification, cDNA synthesis, in vitro transcription reaction for production of biotin-labeled cRNA, hybridization of cRNA with U133A Affymetrix gene chips, scanning of image output files, analysis of gene expression data, and hierarchical and functional clustering algorithms (7). The scanned array images were analyzed by dChip (8). In the dChip analysis, a smoothing spline normalization method was applied before obtaining model-based gene expression indices, also known as signal values. There were no outliers identified by dChip, so all samples were carried on for subsequent analysis. When comparing two (groups of) samples to identify the genes enriched in a given phenotype, we used the lower confidence bound (LCB) of the fold change (FC) between the experiment and the baseline. If the 90% LCB of the FC between the experiment and the baseline was >1.2, the corresponding gene was considered to be differentially expressed. An LCB >1.2 typically corresponds with an “actual” fold change of at least 3 in gene expression. GOTree Machine (GOTM) was used to identify Gene Ontology categories for the input gene set (9).
Electrophoretic mobility shift assays
Nuclear extracts were prepared using Nuclear Extract Kit from Active Motif (Carlsbad, CA). Double-stranded oligonucleotides spanning the tandem NF-κB sites in the VCAM-1 promoter (5′-TGCCCTGGGTTTCCCCTTGAAGGGATTTCCCTCCGCCTC-3′) were labeled with [α-32P] dCTP and Klenow fragment, purified by spun column (Amersham Pharmacia), and incubated with nuclear extracts as described previously (10). To test the effect of antibody on DNA-protein binding, nuclear extracts were preincubated with antibody to p65 (Santa Cruz Biotechnology, Santa Cruz, CA) for 30 min at room temperature. In competition experiments, nuclear extracts were preincubated with 50-fold molar excess of unlabeled wild-type or mutant oligonucleotides (5′-TGCCCTGCCTTTCCGGTTGAAGCCATTTCGGTCCGCC-3′) for 20 min at room temperature, and added to the reaction mixture as described previously (11).
Immunolocalization of p65 NF-κB
For immunolocalization studies, HUVEC or HCAEC were plated onto eight-well chamber slides (Lab-Tek, Christchurch, New Zealand) at a density of 20,000 cells/well. The cells were grown in EGM-2 MV medium for 48 h and fixed in 4% paraformaldehyde at room temperature for 10 min, washed with PBS, and subsequently incubated with primary anti-p65 antibody (Santa Cruz Biotechnology; 1: 100 dilution) for 30 min. Following extensive washes in PBS, the cells were incubated with a FITC-labeled secondary antibody (1:100 dilution) for 30 min. Following additional PBS washes, the slides were mounted in Prolong Gold antifade reagent (Molecular Probes, Eugene, OR) and viewed with confocal fluorescence microscopy. TO-PRO-3 (Molecular Probes) was used for nuclear colocalization.
Assays for total and phospho-IκBα
HUVEC were preconditioned with LPS or TNF-α and then treated with or without thrombin. After thrombin addition (time 0), cytoplasmic fraction was prepared at 15, 30, 45, 60, 120, 180 and 240 min using protein extract kit from Active Motif. Western blots were carried out as described previously (12). In each lane of 10% gel, 120 µg of protein was applied. The specific antibodies against IκBα and phospho-IκBα were purchased from Cell Signaling (Beverly, MA). Western blots were carried out in triplicate, using independent preparations of cytoplasmic extracts. The signals were quantified with NIH Image, and statistical analyses were carried out using Student’s t test.
RESULTS
The effect of preconditioning on thrombin-mediated gene expression in HUVEC
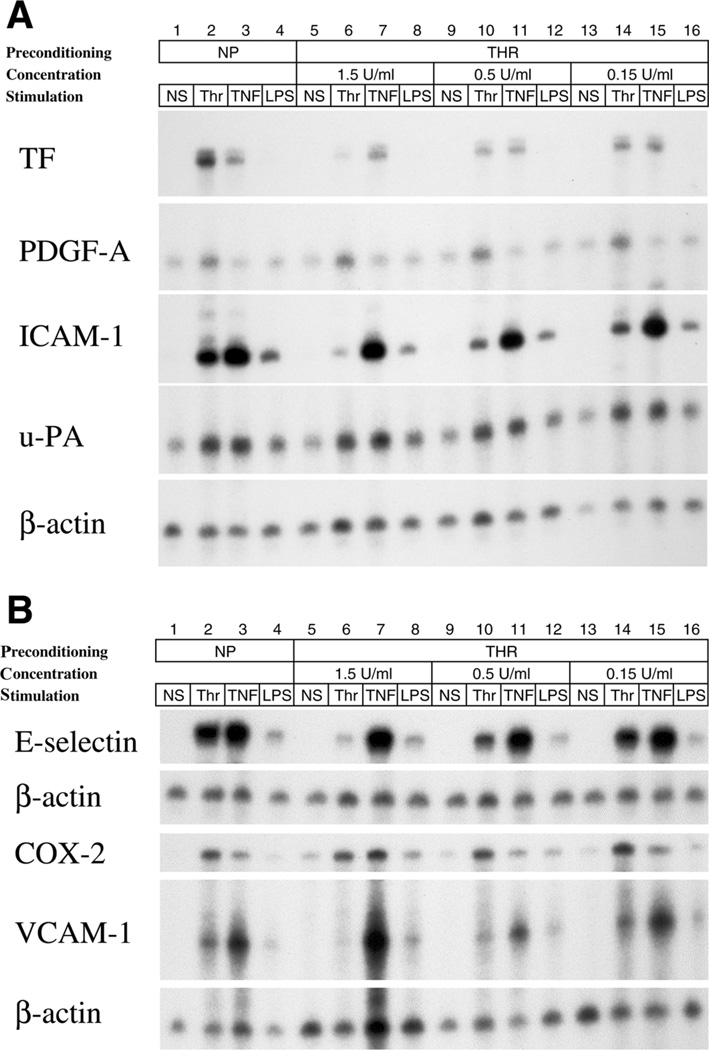
To determine the effect of different sepsis-like preconditioning regimes on thrombin-mediated gene expression, we pretreated HUVEC for 14 h in the presence or absence of high glucose, TNF-α, and/or LPS, and then treated with or without 1.5 U/ml thrombin for 4 h. Alternatively, HUVEC were grown under hypoxic or hyperthermic conditions before thrombin treatment. Cells were harvested for RNA and assayed for gene expression by RNase protection assay. In the absence of preconditioning, thrombin induced the expression of TF, ICAM-1, u-PA, COX-2, E-selectin, VCAM-1, and PDGF-A, but not CD44 (Fig. 1A, 1B; for quantitation, see Supplemental Fig. 1). In the absence of thrombin treatment, preconditioning with TNF-α, and to a lesser extent LPS, increased basal expression of several target genes, including ICAM-1, u-PA, CD44, and COX-2. The effect of LPS was accentuated by concomitant hypoxia.
Figure 1.
The effect of preconditioning on thrombin-mediated expression of candidate genes in endothelial cells. A, B) HUVEC were serum-starved for 6 h, preconditioned with high glucose, high temperature, low oxygen tension, TNF-α, and LPS alone or in combination for 14 h, and then treated with or without 1.5 U/ml thrombin for 4 h. RNase protection assays were carried out for expression of tissue factor (TF), platelet-derived growth factor (PDGF)-A, intercellular adhesion molecule (ICAM)-1, urokinase type plasminogen activator (u-PA), CD44, prostaglandin synthase 2 (COX-2), E-selectin, vascular cell adhesion molecule (VCAM)-1, and β-actin as described in Materials and Methods.
The various preconditioning regimens had different effects on thrombin-mediated gene expression (Fig. 1A, 1B; for quantitation, see Supplemental Fig. 1). Pretreatment with TNF-α or LPS significantly reduced thrombin stimulation of TF, ICAM-1, u-PA, E-selectin, and VCAM-1 but had little or no effect on PDGF-A. In contrast, high glucose, hyperthermia, or hypoxia alone had little or no effect on thrombin response. In cells exposed to a combination of high glucose and hypoxia, there was a trend toward super-induction of thrombin-responsive genes. Interestingly, the data suggest that TNF-α or LPS sensitizes CD44 and COX-2 to the effect of thrombin. In contrast to these sepsis-like regimens, the addition of vascular endothelial growth factor, hepatocyte growth factor, or insulin-like growth factor had no significant effect on thrombin-induced VCAM-1 expression (Supplemental Fig. 2).
To further quantify the above findings, we used a novel TaqMan-based protocol for calculating mRNA copy number per cell (Fig. 2A, 2B). Consistent with the results of RNase protection, TNF-α attenuated subsequent thrombin-mediated induction of TF (from 75-fold to 10-fold), ICAM-1 (from 22-fold to 1.3-fold), VCAM-1 (from 70-fold to 1.2-fold), E-selectin (from 142- fold to 2.9-fold), and u-PA (from 2.9-fold to 1.2-fold). TNF-α had no effect on thrombin stimulation of CD44 and PDGF-A. In contrast to the results of RNase protection assays, TNF-α preconditioning did inhibit thrombin induction of COX-2 (from 10.6-fold to 5-fold). Finally, the same pattern of inhibition was observed with TNF-α doses as low as 0.2 ng/ml (data not shown).
Figure 2.
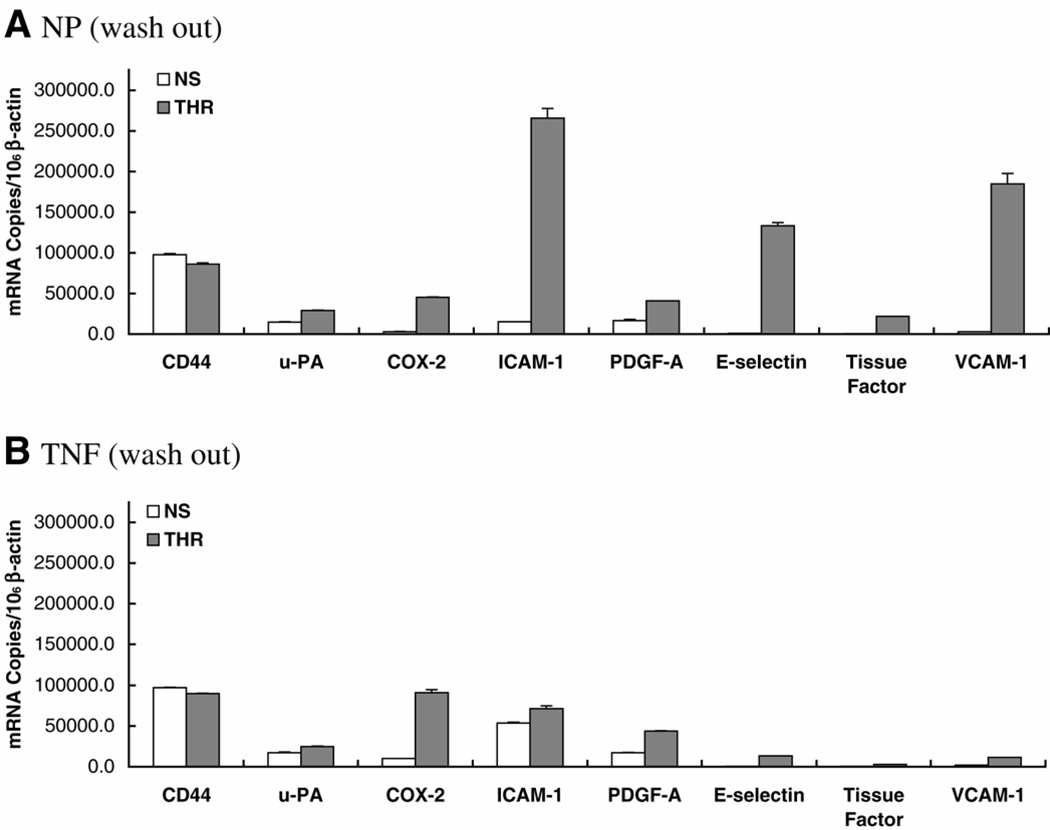
Validation of RNase protection assays using quantitative real-time PCR. Real-time PCR products were detected from the reverse-transcribed cDNA samples using SYBR Green I assay and the ABI Prism 7700 Sequence Detection System. The level of target gene expression was normalized against the β-actin expression in each sample, and the data are presented as mRNA copies per 106 β-actin copies. A) Expression levels in nonpreconditioned HUVEC treated in the absence (NS) or presence (THR) of thrombin for 4 h. B) Expression levels in HUVEC preconditioned with TNF-α for 14 h and treated in the absence (NS) or presence (THR) of thrombin for 4 h. NS, no stimulation; THR, thrombin stimulation; NP, no preconditioning.
DNA microarray analyses of thrombin-responsive gene expression in TNF-α- or LPS-preconditioned HUVEC
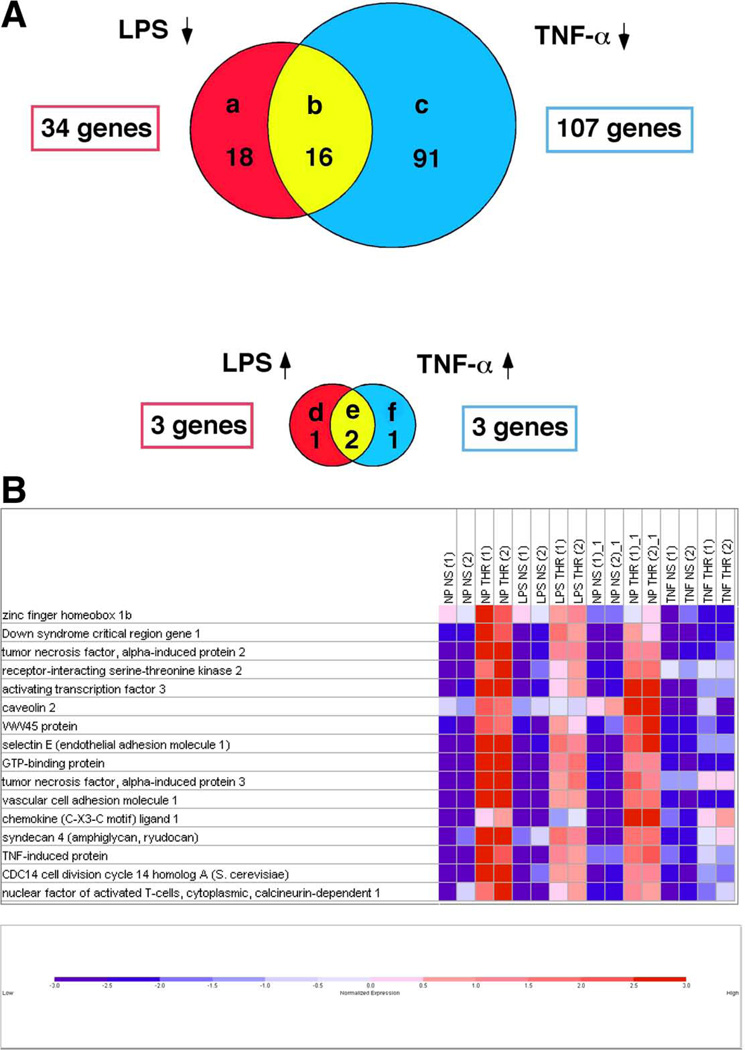
Next, we used DNA microarrays to conduct a global survey of thrombin-responsive genes in preconditioned endothelial cells. Pretreatment with TNF-α alone for 14 h resulted in the induction of 612 genes and repression of 565 transcripts. Pretreatment with LPS alone for 14 h resulted in induction and suppression of 70 and 48 genes, respectively. In the absence of preconditioning, thrombin treatment for 2 h resulted in induction and suppression of 323 and 61 genes, respectively. These latter results are consistent with recent published data from our group (13). Pretreatment of HUVEC with TNF-α or LPS had a significant impact on subsequent thrombin response. Specifically, TNF-α inhibited thrombin-mediated induction of 105 genes (Fig. 3A, group b and c), and resulted in super-induction of 3 genes (Fig. 3A, group e and f) (Supplemental Fig. 3). The remainder of the thrombin responsive genes were unaffected by TNF-α preconditioning (215 genes out of 323 genes). LPS inhibited thrombin-mediated induction of 34 genes (Fig. 3A, group a and b), and resulted in super-induction of 3 thrombin-responsive genes (Fig. 3A, group d and e) (Supplemental Fig. 3). The remainder of the thrombin-responsive genes was unchanged by LPS preconditioning (286 out of 323 genes).
Figure 3.
DNA microarray analyses of thrombin-responsive gene expression in TNF-α or LPS preconditioned endothelial cells. A) Venn diagram of thrombin-stimulated genes whose induction was inhibited by preconditioning with LPS (a), TNF-α (c), or both (b); or up-regulated by preconditioning with LPS (d), TNF-α (f), or both (e). B) Colormap of signal values for the genes in group B. Gene names and Affymetrix IDs are shown next to each row. For each gene, red and blue indicate high and low expression, respectively. For visual purposes, the signal values are normalized to the range [−3,3].
TNF-α and LPS inhibited thrombin stimulation of 16 common genes (Fig. 3A, group b; Fig. 3B). Among these genes are Down syndrome critical region (DSCR)-1, which we recently reported to be a thrombin-responsive autoinhibitory factor (14), numerous genes involved TNF-α signaling, syndecan 4, and caveolin 2. Consistent with the results of the RNase protection assays, TNF-α and LPS inhibited thrombin stimulation of E-selectin, ICAM-1, and VCAM-1, but not PDGF-A or COX-2.
TNF-α and LPS inhibit DNA binding of p65 NF-κB
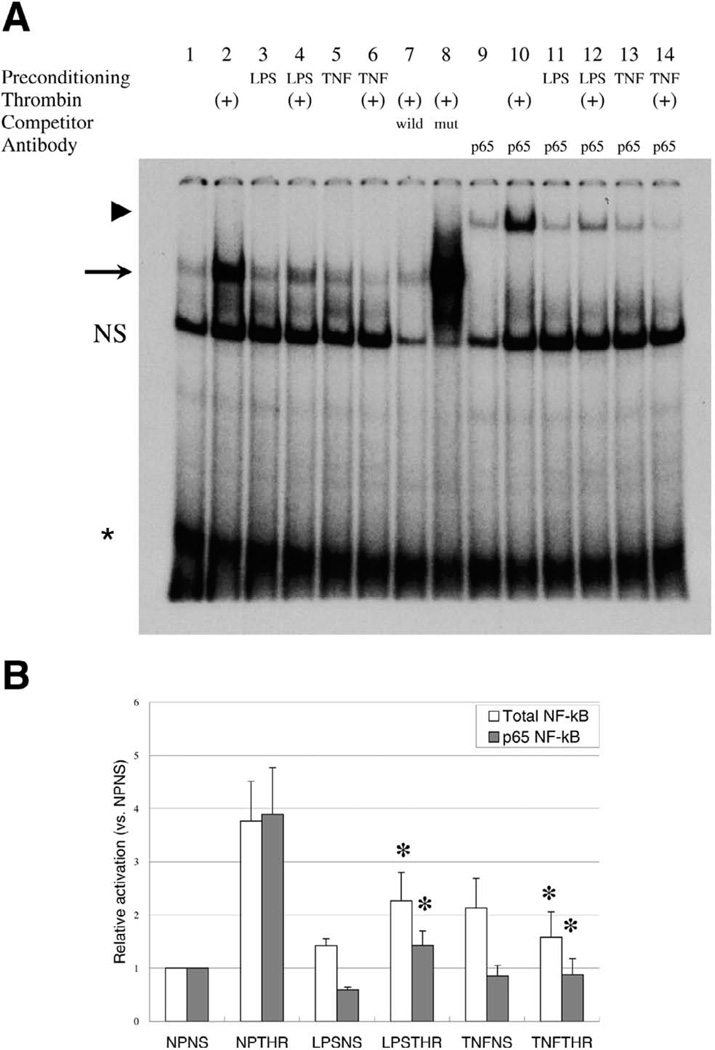
We were interested in determining the mechanism(s) by which pretreatment of HUVEC with TNF-α or LPS altered subsequent thrombin response. NF-κB has been previously implicated in the transcriptional regulation of many of the thrombin-responsive genes that were attenuated by preconditioning with TNF-α or LPS. In duplicate DNA microarray studies, we found that overexpression of a degradation-resistant constitutively active IκBα (IκB S32A/S36A) inhibited thrombin-mediated induction of ICAM-1, VCAM-1, E-selectin, and u-PA, but not CD44, COX-2, and PDGF-B (data not shown). Thus, we focused on the role for this transcription factor in mediating the inhibitory effect of preconditioning on thrombin response. To that end, EMSA were carried out using a radiolabeled double-stranded oligonucleotide probe containing the two adjacent VCAM-1 NF-κB consensus motifs, and nuclear extracts from HUVEC that had been pretreated with TNF-α or LPS and then incubated in the absence or presence of thrombin. In some cases, supershifting anti-p65 NF-κB antibodies were added to the reaction mixture to assay for p65 NF-κB binding. As shown in Fig. 4A and 4B, the addition of thrombin alone resulted in a fourfold increase in total NF-κB and p65 NF-κB binding. Preconditioning with TNF-α alone resulted in increased basal DNA binding of total NF-κB (2.3-fold) and p65 NF-κB (2.2-fold), whereas the effect of LPS was relatively small and limited to total NF-κB (1.3-fold) (Fig. 4A, 4B). More importantly, preconditioning with TNF-α and, to a lesser extent, LPS significantly inhibited thrombin-mediated induction of NF-κB binding. These data were confirmed using ELISA-based assays for p65 NF-κB binding activity (data not shown).
Figure 4.
TNF-α and LPS inhibit DNA binding of p65 NF-κB in endothelial cells. A) Representative electrophoretic mobility shift assay (EMSA) performed with 32 P-labeled NF-κB probe and 12 µg of nuclear extract from control (lanes 1, 3, 5, 9, 11, 13) or thrombin-treated (lanes 2, 4, 6–8, 10, 12, 14) HUVEC preconditioned in the absence (lanes 1, 2, 7, 8, 9, 10) or presence of LPS (lanes 3, 4, 11, and 12) and TNF-α (lanes 5, 6, 13, and 14). To identify specific DNA-protein complexes, nuclear extracts were incubated with wild-type probe (lane 7) or mutated probe (lane 8). The arrow indicates specific NF-κB DNA-protein complex. For supershift analysis, thrombin-treated HUVEC were incubated with antibody against p65 NF-κB (lanes 9–14). The arrowhead indicates the super-shifted complex. B) Quantification of total NF-κB and p65 NF-κB binding. The results show mean ± SE of the specific signal, relative to nonpreconditioned and nonstimulated (NPNS), obtained from three independent experiments. *P < 0.05. NP; no preconditioning; NS, no thrombin stimulation; LPS, LPS preconditioning; TNF, TNF-α preconditioning; THR; thrombin stimulation.
TNF-α inhibits thrombin stimulation of phospho-IκB and nuclear translocation of p65 NF-κB
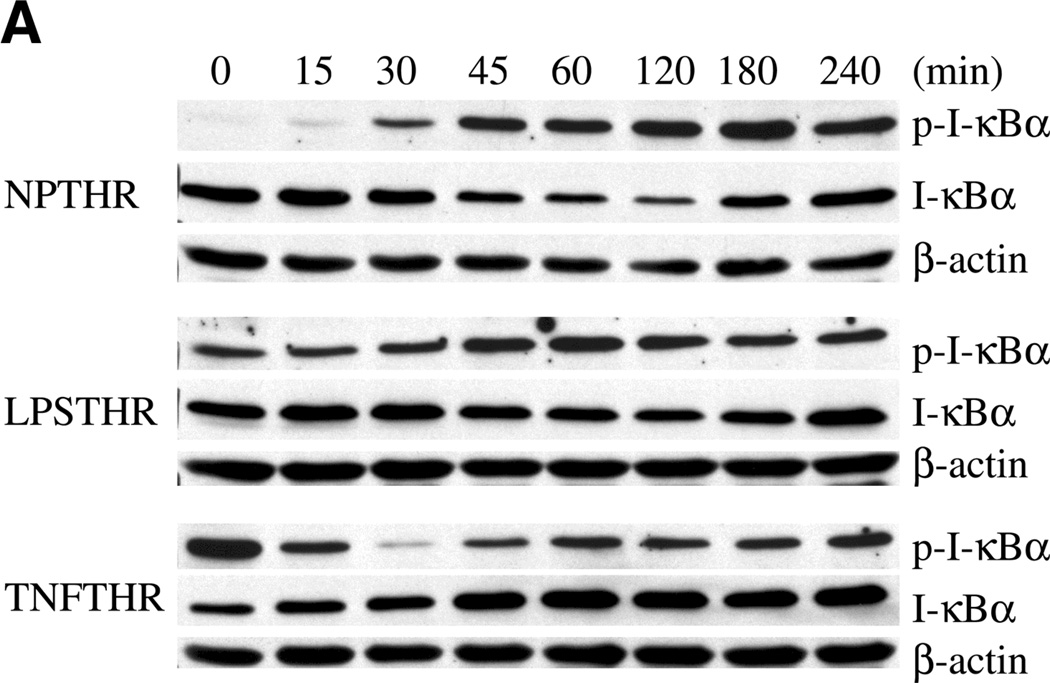
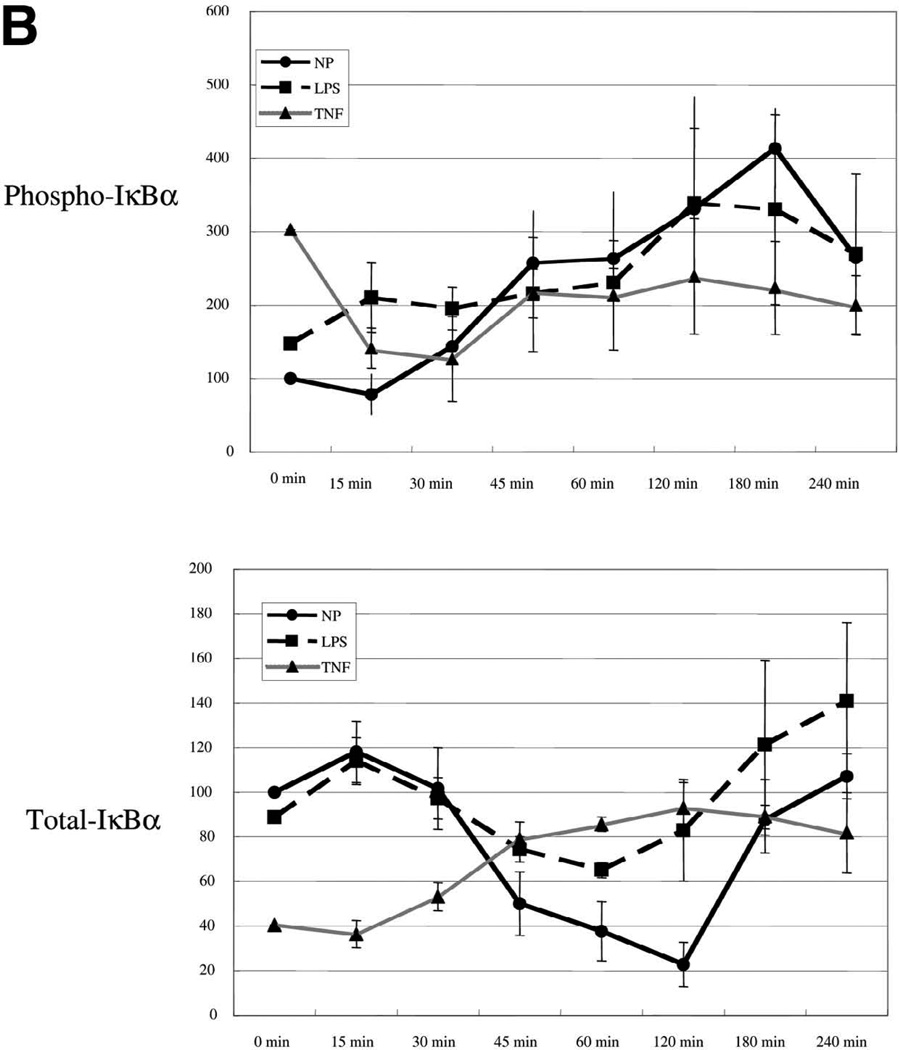
We next wished to study the effect of TNF-α and LPS preconditioning on the IκB-NF-κB signaling axis. Exposure of HUVEC to thrombin alone resulted in time-dependent increase in IκB phosphorylation and reduction in total cytoplasmic IκB, with maximal changes occurring at 180 min and 120 min, respectively (Fig. 5A, 5B). Pretreatment with TNF-α (and to a lesser extent LPS) resulted in increased basal levels of phospho-IκB. Importantly, both agonists blocked the effect of thrombin on total and phospho-IκB. In the case of TNF-α preconditioning, levels of phospho-IκB actually decreased in the thrombin-treated cells, an effect that is likely explained by the removal of TNF-α from the medium.
Figure 5.
TNF-α and LPS inhibit thrombin stimulation of phospho-IκB and p65 NF-κB in endothelial cells. A) Representative Western blot of HUVEC preconditioned with LPS or TNF-α, and then stimulated in the absence or presence of 1.5 U/ml thrombin. After thrombin addition (time 0), cytoplasmic fraction was prepared at 15, 30, 45, 60, 120, 180, and 240 min, and Western blots were performed on the same membrane using specific antibodies against IκBα, phospho-IκBα, and β-actin. In each lane, 120 µg of protein was loaded onto a 10% gel. B) Relative density of both phospho-IκBα and total IκBα at time 0 was adjusted to 100, and relative values of all other bands are plotted accordingly. The data represent mean ± se from three independent experiments.
Based on the results of the EMSA and the Western blots, we predicted that preconditioning with TNF-α or LPS would inhibit nuclear translocation of p65 NF-κB. To test this hypothesis, we carried out immunofluorescent studies in primary endothelial cells. HUVEC and HCAEC treated with thrombin for 2 h demonstrated significant nuclear translocation of p65 NF-κB (compare Fig. 6B and 6C). In endothelial cells preconditioned with TNF-α for 14 h, p65 NF-κB was localized predominantly in the nucleus (compare Fig. 6A and 6G), whereas LPS-primed endothelial cells revealed partial nuclear translocation of p65 NF-κB (compare Fig. 6A and 6D). Following 14 h of preconditioning, TNF-α or LPS was removed and the cells were incubated for 2 h in fresh medium in the absence (control) or presence of thrombin. In the absence of thrombin, a majority of the p65 NF-κB was localized in the cytoplasm after 2 h (Fig. 6F and 6I). A similar pattern of cytoplasmic accumulation was observed in the presence of thrombin (Fig. 6E and 6H). These results suggest that preconditioning with TNF-α or LPS significantly blocks the capacity of thrombin to induce nuclear translocation of p65 NF-κB.
Figure 6.
TNF-α and LPS inhibit thrombi-mediated nuclear translocation of p65 NF-κB in endothelial cells. HCAEC cells cultivated in an eight-well chamber slide were preconditioned for 14 h in the absence (A–C) or presence of TNF-α (D–F) or LPS (G–I). The preconditioning culture medium was then replaced with fresh medium in the absence (C, F, and I) or presence (B, E, and H) of thrombin for 2 h. The cells were fixed, washed, and incubated with monoclonal anti-p65 NF-κB antibody (green). Nuclei are stained by TO-PRO-3 (blue). Shown are the merged images; colocalization of p65 and nucleus causes bright blue color (B). NP; no preconditioning; NS, no thrombin stimulation; LPS, LPS preconditioning; TNF, TNF-α preconditioning; THR; thrombin stimulation. (Magnification ×60, bar=10 µm).
Effect of TNF-α and LPS preconditioning on thrombin signaling is sustained after removal of cytokine from the culture medium
Following a 24 h washout period, in which TNF-α-preconditioned cells were allowed to recover in fresh medium for 24 h before addition of thrombin, the copy number of each target gene approached baseline levels (e.g., in the absence of any preconditioning) (Fig. 7A, 7B). However, the attenuating effect of TNF-α on thrombin-mediated gene induction was still evident, with TF reduced from 65-fold to 6.3-fold; ICAM-1 from 17.6-fold to 1.3-fold; VCAM-1 from 69-fold to 5.8-fold; E-selectin from 164-fold to 29-fold; and u-PA from 2.0-fold to 1.4-fold (Fig. 7B). Induction of COX-2 was reduced from 15-fold to 9-fold. Again, TNF-α had no influence on thrombin’s effect on CD44 or PDGF-A. The inhibitory effect of TNF-α on thrombin-mediated NF-κB binding, IκB phosphorylation, and IκB degradation persisted following the 24 h washout period (data not shown).
Figure 7.
A, B) Effect of preconditioning on inducible gene expression persists following removal of TNF-α from culture medium. A) Same as in Fig. 2 except that preconditioned HUVEC were replaced with fresh medium for 24 h before thrombin treatment. NS, no stimulation; THR, thrombin stimulation; NP, no preconditioning.
Preconditioning with thrombin has little effect on LPS and TNF-α signaling
We next asked whether there is a reciprocal effect of thrombin preconditioning on LPS and TNF-α-mediated gene expression. To that end, HUVEC were preconditioned overnight with 1.5 U/ml thrombin and subsequently exposed to thrombin, LPS, or TNF-α. As shown in Fig. 8A and 8B, preconditioning with thrombin attenuated subsequent thrombin-mediated induction of TF, ICAM-1, E-selectin, and VCAM-1. However, no such effect was seen with PDGF-A, u-PA, or COX-2, arguing against simple consumption or desensitization of PAR-1 (for quantitation, see Supplemental Fig. 4). In contrast to the effects on subsequent thrombin response, preconditioning with thrombin had no effects on TNF-α- or LPS-mediated induction of TF, ICAM-1, E-selectin, or VCAM-1. These results suggest that a change in set point and its effect on subsequent signaling depends on the nature of the preconditioning agent, the agonist being tested and the identity of the target gene.
Figure 8.
Preconditioning with thrombin has little effect on LPS and TNF-α signaling in endothelial cells. A, B) HUVEC cells were serum starved for 6 h, preconditioned in the absence or presence of 1.5 U/ml thrombin for 14 h, and then treated with or without thrombin (1.5 U/ml), TNF-α (20 ng/ml), or LPS (10 µg/ml). RNase protection assays were carried out as in Fig. 1.
DISCUSSION
Thrombin is a multifunctional serine protease that activates protease-activated receptors (PAR) expressed on the surface of multiple cell types, including endothelial cells. PAR-1 is the predominant thrombin receptor on endothelial cells. Activation of PAR-1 leads to a series of posttranscriptional and transcriptional changes, which ultimately contribute to an alteration of structural and/or functional phenotype (reviewed in ref 4). Thrombin has been shown to induce the activity of many transcription factors, including NF-κB, AP1, Egr-1, and the forkhead family of proteins (4). These transcriptional networks, in turn, modulate the expression of many target genes, including cell adhesion molecules, procoagulants, and inflammatory mediators (4).
In the current study, we have shown that inflammatory mediators prime endothelial cells to modulate subsequent thrombin response. TNF-α and LPS alone had the greatest effect on thrombin-mediated gene regulation. An important finding was that for a given regimen, preconditioning inhibited thrombin response of some genes and accentuated the response of others. Moreover, each agonist demonstrated overlapping yet distinct effects on thrombin-responsive gene expression.
Our data point to the importance of NF-κB in mediating the inhibitory effects of preconditioning. We demonstrated that LPS and TNF-α each impair thrombin-mediated phosphorylation of IκB, degradation of cytoplasmic IκB, nuclear mobilization, and DNA binding of p65 NF-κB. The preconditioned endothelial cell is thus hyporesponsive to NF-κB-dependent thrombin signaling. It is interesting to speculate that the priming regimen directly interferes with IκB/NF-κB or with an upstream signal intermediate. One attractive hypothesis is that LPS and TNF-α deplete or inhibit PKC-δ, which is necessary for mediating thrombin stimulation of ICAM-1 and VCAM-1 (11, 15), and potentially other downstream target genes.
We cannot exclude a role for other signal intermediates and/or transcription factors in mediating the effects of preconditioning with inflammatory mediators. For example, preconditioning with TNF-α also inhibited thrombin induction of NF-AT and DSCR-1 (data not shown). Such an effect would be predicted to contribute to the attenuation of thrombin-induced expression of E-selectin, VCAM-1, ICAM-1, and TF. Still other mechanisms may be involved in mediating the positive effects of TNF-α on thrombin-induced COX-2 and CD44, as well as the positive effects of high glucose combined with hypoxia on several thrombin-responsive target genes.
In recent years, a number of other studies have demonstrated the potential for preconditioning regimens to sensitize or desensitize endothelial cells to other agonists. For example, the term “ischemic preconditioning” describes the cytoprotective effect of ischemia-reperfusion or hypoxia on tissues and/or endothelial cells (16). “Transplantation accommodation” occurs when xenoreactive or alloreactive antibodies induce resistance of the endothelium to humoral rejection (17). Repeated challenge of endothelial cells with LPS results in temporary insensitivity to subsequent LPS challenge, a process that has been termed “endotoxin tolerance.” Other preconditioning regimens that have been demonstrated to alter endothelial cell signaling include hypoxia (18), heat shock (19), and IGF-1 (20). Together with our data, these studies support the notion that the history of signal input is a critical determinant of subsequent cell behavior. This is likely to be true under both normal and pathophysiological conditions. At any given point in time, each endothelial cell in the human body has been preconditioned by its past exposure to diverse biomechanical and biochemical forces. The term “set point” refers to the functional sequelae of that input history.
Collectively, these data suggest that the endothelium may be therapeutically primed in such a way as to reduce subsequent activation. A total of five phase 3 clinical trials have demonstrated improved survival in critically ill patients or patients with severe sepsis. These include the use of low tidal volume ventilation (21), activated protein C (22), low dose glucocorticoids (23), intensive insulin therapy (24), and early goal-directed therapy (25). It is possible that each of these regimens exerts its benefit through a protective effect on the endothelium. For example, activated rhAPC may attenuate the response of endothelial cells to inflammatory mediators, early goal-directed therapy may result in favorable hemodynamics at the level of the endothelium, and intensive insulin therapy reverses any deleterious effect of hyperglycemia on endothelial responses. The link between therapeutic efficacy and alteration of endothelial set point is hypothetical and will require further study (26).
Finally, our results have important implications for studying endothelial cells in vitro. In most instances, cultured cells are incubated with agonist at time 0, and assayed at some later point for mRNA, protein, and/or function. Typically, the pretreatment conditions are standardized (e.g., serum-starved medium). We have shown that simple changes in the starting conditions result in complex shifts in subsequent behavior. Moreover, these changes may persist even after removal of the preconditioning regimen. Thus, when designing studies of cultured endothelial cells, it is important to take into consideration (and where necessary, modulate) the pretreatment conditions.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant HL 60585-02.
REFERENCES
- 1.Cines DB, Pollak ES, Buck CA, Loscalzo J, Zimmerman GA, McEver RP, Pober JS, Wick TM, Konkle BA, Schwartz BS. Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood. 1998;91:3527–3561. [PubMed] [Google Scholar]
- 2.Aird WC. The role of the endothelium in severe sepsis and multiple organ dysfunction syndrome. Blood. 2003;101:3765–3777. doi: 10.1182/blood-2002-06-1887. [DOI] [PubMed] [Google Scholar]
- 3.Aird WC. Natural anticoagulant inhibitors: activated Protein C. Best Pract. Res. Clin. Haematol. 2004;17:161–182. doi: 10.1016/j.beha.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Minami T, Sugiyama A, Wu SQ, Abid R, Kodama T, Aird WC. Thrombin and phenotypic modulation of the endothelium. Arterioscler. Thromb. Vasc. Biol. 2003;24:41–53. doi: 10.1161/01.ATV.0000099880.09014.7D. [DOI] [PubMed] [Google Scholar]
- 5.Wu SQ, Minami T, Donovan DJ, Aird WC. The proximal serum response element in the Egr-1 promoter mediates response to thrombin in primary human endothelial cells. Blood. 2002;100:4454–4461. doi: 10.1182/blood-2002-02-0415. [DOI] [PubMed] [Google Scholar]
- 6.Shih SC, Robinson GS, Perruzzi CA, Calvo A, Desai K, Green JE, Ali IU, Smith LE, Senger DR. Molecular profiling of angiogenesis markers. Am. J. Pathol. 2002;161:35–41. doi: 10.1016/S0002-9440(10)64154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitsiades N, Mitsiades CS, Poulaki V, Chauhan D, Fanourakis G, Gu X, Bailey C, Joseph M, Libermann TA, Treon SP. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc. Natl. Acad. Sci. USA. 2002;99:14,374–14,379. doi: 10.1073/pnas.202445099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barash Y, Dehan E, Krupsky M, Franklin W, Geraci M, Friedman N, Kaminski N. Comparative analysis of algorithms for signal quantitation from oligonucleotide microarrays. Bioinformatics. 2004;20:839–846. doi: 10.1093/bioinformatics/btg487. [DOI] [PubMed] [Google Scholar]
- 9.Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minami T, Aird WC. Thrombin stimulation of the vascular cell adhesion molecule-1 promoter in endothelial cells is mediated by tandem nuclear factor-kappa B and GATA motifs. J. Biol. Chem. 2001;276:47,632–47,641. doi: 10.1074/jbc.M108363200. [DOI] [PubMed] [Google Scholar]
- 11.Minami T, Abid MR, Zhang J, King G, Kodama T, Aird WC. Thrombin stimulation of vascular adhesion molecule-1 in endothelial cells is mediated by protein kinase C (PKC)-delta-NF-kappa B and PKC- zeta-GATA signaling pathways. J. Biol. Chem. 2003;278:6976–6984. doi: 10.1074/jbc.M208974200. [DOI] [PubMed] [Google Scholar]
- 12.Abid MR, Guo S, Minami T, Spokes KC, Ueki K, Skurk C, Walsh K, Aird WC. Vascular endothelial growth factor activates PI3K/Akt/forkhead signaling in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2004;24:294–300. doi: 10.1161/01.ATV.0000110502.10593.06. [DOI] [PubMed] [Google Scholar]
- 13.Minami T, Sugiyama A, Wu SQ, Abid R, Kodama T, Aird WC. Thrombin and phenotypic modulation of the endothelium. Arterioscler. Thromb. Vasc. Biol. 2004;24:41–53. doi: 10.1161/01.ATV.0000099880.09014.7D. [DOI] [PubMed] [Google Scholar]
- 14.Minami T, Horiuchi K, Miura M, Abid R, Takabe W, Kohro T, Ge X, Aburatani H, Hamakubo T, Kodama T, Aird WC. VEGF- and thrombin-induced termination factor, down syndrome critical region-1, attenuates endothelial cell proliferation, and angiogenesis. J. Biol. Chem. 2004;279:50,537–50,554. doi: 10.1074/jbc.M406454200. [DOI] [PubMed] [Google Scholar]
- 15.Rahman A, Anwar KN, Uddin S, Xu N, Ye RD, Platanias LC, Malik AB. Protein kinase C-delta regulates thrombin-induced ICAM-1 gene expression in endothelial cells via activation of p38 mitogen-activated protein kinase. Mol. Cell. Biol. 2001;21:5554–5565. doi: 10.1128/MCB.21.16.5554-5565.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad S, Ahmad A, Gerasimovskaya E, Stenmark KR, Allen CB, White CW. Hypoxia protects human lung microvascular endothelial and epithelial-like cells against oxygen toxicity: role of phosphatidylinositol 3-kinase. Am. J. Respir. Cell Mol. Biol. 2003;28:179–187. doi: 10.1165/rcmb.2002-0004OC. [DOI] [PubMed] [Google Scholar]
- 17.Delikouras A, Fairbanks LD, Simmonds AH, Lechler RI, Dorling A. Endothelial cell cytoprotection induced in vitro by allo- or xenoreactive antibodies is mediated by signaling through adenosine A2 receptors. Eur. J. Immunol. 2003;33:3127–3135. doi: 10.1002/eji.200323566. [DOI] [PubMed] [Google Scholar]
- 18.Ishida I, Kubo H, Suzuki S, Suzuki T, Akashi S, Inoue K, Maeda S, Kikuchi H, Sasaki H, Kondo T. Hypoxia diminishes toll-like receptor 4 expression through reactive oxygen species generated by mitochondria in endothelial cells. J. Immunol. 2002;169:2069–2075. doi: 10.4049/jimmunol.169.4.2069. [DOI] [PubMed] [Google Scholar]
- 19.Kohn G, Wong HR, Bshesh K, Zhao B, Vasi N, Denenberg A, Morris C, Stark J, Shanley TP. Heat shock inhibits tnf-induced ICAM-1 expression in human endothelial cells via I kappa kinase inhibition. Shock. 2002;17:91–97. doi: 10.1097/00024382-200202000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Che W, Lerner-Marmarosh N, Huang Q, Osawa M, Ohta S, Yoshizumi M, Glassman M, Lee JD, Yan C, Berk BC. Insulin-like growth factor-1 enhances inflammatory responses in endothelial cells: role of Gab1 and MEKK3 in TNF-alpha- induced c-Jun and NF-kappaB activation and adhesion molecule expression. Circ. Res. 2002;90:1222–1230. doi: 10.1161/01.res.0000021127.83364.7d. [DOI] [PubMed] [Google Scholar]
- 21.ARDSNET. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. The Acute Respiratory Distress Syndrome Network. N. Engl. J. Med. 2000;342:1301–1308. doi: 10.1056/NEJM200005043421801. [DOI] [PubMed] [Google Scholar]
- 22.Bernard GR, Ely EW, Wright TJ, Fraiz J, Stasek JE, Jr, Russell JA, Mayers I, Rosenfeld BA, Morris PE, Yan SB. Safety and dose relationship of recombinant human activated protein C for coagulopathy in severe sepsis. Crit. Care Med. 2001;29:2051–2059. doi: 10.1097/00003246-200111000-00003. [DOI] [PubMed] [Google Scholar]
- 23.Annane D, Sebille V, Charpentier C, Bollaert PE, Francois B, Korach JM, Capellier G, Cohen Y, Azoulay E, Troche G, et al. Effect of treatment with low doses of hydrocortisone and fludrocortisone on mortality in patients with septic shock. JAMA. 2002;288:862–871. doi: 10.1001/jama.288.7.862. [DOI] [PubMed] [Google Scholar]
- 24.van den Berghe G, Wouters P, Weekers F, Verwaest C, Bruyninckx F, Schetz M, Vlasselaers D, Ferdinande P, Lauwers P, Bouillon R. Intensive insulin therapy in the critically ill patients. N. Engl. J. Med. 2001;345:1359–1367. doi: 10.1056/NEJMoa011300. [DOI] [PubMed] [Google Scholar]
- 25.Rivers E, Nguyen B, Havstad S, Ressler J, Muzzin A, Knoblich B, Peterson E, Tomlanovich M. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N. Engl. J. Med. 2001;345:1368–1377. doi: 10.1056/NEJMoa010307. [DOI] [PubMed] [Google Scholar]
- 26.Aird WC. The role of the endothelium in severe sepsis and the multiple organ dysfunction syndrome. Blood. 2003;101:3765–3777. doi: 10.1182/blood-2002-06-1887. [DOI] [PubMed] [Google Scholar]