Abstract
Cycloviruses, small ssDNA viruses of the Circoviridae family, have been identified in the cerebrospinal fluid from symptomatic human patients. One of these species, cyclovirus-Vietnam (CyCV-VN), was shown to be restricted to central and southern Vietnam. Here we report the detection of CyCV-VN species in stool samples from pigs and humans from Africa, far beyond their supposed limited geographic distribution.
Cycloviruses (CyCVs), a recently proposed genus within the Circoviridae family, are non-enveloped single-stranded DNA (ssDNA) viruses with a ≈2 kb circular genome containing two major inversely arranged open reading frames (ORFs) encoding a putative replication-associated protein (Rep) and a capsid (Cap) protein1. An ever growing number of species of CyCVs have been described in mammals (human, chimpanzee, goat, cattle, bat, pig, sheep, and camel), birds (chicken and duck) and insects (dragonflies and wood cockroach)1,2,3,4,5,6. In spite of a high genetic diversity1,5, cross-species transmission seems likely, at least in mammals3,7.
However, in contrast with circoviruses, the pathological significance of CyCV infection remains unclear. CyCVs have been suggested to cause respiratory and neurological infections in humans1,7,8,9. The presence of the virus was demonstrated in serum, stool or cerebrospinal fluid (CSF) of patients with acute flaccid paralysis, paraplegia or suspected central nervous system infections from Pakistan, Tunisia, Malawi and southern and central Vietnam1,7,8. Of particular interest is the description of a high detection rate of the CyCV-Vietnam (CyCV-VN) species in the feces of pigs and chicken from the same area as infected humans7. This observation, together with the high level of conservation between human, swine and chicken CyCV-VN strains suggest that pigs and chicken might act as a reservoir for human infections7. Subsequent investigations on the circulation of CyCV-VN in human CSF specimens from northern Vietnam, Cambodia, Nepal and the Netherlands were negative, suggesting a limited geographic distribution of this CyCV species10.
In this study, the presence of CyCV-VN and CyCV-VN-like strains was demonstrated in human stool samples from Madagascar and swine feces from Cameroon, showing that the geographic distribution extends far beyond southern and central Vietnam as originally thought.
Results
To assess the circulation of CyCV-VN in Africa, we tested, using a CyCV species-specific PCR, 410 human stool samples from Madagascar, 1337 human stool samples from Ghana and 520 pig stool samples from Cameroon. Out of the 410 human samples from Madagascar, three were positive for CyCV-VN-like species and the full genome was amplified, sequenced and characterized (Genbank acc. no. KM392287–KM392289). Ten swine samples from Cameroon were positive (1 from Bamenda, 9 from Douala). The full genome of three positive samples from Douala was obtained (Genbank acc. no. KM392284–KM392286). All human samples from (cases and controls) Ghana remained negative for CyCV-VN.
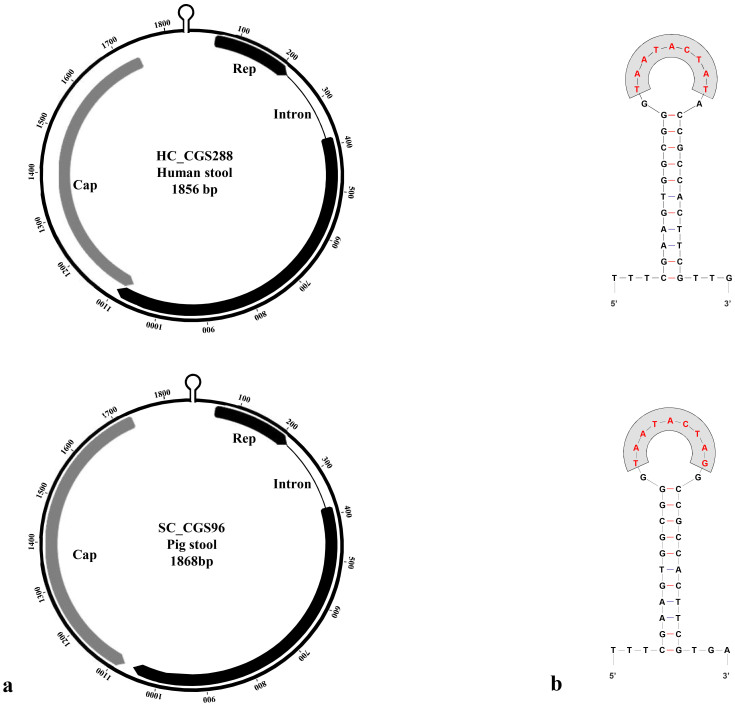
The genomic sequences of human CyCV strains from Madagascar were highly similar (98% identity) to that of human CyCV-VN species7. The genomic organization was identical with a Rep ORF interrupted by a 166 nt long intron with a typical splice donor (GT) and acceptor site (AG) and Cap ORF of the same length and positions (Figure 1). Rep sequence (292 aa) was ≈98% identical to that of CyCV-VN species7 and showed ≈77% and ≈73% identity to the next closely related CyCV species, i.e. Malawian human CSF CyCV (VS5700009) and Tunisian human stool CyCV (TN25), respectively (Table 1)1,8. Cap protein of Madagascan CyCV was 222 aa long like typical CyCV-VN and equally conserved (≈98–99% identity when compared to CyCV-VN) but more divergent compared to Malawian VS5700009 (≈35% identity) and Tunisian TN25 (≈48%) CyCV species (Table 2). In contrast, the swine-derived CyCV strains from Cameroon exhibited a less conserved genome (86% identity to CyCV-VN)7. Although the genome organization was similar (Figure 1), the Rep gene was shorter (earlier STOP codon, at position 1055-57), leading to a Rep protein of 277 residues instead of 2927. As a consequence, Rep and Cap genes did not overlap like in the Madagascan and CyCV-VN strains7. In spite of this, the Rep protein sequence of Cameroonian swine CyCV-VN-like strains was highly conserved (≈94% identity compared to Madagascan and Vietnamese strains). Since CyCVs in the same species are defined as having >85% identity in the Rep protein sequence, these three Cameroonian strains clearly belong to the CyCV-VN group/species (Figure 2). Their Rep sequence identity to Malawian VS5700009 and Tunisian TN25 CyCVs was ≈77% and ≈74%, respectively (Table 1). The Cap gene was 3 nucleotides shorter (deletion of 3 nt after position 1634) and consequently the Cap protein was only 221 residue long. Cap protein was less conserved compared to that of Madagascan and Vietnamese CyCV strains (≈84–85% identity), but was much closer to CyCV-VN Cap proteins than to that of Malawian VS5700009 and Tunisian TN25 CyCV species (≈34% and ≈48%, respectively) (Table 2). Overall, Madagascan human-derived CyCv strains appeared as typical CyCV-VN strains, highly conserved, while Cameroonian swine strains belonged to the same species but were less conserved, especially in the Cap protein sequence.
Figure 1. Genomic organization (a.) and the stem-loop structures (b.) of the cycloviruses discovered in human and animal stool samples from Cameroon and Madagascar.
The locations of two major ORFs, encoding the putative Rep and Cap protein are indicated by arrows. The nonamer sequences (marked in red) within stem-loop structures are also shown.
Table 1. Sequence identity matrix for the replication-associated protein sequence (%). Cameroonian swine (KM392284–KM392286) and Madagascan human cycloviruses (KM392287–KM392289) described in this study are bolded. Vietnamese VN strains are colored in grey, Malawian VS5700009 in light yellow and Tunisian TN25 and TN18 appear in orange1,7,8.
| KM392285 | KM392284 | KM392286 | KM392289 | KM392288 | KM392287 | |
|---|---|---|---|---|---|---|
| KM392285 | 100 | 100 | 93,5 | 93,5 | 93,5 | |
| KM392284 | 100 | 100 | 93,5 | 93,5 | 93,5 | |
| KM392286 | 100 | 100 | 93,5 | 93,5 | 93,5 | |
| KM392289 | 93,5 | 93,5 | 93,5 | 100 | 99,3 | |
| KM392288 | 93,5 | 93,5 | 93,5 | 100 | 99,3 | |
| KM392287 | 93,5 | 93,5 | 93,5 | 99,3 | 99,3 | |
| KF031465 | 93,5 | 93,5 | 93,5 | 97,6 | 97,6 | 97,6 |
| KF031467 | 93,5 | 93,5 | 93,5 | 97,6 | 97,6 | 97,6 |
| KF031468 | 93,5 | 93,5 | 93,5 | 97,6 | 97,6 | 97,6 |
| KF031469 | 93,5 | 93,5 | 93,5 | 97,6 | 97,6 | 97,6 |
| NC_021707 | 93,5 | 93,5 | 93,5 | 97,6 | 97,6 | 97,6 |
| KF031470 | 93,5 | 93,5 | 93,5 | 98,3 | 98,3 | 98,3 |
| KF031471 | 93,5 | 93,5 | 93,5 | 98,6 | 98,6 | 98,6 |
| KF031466 | 93,1 | 93,1 | 93,1 | 97,9 | 97,9 | 97,9 |
| KC771281 | 77,4 | 77,4 | 77,4 | 77,1 | 77,1 | 76,3 |
| NC_021568 | 77,4 | 77,4 | 77,4 | 77,1 | 77,1 | 76,3 |
| GQ404857 | 73,6 | 73,6 | 73,6 | 72,7 | 72,7 | 72,7 |
| GQ404858 | 74,3 | 74,3 | 74,3 | 73,4 | 73,4 | 73,1 |
| HQ738635 | 71,7 | 71,7 | 71,7 | 70,5 | 70,5 | 70,5 |
| NC_014927 | 71,7 | 71,7 | 71,7 | 70,5 | 70,5 | 70,5 |
| HQ738634 | 71,3 | 71,3 | 71,3 | 70,1 | 70,1 | 70,1 |
| HQ738636 | 50,5 | 50,5 | 50,5 | 48,8 | 48,8 | 49,1 |
| NC_014928 | 50,5 | 50,5 | 50,5 | 48,8 | 48,8 | 49,1 |
| GQ404844 | 49,5 | 49,5 | 49,5 | 47,3 | 47,3 | 47,7 |
| GQ404846 | 50,7 | 50,7 | 50,7 | 48,6 | 48,6 | 48,9 |
| GQ404848 | 50,7 | 50,7 | 50,7 | 48,6 | 48,6 | 48,9 |
| GQ404849 | 48,2 | 48,2 | 48,2 | 45,4 | 45,4 | 45,8 |
| GQ404850 | 47,9 | 47,9 | 47,9 | 45,1 | 45,1 | 45,4 |
| HQ738644 | 49,3 | 49,3 | 49,3 | 48,6 | 48,6 | 49,3 |
| NC_014930 | 49,3 | 49,3 | 49,3 | 48,6 | 48,6 | 49,3 |
| HQ738643 | 49,6 | 49,6 | 49,6 | 48,9 | 48,9 | 49,6 |
| KC512919 | 50,7 | 50,7 | 50,7 | 48,8 | 48,8 | 48,8 |
| GQ404847 | 52,7 | 52,7 | 52,7 | 50,2 | 50,2 | 50,5 |
| KC512916 | 50,4 | 50,4 | 50,4 | 48,6 | 48,6 | 48,9 |
| HQ638063 | 50,7 | 50,7 | 50,7 | 50,2 | 50,2 | 49,8 |
| HQ638066 | 50,7 | 50,7 | 50,7 | 49,8 | 49,8 | 49,5 |
| HQ638060 | 50,7 | 50,7 | 50,7 | 50,5 | 50,5 | 50,2 |
| GQ404845 | 52 | 52 | 52 | 49,8 | 49,8 | 50,2 |
| HQ738637 | 51,8 | 51,8 | 51,8 | 51,8 | 51,8 | 51,4 |
| NC_014929 | 51,8 | 51,8 | 51,8 | 51,8 | 51,8 | 51,4 |
| GQ404854 | 51,8 | 51,8 | 51,8 | 50,2 | 50,2 | 50,9 |
| JF938081 | 49,8 | 49,8 | 49,8 | 48,8 | 48,8 | 49,1 |
| HM228874 | 50,7 | 50,7 | 50,7 | 49,8 | 49,8 | 49,5 |
| JF938079 | 50 | 50 | 50 | 48,6 | 48,6 | 48,9 |
| JX185426 | 49,8 | 49,8 | 49,8 | 49,1 | 49,1 | 49,5 |
| NC_023866 | 49,8 | 49,8 | 49,8 | 49,1 | 49,1 | 49,5 |
| JX569794 | 44,8 | 44,8 | 44,8 | 43,7 | 43,7 | 44 |
| NC_020206 | 44,8 | 44,8 | 44,8 | 43,7 | 43,7 | 44 |
| GQ404855 | 52,5 | 52,5 | 52,5 | 50,4 | 50,4 | 50,7 |
| KF726986 | 50 | 50 | 50 | 49,6 | 49,6 | 50 |
| KF726984 | 50 | 50 | 50 | 49,6 | 49,6 | 50 |
| NC_023874 | 50 | 50 | 50 | 49,6 | 49,6 | 50 |
| KF726987 | 50 | 50 | 50 | 49,6 | 49,6 | 50 |
| KF726985 | 50 | 50 | 50 | 49,6 | 49,6 | 50 |
| NC_023869 | 50,7 | 50,7 | 50,7 | 50 | 50 | 50 |
| KM017740 | 40,1 | 40,1 | 40,1 | 39,4 | 39,4 | 39,4 |
| JX260426 | 11 | 11 | 11 | 11 | 11 | 10,7 |
Table 2. Sequence identity matrix for the Capsid protein sequence (%). Cameroonian swine (KM392284–KM392286) and Madagascan human cycloviruses (KM392287–KM392289) described in this study are bolded. Vietnamese VN strains are colored in grey, Malawian VS5700009 in light yellow and Tunisian TN25 and TN18 appear in orange1,7,8.
| KM392288 | KM392289 | KM392287 | KM392285 | KM392286 | KM392284 | |
|---|---|---|---|---|---|---|
| NC_021568 | 35,3 | 35,3 | 35,3 | 34,1 | 34,1 | 34,1 |
| KC771281 | 35,3 | 35,3 | 35,3 | 34,1 | 34,1 | 34,1 |
| KF031470 | 97,7 | 97,7 | 97,3 | 83,8 | 83,8 | 83,8 |
| KF031471 | 97,7 | 97,7 | 97,3 | 83,8 | 83,8 | 83,8 |
| KF031469 | 98,6 | 98,6 | 98,2 | 84,2 | 84,2 | 84,2 |
| KF031468 | 98,6 | 98,6 | 98,2 | 84,2 | 84,2 | 84,2 |
| KF031467 | 98,6 | 98,6 | 98,2 | 84,2 | 84,2 | 84,2 |
| NC_021707 | 98,6 | 98,6 | 98,2 | 84,2 | 84,2 | 84,2 |
| KF031465 | 98,6 | 98,6 | 98,2 | 84,2 | 84,2 | 84,2 |
| KF031466 | 98,2 | 98,2 | 98,6 | 84,2 | 84,2 | 84,2 |
| KM392288 | 100 | 98,6 | 84,7 | 84,7 | 84,7 | |
| KM392289 | 100 | 98,6 | 84,7 | 84,7 | 84,7 | |
| KM392287 | 98,6 | 98,6 | 84,2 | 84,2 | 84,2 | |
| KM392285 | 84,7 | 84,7 | 84,2 | 100 | 100 | |
| KM392286 | 84,7 | 84,7 | 84,2 | 100 | 100 | |
| KM392284 | 84,7 | 84,7 | 84,2 | 100 | 100 | |
| GQ404857 | 48 | 48 | 47,5 | 48 | 48 | 48 |
| GQ404858 | 47,5 | 47,5 | 47,1 | 47,5 | 47,5 | 47,5 |
| HQ738635 | 49,3 | 49,3 | 49,8 | 47,9 | 47,9 | 47,9 |
| NC_014927 | 49,3 | 49,3 | 49,8 | 47,9 | 47,9 | 47,9 |
| HQ738634 | 49,3 | 49,3 | 49,8 | 47,9 | 47,9 | 47,9 |
| KF726987 | 22,8 | 22,8 | 22,4 | 23,8 | 23,8 | 23,8 |
| KF726986 | 22,8 | 22,8 | 22,4 | 23,8 | 23,8 | 23,8 |
| KF726984 | 22,8 | 22,8 | 22,4 | 23,8 | 23,8 | 23,8 |
| NC_023874 | 22,8 | 22,8 | 22,4 | 23,8 | 23,8 | 23,8 |
| KF726985 | 22,8 | 22,8 | 22,4 | 23,8 | 23,8 | 23,8 |
| HQ638060 | 24,9 | 24,9 | 24,9 | 24,5 | 24,5 | 24,5 |
| HQ638066 | 24,5 | 24,5 | 24,5 | 24 | 24 | 24 |
| HQ638063 | 25,3 | 25,3 | 25,3 | 25,8 | 25,8 | 25,8 |
| KC512916 | 24,1 | 24,1 | 24,6 | 24,6 | 24,6 | 24,6 |
| GQ404847 | 25,4 | 25,4 | 25,4 | 24,1 | 24,1 | 24,1 |
| NC_020206 | 19,7 | 19,7 | 19,2 | 19,7 | 19,7 | 19,7 |
| JX569794 | 19,7 | 19,7 | 19,2 | 19,7 | 19,7 | 19,7 |
| GQ404844 | 19 | 19 | 19 | 19,5 | 19,5 | 19,5 |
| GQ404854 | 22,2 | 22,2 | 22,2 | 22,2 | 22,2 | 22,2 |
| HM228874 | 17,3 | 17,3 | 17,3 | 16,5 | 16,5 | 16,5 |
| GQ404845 | 18,7 | 18,7 | 18,7 | 19,2 | 19,2 | 19,2 |
| GQ404855 | 17,2 | 17,2 | 17,2 | 16,8 | 16,8 | 16,8 |
| HQ738636 | 22,6 | 22,6 | 22,2 | 23,6 | 23,6 | 23,6 |
| NC_014928 | 22,6 | 22,6 | 22,2 | 23,6 | 23,6 | 23,6 |
| KC512919 | 23,8 | 23,8 | 23,4 | 22,6 | 22,6 | 22,6 |
| JF938081 | 23,2 | 23,2 | 23,2 | 23,3 | 23,3 | 23,3 |
| HQ738644 | 20,4 | 20,4 | 20,4 | 22,6 | 22,6 | 22,6 |
| NC_014930 | 20,4 | 20,4 | 20,4 | 22,6 | 22,6 | 22,6 |
| HQ738643 | 20,4 | 20,4 | 20,4 | 22,6 | 22,6 | 22,6 |
| GQ404850 | 21,3 | 21,3 | 21,3 | 21,3 | 21,3 | 21,3 |
| GQ404849 | 21,3 | 21,3 | 21,3 | 21,3 | 21,3 | 21,3 |
| JF938079 | 23,5 | 23,5 | 23 | 22,6 | 22,6 | 22,6 |
| NC_023866 | 21,7 | 21,7 | 20,9 | 22,1 | 22,1 | 22,1 |
| JX185426 | 21,7 | 21,7 | 20,9 | 22,1 | 22,1 | 22,1 |
| HQ738637 | 20,9 | 20,9 | 20,9 | 20,9 | 20,9 | 20,9 |
| NC_014929 | 20,9 | 20,9 | 20,9 | 20,9 | 20,9 | 20,9 |
| GQ404848 | 23,8 | 23,8 | 23,4 | 22,1 | 22,1 | 22,1 |
| GQ404846 | 23,4 | 23,4 | 22,9 | 21,6 | 21,6 | 21,6 |
| NC_023869 | 24,2 | 24,2 | 23,7 | 24,7 | 24,7 | 24,7 |
| KM017740 | 18,7 | 18,7 | 18,7 | 17,4 | 17,4 | 17,4 |
| JX260426 | 6,6 | 6,6 | 6,6 | 7,5 | 7,5 | 7,5 |
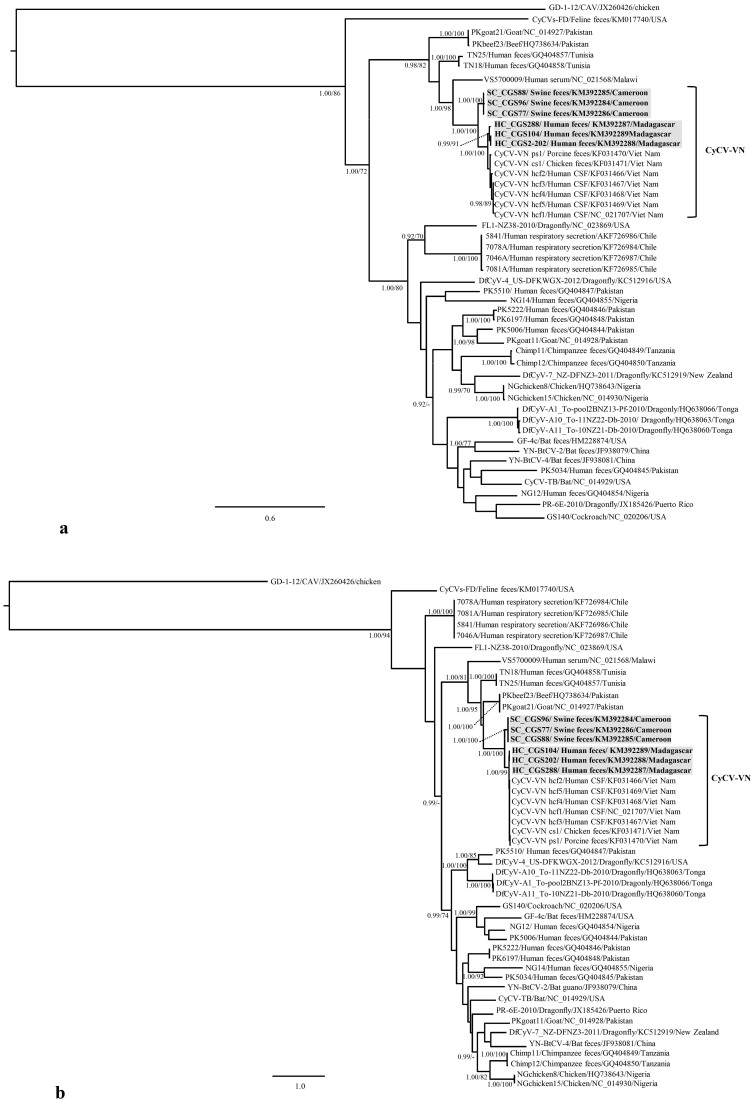
Figure 2. Bayesian phylogenetic trees based on complete amino acid sequences of the Rep (a.) and Cap (b.) protein of the cycloviruses.
Statistical support of grouping from Bayesian posterior probabilities (clade credibilities ≥90%) and 1,000 parallel Maximum Likelihood bootstrap replicates (≥70%) is indicated at the nodes. A Gyrovirus (Chicken anemia virus, GenBank accession number JX260426) was chosen as an outgroup. Taxon information includes the name, source of isolation, GenBank accession number, and country of origin. The cyclovirus strains generated during this study are bolded. The scale bar represents amino acid substitutions per site.
The typical nonamer sequence of the stem loop structure (TAATACTAT) was conserved in Madagascan human CyCV strain genomes and with one nucleotide substitution (TAATACTAG) in Cameroonian swine-derived CyCVs compared to CyCV-VN strains (Figure 1). The sequence of the loop itself was 11 nt long and identical in all these strains (Figure 1). Highly conserved circovirus/cyclovirus Rep motifs WWDGY, DDFYGW and DRYP were present in both Madagascan human and Cameroonian swine strains (1). The dNTP binding site (G-GSK) was intact and the motifs associated with rolling-circle amplification (FTLNN, TPHLQG, CSK) were also identified, with the same alterations (WTLNN and TKHLQG) as in CyCV-VN strains1.
Phylogenetic analysis based on the complete Rep and Cap protein sequences showed that the CyCVs from Cameroon, Madagascar and Vietnam clustered together forming a distinct and highly supported clade/group (CyCV-VN) within the cyclovirus phylogeny (Figure 2). Although, the Cameroonian swine-derived CyCV strains shared a common ancestor with the human-derived CyCVs, they form a distinct lineage within the CyCV-VN clade, suggesting separate evolutionary events.
Discussion
Cycloviruses are recently described members of the Circoviridae family1. CyCVs species have been described in mammals, including humans, birds and insects1,2,3,4,5,6. Recently, two studies reported the detection of CyCVs in the cerebrospinal fluid of symptomatic human patients in Vietnam and Malawi7,8. An extensive specific screening failed to identify the Vietnam CyCV-VN strain in neighboring Asian countries (Cambodia, Nepal) and in the Netherlands, suggesting a very limited distribution to central and southern Vietnam, where it had been detected in human, pig and chicken10.
In order to assess the presence of CyCV-VN on the African continent, we screened a collection of human stool samples from Madagascar and Ghana and of pig stool samples from Cameroon. A total of three human samples and ten swine samples were PCR-positive and confirmed by sequencing. The full genome of the three human CyCV-VN and of three of the swine strains were obtained by primer walking methods.
Phylogenetic analysis showed that both Madagascan and Cameroonian CyCV strains clustered with Vietnam CyCVs forming two distinct sister lineages within CyCV-VN clade and showed distant relationship to previous African cyclovirus strains from Malawi, Tunisia or Nigeria1,3,7,8. It is likely that CyCV-VN can be found in other parts of Africa and Asia and possibly in other continents. Our data in pigs suggest a heterogeneous distribution in Cameroon since most positive cases were concentrated in the Douala region. Further, the 1337 human samples from Ghana were tested negative, suggesting an absence of this viral strain from this area. This apparent unequal circulation might be an explanation for the lack of CyCV-VN detection outside Vietnam in previous studies10.
Our findings further confirm that pigs might play a central role in the epidemiology of cycloviruses in general and for CyCV-VN species in particular7. Besides, the exact pathological significance of CyCV-VN infection in humans is still unknown and further studies are clearly needed to enlighten the transmission route(s), the host tropism and the pathological consequences of these CyCVs.
Overall, the results presented in this study indicate that CyCV-VN and CyCV-VN-like strains circulate in humans and pigs in Africa, far beyond their supposed limited geographic distribution to Vietnam.
Methods
Samples collection
A total of 410 stool samples from healthy children were obtained from Ampasina village, belonging to the township of Andina in the Ambositra region, Madagascar, in 201211. All details including sample preparation are described in ref 11. A total of 1373 human stool samples were retrieved from children up to 15 years of age with diarrhea or vomiting (n = 673) and pediatric controls (n = 664) from a rural area in the Ashanti Region, Ghana. Besides, feces from 520 healthy pigs were sampled in 2012 in the cities of Bamenda (n = 75), Ngaoundere (n = 86), Yaoundé (n = 199) and Douala (n = 160) in Cameroon12.
Ethics Statement
Experiments were conducted in accordance with the ethical principles set out in the Declaration of Helsinki and informed consent was obtained from all subjects. All experimental protocols were approved by the Ministry of Health of the Republic of Madagascar (no. 015 – MSANP/CE, 22 February 2012) and from the Committee on Human Research, Publications and Ethics, School of Medical Science, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana.
DNA extraction, CyCV-VN specific PCR and sequencing
Samples were extracted using Invitek PSP Spin Stool DNA Kit (Stratec Biomedical AG, Birkenfeld, Germany) or QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany), according to manufacturer's instructions. PCR screening of CyCV-VN was performed on extracted nucleic acid using the CyCV-VN-specific diagnostic primers designed based on available CyCV-VN sequences in databases (see Supplementary Table S1 online). In order to further characterize the virus strains and to compare them with previously published data, the full-length genome was successfully amplified by primer walking using specific primers (Table S1 online). PCR were performed using HotStarTaq® Plus Master Mix Kit (Qiagen, Hilden, Germany). The PCR was performed in a 25 μl volume containing 3 μl of sample DNA, 10 μl of HotStarTaq Master Mix, 2 μ ml of each primer (20 pmol), and ddH2O up to 25 μl. The positive and negative controls (extraction and water controls) were included in every PCR run. Amplification conditions were as follows: denaturation at 95°C for 2 min; 45 amplification cycles were performed at 94°C for 20 s, 55°C for 45 s, 72°C for 1 min. Final extension was at 72°C for 7 min. The fragments were sequenced at least twice in each direction by conventional Sangers technology (LGC, Berlin, Germany).
Sequence analysis
Sequence analysis, genomic organization and multiple alignments were performed using Geneious v7.1.7 (Biomatters, Auckland, New Zealand). Secondary DNA structure predictions were performed with the web-based version of mfold (http://mfold.bioinfo.rpi.edu). The trees were constructed by using the Bayesian Markov Chain Monte Carlo sampling method in MrBayes v3.2.213 and maximum likelihood in PhyML v3.114 and included representatives of all cycloviruses species and those six novel human- and swine-derived cycloviruses whose full genome could be determined. The Whelan and Goldman (WAG) amino acid substitution model with gamma-distributed rate variation among sites and a proportion of invariable sites were used.
Supplementary Material
Dataset 1
Acknowledgments
We thank Mathis Petersen, Claudia Poggensee, and Alexandra Bialonski for technical assistance. This article is dedicated to the late Ursula Herrmann (1927–2014) for making this study possible.
Nucleotide sequence accession number The Genbank accession numbers of the cycloviruses described in the study are KM392284-KM392289.
Footnotes
The authors declare no competing financial interests.
Author Contributions R.M.H., H.F., J.M., A.P., H.J., J.B., N.S., D.D., R.M.C., V.S.P., N.G.S., S.P, H.A.M., A.M.D., M.T.K., N.R., E.T. and R.R. collected the samples. M.G., A.P. and D.C. performed laboratory experiments and analyzed the sequences. J.S.C., M.G. and D.C. drafted the manuscript. All authors were involved in reviewing and editing of the manuscript.
References
- Li L. et al. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J. Virol. 84, 1674–1682, 10.1128/JVI.02109-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X. et al. Genetic diversity of novel circular ssDNA viruses in bats in China. J. Gen. Virol. 92, 2646–2653, 10.1099/vir.0.034108-0 (2011). [DOI] [PubMed] [Google Scholar]
- Li L. et al. Possible cross-species transmission of circoviruses and cycloviruses among farm animals. J. Gen. Virol. 92, 768–772, 10.1099/vir.0.028704-0 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K. et al. Dragonfly cyclovirus, a novel single-stranded DNA virus discovered in dragonflies (Odonata: Anisoptera). J. Gen. Virol. 92, 1302–1308, 10.1099/vir.0.030338-0 (2011). [DOI] [PubMed] [Google Scholar]
- Dayaram A. et al. High global diversity of cycloviruses amongst dragonflies. J. Gen. Virol. 94, 1827–1840, 10.1099/vir.0.052654-0 (2013). [DOI] [PubMed] [Google Scholar]
- Padilla-Rodriguez M., Rosario K. & Breitbart M. Novel cyclovirus discovered in the Florida woods cockroach Eurycotis floridana (Walker). Arch. Virol. 158, 1389–1392, 10.1007/s00705-013-1606-x (2013). [DOI] [PubMed] [Google Scholar]
- Tan le V. et al. Identification of a new cyclovirus in cerebrospinal fluid of patients with acute central nervous system infections. MBio 4, e00231–00213, 10.1128/mBio.00231-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S. L. et al. Novel cyclovirus in human cerebrospinal fluid, Malawi, 2010–2011. Emerg. Infect. Dis. 19, 1511–1513, 10.3201/eid1909.130404 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. G., Luchsinger V., Avendaño L. F., Deng X. & Delwart E. Cyclovirus in nasopharyngeal aspirates of Chilean children with respiratory infections. J. Gen. Virol. 95, 922–927, 10.1099/vir.0.061143-0 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan le V. et al. Limited geographic distribution of the novel cyclovirus CyCV-VN. Sci. Rep. 4, 3967, 10.1038/srep03967 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz N. G. et al. Schistosoma mansoni in schoolchildren in a Madagascan highland school assessed by PCR and sedimentation microscopy and Bayesian estimation of sensitivities and specificities. Acta Trop. 134, 89–94, 10.1016/j.actatropica.2014.03.003 (2014). [DOI] [PubMed] [Google Scholar]
- de Paula V. S. et al. Hepatitis E virus genotype 3 strains in domestic pigs, Cameroon. Emerg. Infect. Dis. 19, 666–668, 10.3201/eid1904.121634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F. & Huelsenbeck J. P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- Guindon S. et al. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset 1