Abstract
Dual antiplatelet therapy of aspirin and clopidogrel is pivotal for patients undergoing percutaneous coronary intervention. However, the variable platelets reactivity response to clopidogrel may lead to outcome failure and recurrence of cardiovascular events. Although many genetic and nongenetic factors are known, great portion of clopidogrel variable platelets reactivity remain unexplained which challenges the personalization of clopidogrel therapy. Current methods for clopidogrel personalization include CYP2C19 genotyping, pharmacokinetics, and platelets function testing. However, these methods lack precise prediction of clopidogrel outcome, often leading to insufficient prediction. Pharmacometabolomics which is an approach to identify novel biomarkers of drug response or toxicity in biofluids has been investigated to predict drug response. The advantage of pharmacometabolomics is that it does not only predict the response but also provide extensive information on the metabolic pathways implicated with the response. Integrating pharmacogenetics with pharmacometabolomics can give insight on unknown genetic and nongenetic factors associated with the response. This review aimed to review the literature on factors associated with the variable platelets reactivity response to clopidogrel, as well as appraising current methods for the personalization of clopidogrel therapy. We also aimed to review the literature on using pharmacometabolomics approach to predict drug response, as well as discussing the plausibility of using it to predict clopidogrel outcome.
1. Introduction
Clopidogrel is a second-generation thienopyridine antiplatelet drug which exerts its effect by the inhibition of the platelet's purinergic receptor P2Y12 preventing adenosine diphosphate (ADP) from stimulating it. Clopidogrel is crucial drug for patients enduring high platelets reactivity such as coronary artery disease (CAD), acute coronary syndrome (ACS), and stroke. Some patients may require invasive therapy such as percutaneous coronary intervention (PCI) with stent placed in the occluded artery to ensure enough blood flow through it [1]. PCI patients have to take loading dose of clopidogrel prior to procedure followed by postprocedure dual antiplatelet therapy (DAPT) of low dose aspirin and clopidogrel for duration up to 12 months based on stent type and risk assessment [2]. This DAPT therapy is pivotal to prevent stent thrombosis (ST) and recurrence of ischemic events after PCI. However, some patients may suffer from attenuated platelets inhibition to clopidogrel or clopidogrel high on treatment platelets reactivity (HTPR) which hinders achieving the optimum outcome of DAPT. There are genetic and nongenetic factors contributing to clopidogrel HTPR, however, often challenging therapeutic outcome prediction [3]. Current methods of predicting clopidogrel response do not predict clopidogrel therapeutic outcome adequately. Therefore, investigating new approaches to assess clopidogrel response can help to achieve the desired outcome after PCI. In this review, we aimed to review the literature on clopidogrel variable platelets reactivity and appraise current methods to assess the clopidogrel therapeutic outcome. We also aimed to review the literature on new approaches such as pharmacometabolomics and integrative pharmacometabolomics-pharmacogenetics in assessing clopidogrel therapeutic outcome.
2. Clopidogrel Bioactivation and Clopidogrel HTPR
Clopidogrel is an oral drug which has oral bioavailability of 50% and the maximum peak concentration will be observed within 1 to 2 hours after the administration of the loading dose (600 mg) [4, 5]. The half-life of clopidogrel is from 7 to 8 hours [6]. Almost 50% of clopidogrel dose is excreted in the urine and 46% in the faeces [7]. Of the oral dose, approximately 85% is hydrolysed by esterases into inactive metabolite while the remaining 15% will be activated by the hepatic cytochrome P450 (CYP450) enzymes to the active metabolite through two steps of bioactivation [8]. The hepatic CYP450 enzymes which are involved in the bioactivation process of clopidogrel include the CYP1A2, CYP2B6, and CYP2C19 in the first step and the CYP2B6, CYP2C9, CYP3A4/5, and CYP2C19 in the second step [9–11]. The CYP2C19 enzyme plays vital role in the two bioactivation steps of clopidogrel by participating with 44.9% in the first step and 20.6% in the second step [9, 12]. The CYP3A4 has an essential role in the second step by participating with 39.8% [9]. Clopidogrel has minimum neutropenic side effect compared to ticlopidine (first-generation thienopyridine) [13]. The main side effects of clopidogrel are bleeding, gastrointestinal disorders, and rash, as well as other side effects such as hepatotoxicity and thrombotic thrombocytopenic purpura, albeit they are rare. Therefore, it is well tolerated by patients.
Patients variable platelets inhibition while on clopidogrel was first reported by Järemo et al. in 2002 [14]. In that study, five out of the eighteen PCI patients had weak platelets inhibition in response to clopidogrel loading dose of 300 mg. Since it was first reported, clopidogrel HTPR has been largely documented. It was found to be affecting 15–40% of the patients [4, 15, 16]. Clopidogrel HTPR is associated with poor outcome after PCI. Matetzky and colleagues indicated an association between clopidogrel HTPR and the risk of cardiac events' recurrence among 60 ACS patients undergoing PCI who had taken loading dose of 300 mg followed by daily dose of 75 mg for three months [17]. Substantiating these findings, Geisler et al. (2006) indicated that the primary end point of myocardial infarction, stroke, and death were significantly increased in clopidogrel HTPR patients who were followed up for three months after the PCI [18].
3. Genetic Factors Contributing to Clopidogrel HTPR
There are several identified genetic variabilities which contribute to clopidogrel HTPR. In fact, clopidogrel variable platelets reactivity is highly heritable [19]. As clopidogrel undergoes intestinal absorption, bioactivation by the hepatic CYP450 enzymes, and deactivation by esterases, this process might be affected by several genetic variabilities. Genetic variabilities which may interfere with clopidogrel variable platelets reactivity include the polymorphisms of the CYP2C19 [19], the CYP3A4/5 [20], the CYP2C9 [21], the ATP-Binding Cassette Subfamily B Member 1 (ABCB1) [22], the Paraoxonase-1 (PON1), the Carboxyl Esterase 1 (CES1) [23], and the genetic polymorphism of the P2Y12 receptors [24].
There are 57 active genes of the CYP450 enzymes and 58 pseudogenes in the human genome [25]. For most of the clinically relevant CYP450 enzymes, they are highly affected by genetic polymorphisms [26]. The CYP450 genetic polymorphisms could interfere with gene transcription, gene expression regulation, protein translation, and affinity to substrate [26, 27]. Among the CYP450 reported genetic polymorphisms, the CYP2D6, CYP2C9, and the CYP2C19 were extensively studied and reported, respectively [28]. Of all studied CYP450 genetic polymorphisms, the CYP2C19 polymorphisms were found to be highly associated with clopidogrel variable response, particularly that the enzyme is involved in the two bioactivation steps.
3.1. CYP2C19 Polymorphisms
The CYP2C19 enzyme plays substantial role in the metabolism and bioactivation of drugs such as proton pump inhibitors (PPIs) (omeprazole, lansoprazole, and pantoprazole), selective serotonin reuptake inhibitors (citalopram and sertraline), tricyclic antidepressant (imipramine and amitriptyline), phenytoin, and clopidogrel. The CYP2C19 enzyme is known by its classical reaction; the (S)-mephenytoin 4′-hydroxylation [29]. The gene of the CYP2C19 is highly polymorphic with more than 25 alleles having variable enzymatic activity levels [30]. The star one allele (∗1) is the normal or “wild type,” a homozygous form of this allele's (∗1/∗1) phenotypes for the gained full function of the enzyme. Of all polymorphic alleles of the CYP2C19, two reduced functions alleles were identified as (∗2) and (∗3) alleles have been extensively studied. De Morais et al. (1994) had found that they cause poor metabolism of (S)-mephenytoin metabolism among Japanese [29]. Both the CYP2C19∗2 and CYP2C19∗3 alleles are single nucleotide polymorphism (SNP) of guanine (G) to adenine (A) present at nucleotide 681 in exon 5 and at nucleotide 636 in exon 4, respectively [29, 31]. The CYP2C19∗2 causes aberrant splice site leading to the formation of stop codon which produces truncated, nonfunctional protein which is catalytically inactive [29]. The CYP2C19∗3 causes premature termination codon which leads to the formation of truncated, nonfunctioning protein which is also catalytically inactive [31]. The CYP2C19∗17 is another allele of the CYP2C19 which is associated with increased gene transcription that leads to increased activity of the enzyme [32]. Clinically, individuals are classified into extensive metabolizers (EM), intermediate metabolizers (IM), poor metabolizers (PM), and ultrametabolizer (UM) of drugs based on their CYP2C19 genotype [10]. Table 1 shows subjects' CYP2C19 genotype and their CYP2C19 phenotype. The presence of at least one allele of the CYP2C19∗2 is observed in 15–30%, 33–40%, and 40–50% of the Caucasians, African Americans, and Asians, respectively [33–35]. However, the CYP2C19∗3 allele is present in less than 1% of both Caucasians and African Americans and 7% of Asians.
Table 1.
Phenotype metabolising classes based on CYP2C19 genotype∗.
| Phenotype | Examples of the genotype | Enzyme activity |
|---|---|---|
| Ultrarapid metabolizer (UM) | ∗1/∗17 | Normal or increased enzyme activity |
| ∗17/∗17 | ||
|
| ||
| Extensive metabolizer (EM) | ∗1/∗1 | Normal enzyme activity |
|
| ||
| Intermediate metabolizer (IM) | ∗1/∗2 | Intermediate enzyme activity |
| ∗1/∗3 | ||
| ∗2/∗17 | ||
|
| ||
| Poor metabolizer (PM) | ∗2/∗2 | Low or absent enzyme activity |
| ∗3/∗3 | ||
| ∗2/∗3 | ||
∗This table is adapted and modified with permission from the publisher. Original source: Scott et al. (2013) [10]. Copyright© (2013). Published by Wiley. All rights reserved.
The importance of the CYP2C19 loss of function (LoF) alleles, CYP2C19∗2 and CYP2C19∗3 effect on clopidogrel response, stems not only from their association with clopidogrel HTPR, but also from their association with poor therapeutic outcome [36]. Poor metabolizers (based on CYP2C19 genotype) had lower plasma concentration of clopidogrel active metabolite [21]. Shuldiner and colleagues had found significant association between the CYP2C19∗2 genotype and both the low platelets' inhibition to clopidogrel and the increase of cardiovascular events among PCI patients [19]. Mega and colleagues reported low level of the active metabolite of clopidogrel, reduced platelets inhibition to clopidogrel, and higher risk of ischemic events among patients who are carriers of the CYP2C19 LoF alleles [34]. These results, as well as other findings, encouraged the United States (US) Food and Drug Administration (FDA) to issue labelled warning on clopidogrel box recommending the genotyping of CYP2C19 in order to prevent recurrence of cardiovascular events [12].
3.2. Other Genetic Polymorphisms
Apart from the CYP2C19 polymorphisms, the current literature is consistent about the limited effect of other CYP genetic polymorphisms on the exposure to clopidogrel active metabolite and platelets inhibition. Nevertheless, some controversy exists about some genetic variants. Brandt and colleagues investigated the effect of CYP polymorphisms on the exposure to clopidogrel active metabolite and platelets reactivity [21]. They indicated that the presence of the CYP2C19 or the CYP2C9 LoF variants was associated with reduced exposure to clopidogrel active metabolite and decreased platelets inhibition. However, the CYP1A2, the CYP2B6, the CYP3A4, and CYP3A5 did not have any effect. Park and colleagues indicated similar findings; however, they did not investigate the effect of CYP2C9 variants [37]. Although several studies showed that the CYP2B6 polymorphism did not have an effect, a study indicated that carriers of the CYP2B6 had lower exposure to clopidogrel active metabolite and lower platelets inhibition compared to noncarriers [34]. Although several studies suggested that the association between the CYP3A4 and the CYP3A5 polymorphisms and clopidogrel HTPR is not significant [34, 37–39], others indicated significant association between them [11, 20, 40]. This might be due to the major role of the CYP3A4 and CYP3A5 enzymes in the second step of clopidogrel bioactivation which may substantiate the effect of their genetic polymorphisms on clopidogrel exposure.
The ABCB1 gene is the coding gene for the P-glycoprotein multidrug resistance-1 (MDR1) intestinal transporter, a modulator of clopidogrel absorption [41]. A genetic polymorphism of the ABCB1 causes reduced absorption of clopidogrel [22]. Simon et al. (2009) found that ACS patients who were treated with clopidogrel and were carriers of the two variants alleles of the ABCB1 had higher rate of cardiovascular events at one year of follow-up [35]. Similarly, Mega et al. (2010) indicated that the ABCB1 polymorphism is significantly associated with the risk of deaths due to cardiovascular events or stroke among ACS patients treated with clopidogrel in the “Trial to Assess Improvement in the Therapeutic Outcomes by Optimizing Platelet Inhibition with Prasugrel-Thrombolysis in Myocardial Infarction” (TRITON-TIMI 38) trial [42]. However, other studies failed to indicate the same findings [19, 41].
As the majority of clopidogrel dose is being metabolized by the CES1 enzyme, a reduced function genetic polymorphism of this enzyme can lead to variable response to clopidogrel [23, 43]. Zhu et al. (2013) found that the inhibition of CES1 enzyme and the CES1 reduced function genetic polymorphisms are associated with the reduced hydrolytic metabolism of clopidogrel and the increased concentrations of clopidogrel active metabolite in an in vitro model [43]. Lewis et al. (2013) studied the effect of the CSE1 reduced function genetic variant on clopidogrel exposure and platelets inhibition among 566 healthy volunteers from the Amish “Pharmacogenomics of Anti-Platelet Intervention” (PAPI) study. The study revealed that the level of clopidogrel active metabolite is significantly higher among carriers of the reduced function allele [23]. In addition, the platelets inhibition measured by an ADP-stimulated platelet aggregation was higher in those who were carriers of the reduced function allele. Such findings suggest that the CES1 reduced function genetic polymorphisms may play vital role in the variable platelets inhibitory effect of clopidogrel. Thus, it would be valuable for future investigations to test this role on patients' outcome.
The PON1 is an esterase enzyme which is synthesized in the liver [44]. The PON1, full enzymatic activity, is associated with atheroprotective effect which can be due to its role in enhancing an increased level of the high-density-lipoprotein (HDL) [45–47]. Therefore, patients who were carriers of the PON1 lower activity genetic polymorphism were found to be having an increased oxidative stress and higher risk of cardiovascular events [45]. A presumed role of the PON1 enzyme in the metabolism of clopidogrel and its responsiveness stemmed controversy [46]. Bouman et al. (2011) used an in vitro metabolomics profiling which indicated that the PON1 is imperative enzyme in the hydrolytic cleavage of the intermediate metabolite of clopidogrel (2-oxo-clopidogrel) to the active metabolite of clopidogrel [44]. Accordingly, the PON1 genetic variant of lower enzymatic activity causes lower level of clopidogrel active metabolite. Based on their findings, they conducted a case-cohort study on CAD patients undergoing PCI and a further prospective replication study on another independent sample of 1982 patients with ACS [44]. Both the case-cohort and the prospective replication studies concluded that there was an association between the genetic variant of PON1 and the pharmacokinetics, the pharmacodynamics, and therapeutic outcome of clopidogrel [44]. These findings were challenged by other studies which indicated that this association is not significant [46, 47]. In fact, the effect of PON1 genetic variant on the therapeutic outcome was attributed to the association between this variant and the risk of developing cardiac events [46, 47].
The data on the effect of the P2Y12 genetic variants on clopidogrel response is dialectical. Some studies supported an effect of the P2Y12 polymorphism on platelets activation and clopidogrel response [24, 48, 49], while others refuted this effect [50, 51] or indicated a synergistic effect of it, if coexisted with other genetic variabilities such as CYP2C19 and MDR1 [52, 53]. There are two haplotypes of the P2Y12 gene (H1 and H2) where the H2 represents a gain-of-function haplotype of the receptor which might cause an increase in atherothrombosis and interfere with the response to clopidogrel [48]. The homozygous haplotype of H2 was found to be associated with clopidogrel HTPR [24].
Although the data are conflicting, it could be inferred from current literature that, except for the CYP2C19, most of the investigated genetic polymorphisms have weak or insignificant effect on the clopidogrel exposure and platelets inhibition. However, the coexistence of polymorphisms may have significant effect on platelets inhibition. In fact, the possession of the CYP2C19 polymorphism and other polymorphisms can lead to synergistic effect. For instance, Mega and colleagues concluded that PCI patients who have both ABCB1 and CYP2C19 genetic variants may be vulnerable to the recurrence of cardiac events while they are on clopidogrel treatment [42]. Similarly, some studies indicated that coexistence of CYP2C19 and P2Y12 receptor genetic polymorphisms has higher effect on clopidogrel responsiveness and the clinical outcome than single polymorphism [52, 53]. Furthermore, the use of a drug with enzymatic inhibitory effect in patients who have polymorphism of reduced enzymatic activity for the same enzyme may lower platelets inhibition to clopidogrel. Park et al. (2013) indicated that patients on calcium channel blockers (CCBs) who were carriers of the CYP3A4 (IVS 10 + 12) A allele (GA and AA) had higher platelets reactivity while on clopidogrel compared to the noncarriers-wilt type (GG) [54].
4. Nongenetic Factors Contributing to Clopidogrel HTPR
Other multiple nongenetic factors interfere with clopidogrel response. Clinical factors such as concomitant diseases, drug-drug interaction, patient's compliance, obesity, and age are known to influence clopidogrel response. In addition, lifestyle factors such as smoking and diet can also affect the response.
The presence of comorbid condition such as type 2 diabetes mellitus (T2-DM) increases platelets reactivity in CAD patients which may cause poor therapeutic outcome [55]. In fact, T2-DM patients were found to endure clopidogrel HTPR while on DAPT [56]. However, the interindividual clopidogrel variable platelets reactivity among CAD patients with T2-DM might be due to the concomitance of genetic or nongenetic factors [55, 57]. It had been suggested that the administration of high maintenance dose of clopidogrel or more potent P2Y12 antagonists such as prasugrel can be used to overcome the clopidogrel HTPR in T2-DM [58]. Similar to T2-DM, patients suffering from chronic kidney disease (CKD) had reduced platelets inhibition to clopidogrel [59]. The decreased platelets inhibition in CKD patients was indicated for both clopidogrel and aspirin in patients on DAPT [60]. This reduced response is suggested to be due to the increased baseline platelets reactivity in CKD patients [60, 61]. The association between renal insufficiency and the risk of death and cardiac events after PCI is well established [62]. However, inconsistent data is available, in terms of the risk of bleeding, among PCI patients with underlying CKD [63, 64]. Noteworthily, the concomitance of comorbidities such as T2-DM with moderate to severe CKD was found to be associated with high platelets reactivity in CAD patients on DAPT which might lead to high frequency of poor outcome in this group of patients [65]. Recently, in Japanese population, it has been found that CKD is an independent predictor of PCI outcome, regardless of the CYP2C19 genetic polymorphism status of the patient [66].
As clopidogrel is bioactivated by the CYP450 enzymes, it can be prone to drug-drug interactions, particularly drugs interfering with the CYP450 system. The PPIs are metabolized by the CYP450 enzymes; therefore, they might interact with clopidogrel [67]. However, the data on the extent of PPIs interact with clopidogrel remain controversial which suggests further investigation. In fact, the PPIs inhibit the CYP2C19 enzyme which might reduce the bioactivation of clopidogrel to its active metabolite [67, 68]. Accordingly, this could decrease the antiplatelet effect of clopidogrel which may increase cardiovascular events [68]. The PPIs may not interact to the same extent with clopidogrel [69]. In other words, some studies concluded that omeprazole was associated with the reduced antiplatelet effect of clopidogrel, while esomeprazole and pantoprazole were not [68, 69]. Similarly, lipophilic statins antihyperlipidemic drugs such as atorvastatin and simvastatin were reported to inhibit the antiplatelet effect of clopidogrel [70–72]. These statins interfere with the CYP3A4 enzyme [71]. Thus, it can mitigate the antiplatelet effect of clopidogrel. However, the preventive effect of statins by reducing lipid levels can halt cardiovascular events which might happen consequently to the reduced antiplatelet effect of clopidogrel [70]. Similar to the effect of statins albeit with some controversy in the literature, some drugs such as some CCBs, erythromycin, troleandomycin, and ketoconazole inhibit the CYP3A4 enzyme which will reduce the level of the clopidogrel active metabolite which can lead to reduced antiplatelet effect of clopidogrel [68, 73]. However, the data from some studies did not support this suggestion [74, 75]. For instance, the “Clopidogrel for the Reduction of Events During Observation” (CREDO) trial indicated that the concomitance use of the CCBs does not reduce the efficacy of clopidogrel and there was no evidence of interaction between them [75]. Anticoagulant drugs such as coumarin derivatives may also interfere with platelets inhibition by clopidogrel. Sibbing et al. (2010) indicated a significant reduction in the platelets' inhibitory effect of clopidogrel among patients concomitantly taking phenprocoumon when compared to those who are not taking phenprocoumon. This could be because of the interference with the capacities of the CYP3A4 and the CYP2C9 enzymes which are the main metabolizing enzymes of the phenprocoumon [76]. Sulfonylureas oral hypoglycaemic drugs may also reduce platelets inhibitory effect of clopidogrel [77]. This could be due to the interference with CYP2C9 enzyme, as well [77, 78]. On the other hand, there are also drugs which can increase or decrease the level of clopidogrel active metabolite by the inhibition or induction of the CES1 enzyme [43]. This might lead to clopidogrel variable platelets reactivity [79]. For instance, the phenobarbital, the dexamethasone, and the polycyclic aromatic hydrocarbons are drugs which can interfere with the activity of CES1 leading to variable levels of clopidogrel active metabolite [79].
Patient's compliance is crucial for achieving optimum therapeutic outcome. It has been found that noncompliance contributes significantly to clopidogrel HTPR [80]. Age, lipid levels, and body mass index (BMI) have been found to define nearly 22% of clopidogrel HTPR [19]. An age of more than 65 years old is associated with reduced platelets inhibitory effect of clopidogrel [81]. Furthermore, food, drinks, and smoking may interfere with clopidogrel response. For example, grapefruit can lower platelets inhibitory effect of clopidogrel which could be due to the interference with CYP450 isoforms [82]. In contrast, caffeinated drinks can increase platelets' inhibitory effect of clopidogrel [83]. Similarly, smoking was found to increase platelets' inhibitory effect of clopidogrel [84]. Smoking increases the bioactivation of clopidogrel to its active metabolite by the induction of the CYP1A2 and the CYP2B6 enzymes which are involved in the first step of the bioactivation of clopidogrel. This will increase platelets inhibition effect of clopidogrel.
5. Personalization of Clopidogrel Therapy
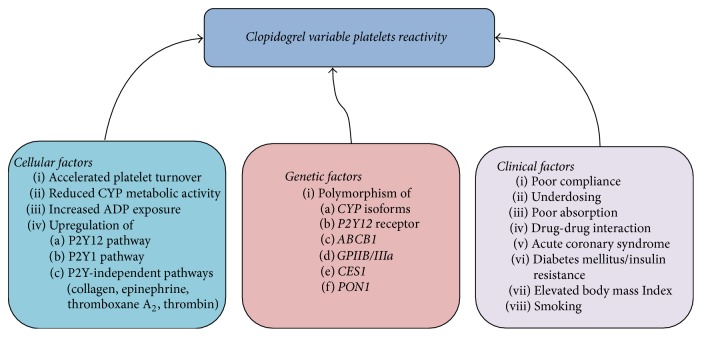
It could be clearly understood that there are multifactorial genetic and nongenetic factors which interfere with clopidogrel variable platelets reactivity as depicted in Figure 1 [85]. However, these known factors failed to explain great portion of the variable platelets reactivity of clopidogrel [86]. For instance, Frelinger et al. (2013) assessed the pharmacokinetics and the pharmacodynamics of 75 mg daily dose of clopidogrel, administered by 160 healthy subjects for 9 days. The 160 subjects were free from nicotine for 6 weeks, prescription drugs for 4 weeks, over-the-counter drugs for 2 weeks, and caffeine and alcohol for 72 hours. All participants were homogenous CYP2C19 EM genotype (∗1/∗1). They were genotyped for the ABCB1, PON1, and CYP3A5 polymorphisms. Of all participants, 45% had clopidogrel HTPR. Despite controlling the aforementioned factors, altogether factors such as age, weight, sex, platelet count, haematocrit, and genetic polymorphisms of ABCB1, PON1, and CYP3A5 explained only 18% of the clopidogrel active metabolite peak plasma concentration (Cmax) and the area under the plasma concentration-time curve (AUCt) variabilities. When Cmax and AUCt were added to these factors, they explained 48% of pharmacodynamics variability. Noteworthily, the results showed that there was no significant association between the polymorphisms of ABCB1, PON1, and CYP3A5 and the pharmacokinetics and pharmacodynamics of clopidogrel. These findings indicated the burden of the unexplained variability of clopidogrel response which consequently affects the therapeutic outcome of patients. This unsolved mystery can be due to unknown genetic polymorphisms or an interaction between several genetic polymorphisms and other nongenetic factors.
Figure 1.
The genetic and nongenetic factors which may contribute to clopidogrel variable platelets reactivity. The figure presents the known genetic and nongenetic factors (cellular and clinical factors) which may contribute to clopidogrel variable platelets reactivity. CYP: cytochrome P450, ADP: adenosine diphosphate, GPIIB/IIIa: Glycoprotein IIB/IIIa, ABCB1: ATP-Binding Cassette Subfamily B Member 1. This figure is adapted and modified with permission from the publisher. Original source: Angiolillo and Ferreiro (2010) [85]. Copyright© (2010) Sociedad Española de Cardiología. Published by Elsevier España SL. All rights reserved.
Personalized therapy is the use of person's genetics, proteomics, and environmental information to prevent, diagnose, and treat disease [87]. It aims to tailor patients therapy based on their genetic and nongenetic information rather than treating the whole population of a disease with the same treatment [88]. Since the discovery of clopidogrel variable platelets reactivity, it has been challenging to achieve optimum personalization of DAPT therapy. However, this optimum antiplatelet therapy could not be achieved without perfect prediction of the therapeutic outcome.
6. Adoption of Pharmacogenetics Biomarkers in Clinical Practice
The biomarker is a biological indicator of disease, physiological state, clinical status, response to drug therapy, or pathogenic process, which can be estimated and appraised for its indicative accuracy [89]. Accordingly, genetic variability which is associated with a biological status can be used as an indicative biomarker of that status. Pharmacogenetics biomarkers have been used to predict drug therapeutic outcome and avoid adverse drug reaction (ADR) prior to drug use. In 2008, the FDA issued table of valid pharmacogenetics biomarkers which contains list of drugs that had FDA label warning of pharmacogenetics testing prior to drug use and this list is frequently updated [90]. As there are several genetic factors which may interfere with clopidogrel variable platelets reactivity, genotyping of these polymorphisms was evaluated for clopidogrel outcome prediction [19, 34]. Apparently, the literature was consistent in this regard. The CYP2C19 polymorphism predominates the effect of other genetic variants. Thus, the FDA considered the CYP2C19 polymorphism valid pharmacogenetics biomarker of clopidogrel efficacy [90].
Although there are consistent literature asserting the association between clopidogrel HTPR and the CYP2C19∗2 and ∗3 LoF alleles, in depth analysis of the data indicated that this association is strong in the PM who are carriers of the homozygous genotypes of the CYP2C19 (∗2/∗2,∗3/∗3) but not for the same extent with the IM who are carriers of the heterozygous genotypes (∗1/∗2,∗1/∗3) [4, 91–93]. Furthermore, patients who are EM but suffering from clopidogrel HTPR would be misclassified as responsive (having optimum clopidogrel platelets inhibition) based on their CYP2C19 genotype [94]. Nasyuhana Sani and colleagues studied the effect of CYP2C19 genotype on clopidogrel platelets reactivity among healthy volunteers from three East Asian ethnicities [92]. They had found that although carriers of one or two alleles of the CYP2C19∗2 had significantly low platelets inhibition while on clopidogrel, low platelets inhibition was found in wild type homozygous carriers CYP2C19 (∗1/∗1) as well. Similarly, Mejin and colleagues had found that there was no significant association between clopidogrel HTPR and CYP2C19 genotype among Malaysian CAD patients [93]. A systematic review and meta-analysis had concluded that, with the exception of stent thrombosis, the CYP2C19 genotype is not significantly associated with cardiovascular events, despite its association with platelets aggregation [95]. Simply put, this can be merely justified by several facts. The CYP2C19∗2 allele contributes to 12% of clopidogrel variable platelets reactivity which means great portion of this variability remains unpredicted by the genotype [19, 96]. The total activity of the CYP2C19 enzyme is affected not only by the genotype but also by the interacting inducer and inhibitor drugs [97]. In other words, some EM subjects may experience poor metabolizing activity of the CYP2C19 due to concomitant administration of inhibitor drugs and vice versa; the PM subjects taking concomitant inducer drugs may experience normal metabolizing activity of the CYP2C19. Besides, although it might be with considerably minimal effect, the synergetic effect of other genetic variabilities such as the PON1, the CYP3A4/5, the ABCB1, the P2Y12 receptors, and the CES1 cannot be neglected [20, 22–24, 40]. Indeed, it is expensive and less practical to genotype all the suspected clopidogrel related genetic variabilities. In addition, the limited role of genetic testing could be due to the fact that most of genetic mutations have inadequate prediction of the outcome [98]. As early mentioned, in the study of Frelinger et al. (2013), the known nongenetic and genetic factors explained 18% of the clopidogrel pharmacokinetics (PK) and 32%–65% of clopidogrel pharmacodynamics (PD). This means great portion of clopidogrel HTPR cannot be identified by CYP2C19 genotyping. Therefore, the Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C19 genotype and Clopidogrel Therapy (CPIC), in its update (2013), did not recommend the CYP2C19 guided therapy for all patients [10].
7. Pharmacokinetics Assessment of Clopidogrel Response
Several studies had evaluated different pharmacokinetics (PK) methods to determine the level of the parent compound clopidogrel and its metabolites as an indicator of clopidogrel platelets inhibition [5, 99, 100]. It was found that the level of clopidogrel active metabolite is correlated with clopidogrel platelets inhibition [86, 101], and the PK of clopidogrel could be predicted by the CYP2C19 genotype [102]. However, applying PK to assess clopidogrel response is limited by several factors. The active thiol metabolite of clopidogrel is not stable which requires adding stabilizing agent within 30 seconds of blood sampling [101]. Some studies used indirect quantification of the active metabolite by quantifying the parent compound and the inactive metabolite [99, 103]. This means an accurate quantification of the active thiol metabolite in blood is difficult. Furthermore, the plasma level of the active metabolite of clopidogrel may not reflect the response in subjects affected by P2Y12 receptor polymorphism, receptor density, fibrinogen, platelets concentration, and accelerated turnover of platelets variabilities [86, 104]. Therefore, the PK assessment can estimate the level of the parent compound and the active metabolite but it may not be optimum method to personalize clopidogrel antiplatelet therapy unless it is done together with other approaches such as pharmacogenetics or platelets function testing (PFT) to provide further information that can help to individualize the therapy.
8. Implementation of Platelets Function Testing (PFT) to Personalize Clopidogrel Therapy
Due to the limited prediction power of the pharmacogenetics biomarkers of clopidogrel platelets inhibition, the suggestion to personalize antiplatelet therapy based on platelets function testing (PFT) gained attraction. This had increased the demand to evaluate the use of PFT assays in the assessment of clopidogrel therapeutic outcome. PFT is one of the important methods to assess the effect of antiplatelet drugs and platelets defects. The PFT assessment of antiplatelet drugs relies on the measurement of platelets inhibition which happened due to the effect of the drug. There are different PFT instruments which are widely used to assess antiplatelet drugs such as the optical light transmission aggregometry (LTA) [105], the VerifyNow® system (VN) (Accumetrics Inc., San Diego, California) [106], the vasodilator stimulated phosphoprotein phosphorylation assay (VASP-P) [107, 108], the multiple electrode platelet aggregometry (MEA) [105], the platelets function analyser (PFA-100) [109–111], the Impact-R assay [110], and the platelet-works® (PW) [110]. The PFT instruments have different techniques, agonist reagents, and testing kits to test different classes of antiplatelet drugs [105, 109]. The PFT agonist reagent induces platelets activation in the sample through stimulation of the drug target. The aggregation of the platelets will be measured by the instrument and it indicates the status of platelets activity after the effect of the drug. Table 2, compares some of the available PFT instruments. For each PFT instrument, there is a specified kit or agonist reagent to test clopidogrel response; it mainly contains ADP as stimulant of P2Y12 receptors. The LTA is the gold standard PFT assay [109]. However, the VASP-P assay is highly specific to P2Y12 receptor. Therefore, it is often considered the gold standard PFT assay for P2Y12 inhibitors. Several studies evaluated the ability of PFT assays to predict major cardiac events after PCI [112, 113]. However, the “Do Platelet Function Assays Predict Clinical Outcome in Clopidogrel-Pretreated Patients Undergoing Elective PCI” (Popular) study had compared the predictability of five different PFT assays [114]. The PFT assays were the LTA, VN, IMPACT-R, PW, and PFA-100. The study concluded that LTA, VN, and PW are the best PFT assessment predictors of atherothrombotic events including stent thrombosis, despite the limitation of low power in predicting bleeding risk. Generally, the use of PFT to tailor clopidogrel therapy has its limitations. The researchers from the “Double Randomization of a Monitoring Adjusted Antiplatelet Treatment versus a Common Antiplatelet Treatment for DES Implantation, and Interruption versus Continuation of Double Antiplatelet Therapy” (ARCTIC) clinical trial had concluded that tailoring antiplatelet therapy based on PFT did not improve outcomes [115]. This can be explained by several confounding factors. For many of PFT assays, the results are interfered by age, gender, ethnicities, diet, platelets count, and haematocrit which lowers their accuracy [109, 110, 116–118]. The results of PFT instruments which utilize platelets aggregation technique for measurement are highly affected by concomitant use of other antiplatelet drugs such as GPIIb-IIIa antagonists [119]. In fact, the simulation nature of PFT by using agonist in an in vitro setting to stimulate platelets activation does not perfectly resemble the physiological process of platelets activation which is affected by many internal pathways [108]. This may lead to PFT results that do not reflect the actual platelets function of the patients. Although the LTA and VASP-P assays are considered gold standard methods to measure platelets function, they are not advocated in clinical practice because they are time consuming and require lab facility and expert handling [120]. On the other hand, the VN-P2Y12 assay and other automated point of care (POC) PFT assays are fast and easy, but they are considerably expensive which limits the routine use of them. Platelets reactivity changes overtime in clopidogrel treated patients which necessitates frequent measurements for precise PFT based personalized therapy [121]. Moreover, there is inconsistency in the identification of clopidogrel responsiveness when the prespecified cut-off points of PFT assays were compared. In a study aimed to evaluate the agreement on clopidogrel HTPR identification between three PFT assays, the LTA, the VASP-P, and the VN-P2Y12, a reduced agreement was reported between the three assays. However, when the PFT results were tested for correlation as continuous variables, there was correlation between the results [122]. This implied that identifying clopidogrel HTPR based on one PFT assay is not enough and may lead to imperfect therapeutic decision. This might be one of the factors that hinders tailoring antiplatelet therapy based on PFT.
Table 2.
Platelets function testing devices.
| PFT device | Specimen | PFT technique | Laboratory requirement/point of care (POC) | Agonist reagents/Cartridges |
|---|---|---|---|---|
| Light transmission aggregometry (LTA) [105] | Platelet rich plasma (PRP) | Optical light transmission aggregometry | Requiring trained lab staff and laboratory facility to do the test | ADP, arachidonic acid, epinephrine |
|
| ||||
| Multiple electrode platelet aggregometry (MEA) [105] | Whole blood | Impedance aggregometry | Semiautomated | ADP, arachidonic acid, collagen |
|
| ||||
| VerifyNow® system (VN) [106] | Whole blood | Optical light transmission aggregometry | Fully automated, POC | P2Y12 kit Aspirin kit GP IIb/IIIa kit |
|
| ||||
| Platelets function analyser (PFA-100) [109–111] | Whole blood | High shear force dynamic flow system | Semiautomated, POC | Collagen with ADP Collagen with epinephrine |
|
| ||||
| Impact-R [110] | Whole blood | Shear stress platelet adhesion | Requiring trained lab staff and laboratory facility to do the test. Extensive sample handling is needed | ADP |
|
| ||||
| Vasodilator-stimulated phosphoprotein phosphorylation assay using flow cytometry (VASP-P) [107, 108] | Whole blood | Flow cytometric fluorescent quantification of intraplatelet VASP | Requiring trained lab staff, intensive lab work, sample preparation | VASP-P2Y12 assay |
|
| ||||
| Platelet works (PW) [110] | Whole blood | Single platelet count | Semiautomated | ADP |
POC: point of care, ADP: adenosine diphosphate, GP IIb/IIIa: glycoprotein IIb/IIIa, and VASP-P: vasodilator-stimulated phosphoprotein phosphorylation. P2Y12 kit: VerifyNow kit designed to test the P2Y12 receptor blockade to assess the response to P2Y12 inhibitors. Aspirin kit: VerifyNow kit designed to test the response to aspirin via arachidonic acid initiated reaction. GP IIb/IIIa kit: VerifyNow kit designed to test the response to GP IIb/IIIa inhibitors. VASP/P2Y12 assay: VASP phosphorylation correlates with the P2Y12 receptor inhibition.
9. The Role of Systems Biology in Personalized Medicine
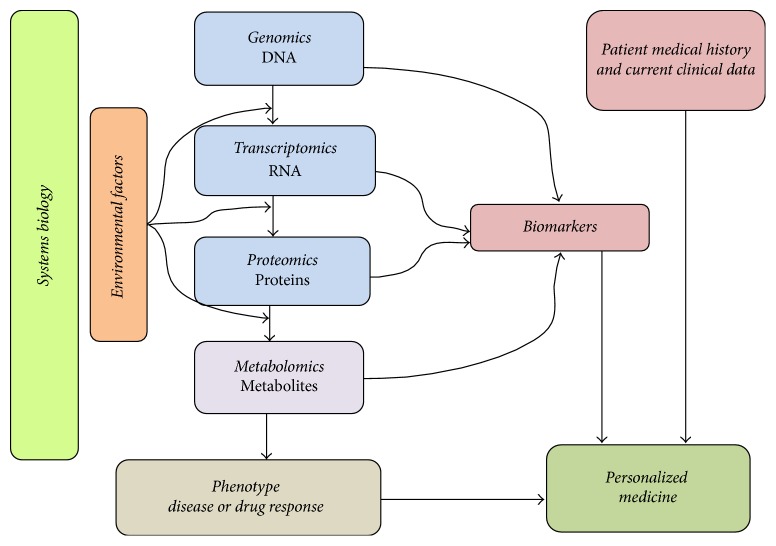
Systems biology is the study of the biochemical, molecular, and supramolecular networks, along with their connections and interactions with the environmental factors in order to investigate an intricate biological perturbation of the living organism [123]. This term is an integrative approach which investigates not only the biological “Omics” such as genomics, transcriptomics, proteomics, and metabolomics, but also the environmental factors such as nutrition, gut microbiota, infections, other drugs, disease state, rest, and sleep which interfere with them. In fact, the conventional biology describes the biological processes in the living system without considering the connections and interactions between them and their environment; however, systems biology is comprehensive in terms of explaining the biological processes, their pathways networks, and other environmental factors which all together contribute to a phenotype such as disease or clinical trait [124, 125]. This will give an extensive understanding of that phenotype by using the multidisciplinary “Omics” data as biomarkers of the phenotype [125]. While the study of the genes and their association with the phenotype is the main interest of genomics, the study of the messenger ribonucleic acid (mRNA) and transcription process is the aim of transcriptomics [126]. The proteomics focuses on peptides and proteins expression [127]. Eventually, as all the substrates in the organism will be subjected to metabolism, the metabolomics focuses on the study of the metabolites [128]. The role of systems biology in the prediction of the phenotype and the personalization of therapy is shown in Figure 2.
Figure 2.
The role of Systems Biology in Personalized Medicine. The figure elucidates how all “Omics” disciplines can provide phenotype biomarkers (disease or drug response biomarkers). Together with patient medical history and current clinical data, these biomarkers can help to personalize patient's therapy. The metabolomics is the ultimate “Omics” that is close to the phenotype, as well as reflecting the perturbation in other “Omics.” This figure is adapted and modified with permission from the publisher. Original source: Louridas and Lourida (2012). Published by Hippokratia. All rights reserved [125].
Apart from pharmacogenomics biomarkers which had been extensively studied, there is scant data on the use of other systems biology approaches such as transcriptomics, proteomics, and metabolomics to find novel biomarkers of clopidogrel variable platelets reactivity and cardiovascular outcome. In a proteomics approach, Caruso et al., 2015, were the first to report an investigation of platelets proteomes after 24 hours of clopidogrel loading dose in 12 ACS patients (6 responders and 6 nonresponders). They indicated significant upregulation of the platelet adhesion molecule cluster differentiation-226 (CD226), downregulation of peroxiredoxin-4, and increased expression of transferrin in patients suffering from clopidogrel HTPR [129]. To the best of our literature review, there is no study which had investigated the use of metabolomics to assess clopidogrel response.
10. Disease and Drug Response Phenotyping Using Metabolomics
Metabolomics (metabonomics in some literature) is the study of the metabolites of the living organisms' metabolome [130]. The metabolites are the ultimate small molecules compounds which are outcomes of the metabolism of the systems biology components and the environmental factors interfering with it [131]. The metabolome of each living system varies with the variations in the systems biology components and the various environmental factors affecting it [130]. Accordingly, the metabolomes of subjects having particular phenotype (disease condition or drug response) cluster in discriminatory way from their controls upon analysing the metabolome of their biological samples [132]. This differentiation usually happens due to the presence of discriminating metabolites in the metabolome which is termed the metabolic fingerprint or metabotype [133]. The identified metabotype can be used as a biomarker of the phenotype. The metabolomics analysis involves analysing biological samples such as urine, serum, plasma, exhaled breath, sputum, cerebrospinal fluids (CSF), and cell culture using spectroscopic instruments such as nuclear magnetic resonance (NMR) or mass spectroscopy (MS) to identify and quantify the metabolites in the sample [134–136]. Metabolomics approach has been used in several studies to find novel biomarkers of diseases. It was used to identify new biomarkers of asthma, chronic obstructive pulmonary disease (COPD), cancer, and other diseases [135, 137, 138]. The use of metabolomics approach to explore and understand different cardiac conditions has been evolved. In an animal model of rats harbouring human renin and angiotensinogen genes, Mervaala and colleagues used metabolomics to test whether angiotensin II alters the metabolic profile of the heart [139]. They concluded that angiotensin II plays key role in the metabolic profile of the heart. Tenori and colleagues investigated the use of NMR profiling of serum and urine to find a metabolic finger print associated with heart failure (HF) and its relationship with New York Heart Association (NYHA) functional classes in humans [140]. The NMR analysis of serum and urine significantly discriminated HF patients from their healthy controls with high accuracy (greater than 86%). However, it was not able to discriminate patients among the NYHA classes of HF.
11. Pharmacometabolomics
Pharmacometabolomics or pharmacometabonomics in some literature is a metabolomics analysis which aims to discover novel biomarkers in the metabolome that predict drug response or toxicity [141, 142]. These novel biomarkers can be used as classifying tool to classify patients to responsive and nonresponsive to drug or may develop and may not develop drug toxicity [143]. Drug response metabotype not only predicts patient's response but also explains response related pathways and monitor patient's outcome during disease management which will improve the personalization of therapy [136, 143, 144]. The term was first proposed by Clayton et al., 2006. They analysed urine samples of rat pre-dose and postdose of 600 mg of paracetamol using proton nuclear magnetic resonance (1H-NMR) spectroscopy to find metabotype which is associated with paracetamol induced hepatotoxicity [145]. It was revealed that high level of predose taurine was associated with low liver damage assessed by mean histology score (MHS) [145]. However, predose low levels of trimethylamine-N-oxide (TMAO) and betaine were associated with increased paracetamol induced liver damage. A repeat of the study in healthy volunteers indicated that high level of predose urinary p-cresol sulfate was associated with low postdose urinary ratios of acetaminophen sulfate to acetaminophen glucoronate (S/G) which is due to the competence between acetaminophen and p-cresol on sulfotransferase enzyme [144]. Thus, the high level of endogenous p-cresol causes an increase in liver amenability for acetaminophen hepatotoxicity and its urine sulfate form can be used as predose predictive biomarker of it. The pharmacometabolomics studies have been evolved in using different biofluids samples to find metabotypes of drug efficacy and toxicity [136, 146–149]. In Table 3, the findings of some pharmacometabolomics studies in humans are summarized.
Table 3.
Examples of pharmacometabolomics studies in humans.
| Study | Drug | Analytical method | Specimen | Main findings |
|---|---|---|---|---|
| Holmes et al., 2006 [136] | Antipsychotic drugs∗ | 1HNMR | CSF | A metabotype of schizophrenia which discriminated antipsychotic drug naive patients from healthy control. This metabotype was alleviated in half of the patients to normal after short term treatment with antipsychotic drugs. |
|
| ||||
| Kim et al., 2010 [146] | Cyclosporine A (CsA) & Tacrolimus (TAC) | 1HNMR | Serum | Time dependent metabolites changes in response to different treatment. The difference in the level of trimethylamine-N-oxide (TMAO) which is associated with graft dysfunction, between 2 groups of immunosuppressant drugs treatment; CsA & TAC were not significant. Lipid metabolites were higher in CsA group increasing the risk of cardiovascular diseases. |
|
| ||||
| Wang et al., 2012 [147] | Methotrexate | 1HNMR | Serum | Identification of 11 metabolites which can discriminate between methotrexate efficacy groups in patients with early rheumatoid arthritis. |
|
| ||||
| Kaddurah-Daouk et al., 2013 [148] | Sertraline | GC-TOF-MS | Serum | Discriminating metabotypes of symptoms reduction between sertraline and placebo in MDD patients on one week and 4 weeks of treatment. Symptoms reduction after one week of sertraline treatment was associated with the reduction of 5-methoxytryptamine levels, while it was associated with lower levels of branched chain amino acids at four weeks of sertraline treatment. |
|
| ||||
| Yerges-Armstrong et al., 2013 [164] | Aspirin | GC-MS | Serum | Identification of aspirin exposure metabotype of 18 metabolites in healthy volunteers who were on aspirin 81 mg for 14 days (HAPI study). Aspirin exposure metabotype was significantly associated with purine metabolic pathway. Inosine and adenosine were significantly higher after aspirin in aspirin HTPR group. |
|
| ||||
| Villaseñor et al., 2014 [149] | Ketamine | LC-QTOF-MS | Plasma | Identification of discriminating metabolites between responders and nonresponders of ketamine among bipolar depression patients. The discriminating metabolites were related to mitochondrial β-oxidation of fatty acid. |
|
| ||||
| Ellero-Simatos et al., 2014 [166] | Aspirin | LC-MS | Serum | Elevated level of baseline serum serotonin is associated with aspirin HTPR based on collagen induced PFT assessment of healthy subjects who were on aspirin 81 mg for 14 days (HAPI study). |
1HNMR: proton nuclear magnetic resonance, GC: gas chromatography, TOF: time of flight, MS: mass spectroscopy, QTOF: quadrupole time of flight, LC: liquid chromatography, CSF: cerebrospinal fluid, CVDs: cardiovascular diseases, CsA: Cyclosporine A, TAC: Tacrolimus, TMAO: trimethylamine-N-oxide, MDD: major depressive disorder, HAPI: Heredity and Phenotype Intervention, and HTPR: high on treatment platelets reactivity.
∗ Antipsychotic drugs: atypical antipsychotic drug such as amisulpride, clozapine, olanzapine, risperidone, quetiapine, and ziprasidone.
11.1. Pharmacometabolomics Spectroscopic Techniques
There are many spectroscopic methods commonly used in metabolomics/pharmacometabolomics studies such as nuclear magnetic resonance (NMR) or 1H-NMR, MS combined with gas or liquid chromatographic extraction (GC, LC) and Fourier transform infrared spectroscopy (FT-IR) [150–152]. Among them, 1H-NMR and MS are widely used [153]. Each of them has its advantages and limitations. The selection of the suitable spectroscopic method in a study depends on the features of each method and the compatibility of the method with the study's aim. The GC or LC combined MS method is more sensitive than 1H-NMR; thus it is highly recommended for metabolomics studies where particular group of compounds is being targeted such as lipid compounds [152, 154]. However, as it requires prior separation using LC or GC, it is time consuming and necessitates more lab work [145]. The 1H-NMR method is fast and does not require extensive sample processing [155]. Besides, prepared 1H-NMR samples can be run in large quantities with automated bulk run of samples [156, 157]. Samples prepared for 1H-NMR analysis can be stored and rerun for further analysis. Therefore, it is preferred choice for untargeted metabolomics analysis of large number of patients' samples.
11.2. Pharmacometabolomics Data Analysis
Pharmacometabolomics data analysis relies on prediction modelling using chemometrics analysis [158]. This includes multivariate analysis through the use of unsupervised and supervised machine learning methods [159]. The commonly used unsupervised method in metabolomics is the principal component analysis (PCA), while the supervised methods include methods such as partial least square (PLS), discriminant function analysis (DFA), and artificial neural networks (ANNs). The PCA provides an overview screening for the systematic variation of the data regardless of prior classification. Thus, it is exploratory analysis and usually followed by the supervised analysis where a prior input classification is introduced. The identification and pathway interpretation of the metabolites include combining knowledge in the pathophysiology of the phenotype with the analytical chemistry of the potential compound [160]. There are available databases which can be used to identify the putative biomarkers in metabolomics analysis such as Human Metabolome Database (HMDB), the Chenomx (Chenomx® Inc., Canada), the MetaboLights, the Birmingham Metabolite Library (BML-NMR), and the Biological Magnetic Resonance Data Bank (BMRB-metabolomics).
12. Pharmacometabolomics Informs Pharmacogenomics
The “pharmacometabolomics informs pharmacogenomics” approach implies that pharmacometabolomics can be used to identify genetic variation which is associated with the variation of drug response or toxicity [161]. Simply put, this concept is based on the fact that the variation in genes or genes' expression may lead to variations in proteins and eventually the metabolites levels which are associated with these pathways will change [162]. Accordingly, a metabotype which is associated with drug response may contain some metabolites associated with the variations in genes or genes expression which are implicated in the variable response. Ji and colleagues were the first to elucidate this concept on the variable response to the selective serotonin reuptake inhibitor (SSRI) escitalopram [163]. From major depressive disorder (MDD) patients on escitalopram whom were included in the Myo-PGRN SSRI study, 20 responders and 20 nonresponders were recruited for plasma pharmacometabolomics analysis. Baseline glycine levels were significantly different (P = 0.005) between the two groups and had negative association with the treatment outcome (remission) [163]. The researchers studied the glycine biosynthesis and metabolism to assign SNPs which are possibly related to these pathways. The selected SNPs were genotyped using the DNA of 512 patients from the same study (Myo-PGRN SSRI). Of the 135 assigned SNPs, the rs10975641 SNP from the glycine dehydrogenase gene was associated with response and remission. Similarly, using the same approach novel genetic variability associated with aspirin HTPR was discovered [164, 165]. Yerges-Armstrong et al. (2013) identified aspirin response metabotype in serum samples of healthy volunteers from the Heredity and Phenotype Intervention (HAPI) Heart study [164]. Aspirin HTPR group had significantly higher postaspirin levels of purine metabolites: inosine and adenosine. Association analysis of SNPs in the purine genes and aspirin HTPR led to the identification of the rs16931294 SNP in the adenosine kinase (ADK) gene region. Aspirin HTPR was significantly associated with the less common G allele rather than the more common A allele of the rs16931294 (β = 0.8, P = 0.00034). Furthermore, the G allele was significantly associated with preaspirin levels of adenosine monophosphate, hypoxanthine, and xanthine and postaspirin levels of inosine and guanosine. Therefore, pharmacometabolomics gained information may help to discover unknown genetic variations and response metabotypes can be used as an economical tool to predict both the response and the possible genetic cause of the response.
13. Conclusion
Clopidogrel had been well established as essential antiplatelet therapy paired with aspirin in DAPT therapy. Due to its high margin of safety, in terms of bleeding and side effects, and its price, clopidogrel remains the preferred P2Y12 antagonists in clinical practice. However, some patients fail to achieve therapeutic outcome while on clopidogrel. Finding precise method for predicting clopidogrel outcome is crucial to guide antiplatelet therapy. Current methods, the CYP2C19 genotyping, PK, and PFT, have their drawbacks. Investigating other systems biology approaches such as proteomics and metabolomics may be the preeminence for clopidogrel personalization. Pharmacometabolomics approach has been used in assessing drug response, as well as identifying novel genetic variations associated with the response. As the perturbation in the metabolome is close to the phenotype, it can be highly reflective of drug's variable response. To date, pharmacometabolomics had never been evaluated in personalizing clopidogrel therapy. An integration of pharmacometabolomics and pharmacogenomics may not only tailor the therapy but also provide better understanding of the mystery behind clopidogrel HTPR.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Torpy J. M., Lynm C., Glass R. M. Percutaneous coronary intervention. The Journal of the American Medical Association. 2004;291(6):p. 778. doi: 10.1001/jama.291.6.778. [DOI] [PubMed] [Google Scholar]
- 2.Levine G. N., Bates E. R., Bittl J. A., et al. 2016 ACC/AHA guideline focused update on duration of dual antiplatelet therapy in patients with coronary artery disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Journal of the American College of Cardiology. 2016;68(10):1082–1115. doi: 10.1016/j.jacc.2016.03.513. [DOI] [PubMed] [Google Scholar]
- 3.Goswami S., Cheng-Lai A., Nawarskas J. Clopidogrel and genetic testing: is it necessary for everyone? Cardiology in Review. 2012;20(2):96–100. doi: 10.1097/crd.0b013e3182455744. [DOI] [PubMed] [Google Scholar]
- 4.Oprea A. D., Popescu W. M. P2Y12 receptor inhibitors in acute coronary syndromes: what is new on the horizon? Cardiology Research and Practice. 2013;2013:15. doi: 10.1155/2013/195456.195456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taubert D., Kastrati A., Harlfinger S., et al. Pharmacokinetics of clopidogrel after administration of a high loading dose. Thrombosis and Haemostasis. 2004;92(2):311–316. doi: 10.1160/TH04-02-0105. [DOI] [PubMed] [Google Scholar]
- 6.Schrör K. Clinical pharmacology of the adenosine diphosphate (ADP) receptor antagonist, clopidogrel. Vascular Medicine. 1998;3(3):247–251. doi: 10.1177/1358836X9800300312. [DOI] [PubMed] [Google Scholar]
- 7.Shepard R. W. Pharmacology: antiplatelet and antithrombin therapy in acute coronary syndromes. The Journal of cardiovascular nursing. 2000;15(1):54–61. doi: 10.1097/00005082-200010000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Shameem R., Hamid M. S., Randhawa A., Spaccavento C., Garatt K. P2Y12 Antagonists: pharmacology, efficacy and patient considerations. Journal of Cardiovascular Disease. 2014;2:91–100. [Google Scholar]
- 9.Kazui M., Nishiya Y., Ishizuka T., et al. Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metabolism and Disposition. 2010;38(1):92–99. doi: 10.1124/dmd.109.029132. [DOI] [PubMed] [Google Scholar]
- 10.Scott S. A., Sangkuhl K., Stein C. M., et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clinical Pharmacology and Therapeutics. 2013;94(3):317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Priyadharsini R., Shewade D. G., Subraja K., et al. Single nucleotide polymorphism of CYP3A5∗3 contributes to clopidogrel resistance in coronary artery disease patients among Tamilian population. Molecular Biology Reports. 2014;41(11):7265–7271. doi: 10.1007/s11033-014-3613-8. [DOI] [PubMed] [Google Scholar]
- 12.Holmes D. R., Jr., Dehmer G. J., Kaul S., Leifer D., O'Gara P. T., Stein C. M. ACCF/AHA clopidogrel clinical alert: approaches to the FDA ‘boxed warning’: a report of the American College of Cardiology Foundation Task Force on clinical expert consensus documents and the American Heart Association endorsed by the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons. Journal of the American College of Cardiology. 2010;56(4):321–341. doi: 10.1016/j.jacc.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Maseneni S., Donzelli M., Taegtmeyer A. B., Brecht K., Krähenbühl S. Toxicity of clopidogrel and ticlopidine on human myeloid progenitor cells: importance of metabolites. Toxicology. 2012;299(2-3):139–145. doi: 10.1016/j.tox.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 14.Järemo P., Lindahl T. L., Fransson S. G., Richter A. Individual variations of platelet inhibition after loading doses of clopidogrel. Journal of Internal Medicine. 2002;252(3):233–238. doi: 10.1046/j.1365-2796.2002.01027.x. [DOI] [PubMed] [Google Scholar]
- 15.Gurbel P. A., Tantry U. S. Drug insight: clopidogrel nonresponsiveness. Nature Clinical Practice Cardiovascular Medicine. 2006;3(7):387–395. doi: 10.1038/ncpcardio0602. [DOI] [PubMed] [Google Scholar]
- 16.Gurbel P. A., Tantry U. S. Clopidogrel resistance? Thrombosis Research. 2007;120(3):311–321. doi: 10.1016/j.thromres.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 17.Matetzky S., Shenkman B., Guetta V., et al. Clopidogrel resistance is associated with increased risk of recurrent atherothrombotic events in patients with acute myocardial infarction. Circulation. 2004;109(25):3171–3175. doi: 10.1161/01.cir.0000130846.46168.03. [DOI] [PubMed] [Google Scholar]
- 18.Geisler T., Langer H., Wydymus M., et al. Low response to clopidogrel is associated with cardiovascular outcome after coronary stent implantation. European Heart Journal. 2006;27(20):2420–2425. doi: 10.1093/eurheartj/ehl275. [DOI] [PubMed] [Google Scholar]
- 19.Shuldiner A. R., O'Connell J. R., Bliden K. P., et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. The Journal of the American Medical Association. 2009;302(8):849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suh J.-W., Koo B.-K., Zhang S.-Y., et al. Increased risk of atherothrombotic events associated with cytochrome P450 3A5 polymorphism in patients taking clopidogrel. Canadian Medical Association Journal. 2006;174(12):1715–1722. doi: 10.1503/cmaj.060664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brandt J. T., Close S. L., Iturria S. J., et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. Journal of Thrombosis and Haemostasis. 2007;5(12):2429–2436. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 22.Su J., Xu J., Li X., et al. ABCB1 C3435T polymorphism and response to clopidogrel treatment in coronary artery disease (CAD) patients: a meta-analysis. PLoS ONE. 2012;7(10) doi: 10.1371/journal.pone.0046366.e46366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis J. P., Horenstein R. B., Ryan K., et al. The functional G143E variant of carboxylesterase 1 is associated with increased clopidogrel active metabolite levels and greater clopidogrel response. Pharmacogenetics and Genomics. 2013;23(1):1–8. doi: 10.1097/FPC.0b013e32835aa8a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staritz P., Kurz K., Stoll M., Giannitsis E., Katus H. A., Ivandic B. T. Platelet reactivity and clopidogrel resistance are associated with the H2 haplotype of the P2Y12-ADP receptor gene. International Journal of Cardiology. 2009;133(3):341–345. doi: 10.1016/j.ijcard.2007.12.118. [DOI] [PubMed] [Google Scholar]
- 25.Ingelman-Sundberg M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends in Pharmacological Sciences. 2004;25(4):193–200. doi: 10.1016/j.tips.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Bains R. K. African variation at Cytochrome P450 genes: evolutionary aspects and the implications for the treatment of infectious diseases. Evolution, Medicine, and Public Health. 2013;2013(1):118–134. doi: 10.1093/emph/eot010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee I., Kim D. Polymorphic metabolism by functional alterations of human cytochrome P450 enzymes. Archives of Pharmacal Research. 2011;34(11):1799–1816. doi: 10.1007/s12272-011-1103-2. [DOI] [PubMed] [Google Scholar]
- 28.Ingelman-Sundberg M., Sim S. C., Gomez A., Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacology & Therapeutics. 2007;116(3):496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 29.De Morais S. M. F., Wilkinson G. R., Blaisdell J., Nakamura K., Meyer U. A., Goldstein J. A. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. Journal of Biological Chemistry. 1994;269(22):15419–15422. [PubMed] [Google Scholar]
- 30.Lee S.-J., Kim W.-Y., Kim H., Shon J.-H., Sang S. L., Shin J.-G. Identification of new CYP2C19 variants exhibiting decreased enzyme activity in the metabolism of S-mephenytoin and omeprazole. Drug Metabolism and Disposition. 2009;37(11):2262–2269. doi: 10.1124/dmd.109.028175. [DOI] [PubMed] [Google Scholar]
- 31.De Morais S. M. F., Wilkinson G. R., Blaisdell J., Meyer U. A., Nakamura K., Goldstein J. A. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Molecular Pharmacology. 1994;46(4):594–598. [PubMed] [Google Scholar]
- 32.Sim S. C., Risinger C., Dahl M.-L., et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clinical Pharmacology and Therapeutics. 2006;79(1):103–113. doi: 10.1016/j.clpt.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Luo H.-R., Poland R. E., Lin K.-M., Wan Y.-J. Y. Genetic polymorphism of cytochrome P450 2C19 in Mexican Americans: a cross-ethnic comparative study. Clinical Pharmacology and Therapeutics. 2006;80(1):33–40. doi: 10.1016/j.clpt.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Mega J. L., Close S. L., Wiviott S. D., et al. Cytochrome P-450 polymorphisms and response to clopidogrel. New England Journal of Medicine. 2009;360(4):354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 35.Simon T., Verstuyft C., Mary-Krause M., et al. Genetic determinants of response to clopidogrel and cardiovascular events. New England Journal of Medicine. 2009;360(4):363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 36.Mega J. L., Simon T., Collet J.-P., et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. The Journal of the American Medical Association. 2010;304(16):1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park J. J., Park K. W., Kang J., et al. Genetic determinants of clopidogrel responsiveness in Koreans treated with drug-eluting stents. International Journal of Cardiology. 2013;163(1):79–86. doi: 10.1016/j.ijcard.2012.09.075. [DOI] [PubMed] [Google Scholar]
- 38.Smith S. M. G., Judge H. M., Peters G., et al. Common sequence variations in the P2Y12 and CYP3A5 genes do not explain the variability in the inhibitory effects of clopidogrel therapy. Platelets. 2006;17(4):250–258. doi: 10.1080/09537100500475844. [DOI] [PubMed] [Google Scholar]
- 39.Fontana P., Hulot J.-S., De Moerloose P., Gaussem P. Influence of CYP2C19 and CYP3A4 gene polymorphisms on clopidogrel responsiveness in healthy subjects. Journal of Thrombosis and Haemostasis. 2007;5(10):2153–2155. doi: 10.1111/j.1538-7836.2007.02722.x. [DOI] [PubMed] [Google Scholar]
- 40.Angiolillo D. J., Fernandez-Ortiz A., Bernardo E., et al. Contribution of gene sequence variations of the hepatic cytochrome P450 3A4 enzyme to variability in individual responsiveness to clopidogrel. Arteriosclerosis, Thrombosis, and Vascular Biology. 2006;26(8):1895–1900. doi: 10.1161/01.ATV.0000223867.25324.1a. [DOI] [PubMed] [Google Scholar]
- 41.Jaitner J., Morath T., Byrne R. A., et al. No association of ABCB1 C3435T genotype with clopidogrel response or risk of stent thrombosis in patients undergoing coronary stenting. Circulation: Cardiovascular Interventions. 2012;5(1):82–88. doi: 10.1161/circinterventions.111.965400. [DOI] [PubMed] [Google Scholar]
- 42.Mega J. L., Close S. L., Wiviott S. D., et al. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. The Lancet. 2010;376(9749):1312–1319. doi: 10.1016/s0140-6736(10)61273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu H.-J., Wang X., Gawronski B. E., Brinda B. J., Angiolillo D. J., Markowitz J. S. Carboxylesterase 1 as a determinant of clopidogrel metabolism and activations. Journal of Pharmacology and Experimental Therapeutics. 2013;344(3):665–672. doi: 10.1124/jpet.112.201640. [DOI] [PubMed] [Google Scholar]
- 44.Bouman H. J., Schomig E., van Werkum J. W., et al. Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nature Medicine. 2011;17(1):110–116. doi: 10.1038/nm.2281. [DOI] [PubMed] [Google Scholar]
- 45.Bhattacharyya T., Nicholls S. J., Topol E. J., et al. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. The Journal of the American Medical Association. 2008;299(11):1265–1276. doi: 10.1001/jama.299.11.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis J. P., Shuldiner A. R. Paraoxonase 1 Q192R variant and clopidogrel efficacy fact or fiction? Circulation: Cardiovascular Genetics. 2012;5(2):153–155. doi: 10.1161/CIRCGENETICS.112.962910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park K. W., Park J. J., Kang J., et al. Paraoxonase 1 gene polymorphism does not affect clopidogrel response variability but is associated with clinical outcome after PCI. PLoS ONE. 2013;8(2) doi: 10.1371/journal.pone.0052779.e52779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fontana P., Dupont A., Gandrille S., et al. Adenosine diphosphate-induced platelet aggregation is associated with P2Y12 gene sequence variations in healthy subjects. Circulation. 2003;108(8):989–995. doi: 10.1161/01.CIR.0000085073.69189.88. [DOI] [PubMed] [Google Scholar]
- 49.Zoheir N., Elhamid S. A., Abulata N., Sobky M. E., Khafagy D., Mostafa A. P2Y12 receptor gene polymorphism and antiplatelet effect of clopidogrel in patients with coronary artery disease after coronary stenting. Blood Coagulation and Fibrinolysis. 2013;24(5):525–531. doi: 10.1097/MBC.0b013e32835e98bf. [DOI] [PubMed] [Google Scholar]
- 50.Bonello L., Bonello-Palot N., Armero S., et al. Impact of P2Y12-ADP receptor polymorphism on the efficacy of clopidogrel dose-adjustment according to platelet reactivity monitoring in coronary artery disease patients. Thrombosis Research. 2010;125(4):e167–e170. doi: 10.1016/j.thromres.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 51.Von Beckerath N., Von Beckerath O., Koch W., Eichinger M., Schömig A., Kastrati A. P2Y12 gene H2 haplotype is not associated with increased adenosine diphosphate-induced platelet aggregation after initiation of clopidogrel therapy with a high loading dose. Blood Coagulation and Fibrinolysis. 2005;16(3):199–204. doi: 10.1097/01.mbc.0000164429.21040.0a. [DOI] [PubMed] [Google Scholar]
- 52.Tang X.-F., Zhang J.-H., Wang J., et al. Effects of coexisting polymorphisms of CYP2C19 and P2Y12 on clopidogrel responsiveness and clinical outcome in patients with acute coronary syndromes undergoing stent-based coronary intervention. Chinese Medical Journal. 2013;126(6):1069–1075. doi: 10.3760/cma.j.issn.0366-6999.20123360. [DOI] [PubMed] [Google Scholar]
- 53.Shalia K. K., Shah V. K., Pawar P., Divekar S. S., Payannavar S. Polymorphisms of MDR1, CYP2C19 and P2Y12 genes in Indian population: effects on clopidogrel response. Indian Heart Journal. 2013;65(2):158–167. doi: 10.1016/j.ihj.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park J. J., Park K. W., Kang J., et al. CYP3A4 genetic status may be associated with increased vulnerability to the inhibitory effect of calcium-channel blockers on clopidogrel. Circulation Journal. 2013;77(5):1289–1296. doi: 10.1253/circj.CJ-12-0682. [DOI] [PubMed] [Google Scholar]
- 55.Angiolillo D. J., Bernardo E., Zanoni M., et al. Impact of insulin receptor substrate-1 genotypes on platelet reactivity and cardiovascular outcomes in patients with type 2 diabetes mellitus and coronary artery disease. Journal of the American College of Cardiology. 2011;58(1):30–39. doi: 10.1016/j.jacc.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 56.Angiolillo D. J., Fernandez-Ortiz A., Bernardo E., et al. Platelet function profiles in patients with type 2 diabetes and coronary artery disease on combined aspirin and clopidogrel treatment. Diabetes. 2005;54(8):2430–2435. doi: 10.2337/diabetes.54.8.2430. [DOI] [PubMed] [Google Scholar]
- 57.Carreras E. T., Hochholzer W., Frelinger A. L., et al. Diabetes mellitus, CYP2C19 genotype, and pharmacodynamic response to clopidogrel dosing: insights from the ELEVATE-TIMI 56 trial. Circulation. 2013;128 doi: 10.1160/TH15-12-0981.A12325 [DOI] [PubMed] [Google Scholar]
- 58.Wiviott S. D., Braunwald E., Angiolillo D. J., et al. Greater clinical benefit of more intensive oral antiplatelet therapy with prasugrel in patients with diabetes mellitus in the trial to assess improvement in therapeutic outcomes by optimizing platelet inhibition with prasugrel–thrombolysis in myocardial infarction 38. Circulation. 2008;118(16):1626–1636. doi: 10.1161/circulationaha.108.791061. [DOI] [PubMed] [Google Scholar]
- 59.Muller C., Caillard S., Jesel L., et al. Association of estimated GFR with platelet inhibition in patients treated with clopidogrel. American Journal of Kidney Diseases. 2012;59(6):777–785. doi: 10.1053/j.ajkd.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 60.Gremmel T., Müller M., Steiner S., et al. Chronic kidney disease is associated with increased platelet activation and poor response to antiplatelet therapy. Nephrology Dialysis Transplantation. 2013;28(8):2116–2122. doi: 10.1093/ndt/gft103. [DOI] [PubMed] [Google Scholar]
- 61.Geisler T., Graß D., Bigalke B., et al. The residual platelet aggregation after deployment of intracoronary stent (PREDICT) score. Journal of Thrombosis and Haemostasis. 2008;6(1):54–61. doi: 10.1111/j.1538-7836.2007.02812.x. [DOI] [PubMed] [Google Scholar]
- 62.Best P. J. M., Lennon R., Ting H. H., et al. The impact of renal insufficiency on clinical outcomes in patients undergoing percutaneous coronary interventions. Journal of the American College of Cardiology. 2002;39(7):1113–1119. doi: 10.1016/S0735-1097(02)01745-X. [DOI] [PubMed] [Google Scholar]
- 63.Latif F., Kleiman N. S., Cohen D. J., et al. In-hospital and 1-year outcomes among percutaneous coronary intervention patients with chronic kidney disease in the era of drug-eluting stents : a report from the EVENT (evaluation of drug eluting stents and ischemic events) registry. JACC: Cardiovascular Interventions. 2009;2(1):37–45. doi: 10.1016/j.jcin.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 64.Best P. J. M., Steinhubl S. R., Berger P. B., et al. The efficacy and safety of short- and long-term dual antiplatelet therapy in patients with mild or moderate chronic kidney disease: results from the Clopidogrel for the Reduction of Events During Observation (CREDO) Trial. American Heart Journal. 2008;155(4):687–693. doi: 10.1016/j.ahj.2007.10.046. [DOI] [PubMed] [Google Scholar]
- 65.Angiolillo D. J., Bernardo E., Capodanno D., et al. Impact of chronic kidney disease on platelet function profiles in diabetes mellitus patients with coronary artery disease taking dual antiplatelet therapy. Journal of the American College of Cardiology. 2010;55(11):1139–1146. doi: 10.1016/j.jacc.2009.10.043. [DOI] [PubMed] [Google Scholar]
- 66.Tabata N., Hokimoto S., Akasaka T., et al. Chronic kidney disease status modifies the association of CYP2C19 polymorphism in predicting clinical outcomes following coronary stent implantation. Thrombosis Research. 2014;134(5):939–944. doi: 10.1016/j.thromres.2014.07.039. [DOI] [PubMed] [Google Scholar]
- 67.Abraham N. S., Hlatky M. A., Antman E. M., et al. ACCF/ACG/AHA 2010 expert consensus document on the concomitant use of proton pump inhibitors and thienopyridines: a focused update of the ACCF/ACG/AHA 2008 expert consensus document on reducing the gastrointestinal risks of antiplatelet therapy and NSAID use. Journal of the American College of Cardiology. 2010;56(24):2051–2066. doi: 10.1016/j.jacc.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 68.Bates E. R., Lau W. C., Angiolillo D. J. Clopidogrel-drug interactions. Journal of the American College of Cardiology. 2011;57(11):1251–1263. doi: 10.1016/j.jacc.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 69.Siller-Matula J. M., Spiel A. O., Lang I. M., Kreiner G., Christ G., Jilma B. Effects of pantoprazole and esomeprazole on platelet inhibition by clopidogrel. American Heart Journal. 2009;157(1):148.e1–148.e5. doi: 10.1016/j.ahj.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 70.Schmidt M., Johansen M. B., Maeng M., et al. Concomitant use of clopidogrel and statins and risk of major adverse cardiovascular events following coronary stent implantation. British Journal of Clinical Pharmacology. 2012;74(1):161–170. doi: 10.1111/j.1365-2125.2012.04169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lau W. C., Waskell L. A., Watkins P. B., et al. Atorvastatin reduces the ability of clopidogrel to inhibit platelet aggregation: a new drug-drug interaction. Circulation. 2003;107(1):32–37. doi: 10.1161/01.cir.0000047060.60595.cc. [DOI] [PubMed] [Google Scholar]
- 72.Neubauer H., Günesdogan B., Hanefeld C., Spiecker M., Mügge A. Lipophilic statins interfere with the inhibitory effects of clopidogrel on platelet function—a flow cytometry study. European Heart Journal. 2003;24(19):1744–1749. doi: 10.1016/s0195-668x(03)00442-1. [DOI] [PubMed] [Google Scholar]
- 73.Farid N. A., Payne C. D., Small D. S., et al. Cytochrome P450 3A inhibition by ketoconazole affects prasugrel and clopidogrel pharmacokinetics and pharmacodynamics differently. Clinical Pharmacology and Therapeutics. 2007;81(5):735–741. doi: 10.1038/sj.clpt.6100139. [DOI] [PubMed] [Google Scholar]
- 74.Olesen J. B., Gislason G. H., Charlot M. G., et al. Calcium-channel blockers do not alter the clinical efficacy of clopidogrel after myocardial infarction: a nationwide cohort study. Journal of the American College of Cardiology. 2011;57(4):409–417. doi: 10.1016/j.jacc.2010.08.640. [DOI] [PubMed] [Google Scholar]
- 75.Good C. W., Steinhubl S. R., Brennan D. M., Lincoff A. M., Topol E. J., Berger P. B. Is there a clinically significant interaction between calcium channel antagonists and clopidogrel?: Results from the clopidogrel for the reduction of events during observation (CREDO) trial. Circulation: Cardiovascular Interventions. 2012;5(1):77–81. doi: 10.1161/circinterventions.111.963405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sibbing D., Von Beckerath N., Morath T., et al. Oral anticoagulation with coumarin derivatives and antiplatelet effects of clopidogrel. European Heart Journal. 2010;31(10):1205–1211. doi: 10.1093/eurheartj/ehq023. [DOI] [PubMed] [Google Scholar]
- 77.Harmsze A. M., Van Werkum J. W., Moral F., et al. Sulfonylureas and on-clopidogrel platelet reactivity in type 2 diabetes mellitus patients. Platelets. 2011;22(2):98–102. doi: 10.3109/09537104.2010.530359. [DOI] [PubMed] [Google Scholar]
- 78.Bell D. S. H. Practical considerations and guidelines for dosing sulfonylureas as monotherapy or combination therapy. Clinical Therapeutics. 2004;26(11):1714–1727. doi: 10.1016/j.clinthera.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 79.Siller-Matula J. M., Trenk D., Krähenbühl S., Michelson A. D., Delle-Karth G. Clinical implications of drug-drug interactions with P2Y12 receptor inhibitors. Journal of Thrombosis and Haemostasis. 2014;12(1):2–13. doi: 10.1111/jth.12445. [DOI] [PubMed] [Google Scholar]
- 80.Serebruany V., Cherala G., Williams C., et al. Association of platelet responsiveness with clopidogrel metabolism: Role of compliance in the assessment of “resistance”. American Heart Journal. 2009;158(6):925–932. doi: 10.1016/j.ahj.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 81.Geisler T., Schaeffeler E., Dippon J., et al. CYP2C19 and nongenetic factors predict poor responsiveness to clopidogrel loading dose after coronary stent implantation. Pharmacogenomics. 2008;9(9):1251–1259. doi: 10.2217/14622416.9.9.1251. [DOI] [PubMed] [Google Scholar]
- 82.Holmberg M. T., Tornio A., Neuvonen M., Neuvonen P. J., Backman J. T., Niemi M. Grapefruit juice inhibits the metabolic activation of clopidogrel. Clinical Pharmacology and Therapeutics. 2014;95(3):307–313. doi: 10.1038/clpt.2013.192. [DOI] [PubMed] [Google Scholar]
- 83.Lev E. I., Arikan M. E., Vaduganathan M., et al. Effect of caffeine on platelet inhibition by clopidogrel in healthy subjects and patients with coronary artery disease. American Heart Journal. 2007;154(4):694.e1–694.e7. doi: 10.1016/j.ahj.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 84.Bliden K. P., DiChiara J., Lawal L., et al. The association of cigarette smoking with enhanced platelet inhibition by clopidogrel. Journal of the American College of Cardiology. 2008;52(7):531–533. doi: 10.1016/j.jacc.2008.04.045. [DOI] [PubMed] [Google Scholar]
- 85.Angiolillo D. J., Ferreiro J. L. Platelet adenosine diphosphate P2Y12 receptor antagonism: benefits and limitations of current treatment strategies and future directions. Revista Espanola de Cardiologia. 2010;63(1):60–76. doi: 10.1016/s0300-8932(10)70010-5. [DOI] [PubMed] [Google Scholar]
- 86.Frelinger A. L., III, Bhatt D. L., Lee R. D., et al. Clopidogrel pharmacokinetics and pharmacodynamics vary widely despite exclusion or control of polymorphisms (CYP2C19, ABCB1, PON1), noncompliance, diet, smoking, co-medications (Including Proton Pump Inhibitors), and pre-existent variability in platelet function. Journal of the American College of Cardiology. 2013;61(8):872–879. doi: 10.1016/j.jacc.2012.11.040. [DOI] [PubMed] [Google Scholar]
- 87.National cancer Institute. National Cancer Institute: Personalized Medicine. Dictionary of Cancer Terms. National Cancer Institute; 2016. http://www.cancer.gov/publications/dictionaries/cancer-terms?cdrid=561717. [Google Scholar]
- 88.Offit K. Personalized medicine: new genomics, old lessons. Human Genetics. 2011;130(1):3–14. doi: 10.1007/s00439-011-1028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Colburn W., DeGruttola V. G., DeMets D. L., et al. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Biomarkers Definitions Working Group. Clinical Pharmacology & Therapeutics. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 90.Food and Drug Administration. Food and Drug Administration (FDA): Table of Pharmacogenomic Biomarkers in Drug Labeling. US Food and Drug Administration; 2015. http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm. [Google Scholar]
- 91.Ahn S. G., Yoon J., Kim J., et al. Genotype- and phenotype-directed personalization of antiplatelet treatment in patients with non-ST elevation acute coronary syndromes undergoing coronary stenting. Korean Circulation Journal. 2013;43(8):541–549. doi: 10.4070/kcj.2013.43.8.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nasyuhana Sani Y., Sheau Chin L., Luen Hui L., et al. The CYP2C19∗1/∗2 genotype does not adequately predict clopidogrel response in healthy Malaysian volunteers. Cardiology Research and Practice. 2013;2013:7. doi: 10.1155/2013/128795.128795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mejin M., Tiong W. N., Lai L. Y. H., et al. CYP2C19 genotypes and their impact on clopidogrel responsiveness in percutaneous coronary intervention. International Journal of Clinical Pharmacy. 2013;35(4):621–628. doi: 10.1007/s11096-013-9783-y. [DOI] [PubMed] [Google Scholar]
- 94.Fontana P., Cattaneo M., Combescure C., Reny J.-L. Tailored Thienopyridine therapy: no urgency for CYP2C19 genotyping. Journal of the American Heart Association. 2013;2(2) doi: 10.1161/jaha.112.000131.e000131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Holmes M. V., Perel P., Shah T., Hingorani A. D., Casas J. P. CYP2C19 genotype, clopidogrel metabolism, platelet function, and cardiovascular events: a systematic review and meta-analysis. JAMA - Journal of the American Medical Association. 2011;306(24):2704–2714. doi: 10.1001/jama.2011.1880. [DOI] [PubMed] [Google Scholar]
- 96.Yin T., Miyata T. Pharmacogenomics of clopidogrel: evidence and perspectives. Thrombosis Research. 2011;128(4):307–316. doi: 10.1016/j.thromres.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 97.Saab Y. B., Zeenny R., Ramadan W. H. Optimizing clopidogrel dose response: a new clinical algorithm comprising CYP2C19 pharmacogenetics and drug interactions. Therapeutics and Clinical Risk Management. 2015;11:1421–1427. doi: 10.2147/tcrm.s83293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hamburg M. A., Collins F. S. The path to personalized medicine. New England Journal of Medicine. 2010;363(4):301–304. doi: 10.1056/nejmp1006304. [DOI] [PubMed] [Google Scholar]
- 99.Patel N. K., Subbaiah G., Shah H., Kundlik M., Shrivastav P. S. Rapid LC-ESI-MS-MS method for the simultaneous determination of clopidogrel and its carboxylic acid metabolite in human plasma. Journal of Chromatographic Science. 2008;46(10):867–875. doi: 10.1093/chromsci/46.10.867. [DOI] [PubMed] [Google Scholar]
- 100.Peer C. J., Spencer S. D., VanDenBerg D. A. H., Pacanowski M. A., Horenstein R. B., Figg W. D. A sensitive and rapid ultra HPLC-MS/MS method for the simultaneous detection of clopidogrel and its derivatized active thiol metabolite in human plasma. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences. 2012;880(1):132–139. doi: 10.1016/j.jchromb.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brandt J. T., Payne C. D., Wiviott S. D., et al. A comparison of prasugrel and clopidogrel loading doses on platelet function: magnitude of platelet inhibition is related to active metabolite formation. American Heart Journal. 2007;153(1):66.e9–66.e16. doi: 10.1016/j.ahj.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 102.Gong I. Y., Crown N., Suen C. M., et al. Clarifying the importance of CYP2C19 and PON1 in the mechanism of clopidogrel bioactivation and in vivo antiplatelet response. European Heart Journal. 2012;33(22):2856–2864. doi: 10.1093/eurheartj/ehs042. [DOI] [PubMed] [Google Scholar]
- 103.Mani H., Toennes S. W., Linnemann B., et al. Determination of clopidogrel main metabolite in plasma: a useful tool for monitoring therapy? Therapeutic Drug Monitoring. 2008;30(1):84–89. doi: 10.1097/ftd.0b013e31815c13fd. [DOI] [PubMed] [Google Scholar]
- 104.Angiolillo D. J., Fernandez-Ortiz A., Bernardo E., et al. Variability in individual responsiveness to clopidogrel. clinical implications, management, and future perspectives. Journal of the American College of Cardiology. 2007;49(14):1505–1516. doi: 10.1016/j.jacc.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 105.Paniccia R., Antonucci E., Maggini N., et al. Assessment of platelet function on whole blood by multiple electrode aggregometry in high-risk patients with coronary artery disease receiving antiplatelet therapy. American Journal of Clinical Pathology. 2009;131(6):834–842. doi: 10.1309/AJCPTE3K1SGAPOIZ. [DOI] [PubMed] [Google Scholar]
- 106.Accumetrics. Verify Now P2Y12 Platelet Reactivity Test: Instruction for Use. Accumetrics; 2013. [Google Scholar]
- 107.Sharma R. K., Voelker D. J., Sharma R., Reddy H. K., Dod H., Marsh J. D. Evolving role of platelet function testing in coronary artery interventions. Vascular Health and Risk Management. 2012;8(1):65–75. doi: 10.2147/VHRM.S28090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Falcão F. J., Carvalho L., Chan M., Alves C. M., Carvalho A. C., Caixeta A. M. Receptores plaquetários P2Y12: importância na intervenção coronariana percutânea. Arquivos Brasileiros de Cardiologia. 2013;101:277–282. doi: 10.5935/abc.20130162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gachet C., Aleil B. Testing antiplatelet therapy. European Heart Journal Supplements. 2008;10:A28–A34. doi: 10.1093/eurheartj/sum081. [DOI] [Google Scholar]
- 110.Paniccia R., Priora R., Liotta A. A., Abbate R. Platelet function tests: a comparative review. Vascular Health and Risk Management. 2015;11:133–148. doi: 10.2147/vhrm.s44469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.SHd. PFA-100 System US Assay Menu. 2012. (Edited by US SHd). [Google Scholar]
- 112.Price M. J., Endemann S., Gollapudi R. R., et al. Prognostic significance of post-clopidogrel platelet reactivity assessed by a point-of-care assay on thrombotic events after drug-eluting stent implantation. European Heart Journal. 2008;29(8):992–1000. doi: 10.1093/eurheartj/ehn046. [DOI] [PubMed] [Google Scholar]
- 113.Marcucci R., Gori A. M., Paniccia R., et al. Cardiovascular death and nonfatal myocardial infarction in acute coronary syndrome patients receiving coronary stenting are predicted by residual platelet reactivity to ADP detected by a point-of-care assay: a 12-month follow-up. Circulation. 2009;119(2):237–242. doi: 10.1161/circulationaha.108.812636. [DOI] [PubMed] [Google Scholar]
- 114.Breet N. J., Van Werkum J. W., Bouman H. J., et al. Comparison of platelet function tests in predicting clinical outcome in patients undergoing coronary stent implantation. JAMA—Journal of the American Medical Association. 2010;303(8):754–762. doi: 10.1001/jama.2010.181. [DOI] [PubMed] [Google Scholar]
- 115.Collet J.-P., Cuisset T., Rangé G., et al. Bedside monitoring to adjust antiplatelet therapy for coronary stenting. New England Journal of Medicine. 2012;367(22):2100–2109. doi: 10.1056/nejmoa1209979. [DOI] [PubMed] [Google Scholar]
- 116.Miller C. H., Rice A. S., Garrett K., Stein S. F. Gender, race and diet affect platelet function tests in normal subjects, contributing to a high rate of abnormal results. British Journal of Haematology. 2014;165(6):842–853. doi: 10.1111/bjh.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim Y. G., Suh J.-W., Park J. J., et al. Different influences of hematocrit on the results of two point-of-care platelet function tests, the verifynow assay and multiple electrode platelet aggregometry. PLoS ONE. 2014;9(11) doi: 10.1371/journal.pone.0114053.e114053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kakouros N., Kickler T. S., Laws K. M., Rade J. J. Hematocrit alters VerifyNow P2Y12 assay results independently of intrinsic platelet reactivity and clopidogrel responsiveness. Journal of Thrombosis and Haemostasis. 2013;11(10):1814–1822. doi: 10.1111/jth.12376. [DOI] [PubMed] [Google Scholar]
- 119.Gurbel P. A., Bliden K. P., Zaman K. A., Yoho J. A., Hayes K. M., Tantry U. S. Clopidogrel loading with eptifibatide to arrest the reactivity of platelets: results of the Clopidogrel Loading With Eptifibatide to Arrest the Reactivity of Platelets (CLEAR Platelets) study. Circulation. 2005;111(9):1153–1159. doi: 10.1161/01.cir.0000157138.02645.11. [DOI] [PubMed] [Google Scholar]
- 120.Jakubowski J. A., Zhou C., Egan B., et al. Modification of the VerifyNow® P2Y12 test BASE channel to accommodate high levels of P2Y12 antagonism. Platelets. 2011;22(8):619–625. doi: 10.3109/09537104.2011.579203. [DOI] [PubMed] [Google Scholar]
- 121.Hochholzer W., Ruff C. T., Mesa R. A., et al. Variability of individual platelet reactivity over time in patients treated with clopidogrel : insights from the ELEVATE–TIMI 56 trial. Journal of the American College of Cardiology. 2014;64(4):361–368. doi: 10.1016/j.jacc.2014.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lemesle G., Landel J.-B., Bauters A., et al. Poor agreement between light transmission aggregometry, Verify Now P2Y12 and vasodilatator-stimulated phosphoprotein for clopidogrel low-response assessment: a potential explanation of negative results of recent randomized trials. Platelets. 2014;25(7):499–505. doi: 10.3109/09537104.2013.840363. [DOI] [PubMed] [Google Scholar]
- 123.Louridas G. E., Kanonidis I. E., Lourida K. G. Systems biology in heart diseases. Hippokratia. 2010;14(1):10–16. [PMC free article] [PubMed] [Google Scholar]
- 124.Kuster D. W. D., Merkus D., Velden J. V. D., Verhoeven A. J. M., Duncker D. J. 'Integrative Physiology 2.0': integration of systems biology into physiology and its application to cardiovascular homeostasis. Journal of Physiology. 2011;589(5):1037–1045. doi: 10.1113/jphysiol.2010.201533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Louridas G. E., Lourida K. G. The new biology: a bridge to clinical cardiology. Hippokratia. 2012;16(2):106–112. [PMC free article] [PubMed] [Google Scholar]
- 126.Cappola T. P., Margulies K. B. Functional genomics applied to cardiovascular medicine. Circulation. 2011;124(1):87–94. doi: 10.1161/CIRCULATIONAHA.111.027300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Piran S., Liu P., Morales A., Hershberger R. E. Where genome meets phenome: rationale for integrating genetic and protein biomarkers in the diagnosis and management of dilated cardiomyopathy and heart failure. Journal of the American College of Cardiology. 2012;60(4):283–289. doi: 10.1016/j.jacc.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 128.Saito K., Matsuda F. Metabolomics for functional genomics, systems biology, and biotechnology. Annual Review of Plant Biology. 2010;61(1):463–489. doi: 10.1146/annurev.arplant.043008.092035. [DOI] [PubMed] [Google Scholar]
- 129.Caruso R., Rocchiccioli S., Gori A. M., et al. Inflammatory and antioxidant pattern unbalance in ‘clopidogrel-resistant’ patients during acute coronary syndrome. Mediators of Inflammation. 2015;2015:12. doi: 10.1155/2015/710123.710123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Goodacre R. Metabolomics of a superorganism. Journal of Nutrition. 2007;137(1) doi: 10.1093/jn/137.1.259S. [DOI] [PubMed] [Google Scholar]
- 131.Guţiu I. A., Andrieş A., Mircioiu C., Radulescu F., Georgescu A.-M., Cioaca D. Pharmacometabonomics, pharmacogenomics and personalized medicine. Romanian Journal of Internal Medicine. 2010;48(2):187–191. [PubMed] [Google Scholar]
- 132.Gladding P. A. Clinical applications of pharmacogenetics: present and near future. Cleveland Clinic Journal of Medicine. 2013;80(8):477–482. doi: 10.3949/ccjm.80a.12111. [DOI] [PubMed] [Google Scholar]
- 133.Semmar N. Metabotype concept: flexibility, usefulness and meaning in different biological populations. In: Roessner D. U., editor. Metabolomics. InTech; 2012. [Google Scholar]
- 134.Lindon J., Nicholson J., Holmes E. The Handbook of Metabonomics and Metabolomics. Amsterdam, The Netherlands: Elsevier BV; 2006. [Google Scholar]
- 135.Ibrahim B., Basanta M., Cadden P., et al. Non-invasive phenotyping using exhaled volatile organic compounds in asthma. Thorax. 2011;66(9):804–809. doi: 10.1136/thx.2010.156695. [DOI] [PubMed] [Google Scholar]
- 136.Holmes E., Tsang T. M., Huang J. T.-J., et al. Metabolic profiling of CSF: evidence that early intervention may impact on disease progression and outcome in schizophrenia. PLoS Medicine. 2006;3(8):1420–1428. doi: 10.1371/journal.pmed.0030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Basanta M., Ibrahim B., Dockry R., et al. Exhaled volatile organic compounds for phenotyping chronic obstructive pulmonary disease: a cross-sectional study. Respiratory Research. 2012;13, article 72 doi: 10.1186/1465-9921-13-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Montuschi P., Paris D., Melck D. Metabolomic analysis by nuclear magnetic resonance spectroscopy of exhaled breath condensate in patient with cystic fibrosis. European Respiratory Journal. 2009;34(supplement 53):p. 63s. doi: 10.1183/09031936.00072408. [DOI] [PubMed] [Google Scholar]
- 139.Mervaala E., Biala A., Merasto S., et al. Metabolomics in angiotensin II-induced cardiac hypertrophy. Hypertension. 2010;55(2):508–515. doi: 10.1161/HYPERTENSIONAHA.109.145490. [DOI] [PubMed] [Google Scholar]
- 140.Tenori L., Hu X., Pantaleo P., et al. Metabolomic fingerprint of heart failure in humans: a nuclear magnetic resonance spectroscopy analysis. International Journal of Cardiology. 2013;168(4):e113–e115. doi: 10.1016/j.ijcard.2013.08.042. [DOI] [PubMed] [Google Scholar]
- 141.Corona G., Rizzolio F., Giordano A., Toffoli G. Pharmaco-metabolomics: an emerging ‘omics’ tool for the personalization of anticancer treatments and identification of new valuable therapeutic targets. Journal of Cellular Physiology. 2012;227(7):2827–2831. doi: 10.1002/jcp.24003. [DOI] [PubMed] [Google Scholar]
- 142.Nicholson J. K., Wilson I. D., Lindon J. C. Pharmacometabonomics as an effector for personalized medicine. Pharmacogenomics. 2011;12(1):103–111. doi: 10.2217/pgs.10.157. [DOI] [PubMed] [Google Scholar]
- 143.Yang Z., Marotta F. Pharmacometabolomics in drug discovery & development: applications and challenges. Metabolomics. 2012;2, article e122 doi: 10.4172/2153-0769.1000e122. [DOI] [Google Scholar]
- 144.Clayton T. A., Baker D., Lindon J. C., Everett J. R., Nicholson J. K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(34):14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Clayton T. A., Lindon J. C., Cloarec O., et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440(7087):1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 146.Kim C., Kim E., Yoo H., et al. Metabonomic analysis of serum metabolites in kidney transplant recipients with cyclosporine a-or tacrolimus-based immunosuppression. Transplantation. 2010;90(7):748–756. doi: 10.1097/tp.0b013e3181edd69a. [DOI] [PubMed] [Google Scholar]
- 147.Wang Z., Chen Z., Yang S., et al. 1H NMR-based metabolomic analysis for identifying serum biomarkers to evaluate methotrexate treatment in patients with early rheumatoid arthritis. Experimental and Therapeutic Medicine. 2012;4(1):165–171. doi: 10.3892/etm.2012.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kaddurah-Daouk R., Bogdanov M. B., Wikoff W. R., et al. Pharmacometabolomic mapping of early biochemical changes induced by sertraline and placebo. Translational Psychiatry. 2013;3, article no. e223 doi: 10.1038/tp.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Villaseñor A., Ramamoorthy A., Santos M. S. D., et al. A pilot study of plasma metabolomic patterns from patients treated with ketamine for bipolar depression: evidence for a response-related difference in mitochondrial networks. British Journal of Pharmacology. 2014;171(8):2230–2242. doi: 10.1111/bph.12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gilany K., Pouracil R. S. M., Sadeghi M. R. Fourier transform infrared spectroscopy: a potential technique for noninvasive detection of spermatogenesis. Avicenna Journal of Medical Biotechnology. 2014;6(1):47–52. [PMC free article] [PubMed] [Google Scholar]
- 151.Savorani F., Rasmussen M. A., Mikkelsen M. S., Engelsen S. B. A primer to nutritional metabolomics by NMR spectroscopy and chemometrics. Food Research International. 2013;54(1):1131–1145. doi: 10.1016/j.foodres.2012.12.025. [DOI] [Google Scholar]
- 152.Scalbert A., Brennan L., Fiehn O., et al. Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics. 2009;5(4):435–458. doi: 10.1007/s11306-009-0168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Alonso A., Marsal S., Julià A. Analytical methods in untargeted metabolomics: state of the art in 2015. Frontiers in Bioengineering and Biotechnology. 2015;3, article 23 doi: 10.3389/fbioe.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Silva Elipe M. V. Advantages and disadvantages of nuclear magnetic resonance spectroscopy as a hyphenated technique. Analytica Chimica Acta. 2003;497(1-2):1–25. doi: 10.1016/j.aca.2003.08.048. [DOI] [Google Scholar]
- 155.Dunn W. B., Bailey N. J. C., Johnson H. E. Measuring the metabolome: current analytical technologies. The Analyst. 2005;130(5):606–625. doi: 10.1039/b418288j. [DOI] [PubMed] [Google Scholar]
- 156.Shulaev V. Metabolomics technology and bioinformatics. Briefings in Bioinformatics. 2006;7(2):128–139. doi: 10.1093/bib/bbl012. [DOI] [PubMed] [Google Scholar]
- 157.Nicholson J. K., Lindon J. C. Systems biology: metabonomics. Nature. 2008;455(7216):1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 158.Bu D. Chemometric Analysis for Spectroscopy: Bridging the Gap between the State and Measurement of a Chemical System 2007, http://www.camo.com/downloads/resources/application_notes/Chemometric%20Analysis%20for%20Spectroscopy.pdf.
- 159.Goodacre R., Vaidyanathan S., Dunn W. B., Harrigan G. G., Kell D. B. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends in Biotechnology. 2004;22(5):245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 160.Pennazza G., Santonico M., Martinelli E., D'Amico A., Natale C. D. Exhaled Biomarkers. 2010. Interpretation of exhaled volatile organic compounds; pp. 115–129. (European Respiratory Society Monograph). [Google Scholar]
- 161.Kaddurah-Daouk R., Weinshilboum R. M. Pharmacometabolomics: implications for clinical pharmacology and systems pharmacology. Clinical Pharmacology and Therapeutics. 2014;95(2):154–167. doi: 10.1038/clpt.2013.217. [DOI] [PubMed] [Google Scholar]
- 162.Raamsdonk L. M., Teusink B., Broadhurst D., et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nature Biotechnology. 2001;19(1):45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- 163.Ji Y., Hebbring S., Zhu H., et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clinical Pharmacology and Therapeutics. 2011;89(1):97–104. doi: 10.1038/clpt.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yerges-Armstrong L. M., Ellero-Simatos S., Georgiades A., et al. Purine pathway implicated in mechanism of resistance to aspirin therapy: pharmacometabolomics-informed pharmacogenomics. Clinical Pharmacology and Therapeutics. 2013;94(4):525–532. doi: 10.1038/clpt.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lewis J. P., Yerges-Armstrong L. M., Ellero-Simatos S., Georgiades A., Kaddurah-Daouk R., Hankemeier T. Integration of pharmacometabolomic and pharmacogenomic approaches reveals novel insights into antiplatelet therapy. Clinical Pharmacology and Therapeutics. 2013;94(5):570–573. doi: 10.1038/clpt.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ellero-Simatos S., Lewis J. P., Georgiades A., et al. Pharmacometabolomics reveals that serotonin is implicated in aspirin response variability. CPT: Pharmacometrics & Systems Pharmacology. 2014;3(7, article e125) doi: 10.1038/psp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]