Abstract
Target-identification and understanding of mechanism-of-action (MOA) are challenging for development of small-molecule probes and their application in biology and drug discovery. For example, although aspirin has been widely used for more than 100 years, its molecular targets have not been fully characterized. To cope with this challenge, we developed a novel technique called quantitative acid-cleavable activity-based protein profiling (QA-ABPP) with combination of the following two parts: (i) activity-based protein profiling (ABPP) and iTRAQ™ quantitative proteomics for identification of target proteins and (ii) acid-cleavable linker-based ABPP for identification of peptides with specific binding sites. It is known that reaction of aspirin with its target proteins leads to acetylation. We thus applied the above technique using aspirin-based probes in human cancer HCT116 cells. We identified 1110 target proteins and 2775 peptides with exact acetylation sites. By correlating these two sets of data, 523 proteins were identified as targets of aspirin. We used various biological assays to validate the effects of aspirin on inhibition of protein synthesis and induction of autophagy which were elicited from the pathway analysis of Aspirin target profile. This technique is widely applicable for target identification in the field of drug discovery and biology, especially for the covalent drugs.
Biologically active small molecules are very useful as probes and drugs for diagnosis and therapeutics. They can be discovered by target-based screenings involving specific proteins and phenotypic screenings using cell- or organism-based assays1. Phenotypic screenings are widely used in traditional and modern biology and pharmacology. A challenging and important issue is the unknown mechanisms of action (MOA) of potential hits found in phenotypic screenings. Small molecules generated by target-based screenings have known binding targets, but it is unknown whether they may have other protein targets in living cells2. Thus, no matter which approach is used for the discovery of biologically active small molecules, it is necessary to perform a target profiling to have a better understanding of their MOA. Among the various approaches of target identification for bioactive small molecules, activity-based protein profiling (ABPP) combining with bio-orthogonal click chemistry is widely utilized both in vitro and in vivo3,4,5,6,7,8,9. ABPP is an ideal strategy that can faithfully recapitulate protein-small molecule interactions in situ (i.e. in live cells), and at the same time enable enrichment of these complexes for subsequent large-scale proteome-wide identification of potential targets10. With the advances of mass spectrometry (MS) technologies, it is feasible to further identify the specific probe-labelling sites on protein targets. For example, the probe is directly incubated with purified proteins identified with ABPP and then labelled proteins are digested and desired peptides are analyzed by MS/MS11. Another promising method for binding site mapping performed in live cells is gel-free ABPP to identify probe-labelled peptides, including the selective enrichment and elution of probe-labelled peptide fragments12,13,14. However, this method discarded the rest of the peptides that are unlabelled. Whereas tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP) employs on-bead trypsin and TEV digestions to simultaneously identify both probe-labelled proteins and their exact sites of probe modification15,16.
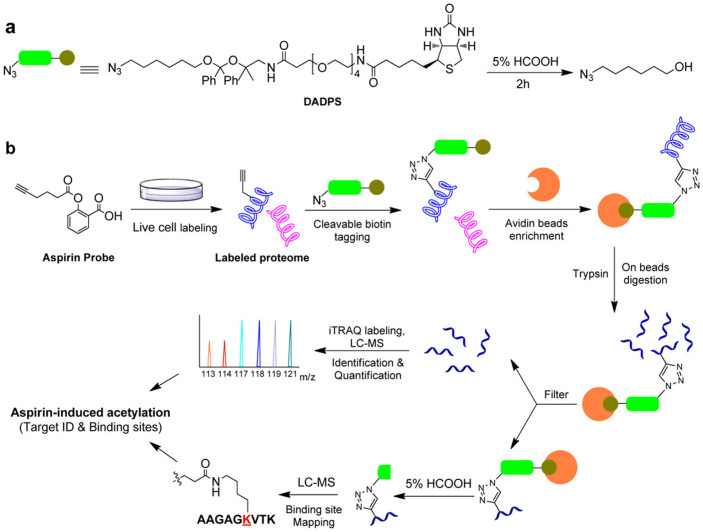
In recent years, quantitative proteomics approaches (SILAC and iTRAQ) have been increasingly applied in ABPP17,18,19,20. The quantitative information generated from such approaches can help differentiate specific from non-specific bindings by comparing enrichment ratios. Meanwhile, a variety of cleavable linkers have been developed and used in ABPP21,22,23,24. Cleavable linkers allow the seized proteins to be liberated after pull down under non-denaturing conditions. In this study, we developed a quantitative acid-cleavable activity-based protein profiling (QA-ABPP) approach that combines iTRAQ with an acid-cleavable linker to profile protein targets and their specific modification sites (Figure 1). In QA-ABPP, the biotin tag (DADPS) containing both an azide group and an acid-cleavable linker reacted with the proteome labelled with an alkynyl ABPP probe. Tagged proteins are then enriched by avidin beads, followed by on-bead trypsin digestion and filtration. These filtrated peptides are labelled with respective iTRAQ reagents, and pooled together for further identification and quantification by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Meanwhile, the alkynyl probe-labelled peptides with a small molecule fragment (237.15 amu; including part of the acid-cleavable linker [143.1 amu] and the aspirin moiety [94.05 amu]) left on the modified amino acids were released from the beads after 5% formic acid treatment for 2 hrs and identified by LC-MS/MS, from which the direct binding site information of the probe was determined. The binding site information acquired by high-resolution mass spectrometry is highly accurate and reliable, as the mass deviation is within 5ppm for most of the identified peptides. By correlating binding site information with quantitative proteomics data, we further corroborated the targets of the probe with exceptionally high confidence.
Figure 1. Overview of quantitative acid-cleavable activity-based protein profiling (QA-ABPP) for protein targets and their binding sites of aspirin.
(a) Structure of acid-cleavable linker and its reaction with formic acid; (b) Overall strategy of QA-ABPP for identification of probe's target and binding sites.
Aspirin, besides its wide application for the reduction of inflammation, pain and fever, was found to lower the rates of heart attack and stroke in patients with cardiovascular disease, and more recently to reduce the incidence of cancer and cancer mortality, especially for gastrointestinal cancers25,26,27,28. To fully understand aspirin's versatility, we designed and synthesized two aspirin-based alkynyl probes (Asp-P1 and Asp-P2) to identify aspirin's protein targets and the exact acetylation sites by virtue of QA-ABPP. By using QA-ABPP, we identified 1110 aspirin-acetylated proteins and 2,775 peptides bearing the acetylation from 870 proteins by our aspirin probes. Aspirin-acetylated amino acid residues were lysine, serine, arginine, histidine, theroine, tyrosine, tryptophan and cysteine. Among which, arginine, histidine, theroine, tyrosine and tryptophan were reported for the first time to be acetylated by aspirin. By correlating the iTRAQ-based quantitative ABPP results with the binding site data, 523 overlapping proteins were confirmed to be modified by aspirin with high confidence. Our results suggest that aspirin may exert its cancer preventative function by acetlylating multiple cellular targets. Furthemore, through pathway analysis and functional validation, our data reveal that aspirin may function by inhibiting protein synthesis and inducing autophagy. Additionally, confocal cell imaging using our aspirin probe was performed to examine the cellular distribution of aspirin-induced acetylation.
Results
Chemical synthesis and labelling profile of aspirin-based probes
Two aspirin probes (Asp-P1 and Asp-P2) were synthesized with different linker lengths to anchor an alkynyl handle (Figure 2a, the synthetic scheme is shown in Supplementary Figure 7). Briefly, salicylic acid was reacted with the designated acyl chlorides to generate Asp-P1 and Asp-P2 probes. In addition, we also synthesized the 4-pentynoate probe (Acetyl-P) (the synthetic scheme is shown in Supplementary Figure 7), which is a reporter for monitoring protein acetylation29. With these probes in hand, we first optimized the labelling concentration of Asp-P1/P2 and Acetyl-P in colon cancer HCT116 cell lines. The three probes were incubated with live cells for 12 hrs, followed by cell lysis. The lysate was then treated with Cy3-azide under click chemistry conditions (CuSO4, TCEP, TBTA) and subsequently detected by fluorescence scannning following SDS-PAGE. The results showed visible Asp-P1/P2 and Acetyl-P fluorescence labelling bands at 1 mM concentration (Supplementary Figure 1)30,31,32. To examine whether the probes can mimic the acetylation effect of Aspirin, we have conducted competition assay by pre-treating the cell lysate with Aspirin before incubating with the probes. Our results showed that Aspirin pre-treatment essentially reduced the labeling signals of the probes, suggesting that both probes largely modify the same proteins as aspirin (Supplementary Figure 2). We then compared the labelling pattern of Asp-P1/P2 and Acetyl-P together with a DMSO-treated negative control at 1 mM concentration for each probe. Asp-P1 and Asp-P2 have approximately the same labelling pattern (Figure 2b), indicating that the slight difference in the lengths of the linkers attached to anchor alkyne handles of the two probes has little effect on their labelling profiles. Surprisingly, Asp-P1/P2 show much stronger labelling signals than Acetyl-P, suggesting that aspirin may induce a significantly higher degree of acetylation than the traditional 4-pentynoate. Considering the expansive and long-term usage of aspirin, it will be highly significant to unravel the acetylation targets of aspirin and the exact sites of this modification.
Figure 2. Synthesis of aspirin probes and their labelling in live cells.
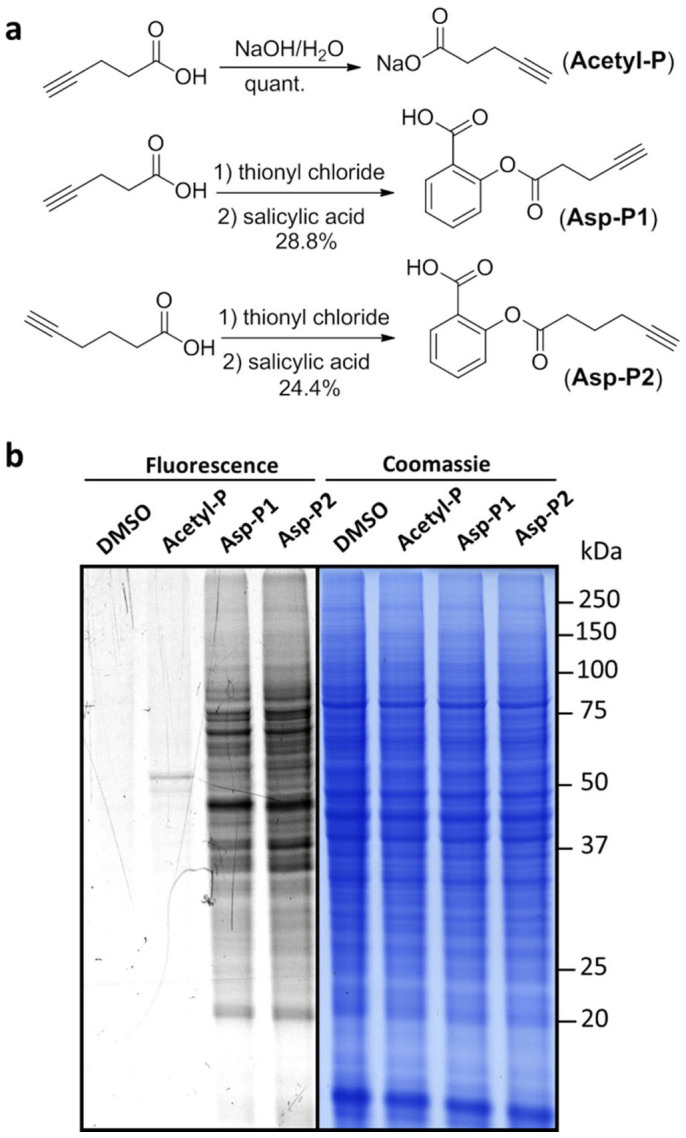
(a) Chemical structures of acetylation probe (Acetyl-P), Aspirin-based probes Asp-P1 and Asp-P2. (b) The in situ fluorescent labelling of HCT116 cells using Acetyl-P, Asp-P1 and Asp-P2 together with a DMSO-treated negative control. Probe-labelled proteomes were visualized by click conjugation to the Cy3-azide tag, SDS gel separation, and fluorescent scanning; the concentrations of all probes were 1 mM.
Identification of protein targets and their modification sites of aspirin-induced acetylation by QA-ABPP
Next, we performed QA-ABPP to simultaneously identify the targets and binding sites of aspirin-induced acetylation using asprin-based probes. Two biological replicates of the asprin probes- and DMSO-treated samples were included to partially overcome biological and experimental variations. After incubating with 1 mM Asp-P1/P2 or DMSO (nagative control) for 12 hrs, HCT116 cells were lysed and reacted with acid-cleavable biotin azide tag (the synthetic scheme is shown in Supplementary Figure 7) before enrichment by pulling down with avidin beads. After washing the beads thoroughly, enriched proteins were digested directly on the beads using trypsin. The resulting peptides were labelled with respective iTRAQ reagents, and pooled together for further identification and quantification by LC-MS/MS. The iTRAQ reporter ions of non-specific binding proteins have equal or similar intensities among the probe-treated and DMSO-treated control samples. Contrastingly, specific target proteins enriched by our probes show highly differential intensities against the DMSO-treated control samples (as illustrated by the significantly higher reporter intensities of 117 and 118, 119 and 121 vs. 113 and 114 shown in Figure. 1b). This technique enables us to discriminate specific protein targets from non-specific and endogenously biotinylated proteins33. Furthermore, the multiplexing nature of iTRAQ-based chemical proteomics approach allows the incorporation of replicates within a single LC-MS/MS analysis, hence increasing the confidence of identifying specific targets while minimizing experimental errors33,34.
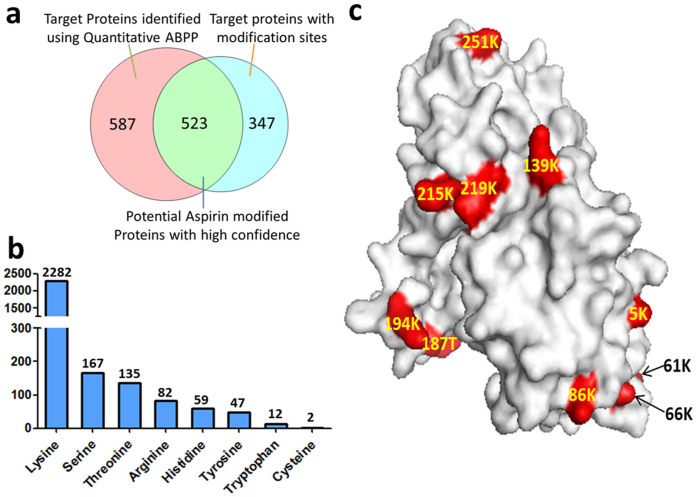
A total of 1194 proteins were successfully identified and quantified using iTRAQ-based quantitative ABPP in our experiment (The full list of the quantified proteins is shown in Supplematary Table 1). To reduce potential false-positive targets, we set a highly stringent differential ratio of 2 as the cut-off threshold to differentiate specific binding targets from non-specific ones. Moreover, the targets must be identified in both Asp-P1/P2 pull down results. Consequently, 1110 proteins were regarded as the specific targets of aspirin using iTRAQ-based quantitative ABPP (The full list of the potential targets is shown in Supplematary Table 2). Since all the protein targets identified with the Asp-P1 probe was also detected with the Asp-P2 probe, the later was used for further identification of aspirin modification sites. Following the pull down by Asp-P2, the washed and filtered beads bearing the binding peptides were cleaved by 5% formic acid for 2 hrs, and the resulting probe-modified peptides were identified using LC-MS/MS. By specifying a variable modification mass shift of 237.15 amu for several potentially modified amino acids, 5011 (redundant) or 2775 (non-redundant) acetylated peptides from 870 proteins were successfully identified (MASCOT peptide score > 30; the full list of the modified redundant and non-redundant peptides was shown in Supplematary Table 3&4; the full list of the modified proteins was shown in Supplematary Table 5). Remarkably, the percentage of the binding site-bearing peptides is extremely high at 70%. In other words, among 7164 (redundant) identified peptides, 5011 (redundant) were aspirin-modified peptides, suggesting that our probe-binding peptide enrichment is highly efficient. In summary, lysine (2282 non-redundant peptide found) is the most common residue to be modified by Asp-P2. We also found 172 peptides with modified serine, 135 peptides with modified threonine, 82 peptides with modified arginine, 59 peptides with modified histidine, 12 peptides with modified tryptophan and 2 peptides with modified cysteine (Figure 3b). Surprisingly, several aspirin-modified residues have never been reported before: such as arginine, histidine, theroine, tyrosine and tryptophan. To assess the degree of false-positive identification of asprin-binding peptides, we performed a database search using the MS data derived from the trypsinized peptides from the probe pull down sample before on-beads acid cleavage, which did not return any modified peptide (MASCOT peptide score >30), thus indicating our data for aspirin-modified peptides are highly accurate and reliable. Although the binding-site information can already comfirm the asprin-modified proteins, we further overlaped the asprin targets identified from quantitative ABPP with the modification site-bearing proteins to obtain highly confident targets. As shown in Figure 3a, a total of 523 asprin targets were finally comfirmed through both methods (The full list of the potential targets is shown in Supplematary Table 6).
Figure 3. Summary of aspirin modified proteins and amino acid residues.
(a) Numbers of proteins identified using quantitative ABPP, proteins with modification sites identified and proteins confirmed with high confidence; (b) Numbers of the aspirin-modified amino acid residues. Numbers on top of the columns are the numbers of peptides modified by Asp-P2; (c) Locations of aspirin-modified residues in the protein GAPDH.
Since we identified a lot of proteins which are modified at multiple sites, next, we analysed the distribution of modified residues. Among the aspirin targets, Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) was found to be acetylated at 25 different residues (Supplematary Table 7). Next, we analyzed the structure of GAPDH and found that all of the aspirin-modified residues were on the surface of the protein, indicating that the asprin modification might only occur when the residue is solvent accessible (Figure 3c).
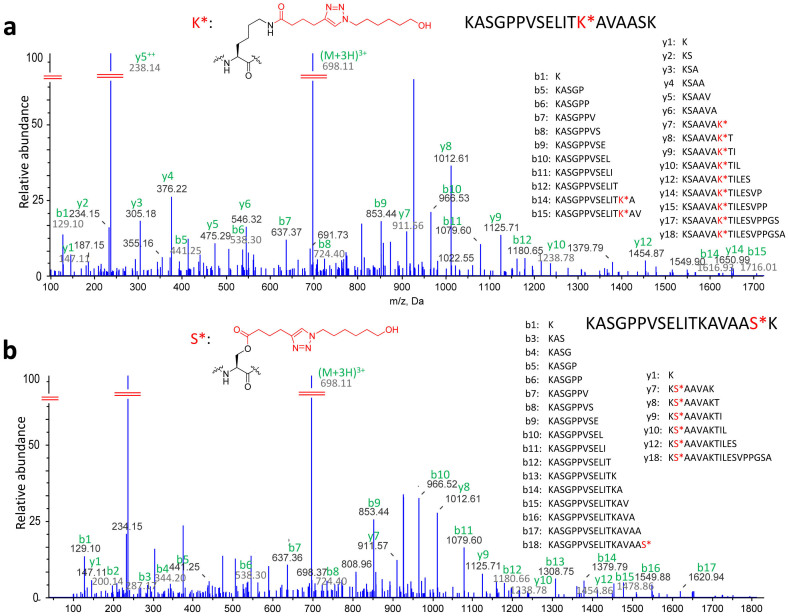
Owing to its crucial role in transcriptional regulation, acetylation is widely reported in histone proteins35,36. For this reason, we went on to examine the aspirin-acetylated histone proteins. As shown in Table 1, four aspirin-acetylated histone proteins have been comfirmed by our QA-ABPP approach. Furthermore, histone H1.4 was found to be extensively modified at 28 different residues. Moreover, we also found a peptide KASGPPVSELITKAVAASK which has been modified at two different sites (Lys46 & Ser51) in Histone H1.4. MS/MS spectra of this peptide are shown in Figure 4a&4b. These results implied that aspirin may regulate transcriptional activities by acetylation of histone proteins.
Table 1. Acetylation sites in Histone proteins identified by QA-ABPP.
| Protein Name | Ratio Asp-P1 | Ratio Asp-P2 | Pep. Exp. Mr. | Pep.Calc. Mr. | Pep. Delta | Peptide Sequence |
|---|---|---|---|---|---|---|
| Histone H1.0 | 3.57 | 4.58 | 1423.871 | 1423.876 | −0.0052 | LVTTGVLK―QTK |
| 1811.9188 | 1811.917 | 0.0022 | SHYK―VGENADSQIK | |||
| 1579.821 | 1579.821 | 0.0005 | TENSTSAPAAK―PK | |||
| 1537.8052 | 1537.81 | −0.0048 | T―ENSTSAPAAKPK | |||
| 1945.0009 | 1945.009 | −0.0082 | YSDMIVAAIQAEKNR― | |||
| Histone H1.4 | 4.81 | 8.82 | 1094.6058 | 1094.608 | −0.0025 | AASGEAK―PK |
| 1293.7408 | 1293.74 | 0.0003 | AASGEAKPK―AK | |||
| 852.5162 | 852.5181 | −0.0018 | AGAAK―AK | |||
| 1219.7419 | 1219.74 | 0.0018 | AK―KPAAAAGAK | |||
| 1077.664 | 1077.666 | −0.0018 | AKK―PAGAAK | |||
| 1814.934 | 1814.928 | 0.0066 | ALAAAGYDVEK―NNSR | |||
| 1434.8107 | 1434.808 | 0.0025 | ASGPPVS―ELITK | |||
| 1962.1123 | 1962.115 | −0.0027 | ASGPPVSELITK―AVAASK | |||
| 1080.626 | 1080.629 | −0.0031 | ATGAATPKK― | |||
| 1067.6078 | 1067.609 | −0.0009 | AVAASK―ER | |||
| 1402.7556 | 1402.757 | −0.0012 | GTGASGSFK―LNK | |||
| 1530.8506 | 1530.852 | −0.0012 | GTGASGSFK―LNKK | |||
| 1774.96 | 1774.958 | 0.0022 | GTLVQT―KGTGASGSFK | |||
| 1421.9341 | 1421.933 | 0.0007 | IKLGLK―SLVSK | |||
| 1222.7038 | 1222.703 | 0.0004 | K―AASGEAKPK | |||
| 1347.8338 | 1347.835 | −0.0012 | K―AKKPAAAAGAK | |||
| 1471.8018 | 1471.803 | −0.0016 | K―ALAAAGYDVEK | |||
| 1562.902 | 1562.903 | −0.0012 | K―ASGPPVSELITK | |||
| 2090.2051 | 2090.21 | −0.0048 | KASGPPVSELITK―AVAASK | |||
| 2090.2051 | 2090.21 | −0.0048 | KASGPPVSELITKAVAAS―K | |||
| 1208.7244 | 1208.724 | 0.0003 | KATGAATPK―K | |||
| 1020.608 | 1020.608 | 0.0001 | K―PAAAAGAK | |||
| 1148.7022 | 1148.703 | −0.0008 | KPAAAAGAK―K | |||
| 1180.7538 | 1180.754 | −0.0005 | LGLK―SLVSK | |||
| 2182.1632 | 2182.163 | −0.0002 | SETAPAAPAAPAPAEK―TPVK | |||
| 2310.2563 | 2310.258 | −0.002 | SETAPAAPAAPAPAEK―TPVKK | |||
| 2438.3482 | 2438.353 | −0.0051 | SETAPAAPAAPAPAEKTPVKKK― | |||
| 1209.7414 | 1209.745 | −0.0031 | SGVSLAALKK― | |||
| 2298.3028 | 2298.295 | 0.0081 | SGVSLAALK―KALAAAGYDVEK | |||
| 1496.8933 | 1496.893 | 0.0006 | SLVSK―GTLVQTK | |||
| Histone H2AX | 2.04 | 3.48 | 1261.629 | 1261.63 | −0.0012 | K―ATQASQEY |
| 1096.5745 | 1096.578 | −0.0033 | K―GHYAER | |||
| 1621.9189 | 1621.915 | 0.0037 | KT―SATVGPKAPSGGK | |||
| 1621.9189 | 1621.915 | 0.0037 | KTSATVGPK―APSGGK | |||
| 1750.0057 | 1750.01 | −0.0044 | KTSATVGPK―APSGGKK | |||
| 2635.624 | 2635.615 | 0.0087 | LLGGVTIAQGGVLPNIQAVLLPK―K | |||
| Histone H2B type 1-O | 2.85 | 4.80 | 1515.796 | 1515.797 | −0.0013 | K―ESYSIYVYK |
| 1314.7312 | 1314.73 | 0.0016 | PDPAK―SAPAPK | |||
| 1758.9292 | 1758.93 | −0.0012 | SRK―ESYSIYVYK |
[a] Ratio Asp-P1/P2 represents the enrichment ratio of aspirin probe versus DMSO control pull downed proteins. [b] The aspirin acetylation sites are highlighted using bold red fonts with underline. [c] The two grey shaded peptides were the same peptide but with different acetylation sites. The detailed mass spectra of these two peptides were shown in Figure 4.
Figure 4. Aspirin modified multiple residues of Histone protein H1.4.
(a) MS/MS sequencing showed the binding of Asp-P2 to Lys46 of histone H1.4. (b) MS/MS sequencing also showed the binding of Asp-P2 to Ser51 of the same peptide. K* and S* represent the Asp-P2 modified Lysine and Serine. The details of mascot scores and other information of these two modified peptides were shown in section 2.11 and 2.12 in Supplementary Information.
Pathway and functional analysis of aspirin-induced protein acetylation
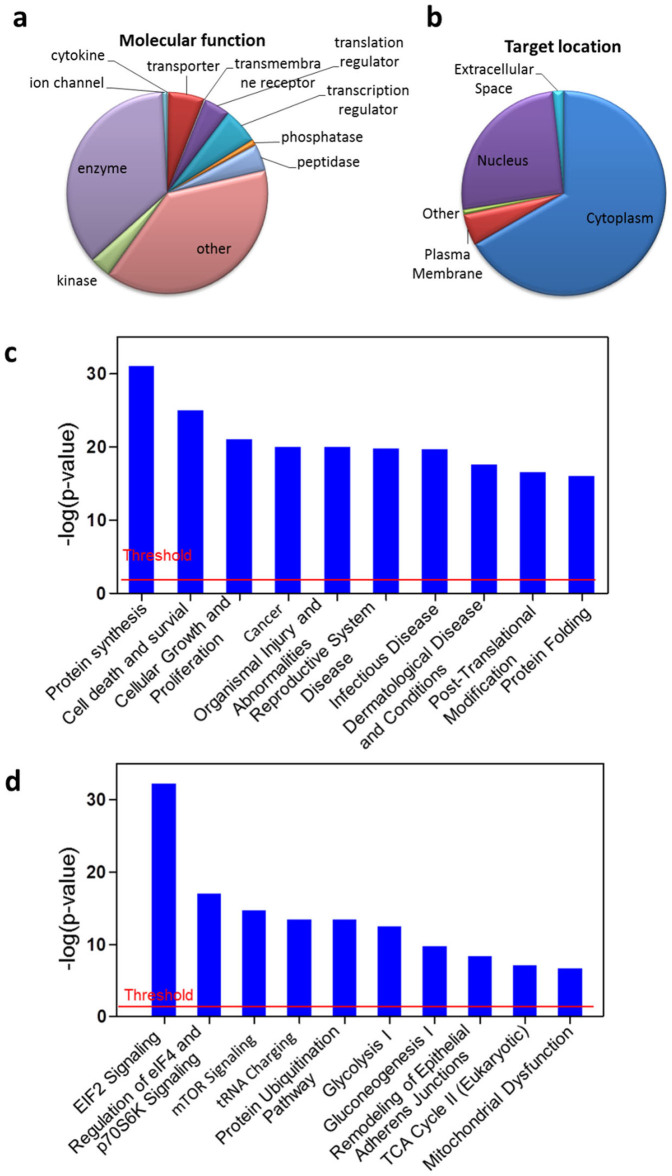
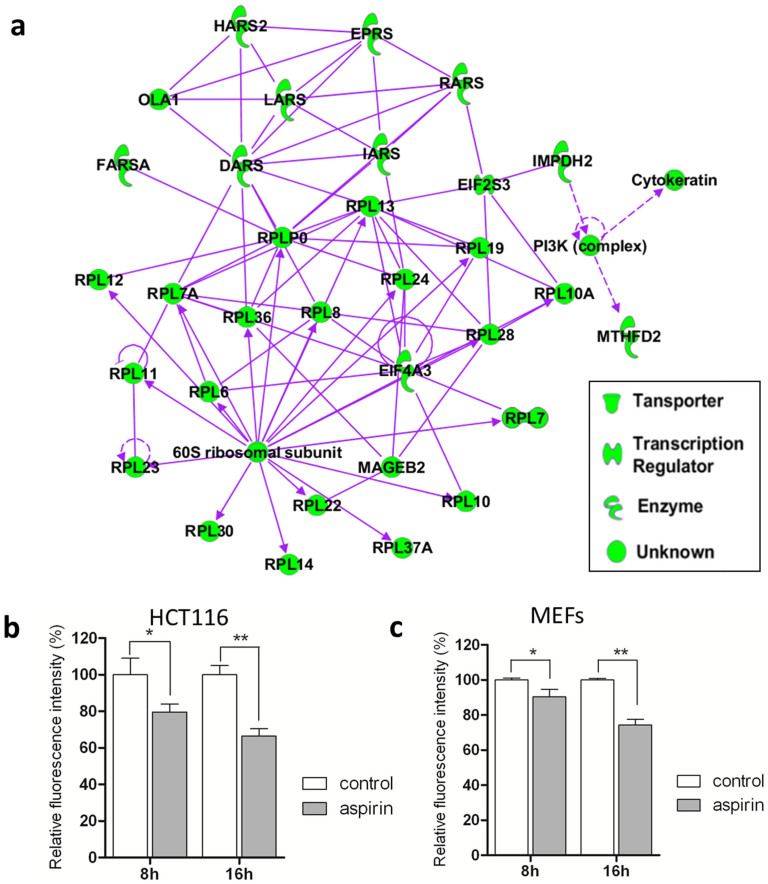
The 523 confirmed aspirin targets are broadly distributed in different parts of the cell and can be categorized into several different molecular functional types: enzyme, kinase, transporter, phosphatase, peptidase, transcription regulator, etc. (Figure 5a&5b). Ingenuity Pathway Analysis™ (IPA) results suggested that aspirin may exert its tumor suppressive effects through multiple cellular pathways including the EIF2, eIF4/p70S6K and mTOR pathways (Figure 5d). Other IPA analysis results were shown in Supplementary Figures 3, 4&5. In particular, mTOR signalling pathway is known to play a critical role in autophagy, while all three pathways are extensively involved in protein synthesis (Figure 5c&Figure 6).
Figure 5. Ingenuity Pathway Analysis™ (IPA) of aspirin-targeted proteins.
(a) Aspirin targets have various biological functions. (b) Aspirin targets located in different cell organelles. (c) Top molecular and cellular functional classes to which the aspirin-targeted proteins are associated. (d) Top canonical pathways to which the aspirin-targeted proteins are associated. The ranking was based on the p values derived from the Fisher's exact test. The high-ranking categories are displayed along the x axis in a decreased order of significance. The y axis displays the - (log) p value. The horizontal line denotes the cut-off threshold for significance (p value <0.05).
Figure 6. Aspirin inhibited de novo protein synthesis.
(a) Ingenuity Pathway Analysis™ (IPA) revealing that aspirin affects gene expression and protein synthesis. All proteins shown as green nodes were identified as the specific targets of aspirin. (b) Aspirin inhibited de novo protein synthesis in HCT116 cells and MEFs. Cells were firstly labelled with 50 μM AHA in L-methionine-free medium with or without aspirin (5 mM) treatment for 8 hrs or 16 hrs. The cells were then harvested, fixed and permeabilized for the click reaction. Finally, cellular fluorescence intensity was analysed using flow cytometry. Data for the relative signal intensity was expressed as the ratio of treated cells to control cells, as mean ± SD from three independent experiments; * P < 0.05, ** P < 0.01, derived from the Student's t-test.
Inhibition of de novo protein synthesis by aspirin
To validate the predicted effects of aspirin on cells as observed from our pathway analysis, we first determined the protein synthesis inhibition (Figure 6a) caused by aspirin treatment. Aspirin-mediated de novo protein synthesis inhibition was detected by reduced AHA (an unnatural amino acid which can be incorporated into the newly synthesized proteins) signal intensity37. We tested AHA signal intensity changes in HCT116 cells and MEFs cells after aspirin treatment. As shown in Figure 6b&6c, aspirin treatment after 16 hrs caused a 40% and 30% reduction in the AHA signal intensity in HCT116 and MEFs cells respectively. This indicates that aspirin is a potential inhibitor of de novo protein synthesis.
Induction of autophagy following mTOR suppression by aspirin
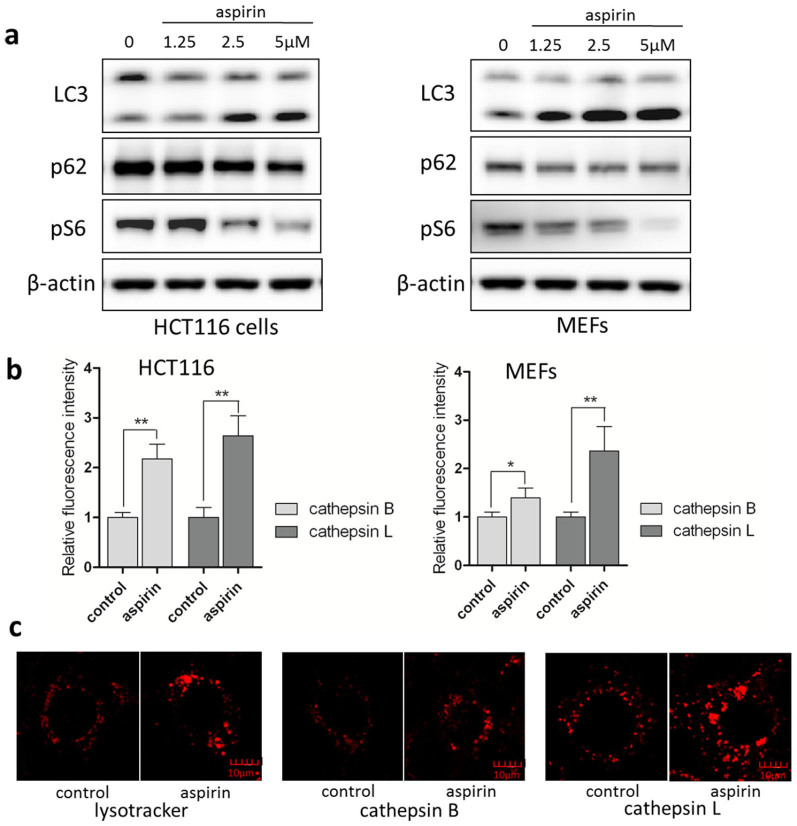
mTOR is a serine/threonine protein kinase serving as the convergent point for many of the upstream stimuli and pathways to regulate cell growth, cell proliferation, cell motility, cell survival, and protein synthesis38,39,40. In mammalian cells, there are two functionally distinct complexes: mTOR complex 1 (mTORC1) and complex 2 (mTORC2). mTORC1 mainly regulates cell growth and protein synthesis by phosphorylating two key translational regulators: eukaryote initiation factor 4E-binding protein (4EBP1) and S6 kinase (S6K)38. At present, it has been well established that mTOR is the key negative regulator of autophagy by suppressing the ULK1 (the mammalian homolog of Atg1 in yeast) complex consisting of ULK1, FIP200 and ATG 1341,42. Our pathway analysis showed that aspirin can influence the mTOR signaling and an aspirin binding peptide GPQTLK*ETSFNQAYGR of mTOR was also observed from our QA-ABPP result. To further evaluate the efficacy of aspirin treatment30, we conducted western blot to determine the changes in mTOR activity and autophagy markers. As shown in Figure 7a, aspirin treatment increased LC3-II level and decreased p62 level in a dose-dependent manner in both the HCT116 and the MEF cells, pointing to an increase in autophagy activity. Meanwhile, the effectiveness of aspirin was verified with the blockage of S6 phosphorylation (Ser235/236) seen in HCT116 cells and MEFs (Figure 7a). Our results show that in HCT116 and MEF cells, aspirin inhibited the downstream effector of mTORC1 (S6). These results are consistent with our targets and pathway analysis where aspirin induced the greatest changes in ribosome biogenesis.
Figure 7. Aspirin induced autophagy and activated lysosomal function in cells.
(a) Aspirin induced autophagy in a dose-dependent manner. HCT116 and MEFs cells were treated with different dosages of aspirin (1.25, 2.5 or 5 mM) for 16 hrs and cell lysates were prepared for western blot. β-actin was used as the loading control. (b) Cells were then loaded with Magic Red Cathepsin B or L reagent for 15 min. Fluorescence intensity of 10,000 cells per sample was measured using flow cytometry. Data were expressed as the ratio of treated cells to control cells for the relative signal intensity, as mean ± SD from three independent experiments; * P < 0.05, ** P < 0.01, derived from the Student's t-test. (c) Aspirin activated lysosomal function in MEFs cells. Cells were treated with aspirin (5 mM) for 16 hrs and then stained with LysoTracker Red DND-99 (50 nM) for 15 min. Scale bar 10 μm.
Activation of lysosomal function by aspirin
Lysosomes are cellular organelles found in all animal cells that contain acid hydrolase enzymes to digest cellular debris, damaged organelles and invaded microorganisms43,44. In the course of autophagy, lysosomes play an essential role in the maturation/degradation stage of autophagy, as the contents in the autophagosomes are eventually degraded in lysosomes after autophagosome-lysosome fusion45. Here, we used two techniques to test the changes of lysosomal activity by aspirin. Firstly, the enzymatic activities of lysosomal cathepsin B and L were measured. Significant increase was found in cells treated with aspirin (Figure 7b&7c). Secondly, a significant increase of LysoTracker staining was observed in MEF cells treated with aspirin (Figure 7c), indicating enhanced acidification of lysosome after aspirin treatment (reduced pH). As reported previously, mTOR suppression can activate lysosomal function in the course of autophagy46,47. Therefore, activation of lysosomal function by aspirin may be due to mTOR signaling inhibition as reflected in our pathway analysis.
Cellular distribution of aspirin probe
To show the cellular distribution of aspirin targets, we used confocal imaging to visualize probe-treated cells (Supplementary Figure 6). MEF cells were treated with Asp-P2, and then fixed with paraformaldehyde before permeabilizing with Triton X-100. Finally, the cells were conjugated to Cy3-azide by click chemistry for fluorescence imaging (colored in red). In cells treated with DMSO, no fluorescence signal was observed. Conversely, Asp-P2-treated cells showed high levels of fluorescence in the whole cell. Thus, our imaging results are consistent with our aspirin target localization results, which show that the aspirin targets are located in various parts of the cells.
Discussion
In this study, quantitative proteomics (iTRAQ) and the acid cleavable linker (DADPS) were combined for the first time to develop QA-ABPP for profiling both a probe's target proteins and its exact binding sites. The usage of DADPS only needs 2-hrs treatment with 5% formic acid to release the probe modified peptides. By using our QA-ABPP and aspirin-based probes, we have identified 1110 aspirin-acetylated proteins and 2,775 peptides bearing modification from 870 proteins by the aspirin probe. Among them, 523 proteins were confidently confirmed as the targets of aspirin. Moreover, we identified 8 different amino acid residues that are modified by aspirin: lysine, serine, arginine, histidine, threonine, tyrosine, tryptophan and cysteine. Besides lysine, serine and cysteine, our study is the first to report the aspirin modification on other amino acid residues. In total, about 82% of the modified residues were lysine, which indicated the approach using anti-acetyl lysine antibody could detect most of the proteins acetylated by aspirin though this approach omitted some acetylations on other residues and inevitably contained some endogenous acetylated proteins27,28. These aspirin-targeted proteins in our study are involved in various cellular processes, such as protein synthesis and autophagy process, which were experimentally proved. Coincidentally, during the preparation of our manuscript, Bateman et al.48 reported the identification of 120 potential aspirin targets, 94 of which were also identified by our approach. However, neither binding site information nor functional studies were reported in their work. Our study not only identified 429 additional aspirin targets, but also validated them with direct aspirin-induced acetylation sites, further supporting with functional studies. Thus, our study provides new molecular insight into this widely used small-molecule medicine. To the best of our knowledge, our study reported and confirmed the most comprehensive modification information for a single small molecule.
It is increasingly accepted that many drugs can target multiple proteins, and it has become a trend in drug research and development49. It is thus not surprising that aspirin, being an effective drug for multiple diseases, can target hundreds of proteins. However, we cannot rule out the possibility that some of the protein targets we identified could be non-specific modifications due to our experimental conditions, although the dosages we adopted in this study are comparable with what have been used in previous studies27,28,30,31,32,48. Future experiments involving different treatment time and dosage may help to further clarify this issue.
QA-ABPP may have wide applications in the field of chemical proteomics. This approach has some advantages over the existing methods. Firstly, by comparing the corresponding enrichment ratios, iTRAQ could differentiate the specific binding targets from the non-specific ones. Secondly, the probe targets proteins in an in vivo setting, more closely reflecting the drug effect under physiological conditions. Thirdly, the drug-modification sites can be directly confirmed with mass spectrometry in a high-throughput manner, avoiding tedious in vitro validation. Fourthly, the information about the drug-modification sites may further our understanding of the MOA of the drug. Lastly, the usage of DADPS only needs 2-hrs treatment with 5% formic acid to release probe-modified peptides. Formic acid is compatible with mass spectrometry and therefore does not need to be removed prior to LC/MS. On the other hand, it is the usage of formic acid that makes our current approach not applicable to compounds that are acid-labile. For these compounds, a potential solution is to change our current acid-cleavable linker to other types of linkers21,22,23,24,29.
Methods
In situ fluorescence labelling using Asp-1/2 and Acetyl-P
HCT116 cells were grown to 80-90% confluence, and switched to medium (1% DMSO) containing Asp-P1/P2 or Acetyl-P (1000 μM) for 12 hrs at 37°C and 5% CO2. Subsequently, the medium was removed, and cells were washed with PBS and detached with trypsin. The cell pellet was resuspended in PBS and washed before sonication in 150 µl of PBS to lyse the cells. The resultant cell lysate was cleared by centrifuging at 13,000 rpm for 30 min. Equal amounts (100 μg) of different treatment samples were used for subsequent fluorescent labelling. For each reaction, Cy3-azide (20 μM), TCEP (1 mM, 100 × fresh stock in water), TBTA ligand (100 μM, 100 × stock in DMSO), and CuSO4 (1 mM, 100 × stock in water) were added to the lysate. The samples were incubated at room temperature for 2 hrs. Next, clicked proteins were precipitated by acetone and air dried. 1× SDS loading buffer (100 μL) was added to dissolve the sample, and 50 μL of sample was separated by SDS PAGE with 12.5% polyacrylamide gel and visualized using a Typhoon 9410 laser scanner (GE Healthcare; Buckinghamshire, UK). Images were analyzed by TotalLab software. For competition assay, the HCT116 cell lysate were pretreated with aspirin (1 mM or 2 mM) for two hrs. Then the lysate were incubated for another 12 hrs together with aspirin probes (500 µM). Probe labeled proteomes were visualized by click conjugation to the Cy3-azide tag followed by SDS-gel separation and fluorescence scanning.
Cell labelling with Asp-1/2 for QA-ABPP study
In the subsequent QA-ABPP study, two biological duplicate of Asp-1/2 treated and two DMSO treated samples were pulled down and digested in parallel. The two biological duplicate of Asp-1/2 treated samples were labelled with iTRAQ reagents and quantified by iTRAQ ratios against the DMSO control samples. Briefly, HCT116 cells were grown to 80-90% confluence in T75 flasks. Used medium was then aspirated and the cells were washed twice with PBS. Asp-1/2 (1000 μM) in 10 ml medium (1% DMSO) was added to the cells in the flasks and incubated for 12 hrs in the CO2 incubator. Culture medium containing 1% DMSO was used as negative control. Subsequently, Asp-1/2 and DMSO-containing media were removed before the cells were washed with PBS and detached with trypsin. The cell pellet was resuspended in PBS, washed and lysed by sonication in PBS. The cell lysates were clarified by centrifugation at 13,000 rpm for 30 min followed by Bradford assay. Equal amount (5 mg) of cell lysates (4 probe treated and 2 DMSO treated samples) were used for subsequent click chemistry to conjugate proteins with the DADPS tag separately. For each reaction, DADPS (20 μM), TCEP (1 mM, 100 × fresh stock in water), TBTA ligand (100 μM, 100 × stock in DMSO), and CuSO4 (1 mM, 100 × stock in water) were added to the cell lysates and incubated at room temperature for 4 hrs. Next, clicked proteins were subjected to precipitation with acetone and air dried. Subsequently, the pellet was dissolved in 1 ml of 0.1% SDS in PBS and incubated with 50 μl of Streptavidin beads (Sigma-Aldrich) under gentle mixing for 2 hrs at room temperature.
On-beads digestion by trypsin
The beads were washed a total of 9 times; thrice with 1% SDS, followed by thrice with urea (6 M) and thrice with PBS. The extensively washed beads were re-suspended in 100 µl 25 mM Triethylammonium bicarbonate (TEAB) and 2 µl TCEP (100 mM stock solution) was added. The beads were placed in a 65°C heat block for 60 min. Next, 1 µl MMTS (200 mM stock solution) was added, and the samples left in the dark and allowed to react for 15 min at room temperature. After reduction and alkylation, trypsin (12.5 ng/µl) was added and incubated at 37°C overnight. The digested peptides were separated from the beads using a filter-spin column (GE Healthcare).
On-beads treatment with formic acid to cleave the probe modified peptide
For the pull down sample using Asp-P2, the washed and filtered beads which bear the binding peptides were further washed three times using 25 mM TEAB to remove the digested unmodified peptides. The beads were then incubated with 5% formic acid for 2 hrs to cleave the acid cleavable biotin tag. The resulting cleaved probe modified peptides were separated from the beads using a filter-spin column and identified using LC-MS/MS.
iTRAQ labelling of the peptides from on-beads digestion by trypsin
iTRAQ labelling was performed using iTRAQ Reagent kit (AB SCIEX; Foster City, CA) based on the vender's instruction manual with minor modifications. The two biological replicates of the negative control pull-down samples were labelled with iTRAQ reagent 113 and 114, respectively. Similarly, two biological replicate of digested pull-down samples by Asp-1/2 probes were labelled with reagent 117, 118 and 119,121 respectively. Briefly, the on-beads digested peptides were dried and reconstituted with equal volume of dissolution buffer (0.5 M TEAB). The peptides were then labelled with the respective iTRAQ reagents and incubated at room temperature for 2 hrs before all the samples were pooled together. The pooled iTRAQ-labelled peptides sample was subjected to strong cation exchange chromatography (SCX) using the iTRAQ Method Development Kit (AB SCIEX). The bound peptides were eluted with 5% ammonium hydroxide (NH4OH) in 30% methanol. The eluate was desalted using a Sep-Pak C18 cartridge (Waters, Milford, MA), dried and then reconstituted with 100 μl of diluent (98% water, 2% acetonitrile (ACN), 0.05% formic acid (FA)).
Nano LC−ESI-MS
The detailed methods for LC-MS/MS was described previously50. Briefly, separation of the iTRAQ labeled peptides or acid cleaved Asp-2 modified peptides was carried out on an Eksigent nanoLC Ultra and ChiPLC-nanoflex (Eksigent, Dublin, CA) in Trap Elute configuration. The samples were desalted with Sep-Pak tC 18 μ Elution Plate (Waters, Miltford, MA, USA) and reconstituted with 50 μl of diluent (98% Water, 2% ACN, 0.1% FA). A volume of 5 μl of the sample was loaded on a 200 μm × 0.5 mm trap column and eluted on an analytical 75 μm × 150 mm column. Both trap and analytical columns were made of ChromXP C18-CL, 3 μm (Eksigent, Germany). Peptides were separated by a gradient formed by 2% ACN, 0.1% FA (mobile phase A) and 98% ACN, 0.1% FA (mobile phase B): 5−12% of mobile phase B (20 min), 12−30% of mobile phase B (90 min), 30−90% of mobile phase B (2 min), 90% of mobile phase B (5 min), 90−5% of mobile phase B (3 min), and 5−5% of mobile phase B (13 min), at a flow rate of 300 nl/min.
The MS analysis was performed on a TripleTOF 5600 system (AB SCIEX, Foster City, CA, USA) in Information Dependent Mode. MS spectra were acquired across the mass range of 400–1250 m/z in high resolution mode (>30000) using 250 ms accumulation time per spectrum. A maximum of 20 precursors per cycle were chosen for fragmentation from each MS spectrum with 100 ms minimum accumulation time for each precursor and dynamic exclusion for 15 s with charge state between 2 to 4. For iTRAQ sample, tandem mass spectra were recorded in high sensitivity mode (resolution >15000) with “adjust CE when using iTRAQ Reagent” on.'
Protein identification and quantification with ProteinPilot™ software
The detailed method of ProteinPilotTM analysis was described previously51. Briefly, the protein identification and iTRAQ quantification were performed with ProteinPilot™ 4.5 (AB SCIEX) which uses the Paragon™ algorithm to perform database searches. The database used was the SwissProt_2013_09 (total sequence 540958). The search parameters used were as follows: Cysteine alkylation with MMTS; Trypsin Digestion; TripleTOF 5600; Biological modifications (All the protein modifications available in the ProteinPilotTM search engine are taken into consideration, which include most if not all the known protein modifications. A whole list of protein modifications is provided as Supplementary information). Redundancy was eliminated by the grouping of identified proteins using the ProGroup algorithm in the software. A decoy database search strategy was used to determine the false discovery rate (FDR) for protein identification. A corresponding randomized database was generated using the Proteomics System Performance Evaluation Pipeline feature in the ProteinPilot™ Software 4.5. In this study, a stringent cut-off threshold with total unused score >1.3 was adopted as the qualification criterion, which corresponded to a protein confidence level of >95% and a false discovery rate (FDR) of 0.33%.
Quantitative iTRAQ data analysis
iTRAQ ratio for each protein was calculated using unique peptides identified only, excluding peptides with miscleavage and peptides missing an iTRAQ reagent label. To reduce the likelihood of selecting the false-positive drug targets, we chose a stringent iTRAQ ratio equivalent to 2 as the cut-off threshold to identify specific protein targets for subsequent experiments. Moreover, the targets must be identified in both Asp-P1/P2 pull down results. Using these criteria, 1110 proteins were identified. The full list of the 1110 potential targets is shown in Supplematary Table 2.'
Asp-2 binding sites mapping
For the identification of drug modification sites, the mass spectra data were converted into Mascot generic format (MGF). The data was searched with Mascot 2.4.0 (Matrix Science). The database used was the SwissProt_2013_09 (total sequence 540958). The search parameters used were as follows: Trypsin Digestion; TripleTOF 5600; Cysteine alkylation of MMTS; Aspirin probe modified residue mass difference 237.24 amu was specified as variable modification at lysine, serine, arginine, histidine, threonine, tyrosine, tryptophan and cysteine residues. Peptide Mass Tolerance was 10 ppm and Fragment Mass Tolerance was 0.4 Da. A decoy database search strategy was used to determine the FDR for peptide identification. We have applied MOWSE score ≥ 30 to filter the identified peptide list, with FDR of 0.15% (Decoy:Normal = 10:6770) for the whole peptide list, and FDR of 0.086% (Decoy:Normal = 4:4645) for the peptides with Aspirin modification.
Pathway analysis of aspirin targets
The specific aspirin targets—identified using the QA-ABPP approach—were analyzed using the Ingenuity Pathway Analysis software (IPA; Ingenuity® Systems, Redwood city, CA). A spreadsheet containing the list of aspirin targets was uploaded into IPA. The software mapped each of the proteins to the repository of information in the Ingenuity Pathways Knowledge base. Molecular networks and canonical pathways regulated by these drugs targets were obtained using IPA core analysis.
Metabolic labelling of newly synthesized proteins with AHA
The cells with 70~80% confluency in a 6-well plate were washed with warm PBS and cultured in L-methionine-free DMEM for 30 min to deplete the intracellular methionine reserves. Following methionine depletion, the cells were labelled with AHA in 10% FBS DMEM (methionine-free) for designated time. After incubation, the cells were then harvested and fixed in 4% formaldehyde in PBS for 15 min and permeabilized with 0.25% Triton™ X-100 in PBS for 20 min at room temperature. Finally, the cells were used for the click reaction. For each reaction, Rhodamine B alkyne (10 μM), TCEP (1 mM, 100 × fresh stock in water), TBTA ligand (100 μM, 100 × stock in DMSO), and CuSO4 (1 mM, 100 × stock in water) were added into the suspended cells. The samples were incubated at room temperature for 2 hrs, and then the reaction cocktail was removed and the cells were washed once with 3% BSA in PBS. After fluorescence tagging, nascent protein synthesis was assessed by flow cytometry, and AHA signal intensity was determined in the FL3 channel. We quantified the fluorescence intensity of the cells and calculated the ratio of the fluorescence intensity of aspirin treated cells to that of the control cells.
Cathepsin B and L activity assay
MEFs and HCT116 cells were both cultured in 24-well plates. After the designated treatment, cells were further loaded with Magic Red cathepsin B or cathepsin L reagents for 15 min. Fluorescence intensities of cells per sample were measured by flow cytometry using the FACS cytometer (BD Biosciences; Franklin Lakes, NJ) or observed under a confocal microscope (Olympus Fluoview FV1000).
Estimation of intralysosomal pH using LysoTracker
The intralysosomal pH was estimated using LysoTracker, following manufacturer's instructions. The fluorescence intensity was observed under a confocal microscope (Olympus Fluoview FV1000) and representative cells were selected and photographed.
Western blotting
Cells were lysed in Laemmli SDS buffer (62.5 mM Tris, pH 6.8, 25% glycerol, 2% SDS, phosphatase inhibitor and proteinase inhibitor cocktails). An equal amount of protein was resolved by SDS-PAGE and transferred onto PVDF membrane. After blocking with 5% non-fat milk, the membrane was probed with designated primary and secondary antibodies, developed with the enhanced chemiluminescence method and visualized with ImageQuant LAS 500 (GE Healthcare).
Cellular imaging of aspirin-induced acetylation
MEFs cells were seeded to a Nunc™ Lab-Tek™ coverglass slide chamber (Thermo Fisher Scientific; Waltham, MA). After 16 hrs aspirin probe (Asp-2, 1 mM) labelling, cells were washed with PBS, then fixed with 4% paraformaldehyde in PBS for 15 min at room temperature before permeabilizing with 0.25% Triton™ X-100 in PBS for 15 min. Cells were washed with PBS and blocked with 1% BSA in PBS for 30 min, then clicked with Cy3 alkyne. The cells were examined and recorded using a confocal microscope (Olympus Fluoview FV1000) and representative cells were selected and photographed.
Other methods
Additional experimental information and results (chemistry and biology) are provided in Supplementary Methods.
Supplementary Material
Supporting Information
Supplematary Table 1
Supplematary Table 2
Supplematary Table 3
Supplematary Table 4
Supplematary Table 5
Supplematary Table 6
Supplematary Table 7
Supplematary Table 8_list of biological modification
Acknowledgments
J.W. and J.Z. were supported by NUS research scholarship. We would like to thank Prof. Kim Chu-Young for helpful discussions and proofreading of this manuscript. We acknowledge the financial support from the Initial Fund of High-level Creative Talents in Hainan Province (Chen Songbi).
Footnotes
The authors declare no competing financial interests.
Author Contributions J.W., C.-J.Z. and Q.L. designed the experiments. J.W., C-J.Z., J.Z., Y.H. and T.K.L. performed the experiments and analysed the data with Y.H., Y.M.L, S.C., S.N. assisted with the data analysis. J.W., C.-J.Z., J.Z., H.-M.S., and Q.L. wrote the manuscript.
References
- Eggert U. S. The why and how of phenotypic small-molecule screens. Nat. Chem. Biol. 9, 206–9 (2013). [DOI] [PubMed] [Google Scholar]
- Ziegler S., Pries V., Hedberg C. & Waldmann H. Target identification for small bioactive molecules: finding the needle in the haystack. Angew. Chem. Int. Ed. Engl. 52, 2744–92 (2013). [DOI] [PubMed] [Google Scholar]
- Speers A. E., Adam G. C. & Cravatt B. F. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 125, 4686–4687 (2003). [DOI] [PubMed] [Google Scholar]
- Ovaa H. et al. Chemistry in Living Cells: Detection of Active Proteasomes by a Two-Step Labeling Strategy. Angew. Chem. Int. Ed. 115, 3754–3757 (2003). [DOI] [PubMed] [Google Scholar]
- Evans M. J. & Cravatt B. F. Mechanism-based profiling of enzyme families. Chem. Rev. 106, 3279–3301 (2006). [DOI] [PubMed] [Google Scholar]
- Paulick M. & Bogyo M. Application of activity-based probes to the study of enzymes involved in cancer progression. Curr. Opin. Genet. Dev. 18, 97–106 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonović M. & Bogyo M. Activity Based Probes as a tool for Functional Proteomic Analysis of Proteases. Expert Rev. Proteomics. 5, 721–730 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher T., Pitscheider M. & Sieber S. A. Natural products and their biological targets: proteomic and metabolomic labeling strategies. Angew. Chem. Int. Ed. 49, 2680–2698 (2010). [DOI] [PubMed] [Google Scholar]
- Willems L. I. et al. Bioorthogonal chemistry: applications in activity-based protein profiling. Acc. Chem. Res. 44, 718–729 (2011). [DOI] [PubMed] [Google Scholar]
- Su Y. et al. Target identification of biologically active small molecules via in situ methods. Curr. Opin. Chem. Biol. 17, 768–75 (2013). [DOI] [PubMed] [Google Scholar]
- Wirth T. & Schmuck K. Duocarmycin analogues target aldehyde dehydrogenase 1 in lung cancer cells. Angew. Chemie 51, 2874–2877 (2012). [DOI] [PubMed] [Google Scholar]
- Okerberg E. S. et al. High-resolution functional proteomics by active-site peptide profiling. Proc. Natl. Acad. Sci. U. S. A. 102, 4996–5001 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam G. C., Burbaum J., Kozarich J. W., Patricelli M. P. & Cravatt B. F. Mapping enzyme active sites in complex proteomes. J. Am. Chem. Soc. 126, 1363–8 (2004). [DOI] [PubMed] [Google Scholar]
- Hekmat O., Kim Y.-W., Williams S. J., He S. & Withers S. G. Active-site peptide “fingerprinting” of glycosidases in complex mixtures by mass spectrometry. Discovery of a novel retaining beta-1,4-glycanase in Cellulomonas fimi. J. Biol. Chem. 280, 35126–35 (2005). [DOI] [PubMed] [Google Scholar]
- Speers A. E. & Cravatt B. F. A tandem orthogonal proteolysis strategy for high-content chemical proteomics. J. Am. Chem. Soc. 127, 10018–9 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E., Speers A. E. & Cravatt B. F. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat. protoc. 2, 1414–25 (2007). [DOI] [PubMed] [Google Scholar]
- Bantscheff M. et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 25, 1035–1044 (2007). [DOI] [PubMed] [Google Scholar]
- Sharma K. et al. Proteomics strategy for quantitative protein interaction profiling in cell extracts. Nat. Methods 6, 741–744 (2009). [DOI] [PubMed] [Google Scholar]
- Ong S. et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc. Natl. Acad. Sci. USA 106, 4617–4622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. et al. Quantitative chemical proteomics approach to identify post-translational modification-mediated protein-protein interactions. J. Am. Chem. Soc. 134, 1982–1985 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf G. C., Heydenreuter W. & Sieber S. A. Chemical proteomics: ligation and cleavage of protein modifications. Curr. Opin. Chem. Biol. 17, 110–7 (2013). [DOI] [PubMed] [Google Scholar]
- Szychowski J. et al. Cleavable biotin probes for labeling of biomolecules via azide-alkyne cycloaddition. J. Am. Chem. Soc. 132, 18351–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonović M., Verhelst S. H. L., Sorum M. T. & Bogyo M. Proteomics evaluation of chemically cleavable activity-based probes. Mol. Cell. Proteomics 6, 1761–70 (2007). [DOI] [PubMed] [Google Scholar]
- Geurink P. P. et al. A cleavable linker based on the levulinoyl ester for activity-based protein profiling. Angew. Chem. Int. Ed. Engl. 49, 6802–5 (2010). [DOI] [PubMed] [Google Scholar]
- Nishihara R. et al. Aspirin use and risk of colorectal cancer according to BRAF mutation status. JAMA 309, 2563–71 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X. et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N. Engl. J. Med. 367, 1596–606 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimuthu S. et al. Aspirin acetylates multiple cellular proteins in HCT-116 colon cancer cells: Identification of novel targets. Int. J. Oncol. 39, 1273–83 (2011). [DOI] [PubMed] [Google Scholar]
- Alfonso L. F., Srivenugopal K. S. & Bhat G. J. Does aspirin acetylate multiple cellular proteins? (Review). Mol. Med. Rep. 2, 533–537 (2009). [DOI] [PubMed] [Google Scholar]
- Yang Y.-Y., Ascano J. M. & Hang H. C. Bioorthogonal chemical reporters for monitoring protein acetylation. J. Am. Chem. Soc. 132, 3640–1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din F. V. N. et al. Aspirin inhibits mTOR signaling, activates AMP-activated protein kinase, and induces autophagy in colorectal cancer cells. Gastroenterology 142, 1504–15.e3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick J. C. H., van Santen M., van den Brink G. R., van Deventer S. J. H. & Peppelenbosch M. P. DNA array analysis of the effects of aspirin on colon cancer cells: involvement of Rac1. Carcinogenesis 25, 1293–8 (2004). [DOI] [PubMed] [Google Scholar]
- Pathi S. et al. Aspirin inhibits colon cancer cell and tumor growth and downregulates specificity protein (Sp) transcription factors. PLoS One 7, e48208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. A quantitative chemical proteomics approach to profile the specific cellular targets of andrographolide, a promising anticancer agent that suppresses tumor metastasis. Mol. Cell. Proteomics 13, 876–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P. L. et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 3, 1154–1169 (2004). [DOI] [PubMed] [Google Scholar]
- Strahl B. D. & Allis C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000). [DOI] [PubMed] [Google Scholar]
- Jenuwein T. & Allis C. D. Translating the histone code. Science 293, 1074–80 (2001). [DOI] [PubMed] [Google Scholar]
- Dieterich D. C. et al. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat. Protoc. 2, 532–40 (2007). [DOI] [PubMed] [Google Scholar]
- Zoncu R., Efeyan A. & Sabatini D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K. & Guan K.-L. Complexity of the TOR signaling network. Trends Cell Biol. 16, 206–12 (2006). [DOI] [PubMed] [Google Scholar]
- Sengupta S., Peterson T. R. & Sabatini D. M. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40, 310–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22, 132–9 (2010). [DOI] [PubMed] [Google Scholar]
- Hosokawa N. et al. Nutrient-dependent mTORC1 Association with the ULK1 – Atg13 – FIP200 Complex Required for Autophagy. Mol Biol Cell 20, 1981–1991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio J. P., Pryor P. R. & Bright N. A. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8, 622–32 (2007). [DOI] [PubMed] [Google Scholar]
- Saftig P. & Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–35 (2009). [DOI] [PubMed] [Google Scholar]
- Eskelinen E.-L. & Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim. Biophys. Acta 1793, 664–73 (2009). [DOI] [PubMed] [Google Scholar]
- Zhou J. et al. Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell Res. 23, 508–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H.-M. & Mizushima N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem. Sci. 39, 61–71 (2013). [DOI] [PubMed] [Google Scholar]
- Bateman L. & Zaro B. An Alkyne–Aspirin Chemical Reporter for the Detection of Aspirin-Dependent Protein Modification in Living Cells. J. Am. Chem. Soc. 135, 14568–14573 (2013). [DOI] [PubMed] [Google Scholar]
- Lu J.-J., Pan W., Hu Y.-J. & Wang Y.-T. Multi-target drugs: the trend of drug research and development. PLoS One 7, e40262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L. A. L. et al. High-performance graphene-titania platform for detection of phosphopeptides in cancer cells. Anal. Chem. 84, 6693–6700 (2012). [DOI] [PubMed] [Google Scholar]
- Ghosh D. et al. Identification of Key Players for Colorectal Cancer Metastasis by iTRAQ Quantitative Proteomics Profiling of Isogenic SW480 and SW620 Cell Lines. J. Proteome Res. 10, 4373–4387 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supplematary Table 1
Supplematary Table 2
Supplematary Table 3
Supplematary Table 4
Supplematary Table 5
Supplematary Table 6
Supplematary Table 7
Supplematary Table 8_list of biological modification