Abstract
Cucumber (Cucumis sativus L.) fruit is a type of fleshy fruit that is harvested immaturely. Early fruit development directly determines the final fruit length and diameter, and consequently the fruit yield and quality. Different cucumber varieties display huge variations of fruit length, but how fruit length is determined at the molecular level remains poorly understood. To understand the genes and gene networks that regulate fruit length in cucumber, high throughout RNA-Seq data were used to compare the transcriptomes of early fruit from two near isogenic lines with different fruit lengths. 3955 genes were found to be differentially expressed, among which 2368 genes were significantly up-regulated and 1587 down-regulated in the line with long fruit. Microtubule and cell cycle related genes were dramatically activated in the long fruit, and transcription factors were implicated in the fruit length regulation in cucumber. Thus, our results built a foundation for dissecting the molecular mechanism of fruit length control in cucumber, a key agricultural trait of significant economic importance.
A fruit is the product of determinate growth from an angiospermous flower or inflorescence, consisting of the seed and its envelope1. Fruits can be classified into dry fruit and fleshy fruit. For dry fruits, such as maize, rice and nuts, the seed is usually the important product. For fleshy fruits, such as grape, tomato and cucurbits, the fruit flesh developed from diverse tissues in the inflorescence is the edible part. Due to its economic and agricultural importance, intensive studies have been performed to explore the molecular, cellular, and physiological events that regulate fruit growth and differentiation2.
Generally, fruit development can be divided into early fruit development and fruit ripening. Early fruit development includes three phases: 1) fruit set, the earliest phase that involves ovary development and fertilization; 2) fruit growth mostly by cell division, which involves seed formation and early embryo development; 3) fruit growth predominantly by cell expansion, during which the fruit reaches its final size and the embryo matures2. Therefore, fruit yield and quality largely depend on cell division and cell expansion during early fruit development.
The cell cycle regulation plays a pivotal role in plant growth and development. Cell division and expansion are tightly controlled by key components in the cell cycle machinery such as the cyclin-dependent protein kinases (CDKs) and the regulatory cyclins3,4. The cell cycle is regulated mainly at two checkpoints, the G1-to-S and G2-to-M transition points. CDKs act as serine threonine kinases in protein complexes, together with cyclins to phosphorylate abundant substrates at the two checkpoints that initiate DNA replication and mitosis, respectively. Five classes of CDKs (A–E) are found in plants5. CDKA is the most numerous class that is constitutively expressed and translated during the cell cycle, while CDKB particularly regulates the S-to-G2 and G2-to-M transitions5,6. Ten groups of cyclins are identified in Arabidopsis7. A-type and B-type cyclins accumulate during the G2 and early M phases, and they regulate the S-to-M and G2-to-M transitions, respectively, while D-type cyclins mainly regulate the phase transition from G1-to-S5,6,8. Recent studies showed that the expression of cyclins and CDKs increased during the early tomato fruit development9,10. Similarly, PaCDKB1 and PaCYCD1 were significantly up-regulated in the early phase of avocado fruit development11, and cell cycle genes and CDKs were highly expressed during the cell division phase of fruit development in cucumber12.
Microtubules are essential components for cell division, cell expansion, and cell morphogenesis13. Stable attachment of microtubules with kinetochores (a specialized protein structure in the centromere) is required for proper chromosome segregation and thus cell division14. Disruption of the orientation of cortical microtubules resulted in defected cell expansion15. Moreover, kinesins, a class of microtubule-based motor proteins, function in both mitosis and meiosis in plants16,17. Four types of kinesins are identified in Arabidopsis. Three kinesins, CsKF1, CsKF2 and CsKF3, have enriched expression in developing fruit and may play important roles during early fruit development in cucumber18.
Expansins are plant cell wall proteins that regulate cell division and plant growth by mediating cell wall extension19,20. Four families of expansins are identified in plants, α-expansins (EXPAs), β-expansins (EXPBs), expansin-like A (EXLA) and expansin-like B (EXLB), among which EXPA is the largest family and EXLB the smallest19,20. Previous studies showed that expansins were highly enriched in growing and ripening fruits, regulating cell enlargement through primary wall synthesis and restructuring21,22.
Cucumber (Cucumis sativus) is a globally cultivated vegetable crop that is of great economic and nutritional importance23. Cucumber fruit is a type of fleshy fruit that is developed from an enlarged ovary. In contrast to most fruits that are consumed when ripe, cucumber fruit is harvested and consumed immaturely (1–2 weeks after anthesis), either as fresh product or as processed food. Therefore, early fruit development directly determines cucumber yield and quality. Fruit development starts with a short fruit set phase, which is followed immediately by a rapid cell division phase until 5 days after anthesis (DAA), and then by an exponential expansion (5–14 DAA) phase18,24,25. Within the two weeks of development, the fruit weight increases around 200 fold in cucumber12,25. Cucumber fruit length concomitants with cell division and expansion, with the most rapid increase during 4–12 DAA and reaching its final size around 12–16 DAA26. Although several studies have been performed on transcriptome profiling or characterization of cell cycle-related genes at different developmental phases (cell division phase versus cell expansion phase)12,18,24,26,27, to the best of our knowledge, no such study has been conducted on cucumber fruits at the same developmental stage but in cultivars with different final fruit lengths. The fruit length of cucumber varies substantially within four geographic groups: the East Asian group, the Eurasian group, the Xishuangbanna group, and the India group. Among them the East Asian group have the longest fruits (over 30 cm), while the Indian group have the shortest (around 4 cm)28. In China, cucumbers can be divided into the northern China type (dark green, dense spines and warts) and the southern China type (light green, sparse spines and warts), and the fruit length varies from as short as 8 cm to as long as 35 cm (Figure S1).
Although fruit length is a key agricultural trait that is of important economic value, the regulatory mechanism of fruit length remains elusive. In order to understand the genes and gene networks that might play a role in controlling the final fruit length in cucumber, we used high throughout RNA-Seq data to compare the transcriptomes of early fruits from two near isogenic lines with different fruit lengths. We obtained 3955 differentially expressed genes between the two lines, in which 2368 genes were significantly up-regulated in the line with long fruit. Moreover, microtubule and cell cycle related genes were dramatically activated in the long fruit, and transcription factors were implicated in the fruit length regulation in cucumber. Thus, our data provide valuable information for further dissection of the molecular mechanism of regulating fruit length in cucumber.
Results
Comparison of gene expression of early fruits from two near isogenic lines with different fruit lengths
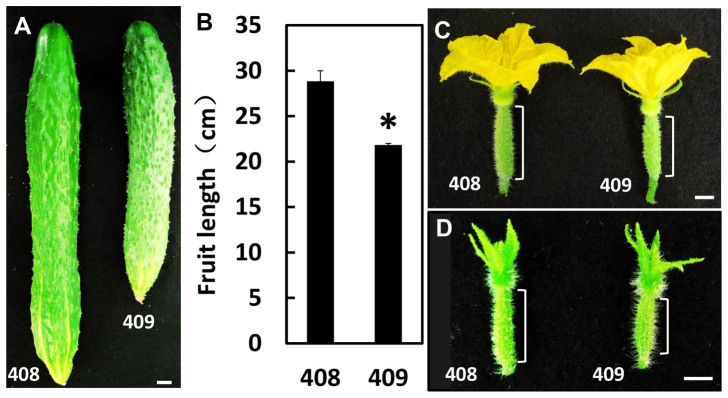
To explore genes and gene networks that control fruit length in cucumber, we performed RNA-Seq analysis of early fruits from two near-isogenic lines 408 and 409. Line 408 is a spontaneous mutant from line 409, and has been stabilized via seven generations of selfing prior to this study. Both 408 and 409 lines belong to the northern China cucumber cultivar, with long dark green fruit covered with dense spines and warts. There is no visible difference between line 408 and 409 in terms of plant morphology and plant height, except that line 408 has long fruits while line 409 has short fruits. At the commercially mature fruit stage, the average fruit length of line 408 is 28.8 cm, which is significantly longer than that (21.8 cm) in line 409 (t-test, p < 0.05) (Figure 1A–1B). The fruit length difference is apparent even as early as at the anthesis stage (Figure 1C). Given that gene expressions usually display significant differences prior to morphologic changes, we chose the very early fruits (around four days before anthesis) for transcriptome analyses (square bracket in Figure 1D).
Figure 1. Morphological characterization of two near-isogenic cucumber lines with different fruit lengths.
(A)–(D) Cucumber line 408 with long fruit (left) and its near-isogenic line 409 with short fruit (right) at commercially mature fruit stage (A)–(B), anthesis stage (C), and 4 days before anthesis (D). The bars in B represent the standard deviation (n = 10). Asterisk indicates that fruit length is significantly different between 408 and 409 (unpaired t test, P < 0.05). Square brackets in C and D show the lengths of early fruits. Samples at the same developmental stage as in D were used for RNA-Seq analyses. Scale bars represent 1 cm.
High-throughput RNA-Seq sequencing generated 14.47 to 16.08 million single-ended reads for each sample, and two biological replicates were performed for each line (Table 1). After low quality regions and adapter sequences being removed, 13.4–14.9 million clean reads were mapped to the cucumber genome using TopHat29 (Table 1). We summarized the expression level of each gene with HT-seq30. After lowly expressed genes being removed, we obtained 20000 genes that had at least 1 RPM (reads per million) in at least two samples. We then used the R package edgeR to identify differentially expressed genes (DEGs)31. Using false discovery rate (FDR) < 0.05 and fold change > 2 as the significance cutoffs, we found 3955 DEGs, in which 2368 genes were significantly up-regulated and 1587 genes were significantly down-regulated in the fruits of line 408 as compared to those of line 409 (Supplemental Table S1).
Table 1. Summary of transcriptome sequencing data.
| Sample | Raw Reads | Clean Reads (Clean/All) | Mapped Reads (Mapped/Clean) | Uniquely mapped reads (Unique/Clean) |
|---|---|---|---|---|
| 408_rep1 | 14535046 | 14331133(98.60%) | 13657318(95.30%) | 13484966(94.10%) |
| 408_rep2 | 16008838 | 15764661(98.47%) | 15039277(95.40%) | 14853164(94.22%) |
| 409_rep1 | 16081734 | 15834155(98.46%) | 15101940(95.38%) | 14929969(94.29%) |
| 409_rep2 | 14466272 | 14249754(98.50%) | 13543924(95.05%) | 13376161(93.87%) |
Validation of RNA-Seq data by quantitative real time RT-PCR assays
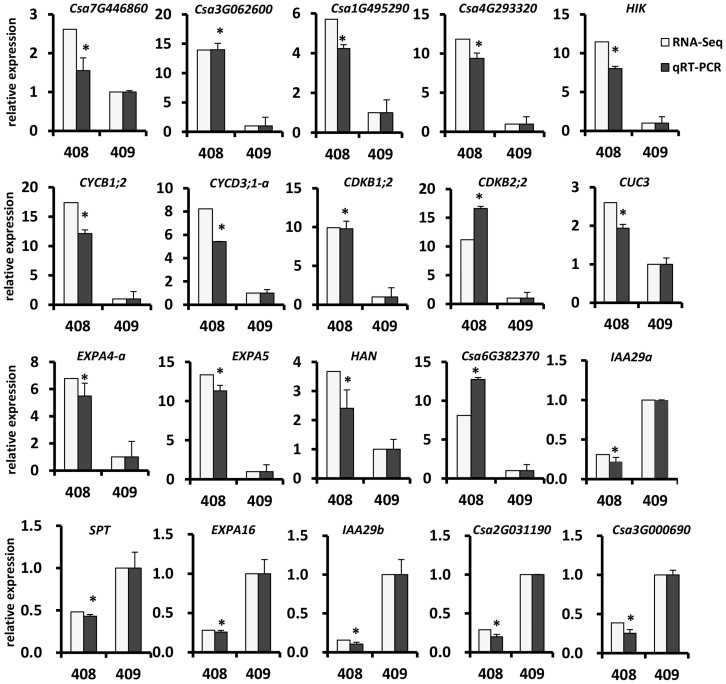
To verify the differentially expressed genes (DEGs) identified by RNA-Seq, we performed quantitative real time RT-PCR (qRT-PCR) assays using independently collected samples that were in the same developmental stage as those used for the RNA-Seq analysis. Among the 20 randomly selected DEGs, 14 genes showed higher expression and 6 genes displayed lower expression in line 408. As shown in Figure 2, all the 20 genes showed the same expression patterns in the qRT-PCR assays as in the RNA-Seq data. The pearson correlation coefficient between qRT-PCR and RNA-Seq data was 0.945 (p = 9.5E-11), indicating that the RNA-Seq data were highly reliable.
Figure 2. Verification of differentially expressed genes by qRT-PCR.
Fourteen DEGs with higher expression and six DEGs with lower expression in line 408 were chosen for qRT-PCR validation. The relative expression level of each gene was expressed as the fold change between two lines in the RNA-Seq data (white bar) and qRT-PCR data (gray bar). The cucumber UBIQUITIN gene was used as an internal control to normalize the expression data. The bars represent the standard deviation (n = 3). Asterisks indicate that the gene transcriptions are significantly different between lines 408 and 409 (unpaired t test, P < 0.05).
Genetic variations between lines 408 and 409
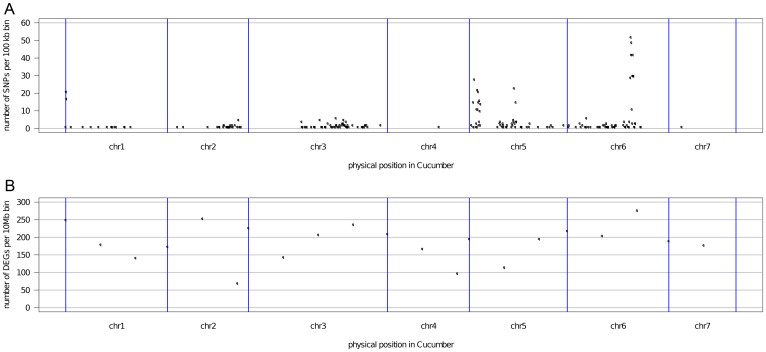
To confirm that lines 408 and 409 have nearly identical genetic background and to identify candidate genetic markers for future gene mapping and functional studies, we used the mapped RNA-Seq reads to obtain single nucleotide polymorphisms (SNPs) between these two lines. Using samtools32 and vcftools33, we obtained only 850 SNPs in the genomes of these two lines, confirming that lines 408 and 409 are near isogenic lines with very few genetic variations. We calculated the density of SNPs in non-overlapping 100 kb bins of each chromosome (Figure 3A) and found four significant peaks on chromosome 1 (Chr1: 1.0 Mb), chromosome 5 (Chr5: 1.4 Mb; 2.3–2.5 Mb), and chromosome 6 (Chr6: 18.1–19.0 Mb). The peak on chromosome 6 has the highest density, with more than 50 SNPs in the neighboring 100 kb bins. Chromosomes 4 and 7 have the lowest SNP density, with only one SNP on each chromosome (Figure 3A). We also calculated the density of DEGs in 10 Mb bins in the genome and found that most DEGs were not located in the SNP rich regions (with the exception of the region near the SNP peak on chromosome 6) (Figure 3B), suggesting that the majority of the SNPs may not affect gene expression directly. 145 SNPs were found to be within or near 70 DEGs (Supplemental Table S2) and thus they may serve as molecular markers for map-based cloning and functional characterization of genes that may regulate fruit length in cucumber.
Figure 3. Genome-wide distributions of SNPs (A) and DEGs (B) in the cucumber genome.
Number of SNPs and DEGs were calculated over non-overlapping 100 kb and 10 MB bins, respectively.
Microtubule and cell cycle related genes are involved in fruit length regulation in cucumber
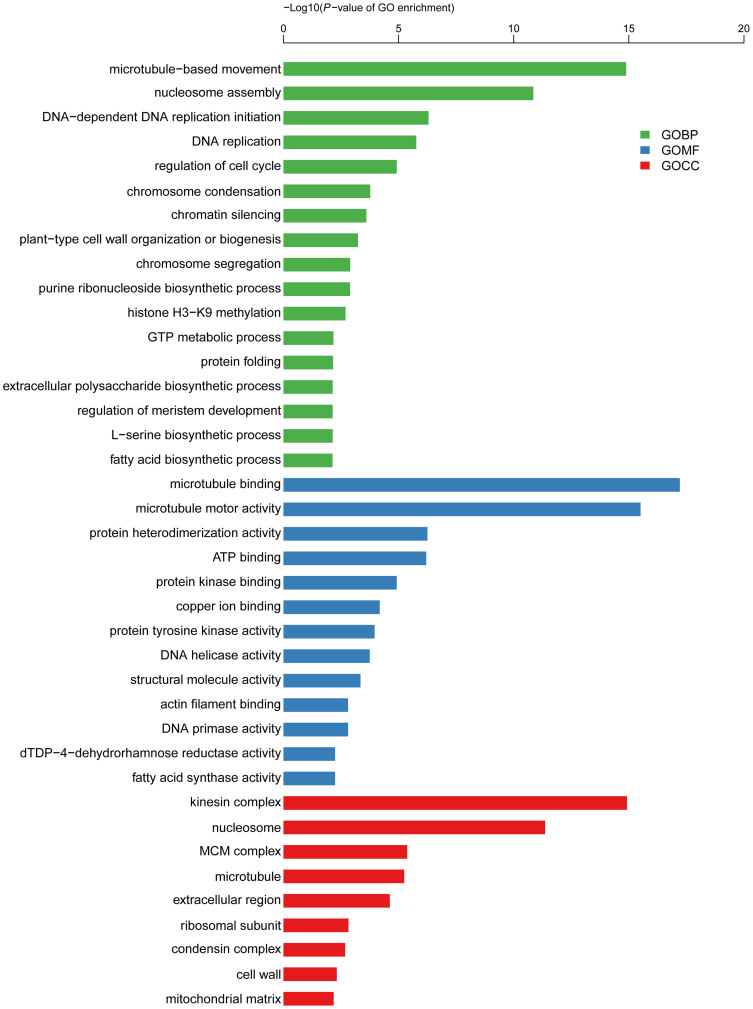
To further understand the function of these DEGs, gene ontology (GO) term enrichment analysis (P ≤ 0.05) was performed. For genes that were up-regulated in line 408, the most significantly enriched GO terms were “microtubule-based movement” (p = 1.3E-15) in the biological process (GOBP) group (green in Figure 4), “microtubule binding” (p = 6.1E-18) in the molecular function (GOMF) group (blue in Figure 4) and “kinesin complex” (p = 1.2E-15) in the cellular component (GOCC) (red in Figure 4), respectively. Because kinesin family proteins are a class of microtubule-based motor proteins that function in mitosis as well as meiosis in both plant and animal systems16,34,35, microtubule-related genes were the highly enriched top three GO categories in the up-regulated DEGs in the 408 line with long fruit. 40 microtubule-related genes that had higher expression in line 408 were listed in Table 2. For example, the microtubule associated protein MAP65 (Csa5G169090) showed a 13 fold (log2FC = 3.72) and the KINESIN 1(Csa1G065980) displayed a 7.6 fold (log2FC = 2.92) higher expression in the line 408 as compared to in line 409 (Table 2). Further, qRT-PCR from independently generated samples verified that five microtubule related genes had significantly higher expression in the fruit of line 408 (top row in Figure 2). In addition, genes involved in nucleosome assembly, DNA replication and cell cycle regulation were highly enriched in the up-regulated DEGs in the line 408 (Figure 4). Further characterization of the DEGs using the MapMan software showed that genes implicated in cell wall (Figure S2A), cell division and cell cycle (Figure S2B) were predominantly up-regulated in the fruit of line 408. In accordance with this finding, 29 cyclin family genes (Table 3) and 15 expansin family genes (Table 4) showed elevated transcription in the 408 line. Moreover, qRT-PCR confirmed that four cyclin genes (CYCB1;2, CYCD3;1-a, CDKB1;2, CDKB2;2) and two expansin genes (EXPA4-a and EXPA5) were significantly induced in the fruit of line 408 as compared to in line 409 (Figure 2), suggesting that the activation of microtubule, cell cycle and cell wall related genes is required for fruit elongation in cucumber.
Figure 4. Significantly enriched Gene Ontology (GO) terms (P < 0.05) in the up-regulated genes in the fruit of line 408 vs 409.
GO terms belong to biological processes (GOBP), molecular functions (GOMF), and cellular components (GOCC) were shown in green, blue, and red, respectively. GO terms were sorted based on p-values.
Table 2. List of selected microtubule-related genes that were differentially expressed in the 408 and 409 cucumber fruit.
| Gene ID | Putative function | Arabidopsishomologue | log2FC | FDR |
|---|---|---|---|---|
| Csa3G062600 | Kinesin motor family protein CsKF2 | AT3G20150 | 3.80 | 1.8E-15 |
| Csa5G169090 | Microtubule associated protein (MAP65/ASE1) family protein | AT5G51600 | 3.72 | 5.6E-11 |
| Csa4G293320 | ATP binding microtubule motor family protein | AT2G37420 | 3.57 | 8.0E-17 |
| Csa1G042550 | ATP binding microtubule motor family protein HIK | AT1G18370 | 3.52 | 4.0E-17 |
| Csa4G064090 | Di-glucose binding protein with Kinesin motor domain | AT1G72250 | 3.44 | 7.9E-14 |
| Csa2G022260 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | AT5G27550 | 3.38 | 2.2E-15 |
| Csa2G173060 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | AT4G14330 | 3.32 | 1.8E-12 |
| Csa1G042790 | beta-6 tubulin (TUB6) | AT5G12250 | 3.32 | 2.1E-02 |
| Csa2G354040 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | AT1G59540 | 3.28 | 1.5E-10 |
| Csa2G427860 | end binding protein 1C | AT5G67270 | 3.24 | 2.0E-08 |
| Csa3G734250 | ATP binding microtubule motor family protein | AT5G02370 | 3.23 | 1.2E-12 |
| Csa5G166470 | ATP binding microtubule motor family protein | AT5G23910 | 3.10 | 1.5E-14 |
| Csa1G025250 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | AT3G44050 | 3.05 | 2.3E-12 |
| Csa2G237130 | phragmoplast orienting kinesin 2 | AT3G19050 | 3.03 | 1.6E-12 |
| Csa6G491670 | end binding protein 1C | AT5G67270 | 3.01 | 6.7E-13 |
| Csa3G777560 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | AT2G36200 | 2.95 | 4.2E-13 |
| Csa1G065980 | kinesin 1 | AT4G21270 | 2.92 | 1.3E-14 |
| Csa4G219360 | P-loop nucleoside triphosphate hydrolases superfamily protein | AT3G10310 | 2.85 | 3.1E-08 |
| Csa2G368830 | tubulin beta-1 chain (TUB1) | AT1G75780 | 2.84 | 1.2E-10 |
| Csa3G176870 | Microtubule associated protein (MAP65/ASE1) family protein | AT5G51600 | 2.83 | 1.2E-12 |
| Csa6G000650 | phragmoplast orienting kinesin 1 | AT3G17360 | 2.79 | 9.9E-14 |
| Csa2G370520 | chromosome-associated kinesin-related | AT5G33300 | 2.62 | 1.3E-10 |
| Csa3G891640 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | AT5G60930 | 2.56 | 3.1E-12 |
| Csa1G015820 | ATP binding microtubule motor family protein | AT1G18550 | 2.52 | 8.0E-11 |
| Csa1G495290 | Kinesin motor family protein CsKF7 | AT3G45850 | 2.51 | 2.8E-10 |
| Csa4G000580 | Tubulin/FtsZ family protein (TUA6) | AT4G14960 | 2.40 | 2.8E-09 |
| Csa3G172990 | phragmoplast-associated kinesin-related protein, putative | AT3G23670 | 2.37 | 1.6E-12 |
| Csa5G583380 | microtubule-associated protein 65-5 | AT2G38720 | 2.17 | 8.9E-08 |
| Csa6G511060 | microtubule-associated proteins 70-5 | AT4G17220 | 2.12 | 7.9E-03 |
| Csa7G197290 | kinesin-like protein 1 | AT3G44730 | 2.04 | 2.5E-07 |
| Csa4G219350 | P-loop nucleoside triphosphate hydrolases superfamily protein | AT3G10310 | 1.85 | 6.3E-06 |
| Csa2G000780 | MAP kinase kinase 6 | AT5G56580 | 1.74 | 1.2E-06 |
| Csa3G821540 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | AT3G10180 | 1.67 | 1.2E-06 |
| Csa6G497050 | tubulin alpha-4 chain (TUA4) | AT1G04820 | 1.65 | 2.5E-06 |
| Csa2G368870 | ATP binding microtubule motor family protein | AT1G20060 | 1.57 | 2.3E-05 |
| Csa6G382990 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | AT1G21730 | 1.41 | 2.6E-05 |
| Csa4G047870 | ATP binding microtubule motor family protein | AT5G66310 | 1.41 | 1.2E-05 |
| Csa7G446860 | ATP binding microtubule motor family protein CsKF1 | AT3G16060 | 1.39 | 2.4E-05 |
| Csa3G414070 | kinesin 3 | AT5G54670 | 1.12 | 4.0E-04 |
| Csa5G153100 | P-loop nucleoside triphosphate hydrolases superfamily protein | AT1G63640 | 1.06 | 1.4E-03 |
Table 3. List of selected cyclin family genes that were differentially expressed in the 408 and 409 cucumber fruit.
| Gene ID | Putative function | Arabidopsis homologue | log2FC | FDR |
|---|---|---|---|---|
| Csa4G425740 | Cyclin family protein CYCB1;2 | AT5G06150 | 4.12 | 3.6E-13 |
| Csa7G071600 | Cyclin -dependent protein kinase 3;2 | AT1G47210 | 4.03 | 1.9E-10 |
| Csa2G354980 | Cyclin -dependent protein kinase 3;2 | AT1G47210 | 3.86 | 4.3E-11 |
| Csa2G022250 | Cyclin family | AT3G05330 | 3.84 | 3.3E-10 |
| Csa6G428000 | Cyclin B1;4 | AT2G26760 | 3.55 | 1.9E-15 |
| Csa6G509580 | Cyclin B2;3 | AT1G20610 | 3.55 | 2.9E-16 |
| Csa4G664480 | Cyclin -dependent kinase B2;2 | AT1G20930 | 3.48 | 2.8E-13 |
| Csa4G658580 | Cyclin B2;4 | AT1G76310 | 3.32 | 6.5E-13 |
| Csa4G198330 | Cyclin -dependent kinase B1;2 | AT2G38620 | 3.31 | 2.3E-10 |
| Csa4G025110 | Cyclin P4;1 | AT2G44740 | 3.13 | 7.6E-04 |
| Csa7G072880 | Cyclin D3;1-a | AT4G34160 | 3.04 | 1.2E-15 |
| Csa7G239600 | Cyclin B3;1 | AT1G16330 | 2.47 | 1.7E-10 |
| Csa3G878930 | mitotic-like Cyclin B3 | AT5G11300 | 2.47 | 1.6E-08 |
| Csa5G603320 | Cyclin D1;1 | AT1G70210 | 2.23 | 9.2E-11 |
| Csa4G641660 | Cyclin D3;2 | AT5G67260 | 2.09 | 2.8E-07 |
| Csa3G199660 | Cyclin D3;1-b | AT4G34160 | 2.06 | 1.5E-08 |
| Csa6G495110 | Cyclin D5;1 | AT4G37630 | 1.79 | 1.5E-06 |
| Csa6G491650 | Cyclin D3;2 | AT5G67260 | 1.69 | 1.9E-05 |
| Csa7G339140 | Cyclin-dependent kinase inhibitor family protein | AT3G24810 | 1.68 | 6.0E-05 |
| Csa4G363410 | Cyclin/Brf1-like TBP-binding protein | AT3G09360 | 1.58 | 4.3E-04 |
| Csa2G356070 | Cyclin D3;2 | AT5G67260 | 1.57 | 1.8E-06 |
| Csa7G435540 | Cyclin A2;4 | AT1G80370 | 1.50 | 2.8E-05 |
| Csa6G491640 | Cyclin D3;3 | AT3G50070 | 1.42 | 3.9E-04 |
| Csa6G078670 | Cyclin P4;1 | AT2G44740 | 1.39 | 2.4E-04 |
| Csa4G642510 | Cyclin D5;1 | AT4G37630 | 1.38 | 1.2E-04 |
| Csa2G379270 | Cyclin A2;1 | AT5G25380 | 1.35 | 3.9E-03 |
| Csa6G499110 | Cyclin D2;1 | AT2G22490 | 1.28 | 4.5E-04 |
| Csa4G000660 | Cyclin D1;1 | AT1G70210 | 1.19 | 2.1E-04 |
| Csa6G121960 | Cyclin D6;1 | AT4G03270 | 1.01 | 3.1E-03 |
Table 4. List of selected expansin family genes that were differentially expressed in the 408 and 409 cucumber fruit.
| Gene ID | Putative function | Arabidopsis homologue | log2FC | FDR |
|---|---|---|---|---|
| Csa5G605760 | expansin A1 | AT1G69530 | 4.76 | 5.3E-29 |
| Csa5G636630 | expansin A5 | AT3G29030 | 3.74 | 2.9E-14 |
| Csa1G009820 | expansin A1 | AT1G69530 | 3.03 | 3.3E-14 |
| Csa6G128580 | expansin B3 | AT4G28250 | 2.88 | 3.1E-10 |
| Csa6G190340 | expansin A4-a | AT2G39700 | 2.76 | 3.5E-09 |
| Csa4G296100 | expansin A4-b | AT2G39700 | 2.29 | 1.1E-07 |
| Csa7G047420 | expansin B3 | AT4G28250 | 2.15 | 3.1E-05 |
| Csa5G182120 | expansin A4-c | AT2G39700 | 2.05 | 1.5E-08 |
| Csa5G332530 | expansin A13 | AT3G03220 | 1.29 | 1.3E-03 |
| Csa5G333040 | expansin A13 | AT3G03220 | 1.23 | 3.4E-03 |
| Csa5G221940 | expansin A8 | AT2G40610 | 1.03 | 1.3E-02 |
| Csa7G019900 | expansin-like A1 | AT3G45970 | −1.03 | 1.2E-02 |
| Csa5G517210 | expansin A16 | AT3G55500 | −1.84 | 5.2E-05 |
| Csa7G019330 | expansin-like A3 | AT3G45960 | −2.35 | 2.3E-04 |
| Csa3G910670 | expansin-like B1 | AT4G17030 | −2.74 | 4.4E-06 |
Transcription factors are implicated in cucumber fruit length control
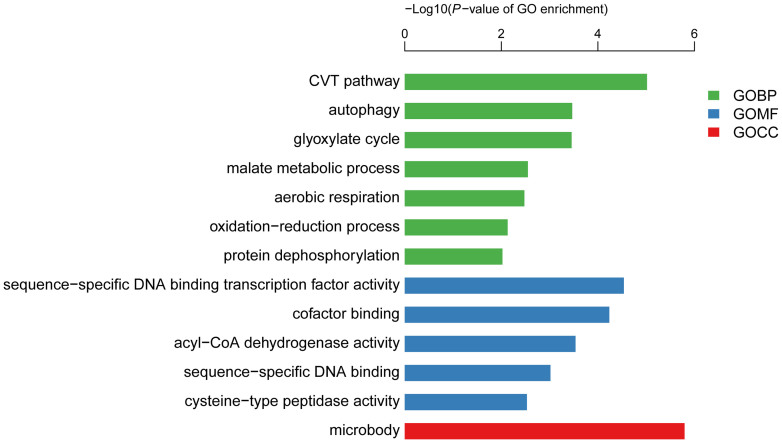
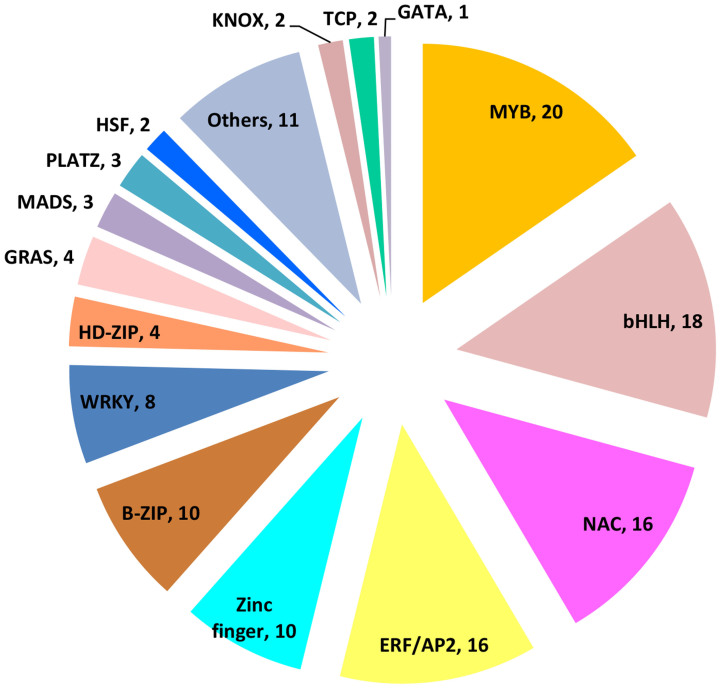
For the down-regulated genes in line 408, the top three enriched GO terms were “sequence−specific DNA binding transcription factor activity” (p = 2.9E-05), “microbody” (p = 1.69E-06) and “CVT pathway” (p = 9.6E-06) (Figure 5). A total of 130 genes that function in transcriptional regulation were found to be repressed in the fruit of line 408 (Supplemental Table 3), and these transcription factors were distributed in different gene families. As shown in Figure 6, there are 20, 18, 16 and 16 genes assigned into the Myb, bHLH, NAC and ERF/AP2 families, respectively, that displayed reduced expression in the 408 line with long fruit, suggesting that these transcription factors may function as negative regulators in fruit elongation in cucumber. The predicted cucumber SPATULA (SPT) (Csa2G356640) showed a 2 fold reduction as detected by RNA-seq and a 2.3 fold decrease as revealed by qRT-PCR (Supplemental Table 3, Figure 2). SPT encodes a basic-helix-loop-helix (bHLH) protein that regulates carpel and fruit development by regulating auxin distribution in Arabidopsis, in coordination with additional bHLH members such as INDEHISCENT (IND) and CRABS CLAW (CRC)36,37,38. Similarly, the putative cucumber FRUITFULL (FUL) (Csa1G039910), a MADS-box gene regulating cell differentiation during fruit development in Arabidopsis and fruit ripening in tomato39,40, displayed 2.3 fold lower expression in the fruit of 408 line (Supplemental Table 3). Moreover, members of other families of transcription factors, such as Csa6G312040 (a zinc finger protein 6), Csa7G413890 (a myb domain protein 2) and Csa5G606310 (a NAC-like protein) showed 96.0, 25.6, and 10.5 fold down-regulation in the fruit of 408 line respectively (Supplemental Table 3), implying that they may play important roles for fruit length regulation in cucumber. Functional categorization by the MapMan software indicated that genes involved in transcription regulation, protein modification and protein degradation were highly enriched in the DEGs, including transcription factors with known function in fruit development (Supplementary Figure S3). For example, HANABA TARANU (HAN), which encodes a GATA-3–like transcription factor with a single zinc finger domain, regulates shoot apical meristem, flower and carpel development together with GATA-3 family genes in Arabidopsis41,42. The putative cucumber HAN gene (Csa6G502700) displayed a 3.7 fold as detected by RNA-seq and 2.4 fold as calculated from qRT-PCR up-regulation in the 408 line with long fruit (Figure 2, Supplemental Table 1). Similarly, the putative cucumber SUPERMAN (SUP) (Csa3G141870), a C2H2 type zinc finger protein that controls carpel numbers and ovule development in Arabidopsis43,44, showed 5.6 fold higher expression in the line 408 as compared to the line 409 (Supplemental Table 1). These data suggest that transcription factors may play a key role in fruit length determination in cucumber.
Figure 5. Significantly enriched Gene ontology (GO) terms (P < 0.05) in the down-regulated genes in the fruit of line 408 vs 409.
GO terms belong to biological processes (GOBP), molecular functions (GOMF), and cellular components (GOCC) were shown in green, blue, and red, respectively. GO terms were sorted based on p-values.
Figure 6. Family assignment of the 130 transcription factors that showed lower expression in the fruit of line 408.
Number of genes assigned to each family is shown behind a comma.
Discussion
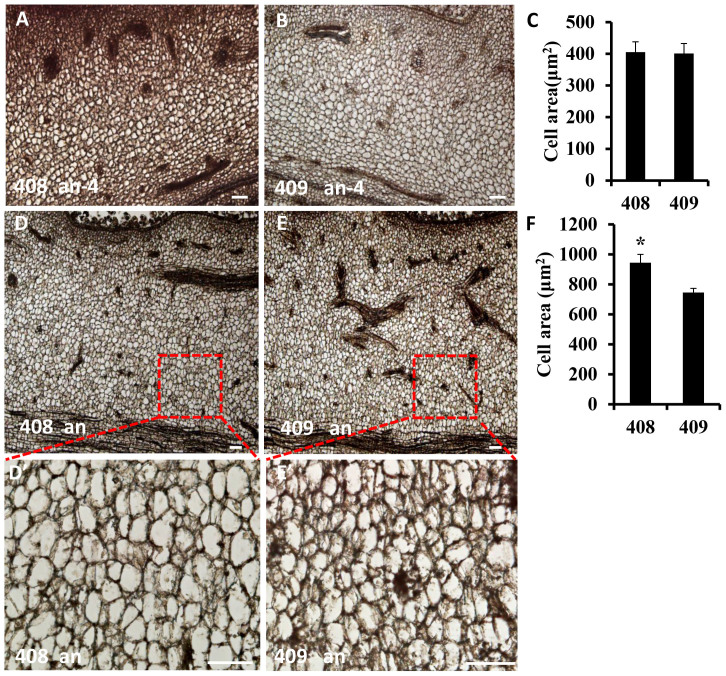
In this study, we provide a comprehensive analysis of genes involved in fruit length control in cucumber. A total of 3955 differentially expressed genes (DEGs) were found, in which 2368 genes were significantly up-regulated and 1587 genes were significantly down-regulated in the 408 line with long fruit (Figure 1, Supplementary Table S1). qRT-PCR confirmed that our RNA-Seq data were highly reliable (Figure 2). Functional categories of the DEGs by GO term enrichment analysis and MapMan showed that microtubule and cell cycle related genes were strongly induced in the long fruit (Figure 4 and supplemental Figure S2). Microtubules have been shown to be essential for proper cell division and cell expansion14,15. Genes that encode both basic structural units of microtubules (alpha tubulins TUAs and beta tubulins TUBs) were found to be up-regulated in the long fruit (Table 2). Moreover, the microtubule associated motor proteins (kinesin and dynein), as well as many other microtubule associated proteins (MAPs) were activated in the 408 line (Table 2). For example, the expressions of kinesin genes CsKF2 (Csa3G062600), CsKF7 (Csa1G495290) and CsKF1 (Csa7G446860) were 13.9, 5.7 and 2.6 fold, respectively, higher in the long fruit (Table 2), which is consistent with previous report that the expression of CsKF2 positively correlated with cell division while the transcription of CsKF1and CsKF7 positively correlated with rapid cell expansion18, suggesting that the long fruit in the 408 line may result from both increased cell number and accelerated cell expansion. In support of this notion, 29 cyclin family genes were all up-regulated in line 408, in which 12 were D-type cyclins, 8 were B-type cyclins and 4 were CDKs (Table 3). Previous evidence showed that D-type and B-type cyclins regulate the phase transition from G1-to-S and G2-to-M respectively, and CDKs regulate cell division through the G1-to-S and G2-to-M checkpoints5,6, and that the expression of cyclins and CDKs positively correlated with rapid cell division during the early fruit development in tomato and cucumber9,10,26. Moreover, 15 expansin family genes were mostly up-regulated in the line 408 with long fruit (Table 4). Previous data also showed that cytoskeleton and cell wall related genes were highly expressed during the peak exponential expansion phase (8 DAA) in cucumber26. Given that the samples we used for RNA-Seq analysis were 4 days before anthesis, a developmental stage with rapid cell division, together with the fact that fruit length but not fruit diameter increased in the line 408 as compared to in the line 409, it is plausible to infer that the long fruit in line 408 may result from both more rapid cell division and accelerated phase progression (from cell division to cell expansion). To test this hypothesis, we compared the cell size at 4 days and 0 day before anthesis between lines 408 and 409 (Figure 7). As expected, there was no noticeable cell size difference between lines 408 and 409 at 4 days before anthesis (Figure 1), suggesting that there were more rapid cell divisions and thus more cells in the line 408 with long fruit (Figure 7A–7C). However, by the day of anthesis, the cell size in line 408 was significantly larger than that in line 409 (Figure 7D–7F), suggesting that line 408 had accelerated phase transition from cell division to cell expansion. In addition, we found that 130 transcription factors were transcriptionally repressed in the fruit of line 408 (Supplemental Table 3), and the majority of them were from MYB, bHLH, NAC and ERF/AP2 gene families (Figure 6). Both AtMYB124 and AtMYB88 encode R2R3 MYB transcription factors, which are expressed during ovule development and have been identified to regulate female reproductive development45. SlFSM1 encodes a SANT/MYB-like domain and is an early fruit-specific gene, and the SlFSM1/FSB1/MYBI complex controls cell expansion and fruit development in tomato46. Furthermore, most known regulators of fruit development such as SPT, IND, CRC, FUL, SUP and HAN were transcription factors that were differentially expressed between lines 408 and 409, and many of them can mediate cell division and expansion36,37,38,40,42. Therefore, we speculate that transcription factors may be the key players for fruit length control in cucumber, while microtubule, CDKs-cyclins and expansins that mediate cell division and cell expansion may be the downstream effects. Further functional studies using genetic transformation of the differentially expressed genes identified in this study, especially transcription factors and the 70 DEGs with SNPs between lines 408 and 409, will shed more light on the precise mechanism of fruit length regulation in cucumber. As such, our results provide a valuable resource for further functional characterization and lay a foundation for molecular breeding for desired fruit length in crops.
Figure 7. Comparison of cell morphology in lines 408 and 409.
(A)–(C) Microscopic longitudinal sections of the young fruits at 4 days before anthesis (an-4) in line 408 (A) and line 409 (B), and the corresponding quantifications of cell size (C). (D)–(F) Microscopic longitudinal sections of the young fruits at anthesis (an) in line 408 (D) and line 409 (F), and the respective quantifications of cell size (F). (D′)–(E′) are the enlarged view of the boxes in D and E, respectively. The bars in C and F represent the standard deviation (n = 3). Asterisk in F indicates that the cell size in line 408 is significantly larger than that in line 409 at anthesis (unpaired t test, P < 0.05). Bar = 50 um.
Methods
Plant materials
The cucumber hermaphrodite line 408 and its near-isogenic line 409 were grown in the Jinliuhuan Experimental Station of Beijing under standard greenhouse conditions. Pest control and water management were performed according to standard practices. Early fruits of about 2–4 cm in length were collected from 408 or 409 lines at the same time on the same day. Three-to-five fruits from different plants were pooled together as one biological sample for each cucumber line. Samples were immediately frozen in liquid nitrogen and stored at −80°C until further use.
RNA extraction and quality test
Frozen fruit samples were grinded in a mortar with liquid nitrogen, and total RNA was isolated using the RNA extraction kit (Huayueyang, China). RNA was checked on 1% agarose gels to avoid possible degradation and contamination, and was then examined by a Nano Photometer spectrophotometer (IMPLEN, CA, USA) for RNA purity. Qubit RNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies, CA, USA) was used to measure RNA concentration, and RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA) was used to evaluate RNA integrity. Only RNA samples that passed the quality tests were chosen for RNA-Seq analyses.
RNA-Seq library construction and sequencing
RNA-Seq library construction was performed using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Ispawich, USA) following manufacturer's instructions and four index codes were added to attribute sequences to different samples47. Briefly, mRNAs were enriched from 3 ug total RNA using magnetic beads with Oligo (dT) (Life technologies, CA, USA), and then fragmented using divalent cations under elevated temperature in the NEB proprietary fragmentation buffer. Double-stranded cDNAs were synthesized using random hexamers and M-MuLV Reverse Transcriptase (RNase H-), followed by DNA Polymerase I and RNase H. After adenylation of the 3′ ends of cDNA fragments, NEBNext adapter oligonucleotides were ligated to cDNA fragments, and the AMPure XP beads system (Beckman Coulter, Beverly, USA) was used to select cDNA fragments of approximately 200 bp in length. cDNA fragments with ligated adapter molecules on both ends were selectively enriched using the NEB Universal PCR Primer and Index primer in a 10-cycle PCR reaction. Products were purified with the AMPure XP beads system and quantified using the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer's instructions. RNA-seq libraries were sequenced on an Illumina HiSeq 2000 platform to generate 100 bp single-ended reads.
Bioinformatics analysis of RNA-Seq data
Raw reads were pre-processed to remove low quality regions and adapter sequences. Clean reads were mapped to the Cucumber genome sequence (http://cucumber.genomics.org.cn, v2i) using TopHat23,29. Read counts of each gene were summarized by the HTSeq-count30. The R package edgeR was used to identify the differentially expressed genes31. The expression of each gene was normalized to reads per million (RPM) to compare among different samples. Lowly expressed genes were removed and only genes with an expression level of at least 1 RPM in at least two samples were kept for further analysis. The genes with at least two folds change in expression between lines 408 and 409, and with a False Discovery Rate (FDR) of less than 0.05 were considered to be differentially expressed.
Sequencing data were deposited to the Gene Expression Omnibus (GEO) database at the National Center for Biotechnology Information (NCBI) with access number GSE60346.
SNP calling using RNA-Seq reads
After clean reads were mapped to the cucumber genome using TopHat, we used the SAMtools32 (version 0.1.18) and vcftools as described in Martin et al33 to detect SNPs between lines 408 and 409. We used “samtools mpileup” with the parameter “-D” to record per-sample read depths and filtered VCF files using an AWK script48. SNPs with a quality value greater than 40 and a read coverage of at least 2 were considered confident SNPs. Genome-wide distributions of SNPs and DEGs on were visualized using R scripts47. Functional annotations of SNP sites were performed using snpEff49.
Gene Ontology (GO) term enrichment analysis
Because GO terms are not well annotated for cucumber genes, we used blast2go50 and interProScan51 to assign GO terms to cucumber genes. The GO term enrichment analysis was conducted for up-regulated and down-regulated genes in line 408, respectively, using the R package TopGO52. Adrian Alexi's improved weighted scoring algorithm and Fisher's exact test were used to determine the significance of GO term enrichment.
MapMan analysis of DEGs
In order to obtain detailed functional categorization of differentially expressed genes between line 408 and line 409, we used the online web tool Mercator53 to obtain the cucumber protein annotation mapping file for MapMan with default parameters and then used the Java software MapMan to assign functional categorizations to DEGs54.
Quantitative real-time RT-PCR
qRT-PCR analyses were performed with independently generated samples from 408 and 409 lines at the same developmental stage. Primers for qRT-PCR were designed using the Primer 5 software and synthesized by Sangon Biotech. cDNAs were reverse transcribed from 3 μg of total RNA using the PrimeScript RT reagent Kit (Takara, Da Lian, China), and qRT-PCR analyses were performed on an ABI PRISM 7500 Real-Time PCR System (Applied Biosystems, USA). The cucumber UBI gene was used as an internal control to normalize the expression data55. Each qRT-PCR experiment was repeated three times. The relative expression of genes was calculated using the 2−ΔΔCt method56 and standard deviation was calculated between three biological replicates. The gene-specific primers are listed in Supplemental Table S4.
Light microscopy of cell morphology
Young fruits at 4 days and 0 day before anthesis were collected from line 408 and line 409, respectively. Fruit samples were fixed in the FAA (50% ethanol, 5% glacial acetic acid, and 3.7% formaldehyde) buffer overnight, and then dehydrated through an ethanol series and embedded in paraplast as described41.
Supplementary Material
Fruit length supplementary data
Table S1
Table S2
Table S3
Acknowledgments
This work was supported by National Basic Research of China 973 program [2012CB113900], National Natural Science Foundation of China [31171399], Chinese Universities Scientific Fund [2013RC030], Earmarked Fund for Beijing Leaf Vegetables Innovation Team of Modern Agro-industry Technology Research System [BLVT-08] and the Chinese Academy of Sciences.
Footnotes
The authors declare no competing financial interests.
Author Contributions L.J., S.Y., Q.W., W.Y. and X.Z. conceived and designed the experiments. L.J., S.Y., M.X., L.Y., X.S. and C.Z. performed the experiments. L.J., S.Y., Y.L., R.L. and X.Z. analyzed the data. L.J., S.Y., Y.L., R.L. and X.Z. wrote the paper. All authors read and approved the final manuscript.
References
- Coombe B. The development of fleshy fruits. Ann Rev Plant Physiol 27, 207–228 (1976). [Google Scholar]
- Gillaspy G., Ben-David H. & Gruissem W. Fruits: A Developmental Perspective. Plant Cell 5, 1439–1451 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inze D. & De Veylder L. Cell cycle regulation in plant development. Annu Rev Genet 40, 77–105 (2006). [DOI] [PubMed] [Google Scholar]
- Van Leene J. et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol 6, 397 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubes J. et al. CDK-related protein kinases in plants. Plant Mol Biol 43, 607–620 (2000). [DOI] [PubMed] [Google Scholar]
- Mironov V. V., De Veylder L., Van Montagu M. & Inze D. Cyclin-dependent kinases and cell division in plants- the nexus. Plant Cell 11, 509–522 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. et al. Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol 135, 1084–1099 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T. & Doerner P. Cell cycle controls: genome-wide analysis in Arabidopsis. Curr Opin Plant Biol 4, 501–506 (2001). [DOI] [PubMed] [Google Scholar]
- Joubes J. et al. Molecular and biochemical characterization of the involvement of cyclin-dependent kinase A during the early development of tomato fruit. Plant Physiol 121, 857–869 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubes J., Walsh D., Raymond P. & Chevalier C. Molecular characterization of the expression of distinct classes of cyclins during the early development of tomato fruit. Planta 211, 430–439 (2000). [DOI] [PubMed] [Google Scholar]
- Sabag M. et al. PaKRP, a cyclin-dependent kinase inhibitor from avocado, may facilitate exit from the cell cycle during fruit growth. Plant Sci 213, 18–29 (2013). [DOI] [PubMed] [Google Scholar]
- Fu F. Q. et al. A role of brassinosteroids in early fruit development in cucumber. J Exp Bot 59, 2299–2308 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L., Li S., Bashline L. & Gu Y. Dissecting the molecular mechanism underlying the intimate relationship between cellulose microfibrils and cortical microtubules. Front Plant Sci 5, 90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H. & Sunkel C. E. Kinetochore-microtubule interactions during cell division. Chromosome Res 12, 585–597 (2004). [DOI] [PubMed] [Google Scholar]
- Bichet A., Desnos T., Turner S., Grandjean O. & Hofte H. BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J 25, 137–148 (2001). [DOI] [PubMed] [Google Scholar]
- Vanstraelen M., Inze D. & Geelen D. Mitosis-specific kinesins in Arabidopsis. Trends Plant Sci 11, 167–175 (2006). [DOI] [PubMed] [Google Scholar]
- Zhou S. et al. Pollen semi-sterility1 encodes a kinesin-1-like protein important for male meiosis, anther dehiscence, and fertility in rice. Plant Cell 23, 111–129 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. Y. et al. Characterization and expression profiling of cucumber kinesin genes during early fruit development: revealing the roles of kinesins in exponential cell production and enlargement in cucumber fruit. J Exp Bot 64, 4541–4557 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D. J. Growth of the plant cell wall. Nat Rev Mol Cell Biol 6, 850–861 (2005). [DOI] [PubMed] [Google Scholar]
- Kende H. et al. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol 55, 311–314 (2004). [DOI] [PubMed] [Google Scholar]
- Brummell D. A., Harpster M. H. & Dunsmuir P. Differential expression of expansin gene family members during growth and ripening of tomato fruit. Plant Mol Biol 39, 161–169 (1999). [DOI] [PubMed] [Google Scholar]
- Rose J. K., Lee H. H. & Bennett A. B. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci U S A 94, 5955–5960 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. et al. The genome of the cucumber, Cucumis sativus L. Nat Genet 41, 1275–1281 (2009). [DOI] [PubMed] [Google Scholar]
- Fu F. Q., Mao W. H., Shi K., Zhou Y. H. & Yu J. Q. Spatio-temporal changes in cell division, endoreduplication and expression of cell cycle-related genes in pollinated and plant growth substances-treated ovaries of cucumber. Plant Biol (Stuttg) 12, 98–107 (2010). [DOI] [PubMed] [Google Scholar]
- Marcelis L. F. M. & Hofman-Eijer L. Cell division and expansion in the cucumber fruit. Journal of horticultural science 68, 665–671 (1993). [Google Scholar]
- Ando K., Carr K. M. & Grumet R. Transcriptome analyses of early cucumber fruit growth identifies distinct gene modules associated with phases of development. BMC Genomics 13, 518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. The expression profiling of the CsPYL, CsPP2C and CsSnRK2 gene families during fruit development and drought stress in cucumber. J Plant Physiol 169, 1874–1882 (2012). [DOI] [PubMed] [Google Scholar]
- Qi J. et al. A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat Genet 45, 1510–1515 (2013). [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. & Salzberg S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Theodor P. P. & Huber W. HTSeq—A python framework to work with high-throughput sequencing data. BioRxiv preprint 10.1101/002824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M. et al. Sequence and ionomic analysis of divergent strains of maize inbred line B73 with an altered growth phenotype. PLoS One 9, e96782 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Toit A. Meiosis: A kinesin on foreign tracks. Nat Rev Mol Cell Biol 14, 400–401 (2013). [DOI] [PubMed] [Google Scholar]
- Ruden D. M., Cui W., Sollars V. & Alterman M. A Drosophila kinesin-like protein, Klp38B, functions during meiosis, mitosis, and segmentation. Dev Biol 191, 284–296 (1997). [DOI] [PubMed] [Google Scholar]
- Girin T. et al. INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell 23, 3641–3653 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M. G., Atkinson A., Bylstra Y. H., Walsh R. & Smyth D. R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128, 1089–1098 (2001). [DOI] [PubMed] [Google Scholar]
- Alvarez J. & Smyth D. R. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386 (1999). [DOI] [PubMed] [Google Scholar]
- Fujisawa M. et al. Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 26, 89–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q., Ferrandiz C., Yanofsky M. F. & Martienssen R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517 (1998). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25, 83–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. et al. HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 16, 2586–2600 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiser J. C., Robinson-Beers K. & Gasser C. S. The Arabidopsis SUPERMAN Gene Mediates Asymmetric Growth of the Outer Integument of Ovules. Plant Cell 7, 333–345 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. et al. Cucumber SUPERMAN has conserved function in stamen and fruit development and a distinct role in floral patterning. PLoS One 9, e86192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkena S., Lee E., Sack F. D. & Lamb R. S. The R2R3 MYB Transcription Factors FOUR LIPS and MYB88 Regulate Female Reproductive Development. Journal of Experimental Botany 63, 5545–5558 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machemer K. et al. Interplay of MYB factors in differential cell expansion, and consequences for tomato fruit development. Plant Journal 68, 337–350 (2011). [DOI] [PubMed] [Google Scholar]
- Wang Z., Gerstein M. & Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10, 57–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M., Wu S., Amand P. S., Stein N. & Poland J. Application of genotyping-by-sequencing on semiconductor sequencing platforms: a comparison of genetic and reference-based marker ordering in barley. PLoS One 8, e76925 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005). [DOI] [PubMed] [Google Scholar]
- Jones P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A., Rahnenfuhrer J. & Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 (2006). [DOI] [PubMed] [Google Scholar]
- Lohse M. et al. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell and Environment 37, 1250–1258 (2014). [DOI] [PubMed] [Google Scholar]
- Thimm O. et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant Journal 37, 914–939 (2004). [DOI] [PubMed] [Google Scholar]
- Wan H. et al. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399, 257–261 (2010). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fruit length supplementary data
Table S1
Table S2
Table S3