Abstract
Dementia is a complex clinical condition characterized by several cognitive impairments that interfere with patient independence in executing everyday tasks. Various neurodegenerative disorders have dementia in common among their clinical manifestations. In addition, these diseases, such as Alzheimer's disease, Parkinson's disease, dementia with Lewy bodies and frontotemporal dementia, share molecular alterations at the neuropathological level. In recent years, the field of neuroepigenetics has expanded massively and it is now clear that epigenetic processes, such as DNA methylation, are mechanisms involved in both normal and pathological brain function. Despite the persistent methodological and conceptual caveats, it has been reported that several genes fundamental to the development of neurodegenerative disorders are deregulated by aberrant methylation patterns of their promoters, and even common epigenetic signatures for some dementia-associated pathologies have been identified. Therefore, understanding the epigenetic mechanisms that are altered in dementia, especially those associated with the initial phases, will allow us not only to understand the etiopathology of dementia and its progression but also to design effective therapies to reduce this global public health problem. This review provides an in-depth summary of our current knowledge about DNA methylation in dementia, focusing exclusively on the analyses performed in human brain.
Introduction
Current demographic trends are leading to a world population comprising a higher proportion of older people. By 2050, for the first time in history, there will be fewer young people than older people.1 This not only poses a major environmental problem2 but will also give rise to an incalculable worldwide socioeconomic burden because of age-related disorders. At present, more than 48 million people worldwide live with dementia and it is estimated that, owing to the lack of preventative therapeutics, there will be more than 130 million patients by 2050.3 The estimated global cost of dementia is US$818 billion for 2016,4 although this figure does not take into account the incalculable social and health costs to relatives and other caregivers.
Dementia is not defined as a specific disease but rather as a collection of cognitive impairments caused by a plethora of disorders. Typically, dementia symptoms include impaired cognitive functions such as loss of memory, language and constructional skills, which are often accompanied by behavioral alterations such as delusions or agitation. Clinical diagnosis of dementia requires the manifestation of two core features characterized by a loss of intellectual ability of sufficient severity to interfere with the patient's independence.5 Some of the diseases that feature dementia are Alzheimer's disease (AD), Parkinson's disease (PD), dementia with Lewy bodies (DLB) and frontotemporal dementia (FTD; Pick's disease).6
Recent investigations have indicated that neuroepigenetic mechanisms are key players in dementia-associated disorders, supporting the hypothesis that the accumulation of epigenetic alterations over the lifespan could be a mechanism underlying these disorders (the ‘Latent Early-life Associated Regulation' (LEARn) model).7, 8 Neuroepigenetic processes are complex mechanisms that regulate the transcriptome of brain cells in serving several functions, such as learning and memory,9 and in the aging process10, 11, 12 and pathological states.13 Epigenetic mechanisms are characterized by their ability to alter transcription, by changing the chromatin structure without modifying the DNA sequence.14 The best understood type of epigenetic modification is DNA methylation,15 which occurs by covalently adding a methyl group from S-adenosyl methionine to the 5′ position of cytosines (5 mC) linked by a phosphodiesterase bond to guanines (CpG dinucleotides). Promoter regions of coding genes (~70% of human genome promoters) are frequently highly enriched in CpGs, forming the CpG islands.16 DNA methylation is generally considered to be a repressive mark17, 18 that principally attracts other proteins associated with gene silencing.19 The addition of methyl groups to the DNA is catalyzed by a set of enzymes called DNA methyltransferases (DNMTs), a group formed by DNMT1, DNMT2 and DNMT3 proteins.20 All of these proteins are strongly expressed in neurons,21 although their function is quite different. While DNMT1 is responsible for maintaining the methylation pattern after cell division,22 DNMT3a and DNMT3b are responsible for the de novo methylation.23, 24 DNA methylation is a reversible process owing to the function of the TET proteins that, through the oxidation of the methyl group, lead the conversion of 5 mC in 5-hydroxymethylcytosines (5 hmC),25 which after successive conversions to 5-formylcytosine and 5-carboxycytosine definitively erase the epigenetic mark. Brain cells are particularly highly enriched in 5 hmC,26 and it has been postulated that it is not merely an intermediate state in the demethylation process but, in fact, a key epigenetic mark in neurological disorders.27, 28
Alterations in the DNA methylation pattern have been seen in other cellular genomes, such as that of the mitochondria. The mitochondrial genome (mtDNA) is small (16.6 kb) and comprises 37 genes. Although it does not contain CpG islands it is highly regulated by DNA methylation processes.29, 30 The existence of both 5 mC and 5 hmC in the mtDNA30 and the presence of mitochondrial DNMT and TET proteins have been confirmed.30, 31 Several laboratories have recently turned their attention to the role of the (hydroxy)methylome of mtDNA and its function regulating gene expression.32 Little is known about brain mtDNA methylation but we do know that increased 5 hmC levels are associated with aging in mice.33
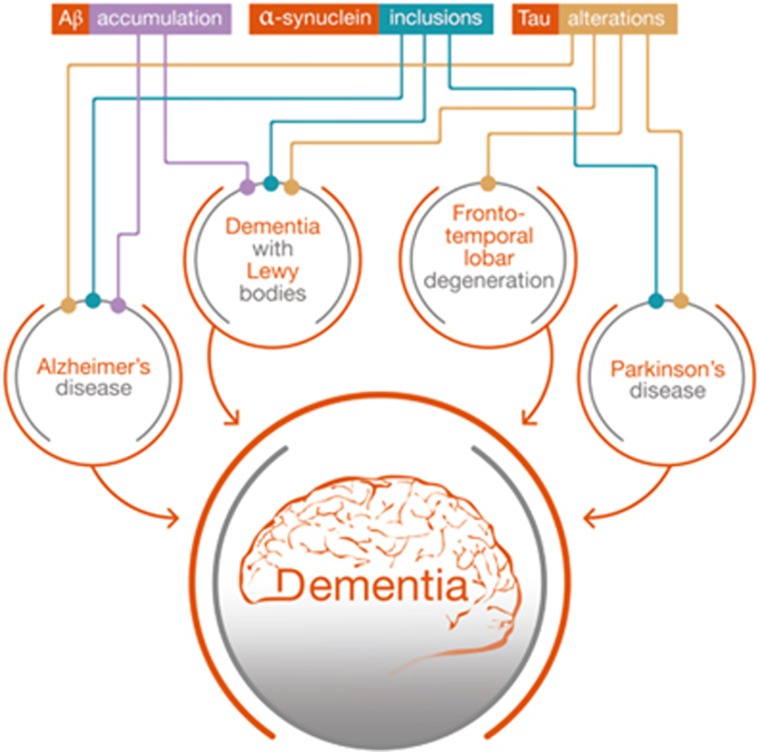
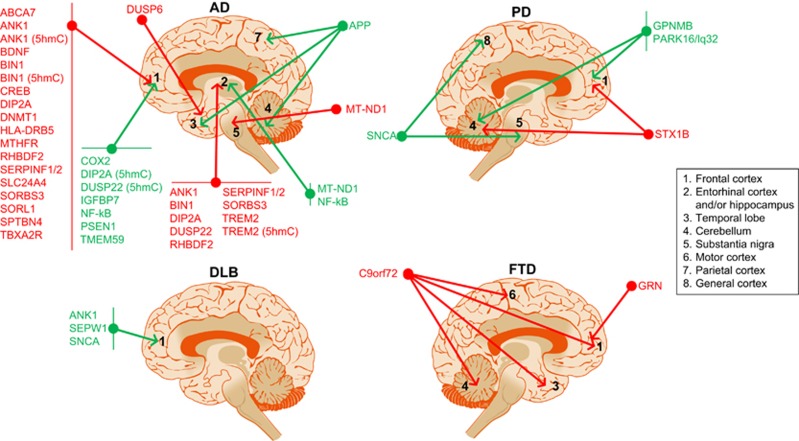
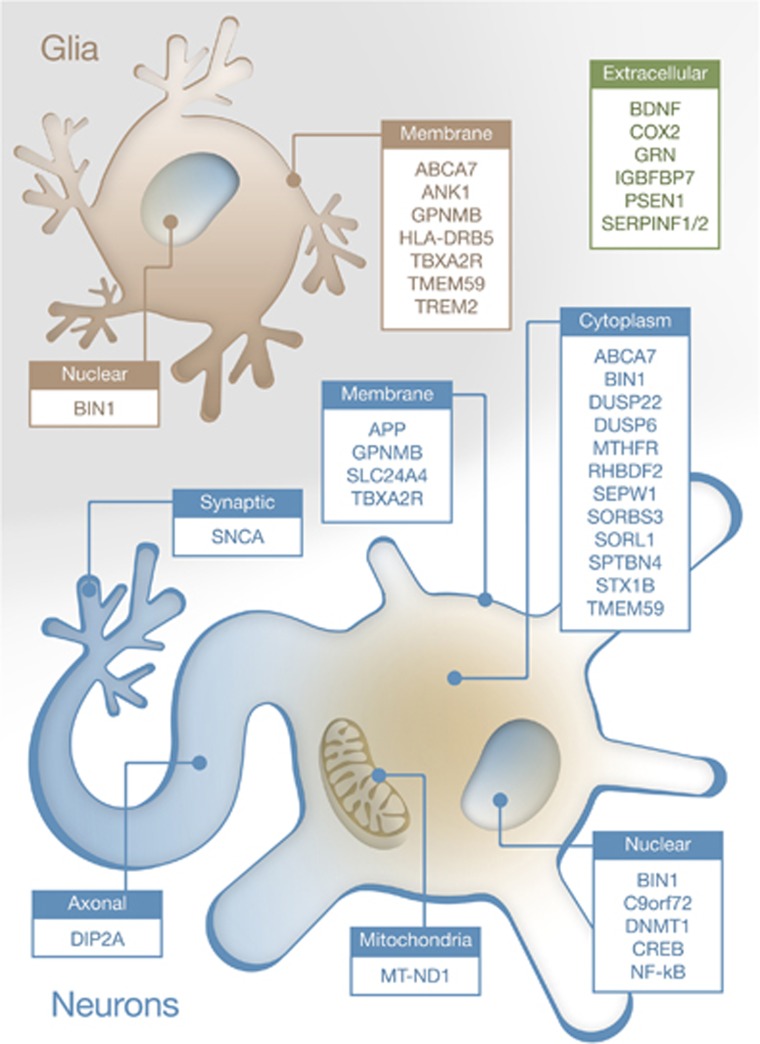
Several disorders with an apparently diverse spectrum of symptoms feature dementia. Surprisingly, those brain alterations exhibit similar molecular alterations, in spite of the observed neuroanatomical differences in the affected areas among these disorders. For example, amyloid plaques, a hallmark of AD, are also formed in DLB, and Tau alterations are common in AD, PD, DLB and FTD.34, 35, 36, 37, 38, 39 These disorders even have similarities at the genetic level, where it has been shown that DLB is equally correlated with AD and PD,40 implying that similar mechanisms are involved (Figure 1). However, it is also possible that different set of alterations or perturbation in brain cells lead to similar response at the genetic level. Our laboratory has recently demonstrated that several dementia-related neurodegenerative disorders (AD, PD, DLB and Down syndrome) share a common set of genes undergoing aberrant DNA methylation shifts.41 These genes, which are involved in functions such as cortex development and general gene regulation, highlight the importance of epigenetic alterations in disorders associated with dementia, despite the limiting number of articles performing cross-disorder profiles. In this review, we will focus on the role of DNA (hydroxy)methylation in dementia-related disorders by summarizing the current knowledge of the role of this particular epigenetic modification in human neurodegeneration. Overall, relevant gene targets for altered DNA methylation in dementia are illustrated in Figure 2, whereas Figure 3 shows the intracellular location of the encoded proteins. The next sections will analyze in detail these epigenetic changes across the broad spectrum of clinical entities associated with dementia.
Figure 1.
Schematic representation of the known connections between Alzheimer's disease, Parkinson's disease, dementia with Lewy bodies and frontotemporal dementia. These disorders not only feature dementia as one of their core clinical manifestations but also share neuropathological alterations such as amyloid-β accumulation, Tau alterations (hyperphosphorylation or loss of function) and α-synuclein aggregation (Lewy bodies).
Figure 2.
Illustration to summarize the major DNA (hydroxy)methylation changes described to date in Alzheimer's disease (AD), Parkinson's disease (PD), dementia with Lewy bodies (DLB) and frontotemporal dementia (FTD) among different brain regions. Red, hypermethylated genes in the disorder; green, hypomethylated genes in the disorder.
Figure 3.
Representation of the known intracellular location of proteins encoded by genes undergoing aberrant DNA methylation in dementia. Nuclear, cytosolic or membrane-associated genes both in neurons (blue) or glia (principally astrocytes and microglia; in pale brown) are illustrated, together with secreted (extracellular) proteins (green).
Alzheimer's disease
AD is the leading form of dementia worldwide. It is an age-related neurodegenerative disorder that severely impairs memory and other cognitive functions. Although clinical manifestations typically occur in patients over 65 years of age, AD develops over decades, and it is believed that the first pathogenic processes start several decades before typical symptoms manifest themselves.42, 43 At the molecular level, AD is characterized by the appearance of two neuropathological hallmarks: senile plaques, comprising insoluble aggregates of amyloid β, and neurofibrillary tangles, which are comprised of abnormally hyperphosphorylated Tau.44, 45 Although the scientific community has amassed considerable knowledge of the molecular alterations and temporal progression of the disease, the events driving the origin of the disorder are unknown. Genetic and non-genetic factors contribute to the etiopathology of AD. To date, almost 300 mutations have been associated with familiar AD, especially those in the APP, PSEN1 and PSEN2 genes.46 However, only ~5% of all AD cases are mainly caused by genetic factors, which draws attention to the need to decipher the non-genetic causative factors of late-onset AD, which is most likely triggered by environmental stimuli that participate in the appearance of AD.
In the past few years, researchers have begun to realize that epigenetic mechanisms are key players in the development/course of AD. Age-associated alterations at the levels of 5 mC and 5 hmC and other epigenetic marks47, 48 have been associated with the progression of AD.49, 50 Several AD-associated genes have been analyzed at the epigenetic level. The APP gene has been described, by using targeted approaches, as hypomethylated in multiple CpG sites on its promoter,51, 52, 53 a concomitant finding with the upregulated levels of APP found in AD brains. These studies used a small cohort, and the findings could not be confirmed in a larger cohort.54 These conflicting results highlight the necessity of performing better and more in-depth studies on the regulation of the APP gene by epigenetic mechanisms. Other APP-related genes, such as APOE, Nicastrin and BACE1 (which is hypomethylated in peripheral blood mononuclear cells of AD patients55), were found to be unaltered in AD brain samples, whereas PSEN1 was hypomethylated and other genes, such MTHFR and DNMT1, were hypermethylated.56 Analyses involving matrix-assisted laser desorption ionization-time of flight mass spectrometry are difficult to interpret, especially because of the small sample sizes used and the high interindividual variance observed, particularly in the PSEN1 and APOE genes. Interestingly, SORL1 and ABCA7, which are genes related to Aβ production, were found to be hypermethylated in AD brains.57 Methylation levels of other genes associated with the amyloid machinery, such as DIP2A, SERPINF1, SERPINF249 and TMEM59,58 are also altered, although the specific meaning of such findings needs to be rigorously addressed. Importantly, the DNA methylation alterations found in De Jager et al.49 correlated with the burden of amyloid plaques identify on the brain tissue, thus associating their finding to the neuropathological state of the subjects.
Other targeted analyses of key molecular players of AD have shown epigenetic alterations in the Tau cascade. Our team demonstrated that DUSP22, a phosphatase-targeting abnormal Tau, presents a hypermethylated promoter in AD that is associated with the lower levels of the protein described in the pathology.59 The levels of methylation of DUSP22 correlated positively with Braak stage of the AD patient. Accordingly, the promoter of the DUSP6 gene, another phosphatase that regulates Tau, was found to be hypermethylated in AD brain60 using a genome-wide approach. Although TAU promoter is not regulated via DNA methylation,54 the data obtained indicate that the molecular cascade associated with Tau hyperphosphorylation or dephosphorylation may be deregulated epigenetically in AD. In another genome-wide study, other molecules associated with Tau alterations, such as BIN1,61 were found to be aberrantly hypermethylated in AD,49, 57 a change correlated with the amyloid plaques burden. Other molecules associated with neuronal function and firing potentials, such as SPTBN4 (spectrin beta 4), are downregulated in human AD brains, a finding that correlates with the hypermethylation status of its promoter.62 SPTBN4 not only participates in the formation of the axon initial segment but also anchors CaMKII to it. CaMKII mislocalization, due to a loss of SPTBN4, is a plausible mechanism for Tau hyperphosphorylation.63 Overall, those sets of data indicate that the two major molecular hallmarks in AD could be regulated via DNA methylation in the early and late phases of the disorder.
Several studies have highlighted the importance of epigenetic processes in the gene expression deregulation occurring in AD, in addition to identifying the AD hallmarks. By analyzing the frontal cortex of human post-mortem samples of AD patients, two independent groups found AD-associated hypermethylation of the promoter of SORBS3, a protein involved in synaptic function.62, 64 Although this effect has been associated with normal aging, this hypermethylation seems to occur faster in AD. Recently, epigenome-wide association studies have identified ANK1 (ankyrin1) as a potential biomarker of AD, as its methylation level is strongly correlated with early stages of the disease and strongly associated with Braak score of the patient. In AD, it is downregulated via hypermethylation of its promoter.41, 49, 65 Little is known about ANK1 function, but it seems to be related to microglia activation and cell motility. Other molecules associated with the immune response show aberrant methylation states. For example, RHBDF2 (related to TNFα function) and HLA-DRB5 are hypermethylated, and nuclear factor-κB (NF-κB) and COX2 (cyclooxygenase-2), which are proinflammatory and associated with inflammatory events in AD, are hypomethylated.41, 49, 57, 66, 67 Mutations of the gene coding for TREM2 (triggering receptor expressed in myeloid cells 2) have been associated with an increased risk of suffering AD.68, 69 TREM2 is expressed in microglia and is associated with microglia activation, phagocytosis and the immune response.70 TREM2 mRNA levels are upregulated in AD hippocampus, its mRNA levels being positively correlated with the AD Braak stages. Interestingly, the increase in mRNA levels are accompanied by a Braak-associated hypermethylation of the TREM2 promoter.71 These results are not those expected from the methylation of promoters, which are normally associated with gene repression, and further research is needed to understand these effects. Overall, synaptic degeneration and immune changes are some of the best known alterations in the pathophysiology of AD,72, 73 and there are evidences that suggests that those are influenced by epigenomic modifications.74
Other molecules have been identified as being epigenetically deregulated in AD. Demyelination processes have been associated with aging but are accelerated in AD, and a recent study has shown that transcriptional alterations in myelination-associated proteins, such as WIF1 and RARRES3, may be mediated via DNA methylation changes.75 SLC2A4, which is thought to be involved in neural development, is also hypermethylated in AD brains.57 Glucose metabolism in brain is altered in this pathological state76 via deregulation of the brain insulin signaling cascade.77, 78 Regarding this pathway, IGFBP7 (insulin-like growth factor binding protein 7) is upregulated in the brains of AD patients probably because of the hypomethylated state of its promoter.79 This protein binds to insulin growth factors and attenuates its protective effects.80 Rao et al.66 demonstrated that the reduction in brain-derived neurotrophic factor previously observed in AD81 was probably due to hypermethylation of its promoter. This epigenetic effect observed in the prefrontal cortex of AD patients was accompanied by a similar one at the promoter of the cAMP response element-binding protein (CREB).66 Both brain-derived neurotrophic factor and CREB are critical proteins for neuronal survival.82, 83 Other authors have also described alterations in genes related to CREB signaling, such as hypermethylation of TBXA2R (thromboxane A2 receptor) gene promoter,62 a G-protein receptor regulating CREB, thereby highlighting the importance of CREB signaling in AD.84
Besides studying the 5 mC levels in AD, researchers have begun to analyze the role of 5 hmC in health and disease. The central nervous system is especially enriched in 5 hmC relative to other body tissues,26 and exhibits a specific pattern across the different brain areas.85 5 hmC is considered to be a key epigenetic mark in brain development and neurological disorders,27, 28 leading reearchers to postulate that age-associated neuronal 5 mC/5mhC changes may be involved in the early and late phases of AD. There are lower 5 hmC levels in several brain regions of late stages of AD patients, such as the neocortex,86 hippocampus,87 entorhinal cortex and cerebellum.88 A genome-wide study has recently found several genes to be associated with neuronal morphology and synaptic function with aberrant 5 hmC levels in the prefrontal cortex of AD patients. Bernstein et al.89 found differential 5 hmC levels of ANK1, DIP2A, BIN1 and DUSP22 genes among those genes described as being epigenetically altered in AD.41, 49, 59, 65 TREM269 also presents alterations in 5 hmC levels in AD brains, insofar as there is a correlated increase in 5 hmC levels in exon 2 and TREM2 mRNA levels.71 However, there are several controversies regarding this epigenetic mark, reflecting the need for further studies to determine the exact role of 5hmC in AD. For example, there are raised levels of 5 hmC in the hippocampus90 and temporal gyrus50 of AD patients. These discrepancies could have arisen as a result of studying different brain populations (i.e. entorhinal cortex vs hippocampus vs temporal gyrus) but also for technical reasons (i.e. immunohistochemistry vs western blotting or dot blot).
Owing to the fact that mitochondrial alterations have been found in AD,91, 92, 93 scientific community have started to analyze the role of the (hydroxy)methylome of mtDNA in brain (mal)function.32 In the context of AD, Blanch et al.94 found a global reduction of 5 mC in mtDNA in mid–late stages of AD (Braak III/IV). Analysis of specific loci showed that the levels of 5 mC in MT-ND1, a protein of the complex I of the respiratory chain, were lower in the entorhinal cortex of AD patients compared with that in age-matched controls. However, no changes were noted in 5 hmC of the mtDNA of AD samples despite that it was described previously as a reduction in 5 hmC levels in aging-associated mtDNA.33 The discrepancy between the Blanch and Dzitoyeva studies could be due to differences between species (humans vs mice, respectively) and the methods used (hydroxyMeDIP assay vs ELISA).
Overall, it is clear that the methylation of DNA is critical to epigenetic processes associated not only with normal brain function and aging but also with AD. Although we still lack information regarding the specific patterns of methylation in specific neuronal populations of key brain nuclei, transcriptomic alterations in AD via epigenetic deregulations could drive the induction of alterations in molecular cascades that will eventually end up with the induction or acceleration of the appearance of AD hallmarks and, consequently dementia and neurodegeneration. The ongoing search for more epigenetic modifications95 will require the development of new technologies to improve our knowledge of the specific role of DNA modification in brain function. Data from a combination of better analyses of human AD samples that take into account the heterogeneity of brain tissue by studying the neuroepigenome via strategies such as single-cell epigenomics,96 and base-resolution whole-genome studies of large cohorts of human samples,97, 98 are sure to be of value for designing early therapeutic interventions aimed at preventing the initial alterations of and, ultimately, AD itself.
Parkinson's disease
PD is a relatively frequent form of neurodegeneration with a prevalence of ~2% worldwide in the over 70 years of age,99 affecting more than four million people100 and with an estimated incidence of 1% in people over 60 years of age.101 PD is characterized by a plethora of symptoms progressing from non-motor changes (olfactory impairments, sleep alterations, constipation and depression)102 to motor alterations (tremor, bradykinesia, rigidity).103 Although the diagnosis of PD is based on the appearance of the motor symptoms, current knowledge indicates that the psychiatric manifestations of the disorder are the first to appear. At the pathophysiological level, PD is characterized by an accumulation of α-synuclein in diverse brain areas, accompanied by a loss of dopaminergic neurons in the substantia nigra and catecholaminergic neurons in the locus coeruleus.15, 102 The brains of PD patients often present Lewy bodies, which are formed by the accumulation of α-synuclein, and by parkin and ubiquitin aggregates.104 Although its cause remains unknown, it has been hypothesized that the disorder is influenced by the interaction of genetic and non-genetic factors, both contributing to a progressive neuronal loss in different brain areas. Monogenic cases of PD, directly affecting ~5–10% of all cases, have been described for mutations in the ATP13A2, DJ1, LRRK2, PARK2, PINK1 and UCHL-1 genes.105, 106 In addition to the autonomic dominant mutations, several risk loci have also been identified.107 However, the majority of PD cases are sporadic, their origin being unknown.
Parkinson's disease dementia (PDD) is a late symptom affecting up to 80% of patients with PD.108 Clinical manifestations of PDD not only include the classic memory impairments but also a myriad of phenotypes, such as alterations of the executive function, deficits in recognition memory, attention processes and visual perception, often accompanied by visual hallucinations and cognitive fluctuations.109 However, the mechanisms underlying PDD are still not understood. Recently published data on the epigenetics of PD have suggested the involvement of this regulatory mechanism in the etiopathology of the disorder, although the exact involvement of DNA methylation in the initiation or progression of PD is still unclear.
Most of the analyses of the role of DNA methylation in PD/PDD have confined themselves to investigating the regulation of genes causing PD. The promoter of SNCA, the gene responsible for the expression of the presynaptic α-synuclein, is hypomethylated at intron 1 in several brain areas such as the substantia nigra and cortex of PD patients.110, 111, 112 This epigenetic regulation of SNCA could explain the higher level of expression of α-synuclein found in PD brains and other neurodegenerative disorders that feature dementia.113 Interestingly, α-synuclein is capable of sequestering DNMT1 from the nucleus, a factor that could explain the global hypomethylation observed in human PD brains.112 Nonetheless, more in-depth studies of SNCA epigenomic regulation are required, as other methylomic studies of PD have not been able to detect differences in SNCA intron 1 methylation levels.41, 114 Besides SNCA, no other genes directly related to PD have been identified as being regulated via methylation of their promoters. Several studies using genome-wide approaches have failed to identify differences in DNA methylation of LRKK2, Parkin, UCHL-1, MAPT and ATP12A2 promoters.41, 115, 116 However, genes such as PARK16/lq32, GPNMB and STX1B, associated with an increased risk of PD, showed altered methylation levels in PD brains.115 Other analyses taking genome-wide approaches identified alterations of several genes at the methylation level.41, 117 Interestingly, methylation levels of the NPAS2 gene, a clock gene, were lower in PD brains.118 Sleep disturbances are a core symptom of PD119 and NPAS2 is involved in sleep regulation.120, 121
Little is known about 5 hmC changes in the brain of PD patients; the only data available have come from the 6-hydroxydopamine-induced rat model of PD. Using that model, Zhang et al.122 were unable to find any difference in the levels of 5 hmC in rat brains.122 Therefore, the potential involvement of 5 hmC in the causes or consequences of PD remains to be determined. Overall, little is known about aberrant DNA (hydroxy)methylation patterns in PD/PDD but the information obtained highlights the need for further research on the neuroepigenomics of PD, as the data collected so far indicate that epigenetic processes may mediate some of the molecular alterations observed in this pathology.
Dementia with Lewy bodies
Besides PD, there are other forms of dementia induced by the formation of Lewy bodies (abnormal aggregates of proteins, principally α-synuclein). DLB is a common form of dementia that differs from PD by the absence of parkinsonism and the progression of the pathology. PD and DLB are classified together as Lewy body dementias and are considered as a continuum rather than as distinct diseases, DLB being an early manifestation of dementia in PD patients.108 DLB is the second most common form of age-related dementia, accounting for an estimated 16–24% of all dementias.123 Although there is still no clear information about its neuropathological changes, DLB shares several pathological marks with AD, PD and PDD: the presence of altered Aβ levels both in DLB and AD has been described, and several mutations associated with AD or PD have also been related to DLB, including mutations in APP, PSEN1/2, SNCA and LRRK2.40, 124 The clinical diagnosis of DLB focuses on the cognitive impairments associated with confusion and attentional deficits, especially those related to visuospatial function.125 These symptoms are often accompanied by psychiatric alterations such as hallucinations, apathy and anxiety.126, 127
Although the molecular driving events responsible for the development of DLB are still unknown, several authors have identified gene networks that are epigenetically deregulated in DLB. Humphries et al.75 demonstrated that DLB brain tissue features DNA methylation alterations in genes related to synaptic function and behavioral response. This is not surprising since cognitive decline is one of the major alterations occurring in DLB.9, 128 Although Aβ overproduction is common in DLB, one study failed to describe DNA methylation changes in human brains of DLB patients in the APP gene53 or other AD-related genes.54 Sleep disturbances are also common in DLB, especially rapid eye movement sleep behavior disorder.129 It is of note that DNA methylation deregulation of the clock genes PER1 and CRY1, which help regulate the circadian cycle and sleep,130 have been reported in DLB blood samples,131 although it will be necessary to analyze the effects on the central circadian clock. Desplats et al.112 described a global hypomethylation state in DLB post-mortem brain samples that probably arises from the reduction in Dnmt1 nuclear levels. The same authors not only described the general hypomethylation state but also reported that the promoter of the SNCA and SEPW1 genes, which are associated with PD,110, 111, 132 was also hypomethylated in DLB. Very recently, our team reported that the DNA methylation level of the promoter of ANK165 was altered in DLB, along with genes related to several pathways, such as MAPK, ErbB, neurotrophin, mTOR, p53 signaling and regulation of the actin cytoskeleton.41
In conclusion, further analysis of the epigenetic alterations in DLB is required. Owing to the molecular alterations common to that disorder, PDD and AD, the identification of epigenetically deregulated genes common to all these disorders41 will provide invaluable knowledge of the early changes in DLB. A better understanding of aberrant DNA methylation, and an analysis of 5 hmC specific to DLB are necessary to enable the better stratification and diagnosis108 of patients, and to accelerate the development of definitive therapeutic approaches.
Frontotemporal dementia
FTD is the second most common form of neurodegenerative disease and the third most common form of dementia, after AD and Lewy body dementias (PDD and DLB). It has an estimated prevalence of 3– 26% worldwide in people older than 65 years of age.133 FTD is a heterogeneous disorder comprising various subtypes, including behavioral-variant frontotemporal dementia (the most common form of the disease, which accounts for ~70% of the cases of FTD) and the primary progressive aphasias, including the semantic-variant primary progressive aphasias and the non-fluent variant primary progressive aphasias.134, 135 Clinical features of FTD vary depending on the subtype of the disorder, although symptoms may converge. In general, FTD is characterized by the presence of cognitive and executive impairments, spatial skills and memory deficits, decline in social conduct and deficits of speech.136 Around 10% of FTD patients develop motor neuron disease and early parkinsonisms.133 All these symptoms are associated with a bilateral frontal and anterior temporal lobe degeneration (frontotemporal lobar degeneration; FTLD), and degeneration of the striatum. At the histological level, FTD is characterized by neuronal loss (especially of large cortical nerve cells) and gliosis accompanied by microvacuolar changes. Other frequently observed alterations are the presence of argyrophilic neuronal inclusions, known as Pick bodies, which are positive for Tau and ubiquitin proteins, and swollen cells.133, 136, 137
Approximately 40% of FTD patients report a history of dementia in the family.138 The most frequent mutations are found in the microtubule-associated protein Tau139 (MAPT) and in the growth factor progranulin (GRN) gene140, 141 (which accounts for >50% of the inherited cases) and on the gene of the chromosome 9 open reading frame 72 (ref. 142) (C9orf72; ~25% of the cases). Other rare mutations appear in genes such as TARDBP, FUS, CHMP2B, TREM2 and VCP.133, 143 However, most FTD cases are sporadic, with molecular alterations of unknown origin.
Although a genome-wide analysis of the methylomic alterations in FTD has not been undertaken, some groups have investigated the role of DNA methylation in the regulation of specific genes associated with the risk of FTD. Two of the most prevalent mutated genes, GRN and C9orf72, have been found in aberrant methylomic state in FTD. The GRN gene is hypermethylated in FTD brains relative to age-matched controls, but notably, also in comparison with AD and PD samples.144 The increased methylation was correlated with lower levels of GRN protein in the frontal cortex of FTD patients. Another of the genes associated with FTD, C9orf72, is epigenetically altered in patients' brains. Expansion of the G4C2 repeat located in the proximal region of C9orf72 is a pathological common cause of FTD.145 Aberrant hypermethylation has been described in the G4C2 repeat,146 and in the CpG island flanking the region of C9orf72.147, 148, 149, 150 The size of the G4C2 repeat is positively correlated with the level of hypermethylation.151 This hypermethylation correlates positively with a reduced neuronal loss, thus pointing out that the downregulation of the genes could have a protective effect in the hippocampus and frontal cortex of FTD patients' brains.152, 153 Banzhaf-Strathmann et al.144 also described the overexpression of DNMT3a in FTD brain samples, which might explain the hypermethylation state of both GRN and C9orf72 genes. Other genes associated with FTD, such as APP, PSEN1 and MAPT, did not show any methylation differences in FTD brain samples compared with age-matched controls.54
The lack of any specific disease-modifying treatment for FTD makes it clear that more in-depth epigenetic analysis is necessary if we are to understand the role of DNA methylation as a potential driving event in this brain disorder. Genome-wide analysis of 5 mC and 5 hmC in different areas of post-mortem brain tissue will yield invaluable insights into potential new gene candidates that are epigenetically deregulated and simultaneously improve patient stratification, helping to differentiate FTD more accurately from other dementias.
Conclusions and future perspectives
Dementia covers a wide spectrum of cognitive abnormalities that arise as a clinical consequence of disorders such as AD, PD, DLB and FTLD. However, our knowledge of its etiology is rather limited and no effective treatments to prevent or revert this condition have been described so far. Dementias are age-associated disorders that, due to the progressive aging of our societies, are likely to increase in incidence.3 Disorders such as AD, PD, DLB and FTLD not only share dementia as one of their core clinical manifestations but also have some molecular alterations in common, implying the existence of similar driving events that may diverge later, during the progression of each disorder. For example Aβ accumulations are considered a hallmark of AD but are also present in DLB; Tau hyperphosphorylation is observed not only in AD but also in PD and DLB; and α-synuclein inclusions are found in PD, DLB and AD.154, 155, 156, 157, 158
The importance of epigenetic mechanisms in learning and memory processes9, 128, 159 has prompted the scientific community to analyze the role of the epigenome in neurological diseases.15 Accordingly, it has been proposed (the LEARn hypothesis) that epigenetic alterations during the lifespan would disturb gene expression over the long term, in an accumulative way, acting as a ‘seed' for the development of disorders such as dementia and neurodegeneration.7 Thus, information about the epigenetic dysregulations of brain function would provide invaluable knowledge about the origin and progression of brain disorders, contributing to the development of effective therapeutic interventions. In this context, our laboratory has recently identified a network of genes undergoing an aberrant methylomic state of their promoters that is common to AD, PD and DLB.41 While this is the first genome-wide demonstration of epigenetic alterations being shared by these disorders, several other alterations common to some of them have been described before (Tables 1 and 2). For example, ANK1, a gene associated with microglia activation, is epigenetically altered in AD41, 49, 65 and DLB.41 Other examples include the gene SNCA, which is aberrantly methylated in PD and DLB,112 and TREM2, a gene mutated in FTD, which is epigenetically altered in AD.71 In addition, α-synuclein can associate with DNMT1, sequestering it in the cytoplasm, resulting in global DNA hypomethylation. This effect of α-synuclein was found not only in PD cases but also in DLB.112 As α-synuclein can also accumulate in AD,154 this mechanism might also contribute to the global DNA hypomethylation seen in the latter. However, more in-depth analysis and genome-wide approaches are essential for understanding the role of aberrant epigenomes in dementia, especially in little understood disorders such as FTD and DLB. This is critical if we are to identify a causative role for DNA methylation in the appearance of neurodegenerative diseases by proving that environmental factors (life experiences, toxins, other disorders) may alter our epigenome in an accumulative way that may eventually end by developing dementia (the LEARn hypothesis7).
Table 1. DNA (hydroxy)methylation alterations in dementia (human brain studies only).
| Gene | Regulation | Disorder | Brain region | Methods | References |
|---|---|---|---|---|---|
| ABCA7 | Increased 5mC | AD | Prefrontal cortex | BS array | 57 |
| ANK1 | Increased 5mC | AD | Entorhinal and prefrontal cortex | WGBS, BS array, pyrosequencing | 41, 49, 65 |
| Reduced 5mC | DLB | Prefrontal cortex | WGBS, BS array, pyrosequencing | 41 | |
| Increased 5hmC | AD | Prefrontal cortex | WG 5 hmC-enriched seq | 89 | |
| APP | Reduced 5mC | AD | Parietal cortex, temporal lobe and cerebellum | MSRE-SB, pyrosequencing | 51, 52, 53 |
| BDNF | Increased 5mC | AD | Prefrontal cortex | MSRE-PCR | 66 |
| BIN1 | Increased 5mC | AD | Entorhinal and prefrontal cortex | BS array | 49, 57 |
| Increased 5hmC | AD | Prefrontal cortex | WG 5 hmC-enriched seq | 89 | |
| C9orf72 | Increased 5mC | FTD | Frontal cortex, temporal cortex, motor cortex, cerebellum | MSRE-PCR, BS-seq, MSRE-SB | 149, 150, 152 |
| COX2 | Reduced 5mC | AD | Prefrontal cortex | MSRE-PCR | 49, 66 |
| CREB | Increased 5mC | AD | Prefrontal cortex | MSRE-PCR | 66 |
| DIP2A | Increased 5mC | AD | Entorhinal and prefrontal cortex | BS array | 49 |
| Reduced 5hmC | AD | Prefrontal cortex | WG 5 hmC-enriched seq | 89 | |
| DNMT1 | Increased 5mC | AD | Prefrontal cortex | BS-MALDI | 56 |
| DUSP22 | Increased 5mC | AD | Hippocampus | BS array, pyrosequencing | 59 |
| Reduced 5hmC | AD | Prefrontal cortex | WG 5 hmC-enriched seq | 89 | |
| DUSP6 | Increased 5mC | AD | Superior temporal gyrus | BS array | 60 |
| GPNMB | Reduced 5mC | PD | Fronta cortex, cerebellum | BS array | 115 |
| GRN | Increased 5mC | FTD | Frontal cortex | BS-MALDI | 144 |
| HLA-DRB5 | Increased 5mC | AD | Prefrontal cortex | BS array | 57 |
| IGFBP7 | Reduced 5mC | AD | Prefrontal cortex | MeDIP | 79 |
| MT-ND1 | Reduced 5mC | AD | Entorhinal cortex and substantia nigra | MeDIP | 94 |
| MTHFR | Increased 5mC | AD | Prefrontal cortex | BS-MALDI | 56 |
| NF-kB | Reduced 5mC | AD | Entorhinal and prefrontal cortex | BS array, MSRE-PCR | 49, 66 |
| PARK16/lq32 | Reduced 5mC | PD | Fronta cortex, cerebellum | BS array | 115 |
| PSEN1 | Reduced 5mC | AD | Prefrontal cortex | BS-MALDI | 56 |
| RHBDF2 | Increased 5mC | AD | Entorhinal and prefrontal cortex | WGBS, BS array, pyrosequencing | 41, 49 |
| SEPW1 | Reduced 5mC | DLB | Frontal cortex | ELISA, MS-PCR | 112 |
| SERPINF1 | Increased 5mC | AD | Entorhinal and prefrontal cortex | BS array | 49 |
| SERPINF2 | Increased 5mC | AD | Entorhinal and prefrontal cortex | BS array | 49 |
| SLC24A4 | Increased 5mC | AD | Prefrontal cortex | BS array | 57 |
| SNCA | Reduced 5mC | PD | Substantia nigra and cortex | BS-seq, ELISA, MS-PCR | 110, 111, 112 |
| Reduced 5mC | DLB | Frontal cortex | ELISA, MS-PCR | 112 | |
| SORBS3 | Increased 5mC | AD | Entorhinal and prefrontal cortex | BS array, pyrosequencing, MS-PCR | 62, 64 |
| SORL1 | Increased 5mC | AD | Prefrontal cortex | BS array | 57 |
| SPTBN4 | Increased 5mC | AD | Frontal cortex | BS array, pyrosequencing | 62 |
| STX1B | Increased 5mC | PD | Fronta cortex, cerebellum | BS array | 115 |
| TBXA2R | Increased 5mC | AD | Frontal cortex | BS array, pyrosequencing, MS-PCR | 62, 64 |
| TMEM59 | Reduced 5mC | AD | Frontal cortex | BS array | 58 |
| TREM2 | Increased 5mC | AD | Hippocampus | BS-seq | 71 |
| Increased 5hmC | AD | Hippocampus | 5hMeDIP | 71 |
Abbreviations: AD, Alzheimer's disease; BS array, bisulfite-modified DNA based arrays; BS-MALDI, bisulfite matrix-assisted laser desorption/ionization; BS-seq, bisulfite sequencing; DLB, dementia with Lewy bodies; ELISA, enzyme-linked immunosorbent assay; FTD, frontotemporal dementia; 5mC, 5-methylcystosine; 5hmC 5-hydroxymethylcytosine; 5hMeDIP; 5-hydroxymethylated DNA immunoprecipitation; MeDIP, methylated DNA immunoprecipitation; MS-PCR, methylation specific PCR; MSRE-PCR, methylation-sensitive restriction enzyme PCR; MSRE-SB, methylation-sensitive restriction enzyme southern blot; PD, Parkinson's disease; WG 5 hmC-enriched seq, whole-genome sequencing analysis of 5-hydroxymethylcytosine-enriched sequences; WGBS, whole-genome bisulfite sequencing.
Table 2. GO analysis using the topGo tool for the genes with described epigenetic alterations in dementia.
| Ontology | GO terms | P-value | Genes |
|---|---|---|---|
| Biological process | Regulation of cellular component organization | 4.4e−09 | ABCA7, ANK1, BIN1, DNMT1, GRN, IGFBP7, MTHFR, PSEN1, SERPINF1, SERPINF2, SNCA, SORBS3, SORL1, SPTBN4, STX1B, TMEM59, TREM2 |
| Vesicle-mediated transport | 9.1e−08 | ABCA7, ANK1, APP, BIN1, C9orf72, GRN, PSEN1, SERPINF2, SNCA, SORL1, SPTBN4, STX1B, TREM2 | |
| Regulation of protein modification process | 2.4e−07 | ABCA7, APP, DNMT1, DUSP22, DUSP6, MTHFR, PSEN1, SERPINF2, SNCA, SORBS3, SORL1, SPTBN4, TMEM59, TREM2 | |
| β-Amyloid formation | 7.0e−07 | ABCA7, PSEN1, SORL1 | |
| Negative regulation of protein metabolic process | 8.2e−07 | ABCA7, APP, DNMT1, DUSP22, DUSP6, PSEN1, SERPINF1, SERPINF2, SNCA, SORL1, TMEM59 | |
| Negative regulation of proteolysis | 1.6e−06 | APP, PSEN1, SERPINF1, SERPINF2, SNCA, SORL1, TMEM59 | |
| Regulation of cellular protein metabolic process | 1.7e−06 | ABCA7, APP, DNMT1, DUSP22, DUSP6, MTHFR, PSEN1, SERPINF1, SERPINF2, SNCA, SORBS3, SORL1, SPTBN4, TMEM59, TREM2 | |
| Regulation of MAPK cascade | 3.5e−06 | ABCA7, DUSP22, DUSP6, PSEN1, SERPINF2, SORBS3, SORL1, SPTBN4, TREM2 | |
| Positive regulation of protein metabolic process | 3.6e−06 | ABCA7, APP, DNMT1, DUSP22, DUSP6, PSEN1, SERPINF2, SNCA, SORBS3, SORL1, SPTBN4, TREM2 | |
| Regulation of protein metabolic process | 4.1e−06 | ABCA7, APP, DNMT1, DUSP22, DUSP6, MTHFR, PSEN1, SERPINF1, SERPINF2, SNCA, SORBS3, SORL1, SPTBN4, TMEM59, TREM2 | |
| Molecular function | Endopeptidase inhibitor activity | 1.7e−04 | APP, SERPINF1, SERPINF2, SNCA |
| Tau protein binding | 1.8e−04 | BIN1, SNCA | |
| Endopeptidase regulator activity | 1.9e−04 | APP, SERPINF1, SERPINF2, SNCA | |
| Peptidase inhibitor activity | 2.0e−04 | APP, SERPINF1, SERPINF2, SNCA | |
| Enzyme binding | 2.7e−04 | ANK1, APP, BIN1, C9orf72, DNMT1, SERPINF2, SNCA, SORL1, SPTBN4, STX1B | |
| Peptidase regulator activity | 4.1e−04 | APP, SERPINF1, SERPINF2, SNCA | |
| Serine-type endopeptidase inhibitor activity | 5.2e−04 | APP, SERPINF1, SERPINF2 | |
| Structural constituent of cytoskeleton | 6.3e−04 | ANK1, SORBS3, SPTBN4 | |
| Spectrin binding | 7.9e−04 | ANK1, SPTBN4 | |
| Tyrosine/serine/threonine phosphatase activity | 2.7e−03 | DUSP22, DUSP6 |
Abbreviations: GO, gene ontology; MAPK, mitogen-activated protein kinase.
The top 10 more significant terms at the biological process and molecular function characterization levels are shown. P-values show the statistical significance of the enrichment of gene ontology terms with analyzed genes.
Epigenetic brain changes are strong correlated with normal aging,11, 160, 161 suggesting that DNA methylation patterns are modified to start senescence programs.162 Interestingly, accelerate epigenetic aging has been described in dementia-related disorders such as AD163 and PD.164 Together with the fact that age is the most important risk factor for many dementia-related disorders, these data suggest a strong association between dementia and an accelerated epigenetic aging.
The young field of neuroepigenomics13 is increasingly identifying new druggable pathways and targets in dementia, demonstrating the importance of the epigenetic component in the healthy brain and in brain disorders.165 The epigenetic discoveries made in the field of dementia in recent years suggest that neuroepigenomic alterations are fundamental in dementia. Several disorders that feature dementia share common aberrant DNA methylation pattern,41 suggesting that these genes could be responsible for the common clinical symptoms presented. However, several methodological and biological drawbacks need to be addressed to properly understand the alterations of the epigenome that occur in dementia. This, together with more in-depth proper cross-disorder studies, will eventually open up the potential for designing effective therapeutic interventions to prevent or revert such pathologies. Until now, all human neuroepigenetic studies have been carried out using post-mortem brain tissue samples. Customarily, grey matter (or often a mixture of white and grey matter) is used for DNA extraction and DNA methylation changes are subsequently analyzed using bisulfite conversion or other techniques.166 Furthermore, the heterogeneity of brain cells167, 168 makes more specific types of preparation essential if we are to understand the specific aberrant methylomes associated with dementia in particular cell populations. This limitations are probably the base of current discrepancies found in the literature on the epigenomics of dementia (different brain sections, even from the same brain area, thus different cellular composition; different ‘life stages' of the patients, etc). Other possible confounders, such geographic distribution of patients,169 sex170 and so on, may be important to explain discrepancies. On the other hand, future studies need to address properly the correlation between neuropathological stage of the disease studied and the changes in (hydroxyl)methylation to properly understand the pathological evolution of the disorder under the epigenomic point of view. Other critical issue is that classically, DNA methylation patterns have been interrogated through sodium bisulfite-treated DNA. This chemical agent transforms ‘unprotected' cytosines to uracils. However, both methylation and hydroxymethylation modifications protect such cytosines to be transformed. Thus, bisulfite-based analysis do not allow to differentiate between 5 mC and 5 hmC, although the data generated on 5 hmC indicates the importance of this modification by itself,161 especially in the brain.171 Recently, a commercially available kit (TrueMethyl Array Kit (Cambridge Epigenetix, Cambridge, UK)) has been developed allowing for the differentiation between 5 mC and 5 hmC.172 Together with the development of new technologies for interrogating the epigenome (i.e. nanopore sequencing173), this kit will eventually increase the current knowledge of the role of 5-hydroxymethylation on dementia, and will force to revise previous published data on DNA methylation on brain disorders, possibly helping to understand current discrepancies. The epigenomic analysis of brain seek to establish strong associations between the (hydroxyl)methylome and the human disorder, but those findings need to be validated to confirm if a specific alteration has a causative role on the disease. Together with the lack of longitudinal studies in human samples that would allow to establish the disorder-causality role of the epigenetic alterations observed, it is clear that the need of better ‘humanized' dynamic models of the pathologies could improve our understanding of dementia. Research with induced pluripotent stem cells differentiated to specific neuronal populations174 or single-cell epigenomics175 will undoubtedly reveal a completely new level of complexity and a set of genes that are altered in dementia-associated pathologies by identifying causal associations between neuronal dysfunctions, gene expression and epigenetic (de)regulation. Recent advances in genetic and epigenetic editing through technologies such CRISPR/Cas9176, 177 will undoubtfully provide a powerful experimental tool that will allow to validate epigenetically altered targets to improve the possible causality of DNA methylome alteration in dementia.
The brain is an extraordinary organ showing high levels of epigenetic features, such as non-CpG methylation.21, 178 The understanding of the role of these non-CpG changes along with the more detailed dissection of the specific role of 5-methylcytosine, 5-hydroxymethylcytosine and newly discovered marks, such as the N6-methyladenine,95, 179 will help the scientific community understand the epigenetic mechanisms underpinning dementia. The recent analysis of human reference epigenomes180 will enable future neuroepigenomics research to exploit the full potential of alterations in DNA methylation patterns in specific genes and cell populations to enable better patient diagnosis, and the stratification and treatment of dementia and other brain disorders.
Acknowledgments
We thank all the members of ME's laboratory and our collaborators for help and stimulating discussions. In particular, we would like to thank Dr Jose V Sanchez-Mut for his work in the laboratory during his PhD, Sebastián Morán and Manuel Castro for their help with the gene network analysis and Dr Joan Carles Balasch for his work on Figures 1 and 3. The work of ME's laboratory is supported by, among other institutions, the E-Rare (ERA-Net for research programs on rare diseases) and EuroRETT (a European network on Rett syndrome, funded by the European Commission under its 6th Framework Program since 2006); the EU Joint Programme—Neurodegenerative Disease Research (JPND; EPI-AD Consortium); Cellex Foundation; and the Health and Science Departments of the Catalan Government (Generalitat de Catalunya). ME is an ICREA Research Professor.
Footnotes
The authors declare no conflict of interest.
References
- United Nations, Department of Economic and Social Affairs, Population Division World Population Ageing (ST/ESA/SER.A/390), 2015.
- Bradshaw CJA, Brook BW. Human population reduction is not a quick fix for environmental problems. Proc Natl Acad Sci USA 2014; 111: 16610–16615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince MJ, Wimo A, Guerchet M, Ali G-C, Wu Y-T, Prina M et al. World Alzheimer Report 2015, The Global Impact of Dementia: An Analysis of Prevalence, Incidence, Cost and Trends. Alzheimer's Disease International (ADI): London, UK, 2015. [Google Scholar]
- Prince MJ, Wu F, Guo Y, Gutierrez Robledo LM, O'Donnell M, Sullivan R et al. The burden of disease in older people and implications for health policy and practice. Lancet 2015; 385: 549–562. [DOI] [PubMed] [Google Scholar]
- American Psychiatric AssociationDiagnostic and Statistical Manual of Mental Disorders: DSM-V, 5th edn. American Psychiatric Publishing: Arlington, VA, USA, 2013. [Google Scholar]
- Sachdev PS, Blacker D, Blazer DG, Ganguli M, Jeste DV, Paulsen JS et al. Classifying neurocognitive disorders: the DSM-5 approach. Nat Rev Neurol 2014; 10: 634–642. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B, Zawia NH. The LEARn model: an epigenetic explanation for idiopathic neurobiological diseases. Mol Psychiatry 2009; 14: 992–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney B, Lahiri DK. Epigenetics of dementia: understanding the disease as a transformation rather than a state. Lancet Neurol 2016; 15: 760–774. [DOI] [PubMed] [Google Scholar]
- Dias BG, Maddox SA, Klengel T, Ressler KJ. Epigenetic mechanisms underlying learning and the inheritance of learned behaviors. Trends Neurosci 2015; 38: 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Agrelo R, Esteller M. Cross-talk between aging and cancer: the epigenetic language. Ann NY Acad Sci 2007; 1100: 60–74. [DOI] [PubMed] [Google Scholar]
- Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci USA 2012; 109: 10522–10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S, Beeri MS, Haroutunian V. Epigenetic determinants of healthy and diseased brain aging and cognition. JAMA Neurol 2013; 70: 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt DJ. The emerging field of neuroepigenetics. Neuron 2013; 80: 624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003; 33(Suppl):245–254. [DOI] [PubMed] [Google Scholar]
- Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol 2009; 8: 1056–1072. [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Grosse I, Zhang MQ. Computational identification of promoters and first exons in the human genome. Nat Genet 2001; 29: 412–417. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 2005; 74: 481–514. [DOI] [PubMed] [Google Scholar]
- Gräff J, Mansuy IM. Epigenetic codes in cognition and behaviour. Behav Brain Res 2008; 192: 70–87. [DOI] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 1997; 88: 471–481. [DOI] [PubMed] [Google Scholar]
- Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol Life Sci 2004; 61: 2571–2587. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci 2014; 17: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem 2004; 279: 48350–48359. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 1998; 19: 219–220. [DOI] [PubMed] [Google Scholar]
- Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem 2005; 280: 13341–13348. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming G-L, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 2011; 145: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 2010; 5: e15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Damayanti NP, Irudayaraj J, Dunn K, Zhou FC. Diversity of two forms of DNA methylation in the brain. Front Genet 2014; 5: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Bernstein A, Chen D, Jin P. 5-Hydroxymethylcytosine: a new player in brain disorders? Exp Neurol 2015; 268: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo AP, Williams SL, Moraes CT. In vivo methylation of mtDNA reveals the dynamics of protein–mtDNA interactions. Nucleic Acids Res 2009; 37: 6701–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci USA 2011; 108: 3630–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellizzi D, D'Aquila P, Scafone T, Giordano M, Riso V, Riccio A et al. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res 2013; 20: 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wijst MGP, Rots MG. Mitochondrial epigenetics: an overlooked layer of regulation? Trends Genet 2015; 31: 353–356. [DOI] [PubMed] [Google Scholar]
- Dzitoyeva S, Chen H, Manev H. Effect of aging on 5-hydroxymethylcytosine in brain mitochondria. Neurobiol Aging 2012; 33: 2881–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippa CF, Schmidt ML, Lee VM, Trojanowski JQ. Antibodies to alpha-synuclein detect Lewy bodies in many Down‘s syndrome brains with Alzheimer's disease. Ann Neurol 1999; 45: 353–357. [DOI] [PubMed] [Google Scholar]
- Galpern WR, Lang AE. Interface between tauopathies and synucleinopathies: a tale of two proteins. Ann Neurol 2006; 59: 449–458. [DOI] [PubMed] [Google Scholar]
- McKeith IG Consensus guidelines for the clinical and pathologic diagnosis ofdementia with Lewy bodies (DLB). Report of the Consortium on DLB International Workshop, 2006, pp 417–423. [DOI] [PubMed]
- Guo JL, Covell DJ, Daniels JP, Iba M, Stieber A, Zhang B et al. Distinct α-synuclein strains differentially promote tau inclusions in neurons. Cell 2013; 154: 103–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan VK, Saykin AJ. Pathways to neurodegeneration: mechanistic insights from GWAS in Alzheimer's disease, Parkinson's disease, and related disorders. Am J Neurodegener Dis 2013; 2: 145–175. [PMC free article] [PubMed] [Google Scholar]
- Hepp DH, Vergoossen DLE, Huisman E, Lemstra AW, Netherlands Brain Bank Berendse HW et al. Distribution and load of amyloid-β pathology in Parkinson disease and dementia with Lewy bodies. J Neuropathol Exp Neurol 2016; 75: 936–945. [DOI] [PubMed] [Google Scholar]
- Guerreiro R, Escott-Price V, Darwent L, Parkkinen L, Ansorge O, Hernandez DG et al. Genome-wide analysis of genetic correlation in dementia with Lewy bodies, Parkinson‘s and Alzheimer's diseases. Neurobiol Aging 2016; 38: 214.e7–214.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Mut JV, Heyn H, Vidal E, Moran S, Sayols S, Delgado-Morales R et al. Human DNA methylomes of neurodegenerative diseases show common epigenomic patterns. Transl Psychiatry 2016; 6: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW et al. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol 2010; 9: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Quiroz YT, Fleisher AS, Chen K, Velez-Pardo C, Jimenez-Del-Rio M et al. Brain imaging and fluid biomarker analysis in young adults at genetic risk for autosomal dominant Alzheimer's disease in the presenilin 1 E280A kindred: a case-control study. Lancet Neurol 2012; 11: 1048–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. The molecular pathology of Alzheimer's disease. Neuron 1991; 6: 487–498. [DOI] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E. Tau in Alzheimer's disease. Trends Cell Biol 1998; 8: 425–427. [DOI] [PubMed] [Google Scholar]
- Cacace R, Sleegers K, van Broeckhoven C. Molecular genetics of early-onset Alzheimer's disease revisited. Alzheimers Dement 2016; 12: 733–748. [DOI] [PubMed] [Google Scholar]
- Lardenoije R, Iatrou A, Kenis G, Kompotis K, Steinbusch HWM, Mastroeni D et al. The epigenetics of aging and neurodegeneration. Prog Neurobiol 2015; 131: 21–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Mut JV, Gräff J. Epigenetic alterations in Alzheimer's disease. Front Behav Neurosci 2015; 9: 135–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L et al. Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci 2014; 17: 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppieters N, Dieriks BV, Lill C, Faull RLM, Curtis MA, Dragunow M. Global changes in DNA methylation and hydroxymethylation in Alzheimer's disease human brain. Neurobiol Aging 2014; 35: 1334–1344. [DOI] [PubMed] [Google Scholar]
- West RL, Lee JM, Maroun LE. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer's disease patient. J Mol Neurosci 1995; 6: 141–146. [DOI] [PubMed] [Google Scholar]
- Tohgi H, Utsugisawa K, Nagane Y, Yoshimura M, Genda Y, Ukitsu M. Reduction with age in methylcytosine in the promoter region −224 approximately −101 of the amyloid precursor protein gene in autopsy human cortex. Brain Res Mol Brain Res 1999; 70: 288–292. [DOI] [PubMed] [Google Scholar]
- Iwata A, Nagata K, Hatsuta H, Takuma H, Bundo M, Iwamoto K et al. Altered CpG methylation in sporadic Alzheimer's disease is associated with APP and MAPT dysregulation. Hum Mol Genet 2014; 23: 648–656. [DOI] [PubMed] [Google Scholar]
- Barrachina M, Ferrer I. DNA methylation of Alzheimer disease and tauopathy-related genes in postmortem brain. J Neuropathol Exp Neurol 2009; 68: 880–891. [DOI] [PubMed] [Google Scholar]
- Marques SCF, Lemos R, Ferreiro E, Martins M, de Mendonça A, Santana I et al. Epigenetic regulation of BACE1 in Alzheimer's disease patients and in transgenic mice. Neuroscience 2012; 220: 256–266. [DOI] [PubMed] [Google Scholar]
- Wang S-C, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer's disease. PLoS One 2008; 3: e2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Chibnik LB, Srivastava GP, Pochet N, Yang J, Xu J et al. Association of Brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA Neurol 2015; 72: 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakulski KM, Dolinoy DC, Sartor MA, Paulson HL, Konen JR, Lieberman AP et al. Genome-wide DNA methylation differences between late-onset Alzheimer's disease and cognitively normal controls in human frontal cortex. J Alzheimer's Dis 2012; 29: 571–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Mut JV, Aso E, Heyn H, Matsuda T, Bock C, Ferrer I et al. Promoter hypermethylation of the phosphatase DUSP22 mediates PKA-dependent TAU phosphorylation and CREB activation in Alzheimer's disease. Hippocampus 2014; 24: 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CT, Roussos P, Garg P, Ho DJ, Azam N, Katsel PL et al. Genome-wide DNA methylation profiling in the superior temporal gyrus reveals epigenetic signatures associated with Alzheimer's disease. Genome Med 2016; 8: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis J, Hansmannel F, Gistelinck M, Mounier A, Van Cauwenberghe C, Kolen KV et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry 2013; 18: 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Mut JV, Aso E, Panayotis N, Lott I, Dierssen M, Rabano A et al. DNA methylation map of mouse and human brain identifies target genes in Alzheimer's disease. Brain 2013; 136: 3018–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Perry G, Troncoso J, Monteiro MJ. α-Calcium-calmodulin-dependent kinase II is associated with paired helical filaments of Alzheimer's disease. qJ Neuropathol Exp Neurol 1996; 55: 954–963. [DOI] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One 2007; 2: e895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunnon K, Smith R, Hannon E, De Jager PL, Srivastava G, Volta M et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer's disease. Nat Neurosci 2014; 17: 1164–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JS, Keleshian VL, Klein S, Rapoport SI. Epigenetic modifications in frontal cortex from Alzheimer's disease and bipolar disorder patients. Transl Psychiatry 2012; 2: e132–e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Sun J, Li S, Wu X, Li L. Oxidative stress induces DNA demethylation and histone acetylation in SH-SY5Y cells: potential epigenetic mechanisms in gene transcription in Aβ production. Neurobiol Aging 2013; 34: 1069–1079. [DOI] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E et al. TREM2 variants in Alzheimer's disease. N Engl J Med 2013; 368: 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M, Wang Y. TREM2 variants: new keys to decipher Alzheimer disease pathogenesis. Nat Neurosci 2016; 17: 201–207. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Rochford CDP, Neumann H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J Exp Med 2005; 201: 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celarain N, Sánchez-Ruiz de Gordoa J, Zelaya MV, Roldán M, Larumbe R, Pulido L et al. TREM2 upregulation correlates with 5-hydroxymethycytosine enrichment in Alzheimer's disease hippocampus. Clin Epigenet 2016; 8: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppner FL, Ransohoff RM, Becher B. Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci 2015; 16: 358–372. [DOI] [PubMed] [Google Scholar]
- Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S et al. Complement and microglia mediate early synapse loss in Alzheimer mousemodels. Science 2016; 352: 712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai L-H et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. Nature 2015; 518: 365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries CE, Kohli MA, Nathanson L, Whitehead P, Beecham G, Martin E et al. Integrated whole transcriptome and DNA methylation analysis identifies gene networks specific to late-onset Alzheimer's disease. J Alzheimers Dis 2015; 44: 977–987. [DOI] [PubMed] [Google Scholar]
- Nordberg A, Rinne JO, Kadir A, Långström B. The use of PET in Alzheimer disease. Nat Rev Neurol 2010; 6: 78–87. [DOI] [PubMed] [Google Scholar]
- Piriz J, Muller A, Trejo JL, Torres-Aleman I. IGF-I and the aging mammalian brain. Exp Gerontol 2011; 46: 96–99. [DOI] [PubMed] [Google Scholar]
- Chua L-M, Lim M-L, Chong P-R, Hu ZP, Cheung NS, Wong B-S. Impaired neuronal insulin signaling precedes Aβ42 accumulation in female AβPPsw/PS1ΔE9 mice. J Alzheimers Dis 2012; 29: 783–791. [DOI] [PubMed] [Google Scholar]
- Agbemenyah HY, Agis-Balboa RC, Burkhardt S, Delalle I, Fischer A. Insulin growth factor binding protein 7 is a novel target to treat dementia. Neurobiol Dis 2014; 62: 135–143. [DOI] [PubMed] [Google Scholar]
- Agis-Balboa RC, Arcos-Diaz D, Wittnam J, Govindarajan N, Blom K, Burkhardt S et al. A hippocampal insulin-growth factor 2 pathway regulates the extinction of fear memories. EMBO J 2011; 30: 4071–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat Rev Neurol 2009; 5: 311–322. [DOI] [PubMed] [Google Scholar]
- Arthur JSC, Fong AL, Dwyer JM, Davare M, Reese E, Obrietan K et al. Mitogen- and stress-activated protein kinase 1 mediates cAMP response element-binding protein phosphorylation and activation by neurotrophins. J Neurosci 2004; 24: 4324–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsky RH, Marini AM. Brain-derived neurotrophic factor in neuronal survival and behavior-related plasticity. Ann NY Acad Sci 2007; 1122: 130–143. [DOI] [PubMed] [Google Scholar]
- Saura CA, Valero J. The role of CREB signaling in Alzheimer's disease and other cognitive disorders. Rev Neurosci 2011; 22: 153–169. [DOI] [PubMed] [Google Scholar]
- Lunnon K, Hannon E, Smith RG, Dempster E, Wong C, Burrage J et al. Variation in 5-hydroxymethylcytosine across human cortex and cerebellum. Genome Biol 2016; 17: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer's disease. PLoS One 2009; 4: e6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouliaras L, Mastroeni D, Delvaux E, Grover A, Kenis G, Hof PR et al. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer's disease patients. Neurobiol Aging 2013; 34: 2091–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condliffe D, Wong A, Troakes C, Proitsi P, Patel Y, Chouliaras L et al. Cross-region reduction in 5-hydroxymethylcytosine in Alzheimer's disease brain. Neurobiol Aging 2014; 35: 1850–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein AI, Lin Y, Street RC, Lin L, Dai Q, Yu L et al. 5-Hydroxymethylation-associated epigenetic modifiers of Alzheimer's disease modulate Tau-induced neurotoxicity. Hum Mol Genet 2016; 25: 2437–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley-Whitman MA, Lovell MA. Epigenetic changes in the progression of Alzheimer's disease. Mech Ageing Dev 2013; 134: 486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS et al. Mitochondrial abnormalities in Alzheimer's disease. J Neurosci 2001; 21: 3017–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow RH, Burns JM, Khan SM. The Alzheimer's disease mitochondrial cascade hypothesis. J Alzheimers Dis 2010; 20 Suppl 2: S265–S279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunnon K, Ibrahim Z, Proitsi P, Lourdusamy A, Newhouse S, Sattlecker M et al. Mitochondrial dysfunction and immune activation are detectable in early Alzheimer's disease blood. J Alzheimers Dis 2012; 30: 685–710. [DOI] [PubMed] [Google Scholar]
- Blanch M, Mosquera JL, Ansoleaga B, Ferrer I, Barrachina M. Altered mitochondrial DNA methylation pattern in Alzheimer disease-related pathology and in Parkinson disease. Am J Phys Anthropol 2016; 186: 385–397. [DOI] [PubMed] [Google Scholar]
- Heyn H, Esteller M. An adenine code for DNA: a second life for N6-methyladenine. Cell 2015; 161: 710–713. [DOI] [PubMed] [Google Scholar]
- Bheda P, Schneider R. Epigenetics reloaded: the single-cell revolution. Trends Cell Biol 2014; 24: 712–723. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009; 462: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods 2014; 11: 817–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringsheim T, Jette N, Frolkis A, Steeves TDL. The prevalence of Parkinson's disease: a systematic review and meta-analysis. Mov Disord 2014; 29: 1583–1590. [DOI] [PubMed] [Google Scholar]
- Marques S, Outeiro TF. Epigenetics in Parkinson's and Alzheimer's diseases. In: Kundu TK (ed). Epigenetics Development and Disease. Sub-cellular Biochemistry, 2013, pp 507–525. [DOI] [PubMed] [Google Scholar]
- de Lau LML, Breteler MMB. Epidemiology of Parkinson's disease. Lancet Neurol 2006; 5: 525–535. [DOI] [PubMed] [Google Scholar]
- Klingelhoefer L, Reichmann H. Pathogenesis of Parkinson disease—the gut–brain axis and environmental factors. Nat Rev Neurol 2015; 11: 625–636. [DOI] [PubMed] [Google Scholar]
- Dickson DW, Braak H, Duda JE, Duyckaerts C, Gasser T, Halliday GM et al. Neuropathological assessment of Parkinson's disease: refining the diagnostic criteria. Lancet Neurol 2009; 8: 1150–1157. [DOI] [PubMed] [Google Scholar]
- Jellinger KA. Formation and development of Lewy pathology: a critical update. J Neurol 2009; 256(Suppl 3):270–279. [DOI] [PubMed] [Google Scholar]
- Farrer MJ. Genetics of Parkinson disease: paradigm shifts and future prospects. Nat Rev Genet 2006; 7: 306–318. [DOI] [PubMed] [Google Scholar]
- Di Fonzo A, Chien HF, Socal M, Giraudo S, Tassorelli C, Iliceto G et al. ATP13A2 missense mutations in juvenile parkinsonism and young onset Parkinson disease. Neurology 2007; 68: 1557–1562. [DOI] [PubMed] [Google Scholar]
- Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet 2014; 46: 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker Z, Possin KL, Boeve BF, Aarsland D. Lewy body dementias. Lancet 2015; 386: 1683–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratwicke J, Jahanshahi M, Foltynie T. Parkinson's disease dementia: a neural networks perspective. Brain 2015; 138: 1454–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowaed A, Schmitt I, Kaut O, Wüllner U. Methylation regulates alpha-synuclein expression and is decreased in Parkinson‘s disease patients' brains. J Neurosci 2010; 30: 6355–6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto L, Takuma H, Tamaoka A, Kurisaki H, Date H, Tsuji S et al. CpG demethylation enhances alpha-synuclein expression and affects the pathogenesis of Parkinson's disease. PLoS One 2010; 5: e15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats P, Spencer B, Coffee E, Patel P, Michael S, Patrick C et al. Alpha-synuclein sequesters Dnmt1 from the nucleus: a novel mechanism for epigenetic alterations in Lewy body diseases. J Biol Chem 2011; 286: 9031–9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründemann J, Schlaudraff F, Haeckel O, Liss B. Elevated alpha-synuclein mRNA levels in individual UV-laser-microdissected dopaminergic substantia nigra neurons in idiopathic Parkinson's disease. Nucleic Acids Res 2008; 36: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boni L, Tierling S, Roeber S, Walter J, Giese A, Kretzschmar HA. Next-generation sequencing reveals regional differences of the α-synuclein methylation state independent of Lewy body disease. Neuromol Med 2011; 13: 310–320. [DOI] [PubMed] [Google Scholar]
- International Parkinson's Disease Genomics Consortium (IPDGC), Wellcome Trust Case Control Consortium 2 (WTCCC2). A two-stage meta-analysis identifies several new loci for Parkinson's disease. PLoS Genet 2011; 7: e1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K, McKnight AJ, Craig D, O'Neill F. Epigenome-wide association study for Parkinson's disease. Neuromol Med 2014; 16: 845–855. [DOI] [PubMed] [Google Scholar]
- Masliah E, Dumaop W, Galasko D, Desplats P. Distinctive patterns of DNA methylation associated with Parkinson disease. Epigenetics 2014; 8: 1030–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Ding H, Zheng Z, Gu Z, Ma J, Chen L et al. Promoter methylation analysis of seven clock genes in Parkinson's disease. Neurosci Lett 2012; 507: 147–150. [DOI] [PubMed] [Google Scholar]
- Iranzo A, Molinuevo JL, Santamaría J, Serradell M, Martí MJ, Valldeoriola F et al. Rapid-eye-movement sleep behaviour disorder as an early marker for a neurodegenerative disorder: a descriptive study. Lancet Neurol 2006; 5: 572–577. [DOI] [PubMed] [Google Scholar]
- Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science 2001; 293: 506–509. [DOI] [PubMed] [Google Scholar]
- Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S et al. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science 2003; 301: 379–383. [DOI] [PubMed] [Google Scholar]
- Zheng T, Lv Q, Lei X, Yin X, Zhang B. Spatial distribution of 5-hydroxymethyl cytosine in rat brain and temporal distribution in striatum. Neurochem Res 2015; 40: 688–697. [DOI] [PubMed] [Google Scholar]
- Vann Jones SA, O'Brien JT. The prevalence and incidence of dementia with Lewy bodies: a systematic review of population and clinical studies. Psychol Med 2014; 44: 673–683. [DOI] [PubMed] [Google Scholar]
- Meeus B, Verstraeten A, Crosiers D, Engelborghs S, Van den Broeck M, Mattheijssens M et al. DLB and PDD: a role for mutations in dementia and Parkinson disease genes? Neurobiol Aging 2012; 33: 629.e5–629.e18. [DOI] [PubMed] [Google Scholar]
- McKeith I, Mintzer J, Aarsland D, Burn D, Chiu H, Cohen-Mansfield J et al. Dementia with Lewy bodies. Lancet Neurol 2004; 3: 19–28. [DOI] [PubMed] [Google Scholar]
- Cummings JL, Mega M, Gray K, Rosenberg-Thompson S, Carusi DA, Gornbein J. The Neuropsychiatric Inventory Comprehensive assessment of psychopathology in dementia. Neurology 1994; 44: 2308–2308. [DOI] [PubMed] [Google Scholar]
- Aarsland D, Ballard C, Larsen JP, McKeith I. A comparative study of psychiatric symptoms in dementia with Lewy bodies and Parkinson's disease with and without dementia. Int J Geriat Psychiatry 2001; 16: 528–536. [DOI] [PubMed] [Google Scholar]
- Fischer A. Epigenetic memory: the Lamarckian brain. EMBO J 2014; 33: 945–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeve BF. REM sleep behavior disorder: updated review of the core features, the REM sleep behavior disorder-neurodegenerative disease association, evolving concepts, controversies, and future directions. Ann NY Acad Sci 2010; 1184: 15–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature 2002; 418: 935–941. [DOI] [PubMed] [Google Scholar]
- Liu H-C, Hu C-J, Tang Y-C, Chang J-G. A pilot study for circadian gene disturbance in dementia patients. Neurosci Lett 2008; 435: 229–233. [DOI] [PubMed] [Google Scholar]
- Stamper C, Siegel A, Liang WS, Pearson JV, Stephan DA, Shill H et al. Neuronal gene expression correlates of Parkinson's disease with dementia. Mov Disord 2008; 23: 1588–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang J, Spina S, Miller BL. Frontotemporal dementia. Lancet 2015; 386: 1672–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden JS, Neary D, Mann DMA. Frontotemporal dementia. Br J Psychiatry 2002; 180: 140–143. [DOI] [PubMed] [Google Scholar]
- Coyle-Gilchrist ITS, Dick KM, Patterson K, Vázquez Rodríquez P, Wehmann E, Wilcox A et al. Prevalence, characteristics, and survival of frontotemporal lobar degeneration syndromes. Neurology 2016; 86: 1736–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden J, Neary D, Mann D. Frontotemporal lobar degeneration: clinical and pathological relationships. Acta Neuropathol 2007; 114: 31–38. [DOI] [PubMed] [Google Scholar]
- Pan X-D, Chen X-C. Clinic, neuropathology and molecular genetics of frontotemporal dementia: a mini-review. Transl Neurodegen 2013; 2: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer JD, Guerreiro R, Vandrovcova J, Uphill J, Reiman D, Beck J et al. The heritability and genetics of frontotemporal lobar degeneration. Neurology 2009; 73: 1451–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Swieten J, Spillantini MG. Hereditary frontotemporal dementia caused by Tau gene mutations. Brain Pathol 2007; 17: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006; 442: 916–919. [DOI] [PubMed] [Google Scholar]
- Zanardini R, Ciani M, Benussi L, Ghidoni R. Molecular pathways bridging frontotemporal lobar degeneration and psychiatric disorders. Front Aging Neurosci 2016; 8: 409–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EGP, Waite A, Rollinson S et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol 2012; 11: 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baizabal-Carvallo JF, Jankovic J. Parkinsonism, movement disorders and genetics in frontotemporal dementia. Nat Rev Neurol 2016; 12: 175–185. [DOI] [PubMed] [Google Scholar]
- Banzhaf-Strathmann J, Claus R, Mücke O, Rentzsch K, van der Zee J, Engelborghs S et al. Promoter DNA methylation regulates progranulin expression and is altered in FTLD. Acta Neuropathol Commun 2013; 1: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011; 72: 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Zhang M, Bruni AC, Maletta RG, Colao R, Fratta P et al. The C9orf72 repeat expansion itself is methylated in ALS and FTLD patients. Acta Neuropathol 2015; 129: 715–727. [DOI] [PubMed] [Google Scholar]
- Xi Z, Zinman L, Moreno D, Schymick J, Liang Y, Sato C et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet 2013; 92: 981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzil VV, Bauer PO, Gendron TF, Murray ME, Dickson D, Petrucelli L. Characterization of DNA hypermethylation in the cerebellum of c9FTD/ALS patients. Brain Res 2014; 1584: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ J, Liu EY, Wu K, Neal D, Suh E, Irwin DJ et al. Hypermethylation of repeat expanded C9orf72 is a clinical and molecular disease modifier. Acta Neuropathol 2014; 129: 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Rainero I, Rubino E, Pinessi L, Bruni AC, Maletta RG et al. Hypermethylation of the CpG-island near the C9orf72 G4C2-repeat expansion in FTLD patients. Hum Mol Genet 2014; 23: 5630–5637. [DOI] [PubMed] [Google Scholar]
- Gijselinck I, Van Mossevelde S, van der Zee J, Sieben A, Engelborghs S, De Bleecker J et al. The C9orf72 repeat size correlates with onset age of disease, DNA methylation and transcriptional downregulation of the promoter. Mol Psychiatry 2015; 21: 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EY, Russ J, Wu K, Neal D, Suh E, McNally AG et al. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol 2014; 128: 525–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan CT, Russ J, Wood EM, Irwin DJ, Grossman M, McCluskey L et al. C9orf72 promoter hypermethylation is neuroprotective: neuroimaging and neuropathologic evidence. Neurology 2015; 84: 1622–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski JQ, Goedert M. Fatal attractions: abnormal protein aggregation and neuron death in Parkinson's disease and Lewy body dementia. Cell Death Differ 1998; 5: 832–837. [DOI] [PubMed] [Google Scholar]
- Iseki E, Togo T, Suzuki K, Katsuse O, Marui W, de Silva R et al. Dementia with Lewy bodies from the perspective of tauopathy. Acta Neuropathol 2003; 105: 265–270. [DOI] [PubMed] [Google Scholar]
- Irwin DJ, Lee VMY, Trojanowski JQ. Parkinson's disease dementia: convergence of α-synuclein, tau and amyloid-β pathologies. Nat Rev Neurosci 2013; 14: 626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendor JT, Logan TP, Edwards RH. The function of α-synuclein. Neuron 2013; 79: 1044–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenkoppele R, Jansen WJ, Rabinovici GD, Knol DL, van der Flier WM, van Berckel BNM et al. Prevalence of amyloid PET positivity in dementia syndromes: a meta-analysis. JAMA 2015; 313: 1939–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder R, Hennion M, Vidal RO, Shomroni O, Rahman R-U, Rajput A et al. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat Neurosci 2015; 19: 102–110. [DOI] [PubMed] [Google Scholar]
- Horvath S, Zhang Y, Langfelder P, Kahn RS, Boks MPM, van Eijk K et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol 2012; 13: R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013; 341: 1237905–1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen P, Shah PP, Nativio R, Berger SL. Epigenetic mechanisms of longevity and aging. Cell 2016; 166: 822–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Bennett DA, Horvath S. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer's disease related cognitive functioning. Aging (Albany NY) 2015; 7: 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Ritz BR. Increased epigenetic age and granulocyte counts in the blood of Parkinson's disease patients. Aging (Albany NY) 2015; 7: 1130–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt DJ, Meaney MJ, Nestler EJ, Akbarian S. Epigenetic Regulation in the Nervous System. Academic Press: New York, NY, USA, 2012. [Google Scholar]
- Kurdyukov S, Bullock M. DNA methylation analysis: choosing the right method. Biology (Basel) 2016; 5; doi:10.3390/biology5010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol 2014; 15: R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Ming G-L, Song H. Decoding neural transcriptomes and epigenomes via high-throughput sequencing. Nat Neurosci 2014; 17: 1463–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Moran S, Hernando-Herraez I, Sayols S, Gomez A, Sandoval J et al. DNA methylation contributes to natural human variation. Genome Res 2013; 23: 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menger Y, Bettscheider M, Murgatroyd C, Spengler D. Sex differences in brain epigenetics. Epigenomics 2010; 2: 807–821. [DOI] [PubMed] [Google Scholar]
- Sun W, Zang L, Shu Q, Li X. From development to diseases: the role of 5hmC in brain. Genomics 2014; 104: 347–351. [DOI] [PubMed] [Google Scholar]
- Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics 2016; 8: 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut IG. New sequencing technologies. Clin Transl Oncol 2013; 15: 879–881. [DOI] [PubMed] [Google Scholar]
- Fernández-Santiago R, Carballo-Carbajal I, Castellano G, Torrent R, Richaud Y, Sánchez-Danés A et al. Aberrant epigenome in iPSC-derived dopaminergic neurons from Parkinson's disease patients. EMBO Mol Med 2015; 7: 1529–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzman O, Tanay A. Single-cell epigenomics: techniques and emerging applications. Nat Rev Genet 2015; 16: 716–726. [DOI] [PubMed] [Google Scholar]
- Kungulovski G, Jeltsch A. Epigenome editing: state of the art, concepts, and perspectives. Trends Genet 2016; 32: 101–113. [DOI] [PubMed] [Google Scholar]
- Mojica FJM, Montoliu L. On the origin of CRISPR-Cas technology: from prokaryotes to mammals. Trends Microbiol 2016; 24: 811–820. [DOI] [PubMed] [Google Scholar]
- Kinde B, Gabel HW, Gilbert CS, Griffith EC, Greenberg ME. Reading the unique DNA methylation landscape of the brain: non-CpG methylation, hydroxymethylation, and MeCP2. Proc Natl Acad Sci USA 2015; 112: 6800–6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 2016; 532: 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A et al. Integrative analysis of 111 reference human epigenomes. Nature 2015; 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]