Abstract
Recent creation of a Unified Microbiome Initiative (UMI) has the aim of understanding how microbes interact with each other and with us. When pathogenic Staphylococcus aureus (S. aureus) infects the skin, the interplay between S. aureus and skin commensal bacteria occurs. Our previous data revealed that skin commensal bacteria can mediate fermentation against the growth of USA300, a community-acquired methicillin-resistant S. aureus MRSA (CA-MRSA). By using a fermentation process with solid media on a small scale, we define poly(ethylene glycol) dimethacrylate (PEG-DMA) as a selective fermentation initiator (SFI) which can specifically intensify the probiotic ability of skin commensal Staphylococcus epidermidis (S. epidermidis) bacteria. At least five SCFAs including acetic, butyric and propionic acids with anti-USA300 activities were produced by PEG-DMA fermentation of S. epidermidis. Furthermore, the S. epidermidis-laden PEG-DMA hydrogels effectively decolonized USA300 in skin wounds in mice. The PEG-DMA and its derivatives may become novel biomaterials to specifically tailor the human skin microbiome against invading pathogens.
Keywords: PEG, Precision microbiome, S. aureus, Selective fermentation, S. epidermidis
Graphical abstract
1 Introduction
The Unified Microbiome Initiative (UMI) seeks to develop and to apply new tools that enable us to gain a deeper understanding of microbiome or nanobiome, a community of extremely small organisms, and its impact on our life [1]. Bacterial interference, in which commensal bacteria used to prevent growth of pathogens, has been shown to be a promising modality for preventing and/or treating infections. Therapeutic application of bacterial interference by active colonization using Staphylococcus epidermidis (S. epidermidis), a commensal bacterium in humans and chickens, was successful in counteracting the infection of Staphylococcus aureus (S. aureus) [2–4]. Previous results in our published papers [5] have demonstrated that a human skin commensal bacterium can fermentatively metabolize glycerol, an endogenous sugar alcohol in the human skin, to produce short-chain fatty acids (SCFAs) with antimicrobial activities against S. aureus [5]. These results demonstrated for the first time that production of SCFAs by fermentation of skin commensal bacteria can be part of skin innate immunity. Different bacterial species make different enzymes that ferment specific carbon sources. Most skin commensal bacteria including S. epidermidis, Propionibacterium acnes (P. acnes) and pathogenic S. aureus can ferment the same carbon source such as glucose to SCFAs [6–8]. To gain maximum chances to survive, commensal bacteria and pathogenic S. aureus in a skin wound may exclude each other via production of SCFAs by fermentation of glucose. When S. aureus survives after the bacterial interference, the infection will proceed to continue to damage the host.
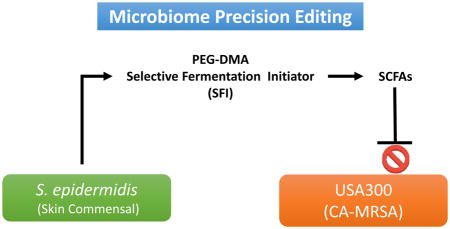
The precision microbiome can refer to the tailoring of treatments of infectious diseases by specifically targeting individual microbial species in the microbiome. Since both commensal and pathogenic bacteria could compete with each other for the same carbon source of fermentation. Our strategy of using precision microbiome approaches is to exclusively trigger the fermentation of S. epidermidis by selective fermentation initiators (SFIs), which will amplify the probiotic activity of S. epidermidis against pathogenic S. aureus. Although an incision and drainage procedure was performed for the majority of methicillin-resistant S. aureus (MRSA)-infected patients, antibiotics are usually given as adjunctive therapy to enhance S. aureus clearing. However, the use of antibiotics for S. aureus treatments is not in compliance with evolutionary medicine since bacteria may fight back by developing the ability to neutralize these antibiotics. Furthermore, the use of antibiotics runs a risk of generating resistant S. aureus and non-specifically kills commensal bacteria.
Phage therapy, as one of precision microbiome approaches, has been used as a manipulation technique, by which pathogens including S. aureus can be targeted with specific bacteriophages, reducing the damage to commensal bacteria of the host [9]. However, many limitations of phage therapy have been reported [10]. One drawback of phage therapy is that bacteriophages could transfer the DNA from a microbe to another. This DNA transfer could be in charge of the transfer of pathogenicity determinants and virulence factors, resulting in the generation of even more resistant microbial strains [11–13]. Also, bacteriophages are self-limiting i.e. they require their hosts to be constantly growing. Furthermore, the use of phage therapy in vivo could elicit a strong antibody response which may eliminate the phages rapidly and thus the use of phages for extended period of time would be impossible [14].
Carbon atoms, as sources of fermentation, form four covalent bonds so they are capable of forming long chains. There are four major categories for classification of carbon-based molecules (CBM) in living things: carbohydrates, lipids, proteins, and nucleic acids. Each type of CMB is made up of specific subunit molecules (monomers) or multiple monomers (polymers). Polyethylene glycol (PEG) containing chains of carbon atoms is a synthetic water-soluble polymer of the common structural formula H(OCH2CH2)nOH. PEG polymers are highly biocompatible and biodegradable molecules which have been approved by the Food and Drug Administration (FDA) in humans for transdermal drug delivery [15]. Many CBMs, in particular carbohydrates, have been used as excellent carbon sources for bacterial fermentation [16]. In this study, we demonstrated for the first time that the poly(ethylene glycol) dimethacrylate (PEG-DMA) can function as a SFI for S. epidermidis. PEG-DMA can exclusively trigger the fermentation of S. epidermidis, but not USA300, a community-acquired MRSA (CA-MRSA), to yield various SCFAs. In addition, to our knowledge, we herein provide the first evidence that the PEG-DMA hydrogel can be used as a 3-dimensional (3-D) scaffold which provides a carbon-rich microenvironment for induction of S. epidermidis fermentation. The S. epidermidis-laden PEG hydrogels containing live probiotic bacteria and their fermentation metabolites may function as probiotic masques or gauze bandages for covering and helping the MRSA-infected wounds heal.
2 Materials and methods
2.1 Ethics statement
Experiments involving mice were performed at the University of California, San Diego (UCSD). The UCSD ethics committee specifically approved this study under an approved Institutional Animal Care and Use Committee (IACUC) protocol (no. S10058).
2.2 Bacterial culture
Bacteria were cultured in tryptic soy broth (TSB) (Sigma, St. Louis, MO, USA). Overnight cultures were diluted 1:100 and cultured to an absorbance at 600 nm [optical density (OD)600]=1.0. Bacteria were harvested by centrifugation at 5,000 g for 10 min, washed with phosphate buffered saline (PBS), and suspended in PBS for further experiments.
2.3 Bacterial fermentation in solid media
For bacterial fermentation in solid media, S. epidermidis (ATCC12228) or USA300 were mixed with 2% molten (w/v) agar (Oxoid. Ltd., London, UK) with/without 20 g/l (2%) PEG-DMA (number-average (Mn)=550; Catalog # 25852-47-5, Sigma) in rich media [10 g/l yeast extract (Biokar Diagnostics, Beauvais, France), 3 g/l TSB, 2.5 g/l K2HPO4 and 1.5 g/l KH2PO4] at 37°C for two days. Agar was cooled to 45°C before bacteria were added to obtain a concentration of 106 colony-forming unit (CFU) on 200 μl agar top in a 96-well V-bottom polypropylene (PP) microtiter plates (Corning®, Corning, NY). Controls include 2% molten agar containing rich media plus PEG-DMA or bacteria alone. Bacterial fermentation in solid media was indicated by adding 0.002% (w/v) phenol red, which changes its color from red-orange to yellow.
2.4 Anti-USA300 overlay assays
Solid media containing 2% molten agar in rich media were cooled to 45°C before USA300 bacteria (105 CFU/200 μl) with/without 2% PEG-DMA were added to a 96-well V-bottom PP microtiter plates. S. epidermidis (ATCC12228) bacteria (105 CFU/20 μl) were spread on top of the USA300-containing solid media, and then cultured at 37°C for two days. The colonies (> 0.005 mm2) of USA300 in solid media were counted by ImageJ software (NIH, Bethesda, MD, USA).
2.5 Gas chromatography mass spectrometry (GC-MS) analysis
S. epidermidis (ATCC12228) (105 CFU/ml) was incubated in rich media (10 ml) in the presence of 2% PEG-DMA for three days. After centrifugation at 5,000 g for 10 min, bacteria in supernatants were further removed by 0.22 micron filters and SCFAs in the fermentation media were detected by ethyl acetate (Residue Analysis OmniSolv OmniSolv, EMD Millipore, Billerica, MA) liquid-liquid extraction after addition of 50 μl of 2H7-butyric acid (1 mg/ml) (C/D/N Isotopes, Quebec, Canada) as an internal standard, acidification with 0.5% ortho-phosphoric acid (Thermo Fisher Scientific Inc., Fair Lawn, NJ) and saturation with sodium chloride (Thermo Fisher Scientific Inc.) followed by GC-MS analysis using an Agilent 5890 Series II GC in conjunction with 5971 MS detector (Agilent Technologies, Inc., Palo Alto, CA). A 70eV electron was utilized for ionization. The levels of acetic, propionic, isobutyric, butyric and isovaleric acids in the fermentation media were quantified by a calibration curve made from six non-zero levels using the Free Fatty Acids Test Standard (Restek Corporation, Bellefonte, PA) which is diluted 500-, 1,000-, 2,000-, 5,000-, and 10,000-fold.
2.6 The minimum bactericidal concentration (MBC) assays
USA300 bacteria (106 CFU/ml) were incubated with butyrate or acetate at various concentrations (0.02–100 mM in PBS) in media on a 96-well microtiter plate (100 μl per well) overnight. The control received PBS alone. After incubation, bacteria were diluted 1:100–1:105 with PBS. MBC was finally examined at a 99.9% (greater than one log10 CFU/ml) killing level and determined by spotting the dilution (5 μl) on an agar plate supplemented with media for the counting of CFUs.
2.7 Fabrication of S. epidermidis-laden PEG-DMA hydrogels
Hydrogels were formed by mixing 10% PEG-DMA or acrylamide (Promega corporation, Madison, WI) with 0.001% ammonium persulfate (APS) (Promega) and 0.1% tetramethylethylenediamine (TEMED) in 500 μl TBS containing phenol red- (0.002%; w/v). To encapsulate bacteria in hydrogels, 500 μl of hydrogel/APS/TEMED media were used to suspend the S. epidermidis (ATCC12228) bacteria (2×108 CFU/ml) and then placed in an incubator at 37°C for three days. The color change of phenol red from red-orange to yellow in hydrogels demonstrated the PEG-DMA fermentation of S. epidermidis.
2.8 In vivo anti-USA300 activity of probiotic PEG-DMA hydrogels
The probiotic PEG-DMA hydrogels (a circle with a diameter of 0.8 cm) were constructed by encapsulating S. epidermidis (ATCC12228) (108 CFU) in hydrogels comprising 10% PEG-DMA, 0.001% APS and 0.1%TEMED in phenol red-free rich media. The PEG-DMA hydrogels without S. epidermidis encapsulation serve as control hydrogels. Two 10 mm wounds were made on the dorsal skin of Institute of Cancer Research (ICR) mice (2–3 month-old females; Harlan Labs, Placentia, CA, USA) following anesthetizing by isoflurane and shaving with electrical clippers. To recapitulate the infection of MRSA, USA300 (108 CFU in 10 μl PBS) was directly applied to the wounds. To determine if the PEG-DMA fermentation of S. epidermidis in hydrogels can mitigate the infection of USA300, a probiotic PEG-DMA hydrogel or a control hydrogel was applied respectively over the wounded sites made on the dorsal skin of the same mice 10 min after USA300 application. Application of PEG-DMA hydrogels without S. epidermidis encapsulation alone or acrylamide gels encapsulated with or without S. epidermidis was also conducted to determine its effect on the USA300 growth in wounds. Three separate experiments with 3 mice per group in each experiment were performed. All experiments using mice were conducted in a biosafety level 2 (BSL-2) facility.
2.9 Bacterial counts in USA300-infected skin
To determine the USA300 counts, the infected wounds were excised 24 h following application of bacteria and hydrogels. The excised skin was homogenized in 500 μl of sterile PBS with a tissue grinder. Bacterial CFUs in the wound were enumerated by plating serial dilutions (1:100–1:105) of the homogenate on a phenol-red supplemented mannitol salt agar (MSA) plate (BD, Sparks, MD, USA). The plate was incubated for 24 h at 37°C to count colonies. Bacteria, which created yellow colonies in MSA plates were recognized as USA300.
2. 10 Statistical analysis
To determine significances between groups, comparisons were conducted using the two-tailed t-test. For all statistical tests, the P-values of <0.05 (*), <0.01 (**), and <0.001 (***) were accepted for statistical significance.
3 Results
3.1 PEG-DMA as a SFI for S. epidermidis
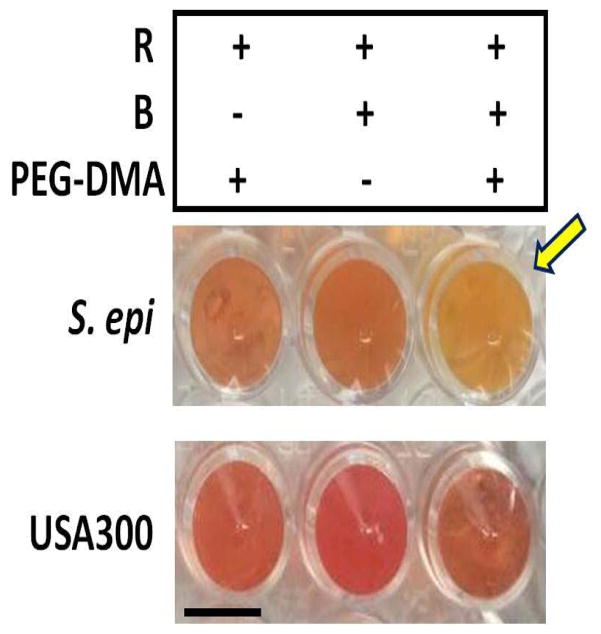
The method of bacterial fermentation in a large volume (>10 ml) of aqueous rich media has been published from our group [5]. Here we establish a fermentation process with solid media on a small (200 μl/well) scale in a 96-well microtiter plate. One of advantages of small-scale fermentation in solid media is to allow us to screen various carbon sources or bacteria in an easier manner. Different microbial species make different enzymes that ferment specific substrates. It has reported that bacteria can use a PEG polymer as a carbon source and fermentatively convert PEG to acetate and ethanol [17]. To examine whether skin commensal bacteria and USA300 differentially utilizes the PEG-DMA for fermentation, two skin commensal bacteria [S. epidermidis (ATCC12228) and P. acnes (ATCC6919)] or USA300 were incubated in the absence or presence of 2% PEG-DMA in solid media containing rich media and molten agar. Controls include solid media with PEG-DMA alone or microbes alone. The PEG-DMA selectively triggered S. epidermidis (Fig. 1), neither USA300 nor P. acnes (data not shown), to undergo fermentation. In the culture of S. epidermidis with PEG-DMA, the phenol red-containing solid media started turning yellow two days after culture. No yellow media in the culture of USA300 or P. acnes (data not shown) with PEG-DMA were detected. To validate the fermentation, S. epidermidis (ATCC12228) or USA300 was cultured in 10 ml phenol red-containing aqueous rich media. The reduction in pH values and 560 nm OD560 for phenol red was quantified. As shown in Fig. S1, a significant decrease in the values of both pH and OD560 was detected when S. epidermidis, but not USA300, was cultured in the presence of PEG-DMA, indicating the PEG-DMA fermentation of S. epidermidis occurs. To examine the PEG-DMA fermentation activity in other S. epidermidis strains, a S. epidermidis strain and a S. aureus strain isolated from human skin were used to test their PEG-DMA fermentation activities in solid media (Fig. S2). Consistently, the PEG-DMA specifically induced the fermentation of S. epidermidis, not S. aureus. The PEG-DMA was thus chosen as a SFI of S. epidermidis.
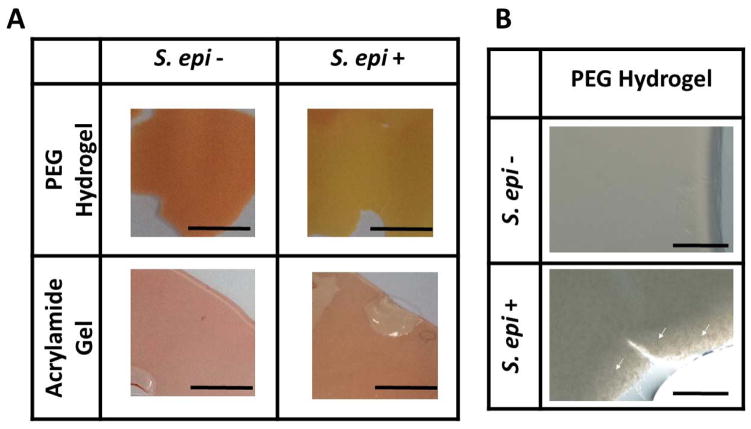
Figure 1.
PEG-DMA induces S. epidermidis, but not USA300, to undergo fermentation. S. epidermidis (ATCC12228) or USA300 (106 CFU on 200 μl agar top) mixed with 2% molten agar were cultured in rich media in the presence or absence of 2% PEG-DMA in a 96-well V-bottom PP microtiter plate at 37°C for two days. Controls included molten agar containing rich media (R) plus PEG-DMA or bacteria (B) alone. Bacterial fermentation in solid media was indicated by adding 0.002% (w/v) phenol red, which changes its color from red-orange to yellow (arrow).
3.2 Suppression of USA300 growth by PEG-DMA fermentation of S. epidermidis
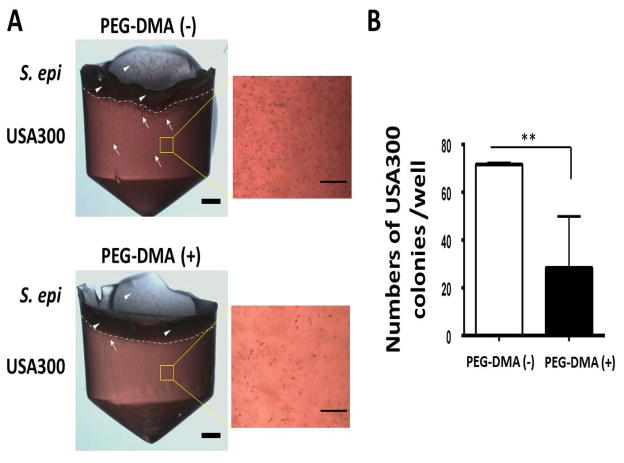
One additional advantage of small-scale fermentation in solid media is that it provides a 3-D scaffold for growing USA300 within solid media. We establish a bacterial overlay assay to investigate whether PEG-DMA is able to enhance the probiotic activities of S. epidermidis for suppression of the growth of USA300. The S. epidermidis bacteria uniformly grow on the top of solid media supplemented with/without PEG-DMA in a well of 96-well microtiter plate. As shown in Fig. 2, the colonies of USA300 with solid media supplemented with PEG-DMA are much less than those in solid media supplemented with PEG-DMA. The results indicate that PEG-DMA fermentation of S. epidermidis grown on the top of solid media may produce SCFAs which were diffused into the solid media. The diffusing SCFAs diminished the growth of USA300 in solid media.
Figure 2.
(A) S. epidermidis counteracts the growth of USA300 in the presence of PEG-DMA in an overlay assay. USA300 bacteria (105 CFU/200 μl; white arrows) were cultured in solid media containing 2% molten agar in rich media with (+)/without (−) 2% PEG-DMA in a 96-well V-bottom PP microtiter plate. S. epidermidis (S. epi) (ATCC12228) bacteria (105 CFU in 20 μl; white arrowheads) were spread on top of the USA300-containing solid media at 37°C for two days. Scale bars=0.1 cm. High magnitude images of USA300 in solid media were displayed. Scale bars=0.5 mm. (B) The numbers of USA300 colonies per well in a 96-well microtiter plate were illustrated as the mean ± standard derivation (SD) of three independent experiments. **P<0.01 (two-tailed t-tests).
3.3 Identification of SCFAs produced by PEG-DMA fermentation by GC-MS
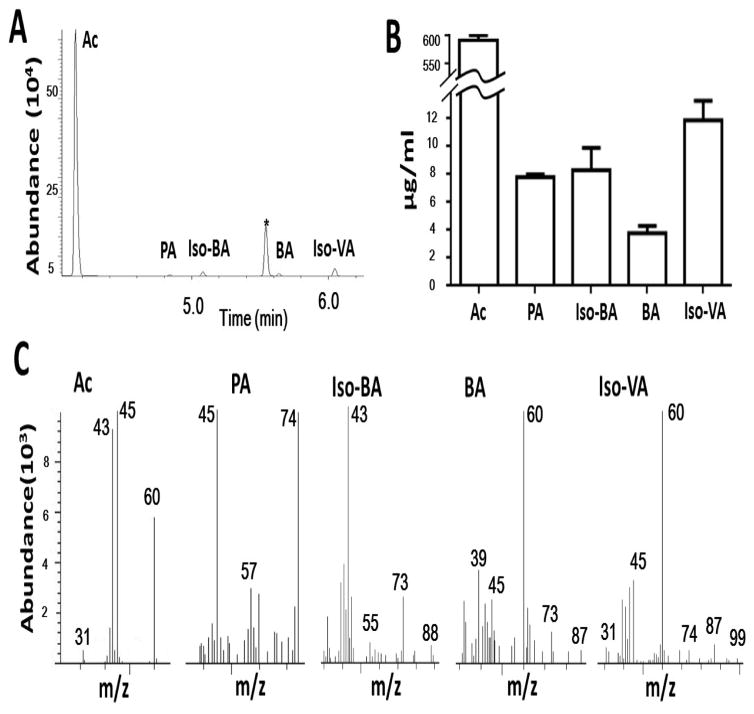
To identify the SCFAs in the ferments, the S. epidermidis bacteria were incubated in phenol red-free rich media in the presence or absence of 2% PEG-DMA for two days. Supernatants of bacterial culture of S. epidermidis mixed with 2H7-butyric acid (an internal standard) were subjected to GC-MS analysis. Extreme low levels (< 0.1 μg/ml) of SCFAs were detected in the supernatants of S. epidermidis culture in the absence of 2% PEG-DMA. Acetic (592.1 μg/ml), propionic (7.8 μg/ml), isobutyric (8.3 μg/ml), butryic (3.8 μg/ml), and isovaleric (11.9 μg/ml) acids are five major SCFAs that were produced by PEG-DMA fermentation of S. epidermidis (Fig. 3A and 3B). Mass spectra of five identified SCFAs were subsequently generated (Fig. 3C). For examples, molecular ions at 31, 43, 45, and 60 m/z values corresponding to [HCO]+, [CH3CO]+, [CH3CO]+, CH3COOH for acetic acid are detected in a MS spectrum. These results demonstrate that S. epidermidis fermentatively metabolized PEG-DMA into SCFAs.
Figure 3.
(A) The ion chromatogram and mass spectrum from GC-MS for identification of SCFAs. Total ion chromatogram for separation of the mixture of SCFAs containing acetic acid (Ac), propionic acid (PA), 2H7-butyric acid (*) (an internal standard), isobutyric acid (Iso-BA), butyric acid (BA) and isovaleric acid (Iso-VA) was displayed by running the GC-MS analysis. (B) The level (μg/ml) of each SCFA was quantified. (C) The mass spectra for AC, PA, Iso-BA, BA, and Iso-VA with their corresponding molecular ions (m/z) were shown.
3.4 Anti-USA300 activities of SCFAs
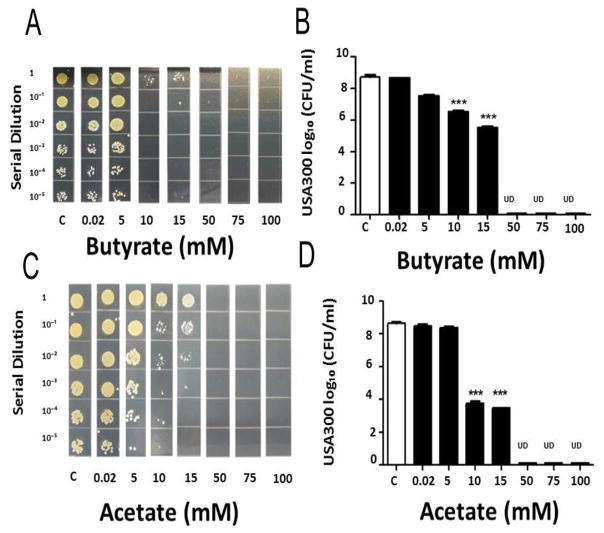
In our previous paper [18], we have demonstrated that propionic acid suppressed the growth of USA300 in vitro and in vivo. The anti-USA300 activity of propionic acid is derived from its ability to reduce the intracellular pH of USA300. Since acetic and butyric acids are two most abundant SCFAs in the media of PEG-DMA fermentation of S. epidermidis (Fig. 3), we examine the anti-USA300 activities of acetic and butyric acids in vitro. To determine MBC values of butyric acid and acetic acid for USA300, butyric acid or acetic acid (0–100 mM) was added into USA300 (106 CFU/ml) culture overnight. The MBC values (>1 log10 inhibition) of butyric acid and acetic acid for USA300 were 10 mM and the concentration of both acids for complete inhibition was 50 mM (Fig. 4). These results demonstrate that PEG-DMA fermentation of S. epidermidis yielded SCFAs with excellent anti-USA300 activities.
Figure 4.
Suppression of USA300 growth by butyric acid and acetic acid. USA300 (106 CFU/ml) was incubated with butyric acid (A, B) or acetic acid (C, D) (0.02–100 mM in PBS) overnight. Incubation of USA300 with PBS served as controls (c). After incubation, USA300 was diluted 1:100–1:105 with PBS, and 5 μl of the dilutions were spotted on an agar plate for MBC assays (A, C). The CFU counts were illustrated as the mean ± SD of three independent experiments (B, D). ***P<0.001 (two-tailed t-tests). UD, undetectable.
3.5 Inhibition of USA300 growth by S. epidermidis-laden PEG-DMA hydrogels
PEG-DMA has been used as a biomaterial for creation of hydrogels [19]. To assess the capability of PEG-DMA for encapsulate the S. epidermidis, PEG-DMA mixed with APS and TEMED was used to suspend the S. epidermidis (ATCC12228) bacteria. Acrylamide mixed with APS, TEMED and S. epidermidis served as a control hydrogel (Fig. 5A). To determine if the encapsulated S. epidermidis in hydrogels (Fig. 5B) can utilize the PEG-DMA as a carbon source for fermentation, PEG-DMA or acrylamide hydrogels were suspended with/without the S. epidermidis for two days in the presence of phenol red. As shown in Fig. 5A, the color change of phenol red from red-orange to yellow was observed in S. epidermidis-laden PEG-DMA, but not acrylamide, hydrogels, demonstrating the probiotic activities of S. epidermidis-laden PEG-DMA hydrogels.
Figure 5.
The fermentation of S. epidermidis in PEG-DMA hydrogels. (A) Hydrogels formed by 10% PEG-DMA (PEG-DMA Hydrogel) or acrylamide (Acrylamide Gel) in phenol red-containing rich media were used to encapsulate the S. epidermidis (2 × 108 CFU/ml; ATCC12228). (B) S. epidermidis indicated with white arrows on the edge of PEG-DMA hydrogels can be visualized in high magnitude images. Hydrogels with (S. epi +) or without (S. epi −) were placed at 37°C for two days. The color change of phenol red from red to yellow in PEG-DMA hydrogels indicated the fermentation of S. epidermidis. Scale bars=0.5 cm; 0.1 cm (high magnitude images).
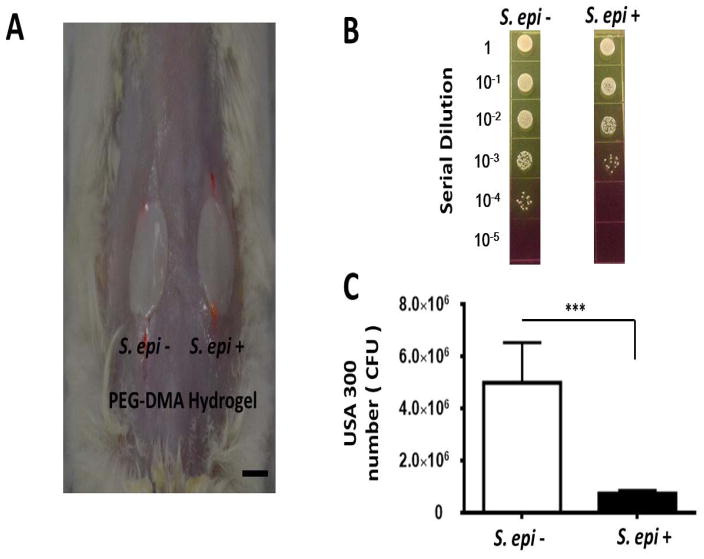
To evaluate the probiotic activities of PEG-DMA hydrogels in vivo, the USA300-infected skin wounds in the dorsal skin of ICR mice were covered by the PEG-DMA hydrogels with or without S. epidermidis encapsulation for 24 h (Fig. 6A). As shown in Fig. 6B, the number (7.5 ± 1.2 × 105 CFU) of USA300 bacteria in the wound covered by a PEG-DMA hydrogel with S. epidermidis was significantly less than that (5.0 ± 1.5 × 106 CFU) in the wound covered by a PEG-DMA hydrogel without S. epidermidis. The results demonstrate that the S. epidermidis-laden PEG-DMA hydrogels possess the probiotic activity against the USA300 skin infection.
Figure 6.
(A) Suppression of USA300 growth in skin wounds by S. epidermidis-laden PEG-DMA hydrogels. The PEG-DMA hydrogels with (S. epi +) or without (S. epi −) S. epidermidis were applied over the wounded sites made on the dorsal skin of the ICR mice 10 min after application of USA300 (106 CFU/1 μl PBS). Scale bar=1.0 cm. (B) Twenty four h after bacterial application, bacterial CFUs in the wound were counted by plating serial dilutions (1:100–1:105) of the wound homogenates on phenol-red supplemented MSA plates. (C) The CFUs of USA300 (yellow colonies) were displayed as the mean ± SD of three separate experiments with 3 mice per group in each experiment. ***P<0.001; two-tailed t-tests.
4 Discussion
PEG-DMA has been widely used as biomaterials for dental and cartilage repair and bone regeneration [20]. It exerts a minimal toxicological response and can be modified to be either bio-inert or biocompatible in vitro [21] and in vivo [22]. Each PEG-DMA monomer can covalently link to up to four other PEG-DMA monomers and the resulting hydrogel polymer forms a covalently crosslinked branched network. PEG-DMA hydrogels have been utilized for encapsulation and stabilization of various compounds [19]. To the best of our knowledge, our findings represent the first report to demonstrate that S. epidermidis, but not USA300, can fermentatively metabolize PEG-DMA to various SCFAs (Fig. 3). If bacterial fermentation occurs within a skin wound, both S. epidermidis and USA300 can use the same carbon source [e.g. glucose] outcompete each other to survive. PEG-DMA can functionally serve as a SFI to specially amplify the fermentation activity of S. epidermidis against USA300. Therefore, PEG-DMA holds great promise for selectively editing the fermentative skin microbiome to reduce the colonization of invading MRSA. It has been documented that a strictly anaerobic, gram-negative, and nonsporeforming bacterium is able to completely degrade a PEG with a molecular weight of 20,000 by fermentation to produce acetaldehyde as an intermediate and acetate and ethanol as final metabolites [23]. The PEG-fermenting strain did not excrete extracellular depolymerizing enzymes and its capability of degrading PEG can be inhibited by ethylene glycol, probably due to blocking of the cellular uptake system. The finding suggested that PEG was taken up into the bacteria and subsequently degraded inside [23]. The depolymerization of PEG inside bacteria can be catalyzed via hydrolysis followed by a modification of the terminal ethylene glycol (EG) unit. Pelobacter propionicus, a gram-negative and strictly anerobic bacterium, can undergo fermentation of a PEG with molecular weights of 1,000 inside the bacteria to produce acetate and propionate [24]. The diol dehydratase and PEG acetaldehyde lyase in fermenting bacteria can degrade PEG [17]. In other laboratories, results obtained with anaerobic bacteria favored PEG degradation outside the bacteria [25]. In fact, an extracellular enzyme which can depolymerize long chain PEGs has been identified [26]. Although the reasons for distinctive capabilities of PEG-DMA fermentation in S. epidermidis and USA300 are not clear, it is possible that two bacteria differentially express the enzymes involved in degradation of PEG-DMA. Future works will include identifying the key enzymes that catalyze the PEG-DMA fermentation in S. epidermidis. Furthermore, besides USA300, one of CA-MRSA bacteria, other MRSA such as MRSA252, a hospital-acquired MRSA [27], non-MRSA, and additional strains of S. epidermidis will be used to validate the action of PEG-DMA as a SFI specific for S. epidermidis.
As shown in Fig. 3, SCFAs are detectable in the media of S. epidermidis PEG-DMA fermentation. For examples, acetic and butyric acids at the concentrations of 592.1 and 3.8 μg/ml corresponding to 9.9 and 0.43 mM were produced respectively in the media of PEG-DMA fermentation (Fig. 3). Numerous SCFAs at low concentrations are measureable in the skin and in the secretions of skin glands [28]. Human sweat contained 0.1% lactic acid, 0.04% citric acid, 0.0096% acetic acid and 0.0062% propionic acid [29]. It is not yet determined what amounts of SCFAs can be produced when PEG-DMA is topically applied onto the skin. The SCFAs produced by fermentation of intestinal microbes in the human colon can locally reach a high level in the 20–140 mM range [30]. The amounts of SCFAs in peripheral circulation are relatively low, ranging from 3 to 7 μM [31]. SCFAs act as class I and II histone deacetylases (HDAC) inhibitors, inhibiting the function of histone deacetylase enzymes, thereby favoring an acetylated state of histones in the cell. Furthermore, treatment of human epidermal keratinocytes (HEKs) with butyric acid resulted in a rapid accumulation of acetylated histone 4 by 8 h, revealing that butyric acid can effectively inhibit the HDAC activity in skin keratinocytes [31, 32]. It has been reported that butyric acid increased histone acetylation states via HDAC inhibition and decreased the production of interleukin (IL)8 by the intestinal epithelium [33]. Furthermore, sodium butyrate can induce the expression of antimicrobial peptides and block the cell internalization of S. aureus [34]. Butyrate and 1,25-dihydroxyvitamin D3 increased the cathelicidin, an antimicrobial peptide and enhanced the antimicrobial activity of keratinocytes against S. aureus [35]. Since SCFAs have documented both antimicrobial and anti-inflammatory effects [28], future studies will include determining whether SCFAs produced by PEG-DMA fermentation regulate the host response to S. aureus infection.
A S. epidermidis-laden PEG-DMA hydrogel can hinder the growth of USA300 bacteria in a skin wound (Fig. 6). The growth suppression of USA300 could be due to the production of anti-USA300 SCFAs in PEG-DMA hydrogels. There is no difference in the USA300 growth in wounds applied with or without a PEG-DMA hydrogel, indicating that PEG-DMA hydrogel itself did not influence the colonization of USA300 in wounds (Fig. S3A, B). Furthermore, no significant change in the USA300 growth was detected when wounds were covered with an acrylamide gel laden with or without S. epidermidis (Fig. S3C, D). These results suggested that PEG-DMA fermentation in a hydrogel may be required for inhibition of the USA300 growth in wounds. In addition, bacterial interference via entry of S. epidermidis into wounds from gels may not be a major factor contributing to inhibition of USA300 growth in wounds. Inhibition of HDACs in skin cells may induce production of cytokines which can regulate the migration of phagocytes (eg. neutrophils) to the S. aureus-infected site [36]. Immunologic tolerance induced by PEG can prevent an immune response against pathogens [37]. Thus, it is worthy to study whether topical application of PEG-DMA onto a skin wound affects the recruitment of phagocytes to the sites of S. aureus infection.
The Center for Disease Control Prevention estimated that more than 90,000 people die from hospital-acquired bacterial infections in the United States (US) each year [38]. CA-MRSA is reported as the most common cause of purulent skin and soft tissue infections in the US [39]. It is assessed that S. aureus accounts for 12 million outpatient visits and 292,000 hospitalizations of which 126,000 are due to MRSA annually in the US alone [38]. Lack of selectivity and induction of resistance are central problems for many antibiotics for treatment of S. aureus infections. Vaccines against S. aureus, although the actions are theoretically specific, have not been used in the clinic and, if available, may become less potent for treatments of patients with significant underlying health matters such as diabetes, surgical intervention, and immune suppression. In contrast, probiotic treatments, which are relatively independent on patients’ health matters, have little or no interference with commensal microbes may complement antibiotics or PEG-DMA on the growth of various skin microbes have been not examined in this study, we believe that PEG-DMA will not systemically induce an imbalance of the human microbiome since it is applied locally onto a skin wound. Besides the function of PEG-DMA as a SFI, we believe that when PEG-DMA is used with antibiotics, it has potential to be an antibiotic adjuvant [39] to augment the fermentation activity of S. epidermidis and reduce the effective doses of antibiotics, decreasing the risk of generating resistant S. aureus and non-specific killing effect of antibiotics on skin commensals. Many bacteria can selectively use substrates from a mixture of different carbon sources via the regulation of carbon catabolite repression (CCR) [40]. In addition, EG, a fermentative metabolite of PEG, can be a carbon source for bacterial fermentation [41]. Thus, when PEG-DMA are applied onto a skin wound, S. epidermidis bacteria may have multiple carbon sources including endogenous glucose for their fermentation. In future studies, we will monitor the fermentation metabolites over time after PEG-DMA application to determine which carbon sources are mainly utilized for S. epidermidis fermentation.
In summary, USA300 and S. epidermidis within a skin wound may compete with each other for the same carbon source of fermentation. Here we employ the precision microbiome approach to identify PEG-DMA as a SFI which can exclusively trigger the fermentation of probiotic S. epidermidis and produce SCFAs with anti-USA300 activities. Antibiotics without selectivity can destroy the probiotic bacteria in the human skin microbiome that helps fight pathogens and maintains homeostasis of microbiome. Our approach to enhance the fermentation activity of skin probiotic bacteria using PEG-DMA as a carbon source emphasizes an undiscovered application of PEG-derived biomaterials for specifically editing the skin microbiome.
Supplementary Material
Acknowledgments
This work was mainly supported by a NIH STTR grant (1R41AR065260-01A1) and partially supported by a MOST (104-2320-B-008-003) grant. We appreciate a Health Sciences Recharge Facility at Department of Pediatrics, UCSD, for GC-MS analysis. We also would like to thank Sunita Keshari for her great effort for editing this manuscript.
Abbreviations
- APS
ammonium persulfate
- BSL-2
biosafety level 2
- CA-MRSA
community-acquired methicillin-resistant S. aureus MRSA
- CBM
carbon-based molecules
- CCR
carbon catabolite repression
- CFU
colony-forming unit
- 3-D
3-dimensional
- EG
ethylene glycol
- FDA
the Food and Drug Administration
- GC-MS
Gas chromatography mass spectrometry
- HDAC
histone deacetylases
- HEKs
human epidermal keratinocytes
- IACUC
Institutional Animal Care and Use Committee
- IL
interleukin
- MBC
the minimum bactericidal concentration
- Mn
number-average
- MRSA
methicillin-resistant S. aureus
- MSA
mannitol salt agar
- OD
optical density
- P. acnes
Propionibacterium acnes
- PBS
phosphate buffered saline
- PEG
polyethylene glycol
- PEG-DMA
poly(ethylene glycol) dimethacrylate
- PP
polypropylene
- S. aureus
Staphylococcus aureus
- SCFAs
short-chain fatty acids
- S. epidermidis
Staphylococcus epidermidis
- SFI
selective fermentation initiator
- TEMED
tetramethylethylenediamine
- TSB
tryptic soy broth
- UCSD
the University of California, San Diego
- SD
standard derivation
- UMI
Unified Microbiome Initiative
- US
the United States
Footnotes
Conflict of interest
The authors declare no financial or commercial conflict of interest.
References
- 1.Biteen JS, Blainey PC, Cardon ZG, Chun M, et al. Tools for the Microbiome: Nano and Beyond. ACS nano. 2016;10:6–37. doi: 10.1021/acsnano.5b07826. [DOI] [PubMed] [Google Scholar]
- 2.Iwase T, Uehara Y, Shinji H, Tajima A, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 3.Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nature medicine. 2009;15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto M. Staphylococcus epidermidis--the ‘accidental’ pathogen. Nature reviews Microbiology. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shu M, Wang Y, Yu J, Kuo S, et al. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PloS one. 2013;8:e55380. doi: 10.1371/journal.pone.0055380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbirato F, Chedaille D, Bories A. Propionic acid fermentation from glycerol: Comparison with conventional substrates. Applied microbiology and biotechnology. 1997;47:441–446. [Google Scholar]
- 7.Robbins GB, Lewis KH. Fermentation of Sugar Acids by Bacteria. Journal of bacteriology. 1940;39:399–404. doi: 10.1128/jb.39.4.399-404.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Safonova TB, Shcherbakova NA, Afanas’eva TI, Sobolev VR. Importance of carbohydrate tests for interspecies differentiation of staphylococci. Zhurnal mikrobiologii, epidemiologii, i immunobiologii. 1978:98–101. [PubMed] [Google Scholar]
- 9.Pincus NB, Reckhow JD, Saleem D, Jammeh ML, et al. Strain Specific Phage Treatment for Staphylococcus aureus Infection Is Influenced by Host Immunity and Site of Infection. PloS one. 2015;10:e0124280. doi: 10.1371/journal.pone.0124280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haq IU, Chaudhry WN, Akhtar MN, Andleeb S, Qadri I. Bacteriophages and their implications on future biotechnology: a review. Virol J. 2012;9 doi: 10.1186/1743-422X-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brabban AD, Hite E, Callaway TR. Evolution of foodborne pathogens via temperate bacteriophage-mediated gene transfer. Foodborne pathogens and disease. 2005;2:287–303. doi: 10.1089/fpd.2005.2.287. [DOI] [PubMed] [Google Scholar]
- 12.O’Shea YA, Boyd EF. Mobilization of the Vibrio pathogenicity island between Vibrio cholerae isolates mediated by CP-T1 generalized transduction. FEMS microbiology letters. 2002;214:153–157. doi: 10.1111/j.1574-6968.2002.tb11339.x. [DOI] [PubMed] [Google Scholar]
- 13.Maiques E, Ubeda C, Tormo MA, Ferrer MD, et al. Role of staphylococcal phage and SaPI integrase in intra- and interspecies SaPI transfer. Journal of bacteriology. 2007;189:5608–5616. doi: 10.1128/JB.00619-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark JR, March JB. Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends in biotechnology. 2006;24:212–218. doi: 10.1016/j.tibtech.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Che J, Okeke CI, Hu ZB, Xu J. DSPE-PEG: a distinctive component in drug delivery system. Current pharmaceutical design. 2015;21:1598–1605. doi: 10.2174/1381612821666150115144003. [DOI] [PubMed] [Google Scholar]
- 16.Dusselier M, Mascal M, Sels BF. Top chemical opportunities from carbohydrate biomass: a chemist’s view of the Biorefinery. Topics in current chemistry. 2014;353:1–40. doi: 10.1007/128_2014_544. [DOI] [PubMed] [Google Scholar]
- 17.Frings J, Schramm E, Schink B. Enzymes Involved in Anaerobic Polyethylene Glycol Degradation by Pelobacter venetianus and Bacteroides Strain PG1. Applied and environmental microbiology. 1992;58:2164–2167. doi: 10.1128/aem.58.7.2164-2167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Dai A, Huang S, Kuo S, et al. Propionic acid and its esterified derivative suppress the growth of methicillin-resistant Staphylococcus aureus USA300. Beneficial microbes. 2014;5:161–168. doi: 10.3920/BM2013.0031. [DOI] [PubMed] [Google Scholar]
- 19.Killion JA, Geever LM, Devine DM, Kennedy JE, Higginbotham CL. Mechanical properties and thermal behaviour of PEGDMA hydrogels for potential bone regeneration application. Journal of the mechanical behavior of biomedical materials. 2011;4:1219–1227. doi: 10.1016/j.jmbbm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Zhang K, Simon CG, Jr, Washburn NR, Antonucci JM, Lin-Gibson S. In situ formation of blends by photopolymerization of poly(ethylene glycol) dimethacrylate and polylactide. Biomacromolecules. 2005;6:1615–1622. doi: 10.1021/bm0500648. [DOI] [PubMed] [Google Scholar]
- 21.Lin-Gibson S, Bencherif S, Cooper JA, Wetzel SJ, et al. Synthesis and characterization of PEG dimethacrylates and their hydrogels. Biomacromolecules. 2004;5:1280–1287. doi: 10.1021/bm0498777. [DOI] [PubMed] [Google Scholar]
- 22.Elisseeff J, McIntosh W, Anseth K, Riley S, et al. Photoencapsulation of chondrocytes in poly(ethylene oxide)-based semi-interpenetrating networks. Journal of biomedical materials research. 2000;51:164–171. doi: 10.1002/(sici)1097-4636(200008)51:2<164::aid-jbm4>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 23.Schink B, Stieb M. Fermentative degradation of polyethylene glycol by a strictly anaerobic, gram-negative, nonsporeforming bacterium, Pelobacter venetianus sp. nov. Applied and environmental microbiology. 1983;45:1905–1913. doi: 10.1128/aem.45.6.1905-1913.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagener S, Schink B. Fermentative degradation of nonionic surfactants and polyethylene glycol by enrichment cultures and by pure cultures of homoacetogenic and propionate-forming bacteria. Applied and environmental microbiology. 1988;54:561–565. doi: 10.1128/aem.54.2.561-565.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dwyer DF, Tiedje JM. Metabolism of polyethylene glycol by two anaerobic bacteria, Desulfovibrio desulfuricans and a Bacteroides sp. Applied and environmental microbiology. 1986;52:852–856. doi: 10.1128/aem.52.4.852-856.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haines JR, Alexander M. Microbial degradation of polyethylene glycols. Applied microbiology. 1975;29:621–625. doi: 10.1128/am.29.5.621-625.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasmussen KJ, Mattsson AH, Pilely K, Asferg CA, et al. Proteome-wide antigen discovery of novel protective vaccine candidates against Staphylococcus aureus infection. Vaccine. 2016;34:4602–4609. doi: 10.1016/j.vaccine.2016.07.016. [DOI] [PubMed] [Google Scholar]
- 28.Burtenshaw JM. The mechanism of self-disinfection of the human skin and its appendages. The Journal of hygiene. 1942;42:184–210. doi: 10.1017/s0022172400035373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobdy E, Murren J. AN-9 (Titan) Current opinion in investigational drugs. 2004;5:628–634. [PubMed] [Google Scholar]
- 30.Garland SH. Short chain fatty acids may elicit an innate immune response from preadipocytes: A potential link between bacterial infection and inflammatory diseases. Med Hypotheses. 2011;76:881–883. doi: 10.1016/j.mehy.2011.02.041. [DOI] [PubMed] [Google Scholar]
- 31.Qian YJ, Jung YS, Chen XB. Delta Np63, a Target of DEC1 and Histone Deacetylase 2, Modulates the Efficacy of Histone Deacetylase Inhibitors in Growth Suppression and Keratinocyte Differentiation. J Biol Chem. 2011;286 doi: 10.1074/jbc.M110.207241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J. Review of the innate immune response in acne vulgaris: Activation of toll-like receptor 2 in acne triggers inflammatory cytokine responses. Dermatology. 2005;211:193–198. doi: 10.1159/000087011. [DOI] [PubMed] [Google Scholar]
- 33.Huang N, Katz JP, Martin DR, Wu GD. Inhibition of IL-8 gene expression in Caco-2 cells by compounds which induce histone hyperacetylation. Cytokine. 1997;9:27–36. doi: 10.1006/cyto.1996.0132. [DOI] [PubMed] [Google Scholar]
- 34.Ochoa-Zarzosa A, Villarreal-Fernandez E, Cano-Camacho H, Lopez-Meza JE. Sodium butyrate inhibits Staphylococcus aureus internalization in bovine mammary epithelial cells and induces the expression of antimicrobial peptide genes. Microb Pathogenesis. 2009;47:1–7. doi: 10.1016/j.micpath.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 35.Schauber J, Oda Y, Buchau AS, Yun QC, et al. Histone acetylation in keratinocytes enables control of the expression of cathelicidin and CD14 by 1,25-dihydroxyvitamin D3. The Journal of investigative dermatology. 2008;128:816–824. doi: 10.1038/sj.jid.5701102. [DOI] [PubMed] [Google Scholar]
- 36.Brogdon JL, Xu Y, Szabo SJ, An S, et al. Histone deacetylase activities are required for innate immune cell control of Th1 but not Th2 effector cell function. Blood. 2007;109:1123–1130. doi: 10.1182/blood-2006-04-019711. [DOI] [PubMed] [Google Scholar]
- 37.Saito T, Kumagai Y, Hiramatsu T, Kurosawa M, et al. Immune tolerance induced by polyethylene glycol-conjugate of protein antigen: clonal deletion of antigen-specific Th-cells in the thymus. Journal of biomaterials science Polymer edition. 2000;11:647–656. doi: 10.1163/156856200743922. [DOI] [PubMed] [Google Scholar]
- 38.Goetghebeur M, Landry PA, Han D, Vicente C. Methicillin-resistant Staphylococcus aureus: A public health issue with economic consequences. The Canadian journal of infectious diseases & medical microbiology = Journal canadien des maladies infectieuses et de la microbiologie medicale / AMMI Canada. 2007;18:27–34. doi: 10.1155/2007/253947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hurley MN, Forrester DL, Smyth AR. Antibiotic adjuvant therapy for pulmonary infection in cystic fibrosis. The Cochrane database of systematic reviews. 2013;6:CD008037. doi: 10.1002/14651858.CD008037.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 41.Gaston LW, Stadtman ER. Fermentation of ethylene glycol by Clostridium glycolicum, spn. J Bacteriol. 1963;85:356–362. doi: 10.1128/jb.85.2.356-362.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Kuo S, Shu M, Yu J, et al. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: implications of probiotics in acne vulgaris. Appl Microbiol Biotechnol. 2014;98:411–424. doi: 10.1007/s00253-013-5394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.