Single-particle cryo-electron microscopy (cryo-EM) has gained its ‘lettres de noblesse’ in structural biology. The synergistic convergence of technological and computational advances now make this a feasible method for determining structures at near-atomic to atomic resolution (∼5–2 Å). The potential impact of cryo-EM on drug discovery is discussed, together with how the method can provide information on conformational variability connected to the function of macromolecular complexes.
Keywords: cryo-electron microscopy, cryo-EM, structural biology, high resolution, conformational variability, drug discovery
Abstract
The invention of the electron microscope has greatly enhanced the view scientists have of small structural details. Since its implementation, this technology has undergone considerable evolution and the resolution that can be obtained for biological objects has been extended. In addition, the latest generation of cryo-electron microscopes equipped with direct electron detectors and software for the automated collection of images, in combination with the use of advanced image-analysis methods, has dramatically improved the performance of this technique in terms of resolution. While calculating a sub-10 Å resolution structure was an accomplishment less than a decade ago, it is now common to generate structures at sub-5 Å resolution and even better. It is becoming possible to relatively quickly obtain high-resolution structures of biological molecules, in particular large ones (>500 kDa) which, in some cases, have resisted more conventional methods such as X-ray crystallography or nuclear magnetic resonance (NMR). Such newly resolved structures may, for the first time, shed light on the precise mechanisms that are essential for cellular physiological processes. The ability to attain atomic resolution may support the development of new drugs that target these proteins, allowing medicinal chemists to understand the intimacy of the relationship between their molecules and targets. In addition, recent developments in cryo-electron microscopy combined with image analysis can provide unique information on the conformational variability of macromolecular complexes. Conformational flexibility of macromolecular complexes can be investigated using cryo-electron microscopy and multiconformation reconstruction methods. However, the biochemical quality of the sample remains the major bottleneck to routine cryo-electron microscopy-based determination of structures at very high resolution.
1. Introduction
Biochemists focus on processes that occur at the molecular level and are therefore invariably fascinated by the small structural details. For instance, visualizing the interaction between a molecule (natural or synthetic) and its protein target has been like the quest for the holy grail for many scientists in the search for enzyme inhibitors. In this context, the drug-discovery process became ‘easier’ and more rational with the complementary use of co-crystallization or ‘soaking’ methods combined with structural biology studies and medicinal chemistry. In this case, co-crystallization applies to the targeted protein and active substances. Note also that active substances are often improperly called ‘drugs’, whereas they are in fact substances that are far from having the characteristics that will admit them to the pharmacopoeia. Nowadays, the active molecule docked in its target can be visualized, either in a solid-state interaction (thanks to co-crystallization or soaking methods and the use of these co-crystals in X-ray diffraction) or in some rare cases in a more dynamic interaction (NMR) with their targets. These techniques have provided medicinal chemists with concrete and highly informative data on the interaction between a molecule and its target protein. The medicinal chemists then orchestrate the synthetic steps leading first to active substances and then to drugs.
Even though crystallography has represented a major advance in drug discovery, the process is still complex. Indeed, a succession of critical and unpredictable steps make it possible to obtain the structures of target proteins: the construction of a plasmid encoding the targets, often excluding overly flexible parts (identified by prediction); the expression of these proteins in efficient expression systems such as bacteria, yeast or insect cells; purification to homogeneity; the identification of crystallization conditions (which are often strictly dependent on the protein sequence); recording the X-ray diffraction data (often on a synchrotron beamline); processing the data and generating electron-density maps; and interpretation of the maps to produce a three-dimensional model of the protein (see, for example, Friedmann et al., 2011 ▸). The aim is to see and understand the structural details that will allow the investigator to build a model showing interactions between different proteins, between parts of the same protein, or between a compound and a protein.
Until the last two decades or so, to aim at the discovery of an optimal inhibitor of an enzyme in the context of drug discovery, successive molecules (obtained from synthesis and derived from some starting candidate) were each analysed using functional tests which provide an understanding of the structure–activity relationship within the chemical series. There is no doubt that modern drug-discovery programmes are now based on the capacity to either measure the molecule/protein target interactions using biophysical methods (see Renaud et al., 2016 ▸) and/or to visualize at the molecular level the position of the compound on its protein target (see reviews by Zuercher et al., 2016 ▸; Brader et al., 2017 ▸). The latter is essentially based on progress in protein crystallization, co-crystallization approaches and crystal X-ray diffraction analyses. These processes have been reviewed at length. They represent an immense step in understanding molecular pharmacology at the closest level, and have made it possible for medicinal chemists to gain information leading them to rationally synthesize series of molecules designed to become drug candidates. Such successful examples are scattered throughout the medicinal chemistry literature and can be found easily (see, for example, Huang et al., 2016 ▸; Gustafsson et al., 2017 ▸; Hazel et al., 2017 ▸; Mills-Davies et al., 2017 ▸).
Why, then, be jubilant at the recent performance of cryo-electron microscopy (cryo-EM)? Let us remember that the data that have been collected and that populate the PDB (Protein Data Bank; http://www.rcsb.org/pdb) were mostly obtained by X-ray crystallography from rigid structures. This rigid state shows the interactions between atoms only in the most stable state, which is not necessarily the most common or the most representative state in a cell. Remember also that what is shown under crystallographic conditions is a protein. This is of course an accomplishment in itself, even if it has become routine to obtain such visualizations of new targets. In recent years, complementary techniques to crystallography have emerged. They are interesting because they reveal proteins in states that are not (or are less) rigid, and are more likely to be those of the proteins in solution. Indeed, cryo-EM is particularly well suited for obtaining structural information on large protein complexes and for systems that exhibit multiple conformational or compositional states. In this respect, there is a groundswell of enthusiasm for this approach (Rupp, 2015 ▸; Vinothkumar, 2015 ▸). We had already sensed this shift several years ago (Jonic & Vénien-Bryan, 2009 ▸). It is certainly no coincidence that the January 2016 issue of Nature Methods designated single-particle cryo-electron microscopy technology the ‘Technology of the Year’ (Eisenstein, 2016 ▸). In the meantime, discussion of the role of cryo-EM in drug discovery as a complement to protein crystallography continues (Merino & Raunser, 2017 ▸; Orlov et al., 2016 ▸; Takizawa et al., 2017 ▸; Wang & Wang, 2017 ▸). Of particular interest, Merk and coworkers have presented data demonstrating that cryo-EM can be used to investigate a broad spectrum of drug–target interactions and of dynamic conformational states (Merk et al., 2016 ▸).
2. What is cryo-electron microscopy?
A number of excellent reviews of electron microscopy are available (Nagayama, 2011 ▸; Oxley et al., 2017 ▸; Schroder, 2015 ▸). The purpose here is not to recapitulate in detail the nature of and the progress in this field, but to highlight recent developments that have been reported. The history of electron microscopy and its application to the observation of biological objects has been marked by major discoveries, each of which has helped to push the resolution limit. These findings relate to sample preparation, microscope performance, image capture or processing of these images. Transmission electron microscopy is an ideal tool for investigating biological specimens, even if at present they are held in a vacuum chamber because electrons do not travel very far in air. These conditions do not yet allow an adequate exploration of the structure of living cells. Spectacular observations of biological material ‘live’ in the electron microscope using a sample holder equipped with microfluidic technologies have been described (Peckys et al., 2015 ▸), but this is not the subject of this commentary. A major advance in electron microscopy was made in the late 1980s, when Jacques Dubochet and coworkers used liquid ethane for the sample preparation of proteins, viruses and macromolecules (Dubochet et al., 1988 ▸). Ultra-rapid freezing to temperatures below −185°C was, and remains, essential for good sample preparation. Indeed, the cooling rate is sufficiently fast to prevent water crystallizing, and rather promotes the formation of vitreous ice (Dubochet et al., 1988 ▸). The sample embedded in vitreous ice is free of stains and artefacts associated with them (typical stains are heavy-metal salts such as uranyl acetate). The sample is then placed in the vacuum chamber of the electron microscope and observed at −180°C. This spectacular progress in sample preparation turned electron microscopy into cryo-EM. For thirty years now, this method of preparation of biological samples has been a major tool in structural biology for the observation of objects in an environment that is less artificial than the solid state observed in a crystal, which was a prerequisite for any high-resolution study. In these fast-freezing conditions, the sample is surrounded by ice in a vitreous state. The density of this vitreous ice is lower than that of proteins or nucleic acids. This difference in density is essential to identify the single particles in the images. In addition, the fast-freezing technique preserves the characteristics of biological structures at atomic resolution and the low temperature provides some protection against the damage induced by electron bombardment. Images are captured during the observation with an electron microscope operating at 200 or 300 kV, and analyses are subsequently performed to reveal the structure of the macromolecules. The projected images of the biological macromolecules originate from multiple random orientations in suspension in the vitreous film. The three-dimensional structure is then calculated by combining and reconstructing projection images into a three-dimensional volume (Fig. 1 ▸, left). Because the information from each image is noisy and incomplete, hundreds of thousands of imaged particles must be aligned and averaged. The quality of the final reconstructed map of the macromolecule thus depends on the accuracy with which the orientations of the particles are determined relative to each other; this is performed using ad hoc software. Over the last forty years, the group of Joachim Frank has developed methods for the image analysis of isolated particles (Frank, 2006 ▸). They demonstrated that by combining images from many particles and identifying the relative orientations of these particles on the electron-microscopy grid, they could increase the signal-to-noise ratio, allowing the calculation of structures in three dimensions. This method is more efficient for large particles because these are more easily visible. There is therefore a lower size limit for the visualization of macromolecules in cryo-EM. Until very recently, it was around 250 kDa. However, it is claimed that in theory, starting from perfect images, one could obtain a final resolution of a reconstruction volume of 3 Å from only 12 000 particles as small as 40 kDa (Henderson, 1995 ▸). Despite the dramatic advances in cryo-EM, until a few years ago the resolution of biological structures was mostly limited to 6 or 8 Å and, at this stage, did not compete with data determined by X-ray crystallography or NMR (Ménétret et al., 2007 ▸; Myasnikov et al., 2005 ▸; Vénien-Bryan et al., 2009 ▸).
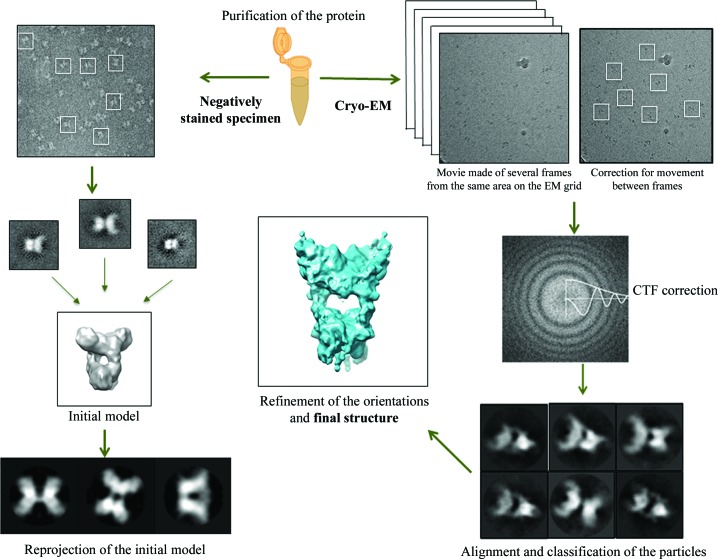
Figure 1.
Schematic representation of three-dimensional single-particle reconstruction. The high purity of the sample is important, as in X-ray crystallography. Initially, the observation of the sample in negative staining is a useful step; as it clearly displays the sample, homogeneity can be checked. The particles are selected from the micrographs, centred and aligned. The classification and averaging images can improve the signal-to-noise ratio, and the class averages can be used to calculate an initial model at low resolution. This is calculated using the common-lines method or the random conical tilt series method (Dubochet et al., 1988 ▸). Then, in the cryo-EM step, the vitrified sample is imaged using a direct electron detector (a film is obtained from multiple images of the same field of the electron-microscope grid). Correcting for movement between the various images of the film is performed. The particles are then windowed and averaged. Determination of the defocus value allows correction of the contrast-transfer function (CTF). After alignment and classification of the data set, the orientation of each projected particle relative to the initial model is assigned. Refinement of the orientation values is performed iteratively until the three-dimensional structure of the macromolecule of interest converges (this figure was adapted from Boutin et al., 2016 ▸).
3. Cryo-electron microscopy and its unique challenges
The two main problems that hinder the determination of biological structures at high resolution using cryo-EM combined with image analysis are (i) the low signal-to-noise ratio (or low contrast) of the captured images, which limits the size of the macromolecules observed (ideally these should be larger than 250 kDa) and (ii) electron beam-induced movement during exposure, which results in degradation of the image quality. The main challenge in cryo-EM is the low signal-to-noise ratio or, in other words, the poor contrast of the image. This low contrast is an inevitable consequence of the low atomic mass of the elements that constitute biological objects (proteins, nucleic acids, carbohydrates, lipids). Exposures above a dose limit of electrons irreversibly break the covalent bonds, resulting in destruction of the very details that we seek to elucidate. Therefore, very low doses of electrons are used when biological structures are observed in cryo-EM. Another difficulty that can thwart attempts to obtain high-resolution information is inherent to the interaction of electrons with the sample. Indeed, in order to obtain detailed biological structural information, the particles must not move more than one ångström, equivalent to the diameter of an H atom. However, more extensive motions appear when the electron beam interacts with a biological sample. This is owing to several things. Firstly, the impact of the electrons on the vitreous ice film in which the biological particles are embedded may cause thermal expansion and the release of constraints at the level of this film. In addition, electron-beam irradiation can break the covalent bonds of biological structures, generating molecules in the gaseous state (such as hydrogen, oxygen, nitrogen and methane; Henderson, 1995 ▸), which escape from the vitreous ice film. These sample movements are inevitably induced by the electron beam and have severely hampered the collection of high-resolution images in the past. Recording an image in low-dose conditions using a conventional medium (film photography or CCD cameras) takes a few seconds, during which the movement of the biological object will degrade the high-resolution data.
4. Why a revolution? Recent technological and methodological innovations
Since 2012, new direct electron detectors (DEDs) have become available. In the past, electron-microscope images were recorded on silver photographic film and then digitized. The advent of image acquisition from CCD (charge-coupled device) detectors thus allowed automated data collection, leading to an increase in the number of images and the size of a data set. However, there is a major limitation for CCD cameras. Incident electrons initially interact with a particular scintillator, which converts them into photons that are then detected. The electron-scattering events in the scintillator give rise to a large ‘cloud’ of photons and electrons, each contributing to the formation of the image. The size of this cloud is larger than the size of the CCD camera pixels; indeed, it can be spread over many pixels. The result is an attenuation of high-resolution signals and thus a limitation of the final resolution.
The new direct electron CMOS (complementary metal-oxide semiconductor) detector overcomes the scintillator step of CCD cameras. A number of technical obstacles had to be overcome to make these new cameras available, the most important of which was the development of a radiation-resistant support to reduce deterioration of the silicon chip. These cameras display a remarkable sensitivity that can significantly increase the image contrast. The easy viewing of ‘small’ proteins of about 100 kDa is now possible. In addition, structural information at high frequencies is preserved. Another remarkable quality of these new DEDs is their fast readout rate: instead of recording one image per second (as for movies and CCD cameras), these new cameras record some movies at a rate of 24–40 frames per second. Thus, we have access to successive images of the same field just like in a video film and the high-speed readout allows the detection of very small movements (on the ångström scale) in these images. The movements of the particles are then corrected from one frame to the other using appropriate software. Each video image is then aligned and stacked to give a final image that is much sharper as each particle is aligned from one frame to the next (Fig. 2 ▸).
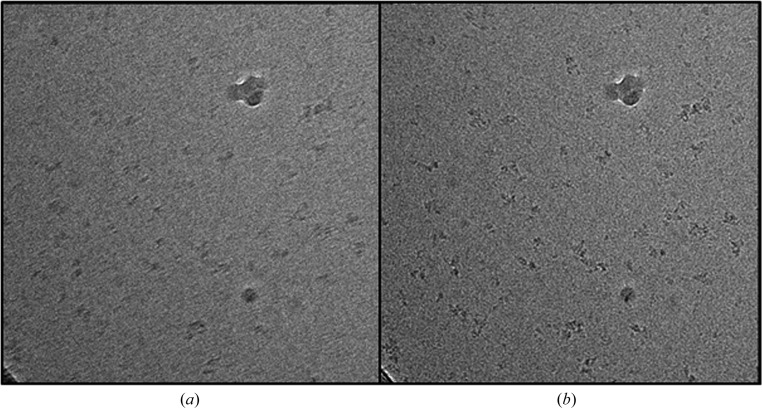
Figure 2.
Motion compensation and recovery of high-resolution information. Averaging of successive frames from a movie of phosphorylase kinase (a 1.3 MDa macromolecule) taken from the same area on the electron-microscope grid before (a) and after (b) alignment. The structural features of the protein are blurred before alignment.
In parallel to the marketing of the new DED cameras, improved image-processing software has emerged: firstly, movie-processing software to perform the correction or the compensation of movements between frames described above (Campbell et al., 2012 ▸; Li et al., 2013 ▸) and, secondly, software for the automated collection of images and software to extract detailed structural information from images captured in an optimal way. For instance, incorporation of the maximum-likelihood approach in three-dimensional calculation has made it possible to determine multiple structures at high resolution of a dynamic structure using one cryo-EM data set (Lyumkis et al., 2013 ▸; Scheres & Chen, 2012 ▸). The maximum-likelihood (ML) method was introduced by Sigworth (1998 ▸). It is a supervised classification method which is based on the Bayes theorem. It makes use of a discriminant function to assign a pixel to the class with the highest likelihood. In other words, it does not give a specific direction to a particle with reference to a similarity test, but rather a set of probabilities of being in a certain direction. Such methods have proven their ability to handle noisy data. Thus, the programs RELION (Scheres & Chen, 2012 ▸) and FREALIGN (Lyumkis et al., 2013 ▸) are now widely used. RELION, with its user-friendly interface, operates mainly without operator intervention, calculates less noisy models in three dimensions and provides quality-control criteria and validation of the final structure with objective criteria and therefore produces more reliable results (Rosenthal & Henderson, 2003 ▸; Scheres & Chen, 2012 ▸; Fig. 1 ▸). Moreover, this software has a powerful classification method that is particularly effective for sorting particles in more homogeneous structural subclasses, which allow the separation of the different conformational states of molecules that are simultaneously present in a preparation deposited on the electron microscope.
5. Recent advances in computational methods for analyzing the conformational variability of macromolecular complexes from cryo-electron microscopy images
In cryo-EM the samples can display different conformational states. Whilst such heterogeneity can limit the resolution of a three-dimensional reconstruction, the identification of distinct states permits investigators to model dynamics and conformational transitions, thereby providing information on biological function. A striking example is that of the AMPA receptor, for which Dürr et al. (2014 ▸) made a film showing large conformational rearrangements of the N-terminal domain and of the ligand binding to the desensitized state. Other noteworthy examples have recently been published, including an analysis of transcribing ribosomes (Abeyrathne et al., 2016 ▸; Behrmann et al., 2015 ▸), understanding the conformational changes necessary for the function of hexameric ATPase p97 (Banerjee et al., 2016 ▸) and a view of the proton-translocation-driven rotational fluctuations of the mitochondrial ATP synthase (Zhou et al., 2015 ▸). In principle, heterogeneous samples (from a biochemical point of view) can also be classified using the same method. Here, the sample may be purified to some extent by image processing rather than during the tedious steps of biochemical purification, but we are still far from this vision.
Conformational variability analysis from cryo-EM images is usually performed by classifying the images into a number of discrete classes representing all of the conformational states present in the specimen. However, recent methods take continuous conformational changes into account (Jonić, 2017 ▸).
Discrete-state methods based on conformational variability analysis in three dimensions can be classified into the following three groups: multi-reference classification, ML classification and classification based on statistical analysis. Supervised multi-reference classification approaches require a good prior knowledge of all conformational states that coexist in the sample (Gao et al., 2004 ▸). ML methods estimate a set of density maps that best describe the given heterogeneous set of images (Lyumkis et al., 2013 ▸; Scheres & Chen, 2012 ▸). Classification based on statistical analysis does not require prior knowledge of conformers, but relies on a preliminary model to assign orientation and translation parameters to each image (Klaholz, 2015 ▸; Penczek et al., 2011 ▸).
Discrete-state classification methods usually contribute to high-resolution reconstructions of single conformations. Within the high-resolution reconstruction strategies, classification methods are used to select a subset of images containing the particles with the most consistent views and conformations (i.e. only those assigned to the classes of highest resolution), and the three-dimensional reconstruction is then computed using only this image subset. In this context, many particle images are thrown away without fully understanding the reasons for their inconsistency with other particles. For instance, a three-dimensional reconstruction at near-atomic resolution is sometimes obtained by collecting 0.5–1.0 million particle images, but retaining only one half after a two-dimensional classification and less than one quarter or even 10% for the final three-dimensional reconstruction (Merk et al., 2016 ▸; Walls et al., 2016 ▸).
However, the classes of lower resolution could come from a mixture of particles with different (possibly unique) conformations that are difficult to represent with the given, usually small, number of classes. Discrete-state methods may provide a limited view of data heterogeneity because of the use of a small number of classes. Continuous-state methods can provide a broader view of data heterogeneity than discrete-state methods (Jonić, 2017 ▸).
While discrete-state methods assume that a few distinct conformational states coexist in the sample, continuous-state methods assume a continuum of conformational states. Currently, only two approaches consider continuous conformational changes explicitly. These are usually referred to as HEMNMA (hybrid electron-microscopy normal-mode analysis; Jin et al., 2014 ▸) and manifold embedding (Dashti et al., 2014 ▸). In the HEMNMA method, the conformational variability is directly analysed and visualized in three-dimensional space. The method analyses EM images using the normal modes of a starting three-dimensional model obtained by EM or X-ray crystallography. The manifold-embedding approach analyses the conformational variability in two-dimensional space determined by one projection direction (using images with virtually the same orientations) and then combines the conformational variability information obtained in different projection directions to visualize it in three dimensions. It is assumed that the effects of orientation changes dominate those of conformational changes. This method does not require knowledge of the number of states/classes.
6. Major breakthroughs: some examples
The first structures at near-atomic resolution were obtained for large complexes with high symmetry, such as viruses with icosahedral symmetry (Yu et al., 2008 ▸; Zhang et al., 2008 ▸). Several studies have recently shown that cryo-EM can be used to obtain near-atomic resolution structures of small complexes (170–150 kDa) with low symmetry (Bartesaghi et al., 2015 ▸; Liao et al., 2013 ▸; Walls et al., 2016 ▸) or no symmetry (Bai et al., 2015 ▸; Wan et al., 2016 ▸), where the best resolution (1.8 Å) was obtained for the 334 kDa glutamate dehydrogenase (Merk et al., 2016 ▸), in which small-molecule inhibitors were localized and the conformational changes induced by the binding of an allosteric inhibitor were defined.
Over the last few years, the number of published structures studied using the methods described above, with a similar structural quality to those obtained by X-ray crystallography, has shown exponential growth. In Supplementary Table S1 we list some of the most exciting cryo-EM studies that can be found in the PDB. With in excess of 30 000 structures since the beginning of 2010, we had to limit our query to proteins and objects with a molecular weight of below 500 kDa. Therefore, this table gives a flavour of the explosion of structures that have been deposited in recent years. Indeed, there are proteins for which several dozen structures have been deposited in the presence of various compounds: substrates and inhibitors, for example. We provide a single entry per protein, complex, particle or (subcellular) structure. It is obvious that those 127 entries represent a huge range of biological structures, some of which were calculated with an outstanding overall resolution (i.e. below 5 Å).
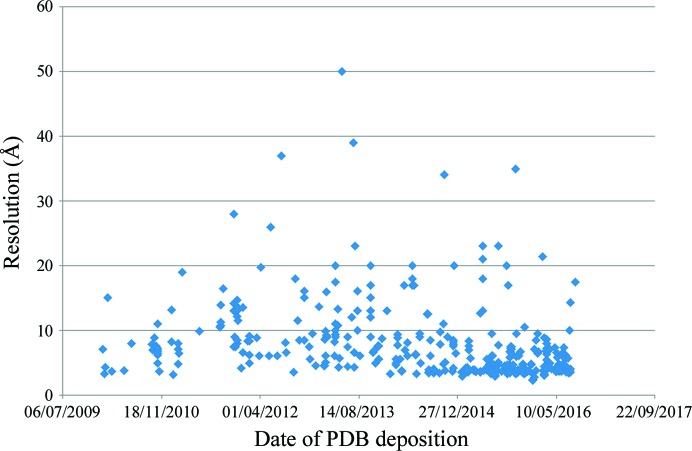
We have also plotted the year of deposition versus the resolution of the structures (Fig. 3 ▸). This figure shows a large increase in high-resolution data from 2012–2013 (with the best resolution being 2.8 Å), but it is also worth pointing out that a fair number of structures at a resolution better than 10 Å were published before 2012, i.e. before the recent advances.
Figure 3.
Representation of the evolution of the cryo-EM resolution as a function of time. The data extracted from the PDB (see Supplementary Table S1 for structures obtained using cryo-EM data) since 2010 were plotted against the corresponding resolution. 16 754 entries (the number of structures) were used to build this figure. Many were similar proteins or structures in different conditions or in the presence of different compounds. The table was constructed as an extract of this initial figure comprising a single entry per deposition.
7. The remarkable case of the ryanodine receptor RyR1
The structure of ryanodine receptor type 1 (RyR1), a large endoplasmic macromolecule of 4 × 500 000 Da, has been determined at 3.8, 6.1 and 4.8 Å resolution (Efremov et al., 2015 ▸; Yan et al., 2015 ▸; Zalk et al., 2015 ▸). This macromolecule regulates the massive outflow of calcium from the endoplasmic reticulum of skeletal or cardiac muscle, which initiates contraction. There are two ryanodine receptor subtypes: RyR1 and RyR2. The former is mainly expressed in skeletal muscle and the latter in the heart. The sequence of the RyR2 gene spans 790 000 base pairs and comprises 105 exons (Otsu et al., 1990 ▸). From a sequence point of view, these two receptors are similar, with 66% amino-acid identity. They are similar enough, at least, for specialists in this area to choose to study RyR1 rather than RyR2, which is a far more relevant target from a pathological prospect but has eluded purification for over a decade (including in the hands of our own group). RyR1 is easier to purify than RyR2, which is the reason why these studies were conducted on RyR1 receptors, the structure of which may also help in understanding the functions of RyR2. This channel is tightly regulated by a series of regulatory proteins, including the proline isomerase calstabin. Until recently, the structures of RyR1 and RyR2 were only partially known (Tung et al., 2010 ▸), making it difficult to fully understand the mechanism of action and regulation at the molecular level. In addition, this lack of structural information complicates the synthesis of new regulatory molecules other than ryanodine and its analogues (Sutko et al., 1997 ▸), the peptide imperatoxine (El-Hayek et al., 1995 ▸) or rapamycin derivatives (Brillantes et al., 1994 ▸). An X-ray crystallographic approach was not appropriate for such a large assembly. The protein from skeletal muscle (non-recombinant) can be purified to high homogeneity using affinity chromatography. The three-dimensional structure of this huge complex (2 × 106 Da) was determined using a relatively low number of particles: 100 000, 230 000 and 245 000 particles, respectively, from 500 to 3400 micrographs with a resolution from 3.8 to 6.1 Å (Efremov et al., 2015 ▸; Yan et al., 2015 ▸; Zalk et al., 2015 ▸). The crystallographic structures of many different domains or subunits of the complex were then positioned in the three-dimensional envelope(s) calculated from the cryo-EM data. This demonstrates the complementarity between cryo-EM and X-ray diffraction in the analyses of complex macromolecules (Wang & Wang, 2017 ▸). Determining the architecture of the multi-protein complex allows a better understanding of the interactions between the different parts and defines the site where some pharmacological regulators interact. The large number of mutations in RyR2 (expressed in the heart; Brini, 2004 ▸) and their association with heart pathologies makes this receptor a target for anti-arrhythmic drug development. It is therefore vital to have access to the detailed structure of this complicated molecular machine in order to elaborate the pharmacological pathways of regulation (Fig. 4 ▸), even though the limitations of these approaches for these particular receptors have already been pointed out (Yuchi & Van Petegem, 2016 ▸).
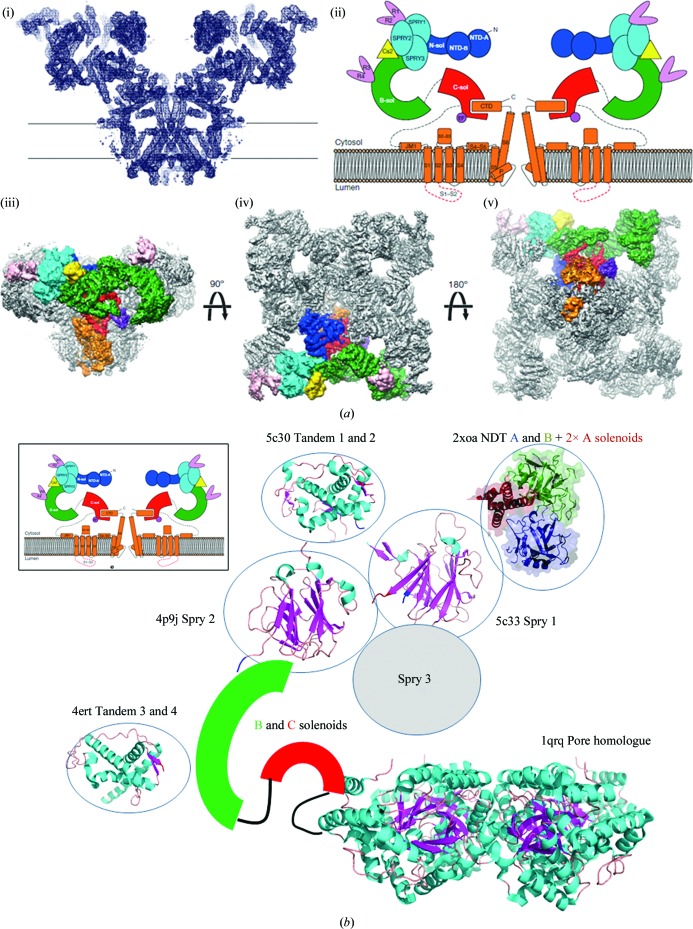
Figure 4.
(a) Structure of ryanodine receptor type 1 (RyR1) from Efremov et al. (2015 ▸). The resolution is 4.8 Å. (i) The molecular architecture of RyR1 determined by cryo-electron microscopy. (ii) Schematic representation of the middle section in the plane of the channel: dark blue, N-terminal domain; cyan, Spry domains 1, 2 and 3; pink, ‘repeat domains’ 1, 2, 3 and 4; green, solenoid connecting domain; red, central solenoid domain; orange, transmembrane and C-terminal domains. Calstabine is coloured yellow. Global representations from cryo-EM perpendicular to the membrane (iii) and views parallel to the membrane from above (iv) and from below (v) are shown. The same colour coding is used as above. (b) Schematic representation of ryanodine receptor type 1. The diagram in (a) is taken from the structures obtained by crystallography and X-ray diffraction. They were placed on the three-dimensional envelope obtained by cryo-electron microscopy. The PDB codes are 5c30 for tandem domains 1 and 2; 2xoa for the N-terminal domain AB part with the two solenoids A; 4p9j for Spry 2; 5c33 for Spry 1; 4ert for tandem domains 3 and 4; and 1qrq for the β-subunit of the voltage-gated potassium channel. This figure was adapted from Boutin et al. (2016 ▸).
Other examples, such as work on complexes such as the nuclear pore (Stuwe et al., 2015 ▸), provide new information on these structures that is in most cases inaccessible to the crystallographer because they are too large and complex.
8. Zooming in on cellular processes at the molecular level using cryo-electron tomography
The methodological developments described above also benefit cellular ultrastructure imaging using cryo-electron tomography (cryo-ET). The principle of tomography is based on the collection of images from samples tilted at different angles containing a number of copies of the macromolecular complex of interest. If the number of different views (projections) of the biological object is sufficient, three-dimensional information on the initial volume can be calculated. This step is called tomographic reconstruction. Subvolumes of the macromolecule of interest are then cut, extracted from the tomogram, aligned and sorted (the subtomogram averaging method). Averaging the different copies of the present structure in different directions can improve the resolution. Thus, a large number of homogeneous subvolumes facilitates the analysis and can provide the molecular characteristics at medium resolution. Therefore, cryo-ET has the capability to obtain nanometre-scale (medium-resolution) information about macromolecular complexes in their native environment, and thus provides a bridge between light microscopy and in vitro structure-determination methods. This is important because many complexes cannot be purified. Knowing both the structure and the location of macromolecular complexes is crucial for understanding their cellular functions. The nuclear periphery of a HeLa cell has been investigated using these methods (Mahamid et al., 2016 ▸). Assisted by the synergistic application of recent technical developments, cryo-ET holds promise for revealing the molecular organization giving rise to cellular function in unperturbed environments. These invaluable methods of investigation in vivo could be used to test new compounds directly on targeted proteins in their natural environment.
9. Perspectives
Although recent developments in cryo-EM have been revolutionary, the development of new pharmacological inhibitors requires structural data at better than 3 Å resolution. Such a resolution currently remains inaccessible for all but a small set of targets. But for how long? The method continues to evolve and improvements are ongoing. Among these are the following.
(i) The development of cameras with improved direct detection of electrons, with thinner sensor chips and with a faster readout rate, together improving the signal-to-noise ratio at high resolution.
(ii) The improvement of the ‘phase plate’ technology. These thin-film phase plates consist of a continuous amorphous carbon film of appropriate thickness with a small central hole placed in the back focal plane of the objective lens of the electron microscope. The zero-order beam passes through the central hole while scattered electrons are phase-shifted by −π/2, resulting in a threefold to fivefold increase in specimen contrast. This will be advantageous for the study of small proteins.
(iii) The motion-compensation methods currently used are still relatively poorly effective for the first frames of the movie pictures. It is precisely at this time that the movements are the largest, but the structural information at the highest resolution disappears quickly because of damage owing to irradiation (Scheres & Chen, 2012 ▸). Solving this problem is crucial and will provide information before the onset of irreparable radiation phenomena.
(iv) Improvements in sample preparation, for example the use of materials that are less likely to move under the electron beam or methods for the easier visualization of small proteins such as the use of specific antibodies (especially Fab fragments) or of fusion proteins in order to more effectively identify proteins on the microscope grid. Such molecules can be used not only as ‘benchmarks’ for better alignment during the classification of objects, but can also help to validate a reconstruction (Wu et al., 2016 ▸). The laboratory of Holger Stark has introduced two techniques to improve complex stability and homogeneity. These techniques are called GraFix (Kastner et al., 2008 ▸) and ProteoPlex (Chari et al., 2015 ▸). GraFix improves both the compositional and structural homogeneity of the samples. ProtoPlex is a high-throughput method that systematically tests the stability of a complex in various buffer conditions. The approach uses a dye that fluoresces in the polar environment created when proteins unfold. This allows the rapid screening of buffer conditions that improve sample quality. The method also accommodates the addition of small molecules, inhibitors, agonistic ligands or antibodies to directly screen for their ability to alter conformational stability.
The immediate challenges that we face are the high costs of the purchase and maintenance of the powerful microscopes that are required to elucidate high-resolution structures. The computation-time requirements of the image-processing pipeline are also important and require supercomputers. Poor access to these instruments (in relation to the growing demand) and their costs call for coordinated funding models and group use. Also, correct sample preparation, image collection and analysis is not as easy as it sounds and appropriate training is required.
Rather than imaging technologies or image-processing methods, we contend that the major bottleneck to routine cryo-EM determination of structures at resolution close to 2 Å is currently the preparation of specimens of adequate quality that take into account intrinsic protein flexibility. There are reasons to think that once these new developments in equipment and software have been implemented in a practical way, maybe in the not-too-distant future, the dream of using cryo-EM to determine protein structures at atomic resolution using only a few thousand particles, as described by Henderson (1995 ▸), will finally become a reality.
10. Conclusion
It is clear to us that the arsenal of methods available to obtain structural data from proteins increases the range of possibilities. It is also clear that, contrary to Callaway’s statement that ‘the revolution will not be crystallized’ (Callaway, 2015 ▸), the crystallographic approach remains the method of choice, and perhaps the easiest route, to continue to obtain the information necessary for the discovery and development of new chemical molecules for medical use targeting proteins of ‘reasonable’ size. This arsenal obviously also includes attractive methodologies such as cryo-EM. What has not changed, however, is the imperative need for high-quality (i.e. purified to homogeneity) proteins or objects to be observed, even if, as mentioned previously, image analysis might be taken as an ultimate purification step. Indeed, the process of purification remains key in this context, and the return to favour of methods such as affinity chromatography (as described in Zalk et al., 2015 ▸) remains one of the steps that will help in obtaining better objects to look at, with a chance of enhancing the resolution of the observations.
One would love to think that today we could analyse everything using cryo-EM, but this faces some practical difficulties, not least the purchase of the microscope.
Supplementary Material
Supplementary Table S1, some of the most exciting cryo-EM studies that can be found in the PDB.. DOI: 10.1107/S2053230X17003740/hv5346sup1.pdf
Acknowledgments
CVB and ZL would like to thank UPMC and CNRS for grants.
Footnotes
This paper is an extension of Boutin et al. [(2016 ▸), Med. Sci. (Paris), 32, 758–767].
References
- Abeyrathne, P. D., Koh, C. S., Grant, T., Grigorieff, N. & Korostelev, A. A. (2016). Elife, 5, e14874. [DOI] [PMC free article] [PubMed]
- Bai, X.-C., Yan, C., Yang, G., Lu, P., Ma, D., Sun, L., Zhou, R., Scheres, S. H. W. & Shi, Y. (2015). Nature (London), 525, 212–217. [DOI] [PMC free article] [PubMed]
- Banerjee, S. et al. (2016). Science, 351, 871–875. [DOI] [PMC free article] [PubMed]
- Bartesaghi, A., Merk, A., Banerjee, S., Matthies, D., Wu, X., Milne, J. L. & Subramaniam, S. (2015). Science, 348, 1147–1151. [DOI] [PMC free article] [PubMed]
- Behrmann, E., Loerke, J., Budkevich, T. V., Yamamoto, K., Schmidt, A., Penczek, P. A., Vos, M. R., Bürger, J., Mielke, T., Scheerer, P. & Spahn, C. M. (2015). Cell, 161, 845–857. [DOI] [PMC free article] [PubMed]
- Boutin, J. A., Li, Z., Vuillard, L. & Vénien-Bryan, C. (2016). Med. Sci. (Paris), 32, 758–767. [DOI] [PubMed]
- Brader, M. L., Baker, E. N., Dunn, M. F., Laue, T. M. & Carpenter, J. F. (2017). J. Pharm. Sci. 106, 477–494. [DOI] [PubMed]
- Brillantes, A. B., Ondrias, K., Scott, A., Kobrinsky, E., Ondriašová, E., Moschella, M. C., Jayaraman, T., Landers, M., Ehrlich, B. E. & Marks, A. R. (1994). Cell, 77, 513–523. [DOI] [PubMed]
- Brini, M. (2004). Biochem. Biophys. Res. Commun. 322, 1245–1255. [DOI] [PubMed]
- Callaway, E. (2015). Nature (London), 525, 172–174. [DOI] [PubMed]
- Campbell, M. G., Cheng, A., Brilot, A. F., Moeller, A., Lyumkis, D., Veesler, D., Pan, J., Harrison, S. C., Potter, C. S., Carragher, B. & Grigorieff, N. (2012). Structure, 20, 1823–1828. [DOI] [PMC free article] [PubMed]
- Chari, A. et al. (2015). Nature Methods, 12, 859–865. [DOI] [PMC free article] [PubMed]
- Dashti, A., Schwander, P., Langlois, R., Fung, R., Li, W., Hosseinizadeh, A., Liao, H. Y., Pallesen, J., Sharma, G., Stupina, V. A., Simon, A. E., Dinman, J. D., Frank, J. & Ourmazd, A. (2014). Proc. Natl Acad. Sci. USA, 111, 17492–17497. [DOI] [PMC free article] [PubMed]
- Dubochet, J., Adrian, M., Chang, J. J., Homo, J. C., Lepault, J., McDowall, A. W. & Schultz, P. (1988). Q. Rev. Biophys. 21, 129–228. [DOI] [PubMed]
- Dürr, K. L., Chen, L., Stein, R. A., De Zorzi, R., Folea, I. M., Walz, T., Mchaourab, H. S. & Gouaux, E. (2014). Cell, 158, 778–792. [DOI] [PMC free article] [PubMed]
- Efremov, R. G., Leitner, A., Aebersold, R. & Raunser, S. (2015). Nature (London), 517, 39–43. [DOI] [PubMed]
- Eisenstein, M. (2016). Nature Methods, 13, 19–22. [DOI] [PubMed]
- El-Hayek, R., Lokuta, A. J., Arévalo, C. & Valdivia, H. H. (1995). J. Biol. Chem. 270, 28696–28704. [DOI] [PubMed]
- Frank, J. (2006). Three-Dimensional Electron Microscopy of Macromolecular Assemblies. Oxford University Press.
- Friedmann, D., Messick, T. & Marmorstein, R. (2011). Curr. Protoc. Protein Sci., Unit 17.4. https://doi.org/10.1002/0471140864.ps1704s66. [DOI] [PMC free article] [PubMed]
- Gao, H., Valle, M., Ehrenberg, M. & Frank, J. (2004). J. Struct. Biol. 147, 283–290. [DOI] [PubMed]
- Gustafsson, R., Jemth, A. S., Gustafsson, N. M., Färnegårdh, K., Loseva, O., Wiita, E., Bonagas, N., Dahllund, L., Llona-Minguez, S., Häggblad, M., Henriksson, M., Andersson, Y., Homan, E., Helleday, T. & Stenmark, P. (2017). Cancer Res. 77, 937–948. [DOI] [PubMed]
- Hazel, P., Kroll, S. H., Bondke, A., Barbazanges, M., Patel, H., Fuchter, M. J., Coombes, R. C., Ali, S., Barrett, A. G. & Freemont, P. S. (2017). ChemMedChem. 12, 372–380. [DOI] [PubMed]
- Henderson, R. (1995). Q. Rev. Biophys. 28, 171–193. [DOI] [PubMed]
- Huang, K.-W., Hsu, K.-C., Chu, L.-Y., Yang, J.-M., Yuan, H. S. & Hsiao, Y.-Y. (2016). J. Med. Chem. 59, 8019–8029. [DOI] [PubMed]
- Jin, Q., Sorzano, C. O., de la Rosa-Trevín, J. M., Bilbao-Castro, J. R., Núñez-Ramírez, R., Llorca, O., Tama, F. & Jonić, S. (2014). Structure, 22, 496–506. [DOI] [PubMed]
- Jonić, S. (2017). Curr. Opin. Struct. Biol. 43, 114–121. [DOI] [PubMed]
- Jonic, S. & Vénien-Bryan, C. (2009). Curr. Opin. Pharmacol. 9, 636–642. [DOI] [PubMed]
- Kastner, B. et al. (2008). Nature Methods, 5, 53–55. [DOI] [PubMed]
- Klaholz, B. P. (2015). Open J. Stat. 5, 820–836.
- Li, X., Mooney, P., Zheng, S., Booth, C. R., Braunfeld, M. B., Gubbens, S., Agard, D. A. & Cheng, Y. (2013). Nature Methods, 10, 584–590. [DOI] [PMC free article] [PubMed]
- Liao, M., Cao, E., Julius, D. & Cheng, Y. (2013). Nature (London), 504, 107–112. [DOI] [PMC free article] [PubMed]
- Lyumkis, D., Brilot, A. F., Theobald, D. L. & Grigorieff, N. (2013). J. Struct. Biol. 183, 377–388. [DOI] [PMC free article] [PubMed]
- Mahamid, J., Pfeffer, S., Schaffer, M., Villa, E., Danev, R., Kuhn Cuellar, L., Forster, F., Hyman, A. A., Plitzko, J. M. & Baumeister, W. (2016). Science, 351, 969–972. [DOI] [PubMed]
- Ménétret, J. F., Schaletzky, J., Clemons, W. M. Jr, Osborne, A. R., Skånland, S. S., Denison, C., Gygi, S. P., Kirkpatrick, D. S., Park, E., Ludtke, S. J., Rapoport, T. A. & Akey, C. W. (2007). Mol. Cell, 28, 1083–1092. [DOI] [PubMed]
- Merino, F. & Raunser, S. (2017). Angew. Chem. Int. Ed. Engl. 56, 2846–2860. [DOI] [PubMed]
- Merk, A., Bartesaghi, A., Banerjee, S., Falconieri, V., Rao, P., Davis, M. I., Pragani, R., Boxer, M. B., Earl, L. A., Milne, J. L. & Subramaniam, S. (2016). Cell, 165, 1698–1707. [DOI] [PMC free article] [PubMed]
- Mills-Davies, N. et al. (2017). Acta Cryst. D73, 9–21. [DOI] [PubMed]
- Myasnikov, A. G., Marzi, S., Simonetti, A., Giuliodori, A. M., Gualerzi, C. O., Yusupova, G., Yusupov, M. & Klaholz, B. P. (2005). Nature Struct. Mol. Biol. 12, 1145–1149. [DOI] [PubMed]
- Nagayama, K. (2011). J. Electron Microsc. (Tokyo), 60, S43–S62. [DOI] [PubMed]
- Orlov, I. et al. (2016). Biol. Cell, 109, 81–93.
- Otsu, K., Willard, H. F., Khanna, V. K., Zorzato, F., Green, N. M. & MacLennan, D. H. (1990). J. Biol. Chem. 265, 13472–13483. [PubMed]
- Oxley, M. P., Lupini, A. R. & Pennycook, S. J. (2017). Rep. Prog. Phys. 80, 026101. [DOI] [PubMed]
- Peckys, D. B., Korf, U. & de Jonge, N. (2015). Sci. Adv. 1, e1500165. [DOI] [PMC free article] [PubMed]
- Penczek, P. A., Kimmel, M. & Spahn, C. M. (2011). Structure, 19, 1582–1590. [DOI] [PMC free article] [PubMed]
- Renaud, J.-P., Chung, C.-W., Danielson, U. H., Egner, U., Hennig, M., Hubbard, R. E. & Nar, H. (2016). Nature Rev. Drug. Discov. 15, 679–698. [DOI] [PubMed]
- Rosenthal, P. B. & Henderson, R. (2003). J. Mol. Biol. 333, 721–745. [DOI] [PubMed]
- Rupp, B. (2015). Trends Biochem. Sci. 40, 419–421. [DOI] [PubMed]
- Scheres, S. H. W. & Chen, S. (2012). Nature Methods, 9, 853–854. [DOI] [PMC free article] [PubMed]
- Schroder, R. R. (2015). Arch. Biochem. Biophys. 581, 25–38. [DOI] [PubMed]
- Sigworth, F. J. (1998). J. Struct. Biol. 122, 328–339. [DOI] [PubMed]
- Stuwe, T., Correia, A. R., Lin, D. H., Paduch, M., Lu, V. T., Kossiakoff, A. A. & Hoelz, A. (2015). Science, 347, 1148–1152. [DOI] [PMC free article] [PubMed]
- Sutko, J. L., Airey, J. A., Welch, W. & Ruest, L. (1997). Pharmacol. Rev. 49, 53–98. [PubMed]
- Takizawa, Y., Binshtein, E., Erwin, A. L., Pyburn, T. M., Mittendorf, K. F. & Ohi, M. D. (2017). Protein Sci. 26, 69–81. [DOI] [PMC free article] [PubMed]
- Tung, C.-C., Lobo, P. A., Kimlicka, L. & Van Petegem, F. (2010). Nature (London), 468, 585–588. [DOI] [PubMed]
- Vénien-Bryan, C., Jonic, S., Skamnaki, V., Brown, N., Bischler, N., Oikonomakos, N. G., Boisset, N. & Johnson, L. N. (2009). Structure, 17, 117–127. [DOI] [PMC free article] [PubMed]
- Vinothkumar, K. R. (2015). Curr. Opin. Struct. Biol. 33, 103–114. [DOI] [PMC free article] [PubMed]
- Walls, A. C., Tortorici, M. A., Bosch, B. J., Frenz, B., Rottier, P. J., DiMaio, F., Rey, F. A. & Veesler, D. (2016). Nature (London), 531, 114–117. [DOI] [PMC free article] [PubMed]
- Wan, R., Yan, C., Bai, R., Huang, G. & Shi, Y. (2016). Science, 353, 895–904. [DOI] [PubMed]
- Wang, H.-W. & Wang, J.-W. (2017). Protein Sci. 26, 32–39. [DOI] [PMC free article] [PubMed]
- Wu, S. et al. (2016). Structure, 20, 582–592.
- Yan, Z., Bai, X.-C., Yan, C., Wu, J., Li, Z., Xie, T., Peng, W., Yin, C.-C., Li, X., Scheres, S. H. W., Shi, Y. & Yan, N. (2015). Nature (London), 517, 50–55. [DOI] [PMC free article] [PubMed]
- Yu, X., Jin, L. & Zhou, Z. H. (2008). Nature (London), 453, 415–419. [DOI] [PMC free article] [PubMed]
- Yuchi, Z. & Van Petegem, F. (2016). Cell Calcium, 59, 209–227. [DOI] [PubMed]
- Zalk, R., Clarke, O. B., des Georges, A., Grassucci, R. A., Reiken, S., Mancia, F., Hendrickson, W. A., Frank, J. & Marks, A. R. (2015). Nature (London), 517, 44–49. [DOI] [PMC free article] [PubMed]
- Zhang, X., Settembre, E., Xu, C., Dormitzer, P. R., Bellamy, R., Harrison, S. C. & Grigorieff, N. (2008). Proc. Natl. Acad. Sci. USA, 105, 1867–1872. [DOI] [PMC free article] [PubMed]
- Zhou, A., Rohou, A., Schep, D. G., Bason, J. V., Montgomery, M. G., Walker, J. E., Grigorieff, N. & Rubinstein, J. L. (2015). Elife, 4, e10180. [DOI] [PMC free article] [PubMed]
- Zuercher, W. J., Elkins, J. M. & Knapp, S. (2016). Cell. Chem. Biol. 23, 173–182. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1, some of the most exciting cryo-EM studies that can be found in the PDB.. DOI: 10.1107/S2053230X17003740/hv5346sup1.pdf