Abstract
Following the first human infection with the influenza A (H10N8) virus in Nanchang, China in December 2013, we identified two additional patients on January 19 and February 9, 2014. The epidemiologic, clinical, and virological data from the patients and the environmental specimen collected from 23 local live poultry markets (LPMs) were analyzed. The three H10N8 cases had a history of poultry exposure and presented with high fever (>38°C), rapidly progressive pneumonia and lymphopenia. Substantial high levels of cytokines and chemokines were observed. The sequences from an isolate (A/Environment/Jiangxi/03489/2013 [H10N8]) in an epidemiologically linked LPM showed highly identity with human H10N8 virus, evidencing LPM as the source of human infection. The HA and NA of human and environmental H10N8 isolates showed high identity (99.1–99.9%) while six genotypes with internal genes derived from H9N2, H7N3 and H7N9 subtype viruses were detected in environmental H10N8 isolates. The genotype of the virus causing human infection, Jiangxi/346, possessed a whole internal gene set of the A/Environment/Jiangxi/10618/2014(H9N2)-like virus. Thus, our findings support the notion that LPMs can act as both a gene pool for the generation of novel reassortants and a source for human infection, and intensive surveillance and management should therefore be conducted.
Influenza A viruses are classified based on their surface proteins, hemagglutinin (HA) and neuraminidase (NA), into sixteen HA subtypes (H1 to H16) and nine NA subtypes (N1 to N9). Most of these subtypes display low pathogenicity in avian species, except for the H5 and H7 viruses, which contain the polybasic amino acid motif at the HA cleavage site and cause severe disease in poultry. Occasionally, some subtypes of avian influenza A virus can jump into humans and cause diseases with a range of clinical symptoms and outcomes, such as conjunctivitis, mild upper respiratory tract disease, severe pneumonia and death1,2,3,4,5,6. Influenza A (H10N8) viruses have circulated mainly in wild birds since their first detection in quails in Italy in 19657. Only two H10N8 isolates were reported in China before the first human H10N8 infection was detected in December 2013; one was identified in an environmental sample obtained from the Dongting Lake wetland in Hunan province in 2007, and the other was isolated from a duck in a live poultry market (LPM) in Guangdong province in 20128,9. The virus isolated from the human case was genetically distinguishable from previous H10N8 viruses, with long genetic distances between their HA and NA genes, and contained the H9N2-derived internal genes10. Notably, as a currently circulating H7N9 virus with the phenotype of low pathogenicity in poultry, the H10N8 virus can cause severe infections in humans, leading to death in 2 out of 3 cases. Here, we compare the host innate response, the progression and features of clinical disease to understand severity-related mechanisms, and we also identify sources of human infections through field investigations of LPMs, including those visited by the patients.
Methods
Patients and diagnostic procedures
After the first human infection with the H10N8 virus was reported, reinforced surveillance of hospitalized patients with severe pneumonia of unexplained origin was conducted from December 1, 2013 to February 28, 2014 in Nanchang City. As a result, two additional human cases were confirmed, according to previously described procedures10.
Clinical and epidemiological data collection
Experiments involving human subjects were performed in accordance with the relevant guidelines and regulations of P. R China. A standardized surveillance and reporting form was used to collect epidemiologic and clinical data, including demographic characteristics, as previously reported10, underlying medical conditions, history of seasonal influenza vaccination, recent exposure to poultry, swine, or other animals, recent visits to a live animal market, clinical signs and symptoms, antiviral treatment, clinical complications, and outcomes were also collected. The institutional review board of the Nanchang Center for Disease Control and Prevention (CDC) approved the experiments, and written informed consent was obtained from the patients and close contacts.
Cytokine and chemokine measurement
Sera from the cases and 13 close contacts were collected and stored at −80°C. We used the cytometric bead array assay (BD Biosciences, San Diego, CA) to measure seven different serum cytokines or chemokines: IFN-γ; IL-6, 8 and 10; IP-10, MCP-1 and MIG.
Field investigations in poultry markets
Samples including fresh poultry feces, sewage, and swabs from culling benches and poultry cages were collected from 23 LPMs located in 5 districts and 4 counties in Nanchang City from December 2013 to February 2014 (Figure 1). The specimens were tested for influenza A virus by targeting the Matrix gene, and viral culture was established via inoculation of 9-day-old specific pathogen-free embryonated chicken eggs (if the test for influenza A was positive).
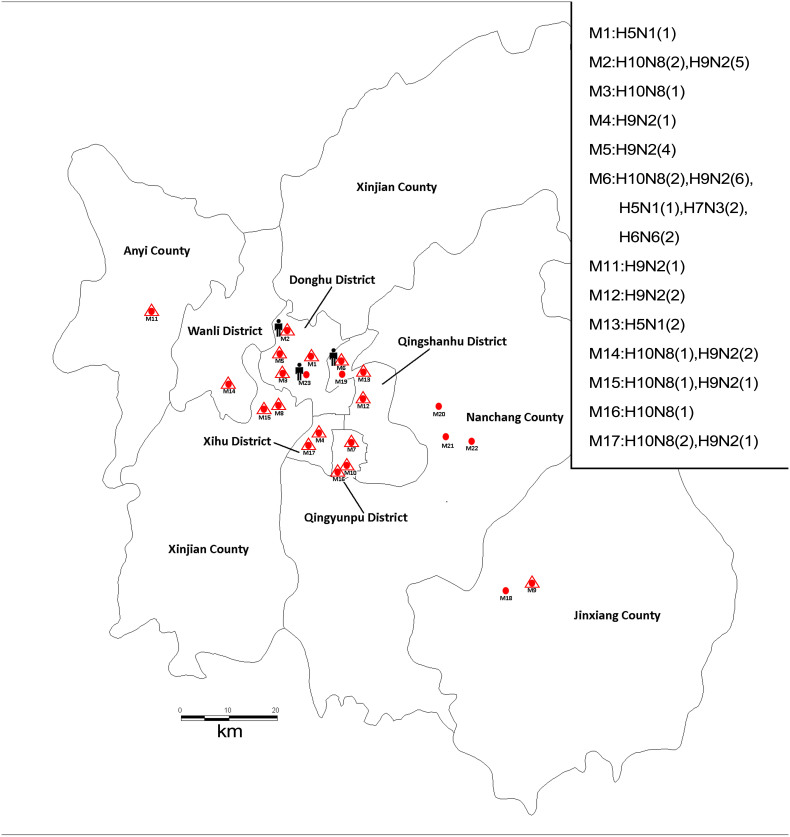
Figure 1. Geographic distribution of the samples from 23 poultry markets in Nanchang, China.
The geographic distribution of the samples was generated using Epi Info 3.5.1 software and modified by ML and YL using Adobe Illustrator CS6 software. Samples including fresh poultry feces, sewage, and swabs from culling benches and poultry cages were collected from 23 LPMs (red dot) located in 5 districts and 4 counties in Nanchang City from December 8, 2013 to February 9, 2014. Fifty-three isolates from 17 markets (red dots surrounded by a triangle) were obtained. The markets where the H10N8, H9N2, H5N1, H7N3 and H6N6 viruses were detected are listed. The markets visited by Cases 1, 2, and 3 were M2, M23 and M6, respectively.
Sequencing and phylogenetic analysis
As previously described10, the full genome of the virus was amplified and sequenced using the automatic Applied Biosystems 3730XL DNA Analyzer (Life Technologies, USA) and deep sequencing. The full genome sequences of the viruses were deposited in the Global Initiative on Sharing All Influenza Data database (GISAID), and the accession numbers are listed in Supplementary Table S1. A maximum likelihood phylogenetic tree for the nucleotide sequences of each gene of selected influenza viruses was constructed using the MEGA5.1 program. The tree topology was evaluated by performing 1,000 bootstrap analyses. The bootstrap values ≥ 0.6 are shown at the major nodes of the phylogenetic trees. The Bioedit 7 program was used for the alignment and analysis of amino acid residues. The percent identity of each virus genes was calculated by using the MegAlign7.1.
Results
Epidemiological and clinical features of human H10N8 infection cases
After the first human infection with H10N8 virus was reported, reinforced surveillance of hospitalized patients with severe pneumonia of unexplained origin was conducted in Nanchang City. As a result, a total of 18 hospitalized patients were reported, and two H10N8 cases were identified; one of the infected patients died. As summarized in Supplementary Figure S1, all three H10N8 cases had visited LPMs 4 to 5 days prior to illness onset; Case 3 had also directly handled chickens and ducks. The interval from illness onset to hospitalization ranged from 2 to 7 days. The disease progressed rapidly, and respiratory failure developed within 2–7 days. Two of these patients died on days 6 and 9 after illness onset. The clinical features of the patients on admission are summarized in Table 1. Generally, the age of the patients ranged from 56 to 75 years, and all were retirees from two different districts of Nanchang City. Cases 1 and 3 suffered from chronic underlying conditions. Similar to Case 1, Cases 2 and 3 mainly presented with cough, sputum and fever at admission as well as a sore throat and poor appetite, respectively. None of the patients had conjunctivitis, rash, diarrhea, or a runny nose. Chest radiography in all cases showed multilobar patchy consolidation and diffuse alveolar opacities on admission (Supplementary Figure S2). Fine crackles in the lower lobe of the left lung were detected in Cases 1 and 2. In addition, pronounced lymphopenia was present in all three cases, with a low proportion of lymphocytes (7%, 8% and 10.8%, respectively; Table 2) and high proportion of neutrophils (76.4%, 87%, and 83.3%, respectively; Table 2). Although the biochemical indices related to the function of the liver, heart and kidney were generally normal, except for a high level of C-reactive protein at admission, the patients' conditions deteriorated rapidly after hospitalization, with the development of severe complications including acute respiratory distress syndrome, septic shock and multi-organ failure (Supplementary Table S2 for Case 2 and Table S3 for Case 3). Oseltamivir was administered at 5 to 7 days after the onset of symptoms, with treatment lasting for 3 days in Case 1, 16 days in Case 2, and one day in Case 3. All patients received glucocorticoid treatment and subsequently received mechanical ventilation. Cases 1 and 3 died at 3 and 2 days after intubation, respectively (Table 2).
Table 1. Clinical features of patients with avian influenza A (H10N8) virus infection on admission.
| Case 1 | Case 2 | Case 3 | |
|---|---|---|---|
| Age/Gender | 73/F | 56/F | 75/M |
| Occupation | retiree | retiree | retiree |
| Location (district) in Nanchang | Donghu | Donghu | Qingshanpu |
| Underlying medical condition | Hypertension, myasthenia gravis, coronary heart disease | No | hypertension |
| Initial symptoms | Cough, sputum, chest tightness | Cough, sputum, sore throat, chest stuffiness | Fever, poor appetite, cough |
| Time from illness onset to admission (days) | 3 | 7 | 2 |
| Clinical characteristics on admission | |||
| Cough | Yes | Yes | Yes |
| Dyspnea | Yes | Yes | No |
| Sputum | Yes | Yes | No |
| Diarrhea | No | No | No |
| Rash | No | No | No |
| Myalgia | Yes | No | No |
| Conjunctivitis | No | No | No |
| Blood pressure (mm Hg) | 150/80 | 139/74 | 166/100 |
| Respiratory rate(breaths per minute) | 21 | 21 | 22 |
| Pulse rate (beats per minute) | 94 | 127 | 100 |
| Crackles | Yes | Yes | No |
| Temperature (°C) | 38.6 | 39.1 | 38.7 |
| Time from illness onset to fever (days) | 2 | 7 | 0 |
| Chest radiograph findings | Bilateral GGO, extensive infiltrates | Bilateral GGO, extensive infiltrates | Bilateral GGO, extensive infiltrates |
GGO, ground-glass opacity.
Table 2. Laboratory values of H10N8 patients on hospitalization and clinical treatments and outcomes.
| Reference range | Case 1 | Case 2 | Case 3 | |
|---|---|---|---|---|
| Laboratory Values | ||||
| White cell count (×109/L). | 4.0–10.0 | 10.34 | 7.90 | 5.07 |
| The proportion of lymphocytes (%) | 20.0–40.0 | 7 | 8 | 10.8 |
| The proportion of neutrophils (%) | 52.0–72.0 | 76.4 | 87 | 83.3 |
| ALT level (U/liter) | 0–40 | 17 | 30 | 53 |
| AST level (U/liter) | 0–40 | 22 | 56 | 69 |
| CRP (mg/liter) | 0–5 | >200 | 115 | 70 |
| Serum glucose(mmol/liter) | 3.9–6.1 | 8.70 | 7.8 | 11.8 |
| Hemoglobulin (g/dl) | 11–16 | 11.7 | 12.3 | 13.5 |
| Platelets (×109/L). | 100–300 | 124 | 145 | 169 |
| Co-infection | NA | No | No | No |
| Complication | NA | |||
| Acute respiratory distress syndrome | Yes | Yes | Yes | |
| Septic shock | Yes | Yes | Yes | |
| Organ failure | Yes | Yes | Yes | |
| Time between illness onset and respiratory failure (days) | NA | 4 | 7 | 2 |
| Time between initiation of mechanical ventilation and death (days) | NA | 3 | NA | 2 |
| Days between onset of symptoms and initiation of oseltamivir(dosage) | NA | 6 (75 mg twice daily) | 7 (75 mg twice daily) | 5 (75 mg twice daily) |
| Duration of antiviral treatment(days) | NA | 3 | 16 | 1 |
| Glucocorticoid treatment (dosage × duration(days) | NA | hydrocortisone (25 mg, q8h × 1) dexamethasone (10 mg, q8h × 1) | methylprednisolone 40 mg × 3 | methylprednisolone 40 mg × 1 |
| Outcome | NA | Died (day 9) | Hospitalized | Died (day 6) |
NA: not applicable.
Cytokine and chemokine responses in H10N8 infection cases
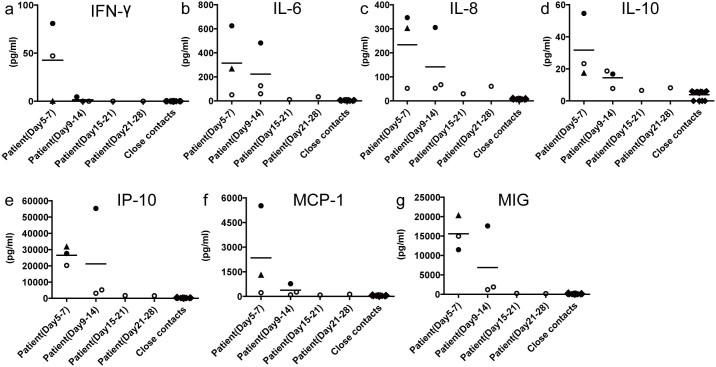
Cytokines and chemokines are important immune modulators released from immune cells in response to infection. Thus, the host cytokine/chemokine response during the acute and convalescent phase was measured. The levels of cytokines and chemokines in acute serum samples from the H10N8 patients were significantly enhanced compared with those in samples from the close contacts (Figure 2). Numerous immune molecules, including IFN-γ, IL-6, anti-inflammatory cytokines (IL-10), inflammatory CC chemokines (MCP-1) and interferon (IFN)-γ responsive chemokines (IP-10, IL-8 and MIG), were strongly induced, especially in the fatal cases (Cases 1 and 3).
Figure 2. Sera chemokine/cytokine levels of patients acutely infected with avian H10N8 virus.
The sera concentrations of the chemokines/cytokines IFN-γ (A), IL-6 (B), IL-8 (C), IL-10 (D), IP-10 (E), MCP-1 (F) and MIG (G) were measured using cytometric bead array assays, according to the manufacturers' instructions. Series of sera were collected on days 5 and 9 after disease onset for Case 1 (filled round), on days 7, 10, 14, 21 and 28 after disease onset for Case 2 (open round) and on day 5 after disease onset for Case 3 (filled triangle); sera were also collected from 13 close contacts. The data was analyzed by GraphPad Prism 5 software package (GraphPad Software) and the data graph was constructed by JZ using Adobe Illustrator CS6 software.
Origin of H10N8 viruses causing human infections
H10N8 RNA was detected in the throat aspiration samples from all patients, and viruses were isolated from those samples in Cases 1 and 3 (denoted as A/Jiangxi-Donghu/346/2013 and A/Jiangxi/09037/2014, respectively). No bacterial or fungal co-infections were detected by blood culture (Table 2).
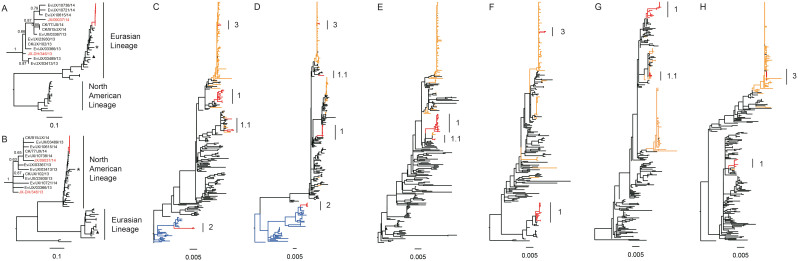
We collected 1,036 samples from 23 poultry markets in Nanchang City between December 8, 2013 and February 9, 2014. A total of 262 samples from FluA-positive specimens were inoculated individually into specific pathogen-free 9-day-old embryonated chicken eggs for virus isolation. Fifty-three isolates were obtained and subtyped by PCR and sequencing, including H10N8 (10), H9N2 (23), H5N1 (4), H7N3 (2), H6N6 (2), mixtures of H10N8 and H9N2 (2) and isolates of FluA-positive and non-H5, 6, 7, 9 and 10 viruses (10) (Figure 1 and Supplementary Table S4). Notably, almost all the observed subtypes were detected in M6, the only wholesale market in Nanchang City that provides poultry to retail markets. To trace the source of the human infections, the H10N8 viruses isolated from Cases 1 and 3 as well as 8 H10N8 and 12 H9N2 viruses isolated from local markets were used for genetic analysis (Supplementary Table S5). The HA and NA genes of the 10 H10N8 viruses shared 99.1–99.9% and 99.4–99.9% identity, respectively (Supplementary Table S6), and formed an independent branch in the corresponding phylogenetic tree that was distinctly different from the two previous H10N8 viruses found in China (A/environment/DongtingLake/Hunan/3–9/2007 and A/duck/Guangdong/E1/2012) (Figure 3). In addition, the eight gene segments of the A/Environment/Jiangxi/03489/2013 virus (Ev/JX/03489, H10N8) isolated from M6 showed the highest similarity with the human H10N8 viruses (99.2–99.9% identity with JX/346 and JX/09037). Cases 3 and 1 visited M6 and M2, respectively, where the poultry was obtained from M6, five days before illness onset. Thus, the epidemiological and virological data indicated that the novel H10N8 virus was most likely transmitted from the wholesale market to retail LPMs, and the human infections were associated with exposure to contaminated poultry markets.
Figure 3. Phylogenetic analyses for eight gene segments of H10N8 viruses.
The maximum likelihood tree for each segment, including HA (A), NA (B), PB2(C), PB1 (D), PA (E), NP (F), MP (G) and NS(H) of the novel H10N8 influenza viruses is shown. The sequences for novel H10N8 viruses and H9N2 viruses in the study, novel H7N9 viruses, other H9N2 viruses and wild birds gene pool are highlighted in red, orange, black and blue, respectively. The major group of H9N2 is labeled as Clade 1, and the minor group is denoted as 1.1 (labeled with a black line). The groups within the duck H7Nx clade are shown as a light blue line and labeled as Clade 2, and the groups within the major H7N9 clade are shown as a yellow line and labeled as Clade 3. Two previous H10N8 viruses, A/environment/DongtingLake/Hunan/3–9/2007 and A/duck/Guangdong/E1/2012, are denoted as a triangle and star, respectively. The phylogenic tree of HA and NA of H10N8 viruses in the study was zoomed out and the name of H10N8 viruses from human or non-human was denoted in red and black, respectively. The sequences of other viruses used for the analysis were downloaded from the Global Initiative on Sharing All Influenza Data (GISAID) database and are shown in black. The regions of the nucleotide sequences used for the phylogenetic analysis were as follows: H10, 1,686 bp; N8, 1,413 bp; PB2, 2,280 bp; PB1, 2,277 bp; PA, 2,151 bp; NP, 1,497 bp; MP, 982 bp and NS, 838 bp.
The internal genes PA, NP and MP shared similarities greater than 99.2%, higher genetic diversity in PB2, PB1 and NS was found in H10N8 viruses with identities of 93.5–99.8%, 89.2–100% and 95.9–100%, respectively (Supplementary Table S6). Based on the phylogenetic trees, the PB2 was grouped into 3 clades, with most falling within the H9N2 lineages circulating among the local LPMs (Clades 1) during the same monitoring period (Figure 3 and Supplementary Figure S3C), and the others were grouped together with the H7N3-like virus circulating in ducks (Clade 2) or the H7N9 virus (Clade 3). PB1 and NS were divided into two groups. Other three internal genes including PA, NP and M formed one group (Figure 3 and Supplementary Figure S3). Based on the different clades of internal genes, 6 genotypes were designated (Table 3). The two human H10N8 isolates belong to genotype JX/346. This genotype contained the whole set of H9N2-derived internal gene segments and was also detected in an epidemiologically-linked wholesale market, M6. Genotypes Ev/JX/3413 and Ev/JX/23930 were observed in the retail LPM (M2) visited by Case 1. Three alternative genotypes, Ev/JX/3366, Ev/JX/3367, and Ev/JX/10615, were also detected in another four retail LPMs. Then, we used this information to speculate on the generation of the diversified genotypes (Figure 4).
Table 3. The genotype of H10N8 and H9N2 viruses isolated from local markets in Nanchang city, China.
| Virus isolate | subtype | sampling site | PB2 | PB1 | PA | NP | MP | NS | Genotype |
|---|---|---|---|---|---|---|---|---|---|
| A/Jiangxi-Donghu/346/2013 | H10N8 | Case 1 | 1 | 1 | 1 | 1 | 1 | 1 | JX/346 |
| A/Jiangxi/09037/2014 | H10N8 | Case 3 | 1 | 1 | 1 | 1 | 1 | 1 | JX/346 |
| A/Environment/Jiangxi/03489/2013 | H10N8 | M6 | 1 | 1 | 1 | 1 | 1 | 1 | JX/346 |
| A/Environment/Jiangxi/03413/2013 | H10N8 | M2 | 3 | 1 | 1 | 1 | 1 | 1 | En/JX/3413 |
| A/Environment/Jiangxi/03366/2013 | H10N8 | M17 | 2 | 2 | 1 | 1 | 1 | 3 | Ev/JX/3366 |
| A/Environment/Jiangxi/03367/2013 | H10N8 | M17 | 1 | 2 | 1 | 1 | 1 | 3 | Ev/JX/3367 |
| A/Environment/Jiangxi/23930/2013 | H10N8 | M2 | 2 | 2 | 1 | 1 | 1 | 1 | Ev/JX/23930 |
| A/Environment/Jiangxi/10615/2014 | H10N8 | M14 | 1 | 2 | 1 | 1 | 1 | 1 | Ev/JX/10615 |
| A/Environment/Jiangxi/10721/2014 | H10N8 | M3 | 1 | 1 | 1 | 1 | 1 | 1 | JX/346 |
| A/Environment/Jiangxi/10738/2014 | H10N8 | M15 | 1 | 2 | 1 | 1 | 1 | 1 | Ev/JX/10615 |
| A/chicken/Jiangxi/102/2013* | H10N8 | Unknown | 2 | 2 | 1 | 1 | 1 | 1 | Ev/JX/23930 |
| A/chicken/Jiangxi/77/2014* | H10N8 | Unknown | 1 | 1 | 1 | 1 | 1 | 1 | JX/346 |
| A/chicken/Jiangxi/B15/2014* | H10N8 | Unknown | 1 | 1 | 1 | 1 | 1 | 1 | JX/346 |
| A/Environment/Jiangxi/03470/2013 | H9N2 | M15 | 1 | 1 | 1 | 1 | 1 | 3 | − |
| A/Environment/Jiangxi/03476/2013 | H9N2 | M5 | 1 | 1 | 1 | 1 | 1 | 3 | − |
| A/Environment/Jiangxi/03481/2013 | H9N2 | M6 | 1 | 1 | 1 | 1 | 1 | 3 | − |
| A/Environment/Jiangxi/03501/2013 | H9N2 | M6 | 1 | 1 | 1 | 1 | 1 | 3 | |
| A/Environment/Jiangxi/20934/2013 | H9N2 | M2 | 1.1 | 1 | 1 | 1 | 1 | 3 | − |
| A/Environment/Jiangxi/03355/2013 | H9N2 | M2 | 1 | 1.1 | 1 | 1 | 1 | 3 | − |
| A/Environment/Jiangxi/03365/2013 | H9N2 | M17 | 1 | 1 | 1 | 1 | 1 | 3 | − |
| A/Environment/Jiangxi/10618/2014 | H9N2 | M14 | 1 | 1 | 1 | 1 | 1 | 1 | − |
| A/Environment/Jiangxi/10623/2014 | H9N2 | M14 | 1 | 1 | 1 | 1 | 1 | 1 | − |
| A/Environment/Jiangxi/10663/2014 | H9N2 | M12 | 1.1 | 3 | 1.1 | 3 | 1.1 | 3 | − |
| A/Environment/Jiangxi/10664/2014 | H9N2 | M12 | 1.1 | 1 | 1.1 | 3 | 1.1 | 3 | − |
| A/Environment/Jiangxi/10726/2014 | H9N2 | M5 | 1.1 | 1 | 1 | 1 | 1 | 3 | − |
Genes in different clades were labeled with different colors and values, 1 represents the clade of A/Environment/Jiangxi/00449/2013 (H9N2)-like virus; 2 is the clade of H7N3 viruses circulating in ducks in South China, represented with A/duck/Zhejiang/2/2011 and A/duck/Jiangxi/7016/2009-like virus and 3 is the clade of A/chicken/Jiangsu/S002/2013(H7N9)-like virus. Star (*) represents that the viruses were from currently-published reports20,21. Minus (−) represents no definition was used for H9N2.
Figure 4. Diversified genotypes of H10N8 viruses in Nanchang, China.
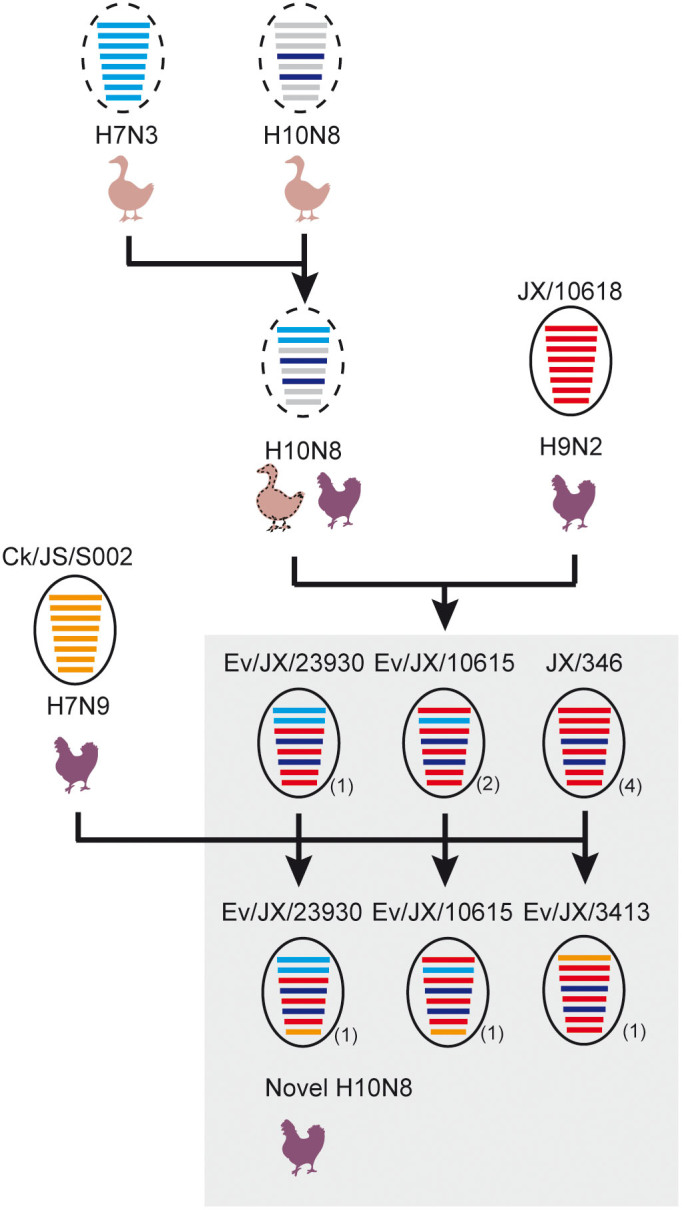
The proposed genotypes for the novel H10N8 viruses were based on phylogeny estimation of the internal genes. The cartoon was created by LY using Adobe Illustrator CS6. The eight gene segments (horizontal bars), from top to bottom, represent PB2, PB1, PA, HA, NP, NA, MP and NS. Genes in different clades were labeled with different colors. The red color represents genes from the Ev/10618/2014(H9N2)-like virus (Clade 1). The blue color represents genes from the H7N3 viruses circulating in ducks in South China (Clade 2) in recent years, and the orange color denotes genes from the CK/JS/S002/2013(H7N9)-like virus (Clade 3). The representative strain is indicated by one circle followed by the total number of isolates in parenthesis. The circle with a dotted line indicates the uncertain data for the virus. Six genotypes of H10N8 viruses with internal genes derived from the H9N2, H7N3 and H7N9 subtype viruses were detected. The possible hosts of the avian influenza viruses, including ducks and chickens, are indicated.
All H10N8 viruses showed genetic markers for mammalian adaptation and virulence in HA (A135T, K137R, S138A), M1 (N30D, T215A), and NS1 (P42S)10,11 but presented an avian signature, specifically glutamic acid (E) at residue 627 in the PB2 protein, with the exception of a mixture of E/K in the throat aspirations from Case 110. Moreover, we identified 13 site mutations in HA and 8 in NA (Table 4), including mutations in Residue 83 located at antigenic site C and Residue 166 and 188 in antigenic site B in the H3 molecule12. Compared with JX/346, Ev/JX/03489 contained 3 amino acid substitutions: M83T and I94S in HA and K143Q in NA. Six amino acid changes were detected in HA (M83T, N119D, N160D, T188I, K414M and F535V) between the two human H10N8 isolates and their NAs shared 100% identity.
Table 4. Amino acid changes in HA and NA proteins of H10N8 viruses in the study.
| Viral protein | Position | Virus isolate | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HA | JX346 | JX09037 | Ev03489 | Ev03413 | Ev03366 | Ev03367 | Ev23930 | Ev10615 | Ev10721 | Ev10738 | |
| 83 | M | T | T | T | T | T | T | T | T | T | |
| 94 | I | I | S | S | I | I | I | I | I | I | |
| 119 | N | D | N | N | D | D | D | N | D | D | |
| 129 | S | S | S | S | S | W | S | S | S | S | |
| 157a | S | S | S | S | S | S | S | N | N | N | |
| 160 | N | D | N | N | N | N | N | N | N | N | |
| 188 | T | I | T | T | T | I | T | I | I | I | |
| 212 | N | N | N | N | N | N | N | N | S | N | |
| 291 | R | R | R | R | K | R | R | R | R | R | |
| 335 | I | I | I | I | I | I | I | I | I | K | |
| 361 | T | T | T | T | A | T | T | T | T | T | |
| 414 | K | M | K | K | K | M | K | M | M | M | |
| 535 | F | V | F | F | F | V | F | V | V | V | |
| NA | |||||||||||
| 8 | I | I | I | I | I | I | I | V | I | I | |
| 20 | L | L | L | I | L | I | L | L | L | L | |
| 111 | R | R | R | K | R | R | R | R | R | R | |
| 143 | K | K | Q | K | K | K | K | K | K | K | |
| 263 | V | V | V | V | V | V | A | V | V | V | |
| 265 | Q | Q | Q | Q | Q | Q | Q | K | Q | Q | |
| 387 | N | N | N | N | N | N | N | N | S | N | |
| 403 | W | W | W | W | R | W | W | W | W | W |
Ev is the abbreviation of environment. The residues located in antigen sites in H3 molecule are highlighted with grey.
Discussion
We previously reported the first human infection with avian influenza A (H10N8) virus in Nanchang City, China10. However, the infection source in this case could not be confirmed because we did not detect the H10N8 viruses in poultry associated with the human case. Subsequently, enhanced active surveillance was initiated to trace the infection source, and an additional two human cases were detected. Furthermore, we identified the epidemiologically linked H10N8 viruses from poultry markets, and all three patients had a clear history of visiting LPMs or making direct contact with live poultry a few days before illness onset. In particular, an H10N8 virus, Ev/03489/2013, isolated from the wholesale market (M6), was highly homologous with H10N8 viruses isolated from Cases 1 and 3. Case 3 visited M6 five days before illness onset, and Case 1 visited a retail poultry market that sold poultry obtained from M6. Thus, all of the available evidence suggests that the LPM was the source of human infection with the H10N8 virus.
The unique trading style of LPMs in Southern China should be noted. This common approach involves grouping birds or different types of poultry from different sources for several days, which are then traded from wholesale markets to retail LPMs. The coexistence of avian influenza viruses from different sources as supported by the finding that various subtype viruses (e.g., H10N8, H9N2, H5N1, H7N3 and H6N6) was detected in the wholesale LPM (M6) and by the detected gene diversity of H10N8, may facilitate interspecies transmission and further gene reassortment. Among the genotypes observed, genotype JX/346 was detected repeatedly and was the only genotype found to cause human infection. This finding might imply that the entire genetic set from H9N2 viruses could be optimal for H10N8 viral fitness, similar to H7N9 viruses13,14,15. Notably, H9N2 and H7N9 viruses have also caused human infections, and extensive genetic reassortments have been reported in these viruses in recent years13,15,16,17,18,19. As a result, increased genetic diversity and the possibility of newly emerging avian influenza A viruses in the future cannot be underestimated.
Recently, three H10N8 isolates from Nanchang City were reported20,21, two belong to genotype JX/346 and one is Ev/JX/23930 according to our analysis. The findings further supported that the diversified H10N8 virus circulated in the poultry in Nanchang City. In addition, a recent serological study in Guangdong indicated that the H10 virus may infect dogs with exposure to poultry22. Therefore, the surveillance of the H10N8 virus should include poultry and mammals. LPMs may also serve as the mixing vessel and source of novel reassortant avian influenza viruses; thus, closure of LPMs is the most important measure for reducing the emergence of novel influenza viruses with potential threats to humans.
Although the initial clinical presentations of the H10N8 cases were generally mild, similar to H7N9 and H5N1 patients23,24, the disease progressed rapidly. In particular, abnormal innate immune responses manifesting as a cytokine storm have been observed in human H5N1 and H7N9 infections, which were attributed to the clinical severity of the infection25,26,27,28,29. Similarly, patterns of elevated cytokines/chemokines and increased levels of C-reactive protein and lactate dehydrogenase were detected in all three human H10N8 cases. Thus, novel avian influenza H10N8 viruses with low pathogenicity in poultry may cause severe infection in humans partially due to their induction of a cytokine storm. Additionally, all cases presented with severe lymphopenia, similar to H7N9 or H5N1 viral infection, implying that the viral infection impaired host immune functions23. Similar to the risk factors related to H7N9 and H5N1 deaths23,24, medical conditions, including coronary heart disease and hypertension, present in two of the fatal cases may have also contributed to the clinical severity.
HA sequencing indicated that the novel H10N8 viruses bound to avian-type receptors preferentially; however, the HA K137R30 substitution might increase the avidity of H10 for human-type receptors. Other mutations associated with mammalian adaptation and virulence, such as HA (A135T and S138A), M1 (N30D and T215A), NS1 (P42S) and the internal genes from the H9N2 virus, may also assist with interspecies transmission of the H10N8 virus and contribute to its severe pathogenesis to humans.
The highly pathogenic avian influenza H5 and H7 viruses have been associated with pandemic potential. However, recently reported human infections with the low-pathogenicity avian influenza H7N9 and H10N8 viruses have raised even greater challenges for pandemic preparedness, as these viruses can spread silently in poultry, which makes it very difficult to monitor their circulation and evolution. Thus, the closure of LPMs and changes in the trading style of poultry are essential to reduce the potential risk of lethal influenza virus infection to humans.
Supplementary Material
supplementary information
Acknowledgments
This work was supported by the National Mega-projects for Infectious Diseases (2014ZX10004002 to Y. Shu, 2013ZX10004-101 to D.L., 2013ZX10004611-003 to J.Z. and 2012ZX10004215 partly to D.W.) and the Chinese National Influenza Center–Centers for Disease Control and Prevention (CDC) collaborative project 5U51IP000334-03 from the CDC China–US Collaborative Program on Emerging and Re-emerging Infectious Diseases. and the National Natural Science Foundation of China (81460302 to H. Chen) and the Major Science and Technological Project of Jiangxi Province (20143ACG70004 to H.C.). We gratefully acknowledge the authors, originating and submitting laboratories of the sequences from GISAID's EpiFlu™ which this research is based.
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.S. and H.C. were co-principal investigators who designed and supervised the study. M.L., H.Y., T.C., J.W., W.X., Y.X., S.C., M.H., X.N., T.G., Y.S., J.L., J.C., Q.L. and S.W. had roles in recruitment and specimen and data collection. H.Y., J.W. and W.X. performed field investigation, specimens and data collection and did data analysis. J.Z., Z.X., W.Z. and J.S. performed the clinical management and analysis. H.B., R.G., J.G., W.H. and Y.Z. performed the viral culture and analysis. X.L., X.Z., X.H.Z. and L.Y. performed the genome sequencing and analysis. J.Z. performed the CAB test and constructed Figure 2. M.L., J.Z., L.Y., T.C., D.W., R.G. and Y.S. drafted the article. L.Y., M.L. and T.C. constructed Figures 1, 3 and 4. D.L., G.W., Z.F. and Y.W. coordinated the study and commented on the report. All authors contributed to the review and revision of the article and have seen and approved the final version.
References
- Pan American Health Organization. Avian influenza virus A (H10N7) circulating among humans in Egypt. EID Weekly Updtes. 2, (2004). Access on 26. Oct. 2014. http://new.paho.org/hq/dmdocuments/2010/Avian_Influenza_Egypt_070503.pdf. [Google Scholar]
- Arzey G. G. et al. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg Infect Dis 18, 814–816, 10.3201/eid1805.111852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R. et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368, 1888–1897, 10.1056/NEJMoa1304459 (2013). [DOI] [PubMed] [Google Scholar]
- Ku A. S. & Chan L. T. The first case of H5N1 avian influenza infection in a human with complications of adult respiratory distress syndrome and Reye's syndrome. J Paediatr Child Health 35, 207–209 (1999). [DOI] [PubMed] [Google Scholar]
- Peiris M. et al. Human infection with influenza H9N2. Lancet 354, 916–917 (1999). [DOI] [PubMed] [Google Scholar]
- Wei S. H. et al. Human infection with avian influenza A H6N1 virus: an epidemiological analysis. Lancet Respir Med 1, 771–778, 10.1016/S2213-2600(13)70221-2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco M. A. et al. Influenza surveillance in birds in Italian wetlands (1992–1998): is there a host restricted circulation of influenza viruses in sympatric ducks and coots? Vet Microbiol 98, 197–208, 10.1016/j.vetmic.2003.10.018 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang H., Xu B., Chen Q., Chen J. & Chen Z. Characterization of an H10N8 influenza virus isolated from Dongting lake wetland. Virol J 8, 42, 10.1186/1743-422X-8-42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao P. et al. Complete genome sequence of an H10N8 avian influenza virus isolated from a live bird market in Southern China. J Virol 86, 7716, 10.1128/JVI.00959-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 383, 714–721, 10.1016/S0140-6736(14)60111-2 (2014). [DOI] [PubMed] [Google Scholar]
- Parry J. H10N8 avian flu virus claims its first known human casualty. BMJ 348, g1360, 10.1136/bmj.g1360 (2014). [DOI] [PubMed] [Google Scholar]
- Caton A. J., Brownlee G. G., Yewdell J. W. & Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31, 417–427 (1982). [DOI] [PubMed] [Google Scholar]
- Liu D., Shi W. & Gao G. F. Poultry carrying H9N2 act as incubators for novel human avian influenza viruses. Lancet 383, 869, 10.1016/S0140-6736(14)60386-X (2014). [DOI] [PubMed] [Google Scholar]
- Mok C. K. et al. Pathogenicity of the novel A/H7N9 influenza virus in mice. MBio 4, 10.1128/mBio.00362-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L. et al. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun 5, 3142, 10.1038/ncomms4142 (2014). [DOI] [PubMed] [Google Scholar]
- Chu Y. C. et al. Continuing evolution of H9N2 influenza viruses endemic in poultry in southern China. Influenza Other Respir Viruses 5 Suppl 1, 68–71 (2011). [PubMed] [Google Scholar]
- Choi Y. K. et al. Continuing evolution of H9N2 influenza viruses in Southeastern China. J Virol 78, 8609–8614, 10.1128/JVI.78.16.8609-8614.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. et al. Genotypic evolution and antigenic drift of H9N2 influenza viruses in China from 1994 to 2008. Vet Microbiol 146, 215–225, 10.1016/j.vetmic.2010.05.010 (2010). [DOI] [PubMed] [Google Scholar]
- Huang Y. et al. Diversified reassortant H9N2 avian influenza viruses in chicken flocks in northern and eastern China. Virus Res 151, 26–32, 10.1016/j.virusres.2010.03.010 (2010). [DOI] [PubMed] [Google Scholar]
- Yang J. et al. Mutations of Novel Influenza A(H10N8) Virus in Chicken Eggs and MDCK Cells. Emerg Infect Dis 20, 1541–1543, 10.3201/eid2009.140257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W. et al. Genesis of the novel human-infecting influenza A(H10N8) virus and potential genetic diversity of the virus in poultry, China. Euro Surveill 19 (2014). [DOI] [PubMed] [Google Scholar]
- Su S. et al. First Evidence of H10N8 Avian Influenza Virus Infections among Feral Dogs in Live Poultry Markets in Guangdong Province, China. Clin Infect Dis 59, 748–750, 10.1093/cid/ciu345 (2014). [DOI] [PubMed] [Google Scholar]
- Wang C. et al. Comparison of patients hospitalized with influenza A subtypes H7N9, H5N1, and 2009 pandemic H1N1. Clin Infect Dis 58, 1095–1103, 10.1093/cid/ciu053 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H. N. et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med 368, 2277–2285, 10.1056/NEJMoa1305584 (2013). [DOI] [PubMed] [Google Scholar]
- de Jong M. D. et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12, 1203–1207, 10.1038/nm1477 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R. et al. Profiles of acute cytokine and antibody responses in patients infected with avian influenza A H7N9. PLoS One 9, e101788, 10.1371/journal.pone.0101788 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opal S. M. & Fedson D. S. The dysfunctional host response to influenza A H7N9: a potential treatment option? Crit Care 18, 135, 10.1186/cc13839 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 381, 1916–1925, 10.1016/S0140-6736(13)60903-4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. et al. Biological features of novel avian influenza A (H7N9) virus. Nature 499, 500–503, 10.1038/nature12379 (2013). [DOI] [PubMed] [Google Scholar]
- Vachieri S. G. et al. Receptor binding by H10 influenza viruses. Nature 511, 475–477, 10.1038/nature13443 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information