Abstract
Membrane-bound fatty acid desaturases (FADs) are of great importance and play multiple roles in plant growth and development. In the present study, 39 full-length FAD genes, based on database searches, were identified in tetraploid upland cotton (Gossypium hirsutum L.) and were phylogenetically clustered into four subfamilies. Genomic localization revealed that 34 genes were mapped on 22 chromosomes, and five genes were positioned on the scaffold sequences. The FAD genes of G. hirsutum in the same subfamily had similar gene structures. The structures of paralogous genes were considerably conserved in exons number and introns length. It was suggested that the FAD gene families in G. hirsutum might be duplicated mainly by segmental duplication. Moreover, the FAD genes were differentially expressed in different G. hirsutum tissues in response to different levels of salt and cold stresses, as determined by qRT-PCR analysis. The identification and functional analysis of FAD genes in G. hirsutum may provide more candidate genes for genetic modification.
Fatty acids, a major source of energy in plants, are the primary constituents of plant membrane phospholipids and store triacylglycerols1. In most of the plant tissues, fatty acid biosynthesis occurs in the plastids by a successive concatenation of two carbon long units resulting in the formation of 16- or 18-carbon long fatty acids that predominate in the cellular membranes2,3. They are over 75% unsaturated and play essential roles like regulating plant growth and responding to variance in environmental signals4,5,6. The unsaturated fatty acids, compared with saturated fatty acid, contain one or more C = C double bonds in their fatty-acyl chains at specific locations4.
Membrane-bound Fatty acid desaturases (FADs), encoded by a large gene family responsible for membrane lipid modification and re-ordering7, are the key enzymes for inducting one or more carbon double bonds at different positions in the hydrocarbon chains of fatty acids8. In the eukaryotic genomes, four large branches of FAD gene family with distinct functions have been identified previously. These are First desaturase, Omega desaturase, Front-end desaturase, and Sphingolipid desaturase9. The predominant First desaturase subfamily members are encoded by Arabidopsis desaturase (ADS) genes and possess double bonds at Δ7 and Δ9 positions of palmitic acid (16:0) or stearic acid (18:0)9,10. In Omega desaturase subfamily, a double bond is found between pre-existing double bond and the acyl end at Δ12 and Δ15 desaturases8. Typically, the Front-end desaturase subfamily contains a cis or trans Δ8 double bond produced by sphingoid long-chain bases at Δ8 desaturases (SLDs)9,11. The sole function of Sphingolipid desaturase subfamily is to modify sphingolipids by causing desaturation at Δ4-position and hydroxylation at the C-4-position. They are also called as dihydroceramide Desaturases (DSDs) due to their highest activity of dihydroceramide12.
Nineteen Gossypium raimondii FAD genes have been separated into above mentioned four branches and named after corresponding encoding genes13. It is known that changing bond number and/or position may form different types of fatty acids14,15. For instance, introducing of a double bond at Δ12 position of oleic acid (18:1) can produce linoleic acid (18:2)16.
To date, several membrane-bound FAD genes from different plant species have been characterized. It is evident by the previous studies that FAD genes are expressed in adverse environmental conditions. For example, in tomato, over-expression of LeFAD3 gene enhanced the tolerance of tomato seedlings against salt stress17, whereas low-expression of LeFAD7 increased tolerance of tomato leaves to high-temperature (45 °C)18. Similarly, over-expression of either FAD3 or FAD8 increased tolerance of tobacco plants to drought19, and over-expression of FAD7 alleviated the damage of cold stress in tobacco plants20. Moreover, over-expression of omega-3 desaturases improved tolerance of various transgenic plants to both drought and chilling stresses20,21,22,23,24. In addition, FAD2 and FAD6 were induced in seedlings of Arabidopsis under salinity stress25,26. In general, the FAD genes regulate the levels of fatty acid unsaturation in membrane lipids thus improving plant tolerance to adverse environments8.
Cotton is an important commercial crop grown worldwide to provide generous supply of natural fiber for textile industry and to provide cottonseeds for food, feed, and bio-fuel productions4. It is cultivated mainly in the tropical and subtropical regions. Temperatures below 15 °C can affect the plant growth and development causing yield losses mainly due to poor germination and high seedling mortality. Gossypium hirsutum L., an allotetraploid species, is one of the most commonly grown cotton species for commercial production4. Originally, it was developed from two ancient diploid cotton species, G. raimondii and G. arboreum, by hybridization and chromosome doubling about 1–2 MYA3. It seems to be an interesting model system not only for the research of genome evolution, but also for studying the roles of polyploid in crop development and domestication with high research value. The fatty acid families from G. raimondii and G. arboreum have already been identified and characterized13. In G. hirsutum, Δ12 desaturase (FAD2) and Δ15 desaturase (FAD3 and FAD7/8) have been studied, mainly focused on cold adaptation at seedling stages1,2,3,7. Furthermore, the FAD2-1 was found to be a seed-specific desaturase responsible for the synthesis of polyunsaturated fatty acids in the cottonseeds27, while the FAD2-2 had low expression levels throughout the seed development2.
In this study, all the FAD genes in G. hirsutum were identified and characterized, and the phylogenetic analysis and structural diversification, as well as the expression profiles of the detected GhFAD genes in response to salt and cold stress regimes across different plant tissues were conducted. This research work may contribute to widen our understanding about the structure and function of FAD gene family in G. hirsutum, which may provide some candidate genes for predictable modification of fatty acid profiles in order to improve the plant vigor and seed nutritional value for cotton breeders1.
Results
Identification of FAD genes in G. hirsutum
Based on the completed genome sequences of G. hirsutum, a genome-wide search for FAD genes was performed by BlastP and tBlastN program, using Arabidopsis and rice FAD genes as the query sequences. Subsequently, all pre-identified proteins were subjected to domain analysis using Pfam and SMART databases for further confirmation, in the presence of FAD protein domain (PF00487). Eventually, a total of 39 FAD genes in G. hirsutum, based on their orthologs with reported counterparts in Arabidopsis28,29,30, were predicted and annotated (Table 1).
Table 1. Basic information regarding the FAD genes in G. hirsutum.
| No. | Gene name | Locus ID | Genomics position | Protein length | Mw(KDa) | PI | Strand |
|---|---|---|---|---|---|---|---|
| 1 | GhFAD7A | Gh_A10G2136 | A10: 100162007-100164622 | 475 | 54.27 | 9.10 | minus |
| 2 | GhFAD7D | Gh_D10G2457 | scaffold4395_D10: 144858-147533 | 450 | 51.15 | 8.91 | minus |
| 3 | GhFAD8.1A | Gh_A04G0783 | A04: 52670441-52672443 | 446 | 50.96 | 8.47 | minus |
| 4 | GhFAD8.1D | Gh_D04G1274 | D04: 41938588-41940588 | 446 | 50.72 | 8.52 | minus |
| 5 | GhFAD8.2D | Gh_D01G2221 | D01: 61104419-61107421 | 435 | 50.40 | 7.42 | plus |
| 6 | GhFAD8.2A | Gh_A01G1961 | A01: 99569646-99572640 | 427 | 49.53 | 8.17 | plus |
| 7 | GhFAD3.1A | Gh_A09G0848 | A09: 55929662-55931410 | 388 | 45.03 | 9.05 | plus |
| 8 | GhFAD3.1D | Gh_D09G0870 | D09: 33787984-33789746 | 388 | 45.11 | 9.05 | plus |
| 9 | GhFAD3.2A | Gh_A13G0564 | A13: 13306656-13308279 | 308 | 35.41 | 8.69 | plus |
| 10 | GhFAD3.3A | Gh_A07G0946 | A07: 17618250-17619951 | 376 | 43.52 | 8.95 | minus |
| 11 | GhFAD3.3D | Gh_D07G1026 | D07: 14269363-14271065 | 376 | 43.55 | 8.69 | minus |
| 12 | GhFAD2.1A | Gh_A13G1850 | A13: 78167608-78168765 | 385 | 44.06 | 9.09 | minus |
| 13 | GhFAD2.1D | Gh_D13G2237 | D13: 58467204-58469315 | 291 | 33.52 | 9.61 | minus |
| 14 | GhFAD2.2D | Gh_D13G2238 | D13: 58471954-58473105 | 383 | 43.89 | 8.95 | minus |
| 15 | GhFAD2.3A | Gh_A11G2814 | A11: 91511504-91512658 | 384 | 44.25 | 8.96 | plus |
| 16 | GhFAD2.3D | Gh_D11G3169 | D11: 64332280-64333434 | 475 | 54.27 | 9.10 | plus |
| 17 | GhFAD2.4A | Gh_A01G2094 | scaffold112_A01: 53713-54846 | 377 | 43.84 | 8.98 | plus |
| 18 | GhFAD2.4D | Gh_D01G1227 | D01: 30322983-30324116 | 377 | 43.66 | 8.95 | minus |
| 19 | GhFAD2.5A | Gh_A01G2091 | scaffold111_A01: 182694-185218 | 205 | 23.76 | 9.48 | plus |
| 20 | GhFAD2.5D | Gh_D01G1226 | D01: 30279978-30281129 | 383 | 44.27 | 8.94 | minus |
| 21 | GhFAD6D | Gh_D13G1979 | D13: 55126909-55130714 | 442 | 51.32 | 9.17 | minus |
| 22 | GhFAD6A | Gh_A13G1619 | A13: 75012308-75016124 | 445 | 51.59 | 9.09 | minus |
| 23 | GhADS5D | Gh_D05G1179 | D05: 10139919-10141857 | 386 | 44.24 | 9.37 | plus |
| 24 | GhADS5A | Gh_A05G3863 | scaffold1234_A05: 29739-31667 | 386 | 44.20 | 9.37 | minus |
| 25 | GhDSD1.1A | Gh_A06G1118 | A06: 70479575-70481245 | 324 | 37.74 | 6.97 | plus |
| 26 | GhDSD1.1D | Gh_D06G1371 | D06: 42795712-42797364 | 324 | 37.72 | 6.95 | plus |
| 27 | GhDSD1.2D | Gh_D10G0219 | D10: 1935044-1936976 | 314 | 36.79 | 8.68 | plus |
| 28 | GhDSD1.2A | Gh_A10G0240 | A10: 2107036-2109095 | 331 | 38.62 | 8.82 | plus |
| 29 | GhSLD1A | Gh_A12G0984 | A12: 61000719-61003073 | 326 | 37.18 | 6.80 | plus |
| 30 | GhSLD1D | Gh_D12G1104 | D12: 37533014-37535355 | 326 | 37.10 | 7.27 | plus |
| 31 | GhSLD2A | Gh_A05G0325 | A05: 3576223-3577566 | 447 | 51.15 | 8.68 | plus |
| 32 | GhSLD2D | Gh_D05G0430 | D05: 3482279-3483622 | 447 | 51.20 | 8.68 | plus |
| 33 | GhSLD3A | Gh_A08G2217 | A08: 103131115-103132458 | 447 | 51.20 | 8.64 | plus |
| 34 | GhSLD3D | Gh_D08G2583 | D08: 65460731-65462074 | 447 | 51.27 | 8.61 | plus |
| 35 | GhSLD4A | Gh_A07G1291 | A07: 30625403-30626746 | 447 | 51.18 | 8.36 | plus |
| 36 | GhSLD4D | Gh_D07G1405 | D07: 23118528-23119871 | 447 | 51.18 | 8.55 | plus |
| 37 | GhSLD5 | Gh_Sca050655G01 | scaffold50655: 42-814 | 218 | 25.04 | 9.03 | plus |
| 38 | GhSLD5D | Gh_D11G0983 | D11: 8582726-8584069 | 447 | 51.37 | 8.94 | plus |
| 39 | GhSLD5A | Gh_A11G0840 | A11: 8503404-8504747 | 388 | 45.03 | 9.05 | plus |
The protein sequences, encoded by these 39 FAD genes in G. hirsutum, were detected. They ranged in length from 205 amino acids of GhFAD2.5A to 475 amino acids of GhFAD7A and GhFAD2.3D, with an average of 388 amino acids approximately. Similarly, the predicted molecular weight (Mw) of these deduced proteins varied from 23.76 kDa to 54.27 kDa, and the theoretical isoelectric point (pI) varied from 6.80 to 9.61.
Phylogenetic relationship among the FAD genes
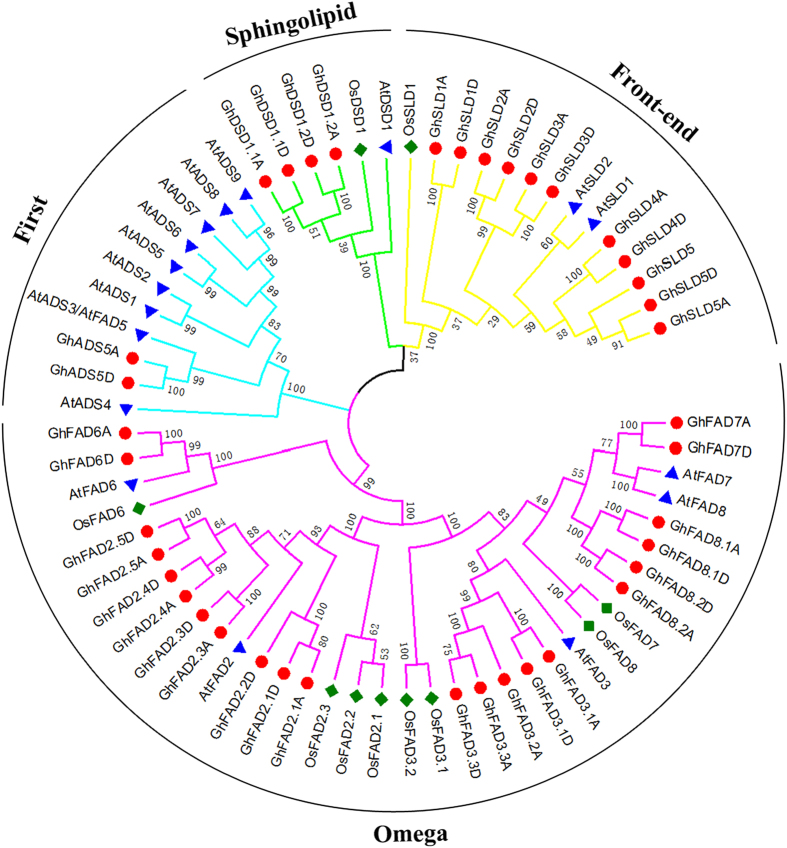
To evaluate the evolutionary relationships among the FAD gene members, all the genes from G. hirsutum (39), Arabidopsis (17), and rice (10) were aligned separately by Neighboring-Joining method to generate an un-rooted phylogenetic tree (Fig. 1). For confirmation, a phylogenetic tree was also constructed by Maximum Evolution method and the results were found almost consistent with Neighbor-Joining method, showing a little differences at some branches (Supplementary Fig. S1). Further, the FAD genes in these three species were divided into four well-defined monophyletic groups, suggesting the existence of a common ancestor before the divergence of monocots and dicots. The First desaturase group encoded by ADS genes was composed of 11 ADS genes, nine from Arabidopsis and two from G. hirsutum. No ADS genes were found in the rice, suggesting some evolutionary role in its genome. The ADS genes were either lost in the rice or were acquired by the dicotyledons after divergence from the last common ancestor. In the Omega desaturase group, there were five genes from Arabidopsis, eight genes from rice, and 22 genes from G. hirsutum. There were 14 SLD genes in the Front-end desaturase group which included 11 gens from G. hirsutum, two from Arabidopsis, and one from rice. The Sphingolipid desaturase subfamily contained only six DSD genes, four DSD genes from G. hirsutum, only one from Arabidopsis and rice each. Interestingly, the G. hirsutum possessed more gene members than Arabidopsis and rice in these three subfamilies except First desaturase in which most of the gene members belonged to Arabidopsis.
Figure 1. Phylogenetic relationships of FAD genes from G. hirsutum, Arabidopsis, and rice.
A Neighbor-Joining (NJ) phylogenetic tree of all detected FAD genes was constructed, using MEGA 5.2 program with bootstrap test (replicated 1000 times). The Omega, First, Sphingolipid, and Front-end subfamily were marked in pink, blue, green, and yellow, respectively.
Though several pairs of paralogous genes in the terminal branches of the G. hirsutum phylogenetic tree were distributed in each subfamily, several other FAD genes were not clustered together with the FAD genes from Arabidopsis and rice. This suggested that these FAD genes might have merged in G. hirsutum genome after diverging from the last common ancestor or have been lost in Arabidopsis and rice. Gene numbers in First and Sphingolipid desaturase groups were lower than others, which harbored two and four GhFAD genes, respectively. Taken together, the presence and the absence of species-specific FAD genes were due to the functional divergence. The FAD genes in G. hirsutum might have undergone rapid expansion during the course of evolution and exhibited a tendency to cluster into the same subgroup due to the conserved functions.
Genomic localization of FAD genes
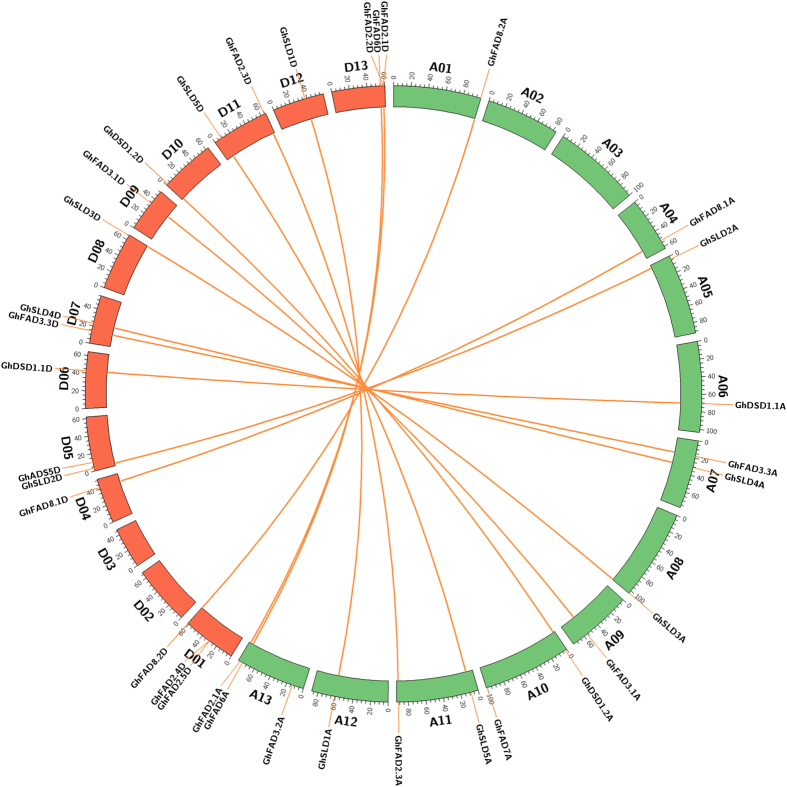
Genomic localization of FAD genes in G. hirsutum notably revealed that 34 genes were mapped on the 22 chromosomes, except for chromosome A02, A03, D02, and D03 (Fig. 2). The rest five genes, GhFAD7D, GhFAD2.4A, GhFAD2.5A, GhADS5A, and GhSLD5 were positioned on the scaffold sequences. As shown in Fig. 2, chromosome A01, A04, A05, A06, A08, A09, A12, D04, D06, D08, D09, D10, and D12 harbored only a single FAD gene each, while two FAD genes were present on each chromosome of A07, A10, A11, D05, D07, and D11. Moreover, there were three FAD genes on chromosome A13, D01, and D13 each.
Figure 2. Genomic localization and paralogs of FAD genes of G. hirsutum.
Chromosomes A and D were indicated with different colors. The paralogous genes belonging to the FAD gene family were connected by orange lines.
Duplication of FAD genes
To date, the mechanism of FAD genes expansion in G. hirsutum remains unclear. We deliberately investigated the relationship between genetic divergence and gene duplication within the FAD gene family of allotetraploid cotton. Total 13 duplicated gene pairs were found in this study, i.e. GhFAD8.2A/GhFAD8.2D, GhFAD8.1A/GhFAD8.1D, GhSLD2A/GhSLD2D, GhDSD1.1A/GhDSD1.1D, GhFAD3.3A/GhFAD3.3D, GhSLD4A/GhSLD4D, GhSLD3A/GhSLD3D, GhFAD3.1A/GhFAD3.1D, GhDSD1.2A/GhDSD1.2D, GhSLDL5A/GhSLD5D, GhFAD2.3A/GhFAD2.3D, GhSLD1A/GhSLD1D, and GhFAD6A/GhFAD6D. They were positioned on individual chromosome and mainly attributed to the segmental duplication events. Meanwhile, no tandem duplication event was found, which showed adjacently positions with no intervening genes instead. This suggested that the expansion of FAD gene family in G. hirsutum was mainly attributed to the segmental duplication events.
Structural organization of FAD genes
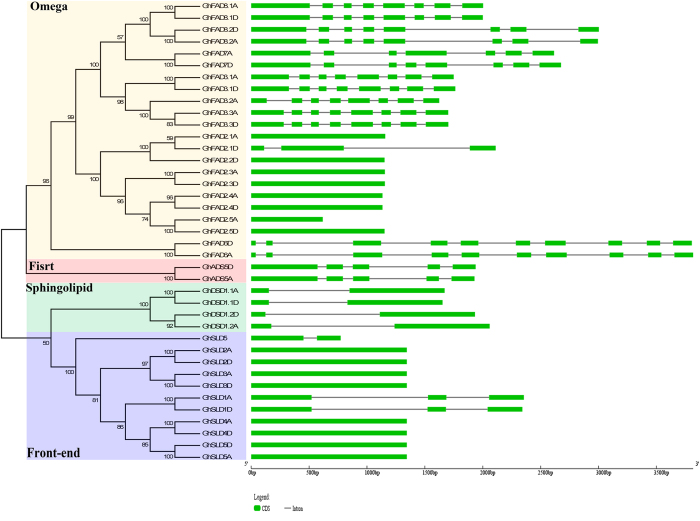
In the current experiment, 39 FAD genes were clustered into four subfamilies (Fig. 3). Generally, the FAD genes in the same subfamily had strikingly similar exon/intron structure than other subfamilies. For instance, in the First desaturase subfamily, GhADS5D and GhADS5A harbored five exons with highly conserved length and four introns in closed size. In contrast, all the genes in the Sphingolipid desaturase subfamily possessed only two same-sized exons. Generally, the genes of the Front-end desaturase subfamily possessed a single exon generally, except GhSLD5 which had two exons and one shorter intron. However, there were three exons in GhSLD1A and GhSLD1D. The similar exon-intron structure agreed well with their close phylogenetic relationship. In contrast to these three relatively conserved subfamilies, the FAD genes in the Omega desaturase subfamily showed a complex distribution of exons and introns. For example, the genes of FAD3, FAD7, and FAD8 sections mostly had up to eight exons of relatively conserved length, except for GhFAD7A which possessed seven exons. While all the FAD genes in FAD2 section had a single exon, except for GhFAD2.1D, which harbored three exons. In addition, GhFAD6D and GhFAD6A were composed of ten exons. Among the paralog pairs, uniformity was observed in the gene structures. Since G.hirsutum is an allotetraploid plant, the essential integrated evolutionary relationship was done by the sequences of the diploid G. raimondii and G. arboreum progenitors (Supplementary Fig. S2). The detailed comparison indicated that the exon-intron structures were identical to the result of phylogenetic relationships and were highly conserved in each subfamily, though parts of the orthologous genes demonstrated certain differences.
Figure 3. Phylogenetic relationship and gene structure of FAD genes in G. hirsutum.
Four subfamilies labeled as Omega, First, Sphingolipid, and Front-end were marked with different color backgrounds. Exons were represented by green boxes and introns by grey lines.
Selective pressure analysis of the duplicated FAD genes
To investigate the selective the duplicated FAD gene pairs, the non-synonymous to synonymous substitution ratios (Ka/Ks) were calculated. Generally, Ka/Ks ratio >1 indicates positive selection, Ka/Ks = 1 indicates neutral selection, while a ratio <1 indicates negative or purifying selection. In G. hirsutum, the Ka/Ks ratios for 13 duplicated pairs were <1 (Table 2), which suggested that they had experienced strong purifying selection pressure contributing largely to the maintenance of function in G. hirtusum FAD gene family.
Table 2. Ka/Ks analysis for the duplicated FAD gene pairs of G. hirsutum.
| Duplicated gene 1 | Duplicated gene 2 | ka | Ks | ka/ks | Purifying selection | Duplicated type |
|---|---|---|---|---|---|---|
| GhFAD8.1A | GhFAD8.1D | 0.011 | 0.056 | 0.199 | Yes | Segmental |
| GhFAD8.2D | GhFAD8.2A | 0.014 | 0.021 | 0.670 | Yes | Segmental |
| GhFAD3.1A | GhFAD3.1D | 0.002 | 0.016 | 0.139 | Yes | Segmental |
| GhFAD3.3A | GhFAD3.3D | 0.010 | 0.032 | 0.320 | Yes | Segmental |
| GhFAD2.3A | GhFAD2.3D | 0.006 | 0.045 | 0.128 | Yes | Segmental |
| GhFAD6D | GhFAD6A | 0.003 | 0.028 | 0.104 | Yes | Segmental |
| GhSLD2A | GhSLD2D | 0.006 | 0.028 | 0.206 | Yes | Segmental |
| GhSLD3A | GhSLD3D | 0.007 | 0.039 | 0.174 | Yes | Segmental |
| GhSLD1A | GhSLD1D | 0.013 | 0.034 | 0.382 | Yes | Segmental |
| GhSLD4A | GhSLD4D | 0.001 | 0.024 | 0.043 | Yes | Segmental |
| GhSLD5D | GhSLD5A | 0.005 | 0.014 | 0.356 | Yes | Segmental |
| GhDSD1.1A | GhDSD1.1D | 0.007 | 0.033 | 0.212 | Yes | Segmental |
| GhDSD1.2A | GhDSD1.2D | 0.004 | 0.054 | 0.074 | Yes | Segmental |
Gene expression patterns in different plant tissues
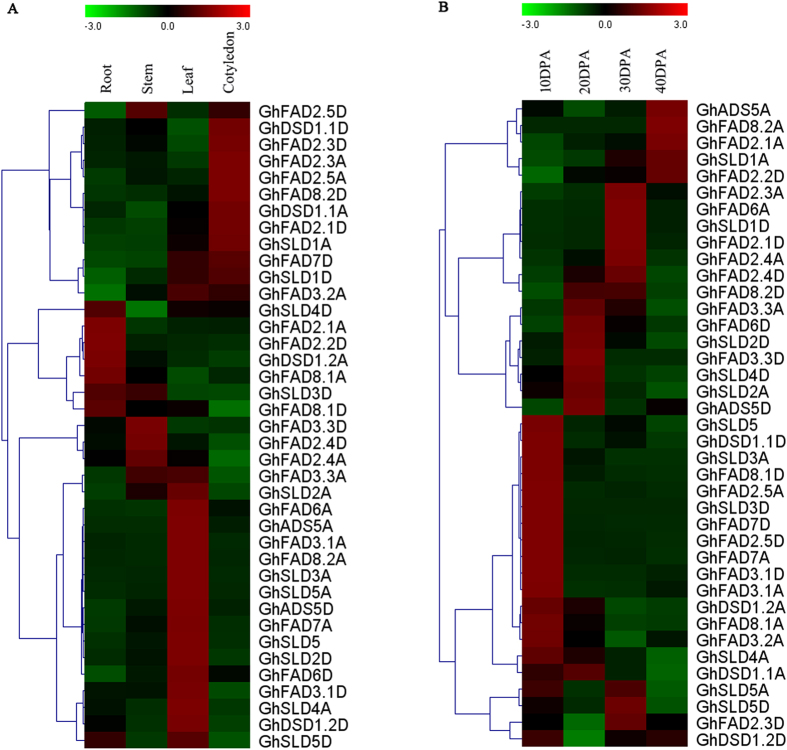
39 FAD genes were multifunctional in different tissues of G. hirsutum plant. In order to evaluate their expression patterns, a comprehensive qRT-PCR analysis was performed and distinct tissue-specific expression patterns were observed as illustrated in Fig. 4A. Eight FAD genes, i.e. GhSLD4D, GhFAD2.1A, GhFAD2.2D, GhDSD1.2A, GhFAD8.1A, GhSLD3D, GhFAD8.1D, and GhSLD5D were preferentially expressed in the roots. In the stems, seven FAD genes, GhFAD2.5D, GhSLD3D, GhFAD3.3D, GhFAD2.4D, GhFAD2.4A, GhFAD3.3A, and GhSLD2A displayed the highest transcript abundance. In the leaves, 20 FAD genes such as GhFAD7D, GhFAD3.2A, and GhSLD5D exhibited high expression levels. Four FAD genes, GhFAD2.1A, GhFAD2.2D, GhDSD1.2A, and GhFAD8.1A, were expressed higher in the roots than other plant tissues, suggesting a potentially specialized function in the roots development or roots activities. While 12 FAD genes, GhFAD2.5D, GhDSD1.1D, GhFAD2.3D, GhFAD2.3A, GhFAD2.5A, GhFAD8.2D, GhDSD1.1A, GhFAD2.1D, GhSLD1A, GhFAD7D, GhSLD1D, and GhFAD3.2A, exhibited specifically high transcript accumulation in the cotyledons.
Figure 4. Expression patterns of 39 FAD genes of G. hirsutum.
(A) Expression patterns in four representative tissues. (B) Expression patterns in developmental ovules. The color scales represented the relative signal intensity values.
There were 18 paralogs among these FAD genes, but only one pairs, GhFAD2.3A/GhFAD2.3D clustered together, which inferred that the expression of the FAD paralogs in G. hirsutum have experienced divergence.
Gene expression patterns in developmental ovules
The expressions of the membrane-bound fatty acid genes in different developmental ovules were shown in Fig. 4B. Eleven FAD genes, GhSLD5, GhDSD1.1D, GhSLD3A, GhFAD8.1D, GhFAD2.5A, GhSLD3D, GhFAD7D, GhFAD2.5D, GhFAD7A, GhFAD3.1D, and GhFAD3.1A shared the characteristic of having maximal expressions in early stage (10day post anthesis, DPA) and lower expressions at later developmental stages. In addition, GhFAD2.4D, GhFAD8.2D, and GhFAD3.3A exhibited specifically high transcript accumulation in ovule development later, which was focused on 20 and 30 DPA. GhADS5A, GhFAD8.2A, GhFAD2.1A, GhSLD1A, and GhFAD2.2D had similar expression behavior of higher expression on 40 DPA stages. However, GhFAD6D, GhSLD2D, GhFAD3.3D, GhSLD4D, GhSLD2A, and GhADS5D were strongly expressed on 20 DPA. Notably, the subfamily of GhFAD2, comprised of GhFAD2.3A, GhFAD2.1D, GhFAD2.4A, and GhFAD2.4D showed strikingly consistent expression patterns in later developmental stages (30 DPA), which clued to the existence of functional conservation.
Gene expression patterns under salt and cold stresses
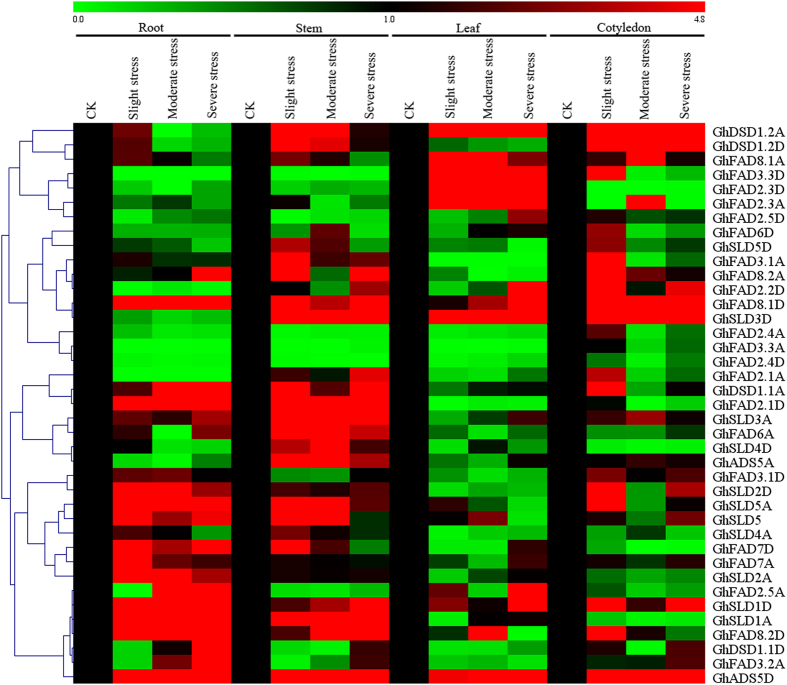
Gene expressions for all the FAD genes were also observed in different tissues under different levels of salt and cold stresses. Results showed that the salt stress caused changes in the expression patterns in the form of induction or suppression (Fig. 5). In the roots, twelve FAD genes, GhFAD8.1D, GhDSD1.1A, GhFAD2.1D, GhSLD2D, GhSLD5A, GhSLD5, GhFAD7D, GhSLD2A, GhSLD1D, GhSLD1A, GhFAD8.2D, and GhADS5D, were up-regulated under different levels of salt stress. In the stems, most of the FAD genes were highly expressed after salt treatment, while GhFAD3.3D, GhFAD2.3D, GhFAD2.5D, GhFAD2.4A, GhFAD3.3A, GhFAD2.4D, and GhFAD2.5A were less expressed over all stages. However, compared with the stems, only few FAD genes were up-regulated in the leaves, but higher expressions of GhFAD3.3D, GhFAD3.1A, GhFAD2.1A, GhDSD1.1A, and GhSLD5A were found under slight salt stress in the cotyledons. Among all, only GhADS5D was highly expressed in all tissues under salt stress. The expressions of GhFAD8.2A in the roots and GhFAD2.2D in the leaves were relatively low at slight stress but showed dramatic increases during severe stress. However, GhDSD1.2A, GhDSD1.2D, GhSLD5A, GhSLD5, GhSLD4A, and GhFAD7D in the stems showed the highest transcript abundance after stress then decreased thereafter. Furthermore, four paralogs, GhDSD1.2A/GhDSD1.2D, GhFAD2.3D/GhFAD2.3A, GhFAD7A/GhFAD7D, and GhSLD1A/GhSLD1D, were clustered together having similar expression patterns.
Figure 5. Expression patterns of 39 FAD genes of G. hirsutum under gradient salt stress.
The color scales represented the relative signal intensity values.
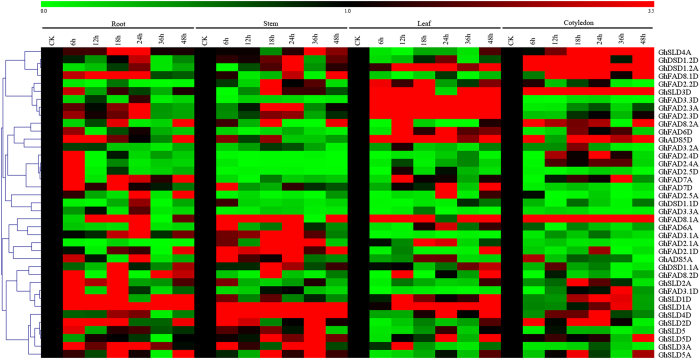
The gene expressions of membrane-bound fatty acids in different plant tissues under different low temperature levels were shown in Fig. 6. In all the tested tissues, the expression level of GhFAD8.1A remained very high under low temperature as compared with other genes, suggesting its potential roles in tissue development and/or function. In the roots, GhFAD2.2D, GhFAD3.2A, and GhFAD2.1A transcripts were largely restricted, whereas the transcripts of GhDSD1.2D, GhDSD1.2A, GhFAD2.5A, GhDSD1.1D, and GhFAD3.3A were increased to their peaks at 24 hours interval after cold treatment. In contrast, GhSLD1A and GhSLD4D showed the highest transcript abundance, whereas GhFAD3.3D, GhFAD6D, GhFAD3.2A, GhFAD2.4D, GhFAD2.4A, GhFAD2.5D, GhFAD7A, GhFAD2.5A, and GhFAD3.3A exhibited low expressions in the stems. In addition, the membrane-bound fatty acid genes were extensively expressed in the leaves and cotyledons, especially for those of GhFAD3.3D, GhFAD2.3A, and GhFAD2.3D in the leaves and GhDSD1.2A, GhSLD3D, and GhFAD8.1A in the cotyledons, which were significantly up-regulated when the salt stress was applied for a longer time. Several fatty acid genes in the cotyledons, such as GhFAD3.3D, GhFAD2.5D, GhFAD7D, and GhFAD3.3A, were suppressed by cold stress application.
Figure 6. Expression patterns of 39 FAD genes of G. hirsutum under gradient cold stress.
The color scales represents the relative signal intensity values.
Discussion
G. hirsutum is a natural allotetraploid produced by the interspecific hybridization of A- and D-genome diploid progenitor species. Based on the genome scans of several plant genomes, the FAD gene family has been systematically investigated in Arabidopsis (17), soybean (10)31, G. raimondii (19), and G. arboreum (20)13. In current work, a total of 39 putative FAD genes were identified in the genomes of G. hirsutum, which contained more FAD genes than other plant species such as Arabidopsis and rice. It might be mainly due to the recent polyploidy and segmental duplication events in G. hirsutum evolutionary history. Coincidentally, the number of FAD genes in G. hirsutum (AD1 allotetraploid) was exactly the sum of those in G. raimondii (19) and G. arboretum (20). Moreover, all these FADs were distinctly classified into four groups as Omega desaturase, First desaturase, Sphingolipid desaturase, and Front-end desaturase consistently. In three subfamilies, Front-end desaturase, Omega desaturase, and Sphingolipid desaturase, G. hirsutum had much more FAD genes than Arabidopsis and rice, which implied that these three subfamilies arose before the divergence of monocots-dicots and might have undergone species specific expansion process. Furthermore, the sizes of the GhFAD proteins varied markedly, as well as the predicted isoelectric points, suggesting that different GhFAD proteins might have functions in different microenvironments.
Gene duplication plays an irreplaceable role in the process of gene family expansion in the genomes. In G. hirsutum, 13 segmental duplicated gene pairs were found, such as five gene pairs in the Front-end desaturase subfamily, two gene pairs in the Sphingolipid desaturase subfamily, and six gene pairs in the Omega desaturase subfamily. These results suggested that the expansion of Sphingolipid desaturase, Front-end desaturase, and Omega desaturase subfamilies were due to the segmental duplication, which was consistent with the previous report13, namely the increasing size of FAD genes might be contributed by the segmental duplication events.
It was noteworthy that paralogous FAD gene pairs demonstrated very similar exon/intron distribution patterns in terms of exon number and intron length. For instance, the FAD genes in the First desaturase subfamily and the Sphingolipid desaturase subfamily of three cotton species were highly conserved and clustered distinctly. However, their expression patterns were greatly divergent. It might have involved in the mechanism of adaptation to different environmental conditions. In the current study, different expression patterns of FAD genes were observed in each tissue. GhADS5A, GhFAD3.1A, and GhSLD3A were expressed markedly at high levels in the leaves but lower in the other tissues, while GhFAD6D was constitutively expressed at high levels in all the tested tissues. The tissue specific FAD gene expression patterns observed in the present study indicated their functional divergence during plant development and growth, which needs further study to identify their actual functions. In addition, most of the FAD2 subfamily such as GhFAD2.3A, GhFAD2.1D, GhFAD2.4A, GhFAD2.4D, GhFAD2.1A, and GhFAD2.2D showed dramatically high abundance in the process of lipid accumulation. It suggested that these genes were seed-specific responsible for the polyunsaturated fatty acids in the seed oil of cultivated cotton. In addition, the genes which were highly expressed on 20 DPA recruited all types of FAD genes for the biogenesis in different positions or lengths. However, the other development stages involved only three subfamilies, which suggested that 20 DPA might be an important stage for ovule development.
Salinity, resulting mainly due to NaCl, is one of the common environmental stresses that afflicts the growth and yield of crops in many regions all over the world6,26,32,33,34. In previous studies, FAD2 and FAD6 were revealed to be required for salt tolerance in Arabidopsis26,32, while LeFAD3 up-regulation could alleviate salt stress in early seedling stage23. In addition, antisense AtFAD7 expression resulted in lower salt tolerance in transgenic tobacco plants6. In this study, low expression level of GhFAD2.4D was found in all tissues treated by salt stress. GhFAD8.1D and GhADS5D genes were significantly up-regulated in the leaves in response to salt stress, which suggested that these FAD genes might adjust to appropriate levels to protect the cell membrane of cotton plant from salt stress. Low temperature is another serious stress, which significantly affects plant growth and development. The FAD2-3 and FAD2-4 genes from G. hirsutum were found to participate in the membrane adaptation to cold stress and induced significantly higher in a previous study7. Similarly, the over-expression of FAD8 could also alleviate the damage of cold stress35. In this study, GhFAD8.1A was highly expressed implying its potential role in the plant tolerance to low temperature stress, which is consistent with the previous findings3,35,36.
Conclusion
In the current work, 39 full-length FAD genes, classified into four well-documented groups in conserved exon/intron structures, have been identified and functionally validated in G. hirsutum. Most GhFAD2 subfamily members showed high abundance in later developmental stages, it might have contributed to the accumulation of oil contents in seeds and can be used as target genes to improve cottonseed quality. qRT-PCR analysis also revealed the corresponding expressions of GhFAD genes under different levels of salt and cold stresses. GhFAD8.1D and GhADS5D exhibited specifically high transcript accumulation in the tested tissues exposed to different levels of cold stress, which suggested that they might be required for salt tolerance. Meanwhile, GhFAD8.1A was likely to enhance the plant tolerance to cold stress according to the significant up-regulation after long periods of treatment. The comprehensive investigation carried out in this study may improve our understanding about the involvement of FAD genes to tolerate adverse environments, and can also provide the candidate genes for functional studies.
Materials and Methods
Sequence retrieval and genome-wide identification
The genome of G. hirsutum was downloaded from MASCOTTON database (http://mascotton.njau.edu.cn/), while the published FAD proteins of Arabidopsis and Oryza sativa (Supplementary Table S1)31,37 were retrieved from The Arabidopsis Information Resource (TAIR release 10, http://www.arabidopsis.org) and the Rice Genome Annotation Project Database (RGAP release 10, http://riceplantbiology.msu.edu/index.shtml), respectively. BlastP and tBlastN programs were applied to search for the initial identification of the candidate FAD genes of G. hirsutum in default parameters, with the queries of Arabidopsis and rice FAD protein sequences. Then, the Pfam (http://pfam.sanger.ac.uk/search)38 and SMART databases (http://smart.embl-heidelberg.de/)39 were used to confirm every putative FADs of G. hirsutum with the domain of Pfam FAD gene family (PF00487). Furthermore, the theoretical molecular weight (Mw) and isoelectric point (pI) of the full-length proteins were predicted by using pI/Mw tool (http://web.expasy.org/protparam/)40.
Phylogenetic construction
Clustal X version 2.041 was used to carry out the multiple sequence alignment of full length protein sequences from three species, G. hirsutum, Arabidopsis, and rice. As soon as a MAGA file was generated, MEGA 5.242 was further applied to construct an unrooted Neighbor-Joining phylogenetic tree with pairwise deletion option and Possion correction model. Bootstrap test was carried out with 1000 replicates for the reliability of interior branches. To confirm the consistency with the Neighbor-Joining method, the phylogenetic tree was also performed with Maximum Evolution method.
Chromosomal locations, gene duplications, and structural analysis
The genes were mapped on G. hirsutum chromosomes using the Circos tool43. Gene duplication of FAD genes in G. hirsutum was indicated by (1) shared aligned sequence covering >80% of the longer gene; (2) similarity of the aligned regions >80%; and (3) only one duplicated event was counted for tightly linked genes44,45. Referring to the different chromosomal locations (Supplementary Table S2), these FAD genes were designated as either tandem duplications or segmental duplications.
Comparing the predicted coding sequences (CDSs) with their corresponding genomic sequences, the exon/intron organization of the individual GhFAD genes were illustrated using the Gene Structure Display Server (GSDS, http://gsds1.cbi.pku.edu.cn/).
Estimating Ka/Ks ratios for duplicated gene pairs
Initially, all the full-length gene sequences of the duplicated FAD gene pairs of G. hirsutum were aligned by Clustal X 2.0 program41 and then the non-synonymous (Ka) and synonymous substitution rates (Ks) were calculated using DnaSp V5.046 software. Finally, the selection pressure of each gene pair was assessed based on the Ka/Ks ratio.
Plant materials and abiotic stress treatment
TM-1, a genetic standard line of G. hirsutum was grown in a temperature-controlled chamber adjusted to 28 °C temperature, 70% relative humidity, and 16/8 hours photoperiod. After five days, half of the germinated cottonseeds were transplanted to black plastic buckets containing MS medium (5 liters). After adaption for ten days in the hydroponics, the MS solutions were adjusted to different salt treatments as 0 (control), 0.3% (slight stress), 0.6% (moderate stress), and 0.9% (severe stress). Three biological replicates were carried out for each treatment. After two weeks of salt treatment, the roots, stems, cotyledons, and leaves were sampled, frozen immediately in the liquid nitrogen, and stored at −80 °C for further analyses.
For cold stress, 30 days old seedlings were transferred to a temperature-controlled chamber adjusted to 10 °C. The plants were harvested at 0, 6, 12, 18, 24, 36, and 48 hours cold intervals to conduct expression analysis. Three biological replicates of each sample were also implemented. All collected samples were immediately frozen in the liquid nitrogen and then stored at −80 °C.
In addition, the flowers of TM-1, planted in the fields in Agricultural station of Zhejiang University, Hangzhou, in 2015, were labeled at 0 DPA. The cotton embryos were harvested at 10, 20, 30, and 40 DPA for analysis of gene expression patterns during the seed development. All collected samples were immediately frozen in the liquid nitrogen and stored at −80 °C.
RNA isolation and qRT-PCR
Total RNA of all collected samples was extracted using the EASYspin Plus Total RNA Extraction Kit (Aidlab, Beijing, China) according to the manufacturer protocol. The NanoDrop 2000 Spectrophotometer was used to determine the quantity and quality of RNAs. Subsequently, using the PrimeScriptTM 1st Strand cDNA Synthesis Kit (TakaRa, Dalian, China) to synthesize first-strand cDNAs. The specific primers were designed according to CDSs (Supplementary Table S3). All melting curves produced by qRT-PCR were shown in Supplementary Fig. S3.
The amplification reactions of qRT-PCR were performed with Lightcycler 96 system (Roche) using SYBR the premix Ex taq (TakaRa) in 20 μL volume with following parameters: initializing denaturation at 95 °C for 30 seconds, following 45 cycles denaturation at 95 °C for 10 seconds; annealing at 53–54 °C for 10 seconds, and extension at 72 °C for 20 seconds. In addition, the default setting for the melting curve stag was chosen. Three biological replicates were maintained for each collected sample. The relative expression levels were calculated according to the 2−ΔΔCt method47. The heatmap for expression profiles were generated with the Mev 4.0 software48 with Pearson correction and complete linkage clustering.
Additional Information
How to cite this article: Feng, J. et al. Genome-wide identification of membrane-bound fatty acid desaturase genes in Gossypium hirsutum and their expressions during abiotic stress. Sci. Rep. 7, 45711; doi: 10.1038/srep45711 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The work was funded by the National High Technology Research and Development Program of China (2013AA102601), China Agriculture Research System (CARS-18-25), and Jiangsu Collaborative Innovation Center for Modern Crop Production.
Footnotes
The authors declare no competing financial interests.
Author Contributions J.F., S.Z., and J.C. conceived all the experiments and analyzed data. J.F. performed experiments, drafted the manuscript and prepared the figures. J.F. and S.Z. wrote and reviewed the manuscript. Y.D., W.L., and Q.H. prepared figures and analyzed data. J.C. performed the experiments. K.D.M. contributed to the manuscript preparation. All authors reviewed the manuscript.
References
- Zhang D. Y. et al. Identification and expression of a new delta-12 fatty acid desaturase (FAD2-4) gene in upland cotton and its functional expression in yeast and Arabidopsis thaliana plants. Plant Physiol. Biochem. 47, 462–471 (2009). [DOI] [PubMed] [Google Scholar]
- Pirtle I. L. et al. Molecular cloning and functional expression of the gene for a cotton Δ-12 fatty acid desaturase (FAD2). Biochimica. et. Biophysica. Atca. 1522, 122–129 (2001). [Google Scholar]
- Yurchenko O. P. et al. Genome-wide analysis of the omega-3 fatty acid desaturase gene family in Gossypium. BMC Plant Biol. 14, 312 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J. & Browse J. Lipid Biosynthesis. Plant Cell 7, 957–970 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somenrille C. Direct tests of the role of membrane lipid composition in low-temperature-induced photoinhibition and chilling sensitivity in plants and cyanobacteria. Proc. Natl. Acad. Sci. USA 92, 6215–6218 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im Y. J., Han O., Chung G. C. & Cho B. H. Antisense expression of an Arabidopsis ω-3 fatty acid desaturase gene reduces salt/drought tolerance in transgenic tobacco plants. Mol. Cells 13, 264–271 (2002). [PubMed] [Google Scholar]
- Kargiotidou A., Deli D., Galanopoulou D., Tsaftaris A. & Farmaki T. Low temperature and light regulate delta 12 fatty acid desaturases (FAD2) at a transcriptional level in cotton (Gossypium hirsutum). J. Exp. Bot. 59, 2043–2056 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T. & Murata N. Regulation of the desaturation of fatty acids and its role in tolerance to cold and salt stress. Curr. Opin. Biotech. 5, 206–210 (2002). [DOI] [PubMed] [Google Scholar]
- Hashimoto K. et al. The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J. Lipid Res. 49, 183–191 (2008). [DOI] [PubMed] [Google Scholar]
- Heilmann I., Pidkowich M. S., Girke T. & Shanklin J. Switching desaturase enzyme specificity by alternate subcellular targeting. Proc. Natl. Acad. Sci. USA 101, 10266–10271 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Markham J. E. & Cahoon E. B. Sphingolipid Δ8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J. 69, 769–781 (2012). [DOI] [PubMed] [Google Scholar]
- Ternes P., Franke S., Zahringer U., Sperling P. & Heinz E. Identification and characterization of a sphingolipid Δ4-desaturase family. J. Biol. Chem. 277, 25512–25518 (2002). [DOI] [PubMed] [Google Scholar]
- Liu W. et al. Characterization of 19 genes encoding membrane-bound fatty acid desaturases and their expression profiles in Gossypium raimondii under low temperature. PloS one 10, e0123281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J. & Cahoon E. B. Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. 49, 611–641 (1998). [DOI] [PubMed] [Google Scholar]
- Wu Q., Liu T., Liu H. & Zheng G. C. Unsaturated fatty acid: metabolism, synthesis and gene regulation. Afr. J. Biotechnol. 8, 1782–1785 (2009). [Google Scholar]
- Panpoom S., Los D. A. & Murata N. Biochemical characterization of a Δ12 acyl-lipid desaturase after overexpression of the enzyme in Escherichia coli. Biochimica et. Biophysica Acta 1390, 323–332 (1998). [DOI] [PubMed] [Google Scholar]
- Wang H. S. et al. A tomato endoplasmic reticulum (ER)-type omega-3 fatty acid desaturase (LeFAD3) functions in early seedling tolerance to salinity stress. Plant Cell Rep. 33, 131–142 (2014). [DOI] [PubMed] [Google Scholar]
- Liu X. Y., Yang J. H., Li B., Yang X. M. & Meng Q. W. Antisense-mediated depletion of tomato chloroplast omega-3 fatty acid desaturase enhances thermal tolerance. J. Integr. Plant Biol. 48, 1096–1107 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang M. et al. Modulated fatty acid desaturation via overexpression of two distinct ω-3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. Plant J. 44, 361–371 (2005). [DOI] [PubMed] [Google Scholar]
- Khodakovskaya M., McAvoy R., Peters J., Wu H. & Li Y. Enhanced cold tolerance in transgenic tobacco expressing a chloroplast omega-3 fatty acid desaturase gene under the control of a cold-inducible promoter. Planta 223, 1090–1100 (2006). [DOI] [PubMed] [Google Scholar]
- Kodama H., Hamada T., Horiguchi C., Nishimura M. & Iba K. Genetic enhancement of cold tolerance by expression of a gene for chloroplast ω-3 fatty acid desaturase in transgenic tobacco. Plant Physiol. 105, 601–605 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama H., Horiguchi C., Nishiuchi T., Nishimura M. & Iba K. Fatty acid desaturation during chilling acclimation is one of the factors involved in conferring low-temperature tolerance to young tobacco leaves. Plant Physiol. 107, 1177–1185 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. et al. Overexpression of endoplasmic reticulum omega-3 fatty acid desaturase gene improves chilling tolerance in tomato. Plant Physiol. Biochem. 47, 1102–1112 (2009). [DOI] [PubMed] [Google Scholar]
- Dominguez T. et al. Increasing omega-3 desaturase expression in tomato results in altered aroma profile and enhanced resistance to cold stress. Plant Physiol. 153, 655–665 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. T. et al. Fatty acid desaturase-6 (Fad6) is required for salt tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 390, 469–474 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang J. T. et al. Arabidopsis fatty acid desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PloS one 7, e30355 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. et al. Evolution of the FAD2-1 fatty acid desaturase 5′ UTR intron and the molecular systematics of Gossypium (Malvaceae). Am. J. Bot. 88, 92–102 (2001). [PubMed] [Google Scholar]
- Miquel M. & Browse J. Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. J. Biol. Chem. 267, 1502–1509 (1992). [PubMed] [Google Scholar]
- Okuley J. et al. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant cell 6, 147–158 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arondel V. et al. Map-based cloning of a gene controlling omega-3 fatty acid desaturation in Arabidopsis. Science 258, 1353–1355 (1992). [DOI] [PubMed] [Google Scholar]
- Chi X. Y. et al. Genome-wide analysis of fatty acid desaturases in soybean (Glycine max). Plant. Mol. Biol. Rep. 29, 769–783 (2011). [Google Scholar]
- An M. et al. Expression of fatty acid desaturase genes and fatty acid accumulation in Chlamydomonas sp. ICE-L under salt stress. Bioresour. Technol. 149, 77–83 (2013). [DOI] [PubMed] [Google Scholar]
- Guerzoni M. E., Lanciotti R. & Cocconcelli P. S. Alteration in cellular fatty acid composition as a response to salt, acid, oxidative and thermal stresses in Lactobacillus helveticus. Microbiology 147, 2255–2264 (2001). [DOI] [PubMed] [Google Scholar]
- Gostinčar C. et al. Expression of fatty-acid-modifying enzymes in the halotolerant black yeast Aureobasidiu pullulans (de Bary) G. Arnaud under salt stress. Stud. Mycol. 61, 51–59 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. W. et al. Characterization of a rice (Oryza sativa L.) gene encoding a temperature-dependent chloroplast omega-3 fatty acid desaturase. Biochem. Biophys. Res. Commun. 340, 1209–1216 (2006). [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan-Nair P. M., Kang I. S., Moon B. Y. & Lee C. H. Effects of low temperature stress on rice (Oryza sativa L.) plastid ω-3 desaturase gene, OsFAD8 and its functional analysis using T-DNA mutants. Plant Cell Tiss. Org. 98, 87–96 (2009). [Google Scholar]
- Li-Beisson Y. H. et al. Acyl-Lipid Metabolism. The Arabidopsis Book e0133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T. & Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjellqvist B., Basse B., Olsen E. & Celis J. E. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 15, 529–539 (1994). [DOI] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. et al. Genome-wide identification and expression profiling of ankyrin-repeat gene family in maize. Dev. Genes Evol. 223, 303–318 (2013). [DOI] [PubMed] [Google Scholar]
- Singh V. K. & Jain M. Genome-wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean. Front. Plant Sci. 6, 918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Saeed A. I. et al. TM4: A Free, Open-Source System for Microarray Data Management and Analysis. Microarray Technologies 34, 374–378 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.