Abstract
Recent large-scale genetic approaches such as genome-wide association studies have allowed the identification of common genetic variations that contribute to risk architectures of psychiatric disorders. However, most of these susceptibility variants are located in noncoding genomic regions that usually span multiple genes. As a result, pinpointing the precise variant(s) and biological mechanisms accounting for the risk remains challenging. By reviewing recent progresses in genetics, functional genomics and neurobiology of psychiatric disorders, as well as gene expression analyses of brain tissues, here we propose a roadmap to characterize the roles of noncoding risk loci in the pathogenesis of psychiatric illnesses (that is, identifying the underlying molecular mechanisms explaining the genetic risk conferred by those genomic loci, and recognizing putative functional causative variants). This roadmap involves integration of transcriptomic data, epidemiological and bioinformatic methods, as well as in vitro and in vivo experimental approaches. These tools will promote the translation of genetic discoveries to physiological mechanisms, and ultimately guide the development of preventive, therapeutic and prognostic measures for psychiatric disorders.
Recent genetic analyses of neuropsychiatric disorders
Schizophrenia, bipolar disorder, major depressive disorder and autism are highly prevalent complex neuropsychiatric diseases across the world populations with varied clinical symptomatology.1 The underlying etiology of these neuropsychiatric disorders remains largely unknown, but a number of family, twin and adoption studies have revealed moderate-to-strong genetic elements in these illnesses.2, 3 Although substantial heritability of neuropsychiatric disorders suggests that genetic approaches may provide valuable information about its biology, early linkage and association analyses in small samples yielded limited success because of the phenotypic heterogeneity and genetic architecture complexity in most psychiatric conditions. Recent large-scale genome-wide association studies (GWASs) through international collaborations suggest that these psychiatric disorders, though all featured genetic heterogeneity,4 share varying degrees of overlap in genetic risk components.5 These studies have significantly boosted our knowledge of reliable genetic variants associated with the diseases.6, 7, 8, 9, 10
Some examples of these GWASs include the Psychiatric Genomics Consortium (PGC2) GWAS of schizophrenia8 as well as Converge consortium GWAS and 23andMe GWAS of major depressive disorder.6, 10 The PGC2 GWAS, which identified 108 independent associated loci for schizophrenia,8 is of epoch-making significance. Most of these identified risk variants shed great light on the pathogenesis of psychiatric illnesses. In the PGC2 schizophrenia GWAS,8 75% of the 108 independent risk loci contained protein-coding genes (40% include one single gene), and a further 8% were within 20 kb of a gene. In fact, many genes within those GWAS risk loci were found to play important roles in neurodevelopment and brain function. For example, GRM3, GRIN2A, SRR and GRIA1 were known to involve in the neurotransmission mediated by glutamate signaling and synaptic plasticity. Some risk loci also contained genes encoding voltage-gated calcium channel subunits (CACNA1C, CACNB2 and CACNA1I) that play pivotal physiological roles. Indeed, genes involved in glutamate signaling, synaptic plasticity and calcium channel activity were also previously implicated in rare genetic variation studies of schizophrenia.11, 12, 13 Moreover, the genes relevant to the major hypotheses for schizophrenia pathogenesis (for example, DRD2, a validated target for antipsychotic drugs) have also been confirmed in the GWASs,8 adding further evidence for these hypotheses. Taken together, the convergence at a broad functional level between studies of common and rare genetic variations suggests that the current genetic approaches are normally reliable.
In addition to its reliability, GWAS is also highly efficient for genetic risk factor identification. It is capable of capturing not only single risk loci7, 8, 10 but also polygenic components in psychiatric diseases. These polygenic components usually involve thousands of common alleles of small effects.14 They are particularly meaningful when determined simultaneously in one study, in which case they could represent biological pathways associated with disease risk. Taking the recent PGC2 schizophrenia GWAS, for example,8 the defined risk loci supported the neuropsychopharmacologic hypotheses that both dopamine D2 receptor and N-methyl-D-aspartate receptor were involved in schizophrenia pathogenesis. Similarly, another pathway analyses based on available GWAS data suggested that malfunction of pathways in histone methylation, neuroimmunology and synaptic signaling transmission likely contributed to the etiology of psychiatric disorders.15 As such, new therapeutic hypotheses can be proposed and ultimately tested based on the underlying molecular mechanisms of genetic risk associations revealed by GWAS.16
Although generally accepted by the scientific community for the reliability and efficiency, the GWAS results are sometimes under debate for the normally small effect sizes of the identified risk loci. This is probably because of the fact that the allele frequencies in patients and controls typically differ by only <2% (the difference between odds ratio is <1.1),6, 7, 8, 9, 10, 17, 18 whereas such variants still yield strong statistical association in GWAS analyses. However, the small effects of single locus at the individual subject level do not mean that the biological implications are necessarily trivial. One example is DRD2, the gene encoding the dopamine D2 receptor. The GWAS variant at this locus was shown to increase the risk of schizophrenia by <10% with an odds ratio of 1.08 at the population level,8 whereas the dopamine D2 receptor has been a primary target of most antipsychotic interventions.19 Therefore, the effect size of a given DNA variant on genetic risk for psychiatric disorders does not always reflect the therapeutic value of its affected molecule or biological pathway, and the GWAS-identified risk loci could provide valuable clues. However, whether a certain GWAS locus is translatable into clinical treatment for psychiatric illnesses still largely depends on the relevance of the proteins and/or transcripts affected by susceptibility genes to common biological pathways.
The idea of translating the bulk of GWAS data to clinical application is compelling, but the actual implementation is not always easy. The vast majority of GWAS risk regions contain numerous high linkage disequilibrium noncoding variants with unknown function, and span wide genomic area of multiple candidate genes.8 For this reason, identifying the specific gene(s) accounting for the clinical association remains a hassle. Considering these characteristics of the current enormous GWAS results, several following important questions should be first asked when utilizing such genetic data to uncover the relevant disease biology. (1) Are there any specific pathogenic gene or transcription products in the particular loci explaining the genetic risk association? (2) Are there any causal variant(s) in certain genomic regions responsible for the disease risk? (3) What are the mechanisms through which the causal variant(s) affect gene expression/function? Moreover, (4) are there physiological consequences of the genetic risk factors? If so, are they related to disease-associated pathological alternations?
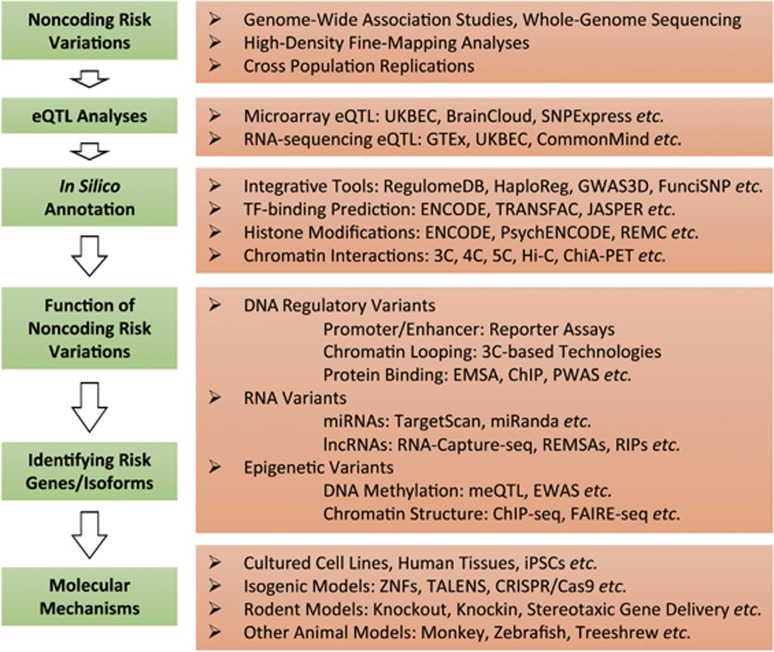
Taken together, GWAS provides valuable information for psychiatric disease research, but efforts are required to tackle possible problems. Here, we have reviewed recent progress in functional evaluation of noncoding variations in psychiatric genetic studies (including GWAS and candidate gene analyses), and have proposed a stepwise pipeline to follow the risk loci identification for prioritizing putative functional variations (Figure 1). Through integrating genomic data, computational approaches and biological assays, we sought to elucidate the function of noncoding GWAS risk loci and their roles in psychiatric illnesses.
Figure 1.
Workflow for functionally analyzing and interpreting noncoding risk loci. 3C, chromosome conformation capture; 4C, circular 3C; 5C, carbon-copy 3C; eQTL, expression quantitative trait locus.
Translating genetic susceptibility loci to risk molecules
Utilization of brain eQTL databases
To understand the roles of noncoding risk loci in disease progression, it is important to predict their functional and physiological impact on human health. Among the numerous risk variants defined by genetic analyses such as GWAS, exome-sequencing or candidate gene studies, nonsynonymous variants are perhaps the easiest to illustrate for the manifestation of protein structure or function alternations.20 Indeed, missense variants in specific risk genes have been found to significantly associate with psychiatric disorders in both GWAS and whole-exome sequencing studies (for example, rare loss-of-function variants in SETD1A).13, 20, 21, 22 However, as discussed above, most psychiatric risk loci are located in noncoding genomic regions with unknown functionality.8 This is probably explained by the accumulating consensus that genetic architecture of psychiatric disorders involves sequence variations that primarily play a role in gene regulation or processing rather than sequences of encoded proteins.23 Therefore, it is necessary to use transcriptomics data combined with genotyping in neural tissues to identify genes or transcripts associated with risk genotypes. These genes and/or transcription products are considered ‘true signals' that are critical in linking the information from genetic associations to functional consequences, mostly through their expression quantitative trait locus (eQTL) effects.
As stated by the name, eQTL provides hints for transcriptome (for example, promoter regulation, enhancer, splicing, microRNA, long noncoding RNA and epigenetic processes) alterations attributed to specific genetic risk factors. As we believe that psychiatric risk variants that do not result in protein coding changes usually manifest as an effect on transcriptomic outcomes, integrative analyses of GWAS and eQTL data are crucial in understanding genetic mechanisms and discovering targetable molecules. In fact, several recent studies have achieved great success following this strategy. For example, the major histocompatibility complex region on human chromosome 6 spanned over hundreds of genes and contained numerous variants conferring risk of schizophrenia in PGC2 GWAS,8 but little was known about the underlying molecular mechanisms of this genomic region. Sekar et al.24 recently identified that the structurally diverse alleles of the complement component 4 (C4) genes were the major contributors. They reported that those alleles caused widely varying levels of C4A and C4B expression in the brain, and the association of common C4 allele with schizophrenia was proportional to its tendency to increase the expression of C4A.24 They further revealed localization of human C4 protein in the neuronal synapses, dendrites, axons and cell bodies, as well as important roles of murine C4 in synapse elimination during postnatal development.24 Overall, this study suggested that excessive dosage of C4 led to increased postnatal synaptic pruning, providing one of the potential mechanisms for the previously observed gray matter loss25 and synaptic structure impairment in schizophrenic brains.26 They illustrated an excellent model for the meticulous identification and characterization of causal, small effect size, common loci found in GWAS. Another interesting example is the series of studies on ANK3 in bipolar disorder. Genetic loci spanning the ANK3 gene were previously reported in several bipolar disorder GWASs.7, 27, 28, 29 A recent study found a loss-of-function variant in a minor isoform of ANK3 that disabled the proper splicing of the protein, resulting in protection against bipolar disorder.30 Intriguingly, another study also described that allelic variation at the bipolar disorder risk single-nucleotide polymorphism (SNP) was correlated with a significant difference in cerebellar expression of a brain-specific ANK3 transcript.31 Although these studies reported different SNPs at ANK3 (and in very low linkage disequilibrium, r2=0.001),30, 31 they together suggested a cis-regulatory transcriptional effect of ANK3 that was relevant to bipolar disorder pathophysiology.
The eQTL data are both informative and easily accessible. There have been a number of eQTL databases with varying sample sizes generated from certain groups (Table 1). These databases present valuable brain transcriptomics (microarray and RNA sequencing) data in post-mortem tissues that has long been an essential substrate for investigating the molecular pathology of psychiatric disorders.32, 33 To date, the major consortia generating brain eQTLs are the GTEx (Genotype-Tissue Expression), the United Kingdom Brain Expression Consortium (UKBEC) and the CommonMind consortium. The original eQTLs in GTEx and UKBEC were derived primarily from microarray technology, but have now switched over to RNA-sequencing approach. The initial version of GTEx (http://www.ncbi.nlm.nih.gov/gtex/GTEX2/gtex.cgi) contained microarray data from studies on human brain cerebellum, frontal cortex, caudal pons and temporal cortex (N=150).34 The newer GTEx Project data are RNA-sequencing results from a variety of human brain regions and are available through the GTEx Portal (http://www.gtexportal.org/home/).35, 36 UKBEC presented microarray eQTL data from 10 human brain regions (www.braineac.org) (N=134).37, 38 They have also generated RNA-sequencing data on substantia nigra and putamen in post-mortem control brains (N=65 and 105, respectively). The CommonMind consortium (http://commonmind.org/WP/) generated data modalities (RNA and DNA sequencing, genotyping) across multiple brain regions (dorsolateral prefrontal cortex (DLPFC), anterior cingulate cortex and superior temporal gyrus) from schizophrenia, bipolar disorder and control samples, totaling a collection of >1000 samples. The CommonMind consortium started the first data release of SNP genotypes and RNA-sequencing results for the DLPFC of over 600 subjects registered at the Mount Sinai, University of Pittsburgh and University of Pennsylvania brain banks since March 2015. Recently, they have published RNA-sequencing analyses of DLPFC from people with schizophrenia (N=258) and controls (N=279), and have reported several schizophrenia-associated eQTL genes.39 Besides, Lieber Institute has published a genome-wide microarray eQTL analysis in human DLPFC of healthy controls (that is, BrainCloud, N=269).32 They also generated RNA-sequencing data with DLPFC and other brain tissues in an expanded sample size including psychiatric patients and healthy individuals, and plan to publically release the RNA-sequencing results upon publication.
Table 1. eQTL studies in human braina.
| Study | No. of individuals | Tissue source | No. of tissue samples | Neuropathology | Age | Gender | Ethnicity | Expression detection methods |
|---|---|---|---|---|---|---|---|---|
| Gibbs et al.34 (part of GTEx pilot) | 150 | Caudal pons | 142 | Neurologically normal controls | 15–101; mean 46.2 | 69% Male and 31% female | Caucasian | Microarray (Illumina Human Ref-8 Expression) |
| Cerebellum | 143 | |||||||
| frontal cortex | 143 | |||||||
| Temporal cortex | 144 | |||||||
| Ramasamy et al.38 (UKBEC) | 134 | Occipital cortex | 129 | Neurologically normal controls | 16–102; mean 59 | 74.5% Male and 25.5% female | Caucasian | Microarray (Affymetrix Huamn ST 1.0) |
| Frontal cortex | 127 | |||||||
| Temporal cortex | 119 | |||||||
| Hippocampus | 122 | |||||||
| Intralobular white matter | 131 | |||||||
| Cerebellar cortex | 130 | |||||||
| Thalamus | 124 | |||||||
| Putamen | 129 | |||||||
| Substantia nigra | 101 | |||||||
| Medulla | 119 | |||||||
| Colantuoni et al.32 (BrainCloud) | 269 | Dorsolateral Pre frontal cortex | 269 | Neurological normal controls | Fetal 80 mean 27.8 | 66% Male and 34% female | 147 African-American; 112 Caucasian; 6 Hispanic; 4 Asian | Microarray (Illumina Human 49K Oligo array) |
| Liu et al.41 | 127 | Prefrontal cortex | 127 | 39 Bipolar disorder; 37 schizophrenia; 11 major depression; 40 controls | 20–65; median 45 | 65% Male and 35% female | Caucasian | Microarray (Affymetrix Human Genome U133A) |
| Myers et al.42 | 193 | Cortex (pooled data from 20% frontal, 70% temporal and 1% parietal) | 193 | Neurological normal controls | 65–100; average 81 | 54% Male and 46% female | Caucasian | Microarray (Illumina Human Refseq-8) |
| Webster et al.43 | 364 | Cortex (pooled from 21% frontal, 73% temporal, 2% parietal and 3% cerebellar) | 188 | Neurological normal controls | 65–100; average 81 | 55% Male and 45% female | Caucasian | Microarray (Illumina Human Refseq-8) |
| Cortex (pooled from 18% frontal, 60% temporal, 10% parietal and 13% cerebellar) | 176 | Patients with late-onset Alzheimer's disease | 68–102; average 84 | 50% Male and 50% female | Caucasian | |||
| Heinzen et al. 40 (SNPExpress) | 93 | Frontal cortex | 93 | Neurological normal controls | 34–90; mean 74 | 59% Male and 41% female | Caucasian | Microarray (Affymetrix Huamn ST 1.0) |
| Zou et al. 44 | ~400 | Cerebellum | 197 | Patients with Alzheimer's disease | Mean±s.d.; 73.6±5.6 | 49% Male and 51% female | Caucasian | Microarray (Illumina HumanHT−12 v4.0) |
| 177 | Patients with other brain pathologies | Mean ± s.d.; 71.7 ± 5.5 | 64% Male and 36% female | Caucasian | ||||
| Temporal cortex | 202 | Patients with Alzheimer's disease | Mean±s.d.; 73.6±5.5 | 47% Male and 53% female | Caucasian | |||
| 197 | Patients with other brain pathologies | Mean±s.d.; 71.6±5.6 | 60% Male and 40% female | Caucasian | ||||
| GTEx, v635, 36 | 72–103 | Anterior cingulate caudate, caudate (basal ganglia), cerebellar hemisphere, cerebellum, cortex, frontal cortex, hippocampus, hypothalamus, nucleus accumbens (basal ganglia), putamen (basal ganglia) | NA | Neurological normal controls | NA | NA | NA | RNA-sequencing (polyA) |
| UKBEC (unpublished) | 65–105 | Substantia nigra, putamen | NA | Neurological normal controls | NA | NA | Caucasian | RNA-sequencing |
| Lieber Institute (unpublished) | >700 | Dorsolateral prefrontal cortex, hippocampus | NA | Patients with bipolar disorder, schizophrenia and major depressive disorder, and neurological normal controls | NA | NA | NA | RNA-sequencing |
| CommonMind consortium39 | 537 | Dorsolateral prefrontal cortex | 537 | 258 Patients with schizophrenia; 279 controls | NA | NA | Caucasian 80.7% African-American 14.7% Hispanic 7.7% East Asian 0.6% | RNA-sequencing (RiboZero) |
Abbreviations: GTEx, Genotype-Tissue Expression; eQTL, expression quantitative trait locus; NA, not available; UKBEC, United Kingdom Brain Expression Consortium.
Websites: GTEx http://www.gtexportal.org/home/.
UKBEC http://www.braineac.org/.
SNPExpress http://igm.cumc.columbia.edu/SNPExpress/.
CommonMind http://commonmind.org/WP/.
Nonexhaustive list of examples.
Cautions needed during eQTL analyses
Although eQTL data provide essential information, cautions are needed when using these expression data sets. First, the eQTLs should be considered ‘authentic' only if they could be replicated across independent samples. A prior report revealed low-to-moderate overlap between eQTL loci across earlier microarray-based eQTL studies32, 34, 40, 41, 42, 43, 44 (the percentage of overlapped eQTL is from 0 to ∼35.4% between pairwise brain studies, as shown in Table 4 of the study by McKenzie et al.45) that might be explained by the different analytical methodologies applied in each study. For example, in the study of Myers et al.,42 the analyses were based on pooled expression data from three different cortical regions (frontal, temporal and parietal) with uncontrolled covariates, such as brain pH value and microarray batch effects. Only a limited number of the significant cis-associations observed in their study survived multiple corrections.42 In parallel, Liu et al.41 performed a brain eQTL analysis focusing on the prefrontal cortex with a statistical procedure optimized for possible confounders. They used surrogate variable analysis46 to adjust for covariate effects and ComBat47 to minimize microarray batch effects before the eQTL analyses. This procedure improved the detection power by removing sources of nongenetic variations from the data, and they have identified an exceedingly large amount of cis-eQTL associations that could stand the strict statistical corrections for multiple testing.41 As such, appropriate analytical methods are obviously pivotal in the eQTL analyses, and methodology improvement are always needed. In line with this idea, CommonMind consortium adjusted for known (for example, RNA integrity, library batch, institution (brain bank), age at death, genetic ancestry, post-mortem interval and sex) and hidden variables detected by surrogate variable analysis (conditional on diagnosis but excluding ancestry) in a recent DLPFC RNA-sequencing eQTL analysis of gene expression in European-ancestry subjects.39 The adjusted expression then underwent eQTL analyses against genotypes, covarying for ancestry and diagnosis. A comparison of the identified eQTLs in the CommonMind study39 with previously reported DLPFC eQTLs32, 34, 36, 38, 48 showed that the CommonMind sample not only captured most eQTLs found in other independent samples, but also discovered a substantial number of genes with previously undetected eQTLs (details shown in Table 1 of the CommonMind study39).
Second, in-depth mRNA characterization are necessary to identify potentially pathogenic transcripts during eQTL analyses. It is well known that multiple mRNA isoforms may arise from one single gene but differ in their expression levels and/or functions in cells or organs. In some cases, psychiatric risk variants may affect only specific transcripts of a given gene, such as AS3MT, ZNF804A, KCNH2 and NRG1.49, 50, 51, 52 Li et al.51 have previously reported a novel truncated AS3MT isoform (AS3MTd2d3) lacking two exons compared with the full-length protein. The AS3MTd2d3 was strongly associated with schizophrenia risk SNPs in the 10q24.32 genomic region. The fact that only AS3MTd2d3 but not the full-length AS3MT was brain enriched, human specific and upregulated during early neuronal differentiation suggested divergent roles between them. Another example is the discovery of a novel truncated ZNF804A splicing variant.52 ZNF804A was previously reported to contain a genome-wide significant risk variant rs1344706 for schizophrenia.53, 54, 55, 56 Researchers later found that such risk variants might contribute to schizophrenia pathology via altered expression of ZNF804A, as significant associations of rs1344706 with ZNF804A gene expression levels was observed in fetal brain samples.57, 58, 59 Taken one step further, Tao et al.52 discovered a novel ZNF804A isoform with an alternative 5′ untranslated region and translation start site using a 5′ RACE (rapid amplification of cDNA ends) assay. This novel truncated ZNF804A isoform, rather than the full-length ZNF804A transcript, was associated with rs1344706 in fetal samples.52 These data suggested that the association between rs1344706 and ZNF804A gene expression might originate from the generation of this novel truncated isoform. Both AS3MT and ZNF804A stories indicate that RNA splicing plays an important role in psychiatric diseases. In fact, RNA alternative splicing has been proposed to be a primary mechanism for genetic variation in disease progression, and many GWAS signals for common traits involved alternative splicing.60 Regarding this, characterizing the transcript structures of a risk gene is pivotal for our general understanding of the disease biology. To study this, the current human mRNA data sets (Ensembl and UCSC) are handy tools. Normally, the diverse transcript structures of a gene can be retrieved in these data sets. However, in the event that novel transcripts exist for a defined gene, one will need to identify them via RNA-sequencing analyses. Specifically, junction-level analyses in RNA-sequencing data sets should be carried out. Junctions are the RNA-sequencing read counts spanning at least two exons, and junction reads between nonadjacent exons (that is, exon-skipping junctions) indicate alternative splicing. Following RNA-sequencing analyses, the transcripts of interest are further investigated using experiments such as RACE and end-to-end PCR. This strategy will provide clues for pathogenic gene/protein product that is vital for future model building based on the biological mechanisms, and development of relevant drugs and therapies.61
Another important issue to consider during eQTL analyses is the temporal and spatial conditions that interact with the identified molecular mechanisms. In a biological system, certain regulatory effects might exist only in some cell types and/or at particular developmental stages, or even under certain biological conditions. Given that some eQTLs and most disease manifestations are tissue specific,38, 62 it is recommended that eQTL analyses be performed in the disease-relevant tissues. For example, eQTLs from adipose tissue have shed great light on obesity-related risk loci,63 and eQTLs from lymphoblastoid cell lines have helped explain genetic risk loci for immunological diseases.64 This idea has also been applied in research for the schizophrenia risk variation at the ZNF804A locus. The genome-wide significant SNP rs1344706 associated with the expression of a truncated ZNF804A isoform at the early stage of human fetal brain development.52 It was therefore important to establish where and when psychiatric risk variants exerted their effects. This easiest way to obtain such information is through publicly available databases. For example, the BrainCloud provides valuable resources of transcriptome expression data from post-mortem DLPFC of normal human subjects across the lifespan (that is, from fetal development through aging). It is thus possible to stratify the subjects based on the developmental stages (for example, prenatal and postnatal) and examine the role of the DNA variant at relevant ages.32 Meanwhile, the UKBEC consortium has produced mRNA expression data for 10 human brain regions from control individuals, establishing a comprehensive data set for regional specificity of gene expression regulation across human brain.38
Next, the varying sample sizes of brain eQTL databases can potentially affect the reliability of eQTL analyses. It is known that the power to detect eQTL is partly a function of sample size. Although it was estimated that a sample size of 100 individuals is sufficient for 80% power in eQTL studies,65 the power to detect an effect across multiple studies is reduced when an expression SNP has a subtle effect size or when multiple genomic loci simultaneously control transcript expression levels. On the other hand, the effect size of expression SNP on gene expression has substantial influence over the power of detecting an eQTL association. For example, Wang et al.66 previously showed that when the eQTL effect size increased from 0.5 phenotypic s.d. (σ) to 4σ, the power of detecting a significant association (P<0.05) increased from 10.5 to 100% (Table 5 in Wang et al.66 study), and the bias of observing authentic effect also decreased (Figure 10 in Wang et al.66 study). As such, larger sample sizes and the expression SNP effect sizes should increase the statistical power for eQTL effect detection.
Last but not least, potential limitations of eQTL analyses should be considered. First, the majority of identified eQTLs are considered to be cis-acting and arbitrarily defined to regulate genes within 1 Mb on a chromosome.67 However, genetic variants can also affect the expression of genes residing further away or even on different chromosomes (defined as trans-eQTLs, although they incur a greater penalty for multiple testing, require greater power for detection and are more prone to false positives that are less replicable than cis-eQTL).68 For example, Fehrmann et al.69 reported independent trans-associated SNPs that affected similar genes, suggesting that independent GWAS associations might influence similar biological pathways. Second, the impact of haplotypes on eQTL effects should be factored in.70 As linkage disequilibrium patterns are usually population specific, associations between variants that tag a haplotype could lead to ambiguous identification of the true casual variant. Third, the targets of eQTL associations could be either coding or noncoding RNAs,71 whereas the latter still remains to be explored. Finally, the mapping of regulatory variants for any complex trait within single or certain populations are often less accurate because of various factors, such as genotyping issues.72, 73, 74 These limitations should be always acknowledged, and eQTL analyses for complex illnesses/conditions should always be conducted across diverse populations.
Collectively, with the help of eQTL analyses, it is possible to identify a molecular mechanism in the brain transcriptome that accounts for the genetic association detected in clinical samples. Such results will guide the subsequent in vitro and in vivo testing for the identified molecules/pathways.61
Differential expression between psychiatric patients and controls
Alongside eQTL analyses, genes that are differentially expressed between patients and healthy controls may play key roles in the pathogenesis of psychiatric disorders and aid in the identification of molecular mechanisms underlying genetic risk loci. That is to say, if a gene (or a transcript) shows strong eQTL association with psychiatric genetic risk loci as well as the illness state, and the risk-associated genotype predicts the same direction of expression difference between cases and controls, it would be an ideal target for further functional studies. Indeed, recent advances in microarray or RNA sequencing techniques have allowed researchers to dissect the roles of gene expression alterations in the pathogenesis of psychiatric disorders including bipolar disorder,75, 76, 77, 78, 79, 80, 81, 82 major depressive disorder83, 84, 85 and schizophrenia.79, 80, 86, 87, 88, 89, 90
However, only a small number of genome-wide significant differentially expressed genes have been reported so far, and only few overlapping differentially expressed genes were reproduced across different studies (for the same illness). This is likely resulted from the multiple confounding factors such as relatively small samples sizes, sample heterogeneity and other technical reasons including instability of RNA and the post-mortem conditions (for example, brain tissue pH changes, coma, respiratory arrest, hypoxia, seizures, dehydration, multiple organ failure and head injury). These confounders may interfere with the relationship between measured gene expression levels and disease status91 to cause difficulty in capturing the desired signals. The following paragraphs will discuss these problems in details.
Among all possible confounding factors, sample size is undoubtedly one major issue that is very difficult to resolve because of the nature of the affected organ in psychiatric disorders. In the past, people applied stringent multiple corrections in genome-wide expression analyses, resulting in identification of extremely significant effects at the cost of substantial false negatives. Such problem becomes harder to resolve in the current gene expression studies that have typically used RNA samples from either brain tissues or peripheral blood.92 Obviously, brain is considered to be the most relevant tissue as its dysfunctions are presumed to be the origin of psychiatric illnesses. However, the brain tissues can only be collected after the participant is deceased, and this significantly affected the total sample size, let alone the number of samples for cases. In fact, this disadvantage is reflected in many of the current brain eQTL resources, in which only healthy controls are utilized. Blood has been an alternative tissue for eQTL analysis. Although it may appear less direct for understanding psychiatric disorders, it is relatively easy to collect. So far, it is generally accepted that blood RNA analyses do provide some clues for the understanding of psychiatric disorders.84, 90
Besides the problems with sample size, technical limitations exist in the current differential expression analytic methods. Microarray analyses with a single probe (or several probes) per gene usually lack ideal resolutions for transcripts. Similarly, insufficient sequencing depth in RNA-sequencing analyses may preclude low-abundant transcripts that are important in the pathogenesis of psychiatric disorders from detection. Taken together, further differential expression studies comparing cases and controls involving larger brain sample sizes and more in-depth RNA-sequencing are necessary.
Resources and bioinformatics analyses to predict regulatory variants
After obtaining the evidence for a link between DNA genotype and gene regulation or processing through eQTL analyses and differential expression analysis, a range of further molecular approaches are required to elucidate the regulatory mechanisms. Public resources and bioinformatics analyses are useful and easily accessible tools that usually provide data suggesting multiple functional component in the genome.
The most well-known and representative bioinformatics resources are the data sets of regulatory elements in noncoding DNA regions obtained through high-throughput sequencing techniques. These regulatory DNA sequences are characterized by an open chromatin for the access of transcription factors (TFs) that modulate gene expression. Specifically, during the transcription of a gene, RNA polymerase II, numerous TFs and accessory molecules are recruited to the gene promoter region. Through binding to the regulatory sequences, transcription initiation complex is formed to start the basal transcription machinery. In this process, the ease of these molecules to access the DNA, which is regulated by posttranslational histone modifications (methylation, acetylation and so on), is the ultimate determinant. Previous studies suggested that a proportion of psychiatric risk variants located within the regulatory sequences (such as promoters and enhancers) influenced gene expression through transcriptional, posttranscriptional and posttranslational (for example, posttranslational modifications of histones or RNA polymerase II as regulation of transcription) mechanisms.93, 94, 95 Importantly, regulatory signals can act over long genomic distances when brought into contact with target promoters by three-dimensional DNA folding. To reveal such mechanisms conveyed by specific genomic variants, computational prediction can be conducted based on recent established large-scale genome-wide data sets (Table 2) such as the Encyclopedia of DNA Elements (ENCODE)96 and the Roadmap Epigenomics Mapping Consortium (REMC).97 These data sets can be routinely mined with multiple computational tools (for example, RegulomeDB,98 HaploReg,99 GWAVA100 and FunciSNP101) for the annotation of DNA variants.
Table 2. Computational tools and resources for the analyses of noncoding risk locia.
| Feature | Description | Significance | Experimental approach | Bioinformatic tools and online resources |
|---|---|---|---|---|
| Open chromatin | Nucleosome-depleted chromatin | DNA sequences harboring regulatory signals | DNase-seq, FAIRE sequencing | ENCODE,96 REMC,97 RegulomeDB,98 HaploReg,99 FunciSNP101 |
| TF-binding prediction | Short DNA consensus recognition sequence characteristic of a particular DNA-binding protein | Computationally predicted TF recognition site | Position weight matrices | TRANSFAC,154 JASPAR,155 MAPPER2,156 GWAS3D,157 DeepSEA158 |
| DNA–protein interaction | Short DNA sequence associated with a DNA-binding protein after precipitation with a specific antibody | Physical protein-nucleic-acid binding | ChIP-seq, DNase footprinting | ENCODE,96 NRCistrome,159 RegulomeDB,98 HaploReg,99 GWAVA100 |
| DNA methylation | Methylation of cytosine residues in CpG dinucleotides | Regulation of gene expression | Methylation array, bisulfite sequencing, MeDIP-seq, MRE-seq | ENCODE,96 REMC,97 MethDB,160, 161 EpiGraph,162 BrainCloudMethyl,163 Fetal brain meQTLs107 |
| DNase I hypersensitive sites | Sensitive to cleavage by the DNase I enzyme | DNA sequences harboring regulatory signals | DNase-seq | ENCODE,96 REMC,97 PsychENCODE,113 DeepSEA158 |
| Histone modifications | Specific posttranslational modifications of particular histone protein residues are associated with various regulatory activities | H3K4me1: promoters and enhancers H3K4me3: promoters H3K27ac: active regulatory region H3K9ac: promoters H3K9me1: active chromatin | ChIP-seq | ENCODE,96 REMC,97 PsychENCODE,113 NRCistrome,159 RegulomeDB,98 HaploReg,99 ChromHMM,164 GWAS3D,157 ChroMoS,165 SEA,166 DeepSEA158 |
| Chromatin interactions | Long-range physical interactions between distal genomic regions | Contact between regulatory motifs, such as tissue-specific enhancers and promoters | 3C, 4C, 5C, Hi-C, ChIA-PET | GWAS3D,157 Hi-C Browser,167 CCSI168 |
| MicroRNA-binding prediction | Short DNA consensus recognition sequence characteristic of a particular microRNA | Computationally predicted microRNA recognition site | Position weight matrices | miRanda,169 Target Scan,170 MicroSNiPer171 |
Abbreviations: 3C, chromosome conformation capture; 4C, circular 3C; 5C, carbon-copy 3C; CCSI, Chromatin Chromatin Space Interaction; ChIA-PET, chromatin interaction analysis by paired-end tag sequencing; ChIP-Seq, chromatin immunoprecipitation followed by next-generation sequencing; DNase-seq, DNase I hypersensitive site sequencing; FAIRE, formaldehyde-assisted isolation of regulatory elements; MeDIP-seq, methylated DNA immunoprecipitation sequencing; MRE-seq, methylation-sensitive restriction enzyme sequencing; NRCistrome, Nuclear Receptor Cistrome; REMC, NIH Roadmap Epigenomics Project; RNA-PET, RNA paired-end tag sequencing; SEA, super-enhancer archive; TF, transcription factor.
Websites: ENCODE https://www.encodeproject.org/.
RegulomeDB http://www.regulomedb.org.
TRANSFAC http://www.gene-regulation.com/index2.
MAPPER2 http://genome.ufl.edu/mapperdb.
GWAS3D http://jjwanglab.org/gwas3d/.
MethDB http://www.methdb.de.
EpiGRAPH http://epigraph.mpi-inf.mpg.de/WebGRAPH/.
BrainCloudMethyl http://braincloud.jhmi.edu/Methylation64/BrainCloudMethyl64bit.htm.
Fetal brain meQTLs http://epigenetics.essex.ac.uk/mQTL/.
PsychENCODE http://psychencode.org/.
ChromHMM http://compbio.mit.edu/ChromHMM/.
Hi-C Browser http://hic.umassmed.edu/welcome/welcome.php.
Target Scan http://www.targetscan.org/vert_71/.
Nonexhaustive list of examples.
Initiated as a follow-up to the Human Genome Project (Genomic Research), ENCODE project aimed to map the functional elements in the genome, usually defined as a segment of the genome having either a biochemical signature (for example, TF-binding site or some other protein-binding site) or a specific chromatin structure (for example, accessible open chromatin), and encodes a product (for example, a protein).96 The initial phase data are available through the UCSC (University of California, Santa Cruz) Genome Browser,102 Table Browser tool103 and the FTP site (ftp://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/). The most recent releases are available from the ENCODE website (https://www.encodeproject.org/). The ENCODE consortium has generated data from both cell lines and human brain tissues. For cell lines, they categorized multiple cell lines into three tiers based on the respective superiority for biological experiments. For example, tier 1 cell lines such as GM12878, H1-hESC and K562 have the highest priority with regard to designing experiments. They later included two cancerous brain cell lines in its 2012 version: glioblastoma (Gliobla) and neuroblastoma (SK-N-SH, and sublines SH-SY5Y and retinoic acid-treated SK-N-SH_RA; also SK-N-BE and clone BE2_C). Though neither of these were in tier 1, they were valuable addition to the database given their nature as brain cells. One major limitation though is that data generated using immortal cell lines may not represent the actual biology in normal cells and tissues. This concern was later addressed when ENCODE added new data from various sources including human brain tissues since the 2012 release. For instance, processed DNase I hypersensitivity data ‘peaks' (regions of statistically significant enrichment based on the signal from the measurement of background abundance in the genome) were available from tissues of nine human brain regions (frontal cortex, cerebellar cortex, cerebellum, globus pallidus, midbrain and middle frontal gyrus), two fetal brains and five primary brain cell types (astrocyte of the cerebellum, astrocyte of the hippocampus, brain microvascular endothelial cell, brain pericyte and choroid plexus epithelial cell). Generally, ENCODE is the first large international collaborative project mapping functional elements in the genome. Its standardized and robust data have greatly contributed to our roadmap from functional annotation prediction to laboratory testing.
Besides mapping the functional elements in the genome, available data sets for epigenome provide another layer of information for gene expression regulation. One major form of epigenetic modification is DNA methylation that involves the enzymatic addition of a methyl group to the carbon-five position on cytosine residues.104 Currently, data from brain tissue are available for allele-skewed methylation (also referred to as methylation quantitative trait loci (meQTL) among other terms), a scenario in which one allele shows significantly different methylation levels compared with another allele at the same base pair location.105, 106 Intriguingly, in two recent genome-wide brain DNA meQTL analyses,107, 108 enrichment of meQTL signals among risk loci in schizophrenia GWAS8 was observed. These data suggested that meQTL might be useful in refining GWAS loci via analyzing discrete sites of DNA methylation in the brain that are associated with schizophrenia (and other psychiatric) risk variants.109
In addition to DNA methylation on cytosine bases, other forms of epigenetic DNA modifications (for example, hydroxymethylation) have been recently reported and require further analyses.110 Meanwhile, mechanisms other than DNA methylation (for example, histone modifications and DNase I hypersensitivity) can also regulate DNA accessibility and gene expression.111 Mapping the epigenome with these information are also potentially helpful for understanding the brain biology and psychiatric illnesses. The NIH REMC97 (http://www.roadmapepigenomics.org/) has undertaken this task and mapped the DNA methylation, DNA accessibility and RNA expression in primary human tissues. Although the sample sizes are tiny (usually one or two samples), there are histone modification and RNA-sequencing data from up to eight adult brain regions (hippocampus middle, substantia nigra, anterior caudate, cingulate gyrus, interior temporal lobe, angular gyrus and dorsolateral prefrontal cortex) and fetal brains. They also possess data of DNA accessibility mediated by DNase I hypersensitivity in fetal brain. Along with human brains, REMC assessed functional elements in stem cells and primary ex vivo tissues. Although such cells are not ideal for epigenome investigation in a living system given the stochastic random epigenetic changes appeared as stem cells divide,112 these data have still provided valuable information.
In summary, these programs have tested the potential impact of DNA sequence variation on several genomic features including histone modifications, chromatin immunoprecipitation followed by next-generation sequencing (ChIP-Seq), DNase I hypersensitive sites, chromatin interactions, evolutionary sequence conservation and TF-binding motifs (measured with targeted biochemical assays and high-throughput sequencing technologies). Mapping these data to a genomic region of interest facilitates the design of further functional assays.
Despite the rapid progress in bioinformatic analyses, many of these computational tools, however, are still not exhaustive, and only limited TFs and cell types have been assayed. Moreover, these programs do not take into account tissue specificity, in which case regulatory variations might influence signals in irrelevant cell types. As a result, the high false negative probability (for example, missing data might lead to the absence of valid results) remains a major challenge. To address this, the PsychENCODE project was found113 to produce a public resource of multidimensional genomic data using samples from ∼1000 healthy and psychiatric disease-affected human post-mortem brains. The key goal of the PsychENCODE project was to look at regulatory elements (for example, TF-binding sites) as was done by the ENCODE Project, but in post-mortem control and psychiatric (schizophrenia, bipolar disorder and autism spectrum disorder) brains. This project is expected to provide an enhanced framework of regulatory genomic elements, to catalog epigenetic modifications and to quantify coding and noncoding RNA and protein expression. The first release made available a few histone modifications and RNA-sequencing data from individuals or from induced pluripotent stem cell (iPSC)-derived neurons on the PsychENCODE Knowledge Portal (https://www.synapse.org/#!Synapse:syn4921369/wiki/235539). Overall, as the knowledge evolves, projects with consideration of specific biological questions are being launched and present useful preliminary reference for designing functional assays.
Collectively, identifying the functional causal variant(s) among psychiatric noncoding risk-associated loci with public bioinformatics tools is a pivotal step to guide functional assays. Regarding potential advantages and limitations discussed above, information in the following aspects should be synthesized: the strength of clinical risk associations, the eQTL effects on gene/isoform expression, the likelihood that the variant perturbs cis-regulatory elements, the relative impact of the variant on reporter gene activity and the evolutionary conservation of the putative regulatory element. Although it is difficult to confidently pinpoint the functional variants underpinning a risk signal, such approaches will prioritize certain variants of interest at associated loci. In addition, using these data to test whether psychiatric risk signals are statistically enriched in regulatory regions of particular cell types provides the information about where these variants are active, and implying relevant biological mechanisms.
Validations of regulatory effects via in vitro functional assays
The eQTL and bioinformatic analyses provide useful tools to predict targeted molecule(s) of psychiatric risk variants. However, this strategy usually only achieves indirect evidence of a molecular association, and experimental testing is necessary to confirm the mechanistic relevance. Thus, we propose that validation of regulatory effects in vitro should be the next step. In recent years, there have been considerable advances in useful techniques for studying noncoding genomic loci and uncovering causal variant(s).
As previously discussed, psychiatric risk variants are enriched in cis-acting eQTLs in brain.93, 94, 95 Underlining the principle that regulatory variations not always affect spatially the closest gene(s), there has been a growing consensus that chromosomal regions frequently fold in order to bring distant regulatory regions in closer proximity to the genes they regulate, such as transcriptional enhancers. These DNA elements are typically located more than 1 kb away from their target genes and regulate transcription through long-range interactions enabled by the formation of chromatin loops.114 The recent PGC2 GWAS has screened the credible risk variants within the 108 genome-wide significant schizophrenia loci in 56 human cell lines and tissues, and found significant enrichment in active enhancers in human brain.8
To verify these long-range chromosomal interactions, chromatin conformation capture (3C) technique is one of the most reliable experimental approaches. The 3C-based techniques involve first formaldehyde crosslinking of interacting sites in cells of interest, and then cutting of DNA with a restriction enzyme and a ligation reaction to join crosslinked DNA fragments to investigate chromosomal interactions at specific candidate loci.115 The 3C-based methods have been utilized in the functional characterizations of GWAS risk-associated loci for several complex traits or diseases such as pigmentation and cancer.116, 117, 118 There have also been several studies using 3C-based methods to interrogate psychiatric risk loci.119, 120 Roussos et al.120 used 3C to elucidate the regulatory roles of a psychiatric risk variant in the intron of CACNA1C. The risk variant was predicted to locate in an enhancer region that interacted with the CACNA1C promoter in human DLPFC and neurons derived from human iPSCs. Using a reporter gene assay, they showed that the risk allele within this enhancer caused lower transcriptional activity, consistent with its association with decreased CACNA1C expression in human cerebellum.121 Although such result seemed to be in the opposite direction from previous studies that showed that the risk allele predicted higher CACNA1C expression in human DLPFC and induced human neurons, such conflict could be because of differential roles of CACNA1C in different brain regions.122, 123 The knowledge of CACNA1C risk variant was further improved by Eckart et al.,124 who showed that risk variants at CACNA1C marked eQTL in the superior temporal gyrus region, and found one SNP rs4765905 showing allele-dependent regulatory function in reporter assays and protein microarrays. Using circular 3C (4C), they revealed interactions of the disease-associated regions (covering the risk SNPs) with CACNA1C promoter. In another study of the psychiatric risk gene MIR137, Duan et al.119 identified a rare enhancer SNP near this gene that conferred risk of schizophrenia and bipolar disorder. The risk allele reduced enhancer activity of its flanking sequence by >50% in human neuroblastoma cells, predicting lower expression of MIR137/MIR2682 that was then also proved with 3C assays (Table 3).
Table 3. Functional genetic variants successfully identified at psychiatric risk locia.
| Disease or phenotype | Locus | Functional variants | Target genes | Key methods | References |
|---|---|---|---|---|---|
| Schizophrenia, bipolar disorder | 1p21.3 | 1:g.98515539A>T | MIR137/MIR2682 | 3C, EMSA, reporter assays | 119 |
| Schizophrenia, bipolar disorder | 2q32.1 | rs1344706 | ZNF804A | eQTL, EMSA, | 52, 136 |
| Schizophrenia | 2q32.1 | rs359895 | ZNF804A | EMSA, reporter assays | 135 |
| Bipolar disorder | 7q21.11 | rs13438494 | PCLO | splicing assays | 142 |
| Bipolar disorder | 7q21.1–q21.2 | rs148754219 | GRM3 | eQTL, EMSA, reporter assays | 137 |
| Schizophrenia | 10q24.32 | VNTR | AS3MT | eQTL, reporter assays | 51 |
| Schizophrenia | 11q23 | rs1076560 | DRD2 | eQTL, splicing assays | 141 |
| Schizophrenia | 12p13.3 | rs2159100/rs12315711 | CACNA1C | 3C, reporter assays | 120 |
| Schizophrenia | 12p13.3 | rs1006737/rs4765905 | CACNA1C | eQTL, 4C, reporter assays, protein arrays | 124 |
Abbreviations: 3C, chromosome conformation capture; 4C, circular 3C; EMSA, electrophoretic mobility shift assay; eQTL, expression quantitative trait locus.
Nonexhaustive list of examples. It should be noted that some of these genetic loci are positive only in candidate gene studies but not in genome-wide association studies (GWASs).
Though being a powerful method for testing chromosomal interaction at specific loci, 3C also has two principle limitations. First, it is unable to distinguish relevant nearby chromatin interactions (within ~20 kb) from background interactions caused by random collisions. Second, 3C can only detect specific interactions between prespecified regions as it relies on PCR primers designed across interacting zones. To address these issues, several variations of the 3C method have been developed. The circular 3C, referred to as 4C, allows screening of the entire genome for sequences in contact with a specific DNA or ‘‘bait' region through inverse PCR with bait primers from a circular intermediate of 3C. However, 4C is also limited because of (1) the inability to identify interactions around the ‘bait' region; (2) the lack of resolution (~100 kb to 1 Mb); (3) the preclusion of certain interactions because of the enzymes used; and (4) the needs for validation of trans-interactions (interchromosomal) and distal cis-interactions (>500 kb from the bait) by independent methods such as fluorescence in situ hybridization. Another ‘upgrade' for 3C is the carbon-copy 3C (also known as 5C) that detects all chromatin interactions across large genomic regions using multiplex PCR in combination with high-throughput sequencing or microarrays.125 Hi-C, a method that could comprehensively detect chromatin interactions in the mammalian nucleus, was also developed.126 In Hi-C, a biotin-labeled nucleotide is incorporated at the ligation junction to enable selective purification of chimeric DNA ligation junctions that is then followed by deep sequencing. The compatibility of Hi-C with next-generation sequencing platforms makes it ideal to detect chromatin interactions on an unprecedented scale. Therefore, Hi-C has the power to explore both biophysical properties and structure of chromatins.126 ChIA-PET (chromatin interaction analysis by paired-end tag sequencing), another variation of 3C originally developed by Fullwood et al.127 to map chromatin interactions bound by estrogen receptor from breast cancer cells treated with estrogen, detects chromatin interactions bound by a defined protein. Several ChIA-PET data sets for CTCF (CCCTC-binding factor), RNA polymerase II and H3K4me2 (a chromatin modification associated with enhancers) are now available in various cell lines.128, 129, 130 However, as a given TF is likely only involved in a subset of chromatin interactions, ChIA-PET data sets do not include all promoter–enhancer interactions. To identify the majority of promoter–enhancer interactions, it is still necessary to use antibodies against the general TFs (such as RNA polymerase II) or chromatin modifications (such as H3K4me1 and H3K4me2) together with deep sequencing.128, 129
These variations of 3C techniques have already been applied in research of several common diseases except for psychiatric disorders. Using the 4C method, Patel et al.131 showed that aberrant TAL1 expression in human T-cell acute lymphoblastic leukemia was mediated by a T-cell acute lymphoblastic leukemia-specific interchromosomal interaction between the TAL1 promoter on chromosome 1 and a regulatory element called TIL16 on chromosome 16.131 Using coined 3C with DNA selection and ligation, a 5C similar technique, Harismendy et al.132 showed that cis-regulatory variants associated with coronary artery disease interacted with IFNA21, located more than 900 kb away. The coronary artery disease risk alleles also disrupted a binding site for STAT1, a well-known effector of interferon signaling. This was confirmed in their following study that treatment of cells with interferon-γ increased the frequency of interaction between the enhancers and IFNA21.132 These studies have shown the great potential of chromatin-interaction approaches. Given that cis-regulatory elements are often highly tissue specific, future chromatin-interaction profiles generated in neuronal cell lines and brain tissues will be an invaluable resource for psychiatric studies.
Once the target gene(s) of a regulatory element has been identified using the 3C and similar technology, the impact of variant(s) on the transactivation of a specific promoter can be tested via standard reporter assay. When the regulatory potential of a certain variant is limited, such assays could map DNA regions harboring regulatory activities and provide hints for its function. Basically, regulatory elements with different alleles of the candidate variants are cloned into a promoter-driven reporter construct (for example, pGL3) and transiently transfected into relevant cell lines. This assay can be used to test the regulatory effects of variants (or haplotypes) located in either the promoter or enhancer regions. Importantly, the effect of the variant(s) might vary depending on the promoter used for reporter expression. The choice of cell type is also critical considering the high tissue- and cell-type specificity of cis-regulatory elements. A recent study compared variable activities of 11 enhancers across 4 mammary epithelial cell lines, and emphasized the importance of choosing appropriate cellular contexts.133
As the majority of regulatory functions are mediated by TFs and similar proteins, another important direction for functional analysis is to assess allele-specific protein binding. Computational prediction of TF binding based on position weight matrices models has been widely used to identify candidate TFs. With this method, quantitative scores are generated for the likelihood of observing a particular nucleotide at a specific position of the candidate TF-binding site. Recent mapping of TFs with DNA by means of ChIP-Seq provides a complementary approach that depicts the genome-wide ‘footprints' created by TFs bound to the DNA at greater sequencing depths. ChIP-Seq can also predict the regulatory status of genomic regions by targeting characteristic histone modifications. For example, promoters and enhancers are typically marked by histone methylations H3K4me3 and H3K4me1, with the additional histone acetylation mark H3K27ac indicating activation and the histone methylation mark H3K27me3 indicating repression. This approach is relatively mature and established, but several limitations still call for cautions during experimental design. First, ChIP assays do not profile more than one TF in each experiment, and the assay resolution is too low to determine the precise binding site. In addition, immunoreactivity assays such as ChIP are potentially compromised in post-mortem human brain because of the impact of post-mortem state (such as post-mortem interval and tissue pH) on epitope fidelity.134 Furthermore, the assay efficiency is highly dependent on the quality of the antibodies used. As a result, ChIP-Seq assay results should be interpreted considering these problems.
Electrophoretic mobility shift assays (EMSAs) can also assess protein binding in vitro, especially the SuperShift EMSA that determines the protein mediating allele-specific binding using antibodies against TFs of interest. In fact, several psychiatric risk loci have been analyzed with the EMSA assays for their impact on TF-binding affinity, such as ZNF804A,135, 136 MIR137 (ref. 119) and GRM3 (ref. 137) (Table 3). Besides, other high-throughput TF-binding methods such as proteome-wide analysis of SNPs (PWAS) using quantitative mass spectroscopy have also been used to screen SNPs for differential TF-binding affinity.138 This technique is very efficient as multiple SNPs and TFs can be analyzed in one experiment. For example, Butter et al.138 applied PWAS to 12 SNPs at the IL2RA locus associated with type 1 diabetes and narrowed down the targets to 4 SNPs showing preferential binding of common TFs. However, the in vitro nature of EMSAs and PWAS gives rise to false positive results, calling for verification by ChIP experiments.
In addition to tests for promoter and enhancer regulatory effects, experimental approaches for alternative splicing have also provided information for the regulatory mechanisms at psychiatric risk loci. One example is the RNA-sequencing analysis using junction data (such as alternative splicing of AS3MT in schizophrenia51) followed by functional verification with in vitro minigene assays. Specifically, a minigene is an artificial gene fragment containing exon(s) and necessary control regions allowing its expression in artificial conditions. The minigene has been used as a splice reporter vector (or exon-trapping vector) to determine the important factors in alternative splicing139 both in vivo and in vitro.140 Using this assay, Cohen et al.141 identified the regulatory mechanism of a schizophrenia risk SNP rs1076560 within DRD2 in Han Chinese (Table 3). The risk SNP was associated with lower D2 short isoform expression in post-mortem brain. Further studies showed that rs1076560 abolished the ability of ZRANB2 to modulate short versus long isoform expression ratios of DRD2 minigene in cultured HEK293 cells, probably by disrupting a binding site for the splicing factor ZRANB2 to diminish binding affinity between DRD2 precursor mRNA and ZRANB2. In another study of PCLO gene (Table 3), Seo et al.142 performed functional minigene analysis of splicing regulatory sequences to characterize the function of rs13438494, a variant in the intron 24 of PCLO that is associated with bipolar disorder in a meta-analysis of GWAS data sets.76 They found that the C allele of rs13438494 reduced the splicing efficiency of the PCLO minigene containing exon 24, intron 24 and exon 25. In addition, prediction analysis using the Human Splice Finder web tool indicated that rs13438494 induced abrogation or creation of enhancer/silencer-binding motifs in this gene. Taken together, rs13438494 altered splicing efficiency by creating or disrupting a splicing motif and associated binding of splicing regulatory proteins.142 Overall, this step provides information on the regulatory mechanism of specific risk loci that usually lead to altered risk gene expression, generation of de novo transcripts or imbalance of the current isoform(s) expression. Such gene expression and/or processing changes are the functional parameters that can be further verified in vitro and in vivo.
Functional analyses of genetic risk in vitro and in vivo
Genes/isoforms
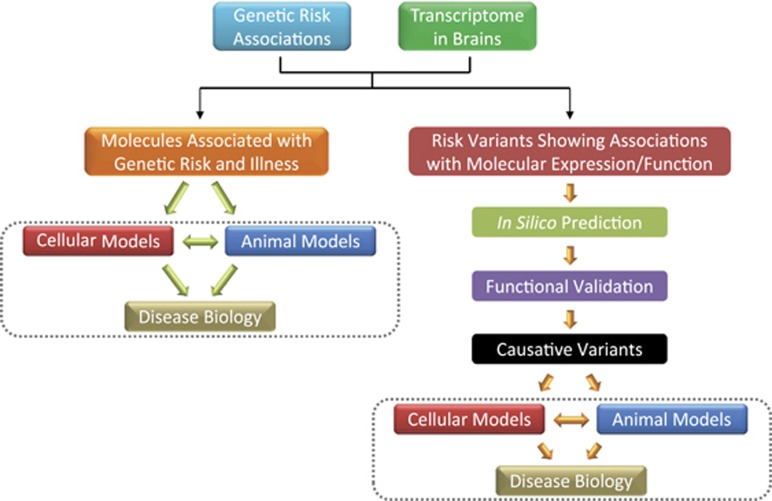
Once the risk genes/transcripts associated with both genetic risk and illness state are located, genetic manipulation of their expression in cultured cells (such as rodent primary cultural neurons) and/or model animals could reveal their influence on neuronal development, brain circuit and disease-related social behaviors (Figure 2). For example, Huffaker et al.49 previously identified a primate-specific isoform (3.1) of the ether-a-go-go related K+ channel (KCNH2) that modulated neuronal firing to be significantly associated with schizophrenia. This risk KCNH2-3.1 transcript was primate specific, brain enriched and highly expressed in patients' hippocampus, whereas the canonical isoform KCNH2-1A was conserved between species, abundant in heart and showed no expression differences between cases and controls. Moreover, rodent primary cortical neurons overexpressed KCNH2-3.1 showed a rapidly deactivating K+ current and a high-frequency, nonadapting firing pattern,49 suggesting a novel function of this specific truncated transcript compared with KCNH2-1A. Later, the same group analyzed the role of KCNH2-3.1 using transgenic mice model, and found that mice overexpressing KCNH2-3.1 had significant alterations in neuronal structure and microcircuit function in the hippocampus and prefrontal cortex. These mice exhibited significant deficits in a hippocampal-dependent object location task and a prefrontal cortex-dependent T-maze working memory task.143 These data further strengthened the contention that KCNH2-3.1 was a risk factor for schizophrenia, and provided information of the neuronal basis of the disease. In recent years, there have also been studies utilizing non-human primates (such as monkeys) to characterize the pathogenic mechanisms of psychiatric risk genes. For example, Liu et al.144 reported that lentivirus-based transgenic cynomolgus monkeys (Macaca fascicularis) expressing human MeCP2 in the brain showed germline transmission of the transgene and exhibited autism-like behaviors. Specifically, these MeCP2 transgenic monkeys had a higher frequency of repetitive circular locomotion and increased stress responses compared with wild-type monkeys. The transgenic monkeys were also socially less interactive, and had a reduced interaction time when paired with other transgenic monkeys in such behavioral tests.
Figure 2.
Roadmap to understand the biology of psychiatric disorders from noncoding risk loci.
Functional variants
When the causative variants of disease is identified, delving into its functional mechanism is a direct and effective approach for verification of the genetic discovery (Figure 2). The most widely used research models are in vitro systems such as human iPSCs and in vivo models with animals. Ideally, it is recommended to create panels of genetically matched iPSCs from patient somatic cells to recapitulate both normal and pathologic human tissue and organ development. These cells then serve as isogenic systems to understand disease mechanisms and to guide drug discovery with higher predictability of prognostic effects.
To study the disease-associated loci for psychiatric disorders with such systems, the common protocol is to generate isogenic human iPSC-derived neurons that differ only at the target SNP locus. This goal is usually achieved using genome-editing systems including designer endonuclease technologies such as zinc-finger nuclease, transcription activator-like effector nuclease (TALEN) and clustered regulatory interspaced short palindromic repeat (CRISPR)/Cas9 endonuclease.145 The consequences of these risk variants on DNA-binding protein occupancy, epigenetic modifications and gene expression in the context of neurodevelopment are then assessed. At present, many in vitro and in vivo studies involving genome-editing tools primarily focused on missense mutations. Sudhof and colleagues146 recently generated two different heterozygous conditional NRXN1 mutations in human embryonic stem cells. NRXN1 encodes neurexin-1, a presynaptic cell adhesion molecule. They found that both heterozygous NRXN1 mutations impaired neurotransmitter release, but had no effect on synapse formation. In another study using TALENs and CRISPR/Cas9, Young-Pearse and colleagues147 disrupted DISC1 near the site of the chromosome translocation found in the Scottish pedigree and found increased WNT signaling in iPSC-derived neural progenitor cells, suggesting its roles in the development of psychiatric disorders. Such studies have also provided implications for future studies on noncoding variations. As an ideal system for studying risk variants in the cellular level, human iPSCs are currently very popular in neuroscience research. However, limited access to clinical samples and difficulties in culturing and manipulating the iPSCs remain the major challenges impeding the establishment of these models.
In addition to the human iPSC systems, researchers also attempted to use model organisms (for example, mouse) to evaluate the functional consequences of the casual variants. The mouse is usually the choice of in vivo mammalian models for its high genome similarity, easy manipulation of genetic background and great capacity to mimic human multifactorial disease phenotypes including neural circuits and social behavior. For example, catechol-O-methyltransferase (COMT) modulates dopamine levels in the prefrontal cortex.148 The human COMT gene contains a polymorphism (Val158Met) that alters its enzyme activity and influences prefrontal cortex function,149, 150 and the Met allele appears to be human specific.151, 152 Recently, Barkus et al.153 introduced the human Met allele into the native mouse COMT gene to produce COMT-Met mice, and developed a mouse model of altered COMT activity comparing with their wild-type littermates. COMT-Met mice had reductions in COMT abundance and activity compared with wild-type controls. When administered with the COMT inhibitor tolcapoe, the attentional performance (assessed with 5-choice serial reaction time task) was only improved in wild-type mice but not in the COMT-Met mice.153 This genetic mutation knock-in mouse model provided an interesting template to study the functions of human risk variant in animals.
Though model animals provide precious information about the physiological impact of a defined genetic variant, this technique should only be used for disease-coding variants occurred in genomic regions that are highly conserved between species and have been (at best) validated using iPSCs. Comparisons between the results from human iPSCs and mouse models will demonstrate the extent to which disease variants converge on common molecular and cellular mechanisms. Meanwhile, cautions should be taken regarding several concerns in translating observations in mice to humans. For example, differences between species often exist in gene function, evolutionary conservation of genome, host responses to environmental changes and genetic backgrounds. In addition, many noncoding variations are not conserved through evolution and could only be studied in vivo using humanized mice. To address these concerns, simultaneous mapping of the genetic variants that already exist and introduction of new genetic variations to the model to ensure proper recapitulation of human genetic landscape are needed in future studies.
Conclusions
Large-scale genetic studies have been successful in identifying multiple genomic loci conferring risk for psychiatric disorders. However, significant obstacles have hampered our ability to pinpoint casual variants, to identify genes/isoforms affected by causal variants and to disentangle the mechanism by which genotype influences phenotype. As opposed to linking rare mutations to Mendelian diseases, revealing the effect of common variants is a tough mission. This review provides a functional pipeline for the identification of candidate causal variants and underlying molecular mechanisms among the noncoding genetic risk loci. Unraveling the complex mechanisms underlying risk associations will ultimately pick up important biological pathways presenting suitable targets for drug development and/or reposition of known therapeutics. Steps toward filling this knowledge gap, as described in this review, will bring us closer to elucidating the genetic bases of psychiatric disorders and offer opportunities for personalized medicine.
Acknowledgments
This work was supported by CAS Pioneer Hundred Talents Program (to ML).
Footnotes
The authors declare no conflict of interest.
References
- Demyttenaere K, Bruffaerts R, Posada-Villa J, Gasquet I, Kovess V, Lepine JP et al. Prevalence, severity, and unmet need for treatment of mental disorders in the World Health Organization World Mental Health Surveys. JAMA 2004; 291: 2581–2590. [DOI] [PubMed] [Google Scholar]
- Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet 2003; 123C: 48–58. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 2003; 60: 1187–1192. [DOI] [PubMed] [Google Scholar]
- Maier W, Lichtermann D, Minges J, Hallmayer J, Heun R, Benkert O et al. Continuity and discontinuity of affective disorders and schizophrenia. Results of a controlled family study. Arch Gen Psychiatry 1993; 50: 871–883. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381: 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converge Consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 2015; 523: 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 2009; 459: 528–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet 2016; 48: 1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry 2012; 17: 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 2014; 506: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014; 506: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia ConsortiumInternational Schizophrenia ConsortiumPurcell SM, International Schizophrenia ConsortiumWray NR, International Schizophrenia ConsortiumStone JL, International Schizophrenia ConsortiumVisscher PM, International Schizophrenia ConsortiumO'Donovan MC et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 2015; 18: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert CR, Xi HS, Wendland JR, O'Donnell P. Translating human genetics into novel treatment targets for schizophrenia. Neuron 2014; 84: 537–541. [DOI] [PubMed] [Google Scholar]
- Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet 2016; 48: 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Bergen SE, Akula N, Song J, Hultman CM, Landen M et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum Mol Genet 2016; doi:10.1093/hmg/ddw181. [DOI] [PMC free article] [PubMed]
- Madras BK. History of the discovery of the antipsychotic dopamine D2 receptor: a basis for the dopamine hypothesis of schizophrenia. J Hist Neurosci 2013; 22: 62–78. [DOI] [PubMed] [Google Scholar]
- Singh T, Kurki MI, Curtis D, Purcell SM, Crooks L, McRae J et al. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci 2016; 19: 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata A, Ionita-Laza I, Gogos JA, Xu B, Karayiorgou M. De novo synonymous mutations in regulatory elements contribute to the genetic etiology of autism and schizophrenia. Neuron 2016; 89: 940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata A, Xu B, Ionita-Laza I, Roos JL, Gogos JA, Karayiorgou M. Loss-of-function variants in schizophrenia risk and SETD1A as a candidate susceptibility gene. Neuron 2014; 82: 773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, Weinberger DR. Schizophrenia genes - famine to feast. Biol Psychiatry 2006; 60: 81–83. [DOI] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N et al. Schizophrenia risk from complex variation of complement component 4. Nature 2016; 530: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Chung Y, He G, Sun D, Jacobson A, van Erp TG et al. Progressive reduction in cortical thickness as psychosis develops: a multisite longitudinal neuroimaging study of youth at elevated clinical risk. Biol Psychiatry 2015; 77: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience 2013; 251: 90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze TG, Detera-Wadleigh SD, Akula N, Gupta A, Kassem L, Steele J et al. Two variants in Ankyrin 3 (ANK3) are independent genetic risk factors for bipolar disorder. Mol Psychiatry 2009; 14: 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet 2008; 40: 1056–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DT, Jiang X, Akula N, Shugart YY, Wendland JR, Steele CJ et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry 2013; 18: 195–205. [DOI] [PubMed] [Google Scholar]
- Hughes T, Hansson L, Sonderby IE, Athanasiu L, Zuber V, Tesli M et al. A loss-of-function variant in a minor isoform of ANK3 protects against bipolar disorder and schizophrenia. Biol Psychiatry 2016; 80: 323–330. [DOI] [PubMed] [Google Scholar]
- Rueckert EH, Barker D, Ruderfer D, Bergen SE, O'Dushlaine C, Luce CJ et al. Cis-acting regulation of brain-specific ANK3 gene expression by a genetic variant associated with bipolar disorder. Mol Psychiatry 2013; 18: 922–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 2011; 478: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman JE, Law AJ, Lipska BK, Hyde TM, Ellis JK, Harrison PJ et al. Genetic neuropathology of schizophrenia: new approaches to an old question and new uses for postmortem human brains. Biol Psychiatry 2011; 69: 140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet 2010; 6: e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013; 45: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015; 348: 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni D, Ryten M, Walker R, Smith C, Imran S, Ramasamy A et al. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem 2011; 119: 275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 2014; 17: 1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 2016; 19: 1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol 2008; 6: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Cheng L, Badner JA, Zhang D, Craig DW, Redman M et al. Whole-genome association mapping of gene expression in the human prefrontal cortex. Mol Psychiatry 2010; 15: 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L et al. A survey of genetic human cortical gene expression. Nat Genet 2007; 39: 1494–1499. [DOI] [PubMed] [Google Scholar]
- Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet 2009; 84: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet 2012; 8: e1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie M, Henders AK, Caracella A, Wray NR, Powell JE. Overlap of expression quantitative trait loci (eQTL) in human brain and blood. BMC Med Genomics 2014; 7: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 2007; 3: 1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007; 8: 118–127. [DOI] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell 2013; 153: 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker SJ, Chen J, Nicodemus KK, Sambataro F, Yang F, Mattay V et al. A primate-specific, brain isoform of KCNH2 affects cortical physiology, cognition, neuronal repolarization and risk of schizophrenia. Nat Med 2009; 15: 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5' SNPs associated with the disease. Proc Natl Acad Sci USA 2006; 103: 6747–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Jaffe AE, Straub RE, Tao R, Shin JH, Wang Y et al. A human-specific AS3MT isoform and BORCS7 are molecular risk factors in the 10q24.32 schizophrenia-associated locus. Nat Med 2016; 22: 649–656. [DOI] [PubMed] [Google Scholar]
- Tao R, Cousijn H, Jaffe AE, Burnet PW, Edwards F, Eastwood SL et al. Expression of ZNF804A in human brain and alterations in schizophrenia, bipolar disorder, and major depressive disorder: a novel transcript fetally regulated by the psychosis risk variant rs1344706. JAMA Psychiatry 2014; 71: 1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet 2008; 40: 1053–1055. [DOI] [PubMed] [Google Scholar]
- Riley B, Thiselton D, Maher BS, Bigdeli T, Wormley B, McMichael GO et al. Replication of association between schizophrenia and ZNF804A in the Irish Case-Control Study of Schizophrenia sample. Mol Psychiatry 2010; 15: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S, Mors O, Borglum AD, Gustafsson O, Werge T, Mortensen PB et al. Expanding the range of ZNF804A variants conferring risk of psychosis. Mol Psychiatry 2011; 16: 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HJ, Norton N, Dwyer S, Moskvina V, Nikolov I, Carroll L et al. Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol Psychiatry 2011; 16: 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guella I, Sequeira A, Rollins B, Morgan L, Myers RM, Watson SJ et al. Evidence of allelic imbalance in the schizophrenia susceptibility gene ZNF804A in human dorsolateral prefrontal cortex. Schizophr Res 2014; 152: 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guella I, Vawter MP. Allelic imbalance associated with the schizophrenia risk SNP rs1344706 indicates a cis-acting variant in ZNF804A. Schizophr Res 2014; 153: 243–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MJ, Bray NJ. Evidence that schizophrenia risk variation in the ZNF804A gene exerts its effects during fetal brain development. Am J Psychiatry 2012; 169: 1301–1308. [DOI] [PubMed] [Google Scholar]
- Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D et al. RNA splicing is a primary link between genetic variation and disease. Science 2016; 352: 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BrainSeq: A Human Brain Genomics Consortium. BrainSeq: neurogenomics to drive novel target discovery for neuropsychiatric disorders. Neuron 2015; 88: 1078–1083. [DOI] [PubMed] [Google Scholar]
- Buonocore F, Hill MJ, Campbell CD, Oladimeji PB, Jeffries AR, Troakes C et al. Effects of cis-regulatory variation differ across regions of the adult human brain. Hum Mol Genet 2010; 19: 4490–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J et al. Genetics of gene expression and its effect on disease. Nature 2008; 452: 423–428. [DOI] [PubMed] [Google Scholar]
- Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007; 448: 470–473. [DOI] [PubMed] [Google Scholar]
- Schliekelman P. Statistical power of expression quantitative trait loci for mapping of complex trait loci in natural populations. Genetics 2008; 178: 2201–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gilbert H, Moreno C, Filangi O, Elsen JM, Le Roy P. Statistical properties of interval mapping methods on quantitative trait loci location: impact on QTL/eQTL analyses. BMC Genet 2012; 13: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Spielman RS. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat Rev Genet 2009; 10: 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013; 45: 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet 2011; 7: e1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier S, Truong V, Brocheton J, Zeller T, Rovital M, Wild PS et al. Genome-wide haplotype analysis of cis expression quantitative trait loci in monocytes. PLoS Genet 2013; 9: e1003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Westra HJ, Karjalainen J, Zhernakova DV, Esko T, Hrdlickova B et al. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet 2013; 9: e1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013; 501: 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Parts L, Glass D, Nisbet J, Barrett A, Sekowska M et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet 2011; 7: e1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet 2012; 8: e1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akula N, Barb J, Jiang X, Wendland JR, Choi KH, Sen SK et al. RNA-sequencing of the brain transcriptome implicates dysregulation of neuroplasticity, circadian rhythms and GTPase binding in bipolar disorder. Mol Psychiatry 2014; 19: 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Higgs BW, Wendland JR, Song J, McMahon FJ, Webster MJ. Gene expression and genetic variation data implicate PCLO in bipolar disorder. Biol Psychiatry 2011; 69: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matigian N, Windus L, Smith H, Filippich C, Pantelis C, McGrath J et al. Expression profiling in monozygotic twins discordant for bipolar disorder reveals dysregulation of the WNT signalling pathway. Mol Psychiatry 2007; 12: 815–825. [DOI] [PubMed] [Google Scholar]
- Seifuddin F, Pirooznia M, Judy JT, Goes FS, Potash JB, Zandi PP. Systematic review of genome-wide gene expression studies of bipolar disorder. BMC Psychiatry 2013; 13: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Vawter MP. Shared gene expression alterations in schizophrenia and bipolar disorder. Biol Psychiatry 2008; 64: 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Xu J, Chen J, Kim S, Reimers M, Bacanu SA et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol Psychiatry 2015; 20: 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MM, Lockstone HE, Huffaker SJ, Wayland MT, Webster MJ, Bahn S. Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Mol Psychiatry 2006; 11: 965–978. [DOI] [PubMed] [Google Scholar]
- Cruceanu C, Tan PP, Rogic S, Lopez JP, Torres-Platas SG, Gigek CO et al. Transcriptome sequencing of the anterior cingulate in bipolar disorder: dysregulation of G protein-coupled receptors. Am J Psychiatry 2015; 172: 1131–1140. [DOI] [PubMed] [Google Scholar]
- Gao L, Gao Y, Xu E, Xie J. Microarray analysis of the major depressive disorder mRNA profile data. Psychiatry Investig 2015; 12: 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Penninx BW, Madar V, Xia K, Milaneschi Y, Hottenga JJ et al. Gene expression in major depressive disorder. Mol Psychiatry 2016; 21: 339–347. [DOI] [PubMed] [Google Scholar]
- Shelton RC, Claiborne J, Sidoryk-Wegrzynowicz M, Reddy R, Aschner M, Lewis DA et al. Altered expression of genes involved in inflammation and apoptosis in frontal cortex in major depression. Mol Psychiatry 2011; 16: 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes MR, Huxley-Jones J, Maycox PR, Lennon M, Thornber A, Kelly F et al. Transcription and pathway analysis of the superior temporal cortex and anterior prefrontal cortex in schizophrenia. J Neurosci Res 2011; 89: 1218–1227. [DOI] [PubMed] [Google Scholar]
- Maycox PR, Kelly F, Taylor A, Bates S, Reid J, Logendra R et al. Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function. Mol Psychiatry 2009; 14: 1083–1094. [DOI] [PubMed] [Google Scholar]
- Mistry M, Gillis J, Pavlidis P. Genome-wide expression profiling of schizophrenia using a large combined cohort. Mol Psychiatry 2013; 18: 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillman SG, Cloonan N, Catts VS, Miller LC, Wong J, McCrossin T et al. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry 2013; 18: 206–214. [DOI] [PubMed] [Google Scholar]
- Sainz J, Mata I, Barrera J, Perez-Iglesias R, Varela I, Arranz MJ et al. Inflammatory and immune response genes have significantly altered expression in schizophrenia. Mol Psychiatry 2013; 18: 1056–1057. [DOI] [PubMed] [Google Scholar]
- Leonard S, Logel J, Luthman D, Casanova M, Kirch D, Freedman R. Biological stability of mRNA isolated from human postmortem brain collections. Biol Psychiatry 1993; 33: 456–466. [DOI] [PubMed] [Google Scholar]
- Mehta D, Menke A, Binder EB. Gene expression studies in major depression. Curr Psychiatry Rep 2010; 12: 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Badner JA, Cheng L, Zhang C, Zhang D, Cox NJ et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol Psychiatry 2013; 18: 340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AL, Jones L, Moskvina V, Kirov G, Gejman PV, Levinson DF et al. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry 2012; 17: 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacanu SA, Chen J, Sun J, Richardson K, Lai CQ, Zhao Z et al. Functional SNPs are enriched for schizophrenia association signals. Mol Psychiatry 2014; 19: 276–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Project Consortium. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 2011; 9: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics ConsortiumRoadmap Epigenomics ConsortiumKundaje A, Roadmap Epigenomics ConsortiumMeuleman W, Roadmap Epigenomics ConsortiumErnst J, Roadmap Epigenomics ConsortiumBilenky M, Roadmap Epigenomics ConsortiumYen A et al. Integrative analysis of 111 reference human epigenomes. Nature 2015; 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012; 22: 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012; 40: D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie GR, Dunham I, Zeggini E, Flicek P. Functional annotation of noncoding sequence variants. Nat Methods 2014; 11: 294–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee SG, Rhie SK, Berman BP, Coetzee GA, Noushmehr H. FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res 2012; 40: e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 2013; 41: D64–D69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 2004; 32: D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths A, Wessler S, Lewontin R, Carroll S. Introduction to Genetic Analysis (W. H. Freeman and Company; 9th edition (February 16, 2007)).
- Tycko B. Mapping allele-specific DNA methylation: a new tool for maximizing information from GWAS. Am J Hum Genet 2010; 86: 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn EL, Schalkwyk LC, Mill J. Allele-specific methylation in the human genome: implications for genetic studies of complex disease. Epigenetics 2010; 5: 578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon E, Spiers H, Viana J, Pidsley R, Burrage J, Murphy TM et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci 2016; 19: 48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Gao Y, Deep-Soboslay A, Tao R, Hyde TM, Weinberger DR et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci 2016; 19: 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliano SA, Ptak C, Mak DY, Shamsi M, Oh G, Knight J et al. Allele-skewed DNA modification in the brain: relevance to a schizophrenia GWAS. Am J Hum Genet 2016; 98: 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013; 341: 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 2008; 132: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatabe Y, Tavare S, Shibata D. Investigating stem cells in human colon by using methylation patterns. Proc Natl Acad Sci USA 2001; 98: 10839–10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psych Encode ConsortiumPsych Encode ConsortiumAkbarian S, Psych Encode ConsortiumLiu C, Psych Encode ConsortiumKnowles JA, Psych Encode ConsortiumVaccarino FM, Psych Encode ConsortiumFarnham PJ et al. The PsychENCODE project. Nat Neurosci 2015; 18: 1707–1712.26605881 [Google Scholar]
- Sexton T, Bantignies F, Cavalli G. Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol 2009; 20: 849–855. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science 2002; 295: 1306–1311. [DOI] [PubMed] [Google Scholar]
- French JD, Ghoussaini M, Edwards SL, Meyer KB, Michailidou K, Ahmed S et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am J Hum Genet 2013; 92: 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praetorius C, Grill C, Stacey SN, Metcalf AM, Gorkin DU, Robinson KC et al. A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway. Cell 2013; 155: 1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med 2015; 373: 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Shi J, Fiorentino A, Leites C, Chen X, Moy W et al. A rare functional noncoding variant at the GWAS-implicated MIR137/MIR2682 locus might confer risk to schizophrenia and bipolar disorder. Am J Hum Genet 2014; 95: 744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Mitchell AC, Voloudakis G, Fullard JF, Pothula VM, Tsang J et al. A role for noncoding variation in schizophrenia. Cell Rep 2014; 9: 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon ES, Grennan K, Busnello J, Badner JA, Ovsiew F, Memon S et al. A rare mutation of CACNA1C in a patient with bipolar disorder, and decreased gene expression associated with a bipolar-associated common SNP of CACNA1C in brain. Mol Psychiatry 2014; 19: 890–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigos KL, Mattay VS, Callicott JH, Straub RE, Vakkalanka R, Kolachana B et al. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Arch Gen Psychiatry 2010; 67: 939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimizu T, Pan JQ, Mungenast AE, Madison JM, Su S, Ketterman J et al. Functional implications of a psychiatric risk variant within CACNA1C in induced human neurons. Mol Psychiatry 2015; 20: 162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckart N, Song Q, Yang R, Wang R, Zhu H, McCallion AS et al. Functional characterization of schizophrenia-associated variation in CACNA1C. PLoS One 2016; 11: e0157086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res 2006; 16: 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 2012; 58: 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 2009; 462: 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 2012; 148: 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res 2012; 22: 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 2011; 43: 630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B, Kang Y, Cui K, Litt M, Riberio MS, Deng C et al. Aberrant TAL1 activation is mediated by an interchromosomal interaction in human T-cell acute lymphoblastic leukemia. Leukemia 2014; 28: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N et al. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature 2011; 470: 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie SK, Coetzee SG, Noushmehr H, Yan C, Kim JM, Haiman CA et al. Comprehensive functional annotation of seventy-one breast cancer risk Loci. PLoS One 2013; 8: e63925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Matevossian A, Jiang Y, Akbarian S. Chromatin immunoprecipitation in postmortem brain. J Neurosci Methods 2006; 156: 284–292. [DOI] [PubMed] [Google Scholar]
- Li M, Luo XJ, Xiao X, Shi L, Liu XY, Yin LD et al. Allelic differences between Han Chinese and Europeans for functional variants in ZNF804A and their association with schizophrenia. Am J Psychiatry 2011; 168: 1318–1325. [DOI] [PubMed] [Google Scholar]
- Hill MJ, Bray NJ. Allelic differences in nuclear protein binding at a genome-wide significant risk variant for schizophrenia in ZNF804A. Mol Psychiatry 2011; 16: 787–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandaswamy R, McQuillin A, Sharp SI, Fiorentino A, Anjorin A, Blizard RA et al. Genetic association, mutation screening, and functional analysis of a Kozak sequence variant in the metabotropic glutamate receptor 3 gene in bipolar disorder. JAMA Psychiatry 2013; 70: 591–598. [DOI] [PubMed] [Google Scholar]
- Butter F, Davison L, Viturawong T, Scheibe M, Vermeulen M, Todd JA et al. Proteome-wide analysis of disease-associated SNPs that show allele-specific transcription factor binding. PLoS Genet 2012; 8: e1002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desviat LR, Perez B, Ugarte M. Minigenes to confirm exon skipping mutations. Methods Mol Biol 2012; 867: 37–47. [DOI] [PubMed] [Google Scholar]
- Cooper TA. Use of minigene systems to dissect alternative splicing elements. Methods 2005; 37: 331–340. [DOI] [PubMed] [Google Scholar]
- Cohen OS, Weickert TW, Hess JL, Paish LM, McCoy SY, Rothmond DA et al. A splicing-regulatory polymorphism in DRD2 disrupts ZRANB2 binding, impairs cognitive functioning and increases risk for schizophrenia in six Han Chinese samples. Mol Psychiatry 2015; 21: 975–982. [DOI] [PubMed] [Google Scholar]
- Seo S, Takayama K, Uno K, Ohi K, Hashimoto R, Nishizawa D et al. Functional analysis of deep intronic SNP rs13438494 in intron 24 of PCLO gene. PLoS One 2013; 8: e76960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr GV, Chen J, Yang F, Ren M, Yuan P, Tian Q et al. KCNH2-3.1 expression impairs cognition and alters neuronal function in a model of molecular pathology associated with schizophrenia. Mol Psychiatry 2016; 21: 1517–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li X, Zhang JT, Cai YJ, Cheng TL, Cheng C et al. Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature 2016; 530: 98–102. [DOI] [PubMed] [Google Scholar]
- Byrne SM, Mali P, Church GM. Genome editing in human stem cells. Methods Enzymol 2014; 546: 119–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak C, Danko T, Zhang Y, Aoto J, Anderson G, Maxeiner S et al. Human neuropsychiatric disease modeling using conditional deletion reveals synaptic transmission defects caused by heterozygous mutations in NRXN1. Cell Stem Cell 2015; 17: 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth P, Han K, Callahan DG, Makovkina E, Muratore CR, Lalli MA et al. Genomic DISC1 disruption in hiPSCs alters Wnt signaling and neural cell fate. Cell Rep 2015; 12: 1414–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunbridge EM, Bannerman DM, Sharp T, Harrison PJ. Catechol-O-methyltransferase inhibition improves set-shifting performance and elevates stimulated dopamine release in the rat prefrontal cortex. J Neurosci 2004; 24: 5331–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lipska BK, Halim N, Ma QD, Matsumoto M, Melhem S et al. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am J Hum Genet 2004; 75: 807–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA 2001; 98: 6917–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotta T, Vidgren J, Tilgmann C, Ulmanen I, Melen K, Julkunen I et al. Kinetics of human soluble and membrane-bound catechol O-methyltransferase: a revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry 1995; 34: 4202–4210. [DOI] [PubMed] [Google Scholar]
- Palmatier MA, Kang AM, Kidd KK. Global variation in the frequencies of functionally different catechol-O-methyltransferase alleles. Biol Psychiatry 1999; 46: 557–567. [DOI] [PubMed] [Google Scholar]
- Barkus C, Korn C, Stumpenhorst K, Laatikainen L, Ballard D, Lee S et al. Genotype-dependent effects of COMT inhibition on cognitive function in a highly-specific, novel mouse model of altered COMT activity. Neuropsychopharmacology 2016; 41: 3060–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V et al. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res 2000; 28: 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res 2014; 42: D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinescu VD, Kohane IS, Riva A. The MAPPER database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res 2005; 33: D91–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Wang LY, Xia Z, Sham PC, Wang J. GWAS3D: Detecting human regulatory variants by integrative analysis of genome-wide associations, chromosome interactions and histone modifications. Nucleic Acids Res 2013; 41: W150–W158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods 2015; 12: 931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol 2011; 12: R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunau C, Renault E, Rosenthal A, Roizes G. MethDB—a public database for DNA methylation data. Nucleic Acids Res 2001; 29: 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoreira C, Hindermann W, Grunau C. An improved version of the DNA Methylation database (MethDB). Nucleic Acids Res 2003; 31: 75–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Walter J, Paulsen M, Lengauer T. CpG island mapping by epigenome prediction. PLoS Comput Biol 2007; 3: e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet 2012; 90: 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 2012; 9: 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manke T, Heinig M, Vingron M. Quantifying the effect of sequence variation on regulatory interactions. Hum Mutat 2010; 31: 477–483. [DOI] [PubMed] [Google Scholar]
- Wei Y, Zhang S, Shang S, Zhang B, Li S, Wang X et al. SEA: a super-enhancer archive. Nucleic Acids Res 2016; 44: D172–D179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Ma W, Songyang Z, Luo Z, Huang J, Dai Z et al. CCSI: a database providing chromatin-chromatin spatial interaction information. Database (Oxford) 2016; 2016: bav124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol 2004; 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Barenboim M, Zoltick BJ, Guo Y, Weinberger DR. MicroSNiPer: a web tool for prediction of SNP effects on putative microRNA targets. Hum Mutat 2010; 31: 1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]