Abstract
Although the importance of cellular forces to a wide range of embryogenesis and disease processes is widely recognized, measuring these forces is challenging, especially in three dimensions. Here, we introduce CellFIT-3D, a force inference technique that allows tension maps for three-dimensional cellular systems to be estimated from image stacks. Like its predecessors, video force microscopy and CellFIT, this cell mechanics technique assumes boundary-specific interfacial tensions to be the primary drivers, and it constructs force-balance equations based on triple junction (TJ) dihedral angles. The technique involves image processing, segmenting of cells, grouping of cell outlines, calculation of dihedral planes, averaging along three-dimensional TJs, and matrix equation assembly and solution. The equations tend to be strongly overdetermined, allowing indistinct TJs to be ignored and solution error estimates to be determined. Application to clean and noisy synthetic data generated using Surface Evolver gave tension errors of 1.6–7%, and analyses of eight-cell murine embryos gave estimated errors smaller than the 10% uncertainty of companion aspiration experiments. Other possible areas of application include morphogenesis, cancer metastasis and tissue engineering.
This article is part of the themed issue ‘Systems morphodynamics: understanding the development of tissue hardware’.
Keywords: cell mechanics, force inference, interfacial tensions, CellFIT-3D, CellFIT, video force microscopy (VFM)
1. Cellular forces
The importance of cellular forces to morphogenesis, wound healing and disease is now widely recognized [1–5]. Experimental studies have shown that irregularities in these forces can give rise to developmental defects and other malformations [6], and computational models have shown that alterations as small as 20% can affect clinical outcomes [7].
In many settings, these driving forces can be treated as equivalent interfacial tensions γi along cell–cell and cell–medium boundaries [8,9] (electronic supplementary material, figure S1), a concept considered as early as the 1960s [10,11] but quickly dropped in favour of the differential adhesion hypothesis [12]. Computer simulations of sorting and other cell movements carried out by the authors, however, showed that a wide range of cell and tissue movements were in fact driven largely by interfacial tensions [13–15] (to which cell–cell adhesions make a counteracting contribution [8]), leading to a new paradigm [2,16–20]. Information about these tensions can be obtained through a variety of experimental techniques, that in generally decreasing length scale include: pipette aspiration, deformable substrates, engineered droplets, laser ablation, atomic force microscopy, optical tweezers, magnetic cytometry, and fluorescence resonance energy transfer (FRET) microscopy [18,21–31]. Unfortunately, these techniques can be expensive, time consuming, invasive, or destructive, and all except substrate deformation, FRET and droplet methods provide information about only a single cell surface at one moment in time.
When force inference techniques for two-dimensional systems [32–36] entered the scene in 2010, they made spatio-temporal maps of cellular forces possible for any available image, including historical ones. Force inference techniques, like CellFIT which is here denoted as CellFIT-2D to distinguish it from the present three-dimensional version, are based on two primary mechanical assumptions: that the tension-carrying boundaries that meet at any triple junction (TJ) [36] produce mechanical equilibrium there, and that these tensions are constant over the span of any given boundary or interface (electronic supplementary material, movie S1). Force inference techniques in this class require only that cell edges be visible, they involve no mechanical intervention, and they produce no additional damage beyond that caused by microscopy. Data from CellFIT-2D revealed tension variations around the perimeters of individual cells, differences between cells in a single population and between populations, elevated tensions along inter-population boundaries [36], and purposeful temporal variations [17].
On the strength of this and other evidence, we propose that cells move and organize primarily by gradual and carefully orchestrated changes in interfacial tensions and protrusion contractions, with the latter acting along the interface between cells and serving as a special case of these tensions. Finite-element studies have shown that force imbalances at TJs cause those junctions to move until TJ angles produce an equilibrium configuration (electronic supplementary material, movie S1) or until a high-tension boundary shortens enough to produce a neighbour change [15]. If these forces were to change systematically and gradually with time, a series of carefully controlled cellular and tissue morphologies could be produced. Modelling studies of whole embryos [37] found that physiological motions were best matched by time-varying driving forces, but the reason for this finding was unclear when those studies were carried out. The finding was contrary to the terraced nature of these forces tacitly assumed by modellers and biologists. In the context of embryogenesis, the context in which the most active research on cell mechanics and morphogenesis was occurring, the underlying mindset was that gene expression or some other process set up conditions for the next developmental step and then it moved ahead on the basis of those conditions. The revised understanding for which we here argue, is that regulatory networks and mechanobiological circuits play a much more active role in controlling the driving forces from moment to moment. If cellular forces do indeed change with time as these studies suggest, then techniques like CellFIT—which can take snapshots of the forces in all of the cells visible in a field of view and do so at multiple sequential times—become particularly useful.
Other kinds of forces—such as intracellular pressures and deviatoric stresses associated with cytoplasm deformation—could be included, as was done in video force microscopy (VFM) [17]. Line tensions along the curvilinear interfaces between trios of cells could also, in principle, be included, but we suspect that their contribution is minimal. The picture of gradual force changes we present should not be confused with models in which increasing forces are applied to elastic systems. Elastic systems would spring back to their initial state should the driving forces be eliminated, though real embryos and their tissues do not. Instead, we contend that embryos and their tissues behave primarily in a plastic manner [38], with their morphogenetic deformations being essentially irreversible due to cell neighbour changes. An apparent exception to this statement is the small elastic component of deformation that can occasionally be observed, as when momentary forces are applied, and a reversible response occurs only because cells do not have time to change neighbours and lock the temporary geometry in place. We have argued elsewhere [28] that viscous, visco-elastic and elastic tissue behaviour can all be produced by cells that have constant edge tensions and viscous cytoplasm.
Interestingly, the TJ force balances on which CellFIT is based do not depend on the mechanical characteristics of the cell membrane system, including whether it is elastic, viscous, governed by rate constants, affected by endocytosis, influenced by adhesion systems or altered by cortical components. In a sense, CellFIT operates one level up from these important details, and simply provides the total relative tension acting along any particular interface, without regard to how it is generated. In addition, intracellular pressures and any stresses from volumetric viscous or elastic components seem to act primarily normal to the interface (except perhaps during laser ablation experiments, which violate other CellFIT assumptions) and likewise, do not affect the TJ force balances [36]. These fortuitous circumstances make CellFIT applicable to a very wide range of biological applications. CellFIT would not be applicable, however, to interfaces where cells are adhered to a substrate, because the forces carried by that substrate would generally not be known. If such forces were known, however, governing equations that include them could be constructed [5] and a suitably adapted version of CellFIT applied. It would also not be applicable to interfaces that contain significant spatial force variations along individual cell faces. One might hope that the quality measures that form part of CellFIT would aid in identifying situations of this kind.
In this article, we outline a series of steps (figure 1) that allow CellFIT-2D to be extended so that cellular forces in three-dimensional aggregates and tissues can be inferred from serial sections. When applied to synthetic sections generated using Surface Evolver (SE) [41], the tensions calculated by CellFIT-3D had errors as small as 1.6%. CellFIT-3D was then applied to eight-cell compaction-stage mouse embryos, and the tensions found had estimated errors lower than the 10% uncertainty associated with accompanying aspiration experiments.
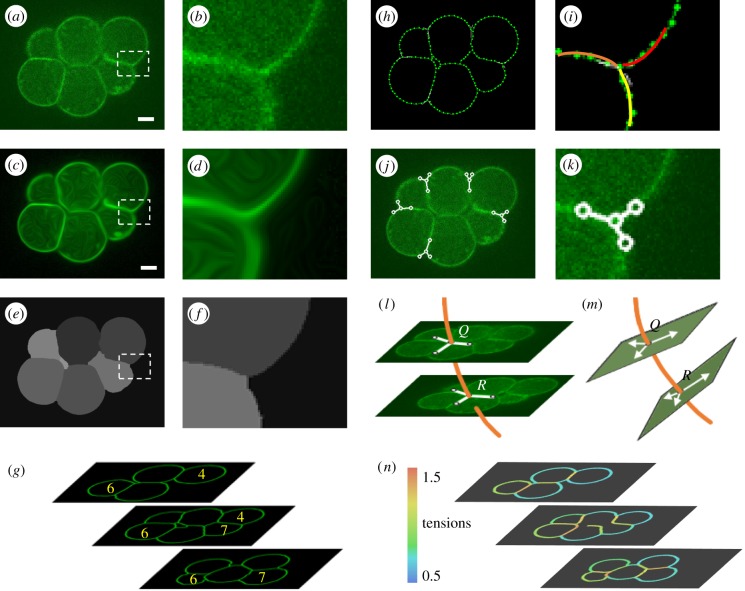
Figure 1.
An overview of CellFIT-3D. A raw confocal image of an eight-cell stage murine embryo is shown in (a) (scale bar, 10 µm), while (b) shows an enlargement of the boxed area. An edge enhancing coherence filter [39] was used to smooth the boundaries (c) and close their gaps (d). Parameters used: Scheme, implicit discretization; total diffusion time = 25; Gaussian sigma = 3. SeedWater [40]-segmented cells are visible in (e) and (f), each denoted by in a different intensity of grey. Corresponding cells in successive images were grouped together (g) and TEs that appeared in multiple images identified. A fine mesh (h) was constructed along each cell boundary and its approach angle to any given triplet determined by circular arc fitting (i). Tangent vectors (j) were calculated automatically and adjusted manually as needed (k). Splines were fit through the triplets that appeared in successive images (l), and dihedral planes and angles calculated (m). Least-squares equations were constructed and solved (see §2i,j) and calculated tensions displayed (n).
2. CellFIT-3D
As in the finite-element models we have used to study embryonic cells for many years and in VFM and CellFIT-2D, we here assume that the subcellular forces generated by various structural protein and adhesion systems can be deemed to generate an equivalent tension tangent to the cell membrane [13,15,17,36,42] (electronic supplementary material, figure S1). The total tension along any given cell–cell or cell–membrane interface i is denoted γi, and it is assumed to be specific to that interface, whether an edge in two dimensions or a face in three dimensions [43].
Furthermore, whenever three cells meet at a particular point and a cutting plane is constructed normal to the TJ between those cells at that point, the vector sum of the membrane tensions in that plane must add to zero. Studies of two-dimensional cellular systems showed that the membrane angles at TJs are unaffected by intracellular pressures or deviatoric stresses associated with reshaping of the viscous cellular cytoplasm [36]. Consequently, TJs can be analysed and used to calculate membrane tensions γi without reference to pressure or viscous forces.
Cellular pressures were not calculated as part of CellFIT-3D, but one could presumably calculate them, as is done in two dimensions, after the interfacial tensions γi are determined [36]. In three dimensions, the local principal curvatures k1 and k2 of cell–cell or cell–medium interfaces change with position unless they are spherical. However, the mean curvature (k1 + k2)/2 would be constant throughout any one of these surfaces if it carries a constant isotropic tension γi, and the pressure difference ΔPi across it would be given by the Young–Laplace equation
| 2.1 |
To determine the spatially varying principal curvatures or even the mean curvature of a surface based on cuts through it, however, is a problem beyond the scope of the present study. For the present, there is probably no reason to calculate intracellular pressures or their differences, as their relevance to development is still unclear and methods to verify them experimentally are limited.
Because of the geometric complexity of three-dimensional systems, it is valuable to distinguish between the curvilinear TJs or triple edges (TEs) that arise between trios of contacting cells or between pairs of cells and the medium, and the points at which those junctional curves pass through individual confocal or other sections. We use the term ‘triplet’ to describe the latter locations where three, but occasionally more, cell boundaries are seen to converge (as in the boxed area of figure 1a). The cell membranes immediately adjacent to the point are considered part of the triplet.
A series of image processing and geometric and mechanical analyses steps are required to implement CellFIT-3D, and they are outlined here.
(a). Image capture
As we will show, the quality of a CellFIT-3D analysis depends on image resolution and section spacing. Reliable tension information can be obtained from data like figure 1a, a portion of a 512 by 512 pixel image from a confocal stack of an eight-cell murine embryo. There were approximately 125 pixels across the diameter of a typical cell, and individual cells were transected an average of 18 times as a result of the 2 µm section spacing.
(b). Image enhancement
The cell boundaries in these images may seem well defined, but at the pixel level (figure 1b) where computational algorithms work, noise, gaps and other anomalies become apparent. An ideal image would have accurate, gap-free white outlines a single pixel wide and a pure black background. A number of image processing algorithms for amending the images toward this ideal were appraised, and an edge enhancing coherence filter [39] that smoothed the boundaries (figure 1c) and largely closed the gaps (figure 1d) was selected. Relevant computer code for this step and a number of the others, as well for as the validation procedures, is available through the indicated references.
(c). Image segmentation
Cells in the processed images were segmented (figure 1e) using a watershed algorithm called SeedWater [40,44]. The algorithm treats the bright pixels at cell edges as if they defined mountain range heights and it begins to flood each mountain-surrounded region from a relatively dark pixel or low-lying point called a seed point. The software chooses these seed points, but users can manually adjust them. Boundaries are found indirectly, based on where the flooded zones contact each other or where they end at the outer mountain ranges. This approach is less sensitive to intracellular noise—spurious hills in low lying regions—and partial gaps than are standard edge tracing techniques. The resulting boundaries may, however, still contain pixel-scale positional noise (figure 1f) and other imperfections.
(d). Grouping of triple edges
In three dimensions, individual TEs may appear as triplets in multiple images and it is necessary to associate these occurrences with each other. As with boundary identification, it is preferable to work with cell cross-sectional areas, and we use a cell-grouping algorithm that considers area overlap across adjacent images, centroid collinearity and other geometric features [45] to group cross-sections associated with the same cell (figure 1g) and in turn to identify triplets belonging to the same TE. If sections are spaced sufficiently close, seed-point proximity can be used to group the outlines. Grouping also makes possible three-dimensional cell models for visualization and verification of mesoscale geometry and topology.
(e). Mesh generation
The pixelated boundaries of sectioned cells may have complex shapes with reversing curvature and may contain noise. In order to obtain good triplet approach angles, and informed by CellFIT-2D [36], we fit cubic splines to the boundary pixels along each edge and use these splines to generate uniformly spaced mesh points (figure 1h).
(f). In-plane angles
CellFIT-2D showed that accurate angles are crucial to the tension calculations, and we found that membrane endpoint directions could be estimated well by fitting separate circular arcs to the last four or five mesh points on each end (figure 1i), as in two dimensions [36]. Standard software exists for arc fitting, the fit is coordinate indifferent, it provides a second-order approximation to the shape of the boundary terminus, and it attenuates noise.
Triplets that appeared in at least three successive images were identified algorithmically, and shown graphically in their corresponding sections (figure 1j). This strategy was more efficient than manually vectorizing triplets of interest. The graphic consisted of a circle at the calculated triplet location and three vectors with circles at their ends in the calculated approach directions. The graphics were automatically overlaid on the raw images and manual angular adjustments were made as needed to best capture the final approach angle at the junction (figure 1k).
(g). Local dihedral angles
To convert the in-plane angles defined by these graphic triplets to true dihedral angles, splines (shown as orange curves in figure 1l) were constructed through sets of grouped triplets. Construction of a reliable spline required a TE to appear in at least three images. Local dihedral planes (figure 1m) were constructed normal to the spline at each triplet, and the boundary vectors were mapped onto them (white arrows) by projection normal to the planes. Finally, a pair of equilibrium equations was constructed for two arbitrary orthogonal directions in each plane. These equations define the ratios that the three boundary tensions γi must have for that section of the TE to be in equilibrium.
(h). Average dihedral angles
The equation pairs generated at various triplets along any one TE can give different force ratios, and the quality of the equations varied. For example, if an image plane is strongly oblique to a TE, the fluorescent signals from one or more of its boundaries may be spread out and grainy. In addition, errors in calculated boundary directions can amplify when projected onto highly tilted dihedral planes, making information from these planes less trustworthy. When more than three equation pairs are available, they are automatically checked for consistency and equations from the ends of the spine, where the dihedral planes tend to be more tilted, eliminated if appropriate. Increasing the number of sections per cell improves the geometry of the spline and increases the number of available equation pairs. However, even when there are seven slices per cell on average (orange curve in electronic supplementary material, figure S3), approximately two thirds of the TEs still appear in two or fewer sections. To amalgamate the multiple equation pairs associated with any one TE into a single set of ratios γi:γj:γk, we recommend a least-squares ratio solver [46].
In contrast to these steps, one could construct point clouds from the meshing points associated with each boundary. One could then fit mathematical surfaces to each cloud, or a portion thereof, and use mathematical descriptions to calculate the TE contact angles. Unfortunately, the shapes that arise are complex, often having reversing concavity and other challenging features, making this approach impractical [47]. One could also take in-plane angles as surrogates for dihedrals and apply CellFIT-2D to individual sections. However, doing so ignores the oblique angles of typical TEs to these sections, and produces tension errors of 50% [47]. Alternatively, for specific TEs, one might use the in-plane angles of the triplet apparently most normal to that TE [21], but numerical tests show that even this approach introduces approximately 5% additional error in the dihedral angle equations, in addition to foregoing the statistical benefits of multi-triplet averages.
(i). Assembly and solution of tension equations
The previous step produces either a pair of equilibrium equations or two tension ratios for each of the n TEs having a sufficient number of useable triplets. In either case, these equations can be assembled into a single matrix equation of the form
| 2.2 |
where G has 2n rows and m columns, and m is the number of tensions γi that appear in one or more of the 2n force-balance equations, and whose values will in due course be found. These equations correspond exactly to the equation pairs that arise in CellFIT-2D [36], and their assembly, solution and evaluation are identical, unaffected by the dimensionality of the host space.
A unique solution to this homogeneous and overdetermined system can be found by constructing and solving the least-squares system
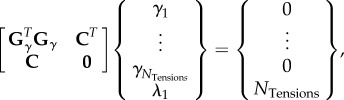 |
2.3 |
or
| 2.4 |
where
| 2.5 |
a choice that selects the particular set of scaled γs having a unit mean.
(j). Solution evaluation
The toolkit developed for assessing CellFIT-2D solutions was found to apply equally well to CellFIT-3D. The overall quality of the equations is assessed using the condition number of G*, a value equal to the ratio of its largest and smallest eigenvalues. This ratio portends the degree to which error in n is amplified in γ* [48], and so indicates the sensitivity of the calculated tensions to angular and other types of error. The condition number will vary with matrix size, and magnitudes substantially higher than those shown here and in other CellFIT analyses [36] may signal structural problems with the equations and reduced solution accuracy.
In contrast, the solution residual
| 2.6 |
provides a measure of how well the pair of equations associated with each TE is satisfied by the least-squares solution γ. The paired components of r indicate the degree to which each TE is out of balance and they are useful for identifying triplets that may have been inaccurately placed during the automated meshing steps and may require manual adjustment. Finally, the scaled cofactors of G*, also known as standard errors [36], indicate the confidence levels associated with individual tensions.
(k). Solution reporting
The calculated tensions can be reported in many different forms, including that shown in figure 1n. As with CellFIT-2D, display of residuals or confidence levels [36,49] might be useful, but doing so in three dimensions is more challenging.
3. Validation
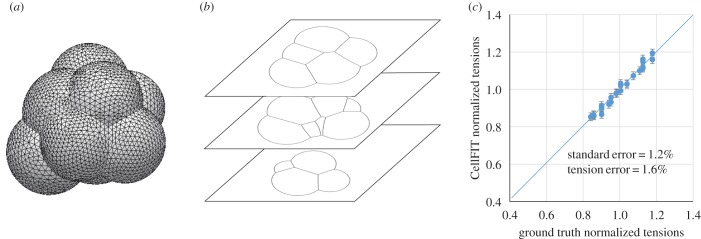
To assess the CellFIT-3D algorithms, they were tested on synthetic sections generated using Surface Evolver [50] (figure 2 and electronic supplementary material) for which ground truth tensions were known. The synthetic data were designed to approximately match the 18 slices per cell of the murine embryo data used here, and the image data were processed as outlined in figure 1. The error in the calculated tensions was found to depend on slice spacing and image resolution, but was insensitive to the number of cells analysed, whether those cells were surrounded by medium or cut cells, and the range of the tension values γI within the model.
Figure 2.
Synthetic data and analysis. A model with eight cells and randomly assigned interfacial tension values varying from each other by 30% was generated using Surface Evolver (a), and used to create synthetic sections (b). The tensions calculated by CellFIT-3D are shown in (c) and error bars indicate their associated standard errors. The RMS tension error relative to ground truth was 1.6%.
Increasing the number of slices per cell increases the number of dihedral planes per TE (electronic supplementary material, figure S3) and allows strongly sloped planes near their ends to be ignored. Seven slices per cell was found to give tension errors of 3.2% in isolated aggregates of eight cells, while 14 slices per cell, often not difficult to obtain experimentally, reduced that error to only 1.6% (figure 2c). In all cases, the number of equations was more than adequate to solve for the unknown tensions (see electronic supplementary material).
Portions of isolated synthetic aggregates of eight and 50 cells were eroded away in order to generate a range of aggregate sizes and conditions—from those fully surrounded by medium, to those with mixed medium and cut cell edges, to fragments containing no complete cells. For aggregates with 14 slices per cell, the errors depended primarily on the image resolution, with 400 and 200 pixels per cell diameter generating errors of 3% and 8%, respectively.
Error was then added to the averaged dihedral angles, and figure 3 shows how adding angular noise to the average dihedral angles affects tension error, residual and standard error. The angular noise was randomly assigned using a Gaussian distribution. Conveniently, all three track similarly with noise, allowing the latter two—which can be calculated from the governing equations associated with any given dataset—to indicate the likely angular error (input noise) and tension error (output uncertainty). A mean angular noise of 5°, a value consistent with typical manual digitizing errors of 5° [36], for example, caused tension errors of 7.1% and residual and standard errors of 9.4% and 5.2%, respectively.
Figure 3.
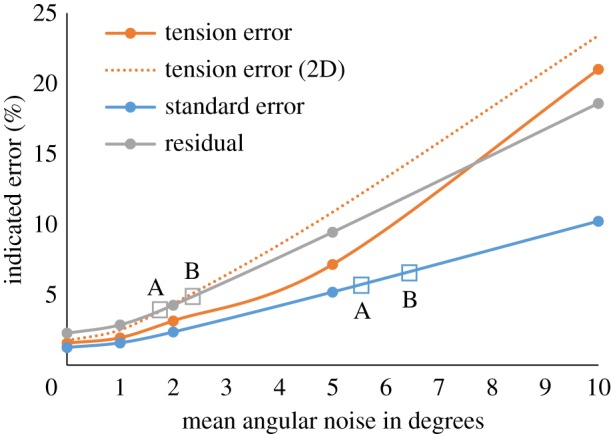
Noise response. Tension error grows as mean angular noise, of the amounts indicated in angular degrees, is introduced into the averaged dihedral angles. The standard error and residual behave similarly, as does the tension error in a 20-cell epithelium analysed using CellFIT-2D [36]. The standard errors and residuals associated with murine embryo analyses (figure 4b and c) are shown as boxes along their corresponding curves, and they indicate, respectively, equivalent angular noise in the averaged dihedral angles of 6–7% and 2–3%. Tension errors can in turn be deduced from these noise levels to be no greater than 7% (see supplementary text).
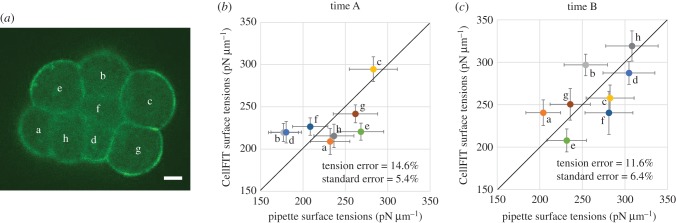
4. Application to murine embryos
CellFIT was then used to investigate eight-cell murine embryos (figure 4a and electronic supplementary materials) undergoing compaction, a process considered to be driven by surface tensions [21,51]. Interfaces were typically seen in at least five slices. Some manual adjustments were necessary in places where the automated meshing was inaccurate. For time A, 90 min after the last four-cell stage blastomeres divided, the CellFIT-3D tensions had residuals of 0.031 and standard errors of 5.4% (figure 4b), while the values for time B (figure 4c), 150 min later, were slightly higher. The corresponding boxes in figure 3 suggest input errors corresponding to noise levels between 2 and 6 and tension errors of 3–9%.
Figure 4.
Analysis of murine embryos. (a) Compaction-stage murine embryos consisting of eight cells were imaged such that cells were nominally 120 pixels in diameter, and had on average 18 sections through each cell (scale bar, 10 µm). Tensions measured using micropipette aspiration at time A (90 min after the last division from four- to eight-cell stage) are shown in (b) and those at time B, 150 min later, in (c). CellFit-3D inferred tensions have been scaled so that their average value is the same as that of the values from the pipette aspiration experiments. Error bars on the experimental values are ±10% (see §4) while CellFIT-3D tensions show their respective standard errors. In this figure, only, tension errors are with respect to aspiration experiments not ground truth.
The surface tensions of all eight cells (figure 4a) were measured by pipette aspiration [21] and the experimental error estimated from repeated tests was 10%. Measurement of tensions on interior surfaces is more challenging, but could be done using optical tweezers or laser ablation, though the latter is destructive and precludes the taking of force measurements at multiple times or locations. The root mean square (RMS) errors between the CellFIT-3D and aspiration values at times A and B (figure 4b,c) were 14.6% and 11.6%, respectively, values consistent with aspiration errors of 10% and standard errors of 5.4% and 6.4%, for times A and B, respectively. The surface tensions of individual cells changed between times A and B, and the time delay between sequential aspiration measurements may have contributed additional error. In contrast, CellFIT typically draws its data from the much narrower window of time required to collect a single set of sections. Several other eight-cell murine embryos were analysed with CellFIT, and similar results were obtained.
5. Discussion
This article demonstrates that cellular forces (interfacial tensions) in three-dimensional aggregates can be inferred from serial sections. In principle, any image sources including confocal sections or histological sections could be used, provided that cell boundaries are well defined and there are at least 6–8 slices per cell. Furthermore, tension errors can in principle be as low as 1.6%, considerably lower than the variability of typical experimental tensometry techniques. Because the assembled CellFIT equations are overdetermined, input and solution quality can be assessed using residuals and standard errors, and TEs with apparently discordant angles or other anomalies can be identified and removed without jeopardizing the tension inference process. These quality measures could also serve to identify situations where membrane tensions are not uniform as assumed in CellFIT, as could be the case should undulating membrane geometries arise, or localized tension sources give rise to nonuniformities in membrane tension.
CellFIT-3D builds on its two-dimensional counterpart, and its three primary challenges—obtaining cell outlines, connecting triplets in successive images, and calculating true dihedral angles—can be overcome using relatively straightforward algorithms. Like its counterpart, CellFIT-3D provides only tension ratios, and should scaled values be required, direct force measurements must be obtained. Fortunately, these ratios are often all that is required [21], as it is these ratios that govern cell shapes and motion patterns, with cytoplasm viscosity and force strength determining motion rates, only [52].
Force inference methods are possible because the relative angles at TEs in equilibrium depend uniquely on membrane forces, even though other mesoscale features (see supplementary text) do not [53,54]. Should four or more cell edges impinge on a single junction or there be a rosette [55], the number of unknowns exceeds the number of equations by more than one, and a unique tension ratio does not exist. However, if the equations and unknowns associated with such junctions are assembled with those from other suitably connected TEs, a unique and trustworthy solution should still be possible [36]. In principle, CellFIT-3D could be extended to calculate intracellular pressures, as in CellFIT-2D [36], but calculating the required surface curvatures is much more difficult in three dimensions, there is currently little experimental interest in these pressures, and their consequences for cell movement remain unclear. The effects of viscous cytoplasmic forces could also be incorporated easily, with their contributions to TJ force balances calculated in a manner similar to that used in VFM.
If the cellular forces that drive cell movements, mesoscale assembly and bulk tissue motions turn out to be time-varying, as recent evidence suggests, then techniques like CellFIT that can provide force maps over a whole field of view and at closely spaced times become particularly useful. Hopefully, in time, the technique presented here will provide new insights into the movements of single or small groups of cells during morphogenesis and diseases like metastasis, formation of mesoscale structures such as acini in various organs and engineered tissues, and bulk tissue movements.
Supplementary Material
Acknowledgements
Animal studies were performed at the European Molecular Biology Laboratory in accordance with Federation for Laboratory Animal Science Associations guidelines and recommendations. Surface Evolver was written by Kenneth Brakke, Susquehanna University, and SeedWater was written by David Mashburn under the supervision of Shane Hutson, Vanderbilt University.
Authors' contributions
The force inference concepts were developed and implemented by G.W.B., J.H.V. and A.E.; the experiments were conducted by J.-L.M. in the lab of T.H.; the Surface Evolver models were developed by S.C. and J.H.V.; the paper was written primarily by G.W.B.
Competing interests
We have no competing interests.
Funding
Funding was provided by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant to G.W.B.
References
- 1.Hamada H. 2015. Role of physical forces in embryonic development. Semin. Cell Dev. Biol. 47–48, 88–91. ( 10.1016/j.semcdb.2015.10.011) [DOI] [PubMed] [Google Scholar]
- 2.Heisenberg CP, Bellaiche Y. 2013. Forces in tissue morphogenesis and patterning. Cell 153, 948–962. ( 10.1016/j.cell.2013.05.008) [DOI] [PubMed] [Google Scholar]
- 3.Wirtz D, Konstantopoulos K, Searson PC. 2011. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer 11, 512–522. ( 10.1038/nrc3080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DuFort CC, Paszek MJ, Weaver VM. 2011. Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 12, 308–319. ( 10.1038/nrm3112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brugues A, et al. 2014. Forces driving epithelial wound healing. Nat. Phys. 10, 683–690. ( 10.1038/nphys3040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guck J, Chilvers ER. 2013. Mechanics meets medicine. Sci. Transl. Med. 5, 212fs41. ( 10.1126/scitranslmed.3007731) [DOI] [PubMed] [Google Scholar]
- 7.Brodland GW, Chen X, Lee P, Marsden M. 2010. From genes to neural tube defects (NTDs): insights from multiscale computational modeling. HFSP J. 4, 142–152. ( 10.2976/1.3338713) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodland GW, Chen HH. 2000. The mechanics of cell sorting and envelopment. J. Biomech. 33, 845–851. ( 10.1016/S0021-9290(00)00011-7) [DOI] [PubMed] [Google Scholar]
- 9.Umetsu D, Aigouy B, Aliee M, Sui L, Eaton S, Julicher F, Dahmann C. 2014. Local increases in mechanical tension shape compartment boundaries by biasing cell intercalations. Curr. Biol. 24, 1798–1805. ( 10.1016/j.cub.2014.06.052) [DOI] [PubMed] [Google Scholar]
- 10.Harris AK. 1976. Is cell sorting caused by differences in the work of intercellular adhesion? A critique of the Steinberg hypothesis. J. Theor. Biol. 61, 267–285. ( 10.1016/0022-5193(76)90019-9) [DOI] [PubMed] [Google Scholar]
- 11.Phillips H, Steinberg MS. 1978. Embryonic tissues as elasticoviscous liquids, I: Rapid and slow shape changes in centrifuged cell aggregates. J. Cell Sci. 30, 1–20. [DOI] [PubMed] [Google Scholar]
- 12.Steinberg MS. 1970. Does differential adhesion govern self-assembly process in histogenesis? Equilibrium configurations and the emergence of a hierarchy among populations of embryonic cells. J. Exp. Zool. 173, 395–434. ( 10.1002/jez.1401730406) [DOI] [PubMed] [Google Scholar]
- 13.Brodland GW. 2002. The differential interfacial tension hypothesis (DITH): a comprehensive theory for the self-rearrangement of embryonic cells and tissues. J. Biomech. Eng. 124, 188–197. ( 10.1115/1.1449491) [DOI] [PubMed] [Google Scholar]
- 14.Brodland GW. 2006. Do lamellipodia have the mechanical capacity to drive convergent extension? Int. J. Dev. Biol. 50, 151–155. ( 10.1387/ijdb.052040gb) [DOI] [PubMed] [Google Scholar]
- 15.Chen HH, Brodland GW. 2000. Cell-level finite element studies of viscous cells in planar aggregates. J. Biomech. Eng. 122, 394–401. ( 10.1115/1.1286563) [DOI] [PubMed] [Google Scholar]
- 16.Lecuit T, Lenne PF. 2007. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 8, 633–644. ( 10.1038/nrm2222) [DOI] [PubMed] [Google Scholar]
- 17.Brodland GW, et al. 2010. Video force microscopy reveals the mechanics of ventral furrow invagination in Drosophila. Proc. Natl Acad. Sci. USA 107, 22 111–22 116. ( 10.1073/pnas.1006591107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campas O, et al. 2014. Quantifying cell-generated mechanical forces within living embryonic tissues. Nat. Methods 11, 183–189. ( 10.1038/nmeth.2761) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levayer R, Hauert B, Moreno E. 2015. Cell mixing induced by myc is required for competitive tissue invasion and destruction. Nature 524, 476–480. ( 10.1038/nature14684) [DOI] [PubMed] [Google Scholar]
- 20.Krieg M, Arboleda-Estudillo Y, Puech P-H, Käfer J, Graner F, Müller DJ, Heisenberg C-P. 2008. Tensile forces govern germ-layer organization in zebrafish. Nat. Cell Biol. 10, 429–436. ( 10.1038/ncb1705) [DOI] [PubMed] [Google Scholar]
- 21.Maitre JL, Niwayama R, Turlier H, Nedelec F, Hiiragi T. 2015. Pulsatile cell-autonomous contractility drives compaction in the mouse embryo. Nat. Cell Biol. 17, 849–855. ( 10.1038/ncb3185) [DOI] [PubMed] [Google Scholar]
- 22.Maitre JL, Berthoumieux H, Krens SF, Salbreux G, Julicher F, Paluch E, Heisenberg CP. 2012. Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science 338, 253–256. ( 10.1126/science.1225399) [DOI] [PubMed] [Google Scholar]
- 23.Thomas G, Burnham NA, Camesano TA, Wen Q. 2013. Measuring the mechanical properties of living cells using atomic force microscopy. J. Vis. Exp. 76, e50497 ( 10.3791/50497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS. 2010. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat. Methods 7, 969–971. ( 10.1038/nmeth.1531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tambe DT, Croutelle U, Trepat X, Park CY, Kim JH, Millet E, Butler JP, Fredberg JJ. 2013. Monolayer stress microscopy: limitations, artifacts, and accuracy of recovered intercellular stresses. PLoS ONE 8, e55172 ( 10.1371/journal.pone.0055172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasza KE, Vader D, Koster S, Wang N, Weitz DA. 2011. Magnetic twisting cytometry. Cold Spring Harb. Protoc. 2011, pdb.prot5599. ( 10.1101/pdb.prot5599) [DOI] [PubMed] [Google Scholar]
- 27.Morimatsu M, Mekhdjian AH, Adhikari AS, Dunn AR. 2013. Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Lett. 13, 3985–3989. ( 10.1021/nl4005145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutson MS, Veldhuis JH, Ma X, Lynch HE, Cranston PG, Brodland GW. 2009. Combining laser microsurgery and finite element modeling to assess cell-level epithelial mechanics. Biophys. J. 97, 3075–3085. ( 10.1016/j.bpj.2009.09.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. 2012. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc. Natl Acad. Sci. USA 109, 12 568–12 573. ( 10.1073/pnas.1204390109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo J, Sachs F, Meng F. 2014. Fluorescence-based force/tension sensors: a novel tool to visualize mechanical forces in structural proteins in live cells. Antioxid. Redox Signal. 20, 986–999. ( 10.1089/ars.2013.5708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugimura K, Lenne PF, Graner F. 2016. Measuring forces and stresses in situ in living tissues. Development 143, 186–196. ( 10.1242/dev.119776) [DOI] [PubMed] [Google Scholar]
- 32.Machado PF, Duque J, Etienne J, Martinez-Arias A, Blanchard GB, Gorfinkiel N. 2015. Emergent material properties of developing epithelial tissues. BMC Biol. 13, p98-015-0200-y. ( 10.1186/s12915-015-0200-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishihara S, Sugimura K, Cox SJ, Bonnet I, Bellaiche Y, Graner F. 2013. Comparative study of non-invasive force and stress inference methods in tissue. Eur. Phys. J. E Soft Matter 36, 9859-13045-8. Epub 2013 Apr 26 ( 10.1140/epje/i2013-13045-8) [DOI] [PubMed] [Google Scholar]
- 34.Brodland GW. 2017. CellFIT-2D. See http://www.civil.uwaterloo.ca/brodland/cellFIT.asp.
- 35.Chiou KK, Hufnagel L, Shraiman BI. 2012. Mechanical stress inference for two dimensional cell arrays. PLoS Comput. Biol. 8, e1002512 ( 10.1371/journal.pcbi.1002512) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brodland GW, Veldhuis JH, Kim S, Perrone M, Mashburn D, Hutson MS. 2014. CellFIT: a cellular force-inference toolkit using curvilinear cell boundaries. PLoS ONE 9, e99116 ( 10.1371/journal.pone.0099116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X, Brodland GW. 2008. Multi-scale finite element modeling allows the mechanics of amphibian neurulation to be elucidated. Phys. Biol. 5, 015003 ( 10.1088/1478-3975/5/1/015003) [DOI] [PubMed] [Google Scholar]
- 38.Brodland GW, Chen DI, Veldhuis JH. 2006. A cell-based constitutive model for embryonic epithelia and other planar aggregates of biological cells. Int. J. Plast. 22, 965–995. ( 10.1016/j.ijplas.2005.05.002) [DOI] [Google Scholar]
- 39.Kroon D. 2010. Image Edge Enhancing Coherence Filter Toolbox. See http://www.mathworks.com/matlabcentral/fileexchange/25449-image-edge-enhancing-coherence-filter-toolbox.
- 40.Mashburn DN, Lynch HE, Ma X, Hutson MS. 2012. Enabling user-guided segmentation and tracking of surface-labeled cells in time-lapse image sets of living tissues. Cytometry A 81, 409–418. ( 10.1002/cyto.a.22034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brakke KA. 2013. The Surface Evolver. See http://facstaff.susqu.edu/brakke/evolver/evolver.html.
- 42.Lecuit T, Lenne PF, Munro E. 2011. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu. Rev. Cell Dev. Biol. 27, 157–184. ( 10.1146/annurev-cellbio-100109-104027) [DOI] [PubMed] [Google Scholar]
- 43.Hutson MS, Brodland GW, Yang J, Viens D. 2008. Cell sorting in three dimensions: topology, fluctuations, and fluidlike instabilities. Phys. Rev. Lett. 101, 148105 ( 10.1103/PhysRevLett.101.148105) [DOI] [PubMed] [Google Scholar]
- 44.Hutson MS.2016. Seedwater Segmenter. See https://my.vanderbilt.edu/shanehutson/software/seedwater-segmenter/.
- 45.Kin Shun Leung T, Veldhuis JH. 2010. Identifying same-cell contours in image stacks: a key step in making 3D reconstruction. Ann. Biomed. Eng. 39, 698–705. ( 10.1007/s10439-010-0198-9) [DOI] [PubMed] [Google Scholar]
- 46.Kwiesielewicz M. 1996. The logarithmic least squares and the generalized pseudoinverse in estimating ratios. Eur. J. Oper. Res. 93, 611–619. ( 10.1016/0377-2217(95)00079-8) [DOI] [Google Scholar]
- 47.Ehsandar A. 2015. Inferring 3D cellular forces from confocal image stacks. PhD Thesis, University of Waterloo, Canada.
- 48.Sewell G. 2014. Computational methods of linear algebra, 3rd edn Hackensack, NJ: World Scientific. [Google Scholar]
- 49.Veldhuis JH, Mashburn D, Hutson MS, Brodland GW. 2015. Practical aspects of the cellular force inference toolkit (CellFIT). Methods Cell Biol. 125, 331–351. ( 10.1016/bs.mcb.2014.10.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brakke KA. 1992. The Surface Evolver. Exp. Math. 1, 141–165. ( 10.1080/10586458.1992.10504253) [DOI] [Google Scholar]
- 51.Turlier H, Maitre JL. 2015. Mechanics of tissue compaction. Semin. Cell Dev. Biol. 47–48, 110–117. ( 10.1016/j.semcdb.2015.08.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brodland GW, Wiebe CJ. 2004. Mechanical effects of cell anisotropy on epithelia. Comput. Methods Biomech. Biomed. Engin. 7, 91–99. ( 10.1080/1025584042000209369) [DOI] [PubMed] [Google Scholar]
- 53.Brodland GW. 2004. Computational modeling of cell sorting, tissue engulfment, and related phenomena: A review. Appl. Mech. Rev. 57, 47–76. ( 10.1115/1.1583758) [DOI] [Google Scholar]
- 54.Davies JT, Rideal EK. 1963. Interfacial phenomena, 2nd edn New York: Academic Press. [Google Scholar]
- 55.Blankenship JT, Backovic ST, Sanny JS, Weitz O, Zallen JA. 2006. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. Cell 11, 459–470. ( 10.1016/j.devcel.2006.09.007) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.