Abstract
The simple, rapid magnetic manipulation of paramagnetic particles (PMPs) paired with the wide range of available surface chemistries has strongly positioned PMPs in the field of analyte isolation. One recent technology - Sliding Lid for Immobilized Droplet Extractions (SLIDE) - presents a simple, rapid alternative to traditional PMP isolation protocols. Rather than remove fluid from PMP-bound analyte, SLIDE directly removes the PMPs from the fluid. SLIDE collects the PMPs on a hydrophobic, removable surface, which allows PMPs to be captured from one well and then transferred and released into a second well. Despite several key advantages, SLIDE remains limited by its passive magnetic manipulation that only allows for a one-time capture-and-release of PMPs – preventing wash steps and limiting purity. Furthermore, the strategy employed by SLIDE constrains the position of the wells, thereby limiting throughput and integration into automated systems. Here, we introduce a new, mechanically and operationally simplistic magnetic manipulation system for integration with the SLIDE technology to overcome the previously stated limitations. This magnetic system is compatible with nearly any plate design, can be integrated into automated workflows, enables high-throughput formats, simplifies mechanical requirements, and is amenable to a range of analytes. Using this magnetic system, PMPs can be collected, released, and re-suspended throughout multiple wells regardless of proximity. We demonstrate this system’s capabilities to isolate whole cells, mRNA, and DNA, demonstrating up to a 28-fold improvement of purity via the multi-wash protocols enabled by this magnetic technology.
Graphical Abstract
Introduction
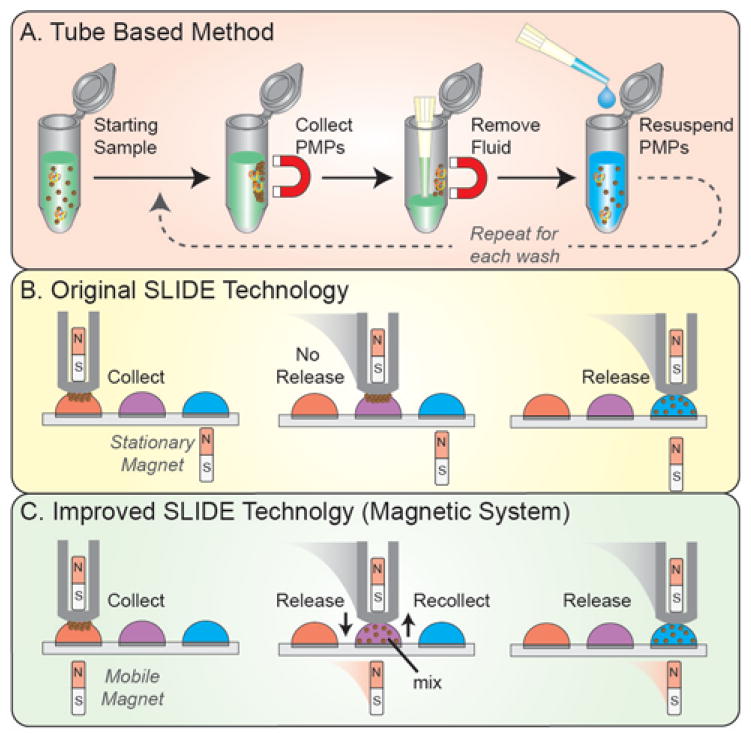
Isolation via paramagnetic particles (PMP) is becoming increasingly popular for sample preparation and cell manipulation. Due to a wide range of analyte-specific surface chemistries (silica, antibodies, streptavidin, etc.), PMPs enable isolation of numerous analytes, including RNA, DNA, whole cells, and proteins. Leveraging magnetic forces, the PMP-bound analytes can be isolated from diverse backgrounds and purified via a bind, wash, and elute protocol1. In tube-based PMP isolation protocols – one of the most common isolation strategies - sample, buffer (e.g., lysis buffer), and PMPs are incubated in a microtube (Fig. 1A). Following PMP-analyte binding, a magnet collects the PMP-bound analyte against the side of the tube as the unbound material and excess sample is removed by pipette. The collected sample is then washed several times by adding wash buffer, mixing via pipette or vortexing, re-collecting the PMPs, then removing the wash buffer. After washing, an elution buffer is often added, facilitating dissociation between the PMPs and the target analyte. Unfortunately, tube-based techniques are time-consuming and prone to sample loss2. Thus, several alternative approaches – categorized, herein, as exclusion-based sample preparation (ESP) – have been developed3–10.
Figure 1.
Sample preparation technology comparison. (A) The tube based approach to sample preparation requires several lengthy steps to collect, wash, and purify the PMPs. (B) The SLIDE technology requires one simple sliding motion purify the PMPs. However, the release magnet (bottom) is stationary so the PMPs cannot be released or recollected in wash wells – a necessary step for improving purity. (C) The new magnetic system facilitates a mobile release magnet to enable release and recollection of PMPs in any well.
ESP takes an alternative approach to isolating PMPs by removing PMPs from the sample rather than the sample from immobilized PMPs (e.g., tube-based approaches). To remove PMPs from the sample, ESP technologies leverage interfaces (aqueous-aqueous, aqueous-air, aqueous-oil), which in turn reduce loss of the original sample and yield higher purity despite fewer, less-rigorous wash steps. Reducing the number of washes becomes of particular importance when isolating low affinity targets, as wash steps can cause loss of target analyte11. Additionally, because the PMPs are removed from the sample, there is no fluid transfer, no dilution, and negligible loss of the sample, enabling repeated re-interrogation of the sample with ESP.
One form of ESP – Sliding Lid for Immobilized Droplet Extractions (SLIDE) – provides improvements in speed and simplicity by collecting, washing, and releasing PMP-bound analyte in a single motion12 (Fig. 1B). To accomplish this, the SLIDE platform leverages opposing magnetic fields, where a fixed release magnet (lower magnet) repels a floating collection magnet (upper magnet), thereby establishing magnetic dominance to attract the PMPs. However, SLIDE’s requirement for release magnets creates limitations. First, the stationary release magnets prevent PMPs from being recollected after release, thereby limiting the SLIDE processes to a single step (i.e., capture and release) prohibiting consecutive washes (i.e., releasing and recollecting PMPs). Secondly, the existence of adjacent magnetic fields limits the proximity with which wells can be spaced, limiting both well density and throughput. Furthermore, while PMPs are under the influence of a strong magnetic field, mixing or resuspending the PMPs is not straightforward; the plate must be physically separated, adding complexity and requiring additional functionalities. The original SLIDE platform presents a simplistic approach to sample preparation, although SLIDE’s limitations reduce its utility and complicate its integration into automated systems.
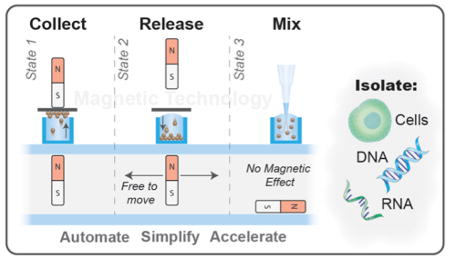
Here we present a new automated magnetic PMP manipulation system amenable to a wide range of protocols and micro-plate formats, which has no limitations on well proximity (provided the wells are sufficiently far apart that the droplets do not connect), and operates with the functional simplicity of SLIDE (Fig. 1C). This magnetic system consists of two components, a magnetic box and a magnetic head, which work in conjunction via a mechanically controlled capture magnet and a free-floating release magnet. In each well, the PMPs can be released, re-captured, and mixed with negligible influence of magnetic fields. Ultimately, this technique enables rapid isolation of a variety of analytes with improved sample purity. Here, we characterize the magnetic system, describe design criteria and considerations, and demonstrate this system’s amenability to a range of PMPs and sample types by isolating whole cells, RNA, and DNA.
Experimental
Automated ESP system Operation
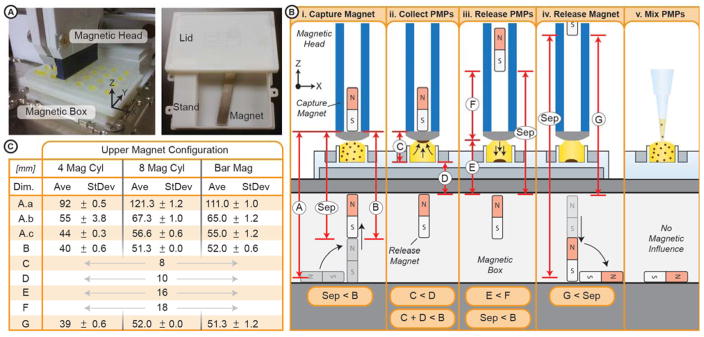
The automated ESP system integrates several key components including: custom microplates, consumable strips, a PIPETMAX™ automated liquid handler (Gilson), and the new magnetic system. The consumables used in the automated ESP platform were developed for a manual commercialized ESP embodiment known as EXTRACTMAN™ (Gilson). The consumables include custom microplates and hydrophobic polypropylene (PP) strips. The microplate consists of four rows of wells, each row comprising: a sample well (~450 μL), a large wash well (~275 μL), three small wash wells (110 μL), and a capture/elution well (110 μL or 15 μL depending on the plate). The consumable PP strips are used as a PMP transfer surface. All experiments are performed on a PIPETMAX™ automated liquid handler (Gilson), which is equipped with a standard 8-channel pipette and a custom magnetic head (SI Fig. 1). The magnetic head - a retrofitted 8-channel pipette - houses one of three magnetic configurations: four cylindrical magnets, eight cylindrical magnets, or one bar magnet. The axially polarized cylindrical magnets (#D4X-N52, K&J Magnetics) are spaced 9 mm apart (matching 96-well format) with the poles oriented in the same direction. The bar magnet (3″ L, 0.5″ W, 0.125″ t) is magnetized through the width (#52PM02 (Custom ordered from K&J). The height of the magnets are robotically controlled to achieve three magnetic states: (i) The magnetic field from the magnetic head dominates, causing the PMPs to collect on the head; (ii) The magnetic field from the box dominates, pulling the PMPs off of the head and into the well; (iii) The well is devoid of a substantial magnetic field, enabling mixing/resuspension of the PMPs. The PP strip is affixed to the bottom of the magnetic head and the EXTRACTMAN™ plate is positioned on top of the magnetic box. Each well is filled to form a convex meniscus (i.e., the fluid protruded above the well top), which enables fluidic contact between the well and the magnetic head. Like the previously published SLIDE, once fluidic contact is made, PMPs are magnetically transferred to the PP strip. However, due to the non-wetting nature of this strip and low area of liquid contact, minimal liquid is transferred along with the PMPs. With the added capabilities of the introduced magnetic system, PMPs are able to be washed in the wash wells (by releasing, pipette-mixing, and recollecting) and then released in the elution well. Only the magnets within the head are mechanically manipulated, whereas the magnet within the magnetic box responds to the magnetic field imposed by the magnetic head. As a result, the magnet in the magnetic box follows (tracks) the position of the head, enabling full control of the PMPs.
Magnetic Box Design, Fabrication, and Function
The magnetic box comprises a hollow stand and lid, that are 3D printed13 in Accura 25 (Midwest Prototyping) and screwed together, forming a hollow compartment (Fig. 2A). The footprint of the stand meets the dimensional standards set by the Society for Biomolecular Sciences (SBS standards) and the lid is sized to receive a standard microplate. The hollow compartment, is 3.1″ × 3.5″ × 0.75″ (W x L x H) - sized to house a bar magnet. The EXTRACTMAN™ plate is 11 mm tall, with a well depth of 4 mm. Each well is filled with the prescribed volumes to form a convex meniscus; the height of the meniscus (from the well top), ranges from 1 to 2.5 mm, dependent on fluid properties (e.g., surface tension).
Figure 2.
System Overview. (A) Image of the automated system, where a well plate is set on top of the box (left image) and image of the box open (right image). (B) Schematic of the operation of the magnetic box. (Panel i) Once the capture magnet is close enough to the release magnet, the release magnet is attracted to its capture position. (Panel ii) The capture magnet is closer to the magnetic PMPs, thus the PMPs are attracted to the capture magnet. At this point the head can be transferred to a new well. (Panel iii) The capture magnet is raised, such that the release magnet is still within magnetic range of the capture magnet, but the PMPs are closer, thereby attracted to, the release magnet. (Panel iv) The capture magnet is moved further away, thereby releasing the release magnet. The release magnet falls away from the plate. (Panel v) Once the release magnet falls away, there is minimal magnetic influence on the PMPs, enabling re-suspension of the PMPs via mixing. (C) A list of dimensions corresponding to the schematic.
PMP movement is controlled (captured) by tuning the proximity of each magnet to establish a magnetic dominance over the PMPs resulting in their movement (patent pending). In this system, the capture magnet (within the head) is controlled robotically, the release magnet (in the magnetic box) is able to move freely within the hollow compartment but responsive to the capture magnet, and the PMPs are free to move between the well and the strip. A typical PMP transfer cycle was performed as follows (Fig. 2B & 2C). Before the system begins, due to the absence of any capture magnets, the release magnet begins on its side. Step one: pick up the release magnet (Fig. 2C-i) and attract it to the top of the compartment. To do this, the separation distance (Sep) between the capture and release magnets must be less than the attractive distance of the magnets (Dim. B) (i.e., the attractive force between two magnets must overcome the gravitational force on the release magnet). Step two: collect the PMPs against the capture surface (consumable strip) (Fig. 2C-ii). To do this, the distance between the capture magnet and the bottom of the well (Dim. C) must be less than the distance between the release magnet and the bottom of the well (Dim. D) (i.e., the furthest PMP from the capture magnet must be closer to the capture magnet than the release). Step three: transfer the PMPs to a desired well then release them (Fig. 2C-iii). To release the PMPs, the distance between the capture surface and the release magnet (Dim. E) must be less than the distance between the capture surface and the capture magnet (Dim. F). Meanwhile, to assure the release magnet is held against the top of the compartment, the distance between the two magnets (Sep) must remain less than the attractive distance (Dim. B). Step four: remove magnetic influence by releasing the release magnet (Fig. 2C-iv). To do this, the distance between the magnets (Sep) must exceed the attractive distance (Dim. G). Importantly, the responsive distance – the distance below which the release magnet has any response to the capture magnet – is much greater than the attractive distance. Thus, to assure complete removal of magnetic influence, the separation must exceed the responsive distance (Dim. A.a – A.c, varies based on magnet configuration) (see results section for more details).
Biological Validation
Both target analytes and PMPs can influence the operation of a technology such as this where the attractive force between a PMP and a magnet is a function of the PMP size and material14. To validate the technology, we isolate whole cells, RNA, and DNA using a variety of PMPs. LNCaP whole cells are isolated from a background of peripheral blood mononuclear cells using 25 μg of Dynabeads M270 PMPs (14311D, Life Technologies) that were pre-conjugated to an anti-EpCAM antibody. Cells were identified through immuno staining and enumerated via fluorescent microscopy. RNA and DNA are isolated from sample containing 2000 LNCaP cells. The RNA sample is lysed with reagents provided in the Dynabeads mRNA DIRECT Purification Kit (#61011, Thermo-Fisher) and isolated with 10 μL of Dynabeads® Oligo(dT)25 PMPs. The DNA sample is lysed with Buffer RLT Plus (#1053393, Qiagen) and isolated with 5 μL of MagneSil® KF PMPs (MD1441, Promega). RNA and DNA are quantified by RT-qPCR and qPCR, respectively. Extensive protocols and additional details can be found in the Supporting Information. Data is expressed as averages of the technical replicates (n=3), error bars represent standard deviation, and p-values are calculated using two-tailed heteroscedastic t-test (statistical significance is defined as p < 0.05).
Results
Magnetic Box dimensions
The dimensions of the presented magnetic system are paramount to achieve precise magnetic dominance over a well of PMPs. Several PMP manipulation cycles often occur in an isolation protocol, each cycle consisting of four basic steps: collect, transfer, release, and (optional) mixing. During collection, increasing the strength of the capture magnets (by changing the magnetic configuration) significantly lengthens the attractive distance: four cylinder magnets (40 mm), eight cylinder magnets (51 mm), or one bar magnet (52 mm) (Fig. 2C, Dim. B). Similarly, strengthening the release magnet has the same effect on release; however, to minimize weight while maximizing the magnetic strength we utilize a bar magnet. Furthermore, to successfully release the PMPs from the capture surface, a buffer distance of ~2 mm is necessary to assure the magnetic dominance overcomes any foreign forces on the PMPs15 (e.g., sticky sample) (i.e., {Dim. E + Buffer Distance} < Dim. F). To facilitate mixing, the PMPs must be devoid of a magnetic field. Accordingly, the spacing between the capture and release magnets must be sufficiently large to release the release magnet. For instance, with four cylindrical magnets in the head, if Sep is below 92 mm (Dim. A.c), the release magnet will still react (stand up) to the capture magnet. Therefore, spacing must exceed 92 mm to release the release magnet, causing it to fall on its side and release the PMPs from the magnetic field.
Whole Cell Isolation
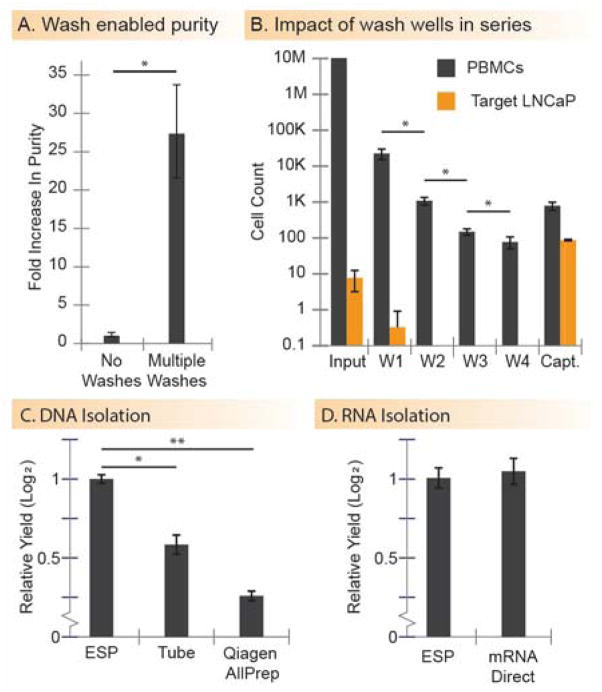
Purity and capture efficiency are the two most important measurements when isolating rare cells from complex or high-background cell-populations. One way to improve purity is through the addition of wash steps. The presented magnetic system is capable of capturing and releasing PMPs in a serial manner, enabling multiple washes. When isolating ~100 target LNCaP cells from a background of ~10 million peripheral blood mononuclear cells (PBMCs), the addition of four washes enables a ~28 fold improvement of purity over the original SLIDE platform (Fig. 3A, p = .016). In an isolation procedure consisting of four washes, we captured ~90% of target cells and reduced the background population of PBMCs by four orders of magnitude (Fig. 3B, bars represent cell counts in each respective well). PBMCs were removed from the sample via a series of four washes, which was quantified by counting the PBMCs remaining in each wash well. The efficacy of the washes decreased with additional wash (p < .05 for each case). Some PBMCs remained in the captured sample, suggesting that there is a lower limit to which PBMCs can be depleted. The PBMC fraction that made it into the final well of captured cells consisted primarily of cells that non-specifically adhered to the M270 PMPs (cell surface was covered with PMPs), as opposed to non-specific carryover (i.e., fluid carried over in the interstitial space of the PMP aggregate). Roughly 90% of target cells were captured, with little or no target cells lost in the wash wells (<5% in all replicates, N=3). The ~10% uncaptured target cells (remaining in the input) typically did not adhere to the PMPs. Based on observations, we speculate that the lack of adhesion is a result of improper (e.g., insufficient, or mutated) EpCAM expression on the cell surface, potentially a result of repetitive cell passages or cell cycle. Based on prior results, it is unlikely that the loss of cells is caused by depletion of beads or improper coating of the antibody.
Figure 3.
Data collected using the magnetic system. (A) The washing capabilities enabled by the magnetic box yield significantly higher purity over simply capturing and releasing cells (n = 3, p = .016). (B) Target cells are captured from a background of PBMCs using PMPs. The isolate is released in a series of four washes (W1-W4), to remove PBMC contaminant cells, then released in a final capture well (Capt.). The contents of each well (input, wash, and capture wells) are enumerated and the counts are shown (n = 3). (C and D) DNA and RNA are isolated using the magnetic box and compared to traditional methods. (n = 3). In all cases, error bars represent the standard deviation. (* indicates p < 0.05; ** indicates p < 0.01)
Nucleic Acid Isolation
To test this system’s compatibility with different types of PMPs and sample consistencies (i.e., sticky or viscous), we isolate RNA and DNA using application specific PMPs. For DNA extraction, the magnetic system demonstrates a higher recovery and tighter error bars as compared to traditional tube-based approaches (~0.8 fold increase, p = 0.033) and Qiagen AllPrep spin-columns (~2 fold increase, p = 0.005) (Fig. 3C). For RNA isolation, using reagents from the mRNA Direct Kit, the automated ESP demonstrates equivalent yield (no statistical difference, p = 0.74) to the manufacturer’s tube-based protocol (Fig. 3D). Importantly, the RNA and DNA are amplified via RT-qPCR-based reactions, confirming adequate purity for sensitive downstream analytical techniques.
Discussion
Fundamentally, this magnetic system is nothing more than a magnet in a box paired with a controllable capture magnet; yet, this simple infrastructure enables complex PMP manipulation and facilitates a wide range of assay workflows. While many PMP based assays are performed in tubes1, where a magnet is placed on the side of the tube to collect the PMPs or manually removed to re-suspend the PMPs, automation of these approaches can be complex. These traditional techniques have been adapted in automated platforms for use on both tubes and microplates, where the magnets are placed on the bottom, side, or even the inside of a well. However, to release the PMPs back into solution, the magnet and the tube/plate must be mechanically separated – adding complexity and required functionality in an automated platform – and the increased contact between the collector and the liquid is prone to higher carryover. Furthermore, when exchanging fluids in traditional tube-based approaches, complete fluid removal can be difficult; perpetually leaving potential contaminants with the PMPs. SLIDE provides a gentle, simple, and clean (low carryover) alternative for PMP isolation. The presented system overcomes the limitations of the original SLIDE technology, enabling adaptation into an automated platform and limitless manipulations of PMPs in and out of series of wells. The magnetic box is entirely self-contained – no mechanical or electrical manipulation required - making the system amenable to nearly any automated platform or plate format – including commercially available microtiter plates. However, to fully adapt the magnetic box to a new platform or plate, several design considerations must be taken into account.
Selection of the release magnet can be tuned to specific applications. The single bar magnet provided a high strength-to-weight ratio, and spanned the entire width of the plate making it adaptable to most plate configurations. Because beads collect in a region tied to the thickness of the magnet16 (.125″), this thin magnet enabled us to use plates with a 384-well format. Alternatively, arrays of magnets can also be used if only specific wells need to be magnetized. Regardless of the configuration of the release magnet (bar, array, etc.), it is important that the size of the compartment is sufficiently wide, such that the magnet can move freely and orient itself without getting stuck.
Advantages of this automated magnetic system become evident through testing various biological situations. The variety of analytes serve to demonstrate the versatility of this platform in extracting PMPs and bound analyte from various buffers, backgrounds, volumes, and washes, while using different mixing capabilities. The range of commercial PMPs tested demonstrates the system’s compatibility with PMPs of various sizes, surface chemistries, and magnetic moments. One important consideration is that carryover of detergents from many lysis buffers will inhibit PCR and thus, the PMPs must be adequately washed. In this system, PMPs can be transferred through a series of wash wells and mixed, as necessary, to ensure adequate washing and removal of lysis/binding buffer. In the cell capture experiments, contaminant cells are depleted by a combination of multiple washes with mixing. The success, as benchmarked against existing methods, demonstrates this platform’s utility in isolating a wide range of analytes, as well is its adaptability to various protocols and process configurations.
Conclusion
Presented here is a magnetic system that is both mechanically and operationally simplistic. Using the design criteria described herein, this magnetic technology is adaptable to nearly any plate configuration, including commercially available microplates. Both magnets are mobile, meaning there are no interfering magnetic fields, allowing PMPs to be resuspended or transferred in any sequence. Importantly, this technology is not reliant on any mechanical or electrical actuation (only that of the capture magnet) making it easy to integrate into any automated system where the capture magnets are robotically controlled. Using a two magnet system we are able to manipulate PMPs in several different operational modes including: 1) collecting PMPs at the surface of the well, 2) moving them over a neighboring well, 3) pulling them into the well, 4) releasing them for mixing as desired, and 5) recollecting them with ease to repeat this process any number of desired times. We have demonstrated the utility of this system by isolating DNA and mRNA with equivalent or enhanced yield to traditional assays, as well as isolation of whole cells with significant improvement in purity which was enabled through this new magnetic system.
Supplementary Material
Acknowledgments
DJB received funding from University of Wisconsin Carbone Cancer Center Support Grant (NIH P30 CA014520); SMB and HMP received funding from NIH R01 CA181648-01A1; DJG, HMP, SMB received funding from Prostate Cancer Foundation Challenge Award; MCR was funded by NIH T32 GM08349
Footnotes
Conflict of Interest:
The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: D. J. G., S. M. B., and D. J. B. hold equity in Salus Discovery, LLC; D. J. G. and D. J. B. hold equity in Tasso, Inc.; D. J. B. holds equity in Stacks to the Future, LLC, Onexion Biosystems, LLC and Lynx Biosciences, LLC.
Supporting Information. Cell culture, cell isolation, RNA isolation, and DNA isolation protocols for the biological validation.
References
- 1.Berensmeier S. Appl Microbiol Biotechnol. 2006;73:495–504. doi: 10.1007/s00253-006-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moussavi-Harami SF, Annis DS, Ma W, Berry SM, Coughlin EE, Strotman LN, Maurer LM, Westphall MS, Coon JJ, Mosher DF. J Proteome Research. 2013;12:3393–3404. doi: 10.1021/pr400225p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry SM, Alarid ET, Beebe DJ. Lab Chip. 2011;11:1747–1753. doi: 10.1039/c1lc00004g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bordelon H, Adams NM, Klemm AS, Russ PK, Williams JV, Talbot HK, Wright DW, Haselton FR. ACS Appl Mater Interfaces. 2011;3:2161–2168. doi: 10.1021/am2004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casavant BP, Guckenberger DJ, Berry SM, Tokar JT, Lang JM, Beebe DJ. Lab Chip. 2013;13:391–396. doi: 10.1039/c2lc41136a. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Abolmatty A, Faghri M. Microfluid Nanofluid. 2011;10:593–605. [Google Scholar]
- 7.Okochi M, Tsuchiya H, Kumazawa F, Shikida M, Honda H. J Biosci Bioeng. 2010;109:193–197. doi: 10.1016/j.jbiosc.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Shikida M, Takayanagi K, Honda H, Ito H, Sato K. J Micromech Microeng. 2006;16:1875. [Google Scholar]
- 9.Sur K, McFall SM, Yeh ET, Jangam SR, Hayden MA, Stroupe SD, Kelso DM. J Mol Diagn. 2010;12:620–628. doi: 10.2353/jmoldx.2010.090190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas PC, Strotman LN, Theberge AB, Berthier E, O’Connell R, Loeb JM, Berry SM, Beebe DJ. Anal Chem. 2013;85:8641–8646. doi: 10.1021/ac4012914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry SM, Chin EN, Jackson SS, Strotman LN, Goel M, Thompson NE, Alexander CM, Miyamoto S, Burgess RR, Beebe DJ. Anal Biochem. 2014;447:133–140. doi: 10.1016/j.ab.2013.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casavant BP, Guckenberger DJ, Beebe DJ, Berry SM. Anal Chem. 2014:6355–6362. doi: 10.1021/ac500574t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Au AK, Lee W, Folch A. Lab Chip. 2014;14:1294–1301. doi: 10.1039/c3lc51360b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatch GP, Stelter RE. J Magn Magn Mater. 2001;225:262–276. [Google Scholar]
- 15.Gaillard C, Strauss F. Tech Tips Online. 1998;3:63–65. [Google Scholar]
- 16.Gassner AL, Abonnenc M, Chen HX, Morandini J, Josserand J, Rossier JS, Busnel JM, Girault HH. Lab Chip. 2009;9:2356–2363. doi: 10.1039/b901865d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.