Abstract
The microRNA (miRNA) pathway is well established to be involved in host-pathogen interactions. As key insect pollinators, bees are suffering from widely spreading viruses, especially honeybees and bumblebees. In order to better understand bee-virus interaction, we comparatively analyzed the involvement of the bumblebee miRNA pathway upon infection by two different viruses. In our setup, an avirulent infection is induced by slow bee paralysis virus (SBPV) and a virulent infection is induced by Israeli acute paralysis virus (IAPV). Our results showed the increased expressions of dicer-1 and ago-1 upon SBPV infection. There were 17 and 12 bumblebee miRNAs differentially expressed upon SBPV and IAPV infections, respectively. These results may indicate the involvement of the host miRNA pathway in bumblebee-virus interaction. However, silencing of dicer-1 did not influence the genome copy number of SBPV. Target prediction for these differentially expressed miRNAs showed their possible involvement in targeting viral genomic RNA and in the regulation of networks in bumblebee. Our study opens a new insight into bee-virus interaction meditated by host miRNAs.
RNA interference (RNAi) is a conserved mechanism for posttranscriptional gene silencing to regulate gene expression and defense against viral infection in insects. Currently, three sub-pathways are recognized under the RNAi pathway based on the produced small RNAs, including: small interfering RNA (siRNAs), microRNAs (miRNA), and Piwi-interacting RNAs (piRNAs) pathways. In bees, two aspects of the siRNA-pathway-mediated antiviral response has been considered: virus derived siRNAs and utilization of this pathway to control virus through virus-specific dsRNA1. Our previous study in the bumblebee Bombus terrestris showed that Israeli acute paralysis virus (IAPV) replicated very fast and acted as a very virulent virus whereas slow bee paralysis virus (SBPV) also replicated fast but induced no mortality after injection; furthermore, both IAPV and SBPV infections could induce the expression of dicer-2, while IAPV, but not SBPV, infection triggered the production of predominantly 22 nt-long virus-derived siRNAs2.
As another important sub-pathway of RNAi in insects, the miRNA pathway has been documented to be involved in different aspects of development, such as formation of germ cells, wing and muscle, neurogenesis, apoptosis, and phenotypic plasticity3, while this pathway is also well established to be involved in host-pathogen interactions3,4. The canonical biogenesis of miRNAs initiates in the nucleus where monocistronic, bicistronic or polycistronic transcripts are produced. These contain stem-loop structures known as the primary miRNA (pri-miRNA). The pri-miRNA is cleaved by Drosha and Pasha to liberate the precursor miRNA (pre-miRNA). After exportation to the cytoplasm, the hairpin head of the pre-miRNA stem-loop is cut by Dicer-1 to yield a miRNA duplex. The miRNA duplex is then loaded into RNA induced silencing complex (RISC), which typically includes Argonaut 1 (Ago-1), resulting in the degradation of one of the strands. Then, the mature miRNA binds to the target mRNA and leads to mRNA degradation or translational repression5. Besides, there are also non-canonical pathways of miRNA biogenesis, which are Drosha-independent but can be Dicer-dependent or Dicer-independent6. The production and regulatory effects of miRNAs on insect-virus interactions could be complex. The first layer of complexity relates to the origin of miRNAs, which could be derived from the host or the virus. The second layer of complexity arises from the two-way interplay, meaning host-encoded miRNAs can target genes from both the host and the virus, and vice versa for virus encoded miRNAs7.
Differential expression of miRNAs has been associated with honeybee development and social behaviors8,9,10,11. Recently, with the genome sequencing of two bumblebee species, Bombus terrestris and B. impatiens, two datasets of miRNAs have been annotated for both species12. However, the regulatory effect of the miRNA pathway on bee-virus interaction is still unknown. Thus, in the current report, we comparatively analyzed the involvement of bumblebee miRNAs upon infections of an avirulent virus SBPV and a virulent virus IAPV. First, we analyzed the expression of the core genes (dicer-1 and ago-1) of the miRNA pathway upon virus infection. Secondly, through small RNA sequencing, we analyzed the miRNA profiles following viral infections. To have a further insight into miRNA-mRNA interaction, we predicted the possible targets for these miRNAs. Finally, we silenced dicer-1 to analyze the outcome of SBPV infection. Our study showed different features of the miRNA pathway response between the infection by the avirulent and the virulent virus, which may be associated with the virulence of the bee viral infections. In addition, these initial data could be the starting point to understand the function of miRNAs in cell regulatory systems of bee-virus interactions.
Results and Discussion
Significant effects of SBPV and IAPV infections on the expressions of dicer-1 and ago-1
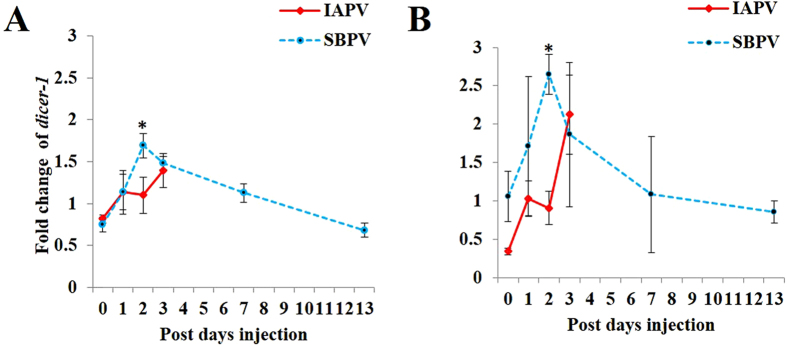
Aside from some non-canonical pathways of miRNA biogenesis, both Dicer-1 and Ago-1 are the core components of miRNA biogenesis and determine the pathway’s activity on gene regulation6. Here, we measured the expression of B. terrestris dicer-1 and ago-1 upon viral infections. In Niu et al.2, we showed that SBPV and IAPV replicated fast, but the two viruses differed largely in causing mortality in bees: SBPV as an avirulent virus and IAPV as an extremely virulent virus2. In the current study, we have separated PBS controls for the two viruses, thus we used independent t-test to separate means of the expressions of dicer-1 and ago-1. For IAPV (four time points) and SBPV (six time points) the Bonferroni corrected p values were 0.0125 and 0.0083, respectively. Our results showed that only a noticeable increase of both dicer-1 and ago-1 was observed upon IAPV at 3 days post infection (dpi) (Fig. 1), while both dicer-1 (t-test: t = 5.861, df = 8, p < 0.001) and ago-1 (t-test: t = 5.370, df = 4.578, p = 0.004) were significantly upregulated at 2 dpi of SBPV (Fig. 1 and Table S1).
Figure 1. Fold changes of two core gene expressions in the miRNA pathway upon IAPV and SBPV infection.
(A) Fold change of dicer-1 expression upon viral infections; (B) Fold change of ago-1 expression upon viral infections. The fold changes of gene expression were equal to the ratio of the relative expression of each gene in virus infected samples over the relative expression of this gene in control samples (PBS injected bees) (n = 4~5). The relative expression of each gene was calculated based on internal reference gene PPIA. The error bar represented the standard error of mean. The level of significance was calculated by t-test, and p < 0.0125 for the group of IAPV and p < 0.0083 for the group of SBPV was labeled with asterisks (*), respectively.
Small RNA sequencing reveals differentially expressed miRNAs upon SBPV and IAPV infections
Above we presented indications that the expression of core components of the miRNA pathway could be altered upon viral infections in B. terrestris, especially after SBPV infection, which implied the potential involvement of the miRNA pathway in host-virus interaction. To explore this possibility further, we used small RNA sequencing to find out which miRNAs could be influenced. Currently, there are 217 miRNAs described for the honeybee Apis mellifera in miRBase, while in the two bumblebee species, B. terrestris and B. impatiens, 130 and 115 miRNAs have been identified, respectively12. Our results revealed that 17 and 12 host miRNAs were differentially expressed upon infection with SBPV and IAPV, respectively (Table 1, Table S2 and S3). From the 17 miRNAs differentially expressed in bumblebees infected with SBPV, 10 miRNAs were upregulated and 7 were downregulated. In bumblebees infected with IAPV, 12 miRNAs were differentially expressed, which included 7 upregulated and 5 downregulated miRNAs. Among all these differentially expressed miRNAs, 3 miRNAs (bte-miR-277, bte-bantam, and bte-miR-263a) were upregulated and 3 (bte-miR-3759, bte-miR-11, and bte-miR-24) were downregulated in both viral infections.
Table 1. Differentially expressed miRNAs upon virus infection.
| miRNA name | fold change | Targets of homologous miRNA in insects and its relative prediction in Bombus terrestris | ||
|---|---|---|---|---|
| SBPV/PBS | IAPV/PBS | Homologous miRNA functions (target) in insects | Prediction of homologous targets or related targets in Bombus terrestris | |
| bte-miR-13b | 2.26 | Juvenile hormone signaling pathway18 | insulin-like growth factor 2 mRNA-binding protein 1-like (LOC100647990); ecdysone-induced protein 75B, isoforms C/D-like, (LOC100644185) | |
| bte-miR-927a | 1.75 | |||
| bte-miR-277 | 1.60 | 1.45 | Branched-chain amino acid catabolism33 Neurodegeneration34 | insulin receptor substrate 1-like (LOC100644779); insulin-degrading enzyme-like (LOC100644699) |
| bte-miR-263b | 1.54 | Apoptosis14 | apoptosis-inducing factor 1, mitochondrial-like (LOC100651200); cell division cycle and apoptosis regulator protein 1-like (LOC100651179); apoptosis 2 inhibitor-like (LOC100647685); apoptosis regulator R1-like (LOC100649633); PRKC apoptosis WT1 regulator protein-like, transcript variant 1 (LOC100649375) | |
| bte-bantam | 1.49 | 1.28 | Apoptosis13 Stem cell regulation35; Cell proliferation36 Tumor suppressor37 | apoptosis 2 inhibitor-like (LOC100650673); SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1-like (LOC100645044); disks large 1 tumor suppressor protein-like (LOC100643982); fat-like cadherin-related tumor suppressor homolog (LOC100650960); cadherin-related tumor suppressor-like (LOC100651028) |
| bte-miR-9a | 1.36 | Wing development38 Transcription factor of senseless39,40; | neurogenic locus Notch protein-like (LOC100645932); strawberry notch-like (LOC100649094) | |
| bte-miR-31a | 1.32 | |||
| bte-miR-263a | 1.26 | 1.42 | Apoptosis14 | apoptosis-inducing factor 1, mitochondrial-like (LOC100651200); cell division cycle and apoptosis regulator protein 1-like (LOC100651179); apoptosis-inducing factor 3-like, transcript variant 1 (LOC100649657) |
| bte-miR-10 | 1.26 | |||
| bte-miR-750 | 1.22 | |||
| bte-miR-305 | 1.30 | Intestinal stem cells20 | insulin-like growth factor-binding protein complex acid labile subunit-like (LOC100651887); insulin-degrading enzyme-like (LOC100644699); insulin-like peptide receptor-like (LOC100645413); insulin-like receptor-like (LOC100650952); insulin-like growth factor-binding protein complex acid labile subunit-like (LOC100643961) | |
| bte-miR-13a | 1.30 | juvenile hormone signaling pathway18 | insulin-like growth factor-binding protein complex acid labile subunit-like (LOC100647673); insulin-like receptor-like (LOC100651348); ecdysone-induced protein 75B, isoforms C/D-like (LOC100644185) | |
| bte-miR-278 | 1.28 | Energy homeostasis16 P450s 17 | insulin-like peptide receptor-like (LOC100645413); insulin gene enhancer protein ISL-1-like (LOC100644486); cytochrome P450 18a1-like (LOC100642897); cytochrome P450 49a1-like (LOC100646342); cytochrome P450 4C1-like (LOC100651255); cytochrome P450 4g15-like (LOC100652170); cytochrome P450 6a13-like (LOC100646677); cytochrome P450 6a2-like (LOC100646434), (LOC100647785), (LOC100651291), (LOC100647041); cytochrome P450 6k1-like (LOC100642816), (LOC100642936), (LOC100643678), (LOC100647803), (LOC100648391), (LOC100648995), (LOC100650427); cytochrome P450 9e2-like (LOC100647566), (LOC100648545), (LOC100649871), (LOC100649988) | |
| bte-miR-375 | 1.22 | |||
| bte-miR-252a | 0.78 | Dengue virus replication21 | 3′UTR of SBPV | |
| bte-miR-316 | 0.72 | |||
| bte-miR-276 | 0.67 | Olfactory41 | putative odorant receptor 13a-like (LOC100647497); odorant receptor Or2-like (LOC100644343); putative odorant receptor 82a-like (LOC100646456); odorant receptor 47b-like (LOC100646577); putative odorant receptor 13a-like (LOC100646817); putative odorant receptor 63a-like (LOC100644217) | |
| bte-miR-3718a | 0.66 | |||
| bte-miR-3759 | 0.57 | 0.76 | ||
| bte-miR-11 | 0.56 | 0.61 | Apoptosis15 | DNA damage-binding protein 1-like (LOC100650523) |
| bte-mc-24 | 0.44 | 0.68 | ||
| bte-mc-753 | 0.75 | |||
| bte-miR-283 | 0.71 | |||
Table 1 summarizes the potential functions of the differentially abundant miRNAs based on the function of homologous miRNAs demonstrated in others insects. Two reappearing functions (apoptosis and energy homeostasis), known to be regulated by miRNAs, struck our attention as they have also been described to be important factors in virus-host interactions.
Apoptosis
Homologous miRNAs of bte-miR-263a, bte-bantam, and bte-miR-11, three commonly differentially regulated miRNAs during SBPV and IAPV infections, have been shown to be involved in apoptosis13,14,15. In addition, a homolog of bte-miR-263b, which is upregulated by SBPV infection but not by the infection of IAPV, has been linked with apoptosis14.
Energy homeostasis (insulin-associated)
A homolog of bte-miR-278, upregulated by IAPV infection, is involved in energy homeostasis resulting in the regulation of levels of insulin16 and regulation of the detoxification enzyme P450s17. Homologs of bte-miR-13a and -13b upregulated by IAPV and SBPV, respectively, regulate the expression of juvenile hormone (JH)18. JH plays an import role in regulation of insect growth and molting, which are also associated with ecdysone and insulin signaling pathways19. The homolog of another miRNA regulated by IAPV infection, bte-miR-305, was shown to regulate Notch and insulin pathways in the intestinal stem cells of the Drosophila melanogaster gut20.
Validation of reference miRNAs upon different time points of viral infections by RT-qPCR
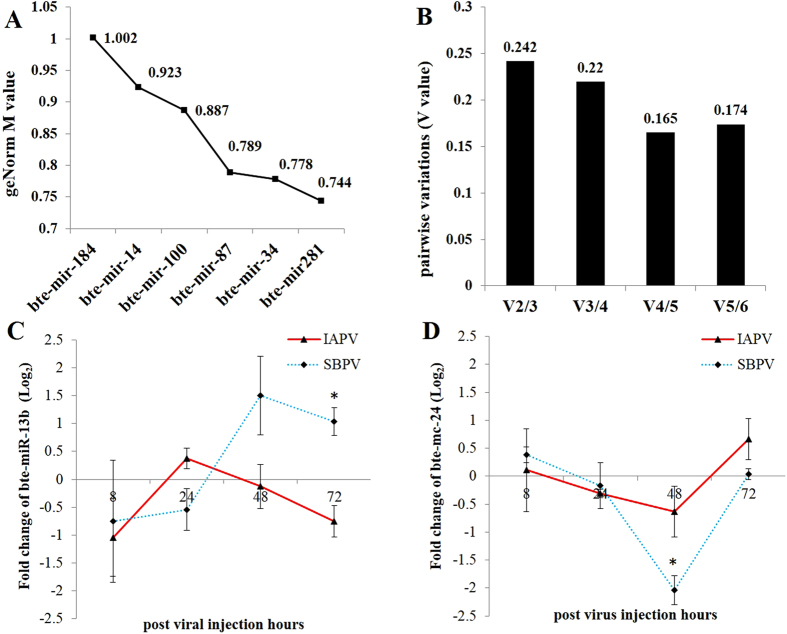
Until now, there have not been stable reference miRNAs known to normalize the data upon viral infections and non-infection in bumblebees. With the pre-selection criteria, based on primers efficiency and specificity (Table S4 and Figure S1), we eventually validated six miRNAs, including bte-miR-14, bte-miR-100, bte-miR-34, bte-miR-87, bte-miR-184 and bte-miR-281, in the same samples used to validate several key differentially expressed miRNAs indicated by small RNA sequencing. By geNorm, the results revealed that all these candidate reference miRNAs showed an M value of less than 1 except bte-miR-184, which showed an M value of 1.002 (Fig. 2A). Pairwise variations indicated the lowest V value of 0.165 (V4/5) upon the combination of the four most stable reference miRNAs (Fig. 2B). These results suggested the optimal references within these candidate reference miRNAs were the combination of four miRNAs, bte-miR-281 + bte-miR-34 + bte-miR-87 + bte-miR-100, for data analysis of miRNA expressions upon different injection time points of the two viruses.
Figure 2. Validation of reference miRNAs upon different time points of IAPV and SBPV infections by RT-qPCR.
(A) M value of the candidate reference miRNAs analyzed in samples of different time points of IAPV and SBPV infections by geNorm. (B) Pairwise variations (V value) for the combination of the candidate reference miRNAs by geNorm. (C,D) The fold changes of bte-miR-13b and bte-miR-24 expressions were calculated by the ratio of the relative expression of miRNA in virus infected samples over the relative expression of this miRNA in control samples (PBS injected bees) (n = 3). The relative expression of miRNA was calculated based on the combination of four optimal reference miRNAs: bte-miR-281 + bte-miR-34 + bte-miR-87 + bte-miR-100. The error bar represents the standard error of mean. The level of significance was calculated by t-test and p < 0.0125 was labeled with asterisks (*).
To test the optimal reference miRNAs, we further evaluated the abundance of bte-miR-13b and bte-mc-24, the most potential miRNAs indicated by sequencing data to be involved in virus-host interaction, upon different time points after injection of the two viruses by RT-qPCR. The relative expressions of the miRNAs were normalized by four miRNAs selected above (miR-281 + bte-miR-34 + bte-miR-87 + bte-miR-100). At each time point, we separated PBS controls for two viruses, thus we used independent t-test to separate means but with Bonferroni correction as four comparisons for each virus were conducted, i.e. at four time points between virus injections and PBS injections. Therefore, the strict p value of 0.0125 was used to judge the statistical difference of miRNAs expressions. The fold change (log2 transformed) of miRNAs was calculated as the division of miRNAs expression in virus-injected samples by the very time points of PBS injected samples. The results showed that bte-miR-13b was upregulated at 48 and 72 h post-injection of SBPV, but only with significant difference at 72 h (p = 0.010) (Fig. 2C). The significant downregulation of bte-mc-24 (p < 0.001) was detected at 48 h post-injection of SBPV (Fig. 2D). These results were consistent with sequencing data and thereby these evaluated reference miRNAs could be used for further miRNA analysis upon virus infections.
In silico target prediction of differentially expressed miRNAs shows a possible host-virus interaction network mediated by miRNAs
The current understanding is that one miRNA may target hundreds of genes, while a gene may be regulated by multiple miRNAs. Based on RNA22, we identified a total of 7465 genes possibly targeted by 130 miRNAs in B. terrestris. Among them, 7216 genes were possibly targeted by 23 differentially expressed miRNAs upon virus infection. Some predicted targets of these 23 miRNAs could be visualized in the GO and KEGG enrichments by ClueGO in two groups: 17 miRNAs differentially expressed by SBPV infection, and 12 miRNAs differentially expressed by IAPV infection. The analysis showed that there were 32 and 21 groups of GO/KEGG pathways in the network of SBPV (Figure S2) and IAPV (Figure S3). A number of 11 GO/KEGG pathways were common in the two datasets, which is consistent with the 6 common miRNAs influenced by SBPV and IAPV infections. Generally, these pathways can be associated with certain level of molecular activities in the view of virus-host interaction, such as DNA/RNA, protein, metabolism, cell activity, host disease, and host antiviral immunity (Table 2). These visualizations of miRNA-mRNA-GO/KEGG enrichments may show a possible host-virus interaction network regulated by virus-influenced miRNAs. Moreover, these potential targets reported in the homologous miRNAs, some of them also show an overlap with our predictions (Table 1). A future direction of research could be the role of apoptosis and insulin associated energy homeostasis in host-virus interaction based on miRNAs.
Table 2. The most significant enriched GO/KEGG from the targets of differentially expressed miRNAs upon virus infection.
| Diff. exp. miRNAs upon SBPV infection | Diff. exp. miRNAs upon IAPV infection | ||
|---|---|---|---|
| GO/KEGG ID | name | GO/KEGG ID | name |
| KEGG:00250 | Alanine, aspartate and glutamate metabolism | KEGG:00250 | Alanine, aspartate and glutamate metabolism |
| GO:0005524 | ATP binding | GO:0005524 | ATP binding |
| KEGG:04360 | Axon guidance | KEGG:04360 | Axon guidance |
| GO:0005975 | carbohydrate metabolic process | GO:0005975 | carbohydrate metabolic process |
| GO:0003924 | GTPase activity | GO:0003924 | GTPase activity |
| KEGG:05410 | Hypertrophic cardiomyopathy (HCM) | KEGG:05410 | Hypertrophic cardiomyopathy (HCM) |
| GO:0048519 | negative regulation of biological process | GO:0048519 | negative regulation of biological process |
| GO:0000166 | nucleotide binding | GO:0000166 | nucleotide binding |
| GO:0006082 | organic acid metabolic process | GO:0006082 | organic acid metabolic process |
| KEGG:03008 | Ribosome biogenesis in eukaryotes | KEGG:03008 | Ribosome biogenesis in eukaryotes |
| KEGG:05202 | Transcriptional misregulation in cancer | KEGG:05202 | Transcriptional misregulation in cancer |
| GO:0016887 | ATPase activity | GO:0006520 | cellular amino acid metabolic process |
| GO:0016830 | carbon-carbon lyase activity | GO:0005856 | cytoskeleton |
| KEGG:04110 | Cell cycle | GO:0003677 | DNA binding |
| KEGG:04713 | Circadian entrainment | GO:0006259 | DNA metabolic process |
| GO:0006732 | coenzyme metabolic process | GO:0004386 | helicase activity |
| GO:0005794 | Golgi apparatus | GO:0008237 | metallopeptidase activity |
| KEGG:04390 | Hippo signaling pathway | KEGG:04145 | Phagosome |
| GO:0016788 | hydrolase activity, acting on ester bonds | GO:0004672 | protein kinase activity |
| KEGG:04630 | Jak-STAT signaling pathway | GO:0038023 | signaling receptor activity |
| GO:0000278 | mitotic cell cycle | KEGG:04530 | Tight junction |
| GO:0002009 | morphogenesis of an epithelium | ||
| GO:0001882 | nucleoside binding | ||
| GO:0031090 | organelle membrane | ||
| GO:0006508 | proteolysis | ||
| KEGG:00230 | Purine metabolism | ||
| KEGG:04015 | Rap1 signaling pathway | ||
| GO:0010646 | regulation of cell communication | ||
| GO:0010033 | response to organic substance | ||
| KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | ||
| GO:0004888 | transmembrane signaling receptor activity | ||
| KEGG:05203 | Viral carcinogenesis | ||
Notes: pathways with bold are the same GO/KEGG appeared in both SBPV and IAPV dataset. These GO/KEGG could be associated with certain biological functions in the view of host-virus interaction. Network of SBPV influenced miRNAs-targets-GO/KEGG: In DNA/RNA associated GO/KEGG, (GO:0005524), (GO:0003924), (GO:0000166), (GO:0016887), (GO:0001882), (KEGG:00230). In protein associated GO/KEGG, (KEGG:00250), (KEGG:03008), (GO:0005794), (GO:0006508). In metabolism associated GO/KEGG, (GO:0005975), (GO:0006082), (GO:0016830), (GO:0006732). In cell activity associated GO/KEGG, (KEGG:04110), (GO:0000278), (KEGG:04015), (GO:0010646), (GO:0010033), (KEGG:04550), (GO:0004888). In host disease associated GO/KEGG, (KEGG:05410), (KEGG:05202), (KEGG:05203). In host antiviral immunity associated GO/KEGG, (KEGG:04390), (KEGG:04630). Network of IAPV influenced miRNAs-targets-GO/KEGG: In DNA/RNA associated GO/KEGG, (GO:0005524), (GO:0003924), (GO:0000166), (GO:0003677), (GO:0006259), (GO:0004386). In protein associated GO/KEGG, (KEGG:00250), (KEGG:03008), (GO:0006520), (GO:0008237), (GO:0004672). In metabolism associated GO/KEGG, (GO:0005975), (GO:0006082). In cell activity associated GO/KEGG, (GO:0005856), (GO:0038023), (KEGG:04530). In host disease associated GO/KEGG, (KEGG:05410), (KEGG:05202). In host antiviral immunity Phagosome (KEGG:04145).
In addition, we also explored if host miRNAs could directly target viral genomic RNAs using RNAhybrid and RNA22. The results identified a total of 24 target sites on SBPV and IAPV genomes by host miRNAs (Table 3). The predicted target sites were located in the UTRs as well as ORFs (open reading frame). Of these 24 target sites, 16 were predicted to be targeted by differentially expressed miRNAs upon virus infections. We then applied a Chi square test to analyze whether these predictions, mainly linked with differential miRNAs, acted just by chance. We found a significant difference between the number of differentially expressed bumblebee miRNAs by each virus in targeting its own viral genomic RNAs and all bumblebee miRNAs in targeting viral genomic RNAs for both SBPV (χ2 = 14.816, p < 0.001) and IAPV (χ2 = 13.422, p < 0.001) analyses. This could suggest that the direct host miRNA-virus interplay might also play a role in bumblebee-SBPV/IAPV interaction. Intriguingly, a homolog of bte-miR-252a downregulated by SBPV, was shown to have a direct interaction with dengue virus (DENV) via targeting the DENV E protein transmembrane region21. Through RNAhybrid and RNA22, we also predicted that bte-miR-252a could possibly target the 3′UTR of SBPV (Table 3).
Table 3. Predicted targets of host miRNAs in the viral genomes.
| Target | miRNA | MFE (Kcal/mol) | Binding Site Start Position | Target Gene Annotation |
|---|---|---|---|---|
| SBPV | bte-miR-276 | −21.8 | 7261 | Polyprotein_ORF |
| SBPV | bte-miR-276 | −20.7 | 2285 | Polyprotein_ORF |
| IAPV | bte-bantam | −25 | 4919 | Polymerase Polyprotein_ORF |
| IAPV | bte-mc-24 | −28.5 | 6219 | Polymerase Polyprotein_ORF |
| IAPV | bte-mc-24 | −28.1 | 6554 | UTR |
| IAPV | bte-mc-24 | −25 | 328 | 5′UTR |
| SBPV | bte-miR-252a | −29.6 | 9370 | 3′UTR |
| SBPV | bte-miR-9a | −25.1 | 3777 | Polyprotein_ORF |
| IAPV | bte-miR-11 | −21.8 | 7752 | Structural Polyprotein_ORF |
| IAPV | bte-miR-283 | −20.8 | 1145 | Polymerase Polyprotein_ORF |
| IAPV | bte-mc-24 | −22.5 | 9474 | 3′UTR |
| IAPV | bte-mc-753 | −22.4 | 7878 | Structural Polyprotein_ORF |
| SBPV | bte-miR-263b | −21.9 | 3705 | Polyprotein_ORF |
| SBPV | bte-miR-276 | −22.9 | 2500 | Polyprotein_ORF |
| SBPV | bte-miR-3759 | −23.3 | 7714 | Polyprotein_ORF |
| SBPV | bte-mc-24 | −20.1 | 9239 | 3′UTR |
| SBPV | bte-mc-753* | −26.3 | 7748 | Polyprotein_ORF |
| IAPV | bte-miR-263b* | −24.2 | 8501 | Structural Polyprotein_ORF |
| IAPV | bte-miR-9a* | −20.2 | 5318 | Polymerase Polyprotein_ORF |
| IAPV | bte-miR-252a* | −22.2 | 3819 | Polymerase Polyprotein_ORF |
| IAPV | bte-miR-252a* | −21.8 | 7091 | Structural Polyprotein_ORF |
| IAPV | bte-miR-263b* | −20.1 | 6255 | Polymerase Polyprotein_ORF |
| IAPV | bte-miR-750* | −21.9 | 1993 | Polymerase Polyprotein_ORF |
| SBPV | bte-miR-305* | −22.7 | 785 | Polyprotein_ORF |
*Not differentially expressed due to that particular viral infection.
Further analyses are needed to elucidate these direct miRNAs-virus interaction, which may provide us with an alternative strategy to use miRNAs to control the viruses or in a combination with the strategy of virus specific small interfering RNA (siRNA) and dsRNA through the siRNA pathway1.
Depletion of dicer-1 by RNAi does not lead to an altered genome copy number of SBPV
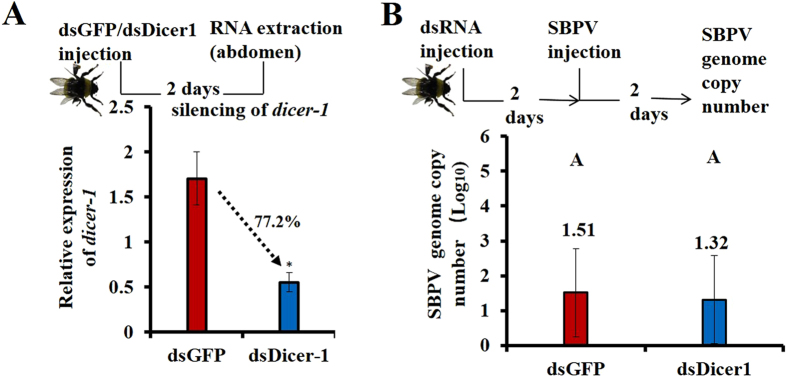
Although our results showed an altered expression of dicer-1 and ago-1 and differential abundance of miRNAs upon virus infections, the role of the miRNA pathway itself on the outcome of viral pathogenicity is not clear. Therefore, we used RNAi to silence dicer-1 to detect its influence on the outcome of SBPV genome copy number (gcn), which was the avirulent infection and significantly influenced the expression of dicer-1, ago-1 and miRNAs. Our result showed a ~77% depletion of dicer-1 transcripts after two days of injection of dsRNA (Fig. 3A). However, silencing of dicer-1 did not result in a significant difference in SBPV gcn (Fig. 3B). In our previous study, we also found that silencing of dicer-2 had no effect on IAPV and SBPV viral gcn, although silencing efficiency in dicer-2 depleted insects was reduced2. However, the answer to whether these results are associated with incomplete silencing of these key components from the RNAi pathway demands further study. In addition, it has been noted that various bumblebee tissues show different susceptibilities to viral infection22, but yet the virus-bumblebee interactions at the level of tissues rather than whole body or body parts awaits further studies. Recent studies showed that RNAi and JAK-STAT showed a cross-talk activity during viral infections23,24. In our previous study, we also detected the association between the RNAi and JAK/STAT pathways upon the infection of bee viruses in bumblebee25. Therefore, silencing of genes of the RNAi pathway could influence different (especially immune) pathways.
Figure 3. Silencing of dicer-1 and its effect on the genome copy number of SBPV.
(A) The injection of dsDicer-1 (n = 5) to target dicer-1 led to 77.2% reduction of its transcript levels compared with injection of the negative control dsGFP (n = 5). (B) Pre-silencing of dicer-1 led to no difference of genome copy number of SBPV (n = 12).
In conclusion, we found that an avirulent virus infection induced by SBPV and a virulent infection induced by IAPV in bumblebee B. terrestris could alter the miRNA pathway core genes’ expression and the abundance of miRNAs. It seems that based on the level of induction of dicer-1 and ago-1 and the number of differentially expressed miRNAs, the miRNA regulation was more disturbed or influenced after SBPV infection compared with IAPV infection. The potential target predictions and GO and KEGG enrichments were produced for initial visualization purposes and still lack any biological evidence, while it reflected the potential of bumblebee miRNAs that could be involved in various aspects of molecular activities in the view of virus-host interaction. In addition, some differentially expressed miRNAs upon virus infection may directly target the viral genome based on our prediction. Further, the selected reference miRNAs could be used for miRNA expression analysis through RT-qPCR. In summary, our study opens a new insight into bee-virus interactions meditated by the miRNA pathway. These initial data and observations could direct and facilitate our further studies to search a host miRNA-based strategy to tackle bee viruses.
Methods
Insects and virus inoculation
The colonies of B. terrestris were obtained from Biobest NV (Westerlo, Belgium). The bees used in this study were screened by RT-PCR (Table S5) to make sure that they were free of SBPV and IAPV. Newly emerged workers within 24 hours were genuinely collected from various colonies (within each experiment a maximum of 2 newly emerged workers were collected from a single colony, this was to ensure a random genetic background and to avoid overlap between treatment and genetic background) to keep the homogeneity of bees used in this study and kept in micro-colonies fed with pollen and sugar water ad libitum for further experiments. All the micro-colonies were maintained in an incubator (Panasonic, Japan) at 29–31 °C, 60–65% relative humidity, and continuous darkness26.
Virus inocula were produced as described previously26. The virus particles were counted by using transmission electron microscopy. To inoculate bees, we injected 20,000 and 20 particles of SBPV and IAPV into five to eight days old workers on the side of the soft white-like cuticle between the 1st and 2nd abdominal segments by a nanoinjector (Eppendorf, Germany) according to a previous study2. PBS (10 mM phosphate buffer pH 7.0/0.02% diethyl dithiocarbamate) injected bumblebees served as control, which was the same solution used to purify viral samples. Although both viruses replicate very fast, we define SBPV as an avirulent virus infection while IAPV represent a highly virulent infection as the injection of SBPV causes no mortality and IAPV causes extremely high mortality after 3 days and 100% mortality around 8 days2.
RNA isolation, cDNA synthesis, and qPCR
A total of 1.5~2 ml RLT buffer was initially used to homogenize the individual bumblebee by mortar and the supernatant was centrifuged three times to remove debris. Thereafter, the protocol of the RNeasy mini kit (Qiagen, Germany) was followed. The TURBO DNA-free™ kit (Ambion, USA) was used to remove the possible genomic DNA contamination in RNA samples. The quantity and quality of RNA samples were checked by Nanodrop and electrophoresis on 1.5% agarose gel. An amount of 2 μg total RNA was used to synthesize the cDNA by SuperScript® II Reverse Transcriptase (Invitrogen, USA) using oligo (dT) primers. To make sure there was no genomic DNA contamination, we checked cDNA samples by PCR with exon spanning primers for RPL23 (Table S5). The cDNA should produce an amplicon of 143 bp whereas the presence of genomic DNA will produce an extra amplicon of 452 bp. Next, qPCR was performed on a CFX96™ Real-Time PCR Detection with GoTaq® qPCR master (Promega, USA). Each reaction was performed in duplicate. The amplification specificities of primers used in this study were checked by both electrophoresis of the RT-PCR products and analysis of the dissociation curve by qPCR. A 10-fold serial dilution of cDNA was applied to calculate the amplification efficiency (Table S5). In addition, the RT-PCR products were sequenced in order to confirm their primers’ amplification specificities.
Core gene expression of the miRNA pathway
We collected RNA samples at 8 h, and 1, 2, 3, 7 and 13 dpi with SBPV and 8 h, and 1, 2, and 3 dpi with IAPV, to analyze gene expressions (n = 4~5). These time points were ascertained from previous study in consideration of the viral replication and viral caused mortality2. For each bumblebee individual, the whole body of each individual bee was used to extract RNA. PBS injected bees were also collected at all the different time points to serve as non-infected controls. Four to five bumblebee individuals were included in each time point for virus and PBS. The relative expression of key components, dicer-1 and ago-1, of the miRNA pathway were normalized by internal control peptidylprolyl isomerase A (PPIA)26. The fold change of gene expression at each time point was given as the ratio of the relative gene expressions of the virus treated samples over the PBS controls collected at the same time point.
Small RNA sequencing and target prediction of miRNAs
Small RNA sequencing was performed on RNA samples of SBPV and IAPV at 2 dpi, representing a fast replicating stage of IAPV and SBPV2. The PBS injected bees were included as control. For each bumblebee individual, the whole body of each individual bee was used to extract RNA. Four bumblebee individuals for each treatment and control were sequenced by the NXTGNT sequencing platform from the Ghent University. In detail, concentration and quality of the total extracted RNA was checked using the Quant-iTTM RiboGreen® RNA assay kit (Invitrogen, USA) and the RNA 6000 pico chip (Agilent Technologies, USA). Subsequently, 1 μg of total RNA was used to start the library preparation using the TailorMix miRNA Sample Preparation Kit v7 (SeqMatic, USA). Library preparation was carried out according to the manufacturer’s instructions. Then tRNA was added as carrier to minimize the loss of RNA via tube interaction. Libraries were quantified by qPCR, according to Illumina’s protocol ‘Sequencing Library qPCR Quantification protocol guide’ (version February 2011). A high sensitivity DNA chip (Agilent Technologies, USA) was used to check the libraries’ size distribution and quality. Single-end index 50 bp sequencing was performed on an Illumina MiSeq sequencer by loading 7 pM of each sample on the flowcell. A 10% PhiX spike-in was added as control. An average sequencing depth of 1.2 million reads in each sample was performed.
Ambiguous and low quality bases and adaptor sequences were trimmed from the sequencing reads using CLC Genomics Workbench 7.0.2. No ambiguous bases were allowed and a quality setting of 0.05 was applied. Reads smaller than 15 bp after trimming or reads containing more than 10% of bases with Phred quality score lower than 20 were filtered with CLC Genomics Workbench 7.0.2 and fastX-toolkit 0.0.13.1, respectively. The miRNA reads were counted by CLC Genomics Workbench 7.0.2 based on annotated bumblebee miRNA dataset12. Differential expression analysis between virus-infected and non-infected bees was performed in the R Bioconductor-package limma on quantile normalized data. The results were corrected by multiple testing using a Benjamini-Hochberg False Discovery Ratio at a cut-off value of 0.05. To select differentially expressed miRNAs, we used a strict criteria based on miRNA counts (greater than 100), fold change (larger than 20% difference), and adjusted p value (less than 0.01).
In this study, we screened the B. terrestris annotated mRNAs to identify potential miRNA binding sites using the core algorithm of improved version RNA22 v227. This tool is publicly available from Computational Medicine Centre of Thomas Jefferson University (https://cm.jefferson.edu/rna22). RNA22 is a pattern based target prediction program which first searches for reverse complementary sites of patterns within given mRNA sequences and identifies the hot spots. In the next step, the algorithm searches for miRNAs that are likely to bind to these sites. We allowed maximum one mismatch in the seed region and minimum 12 nt matches in the entire binding site. We set the sensitivity and specificity thresholds at 63% and 61%, respectively.
To further view the potential targets of miRNAs with differential expressions upon viral infections, we built a network of enrichment of Gene Ontology (GO) terms or Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways by ClueGO28. Briefly, ClueGO is a Cytoscape plug-in App to facilitate the visualization of functionally grouped terms in the form of networks and charts. We used a maximum of 25 predicted targets per miRNAs (folding energy cut-off: −20 Kcal/mol) as input. The GO term retrieved from Uniprot (ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/UNIPROT/) and KEGG annotation (http://www.genome.jp/kegg/kegg2.html) of B. terrestris were input with ClueGO main id, Entrez gene ID. The default parameters of ClueGO 2.1.7 were used for network construction. In details, the GO term level was defined with minimum 3 and maximum 8; in each term cluster, it required a minimum of 3 genes with a minimum of 4.0% clustered genes over the total genes in the term. The enrichment of predicted target genes of miRNAs to GO term/KEGG pathway was tested based on the hypergeometric distribution. A further multiple testing was followed by Bonferroni step-down. Finally, ClueGO created a binary gene-term matrix with the selected terms and their associated genes. Based on this matrix, a term-term similarity matrix was calculated using chance corrected kappa statistics to determine the association strength between the terms. In the current analysis, we used a cut-off value 4.0 for kappa score, to represent the most significant (leading) terms in associated functional groups, which could be visualized in the network.
In addition, we also predicted the potential targets of B. terrestris miRNAs in SBPV and IAPV genomic RNA by RNA2227 and RNAhybrid29, which use different prediction assumptions and thus made our prediction more reliable. RNAhybrid is a tool for finding the normalized minimum free energy of hybridization for miRNA and their mRNA target genes. The small RNA sequence is hybridized to the best fitting part of the mRNA. We did not allow G:U pairing in the seed region (nucleotide 2–8) and forced miRNA-target duplexes to have a helix in this region. Maximum 5 nt were approved as unpaired nucleotides in either side of an internal loop. The outcomes of the two tools were merged, and miRNA binding sites that were predicted by both tools were considered highly confident.
Validation and expression analysis of miRNAs by RT-qPCR
The same samples collected at 8, 24, 48, 72 h (n = 3) for both virus injections and PBS controls were used to further analyze the expressions of miRNAs. An amount of 2 μg total RNA was used to prepare cDNA by miScript® II RT kit (QIAGEN, Germany). To quantify mature miRNAs in the study, the 5x miScript HiSpec buffer was used. The total volume of 20 μl reaction system was mixed by adding buffer, nucleic acids mix, reverse transcriptase mix, RNA template and RNase-free water, and then followed the protocol of the kit. For RT-qPCR, an volume of 20 μl reaction system was mixed by adding SYBR green PCR master mix, universal primer (provided by the kit), miRNA specific primer (Table S4), template cDNA prepared above and RNase free water, by following the protocol of miScript SYBR® Green PCR kit (QIAGEN, Germany). Each reaction contained two technical replicates. To check the efficiency and specificity of miRNA primers, we evaluated the miRNAs’ amplification efficiencies by using a 10-fold serial dilution of mixture cDNA and the melting curves in all tested cDNA samples.
The validation of references as the same principle of mRNA expressions through RT-qPCR, was also suggested before the detection of miRNAs’ expressions30,31. To select good candidate reference miRNAs, we selected those with high counts (>100) and low expression variabilities indicated by sequencing data. Some miRNAs with significant changes upon viral infections revealed by the small RNA sequencing (section 2.4) were chosen to analyze their expressions in samples described above. The analysis of stable reference miRNAs was calculated by plug-in software geNorm in qBasePLUS and the relative expressions of miRNAs were normalized by selected optimal reference miRNAs through qBasePLUS.
Silencing Dicer-1 and detection of its effect on viral genome copy number
The same protocol of RNAi to silence a specific gene was performed according to our previous study2. A fragment of dicer-1 was amplified by PCR with target gene sequence specific primers plus T7 promoters (Table S5). This partial DNA template was purified by E.Z.N.A.® Cycle-Pure Kit (Omega, USA), and was sequenced in order to confirm the identity of the amplification. Then, 1 μg of template was used to synthesize dsRNA according to the guideline of MEGAscript® RNAi Kit (Invitrogen, USA). The concentration and quality of dsRNA were verified by Nanodrop and electrophoresis on 1.5% agarose gel. A total of 20 μg (20 μl) of dsRNA was injected into five to eight days old workers, and the same dose of dsGFP served as negative control. We used RNAi approach to investigate whether silencing of dicer-1 influences the genome copy number (gcn) of SBPV. First we silenced dicer-1 by injection of dsDicer1 (dsGFP serves as a control). After two days, the second injections were performed to inoculate bees with SBPV. Subsequently, in each sample, the abdomen of individual bumblebees was used for RNA extractions. To only measure the dicer-1 silencing efficiency, five bumblebee individuals were used. For detecting the genome copy number of SBPV upon silencing of dicer-1, 12 bumblebee individuals were included. The whole experiment was repeated twice.
The measurement of SBPV gcn was based on a standard curve made from the Cq values (x) detected from a dilution of DNA templates of partial SBPV genome converting to corresponding gcn (y)2. In detail, a part of the SBPV genome (Table S5) was amplified and purified by E.Z.N.A.® Cycle-Pure Kit (Omega, USA). The partial sequence was confirmed by Sanger sequencing (LGC genomics, Germany). The concentration of purified templates was measured by Quant-iTTM PicoGreen® dsDNA assay kit (Invitrogen, USA). The concentration was converted to gcn per μl by the online tool (URL: http://cels.uri.edu/gsc/cndna.html; Accessed date: 20/June/2014). A 10 fold serial dilution of templates was made to obtain a standard curve for each virus by qPCR. The equation is y = -0.2926x + 9.4426 (R2 = 0.9996). The normalized gcn of each sample was represented by the ratio of the gcn calculated based on the standard curve and the normalization factor from the internal reference gene PPIA26 with the framework of qBase32.
Additional Information
How to cite this article: Niu, J. et al. Infections of virulent and avirulent viruses differentially influenced the expression of dicer-1, ago-1, and microRNAs in Bombus terrestris. Sci. Rep. 7, 45620; doi: 10.1038/srep45620 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
Jinzhi Niu is recipient of a doctoral grant from the China Scholarship Council (CSC) and China Postdoctoral Science Foundation Grant (2016M600715). The authors acknowledge support of the Fund for Scientific Research-Flanders (FWO-Vlaanderen, Belgium) and the Foundation Project of Southwest University (SWU114049). SA and KE were supported by a National Health and Medical Research Council (APP1062983) and Horticulture Innovation Australia (VG13111).
Footnotes
The authors declare no competing financial interests.
Author Contributions J.N. and I.M. conceived the experiments, J.N. conducted the experiments, D.D.C. and D.D. analyzed the differential expressions of microRNAs, K.E. and S.A. predicted the targets of microRNAs. J.N., I.M. and G.S. analyzed the results. All authors reviewed the manuscript.
References
- Niu J., Meeus I., Cappelle K., Piot N. & Smagghe G. The immune response of the small interfering RNA pathway in the defense against bee viruses. Curr. Opin. Insect Sci. 6, 22–27 (2014). [DOI] [PubMed] [Google Scholar]
- Niu J. et al. In vivo study of Dicer-2-mediated immune response of the small interfering RNA pathway upon systemic infections of virulent and avirulent viruses in Bombus terrestris. Insect Biochem. Mol. Biol. 70, 127–137 (2016). [DOI] [PubMed] [Google Scholar]
- Asgari S. MicroRNA functions in insects. Insect Biochem. Mol. Biol. 43, 388–397 (2013). [DOI] [PubMed] [Google Scholar]
- Hussain M. & Asgari S. MicroRNAs as mediators of insect host–pathogen interactions and immunity. J. Insect Physiol. 70, 151–158 (2014). [DOI] [PubMed] [Google Scholar]
- Lucas K. & Raikhel A. S. Insect MicroRNAs: Biogenesis, expression profiling and biological functions. Insect Biochem. Mol. Biol. 43, 24–38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.-S. & Lai Eric C. Alternative miRNA Biogenesis Pathways and the Interpretation of Core miRNA Pathway Mutants. Mol. Cell 43, 892–903 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari S. Regulatory role of cellular and viral microRNAs in insect–virus interactions. Curr. Opin. Insect Sci. 8, 104–110 (2015). [DOI] [PubMed] [Google Scholar]
- Weaver D. et al. Computational and transcriptional evidence for microRNAs in the honey bee genome. Genome Biol. 8, R97 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behura S. K. & Whitfield C. W. Correlated expression patterns of microRNA genes with age-dependent behavioural changes in honeybee. Insect Mol. Biol. 19, 431–439 (2010). [DOI] [PubMed] [Google Scholar]
- Greenberg J. K. et al. Behavioral plasticity in honey bees is associated with differences in brain microRNA transcriptome. Genes Brain Behav. 11, 660–670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F. et al. Next-generation small RNA sequencing for microRNAs profiling in Apis mellifera: comparison between nurses and foragers. Insect Mol. Biol. 21, 297–303 (2012). [DOI] [PubMed] [Google Scholar]
- Sadd B. et al. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 16, 76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D. R., Stark A., Russell R. B. & Cohen S. M. Bantam Encodes a Developmentally Regulated microRNA that Controls Cell Proliferation and Regulates the Proapoptotic Gene hid In Drosophila. Cell 113, 25–36 (2003). [DOI] [PubMed] [Google Scholar]
- Hilgers V., Bushati N. & Cohen S. M. Drosophila microRNAs 263a/b Confer Robustness during Development by Protecting Nascent Sense Organs from Apoptosis. PLoS Biol. 8, e1000396 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott M., Islam A., Lopez-Bigas N. & Frolov M. V. mir-11 limits the proapoptotic function of its host gene, dE2f1. Genes Dev. 25, 1820–1834 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman A. A. & Cohen S. M. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 20, 417–422 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z. et al. MiR-278-3p regulates pyrethroid resistance in Culex pipiens pallens. Parasitol. Res. 114, 699–706 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano J., Montañez R. & Belles X. MiR-2 family regulates insect metamorphosis by controlling the juvenile hormone signaling pathway. Proc. Natl. Acad. Sci. USA 112, 3740–3745 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth C. K. et al. Juvenile hormone regulates body size and perturbs insulin signaling in Drosophila. Proc. Natl. Acad. Sci. USA 111, 7018–7023 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foronda D., Weng R. F., Verma P., Chen Y. W. & Cohen S. M. Coordination of insulin and Notch pathway activities by microRNA miR-305 mediates adaptive homeostasis in the intestinal stem cells of the Drosophila gut. Genes Dev. 28, 2421–2431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H. et al. miR-252 of the Asian tiger mosquito Aedes albopictus regulates dengue virus replication by suppressing the expression of the dengue virus envelope protein. J. Med. Virol. 86, 1428–1436 (2014). [DOI] [PubMed] [Google Scholar]
- Wang H., Meeus I. & Smagghe G. Israeli acute paralysis virus-associated paralysis symptoms, viral tissue distribution and Dicer-2 induction in bumblebee workers (Bombus terrestris). J. Gen. Virol. 97, 1981–1989 (2016). [DOI] [PubMed] [Google Scholar]
- Paradkar P. N., Duchemin J.-B., Voysey R. & Walker P. J. Dicer-2-Dependent Activation of Culex Vago Occurs via the TRAF-Rel2 Signaling Pathway. PLoS Negl Trop Dis 8, e2823 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradkar P. N., Trinidad L., Voysey R., Duchemin J.-B. & Walker P. J. Secreted vago restricts west nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. USA 109, 18915–18920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J., Meeus I. & Smagghe G. Differential expression pattern of Vago in bumblebee (Bombus terrestris), induced by virulent and avirulent virus infections. Sci. Rep. 6, 34200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J., Cappelle K., de Miranda J. R., Smagghe G. & Meeus I. Analysis of reference gene stability after Israeli acute paralysis virus infection in bumblebees Bombus terrestris. J. Invertebr. Pathol. 115, 76–79 (2014). [DOI] [PubMed] [Google Scholar]
- Miranda K. C. et al. A Pattern-Based Method for the Identification of MicroRNA Binding Sites and Their Corresponding Heteroduplexes. Cell 126, 1203–1217 (2006). [DOI] [PubMed] [Google Scholar]
- Bindea G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J. & Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34, W451–W454 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagias K., Podolska A. & Pocock R. Reliable reference miRNAs for quantitative gene expression analysis of stress responses in Caenorhabditis elegans. BMC Genomics 15, 222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X. et al. Selection of suitable inner reference genes for normalisation of microRNA expression response to abiotic stresses by RT-qPCR in leaves, flowers and young stems of peach. Sci. Hortic. 165, 281–287 (2014). [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F. & Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8, R19 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esslinger S. M. et al. Drosophila miR-277 controls branched-chain amino acid catabolism and affects lifespan. RNA Biol. 10, 1042–1056 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H., Poidevin M., Li H., Chen D. & Jin P. MicroRNA-277 Modulates the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats. PLoS Genet. 8, e1002681 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. et al. The Bantam microRNA Is Associated with Drosophila Fragile X Mental Retardation Protein and Regulates the Fate of Germline Stem Cells. PLoS Genet. 5, e1000444 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becam I., Rafel N., Hong X., Cohen S. M. & Milán M. Notch-mediated repression of bantam miRNA contributes to boundary formation in the Drosophila wing. Development 138, 3781–3789 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang Y. & Lai Z.-C. Mob as tumor suppressor is regulated by bantam microRNA through a feedback loop for tissue growth control. Biochem. Biophys. Res. Commun. 439, 438–442 (2013). [DOI] [PubMed] [Google Scholar]
- Biryukova I., Asmar J., Abdesselem H. & Heitzler P. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Dev. Biol. 327, 487–496 (2009). [DOI] [PubMed] [Google Scholar]
- Cassidy Justin J. et al. miR-9a Minimizes the Phenotypic Impact of Genomic Diversity by Buffering a Transcription Factor. Cell 155, 1556–1567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang F., Lee J.-A. & Gao F.-B. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 20, 2793–2805 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H. et al. MicroRNA-276a Functions in Ellipsoid Body and Mushroom Body Neurons for Naive and Conditioned Olfactory Avoidance in Drosophila. J. Neurosci. 33, 5821–5833 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.