Abstract
Francisella tularensis, the causative agent of tularemia, is a Gram-negative bacterium that infects a variety of cell types including macrophages, and propagates with great efficiency in the cytoplasm. Iron, essential for key enzymatic and redox reactions, is among the nutrients required to support this pathogenic lifestyle and the bacterium relies on specialized mechanisms to acquire iron within the host environment. Two distinct pathways for iron acquisition are encoded by the F. tularensis genome- a siderophore-dependent ferric iron uptake system and a ferrous iron transport system. Genes of the Fur-regulated fslABCDEF operon direct the production and transport of the siderophore rhizoferrin. Siderophore biosynthesis involves enzymes FslA and FslC, while export across the inner membrane is mediated by FslB. Uptake of the rhizoferrin- ferric iron complex is effected by the siderophore receptor FslE in the outer membrane in a TonB-independent process, and FslD is responsible for uptake across the inner membrane. Ferrous iron uptake relies largely on high affinity transport by FupA in the outer membrane, while the Fur-regulated FeoB protein mediates transport across the inner membrane. FslE and FupA are paralogous proteins, sharing sequence similarity and possibly sharing structural features as well. This review summarizes current knowledge of iron acquisition in this organism and the critical role of these uptake systems in bacterial pathogenicity.
Keywords: Siderophore, FeoB, intracellular pathogen, FslE, FupA, TonB-independent, Francisella tularensis
Introduction
Francisella tularensis, the etiological agent of the zoonosis tularemia, is a Gram-negative gamma-proteobacterium with a small genome of 1.89 Mb (Sjöstedt, 2007). The species is further differentiated into three subspecies, of which tularensis causes a more severe disease than holarctica, while mediasiatica is less well-studied. The closely related species F. novicida, considered to be of a more ancestral lineage (Svensson et al., 2005) and with a genome sequence identity of ~98% (Larsson et al., 2009) is an opportunistic human pathogen, but can cause a virulent tularemia-like disease in mice.
In the laboratory, studies have largely focused on virulent strain Schu S4 of the tularensis subspecies, the attenuated live vaccine strain (LVS) derived from a holarctica isolate and the strain U112 of F. novicida. These three isolates share many biological attributes although their genetic and functional differences significantly impact virulence (Jones et al., 2012; Celli and Zahrt, 2013; Kingry et al., 2014). For this reason, all three strains are referred to in this review as F. tularensis unless specifically identified in order to highlight particular differences.
F. tularensis is a facultative intracellular pathogen infecting a wide variety of cells, including mammalian and arthropod cells (Ozanic et al., 2015). Following uptake into the macrophage, the bacteria at first reside within a phagosome, but then rapidly escape into the cytoplasm. Phagosomal escape is dependent on the igl operon and associated genes in the Francisella Pathogenicity Island (FPI) that encode components of a putative Type VI secretion system (Barker et al., 2009; de Bruin et al., 2011). The bacteria replicate to high numbers in the cytoplasm resulting finally in apoptotic death of the host cell. Adaptation to the specialized intracellular lifestyle is associated with evolutionary loss of genes for many metabolic pathways (Rohmer et al., 2007; Larsson et al., 2009), but F. tularensis has retained or evolved mechanisms to efficiently acquire essential nutrients within the intracellular niche of the different cell types that it infects (Meibom and Charbit, 2010).
Mice have been extensively used to model animal infection (Lyons and Wu, 2007). Phagocytic cells are thought to be the first infected (Hall et al., 2008); subsequently infection is disseminated to other tissues in the body. F. tularensis exercises several strategies to evade immune responses and is able to replicate to high levels in the liver, spleen and lungs before the immune system is provoked to respond with a destructive cytokine storm (Sharma et al., 2011; Jones et al., 2012).
Iron and francisella
Francisella requires iron for essential cellular functions. Early studies reported that infection with F. tularensis induces an iron-withholding response typical of the innate nutritional immunity defense mechanism (Pekarek et al., 1969). However, the intracellular pathogen manipulates host cell iron metabolism to support growth; LVS induces infected macrophages to increase iron flow through the cell by enhanced expression of the transferrin receptor TfR1 for uptake of iron and in parallel, increased expression of Dmt1 that moves endosomal iron into the cytoplasm and a slight increase in ferroportin that promotes outflow of iron from the cell (Pan et al., 2010). A functional Nramp1protein that also transports endosomal iron into the cytoplasm restricts growth of endosome-resident bacteria but enhances Francisella growth (Kovářová et al., 2000, 2002), highlighting the importance of cytoplasmic iron availability for pathogenesis. However, the nature of the host iron sources accessed by the organism remains to be characterized. Iron-limitation restricts growth of bacteria in culture (Deng et al., 2006; Sullivan et al., 2006) as well as within the macrophage (Fortier et al., 1995).
As might be predicted, growth of F. tularensis is inhibited by gallium, which competes with ferric iron for uptake and also interferes with iron-dependent biological processes (Olakanmi et al., 2010; Lindgren and Sjöstedt, 2016). Inhibition of the iron-associated enzymes catalase and superoxide dismutase leads to increased susceptibility to oxidative stress (Bakshi et al., 2006; Lindgren et al., 2007; Olakanmi et al., 2010; Binesse et al., 2015).
Iron metabolism appears to differ among F. tularensis isolates. LVS expresses higher levels of bacterioferritin as compared to Schu S4 (Hubálek et al., 2003, 2004). Consistent with these findings, isolates of the holarctica subspecies have greater iron stores than tularensis isolates, and since iron is closely associated with generation of reactive oxygen species, holarctica strains are more susceptible to oxidative stress (Lindgren et al., 2011).
Fur and iron regulation of genes
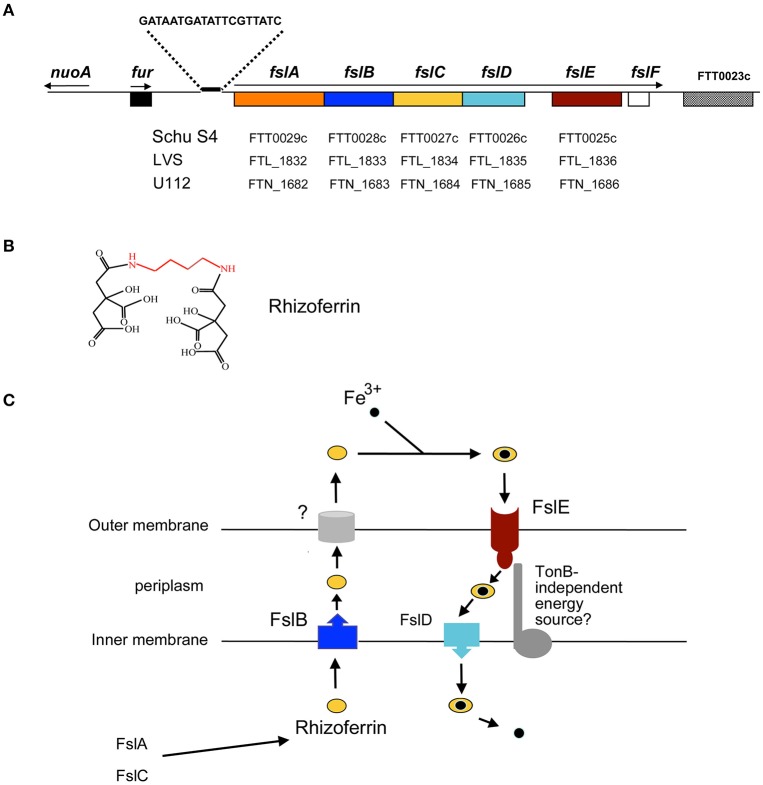
A fur ortholog is encoded in the F. tularensis genome and the predicted Fur protein contains elements known to be important for Fur function (Pérard et al., 2016). The fur gene is adjacent to the fsl operon encoding components of a siderophore- mediated iron uptake pathway, and a canonical Furbox is located upstream of the first gene of the operon, fslA (Figure 1A). Expression of the fsl operon is induced in iron-limiting media (Deng et al., 2006; Sullivan et al., 2006; Buchan et al., 2008). Loss of the fur gene results in deregulated transcription of the fsl operon and increased siderophore production (Buchan et al., 2008; Ramakrishnan et al., 2008). Expression of the inner membrane ferrous iron transporter feoB is also upregulated in a fur mutant (Pérez and Ramakrishnan, 2014).
Figure 1.
Siderophore-mediated iron acquisition in F. tularensis. (A) The fur-fsl locus of the F. tularensis subsp. tularensis chromosome is depicted and the corresponding gene designations in the Schu S4, LVS, and U112 genomes are indicated. The Furbox upstream of the fslABCDEF operon is shown. Arrows indicate the direction and extent of transcribed regions. (B) A schematic of the F. tularensis siderophore, rhizoferrin comprising two citrate molecules linked by amide bonds to a putrescine backbone (in red). (C) Current model for siderophore-mediated iron acquisition in F. tularensis and the roles of the fsl operon products. FslA and FslC encode enzymes for biosynthesis of the siderophore. The siderophore is released into the extracellular medium by the action of FslB and an as yet unknown outer membrane component. The outer membrane siderophore receptor FslE, and FslD in the inner membrane mediate uptake of the ferric-siderophore complex. It is not known if a TonB analog facilitates FslE function.
Exposure to iron limitation was also shown to increase virulence of an F. tularensis isolate, suggesting that pathogenicity is influenced by iron levels (Bhatnagar et al., 1995). Microarray analysis of RNA indicated that besides the fsl operon, transcription of the igl operon was increased under iron limitation (Deng et al., 2006). Proteomic analysis also confirmed that iron limitation results in increased levels of the IglC protein (Lenco et al., 2007). However, although an fslB-lacZ reporter could be repressed by overexpression of Fur, an iglB-lacZ reporter was not similarly repressible (Buchan et al., 2008), suggesting that a mechanism besides Fur regulates the igl genes in response to iron levels in F. tularensis.
Siderophore-mediated iron acquisition
A “growth inducing substance (GIS)” that promoted growth of F. tularensis bacteria from small inocula was reported in the 1960s (Halmann and Mager, 1967; Halmann et al., 1967); in all likelihood this substance was the siderophore now identified as the polycarboxylate rhizoferrin (Drechsel et al., 1991; Thieken and Winkelmann, 1992; Sullivan et al., 2006). Rhizoferrin is structurally simple, comprising 2 citrate moieties linked through amide bonds to a putrescine backbone (Figure 1B). Originally identified as a fungal siderophore (Drechsel et al., 1991; Thieken and Winkelmann, 1992), rhizoferrin was subsequently also isolated from a strain of the bacterium Ralstonia pickettii (Münzinger et al., 1999). The identification of the siderophore made by F. tularensis as rhizoferrin (Sullivan et al., 2006), and the subsequent identification of the Legionella pneumophila siderophore legiobactin as also rhizoferrin (Burnside et al., 2015) suggests that this siderophore may be more widely prevalent in bacteria than suspected. Structurally related siderophores made by bacteria include staphyloferrin A (Konetschny-Rapp et al., 1990) and corynebactin (Zajdowicz et al., 2012), where D-ornithine and lysine, respectively, constitute the siderophore backbone in place of the putrescine present in rhizoferrin. The R. pickettii rhizoferrin was shown by CD spectroscopy to be an S-S enantiomer in contrast to the R-R fungal molecule (Münzinger et al., 1999). Whether all bacterial rhizoferrins adopt the S-S conformation is not clear, but bacteria making rhizoferrin are capable of utilizing the fungal form for iron uptake (Münzinger et al., 1999; Kiss et al., 2008).
Genes for synthesis and transport of Francisella rhizoferrin are located on the siderophore operon fslABCDEF (also designated figABCDEF; (Deng et al., 2006; Sullivan et al., 2006; Milne et al., 2007; Buchan et al., 2008; Ramakrishnan et al., 2008); Figures 1A,C). Analysis of individual mutants as well as complementation of a strain carrying a complete deletion of the fslA-F genes helped to determine the roles played by the different genes in siderophore-mediated iron acquisition, as detailed below.
fslA and fslC and share homology with genes found in siderophore biosynthetic loci of other bacteria. FslA is similar to the aerobactin synthetases IucA/IucC and a member of the non-ribosomal peptide synthetase-independent siderophore (NIS) synthetases, enzymes that assemble non-peptide siderophores using dicarboxylic acids and diamines or amino-alcohols (Challis, 2005). FslC is predicted to be a member of the pyridoxal phosphate-dependent decarboxylases. Mutant analysis demonstrated that both fslA and fslC are required for Francisella rhizoferrin production (Deng et al., 2006; Sullivan et al., 2006; Lindgren et al., 2009; Thomas-Charles et al., 2013). Rhizoferrin biosynthesis in F. tularensis, involving just two dedicated enzymes, may be the simplest siderophore biosynthetic pathway identified thus far.
fslB encodes a transporter of the Major Facilitator superfamily (MFS) and deletion of this gene in F. novicida results in reduced levels of siderophore activity in the culture medium (Kiss et al., 2008). Additionally, detection of siderophore activity in culture supernatants of an LVS ΔfslA-F mutant required complementation with the fslB gene in addition to the biosynthetic genes fslA and fslC (Pérez et al., 2016). These observations support a role for FslB in export of the siderophore across the cytoplasmic membrane. How the siderophore is channeled through the outer membrane into the extracellular space is currently not known.
fslD encodes an inner membrane MFS protein and deletion of this gene was found to have little effect on siderophore production (Kiss et al., 2008). The role of this protein in siderophore-mediated iron uptake across the cytoplasmic membrane was deduced on the basis of complementation studies: a Schu S4 ΔfslA-F mutant is able to transport 55Fe3+ complexed to siderophore only when complemented with the fslD gene in addition to the fslE receptor gene (see below; Pérez et al., 2016).
fslE, the fifth gene in the operon, encodes an outer membrane protein unique to the Francisella genus Larsson et al., 2005; Huntley et al., 2007). ΔfslE mutants are impaired for growth in iron-limiting media and are unable to utilize exogenous siderophore for growth (Kiss et al., 2008; Ramakrishnan et al., 2008). In transport assays, fslE mutants proved incapable of siderophore-mediated 55Fe3+ uptake, establishing a role for FslE as receptor for the siderophore (Ramakrishnan et al., 2012). FslE can also transport the iron mimic gallium in complex with rhizoferrin as shown by the resistance of fslA and fslE mutants to gallium (Pérez et al., 2016).
The last gene of the fsl operon, fslF, varies structurally among the tularensis and the novicida species, being truncated in the tularensis isolates. Studies with a ΔfslF mutant in Schu S4 indicate that the gene does not influence iron transport in F. tularensis (Pérez et al., 2016).
Siderophore-mediated iron transport by outer membrane receptors in Gram-negative bacteria is typically dependent on the proton motive force transduced by the TonB-ExbB-ExbD complex (Noinaj et al., 2010). The Francisella genome, however, does not encode orthologs of tonB, exbB, and exbD, implying that alternative mechanisms must facilitate siderophore -iron uptake.
Ferrous iron uptake
The F. tularensis genome encodes an inner membrane ferrous iron transport system comprising unlinked genes feoA and feoB. ΔfeoB mutants of LVS and Schu S4 are deficient for growth on iron-limiting media (Thomas-Charles et al., 2013; Pérez and Ramakrishnan, 2014; Pérez et al., 2016). 55Fe uptake assays demonstrated that the F. tularensis ΔfeoB mutants are completely deficient in ferrous iron uptake (Pérez and Ramakrishnan, 2014; Pérez et al., 2016), implying that the Feo system is the sole ferrous iron transporter across the inner membrane (Figure 2). It is likely that FeoA supports FeoB function as seen in Salmonella Typhimurium (Kim et al., 2012) and Vibrio cholerae (Weaver et al., 2013; Stevenson et al., 2016).
Figure 2.
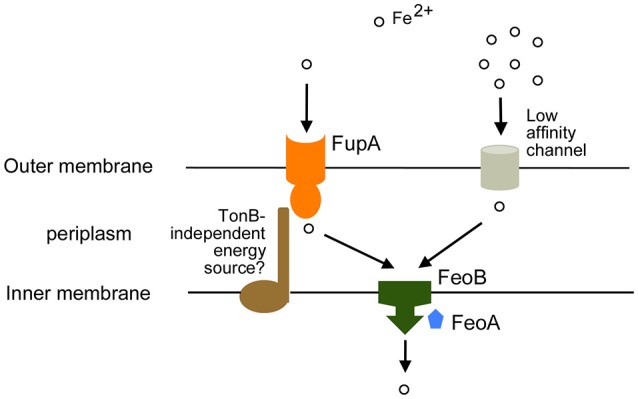
Ferrous iron acquisition in F. tularensis. Transport of ferrous iron relies on the high affinity transporter FupA and uncharacterized low affinity channels in the outer membrane. Inner membrane transport is dependent on the FeoB transporter with likely involvement of FeoA. We raise the possibility that an accessory protein may facilitate FupA function.
Given the soluble nature of ferrous iron, the general assumption has been that it diffuses into the periplasmic space through non-specific porin proteins in the outer membrane. However, growth and 55Fe transport assays indicate that F. tularensis is capable of high-affinity uptake of ferrous iron mediated by the specific outer membrane protein FupA (Ramakrishnan et al., 2012). FupA was initially characterized as a virulence factor in Schu S4 (Twine et al, 2005) and found to influence bacterial intracellular replication (Twine et al, 2005; Asare et al., 2010). An involvement in iron acquisition was established by the finding that Schu S4 ΔfupA mutant grew poorly under iron limitation, had lowered internal iron levels and was deregulated for siderophore production (Lindgren et al., 2009). A link to ferrous iron uptake was indicated by the finding that ferrous and ferric iron supplements supported growth of a fupA mutant to different extents on agar plates (Ramakrishnan et al., 2012). 55Fe transport assays clearly demonstrated that the fupA mutant was unable to transport ferrous iron at limiting concentrations (~0.1 μM) although low affinity ferrous iron transport at high concentrations (~3 μM) could still be observed, and siderophore-iron uptake was not perturbed (Ramakrishnan et al., 2012). These findings suggest that FupA serves as a F. tularensis adaptation to efficiently acquire ferrous iron even in low abundance settings when general diffusion-based transport across the outer membrane might prove inadequate (Figure 2).
FslE and FupA: related high-affinity iron transport proteins
FupA and FslE are paralogs belonging to a family of proteins unique to Francisella (Larsson et al., 2005). Both proteins have been localized to the outer membrane of F. tularensis (Huntley et al., 2007; Ramakrishnan and Sen, 2014) and share a global 54% identity and 69% similarity in amino acid sequence. FupA with 557 amino acid residues is larger than FslE (509 residues). The Hidden Markov Model-based PRED-TMBB program (Bagos et al., 2004a,b) predicts that both FupA and FslE fold as β-barrels in the outer membrane with amino-terminal periplasmic domains. FupA is predicted to form a 16-stranded barrel with a periplasmic domain of 201 residues while FslE could form a 14-stranded barrel with a 152 residue periplasmic domain. This structure is reminiscent of typical TonB-dependent transporters (Noinaj et al., 2010). The greatest similarity between FslE and FupA is in the predicted β-barrel domains.
The fupA gene is located adjacent to a paralog fupB on the chromosome. Infrequent recombinational deletion events have been observed leading to formation of fupA/B hybrid genes (Twine et al, 2005; Rohmer et al., 2006); such recombination accounts for a significant reduction in virulence as seen in LVS, which can be reversed by restoration of the full length fupA gene (Salomonsson et al., 2009). The FupA/B hybrid protein encoded by LVS is less efficient at high-affinity ferrous iron uptake than FupA, but gains siderophore-iron uptake capability (Sen et al., 2010; Ramakrishnan and Sen, 2014). The structural and functional overlap in protein function raises the intriguing possibility that a common mechanism may underlie transport by FslE and FupA.
fupA expression is independent of iron and fur regulation, suggesting that FupA may have functions in addition to iron transport. fupA mutants have increased resistance to copper, and transport assays indicated that copper competes with ferrous iron for transport (Pérez et al., 2016). A role for FupA in maintenance of outer membrane integrity has also been proposed (Nallaparaju et al., 2011). Alhough, the fsl operon is regulated in response to iron levels, it was reported recently that calcium and magnesium limitation also result in increased fslE transcription (Wu et al., 2016). These observations are consistent with the idea that the high affinity iron transport proteins in the outer membrane of F. tularensis may assume roles in transport of additional substrates under stress.
Interestingly, FslE appears structurally different from rhizoferrin receptors of other bacteria. The LbtU siderophore receptor of L. pneumophila is not predicted to have a distinct periplasmic domain (Chatfield et al., 2011). RumA, the rhizoferrin receptor in Morganella morganii is a TonB-dependent transporter (Kühn et al., 1996). Mechanisms for rhizoferrin transport thus appear to have evolved independently in different bacteria.
Iron uptake and pathogenesis
Analysis of the transcriptome of Schu S4-infected macrophages demonstrated that the fsl genes are among the most highly induced genes in the intracellular niche (Wehrly et al., 2009). Similar results were obtained using LVS infected hepatocytes (Thomas-Charles et al., 2013), suggesting that the siderophore uptake system contributes to survival within different tissue types. fsl mutants were identified in a negative selection screen of a U112 transposon mutant library in mice (Weiss et al., 2007) and a signature-tagged mutant screen identified fslA and feoB to be important for pulmonary infection by LVS (Su et al., 2007). Nevertheless, individual mutants in the fsl, fupA, and feoB genes are capable of intracellular growth although the reduced growth of the feoB mutants in hepatocytes suggests that ferrous iron is likely the major iron source within these cells (Lindgren et al., 2009; Ramakrishnan et al., 2012; Thomas-Charles et al., 2013; Pérez and Ramakrishnan, 2014; Pérez et al., 2016). Both iron uptake pathways appear to contribute to utilization of iron from heme for growth (Lindgren et al., 2015), suggesting that iron needs to be released from the heme for use by the bacteria. Screens with U112 mutants have implicated additional genes in iron acquisition, but they have not been definitively characterized (Crosa et al., 2009).
A Schu ΔfslE ΔfupA mutant deficient for both siderophore and high affinity ferrous iron uptake grows slowly, is attenuated for growth in macrophages and completely avirulent in mice (Ramakrishnan et al., 2012). ΔfslA ΔfeoB mutants of LVS and Schu S4 deficient for siderophore biosynthesis and for all ferrous iron uptake have an even more severe defect, with dependence on extraneous siderophore for growth, loss of all capacity for intracellular growth and complete loss of virulence (Pérez and Ramakrishnan, 2014; Pérez et al., 2016). These findings indicate that both subspecies of F. tularensis have a similar repertoire of iron uptake mechanisms, limited to just the fsl and feo-mediated mechanisms. Interestingly, although the ΔfslE ΔfupA and the ΔfslA ΔfeoB mutants are avirulent in mice, exposure to these strains protects from subsequent challenge with the virulent strain (Ramakrishnan et al., 2012; Pérez et al., 2016), making them good candidates for further exploration as live vaccines.
Conclusions
With a reduced genome, F. tularensis has evolved to efficiently support its lifestyle as an intracellular pathogen with a minimal set of two iron acquisition pathways. Host iron sources utilized and mechanisms regulating the transport proteins FslE and FupA are interesting questions for future investigations.
Author contributions
The author confirms being the sole contributor of this work and approved it for publication.
Funding
Work in the author's lab has been supported by grants AI056227, AI067823, and AI119471 from the National Institute of Allergy and Infectious Diseases (NIH) and by intramural support from the School of Medicine, University of Virginia.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Asare R., Akimana C., Jones S., Abu Kwaik Y. (2010). Molecular bases of proliferation of Francisella tularensis in arthropod vectors. Environ. Microbiol. 12, 2587–2612. 10.1111/j.1462-2920.2010.02230.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagos P. G., Liakopoulos T. D., Spyropoulos I. C., Hamodrakas S. J. (2004a). A hidden markov model method, capable of predicting and discriminating β barrel outer membrane proteins. BMC Bioinformatics 5:29. 10.1186/1471-2105-5-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagos P. G., Liakopoulos T. D., Spyropoulos I. C., Hamodrakas S. J. (2004b). PRED-TMBB: a web server for predicting the topology of β barrel outer membrane proteins. Nucleic Acids Res. 32, W400–W404. 10.1093/nar/gkh417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi C. S., Malik M., Regan K., Melendez J. A., Metzger D. W., Pavlov V. M., et al. (2006). Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J. Bacteriol. 188, 6443–6448. 10.1128/JB.00266-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J. R., Chong A., Wehrly T. D., Yu J. J., Rodriguez S. A., Liu J., et al. (2009). The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol. Microbiol. 74, 1459–1470. 10.1111/j.1365-2958.2009.06947.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar N. B., Elkins K. L., Fortier A. H. (1995). Heat stress alters the virulence of a rifampin-resistant mutant of Francisella tularensis LVS. Infect. Immun. 63, 154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binesse J., Lindgren H., Lindgren L., Conlan W., Sjöstedt A. (2015). Roles of reactive oxygen species-degrading enzymes of Francisella tularensis SCHU S4. Infect. Immun. 83, 2255–2263. 10.1128/IAI.02488-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan B. W., McLendon M. K., Jones B. D. (2008). Identification of differentially regulated Francisella tularensis genes by use of a newly developed Tn5-based transposon delivery system. Appl. Environ. Microbiol. 74, 2637–2645. 10.1128/AEM.02882-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside D. M., Wu Y., Shafaie S., Cianciotto N. P. (2015). The Legionella pneumophila siderophore legiobactin is a polycarboxylate that is identical in structure to rhizoferrin. Infect. Immun. 83, 3937–3945. 10.1128/IAI.00808-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J., Zahrt T. C. (2013). Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb. Perspect. Med. 3:a010314. 10.1101/cshperspect.a010314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis G. L. (2005). A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. ChemBioChem 6, 601–611. 10.1002/cbic.200400283 [DOI] [PubMed] [Google Scholar]
- Chatfield C. H., Mulhern B. J., Burnside D. M., Cianciotto N. P. (2011). Legionella pneumophila LbtU acts as a novel, tonb-independent receptor for the legiobactin siderophore. J. Bacteriol. 193, 1563–1575. 10.1128/JB.01111-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa L. M., Crosa J. H., Heffron F. (2009). Iron Transport in Francisella in the absence of a recognizable TonB protein still requires energy generated by the proton motive force. Biometals 22, 337–344. 10.1007/s10534-008-9170-7 [DOI] [PubMed] [Google Scholar]
- de Bruin O. M., Duplantis B. N., Ludu J. S., Hare R. F., Nix E. B., Schmerk C. L., et al. (2011). The biochemical properties of the Francisella Pathogenicity Island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU Suggest roles in Type VI secretion. Microbiology 157, 3483–3491. 10.1099/mic.0.052308-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng K., Blick R. J., Liu W., Hansen E. J. (2006). Identification of Francisella tularensis genes affected by iron limitation. Infect. Immun. 74, 4224–4236. 10.1128/IAI.01975-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel H., Metzger J., Freund S., Jung G., Boelaert J. R., Winkelmann G. (1991). Rhizoferrin- a novel siderophore from the fungus Rhizopus microsporus Var. Rhizopodiformis. Biometals 4, 238–243. [Google Scholar]
- Fortier A. H., Leiby D. A., Narayanan R. B., Asafoadjei E., Crawford R. M., Nacy C. A., et al. (1995). Growth of Francisella tularensis LVS in Macrophages: the acidic intracellular compartment provides essential iron required for growth. Infect. Immun. 63, 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. D., Woolard M. D., Gunn B. M., Craven R. R., Taft-Benz S., Frelinger J. A., et al. (2008). Infected-host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis Schu S4, LVS, or U112. Infect. Immun. 76, 5843–5852. 10.1128/IAI.01176-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halmann M., Mager J. (1967). An endogenously produced substance essential for growth initiation of Pasteurella tularensis. J. Gen. Microbiol. 49, 461–468. 10.1099/00221287-49-3-461 [DOI] [Google Scholar]
- Halmann M., Magda B., Mager J. (1967). Nutritional requirements of Pasteurella tularensis for growth from small inocula. J. Gen. Microbiol. 49, 451–460. 10.1099/00221287-49-3-451 [DOI] [Google Scholar]
- Hubálek M., Hernychová L., Brychta M., Lenčo J., Zechovská J., Stulík J. (2004). Comparative proteome analysis of cellular proteins extracted from highly virulent Francisella tularensis ssp. tularensis and less virulent, F. tularensis ssp. holarctica and F. tularensis ssp. mediaasiatica. Proteomics 4, 3048–3060. 10.1002/pmic.200400939 [DOI] [PubMed] [Google Scholar]
- Hubálek M., Hernychová L., Havlasová J., Kasalová I., Neubauerová V., Stulík J., et al. (2003). Towards proteome database of Francisella tularensis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 787, 149–177. 10.1016/S1570-0232(02)00730-4 [DOI] [PubMed] [Google Scholar]
- Huntley J. F., Conley P. G., Hagman K. E., Norgard M. V. (2007). Characterization of Francisella tularensis outer membrane proteins. J. Bacteriol. 189, 561–574. 10.1128/JB.01505-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. L., Napier B. A., Sampson T. R., Llewellyn A. C., Schroeder M. R., Weiss D. S. (2012). Subversion of host recognition and defense systems by Francisella spp. Microbiol. Mol. Biol. Rev. 76, 383–404. 10.1128/MMBR.05027-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Lee H., Shin D. (2012). The FeoA protein is necessary for the FeoB transporter to import ferrous iron. Biochem. Biophys. Res. Commun. 423, 733–738. 10.1016/j.bbrc.2012.06.027 [DOI] [PubMed] [Google Scholar]
- Kingry L. C., Petersen J. M. (2014). Comparative review of Francisella tularensis and Francisella novicida. Front. Cell. Infect. Microbiol. 4:35. 10.3389/fcimb.2014.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss K., Liu W., Huntley J. F., Norgard M. V., Hansen E. J. (2008). Characterization of fig operon mutants of Francisella novicida U112. FEMS Microbiol. Lett. 285, 270–277. 10.1111/j.1574-6968.2008.01237.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konetschny-Rapp S., Jung G., Meiwes J., Zähner H. (1990). Staphyloferrin a: a structurally new siderophore from staphylococci. Eur. J. Biochem. 191, 65–74. 10.1111/j.1432-1033.1990.tb19094.x [DOI] [PubMed] [Google Scholar]
- Kovářová H., Halada P., Man P., Golovliov I., Kovářová Z., Špaček J., et al. (2002). Proteome Study of Francisella tularensis Live vaccine strain-containing phagosome in Bcg/Nramp1 congenic macrophages: resistant allele contributes to permissive environment and susceptibility to infection. Proteomics 2, 85–93. [DOI] [PubMed] [Google Scholar]
- Kovářová H., Hernychova L., Hajduch M., Sirova M., Macela A. (2000). Influence of the Bcg locus on natural resistance to primary infection with the facultative intracellular bacterium Francisella tularensis in mice. Infect. Immun. 68, 1480–1484. 10.1128/IAI.68.3.1480-1484.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn S., Braun V., Köster W. (1996). Ferric rhizoferrin uptake into Morganella morganii: characterization of genes involved in the uptake of a polyhydroxycarboxylate siderophore. J. Bacteriol. 178, 496–504. 10.1128/jb.178.2.496-504.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P., Elfsmark D., Svensson K., Wikström P., Forsman M., Brettin T., et al. (2009). Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog. 5:e1000472. 10.1371/journal.ppat.1000472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P., Oyston P. C., Chain P., Chu M. C., Duffield M., Fuxelius H. H., et al. (2005). The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37, 153–159. 10.1038/ng1499 [DOI] [PubMed] [Google Scholar]
- Lenco J., Hubalek M., Larsson P., Fucikova A., Brychta M., Macela A., et al. (2007). Proteomics analysis of the Francisella tularensis LVS response to iron restriction: induction of the F. tularensis pathogenicity island proteins IglABC. FEMS Microbiol. Lett. 269, 11–21. 10.1111/j.1574-6968.2006.00595.x [DOI] [PubMed] [Google Scholar]
- Lindgren H., Sjöstedt A. (2016). Gallium potentiates the antibacterial effect of gentamicin against Francisella tularensis. Antimicrob. Agents Chemother. 60, 288–295. 10.1128/AAC.01240-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H., Honn M., Golovlev I., Kadzhaev K., Conlan W., Sjostedt A. (2009). The 58-kilodalton major virulence factor of Francisella tularensis Is required for efficient utilization of iron. Infect. Immun. 77, 4429–4436. 10.1128/IAI.00702-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H., Honn M., Salomonsson E., Kuoppa K., Forsberg Å., Sjö¨stedt A. (2011). Iron content differs between Francisella tularensis Subspecies tularensis and Subspecies holarctica strains and correlates to their susceptibility to H(2)O(2)-induced killing. Infect. Immun. 79, 1218–1224. 10.1128/IAI.01116-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H., Lindgren L., Golovliov I., Sjöstedt A. (2015). Mechanisms of heme utilization by Francisella tularensis. PLoS ONE 10:e0119143. 10.1371/journal.pone.0119143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H., Shen H., Zingmark C., Golovliov I., Conlan W., Sjöstedt A. (2007). Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect. Immun. 75, 1303–1309. 10.1128/IAI.01717-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons R. C., Wu T. H. (2007). Animal models of Francisella tularensis infection. Ann. N. Y. Acad. Sci. 1105, 238–265. 10.1196/annals.1409.003 [DOI] [PubMed] [Google Scholar]
- Meibom K. L., Charbit A. (2010). Francisella tularensis metabolism and its relation to virulence. Front. Microbiol. 1:140. 10.3389/fmicb.2010.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne T. S., Michell S. L., Diaper H., Wikström P., Svensson K., Oyston P. C., et al. (2007). A 55 kDa hypothetical membrane protein is an iron-regulated virulence factor of Francisella tularensis subsp. Novicida U112. J. Med. Microbiol. 56(Pt 10), 1268–1276. 10.1099/jmm.0.47190-0 [DOI] [PubMed] [Google Scholar]
- Münzinger M., Taraz K., Budzikiewicz H., Drechsel H., Heymann P., Winkelmann G., et al. (1999). S,S-Rhizoferrin (Enantio-Rhizoferrin) - a siderophore of Ralstonia (Pseudomonas) pickettii DSM 6297 - the optical antipode of R,R-rhizoferrin isolated from fungi. Biometals 12, 189–193. 10.1023/A:1009259118034 [DOI] [Google Scholar]
- Nallaparaju K. C., Yu J. J., Rodriguez S. A., Zogaj X., Manam S., Guentzel M. N., Seshu J., et al. (2011). Evasion of IFN- γ signaling by Francisella novicida is dependent upon Francisella outer membrane protein, C. PLoS ONE 6:e18201. 10.1371/journal.pone.0018201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj N., Guillier M., Barnard T. J., Buchanan S. K. (2010). TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60. 10.1146/annurev.micro.112408.134247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olakanmi O., Gunn J. S., Su S., Soni S., Hassett D. J., Britigan B. E. (2010). Gallium disrupts iron uptake by intracellular and extracellular Francisella strains and exhibits therapeutic efficacy in a murine pulmonary infection model. Antimicrob. Agents Chemother. 54, 244–253. 10.1128/AAC.00655-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanic M., Marecic V., Abu Kwaik Y., Santic M. (2015). The divergent intracellular lifestyle of Francisella tularensis in evolutionarily distinct host cells. PLoS Pathog. 11:e1005208. 10.1371/journal.ppat.1005208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Tamilselvam B., Hansen E. J., Daefler S. (2010). Modulation of iron homeostasis in macrophages by bacterial intracellular pathogens. BMC Microbiol. 10:64. 10.1186/1471-2180-10-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarek R. S., Bostian K. A., Bartelloni P. J., Calia F. M., Beisel W. R. (1969). The effects of Francisella tularensis infection on iron metabolism in man. Am. J. Med. Sci. 258, 14–25. 10.1097/00000441-196907000-00003 [DOI] [PubMed] [Google Scholar]
- Pérard J., Covés J., Castellan M., Solard C., Savard M., Miras R., Galop S., et al. (2016). Quaternary structure of fur proteins, a new subfamily of tetrameric proteins. Biochemistry 55, 1503–1515. 10.1021/acs.biochem.5b01061 [DOI] [PubMed] [Google Scholar]
- Pérez N. M., Ramakrishnan G. (2014). The reduced genome of the Francisella tularensis live vaccine strain (LVS) encodes two iron acquisition systems essential for optimal growth and virulence. PLoS ONE 9:e93558. 10.1371/journal.pone.0093558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez N., Johnson R., Sen B., Ramakrishnan G. (2016). Two parallel pathways for ferric and ferrous iron acquisition support growth and virulence of the intracellular pathogen Francisella tularensis Schu S4. Microbiologyopen 5, 453–468. 10.1002/mbo3.342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan G., Sen B. (2014). The FupA/B protein uniquely facilitates transport of ferrous iron and siderophore-associated ferric iron across the outer membrane of Francisella tularensis live vaccine strain. Microbiology 160(Pt 2), 446–457. 10.1099/mic.0.072835-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan G., Meeker A., Dragulev B. (2008). Fsle is necessary for siderophore-mediated iron acquisition in Francisella tularensis Schu S4. J. Bacteriol. 190, 5353–5361. 10.1128/JB.00181-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan G., Sen B., Johnson R. (2012). Paralogous outer membrane proteins mediate uptake of different forms of iron and synergistically govern virulence in Francisella tularensis tularensis. J. Biol. Chem. 287, 25191–25202. 10.1074/jbc.M112.371856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L., Brittnacher M., Svensson K., Buckley D., Haugen E., Zhou Y., et al. (2006). Potential source of Francisella tularensis live vaccine strain attenuation determined by genome comparison. Infect. Immun. 74, 6895–6906. 10.1128/IAI.01006-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L., Fong C., Abmayr S., Wasnick M., Larson Freeman T. J., Radey M., et al. (2007). Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 8:R102. 10.1186/gb-2007-8-6-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonsson E., Kuoppa K., Forslund A. L., Zingmark C., Golovliov I., Sjöstedt A., et al. (2009). Reintroduction of two deleted virulence loci restores full virulence to the live vaccine strain of Francisella tularensis. Infect. Immun. 77, 3424–3431. 10.1128/IAI.00196-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen B., Meeker A., Ramakrishnan G. (2010). The fslE Homolog, FTL-0439 (fupA/B), Mediates siderophore-dependent iron uptake in Francisella tularensis LVS. Infect. Immun. 78:10. 10.1128/IAI.00503-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J., Mares C. A., Li Q., Morris E. G., Teale J. M. (2011). Features of sepsis caused by pulmonary infection with Francisella tularensis Type A strain. Microb. Pathog. 51, 39–47. 10.1016/j.micpath.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostedt A. (2007). Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann. N. Y. Acad. Sci. 1105, 1–29. 10.1196/annals.1409.009 [DOI] [PubMed] [Google Scholar]
- Stevenson B., Wyckoff E. E., Payne S. M. (2016). Vibrio cholerae FeoA, FeoB, and FeoC interact to form a complex. J. Bacteriol. 198, 1160–1170. 10.1128/JB.00930-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Yang J., Zhao D., Kawula T. H., Banas J. A., Zhang J. R. (2007). Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 75, 3089–3101. 10.1128/IAI.01865-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J. T., Jeffery E. F., Shannon J. D., Ramakrishnan G. (2006). Characterization of the siderophore of Francisella tularensis and role of fslA in siderophore production. J. Bacteriol. 188, 3785–3795. 10.1128/JB.00027-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson K., Larsson P., Johansson D., Byström M., Forsman M., Johansson A. (2005). Evolution of Subspecies of Francisella tularensis. J. Bacteriol. 187, 3903–3908. 10.1128/JB.187.11.3903-3908.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieken A., Winkelmann G. (1992). Rhizoferrin: a complexone type siderophore of the Mucorales and Entomophthorales (Zygomycetes). FEMS Microbiol. Lett. 94, 37–41. 10.1111/j.1574-6968.1992.tb05285.x [DOI] [PubMed] [Google Scholar]
- Thomas-Charles C. A., Zheng H., Palmer L. E., Mena P., Thanassi D. G., Furie M. B. (2013). FeoB-mediated uptake of iron by Francisella tularensis. Infect. Immun. 81, 2828–2837. 10.1128/IAI.00170-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twine S., Byström M., Chen W., Forsman M., Golovliov I., Johansson A., et al. (2005). A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58-kilodalton protein is attenuated for virulence and is an effective live vaccine. Infect. Immun. 73, 8345–8352. 10.1128/IAI.73.12.8345-8352.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver E. A., Wyckoff E. E., Mey A. R., Morrison R., Payne S. M. (2013). FeoA and FeoC are essential components of the Vibrio cholerae ferrous iron uptake system, and FeoC interacts with FeoB. J. Bacteriol. 195, 4826–4835. 10.1128/JB.00738-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrly T. D., Chong A., Virtaneva K., Sturdevant D. E., Child R., Edwards J. A., et al. (2009). Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell. Microbiol. 11, 1128–1150. 10.1111/j.1462-5822.2009.01316.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D. S., Brotcke A., Henry T., Margolis J. J., Chan K., Monack D. M. (2007). In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U.S.A. 104, 6037–6042. 10.1073/pnas.0609675104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Ren G., Gunning W. T., III., Weaver D. A., Kalinoski A. L., Khuder S. A., et al. (2016). FmvB: a Francisella tularensis magnesium-responsive outer membrane protein that plays a role in virulence. PLoS ONE 11:e160977. 10.1371/journal.pone.0160977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajdowicz S., Haller J. C., Krafft A. E., Hunsucker S. W., Mant C. T., Duncan M. W., et al. (2012). Purification and structural characterization of siderophore (Corynebactin) from Corynebacterium diphtheriae. PLoS ONE 7:e34591. 10.1371/journal.pone.0034591 [DOI] [PMC free article] [PubMed] [Google Scholar]