Abstract
High-throughput screening (HTS) is the primary driver to current drug discovery efforts. New therapeutic agents that enter the market are a direct reflection of the structurally simple compounds that make up screening libraries. Unlike medically relevant natural products (e.g., morphine), small molecules currently being screened have low fraction sp3 character and few, if any, stereogenic centers. Although simple compounds have been useful in drugging certain biological targets (e.g., protein kinases), more sophisticated targets (e.g., transcription factors) have largely evaded the discovery of new clinical agents from screening collections. Here, we describe a tryptoline ring distortion strategy that enabled the rapid synthesis of 70 complex and diverse compounds from yohimbine 1, an indole alkaloid. The compounds that were synthesized had architecturally complex and unique scaffolds, unlike 1 and other scaffolds. These compounds were subjected to phenotypic screens and reporter gene assays leading to the identification of new compounds that possess various biological activities, including: antiproliferative activities against cancer cells with functional hypoxia inducible factors, nitric oxide inhibition, inhibition and activation of the antioxidant response element (ARE). This tryptoline ring distortion strategy can begin to address diversity problems in our screening libraries while occupying biologically relevant chemical space in areas critical to human health.
Keywords: drug discovery, tryptoline, ring distortion, yohimbine, biological screening
Graphical abstract
Introduction
Through the 1980’s, natural products were the primary source of new drug molecules as efforts to discover new biologically active compounds involved screening collections of natural products or crude extracts.1 These were highly fruitful expeditions that gave us bioactive entities, such as morphine, vancomycin and taxol, which all possess exquisite biological activities due to their complex molecular structures interacting with their corresponding biological target in a highly specific and selective fashion. Due to advances in molecular biology, solid-phase synthesis and screening technologies, the 1990’s led to a paradigm shift from screening natural products/extracts in whole-cell (phenotype) screens to large collections of relatively simple organic compounds (containing low sp3-character and essentially no stereochemistry) and target-based, high throughput screens (HTS).2,3
HTS has become the primary driver in drug discovery and has led to several FDA-approved drugs, including: gefitinib, erlotinib, sorafenib, tipranavir, tolvaptan and lapatinib.2 Despite success in drugging certain protein targets (tyrosine kinases), current screening libraries have been ineffective at targeting more sophisticated biological targets (i.e., protein-protein interactions4–6, transcription factors7,8) and disease areas (i.e., antibiotic-resistant bacteria3). The inability of current screening libraries to produce compounds that effectively drug “undruggable” biological targets are attributed to an underrepresentation of chemical diversity, or chemical space.3,9–11 As such, complex and diverse small molecules are of significant interest to drug discovery.
Several elegant synthesis strategies have been developed to address chemical diversity deficiencies through the rapid synthesis of complex molecules for biological screening; however, the two most well-known approaches include: diversity-oriented synthesis9,12–17 (DOS; pioneered by Schreiber and co-workers) and biology-oriented synthesis18–23 (BIOS; led by Waldmann and co-workers). Additionally, alternative and elegant syntheses of natural product-inspired scaffolds has been an area of significant interest in recent years.24–26 These strategies are aimed at capturing various structural features of natural products (i.e., fused ring systems, stereochemical complexity) into small molecules that can be rapidly accessed for biological screening. DOS and BIOS have led to several interesting biological discoveries in diverse disease areas and are promising approaches to the identification of therapeutic leads and probe molecules to interrogate biological systems.20,22,23,27–29
An alternative “ring distortion” approach utilizes available natural products containing a complex and fused ring system as synthetic starting points. Through rapid synthetic sequences from the selected natural product, the strategic goal is to dramatically alter the complex ring system inherent to the natural product thus generating complex small molecules with unique and diverse molecular architectures.30 Ring distortion involves the altering of complex ring systems through various chemical reactions that enable ring cleavage, expansion, rearrangement and fusion.30–32 The Hergenrother lab has carried out ring distortion campaigns on gibberellic acid30, adrenosterone30, quinine30, abietic acid31 and pleuromutilin.32 These have been largely successful due to the utility of strategically useful ketone- and alkene-based chemistries in addition to complex molecular frameworks that enable dramatic ring rearrangement reactions for ring distortion.
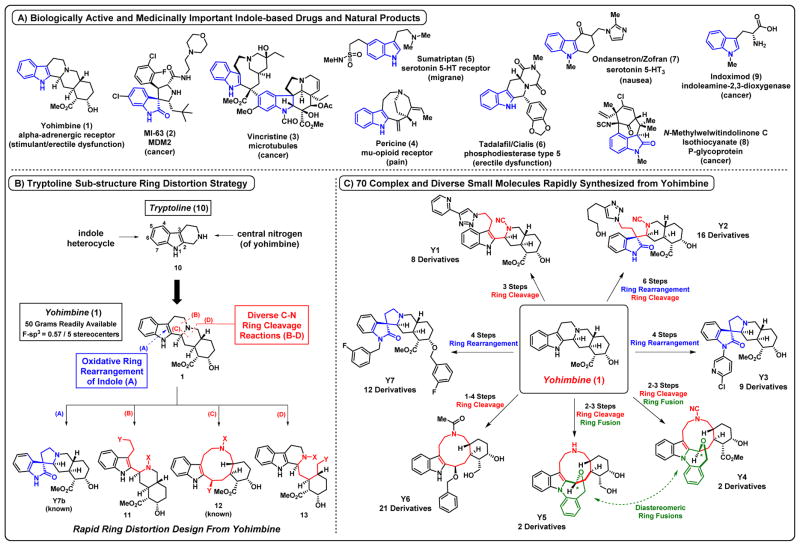
Indole-containing compounds span an array of chemical space from complex natural products to very simple compounds (Fig 1A, indoles 1–9). Interestingly, simple and complex indole-containing compounds demonstrate diverse biological activities due to the numerous biological targets that bind various indoles, making the indole nucleus a privileged scaffold that occupies an interesting and biologically relevant chemical space. Yohimbine 1 is a complex indole alkaloid isolated from the bark of Pausinystalia trees and the Rauwolfia root that acts as an α2-adrenergic receptor antagonist.33 This alkaloid is highly abundant and sold over-the-counter for its fat-burning/stimulatory and aphrodisiac properties. We became interested in 1 as a starting point for ring distortion due to its highly complex ring system fused to an indole nucleus. Multiple synthetic reactions of 1 as a starting material have been reported in the literature, including: oxidation of the secondary alcohol34, allylation of the 3-position of the indole nucleus35, alkylation of the indole nitrogen36, hydroxylation of the indole nitrogen37,38, reduction of the 2,3-positions of the indole heterocycle37,38, and ethylene glycol protection of the 2,3-positions of the indole nucleus.39 Our ring distortion strategy involved two known transfromations, including: cyanogen bromide ring cleavage with alcohols40 and transformation of 1 into a spirooxindole-containing scaffold.41 Yohimbine 1 provides an outstanding platform to develop an innovative ring distortion strategy involving the basic reactivity of the indole heterocycle to rapidly generate complex and diverse small molecules for biological screening in diverse disease areas (i.e., cancer, inflammation, drug-resistant bacteria).
Figure 1.
A) Biologically active indole-containing and indole-related molecules. B) Tryptoline ring distortion strategy applied to the indole alkaloid Yohimbine. C) Complex and diverse small molecules rapidly synthesized from Yohimbine.
Results and Discussion
Tryptoline-Based Ring Distortion Strategy
We devised a ring distortion strategy based on two structural features connected through the tryptoline 10 sub-structure of 1, which include: 1.) the centrally positioned basic nitrogen with three unequivocal C-N bonds (bonds B, C and D; Fig 1B), ideal for dramatic and diverse ring cleavage reactions to afford 11–13 and 2.) oxidative rearrangement of the 2,3-disubstituted indole nucleus to give ring-rearranged product Y7b (bond A; Fig 1B). Our goals were to develop short synthetic sequences that involve ring distortion reactions directly from 1 to enable optimal choreographing of ring cleavage, scrambling, fusion or rearrangement reactions in tandem with subsequent diversification reactions (i.e., click chemistry). This tryptoline ring distortion strategy was developed to generate several complex and diverse scaffolds totaling 70 small molecules in rapid synthetic sequences from 1 (Fig 1C).
Ring Distortion of the Indole Alkaloid Yohimbine
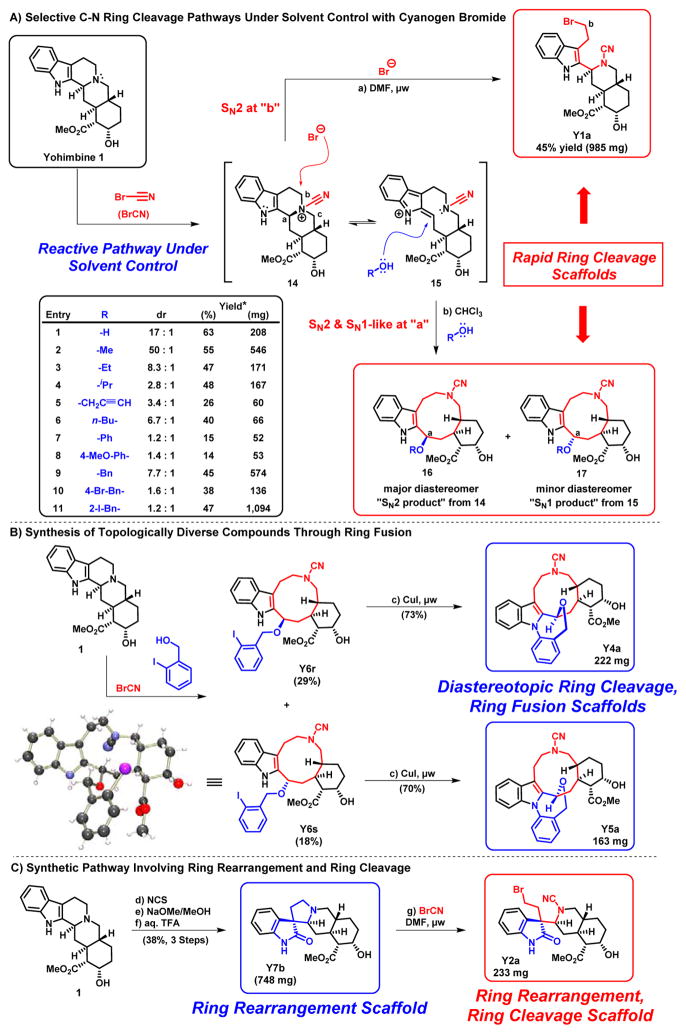
Initially, we targeted the C-N bonds of yohimbine’s central amine for regioselective ring cleavage reactions. Using cyanogen bromide (BrCN), we were able to activate the basic nitrogen of 1 to give intermediate 14 (Scheme 1), from which we generated numerous diverted ring-distorted compounds. Two of the unequivocal C-N bonds of 1 could be selectively cleaved with cyanogen bromide through careful selection of solvent.
Scheme 1.
Summary of ring distortion from 1. Reagents and Conditions: a) BrCN (3.0 eq.), DMF, μw, 100°C, 2.75 min, 45%; b) BrCN (3.0 eq.), ROH:CHCl3 (3:1), rt, 16 h, 14 – 63% (11 examples); c) Copper(I) Iodide (0.35 eq.), K2CO3 (1.5 eq.), DMEDA (0.7 eq.), MeCN, μw, 160 °C, 13 min, 70 – 73%; d) NCS (1.1 eq.), CH2Cl2, −41 °C, 0.75 h, crude; e) 0.5 M NaOMe/MeOH, rt, 2.5 h, 66%; f) 10% TFA (aq), reflux, 1 h, 58%; g) BrCN (3.0 eq.), DMF, rt to 60 °C, 22 h, 52%. Notes: DMEDA = N,N′-dimethylethylenediamine, NCS = N-chlorosuccinimide. *Indicates combined percent yield and isolated yields of ether diastereomers.
Cyanogen bromide C-N ring cleavage of 1 proceeds through an SN2 pathway in dimethylformamide (DMF; Scheme 1A). The liberated bromide anion generated upon formation of 14 attacks the most accessible “b” carbon adjacent to the activated central nitrogen of 14 in only 3 minutes using microwave irradiation resulting in Y1a as the only observed product. We were able to successfully carry this reaction out on a 985 milligram scale (45% yield), which was crucial for subsequent analogue synthesis.
Contrasting to the SN2 pathway, a 3:1 chloroform:alcohol solvent mixture results in an SN1-like pathway in which the alcohol is a nucleophile and attacks the “a” carbon of 14. This reaction has been previously described40; however, we extended this reaction by utilizing diverse alcohol solvent/nucleophiles, which react with intermediates 14 and 15 to yield diastereomeric mixtures of ethers 16 (“SN2 product”; inverted stereochemistry at “a”) and 17 (“SN1 product”; retention of stereochemistry at “a”). Diastereomers produced from this reaction can be readily separated via chromatography and tested in biological screens. Interestingly, less sterically encumbered alcohols (i.e., methanol) gave “SN2 products” (i.e., 16) as the major diastereomer (3.4:1 to 50:1 drs; Scheme 1A), which can be rationalized by considering that 14 and 15 can permit less hindered nucleophiles to undergo “back-side attack” at carbon “a” of structure 14. Alcohols with more steric bulk (i.e., 2-iodobenzyl alcohol) gave ~1:1 mixtures of diastereomeric products, which can be explained through an SN1-like mechanism involving nucleophilic attack of both faces on resonance-stabilized carbocation 15. This ring-cleavage reaction proceeded in 14–63% yield (11 examples yielded 19 ethers) and the major diastereomers were isolated on scales between 6.9 and 576 milligrams, which is sufficient for additional ring distortion reactions, analogue synthesis and biological screening.
We took advantage of the lack of diastereocontol of cyanogen bromide/alcohol ring cleavage to generate diverted ring fusion products Y4a and Y5a (Scheme 1B), which have unique complex molecular architectures. Yohimbine was initially subjected to cyanogen bromide ring cleavage in chloroform:2-iodobenzyl alcohol to give a 1.2:1 diastereomeric mixture of Y6r and Y6s (47% combined yield). The diastereomeric mixture was readily separated and each diastereomer was taken forward and subjected to copper(I) iodide-catalyzed intramolecular C-N coupling between the aryl iodide and indole nitrogen to give diverted ring fusion products Y4a (70%) and Y5a (73%) on 163–222 milligram scale.
Indole heterocycles can undergo oxidation with subsequent alkyl migration, translating to a ring rearrangement of 1. An oxidative rearrangement such as this allows us to take advantage of the intrinsic reactivity of the indole ring, which was a critical aspect of our tryptoline ring distortion strategy. A three-step, oxidative-rearrangement of yohimbine to afford ring-rearranged spirooxindole Y7b (Scheme 1C) was previously reported.41 Although initial attempts to repeat the literature chlorination reaction with hypochlorite failed in our hands, this was remedied by using N-chlorosuccinimide (NCS) to afford clean chlorination at the 3-position of the indole of 1. This was followed by treatment with sodium methoxide (NaOMe) to induce stereospecific alkyl migration and expulsion of the 3-chloro group. Final treatment with trifluoroacetic acid (TFA) afforded ring-rearranged product Y7b (38%/3 steps). We were able to generate 748 milligrams of Y7b in one run through this three step route during these investigations without focused scale-up efforts.
We subjected Y7b to cyanogen bromide ring cleavage in DMF to afford Y2a as a single product in 52% yield (233 milligrams; Scheme 1C; X-ray in Supporting Information). The propensity of bromide to attack the “b” carbon of Y7b can be rationalized by an SN2 attack of the least sterically encumbered carbon atom of the respective cyanamide intermediate (not shown). Despite many attempts, ring cleavage at the “D” bond (Fig 1B) of 1 was evaded; however, future developments are aimed to address this to generate interesting structures similar to 13.
The rapid, tryptoline-based ring distortion pathways developed during these investigations were highly efficient in the generation of several new scaffolds in 1 to 4 steps from 1. During these investigations, multiple C-N bonds to the central nitrogen atom of 1 were selectively cleaved with cyanogen bromide, an oxidative rearrangement of the indole heterocycle of 1 was utilized and C-N bond coupling reactions were used in ring fusion reactions to generate diverted complex scaffolds. Overall, this tryptoline ring-distortion platform enabled the rapid synthesis of novel complex and diverse chemotypes that are important to biological screening and drug discovery efforts.
Diversification of Ring Distorted Compounds
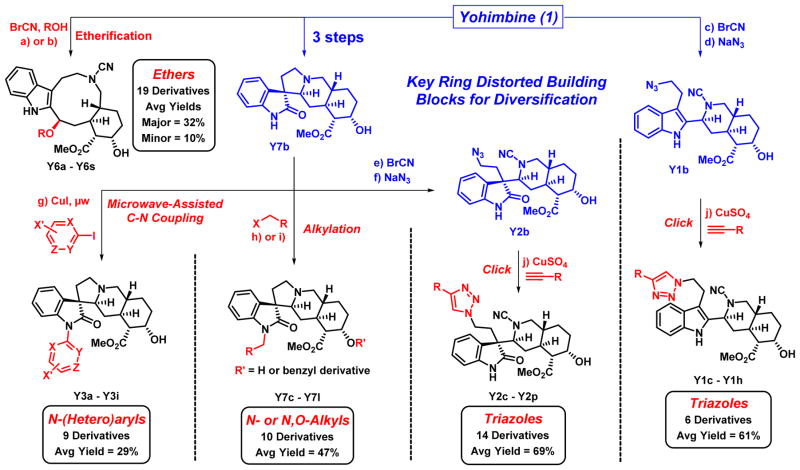
Following tryptoline ring distortion of 1, our goals were to decorate newly synthesized complex and diverse scaffolds using more traditional diversification strategies. This allows for the synthesis of sub-libraries for each scaffold enabling further exploration of chemical space during biological screening endeavors. Diversification of yohimbine-derived architectures found in Scheme 1 was ideal as these complex small molecules are endowed with multiple functional groups serving as potential synthetic handles for library generation.
Reactions that lead to ring distortion AND chemical diversification (sub-libraries) in the same step are highly useful in this approach
For example, subjecting 1 to cyanogen bromide and 11 different alcohols (including water; Scheme 1A) led to 19 total ring-cleaved ethers (Y6a-Y6s; Scheme 2). The library potential for this reaction was enhanced as two diastereomers are typically produced from this reaction. In 8 of the 11 examples, both diastereomers were isolated (pure) for biological screening and additional synthesis efforts.
Scheme 2.
Library synthesis to afford sub-libraries of yohimbine ring-distorted compounds. Reagents and Conditions: a) BrCN (3.0 eq.), THF:H2O (2.5:1), rt, 23 h, 57%; b) BrCN (3.0 eq), alcohol:CHCl3 (1:3), rt (or reflux for Y6j-Y6o), 6.5 h (major diastereomer yields: 7 – 55%; minor diastereomer yields: 7 – 21%); c) BrCN (3.0 eq.), DMF, μw, 100 °C, 2.75 min, 56%; d) NaN3 (3.0 eq.), DMF, rt to 75 °C, 22 h, 91%; e) BrCN (3.0 eq), DMF, rt to 60 °C, 22 h, 52%; f) NaN3 (3.0 eq), DMF, rt to 80 °C, 16.5 h, 99%; g) Aryl iodide (1.1 eq.), DMEDA (0.70 eq.), CuI (0.35 eq.), MeCN, μw, 160 °C, 13 min, 20 – 37%; h) alkyl halide (1.9 eq.), K2CO3 (3.8 eq.), DMF, rt, 15 h, 44 – 80%; i) NaH (4.3 eq.), THF, 0 °C; then benzyl bromide (2.0 eq.), THF, 0 °C to rt, 16 h, 29 – 66%; j) alkyne (3.0 eq.), anhydrous CuSO4 (0.5 eq.), sodium ascorbate (1.5 eq.), tBuOH:H2O (1:2), CH2Cl2, rt, 2 h, Y1 yields: 19 – 78%, Y2 yields: 50 – 78%. BrCN = cyanogen bromide, DMEDA = N,N′-dimethylethylenediamine.
Due to the synthetic utility of cyanogen bromide in ring cleavage reactions, multiple ring-cleaved compounds contained primary bromides (Y1a and Y2a) which were subjected to sodium azide displacement (SN2) to yield azides Y1b and Y2b, respectively (Scheme 2). Each azide was then subjected to copper(I)-catalyzed click reactions with a diverse panel of available terminal alkynes to generate two diverse triazole sub-libraries (Y1c-Y1h, 6 examples, 61% average yield; Y2c-Y2p, 14 examples, 69% average yield).
We found ring-rearranged Y7b to be very useful in diverted library synthesis due to the amide and hydroxyl functional groups readily available as synthetic handles. Ten mono- or dialkylated products of Y7b were synthesized with available alkyl halides (Y7c-Y7l, 47% average yield; Scheme 2). Y7b was also diversified using copper(I)-catalyzed C-N coupling between the amide nitrogen and nine diverse (hetero)aryl iodides (29% average yield).
Other synthetic modifications were made to yohimbine-derived small molecules, including: 1) lithium aluminum hydride reductions of cyanamide/ester functional groups and 2) acylation (Supporting Information). These efforts resulted in 70 complex and diverse compounds synthesized from yohimbine using rapid and diverted synthetic pathways. During these investigations, we were able to synthesize each of the analogues in these sub-libraries on a 6.0 to 576 milligram scale, which is sufficient for extensive biological investigations.
Biological Investigations of Yohimbine-Derived Small Molecules
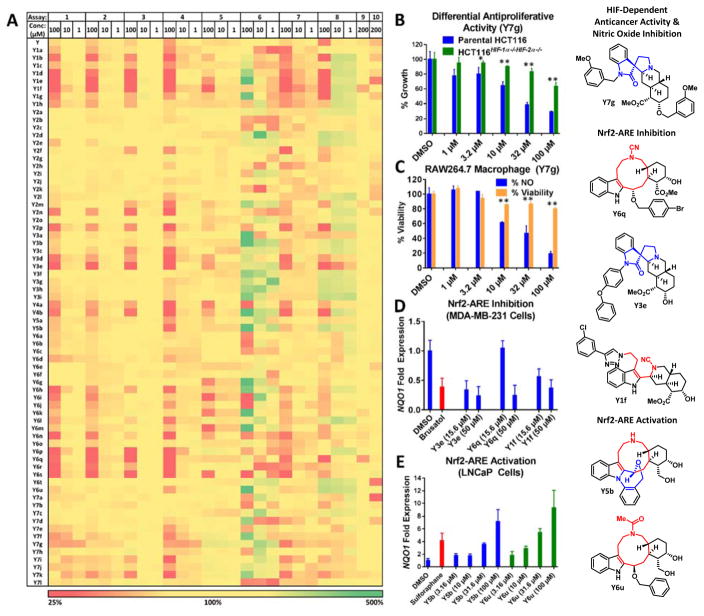
We screened our yohimbine ring distortion library in several phenotypic and focused pathway-specific screens related to cancer and inflammation as well as pathogenic bacteria (Fig 2A). Specifically, we aimed to identify selective antiproliferative agents that preferentially act on cancer cells with functional hypoxia-inducible factors (HIF) pathways. HIF transcription factor mediated signaling is known to be activated in cancer and linked to angiogenesis, cell growth, survival and metastasis. We employed isogenic HCT116 colorectal cancer cell lines, including parental HCT116 and HCT116 double knockout cells lacking HIF-1α and HIF-2α,42,43 and screened our library for differential antiproliferative activity. Of the 70 compounds tested, Y7g stood out as the most selective antiproliferative agent upon dose-response analysis (Fig 2B), suggesting that this compound could be a starting point for the development of selective and more potent HIF inhibitors. Chronic inflammation is a known factor in the etiology of colorectal cancer and, intriguingly, the same compound (Y7g) also showed the most selective anti-inflammatory activity in RAW264.7 macrophage cells based on the inhibition of nitric oxide (NO) production (Fig 2C). Cell viability was almost unaffected in this cell type, further supporting the cancer cell selectivity of Y7g.
Figure 2.
A) Heat map matrix summarizing primary biological screening results of the yohimbine ring distortion library (biological activity scale key: red = inhibition; yellow = no change; green = activation), including the structures of six validated hit compounds. Rows correspond to compounds tested. Columns correspond to primary biological activity screens (1–10) and concentrations tested (200, 100, 10 and 1 μM). 1) Parental HCT116 and 2) HCT116HIF-1α−/−HIF-2α−/− cell viability; 3) RAW264.7 cell viability; 4) RAW264.7 NO production; 5) LNCaP cell viability; 6) LNCaP cell ARE activity; 7) MDA-MB-231-ARE-luc cell viability; 8) MDA-MB-231-ARE-luc cell ARE activity; 9) S. aureus and 10) A. baumannii growth. B) E) Validation experiments of hit compounds. B) HIF-dependent anticancer activity. Cell viability of parental HCT116 and HCT116HIF-1α−/−HIF-2α−/− cells was determined after 48 h exposure using MTT assay (p-values: ≤ 0.01 **, ≤ 0.05 *; pairwise student t-test comparing cell viability between parental HCT116 and HCT116HIF-1α−/−HIF-2α−/− cells at a given concentration). C) NO production and cell viability. Production of NO and cell viability of RAW264.7 cells were determined after 24 h exposure using Griess reagent and MTT assay, respectively (p-values: ≤ 0.01 **, ≤ 0.05 *; pairwise student t-test comparing relative NO production and viability of RAW264.7 cells at a given concentration). D) MDA-231-ARE-luc cell ARE activity and E) LNCaP cell ARE activity. ARE inhibition in MDA-MB-231-ARE-luc and ARE activation in LNCaP cells after 24 h exposure were validated by effects on endogenous NQO1 transcript levels measured by RT-qPCR. ACTB expression was used as an internal control for normalization. All validation data are presented relative to vehicle control (0.5% DMSO). Brusatol (125 nM) and sulforaphane (10 μM) served as positive controls for MDA-MB-231-ARE-luc ARE inhibition and LNCaP ARE activation assays, respectively.
We then assessed the potential of the library compounds to modulate the activity of another key transcription factor, NF-E2-related factor 2 (Nrf2), which acts on the antioxidant response element (ARE) in the promoter region upstream of antioxidant and phase II detoxification enzymes. Activators of the Nrf2-ARE pathway have potential as cancer preventive agents because of their ability to increase antioxidant status of the cell and protect from oxidative damage that could lead to cancer. Inhibitors in turn may increase the susceptibility of cancer cells to chemotherapeutic agents and overcome drug resistance since Nrf2 is commonly activated in cancer.44 To screen for Nrf2 inhibitors, we used MDA-MB-231 breast cancer cells (a model for invasive, triple negative breast cancer) stably transfected with the ARE-luciferase reporter, where the promoter region is derived from the human NQO1 gene.45 As a control, we monitored cell viability to ensure the identification of false positives in the reporter gene assay that are a result of concomitant reduced cell viability. We selected Y3e, Y6q and Y1f for validation by RT-qPCR, assessing the effects of the compounds on the transcription of the endogenous Nrf2 target gene, NQO1, in comparison with brusatol.46 All three compounds downregulated NQO1 transcription (Fig 2D).
To identify activators of Nrf2 we transiently transfected LNCaP androgen-sensitive prostate cancer cells with ARE-luciferase reporter based on the human NQO1 promoter47–49 and then challenged them with our yohimbine library. As a control, we monitored LNCaP viability in parallel (Fig 2E). While many compounds activated the ARE at 100 μM potentially as a stress response, Y5b and Y6u were also active at lower concentrations when we validated the hits by RT-qPCR for levels of endogenous NQO1. These compounds were only slightly less potent than the positive control, sulforaphane.
We also screened our yohimbine library against Staphylococcus aureus (Gram-positive pathogen) and Acinetobacter baumannii (Gram-negative); however, only a few compounds demonstrated weak antibacterial activities (i.e., partial growth inhibition) against A. baumannii at 200 μM. We did find the antibacterial screen insightful to cross-reference activity profiles of validated hit compounds with selective antiproliferative, NO inhibition and Nrf2-ARE modulation. Of the six validated hit compounds identified during these biological investigations (Y7g, Y6q, Y3e, Y1f, Y5b, Y6u; Figure 2), only Y1f and Y6q demonstrated slight (~20%) growth inhibition activities at 200 μM against S. aureus. Interestingly, we note that 1 does not possess any of these activities inherently, illustrating the potential for yohimbine, and other natural products, to reach beyond their own biological capabilities using this ring distortion approach. Overall, yohimbine ring-distorted small molecules cover a biologically relevant chemical space as evident by the diverse array of active compounds we identified in a relatively small library of 70 compounds.
Indole alkaloids possess an array of complex molecular architectures that is able to produce diverse biological activities as this class of natural products has intrigued scientists for decades.50–54 Total synthesis efforts date back to Woodward’s elegant construction of (±)-reserpine52 reported in 1956 and continue to inspire the development of new synthetic methods and approaches53 today, including Jacobsen’s 2013 total synthesis of (+)-reserpine.54
As alternative synthesis approaches have emerged to drive biological discoveries, indoles have remained a considerable interest as unnatural indole small molecules have demonstrated impressive biological activities in diverse disease areas of importance to human health. Synthetic efforts have led to the discovery of indoline compounds capable of resensitizing methicillin-resistant Staphylococcus aureus to methicillin55,56 while indole-inspired BIOS has led to the identification of inhibitors of protein phosphatases, which are important drug targets for diabetes and cancer.19 BIOS efforts have also led to the discovery of spirooxindole small molecules that influence neuronal network formation and demonstrate neurite outgrowth-promoting activities.22
Here, we utilized the complex indole alkaloid, yohimbine 1, as a starting point for diverted ring distortion by combining the inherent reactivity of the indole nucleus with the tryptoline scaffold as a complementary approach to DOS and BIOS, termed “Complexity-to-Diversity” (CtD). New ring-distorted compounds from 1 were advanced through further derivatization and subjected to initial biological screens leading to the discovery of Y7g which demonstrated dual HIF-dependent anticancer and anti-inflammatory activities while five additional ring distorted compounds (Y6q, Y3e, Y1f, Y5b, Y6u; Figure 2) were found to selectively modulate the ARE pathway in cancer cell lines.
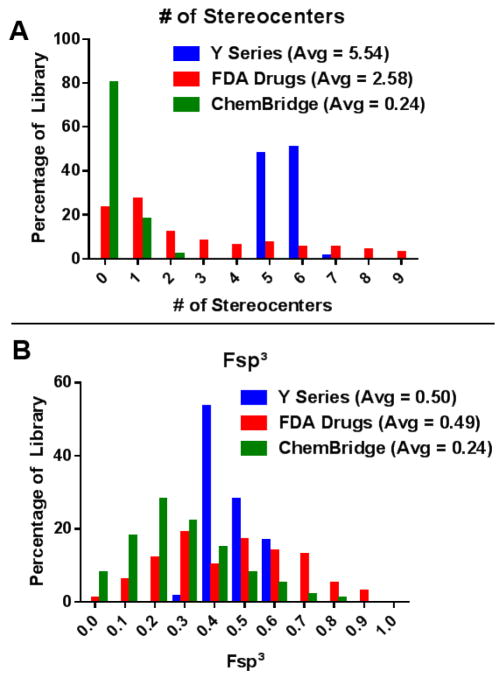
We compared the yohimbine ring distortion library to a ChemBridge (commercial) Library31 and the 100 top-selling, FDA-approved drugs from 2013 by analyzing the number of stereocenters per compound and fraction sp3 (Fsp3) character for compounds in each library (Fig 3). Each compound from our yohimbine-derived library averages 5.54±0.53 stereocenters, which is considerably higher than the FDA-approved drugs (2.58±2.61 stereocenters per compound) and the ChemBridge Library (0.24 stereocenters per compound). The Fsp3 character for the yohimbine-derived library is 0.50±0.08 which is similar to the FDA-approved drugs (Fsp3 = 0.49±0.21) and higher than the ChemBridge library (Fsp3 = 0.24).
Figure 3.
A) Analysis of the stereochemical complexity of the yohimbine ring distortion library (Y Series) compared to FDA Drugs and a ChemBridge library. B) Comparison of the Fsp3 character of the yohimbine library (Y Series), FDA Drugs and a ChemBridge library.
These combined efforts led to the identification of multiple biologically active, ring-distorted small molecules from 1 demonstrating the potential for this ring distortion approach to discover new biologically active compounds. This complex and diverse library of small molecules occupies biologically relevant chemical space. Future work will include developing improved analogues of the hit compounds identified and validated during these studies.
Conclusions
In conclusion, we have developed a new tryptoline-based ring distortion strategy that utilized the indole heterocycle of yohimbine to carry out diverse ring cleavage, ring rearrangement and ring fusion reactions enabling the rapid synthesis of multiple scaffolds totaling 70 complex and diverse small molecules. The resulting library was screened for biological activity and several hit compounds were identified in an array of assays, including Y7b which demonstrated HIF-dependent anticancer and anti-inflammatory activities. This tryptoline-based ring distortion has clear potential for the discovery of new biologically active small molecules. Hit-to-lead optimization of active compounds identified during these investigations are currently underway and aim to address multiple disease areas critical to human health.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the College of Pharmacy at the University of Florida for start-up funds (R.W.H.) and the NIH (R01CA172310, H.L.; R50CA211487, R.R.) to support this work. We thank Professors Donna Zhang for MDA-MB-231-ARE-luc cells and Jeffrey Johnson for ARE-luc plasmid. We also thank Dr. Khalil Abboud and The Center for X-Ray Crystallography at the University of Florida for obtaining an X-ray of Y2a and Y6s. All high resolution mass spectra (HRMS) were obtained from the Chemistry Department at the University of Florida.
Footnotes
Supporting Information. All experimental details concerning procedures for chemical synthesis/biological screens/validation of biologically active hit compounds, full characterization data (1H, 13C NMR, HRMS, melting point) for all new compounds with 2-D NMR spectra and X-ray data for select compounds can be found in the Supporting Information associated with this manuscript.
References
- 1.Newman DJ, Cragg GM. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swinney DC, Anthony J. Nat Rev Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 3.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 4.Dömling A. Curr Opin Chem Biol. 2008;12:281–291. doi: 10.1016/j.cbpa.2008.04.603. [DOI] [PubMed] [Google Scholar]
- 5.Villoutreix BO, Kuenemann MA, Poyet JL, Bruzzoni-Giovanelli H, Labbé C, Lagorce D, Sperandio O, Miteva MA. Mol Inform. 2014;33:414–437. doi: 10.1002/minf.201400040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbi-Verge C, Kim PM. Cell Commun Signal. 2016;14:12. doi: 10.1186/s12964-016-0131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grivas PD, Kiaris H, Papavassiliou AG. Trends Mol Med. 2011;17:537–538. doi: 10.1016/j.molmed.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Grivas PD, Papavassiliou AG. Cancer. 2013;119:1120–1128. doi: 10.1002/cncr.27908. [DOI] [PubMed] [Google Scholar]
- 9.Schreiber SL. Nature. 2009;457:153–154. doi: 10.1038/457153a. [DOI] [PubMed] [Google Scholar]
- 10.Bauer RA, Wurst JM, Tan DS. Curr Opin Chem Biol. 2010;14:308–314. doi: 10.1016/j.cbpa.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dandapani S, Marcaurelle LA. Nat Chem Biol. 2010;6:861–863. doi: 10.1038/nchembio.479. [DOI] [PubMed] [Google Scholar]
- 12.Schreiber SL. Science. 2000;287:1964–1969. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 13.Hung AW, Ramek A, Wang Y, Kaya T, Wilson JA, Clemons PA, Young DW. Proc Natl Acad Sci. 2011;108:6799–6804. doi: 10.1073/pnas.1015271108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopp F, Stratton CF, Akella LB, Tan DS. Nat Chem Biol. 2012;8:358–365. doi: 10.1038/nchembio.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer RA, Wenderski TA, Tan DS. Nat Chem Biol. 2013;9:21–29. doi: 10.1038/nchembio.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beckmann HS, Nie F, Hagerman CE, Johansson H, Tan YS, Wilcke D, Spring DR. Nat Chem. 2013;5:861–867. doi: 10.1038/nchem.1729. [DOI] [PubMed] [Google Scholar]
- 17.Grossman A, Bartlett S, Janecek M, Hodgkinson JT, Spring DR. Angew Chem Int Ed. 2014;53:13093–13097. doi: 10.1002/anie.201406865. [DOI] [PubMed] [Google Scholar]
- 18.Antonchick AP, Gerding-Reimers C, Catarinella M, Schürmann M, Preut H, Ziegler S, Rauh D, Waldmann H. Nat Chem. 2010;2:735–740. doi: 10.1038/nchem.730. [DOI] [PubMed] [Google Scholar]
- 19.Wetzel S, Bon RS, Kumar K, Waldmann H. Angew Chem Int Ed. 2011;50:10800–10826. doi: 10.1002/anie.201007004. [DOI] [PubMed] [Google Scholar]
- 20.Basu S, Ellinger B, Rizzo S, Deraeve C, Schürmann M, Preut H, Arndt HD, Waldmann H. Proc Natl Acad Sci. 2011;108:6805–6810. doi: 10.1073/pnas.1015269108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Hattum H, Waldmann H. J Am Chem Soc. 2014;136:11853–11859. doi: 10.1021/ja505861d. [DOI] [PubMed] [Google Scholar]
- 22.Antonchick AP, López-Tosco S, Parga J, Sievers S, Schürmann M, Preut H, Höing S, Schöler HR, Sterneckert J, Rauh D, Waldmann H. Chem Biol. 2013;20:500–509. doi: 10.1016/j.chembiol.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Švenda J, Sheremet M, Kremer L, Maier L, Bauer JO, Strohmann C, Ziegler S, Kumar K, Waldmann H. Angew Chem Int Ed. 2015;54:5596–5602. doi: 10.1002/anie.201500112. [DOI] [PubMed] [Google Scholar]
- 24.Balthaser BR, Maloney MC, Beeler AB, Porco JA, Synder JK. Nat Chem. 2011;3:969–973. doi: 10.1038/nchem.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLeod MC, Singh G, Plampin JN, III, Rane D, Wang JL, Day VW, Aubé J. Nat Chem. 2014;6:133–140. doi: 10.1038/nchem.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aquino C, Sarkar M, Chalmers MJ, Mendes K, Kodadek T, Micalizio GC. Nat Chem. 2011;4:99–104. doi: 10.1038/nchem.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan DS. Nat Chem Biol. 2005;1:74–84. doi: 10.1038/nchembio0705-74. [DOI] [PubMed] [Google Scholar]
- 28.Goess BC, Hannoush RN, Chan LK, Kirchhausen T, Shair MD. J Am Chem Soc. 2006;128:5391–5403. doi: 10.1021/ja056338g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins I, Jones AM. Molecules. 2014;19:17221–17225. doi: 10.3390/molecules191117221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huigens RW, III, Morrison KC, Hicklin RW, Flood TA, Richer MF, Hergenrother PJ. Nat Chem. 2013;5:195–202. doi: 10.1038/nchem.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rafferty RJ, Hicklin RW, Maloof KA, Hergenrother PJ. Angew Chem Int Ed. 2014;53:220–224. doi: 10.1002/anie.201308743. [DOI] [PubMed] [Google Scholar]
- 32.Hicklin RW, López-Sliva TL, Hergenrother PJ. Angew Chem Int Ed. 2014;53:9880–9883. doi: 10.1002/anie.201404765. [DOI] [PubMed] [Google Scholar]
- 33.Friesen K, Palatnick W, Tenebein M. J Emerg Med. 1993;11:287–288. doi: 10.1016/0736-4679(93)90048-c. [DOI] [PubMed] [Google Scholar]
- 34.Albright J, Goldman L. J Org Chem. 1965;30:1107–1110. doi: 10.1021/jo01015a038. [DOI] [PubMed] [Google Scholar]
- 35.Kagawa N, Malerich J, Rawal V. Org Lett. 2008;10:2381–2384. doi: 10.1021/ol8006277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhat UG, Winter MA, Pearce HL, Beck WT. Mol Pharm. 1995;48:682–689. [PubMed] [Google Scholar]
- 37.Somei M, Noguchi K, Yamada F. Heterocycles. 2001;55:1237–1240. [Google Scholar]
- 38.Somei M, Noguchi K, Yoshino K, Mori K, Asada M, Yamada F, Tanaka Y, Shigenobu K, Koike K. Heterocycles. 2006;69:259–269. [Google Scholar]
- 39.Takayama H, Misawa K, Okada N, Ishikawa H, Kitajima M, Hatori Y, Murayama T, Wongseripipatana S, Tashima K, Matsumoto K, Horie S. Org Lett. 2006;8:5705–5708. doi: 10.1021/ol062173k. [DOI] [PubMed] [Google Scholar]
- 40.Albright JD, Goldman L. J Am Chem Soc. 1969;91:4317–4318. [Google Scholar]
- 41.Stahl R, Borschberg H. Helv Chim Acta. 1996;79:1361–1378. [Google Scholar]
- 42.Burkitt K, Chun SY, Dang DT, Dang LH. Mol Cancer Ther. 2009;8:1148–1156. doi: 10.1158/1535-7163.MCT-08-0944. [DOI] [PubMed] [Google Scholar]
- 43.Bousquet MS, Ma JJ, Ratnayake R, Havre PA, Yao J, Dang NH, Paul VJ, Carney TJ, Dang LH, Luesch H. ACS Chem Biol. 2016;11:1322–1331. doi: 10.1021/acschembio.5b00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Pharmacol Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du Y, Villeneuve NF, Wang XJ, Sun Z, Chen W, Li J, Lou H, Wong PK, Zhang DD. Environ Health Perspect. 2008;116:1154–1161. doi: 10.1289/ehp.11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, Zhang DD. Proc Natl Acad Sci. 2011;108:1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yates MS, Tauchi M, Katsuoka F, Flanders KC, Liby KT, Honda T, Gribble GW, Johnson DA, Johnson JA, Burton NC, Guilarte TR, Yamamoto M, Sporn MB, Kensler TW. Mol Cancer Ther. 2007;6:154–162. doi: 10.1158/1535-7163.MCT-06-0516. [DOI] [PubMed] [Google Scholar]
- 48.Moehlenkamp JD, Johnson JA. Arch Biochem Biophys. 1999;363:98–106. doi: 10.1006/abbi.1998.1046. [DOI] [PubMed] [Google Scholar]
- 49.Ratnayake R, Liu Y, Paul VJ, Luesch H. Cancer Prev Res. 2013;6:989–999. doi: 10.1158/1940-6207.CAPR-13-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Conner SE, Maresh JJ. Nat Prod Rep. 2006;23:532–547. doi: 10.1039/b512615k. [DOI] [PubMed] [Google Scholar]
- 51.Kaushik NK, Kaushik N, Attri P, Kumar N, Kim CH, Verma AK, Choi EH. Molecules. 2013;18:6620–6662. doi: 10.3390/molecules18066620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodward RB, Bader FE, Bickel H, Frey AJ, Kierstead RW. J Am Chem Soc. 1956;78:2023–2025. [Google Scholar]
- 53.Yang Y, Bai Y, Sun S, Dai M. Org Lett. 2014;16:6216–6219. doi: 10.1021/ol503150c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajapaksa NS, McGowan MA, Rienzo M, Jacobsen EN. Org Lett. 2013;15:706–709. doi: 10.1021/ol400046n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Podoll JD, Liu Y, Chang L, Walls S, Wang W, Wang X. Proc Natl Acad Sci. 2013;110:15573–15578. doi: 10.1073/pnas.1310459110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu W, Wang W, Wang X. Angew Chem Int Ed. 2015;54:9546–9549. doi: 10.1002/anie.201503736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.