Abstract
The objective of this study was to determine the minimal inhibitory concentration of colistin for Escherichia coli from food animals and the possible underlying colistin resistance mechanisms. During 2007–2014, 4,438 E. coli isolates of food animal origins were collected. The susceptibility of colistin was tested by the agar dilution method. Mutations in pmrA, pmrB, and mgrB and the presence of mcr-1 gene were determined by PCR and DNA sequencing. Complementation experiments were carried out to evaluate the contribution of the mutations to colistin resistance. There was a high frequency of colistin resistance in E. coli from pigs on farm (24.1%) and at slaughter (24.3%) in 2013–2014, followed by chickens on farm (14.0%) and at slaughter (9.5%). The resistance frequency of E. coli in cow isolates was the lowest (0.9%). MIC distribution for colistin showed that most isolates (75.2%) were distributed at 0.25 mg/L–0.5 mg/L, followed by 4 mg/L–8 mg/L (16.8%). Compared with the isolates from pigs and chickens recovered during 2013–2014, E. coli isolates collected during 2007–2008 (5.5%) and 2010–2011 (12.4%) showed significantly lower frequency of colistin resistance (P < 0.05). DNA sequencing and complementation experiments failed to detect any insertion inactivation or mutation in pmrA, pmrB, and mgrB associated with colistin resistance. However, 91.0% colistin-resistant isolates were positive for mcr-1. The high frequency of colistin resistance and mcr-1 gene among E. coli isolates from food animals in China urged the need to minimize potential risks of colistin resistance development and the spread of mcr-1 gene.
Keywords: Escherichia coli, colistin, food animals, resistance, mcr-1
Introduction
The rising prevalence of multidrug-resistant (MDR) gram-negative Enterobacteriaceae (GNB), especially carbapenem- resistant, has resulted in a renewed interest in polymyxins, especially polymyxin E (colistin), for the management of gram-negative infections in many countries (Falagas and Michalopoulos, 2006; Cassir et al., 2014). Despite their relatively recent reintroduction in clinical practice, reports on colistin resistant isolates are on the rise (Falagas et al., 2010; Olaitan et al., 2014a). Resistance to polymyxins has been traditionally regarded as occurring via mutations in genes regulating the synthesis of LAra4N (Falagas et al., 2010; Olaitan et al., 2014b). However, we recently described for the first time the emergence of plasmid-mediated colistin resistance gene, mcr-1, which now has been identified in several Enterobacteriaceae species from various sources (environment, food, animal and humans) (Liu et al., 2016).
Colistin has been used in veterinary medicine through prophylactic or metaphylactic practices, but the prevalence of colistin resistance in bacteria isolated from animals in many countries was still low (Kempf et al., 2013; Wasyl et al., 2013; Quesada et al., 2014). In China, colistin has been widely used in veterinary medicine, especially in swine and poultry for many years. We previously detected high prevalence of mcr-1 among E. coli isolates from pigs at slaughter in Guangzhou (Liu et al., 2016). Soon after, mcr-1 gene has been reported to be present in Enterobacteriaceae from animals, food and humans worldwide (Schwarz et al., 2001; Quan et al., 2017; Wang et al., 2017a). However, little is known about the prevalence of colistin resistance and mcr-1 gene among commensal E. coli isolates from other food animals in China. The aim of this study was to investigate the frequency of colistin resistance among commensal E. coli isolates from farm animals (chicken, cattle, and pig) and food animals at slaughter recovered from 12 provinces of China and to determine the possible underlying mechanisms among part of colistin-resistant isolates.
Materials and Methods
Origin of E. coli Isolates
Cloacal samples from chickens (laying hens, chickens, and broilers) and rectal swabs from pigs (piglets, weaned pigs, fattening pigs, and sows) and cattle were collected from 107 food animal farms located in different geographic areas of China (Guangdong, Henan, Jiangxi, Ningxia, Jilin, Qinghai, Sichuan, Shanghai, Jiangsu, Shandong, Beijing, and Neimeng) from May 2013 to August 2014 (Table 1). Animals were randomly selected for sampling on each farm based on their age and stage of production. Ten to thirty samples per stage of production per farm were collected. In addition, cecal contents of chickens from seven farmers markets and two live-bird markets and rectal swabs of pigs from two live pig markets and eight abattoirs located in Guangdong, Henan, Shandong, Liaoning, and Sichuan province were collected at slaughter between April 2013 and August 2014. No more than five animal samples per farm were analyzed. All samples were seeded on MacConkey agar plates and were incubated at 37°C for 24 h. One presumptive colony with typical E. coli morphology and size was selected and then inoculated on eosin-methylene blue agar. After incubation, suspected E. coli colony was identified using classical biochemical methods. In addition, 349 E. coli isolates (91 were from chicken during 2007–2008, 86 from chicken during 2010–2011, and 172 from pigs during 2010–2011) from healthy food animals mentioned in our previous study were also included in this study for comparison (Yang et al., 2014).
Table 1.
Prevalence of colistin resistance among Escherichia coli isolates of different origins.
| Animals | Farm number | Samples | Number of isolates | Number of colistin-resistant isolates (%) |
|---|---|---|---|---|
| 2013–2014 | ||||
| Laying hens | 21 | 357 | 295 | 25 (8.5) |
| Broilers | 43 | 886 | 611 | 102 (16.7) |
| Chickens | 6 | 90 | 67 | 9 (13.4) |
| All farm chickens | 47 | 1333 | 973 | 13 (14.0) |
| Chickens at slaughter | 456 | 325 | 31 (9.5) | |
| Piglets | 15 | 275 | 246 | 57 (23.2) |
| Weaned pigs | 12 | 180 | 150 | 97 (64.7) |
| Fattening pigs | 32 | 713 | 664 | 141 (21.2) |
| Sows | 24 | 361 | 332 | 26 (7.8) |
| All farm pigs | 46 | 1529 | 1392 | 335 (24.1) |
| Pigs at slaughter | 1200 | 1063 | 258 (24.3) | |
| Cows | 13 | 370 | 336 | 3 (0.9) |
| Total | 4888 | 4089 | 763 (18.7) | |
| 2007–2008 | ||||
| Farm chickens | 91 | 5 (5.5) | ||
| 2010–2011 | ||||
| Farm chickens | 86 | 10 (11.6) | ||
| Farm pigs | 172 | 22 (12.8) |
Antimicrobial Susceptibility Testing
The minimal inhibitory concentration (MIC) of colistin was determined by the agar dilution method according to the protocols recommended in M100-S25 of the (Clinical and Laboratory Standards Institute, 2013). For isolates from pigs at slaughter, MICs of ampicillin, cefotaxime, imipenem, gentamicin, amikacin, neomycin, apramycin, florfenicol, tetracycline, ciprofloxacin, and fosfomycin were also determined. The results were interpreted according to epidemiological cut-off (ECOFF) values recommended by EUCAST1 (colistin, florfenicol, and neomycin) and the interpretative criteria recommended by CLSI (M100-S25) (ampicillin, cefotaxime, gentamicin, amikacin, fosfomycin, and ciprofloxacin) (Clinical and Laboratory Standards Institute, 2013).
Statistical significance for the comparison of resistance prevalence data was determined by the χ2 test. P values less than 0.05 were considered statistically significant.
PCR Amplification and Sequencing
A total of 200 colistin-resistant E. coli isolates of different origins (127 from pigs, 70 from chickens, and 3 from cows) were randomly selected for PCR amplification of mcr-1 (Liu et al., 2016). In addition, 50 of them were randomly selected for sequencing for genes encoding PmrA, PmrB, and MgrB. pmrA were amplified using primers described previously (Quesada et al., 2014). The primers used for amplification of entire mgrB and pmrB genes were as follows: EmgrB-F (5′- CCGCTGAGTAATAATCCTAT -3′) and EmgrB-R (5′- TACAACCAAAGACGCAAT -3′), EpmrB-F (5′- ATAAGCTGAAACGGATGGC -3′) and EpmrB-R (5′- CATAATAATCAGGGCGAAAGT -3′). PCR products of pmrA, pmrB, and mgrB were sequenced and the nucleotides and deduced protein sequences were analyzed at the National Center for Biotechnology Information web site2. In addition, the pmrA, pmrB, and mgrB sequences of five colistin-susceptible E. coli isolates were determined as control.
Complementation Experiments
The wild-type mgrB and pmrB genes from an E. coli reference strain ATCC 25922 (colistin MIC of 0.25 μg/ml) were amplified by PCR using primers EmgrB-F/EmgrB-R and EpmrB-F/EpmrB-R, respectively. The non-coding mdh sequence was amplified by PCR using primers described previously (Jayol et al., 2014). The PCR products were cloned into the plasmid pCR-BluntII-TOPO (Invitrogen) encoding resistance to kanamycin and zeocin. The resulting plasmids pTOPO-mgrB, pTOPO-pmrB, and pTOPO-mdh were separately transformed into E. coli TOP10 strains by electroporation. Transformants were selected by on Mueller–Hinton agar supplemented with 50 mg/L of kanamycin. The recombinant plasmids were isolated and transformed into electrocompetent colistin-resistant E. coli with mgrB mutation (pTOPO-mgrB) or E. coli with pmrB mutation (pTOPO-pmrB) via electroporation. The transformants were selected on Mueller–Hinton agar supplemented with zeocin (25 mg/L) and the presence of the cloned gene was confirmed by PCR. The colistin MICs of the transformants were determined by the agar dilution method.
Results
Antimicrobial Susceptibility
Overall, 4089 commensal E. coli collected from 973 chickens on farm, 1392 pigs on farm, 325 chickens at slaughter, 1063 pigs at slaughter, and 336 cows on farm during 2013–2014 were recovered from 4888 samples. Among them, 763 (18.7%) isolates showed resistance to colistin (MIC ≥ 4 mg/L) (Table 1). The MIC values were shown in Figure 1. MICs of most isolates (75.2%) were distributed at 0.25 mg/L–0.5 mg/L, followed by 4 mg/L–8 mg/L (16.8%). Only MICs of 6.1% isolates and 1.9% isolates were distributed at 1 mg/L–2 mg/L and ≥16 mg/L, respectively.
FIGURE 1.
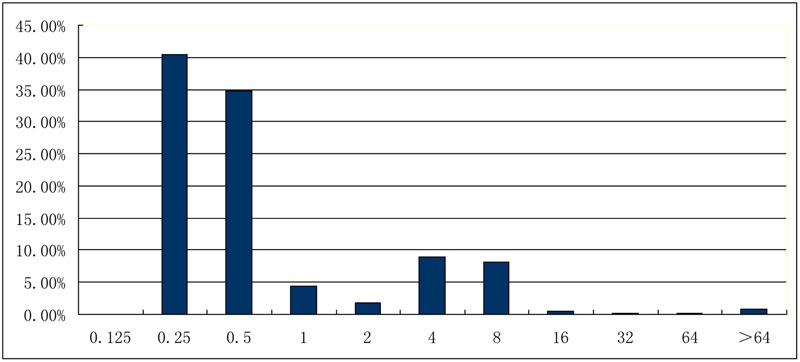
Distribution of minimal inhibitory concentrations (mg/L) of Escherichia coli isolates toward colistin.
There was a high frequency of colistin resistance in E. coli from pigs on farm (24.1%) and at slaughter (24.3%), followed by chickens on farm (14.0%) and at slaughter (9.5%). The resistance frequency of cow isolates was the lowest (0.9%). Compared with the isolates recovered during 2013-2014, E. coli isolates collected during 2007–2008 and 2010–2011 showed significantly lower frequency of colistin resistance (P < 0.05, Table 1).
Of the 258 colistin resistant isolates from pig at slaughter, 76.7% showed resistance to 3–9 other antimicrobial agents, including tetracycline (93.8%), ampicillin (79.5%), florfenicol (64.3%), cefotaxime (13.2%), neomycin (56.6%), gentamicin (29.1%), ciprofloxacin (28.3%), apramycin (12.4%), fosfomycin (7.8%), and amikacin (0.4%) (Table 2). The frequencies of antimicrobial resistance to other antimicrobial agents among colistin resistant isolates were significantly higher than those of colistin susceptible isolates (P < 0.01), except to amikacin and gentamicin. All of the isolates were susceptible to imipenem.
Table 2.
Comparison of antimicrobial susceptibility of colistin-susceptible isolates and colistin-resistant isolates from pigs at slaughter.
| Antimicrobial agents | Colistin-susceptible isolates (n = 805) (%) | Colistin-resistant isolates (n = 258) (%) | chi-square value | P-value |
|---|---|---|---|---|
| Ampicillin | 65.30 | 79.50 | 18.126 | < 0.0001 |
| Cefotaxime | 6.60 | 13.20 | 11.307 | 0.0008 |
| Amikacin | 0.90 | 0.90 | 0 | 1.000 |
| Gentamicin | 23.90 | 29.10 | 2.8293 | 0.093 |
| Apramycin | 4.5 | 12.40 | 20.5248 | < 0.0001 |
| Neomycin | 32.60 | 56.60 | 47.52 | < 0.0001 |
| Tetracycline | 84.80 | 93.80 | 293.998 | < 0.0001 |
| Florfenicol | 47.50 | 64.30 | 22.31 | < 0.0001 |
| Fosfomycin | 2.10 | 7.80 | 18.5003 | < 0.0001 |
| Ciprofloxacin | 17.60 | 28.30 | 13.7470 | 0.0002 |
mcr-1 Detection and Sequence of pmrA, pmrB, and mgrB Genes
Of the 200 randomly seiected colistin-resistant isolates, 182 (91.0%) were positive for mcr-1. The sequences of the pmrA, pmrB, and mgrB genes known to be involved in polymyxin resistance were determined in 50 isolates. For PmrA, no amino acid substitution was observed among the 50 isolates except one isolate that had the G144S substitution. However, G144S substitution was found to be present in colistin-susceptible isolates (Quesada et al., 2014). For MgrB, 4 isolates from different regions possessed D31G substitution. For PmrB, one isolate had two amino acid substitutions (T246I and D282N). However, the MICs of colistin remained unchanged upon transformation with plasmid pTOPO-mgrB or pTOPO-pmrB.
Discussion
Despite the frequent use of colistin in animal farming for over 50 years, the occurrence of colistin resistance among E. coli strains isolated from food animals remains low (<1%) (Kempf et al., 2013; Kieffer et al., 2015). However, in this study, we found a very high prevalence of colistin resistance (18.7%) among commensal E. coli isolates from food animals, especially pigs. The frequency of resistance in commensal intestinal E. coli is considered to be a good marker for the selection pressure exerted by antibiotic use in the host animals and the resistance problems to be predicted in pathogenic bacteria (van den Bogaard and Stobberingh, 2000). This high prevalence of colistin resistance may be due to the increasing use of colistin in food animals in recent years. Our previous studies showed that most E. coli strains from chicken and pigs in China showed resistance to fluoroquinolones and florfenicol, and over 20% isolates exhibited resistance to third-generation cephalosporins, amikacin and fosfomycin (Chen et al., 2014; Rao et al., 2014). Thus, in recent years, the lack of effective drugs against E. coli might be attributed by the increased consumption of colistin in veterinary medicine, especially in piglets which are frequently treated with colistin sulphate for colibacillosis. This high selective pressure might result in the highest prevalence (64.7%) of colistin resistance among E. coli isolates from weaned piglets found in this study. Compared with pig and chicken isolates, the prevalence of colistin resistance among E. coli from cows was very low (0.9%) which might be associated with the infrequent use of this drug on dairy-farm.
To determine whether there was an increase of colistin resistance from 2007 to 2014, E. coli isolates collected in our previous study were reviewed for colistin resistance. By comparison, colistin resistance among E. coli isolated from chicken raised nearly three times from 2007/2008 to 2013/2014 and that among E. coli isolated from pigs raised nearly two times from 2010/2011 to 2013/2014. Though we could not obtain the amount of colistin consumption on each farm sampled in this study, data from China Veterinary Drug Association showed that the volume of colistin sales increased significantly from 2011 to 2013 (China Veterinary Drug Association, 2014). Taken together, our results revealed that colistin resistance in food animals was correlated with the consumption of colistin.
Interestingly, MICs of most colistin- resistant isolates were 4 or 8 mg/L. The emergence and spread of E. coli with low level of colstin resistance (MIC = 4 or 8) might lead to the treatment failure of diarrhea with standard colistin dosage (2–20 mg/kg, ppm). Thus, farmers have to illegally use medicated feed added with increased dosage of colistin (80–100 ppm) to prevent diarrhea in piglets (personal communication).
Colistin resistance among commensal E. coli isolates recovered from pigs at slaughter was also worrisomely high. These resistant bacteria might contaminate meat during slaughtering procedures and transfer to humans by food chain or to workers via direct animal contact as indicated in some previous studies (Angulo et al., 2004; Liebana et al., 2013). Recently, several studies suggested the possibility of the transference MCR-1-producing Enterobacteriaceae to humans via food chain (Campos et al., 2016; Carnevali et al., 2016; Figueiredo et al., 2016; Wang et al., 2017b). Hence it is urgent to limit the usage of polymyxins (colistin) in veterinary medicine especially as feed additives in China. Fortunately, following our discovery of mcr-1, the Chinese Government has banned the use of colistin in animal feed since Nov 1, 2016 (Walsh and Wu, 2016).
Similar to our previous results, colistin resistance is mainly caused by mcr-1 gene. Quesada et al. have recently found mutations in PmrAB that confer resistance to polymyxins in E. coli (Quesada et al., 2014). However, in this study, we failed to detect any meaningful mutation in pmrAB and mgrB conferring resistance to colistin. Further studies are needed to understand the possible mechanism mediating colistin resistance among mcr-1-negative isolates.
Conclusion
We have detected a high prevalence of colistin resistance and mcr-1 gene in E. coli from food animals. Though colistin exhibited high antimicrobial activities against GNB, including E. coli, A. baumannii, Pseudomonas aeruginosa isolates, and K. pneumonia in human (Chen et al., 2015), the frequent presence of mcr-1-positive E. coli and in food animals might be a threat to human. As colistin is the last therapeutic option against infections caused by MDR GNB, careful monitoring of the evolution of colistin resistance and the spread of mcr-1 gene in isolates from humans in China is urgently needed.
Author Contributions
Conceived and designed the experiments: J-HL, XH, and ZZ. Performed the experiments: XH, LY, XC, CZ, XY, YL, SW, ZG, and LY. Analyzed the data: J-HL, XH, LY, YL, and ZZ. Wrote the paper: J-HL and XH.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by the National Key Basic Research Program of China (No. 2013CB127200) and the Guangdong Natural Science Foundation (No. S2012030006590).
Footnotes
References
- Angulo F. J., Nargund V. N., Chiller T. C. (2004). Evidence of an association between use of anti-microbial agents in food animals and anti-microbial resistance among bacteria isolated from humans and the human health consequences of such resistance. J. Vet. Med. B Infect. Dis. Vet. Public Health 51 374–379. 10.1111/j.1439-0450.2004.00789.x [DOI] [PubMed] [Google Scholar]
- Campos J., Cristino L., Peixe L., Antunes P. (2016). MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill. 21 1–5. 10.2807/1560-7917.ES.2016.21.26.30270 [DOI] [PubMed] [Google Scholar]
- Carnevali C., Morganti M., Scaltriti E., Bolzoni L., Pongolini S., Casadei G. (2016). Occurrence of mcr-1 in colistin-resistant Salmonella enterica isolates recovered from humans and animals in Italy, 2012 to 2015. Antimicrob. Agents Chemother. 60 7532–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassir N., Rolain J. M., Brouqui P. (2014). A new strategy to fight antimicrobial resistance: the revival of old antibiotics. Front. Microbiol. 5:551 10.3389/fmicb.2014.00551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wang Z., Li H., Wang Q., Zhao C., He W., et al. (2015). In vitro activities of 16 antimicrobial agents against Gram-negative bacteria from six teaching hospitals in China. Jpn. J. Infect. Dis. 68 263–267. 10.7883/yoken.JJID.2014.202 [DOI] [PubMed] [Google Scholar]
- Chen X., Zhang W., Yin J., Zhang N., Geng S., Zhou X., et al. (2014). Escherichia coli isolates from sick chickens in China: changes in antimicrobial resistance between 1993 and 2013. Vet. J. 202 112–115. 10.1016/j.tvjl.2014.06.016 [DOI] [PubMed] [Google Scholar]
- China Veterinary Drug Association (2014). Annual Report on Development of Veterinary Medicine Industry in China (2012). Beijing: China Agriculture Press. [Google Scholar]
- Clinical and Laboratory Standards Institute (2013). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Test for Bacteria Isolated from Animals; Second Informational Supplement. CLSI Document VET01-S23. Wayne, PA: CLSI. [Google Scholar]
- Falagas M. E., Michalopoulos A. (2006). Polymyxins: old antibiotics are back. Lancet 367 633–634. 10.1016/S0140-6736(06)68241-X [DOI] [PubMed] [Google Scholar]
- Falagas M. E., Rafailidis P. I., Matthaiou D. K. (2010). Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist. Updat. 13 132–138. 10.1016/j.drup.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Figueiredo R., Card R. M., Nunez J., Pomba C., Mendonça N., Anjum M. F., et al. (2016). Detection of an mcr-1-encoding plasmid mediating colistin resistance in Salmonella enterica from retail meat in Portugal. J. Antimicrob. Chemother. 71 2338–2340. 10.1093/jac/dkw240 [DOI] [PubMed] [Google Scholar]
- Jayol A., Poirel L., Brink A., Villegas M. V., Yilmaz M., Nordmann P. (2014). Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 58 4762–4766. 10.1128/AAC.00084-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf I., Fleury M. A., Drider D., Bruneau M., Sanders P., Chauvin C., et al. (2013). What do we know about resistance to colistin in Enterobacteriaceae in avian and pig production in Europe? Int. J. Antimicrob. Agents 42 379–383. 10.1016/j.ijantimicag.2013.06.012 [DOI] [PubMed] [Google Scholar]
- Kieffer N., Poirel L., Nordmann P., Madec J. Y., Haenni M. (2015). Emergence of colistin resistance in Klebsiella pneumoniae from veterinary medicine. J. Antimicrob. Chemother. 70 1265–1267. [DOI] [PubMed] [Google Scholar]
- Liebana E., Carattoli A., Coque T. M., Hasman H., Magiorakos A. P., Mevius D., et al. (2013). Public health risks of enterobacterial isolates producing extended-spectrum beta-lactamases or AmpC beta-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin. Infect. Dis. 56 1030–1037. 10.1093/cid/cis1043 [DOI] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet infect. Dis. 16 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Olaitan A. O., Diene S. M., Kempf M., Berrazeg M., Bakour S., Gupta S. K., et al. (2014a). Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int. J. Antimicrob. Agents 44 500–507. 10.1016/j.ijantimicag.2014.07.020 [DOI] [PubMed] [Google Scholar]
- Olaitan A. O., Morand S., Rolain J. M. (2014b). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J., Li X., Chen Y., Jiang Y., Zhou Z., Zhang H., et al. (2017). Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect. Dis. 17 400–410. 10.1016/S1473-3099(16)30528-X [DOI] [PubMed] [Google Scholar]
- Quesada A., Porrero M. C., Tellez S., Palomo G., Garcia M., Dominguez L. (2014). Polymorphism of genes encoding PmrAB in colistin-resistant strains of Escherichia coli and Salmonella enterica isolated from poultry and swine. J. Antimicrob. Chemother. 70 71–74. 10.1093/jac/dku320 [DOI] [PubMed] [Google Scholar]
- Rao L., Lv L., Zeng Z., Chen S., He D., Chen X., et al. (2014). Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003-2012. Vet. Microbiol. 172 534–541. 10.1016/j.vetmic.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Schwarz S., Kehrenberg C., Walsh T. R. (2001). Use of antimicrobial agents in veterinary medicine and food animal production. Int. J. Antimicrob. Agents 17 431–437. 10.1016/S0924-8579(01)00297-7 [DOI] [PubMed] [Google Scholar]
- van den Bogaard A. E., Stobberingh E. E. (2000). Epidemiology of resistance to antibiotics. Links between animals and humans. Int. J. Antimicrob. Agents 14 327–335. 10.1016/S0924-8579(00)00145-X [DOI] [PubMed] [Google Scholar]
- Walsh T. R., Wu Y. (2016). China bans colistin as a feed additive for animals. Lancet Infect. Dis. 16 1102–1103. 10.1016/S1473-3099(16)30329-2 [DOI] [PubMed] [Google Scholar]
- Wang Y., Tian G. B., Zhang R., Shen Y., Tyrrell J. M., Huang X., et al. (2017a). Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 10.1016/S1473-3099(16)30527-8 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang R., Li J., Wu Z., Yin W., Schwarz S., et al. (2017b). Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2:16260 10.1038/nmicrobiol.2016.260 [DOI] [PubMed] [Google Scholar]
- Wasyl D., Hoszowski A., Zajac M., Szulowski K. (2013). Antimicrobial resistance in commensal Escherichia coli isolated from animals at slaughter. Front. Microbiol. 4:221 10.3389/fmicb.2013.00221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Zeng Z., Rao L., Chen X., He D., Lv L., et al. (2014). The association between occurrence of plasmid-mediated quinolone resistance and ciprofloxacin resistance in Escherichia coli isolates of different origins. Vet. Microbiol. 170 89–96. 10.1016/j.vetmic.2014.01.019 [DOI] [PubMed] [Google Scholar]


