Abstract
Cancers are often initiated by genetic events that activate proto-oncogenes or inactivate tumor suppressor genes. These events are also critical for sustained tumor cell proliferation and survival, a phenomenon described as oncogene addiction. In addition to this cell intrinsic role, recent evidence indicates that oncogenes also directly regulate immune responses, leading to immunosuppression. Expression of many oncogenes, or loss of tumor suppressors, indeed induces the expression of immune checkpoints including PD-L1, which regulate the immune response. Here, we discuss how oncogenes, and in particular MYC, suppress immune surveillance and how oncogene-targeted therapies may restore the immune response against tumors.
Oncogenic MYC Addiction and the Immune System
Proto-oncogenes physiologically regulate normal cell proliferation, growth, differentiation, and apoptosis [1]. The activation, amplification, or mutation of oncogenes combined with the mutation or loss of expression of tumor suppressor genes cooperate to initiate tumorigenesis [1]. The c-MYC (MYC, see Abbreviations) oncogene is a transcription factor that normally regulates the expression of up to thousands of genes that in turn regulate proliferation, growth, differentiation, stemness, and metabolism [2–5]. MYC is genetically activated and/or overexpressed in most types of human cancer, thereby driving autonomous proliferation, growth and self-renewal [6–10], blocking differentiation [11,12], and inducing genomic destabilization [13]. Thus, MYC is thought to be a central driver of normal and malignant cellular growth and proliferation.
Oncogenes are not only critical in the process of the initiation of tumorigenesis; their sustained activation is required for the maintenance of a neoplastic state. The inactivation of a single driver oncogene can often result in dramatic regression of a cancer, a phenomenon that is referred to as “oncogene addiction.” Oncogene addiction has been most studied in experimental transgenic mouse models whereby oncogenes, such as MYC, can be conditionally expressed, using the Tet System. Using such model systems, it has been shown by many investigators that cancers induced by MYC [6,7,14,15], the GTPase RAS [16,17], BCR-ABL (breakpoint cluster region – Abelson kinase fusion) [18], the human epidermal growth factor receptor HER2/neu [19,20], the tyrosine kinase receptor MET [21] as well as many other oncogenes [22–24] undergo rapid and sustained regression upon oncogene inactivation.
Until recently, it was thought that oncogenes, including MYC, contributed to normal and pathological cellular proliferation and growth only through tumor cell specific, host-independent mechanisms. However, when examined in conditional transgenic mouse models, immune cells were found to be critical for the regression that occurs upon oncogene withdrawal, suggesting that oncogenes play an important role through the regulation of host immune-mediated mechanisms in models of lymphoma [25]. Similarly, it was found that host immune mechanisms regulate angiogenesis and tumor growth in pancreatic islet tumors [26]. In agreement with this hypothesis, MYC regulates the expression of immune checkpoints, including Cluster of Differentiation 47 (CD47, also known as Integrin Associated Protein, which allows cells to avoid phagocytosis [27]) and programmed death ligand 1 (PD-L1, also known as CD274 and B7-H1) [28]. Collectively, these findings indicate that oncogenes cause cancer not just through influencing cell growth and death pathways in the tumor cells, but also through their influence on immune evasion and immunosuppression.
During the evolution of a tumor, it is thought that cancer cells go through a process termed immune editing, allowing tumor cells to bypass immune surveillance mechanisms, either through the establishment of an immunosuppressive environment and/or through avoidance of recognition by immune cells [29]. Some tumor cells are immuno-stimulatory and are eliminated, whereas others avoid detection and continue to grow, forming the remaining, edited tumor. Furthermore, tumor micro-environments are often generally immunosuppressive, and although many mechanisms contribute to this phenomenon, changes in the expression of immune checkpoints in cancers appear to play an important role [30]. With recent advances in checkpoint blockade therapies, it is now clear that even “edited” tumors can be targeted by the immune system in a clinically effective manner through the alteration of the immunosuppression induced by tumor cells and their environment.
Here, we will discuss how oncogenes regulate key immune checkpoints. This could have significant implications for understanding the mechanisms by which oncogenes cause cancer and subsequently for therapy of cancer. We will suggest how and why oncogenes physiologically regulate the immune response and how and why abnormal oncogene activation can disrupt the immune response and contribute to tumorigenesis.
Oncogenes Regulate the Immune Response
Many studies report that oncogenes regulate components of the immune response, suggesting that more generally this is a mechanism of tumorigenesis. In some cases, these other oncogenes may work via MYC. The immune checkpoint regulators are key components of the immune response that can be regulated by oncogenic pathways [31], as has been seen particularly in MYC-driven tumors and lymphomas. PD-L1 is an immune checkpoint that suppresses the immune system so that PD-L1 binding to its receptor sends T cells a signal to disengage [32,33]. PD-L1 has a role in the malignant progression of many types of cancer including lymphoma [34]. There are multiple oncogenic signaling pathways used to dampen the immune response and encourage immune privilege within the tumor [30]. In the next section, we focus on what is understood about how oncogenes regulate PD-L1.
Several Oncogenes and Tumor Suppressors Regulate PD-L1 Expression
The gain of expression of oncogenes or the loss of expression of tumor suppressors has been shown to regulate immune checkpoints [35–38]. The upregulation of PD-L1 and related immunosuppressive pathways in mouse models of epidermal growth factor receptor (EGFR)-driven lung cancer was accompanied by decreases in cytotoxic tumor infiltrating lymphocytes (CTLs) and increases in T cell exhaustion [36]. The expression of mutant EGFR induced PD-L1, whereas EGFR inhibitors reduced PD-L1 expression [36]. Additionally, PD-L1 is upregulated by oncogenic signaling pathways associated with the resistance to the serine/threonine-protein kinase BRAF inhibitor vemurafenib [37]. Thus, multiple signaling pathways regulate the expression of PD-L1.
The WNT pathway and phosphinositide-3 kinase (PI3K) pathway regulate immune checkpoints. In autochthonous mouse melanoma models, WNT and CTNNB1 (encoding the β-catenin protein) signaling suppresses T cells, conferring resistance to anti-PD-L1/anti-CTLA-4 therapy [39]. The activation of WNT/β-catenin is associated with the absence of a T cell signature and T cell infiltration in the tumor. Cyclin-dependent kinase 5 (CDK5) disruption reduces PD-L1 in a mouse model of medulloblastoma, increasing anti-tumor immunity via CD4+ T cell-mediated tumor rejection [40]. The transformation of T lymphocytes through the expression of the oncogenic tyrosine kinase NPM-ALK (fusion of nucleophosmin – anaplastic lymphoma kinase) led to upregulation of signal transducer and activator of transcription 3 (STAT3) and mechanistic target of rapamycin (mTORC1) along with cell surface expression of PD-L1 [38], suggesting that upregulation of PD-L1 is an early step in carcinogenesis. The AKT/mTOR pathway activates PD-L1 in vitro and in vivo in human lung cancer [41]. Both oncogenic and IFNγ-inducible expression of PD-L1 depended on mTOR, and in human lung adenocarcinomas and squamous cell carcinoma, cell surface expression of PD-L1 was correlated with mTOR activation. The same study found that in mouse models of lung cancer, combination of an mTOR inhibitor with an anti-PD1 antibody elevated levels of infiltrating lymphocytes and reduced regulatory T cell levels. Thus, both the WNT and PI3K pathways are important regulators of PD-L1.
Loss of tumor suppressors such as phosphatase and tensin homolog (PTEN), a key regulator of the PI3K pathway, and p53, a regulator of apoptosis, has been associated with increased expression of PD-L1. The loss of PTEN increased PD-L1 expression, eliciting immuno-resistance in glioma [35]. The loss of PTEN also elevated PD-L1 in melanoma alongside secretion of immunosuppressive factors [42] in short-term culture of patient-derived melanoma cells [42]. In human lung tumors, PD-L1 expression was significantly associated with aberrant p53 expression, which in turn associated with reduced infiltrating immune cells [43]. In hepatocellular carcinoma, PD-L1 expression was negatively correlated to p53 expression [44]. In human lung adenocarcinoma, genetic events in p53 correlated with higher PD-L1 [45]. Thus, dysregulation of tumor suppressors correlates with changes in PD-L1 expression.
From the studies discussed here, it is clear that PD-L1 is regulated by many oncogenes and tumor suppressors, suggesting that the onset and maintenance of tumorigenesis is directly linked with increased levels of PD-L1 expression
Oncogene Inactivation Restores the Immune Response
Cancers are often “addicted” to driver oncogenes, such that inactivation of a driver oncogene can result in dramatic tumor regression [6]. Oncogene addiction was largely thought to occur based on tumor cell-intrinsic pathways. However, recent observations indicate that the immune system plays a role in tumor regression following oncogene inactivation [46,47]. Experimentally, inactivation of the MYC or BCR-ABL oncogenes in genetically engineered conditional mouse hematopoietic tumors failed to induce sustained tumor regression in the absence of an intact host immune system [25]. CD4+ helper T cells were found to be required to remodel the tumor microenvironment and reduce the numbers of malignant cells left following treatment, a parameter called minimal residual disease (MRD). In the absence of CD4+ T cells, oncogene inactivation failed to shut down angiogenesis in the host or elicit cellular senescence in tumor cells [25]. Thus, oncogene addiction occurs both through tumor-intrinsic mechanisms, such as induction of apoptosis and inhibition of proliferation, and host immune-dependent mechanisms.
In this context, the role of MYC in the regulation of immune checkpoints was critical for the observed phenotype. Indeed, preventing MYC inactivation from suppressing CD47 or PD-L1 via constitutive expression of these checkpoints in mouse tumor cells blocked sustained tumor regression [27]. For both genes, MYC inactivation decreased mRNA and surface protein expression, although other immune receptors were unaffected. Mechanistically, MYC was shown to bind to the promoters of CD47 and PD-L1 as measured by chromatin immune-precipitation followed by sequencing (ChIP-Seq) [27]. MYC binding was generally seen only at relatively high levels of MYC expression, suggesting that this may represent promoter “invasion.” Notably, MYC only modestly induced these immunomodulatory targets on its own but knocking down expression of CD47 or PD-L1 prevented MYC-induced tumors from establishing in immunocompetent hosts [27]. In terms of immune response, MYC’s regulation of PD-L1 and CD47 was shown to be casually involved in the recruitment of T cells and macrophages. This recruitment modified the tumor microenvironment, ultimately influencing both angiogenesis and induction of cellular senescence [25,28].
To date, no drugs have been identified to directly target MYC. Bromodomain and extraterminal protein (BET) inhibitors can block MYC transcriptional function and can reduce MYC expression [48,49]. However, these inhibitors do not only work through MYC [48–50]. The BET inhibitor JQ1 [48,51] decreased expression of PD-L1 and CD47 but not of other immune receptors [28,52] and was associated with the recruitment of T cells [52]. Thus, drugs that target MYC associated pathways may be useful to alter the expression of immune checkpoints.
General Mechanistic and Therapeutic Implications
MYC’s regulation of the expression of the immune checkpoints CD47 and PD-L1 suggests a direct role in coordinating the immune response (Figure 1). Several questions arise: First, is MYC a general regulator the immune response? This remains to be shown since there are a multitude of immune checkpoints, chemokines, and host immune effector mechanisms that MYC regulates. Other oncogenes can regulate the immune response against tumors, as described above, including BRAF and β-catenin [39,53,54]. These could work through MYC, or they could work through parallel pathways. The inactivation of several different oncogenes has been shown to restore an immune response, including the inactivation of HER2 [55], BRAF [53,56,57], sonic hedgehog (SHH) [58], β-catenin, [59], EGFR [60] and the zinc finger protein SNAI1 (Snail) [61]. Thus, it is likely that many oncogenes coordinate to regulate the immune response. Since oncogene inactivation often induces tumor cell death [6], it is of great interest to determine whether cell death following oncogene withdrawal may also, albeit indirectly, activate the immune system through effects on immunogenic or tolerogenic cell death [62].
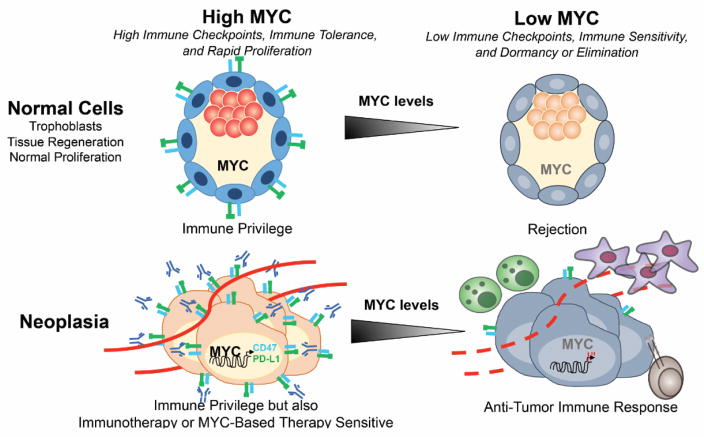
Figure 1.
Proto-oncogene and Oncogene Mediated Regulation of Immune Privilege We suggest a model where in normal physiological situations requiring rapid proliferative expansion, such as during development, tissue regeneration, and immune response (left, top panel) or during pathological conditions such as tumor growth (left, bottom panel), oncogenes such as MYC induce immune checkpoint expression, thereby allowing immunologic privilege. However, oncogene inactivation can elicit immune recognition, which in normal cells can be associated with quiescence (right, top panel) but in tumor cells can be associated with immune response and tumor elimination (right, bottom panel). Further, MYC inactivation in tumors appears to engage the immune system to elicit cellular senescence in tumor cells and to collapse the vascular endothelial cells. MYC-driven tumors necessarily might express high levels of immune checkpoints suppressing immune response and thereby evade immune surveillance, but this oncogene addiction through immune checkpoints may also render them highly susceptible to immunotherapy.
In addition to effects on the expression of immune checkpoints, oncogenes can regulate other immune-associated molecules including cytokines. MYC has been shown to regulate many secreted molecules. MYC regulates thrombospondin-1, which causally regulates angiogenesis and cellular senescence [25]. MYC also regulates Type 1 IFN [63], which influences innate and adaptive immunity [64]. Thus, expression of MYC could engender a non-immunogenic phenotype to cancer cells through multiple mechanisms, including cell surface checkpoint proteins [28] and secreted factors [63].
Second, do oncogenes such as MYC regulate immune checkpoints in a way that would make the tumor highly susceptible to immune therapy? MYC and other oncogenes, by inducing expression of immune checkpoints, may make tumors more susceptible to immune checkpoint-based therapies. However, this remains to be determined. Generally, tumors are all thought to undergo immune editing to evade immune surveillance [65,66]. The immune system would eliminate many nascent tumor cells, thereby evolutionarily selecting relative immune resistant and thus “edited” cells. However, if MYC up regulates immune checkpoints, this could bypass the requirement for tumor to find other ways “edit” the induced expression of other immune resistance mechanisms. MYC activation would enable tumors to proliferatively expand; neoantigens may be generated but not able to elicit an immune response. Acute inactivation of the MYC oncogene then would necessarily suppress immune checkpoints and restore an immune response against the tumor. Thus, it seems likely that tumors that express high levels of MYC may be particularly susceptible to immune checkpoint therapy, but this remains to be seen.
Third, do oncogenes and tumor suppressors physiologically regulate the expression of immune checkpoints? MYC and other oncogenes may physiologically regulate immune checkpoints to enable normal proliferation in a manner that prevents an immune response. During normal development, tissue regeneration, and the generation of an immune response, normal cells are required to undergo rapid proliferative expansion. Rapidly proliferating cells may be at risk of an immune response, as they upregulate NKG2D and other stress ligands [67,68]. During certain physiologic circumstances, an immune response against the rapidly proliferating cells must be suppressed (Figure 1). MYC may couple the requirement of normal cellular proliferation with the ability to prevent rapidly proliferating cells from eliciting an autoimmune response.
One physiological circumstance in which MYC may serve to induce proliferation but prevent an immune response would be during the invasion of the uterine wall by the trophoblast layer during embryonic development [69]. In this important step in development, healthy cells invade into another tissue. Tightly regulated programming must prevent an immune response, especially as the embryo-derived trophoblast is not “self.” MYC is highly expressed in the trophoblast [69], a highly proliferative tissue that even is described as “pseudomalignant” [70]. The trophoblast uses multiple pathways, including the repression of polymorphic human leukocyte antigens (HLA) molecules and the induction of PD-L1, to prevent an immune response from the mother [71]. Notably, MYC inhibition of pre-implantation blastocysts induces dormancy [72]. Thus, MYC and for that matter other oncogenes might regulate immune checkpoints as part of normal developmental programs to protect rapidly proliferating cells from immunological rejection.
MYC and other oncogenes may induce immune checkpoints and regulate other immune modulators as part of a physiologic mechanism to ensure that normally proliferating cells remain immune privileged (Figure 1). Cancers may coopt this physiologic mechanism through specific oncogenes like MYC that not only drive proliferation but also serves as a secondary “hit” enabling tumors to bypass immune surveillance. Hence, tumorigenesis may recapitulate yet distort a program that normally helps immunologically define self from nonself.
Concluding Remarks
If MYC physiologically regulates the immune response, then there are many predictions. The activation of MYC in a cell in the absence of immune checkpoints such as CD47 and PD-L1 should elicit an immune response and elimination of normal cells. MYC-induced tumors may express many neoantigens that will be uncovered upon MYC inactivation. Many other immune regulators [30], checkpoints, and secreted factors, are likely to be similarly regulated by MYC. Other oncogenes that regulate MYC gene expression, protein activation, or stability, may similarly regulate immune checkpoints. Drugs that target the MYC pathway could be effective at eliciting an immune response against tumors.
MYC has been already established to regulate the tumor microenvironment and host immune system [25,28]. The challenge now is to understand how MYC and other oncogenes specifically reshape the immune landscape. This insight should lead to better therapeutic strategies for cancer. In particular, identifying the specific mechanisms and immune components regulated by MYC and other oncogenes should lead to better experimental strategies to use to restore the immune response against tumors and to remodel the tumor microenvironment [73,74].
The realization that oncogenes such as MYC function as central regulators of normal and pathological growth and proliferation, but also of the host immune response, should galvanize renewed interest in developing drugs against this otherwise difficult therapeutic target.
Trends Box.
Oncogenes such as MYC may drive tumor growth both through their intrinsic influence on cellular proliferation but also through their regulation of immune checkpoints that enables evasion from immune surveillance.
The MYC oncogene causally regulates immune checkpoint expression. Other oncogenes including EGFR, STAT3, BRAF, β-catenin, and AKT/mTOR, as well as loss of tumor suppressor genes such as PTEN, appear to regulate PD-L1 expression.
Targeted inactivation of an oncogene with therapy may restore an immune response against tumors and remodel the tumor microenvironment.
Outstanding questions box.
Do oncogenes generally regulate expression of immune checkpoints, and if so, through what mechanisms?
Do oncogenes physiologically regulate immune checkpoints to prevent an immune response against proliferating cells?
Do oncogenes predict sensitivity to immune therapy and can targeted oncogene therapy be used to restore an immune response against tumors?
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 7.Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 8.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 9.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. discussion 3080. [DOI] [PubMed] [Google Scholar]
- 10.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci U S A. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evan G, Harrington E, Fanidi A, Land H, Amati B, Bennett M. Integrated Control of Cell Proliferation and Cell Death by the c-myc Oncogene. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences. 1994;345:269–275. doi: 10.1098/rstb.1994.0105. [DOI] [PubMed] [Google Scholar]
- 12.Eisenman RN. Deconstructing myc. Genes Dev. 2001;15:2023–2030. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- 13.Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci U S A. 1999;96:3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marinkovic D, Marinkovic T, Mahr B, Hess J, Wirth T. Reversible lymphomagenesis in conditionally c-MYC expressing mice. Int J Cancer. 2004;110:336–342. doi: 10.1002/ijc.20099. [DOI] [PubMed] [Google Scholar]
- 15.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 16.Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O’Hagan R, Pantginis J, Zhou H, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 17.Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA, Koretsky A, Varmus HE. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15:3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huettner CS, Zhang P, Van Etten RA, Tenen DG. Reversibility of acute B-cell leukaemia induced by BCR-ABL1. Nat Genet. 2000;24:57–60. doi: 10.1038/71691. [DOI] [PubMed] [Google Scholar]
- 19.Xie W, Chow LT, Paterson AJ, Chin E, Kudlow JE. Conditional expression of the ErbB2 oncogene elicits reversible hyperplasia in stratified epithelia and up-regulation of TGFalpha expression in transgenic mice. Oncogene. 1999;18:3593–3607. doi: 10.1038/sj.onc.1202673. [DOI] [PubMed] [Google Scholar]
- 20.Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, Innocent N, Cardiff RD, Schnall MD, Chodosh LA. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2:451–461. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- 21.Wang R, Ferrell LD, Faouzi S, Maher JJ, Bishop JM. Activation of the Met receptor by cell attachment induces and sustains hepatocellular carcinomas in transgenic mice. J Cell Biol. 2001;153:1023–1034. doi: 10.1083/jcb.153.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewald D, Li M, Efrat S, Auer G, Wall RJ, Furth PA, Hennighausen L. Time-sensitive reversal of hyperplasia in transgenic mice expressing SV40 T antigen. Science. 1996;273:1384–1386. doi: 10.1126/science.273.5280.1384. [DOI] [PubMed] [Google Scholar]
- 23.Gunther EJ, Moody SE, Belka GK, Hahn KT, Innocent N, Dugan KD, Cardiff RD, Chodosh LA. Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev. 2003;17:488–501. doi: 10.1101/gad.1051603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felsher DW. Reversibility of oncogene-induced cancer. Curr Opin Genet Dev. 2004;14:37–42. doi: 10.1016/j.gde.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Rakhra K, Bachireddy P, Zabuawala T, Zeiser R, Xu L, Kopelman A, Fan AC, Yang Q, Braunstein L, Crosby E, et al. CD4(+) T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell. 2010;18:485–498. doi: 10.1016/j.ccr.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soucek L, Lawlor ER, Soto D, Shchors K, Swigart LB, Evan GI. Mast cells are required for angiogenesis and macroscopic expansion of Myc-induced pancreatic islet tumors. Nat Med. 2007;13:1211–1218. doi: 10.1038/nm1649. [DOI] [PubMed] [Google Scholar]
- 27.Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD, Jr, van Rooijen N, Weissman IL. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gutgemann I, Eilers M, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016 doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases--elimination, equilibrium and escape. Curr Opin Immunol. 2014;27:16–25. doi: 10.1016/j.coi.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pardoll D. Cancer and the Immune System: Basic Concepts and Targets for Intervention. Semin Oncol. 2015;42:523–538. doi: 10.1053/j.seminoncol.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaguchi T, Kawakami Y. Cancer-induced heterogeneous immunosuppressive tumor microenvironments and their personalized modulation. Int Immunol. 2016;28:393–399. doi: 10.1093/intimm/dxw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5:1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 33.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Wang J, Li C, Ke XY. Contribution of PD-L1 to oncogenesis of lymphoma and its RNAi-based targeting therapy. Leuk Lymphoma. 2012;53:2015–2023. doi: 10.3109/10428194.2012.673228. [DOI] [PubMed] [Google Scholar]
- 35.Parsa AT, Waldron JS, Panner A, Crane CA, Parney IF, Barry JJ, Cachola KE, Murray JC, Tihan T, Jensen MC, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13:84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 36.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, Mikse OR, Cherniack AD, Beauchamp EM, Pugh TJ, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3:1355–1363. doi: 10.1158/2159-8290.CD-13-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atefi M, Avramis E, Lassen A, Wong DJ, Robert L, Foulad D, Cerniglia M, Titz B, Chodon T, Graeber TG, et al. Effects of MAPK and PI3K pathways on PD-L1 expression in melanoma. Clin Cancer Res. 2014;20:3446–3457. doi: 10.1158/1078-0432.CCR-13-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q, Wei F, Wang HY, Liu X, Roy D, Xiong QB, Jiang S, Medvec A, Danet-Desnoyers G, Watt C, et al. The potent oncogene NPM-ALK mediates malignant transformation of normal human CD4(+) T lymphocytes. The American journal of pathology. 2013;183:1971–1980. doi: 10.1016/j.ajpath.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 40.Dorand RD, Nthale J, Myers JT, Barkauskas DS, Avril S, Chirieleison SM, Pareek TK, Abbott DW, Stearns DS, Letterio JJ, et al. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science. 2016;353:399–403. doi: 10.1126/science.aae0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lastwika KJ, Wilson W, 3rd, Li QK, Norris J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, et al. Control of PD-L1 Expression by Oncogenic Activation of the AKT-mTOR Pathway in Non-Small Cell Lung Cancer. Cancer Res. 2016;76:227–238. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- 42.Dong Y, Richards JA, Gupta R, Aung PP, Emley A, Kluger Y, Dogra SK, Mahalingam M, Wajapeyee N. PTEN functions as a melanoma tumor suppressor by promoting host immune response. Oncogene. 2014;33:4632–4642. doi: 10.1038/onc.2013.409. [DOI] [PubMed] [Google Scholar]
- 43.Cha YJ, Kim HR, Lee CY, Cho BC, Shim HS. Clinicopathological and prognostic significance of programmed cell death ligand-1 expression in lung adenocarcinoma and its relationship with p53 status. Lung Cancer. 2016;97:73–80. doi: 10.1016/j.lungcan.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Kan G, Dong W. The expression of PD-L1 APE1 and P53 in hepatocellular carcinoma and its relationship to clinical pathology. Eur Rev Med Pharmacol Sci. 2015;19:3063–3071. [PubMed] [Google Scholar]
- 45.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, Behrens C, Kadara H, Parra ER, Canales JR, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Restifo NP. Can antitumor immunity help to explain “oncogene addiction”? Cancer Cell. 2010;18:403–405. doi: 10.1016/j.ccr.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Felsher DW. MYC Inactivation Elicits Oncogene Addiction through Both Tumor Cell-Intrinsic and Host-Dependent Mechanisms. Genes Cancer. 2010;1:597–604. doi: 10.1177/1947601910377798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ., 3rd Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao W, Yue P, Khuri FR, Sun SY. The BET bromodomain inhibitor, JQ1, facilitates c-FLIP degradation and enhances TRAIL-induced apoptosis independent of BRD4 and c-Myc inhibition. Oncotarget. 2015;6:34669–34679. doi: 10.18632/oncotarget.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu H, Bengsch F, Svoronos N, Rutkowski MR, Bitler BG, Allegrezza MJ, Yokoyama Y, Kossenkov AV, Bradner JE, Conejo-Garcia JR, et al. BET Bromodomain Inhibition Promotes Anti-tumor Immunity by Suppressing PD-L1 Expression. Cell Rep. 2016;16:2829–2837. doi: 10.1016/j.celrep.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilmott JS, Long GV, Howle JR, Haydu LE, Sharma RN, Thompson JF, Kefford RF, Hersey P, Scolyer RA. Selective BRAF inhibitors induce marked T-cell infiltration into human metastatic melanoma. Clin Cancer Res. 2012;18:1386–1394. doi: 10.1158/1078-0432.CCR-11-2479. [DOI] [PubMed] [Google Scholar]
- 54.Sweis RF, Spranger S, Bao R, Paner GP, Stadler WM, Steinberg G, Gajewski TF. Molecular Drivers of the Non-T-cell-Inflamed Tumor Microenvironment in Urothelial Bladder Cancer. Cancer Immunol Res. 2016;4:563–568. doi: 10.1158/2326-6066.CIR-15-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor C, Hershman D, Shah N, Suciu-Foca N, Petrylak DP, Taub R, Vahdat L, Cheng B, Pegram M, Knutson KL, et al. Augmented HER-2 specific immunity during treatment with trastuzumab and chemotherapy. Clin Cancer Res. 2007;13:5133–5143. doi: 10.1158/1078-0432.CCR-07-0507. [DOI] [PubMed] [Google Scholar]
- 56.Steinberg SM, Zhang P, Malik BT, Boni A, Shabaneh TB, Byrne KT, Mullins DW, Brinckerhoff CE, Ernstoff MS, Bosenberg MW, et al. BRAF inhibition alleviates immune suppression in murine autochthonous melanoma. Cancer Immunol Res. 2014;2:1044–1050. doi: 10.1158/2326-6066.CIR-14-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khalili JS, Liu S, Rodriguez-Cruz TG, Whittington M, Wardell S, Liu C, Zhang M, Cooper ZA, Frederick DT, Li Y, et al. Oncogenic BRAF(V600E) promotes stromal cell-mediated immunosuppression via induction of interleukin-1 in melanoma. Clin Cancer Res. 2012;18:5329–5340. doi: 10.1158/1078-0432.CCR-12-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otsuka A, Dreier J, Cheng PF, Nageli M, Lehmann H, Felderer L, Frew IJ, Matsushita S, Levesque MP, Dummer R. Hedgehog pathway inhibitors promote adaptive immune responses in basal cell carcinoma. Clin Cancer Res. 2015;21:1289–1297. doi: 10.1158/1078-0432.CCR-14-2110. [DOI] [PubMed] [Google Scholar]
- 59.Holtzhausen A, Zhao F, Evans KS, Tsutsui M, Orabona C, Tyler DS, Hanks BA. Melanoma-Derived Wnt5a Promotes Local Dendritic-Cell Expression of IDO and Immunotolerance: Opportunities for Pharmacologic Enhancement of Immunotherapy. Cancer Immunol Res. 2015;3:1082–1095. doi: 10.1158/2326-6066.CIR-14-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumai T, Matsuda Y, Oikawa K, Aoki N, Kimura S, Harabuchi Y, Celis E, Kobayashi H. EGFR inhibitors augment antitumour helper T-cell responses of HER family-specific immunotherapy. Br J Cancer. 2013;109:2155–2166. doi: 10.1038/bjc.2013.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 62.Green DR, Ferguson T, Zitvogel L, Kroemer G. Immunogenic and tolerogenic cell death. Nat Rev Immunol. 2009;9:353–363. doi: 10.1038/nri2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schlee M, Holzel M, Bernard S, Mailhammer R, Schuhmacher M, Reschke J, Eick D, Marinkovic D, Wirth T, Rosenwald A, et al. C-myc activation impairs the NF-kappaB and the interferon response: implications for the pathogenesis of Burkitt’s lymphoma. Int J Cancer. 2007;120:1387–1395. doi: 10.1002/ijc.22372. [DOI] [PubMed] [Google Scholar]
- 64.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 65.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 66.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 67.Marcus A, Gowen BG, Thompson TW, Iannello A, Ardolino M, Deng W, Wang L, Shifrin N, Raulet DH. Recognition of tumors by the innate immune system and natural killer cells. Adv Immunol. 2014;122:91–128. doi: 10.1016/B978-0-12-800267-4.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev. 2010;235:267–285. doi: 10.1111/j.0105-2896.2010.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pfeifer-Ohlsson S, Scott Goustin A, Rydnert J, Wahlström T, Bjersing L, Stehelin D, Ohlsson R. Spatial and temporal pattern of cellular myc oncogene expression in developing human placenta: Implications for embryonic cell proliferation. Cell. 1984;38:585–596. doi: 10.1016/0092-8674(84)90513-0. [DOI] [PubMed] [Google Scholar]
- 70.Rydnert J, Pfeifer-Ohlsson S, Goustin AS, Ohlsson R. Temporal and spatial pattern of cellular myc oncogene expression during human placental development. Placenta. 1987;8:339–345. doi: 10.1016/0143-4004(87)90061-0. [DOI] [PubMed] [Google Scholar]
- 71.Petroff MG, Chen L, Phillips TA, Azzola D, Sedlmayr P, Hunt JS. B7 family molecules are favorably positioned at the human maternal-fetal interface. Biol Reprod. 2003;68:1496–1504. doi: 10.1095/biolreprod.102.010058. [DOI] [PubMed] [Google Scholar]
- 72.Scognamiglio R, Cabezas-Wallscheid N, Thier MC, Altamura S, Reyes A, Prendergast AM, Baumgartner D, Carnevalli LS, Atzberger A, Haas S, et al. Myc Depletion Induces a Pluripotent Dormant State Mimicking Diapause. Cell. 2016;164:668–680. doi: 10.1016/j.cell.2015.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Casey SC, Li Y, Felsher DW. An essential role for the immune system in the mechanism of tumor regression following targeted oncogene inactivation. Immunol Res. 2014;58:282–291. doi: 10.1007/s12026-014-8503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Casey SC, Li Y, Fan AC, Felsher DW. Oncogene withdrawal engages the immune system to induce sustained cancer regression. J Immunother Cancer. 2014;2:24. doi: 10.1186/2051-1426-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]