Abstract
The annual growth cycle of trees is the result of seasonal cues. The onset of winter triggers an endodormant state preventing bud growth and, once a chilling requirement is satisfied, these buds enter an ecodormant state and resume growing. MADS-box genes with similarity to Arabidopsis SHORT VEGETATIVE PHASE (SVP) [the SVP-like and DORMANCY ASSOCIATED MADS-BOX (DAM) genes] have been implicated in regulating flowering and growth-dormancy cycles in perennials. Here, we identified and characterized three DAM-like (MdDAMs) and two SHORT VEGETATIVE PHASE-like (MdSVPs) genes from apple (Malus × domestica ‘Royal Gala’). The expression of MdDAMa and MdDAMc indicated they may play a role in triggering autumn growth cessation. In contrast, the expression of MdDAMb, MdSVPa and MdSVPb suggested a role in maintaining bud dormancy. Consistent with this, ectopic expression of MdDAMb and MdSVPa in ‘Royal Gala’ apple plants resulted in delayed budbreak and architecture change due to constrained lateral shoot outgrowth, but normal flower and fruit development. The association of MdSVPa and MdSVPb expression with floral bud development in the low fruiting ‘Off’ trees of a biennial bearing cultivar ‘Sciros’ suggested the SVP genes might also play a role in floral meristem identity.
Keywords: apple, DAM, SVP, dormancy, ectopic expression, biennial bearing
Introduction
The annual growth cycle of trees in temperate zones consists of dormant and active growth phases. External environmental factors such as cold or drought stress trigger the transition to dormant phase, resulting in shoot growth cessation and bud set. Subsequently, internal signals in the bud are induced to prevent its growth during the endodormant state (Lang et al., 1987; Horvath et al., 2003). These buds shift to the ecodormant state after satisfying the chilling requirement, where buds are capable of resuming growth (Lang et al., 1987).
Recent studies in Populus revealed striking similarities between genes and pathways that regulate flowering time in annuals and growth and dormancy cycles in perennials, as reviewed by Ding and Nilsson (2016); homologs of circadian-clock and photoperiod pathway components phytochromes and CONSTANS (CO) impact on growth cessation, homologs of key flowering genes FLOWERING LOCUS T (FT), bZIP transcription factor FD and floral meristem identity gene APETALA1 (AP1) have all been shown to regulate growth, while a homolog of TERMINAL FLOWER1 (TFL1) affects chilling-requirement and dormancy release. A role of hormones, particularly abscisic acid (ABA) has been well established in both bud and seed dormancy (Rohde and Bhalerao, 2007; Horvath et al., 2008; Graeber et al., 2012) and commonalities in genes that regulate bud and seed dormancy were reported in peach (Leida et al., 2012a; Wang et al., 2016). A novel regulator, putative APETALA2/Ethylene responsive transcription factor EARLY BUD-BREAK 1 (EBB1) was also identified as a determinant of the time of bud break in Populus (Yordanov et al., 2014) and subsequently associated with bud break regulation in apple (Wisniewski et al., 2015). Ectopic expression of a peach C-repeat binding factor (CBF/DREB) gene PpCBF1 in apple resulted in unexpected short-day induced dormancy and delayed budbreak in spring (Wisniewski et al., 2011; Artlip et al., 2014), demonstrating a role of a cold-tolerance gene in regulation of growth and dormancy in trees.
Another class of genes clearly associated with regulation of flowering in annuals and dormancy in perennials are MADS-box genes with similarity to Arabidopsis SHORT VEGETATIVE PHASE (SVP). Deletion of six DORMANCY ASSOCIATED MADS-BOX (DAM) genes in the EVERGROWING (EVG) locus resulted in a complete lack of dormancy in the peach (Prunus persica) evergrowing (evg) mutant (Bielenberg et al., 2004, 2008). Peach DAM gene expression tracks seasonal light and temperature cycles, integrating environmental cues that regulate the transition into and out of endodormancy (Li et al., 2009). DAM1, DAM 2 and DAM 4 showed elevated expression over summer and early autumn, which coincided with the timing of short-day induced growth cessation. Therefore, these three DAM genes were inferred as the most likely candidates for the cause of the non-dormant bud set phenotype seen in the evg mutant trees (Li et al., 2009). DAM3 did not show any distinct expression pattern throughout the annual cycle, but DAM5 and DAM6 expression was elevated over the winter, suggesting that they may play a role in the maintenance of bud endodormancy (Li et al., 2009). Correspondingly, overexpression of the Japanese apricot (Prunus mume) homolog of DAM6 in poplar caused early growth cessation and early terminal bud set, confirming the growth inhibitory function (Sasaki et al., 2011). The growth inhibitory function was also confirmed in kiwifruit. Overexpression of Actinidia chinensis SVP3 retarded reproductive development in transgenic A. eriantha (Wu et al., 2014) and overexpression of SVP2 delayed budbreak in transgenic A. deliciosa (Wu et al., 2017).
In Rosaceae, DAM genes form a subclade separate from true Arabidopsis SVP-like and potato StMADS11-like subclades (Velasco et al., 2010; Wells et al., 2015; Niu et al., 2016; Porto et al., 2016). In peach, the SVP gene family is expanded to six DAM genes, tandemly arranged on peach chromosome 1 (Jimenez et al., 2009; Illa et al., 2011), one true SVP ortholog PpeMADS57 and the less similar PpeMADS58, located on chromosomes 6 and 8, respectively (Wells et al., 2015). A similar expansion of six tandemly arranged DAM genes was found on Japanese apricot chromosome 2, but two SVP orthologs are located on chromosomes 1 and 6 (Sasaki et al., 2011; Xu et al., 2014). In pear, three DAM and one SVP orthologs have been identified (Ubi et al., 2010; Niu et al., 2016). Several studies attempted to identify and characterize apple DAM genes after the apple genome sequence release (Velasco et al., 2010). The presence of DAM and SVP orthologs was confirmed (Mimida et al., 2015; Wisniewski et al., 2015; Kumar G. et al., 2016; Porto et al., 2016), with some discrepancies in nomenclature used in reports and the possible presence of misannotated genes or pseudogenes (Mimida et al., 2015; Porto et al., 2016). Despite the nomenclature discrepancies, apple DAM genes appear to be arranged in tandem on chromosomes 8 and 16, in regions syntenic to the peach chromosome 1, where six peach DAM genes are located (Illa et al., 2011; Mimida et al., 2015; Porto et al., 2016). Two SVP orthologs are located on apple chromosomes 4 and 11, respectively. Ectopic expression of the SVP ortholog MdJOINTLESSa (MdJa) restored the JOINTLESS-deficient tomato mutant (Nakano et al., 2015), demonstrating similar molecular activity between SVP-like genes, while differential seasonal expression of apple DAM and SVP genes at different stages of apical bud development suggested roles in bud dormancy (da Silveira Falavigna et al., 2013; Mimida et al., 2015; Porto et al., 2016). Seasonality and differential expression patterns were also demonstrated in bark tissue of apple (Wisniewski et al., 2015). In addition, sharp spikes in expression in bark and vegetative buds of a transgenic PpCBF1 apple line with altered cold acclimation, dormancy, and growth suggested for distinct roles in regulation of bud dormancy and potentially cambial dormancy (Wisniewski et al., 2015).
So far there has been no attempt at functional characterization of apple DAM and SVP genes by transgenic overexpression in apple. Similarly, little is known about the role of these genes in floral induction, flower and fruit development and if they contribute to biennial bearing, i.e., limited flowering after a year with heavy fruit load, which has been a major constraint on fruit production in many apple cultivars.
In this study, DAM and SVP homologs were identified and tested for expression in apple tissues and in apical buds during the growth and dormancy cycle. Potential roles in regulation of flowering were examined using apical buds of fruit-bearing and non-bearing trees. To examine whether apple DAM and SVP genes play the same or similar roles in apple, a full-length apple DAM cDNA (MdDAMb) and a full-length SVP cDNA (MdSVPa) were characterized using ectopic overexpression in transgenic apple. Their role in growth retardation is discussed.
Materials and Methods
Apple Tissue Sample Collections
Apple (Malus × domestica ‘Royal Gala’) samples were collected from 3-year-old plants grown in a glasshouse at ambient conditions. Three plants were used for sampling. Root, stem, leaf, flower and fruit tissue sampling was performed during the spring to autumn season of 2008 in a glasshouse at Plant & Food Research, Mt Albert, Auckland, New Zealand. Flowers were collected 5 days after bloom (October 2008) and fruit 132 days after bloom. The remaining tissues were sampled at the beginning of summer (December 2008), except for apical buds which were sampled in early autumn (March 2008). Apical bud samples were collected from fruit bearing and non-fruit bearing ‘Sciros’/Pacific RoseTM apple trees grown in the field under standard orchard management. In total, 10 fruit bearing and 10 non-fruit bearing plants were used. Because of the bud tissue availability and the duration of the experiment, buds from multiple trees had to be used as one biological replicate and two replicates were collected at each sampling date. The sampling was performed at approximately four-weekly intervals from early summer 2011 to late spring 2012 at the Plant & Food Research orchard near Havelock North, New Zealand. The beginning of leaf drop was the last week of May 2012, 50% of budbreak was recorded at the second week of September 2012 and 80% full-bloom in the first week of October 2012.
Identification and Phylogenetic Study of ‘Royal Gala’ SVP and DAM-like Genes
The full-length apple ‘Royal Gala’ DAM-like (MdDAMb) and SVP-like (MdSVPa) transcripts were identified from the Plant & Food Research Malus EST database (Newcomb et al., 2006) by BLAST alignment (Altschul et al., 1990). The remaining DAM-like and SVP-like coding sequences were amplified using oligonucleotide primers designed from ‘Royal Gala’ partial sequences and previously reported apple DAM/SVP/JOINTLESS genes (Supplementary Table S1). The predicted full length amino acid sequences were aligned with the full length of amino acid sequences of Arabidopsis SVP (AT2G22450) and AGL24 (AT4G24540), potato StMADS11 (AF008652), apple MdDAM1 (KT582786), MdDAM2 (KT582787), MdDAM4 (KT582789), MdJb (LC004730), apricot PmDAM6 (AB437345), leafy spurge EeDAM2 (EU339320), three pear DAM (PpyDAM1, PpyDAM2 and PpyDAM3) and one pear SVP (PpySVP) (Niu et al., 2016), six peach DAM (PpeDAM1, PpeDAM2, PpeDAM3, PpeDAM4, PpeDAM5 and PpeDAM6) (Bielenberg et al., 2006) and one peach SVP (PpeMADS57) (Wells et al., 2015), trifoliate orange PtSVP (FJ373211), kiwifruit SVP genes (AcSVP1, AcSVP2, AcSVP3 and AcSVP4) (Wu et al., 2012). The alignments were performed using Clustal W (opening = 10, extension = 0.2). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 7.0 (Kumar S. et al., 2016), minimum evolution phylogeny tested with 1000 bootstrap replicates and above 50% of the cut-off value for condensed tree.
RNA Extraction and Quantitative RT-PCR (qRT-PCR) Analysis
Apple bud and tissue RNA was extracted using the Spectrum Plant Total RNA kit (Sigma–Aldrich) according to the manufacturer’s instructions. Total RNA (5 μg) was treated with DNaseI (Ambion) and reverse-transcribed at 37°C using the BluePrint® Reagent kit for RT-PCR (TaKaRa). Amplification and quantification were carried out using the LightCycler® 480 System, LightCycler 480 SYBR Green I Master Mix and LightCycler 480 software version 1.5 (Roche Diagnostics). qPCR conditions were 5 min at 95°C, followed by 40 cycles of 5 s at 95°C, 5 s at 60°C, and 10 s at 72°C, followed by 65–95°C melting curve detection. Relative abundance was calculated using the ΔΔCT method and efficiency was corrected based on standard curves generated from cDNA serial dilutions. Oligonucleotide primers used in this study (Supplementary Table S1) were designed to amplify products between 150–300 bp in size. The reference gene MdSGL29F (GenBank accession number CN892118) was chosen because of demonstrated low-variance across various tissues, development and a range of treatments (Bowen et al., 2014). Other reference genes were considered and demonstrated similar patterns across the samples (Supplementary Figure S1). Three biological replicates (individual trees) were used for tissue-specific expression experiments. Two biological replicates were used for seasonal bud expression experiments. Each single transgenic plant (single line) was used to evaluate transgene expression levels; young leaf tissue was used for this experiment. Four technical replicates were used for each biological replicate.
Plasmid Preparation and Plant Transformation
Full length cDNA sequences of MdSVPa and MdDAMb were cloned into a binary vector pART277 carrying the CaMV 35S promoter, derived from the binary vector pART27 (Gleave, 1992). A construct of the 35S promoter-driven uidA (GUS) gene was used as a control. Agrobacterium tumefaciens strain LBA4404 containing the above binary vectors were transformed into the leaf disks of M. × domestica ‘Royal Gala’ plants, following a previously reported method (Yao et al., 1995). Transgenic plants were transferred to soil and grown under light and temperature conditions replicating the ambient environment in a containment glasshouse at Plant & Food Research, Auckland, New Zealand. Transgenic MdDAMb and MdSVPa plants and controls were carefully monitored at all stages of plant development.
Results
Apple ‘Royal Gala’ DAM and SVP Genes
Interrogation of the apple EST database (Newcomb et al., 2006) identified a full-length ‘Royal Gala’ SVP-like and a full-length ‘Royal Gala’ DAM-like sequence, previously annotated as MdMADS16 (GeneBank accession number HM122599) and MdMADS20 (GeneBank accession number HM122604), respectively. The predicted amino acid sequence of ‘Royal Gala’ MdMADS16 is identical to the previously described ‘Fuji’ MdSVPa and corresponds to MdJa (Nakano et al., 2015) and gene model MDP0000233948 on chromosome 11. The predicted amino acid sequence of ‘Royal Gala’ MdMADS20 is identical to previously reported ‘Fuji’ MdDAMb (Mimida, GeneBank accession number ADL36743), and corresponds to gene model MDP0000255146 on chromosome 16 (Supplementary Table S2).
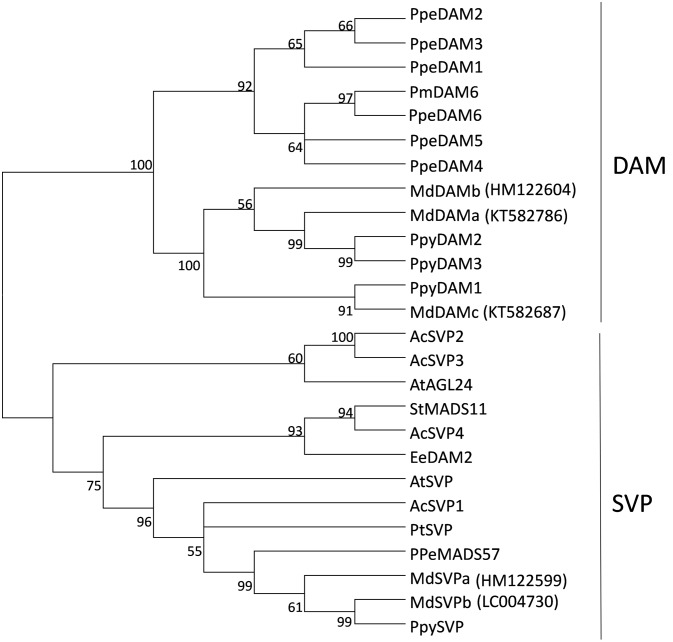
Further interrogation of ‘Royal Gala’ transcripts identified partial sequences that were highly similar to previously reported MdDAMa/MdDAM1, MdDAMc/MdDAM2 and MdSVPb/MdDAM3/MdJb sequences (Mimida et al., 2015; Nakano et al., 2015; Wisniewski et al., 2015; Porto et al., 2016). Amplification of full-length sequences from ‘Royal Gala’ tissues confirmed that the ‘Royal Gala’ sequences were identical or almost identical to previous reports. However, we could not identify a ‘Royal Gala’ MdDAMd/MdDAM3, already suggested to be either a pseudogene or a result of misannotation (Mimida et al., 2015; Porto et al., 2016), nor MdDAM4 (KT582789) (Porto et al., 2016). To prevent confusion, the nomenclature proposed by Mimida et al. (2015) was chosen (Supplementary Table S2) to reflect the relationship to DAM and true SVP homologs. Phylogenetic study of predicted full-length protein sequences confirmed the placement of ‘Royal Gala’ MdDAM and MdSVP proteins in the DAM and SVP subclades (Figure 1).
FIGURE 1.
Phylogenetic tree of DAM and SVP predicted proteins. The tree was built from the full-length amino acid sequence alignment using the neighbor-joining method and the bootstrap test (1000 replicates). Bootstrap values above 50% are shown next to the branches. Ac, Actinidia chinensis, At, Arabidopsis thaliana, Ee, Euphorbia esula, Pm, Prunus mume, Ppe, Prunus persica, Ppy, Pyrus pyrifolia, Pt, Poncirus trifoliata, St, Solanum tuberosum, Md, Malus × domestica ‘Royal Gala’ sequences, with indicated accession numbers.
Tissue-Specific Expression Profiles of MdDAM and MdSVP Genes
To investigate whether MdDAM genes and MdSVP genes were expressed in a tissue-specific manner, qPCR was performed on extracts from various apple tissue samples. Transcripts of MdDAM and MdSVP were predominately found in vegetative tissues and apical buds (Figure 2). MdDAMa and MdDAMc were expressed in apical buds, root and stem. Transcripts of MdDAMb were predominantly found in leaf, stem, but also in root, flower and fruit. MdSVPa transcripts were detected in apical buds, root, stem and leaf tissues and a similar pattern but lower relative expression, particularly in apical buds, was detected for MdSVPb.
FIGURE 2.
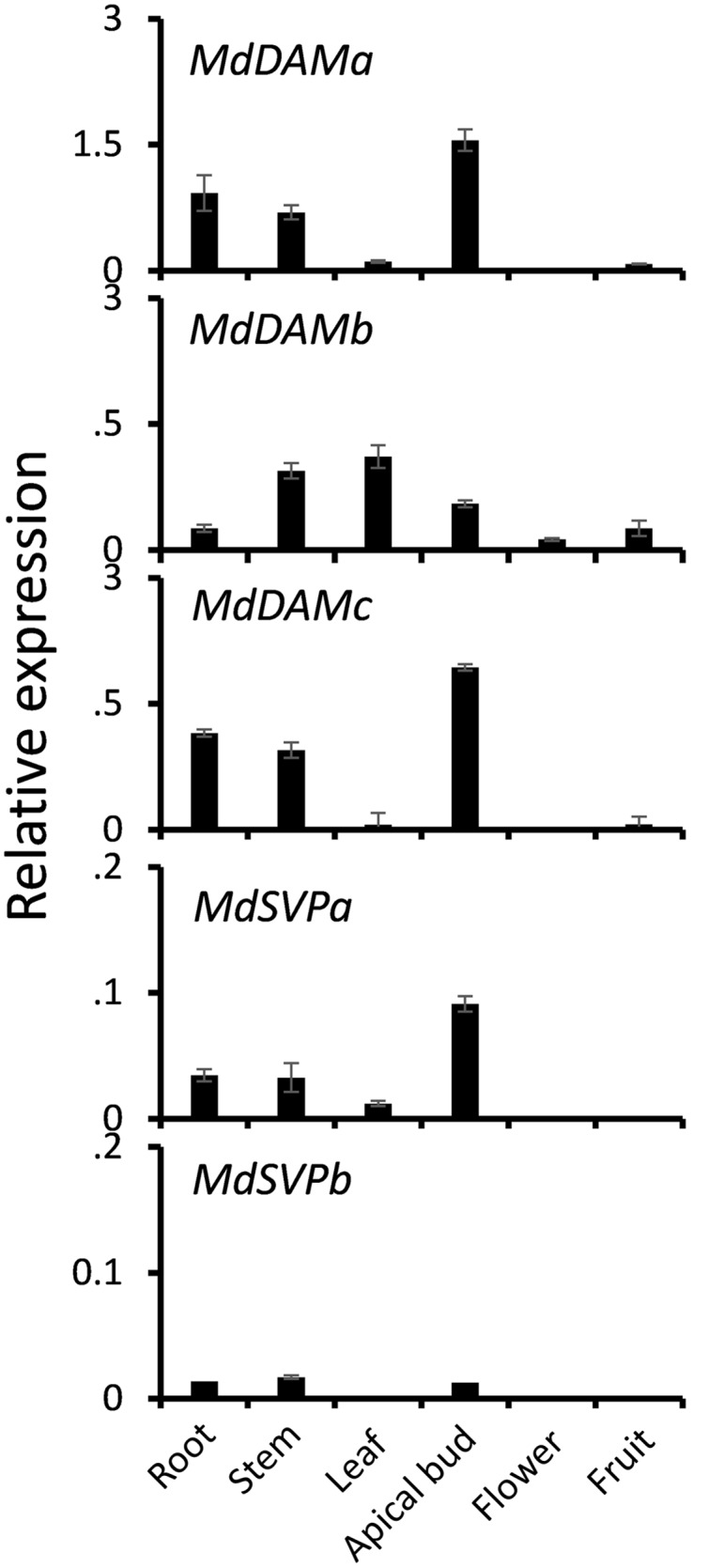
Relative expression of MdDAM and MdSVP genes in root, stem, leaf, apical bud, flower, and fruit of apple. The level of expression was normalized to apple reference gene, MdSGL29F. Error bars represent ± SE of three biological replicates.
Distinct Seasonal Expression Patterns in Apical Buds over a Growth and Dormancy Cycle
Expression analysis was performed for MdSVP and MdDAM genes in apical buds from ‘Sciros’ apple trees harvested over a full growing season, including autumn growth cessation and winter dormancy. This cultivar was chosen for its well-established alternating cropping pattern (Bertelsen and Tustin, 2002), with plants in the light and heavy flowering year of a biennial bearing cycle growing in the same orchard plot. Buds were harvested from fruit-bearing (‘On’) and non-bearing (‘Off’) trees to ascertain potential roles of these genes in flowering. Distinct patterns were observed in buds monitored between establishment of dormancy to reactivation of growth in spring (Figure 3A) and during the full bloom and fruit development stage to leaf drop, which coincide with transitions between meristem identities and subsequent floral organ development in ‘Off’ trees (Figure 3B). Transcripts of MdDAMa and MdDAMc were mostly absent in winter and spring months. MdDAMb was elevated in late winter, peaking in early spring, before visible budbreak. MdSVPa transcript was elevated during dormancy in winter months. Similar mid-winter accumulation was observed for MdSVPb (Figure 3A). MdDAMc transcripts accumulated in summer months and declined in autumn, while MdDAMa accumulated in summer and peaked in autumn (Figure 3B). Small differences, but similar general trends, were detected in buds collected from ‘On’ and ‘Off’ trees for all MdDAM genes. Steady MdDAMa and MdDAMc accumulation with single prominent peaks in April and February, respectively, were detected in ‘Off’ trees, while expression remained elevated over a longer period in ‘On’ trees (Figure 3B). More prominent differences in accumulation between ‘On’ and ‘Off’ trees were detected for MdSVP genes. Summer accumulation, with a peak in February, was observed for MdSVPa and MdSVPb in ‘Off’ trees (Figure 3B) and a faster accumulation rate of MdSVPa in winter and MdSVPb in spring were detected in ‘On’ trees (Figure 3A).
FIGURE 3.
Relative expression of MdDAM and MdSVP genes in apple apical buds during the season. (A) MdDAM and MdSVP genes expression during winter dormancy and active growth in spring. (B) MdDAM and MdSVP genes expression from full bloom to leaf drop. The solid line represents samples collected from the fruit bearing (‘On’) trees. The dotted line represents samples collected from non-fruit bearing (‘Off’) trees. The level of expression was normalized to apple MdSGL29F. Data points represent the mean ± SE of two biological replicates. 50% budbreak is indicated by a vertical line. Stages representing predicted common terminal meristem identity in ‘Off’ trees are presented below. The timing of transitions between meristem identities was adopted from Foster et al. (2003): competent vegetative meristems (VM) transition to inflorescence meristems (IM) around 100 days after full bloom (DAFB) and to terminal floral meristem (FM) identity between 100 and 141 DAFB.
Ectopic Overexpression of MdDAMb and MdSVPa Affects Apple Plant Development
For further functional characterization of apple DAM and SVP genes, ectopic overexpression of MdDAMb and MdSVPa was performed. Their full-length cDNAs driven by the CaMV 35S promoter were transformed into ‘Royal Gala,’ using an established apple transformation protocol (Yao et al., 1995). Initially, a very low transformation efficiency and retarded growth were observed with both 35S:MdDAMb and 35S:MdSVPa, but not the control 35S:GUS construct. After multiple transformation experiments, four MdDAMb and six MdSVPa transgenic lines were established (Figure 4A) and grown alongside four 35S:GUS control lines for 2 years. Transgenic MdDAMb and MdSVPa plants had a strong apical dominance phenotype, with one main upright stem and constrained lateral shoot outgrowth during the first growing season (Figure 4B). Pruning of shoot tips and bending of the stems induced lateral shoot growth (Figure 5), but the timing of spring budbreak in transgenic MdDAMb and MdSVPa lines was significantly delayed in both years (Figure 6). No early leaf senescence and no early bud set could be detected in transgenic MdDAMb and MdSVPa lines over the 2 years of phenotypic analysis.
FIGURE 4.
Appearance of transgenic MdDAMb and MdSVPa ‘Royal Gala’ lines at the first year spring in the glasshouse. (A) Relative expression of MdDAMb and MdSVPa in leaf tissue of transgenic ‘Royal Gala’ lines. The expression of each gene was normalized against apple MdSGL29F. Error bars represent SE for four technical replicates. (B) Phenotypes of constitutive expression of MdDAMb and MdSVPa in ‘Royal Gala’ compared with controls.
FIGURE 5.
Appearance of transgenic MdDAMb and MdSVPa ‘Royal Gala’ lines at the second year spring in the glasshouse. All plants were pruned to induce lateral shoot growth.
FIGURE 6.
Delayed budbreak in transgenic MdDAMb and MdSVPa ‘Royal Gala’ lines. Budbreak was recorded as a number of days from 100% leaf drop in late autumn to the first visible leaf in spring and presented as the mean ± SE of individual transgenic lines.
Ectopic Overexpression of MdDAMb and MdSVPa Does Not Affect Apple Flower and Fruit Development
The first flowering was observed in the spring of the third year after transferring of plants into the glasshouse. The time of flowering (appearance of visible flower buds) was similar between transgenic MdDAMb, MdSVPa and control lines (data not shown). Flowers on less branched MdSVPa lines were mostly harbored on the main stem, in contrast to control plants where most flowers were found on lateral branches (Figures 7A,B), but the duration of flower and fruit development, as well as flower and fruit morphology appeared unaffected (Figures 7C,D).
FIGURE 7.
Flowers and fruits on 35S:MdSVPa transgenic lines versus controls. (A) The flowers of 35S:MdSVPa transgenic lines harbored on the main stem. (B) Typical position of flowers on control plants. (C,D) Fruit on 35S:MdSVPa transgenic lines and controls.
Discussion
In temperate horticultural trees, adequate timing of the onset and duration of winter dormancy are essential to avoid unfavorable winter conditions and ensure flowering in the following season. Despite the importance of dormancy in apple production, very little is known about the molecular processes underlying seasonal growth cycles. MADS-box genes with similarity to SVP were advanced as potential regulators for apple dormancy control (Mimida et al., 2015; Wisniewski et al., 2015; Porto et al., 2016), but the functional characterization using transgenic plants has not yet been reported. The objective of the present study was to functionally characterize apple DAM and SVP genes in transgenic apple ‘Royal Gala’ and perform expression analyses which would shed light on potential roles during apple vegetative and flower development.
Expression of three DAM and two SVP genes have been confirmed in tissues of apple cultivar ‘Royal Gala’ and further tested in apical buds of a biennial bearing apple cultivar ‘Sciros.’ The predominantly vegetative expression and distinct seasonal profiles suggested conserved roles in vegetative growth, but divergent functions in apple bud development and growth-dormancy cycles. The progressive upregulation of MdDAMa and MdDAMc during summer and autumn coincided with cessation of terminal growth, and displayed a pattern whereby MdDAMc accumulation peaks and declines preceded those of MdDAMa. This is highly similar to the pattern reported for other apple cultivars (Mimida et al., 2015; Wisniewski et al., 2015). These seasonal expression profiles concurred with patterns of peach DAM1, DAM 2 and DAM 4, predicted to be the most likely candidates causing the non-dormant bud phenotype in the evg mutant (Li et al., 2009). On the other hand, expression of MdDAMb was elevated in apical buds just before budbreak, resembling the expression patterns of peach DAM5 and DAM6, which were regulated by cold exposure and inversely correlated with budbreak rate (Li et al., 2009; Jiménez et al., 2010; Leida et al., 2012b). Similarly, both MdSVPa and MdSVPb accumulated in apple buds during winter, reaching their peak during the coldest time of the year, followed by a transient decline before budbreak. At this stage, it is unclear if any of the apple DAM and SVP genes directly responds to cold exposure or if their expression reflects developmental events in the bud. Direct binding of cold response C-repeat binding factors (CBF) to C-repeat (CRT)/dehydration-responsive elements (DRE) in the promoter regions of apple DAM and SVP genes has been proposed (Wisniewski et al., 2015) and CBF-mediated enhanced DAM expression was also recently demonstrated in pear (Niu et al., 2016). CRT/DRE motifs have been identified upstream of the coding regions of MdDAMa, MdDAMc and MdSVPb (Mimida et al., 2015; Wisniewski et al., 2015), at least two CRT/DRE motifs (A/GCCGAC) are located within 2 kb upstream of MdSVPa transcription start site and a putative CRT/DRE motif has been identified in the upstream region of MdDAMb, but requires confirmation because of the poor sequence quality in that genomic region. The significance of the CBF pathway and the cross talk between cold and dehydration signaling (Shinozaki and Yamaguchi-Shinozaki, 2000) could therefore be of key importance in convergence of signals that regulate DAM/SVP expression and distinct aspects of dormancy and growth, but the molecular mechanisms underlying distinct expression of DAM and SVP genes in apple remain to be determined.
In Arabidopsis, SVP functions as a repressor of the floral transition during the vegetative phase and later it contributes to the specification of floral meristems, acting to supress floral organ development (Gregis et al., 2009). Hence, removal of SVP is necessary to enable flower organ development, consistent with general absence of apple DAM and SVP transcripts in mature flower and fruit tissues. To address their potential roles in early apple flower development, the predictable biennial bearing pattern of ‘Sciros’ was utilized (Bertelsen and Tustin, 2002). Low flowering and reduced cropping in ‘Off’ years results from the lack of floral induction on fruit-bearing trees in ‘On’ years and is accompanied by differential expression of several flowering regulator genes (Guitton et al., 2016). MdDAM genes in this study showed similar patterns regardless of fruit load, suggesting that DAM gene expression was not principally affected by the physiological states of apical buds. MdSVPa and MdSVPb were upregulated in buds of ‘Off’ trees in February, coinciding with the predicted prevalence of floral meristems in buds of non-bearing trees between 100 and 141 days after full bloom (DAFB) (Foster et al., 2003), indicating potential roles of MdSVPa and/or MdSVPb in floral meristem identity similar to Arabidopsis (Gregis et al., 2009). However, these roles may be minor, considering the normal flower development recorded in transgenic apple lines.
For the functional study, we successfully generated transgenic lines overexpressing a DAM and an SVP gene from apple. The low regeneration efficiency was consistent with previous reports (Sasaki et al., 2011; Wu et al., 2017), suggesting that overexpression of SVP-like genes strongly inhibits outgrowth of shoots in tissue culture, which may explain the limited number of reported transgenic approaches to functionally characterize SVP/DAM genes in woody perennial species. To our knowledge, this is the first report of homologous overexpression of SVP-like genes in a horticultural tree. The role in growth inhibition was corroborated by constrained axillary shoot growth and significantly delayed spring budbreak in transgenic MdDAMb and MdSVPa lines. The normal timing of leaf senescence, growth cessation and apical bud set suggested against a role in establishment of dormancy and the normal flower and fruit development suggested no or perhaps only minor roles in floral meristem and floral organ development. Previous studies demonstrated a general growth inhibitory function of DAM and SVP genes, but also a role in dormancy onset. Overexpression of Japanese apricot DAM6 in transgenic poplar resulted in premature growth cessation and terminal bud set in autumn and delayed shoot outgrowth in spring (Sasaki et al., 2011). However, in kiwifruit, SVP2 performed as a growth repressor once dormancy has been established, but appeared insufficient to suppress growth in permissive conditions (Wu et al., 2017). The current study suggests that MdDAMb and MdSVPa genes play similar growth inhibition roles in apple, without the capacity to promote growth cessation, consistent with their seasonal expression patterns. Therefore, MdSVPa and MdDAMb may maintain growth suppression upon establishment of dormancy, whilst MdDAMa and MdDAMc remain as potential candidates for regulation of growth cessation in autumn.
In summary, apple DAM and SVP-like genes likely play key roles in regulating growth and dormancy cycles in apple. This knowledge can help develop new varieties with budbreak and flowering dates which ensure optimal growth and adequate production in different growing regions and environments. The transgenic lines generated over the course of this study will help elucidate the genes, pathways and biochemical aspects underlying the growth inhibitory functions of MdDAMb and MdSVPa.
Author Contributions
RW designed and performed experiments and prepared the manuscript. ST and SK provided technical assistance to RW. SDT designed the study of biennial bearing apple. RH and AA provided bioinformatics expertise and intellectual input. RM revised the manuscript. EV-G was involved in design of the study and planning the content, editing, and finalizing the manuscript. All authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewers JPL, TA and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
Authors wish to thank Tim Holmes for photography, Sara Uhrle for RNA extractions, Monica Dragulescu and Wade Wadasinghe for maintenance of plants in the glasshouse, Toshi Foster, Cath Kingston and Bianca Wohlers for critical reading of the manuscript.
Footnotes
Funding. This work was funded by the New Zealand Ministry of Business, Innovation and Employment, contract C10X0816.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00477/full#supplementary-material
Relative expression of MdDAM and MdSVP genes in apple apical buds during the season. (A) MdDAMb expression during winter dormancy and active growth in spring. (B) MdDAMb expression from full bloom to leaf drop. The level of expression was normalized to three previously characterized apple reference genes MDP0000173025, MDP0000326399 and MdActin (GenBank accession number CN938023) (Espley et al., 2007; Bowen et al., 2014) and compared to reference gene SGL29L used in this study (Figure 3). Data points represent the mean ± SE of two biological replicates.
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Artlip T. S., Wisniewski M. E., Norelli J. L. (2014). Field evaluation of apple overexpressing a peach CBF gene confirms its effect on cold hardiness, dormancy, and growth. Environ. Exp. Bot. 106 79–86. 10.1016/j.envexpbot.2013.12.008 [DOI] [Google Scholar]
- Bertelsen M. G., Tustin D. S. (2002). Suppression of flower bud formation in light cropping trees of ‘Pacific Rose’ apple using gibberellin sprays. J. Hortic. Sci. Biotechnol. 77 753–757. 10.1080/14620316.2002.11511568 [DOI] [Google Scholar]
- Bielenberg D. G., Wang Y., Fan S., Reighard G. L., Scorza R., Abbott A. G. (2004). A deletion affecting several gene candidates is present in the Evergrowing peach mutant. J. Hered. 95 436–444. 10.1093/jhered/esh057 [DOI] [PubMed] [Google Scholar]
- Bielenberg D. G., Wang Y., Li Z. G., Zhebentyayeva T., Fan S. H., Reighard G. L., et al. (2008). Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 4 495–507. 10.1007/s11295-007-0126-9 [DOI] [Google Scholar]
- Bielenberg D. G., Wang Y., Reighard G. L., Abbott A. G. (2006). Sequencing and analysis of the peach EVG locus. Acta Hortic. 713 73–82. 10.17660/ActaHortic.2006.713.6 [DOI] [Google Scholar]
- Bowen J., Ireland H. S., Crowhurst R., Luo Z., Watson A. E., Foster T., et al. (2014). Selection of low-variance expressed Malus × domestica (apple) genes for use as quantitative PCR reference genes (housekeepers). Tree Genet. Genomes 10 751–759. 10.1007/s11295-014-0720-6 [DOI] [Google Scholar]
- da Silveira, Falavigna V., Porto D. D., Buffon V., Margis-Pinheiro M., Pasquali G.,et al. (2013). Differential transcriptional profiles of dormancy-related genes in apple buds. Plant Mol. Biol. Rep. 32 796–813. [Google Scholar]
- Ding J., Nilsson O. (2016). Molecular regulation of phenology in trees — because the seasons they are a-changin’. Curr. Opin. Plant Biol. 29 73–79. 10.1016/j.pbi.2015.11.007 [DOI] [PubMed] [Google Scholar]
- Espley R. V., Hellens R. P., Putterill J., Stevenson D. E., Kutty-Amma S., Allan A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49 414–427. 10.1111/j.1365-313X.2006.02964.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T., Johnston R., Seleznyova A. (2003). A morphological and quantitative characterization of early floral development in apple (Malus × domestica Borkh.). Ann. Bot. 92 199–206. 10.1093/aob/mcg120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave A. P. (1992). A versatile binary vector system with a T-DNA organizational-structure conducive to efficient integration of cloned dna into the plant genome. Plant Mol. Biol. 20 1203–1207. 10.1007/BF00028910 [DOI] [PubMed] [Google Scholar]
- Graeber K. A. I., Nakabayashi K., Miatton E., Leubner-Metzger G., Soppe W. J. J. (2012). Molecular mechanisms of seed dormancy. Plant Cell Environ. 35 1769–1786. 10.1111/j.1365-3040.2012.02542.x [DOI] [PubMed] [Google Scholar]
- Gregis V., Sessa A., Dorca-Fornell C., Kater M. M. (2009). The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 60 626–637. 10.1111/j.1365-313X.2009.03985.x [DOI] [PubMed] [Google Scholar]
- Guitton B., Kelner J., Celton J., Sabau X., Renou J., Chagné D., et al. (2016). Analysis of transcripts differentially expressed between fruited and deflowered ‘Gala’ adult trees: a contribution to biennial bearing understanding in apple. BMC Plant Biol. 16:55 10.1186/s12870-016-0739-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath D. P., Chao W. S., Suttle J. C., Thimmapuram J., Anderson J. V. (2008). Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genomics 9:536 10.1186/1471-2164-9-536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath D. P., Schaffer R., West M., Wisman E. (2003). Arabidopsis microarrays identify conserved and differentially expressed genes involved in shoot growth and development from distantly related plant species. Plant J. 34 125–134. 10.1046/j.1365-313X.2003.01706.x [DOI] [PubMed] [Google Scholar]
- Illa E., Sargent D. J., Girona E. L., Bushakra J., Cestaro A., Crowhurst R., et al. (2011). Comparative analysis of rosaceous genomes and the reconstruction of a putative ancestral genome for the family. BMC Evol. Biol. 11:9 10.1186/1471-2148-11-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez S., Lawton-Rauh A. L., Reighard G. L., Abbott A. G., Bielenberg D. G. (2009). Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant Biol. 9:81 10.1186/1471-2229-9-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez S., Reighard G., Bielenberg D. (2010). Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Mol. Biol. 73 157–167. 10.1007/s11103-010-9608-5 [DOI] [PubMed] [Google Scholar]
- Kumar G., Arya P., Gupta K., Randhawa V., Acharya V., Singh A. K. (2016). Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malus × domestica). Sci. Rep. 6:20695 10.1038/srep20695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. A., Early J. D., Martin G. C., Darnell R. L. (1987). Endodormancy, paradormancy, and ecodormancy – physiological terminology and classification for dormancy research. HortScience 22 371–377. [Google Scholar]
- Leida C., Conejero A., Arbona V., Gómez-Cadenas A., Llácer G., Badenes M. L., et al. (2012a). Chilling-dependent release of seed and bud dormancy in peach associates to common changes in gene expression. PLoS ONE 7:e35777 10.1371/journal.pone.0035777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leida C., Conesa A., Llacer G., Badenes M. L., Rios G. (2012b). Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytol. 193 67–80. 10.1111/j.1469-8137.2011.03863.x [DOI] [PubMed] [Google Scholar]
- Li Z. G., Reighard G. L., Abbott A. G., Bielenberg D. G. (2009). Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 60 3521–3530. 10.1093/jxb/erp195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimida N., Saito T., Moriguchi T., Suzuki A., Komori S., Wada M. (2015). Expression of DORMANCY-ASSOCIATED MADS-BOX (DAM)-like genes in apple. Biol. Plant. 59 237–244. 10.1038/srep20695 [DOI] [Google Scholar]
- Nakano T., Kato H., Shima Y., Ito Y. (2015). Apple SVP family MADS-box proteins and the tomato pedicel abscission zone regulator JOINTLESS have similar molecular activities. Plant Cell Physiol. 56 1097–1106. 10.1093/pcp/pcv034 [DOI] [PubMed] [Google Scholar]
- Newcomb R. D., Crowhurst R. N., Gleave A. P., Rikkerink E. H., Allan A. C., Beuning L. L., et al. (2006). Analyses of expressed sequence tags from apple. Plant Physiol. 141 147–166. 10.1104/pp.105.076208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Q., Li J., Cai D., Qian M., Jia H., Bai S., et al. (2016). Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 67 239–257. 10.1093/jxb/erv454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porto D. D., Da Silveira Falavigna V., Arenhart R. A., Perini P., Buffon V., Anzanello R., et al. (2016). Structural genomics and transcriptional characterization of the Dormancy-Associated MADS-box genes during bud dormancy progression in apple. Tree Genet. Genomes 12 1–15. 10.1007/s11295-016-1001-3 [DOI] [Google Scholar]
- Rohde A., Bhalerao R. P. (2007). Plant dormancy in the perennial context. Trends Plant Sci. 12 217–223. 10.1016/j.tplants.2007.03.012 [DOI] [PubMed] [Google Scholar]
- Sasaki R., Yamane H., Ooka T., Jotatsu H., Kitamura Y., Akagi T., et al. (2011). Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot (Prunus mume). Plant Physiol. 157 485–497. 10.1104/pp.111.181982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3 217–223. 10.1016/S1369-5266(00)00067-4 [DOI] [PubMed] [Google Scholar]
- Ubi B. E., Sakamoto D., Ban Y., Shimada T., Ito A., Nakajima I., et al. (2010). Molecular cloning of dormancy-associated MADS-box gene homologs and their characterization during seasonal endodormancy transitional phases of Japanese pear. J. Am. Soc. Hortic. Sci. 135 174–182. [Google Scholar]
- Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., Kalyanaraman A., et al. (2010). The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 42 833–839. 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- Wang D., Gao Z., Du P., Xiao W., Tan Q., Chen X., et al. (2016). Expression of ABA metabolism-related genes suggests similarities and differences between seed dormancy and bud dormancy of peach (Prunus persica). Front. Plant Sci. 6:1248 10.3389/fpls.2015.01248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells C. E., Vendramin E., Tarodo S. J., Verde I., Bielenberg D. G. (2015). A genome-wide analysis of MADS-box genes in peach [Prunus persica (L.) Batsch]. BMC Plant Biol. 15:41 10.1186/s12870-015-0436-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski M., Norelli J., Artlip T. (2015). Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 6:85 10.3389/fpls.2015.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski M., Norelli J., Bassett C., Artlip T., Macarisin D. (2011). Ectopic expression of a novel peach (Prunus persica) CBF transcription factor in apple (Malus × domestica) results in short-day induced dormancy and increased cold hardiness. Planta 233 971–983. 10.1007/s00425-011-1358-3 [DOI] [PubMed] [Google Scholar]
- Wu R., Wang T., Mcgie T., Voogd C., Allan A. C., Hellens R. P., et al. (2014). Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time. J. Exp. Bot. 65 4985–4995. 10.1093/jxb/eru264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. M., Walton E. F., Richardson A. C., Wood M., Hellens R. P., Varkonyi-Gasic E. (2012). Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J. Exp. Bot. 63 797–807. 10.1093/jxb/err304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R.-M., Wang T., Warren B. A. W., Allan A. C., Macknight R. C., Varkonyi-Gasic E. (2017). Kiwifruit SVP2 gene prevents premature budbreak during dormancy. J. Exp. Bot. 10.1093/jxb/erx014 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Zhang Q., Sun L., Du D., Cheng T., Pan H., et al. (2014). Genome-wide identification, characterisation and expression analysis of the MADS-box gene family in Prunus mume. Mol. Genet. Genomics 289 903–920. 10.1007/s00438-014-0863-z [DOI] [PubMed] [Google Scholar]
- Yao J.-L., Cohen D., Atkinson R., Richardson K., Morris B. (1995). Regeneration of transgenic plants from the commercial apple cultivar Royal Gala. Plant Cell Rep. 14 407–412. 10.1007/bf00234044 [DOI] [PubMed] [Google Scholar]
- Yordanov Y. S., Ma C., Strauss S. H., Busov V. B. (2014). EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proc. Natl. Acad. Sci. U.S.A. 111 10001–10006. 10.1073/pnas.1405621111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative expression of MdDAM and MdSVP genes in apple apical buds during the season. (A) MdDAMb expression during winter dormancy and active growth in spring. (B) MdDAMb expression from full bloom to leaf drop. The level of expression was normalized to three previously characterized apple reference genes MDP0000173025, MDP0000326399 and MdActin (GenBank accession number CN938023) (Espley et al., 2007; Bowen et al., 2014) and compared to reference gene SGL29L used in this study (Figure 3). Data points represent the mean ± SE of two biological replicates.