Abstract
The phylum candidatus Saccharibacteria formerly known as Candidate Division TM7 is a highly ubiquitous phylum with 16S rRNA gene sequences reported in soils, sediments, wastewater and animals, as well as a host of clinical environments. Here, the application of two taxon-specific primers on environmental and human-associated samples using bar-coded tag pyrosequencing revealed two new clades for this phylum to exist and we propose that the division consists of 2 monophyletic and 2 polyphyletic clades. Investigation into TM7 ecology revealed that a high proportion (58%) of phylotypes were sample specific, few were widely distributed and of those most widely distributed all belonged to subdivision 3. Additionally, 50% of the most relatively abundant phylotypes observed were also subdivision 3 members. Community analysis showed that despite the presence of a high proportion of unique phylotypes, specific groups of samples still harbor similar TM7 communities with samples clustering together. The lack of relatively abundant phylotypes from subdivisions 1, 2 and 4 and the presence of very few cosmopolitan members' highlights not only the site specific nature of this phylum but provides insight into why the majority of studies into TM7 have been biased towards subdivision 3.
The Candidatus Saccharibacteria phylum has been recently coined following complete genome sequencing of several Candidate Division TM7 members from wastewater1. TM7 is a well-known, ubiquitous bacterial phylum described from 16S rRNA gene sequence and genome data only1,2. First identified in a German peat bog3, TM7 16S rRNA gene sequences have subsequently been shown to be present in soil, seawater, activated sludge and many animal and human-associated sources2,4,5,6,7,8. The widespread nature of the phylum has gained interest from both environmental and clinical research groups leading to the design of TM7-specific FISH probes and PCR primers2,9. More recently metagenomics and single-cell genomics approaches have been applied to investigate TM71,7,10. Still, due to a lack of cultured isolates and a paucity of 16S rRNA gene sequences in repositories knowledge on the biology of this enigmatic group is in its infancy11,12.
There have been several reports on the phylogenetic order within the TM7 group, which have proposed 2 or 3 class level clades2,4,7. Three subdivisions proposed by Hugenholtz et al. (2001) and Marcy et al. (2007) displayed a bias in diversity towards subdivision 1. More recently, Dinis et al. 2011 who correlated each clade to sequence origin, reported 2 monophyletic subdivisions; one group consisting of bacteria from environmental sources and the other encompassing clinical and environmental clusters. However, this data disagrees with the other phylogenetic reconstructions of TM7 with sequences from various environmental samples distributed throughout the entire phylum2,6.
A major problem in establishing a reliable taxonomic structure is a lack of available DNA sequences. The RDP contains a little over 4500 TM7 16S rRNA sequences, a small amount considering the highly ubiquitous nature of TM7 with an apparent relative abundance between 1 and 10%2,4,9,13,14,15,16. One way to overcome this lack of sequence data is to survey environments where they are present using amplicon pyrosequencing. Bar coded sequencing technology enables millions of sequences to be obtained in a timely and effective manner17,18. One drawback to these methods is that to obtain more TM7 phylum sequences, highly taxon-specific PCR primers that generate <600 bp amplicons (depending on technology used) are required. TM7 taxon-specific PCR primers and probes were first reported in 2001 and have been used since in a number of studies4,8,19,20. In 2003, Brinig et al., required a more specialised set of primers and probes to investigate TM7 within the human oral cavity and therefore designed phylum-specific probes based on sequences of oral origin only9.
Here, we proposed that currently available TM7 primers were not highly specific to the phylum and thus we describe the development and validation of a TM7 taxon-specific PCR primer set that covers a high percentage of the candidate phylum. We applied this primer set in a 454 bar coded tag pyrosequencing survey on a collection of samples from both environmental and human sources. The resulting sequence data was inserted into an ARB parsimony tree containing full-length reference sequences to visualise the phylogeny of the division, which was then used to investigate the distribution and abundance of TM7 communities from within several diverse environments.
Results
Primer selection and validation
Four potential PCR primers were selected based on results from the RDP Probe Match function (Table 1). When combined in silico, primers TM7-590F and TM7-965R exhibited 94.7% and 92.5% coverage of the TM7 phylum respectively, with no correlating matches to other phyla when paired (Tables S2 and S3). By comparison, previously designed primers exhibited between 41.9–93.4% coverage of the phylum with specificities ranging between 73.9–99.8%. Following optimisation of PCR conditions using DNA from TM7 environmental clone AY540773 (PCF39)8, an optimal annealing temperature of 61°C and an optimal magnesium chloride concentration of 2 mM was determined for each primer set. The amplicon from primer pair TM7-590F/TM7-965R was also of an appropriate length for 454 pyrotag sequencing, being approximately 375 bp long. Unfortunately, primer pair TM7-211F/TM7-590F (synthesized as a reverse primer) yielded non-specific amplicons under most PCR conditions while TM7-211F/TM7-965R produced a fragment too long (approximately 750 bp) for pyrosequencing.
Table 1. Candidate Division TM7-specific PCR primers.
| Name | 5′ – 3′ Sequence |
|---|---|
| TM7-211F | GAGCGGCGGACGGCTGAG |
| TM7-686R | CTACGCAACYCTTTACRCCC |
| TM7-590F* | GWAAAGAGTWGCGTAGGYGG |
| TM7-965R* | WTRCTTAACGCGTTAGCTTCGCT |
*Primers used for pyrosequencing.
Analysis of pyrosequencing data and phylogenetic analysis
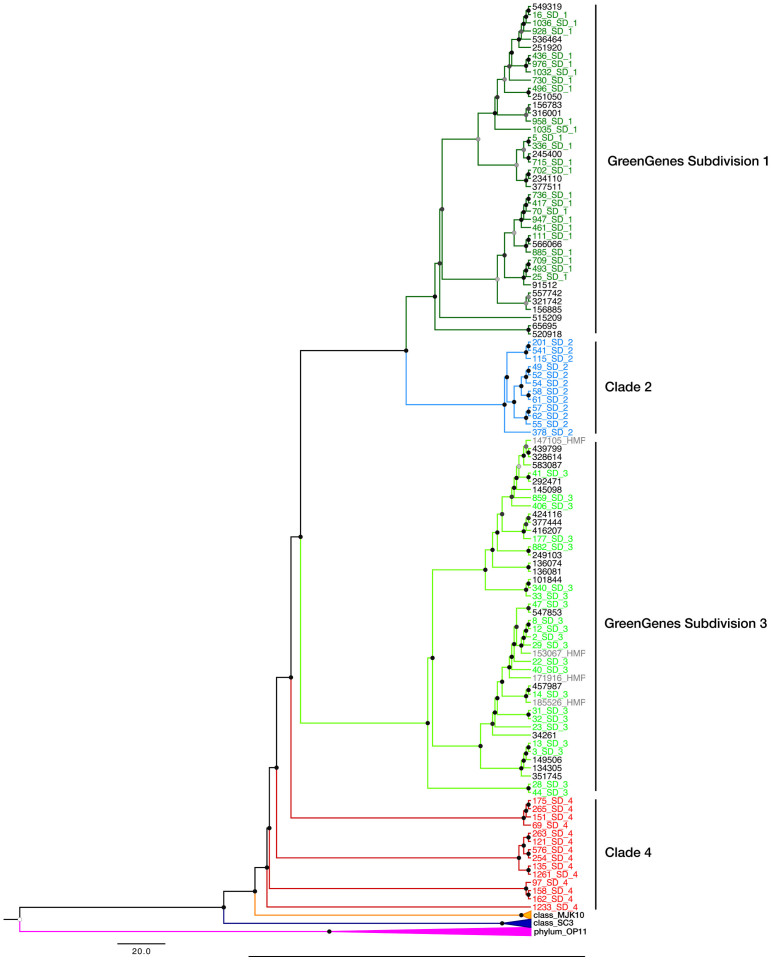
The collective dataset produced 1299 species-level OTUs (0.02 dissimilarity clustering) spanning the V6–V9 regions21. This is compared to 3145 TM7 member OTUs in the RDP as of release 10 update 2911. The evolutionally dendogram was produced from 53 TM7 full-length 16S rDNA reference sequences from the GreenGenes database including 4 from the human microbiome project (HMP). One representative sequence (100% sequence identity) for each of our phylotypes was then inserted in the ARB parsimony tree to visualise clustering of sequences. For ease of interpretation only reference sequences that highlighted clustering of the divisions was included in the final phylogenetic dendogram (Figure 1). At the same time a maximum-likelihood tree was also produced using the GreenGenes sequences to gain a closer image of the similarity of our sequences against those in the database (Figure S1). Since this similarity tree had an abundance of terminal branches, resulting in poor visualisation, representative OTU sequences clustered at 0.08 dissimilarity was used (92% similarity). This reduced the number of terminal branches for ease of interpretation and generated 495 OTUs.
Figure 1. Evolutionary dendogram of the Candidatus Saccharibacteria phylum.
Phylogeny constructed with 72 new sequences (OTUs defined at 0.08 dissimilarity), as well as 57 sequences from GreenGenes and the HMP representing previously defined class level clades 1, 3, MJK10, SC3 and the outgroup OP11. The reference sequences from GreenGenes were prefixed with the subdivision affiliation assigned by the GreenGenes taxonomy; Subdivision 1 (dark green), Subdivision 3 (light green), Class MJK10 (yellow), SC3 (dark blue) or OP11 (pink). The branches of the trees are colored according to the 4 proposed clades; 1 (dark green), 2 (blue), 3 (light green), 4 (red). Bootstrap confidence was calculated from 1000 replicates and displayed by circles at each node. The color of the circles, ranging from white to black corresponds to the bootstrap confidence values (between 50 and 100%).
The evolutionary dendogram consisted of up to 4 clades, with all GreenGenes sequences falling within Subdivisions 1 and 3 only. Two additional clades were observed outside the 2 major subdivisions that consisted solely of the TM7 sequences recovered here. Clades 1 and 2 were monophyletic whilst 3 and 4 formed paraphyletic groups (Figures 1 and S1). Clade 4 appears to be the highest rank of the 4 subgroups, with clades 1, 2 and 3 as daughter groups. Of all 1299 species-level OTUs observed in this TM7 dataset, approximately 0.3% belonged to MJK10, 35.8% to subdivision or clade 1, 22.2% to clade 2, 27.5% to subdivision or clade 3, and 14.3% to clade 4 (Table 2; Figure 3). All sequences affiliated with previously designated subdivisions 1 and 3 were concurrent with those defined by GreenGenes as all the downloaded reference sequences resided within their expected clades12,22. For MJK120, only 7 sequences belonged to this group, yet it was basal to all the other clades within TM7 (Figures 1 and S1). For the purpose of this study we then grouped MJK10 with clade 4.
Table 2. Total number of representative OTUs identified from the four proposed subdivisions and the relative abundance or total reads obtained.
| Subdivision | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Total OTUs | 1,228 | 179 | 159 | 119 |
| Total Reads | 15,652 | 8,888 | 13,224 | 2,521 |
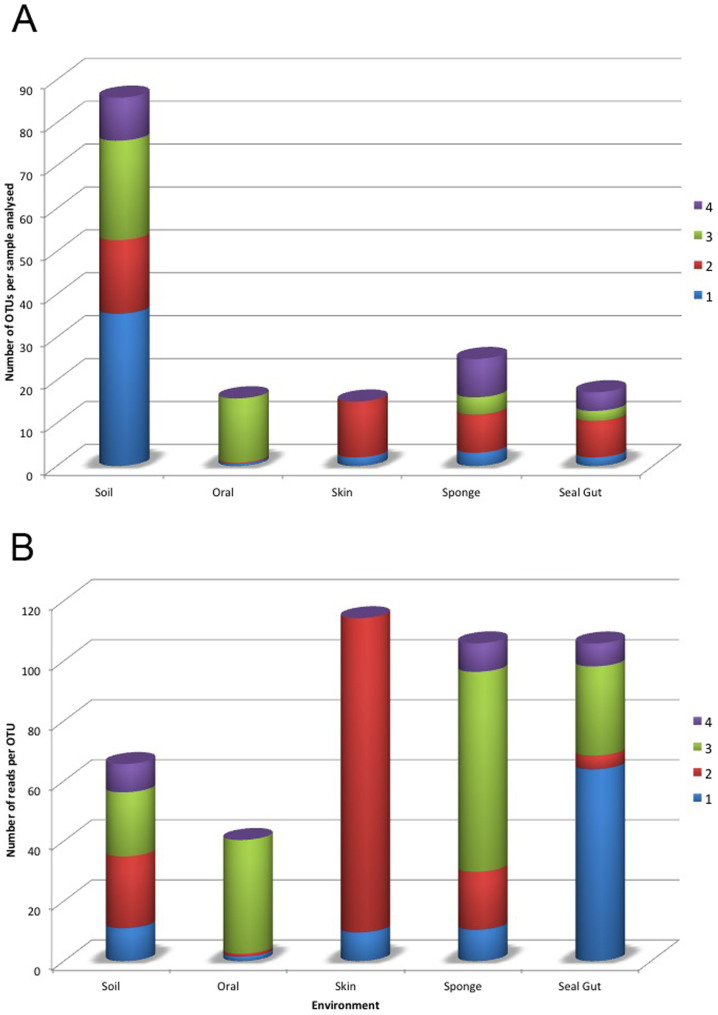
Figure 2.
Bar charts comparing the relative abundance of TM7 phylotypes per individual sample from the 4 proposed clades (A) OTUs recovered per sample. (B) Total reads obtained per sample from each subdivision. Environmental samples contained TM7 sequences from all 4 proposed clades with soil harboring the greatest number of phylotypes per sample analysed. OTUs from clades 2 and 4 were distributed widely, yet no reliable 16S rRNA gene sequences were present in the databases for these phylotypes. In most cases, a greater distribution and abundance of subdivision 3 OTUs was found in environmental samples.
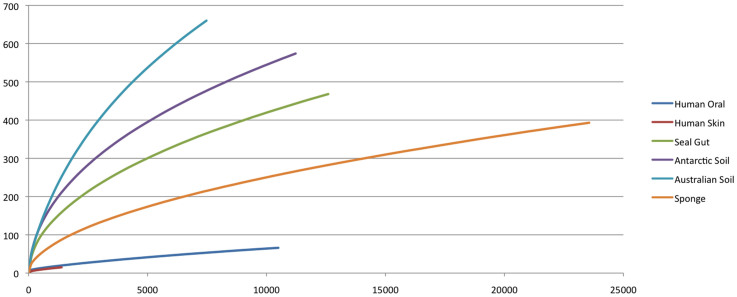
Figure 3. Rarefaction curves for species-level TM7 OTUs (0.02 similarity) recovered using new taxon-specific primers and 454 bar coded tag pyrosequencing.
Each group of samples was sequenced to various depths with human oral samples sequenced to asymptote. The higher species richness of environmental samples, particularly soil and seal faecal samples is highlighted with the Australian soil requiring much more sequencing then the other environments.
The total number of OTUs identified within clade 1 was far greater (1,228) than subdivisions 2,3 and 4 combined indicating the greater richness of this subdivision (Table 2; Figure 2). Yet the number of total reads recovered for subdivision 1 and 3 were similar. For subdivisions 2 and 4 significantly fewer total reads were obtained despite recovering a similar number of OTUs as was recovered in subdivision 3. The distribution of the reads was consistent with the proportion of samples submitted from each environment (Figure 2). The oral samples consisted almost exclusively of sequences falling within subdivision 3, while the skin sample consisted almost exclusively of a new cluster (clade 2) (Figures 1 and S1). The remainder of sequences from the environmental samples, as well as those from seal faeces, were distributed throughout the phylum (Table 2; Figure 2). Besides the affiliation of oral samples to subdivision 3 and the skin sample sequences to clade 2, no other source of TM7 OTUs showed any correlation to a particular subgroup of the phylum. Rarefaction curves highlighted the greater species richness of environmental samples over human host samples (Figure 3). Soil samples contained the greatest species richness with the Australian soil group requiring more sequencing to reach an asymptote. By comparison, the human host samples appeared to be adequately sequenced reaching an asymptote in this study.
The relative abundance of a particular OTU varied significantly between samples with 58% specific to one individual sample only. Analysis of similarities (ANOSIM) highlighted that despite this variability, groups of samples from a specific environment were significantly more similar to each other then to the other environments analysed with an overall significance level of 0.001 and a Global R value 0.488 (Table 4). Those groups that were most similar were Australian soil and sponge samples as well as oral and sponge samples with a 0.001 significance level combined with Global R values of 0.443 and 0.439, respectively. While oral and Australian soil samples exhibited a 0.939 Global R value at a significance level of 0.003.
Table 3. Most prevalent OTUs, their proposed subdivision affiliation and their relative abundance as determined by (A) total number of TM7 reads recovered or (B) those shared OTUs present in 6 or more samples analysed.
| A | |||||
|---|---|---|---|---|---|
| OTU | Total reads | Positive samples | Clade | % Similarity TM7; Accession number | Reference; Sample type |
| 343 | 2536 | 2 | 3 | 98; AF269025 | Hugenholtz et al., 2001; wastewater treatment plant |
| 9 | 2175 | 4 | 2 | 92; EU056419 | Kuehbacher, et al., 2008; human intestine active in inflammatory bowel disease |
| 1 | 1556 | 5 | 3 | 100; HM215448 | Dinis et al., 2011; human tooth surface scraping |
| 49 | 1305 | 4 | 2 | 100; JN713533 | Dewhirst et al., 2009; canine oral cavity subgingival plaque |
| 364 | 1250 | 3 | 1 | 94; JF03514 | Zhang et al., 2012; root and rhizophere soil |
| 504 | 1040 | 5 | 3 | 97; FJ629383 | Lou et al., 2009; agricultural soil with toluene |
| 336 | 786 | 5 | 1 | 100; AY540773 | Ferrari et al., 2005; soil |
| 360 | 581 | 2 | 1 | 97; FJ542866 | Rattray et al., 2010; earthworm gut |
| 18 | 552 | 3 | 3 | 100; GU410601 | Dewhirst et al., 2010; human oral cavity |
| 309 | 545 | 7 | 3 | 100; EU056483 | Kuehbacher et al., 2008; human intestine active in inflammatory bowel disease |
Table 4. Analysis of similarity (ANOSIM) between TM7 communities from within sample groups.
| Groups Observed | R Statistic | Significance level |
|---|---|---|
| Oral: Seal Gut | 0.54 | 0.005 |
| Oral: Polar Soil | 0.844 | 0.029 |
| Oral; Soil | 0.939 | 0.003 |
| Oral: Sponge | 0.439 | 0.001 |
| Seal Gut: Polar Soil | 0.083 | 0.276 |
| Seal Gut: Sponge | 0.228 | 0.030 |
| Seal Gut: Soil | 0.278 | 0.010 |
| Polar Soil: Soil | 0.439 | 0.012 |
| Polar Soil: Sponge | 0.393 | 0.008 |
| Soil: Sponge | 0.443 | 0.001 |
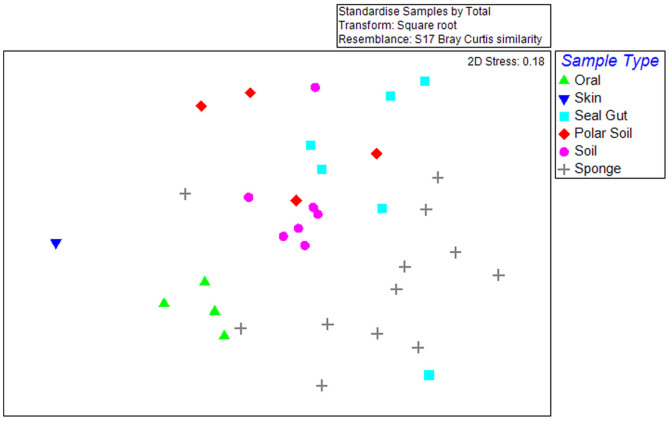
We used non-metric multidimensional scaling (nMDS) analysis (which clusters samples based on OTU composition) to visualize clusters of samples containing highly similar TM7 phylotype compositions (Figure 4). The nMDS plot also showed that host-specific and/or site-specific groups of samples did harbour similar communities with samples clustering together. However, the level of similarity observed was less than 10% for all sample groups investigated apart from the human oral group. We also carried out SIMPER analysis that confirmed oral samples to be the most closely related TM7 communities with 29.2% similarity between samples observed. Groups of different environmental and animal host samples exhibited less similarity to each other with 6.2% for polar soil, 6.8% for seal faeces and 8.6% for Australian soils. Sponge samples contained the greatest variability between samples with just 2.9% similarity observed within this group (Figure 4). The high variation between marine sponge samples observed may have been due to the species of sponge analyzed as this information was not known.
Figure 4. nMDS plots showing the community similarity between sample groups.
Despite the presence of very few abundant phylotypes in more than one sample, TM7 communities from various hosts or environments were more closely related to each other than to the other environments analysed with clustering observed. At a level of 20% similarity most groups did not completely cluster, highlighting the variable nature of this enigmatic phylum.
Only 42% of the species-level OTUs identified in this study was present in two or more of the 35 samples analyzed. Of these, the most widely distributed 8 OTUs identified were found to cluster within subdivisions 1 and 3, as well a new cluster (clade 2), yet these sequences were only detected in just 17–20% of samples analysed (Figure 5A, Table 3). While present in the highest number of samples investigated these phylotypes exhibited a low level (0.05–2% total reads) of relative abundance throughout the entire dataset. By comparison, the 10 phylotypes that contributed to the greatest number (2–9%) of total TM7 reads were all affiliated with the well-characterised subdivision 3 (Figure 5B, Table 3). The most dominant phylotype observed (OTU343) contributed 2,536 total reads or 9% relative abundance, but, the majority of these reads (2,531) were recovered from a single seal faecal sample only. This trend was observed repeatedly for the most dominant and widely distributed phylotypes with OTU336, from subdivision 1, detected in 5 samples. However, its relative abundance was 2.9% (786 total reads) and of these 694 reads was recovered from a single sponge sample.
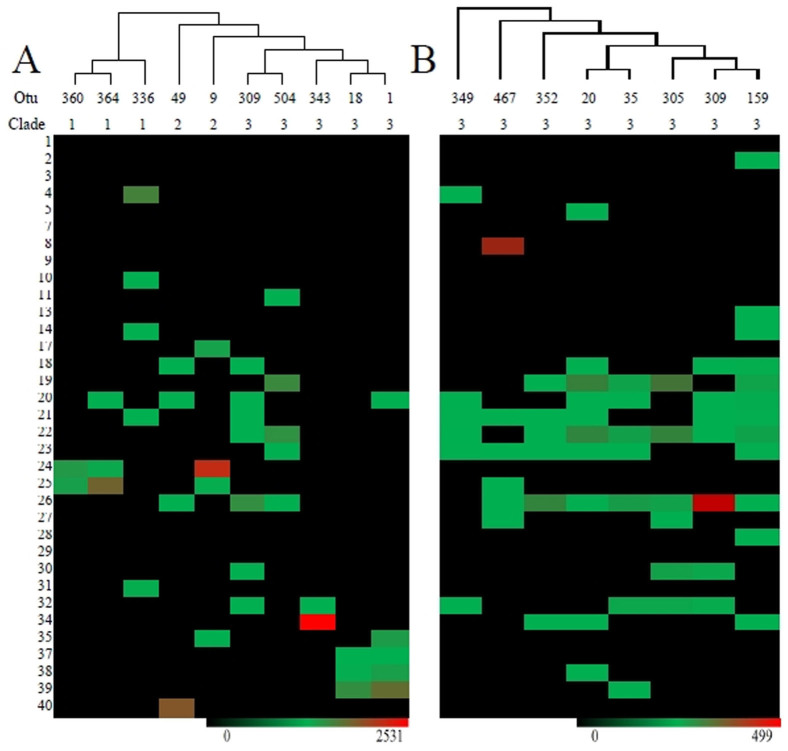
Figure 5. Heatmap visualisation of the most dominant TM7 OTUs identified along with their proposed clade affiliation (A) the most highly abundant phylotypes and (B) the most widely distributed phylotypes observed.
Each gradient key displays the highest number of total reads obtained for each group of phylotypes with many OTUs present in relatively high abundance for one sample only (red/brown). The most widely distributed phylotypes all clustered with subdivision 3 sequences, all in relatively low abundance (<0.02% total reads) and were found in soils, sponge, seal gut and human oral samples. The most abundant phylotypes, representing 2–9% of total reads were present within clades 1, 2 or 3. Those affiliated with clade 1 were detected in Antarctic and Australian soils, with OUT 336 also found in seal gut and sponge samples. Clade 2 OTUs were primarily affiliated with skin or oral samples and soils. Clade 3 OTUs were affiliated with a range of samples, with OTU's 1 and 18 almost exclusively detected in oral human samples, OTU 309 was found in soils and seal gut samples, while OTU 343 was only detected in seal gut samples. For details on each sample and each OTUs closest match in the NCBI database see Tables S1 and 3.
Discussion
We have validated a highly specific set of Candidatus Saccharibacteria phylum specific PCR primers suitable for 454 bar coded tag pyrosequencing. The application of this primer pair resulted in a significant increase in the number of 16S rRNA gene sequences currently available in databases and revealed that two new clades may exist for this phylum2,4,7,10. The evolutionary tree showed that these novel sequences did indeed cluster outside well-described TM7 subdivisions 1 and 32. Clades 1 and 2 appeared to be monophyletic, while clade 3, (which was previously reported as a monophyletic clade 24 and clade 4, appeared to form polyphyletic groups. We recognise that as the TM7 sequences recovered here were short (375 bp) and a more reliable taxonomy of this phylum is now required consisting of full-length sequences from clades 2 and 4.
There have been several reports on TM7 phylogeny with an association of human derived samples to one subdivision and environmental members to another subdivision4,9. Using our data and all currently available Greengenes and HMP full-length TM7 16S rDNA gene sequences we highlight human-associated samples to be almost exclusively associated with subdivision 3 (previously known as clade 2 in Dinis et al., 2011), while environmental OTUs were distributed throughout the entire phylum (Figure 1). Thus, the evolutionary tree appears to be most similar to the phylogeny constructed by Hugenholtz et al., 2001. Additionally, the affiliation of the majority of sequences from the skin sample to a new cluster (clade 2), and the lack of reliable reference sequences to both clades 2 and 4 in the databases reveals that there are still advantages of using up-to-date PCR primers in microbial ecology investigations.
Further analysis into the diversity and abundance of TM7 communities recovered revealed that while TM7 members are reported to be ubiquitous, many individual samples analysed here harbored a high proportion of unique phylotypes. This sample-specific distribution of phylotypes was widespread and was surprizing as very few cosmopolitan OUT's were identified. Of the most prevalent 8 OTUs detected (in 15–20% of samples only), all 8 were associated with subdivision 3 (Figure 4;Table 3), which was consistent with subdivision 3 being reported on extensively in environmental samples, particularly sewage treatment plants2,23,24. While these 8 OTUs were found in the greatest number of samples, half of the OTUs were detected in low relative abundance per sample, contributing to less than 2% of total reads recovered. This is much lower than for members from the ubiquitous marine bacterium clade SAR11 who were recently reported to be present in 50%–100% of marine samples analyzed25.
The 10 most dominant TM7 sequences identified here were distributed within clades 1, 2 and 3 and those associated with clades or subdivisions 1 and 3 were identified previously in wastewater treatment plants, earthworm gut, soils, as well as human and animal clinical samples2,26,27. Subdivision 3 OTU's 309 and 9, distributed in soils, oral and seal faecal samples were most closely related (100 and 92% similarity) to 16S rRNA gene sequences associated with active inflammatory bowel disease6. These most abundant phylotypes were also detected at high relative abundance in one sample only (Figure 5, Table 3). For example, subdivision 1 OTU336 exhibiting 100% similarity to PCF39, a microcolony forming TM7 bacteria from soil8, was detected in sponge, seal faecal and soil samples, however, the majority of reads were detected in one sponge sample only.
The similarity of TM7 communities between groups of samples showed that despite a high proportion of phylotypes being unique to one individual sample, community diversity and composition from the same host or environment were still more similar to each other (0.001 significance level and Global R value 0.488) than to the other groups of samples investigated. Host species have recently been suggested to play a significant role in predicting the bacterial community present, with unique communities reported to be host-specific in amphibians28 and seals29. The level of similarity we observed between groups was low between 2–29% and may be due to both the high proportion of unique phylotypes present and the lack of adequate sequence coverage for environmental samples. In soil, there was greater species richness for subdivision 1, as reported previously, but a higher abundance of subdivision 3 members. Currently, all metagenomic and genome reports on TM7 have focused on subdivision 3 members from wastewater or oral samples, including the most recent publication of the complete genome for 4 subdivision 3 members from wastewater1,7,10. We hypothesize that the greater abundance and distribution of subdivision 3 members has contributed to a sampling bias that we are now observing for this group.
We reveal Candidatus Saccharibacteria to be a diverse phylum, consisting of a high proportion (58%) of site and sample-specific phylotypes distributed throughout 2 monophyletic and 2 polyphyletic clades. The presence of very few cosmopolitan members highlights the site-specific nature of this phylum and provides insight into why members from this group have yet to be recovered into pure culture. We hypothesize that the lack of relatively abundant phylotypes from clades 1, 2 and 4 may have contributed to why the majority of investigations into TM7 have been biased towards subdivision 3 members and highlights the future requirement for targeted investigations across all clades within this enigmatic group.
Methods
Sample sources and DNA extraction
Thirty-five diverse environmental, animal and human-associated samples from Australia and Antarctica were obtained (Table S1). For the 12 Australian and Antarctic soil samples, 300 mg was extracted using the FastDNA SPIN Kit for soil (FastDNA SPIN kit for soil, MP Biomedicals, Australia) as described18. For the 5 human samples, the prepGEM Tissue (ZyGEM Corporation, New Zealand) DNA extraction method was used as described30 and extracted gDNA was preserved with 0.5 M EDTA (0.2 μl). For the 12 marine sponge and 6 seal faecal samples analysed gDNA was supplied following extraction using the QIAamp stool kit29.
Design and in silico testing of TM7-specific PCR primers
For effective targeting of the TM7 via 454 pyrosequencing, oligonucleotide primers with a high specificity for the phylum were required. To design new primers all available TM7 phylum sequences were downloaded from the GreenGenes online resource (http://greengenes.lbl.gov/cgi-bin/nphindex.cgi)12 and were aligned with Clustal X v1.83.131. Alignments were then visually scanned for conserved regions homologous to only TM7 sequences. An in silico analysis of primer specificity was performed with candidate primer sequences as well as previously published primer pairs using the RDP's probe match function (http://rdp.cme.msu.edu/)32 (Tables S2 and S3). Finally, 4 candidate primers were designed, synthesized (Sigma-Aldrich, Australia) and evaluated for suitability for 454 bar coded tag pyrosequencing. Naming of each primer was based on the Escherichia coli numbering of the 16S rRNA gene.
Optimisation of the PCR conditions for all candidate primers
GoTaq Flexi hot-start polymerase (Promega, Australia) was used for all PCRs. A gradient PCR assay was used to optimize the PCR protocol for all candidate primers using various primer pairs (Table 1). Reactions (50 μl) were set up over a range of annealing temperatures (58–65°C) and magnesium chloride concentrations (1.5–4 mM) to amplify the 16S rRNA gene from positive and negative controls. The positive control used was a TM7 16S rRNA gene fragment (AY540773) obtained in our laboratory in a previous study8 and Escherichia coli gDNA was used as the negative control. The initial master mix contained 1.5–4 mM MgCl2, 800 μM dNTPs, 5 μg BSA, 10 pmol each primer and approximately 10 ng of DNA was used in each 50 μl PCR reaction. The PCR program consisted of initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 58–65°C for 30 s and 72°C for 30 s for every 500 base pairs of sequence to be amplified. All reactions were terminated with a final step of 72°C for 5 min. Reactions was performed using a Bio-Rad MyCycler Thermocycler (Bio-Rad, Australia). Products were visualized on a 2% agarose gel. Inspecting the electrophoresed products and selecting the corresponding band that produced the brightest single product of the correct size was used to determine the optimal annealing temperature and magnesium chloride concentration.
454 bar coded amplicon pyrosequencing
The Roche 454 FLX Titanium platform was used for amplicon pyrosequencing obtaining reliable read lengths of ~400 bp. At the time this was the most reliable technology available for bar coded sequencing. All 35 environmental and human samples tested positive by our TM7-specific PCR and were diluted to 10 ng/μl and sent for tag-encoded pyrosequencing. The sequencing was performed at the Research and Testing Laboratory (Lubbock, USA) (www.researchandtesting.com) using the forward candidate primer TM7-590F attached to biotinylated adapters. Bar-coded pyrosequencing involved an initial PCR stage with fusion primers containing custom unique 8-mer barcodes linked to the PCR primers. Subsequently, emulsion PCR (emPCR) was performed and then following quality checking of amplified products, the samples were titrated and mixed equally based on amplicon concentration then applied to the picotitre plate for pyrosequencing33. Standard flowgram format (Sff) files were generated from the raw data output from the instrument including the sequence data, quality files and flowgrams.
Processing pipeline for raw pyrosequencing data
Sequence data were processed with the mothur software package34. Reads were denoised with the PyroNoise algorithm using the author's defined parameters, flowgrams with values below 360 were removed and those above 720 were truncated to this value35, trimmed to remove short (<200 bp) reads and pre-clustered at 1% dissimilarity to account for the per-base error rate in 454 FLX Titanium sequencing. NAST alignment against the SILVA database was performed36 followed by chimaera removal using the UCHIME algorithm37. Distances between sequences were calculated and then clustering was done at 98% similarity to obtain species level OTUs as defined for the V6–V9 region21,37. Representatives of each OTU were taxonomically identified using the GreenGenes 2011 taxonomy and analysed using the Basic Local Alignment Search Tool (BLAST) in GenBank22.
Analysis of data and phylogenetic tree construction
Representative TM7 16S rDNA gene sequences present in GreenGenes were collated with TM7 sequences from the Human Microbiome Project (http://www.hmpdacc.org/) and imported into the ARB software environment38. The phylogenetic placements of 16S rRNA sequences were determined using the neighbour-joining method within ARB as described previously (substitution correction model: Olsen correction)38,39. The partial length TM7 sequences obtained in this study were then aligned to the Greengenes ARB sequence database, and inserted into the evolutionary tree with the parsimony insertion tool. The OP11 phylum was retained in the phylogeny as an outgroup. Those clades that contained reference sequences from GreenGenes or the HMP were labelled as GreenGenes Subdivision 1 or 3 (which represents the subdivision defined by Greengenes). To estimate confidence of the resulting tree topology, bootstrap resampling analysis for 1000 replicates was carried out. All GreenGenes reference sequences were labelled within the tree with their accession number in black and those with representatives from the human microbiome project were labelled as accession number_HMP in grey font. The reference sequences obtained in this study were labelled with a number followed by their proposed affiliation to clades 1–4. Sequences clustering within the GreenGenes subdivisions 1 and 3 were labelled in green, while those affiliated with 2 new clusters were labelled blue (clade 2) or red (clade 4). A similarity tree was also constructed to visualize sequences in closer detail using the FastTree software application40. This generated a maximum-likelihood tree and included were reference sequences from GreenGenes, including class level subdivisions 1, 3 and MJK10, as well as an unclassified clade also with several outgroup species, Escherichia albertii str 19982 (AY696662), Bacillus acidicola str. TSAS-1 (GQ389780) and Actinomyces catuli str. CCUG 41709 (AJ276805). Community dissimilarity was determined by first generating a maximum-likelihood tree from unique sequences using the FastTree program. This involved using a sequence alignment and first generating a fast heuristic-style neighbour-joining tree then, through a combination of nearest-neighbour interchanges (NNI) and subtree-prune-regraft moves (SPR), a minimum-evolution (ME) tree was created. This ME tree was then rearranged using a general-time-reversible model, into a maximum likelihood tree with optimized topology and branch-lengths. Bootstrap replications (1000) were performed and bootstrap confidence values were displayed on the ARB evolutionary and the similarity trees with circles at the nodes colored in a gradient from white to black to indicate bootstrap values between 50 and 100% respectively.
Statistical analyses
Multivariate statistical analyses were performed using the Plymouth Routines in Multivariate Ecological Research (Primer-E v6) program41. These analyses were based on sample-by-OTU abundance matrices produced in the Mothur software package. Data was standardized, square root transformed and a resemblance matrix using the Bray-Curtis dissimilarity algorithm was calculated. Similarities between sample groups were visualized using non-metric multidimensional scaling (nMDS) plots. NMDS plots, SIMPER and ANOSIM analysis, which looks at the similarity between samples was used as well as generation of rarefaction curves to compare TM7 communities between and within sample groups. Rarefaction data were generated via a sampling without replacement method using the Mothur package.
Deposition of pyrosequencing data into NCBI
The TM7 phylum library of pyrosequencing data was deposited in the NCBI-NIH Sequence Read Archive (SRA) under the accession SRA054930. Accession numbers also submitted for each TM7 phylotype identified here under accession numbers KF578542-KF579840.
Supplementary Material
Supplementary information
Acknowledgments
The authors would like to thank Dr Andrew Bissett from CSIRO Canberra for sharing his knowledge on using ARB. We would also like to thank researchers from UNSW, Macquarie University and the Australian Antarctic Division for the supply of gDNA from a range of samples; Drs Nathan Fenning for the supply of clinical gDNA, Tiffanie Nelson for seal gut gDNA, Chris Fan Lu for sponge gDNA, Rachael Anderson for Antarctic soil gDNA and Malcolm Walter, Karthikeyan Gunasekaran, Jason Lowe for Australian soil gDNA samples. This work was fully supported by UNSW internal grant funding schemes.
Footnotes
The authors declare no competing financial interests.
Author Contributions B.F. wrote the manuscript and prepared many of the figures. T.W. carried out the laboratory work and prepared figures 1 and S1. M.J. prepared figure 5. B.N. offered input into the design of the experiments and towards the figures presented in the manuscript. All authors reviewed the manuscript.
References
- Albertsen M. et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nature Biotechnol. 31, 533–538, 10.1038/nbt.2579 (2013). [DOI] [PubMed] [Google Scholar]
- Hugenholtz P., Tyson G. W., Webb R. I., Wagner A. M. & Blackall L. L. Investigation of Candidate Division TM7, a Recently Recognised Major Lineage of the Domain Bacteria with No Known Pure-Culture Representatives. Appl. Environ. Microbiol. 67, 411–419 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheims H., Sproer C., Rainey F. A. & Stackebrandt E. Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology 142 (Pt 10), 2863–2870 (1996). [DOI] [PubMed] [Google Scholar]
- Dinis J. M. et al. In search of an uncultured human-associated TM7 bacterium in the environment. PLoS ONE 6, e21280, 10.1371/journal.pone.0021280 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 3, REVIEWS0003 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehbacher T. et al. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J. Med. Microbiol. 57, 1569–1576, 10.1099/jmm.0.47719-0 (2008). [DOI] [PubMed] [Google Scholar]
- Marcy Y. et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. U S A 104, 11889–11894, 10.1073/pnas.0704662104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari B., Binnerup S. J. & Gillings M. R. Microcolony cultivation on a soil substrate membrane system recovers previously unculturable bacteria. Appl. Environ. Microbiol. 71, 8174–8720 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinig M. M., Lepp P. W., Ouverney C. C., Armitage G. C. & Relman D. A. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl. Environ. Microbiol. 69, 1687–1694 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M. et al. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl. Environ. Microbiol. 73, 3205–3214, 10.1128/AEM.02985-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. R. et al. The Ribosomal Database Project (RDP-II): Introducing myRDP Space and Quality Controlled Public Data. Nucleic Acids Res. 35, D169–D172 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072, 10.1128/AEM.03006-05 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanga M., Paster B. J. & Guerrero R. The taxophysiological paradox: changes in the intestinal microbiota of the xylophagous cockroach Cryptocercus punctulatus depending on the physiological state of the host. Int. Microbiol. 12, 227–236 (2009). [PubMed] [Google Scholar]
- Elinav E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757, 10.1016/j.cell.2011.04.022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouverney C. C., Armitage G. C. & Relman D. A. Single-Cell Enumeration of an Uncultivated TM7 Subgroup in the Human Subgingival Crevice. Appl. Environ. Microbiol. 69, 6294–6298, 10.1128/aem.69.10.6294-6298.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Kong Y. & Nielsen P. H. r. In situ detection of protein-hydrolysing microorganisms in activated sludge. FEMS Microbiol. Ecol. 60, 156–165, 10.1111/j.1574-6941.2007.00279.x (2007). [DOI] [PubMed] [Google Scholar]
- Sogin M. L. in Uncultivated Microorganisms Vol. 10, Microbiology Monographs (ed Epstein, S.) Ch. 2, 19–34 (Springer, 2009). [Google Scholar]
- Winsley T., van Dorst J. M., Brown M. V. & Ferrari B. C. Capturing greater 16S rRNA gene sequence diversity within the domain Bacteria. Appl. Environ. Microbiol. 78, 5938–5941, 10.1128/AEM.01299-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari B. C., Winsley T., Gillings M. & Binnerup S. Cultivating previously uncultured soil bacteria using a soil substrate membrane system. Nat. Protoc. 3, 1261–1269 (2008). [DOI] [PubMed] [Google Scholar]
- Luo C., Xie S., Sun W., Li X. & Cupples A. M. Identification of a novel toluene-degrading bacterium from the candidate phylum TM7, as determined by DNA stable isotope probing. Appl. Environ. Microbiol. 75, 4644–4647, 10.1128/AEM.00283-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Morrison M. & Yu Z. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J. MIcrobiol. Meth. 84, 81–87, 10.1016/j.mimet.2010.10.020 (2011). [DOI] [PubMed] [Google Scholar]
- McDonald D. et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618, 10.1038/ismej.2011.139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere D. et al. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 3, 700–714, 10.1038/ismej.2009.2 (2009). [DOI] [PubMed] [Google Scholar]
- Kong G. K., Adams J. J., Cappai R. & Parker M. W. Structure of Alzheimer's disease amyloid precursor protein copper-binding domain at atomic resolution. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 63, 819–824, 10.1107/S1744309107041139 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. V. et al. Global biogeography of SAR11 marine bacteria. Mol. Syst. Biol. 8, 595 10.1038/msb.2012.28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst F. E. et al. The canine oral microbiome. PLoS ONE 7, e36067 10.1371/journal.pone.0036067 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray R. M., Perumbakkam S., Smith F. & Craig A. M. Microbiomic comparison of the intestine of the earthworm Eisenia fetida fed ergovaline. Curr. Microbiol. 60, 229–235, 10.1007/s00284-009-9530-8 (2010). [DOI] [PubMed] [Google Scholar]
- McKenzie V. J., Bowers R. M., Fierer N., Knight R. & Lauber C. L. Co-habiting amphibian species harbor unique skin bacterial communities in wild populations. ISME J. 6, 588–596 10.1038/ismej.2011.129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T. M., Rogers T. L., Carlini A. R. & Brown M. V. Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environ. Microbiol. 15, 1132–1145, 10.1111/1462-2920.12022 (2013). [DOI] [PubMed] [Google Scholar]
- Ferrari B. C., Power M. L. & Bergquist P. L. Closed-tube DNA extraction using a thermostable proteinase is highly sensitive, capable of single parasite detection. Biotechnol. Lett. 29, 1831–1837 (2007). [DOI] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948, 10.1093/bioinformatics/btm404 (2007). [DOI] [PubMed] [Google Scholar]
- Cole J. R. et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35, D169–172, 10.1093/nar/gkl889 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd S. E., Sun Y., Wolcott R. D., Domingo A. & Carroll J. A. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog. Dis. 5, 459–472, 10.1089/fpd.2008.0107 (2008). [DOI] [PubMed] [Google Scholar]
- Schloss P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541, 10.1128/AEM.01541-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C. et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Meth. 6, 639–641, 10.1038/nmeth.1361 (2009). [DOI] [PubMed] [Google Scholar]
- Caporaso J. G. et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267, 10.1093/bioinformatics/btp636 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C. & Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200, 10.1093/bioinformatics/btr381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W. et al. ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371, 10.1093/nar/gkh293 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T. et al. Characterization of filamentous bacteria, belonging to candidate phylum KSB3, that are associated with bulking in methanogenic granular sludges. ISME J. 1, 246–255, 10.1038/ismej.2007.28 (2007). [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S. & Arkin A. P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490, 10.1371/journal.pone.0009490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K. R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143 (1993). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information