Abstract
Background
Porcine reproductive and respiratory syndrome virus (PRRSV) has caused several outbreaks in China since 2006. However, the genetic diversity of PRRSV in China has greatly increased by rapid evolution or recombination events. Modified live-attenuated vaccines are widely used to control this disease worldwide. Although the risk and inefficacy of the vaccine has been reported, the genetic diversity between epidemic field strains and vaccine strains in China has not been completely elucidated.
Methods
A total of 293 clinical samples were collected from 72 pig farms in 16 provinces of China in 2015 for PRRSV detection. A total of 28 infected samples collected from 24 pig farms in nine provinces were further selected for immunohistochemical analysis and whole genome sequencing of PRRSV. Phylogenetic analysis and recombination screening were performed with the full genome sequences of the 28 strains and other 623 reference sequences of PRRSV.
Results
Of 293 clinical samples, 117 (39.93%) were positive for PRRSV by RT-PCR. Phylogenetic results showed that the 28 strains were nested into sublineage 10.5 (classic highly pathogenic [HP]-PRRSV), sublineage 10.6 (HP-PRRSV-like strains and related recombinants), sublineage 10.7 (potential vaccine JXA1-R-like strains), and lineage 9 (NADC30-like strains and recombinants of NADC30-like strains), respectively, suggesting that multiple subgenotypes of PRRSV currently circulate in China. Recombination analyses showed that nine of 28 isolates and one isolate from other laboratory were potential complicated recombinants between the vaccine JXA1-R-like strains and predominant circulating strains.
Conclusions
These results indicated an increase in recombination rates of PRRSV under current vaccination pressure and a more pressing situation for PRRSV eradication and control in China.
Electronic supplementary material
The online version of this article (doi:10.1186/s12985-017-0735-3) contains supplementary material, which is available to authorized users.
Keywords: PRRSV, Vaccine JXA1-R, Predominant circulating strains, Recombination
Background
Porcine reproductive and respiratory syndrome (PRRS) is an economically critical swine disease worldwide that causes severe reproductive failure in sows, poor viability of piglets, and respiratory disease with secondary infection in growing pigs. The agent of PRRS, PRRS virus (PRRSV), belongs to the Arterivirus genus and has a single-stranded positive-sense RNA genome of approximately 15 kb, which is organized into nine overlapping open reading frames (ORFs) [1–4]: ORF1a, ORF1b, ORF2a, ORF2b, and ORFs 3–7. ORF1a and ORF1b, which comprise 80% of the genome, encode the viral nonstructural proteins involved in genome transcription and replication [5], whereas ORF2a, ORF2b, and ORFs 3–7 encode the viral structural proteins GP2, E, GP3, GP4, GP5, M, and N, respectively [6–8]. The two genotypes of PRRSV, the European type (Type 1) and North American type (Type 2), share only about 60% nucleotide similarities at the genomic level and are represented by Lelystad virus and VR-2332, respectively [9–11].
The North American type PRRSV has been a major viral genotype in China [12–14], but was not a major concern until 2006, when highly pathogenic PRRSV (HP-PRRSV), characterized by a 30-aa deletion in Nsp2, emerged [15]. Having evolved from a less-pathogenic variant in China, HP-PRRSV has widely spread and continues to cause high mortality and huge economic losses to the swine industry in China [15, 16]. In a second major outbreak in China and other Southeast Asian countries in 2009 and 2010, the HP-PRRSV strain was later shown to be a recombinant between two Chinese PRRSV strains [17, 18]. NADC30 was isolated in the US in 2008 and in China in 2013 [19]. Associated with the importation of NADC30-like strains or recombination events between NADC30-like and domestic strains in China, a third outbreak occurred from 2013 to 2015 [19–21].
Imported and domestic modified live-attenuated vaccines (MLVs) have been widely used to safeguard against PRRSVs since the outbreaks in China. However, evidence of the risk and inefficacy of MLVs to prevent the spread of the latest PRRSV strain has begun to accumulate [21–24]. In order to investigate the epidemiological and evolutionary characteristics of PRRSV in 2015, a total of 293 clinical samples were collected from pigs in 16 provinces in China and the full-length sequence of 28 PRRSV isolates were sequenced and analyzed.
Methods
Sample collection
A total of 293 clinical lung samples were collected from pigs from different swine herds that experienced high fever and reproductive and respiratory syndrome in 16 provinces of China in 2015. A portion of each sample was homogenized for RNA extraction and stored at −70 °C. Meanwhile, the clinical symptoms, morbidity, mortality, and changes in autopsy tissues of pigs were examined.
RNA extraction and reverse transcriptase polymerase chain reaction (RT-PCR)
Total RNA was extracted from tissue homogenates using TRIzol reagent (Life Technologies, New York, NY, USA). cDNA was constructed from 7 μL of RNA using M-MLV reverse transcriptase (Promega, Madison, WI, USA). Two microliters of cDNA was used as a template for subsequent PCR analysis. Specific PRRSV nucleotide sequences of 143 bp were amplified with the primers 5′-AGC TGT GCC ARA TGY TGG-3′ and 5′-GGT RAA GTG ATG YCT GAC-3′ targeting ORF7. The cycling conditions were 95 °C for 1 min, followed by 35 cycles at 95 °C for 30 s, 56 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 2 min.
Immunohistochemical (IHC) analysis
The presence of PRRSV antigens in lung tissues was examined by IHC analysis. Briefly, the lung tissues were fixed in 10% neutral buffered formalin for 48 h, sectioned, and stained with monoclonal antibodies specific for the N protein of PRRSV for detection of antigen-positive cells, as previously described [25].
Full-length sequencing of PRRSV
The 28 PRRSV infected samples (Table 1) were chosen for further whole genome sequencing. Viral RNA purification from lung tissues and cDNA synthesis were performed as described previously [26], and PCR amplification of 14 fragments covering the PRRSV genome was performed with primers described in a previous report with minor modifications [27] (Additional file 1: Table S1). The quality of the PCR products was confirmed by electrophoresis on a 1% agarose gel and then purified using the PureLink Quick Gel Extraction Kit (Life Technologies). Then, the PCR products were quantified, cloned into the pMD18-T vector (Takara Bio, Inc., Shiga, Japan), and sequenced with an automated genome sequencer (Genetic Analyzer 3730XL; Applied Biosystems, Foster City, CA, USA).
Table 1.
Information of the 28 PRRSV isolates in China
| No. | Isolate | Province | Accession no. | Clinical background | Herd size(sows) | Vaccination history of MLV | |
|---|---|---|---|---|---|---|---|
| Morbidity (%) | Mortality (%) | ||||||
| 1 | 15GD1 | Guangdong | KX815407 | 40.2 | 21.4 | 700 | No |
| 2 | 15GD2 | Guangdong | KX815408 | 63.6 | 62.9 | 3000 | JXA1-R,TJM-F92 |
| 3 | 15GD3 | Guangdong | KX815409 | ||||
| 4 | 15GD4 | Guangdong | KX815410 | 41.6 | 7.2 | 2400 | No |
| 5 | 15HEB1 | HeBei | KX815411 | 80.8 | 86.5 | 1400 | No |
| 6 | 15HEB3 | HeBei | KX815412 | 62.7 | 53.4 | 450 | JXA1-R |
| 7 | 15HEN1 | Henan | KX815413 | 71.7 | 72.5 | 1000 | No |
| 8 | 15HEN3 | Henan | KX815414 | 29.6 | 3.4 | 1300 | No |
| 9 | 15HEN4 | Henan | KX815415 | 33.2 | 7.6 | 420 | No |
| 10 | 15HUN1 | Hunan | KX815416 | 65.4 | 54.1 | 3400 | No |
| 11 | 15HUN2 | Hunan | KX815417 | ||||
| 12 | 15HUN3 | Hunan | KX815418 | 23 | 61.4 | 200 | R98 |
| 13 | 15JX1 | Jiangxi | KX815419 | 80 | 48.8 | 700 | No |
| 14 | 15JX2 | Jiangxi | KX815420 | 67.4 | 26.6 | 390 | No |
| 15 | 15JX3 | Jiangxi | KX815421 | 2.9 | 2.2 | 400 | No |
| 16 | 15JX4 | Jiangxi | KX815422 | 12.3 | 54.9 | 1500 | RespPRRS |
| 17 | 15LN1 | Liaoning | KX815423 | 51.1 | 23.8 | 320 | No |
| 18 | 15LN2 | Liaoning | KX815424 | 35.4 | 2.3 | 250 | No |
| 19 | 15LN3 | Liaoning | KX815425 | 94.7 | 87.1 | 600 | No |
| 20 | 15SC1 | Sichuan | KX815426 | 12.9 | 56.2 | 500 | JXA1-R |
| 21 | 15SC2 | Sichuan | KX815427 | 33.8 | 4.8 | 800 | No |
| 22 | 15SC3 | Sichuan | KX815428 | 75 | 90.8 | 1500 | No |
| 23 | 15SN1 | Shaanxi | KX815429 | 50.6 | 31.6 | 350 | TJM-F92 |
| 24 | 15SN2 | Shaanxi | KX815430 | ||||
| 25 | 15SN3 | Shaanxi | KX815431 | ||||
| 26 | 15ZJ1 | Zhejiang | KX815432 | 28.4 | 16 | 660 | CH-1R |
| 27 | 15ZJ2 | Zhejiang | KX815433 | 65 | 35.9 | 1400 | JXA1-R |
| 28 | 15ZJ3 | Zhejiang | KX815434 | 57.6 | 32.3 | 570 | No |
Phylogenetic analysis
A total of 651 genomic sequences, which included 28 sequences from the present study and all available sequences of type 2 PRRSVs (n = 623) retrieved from the GenBank database in March 2017 (https://www.ncbi.nlm.nih.gov/genbank/) were aligned using the MUSCLE multiple alignment program (v3.8.31) [28] under default settings with minor manual adjustments afterward. Phylogenetic trees were constructed using the neighbor-joining algorithm with the Kimura 2-parameter substitution model and bootstrap tests of 1000 replicates using the MEGA6 sequence alignment tool [29]. Interactive tree of life (http://itol.embl.de) [30] was employed to display and annotate the phylogenetic trees. The genetic distances of intra- and inter- clusters were computed using MEGA 6 with Kimura 2-parameter model and 1000 bootstrap replicates. Sub-data sets with a diversity level of >10% were further divided into smaller monophyletic clusters and sublineages within lineages were identified with a cut off of 1.4%.
Recombination analyses
To accurately identify potential recombinants, sequences of the 28 strains identified in this study were combined with up to 623 isolate sequences for recombination screening. Recombination events were only considered significant when supported by at least three of the following five methods: RDP [31], GENECONV [32], MAXCHI [33], CHIMAERA [34], and 3SEQ [35] implemented in RDP v4.16 [36]. In order to check the recombination signals and estimate the breakpoint locations, sequences of potential recombinants isolated in this study or similar sequences of the parental lineage and those retrieved from the GenBank database were chosen for similarity plot analysis implemented in SIMPLOT v3.5.1 [37], with a window of 500 bp and step size of 10 bp. Furthermore, recombination events were confirmed with a neighbor-joining phylogenetic tree.
Only the strains that fit all of the following three criteria, as modified from a previous report [23], were recognized as JXA1-R (JXA1 P80)-like strains: (1) nested in the phylogenetic tree within the same cluster as the vaccine strain JXA1-R, (2) sharing the highest sequence identity with the JXA1-R strain (>99%), and (3) presenting amino acids (aa) unique to JXA1-R strain derivatives.
Results
Detection of PRRSV in the clinical samples
A total of 293 clinical samples were collected from different pig farms in 16 provinces in China in 2015. Of these samples, 117 (39.93%) were positive for PRRSV detection by RT-PCR, which was similar to that report in 2010 (45.2%) [17], suggesting that PRRSV was still prevalent in China in 2015. Representative sick pigs exhibited high and continuous fever, labored breathing, blue ears, blue nose, erythematous blanching rashes, pimples in the ears or on the back, and loss of appetite (Fig. 1a, b). Pathological examination results showed hyperplasia, edema, and hemorrhage in the lungs (Fig. 1c), and severe hemorrhagic spots in the lymph nodes (Fig. 1d). IHC results indicated obvious PRRSV antigen in the lungs of diseased piglets (Fig. 1e), as compared with healthy piglets (Fig. 1f).
Fig. 1.
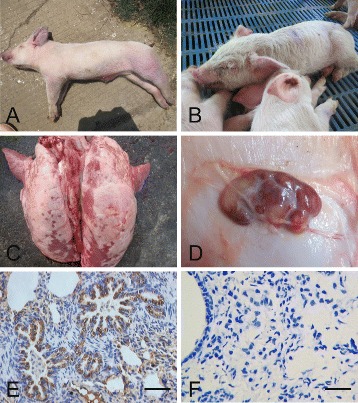
Clinical symptoms, gross lesions, and PRRSV detection by immunohistochemical analysis. The representative sick pigs exhibited blue ears, blue noses (a), and conjunctivitis (b); hyperplasia and edema in the lungs (c), and hemorrhagic spots in the lymph nodes (d). There were obvious PRRSV antigens in the lung tissue of diseased piglets (e) as compared to healthy piglets (f). Bar = 50 μm
The full length sequences of 28 PRRSV strains from 24 pig farms in nine provinces were amplified and sequenced for further analysis. Strain names, areas, accession numbers, and clinical backgrounds are summarized in Table 1.
Phylogenetic analysis
As shown in Fig. 2, the phylogenetic tree showed that the 28 isolates and another 623 isolates of PRRSV were clustered into 10 lineages, which generally have intralineage diversities of less than 10% (Table 2). The pairwise homology of the 28 isolates ranged from 76.3 to 99.9%, and the 28 isolates were divided into two lineages: 9 and 10 (Additional file 2: Table S2; Fig. 2). Meanwhile, the vaccine strain (ATP) and its parental strain (JA142) belonged to lineage 1. MLV RespPRRS/Repro and its parental, the prototype north American strains VR-2332, belonged to lineage 2. Some local endemic strains in Hong Kong and Taiwan, like isolates HK2, were clustered into lineage 3, and isolate CA-2, a virulent Korean strain, was clustered into lineage 4. MN184 variants, virulent MN414, and NADC31 were nested within lineage 5, 6, and 7, respectively. QYYZ-like strains and it recombinants isolated from southern China were clustered into lineage 8.
Fig. 2.
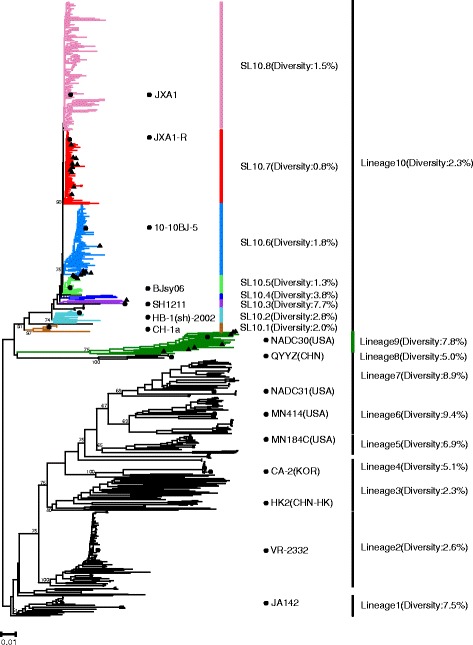
Phylogenetic tree of 28 strains isolated in this study and other 623 PRRSV strains. Lineage 9 is highlighted in green. Sublineages 10.1–10.8 are highlighted in sandy brown, light blue, purple, blue, light green, deep sky blue, red, and pink, respectively. The representative strains in lineages or sublineages are labeled with black circles (●) and the 28 strains isolated in this study are labeled with black triangles (▲). In sublineage 10.7, 10 isolates (15ZJ2, 15ZJ3, 15HUN3, 15LN2, 15JX2, 15JX3, 15JX4, 15HEN3, 15SC1, and 15SC2) from the present study were identified as potential vaccine JXA1-R-like strains with a 30-aa deletion in Nsp2
Table 2.
Pairwise interlineage genetic distance comparisons for complete genome sequences of type 2 PRRSVs
| % Difference for lineage | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| 1 | ||||||||||
| 2 | 10.8 | |||||||||
| 3 | 17.4 | 14.9 | ||||||||
| 4 | 18.3 | 15.4 | 19.7 | |||||||
| 5 | 17.7 | 16.0 | 19.5 | 16.8 | ||||||
| 6 | 19.1 | 17.9 | 20.9 | 20.6 | 15.2 | |||||
| 7 | 19.9 | 18.5 | 21.3 | 18.9 | 12.9 | 13.4 | ||||
| 8 | 15.8 | 14.1 | 18.7 | 20.8 | 20.5 | 21.9 | 21.9 | |||
| 9 | 18.0 | 16.9 | 20.8 | 20.5 | 16.2 | 13.2 | 16.6 | 21.1 | ||
| 10 | 10.6 | 11.8 | 18.3 | 18.8 | 18.7 | 20.1 | 20.3 | 13.9 | 18.5 | |
Seven strains (15LN1, 15ZJ1, 15HEN4, 15JX1, 15HEN1, 15SC3, and 15LN3) in this study were nested in lineage 9 together with NADC30-like strains and its recombinants. A total of 350 isolates mainly from China were clustered into lineage 10, which was divided into eight sublineages (10.1–10.8).
CH-1a (isolated in 1996) was located in sublineage 10.1 and HB-1(sh)/2002 (isolated in 2002) was nested in sublineage 10.2. 15HEB1 isolated in this study and recombinant SH1211 were nested within sublineage 10.3. The early HP-PRRSV isolates [15, 26] were clustered in sublineage 10.5 and 10.8. Four strains (15SN1, 15SN2, 15SN3, and 15GD4) isolated in this study and BJsy06 (isolated in 2006) were located in sublineage 10.5. HP-PRRSV JXA1, isolated in the first outbreak in 2006 [15], was clustered in subgroup 10.8. The isolates identified in this study (15GD2, 15GD3, 15HUN1, and 15HUN2) were potential recombinants associated with 2009–2010 HP-PRRSV-like strains and were located in sublineage 10.6. Sublineage 10.7 consisted of JXA1 derivatives by serial passaging in MARC-145 cells [38, 39] and potential vaccine JXA1-R (JXA1 P80)-like strains. In sublineage 10.7, 10 isolates (15ZJ2, 15ZJ3, 15HUN3, 15LN2, 15JX2, 15JX3, 15JX4, 15HEN3, 15SC1, and 15SC2) from the present study were identified as potential vaccine JXA1-R-like strains with a 30-aa deletion in Nsp2.
These results indicated that multiple subgenotypes of PRRSV co-existed in China in 2015, which included classic HP-PRRSV (sublineages 10.5 and 10.8), 2009–2010 HP-PRRSV-like strains and its recombinants (sublineage 10.6), potential vaccine JXA1-R-like strains (sublineage 10.7), and NADC30-like strains and its recombinants strains (lineage 9).
Distribution of JXA1-R vaccine-like strains in China
In sublineage 10.7 with vaccine strain JXA1-R as shown in Fig. 2, there were 44 JXA1-R-like strains, which included the 10 potential JXA1-R-like strains in this study and seven reported strains in China (NT1, NT2, NT3, 11NZ-GD, 11XX-GD, 11SH1-GD, and 11SH-GD) [23, 27]. These 27 Chinese strains were deposited in the GenBank database because they shared the highest nucleotide identities with JXA1-R strains (>99%) (Additional file 3: Table S3) and contained 6–11 amino acids unique to JXA1-R (Additional file 4: Figure S1). The geographical distribution results revealed that those potential JXA1-R-like strains were detected in all 16 provinces or autonomous cities (regions) with a wide geographical range in China (Fig. 3).
Fig. 3.
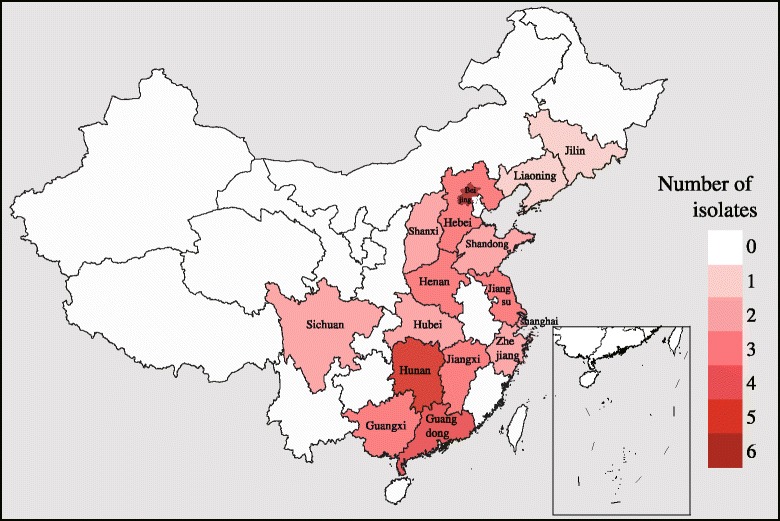
Distribution of 44 potential JXA1-R-like strains in China. Distribution if presented as the number of isolates per province or autonomous city (region), from most cases (6, dark red) to no case (0, white)
Recombination analyses
All recombination events in RDP were detected and confirmed by SIMPLOT and phylogenetic trees (Additional file 5: Figure S2). As shown in Fig. 4, nine of 28 isolates and one isolates from another laboratory were the result of recombination events between the JXA1-R and predominant circulating strains. Three kinds of recombination events were involved: inner-lineage, inter-lineage, and both inner-lineage and inter-lineage (Table 3). First, 15GD2, 15GD3, 15HUN1, 15HUN2, and 10FS1-GD were the products of inner-lineage recombination events between 2009 and 2010 HP-PRRSV-like strains (sublineage 10.6) and JXA1-R vaccine strains (sublineage 10.7). The genomes of 15GD2 and 15GD3 shared the same recombination pattern, as did the genomes of 15HUN1 and 15HUN2. Second, 15JX1, 15HEN1, and 15SC3 were inter-lineage recombination events between strains NADC30 (lineage 9) and JXA1-R. Third, 15HEB1, and 15LN3 were mosaic recombination events between strains NADC30, 2009–2010 HP-PRRSV, and JXA1-R. These results indicate that the recombination events in China were relatively complicated. Among them, Nsp2 or Nsp9 exhibited the complicated recombination events, involving eight breakpoints from six recombination events (Fig. 5).
Fig. 4.
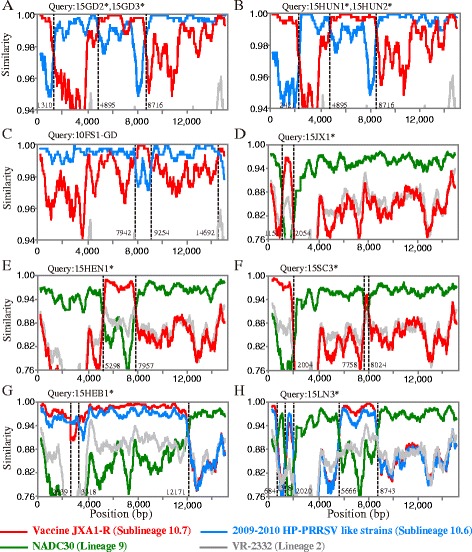
SimPlots for all putative recombinants analyzed in this study. Comparisons of genetic similarity between recombinant and parental strains were made using SimPlot. The results are shown for strains 15GD2*, 15GD3* (a), 15HUN1*, 15HUN2* (b), 10FS1-GD (c), 15JX1* (d), 15HEN1* (e), 15SC3* (f), 15HEB1* (g), and 15LN3* (h). Strains isolated in this study are marked with asterisks (*). The vertical axis represents the nucleotide sequence similarity between the putative recombinant strains, vaccine JXA1-R (sublineage 10.7, red), 2009-2010 HP-PRRSV-like strains (sublineage 10.6, deep sky blue), strain NADC30 (lineage 9, green), and strain VR-2332 (lineage 2, grey). The specific parental 2009-2010 HP-PRRSV strains of the recombinant strains are shown in Table 3. The horizontal axis shows the relative nucleotide position in reference to the full-length genome of VR-2332. Recombination breakpoints are shown as black dotted lines with the location indicated at the bottom
Table 3.
PRRSV recombination events documented in this study
| Recombination set | Year | Location | Parental strains | Breakpoint position (nt) |
|---|---|---|---|---|
| Set1 ( innerlineages ) | ||||
| ( SL10.7 ± 10.6 ) | ||||
| 15GD2*,15GD3* | 2015 | Guangdong | 15GD1 + JXA1-R | 1310, 4895, 8716 |
| 15HUN1*,15HUN2* | 2015 | Hunan | 15GD1 + JXA1-R | 2421, 4895, 8716 |
| 10FS1-GD | 2010 | Guangdong | 10HD-GD + JXA1-R | 7942, 9254, 14692 |
| Set2 ( interlineages ) | ||||
| ( L9 ± SL10.7 ) | ||||
| 15JX1* | 2015 | Jiangxi | NADC30 + JXA1-R | 1151, 2054 |
| 15HEN1* | 2015 | Henan | NADC30 + JXA1-R | 5298, 7957 |
| 15SC3* | 2015 | Sichuan | NADC30 + JXA1-R | 2004, 7758, 8024 |
| Set3 ( inner & interlineages ) | ||||
| ( L9 ± SL10.7 ± 10.6 ) | ||||
| 15HEB1* | 2015 | Hebei | NADC30 + JXA1-R + 10-10JX | 2639, 3318, 12171 |
| 15LN3* | 2015 | Liaoning | NADC30 + JXA1-R + 10-10JX | 684, 1318, 2020, 5666, 8743 |
*Strains isolated in this study; Breakpoint position in reference to VR-2332 (accession number U87392)
Fig. 5.
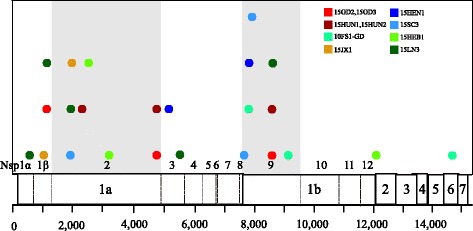
Recombination breakpoints identified across the full-length genome. The putative recombinants are listed in the key, with the colored lines matching each corresponding breakpoint. Most breakpoints were localized in Nsp2 and Nsp9, and the backgrounds of the two regions are highlighted in grey
Discussion
PRRSV is among the most economically important viruses in the swine industry worldwide [40, 41]. In 2006, a HP-PRRSV with a unique discontinuous 30-aa deletion in Nsp2 emerged and caused the most severe viral diseases in swine in China to date [15]. The genetic diversity of this HP-PRRSV has greatly increased by rapid evolution and recombination events [14, 17–19]. The current commercial PRRSV vaccines cannot provide complete protection against this disease [21]. In this study, 117 (39.93%) of 293 clinical samples were positive for PRRSV by RT-PCR, similar to that reported in 2010 (45.2%) [17]. Full-length sequences of 28 PRRSV isolates showed that they belonged to two lineages together with HP-PRRSV isolated in China. Among them, 10 JXA1-R-like isolates and 44 other Chinese strains were possibly derived from JXA1-R strains by recombination events of different strains. These findings indicated that the mosaic recombinant strains associated with the vaccine strains JXA1-R and HP-PRRSV were the predominant circulating strains of PRRSV in China.
The virulence reversion of the vaccine strain JXA1-R has been described [23, 27], but no recombination of this strain has yet been reported. Recombination analyses showed that 10 strains, which included nine strains from the present study and one isolate retrieved from the GenBank database, evolved through complicated recombination events between vaccine JXA1-R-like strains and predominant circulating strains in China. Phylogenetic analysis showed that the nine strains isolated in this study were positioned within major clusters with recombinants from other laboratories (Fig. 2). Furthermore, all breakpoints of the recombinants isolated in this study were not adjacent to the corresponding primer binding sites, suggesting that all of the recombinant strains isolated in this study are circulating recombinant strains rather than laboratory artifacts.
Most of the breakpoints of recombination events in this study were located in Nsp2 or Nsp9. Similarly, Liu et al. [11] conducted in vivo experiments to identify recombination events with Nsp2, ORF3, and ORF5, which demonstrated that Nsp2 was the most complicated region for recombination. Furthermore, all breakpoints of the recombinant strain Em2007 (lineage 10) were located in Nsp2 or Nsp9 [22], as were two of the three breakpoints of 2009–2010 HP-PRRSV [18]. Thus, it is possible that Nsp2 or Nsp9 is the hot region for breakpoint of recombination events.
Geographically, previous vaccine-like strains or recombinants related with vaccine in China were located in limited ranges and represented by single sequences. However, the results of the present study revealed that JXA1-R-like strains were detected in all 16 provinces or autonomous cities (regions) in China, denoting a wide geographical range. In 2010, only two JXA1-R-like strains (GX1002 or GX1003) were examined. Since then, the emergence of JXA1-R-like strains has greatly increased, as shown by the data presented in Table 4. These findings imply that HP-PRRSV vaccine-like strains were the predominant strains circulating in the field followed by the PRRSV vaccination.
Table 4.
Time of emergence of vaccine JXA1-R-like strains in China
| Isolation year | No. Isolation | Isolate | Reference |
|---|---|---|---|
| 2010 | 2 | GX1002, GX1003 | [45], GenBank |
| 2011 | 6 | GX1001, NVDC-HeB1-2011, 11NZ-GD, 11SH1-GD, 11SH-GD, 11XX-GD | [27, 46], GenBank |
| 2012 | 10 | NT1, NT2, NT3, JL-04/12, NVDC-BJ3-2012, NVDC-BJ4-2012, NVDC-BJ5-2012, NVDC-BJ6-2012, NVDC-BJ9-2012, NVDC-SD2-2012 | [23, 47], GenBank |
| 2013 | 7 | NVDC-SDXX-2013, NVDC-HBCZ-2013, NVDC-SXJC-2013, HNyc13, HEB-2013, HEB 20130008-14, NVDC-BJPG-2013 | GenBank |
| 2014 | 8 | HNxa14, HUN-2014, HB2014001, HENZK-1, NVDC-shh01-2014, NVDC-SHH02-2014, NVDC-HuNCS-2014, NVDC-13SXJC-2014 | GenBank |
| 2015 | 11 | HENPDS-2, 15ZJ2, 15ZJ3, 15HUN3, 15LN2, 15JX2, 15JX3, 15JX4, 15HEN3, 15SC1, 15SC2 | GenBank, this study |
Recombination events between field PRRSV strains could result in greater PRRSV pathogenicity, such as strain JL580 from the recombination between NADC30 and HP-PRRSV 09HEN1, which has been confirmed as highly pathogenic [19]. In this study, the PRRSV-positive piglets exhibited severe clinical symptoms and gross lesions typical of HP-PRRS. Morbidity (63.6%–94.7%) and mortality (48.8%–90.8%) rates were relatively high among piglets infected the recombinant strains (Table 1, Additional file 6: Figure S3), suggesting that the recombinant strains associated with JXA1-R might be highly pathogenic. Of course, the pathogenicity of these recombinant JXA1-R-like strains should be examined in future studies.
In order to control this disease, at least seven MLV PRRSV vaccine strains (JXA1-R, HuN4-F112, TJM-F92, GDr180, RespPRRS/Repro, CH-1R, and R98) have been widely used in China [38, 42, 43]. In order to determine the original of these isolates, we compared 28 sequences identified in this study with all available genomic sequences (n = 623) of type 2 PRRSV in the GenBank database. The sublineage 10.7 include up to 65 field samples with a wide geographical range, in which the intralineage diversity was the lowest (0.8%), and found nine JXA1-R-like isolates out of the 28 isolates based one the three criteria reported previously [23]. Of course, it was possible that these recombinants might be also derived from other HP-PPRSV vaccine strains, such as HuN4-F112, TJM-F92, and GDr180, because those vaccine strains are derived from HP-PRRSV and there is no published complete sequences of those vaccine strains. The improper use of vaccines may be associated with the emergence of mosaic recombinant strains [44]. Hence, immunization schedules should be optimized to prevent future outbreaks of this infectious disease.
Conclusion
The findings of this study demonstrated that HP-PRRSV vaccine like strains (JXA1-R-like strains) are widely circulated in China and mosaic recombinant strains associated with the vaccine strains and predominant circulating strains of PRRSV emerged in different provinces of China under current vaccination pressure. PRRSV eradication and control is a pressing situation in China.
Additional files
Primers used in generating overlapping amplicons spanning PRRSV genomes. (DOCX 13 kb)
Homology between the 28 isolates sequenced in this study. (DOCX 19 kb)
Sequence identity of the JXA1-R-like strains with JXA1 derivatives and representative strains. (DOCX 20 kb)
Comparisons of partial ORF1a, ORF1b, and GP4 deduced amino acid sequences. *Unique and identical amino acids among the JXA1 derivatives. (PDF 491 kb)
Phylogenetic analysis of parental regions of putative 10 recombinant strains. The parental vaccine JXA1-R group (sublineage 10.7) is shown in red, the 2009–2010 HP-PRRSV-like group (sublineage 10.6) is shown in deep sky blue, and the NADC30-like strain (lineage 9) is shown in green. Putative recombinant strains are labeled with black triangles (▲). (PDF 502 kb)
Morbidity and mortality rates among farms with pigs infected with recombinant JXA1-R-like strains. (PDF 87 kb)
Acknowledgements
This work was mainly supported by the National Natural Science Foundation (31230071,31672565), a grant from the Ministry of Agriculture (CARS-36) for development of techniques to control swine diseases, and the priority academic program development of Jiangsu higher education institutions (PAPD).
Authors’ contributions
HZ and PJ designed the study. HZ, QH, LZ, and ZZ performed the experiments. HZ, QH, HS, YW, and PJ analyzed the data. HZ and PJ wrote the manuscript. All authors have read and approved the submitted manuscript.
Competing interests
The authors declare that they have no competing interests.
Ethics approval
All animal protocols were approved by the Animal Care and Ethics Committee of Nanjing Agricultural University (permit number: IACECNAU 20121001) and followed the Guiding Principles for Biomedical Research Involving Animals.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- HP-PRRSV
Highly pathogenic porcine reproductive and respiratory syndrome virus
- IHC
Immunohistochemistry
- MLVs
Modified live-attenuated vaccines
- ORFs
Open reading frames
- PCR
Polymerase chain reaction
- PRRS
Porcine reproductive and respiratory syndrome
- PRRSV
Porcine reproductive and respiratory syndrome virus
- RT-PCR
Reverse transcription polymerase chain reaction
Contributor Information
Huajian Zhao, Email: zhjzhaohuajian@163.com.
Qinggong Han, Email: hanqinggong7606@163.com.
Lei Zhang, Email: 13673737467@163.com.
Zhiyong Zhang, Email: zhangzy1888@163.com.
Yufeng Wu, Email: yfwu@njau.edu.cn.
Hong Shen, Email: hongshenks@hotmail.com.
Ping Jiang, Phone: +86 25 84395504, Email: jiangp@njau.edu.cn.
References
- 1.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 2.Hanada K. The origin and evolution of porcine reproductive and respiratory syndrome viruses. Mol Biol Evol. 2005;22:1024–1031. doi: 10.1093/molbev/msi089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murtaugh MP, Stadejek T, Abrahante JE, Lam TTY, Leung FCC. The ever-expanding diversity of porcine reproductive and respiratory syndrome virus. Virus Res. 2010;154:18–30. doi: 10.1016/j.virusres.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Marthaler D, Rovira A, Rossow S, Murtaugh MP. Emergence of a virulent porcine reproductive and respiratory syndrome virus in vaccinated herds in the United States. Virus Res. 2015;210:34–41. doi: 10.1016/j.virusres.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Snijder EJ, van Tol H, Roos N, Pedersen KW. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J Gen Virol. 2001;82:985–994. doi: 10.1099/0022-1317-82-5-985. [DOI] [PubMed] [Google Scholar]
- 6.Bautista EM, Faaberg KS, Mickelson D, McGruder ED. Functional properties of the predicted helicase of porcine reproductive and respiratory syndrome virus. Virology. 2002;298:258–270. doi: 10.1006/viro.2002.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stadejek T, Stankevicius A, Storgaard T, Oleksiewicz MB, Belak S, Drew TW, Pejsak Z. Identification of radically different variants of porcine reproductive and respiratory syndrome virus in Eastern Europe: towards a common ancestor for European and American viruses. J Gen Virol. 2002;83:1861–1873. doi: 10.1099/0022-1317-83-8-1861. [DOI] [PubMed] [Google Scholar]
- 8.Wootton S, Yoo D, Rogan D. Full-length sequence of a Canadian porcine reproductive and respiratory syndrome virus (PRRSV) isolate. Arch Virol. 2000;145:2297–2323. doi: 10.1007/s007050070022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson EA, Christopher-Hennings J, Drew T, Wensvoort G, Collins JE, Benfield DA. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J Clin Microbiol. 1993;31:3184–3189. doi: 10.1128/jcm.31.12.3184-3189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng XJ, Paul PS, Halbur PG, Lum MA. Phylogenetic analyses of the putative M (ORF 6) and N (ORF 7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): implication for the existence of two genotypes of PRRSV in the U.S.A. and Europe. Arch Virol. 1995;140:745–755. doi: 10.1007/BF01309962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu D, Zhou R, Zhang J, Zhou L, Jiang Q, Guo X, Ge X, Yang H. Recombination analyses between two strains of porcine reproductive and respiratory syndrome virus in vivo. Virus Res. 2011;155:473–486. doi: 10.1016/j.virusres.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Liu T, Zhu C, Jin Y, Zhang Y. Genetic variation of Chinese PRRSV strains based on ORF5 sequence. Biochem Genet. 2006;44:421–431. doi: 10.1007/s10528-006-9039-9. [DOI] [PubMed] [Google Scholar]
- 13.Zhou YJ, Hao XF, Tian ZJ, Tong GZ, Yoo D, An TQ, Zhou T, Li GX, Qiu HJ, Wei TC, Yuan XF. Highly virulent porcine reproductive and respiratory syndrome virus emerged in China. Transbound Emerg Dis. 2008;55:152–164. doi: 10.1111/j.1865-1682.2008.01020.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Z, Ni J, Cao Z, Han X, Xia Y, Zi Z, Ning K, Liu Q, Cai L, Qiu P, et al. The epidemic status and genetic diversity of 14 highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) isolates from China in 2009. Vet Microbiol. 2011;150:257–269. doi: 10.1016/j.vetmic.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, et al. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One. 2007;2:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An TQ, Tian ZJ, Xiao Y, Li R, Peng JM, Wei TC, Zhang Y, Zhou YJ, Tong GZ. Origin of highly pathogenic porcine reproductive and respiratory syndrome virus, China. Emerg Infect Dis. 2010;16:365–367. doi: 10.3201/eid1602.090005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu X, Chen N, Wang L, Wu J, Zhou Z, Ni J, Li X, Zhai X, Shi J, Tian K. New genomic characteristics of highly pathogenic porcine reproductive and respiratory syndrome viruses do not lead to significant changes in pathogenicity. Vet Microbiol. 2012;158:291–299. doi: 10.1016/j.vetmic.2012.02.036. [DOI] [PubMed] [Google Scholar]
- 18.Shi M, Holmes EC, Brar MS, Leung FCC. Recombination is associated with an outbreak of novel highly pathogenic porcine reproductive and respiratory syndrome viruses in China. J Virol. 2013;87:10904–10907. doi: 10.1128/JVI.01270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao K, Ye C, Chang X, Jiang C, Wang S, Cai X, Tong G, Tian Z, Shi M, An T. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J Virol. 2015;89:10712–10716. doi: 10.1128/JVI.01446-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L, Wang Z, Ding Y, Ge X, Guo X, Yang H. NADC30-like strains of porcine reproductive and respiratory syndrome virus, China. Emerg Infect Dis. 2015;21:2256–2257. doi: 10.3201/eid2112.150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Zhuang J, Wang J, Han L, Sun Z, Xiao Y, Ji G, Li Y, Tan F, Li X, Tian K. Outbreak investigation of NADC30-like PRRSV in South-East China. Transbound Emerg Dis. 2016;63:474–479. doi: 10.1111/tbed.12530. [DOI] [PubMed] [Google Scholar]
- 22.Li B, Fang L, Xu Z, Liu S, Gao J, Jiang Y, Chen H, Xiao S. Recombination in vaccine and circulating strains of porcine reproductive and respiratory syndrome viruses. Emerg Infect Dis. 2009;15:2032–2035. doi: 10.3201/eid1512.090390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Xia T, Zhou Y, Yu L, Yang S, Huang Q, Li L, Gao F, Qu Z, Tong W, Tong G. Characterization of three porcine reproductive and respiratory syndrome virus isolates from a single swine farm bearing strong homology to a vaccine strains. Vet Microbiol. 2015;179:242–249. doi: 10.1016/j.vetmic.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 24.Lu WH, Tun HM, Sun BL, Mo J, Zhou QF, Deng YX, Xie QM, Bi YZ, Leung FC, Ma JY. Re-emerging of porcine respiratory and reproductive syndrome virus (lineage 3) and increased pathogenicity after genomic recombination with vaccine variant. Vet Microbiol. 2015;175:332–340. doi: 10.1016/j.vetmic.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 25.Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Andrews JJ, Lum MA, Rathje JA. Comparison of the antigen distribution of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol. 1996;33:159–170. doi: 10.1177/030098589603300205. [DOI] [PubMed] [Google Scholar]
- 26.Zhou L, Chen S, Zhang J, Zeng J, Guo X, Ge X, Zhang D, Yang H. Molecular variation analysis of porcine reproductive and respiratory syndrome virus in China. Virus Res. 2009;145:97–105. doi: 10.1016/j.virusres.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 27.Zhang M, Xie J, Sun L, Cao Z, Gu H, Deng S, Chen Y, Cao Z, Tang F, Su S, Zhang G. Phylogenetic analysis and molecular characteristics of 17 porcine reproductive and respiratory syndrome virus isolates in Southern China from 2010 to 2011. Microb Pathogenesis. 2013;65:67–72. doi: 10.1016/j.micpath.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 32.Padidam M, Sawyer S, Fauquet CM. Possible emergence of new geminiviruses by frequent recombination. Virology. 1999;265:218–225. [DOI] [PubMed]
- 33.Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 34.Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A. 2001;98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boni MF, Posada D, Feldman MW. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics. 2007;176:1035–1047. doi: 10.1534/genetics.106.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han W, Wu J, Deng X, Cao Z, Yu X, Wang C, Zhao T, Chen N, Hu H, Bin W, et al. Molecular mutations associated with the in vitro passage of virulent porcine reproductive and respiratory syndrome virus. Virus Genes. 2009;38:276–284. doi: 10.1007/s11262-008-0322-1. [DOI] [PubMed] [Google Scholar]
- 39.Yu X, Chen N, Deng X, Cao Z, Han W, Hu D, Wu J, Zhang S, Wang B, Gu X, Tian K. Genomic sequencing reveals mutations potentially related to the overattenuation of a highly pathogenic porcine reproductive and respiratory syndrome virus. Clin Vaccine Immunol. 2013;20:613–619. doi: 10.1128/CVI.00672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pejsak Z, Stadejek T, Markowska-Daniel I. Clinical signs and economic losses caused by porcine reproductive and respiratory syndrome virus in a large breeding farm. Vet Microbiol. 1997;55:317–322. doi: 10.1016/S0378-1135(96)01326-0. [DOI] [PubMed] [Google Scholar]
- 41.Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc. 2005;227:385–392. doi: 10.2460/javma.2005.227.385. [DOI] [PubMed] [Google Scholar]
- 42.Leng X, Li Z, Xia M, He Y, Wu H. Evaluation of the efficacy of an attenuated live vaccine against highly pathogenic porcine reproductive and respiratory syndrome virus in young pigs. Clin Vaccine Immunol. 2012;19:1199–1206. doi: 10.1128/CVI.05646-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian Z, An T, Zhou Y, Peng J, Hu S, Wei T, Jiang Y, Xiao Y, Tong G. An attenuated live vaccine based on highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) protects piglets against HP-PRRS. Vet Microbiol. 2009;138:34–40. doi: 10.1016/j.vetmic.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 44.Xie J, Cui T, Cui J, Chen Y, Zhang M, Zhou P, Deng S, Su S, Zhang G. Epidemiological and evolutionary characteristics of the PRRSV in Southern China from 2010 to 2013. Microb Pathogenesis. 2014;75:7–15. doi: 10.1016/j.micpath.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 45.Shi K, Tao C, Lin C, Li G, Li J. Complete genome sequence of a porcine reproductive and respiratory syndrome virus variant with a new deletion in the 5′ untranslated region. Genome Announc 2013;1(1):e00090–12. [DOI] [PMC free article] [PubMed]
- 46.Shi K, Mo S, Huang H, Lin C, Tao C, Li J, Li G. Complete genome sequence of a highly pathogenic porcine reproductive and respiratory syndrome virus variant isolated from a backyard piglet. Genome Announc. 2013;1(1):e00123–12. [DOI] [PMC free article] [PubMed]
- 47.Wang F, Guo L, Yang Y, Song N, Chen L, Cheng S, Wen Y. Isolation and sequence analysis of highly pathogenic porcine reproductive and respiratory syndrome virus from swine herds in the Jilin Province of China. Indian J Virol. 2013;24:90–92. doi: 10.1007/s13337-012-0124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in generating overlapping amplicons spanning PRRSV genomes. (DOCX 13 kb)
Homology between the 28 isolates sequenced in this study. (DOCX 19 kb)
Sequence identity of the JXA1-R-like strains with JXA1 derivatives and representative strains. (DOCX 20 kb)
Comparisons of partial ORF1a, ORF1b, and GP4 deduced amino acid sequences. *Unique and identical amino acids among the JXA1 derivatives. (PDF 491 kb)
Phylogenetic analysis of parental regions of putative 10 recombinant strains. The parental vaccine JXA1-R group (sublineage 10.7) is shown in red, the 2009–2010 HP-PRRSV-like group (sublineage 10.6) is shown in deep sky blue, and the NADC30-like strain (lineage 9) is shown in green. Putative recombinant strains are labeled with black triangles (▲). (PDF 502 kb)
Morbidity and mortality rates among farms with pigs infected with recombinant JXA1-R-like strains. (PDF 87 kb)


