Abstract
Clinical isolates of the fungal human pathogen Candida albicans are invariably diploid and heterozygous, impeding genetic study. Recent isolation of C. albicans haploids opens opportunities to apply technologies unfeasible in diploids. However, doubts remain on whether the haploids, derived from chromosome loss, can represent the diploids. Here, we use C. albicans haploids to investigate biofilm, a key virulence attribute. We conducted the first comprehensive characterization of biofilm formation of the haploids in comparison with the diploids. We demonstrate that the haploids form biofilms with essentially the same characteristics as the diploids. Screening a haploid mutant library has uncovered novel GTPase-related genes as biofilm regulators, including IRA2 that encodes an activator of the Ras GTPase. IRA2-deletion mutants develop poorly constructed biofilm in both haploid and diploid C. albicans. Our results demonstrate that the haploids are a valid model for C. albicans biofilm research and a powerful tool for uncovering novel regulators.
Candida albicans is one of the most prevalent fungal pathogens in humans which causes both superficial infections and systemic diseases1,2. Candida species rank as the fourth leading cause of hospital-acquired bloodstream infection and are associated with high morbidity and mortality rates2. C. albicans forms surface-attached communities known as biofilms, which is a major contributory factor to therapeutic failure due to higher resistance of biofilm mode of growth to antifungal drugs and the host immunity3,4,5.
C. albicans possesses an excellent ability to form biofilm communities on medical devices and tissue surfaces, which are extremely difficult to eradicate, giving dire consequences to the patients. Due to its medical importance biofilm mode of growth of C. albicans has been extensively studied by various research groups. Last decade, there has been a major drive to develop novel strategies, particularly to combat Candida biofilms by academic and pharmaceutical investigators. Therefore, understanding mechanisms governing the biofilm formation and its properties will greatly enhance devising new anti-biofilm therapies against this recalcitrant fungal pathogen. However, a significant difficulty in the study of C. albicans biology, including biofilm formation, is the diploid nature of the genome, which hinders the application of forward genetics approaches in identifying novel genes and drug targets. To date, ~4,500 open reading frames in the C. albicans genome remain uncharacterized.
Historically, C. albicans was regarded as an obligate diploid organism with no haploid state6,7. Hickman et al recently discovered haploid strains of C. albicans8. This ground-breaking discovery has a profound impact on the current understanding of C. albicans biology and pathogenicity. In addition, the availability of C. albicans haploid immediately offers high promise to enable, for the first time, applying powerful genetic and molecular approaches previously very difficult or impossible to do in diploid cells9. However, as the haploid strains were generated through loss of one set of chromosomes from a highly heterozygous diploid parent, many recessive mutations are unmasked resulting in a genetic background different from their parent. Indeed, the haploid strains show reduced growth rate and diminished virulence compared to their diploid parent, although they do exhibit several key characteristics defining this species, such as the yeast-hyphae transition, the white-opaque switching, and chlamydospore formation8. These issues have raised doubts on the suitability of using the haploid strains as research tools, because findings derived from the haploid may not be reproduced in the diploid.
In this work, we performed proof-of-concept experiments. We first comprehensively compared C. albicans haploid and diploid strains for their ability to form biofilm, a trait of high importance for life-threatening systemic infection but yet uncharacterized in C. albicans haploid. We then constructed a haploid gene-deletion library for genes encoding uncharacterized GTPases and their regulators, and screened the library to look for novel regulators of biofilm formation. We found that the haploid strains are fully capable of forming biofilms, albeit at a slower rate compared to the diploid strains. The haploid biofilm matured and reached a comparable level to the diploid biofilm after three days. The matured haploid biofilm exhibited essentially the same compositional and architectural characteristics as the diploid biofilm. In addition, by screening the haploid mutant library, we identified IRA2 as a novel positive regulator of biofilm development in both C. albicans haploids and diploids. This study, therefore, demonstrates that C. albicans haploids can serve as a valid model for the study of biofilm formation and for rapid identification of novel gene functions.
Results
C. albicans haploid forms biofilm in vitro
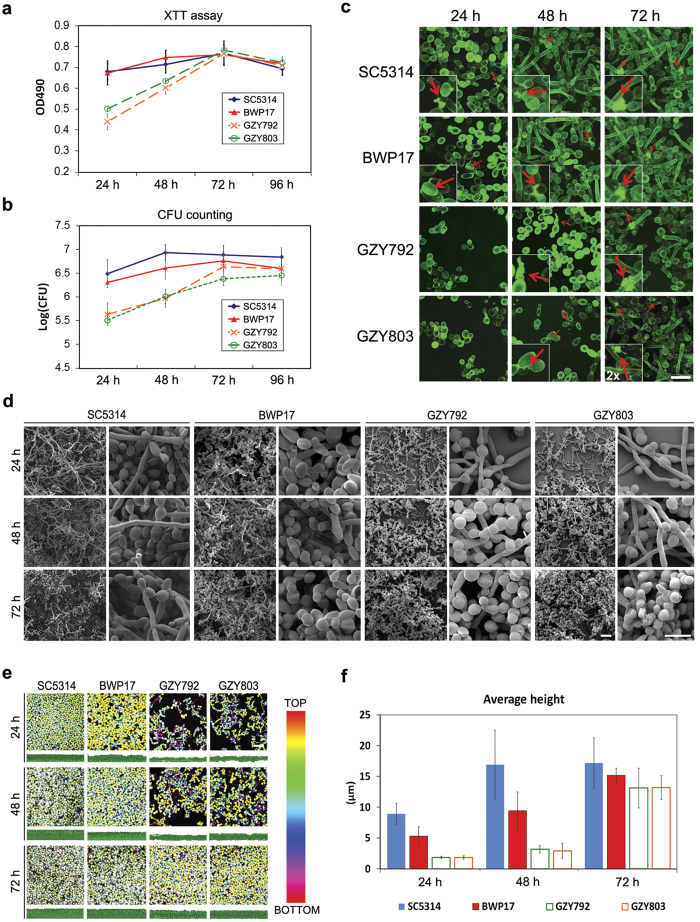
To determine whether C. albicans haploids can serve as a valid model to study biofilm formation, we carried out the first comprehensive characterization of C. albicans haploid biofilms in comparison to their diploid counterparts. We assessed the biofilm forming ability of two haploid strains, GZY792 and GZY8038, and two common laboratory diploid strains, SC5314 and BWP17. These strains were cultured according to the published protocol10 to allow the development of biofilm. The biofilms formed by each strain were then quantified at 24 h intervals by both the XTT reduction assay (Fig. 1a) and colony forming unit (CFU) counting method (Fig. 1b). The haploid strains were capable of forming mature biofilm communities albeit at a slower rate compared to the diploids (Fig. 1a,b). At 24 h after initial adhesion, haploid cells grew and expanded into microcolonies, and appearance of extracellular materials (ECM) was observed at 48 h. For the diploid strains, ECM was observed at 24 h (Fig. 1c). Scanning electron microscopy (SEM) and confocal laser scanning microscopy (CSLM) imaging revealed that haploid strains demonstrated mature biofilm architecture by 72 h, whereas diploid strains exhibited the same by 48 h (Fig. 1d,e). Mature haploid biofilms and diploid biofilms were structurally similar with spatial arrangements of yeast and hyphal cells embedded in ECM giving the classical three-dimensional appearance of C. albicans biofilms. The biofilm comprised of stacks of yeast and hyphal cells with 10–20 μm in height, supported by extracellular matrix, although diploid biofilms contained more hyphal cells by 24 h than haploid biofilms (Fig. 1c–e). We also measured the height of biofilms formed by haploid and diploid strains at different time points. At 24 and 48 h, the average heights of haploid biofilms were substantially less than those of the diploid biofilms. However, the haploid strains could construct biofilms by 72 h with an average height around 13 μm, which was comparable with that of diploid biofilms (Fig. 1f). Taken together, we conclude that the haploid strains are capable of forming biofilms characteristic of the diploid biofilm in vitro.
Figure 1. Comparison of biofilms formed by C. albicans haploid and diploid in vitro.
(a,b) Quantification of the biomass of biofilms formed by diploid strains (SC5314 and BWP17) and haploid strains (GZY792 and GZY803) at different time points with XTT reduction assay (a) and CFU counting method (b). Cells were cultured in GMM medium (supplemented with required amino acids when necessary) at 30 °C overnight and re-inoculated to allow the development of biofilm at 37 °C. The assays were performed in triplicates, and the means were used to generate the curve with standard error. (c) Visualization of extracellular materials (ECM, indicated by arrows) on haploid and diploid biofilms formed at 24 h, 48 h, and 72 h. ConA-Alexa 488 (green) was used to stain the glucose and mannose residues in the fungal cell wall and extended ECM. Bar, 15 μm. (d) Visualization of haploid and diploid biofilms formed at 24 h, 48 h, and 72 h with scanning electron microscopy. Bar, 15 μm. (e) Confocal imaging of haploid and diploid biofilms formed at 24 h, 48 h, and 72 h. For each time point, the upper panels show the top view (with the depth of biofilm color-coded) and the bottom panels show the side view of the biofilm. (f) Comparison of the average heights of haploid and diploid biofilms formed at 24 h, 48 h, and 72 h. Three different sections of each biofilm confocal image were taken for height estimation using BioImage_L to calculate the average height.
C. albicans haploid maintains its ploidy during biofilm formation
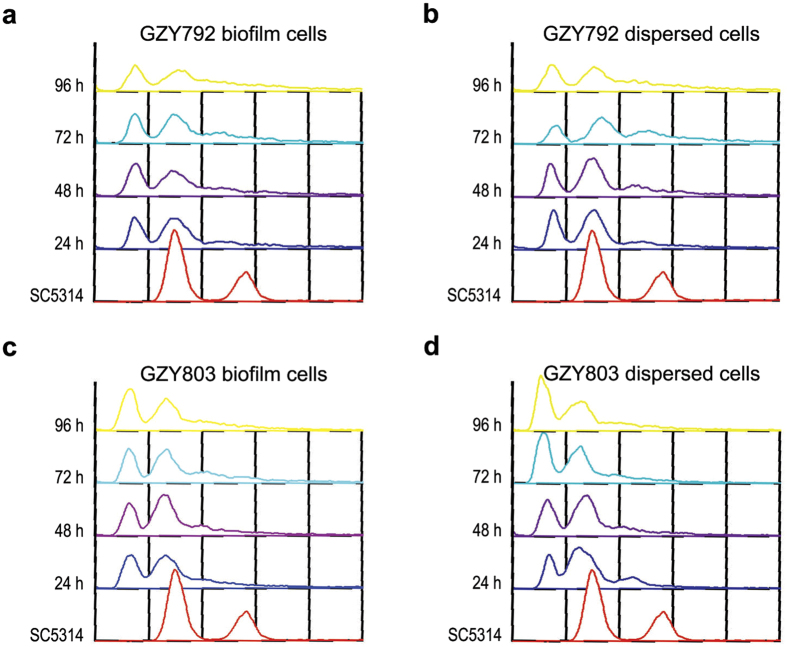
C. albicans haploids are naturally unstable and tend to auto-diploidize under various culture conditions8. Although the haploid tool strains (GZY792 and GZY803) we used for biofilm formation are substantially stable to sustain molecular genetic manipulations8,9, it remains unclear whether they are also stable during biofilm formation. To address this issue, we took biofilm samples as well as the dispersed cells at different time points during the haploid biofilm development for ploidy assessment by flow cytometry (Fig. 2a–d). Both biofilm and dispersed cells of GZY792 and GZY803 maintained the haploid state during the whole course of biofilm formation (up to 96 h), indicating that the C. albicans haploid tool strains are stable in ploidy during the development of biofilms.
Figure 2. Ploidy examination of haploid biofilms and dispersed cells.
Haploid strains GZY792 (a,b) and GZY803 (c,d) were cultured at 30 °C overnight and re-inoculated to allow the development of biofilm at 37 °C. Samples were taken from both biofilm (a,c) and dispersed cells (b,d) at the time points as indicated for ploidy examination by flow cytometry analysis. SC5314 was used as the standard for diploidy.
C. albicans haploids form biofilm ex vivo
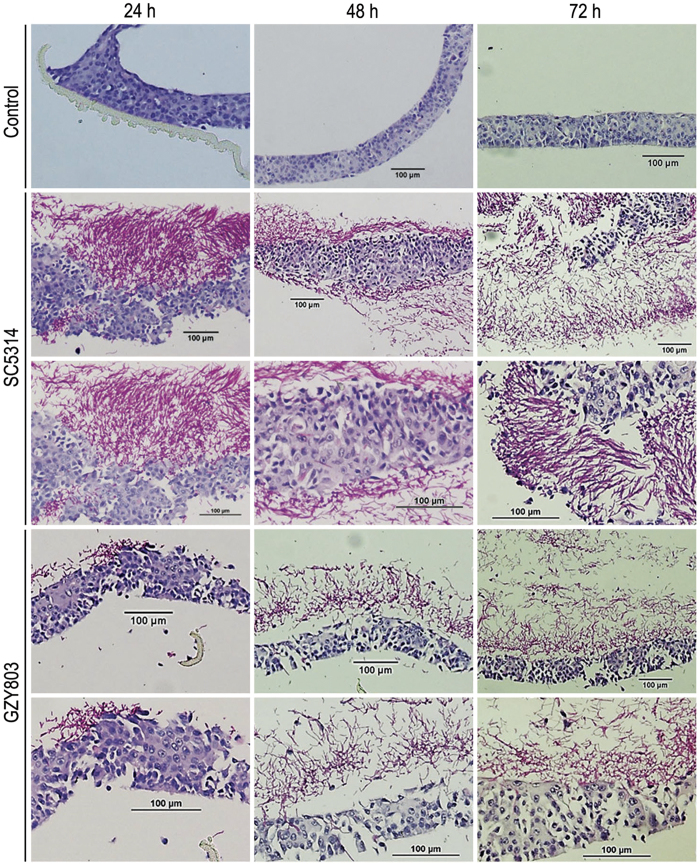
In order to ascertain the comparative biofilm formation ability, reconstituted human oral epithelial (RHOE) model was used to examine the ex vivo biofilm formation of haploid GZY803 cells in comparison to diploid SC5314 cells (Fig. 3). Consistent with the observations on in vitro biofilm formation, the haploid strain was able to form comparable biofilms on RHOE, albeit at a slower rate compared to the diploid strain. The diploid strain formed a thick layer of mature biofilm on the oral epithelial surface by 48 h, whereas the haploid strain formed the same by 72 h. Fungal invasion into the tissues could be detected as early as in 24 h biofilm, which was more obvious in diploid strains. Tissue invasion by GZY803 cells became obvious by 48–72 h. Together, these results further support the validity of using C. albicans haploids as a new tool to study biofilm formation.
Figure 3. Comparison of biofilms formed by C. albicans haploid and diploid on ex vivo reconstituted human oral epithelial cells.
Reconstituted human oral epithelia were infected with the diploid SC5314 and haploid GZY803, respectively, and incubated at 37 °C with 5% CO2. Samples were taken at the time points as indicated and processed for acid-Schiff staining to visualize C. albicans biofilm formation and tissue invasion.
Screen of a haploid mutant library for new biofilm regulators
Next, we utilized this novel haploid C. albicans biofilm model to identify novel regulators of biofilm formation. GTPases are modulators of hyphal formation in C. albicans11,12, which could influence biofilm forming ability13. However, little is known about the role of GTPases in regulating C. albicans biofilm formation. Our group recently established an efficient protocol to rapidly generate specific gene knock out mutants in C. albicans haploids9, which greatly facilitates the discovery of novel gene functions. Using this new methodology, we constructed from the haploid strain GZY803 a gene-deletion library for 35 putative GTPases and GTPase regulators with unknown functions and screened the mutants for defects in biofilm formation using the newly established haploid biofilm model. Biofilm was allowed to develop in GMM medium supplemented with required amino acids at two different temperatures, 30 °C and 37 °C, to select temperature-independent regulators from the mutant library. Biofilm formation was evaluated by XTT assay at 72 h after initial adhesion, since the parental haploid strain (GZY803) and most diploid strains5 were capable of forming mature biofilm by 72 h. Readings taken for biofilms formed by each mutant was normalized to the parent strain GZY803. Foregoing experiments revealed that deletion of IRA2, ARL1, LRG1, AGE2 or the uncharacterized ORF19.3216 gene significantly affected the biofilm formation in a temperature-independent manner (Fig. 4). Interestingly, deletion of IRA2 or ORF19.3216 had inhibitory effect on biofilm formation while deletion of ARL1, LRG1, AGE2 seemed to promote the development of biofilm. Similar results were obtained when biofilm was allowed to develop in RPMI, another medium commonly used for C. albicans studies (Fig. S1), suggesting that the observed phenotypes are independent of the culture medium.
Figure 4. Comparison of biofilms formed in vitro by different C. albicans haploid mutants with their parental strain GZY803.
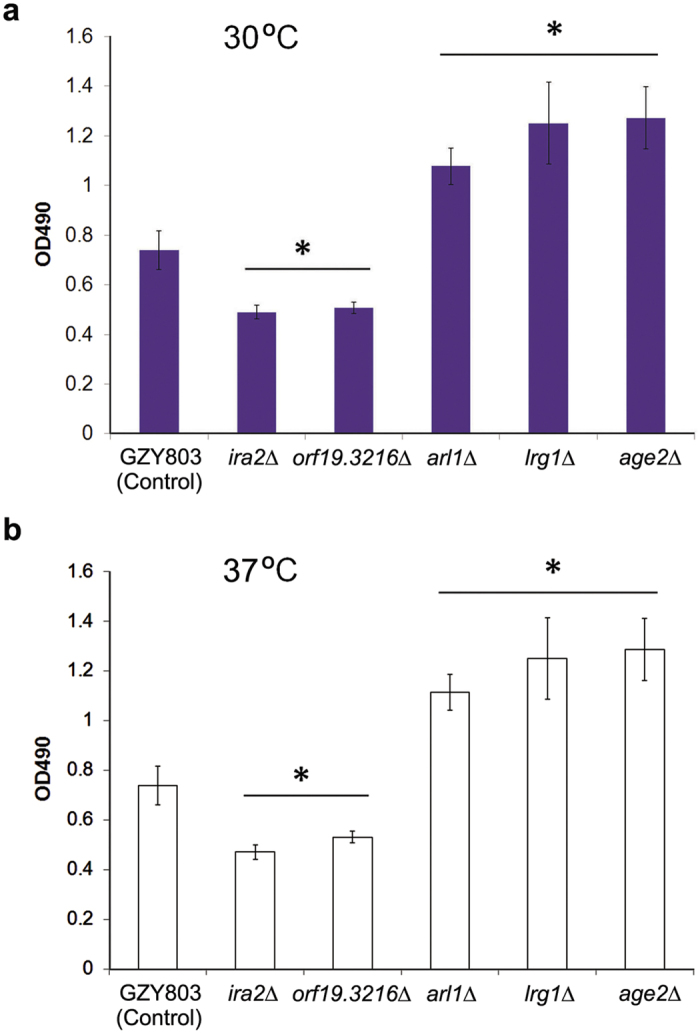
(a,b) Haploid mutants ira2Δ, orf19.3216Δ, arl1Δ, lrg1Δ, age2Δ and the parental strain GZY803 were cultured at 30 °C overnight and re-inoculated to allow the development of biofilm at either 30 °C (a) or 37 °C (b) for 72 h. The biomass of biofilm formed by each strain was quantified with XTT reduction assay. Each sample was tested in triplicates and the means were used to generate the bar with standard error. (*): p-value < 0.05.
Confirmation of IRA2 as a biofilm regulator in C. albicans haploids
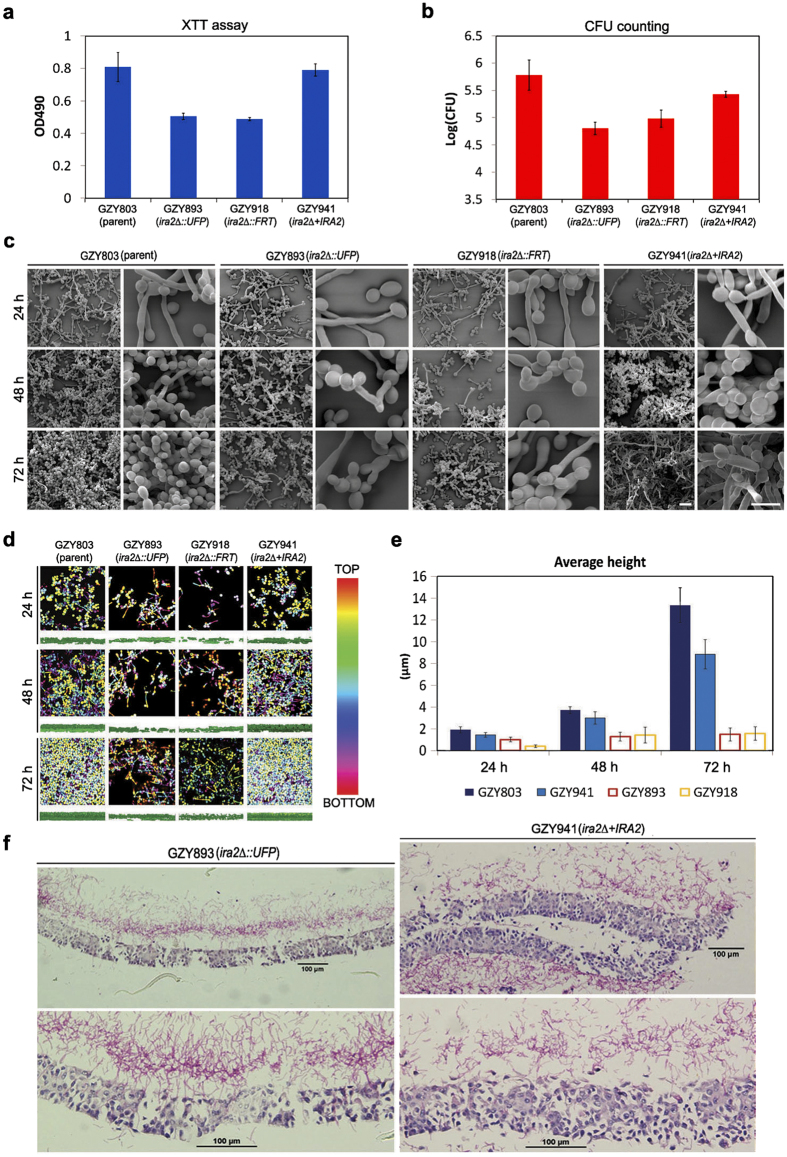
Ira2 is a putative GTPase-activating protein of Ras1, an activator of the cAMP/protein kinase A pathway that critically controls the hyphal growth and biofilm formation. However, the function of C. albicans IRA2 has not been characterized in details, although a previous study had demonstrated that the ira2Δ/Δ is not defective in both yeast and hyphal growth14. We found that the ira2Δ haploid mutant has a similar growth rate like its wild-type parent (GZY803) under biofilm culture conditions (Fig. S2a). In agreement with the previous study14, the ira2Δ haploid mutant is also not defective in morphogenesis as it exhibited normal yeast cell morphology and produced hyphae indistinguishable from those of the wild-type parent at 37 °C upon serum induction (Fig. S2b). These findings suggest that IRA2 might have a direct role in regulating biofilm formation instead of acting indirectly via the Ras pathway that regulates the hyphal development. To confirm that IRA2 is a biofilm regulator in the haploid context, we examined the biofilm formation of both ura+ ira2Δ (GZY893) and ura– ira2Δ (GZY918) strains at 24 h, 48 h, and 72 h relative to their parental strain GZY803 and a rescued strain GZY941. Strikingly, at the mature stage of biofilm formation (72 h), both ira2∆ strains exhibited about 10-fold reduction in CFU compared to the parental strain, which is consistent with the results of the XTT reduction assay (Fig. 5a,b). SEM and CSLM imaging showed that the ira2∆ strains formed a meagre biofilm, being thinner and less dense than the biofilm formed by the parental strain (Fig. 5c,d). Estimation of biofilm average height from CSLM was also consistent with the findings (Fig. 5e). When a wild-type copy of IRA2 was reintroduced into the ira2∆ mutant, the resulting rescued strain (GZY941) largely restored the biofilm formation features of the haploid strain as demonstrated by XTT, CFU and CSLM results (Fig. 5a–e), suggesting that the biofilm formation defects observed in ira2∆ mutants are indeed due to the loss of IRA2 gene function. Consistently, the RHOE model also showed a significantly reduced ability of the ira2∆ strain to form biofilm in comparison with the rescued strain (Fig. 5f). This comprehensive set of data firmly established an important regulatory role for IRA2 in regulating biofilm formation in C. albicans haploids.
Figure 5. Examination of biofilms formed by haploid ira2Δ mutants.
(a,b) Quantification of the biomass of biofilms formed in vitro by haploid ira2Δ mutants (GZY893 and GZY918) and the rescued strain (GZY941) with XTT reduction assay (a) and CFU counting (b). Cells were cultured at 30 °C overnight and re-inoculated to allow the development of biofilm at 37 °C for 72 h. The parental strain GZY803 was used as the control. The assays were performed in triplicates and the means were used to generate the bar with standard error. (c) Visualization of biofilms formed by GZY803, GZY893, GZY918 and GZY941 at 24 h, 48 h, and 72 h with scanning electron microscopy. Bar, 15 μm. (d) Confocal imaging of biofilms formed by GZY803, GZY893, GZY918 and GZY941 at 24 h, 48 h, and 72 h. For each time point, the upper panels show the top view (with the depth of biofilm color-coded) and the bottom panels show the side view of the biofilm. Bar, 15 μm. (e) Comparison of the average heights of biofilms formed by GZY803, GZY893, GZY918 and GZY941 at 24 h, 48 h, and 72 h. Three different sections of each biofilm confocal image were taken for height estimation using BioImage_L to calculate the average height (with standard error). (f) Comparison of ex vivo biofilms formed by haploid ira2Δ mutant and its rescued strain. Reconstituted human oral epithelia was infected with the GZY893 and GZY941, respectively, and incubated at 37 °C for 72 h. Samples were taken and processed for acid-Schiff staining to visualize C. albicans biofilm formation and tissue invasion.
IRA2 as a biofilm regulator in C. albicans diploids
The above results demonstrated that IRA2 is a positive biofilm regulator in the haploid background. To examine whether the new biofilm regulators identified from the haploid biofilm model play the same role in C. albicans diploids, we constructed an ira2∆/∆ mutant (GZY921) from the diploid strain BWP17 for biofilm formation assays. Previous studies have established BCR1 as a key biofilm regulator since the bcr1∆/∆ mutant only shows defects in biofilm formation but not in hyphal morphogenesis15. Therefore, we included bcr1∆ haploid (GZY1095) and bcr1∆/∆ diploid (GZY1094) mutants as controls in the experiments. Our XTT and CFU results reflected a consistent observation of the biofilm formation between haploid and diploid mutants (Fig. 6). Both bcr1∆ haploid and bcr1∆/∆ diploid strains were defective in forming mature biofilm, thus serving as good negative controls. Similarly, ira2∆ haploid and ira2∆/∆ diploid strains both showed a reduction in biofilm formation compared to the haploid GZY803 or diploid BWP17 parental strains, respectively. Again, the defective biofilm formation of ira2 haploid and diploid mutants could be largely rescued by re-integration of a wild-type IRA2 gene back to the mutants, further confirming the important role of IRA2 in biofilm formation. In summary, our results demonstrate that the C. albicans haploid strains we developed are powerful tools in the discovery of novel regulators of traits as complex as biofilm formation in this fungal pathogen of humans.
Figure 6. Quantification of biofilms formed by haploid ira2Δ and diploid ira2Δ/Δ mutants.
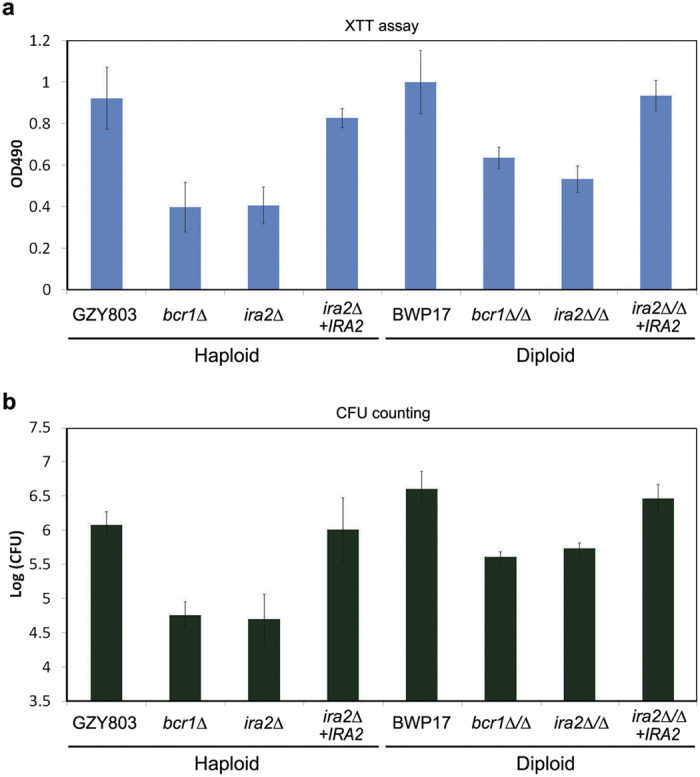
The biomass of biofilms formed in vitro by haploid ira2Δ (GZY893) and its rescued strain (ira2Δ+IRA2, GZY941), as well as diploid ira2Δ/Δ (GZY921) and its rescued strain (ira2Δ/Δ+IRA2, GZY1022) were quantified with XTT reduction assay (a) and CFU counting (b). GZY803 with bcr1Δ (GZY1095), and BWP17 with bcr1Δ/Δ (GZY1094), were used as the positive and negative controls for haploid and diploid, respectively. Cells were cultured at 30 °C overnight and re-inoculated to allow the development of biofilm at 37 °C for 72 h.
Discussion
The discovery of C. albicans haploids was revolutionary, as this fungus had long been thought to be an obligate diploid6,7. And, the diploid nature of this organism has impeded application of some powerful genetic approaches routinely used in organisms with a haploid phase in their life cycle16, thus greatly hindering progress in C. albicans research. There have been doubts on using the haploids to study C. albicans, because the haploids carry many unmasked recessive mutations compared to their diploid parent. In this study, we demonstrate the successful use of C. albicans haploids to discover new regulators of biofilm formation. We performed the first detailed characterization of C. albicans haploid biofilms, and then used the haploids in genetic screens to find novel regulators.
We developed and characterized C. albicans haploid biofilms using two models. One was an in vitro model using multi-well tissue culture plates, and the other was the ex vivo RHOE model which is known to simulate clinical conditions17. We observed that the haploid strains followed the same stages as the diploids to develop mature biofilms (Fig. 1), and were able to stably maintain their ploidy during the whole course of biofilm development (Fig. 2). The only difference was that the haploids showed less initial adhesion to the substrate and needed more time to reach a stable mature biofilm community. For most C. albicans diploid strains, microcolony formation, sign of early biofilm development stage, is at 3–4 h5. Intermediate biofilm development, marked by the emergence of early extracellular material, is at 12 h after initial adhesion. Maturation of diploid biofilm is observed at 38–72 h5. The C. albicans haploid cells developed the microcolony and intermediate biofilm at 24 h and 48 h, respectively, later than the diploid strains. Nevertheless, the haploid biofilm still reached the mature stage at 72 h. The mature haploid biofilm comprised of metabolically active cells encased in extracellular materials (Fig. 1). This level of complexity in terms of yeast and hyphal architecture of the haploid biofilms was comparable to that of mature diploid biofilms.
RHOE is a well-recognized model for the study of host-microbial interaction and oral candidiasis17,18. Corroborating the results of the in vitro model, the haploid strains formed comparable biofilms to that of diploids on the epithelium at 72 h (Fig. 3). However, we noted a delay in tissue invasion by the haploid cells. The diploid cells had invaded the epithelia by 24 h, whereas the haploid cells showed no clear invasion until 48 h. This indicates that the haploid strain is capable of invading the epithelium in spite of some delay. The delay is probably attributable to the decreased adhesion properties and slower growth of the haploid cells than the diploids19. Overall, despite slower development in early stages, the haploid strains eventually formed mature biofilms exhibiting essentially the same cell composition and architecture as the biofilms formed by the diploid strains. Also, we did not detect autodiploidization in the process of haploid biofilm formation (Fig. 2). Thus, our results demonstrate that C. albicans haploid strains can be utilized to assess phenotypic features in mature biofilms under appropriate experimental settings.
GTPases are molecular switches involved in the regulation of numerous biological processes. In C. albicans, small GTPases such as Cdc42 and Ras1 have been extensively investigated for their roles in regulating hyphal growth and tissue invasion11,12,20,21,22,23. To unravel novel gene functions, we have constructed a haploid gene-deletion library covering mostly genes encoding uncharacterized GTPases and their regulators listed in the Candida Genome database (manuscript in preparation). To identify new regulators of biofilm formation in C. albicans, we made use of this library to screen for mutants exhibiting altered ability for biofilm formation in this study. Initial screening revealed that deletion of IRA2, LRG1, ARL1, AGE2 or ORF19.3216 in the haploid significantly affected biofilm formation at both 30 and 37 °C (Fig. 4). None of these genes have been characterized in C. albicans, although some of their homologues have been studied in other fungi such as Saccharomyces cerevisiae (Sc). ScIRA2 encodes a GTPase-activating protein (GAP) that negatively regulates the small GTPase Ras2 which activates the cAMP/protein kinase A (PKA) pathway24. C. albicans has two Ras proteins, Ras1 and Ras2, that play antagonistic roles in regulating the cAMP/PKA pathway25 which governs both hyphal growth and biofilm development26,27,28. Furthermore, ScIRA2 is a key modulator for cellular response against heat shock, starvation, and dormant cell formation24,29. The observed involvement of IRA2 in biofilm formation may help to explain the molecular mechanism underlying the tolerance of C. albicans biofilm against extreme conditions. ScLRG1 encodes a GAP of Rho1 involved in negative regulation of the Pkc1-mediated cell wall integrity signaling pathway30, but its role in biofilm has not been examined. ScARL1 encodes a soluble GTPase having a role in regulation of membrane trafficking31, and ScAge2 is an ADP-ribosylation factor GAP effector playing a part in Trans-Golgi-Network transport31,32. Our observations suggest that membrane trafficking may have an important role in biofilm development33. Consistently, the cAMP pathway activates directional membrane trafficking during hyphal development34,35 which is required for biofilm formation in C. albicans36.
To confirm that IRA2 is indeed required for proper biofilm formation, we used two biofilm models and a range of assays, including XTT reduction assay, CFU counting, microscopic imaging and analysis, and confocal computational analysis, to assess biofilm formation of the haploid ira2Δ mutant. The results consistently demonstrated defective biofilm formations in the mutant, which could be rescued by reintroducing a copy of wild-type IRA2 back to the mutant (Fig. 5). When we deleted both copies of IRA2 in the diploid BWP17 strain, the same defects on biofilm formation were also observed in the diploid ira2Δ/Δ mutant (Fig. 6), suggesting that the biofilm defects of the haploid ira2Δ mutant is directly attributed to IRA2 gene function rather than a phenotype peculiar to the genetic background of the strain. This comprehensive set of data indicates that IRA2 plays an important role in the regulation of C. albicans biofilm formation.
In addition to ira2Δ, four other mutants (orf19.3216Δ, arl1Δ, lrg1Δ, age2Δ) have also been found to significantly affect C. albicans biofilm formation in the initial screening assays (Fig. 4). The enhanced biofilm formation of arl1Δ and lrg1Δ mutants might be due to their abnormal morphogenesis during yeast and/or hyphal growth (Fig. S2b), while the enhanced biofilm formation of age2Δ at 37 °C is likely a result of its accelerated growth rate (Fig. SFig. S2a). Interestingly, the orf19.3216Δ mutant shows a similar growth rate as the wild-type parental strain at both 30 °C and 37 °C, and exhibits normal yeast and hyphal morphologies (Fig. S2), rendering it a good candidate of another previously unknown biofilm regulator. However, the roles of these genes in regulating C. albicans biofilm formation remains to be further investigated.
In summary, we have demonstrated that in spite of genetic differences, C. albicans haploids can be used as a valid model for the study of complex traits, such as biofilm formation, that characterize C. albicans diploids. Importantly, the feasibility to carry out genetic screens using the haploid strains can greatly accelerate discovery of new genes or pathways and speed up identification of novel antifungal drug targets.
Methods
Strains, plasmids, and growth conditions
Construction of yeast strains and plasmids used in this study are described in Supplemental Table 1 and 2, respectively. Recombinant DNA manipulations were performed according to standard methods. E.coli XL1 blue (Stratagene) was used as the host strain for recombinant plasmids and cultured in LB broth (0.5% yeast extract, 1% tryptone, and 0.5% NaCl, pH 7.0) supplemented with ampicillin (100 μg/ml). Transformation of C. albicans was performed according to the protocol of the Fast Yeast Transformation Kit (G-Biosciences). Targeted gene deletion was performed by transforming the host cells with gene deletion cassette and verified by colony PCR as described9. Loop out of URA3 via FLP-mediated excision followed the previous protocols37. Yeast cells were routinely grown at 30 °C in YPD (2% yeast extract, 1% peptone, and 2% glucose), or GMM (glucose minimal medium, 6.79 g/l yeast nitrogen base without amino acids, and 2% glucose) supplemented with appropriate amino acids (uridine 80 μg/ml, arginine 40 μg/ml, and histidine 40 μg/ml) and 5-FOA (1 mg/ml) when necessary. Solid medium plates were prepared by the addition of 2% agar.
In vitro biofilm development
A single colony of each yeast strain was inoculated into GMM medium (supplemented with required amino acids) and grown at 30 °C overnight. The biofilm was developed according to a previous published protocol10. In brief, cell suspension was prepared in GMM (supplemented with required amino acids) with the overnight culture at MacFarland standard 0.375 (equivalent to 107 cells/ml). Aliquots of 100 μl of cell suspension were transferred into wells of pre-sterilized flat-bottomed 96-well microtiter plates (Greiner bio-one). The plates were incubated at 30 °C or 37 °C (depending on the nature of the experiment) for 1.5 h with a shaking speed of 80 rpm. After this adhesion phase, non-adhered cells were removed and the adhered cells were replenished with 200 μl of fresh GMM (supplemented with required amino acids). The plates were incubated under the same conditions for up to 96 h, and dispersed cells and biofilm samples were taken every 24 h for evaluation using standard methodologies.
Biofilm quantification
The biofilm was quantified using 2, 3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide (XTT) reduction assay and colony forming unit (CFU) counting method. After culture medium was aspirated, biofilms were washed with 100 μl of phosphate buffered saline (PBS) once to remove non-adhered cells. XTT assay was performed according to a previously optimized protocol10. Briefly, biofilms were incubated with XTT solution (containing 4 μM menadione and 0.2 mg/ml XTT in PBS) at 37 °C and kept in dark for 20 min. The solutions were transferred to a new plate and colorimetric changes were measured at 490 nm using a calibrated spectrophotometer (Multiskan™ GO, Thermo Scientific). In parallel, some biofilms were taken for enumeration of cells by CFU counting method as previously described38. Dilution series were prepared from the biofilm suspensions and colonies were counted after incubation at 30 °C for 2 days.
Ex vivo biofilm development
C. albicans biofilm formation on ex vivo reconstituted human oral epithelium (RHOE) was performed as described previously39. Upon arrival, the RHOE was transferred to a 24-multiwell plate (Nunc). The tissues was recovered by incubating at 37 °C overnight in serum-free, chemically defined medium MCDB 153 (Skinethic Laboratory) containing 5 mg/ml insulin, 1.5 mM CaCl2, 25 mg/ml gentamicin and 0.4 mg/ml hydrocortison. The tissues was then infected with 200 μl of 108 cells/ml C. albicans cells in GMM medium and incubated at 37 °C in 5% CO2 for up to 72 h. Samples were harvested at desired time points, washed gently to remove non-adherent C. albicans cells and fixed in 4% paraformaldehyde (Sigma) in PBS for 1 h at room temperature. The fixed tissues were then taken and processed for acid-Schiff (PAS) staining to visualize the C. albicans biofilm formation and tissue invasion.
Scanning electron microscopy
Scanning electron microscopy was used to examine the topographic features of the C. albicans biofilms formed on coverslips (Thermo Scientific) under similar environmental conditions as described above. The biofilms on coverslips were washed with PBS once and fixed with 2.5% glutaraldehyde (Sigma) overnight at 4 °C. The coverslips were osmicated with 1% osmium tetraoxide for 30 min and dehydrated with a series of ethanol alcohol (70% for 1 h, 95% for 10 min and 100% for 10 min). Coverslips were air dried in a desiccator prior to sputter coating with gold. Afterwards, specimens were mounted on the aluminium stubs and coated with gold in a low-pressure atmosphere with an ion sputter coater (JFC1 100, Jeol). The topographic features of the biofilm were visualized with a scanning electron microscope (Philips XL30CP).
Confocal laser scanning microscopy
C. albicans biofilms were developed on Thermanox (Nunc) 8 well plates under similar environmental conditions as described above. For three dimensional reconstruction of biofilm, live cells in biofilms were stained with SYTO9 and dead cells with propidium iodide (Invitrogen) for 15 min in dark as previously described38. The biofilms were observed with an Olympus Fluoview FV1000 TIRF confocal microscope. Z-sections were collected. Images were analysed using Olympus FV10-ASW Viewer and Fiji software40. Estimation of the biofilm biovolume was supported by bioImage_L developed by Chávez de41.
Extracellular matrix staining
C. albicans biofilms were developed on 8 well chamber glass (Nunc) as described above and samples were taken at different time point. The cells were stained with Concanavalin-Alexa Flour® 488 (Green) (Invitrogen, 100 μg/ml in PBS) by incubating in dark for 20 min as described5. The images were taken with an Olympus Fluoview FV1000 TIRF confocal microscope. Z-sections were collected. Images were analysed using Olympus FV10-ASW Viewer and Imaris software (Bitplane).
Ploidy analysis by flow cytometry
Ploidy of C. albicans was analyzed by flow cytometry as previously described9. Briefly, adhered and dispersed C. albicans cells were collected at different time points during the development of biofilm, washed once with PBS, and fixed with 70% ethanol at 4 °C overnight. Cells were then treated with RNase A (5 mg/ml) at 37 °C for 6–7 h or overnight. After washed once with PBS, cells were further incubated with propidium iodine (5 mg/ml) in dark at 4 °C overnight. The stained samples were sonicated once to separate clustered cells, and subjected to DNA content analysis using BD FACSCalibur. Data acquisition was performed with 10,000 cells for each sample by using the CellQuest Pro software. The acquired data were then analyzed using WinMDI (version 2.8) software. Diploid cells (SC5314) were used as the control to optimize the instrument settings and calculate sample ploidy value.
Statistical analysis
Statistical analysis was performed using Kruskal-Wallis test, and for any pairwise comparison, Mann-Whitney U test with Bonferroni correction. Analysis was done using SPSS software (Version 16.0, SPSS Inc.). The level of significance was set at P < 0.05.
Additional Information
How to cite this article: Seneviratne, C. J. et al. New “haploid biofilm model’’ unravels IRA2 as a novel regulator of Candida albicans biofilm formation. Sci. Rep. 5, 12433; doi: 10.1038/srep12433 (2015).
Supplementary Material
Acknowledgments
This study was supported by NMRC-NIG (R-221-000-073-511), NUS-Start-up (R-221-000-064-133) to CJS and funding from Agency for Science, Technology, and Research (A*STAR) to YW.
Footnotes
The authors declare no competing financial interests.
Author Contributions C.J.S. and Y.W. conceived, designed, provided general guidance, and co-wrote the manuscript. G.Z. constructed most of the yeast strains, and co-wrote the manuscript. T.T. performed the biofilm experiments and co-wrote the manuscript. F.Y.C. and Y.M.W. assisted strain construction and flow cytometry experiment. S.S.W. and L.P.S. performed the RHOE experiments. H.W. and J.G. generated lrg1∆ and age2∆ mutants, respectively.
References
- Ashman R. B., Farah C. S., Wanasaengsakul S., Hu Y., Pang G. & Clancy R. L. Innate versus adaptive immunity in Candida albicans infection. Immunology and cell biology 82, 196–204 (2004). [DOI] [PubMed] [Google Scholar]
- Pfaller M. A. & Diekema D. J. Epidemiology of invasive candidiasis: a persistent public health problem. Clinical microbiology reviews 20, 133–163 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneviratne C. J., Jin L. & Samaranayake L. P. Biofilm lifestyle of Candida: a mini review. Oral diseases 14, 582–590 (2008). [DOI] [PubMed] [Google Scholar]
- Ramage G., Saville S. P., Thomas D. P. & Lopez-Ribot J. L. Candida biofilms: an update. Eukaryot Cell 4, 633–638 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra J., Kuhn D. M., Mukherjee P. K., Hoyer L. L., McCormick T. & Ghannoum M. A. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. Journal of bacteriology 183, 5385–5394 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA 101, 7329–7334 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S. M. & Johnson A. D. Genetics of Candida albicans, a diploid human fungal pathogen. Annual review of genetics 41, 193–211 (2007). [DOI] [PubMed] [Google Scholar]
- Hickman M. A. et al. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature 494, 55–59 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G., Wang Y. M., Chan F. Y. & Wang Y. One-step targeted gene deletion in Candida albicans haploids. Nat Protoc 9, 464–473 (2014). [DOI] [PubMed] [Google Scholar]
- Seneviratne C. J., Jin L. J., Samaranayake Y. H. & Samaranayake L. P. Cell density and cell aging as factors modulating antifungal resistance of Candida albicans biofilms. Antimicrobial agents and chemotherapy 52, 3259–3266 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana M., Hopkins J. & Arkowitz R. A. Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot Cell 4, 588–603 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausauer D. L., Gerami-Nejad M., Kistler-Anderson C. & Gale C. A. Hyphal guidance and invasive growth in Candida albicans require the Ras-like GTPase Rsr1p and its GTPase-activating protein Bud2p. Eukaryot Cell 4, 1273–1286 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie G. S. & Douglas L. J. Role of dimorphism in the development of Candida albicans biofilms. Journal of medical microbiology 48, 671–679 (1999). [DOI] [PubMed] [Google Scholar]
- Shapiro R. S. et al. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol 19, 621–629 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J. & Mitchell A. P. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol 15, 1150–1155 (2005). [DOI] [PubMed] [Google Scholar]
- Samaranayake D. P. & Hanes S. D. Milestones in Candida albicans gene manipulation. Fungal Genet Biol 48, 858–865 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller M., Zakikhany K., Naglik J. R., Weindl G. & Hube B. Models of oral and vaginal candidiasis based on in vitro reconstituted human epithelia. Nat Protoc 1, 2767–2773 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. B., Cheng G., Chandra J., Mukherjee P., Ghannoum M. A. & Hoyer L. L. RT-PCR detection of Candida albicans ALS gene expression in the reconstituted human epithelium (RHE) model of oral candidiasis and in model biofilms. Microbiology 150, 267–275 (2004). [DOI] [PubMed] [Google Scholar]
- Gancedo J. M. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS microbiology reviews 25, 107–123 (2001). [DOI] [PubMed] [Google Scholar]
- Atre A. N., Surve S. V., Shouche Y. S., Joseph J., Patole M. S. & Deopurkar R. L. Association of small Rho GTPases and actin ring formation in epithelial cells during the invasion by Candida albicans. FEMS immunology and medical microbiology 55, 74–84 (2009). [DOI] [PubMed] [Google Scholar]
- Zheng X. D., Lee R. T., Wang Y. M., Lin Q. S. & Wang Y. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J 26, 3760–3769 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H. M. & Wang Y. RA domain-mediated interaction of Cdc35 with Ras1 is essential for increasing cellular cAMP level for Candida albicans hyphal development. Mol Microbiol 61, 484–496 (2006). [DOI] [PubMed] [Google Scholar]
- Brand A. C., Morrison E., Milne S., Gonia S., Gale C. A. & Gow N. A. Cdc42 GTPase dynamics control directional growth responses. Proc Natl Acad Sci USA 111, 811–816 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Tamanoi F., Kaziro Y., Matsumoto K. & Toh-e A. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol Cell Biol 10, 4303–4313 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Fang H. M., Wang Y. M., Zeng G. S., Zheng X. D. & Wang Y. Ras1 and Ras2 play antagonistic roles in regulating cellular cAMP level, stationary-phase entry and stress response in Candida albicans. Mol Microbiol 74, 862–875 (2009). [DOI] [PubMed] [Google Scholar]
- Inglis D. O. & Sherlock G. Ras signaling gets fine-tuned: regulation of multiple pathogenic traits of Candida albicans. Eukaryot Cell 12, 1316–1325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D. A. & Sundstrom P. The Ras/cAMP/PKA signaling pathway and virulence in Candida albicans. Future microbiology 4, 1263–1270 (2009). [DOI] [PubMed] [Google Scholar]
- Yi S. et al. Alternative mating type configurations (a/alpha versus a/a or alpha/alpha) of Candida albicans result in alternative biofilms regulated by different pathways. PLoS biology 9, e1001117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. et al. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell 60, 803–807 (1990). [DOI] [PubMed] [Google Scholar]
- Lorberg A., Schmitz H. P., Jacoby J. J. & Heinisch J. J. Lrg1p functions as a putative GTPase-activating protein in the Pkc1p-mediated cell integrity pathway in Saccharomyces cerevisiae. Mol Genet Genomics 266, 514–526 (2001). [DOI] [PubMed] [Google Scholar]
- Rosenwald A. G. et al. ARL1 and membrane traffic in Saccharomyces cerevisiae. Yeast 19, 1039–1056 (2002). [DOI] [PubMed] [Google Scholar]
- Poon P. P., Nothwehr S. F., Singer R. A. & Johnston G. C. The Gcs1 and Age2 ArfGAP proteins provide overlapping essential function for transport from the yeast trans-Golgi network. J Cell Biol 155, 1239–1250 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanisamy S. K., Ramirez M. A., Lorenz M. & Lee S. A. Candida albicans PEP12 is required for biofilm integrity and in vivo virulence. Eukaryot Cell 9, 266–277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A., Lane R., Beniston R., Chapa-y-Lazo B., Smythe C. & Sudbery P. Hyphal growth in Candida albicans requires the phosphorylation of Sec2 by the Cdc28-Ccn1/Hgc1 kinase. EMBO J 29, 2930–2942 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. X., Wang H., Wang Y. M. & Wang Y. Novel mechanism coupling cyclic AMP-protein kinase A signaling and golgi trafficking via Gyp1 phosphorylation in polarized growth. Eukaryot Cell 13, 1548–1556 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. H., Kabrawala S., Fox E. P., Nobile C. J., Johnson A. D. & Bennett R. J. Genetic control of conventional and pheromone-stimulated biofilm formation in Candida albicans. PLoS Pathog 9, e1003305 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morschhauser J., Michel S. & Staib P. Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol Microbiol 32, 547–556 (1999). [DOI] [PubMed] [Google Scholar]
- Seneviratne C. J., Silva W. J., Jin L. J., Samaranayake Y. H & Samaranayake L. P. Architectural analysis, viability assessment and growth kinetics of Candida albicans and Candida glabrata biofilms. Archives of oral biology 54, 1052–1060 (2009). [DOI] [PubMed] [Google Scholar]
- Jayatilake J. A., Samaranayake Y. H., Cheung L. K. & Samaranayake L. P. Quantitative evaluation of tissue invasion by wild type, hyphal and SAP mutants of Candida albicans, and non-albicans Candida species in reconstituted human oral epithelium. Journal of oral pathology & medicine : official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology 35, 484–491 (2006). [DOI] [PubMed] [Google Scholar]
- Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez de Paz L. E. Image analysis software based on color segmentation for characterization of viability and physiological activity of biofilms. Applied and environmental microbiology 75, 1734–1739 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.