Abstract
Collective cell migration plays a crucial role in several biological processes, such as embryonic development, wound healing, and cancer metastasis. Here, we focused on collectively migrating Madin-Darby Canine Kidney (MDCK) epithelial cells that follow a leader cell on a collagen gel to clarify the mechanism of collective cell migration. First, we removed a leader cell from the migrating collective with a micromanipulator. This then caused disruption of the cohesive migration of cells that followed in movement, called “follower” cells, which showed the importance of leader cells. Next, we observed localization of active Rac, integrin β1, and PI3K. These molecules were clearly localized in the leading edge of leader cells, but not in follower cells. Live cell imaging using active Rac and active PI3K indicators was performed to elucidate the relationship between Rac, integrin β1, and PI3K. Finally, we demonstrated that the inhibition of these molecules resulted in the disruption of collective migration. Our findings not only demonstrated the significance of a leader cell in collective cell migration, but also showed that Rac, integrin β1, and PI3K are upregulated in leader cells and drive collective cell migration.
Collective cell migration plays a pivotal role in many biological events as it is observed in embryogenesis, wound healing, and collective cancer metastasis1,2,3. Previous studies have shown the presence of specialized “leader” cells or “tip” cells at the leading edges of colonies of collectively migrating cells4,5,6,7,8,9. It has also been shown that emerging leader cells and the subsequent migration of leader cells is accompanied by nearby cells called “follower” cells that cause collective cell migration. However, the mechanisms by which leader cells migrate in front of follower cells and the differences between leader cells and follower cells are still unclear.
We previously demonstrated that Madin-Darby canine kidney (MDCK) cells cultured on a soft collagen gel exhibit more cohesive movement as opposed to cultures on a stiff glass substrate5. Moreover, leader cells extend large lamellipodia and exhibit clear front-rear polarity. It is apparent that leader cells play an important role in the cohesive movement of MDCK cells; however, the specific characteristics of leader cells and the relationship between leader cells and their neighboring “follower” cells have not been widely investigated. Therefore, we analyzed leader cells emerging from MDCK cell cultures on a soft collagen gel.
It is well-known that a large number of molecules contribute to cell migration10. Of these molecules, Rac, a small GTPase protein, is a key regulator of actin dynamics11 and cell migration12. Previous reports showed that Rac plays a crucial not only in single cell migration but also in collective cell migration13,14,15,16. In addition, another group demonstrated that Rac activity in Drosophila oogenesis is strictly regulated to movement towards the direction of the collectively migrating border cells14,17. Neural crest (NC) cell migration is one well-characterized model of in vivo collective cell migration18. In this system, NC cells are sensitive to chemokine stromal-derived factor1 (SDF1) and collectively migrate towards the source of SDF1. When NC cells move as a cell mass, cell-cell contact between each NC cell regulates Rac1 activity and promotes directional migration15.
Integrins are transmembrane hetero-dimeric receptors for extracellular matrix (ECM) proteins, i.e., collagen and laminin, which regulate persistent cell migration and cancer invasion19,20. Previous studies showed that integrin β1 is expressed in pro-migratory cells at the leading edge of primary melanoma explants cultured in 3D collagen21. The relationship between Rac and integrins has been discussed. Because Rac guanine-nucleotide exchange factor (GEF) Tiam1 is recruited to integrin β1 complexes through adaptor protein 14-3-3ζ22, Rac is known as a downstream signaling molecule of integrin β1. In addition, conversely, some studies showed that integrins are also regulated by Rac12,23. Phosphoinositide 3-kinase (PI3K) is a major contributor to cell migration, polarity, and survival24,25,26. PI3K also regulates Rac activity by producing PtdIns(3,4,5)P3, which activates Rac GEFs24. Recently, the spatial distribution of active PI3K, active Rac, and integrin α5 and their relationship in cells with one end free under the stimulation of platelet-derived growth factor (PDGF) was discussed13. However, the contribution of these molecules to collective migration of MDCK cells remains unclear.
Here, we demonstrate that leader cells are essential for the collective migration of MDCK cells. Furthermore, we showed that Rac, integrin β1, and PI3K are upregulated in leader cells and that the inhibition of these molecules disrupts collective migration. Finally, we showed that these molecules relate to each other, forming a signaling pathway in leader cells. The findings of this study, in part, reveal the mechanisms of collective cell migration and cohesive cancer cell invasion.
Results
Elimination of a leader cell disrupts collective cell migration
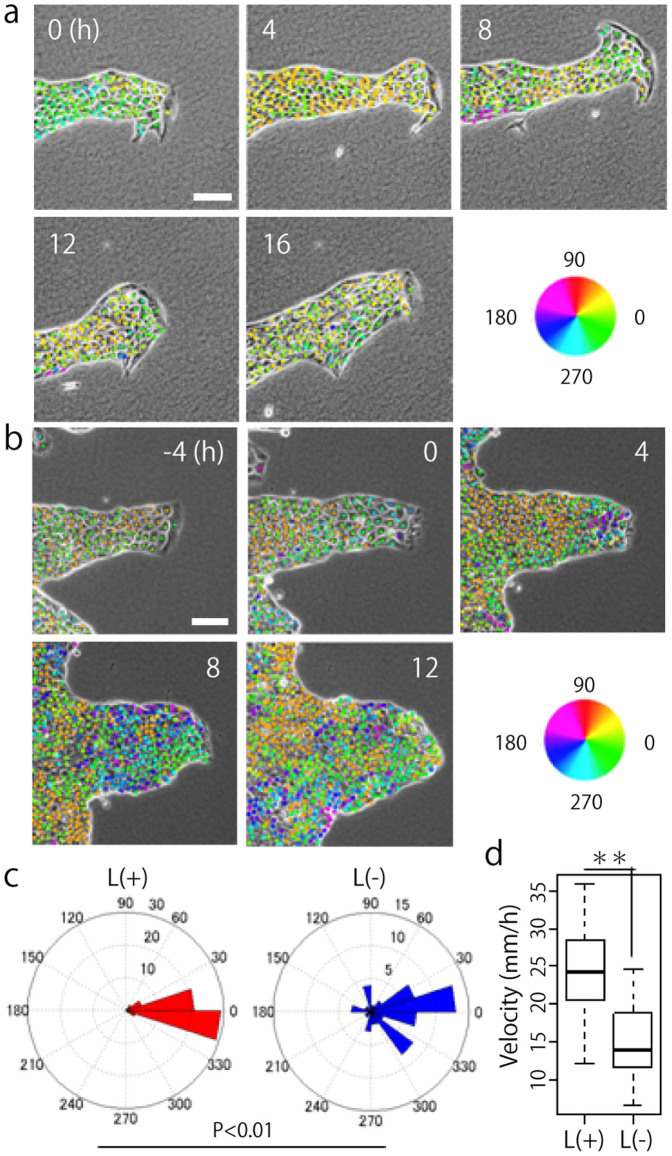
Time-lapse imaging was used to examine MDCK cells in which a leader cell was observed when cultured on a collagen gel. A single leader cell in a colony, exhibiting a “finger-like” morphology was distinguishable (Fig. 1a and Movie 1). A mass of cells directed by a leader cell was observed to migrate in a single direction for 16 h. The hue of the dots in the figure represents the direction of migration of each cell (Fig. 1). To investigate the role of a leader cell in collective cell migration, we attempted to remove the leader cell on a collagen gel by micromanipulation and performed time-lapse imaging for 16 h to compare the movement of each cell with control data. The time at which the leader cell was killed was designated as 0 h, and after the death of the leader cell, the migration of follower cells in the same direction stopped and their movement gradually became random and slower (Fig. 1b, c, d and Movie 2). Thus, our results indicate that leader cells play a crucial role in the collective migration of MDCK cells on collagen gel.
Figure 1. Leader cells play a crucial role in collective migration of MDCK cells.
(a) MDCK cells moving collectively as directed by a leader cell. The migratory direction of each cell is shown by colored dots. (b) The direction of the follower cells moving after removal of the leader cell at 0 h. (a and b) Scale bar: 100 μm. (c) Quantification of migration direction (N = 3). In each experiment, 20 follower cells were manually tracked. L(+): before and L(−): after a leader was killed. P < 0.01 by Watson's two-sample test of homogeneity. (d) Quantification of migratory velocity. **P < 0.01 by Student's t-test. The mean value ± S.D.
Rac, integrin β1, and PI3K are upregulated in leader cells
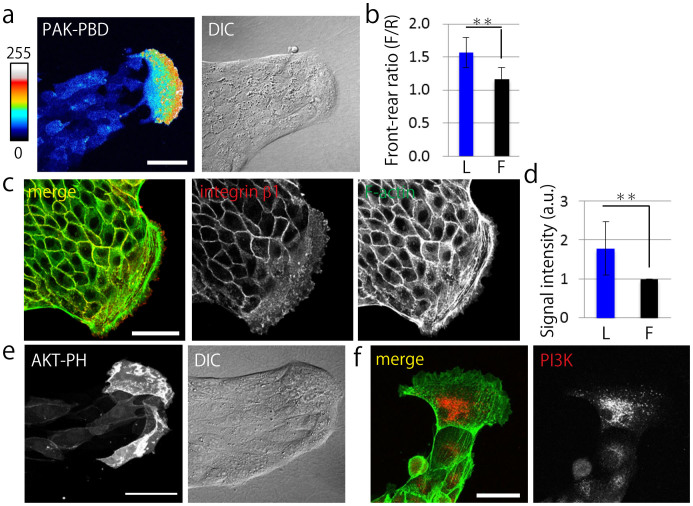
Next, we determined the difference between leader cells and follower cells by evaluating the expression of proteins involved in cell migration and polarization. To investigate the distribution of active Rac, we developed a MDCK PAK-PBD-AcGFP transgenic cell line (See Methods). The typical collective migration of MDCK cells expressing PAK-PBD-AcGFP showed that PAK-PBD probe was clearly localized at the leading edge of a leader cell (Fig. 2a). Comparison of the intensity of the localized probe in leader cells and follower cells by using image analysis (Fig. 2b) showed that the leader cells had more apparent polarity of active Rac than the follower cells. Immunofluorescence staining indicated that integrin β1 was highly localized in the lamellipodia of leader cells (Fig. 2c, Supplementary Fig. 1a) and the signal was brighter in leader cells than in follower cells (Fig. 2d). Moreover, to examine integrin β1 activation in the lamellipodia of leader cells, we performed immunofluorescence staining of phosphorylated FAK, because FAK phosphorylation is induced by integrin β1 activation27,28. We observed that phosphorylated FAK (Y397 and Y925) was clearly localized in the leading edge of the leader cells (Supplementary Fig. 1b and c) and co-localized with integrin β1 (Supplementary Fig. 1c). We also investigated the localization of AKT-PH-GFP as an indicator of active PI3K29. Collective cell migration was observed and AKT-PH-GFP probe was clearly localized at the leading edge of a leader cell (Fig. 2e), which showed a localization pattern similar to that reported previously30. In addition, fluorescence live cell imaging of MDCK AKT-PH-GFP cells showed translocation of the AKT-PH-GFP probe and a brighter fluorescence signal was observed in the ruffling sites of the lamellipodia of leader cells (Supplementary Fig. 2a and b). Moreover, PI3K expression was observed by immunofluorescence staining (Fig. 2f). To compare the signal intensity of PI3K in leader cells with that in follower cells, we conducted staining of GAPDH as a cytoplasmic staining control (Supplementary Fig. 3a). We found that PI3K expression was higher in the leader cells (Fig. 2f, Supplementary Fig. 3a and b). Further, we stained phosphorylated Akt to investigate PI3K activation, which revealed the localization of phosphorylated Akt at the leading edge of the leader cell (Supplementary Fig. 3c). These findings demonstrate that Rac, integrin β1, and PI3K were clearly localized and upregulated in the leader cells.
Figure 2. Rac, integrin β1, and PI3K are upregulated in leader cells.
(a) Fluorescent image of the typical finger-like structure exhibited by MDCK cells expressing PAK-PBD-AcGFP. DIC: Differential interference contrast image. Fluorescent intensity is represented with colors from hot to cold. (b) Quantification of the ratio of the mean fluorescent intensity of PAK-PBD-AcGFP determined in a region of the leading front (F) divided by the signal intensity of the trailing rear (R) (N = 15). (c) Fluorescent images of integrin β1 stained by AIIB2. Red: integrin β1. Green: F-actin. (d) Quantification of the fluorescent intensity of integrin β1 at the bottom of cells. L: leader cells, F: follower cells. The signal intensity of leader cells was normalized by that of the follower cells (N = 8). (e) Fluorescent images of collectively migrating MDCK cells expressing AKT-PH-GFP. (f) Fluorescent images of PI3K. Green: F-actin, Red: PI3K. (a, c, e, and f) Scale bars: 50 μm. (b and d) The mean value ± S.D. **P < 0.01 by Student's t-test.
Rac in a leader cell is activated by integrin β1 and PI3K
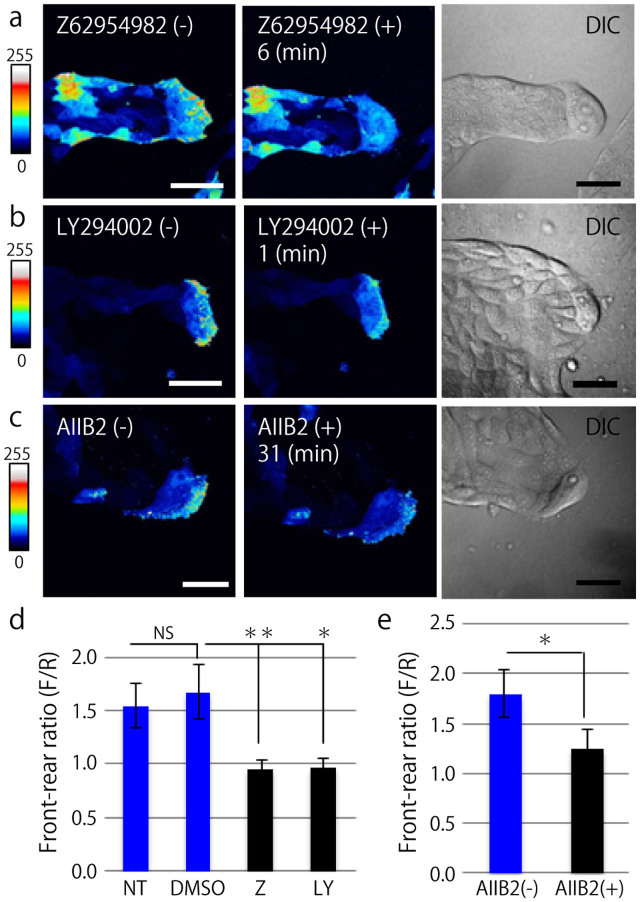
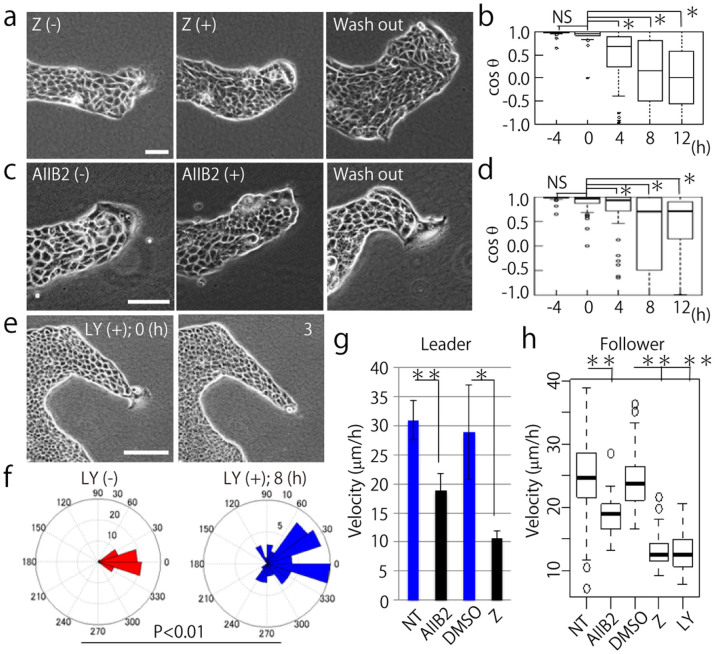
The interaction of Rac, integrin β1, and PI3K was evaluated using live-cell imaging of MDCK cells expressing PAK-PBD-AcGFP. For a positive control, Rac1 in MDCK cells undergoing collective migration was inhibited using a Rac1 inhibitor (Z62954982); as a result, the high fluorescent signal at the leading edge of the leader cell was immediately decreased (Fig. 3a), indicating that active Rac1 downregulation could be observed using the PAK-PBD-AcGFP probe. We also verified the effects of DMSO treatment as a negative control, where the fluorescent signal was not altered (Supplementary Fig. 4). Next, PI3K and integrin β1 activity was inhibited using an inhibitor (LY294002) and an inhibitory antibody (AIIB2), respectively. In the case of PI3K inhibition, the polarized signal of active Rac rapidly disappeared (Fig. 3b), while the disappearance of the active Rac signal because of integrin β1 inhibitory antibody took approximately 30 min (Fig. 3c). The mean value of the intensity ratio was calculated by dividing the intensity of the signal at the migrating front of a leader cell by one in the rear (Fig. 3d and e); consequently, we determined that Rac in a leader cell was regulated by both integrin β1 and PI3K.
Figure 3. Rac in a leader cell is regulated by PI3K and integrin β1.
(a, b, and c) Representative images of MDCK cells expressing PAK-PBD-AcGFP before (left) and after inhibitor treatment (center). The right panels show the DIC images before inhibitor treatment. Scale bars: 50 μm. (a) Z62954982 (Rac1 inhibitor, 100 μM). (b) LY294002 (PI3K inhibitor, 10 μM). (c) AIIB2 (integrin β1 inhibitory antibody, 1.5 μg/ml). (d) Quantification of the ratio of the mean fluorescent signal of PAK-PBD-AcGFP determined in the leading front area (F) divided by the fluorescent intensity of the trailing rear (R). NT: before inhibitor treatment (non-treated), DMSO: negative control, Z: 6 min after treatment of Z62954982, LY: 1 min after treatment of LY294002 (at least N = 3). (e) Quantification of the front-rear signal intensity ratio of MDCK cells expressing PAK-PBD-AcGFP before (AIIB2(−)) and 31 min after (AIIB2(+)) AIIB2 treatment (N = 3). (d and e) The mean value ± S.D. NS: non-significant, **P < 0.01, *P < 0.05 by Student's t-test.
The activation of PI3K in a leader cell is mediated by Rac1 but not by integrin β1
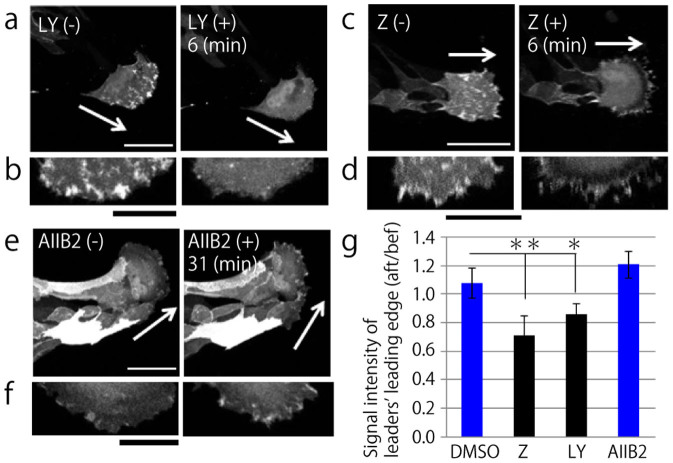
Since PI3K is known to be regulated by integrins24 and has a positive feedback loop with Rac31,32, we investigated PI3K signaling by using live-imaging of MDCK cells expressing AKT-PH-GFP. PI3K inhibition with LY294002 was performed to verify the experimental condition. Before inhibition of PI3K, the AKT-PH-GFP probe was localized to the migrating edge of a leader cell; however, following treatment with the inhibitor, active PI3K signal disappeared (Fig. 4a and b). We also verified that DMSO treatment did not influence active PI3K localization (Supplementary Fig. 5). Next, we elucidated the influence of Rac or integrin β1 inhibition on active PI3K localization. Rac1 inhibition resulted in the disappearance of the fluorescent signal for active PI3K (Fig. 4c and d). In contrast, treatment with AIIB2 to inhibit integrin β1 produced no effect (Fig. 4e and f). Comparison of the fluorescent intensity of the leading edge in leader cells before and after inhibitor treatment (Fig. 4g) showed that Rac and PI3K formed a positive feedback loop, whereas integrin β1 and PI3K regulated Rac activity individually.
Figure 4. PI3K in a leader cell is also regulated by Rac1 but not by integrin β1.
(a, c, and e) Images obtained by live imaging of MDCK cells expressing AKT-PH-GFP, before inhibitor treatment (left panel) and after inhibitor treatment (right panel). Scale bars: 50 μm. (a) LY294002 (PI3K inhibitor, 10 μM). (c) Z62954982 (Rac1 inhibitor, 100 μM). (e) AIIB2 (integrin β1 inhibitory antibody, 1.5 μg/ml). (b, d, and f) Magnified images of the leading edges in a leader cell are shown (a, c, and e, respectively). Left: before inhibitor treatment, right: after inhibitor added. Scale bars: 25 μm. (g) Quantification of the ratio of the mean fluorescent intensity of AKT-PH-GFP described in the leading front area after the inhibitor was added (aft) divided by the fluorescent intensity of the same region before inhibitor treatment (bef). DMSO: negative control, Z: 6 min after Z62954982 treatment, LY: 6 min after LY294002, AIIB2: 31 min after AIIB2 treatment. **P < 0.01, *P < 0.05 by Student's t-test. The mean value ± S.D.
Rac, integrin β1, and PI3K are essential for collective cell migration
Next, we investigated how the inhibition of Rac, integrin β1, and PI3K affects collective motion. Time-lapse imaging was performed to characterize the effect of each inhibitor on cell morphology and migration direction and velocity. First, we performed Rac1 inhibition by using an inhibitor Z62954982, which resulted in the shrinking of leader cells. However, after removing the Rac1 inhibitor, the leader cell returned to its previous state with extended lamellipodia and led to collective cell migration (Fig. 5a and Movie 3). After blocking Rac1 function, the migratory direction of the follower cells became diverse (Fig. 5b and Supplementary Fig. 6a). Blockage of integrin β1 function showed similar effects (Fig. 5c and d, Supplementary 6b, and Movie 4), but in case of inhibited PI3K function, death or shrinkage of the leader cell was observed (Fig. 5e, Supplementary Fig. 7a, and Movie 5). On performing annexin V and Hoechst 33342 staining, we found that the death of the leader cells was due to apoptosis (Supplementary Fig. 7b and c). Although, apoptosis occurred particularly in the leader cells, it was not dose-dependent (Supplementary Fig. 7d and e). Moreover, we measured cell survival on collagen-coated glass surface because the cells cultured on the glass showed elongated shape and morphology, similar to the leader cells cultured on collagen gel. We concluded that the apoptosis of the leader cells on a collagen gel does not depend on its elongated shape (Supplementary Fig. 8a and b). After PI3K inhibition, follower cells moved more randomly (Fig. 5f). The migratory velocity of both leader and follower cells decreased by the inhibition of Rac1, integrin β1, and PI3K (Fig. 5g and h). In addition, the velocity of the follower cells after inhibition was widely distributed compared to the velocity of the inhibited leader cells. No effect on collective migration was observed in the cells treated with DMSO as a negative control (Supplementary Fig. 9a, b, and c).
Figure 5. Inhibition of Rac1, integrin β1, and PI3K disrupts collective migration of MDCK cells.
(a, c, and e) Time-lapse phase contrast images of migrating MDCK cells. (−) indicates before and (+) represents after inhibition. (a) Rac1 inhibition with Z62954982 (100 μM). (c) integrin β1 inhibition with AIIB2 (1.5 μg/ml). (e) PI3K inhibition with LY294002 (10 μM). Wash out means washing or removal of the inhibitor with fresh medium. Scale bars: 100 μm. (b and d) The angles between the migration direction of leader cells and follower cells treated with inhibitors are shown by cosθ. (b) Z62954982 (100 μM) and (d) AIIB2 (1.5 μg/ml) was treated at time 0. NS: non-significant, *P < 0.01 by Wilcoxon rank sum test. Twenty follower cells were randomly chosen in each experiment (N = 3). The mean value ± S.D. (f) Migratory direction of follower cells before and after PI3K inhibitor treatment. Twenty follower cells were analyzed from each experiment (N = 3). NS: non-significant, *P < 0.01 by Watson's two-sample test of homogeneity. (g) The migration velocity of leader cells under inhibitor treatment. NT: before treatment. AIIB2: integrin β1 inhibitor (1.5 μg/ml), DMSO: negative control, Z: Rac1 inhibitor (Z62954982, 100 μM) (at least N = 3). **P < 0.01, *P < 0.05. The mean value ± S.D. (h) The migration velocity of follower cells with each inhibitor. The inhibitor concentrations were the same as (g), and the PI3K inhibitor (LY294002) concentration used was 10 μM. Twenty follower cells were randomly chosen from 1 image (N = 3). **P < 0.01. The mean value ± S.D.
After that, we investigated whether the finger-like morphology was shaped or not under conditions of Rac1, integrin β1, and PI3K inhibition. To generate colonies of identical size on a collagen gel, we developed the ring assay (Supplementary Fig. 10a and Methods). When the evaluated proteins were inhibited from the beginning of culturing, immediately after the rings were removed, the number of leader cells at the top of the fingers decreased (Supplementary Fig. 10b). The colonies formed by the ring became smaller and smoother than the DMSO control (Supplementary Fig. 10c). These results demonstrate that Rac1, integrin β1, and PI3K were essential for collective cell migration.
Migration of leader cells is dependent on Rac1 activity
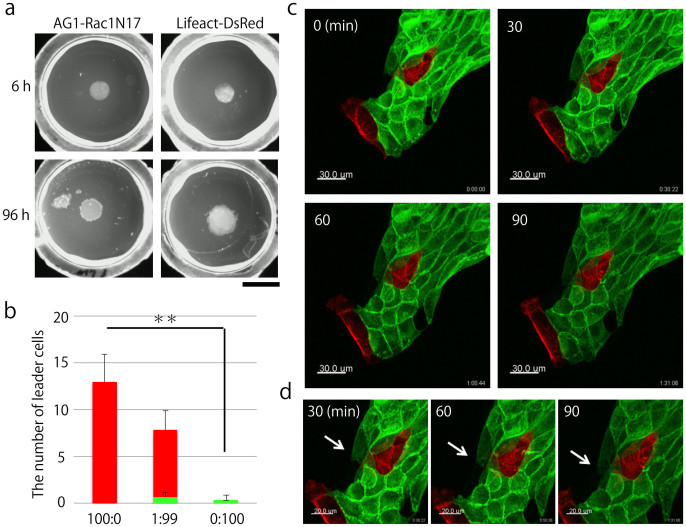
Finally, we investigated the role of Rac1 in collective migration by using a dominant-negative form of Rac1 (Rac1N17). We developed MDCK AG1-Rac1N17 cells and MDCK Lifeact-DsRed cells that were used as plasmid control cells (Supplementary Fig. 11a). The colony edge of MDCK AG1-Rac1N17 cells cultured on a collagen gel was dense because AG1-Rac1N17-expressing cells could not efficiently migrate on a collagen gel compared with MDCK Lifeact-DsRed cells (Fig. 6a and Supplementary Fig. 11b). First, we used ring-assay and counted the number of leader cells generated from MDCK Lifeact-DsRed cells per dish. On an average, approximately thirteen leader cells emerged from the colonies (13 ± 2.94, N = 4) (Fig. 6b). Next, we mixed MDCK AG1-Rac1N17 cells with MDCK Lifeact-DsRed cells at a ratio of 99:1 and cultured these cells on a collagen gel. Although approximately seven leader cells (7.17 ± 2.04, N = 6) originating from Lifeact-DsRed-expressing cells emerged per dish, few leader cells (0.667 ± 0.516, N = 6) were also derived from the MDCK AG1-Rac1N17 cells (Fig. 6b). When we cultured only MDCK AG1-Rac1N17 cells on a collagen gel, we rarely found AG1-positive leader cells (0.333 ± 0.516, N = 6) (Fig. 6b). Finally, we performed fluorescent live imaging of mixed culture cells. We observed that a leader cell derived from a Lifeact-DsRed-expressing cell led follower cells (Fig. 6c, upper panels). Moreover, we also observed that an AG1-Rac1N17-expressing follower cell elongated and migrated behind the leader cell (Fig. 6c, lower panels). These results showed that migration of a leader cell was dependent on Rac1 activity.
Figure 6. Activation of Rac1 in leader cells, but not in follower cells, is crucial for collective migration.
(a) Expansion of MDCK AG1-Rac1N17 and MDCK Lifeact-DsRed cells cultured on collagen gels. Upper panels show the colony morphologies observed immediately after the rings were removed, while lower panels show the morphologies observed 96 h after the rings were removed. Bar: 1 cm. (b) The number of leader cells, each distinguished by a different color. Red represents MDCK Lifeact-DsRed cells and green represents MDCK AG1-Rac1N17 cells. MDCK Lifeact-DsRed and MDCK AG1-Rac1N17 cells were mixed at the following ratios: 100:0 (N = 4), 1:99 (N = 6), and 0:100 (N = 6). The number of total seeded cells was 105 cells. **P < 0.01 by Welch's t-test. Results are shown as mean value ± S.D. (c) Live cell imaging of mixed culture cells at the following ratio: MDCK Lifeact-DsRed cells:MDCK AG1-Rac1N17 cells = 1:99. The number of total seeded cells was 105 cells. Bar: 30 μm. (d) Magnified images of (c). Arrows indicate an elongating follower cell expressing AG1-Rac1N17. Bar: 30 μm.
Discussion
In this study, we demonstrated a role for leader cells in the collective cell migration of MDCK cells and the activated signaling in leader cells. Though collective cell migration is a requisite process for maintaining life, analysis of the mechanism of this cohesive migration is insufficient.
Several marker proteins that determine the heterogeneity of front cells and rear cells were previously characterized4. One of these proteins, integrin β1, was evaluated in a series of studies addressing collective cancer invasion8,21,33. Although in previous reports, integrin β1 was highly expressed in the front cells of cancer collectives, the mechanism for integrin β1's role in front cells was not well understood. Here, we showed that Rac plays a pivotal role in leader cells, and it is regulated by integrin β1. We hypothesized that PI3K was an intermediate protein between integrin β1 and Rac; however, PI3K regulates Rac independently from integrin β1 in a leader cell. Tiam1, which is a Rho-GEF for Rac, was shown to activate Rac by a PI3K-independent mechanism34. In other studies, Rac and PI3K were shown to form feed-forward loops31,32. In our results, blockage of Rac1 decreased the active PI3K signal, and vice versa. Another study showed that Ras is essential for directional migration of Dictyostelium through regulation of PI3K35. Although we did not elucidate the upper signals of PI3K in this study, Ras might be an appropriate candidate for PI3K activator. Taken together, our results suggest that the feed-forward loop between Rac and PI3K is established in a leader cell, and that it drives leader-cell migration.
Inhibition of Rac1 and integrin β1 causes morphological changes in leader cells, and blockage of PI3K occasionally results in leader cell apoptosis. In addition, apoptosis by inhibiting PI3K is specifically observed in leader cells. PI3K is a well-characterized contributor to cell survival26. Recently, an anti-apoptosis protein, Bcl-2, was shown to be highly expressed in the leading cells of cancer collectives8. As such, proteins involved in cell survival may not only regulate survival, but also leader cell migration.
In our model, leader cells were represented by large lamellipodia, and active Rac and active PI3K were clearly localized in the leading edge of the leader cell without any additional growth factor stimuli13. However, the question as to how leader cells emerged from the colony still remained. When we killed a leader cell using a micromanipulator, another cell from the population of followers sometimes became a leader cell and led neighboring cells (data not shown). For example, in the case of Drosophila oocyte border cell migration, a leading border cell took the place of leading position by other border cells36. However, in our model, a leader cell had fibroblast-like large cell body and was not replaced by another cell. Therefore, on becoming a leader cell, a feedback loop might be responsible for the maintenance of leader cell characteristics in our system.
Recently, M. Reffay et al. showed that collectively migrating leader cells exert traction force via RhoA and phosphorylated MLC and by using this force leader cells drag finger-like morphology of follower cells16. Moreover, in that study, the data illustrated that activated Rac1 is not a primary contributor to the migration of leader cells in the collective migration16. Therefore, our results may be seen as controversial. In our study, we inhibited Rac1 by using a pharmacological inhibitor, Z62954982, which is a novel inhibitor that is characterized as more powerful than NSC2376637. Moreover, to further assess the role of Rac1 in leader cells and collective migration, we developed and characterized the MDCK AG1-Rac1N17 cell line in our system. Compared with MDCK Lifeact-DsRed cells, MDCK AG1-Rac1N17 cells rarely became leader cells (Fig. 6b). Moreover, fluorescent live cell imaging showed that a leader cell expressing Lifeact-DsRed led follower cells expressing AG1-Rac1N17 (Fig. 6c). From these results, we determined that the migration of leader cells is highly dependent on Rac1 activity, at least in our system. In addition to our results, T. Omelchenko, et al., who were the first to find leader cells, showed that treatment of Y27632 or expression of dominant-negative RhoA induced many edge cells to form a leader-like morphology7. This result is consistent with our results. Considering the results of our study and that of previous studies7,16, migration of leader cells may be regulated by several small GTPases in a highly complex manner.
Recent studies showed that polarity of collectively migrating cells is established through cell-cell contact or cell-cell adhesion13,15,38,39,40. Based on this notion, because leading cells have a free edge, leader cells may extend lamellipodia towards the opposing side of cell-cell contact. The outer cells of the finger-like shapes also have a large free edge, which may suggest that there is a restriction mechanism for extending the lamellipodia of followers towards another direction other than that of migrating leader cells. Recently, M. Reffay et al. clearly showed one of the restriction mechanisms by using laser ablation to cut the thick actin cables along the side of the finger-like structures16. Additionally, it is well known that in collectively migrating epithelial sheets, the cells behind the leading edge extend cryptic lamellipodia41. Thus, it is important to clarify how these cryptic lamellipodia are regulated in order to understand the origin of cell collectiveness. Using a light-mediated Rac activation technique, it was shown that focal activation of Rac is crucial for correct migration of Drosophila border cells17, and cell-cell communication for regulating particular activation of Rac is controlled by Rab11 and Moesin14. On the other hand, in a study using a single cell, membrane tension was shown to play an important role in preventing the extension of an additional front42. Therefore, both protein interaction and mechanical stimuli may control the formation of the migrating edge in the scale of the “finger”.
The mechanism for collectively persistent migration is still unknown. Two specific models, a chemotactic model and a mechanical model, have been discussed43,44,45,46,47. The chemotactic model is actively studied using in vivo models, such as a lateral line of Zebrafish48,49,50 or the neural crest of Xenopus15. In these systems, the characterized chemokine, i.e. SDF1, guides groups of cells. The mechanical model is examined using in vitro experimental models to show in expanding epithelial monolayers how each cell migrates along orientations of maximum principal stress43. In our study, we showed that elimination of a leader cell from the migrating collective disrupts collective cell migration. Furthermore, inhibition of Rac1, integrin β1, and PI3K, which were shown to be highly localized in leader cells, also disrupted collective cell migration. These results may indicate that pulling forward by the migrating leader cells is a mechanical trigger for subsequent migration of follower cells16,51. Although the migration velocity of follower cells also decreased with the inhibitor, some population of follower cells still moved faster than the leader cells. Without dragging by a leader cell, these migrating cells appeared to lose directionality, and collective cell migration was completely disrupted.
In summary, we showed the importance of leader cells in collective cell migration of MDCK cells on a collagen gel. Furthermore, Rac, integrin β1, and PI3K were upregulated and shown to be related to each other in a leader cell as well as involved in collective cell migration. These results partially clarified the mechanisms of cohesive cell movement and collective cancer invasion led by leader cells.
Methods
Cell culture and ring assay on a collagen gel
MDCK cells were purchased from Riken Cell Bank and cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma, St. Louis, MO) containing 1% antibiotic/antimycotic solution (Sigma) and 10% fetal bovine serum (FBS; Equitech-Bio, Inc., Kerrville, TX). Cells were maintained in a humidified incubator at 37°C and 5% CO2. For time-lapse imaging, immunofluorescence staining, and live cell imaging, we prepared a 1.6 mg/ml collagen type I gel with Cellmatrix type I-P (Nitta Gelatin Inc., Osaka, Japan). To form a circular monolayer colony, a hand-made plastic ring (diameter, 5 mm) was placed on a collagen gel. The cell suspension obtained after trypsinization was dropped within the ring and incubated for 6 h. The ring was removed, and unattached cells were washed away (Supplementary Fig. 8a). After 2–3 days, several leader cells were observed in each dish.
Reagents
Rac1 inhibitor Z6295498237 (Calbiochem, La Jolla, CA) and PI3K inhibitor LY294002 (Sigma) were reconstituted in dimethyl sulfoxide (DMSO) for use in experiments. A monoclonal antibody, AIIB2, obtained from the Developmental Studies Hybridoma Bank at the University of Iowa was used as a function-blocking antibody against integrin β1. Alexa Fluor-488 phalloidin (Invitrogen, Carlsbad, CA) was used for immunofluorescence staining of F-actin. The PI3K antibody was purchased from BD (610045; BD Biosciences, San Jose, CA). AlexaFluor-546 goat anti-mouse IgG (A-11003; Invitrogen) and AlexaFluor-546 goat anti-rat IgG (H + L; Invitrogen) were used as secondary antibodies.
Construction of plasmids and developing cell lines
To construct a PAK-PBD encoding plasmid, we used a similar procedure described in the previous study32,52. Briefly, mRNA was extracted from human lung adenocarcinoma cell line A549 (American Type Culture Collection, Manassas, VA). Complementary DNA (cDNA) pool of the A549 cell was obtained by RT-PCR using the ReverTra Ace® qPCR RT kit (Toyobo, Osaka, Japan), PAK-PBD encoding DNA was amplified by PCR using the cDNA pool and KOD-FX (Toyobo). The primer set used for the PCR is as follows: 5′-AAGAATTCATGAATAAAAAGAAAGAGAAAGAG-3′ (forward) and 5′-AAGGTACCAGCTGACTTATCTGTAAAG-3′ (reverse). The obtained PCR product and a pAcGFP-N3 vector (TaKaRa Bio, Shiga, Japan) were digested using EcoRI and KpnI and purified by electrophoresis and commercially available Recochips (TaKaRa Bio). The digested DNA was ligated using the DNA Ligation Kit Mighty Mix (TaKaRa Bio), and introduced to competent Escherichia coli cells (HST08 Premium; TaKaRa Bio). After cloning of PAK-PBD into the vector, we identified the plasmid sequence by DNA sequencing. To fuse the fluorescent protein to the Rac1N17-encoding plasmid, Rac1N17 DNA was amplified by PCR using the human Rac1N17-encoding plasmid (generously gifted by Dr. Michiyuki Matsuda, iCeMS, Kyoto University) and KOD-FX (Toyobo). The following primer set was used for PCR: 5′- AAGGTACCATGCAGGCCATCAAGTGTG-3′ (forward) and 5′- GGAATTCTTACAACAGCAGGCATTTTC-3′ (reverse). The PCR product and a phmAG1-MClinker (TaKaRa Bio) were digested using EcoRI and KpnI. Further methods were followed by plasmid construction, which was same as PAK-PBD-AcGFP. Lifeact-TagGFP2 pCMV was purchased from ibidi (ibidi LLC, Verona, WI) and cloned into a pDsRed-Monomer-N1 (TaKaRa Bio). An AKT-PH-GFP plasmid was generously gifted by Dr. Tamas Balla (National Institute of Child Health & Human Development, National Institutes of Health). MDCK cells were transfected with PAK-PBD-AcGFP, AKT-PH-GFP, AG1-Rac1N17, and Lifeact-DsRed plasmids by using the Lipofectamine 2000 reagent (Invitrogen). Transfected cells were selected by culturing with G418 (Promega, Madison, WI) containing DMEM and picking the cells using a micropipette. Cell populations with approximately 70% fluorescent protein-positive cells were subsequently used as cell lines for transgenic MDCK cells.
Time-lapse imaging and leader cell removal by micromanipulation
Time-lapse observation was performed using a phase-contrast microscope (TE300, Nikon, Tokyo, Japan) equipped with a 10× objective lens. To maintain the temperature at 37°C, the microscope was kept in an acrylic resin box. Images were captured every 5 min for 8 h without inhibitor, and after inhibitor treatment, images were captured for 12 h. To isolate a leader cell during collective cell migration, borosilicate glass (Sutter Instrument Company, Novato, CA) was pulled using a micropipette puller (Model P-37; Sutter Instrument Company), attached to a micromanipulator (MP-285; Sutter Instrument Company), and used to impale the leader cell nucleus observed by phase-contrast microscopy.
Manual cell tracking
To quantify the velocity and direction of migrating cells, we manually tracked the nucleus of each cell using the Image J software and the Manual Tracking plug-in (National Institutes of Health, Bethesda, MD). Follower cells were defined as the cells within a distance of 200 μm from the leader cell nuclei, and 20 individual follower cells were randomly chosen in independent experiments.
Immunofluorescence staining
Cells cultured on a glass-bottom dish covered with a collagen gel were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) and permeabilized with 0.5% Triton X-100 in PBS. Blocking was performed using 0.5% bovine serum albumin (BSA)/PBS. The cells were incubated with a specific primary antibody, and reacted with a secondary antibody or Alexa Fluor-488 phalloidin (Invitrogen). Fluorescence images were captured via confocal laser scanning microscopy (C1 Confocal Imaging System; Nikon). Max-intensity z-projected images were obtained and analyzed using the Image J software.
Imaging of colony morphology
To image the morphology of the colonies growing on collagen gels, we performed Hoechst33342 (Sigma) staining and used a transilluminator to capture the images. Colonies were fixed and permeabilized as described under immunofluorescence staining.
Fluorescent live cell imaging
MDCK PAK-PBD-AcGFP and MDCK AKT-PH-GFP on a glass bottom dish covered with a collagen gel cultured with media without phenol red (D5921; Sigma) containing 25 mM HEPES (Sigma). To cut down on evaporation of media, we dropped mineral oil (M5310; Sigma) on it. Fluorescent images and transmitted light images were obtained using an A1R confocal imaging system (Nikon) every 5 min. Inhibitors mixed with DMEM were added by micropipette and 1 min after inhibitor treatment, we resumed capturing images. Fluorescent intensity was analyzed using Image J software. GFP-positive cells were selected, and the front-rear signal intensity was calculated using line-plotted intensity data for analysis of MDCK PAK-PBD-AcGFP. We mixed MDCK Lifeact-DsRed cells with MDCK AG1-Rac1N17 cells at a ratio of 1:99 and performed fluorescent live cell imaging. Images were edited using IMARIS software (Carl Zeiss, Oberkochen, Germany).
Statistical analysis
Each experimental condition was repeated at least 3 times. The error bars represent the mean ± S.D. The angles between the migration direction of the leaders and followers were described as cosθ and illustrated by box-plot using R software (R Development Core Team, Vienna, Austria) (Fig. 5b and d). The migration direction of the follower cells was graphed by rose-diagram using Matlab software (Fig. 1e and 5f). Comparison of the migration velocity and fluorescent intensity was performed with a Student's t-test using the R software. Comparison of the migration direction was performed using the Wilcoxon rank sum test (Fig. 5b and d) or Watson's two-sample test of homogeneity (Fig. 1e and 5f) using the R software. The number of leader cells generated from the mixed colony was tested by Welch's t-test using R software (Fig. 6b). Two data sets were considered significantly different if the P-value was smaller than 0.05.
Supplementary Material
Supplementary Information
Movie 1
Movie 2
Movie 3
Movie 4
Movie 5
Acknowledgments
This research was supported by JSPS KAKENHI Grant Numbers 26106702, 25127701, 26430104 to H.H.; 24106502 to T.M.; 25287106 to K.K. This study was also partly supported by special expenditures for “Reverse Translational Research from Advanced Medical Technology to Advanced Life Science” to H.H. by the Ministry of Education, Culture, Sports, Science and Technology, Japan. The authors are grateful to Mr. K. Umeno for his preliminary technical support.
Footnotes
The authors declare no competing financial interests.
Author Contributions N.Y., T.M., K.K. and H.H. conceived and designed the experiments. N.Y. performed the experiments. N.Y. analyzed the data. N.Y. contributed reagents/materials/analysis tools. N.Y., T.M. and H.H. wrote the paper.
References
- Friedl P. & Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol 10, 445–57 (2009). [DOI] [PubMed] [Google Scholar]
- Friedl P., Hegerfeldt Y. & Tusch M. Collective cell migration in morphogenesis and cancer. Int J Dev Biol 48, 441–9 (2004). [DOI] [PubMed] [Google Scholar]
- Rorth P. Collective cell migration. Annu Rev Cell Dev Biol 25, 407–29 (2009). [DOI] [PubMed] [Google Scholar]
- Cheung K. J., Gabrielson E., Werb Z. & Ewald A. J. Collective invasion in breast cancer requires a conserved Basal epithelial program. Cell 155, 1639–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga H., Irahara C., Kobayashi R., Nakagaki T. & Kawabata K. Collective movement of epithelial cells on a collagen gel substrate. Biophys J 88, 2250–6 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A. A. & Friedl P. Determinants of leader cells in collective cell migration. Integr Biol (Camb) 2, 568–74 (2010). [DOI] [PubMed] [Google Scholar]
- Omelchenko T., Vasiliev J. M., Gelfand I. M., Feder H. H. & Bonder E. M. Rho-dependent formation of epithelial “leader” cells during wound healing. Proc Natl Acad Sci U S A 100, 10788–93 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J. et al. Significance of cancer-associated fibroblasts in the regulation of gene expression in the leading cells of invasive lung cancer. J Cancer Res Clin Oncol 139, 379–88 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poujade M. et al. Collective migration of an epithelial monolayer in response to a model wound. Proc Natl Acad Sci U S A 104, 15988–93 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A. J. et al. Cell migration: integrating signals from front to back. Science 302, 1704–9 (2003). [DOI] [PubMed] [Google Scholar]
- Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D. & Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70, 401–10 (1992). [DOI] [PubMed] [Google Scholar]
- Keely P. J., Westwick J. K., Whitehead I. P., Der C. J. & Parise L. V. Cdc42 and Rac1 induce integrin-mediated cell motility and invasiveness through PI(3)K. Nature 390, 632–6 (1997). [DOI] [PubMed] [Google Scholar]
- Ouyang M. et al. N-cadherin regulates spatially polarized signals through distinct p120ctn and beta-catenin-dependent signalling pathways. Nat Commun 4, 1589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel D., Wang X., Laflamme C., Montell D. J. & Emery G. Rab11 regulates cell-cell communication during collective cell movements. Nat Cell Biol 15, 317–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E. et al. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell 19, 39–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reffay M. et al. Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nat Cell Biol 16, 217–23 (2014). [DOI] [PubMed] [Google Scholar]
- Wang X., He L., Wu Y. I., Hahn K. M. & Montell D. J. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol 12, 591–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Fontaine C. et al. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature 456, 957–61 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissoglu K. & Schwartz M. A. Integrin signalling in directed cell migration. Biol Cell 98, 547–55 (2006). [DOI] [PubMed] [Google Scholar]
- Guo W. & Giancotti F. G. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol 5, 816–26 (2004). [DOI] [PubMed] [Google Scholar]
- Hegerfeldt Y., Tusch M., Brocker E. B. & Friedl P. Collective cell movement in primary melanoma explants: plasticity of cell-cell interaction, beta1-integrin function, and migration strategies. Cancer Res 62, 2125–30 (2002). [PubMed] [Google Scholar]
- O'Toole T. E., Bialkowska K., Li X. & Fox J. E. Tiam1 is recruited to beta1-integrin complexes by 14-3-3zeta where it mediates integrin-induced Rac1 activation and motility. J Cell Physiol 226, 2965–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiosses W. B., Shattil S. J., Pampori N. & Schwartz M. A. Rac recruits high-affinity integrin alphavbeta3 to lamellipodia in endothelial cell migration. Nat Cell Biol 3, 316–20 (2001). [DOI] [PubMed] [Google Scholar]
- Cain R. J. & Ridley A. J. Phosphoinositide 3-kinases in cell migration. Biol Cell 101, 13–29 (2009). [DOI] [PubMed] [Google Scholar]
- Kolsch V., Charest P. G. & Firtel R. A. The regulation of cell motility and chemotaxis by phospholipid signaling. J Cell Sci 121, 551–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke T. F., Hornik C. P., Segev L., Shostak G. A. & Sugimoto C. PI3K/Akt and apoptosis: size matters. Oncogene 22, 8983–98 (2003). [DOI] [PubMed] [Google Scholar]
- Choma D. P., Milano V., Pumiglia K. M. & DiPersio C. M. Integrin alpha3beta1-dependent activation of FAK/Src regulates Rac1-mediated keratinocyte polarization on laminin-5. J Invest Dermatol 127, 31–40 (2007). [DOI] [PubMed] [Google Scholar]
- Bryant D. M. et al. A Molecular Switch for the Orientation of Epithelial Cell Polarization. Dev Cell 31, 171–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant G. et al. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science 287, 1037–40 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F. et al. Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat Cell Biol 4, 513–8 (2002). [DOI] [PubMed] [Google Scholar]
- Yang H. W. et al. Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol Cell 47, 281–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S. et al. Rac and Cdc42 play distinct roles in regulating PI(3,4,5)P3 and polarity during neutrophil chemotaxis. J Cell Biol 160, 375–85 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T. et al. TRIM27/MRTF-B-Dependent Integrin β1 Expression Defines Leading Cells in Cancer Cell Collectives. Cell Rep 7, 1156–67 (2014). [DOI] [PubMed] [Google Scholar]
- Lambert J. M. et al. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat Cell Biol 4, 621–5 (2002). [DOI] [PubMed] [Google Scholar]
- Sasaki A. T., Chun C., Takeda K. & Firtel R. A. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. Journal of Cell Biology 167, 505–518 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A. et al. Two distinct modes of guidance signalling during collective migration of border cells. Nature 448, 362–5 (2007). [DOI] [PubMed] [Google Scholar]
- Ferri N., Corsini A., Bottino P., Clerici F. & Contini A. Virtual screening approach for the identification of new Rac1 inhibitors. J Med Chem 52, 4087–90 (2009). [DOI] [PubMed] [Google Scholar]
- Weber G. F., Bjerke M. A. & DeSimone D. W. A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev Cell 22, 104–15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo-Carcedo C. et al. Collective cell migration requires suppression of actomyosin at cell-cell contacts mediated by DDR1 and the cell polarity regulators Par3 and Par6. Nat Cell Biol 13, 49–58 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortier J. G., Martin S., Meyer D., Rosa F. M. & David N. B. Collective mesendoderm migration relies on an intrinsic directionality signal transmitted through cell contacts. Proc Natl Acad Sci U S A 109, 16945–50 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui R. & Fenteany G. Multiple rows of cells behind an epithelial wound edge extend cryptic lamellipodia to collectively drive cell-sheet movement. J Cell Sci 118, 51–63 (2005). [DOI] [PubMed] [Google Scholar]
- Houk A. R. et al. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell 148, 175–88 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambe D. T. et al. Collective cell guidance by cooperative intercellular forces. Nat Mater 10, 469–75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Picamal X. et al. Mechanical waves during tissue expansion. Nature Physics 8, 628–U66 (2012). [Google Scholar]
- Trepat X. et al. Physical forces during collective cell migration. Nature Physics 5, 426–430 (2009). [Google Scholar]
- Ng M. R., Besser A., Danuser G. & Brugge J. S. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J Cell Biol 199, 545–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D. et al. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell 157, 1146–59 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkiteswaran G. et al. Generation and dynamics of an endogenous, self-generated signaling gradient across a migrating tissue. Cell 155, 674–87 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas P. & Gilmour D. Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Dev Cell 10, 673–80 (2006). [DOI] [PubMed] [Google Scholar]
- Dona E. et al. Directional tissue migration through a self-generated chemokine gradient. Nature 503, 285–9 (2013). [DOI] [PubMed] [Google Scholar]
- Reffay M. et al. Orientation and polarity in collectively migrating cell structures: statics and dynamics. Biophys J 100, 2566–75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynov V. S. et al. Localized Rac activation dynamics visualized in living cells. Science 290, 333–7 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Movie 1
Movie 2
Movie 3
Movie 4
Movie 5