Abstract
Background and Aims Root border cells and border-like cells (BLCs), the latter originally described in Arabidopsis thaliana, have been described as cells released at the root tips of the species in which they occur. BLCs are thought to provide protection to root meristems similar to classical root border cells. In addition, four defensin peptides (Hc-AFP1–4) have previously been characterized from Heliophila coronopifolia, a South African semi-desert flower, and found to be strongly antifungal. This provided an opportunity to evaluate if the BLCs of H. coronopifolia indeed produce these defensins, which would provide evidence towards a defence role for BLCs.
Methods Fluorescence microscopy, using live-cell-imaging technology, was used to characterize the BLCs of H. coronopifolia. Quantitative real-time PCR (qRT-PCR) analysis and immunofluorescence microscopy was used to characterize these defensin peptides.
Key Results BLCs originated at the root apical meristem and formed a protective sheath at the tip and along the sides as the root elongated in solid medium. BLCs have a cellulose-enriched cell wall, intact nuclei and are embedded in a layer of pectin-rich mucilage. Pectinase treatments led to the dissolution of the sheath and dissociation of the root BLCs. Hc-AFP1–4 genes were all expressed in root tissues, but Hc-AFP3 transcripts were the most abundant in these tissues as measured by qRT-PCR. A polyclonal antibody that was cross-reactive with all four defensins, and probably recognizing a general plant defensin epitope, was used in fluorescence microscopy analysis to examine the presence of the peptides in the root tip and BLCs. Data confirmed the peptides present in the root tip tissues, the mucilage sheath and the BLCs.
Conclusions This study provides a link between defensin peptides and BLCs, both embedded in a protective pectin mucilage sheath, during normal plant growth and development. The presence of the Hc-AFP3 defensin peptides in the BLCs suggests a role for these cells in root protection.
Keywords: Brassicaceae, Heliophila coronopifolia, root border-like cells, pectin-rich mucilage, defensins, pectinase, Hc-AFP
INTRODUCTION
The root apical meristem facilitates the elongation of growing roots for soil anchorage, nutrient assimilation and water absorption in plants (Raven et al., 2013). The region of the root tip, near the apical meristem, produces the root cap, consisting of peripheral root cells and mucilage. This region is crucial in the detection of external stimuli (i.e. water, nutrients, pathogens, etc.) and helps to guide the growing root (Hawes et al., 2003; Iijima et al., 2003; Haichar et al., 2014). During these events, the root body produces numerous cells, in a dynamic process that appears to be linked to plant growth (Hawes and Pueppke, 1986; Wen et al., 2014). These cells, termed border cells, function by acting as a protective sheath (consisting of cells embedded in mucilage) around the root tip (Hawes and Lin, 1990). Border cells are defined as individually released cells, produced at the root periphery, which disperse in suspension after a few seconds when the root tip is placed in water (Hawes et al., 1998). These border cells have been shown to be viable and metabolically active (Hawes and Pueppke, 1986; Hawes and Brigham, 1992) and express a unique pattern of mRNA transcripts and proteins as compared with those produced by the root tip (Brigham et al., 1995). Border cells secrete abundant mucilage which is composed of a diverse mixture of proteins, polysaccharides, secondary metabolites and extracellular DNA which is believed to retard and entrap pathogens, thereby preventing root infection (Hawes and Brigham, 1992; Wen et al., 2007, 2009; Driouich et al., 2013). These cells have also been shown to protect plants from abiotic stress, specifically aluminium (and other metals) toxicity (Miyasaka and Hawes, 2001; Kopittke et al., 2011, 2012) and are believed to mediate their protective functions by modifying the immediate environment around the root tip (Brigham et al., 1995; Gunawardena et al., 2005). The release of root border cells, embedded in mucilage, is believed to act as a lubricating layer which supports root growth in the soil (Hawes et al., 2003). Border cells are also believed to provide an interface between the broader plant–root–rhizosphere environment as has been observed in Oryza sativa, Pisum sativum and Nicotiana tabacum (Hawes et al., 2003).
Work on border cells has been conducted with a focus on agricultural applications, but as the model plant Arabidopsis thaliana and most other Brassicaceae species lack classical border cells (Hawes et al., 1998) this has complicated progress in this area. However, more recent studies have demonstrated that many Brassicaceae species produce rows of linearly attached cells at the extremity of the root tip (Vicré et al., 2005; Driouich et al., 2007). These ‘rows of cells’, by analogy, have been considered similar from a functional perspective to classical border cells and have thus been termed ‘border-like cells’ (BLCs) (Vicré et al., 2005). In A. thaliana it has been shown that these cells are viable and present the characteristics of metabolically active cells, but probably secrete much less mucilage and remain attached to the root for much longer than border cells (Vicré et al., 2005). It has also been shown that homogalacturonan polymers are responsible for the attachment of BLCs to each other, whereas homogalacturonan-deficient mutants produce individual ‘BLCs’ which stay trapped around the root tip by releasing copious amounts of mucilage, similarly to the well-characterized border cells (Driouich et al., 2007). It has recently been proposed that BLCs function similarly to border cells (Driouich et al., 2010, 2013), but supporting information is required to confirm this. As border cells have been proposed to function in plant defence, and specifically in protecting the root tip (Hawes et al., 2000), it is highly probable that BLCs act in the same manner. Interestingly, arabinogalactan proteins have been found in the mucilage secreted by both border cells and BLCs (Cannesan et al., 2012; Vicré et al., 2005) and have been shown to inhibit the germination of zoospores of the oomycete Aphanomyces euteiches (Cannesan et al., 2012; Driouich et al., 2013). A recent study on BLCs from A. thaliana and flax has also shown that BLCs have the capacity to trigger specific defence responses after elicitation with flagelin 22 (Plancot et al., 2013). Hence studying border cells and BLCs in novel species offers a means to understand specific defence responses to biotic and abiotic stresses in soil rhizosphere environments.
The family Brassicaceae (mustard), which includes A. thaliana and several important crop species such as cabbage, canola and mustard, remains relatively unexplored when considering the significant species, genetic and physiological diversity (Kiefer et al., 2014). In addition to A. thaliana as a model system for general plant biology, other tribes, genera and species of the mustard family are being studied as models in evolutionary biology (Kagale et al., 2014; Kiefer et al., 2014). The tribe Heliophileae consists of approx. 90 Heliophila species, which are all endemic to specific areas of southern Africa (Mandáková et al., 2012).
In this study, we examined whether H. coronopifolia, a Brassicaceae species endemic to South Africa (De Beer and Vivier, 2011) produces BLCs. The root tip area and its associated BLCs were characterized under various culture conditions, and shown to indeed produce BLCs. This species is of interest as four defensin peptide-encoding genes (Hc-AFP1–4) have been isolated and characterized from it; the encoded peptides were confirmed to have strong antifungal activities (De Beer, 2008; De Beer and Vivier, 2011). The aim was therefore to characterize the development and turnover of the BLCs in this species and to examine whether these cells produce the Hc-AFP defensins. A quantitative real-time PCR (qRT-PCR) analysis confirmed that Hc-AFP-1–4 transcripts were expressed in root tissues and an immunofluorescence protocol optimized for H. coronopifolia roots revealed that the root tip, the mucilage and the BLCs contain defensins, providing evidence that BLCs and the secreted mucilage contribute to a protective role in the sensitive root tip zone.
MATERIALS AND METHODS
Plant growth and tissue culture conditions
Seeds of H. coronopifolia were purchased from Silverhill Seeds (Kenilworth, Cape Town, South Africa). After 2 weeks of vernalization at 4 °C, seeds were surface sterilized (Balcells, 1991). Briefly, seeds were washed for 2 min in 70 % (v/v) aqueous ethanol, followed by 5 min immersion in a solution of calcium hypochlorite (5 %, w/v) and sodium dodecyl sulphate (SDS; 0–5 %, w/v), followed by a series of rinses (3×) in sterile distilled water. Sterilized seeds were transferred to rectangular Petri plates containing MS media (Murashige and Skoog, 1962) solidified with phytagel (0–3 %, w/v). Plates were sealed with parafilm and positioned vertically (using a support) to encourage roots to grow downwards along the surface of the phytagel and not penetrate the MS–phytagel matrix. To observe the root tips growing in liquid media, 5-d-old seedlings were transferred to a specialized test tube (custom made by the Department of Chemistry, Stellenbosch) with a narrower section at the base to maintain seedlings upright while allowing the root tip and body to remain in contact with the liquid MS. All seedlings were grown in a controlled plant tissue culture growth room (16-h light/8-h dark cycle, 23 °C). The root tips and associated cells were used to confirm the presence of BLCs in this species and thereafter to follow the development and characteristics of the BLCs under controlled conditions. It was found that the best period to observe root BLCs was 5–10 d following germination and, unless stated otherwise, all further experiments used root tips from seedlings in this period of development.
Microscopy
The appearance and formation of root growth, root cap formation and root BLCs were evaluated using live-cell-imaging video microscopy with an Olympus Cell-R system coupled to an Olympus IX 81 inverse fluorescence microscope equipped with an F-view-II cooled CCD camera (Soft Imaging System, Berlin, Germany). The system is illuminated using a xenon lamp (Olympus Biosystems, Tokyo, Japan), and uses excitation filters at selected wavelengths (λ 360, 472 and 572 nm). Emission spectra were collected using a ‘UBG triple-band pass emission filter cube’ (Chroma, Bellows Falls, VT, USA). Images were z-stacked using Cell-R imaging software, with each set of images optimized for exposure and background subtraction, and simultaneously captured/compiled for signal processing. Parameters for Syto-9 staining (Krause et al., 2008) were as follows: λex = 485 nm and λem = 498 nm, filtered at 472 nm which measures DNA intercalation (at a concentration of 8.35 μm). Parameters for calcofluor white staining (Monheit et al., 1984) were λex = 355 nm and λem of between 300 and 440 nm with an excitation filter of 360 nm after activation using KOH. Calcofluor white is used for cell morphology and labelling of cell wall beta-glucans (Monheit et al., 1984). Staining with calcofluor white was done as described by Andème-Onzighi et al. (2002). For the video microscopy study evaluating the influence of different growth and media conditions, seedlings were either imaged directly on MS agar plates, or transferred to glass slides and covered with liquid MS. An image was acquired every 15 min over a 24-h period for growth on solid media, whereas the assay continued for 36 h when liquid media was tested (see Supplementary Data, Videos S1 and S2).
RNA extraction
For each biological repeat, about 50 roots (last 5 cm with the root tip) of 2-week-old H. coronopifolia were collected from culture plates and ground to a fine powder under liquid nitrogen using a mortar and pestle, under RNase-free conditions. Two spatula scoops of ground powder (approx. 100 mg) were placed in an Eppendorf tube containing 300 μL phenol (Tris-buffered pH 8·0) and 700 μL of an extraction buffer composed of Tris/HCl (0·1 m, pH 8·0), SDS (1·5 %, w/v), LiCl (300 mm), Na2-EDTA (10 mm), Na-deoxycholate (1 %, w/v), Nonidet P-40 (1 %, w/v), thiourea (5 mm) and Na-metabisulfite (1 %, w/v). The RNA extraction protocol was adapted from Joubert et al. (2013) (for tobacco, including two chloroform precipitation steps and using ethanol instead of isopropanol). RNA integrity was confirmed using agarose gel electrophoresis [1·6 % (w/v) agarose, 1× gel red) in formaldehyde buffer (120 V, 45 min). A ‘RiboRuler High Range RNA ladder’ (Fermentas, Fitchburg, WI, USA) was used for molecular weight determination.
Preparation of cDNA from RNA
RNA was treated with recombinant DNase I (RNAse free kit, Roche, Mannheim, Germany) in 1× buffer, to digest all genomic DNA present. The reaction consists of 20 μL of purified nucleic acid, buffer, enzyme and DEPC-treated water [1 % (v/v) DEPC added to milliQ water, incubated at 37 °C overnight and autoclaved before use] in a final volume of 100 μL for 2 h at 37 °C. A PCR was used to verify the complete digestion of genomic DNA: the reaction involved 0·2 μL of GoTaq (Taq DNA polymerase) (Promega), 2 mm MgCl2, 200 μm dNTPs, 1× Green GoTaq Flexi buffer, 250 nm psbA primers (see Supplementary Data, Table S1 for all primer details) and DEPC-dH2O. The reaction programme using a Biometric thermocycler (Applied Biosystems, Foster City, CA, USA) was : 94 °C, 5 min; 40 cycles of 94 °C, 30 s, 50 °C, 30 s and 72 °C, 2 min); 72 °C, 7 min. Verification of the PCR was performed using agarose gel electrophoresis and molecular weights were determined using a FastRule Low Range DNA Ladder. The cDNA synthesis was performed using DNA-free RNA and the ‘SuperScript III Platinum’ kit (Two-Step qRT-PCR Kit) (Invitrogen Life Technologies, Carlsbad, CA, USA) following the manufacturer’s instructions.
qRT-PCR
All primers used are reported in Table S1. Analyses were performed from cDNA isolated from root tissue (this study) as well as cDNA produced and evaluated previously from aerial organs (De Beer, 2008; De Beer and Vivier, 2011) as a positive control. The constitutively expressed elongation factor encoding gene (EF1α) was used as an internal control (‘housekeeping’), whereas λ DNA (Roche) and its specific primers (Table S1) was included for normalization according to the linear regression efficiency (LRE) method (Rutledge, 2011). qRT-PCR was performed using four biological repeats of the samples: Tissue powder from each biological repeat was split into three different tubes (technical repeats) for each extraction; when cDNA quality was confirmed, the three technical repeats were pooled and the qRT-PCR was conducted in triplicate for each sample. qRT-PCR was performed with the Kapa Sybr Fast qPCR kit (Kapa Biosystems, Cape Town, South Africa). The reactions were performed in a ABI7500 Real-Time PCR (Applied Biosystems) system using the following programme: 95 °C, 3 min; 40 cycles of 95 °C, 15 s, 60 °C, 32 s, and a dissociation curve from 60 to 95 °C. Data from the proprietary software 7500 System Software were exported to Microsoft Excel and quantification of LRE was done using LRE Analyzer ver. 0.8.7 (Rutledge, 2011). The LRE Analyzer software normalized the data with λ DNA and expressed the result as a number of target molecules, which was converted into a percentage when the data were tabulated. The different biological repeats were analysed independently and normalized using the internal control EF1α. For statistical purposes the data represent the standard deviation of the mean for three biological replicates with three technical repeats.
Immunofluorescent labelling of defensins using a polyclonal anti-Hc-AFP2 antibody
An Hc-AFP2 polyclonal primary antibody (obtained and tested by Prof. A. Poole, University of the Western Cape, South Africa) was obtained from antiserum produced in mice from purified Hc-AFP2 peptides (De Beer, 2008; Barkhuizen, 2013). The Hc-AFP2 peptide was previously produced using an Escherichia coli-based peptide expression system, producing a peptide of approx. 5·5 kDa, and then functionally characterized (Kraemer et al., 2011; Barkhuizen, 2013). The Hc-AFP2 polyclonal antibody was shown to cross-react against all four defensins (Hc-AFP1–4), as well as VvAMP1, an antimicrobial peptide from grapevine (Vitis vinifera) (De Beer and Vivier, 2008) by using Western and dot blotting (see Supplementary Data). Pre-bleed serum was similarly tested using dot blotting (De Beer, 2008; Barkhuizen, 2013). Given that the Hc-AFP2 polyclonal primary antibody does cross-react with other defensins, it is likely that the epitope is conserved among these peptides: such as the α-helix plus three β-sheets common tertiary motif (Fant et al., 1998). Consequently, the use of this antibody serum in labelling experiments must be used in conjunction with qRT-PCR to evaluate peptide expression. This polyclonal antibody was used in subsequent immunocytochemistry protocols. The immunofluorescence protocol was evaluated for several parameters before an optimal set of conditions was found for roots of H. coronopifolia. The optimized method was as follows. Seedling roots (5–10 d post-germination) were transferred onto poly-lysine- (0·02 g poly-l-lysine in 396 mL water; 4 mL 1 m Tris pH 8) coated slides. After 5 min to allow roots to adhere to the slides, blocking buffer [skimmed milk in 0·1 m phosphate-buffered saline (PBS)] was added to the immobilized roots for 30 min. The polyclonal primary antibody, in the presence of pectinase (1 : 20) was incubated overnight for 12 h at 4 °C (see Supplementary Data Table S2 for details of the primary antibodies used). After draining off the excess antibody solution and gentle rinsing of the samples with 1·5 mL PBS, the anti-mouse secondary antibody was added and incubated for 30 min at room temperature (23 °C) in the dark. Gentle rinsing in 0·1 m PBS (3×) was used to remove/reduce non-specific binding of secondary antibodies.
On average 16 roots were used for analyses and the experiments were independently replicated at least four times. Representative images are reported after visual inspection from the independent experiments. Images were post-treated with background subtraction. The background subtraction value was defined by removing any signal from controls (PBS with and without pectinase in place of the primary antibody) (see Supplementary Data Fig. S1). Thirty roots were used to define the background subtraction value and all subsequent experiments were conducted with three negative control roots to cross-check whether this value remained applicable for the background subtraction. An additional control using a mammalian primary antibody (non-plant negative control), p-p38 (D-8) (sc-7973, Santa Cruz Biotechnology, Dallas, TX, USA), which binds phosphorylated Tyr 182 of mouse, rat and human origin was used.
RESULTS
Formation of root border-like cells in Heliophila coronopifolia
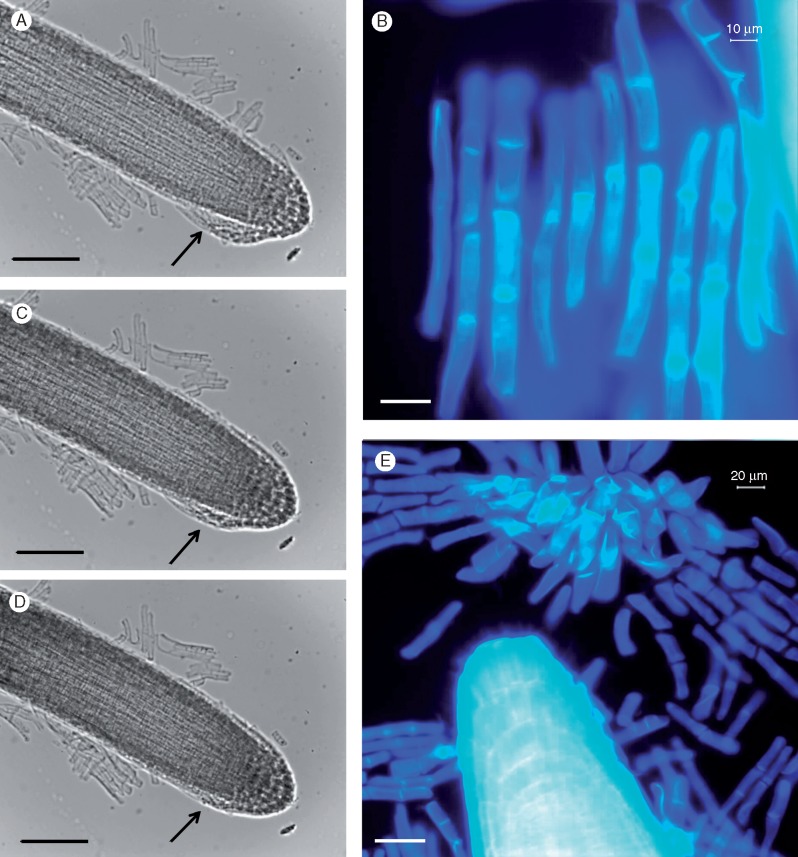
Microscopic observation of the root tip, stained with calcofluor white, revealed detached rows of root cells which are characteristic of BLCs found in members of the Brassicaceae including A. thaliana (Fig. 1A, B). These cells were further investigated and followed using fluorescence microscopy coupled with live-cell-imaging ‘video microscopy’ technology. BLCs were first noticeable (using microscopy) in 3-d-old seedlings where the root had grown approx. 5 mm in length. The formation of root BLCs are shown in Fig. 1(C–E) (images extracted from live-cell video microscopy analyses performed over a 36-h period). The full movie videos are provided in the Supplementary Data as Videos S1 and S2. These images are representative of an extensive set of observations from about 30 roots per experiment (three biological experiments were performed comprising approx. 100 plants in total). The BLCs appear to originate from cell layers present at the root tip and appeared as ‘organized rows’ of cells, which successively break away from the root (Fig. 1C–E). The BLCs continue to remain attached to each other and within the vicinity of the root cap. This is typically observed for BLCs, such as in A. thaliana, in contrast to what is seen in root border cells (Driouich et al., 2010).
Fig. 1.
Root tip and detached border-like cells (BLCs) stained with calcofluor white which stains cellulose-enriched cell walls (A) and BLCs at higher magnification (B). The appearance and formation of border-like cells (BLCs) (C–E) at the root tip of Heliophila coronopifolia viewed under phase contrast microscopy (frames C–E are at 15-min intervals). The arrows indicate attached BLCs (micrographs extracted from a live-cell imaging experiment, see Video S1). Scale bars: 40 μm (A), 20 μm (B), 200 μm (C–E).
Following the confirmation that H. coronopifolia produces BLCs, the characteristics of the root tip cells, the root cap and the BLCs were further investigated using various stains visible with fluorescence microscopy. The root tip cells stained with calcofluor white (Fig. 1A, B) (which indicates the presence of β-glucans including cellulose) and Syto-9 (Fig. 2A) (which binds RNA and DNA revealing nuclei). The cells constituting the root cap (including the BLCs) were also stained with calcofluor white (Fig. 1B) and Syto-9 (Fig. 2). The presence of intact nuclei of BLCs was also assessed with Syto-9 24 h after their separation from the root tip (Fig. 2A, B) staining. A mucilaginous matrix was faintly visible (Fig. 2A) surrounding the root tip and associated BLCs. The root cap with BLCs was observed to detach from the root tip (Fig. 1A, B), suggesting that these cells (i.e. BLCs) are extensions of the root cap. Furthermore, BLCs showed a similar pattern of growth to those cells formed at the root meristem, i.e. they elongate after formation. At the root meristem and cap they have a round shape and then grow in length to acquire an elongated mature form (Fig. 1A, B). The live-cell-imaging study (see Videos S1 and S2) did not show the BLCs undergoing cell division during the 24–36 h of recording observations.
Fig. 2.
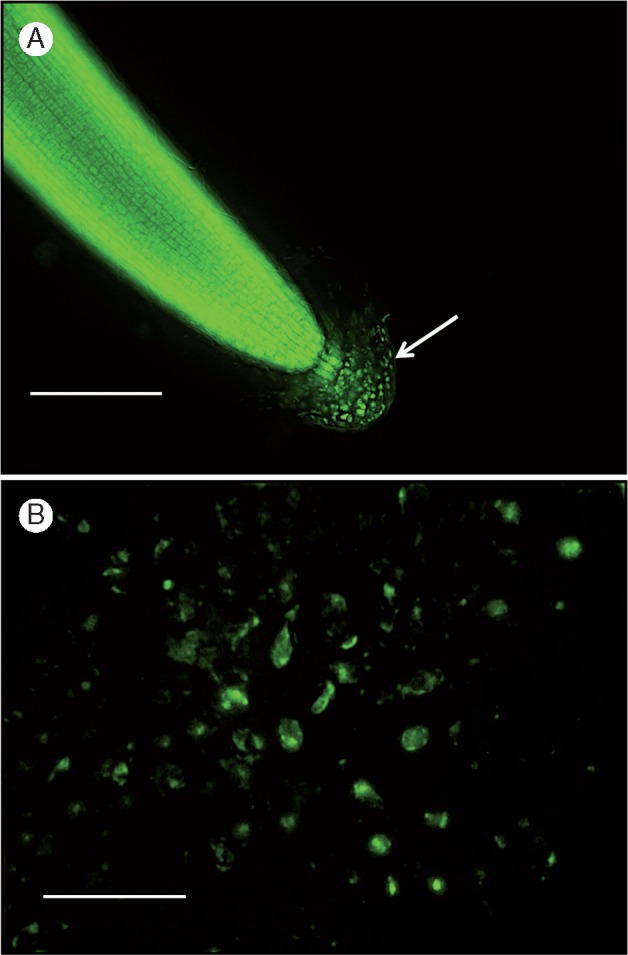
Characterization of root border-like cells (BLCs) of Heliophila coronopifolia using the fluorescent dye Syto-9, which binds to cellular RNA and DNA. (A) shows root cap and BLCs. (B) shows BLCs. Arrows indicate the root cap with the BLCs. Scale bars: 200 μm (A), 800 μm (B).
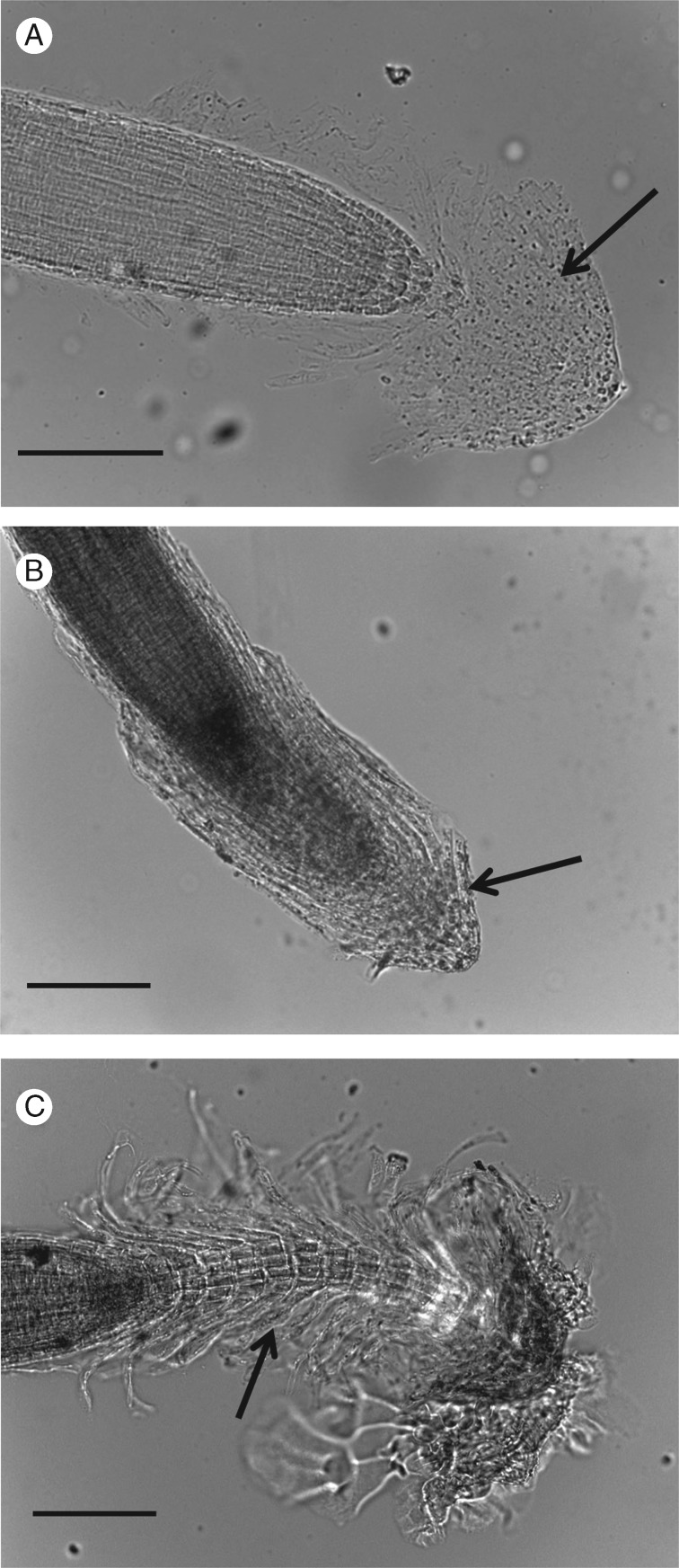
Influence of root environment on BLCs and root cap organization
To evaluate if the root environment influenced the morphology and general appearance of the cells at the root tip (and associated BLCs) seedlings of H. coronopifolia were grown and observed in two different growth media (Fig. 3). When roots of H. coronopifolia were placed in a liquid environment (i.e. liquid MS media), the root cap (and associated BLCs) detached from the root tip and mucilage (Fig. 3A). The BLCs were found to spread around the root tip and cap (Fig. 3B). The root cap with associated BLCs was found to extend into the watery milieu surrounded by a faintly visible mucilage matrix (Fig. 3A). Interestingly, the roots placed in liquid media did not appear to elongate, although BLCs were still formed. In contrast, when H. coronopifolia seedlings were placed in solid MS media, the root cap and BLCs remained closely associated with the main root and formed a protective sheath that surrounded the growing tip (Fig. 3B). Moreover, an interesting observation was made in older plants where the roots were left to grow for more than 15 d (Fig. 3C). These show a stacking of root caps (and associated BLCs) that formed as the roots grew (Fig. 3C). These structures remained attached to each other and suggest that the root cap structure and associated BLCs are continuously regenerated in the growing root (Fig. 3C).
Fig. 3.
The influence of growth conditions on the morphology of the root cap and associated border-like cells (BLCs) of Heliophila coronopifolia. Images of the root tip (and BLCs) of H. coronopifolia growing in liquid MS media (A, B) and solid MS media (C). Images of the root tip (and BLCs) of H. coronopifolia after 15 d of growth in solid MS media (C). Arrows show BLCs. Scale bars: 200 μm.
A pectinase treatment disrupted the mucilage sheath
The composition of the observed mucilage that encapsulates the BLCs of H. coronopifolia was suspected to be pectin. As the root mucilage of the model Brassicaceae species, A. thaliana, was found to be enriched in pectin (particularly in homogalacturonan and xylogalacturonans) (Durand et al., 2009), a series of pectinase treatments was performed in an attempt to improve the accessibility of the BLCs to antibody probes. Seedling roots were incubated in a PBS solution containing pectinase at 1 : 20 aqueous dilution followed by calcofluor white staining (see Supplementary Data Fig. S2). The pectinase treatment disrupted the mucilage matrix in this species. In comparison to the control assay where pectinase was omitted (Fig. S2a), a high concentration of pectinase (1 : 2 dilution: 14.25 U) resulted in complete disorganization of the root cap and disruption of the root cells (Fig. S2b), suggesting pectins are important for tissue cohesion. A 1 : 20 dilution of the pectinase (1·425 U) was found to degrade the mucilage sheath without affecting the morphology of the root tip and BLCs (Fig. S2c), whereas lower concentrations of pectinase (0·57 and 0·285 U) (Fig. S2d) did not significantly affect the integrity of the mucilage matrix.
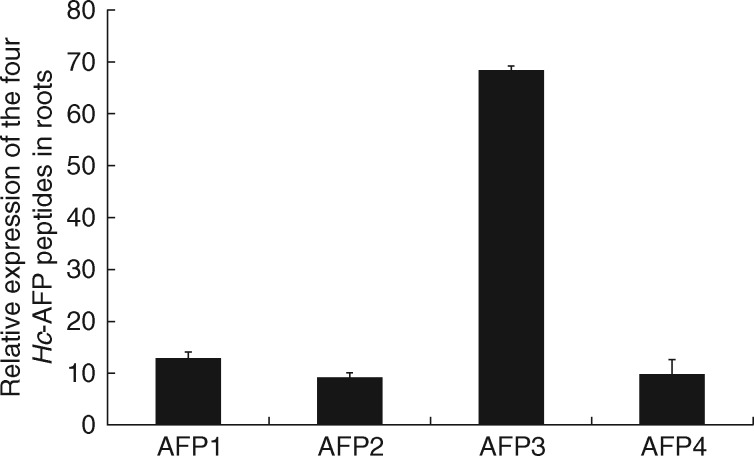
Expression of defensins in the roots and root BLC of H. coronopifolia
A qRT-PCR analysis confirmed that all four Heliophilia antifungal peptides, Hc-AFP1–4 (De Beer and Vivier, 2011), were expressed in the root tissues. Three of the peptide encoding genes showed basal levels of relative expression (Hc-AFP1 with 13 %, Hc-AFP2 with 9 % and Hc-AFP4 with 10 % expression), whereas the Hc-AFP3 transcripts were the most abundant (68 % of the relative expression) in the root tissues (Fig. 4).
Fig. 4.
Relative gene expression of the Hc-AFP1–4 transcripts in the root tip and associated border-like cells (BLCs) of Heliophila coronopifolia measured using qRT-PCR. Expression was confirmed using the LRE method (Rutledge, 2011) as well as the housekeeping gene Hc-EF1α (De Beer and Vivier, 2011; Pabinger et al., 2014). Data represent the mean and standard deviation for three independent biological repeats performed in triplicate.
Determining the presence and distribution of Hc-AFP peptides in living roots of H. coronopifolia using immunofluorescence microscopy
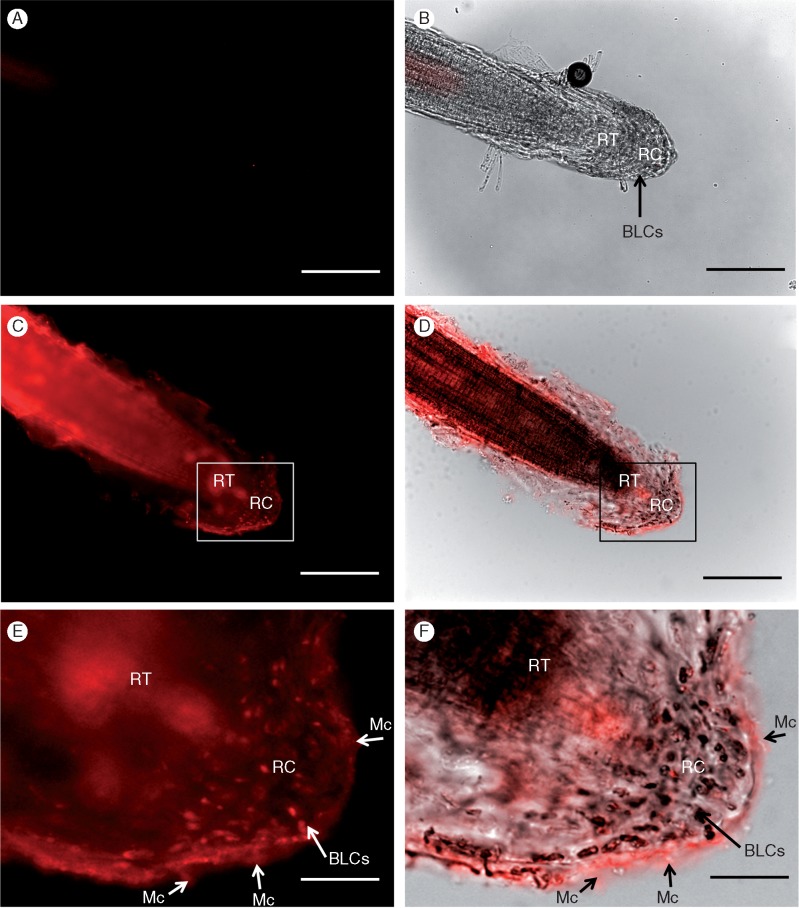
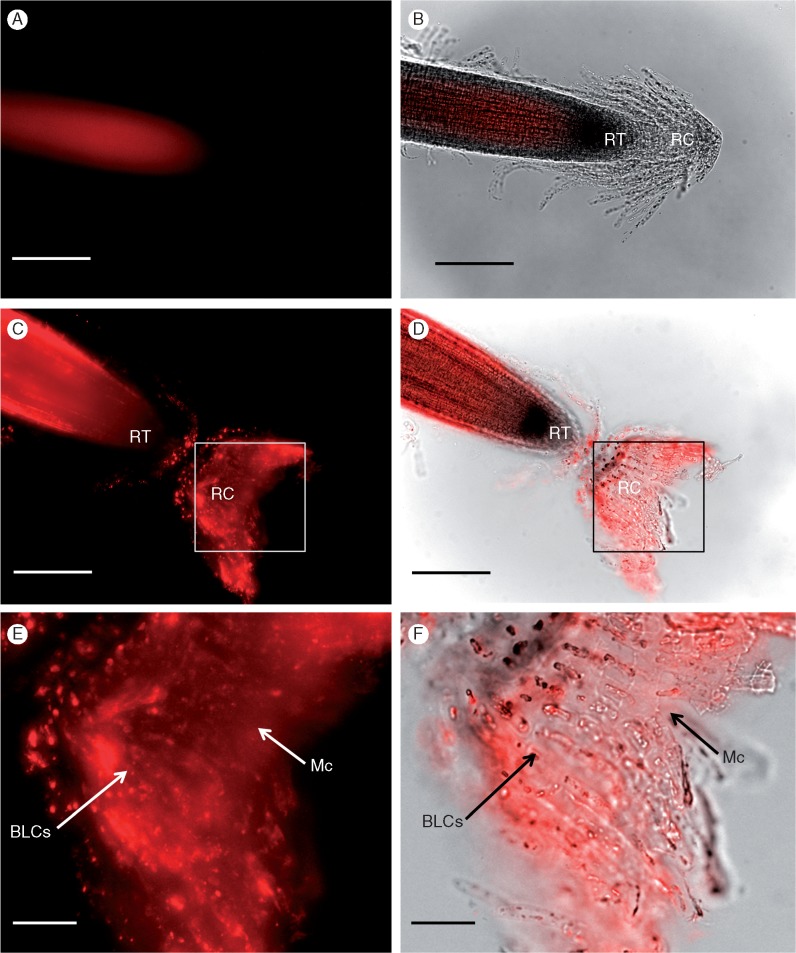
The treatment of H. coronopifolia roots with primary anti-Hc-AFP2 antibodies produced signal throughout the root tip, the mucilage matrix and the BLCs (Fig. 5). The control roots (where the primary antibody is replaced with PBS or with a mammalian primary antibody, i.e. a non-plant-negative control) showed no signal after background subtraction (Supplementary Data Fig. S1a-b and Fig. 5A, B). anti-Hc-AFP2 polyclonal antibody signal was prominent in the root body itself (Fig. 5C), which was confirmed using an overlay of fluorescence with phase contrast imaging (Fig. 5D). Signal was visible at the outer layers of the root and the mucilage surrounding the root cap (Fig. 5C–E). Higher magnification images of the matrix confirmed binding of anti-Hc-AFP2 antibodies to the cells therein (Fig. 5E, F). To improve access of the primary polyclonal antibody to the Hc-AFP peptides in the BLCs, the procedure was repeated after pectinase treatment. As previously stated, a 1 : 20 dilution (1·425 U) of pectinase was found to maximize mucilage dissolution while minimizing any damage of the root cap integrity (Fig. S2). As seen in Fig. 5(C, D), a pectinase treatment coupled with gentle rinsing caused an inversion of the root cap exposing the underlying– adjacent BLCs. Clear signals were now obtained at the root tip (as in Fig. 6), as well as from the BLCs themselves, further confirming cell labelling. Higher magnification images of the BLCs and root cap (Fig. 6E, F) confirmed a signal in the mucilage (Fig. 6E) and with the associated BLCs (Fig. 6F). Similarly, a signal associated with cellular structures was seen along the length of the root surface (Fig. 6C, D) in addition to the BLCs that elongate along the sides of the tip. These data confirmed the presence of Hc-AFP defensins in the root tip, mucilage and root BLCs of H. coronopifolia.
Fig. 5.
Immunolabelling of the root cap and associated border-like cells (BLCs) of Heliophila coronopifolia with the anti-Hc-AFP2 polyclonal antibodies using a fluorescent secondary antibody (without pectinase treatment). (A, B) Controls with a mammalian primary antibody (non-plant negative control) (p-p38, see Table S2 for details on antibodies). Images with anti-Hc-AFP2 label; (C, D) the root tip with its BLCs and images, and (E, F) BLCs at higher magnification. Images with immunofluorescence mode (A, C, E), images with overlayed transmission and immunofluorescence mode (B, D, F). Arrows show specific labelling with primary antibody. RT, root tip; RC, root cap; BLCs, border-like cells; Mc, mucilage. Scale bars: 200 μm (A–D), 400 μm (E, F).
Fig. 6.
Immunolabelling of the root cap and associated border-like cells (BLCs) of Heliophila coronopifolia with the anti-Hc-AFP2 polyclonal antibody using a fluorescent secondary antibody, after pre-treatment with pectinase. (A, B) Controls with a mammalian primary antibody (non-plant negative control) (p-p38, see Table S2 for details on antibodies). (C, D) The root tip with its BLCs and (E, F) BLCs at higher magnification. Images with immunofluorescence mode (A, C, E), and images with overlayed transmission and immunofluorescence mode (B, D, F). Arrows show specific labelling with primary antibody. RT, root tip; RC, root cap; BLCs, border-like cells; Mc, mucilage. Scale bars: 200 μm (A–D), 400 μm (E, F).
DISCUSSION
Four defensin peptides (Hc-AFP1–4) have been isolated from and characterized in H. coronopifolia (De Beer and Vivier, 2011). Defensins occur naturally in a variety of plant species, are usually constitutively expressed and are commonly associated with antifungal activity (Broekaert et al., 1995). They are small (approx. 5 kDa), basic peptides that possess stabilizing cysteine bonds (CSCSαβ) and have a tertiary structure composed of an α-helix and three antiparallel β-sheets (Fant et al., 1998). Although the main functions of defensins are believed to be antimicrobial, other interesting roles, such as providing tolerance to Zn (Hawes et al., 1998; Shahzad et al., 2013), have been uncovered. The Hc-AFP1–4 peptides have strong antifungal activities against the necrotrophic fungus Botrytis cinerea as well as Fusarium solani, a soil-borne fungal pathogen (De Beer and Vivier, 2011). Antifungal activities of the Hc-AFP1–4 peptides have included hyphal tip swelling, hyper-branching, and disruption and lysis of the fungal membranes through permeabilization (De Beer and Vivier, 2011). The Hc-AFP1–4 genes have previously been shown to be differentially expressed between vegetative and reproductive organs of H. coronopifolia (De Beer and Vivier, 2011). This study confirms that although all four peptide-encoding genes are expressed in root cells, Hc-AFP3 transcripts were the most abundantly expressed.
Furthermore H. coronopifolia produces BLCs, similar to other Arabidopsis and some other Brassicaceae species, providing motivation to investigate whether these specialized cells produce Hc-AFP1–4 peptides. The results obtained suggest that root BLCs act both in a mechanical protectant role at the root tip, as well as producing compounds that can participate in defence against pathogens, as outlined below.
The behaviour of the root BLCs of H. coronopifolia in different growth media suggests a mechanically protective role for the growing root tip
Heliophila coronopifolia’s BLCs have been shown to form at the root tip and then elongate to form organized stacks of cells embedded in mucilage. A pectin-rich mucilage matrix was also observed in A. thaliana, Pisum sativum and Brassica napus (Durand et al., 2009; Cannesan et al., 2012). The BLCs of A. thaliana possess significant numbers of mitochondria, endoplasmic reticula, Golgi stacks and secretory vesicles visible under electron microscopy (Vicré et al., 2005) and so the mucilage is therefore considered to be at least partly produced from these cells. In H. coronopifolia grown in liquid media the BLCs were not so obviously stacked together and appeared loosely attached to the matrix and root cap, whereas the roots grown in solidified media had a more organized structure with the BLCs remaining attached to each other and covering the growing root tips. These differences in growth morphology could be due to the fact that the mucilage dissolves more easily in liquid than solid media, as is shown to occur in the many species that produce root border cells (Hawes et al., 2000, 2003). It is also possible that less mucilage is formed under these laboratory culture conditions, as the roots are not in an environment that produces mechanical stress on the root tip. Interestingly, Iijima et al. (2003) showed that in maize the number of border cells produced was related to the compactness of the soil. The observations made on the solid growth medium confirm that the root cap, associated BLCs and mucilage thus also probably function in lubrication, protection and the regeneration processes of growing roots in H. coronopifolia as suggested by Driouich et al. (2010) for other species. In H. coronopifolia older roots showed stacking of root caps with BLCs that formed and remained attached to the growing root tip. The mechanical shearing forces in the gel medium are probably not severe enough to cause the structures to become dislodged as would most likely occur in soil conditions. Published data show that these cells have the capacity to remain intact and viable even after separation from the root tip (Vicré et al., 2005; Cannesan et al., 2012; Plancot et al., 2013); this is also believed to occur in H. coronopifolia.
Defensin peptides are produced in the root tip and BLCs of H. coronopifolia
A number of studies have advanced understanding of how border cells protect the root apex (Vicré et al., 2005; Cannesan et al., 2012; Plancot et al., 2013). Root border cells secrete mucilage which provides protection against biotic and abiotic stress (Hawes et al., 2000; Miyasaka and Hawes, 2001). A thorough review on the role of root exudates in plant defence is provided by Baetz and Martinoia (2014). Although there are only a few studies focused on BLCs in this regard, they all strongly suggest that these cells are crucial in protecting the root tip (Cannesan et al., 2012; Plancot et al., 2013). The root cap and associated BLCs were reported to be able to entrap and inhibit germination of oomycete zoospores (Cannesan et al., 2012). The role of arabinogalactan proteins isolated from the BLC mucilage was shown to play an important role in that inhibitory process (Cannesan et al., 2012). BLCs from Linum sativum were shown to respond to elicitation, providing support for the idea that these cells participate in and respond by supporting induced plant defence (Plancot et al., 2013). In the case of H. coronopifolia the combined immunocytochemical and gene expression analysis showed that the root tip and its associated BLCs produce and probably secrete antifungal peptides. Given that A. thaliana is believed to harbour over 300 putative defensins (De Coninck et al., 2013) and a full understanding of the plant peptidome (Tavormina et al., 2015) is still very much ongoing, it is certainly possible that H. coronopifolia contains many more defensins than have hitherto been isolated. As defensins are considered to be part of the innate defence of plants, it is logical to suggest that BLCs also participate in providing a protective environment even before pathogens are sensed and a response is activated. In this study, immunomicroscopy combined with gene expression confirmed the presence of defensin peptides in the root tissues, the BLCs, as well as the mucilage (Figs 4 and 5); this is in line with the known apoplastic localization of these peptides (De Beer, 2008). Given these data, it is also likely that only one peptide, Hc-AFP3, is produced in roots, given the basal expression levels found for the other three defensins. These peptides (present in the mucilage) could therefore be part of the root extracellular trap described by Driouich et al. (2013) functioning to protect roots against pathogens. It has similarly been shown in mammalian neutrophils where defensins were found in the extracellular matrix controlling the microbial trapping activities (Saitoh et al., 2012; Nguyen et al., 2014). Given the known antifungal roles and characteristics previously described for these peptides, their presence in BLCs also provides support for their role in preformed defence. It is also interesting to note that blast sequence analysis confirmed that Hc-AFP4 had high homology (89 %) to the A. halleri defensin PDF1.1 (Mirouze et al., 2006). It has recently been demonstrated that PDF1.1 plays an important role in tolerance to zinc toxicity (Shahzad et al., 2013; Nguyen et al., 2014; Wen et al., 2014). Given that border cells have also been shown to be involved in aluminium tolerance (Miyasaka and Hawes, 2001; Zhu et al., 2003), as well as tolerance to arsenic and other metals (i.e. zinc, copper, nickel) (Kopittke et al., 2011, 2012), it is possible that a similar function could be ascribed to H. coronopifolia BLCs. It would be interesting to test whether the Hc-AFP1–4 peptides contribute to metal ion tolerance which would suggest a role for them (and the cells that produce them) in alleviating both abiotic and biotic stresses, as described for PDF1 (Nguyen et al., 2014). In conclusion, this study has produced evidence for BLC involvement in preformed root defence. Further investigation is warranted, with elucidation of the mechanisms involved being of considerable importance for vegetable crop species in the family Brassicaceae.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Video S1: live-cell imaging of a growing root in liquid MS medium over time. The frames were taken every 15 min over a 36-h period. Video S2: live-cell imaging of a growing root in solid MS medium over time. The frames were taken every 15 min over a 24-h period. Table S1: list and details of the primers used for the q-RT-PCR analysis of the Hc-AFP genes. Table S2: list of antibodies used for the immunofluorescence protocol optimization and the observation of the Hc-AFP1–4 peptides. Figure S1: controls for the immunofluorescence experiments showing root tip incubated with phosphate buffer in place of the primary antibody and control showing root tip incubated with phosphate buffer and pectinase without primary antibody after background subtraction. Figure S2: root tips and their BLCs stained with calcofluor after 12 h of incubation in 0·1 m PBS with different concentrations of pectinase.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by funding from Stellenbosch University, Wine Industry Network of Expertise and Technology (Winetech) and the South African Technology and Human Resources for Industry Programme (THRIP). The Central analytical facility (CAF) of Stellenbosch University, in particular Dr Ben Loos and Ms Lize Engelbrecht, is gratefully thanked for their expertise and technical support. Dr Abré de Beer is thanked for his advice on antifungal peptides and Ms Varsha Premsagar is thanked for her technical assistance. Dr Marc-Antoine Cannesan, University of Rouen, is thanked for his preliminary observations of border-like cells in this species. Thanks are also due to the University of Rouen, the GRR-Végétal-Agronomie- Sols-Innovation of Haute Normandie, Le Fonds Européen de Développement Regional (FEDER) for financial support to A.D.
LITERATURE CITED
- Andème-Onzighi C, Sivaguru M, Judy-March J, Baskin TI, Driouich A.. 2002. The reb1-1 mutation of Arabidopsis alters the morphology of trichoblasts, the expression of arabinogalactan proteins and the organization of cortical microtubules. Planta 215: 949–958. [DOI] [PubMed] [Google Scholar]
- Baetz U, Martinoia E.. 2014. Root exudates: the hidden part of plant defense. Trends in Plant Science 19: 90–97. [DOI] [PubMed] [Google Scholar]
- Balcells L. 1991. A method for sterilizing Arabidopsis seed In D Flanders, C Dean, eds. Arabidopsis: The Complete Guide. Norwich: Agriculture and Food Research Plant Molecular Biology Arabidopsis Programme, Cambridge Laboratory, 1·2–2·2. [Google Scholar]
- Barkhuizen H. 2013. Mode of action studies of defensin peptides from native South African Brassicaceae species. Master of Science thesis, Stellenbosch University, South Africa.
- Brigham LA, Woo HH, Nicoll M, Hawes MC.. 1995. Differential expression of proteins and mRNAs from border cells and root tips of pea. Plant Physiology 109: 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert W, Terras F, Cammue B, Osborn R.. 1995. Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiology 108: 1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannesan MA, Durand C, Burel C, et al. 2012. Effect of arabinogalactan proteins from the root caps of pea and Brassica napus on Aphanomyces euteiches zoospore chemotaxis and germination. Plant Physiology 159: 1658–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beer A. 2008. Isolation and characterization of antifungal peptides from plants. Dissertation of Doctor of Philosophy, Stellenbosch University, South Africa.
- De Beer A, Vivier M.. 2008. Vv-AMP1, a ripening induced peptide from Vitis vinifera shows strong antifungal activity. BMC Plant Biology 8: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beer A, Vivier M.. 2011. Four plant defensins from an indigenous South African Brassicaceae species display divergent activities against two test pathogens despite high sequence similarity in the encoding genes. BMC Research Notes 4: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coninck B, Carron D, Tavormina P, et al. 2013. Mining the genome of Arabidopsis thaliana as a basis for the identification of novel bioactive peptides involved in oxidative stress tolerance. Journal of Experimental Botany 64: 5297–5307. [DOI] [PubMed] [Google Scholar]
- Driouich A, Durand C, Vicré-Gibouin M.. 2007. Formation and separation of root border cells. Trends in Plant Science 12: 14–19. [DOI] [PubMed] [Google Scholar]
- Driouich A, Durand C, Cannesan MA, Percoco G, Vicré-Gibouin M.. 2010. Border cells versus border-like cells are they alike. Journal of Experimental Botany 61: 3827–3831. [DOI] [PubMed] [Google Scholar]
- Driouich A, Follet-Gueye ML, Vicré-Gibouin M, Hawes MC.. 2013. Root border cells and secretions as critical elements in plant host defense. Current Opinion in Plant Biology 16: 489–495. [DOI] [PubMed] [Google Scholar]
- Durand C, Vicré-Gibouin M, Follet-Gueye ML, et al. 2009. The organization pattern of root border-like cells of Arabidopsis is dependent on cell wall homogalacturonan. Plant Physiology 150: 1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fant F, Vranken W, Broekaert W, Borremans F.. 1998. Determination of the three-dimensional solution structure of Raphanus sativus antifungal protein 1 by 1H NMR. Journal of Molecular Biology 279: 257–270. [DOI] [PubMed] [Google Scholar]
- Gunawardena U, Rodriguez M, Straney D, Romeo JT, VanEtten HD, Hawes MC.. 2005. Tissue-specific localization of pea root infection by Nectria haematococca. Mechanisms and consequences. Plant Physiology 137: 1363–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haichar FZ, Santaella C, Heulin T, Achouak W.. 2014. Root exudates mediated interactions belowground. Soil Biology and Biochemistry 77: 69–80 [Google Scholar]
- Hawes MC, Brigham LA.. 1992. Impact of root border cells on microbial populations in the rhizosphere. Advances in Plant Pathology 8: 119–142. [Google Scholar]
- Hawes MC, Lin E.. 1990. Correlation of pectolytic enzyme activity with the programmed release of cells from root. Plant Physiology 94: 1855–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes MC, Pueppke SG.. 1986. Sloughed peripheral root cap cells: yields from different species and callus formation from single cells. American Journal of Botany 73: 1466–1473. [Google Scholar]
- Hawes MC, Brigham LA, Wen F, Woo HH, Zhu Y.. 1998. Function of root border cells in plant health: pioneers in the rhizosphere. Annual Reviews in Phytopathology 36: 311–327. [DOI] [PubMed] [Google Scholar]
- Hawes MC, Gunawardena U, Miyasaka S, Zhao X.. 2000. The role of root border cells in plant defense. Trends in Plant Science 5: 128–133. [DOI] [PubMed] [Google Scholar]
- Hawes MC, Bengough G, Cassab G, Ponce G.. 2003. Root caps and rhizosphere. Journal of Plant Growth Regulation 21: 352–367 [Google Scholar]
- Iijima M, Barlow PW, Bengough G.. 2003. Root cap structure and cell production rates of maize (Zea mays) roots in compacted sand. New Phytologist 160: 127–134. [DOI] [PubMed] [Google Scholar]
- Joubert DA, de Lorenzo G, Vivier MA.. 2013. Regulation of the grapevine polygalacturonase-inhibiting protein encoding gene: expression pattern, induction profile and promoter analysis. Journal of Plant Research 126: 267-281. [DOI] [PubMed] [Google Scholar]
- Kagale S, Robinson SJ, Nixon J, et al. 2014. Polyploid evolution of the Brassicaceae during the Cenozoic era. The Plant Cell 26: 2777–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer M, Schmickl R, German DA, et al. 2014. BrassiBase: Introduction to a novel knowledge database on Brassicaceae evolution. Plant and Cell Physiology 55: e3(1–9). [DOI] [PubMed] [Google Scholar]
- Kopittke PM, Menzies NW, de Jonge MD, et al. 2011. In situ distribution and speciation of toxic copper, nickel, and zinc in hydrated roots of cowpea. Plant Physiology 156: 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopittke PM, de Jonge MD, Menzies NW, et al. 2012. Examination of the distribution of arsenic in hydrated and fresh cowpea roots using two- and three-dimensional techniques. Plant Physiology 159: 1149–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer BF, Campbell RA, Schwertz H, et al. 2011. Novel anti-bacterial activities of β-defensin 1 in human platelets: suppression of pathogen growth and signaling of neutrophil extracellular trap formation. PLoS Pathogens 7: e1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Rösch P, Radt B, Popp J.. 2008. Localizing and identifying living bacteria in an abiotic environment by a combination of raman and fluorescence microscopy. Analytical Chemistry 80: 8568–8575. [DOI] [PubMed] [Google Scholar]
- Mandáková T, Mummenhoff K, Al-Shehbaz IA, Mucina L, Mühlhausen A, Lysak MA.. 2012. Whole-genome triplication and species radiation in the southern African tribe Heliophileae (Brassicaceae). Taxon 61: 989–1000. [Google Scholar]
- Mirouze M, Sels J, Richard O, et al. 2006. A putative novel role for plant defensins: a defensin from the zinc hyper-accumulating plant, Arabidopsis halleri, confers zinc tolerance. The Plant Journal 47: 329–342. [DOI] [PubMed] [Google Scholar]
- Miyasaka SC, Hawes MC.. 2001. Possible role of root border cells in detection and avoidance of aluminum toxicity. Plant Physiology 125: 1978–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monheit JE, Cowan DF, Moore DG.. 1984. Rapid detection of fungi in tissues using calcofluor white and fluorescence microscopy. Archives in Pathology Laboratory Medicine 108: 616–618. [PubMed] [Google Scholar]
- Murashige T, Skoog F.. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum 15: 473–497. [Google Scholar]
- Nguyen NN, Ranwez V, Vile D, et al. 2014. Evolutionary tinkering of the expression of PDF1s suggests their joint effect on zinc tolerance and the response to pathogen attack. Frontiers in Plant Sciences 11: (5)70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabinger S, Rödiger S, Kriegner A, Vierlinger K, Weinhäusel A.. 2014. A survey of tools for the analysis of quantitative PCR (qPCR) data. Biomolecular Detection and Quantification 1: 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plancot B, Sanraella C, Jaber R, et al. 2013. Deciphering the responses of root border-like cells of Arabidopsis and Flax to pathogen-derived elicitors. Plant Physiology 163: 1584–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven PH, Evert RF, Eichhorn SE.. 2013. Biology of plants. New York: Worth Publishers. [Google Scholar]
- Rutledge RG. 2011. A Java program for LRE-based real-time qPCR that enables large-scale absolute quantification. PLoS One 6: e17636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Komano J, Saitoh Y, et al. 2012. Neutrophil extracellular traps mediate a host defense response to human immunodeficiency virus-1. Cell Host & Microbe 12: 109–116. [DOI] [PubMed] [Google Scholar]
- Shahzad Z, Ranwez V, Fizames C, et al. 2013. Plant Defensin type 1 (PDF1): protein promiscuity and expression variation within the Arabidopsis genus shed light on zinc tolerance acquisition in Arabidopsis halleri. New Phytologist 200: 820–833. [DOI] [PubMed] [Google Scholar]
- Tavormina P, De Coninck B, Nikonorova N, De Smet I, Commue BPA.. 2015. The plant peptidome: an expanding repertoire of structural features and biological functions. The Plant Cell 27: 2095–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicré M, Santaella C, Blanchet S, Gateau A, Driouich A.. 2005. Root border-like cells of Arabidopsis: microscopical characterization and role in the interaction with rhizobacteria. Plant Physiology 138: 998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F, VanEtten HD, Tsaprailis G, Hawes MC.. 2007. Extracellular proteins in pea root tip and border cell exudates. Plant Physiology 143: 773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F, White GJ, Van Etten HD, Xiong Z, Hawes MC.. 2009. Extracellular DNA is required for root tip resistance to fungal infection. Plant Physiology 151: 820–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F, Brigham LA, Curlando-Riviera G, Xiong Z, Hawes MC.. 2014. Altered growth and root tip morphology in Pisum sativum L. in response to altered expression of a gene expressed in border cells. Plant and Soil 377: 179–187. [Google Scholar]
- Zhu MY, Ahn SJ, Matsumoto H.. 2003. Inhibition of growth and development of root border cells in wheat by Al. Physiologia Plantarum 117: 359–367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.