Abstract
Background and Aims The Nictaba family groups all proteins that show homology to Nictaba, the tobacco lectin. So far, Nictaba and an Arabidopsis thaliana homologue have been shown to be implicated in the plant stress response. The availability of more than 50 sequenced plant genomes provided the opportunity for a genome-wide identification of Nictaba-like genes in 15 species, representing members of the Fabaceae, Poaceae, Solanaceae, Musaceae, Arecaceae, Malvaceae and Rubiaceae. Additionally, phylogenetic relationships between the different species were explored. Furthermore, this study included domain organization analysis, searching for orthologous genes in the legume family and transcript profiling of the Nictaba-like lectin genes in soybean.
Methods Using a combination of BLASTp, InterPro analysis and hidden Markov models, the genomes of Medicago truncatula, Cicer arietinum, Lotus japonicus, Glycine max, Cajanus cajan, Phaseolus vulgaris, Theobroma cacao, Solanum lycopersicum, Solanum tuberosum, Coffea canephora, Oryza sativa, Zea mays, Sorghum bicolor, Musa acuminata and Elaeis guineensis were searched for Nictaba-like genes. Phylogenetic analysis was performed using RAxML and additional protein domains in the Nictaba-like sequences were identified using InterPro. Expression analysis of the soybean Nictaba-like genes was investigated using microarray data.
Key Results Nictaba-like genes were identified in all studied species and analysis of the duplication events demonstrated that both tandem and segmental duplication contributed to the expansion of the Nictaba gene family in angiosperms. The single-domain Nictaba protein and the multi-domain F-box Nictaba architectures are ubiquitous among all analysed species and microarray analysis revealed differential expression patterns for all soybean Nictaba-like genes.
Conclusions Taken together, the comparative genomics data contributes to our understanding of the Nictaba-like gene family in species for which the occurrence of Nictaba domains had not yet been investigated. Given the ubiquitous nature of these genes, they have probably acquired new functions over time and are expected to take on various roles in plant development and defence.
Keywords: Lectin, Nictaba, comparative genomics, evolution, legume, Glycine max, food crop, phylogeny, duplication
INTRODUCTION
Almost 50 years ago, gene duplication was first considered as the driving force behind evolution by Ohno (1970). Over the years, this has been confirmed by various researchers and gene duplication is now considered to be of great importance for evolution in general. Whole-genome duplications (WGDs) in particular are acknowledged as foremost players in evolution, resulting in expanded biological complexity (Lynch and Conery, 2000; Otto and Whitton, 2000; Wendel, 2000; Van de Peer et al., 2009; Lynch, 2013). Following a WGD event, retained duplicated genes often undergo sub- or neofunctionalization due to increased genetic redundancy (Fawcett et al., 2009). Whole-genome duplication events are common in plants and at least two WGDs resulted in the diversification of seed plants and angiosperms (Jiao et al., 2011). In addition to WGDs, other types of local duplication events (gene-scale duplications) also contribute to gene expansion and generation of new functions for homologous genes. Segmental duplication involves duplicative transpositions of relatively small DNA regions, while tandem duplication mainly occurs through unequal crossing over between chromosomes (Zhang, 2003; Cannon et al., 2004; Leister, 2004; Freeling, 2009). Ultimately, polyploid plants tend to diploidize and this process is associated with chromosomal rearrangements and gene and chromosome loss (Lynch and Conery, 2000).
The Leguminosae or Fabaceae, also known as the legume family, is an interesting family to study the contributions of duplication events to plant evolution. It is the third largest family of flowering plants and includes several crops that are of high economic value as major protein sources for humans and animals. Moreover, the genome sequences of multiple members of the legume family are available, including Medicago truncatula (barrel clover), Cicer arietinum (chickpea), Lotus japonicus (Japanese trefoil), Glycine max (soybean), Cajanus cajan (pigeon pea), Phaseolus vulgaris (common bean), Vigna radiata (mung bean) and Lupinus angustifolius (lupin) (Sato et al., 2008; Schmutz et al., 2010, 2014; Varshney et al., 2011, 2013; Young et al., 2011; Jain et al., 2013; Yang et al., 2013; Kang et al., 2014). Within the legume family, different polyploidy events have occurred. Analysis of the soybean genome, for example, revealed that three rounds of WGD contributed to the current G. max genome: a common WGD of all rosids [130–240 million years ago (Mya)], a legume-specific WGD ∼59 Mya and a more recent Glycine-specific WGD event 13 Mya (Shoemaker et al., 2006; Schmutz et al., 2010; Severin et al., 2011; Cannon et al., 2015). These duplication events gave rise to a soybean genome in which 75 % of its genes are present in multiple copies (Roulin et al., 2013).
The wealth of many completely sequenced genomes has allowed the analysis of gene family expansion across species. This comparative analysis of gene families has facilitated insights into how proteins can confer adaptation. Protein domains are the functional and structural components of proteins. Evolutionarily, they are well conserved across taxa and are frequently rearranged within and/or between proteins and even genomes. Protein domain rearrangements are driven by evolutionary events such as duplication, fusion, fission and domain loss, and play an essential role in the evolution and expansion of multi-domain proteins (Kummerfeld and Teichmann, 2005; Weiner et al., 2006; Moore et al., 2008; Moore and Bornberg-Bauer, 2012). Therefore, protein domains are considered discrete evolutionary units and could be related to plant adaptation and tolerance to variable environmental conditions (Yang and Bourne, 2009; Sharma and Pandey, 2016). The plant lectin family comprises all proteins that specifically bind carbohydrates. This protein–carbohydrate interaction is involved in a variety of essential processes in the plant (Van Damme et al., 2008; Lannoo and Van Damme, 2014). Plant lectins can be further divided into distinct subfamilies, specified by their conserved carbohydrate recognition domain (Van Damme et al., 2008). One of these families, the Nictaba-like family, groups all proteins that contain a protein domain that shows homology with the Nicotiana tabacum agglutinin, which is abbreviated as Nictaba and also known as the tobacco lectin. Nictaba homologues were shown to be ubiquitous in plants, including some crop species (Delporte et al., 2015). However, the tobacco lectin is the best characterized member of this lectin family at genetic and biological levels. It is believed that Nictaba acts as a signalling molecule in response to stress and triggers gene expression through interaction with histones (Delporte et al., 2014), yet the biological function of lectin homologues has not yet been uncovered. Recently, the distribution and expansion of Nictaba homologues in soybean were analysed, and the results indicated that both tandem and segmental duplications were responsible for the expansion of this family in soybean (Van Holle and Van Damme, 2015). Although a survey of Nictaba-like genes in the plant kingdom has been performed in the past, the number of plant species included has been limited and few phylogenetic conclusions have been drawn (Delporte et al., 2015). Further investigation of the genetic diversity of the family of Nictaba-like genes in crop species will yield new insights into its evolutionary relationships. In this study, bioinformatics methods were employed for the identification and comparison of the Nictaba-like gene family in six legume species (soybean, barrel clover, Japanese trefoil, common bean, pigeon pea, chickpea), two Solanaceae species (potato and tomato), cacao, coffee, three Poaceae species (rice, maize and sorghum) and two additional monocots (banana and oil palm). Using a multidisciplinary analysis, new insights are generated and the phylogenetic relationships, domain organization, duplication modes, chromosome distribution and expression analysis of this family of putative lectin genes across different species are discussed, with a special focus on the Nictaba-like lectin (NLL) genes from soybean, further referred to as GmNLLs. The results provide useful information to help us understand the role of Nictaba-like genes in plant growth and development.
MATERIALS AND METHODS
Data retrieval and sequence analysis
Putative NLL genes in the different plant genomes were identified by BLASTp searches using the protein sequence of Nictaba (AAK84134.1) against the corresponding translated genome sequence. Phytozome v10.3 (http://phytozome.jgi.doe.gov/) was used for the following plant genomes: Zea mays (v6a), Oryza sativa (filtered MSU release 7.0), Glycinemax (Wm82.a2.v1), Phaseolusvulgaris (v1.0), Medicago truncatula (Mt4.0v1), Musa acuminata (v1) (banana), Theobroma cacao (v1.1) (cacao) and Sorghum bicolor (MIPS v3.1) (Goodstein et al., 2012). BLASTp searches against the translated genomes of Lotus japonicus (v3.0), Cicer arietinum (v1.0) and Cajanus cajan (v1.0) were carried out with the BLAST tool available from the legume Information System website (http://legumeinfo.org/), while the Solanum lycopersicum (ITAG release 2.40) and Solanum tuberosum (PGSC DM v3.4) BLASTp searches were executed on the Sol Genomics Network (https://solgenomics.net/tools/blast/) website. The Coffee Genome Hub website (http://coffee-genome.org/coffeacanephora) and the Kyoto Encyclopedia of Genes and Genomes website (http://www.kegg.jp/kegg-bin/show_organism?org=egu) were used to perform BLASTp searches for Coffea canephora and Elaeis guineensis (oil palm), respectively. The top hit of each BLASTp search was used as a template for a second BLASTp search to retrieve more candidate sequences. BLAST searches using the nucleotide sequence of Nictaba against the genome sequences did not yield any additional sequences. The availability of a Pfam ID (PF14299) enabled the search of the Pfam database for more candidate sequences (Finn et al., 2016). Protein sequences encoded by all potential Nictaba-like genes were downloaded and scanned with InterPro (http://www.ebi.ac.uk/interpro/) (Mitchell et al., 2015) to verify the presence of the Nictaba domain and identify any additional annotated protein domains. Only those sequences containing one or more lectin domains were considered for further analysis. Next to the amino acid sequences, the chromosomal localization of the NLL genes was downloaded from the different databases.
Homologue identification
Tandem duplications of all NLL genes within one species and segmental duplications across the different legumes were assessed as described previously (Van Holle and Van Damme, 2015).
Phylogenetic analysis
Maximum likelihood phylogenetic trees were constructed with the protein sequences of the lectin domains. Sequences were aligned with MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/) using the default parameters (Edgar, 2004) and blocks of conserved aligned sequences were generated with trimAl using the automated1 option (Capella-Gutiérrez et al., 2009). For protein sequences containing multiple Nictaba domains, all domains were separately included in the alignment. Maximum likelihood-based phylogenetic trees were built with RAxML v8.2.4 using the GTRGAMMA model, with automatic determination of the protein substitution model, random number seed, using distinct starting trees. Subsequent bootstrap analysis was performed to assess the robustness of the phylogenetic trees (Stamatakis, 2014). The FigTree v1.4.2 software (http://tree.bio.ed.ac.uk/software/figtree/) was used to visualize the phylogenetic trees.
Molecular modelling
Homology modelling of Nictaba and one selected Nictaba-like lectin (GmNLL1 or Glyma.06G221100) from soybean was performed using YASARA Structure (Krieger et al., 2002). Different models were built from the X-ray coordinates of the carbohydrate-binding module (CBM) of the glycoside hydrolase family 10 protein from Prevotella bryantii B14 (PDB code 4MGQ) and Bacteroides intestinalis (PDB code 4QPW) (Zhang et al., 2014), and the CBM4-2 of the xylanase from Rhodothermus marinus (PDB code 1K42) (Simpson et al., 2002). Finally, a hybrid model of the proteins was built using the different previous models. PROCHECK was used to assess the geometric quality of the three-dimensional models (Laskowski et al., 1993). In this respect, all residues of the Nictaba model were correctly assigned in the allowed regions of the Ramachandran plot except for three residues (Glu2, Pro71, Arg112). Similarly, three residues of the GmNLL1 model (Leu57, Leu140, Thr163) were found to occur in the non-allowed region of the Ramachandran plots. Using ANOLEA to evaluate the models, only one residue of Nictaba out of 165 and 14 residues of the GmNLL1 out of 163 exhibited an energy higher than the threshold value (Melo and Feytmans, 1998). The residues were mainly located in the loop regions connecting the β-sheets in the models. The calculated QMEAN6 score of Nictaba and Glyma.06G221100 were 0·36 and 0·41, respectively (Arnold et al., 2006; Benkert et al., 2011). Molecular cartoons were drawn with the UCSF Chimera package (Pettersen et al., 2004).
Online tools and database resources
Selected Nictaba-related sequences were screened for the presence of transmembrane domains using the TMHMM server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and the SignalP 4·1 server (http://www.cbs.dtu.dk/services/SignalP/) was used to predict the presence of a signal peptide (Krogh et al., 2001; Petersen et al., 2011). Coding sequences and genomic sequences of GmNLL genes were downloaded from Phytozome (https://phytozome.jgi.doe.gov/biomart) and the Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn/) was used to determine and visualize the intron/exon organization of the genes (Hu et al., 2015). Microarray data (Libault et al., 2010) were visualized in a heat map using the BAR HeatMapper Plus Tool (http://bar.utoronto.ca/ntools/cgi-bin/ntools_heatmapper_plus.cgi) and logos of the Nictaba domain sequences from soybean were generated with WebLogo3 (http://weblogo.berkeley.edu/logo.cgi) (Crooks et al., 2004).
RESULTS
Genome-wide identification of Nictaba homologues in soybean and other food crops
Nictaba-related genes are characterized by the presence of a carbohydrate recognition domain with sequence similarity to Nictaba and have previously been identified in a limited number of plants (Lannoo et al., 2008; Delporte et al., 2015). In this study, a total of 360 putative NLL genes were identified in 15 crop genomes using a combination of BLASTp, InterPro analysis and hidden Markov models (Table 1). In total, 139 NLL genes were identified in six legume species (G. max, P. vulgaris, C. cajan, L. japonicus, M. truncatula and C. arietinum) and 74 genes were found in tomato and potato. The T. cacao and C. canephora genomes retained 27 and 19 NLL genes, respectively. In the three Poaceae species (O. sativa, Z. mays and S. bicolor), 53 NLL genes were identified. The M. acuminata genome contained 23 NLL genes and an additional 25 NLL genes were found in the E. guineensis genome. The M. truncatula genome contained the highest number (44) of Nictaba-related genes. A variable number of genes (13–31) was identified in genomes of the other plants. Overall, the chromosome number or genome size was not correlated with the number of retrieved Nictaba-related genes. The M. truncatula genome amounted to 470 Mb over eight chromosomes and contain 44 NLL genes, while the soybean genome was more than double in size and in chromosome number but contained only 31 NLL genes. The sequence characteristics of all NLL genes from soybean are listed in Supplementary Data Table S1. The translated Nictaba-like protein sequences vary from 163 to 341 amino acids, and are mostly encoded by three exons.
Table 1.
Distribution of NLL genes in different crop species
| Lineage | Species | Genome size | Chromosome number | Number of genes |
|---|---|---|---|---|
| Eukaryota | ||||
| Monocots | ||||
| Poaceae | O. sativa | 480 Mb | 12 | 20 |
| Z. mays | 2400 Mb | 10 | 16 | |
| S. bicolor | 732 Mb | 10 | 17 | |
| Musaceae | M. acuminata | 523 Mb | 11 | 23 |
| Arecaceae | E. guineensis | 1·8 Gb | 16 | 25 |
| Dicots | ||||
| Fabaceae | ||||
| Phaseoleae | G. max | 1115 Mb | 20 | 31 |
| P. vulgaris | 625 Mb | 11 | 17 | |
| C. cajan | 833 Mb | 11 | 13 | |
| Lotaea | L. japonicus | 470 Mb | 6 | 21 |
| Trifolieae | M. truncatula | 470 Mb | 8 | 44 |
| Cicereae | C. arietinum | 740 Mb | 8 | 13 |
| Malvaceae | T. cacao | 331 Mb | 10 | 27 |
| Solanaceae | S. lycopersicum | 950 Mb | 12 | 31 |
| S. tuberosum | 840 Mb | 12 | 43 | |
| Rubiaceae | C. canephora | 710 Mb | 11 | 19 |
Phylogenetic analysis demonstrates that all NLL genes have a common ancestor
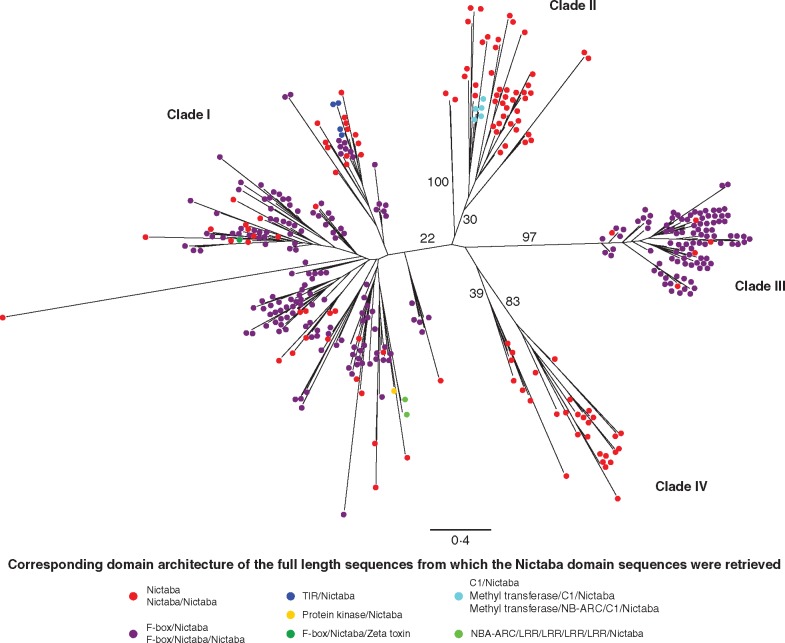
To unravel the evolutionary relationships between the Nictaba homologues in the different plant species, a maximum likelihood phylogenetic tree was constructed using the amino acid sequences encoding the Nictaba domain from G. max, P. vulgaris, C. cajan, L. japonicus, M. truncatula, C. arietinum, T. cacao, S. lycopersicum, S. tuberosum, C. canephora, O. sativa, Z. mays, S. bicolor, M. acuminata and E. guineensis (Fig. 1).
Fig. 1.
Maximum likelihood tree constructed with RAxML and based on all Nictaba domain sequences retrieved from the 15 genomes under study. Concatenated alignments of all Nictaba domain sequences were used in the RAxML analysis. Distances are proportional to evolutionary distances and are specified by the scale bar (0·4), and the numbers refer to percentage bootstrap values. Coloured circles mark the different domain architectures of the full-length Nictaba-related sequences.
The phylogenetic analysis of the Nictaba homologues included only the Nictaba domain sequences since the complete protein sequences differ too much in length and domain organization, making it difficult to generate a suitable alignment. RAxML analysis of the Nictaba-related protein sequences generated a phylogenetic tree that contained four clades. Although clade I can be further divided into multiple subclades, they were all classified as clade I due to the low bootstrap values among the subclades. All clades contain sequences from both monocots and dicots, indicating a close phylogenetic relationship. Remarkably, clade IV is the only clade in which no sequences from the Poaceae were found, suggesting that this group of genes evolved independently from the NLL sequences from grasses (Supplementary Data Fig. S1, Supplementary Data Table S2). Based on the phylogenetic tree, all NLL genes have probably diverged from a common ancestor protein in Angiospermae.
Due to the high complexity of phylogeny, a separate phylogenetic analysis of NLL proteins from soybean was performed using the Nictaba domain sequences from all soybean NLL proteins. As shown in Supplementary Data Fig. S2, the NLL genes from soybean can be categorized into seven clades. Subclade A is the largest group, containing 11 members, followed by subclades B and F, encompassing six and five members, respectively.
Structural features of the NLL genes are related to their phylogenetic relationships
Considering the chromosomal localization of the NLL genes under study, most of the genes are unevenly distributed over the chromosomes and some gene clusters are observed (Fig. 2). Remarkably, in Z. mays, the 16 NLL genes are located on only two out of ten chromosomes (chromosome 2 and chromosome 5). Interestingly, the distribution patterns of the Nictaba-related genes from S. lycopersicum and S. tuberosum show high similarity.
Fig. 2.
Chromosomal distribution of NLL genes across different species. All chromosomes are visualized in distinct colours and chromosome zero is defined by the scaffolds that could not be mapped on any of the chromosomes.
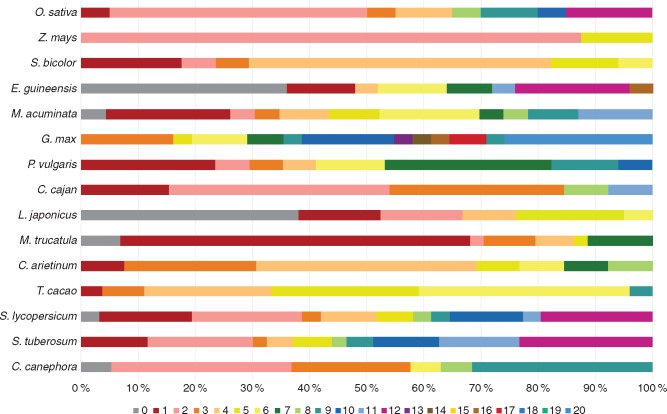
Comparison of the domain architecture also highlighted structural diversity between the NLL genes from different species. Next to the Nictaba protein domain, eight additional annotated protein domains could be identified (Table 2). Combinations of the Nictaba domain with a second Nictaba domain, an F-box domain, a protein kinase domain, a ζ (zeta) toxin domain, a TIR (Toll/interleukin-1 receptor) domain, a C1 domain, a methyltransferase domain, an NB-ARC (nucleotide-binding adaptor shared by APAF-1, R proteins and CED-4) domain and/or leucine-rich repeats (LRRs) result in ten different domain architectures in the species under study. In most cases, the multi-domain architecture involves the presence of an F-box domain N-terminally of the Nictaba domain. Furthermore, F-box Nictaba is the most abundant domain architecture in all plants studied here. The F-box Nictaba domain architecture and the single Nictaba domain architecture are the only domain combinations that were identified in every genome. The TIR Nictaba domain organization is unique for the Solanaceae, and the combination of the NB-ARC domain, LRRs and the Nictaba domain could only be identified in monocots. In some species, rare combinations were identified such as the protein kinase domain combined with the Nictaba domain, the combination of C1 domains with the Nictaba domain in T. cacao and the F-box Nictaba ζ toxin combination in M. truncatula. A gene encoding a protein with two tandem-arrayed Nictaba domains was identified in three species belonging to non-related families: S. bicolor, G. max and S. tuberosum. All translated NLL protein sequences were further investigated for the presence of signal peptides and transmembrane domains. None of the sequences contained a signal peptide or transmembrane domains, suggesting these proteins are all targeted to the cytosol.
Table 2.
Domain architectures in each of the explored plant species
| Domain architecture | O. sativa | Z. mays | S. bicolor | M. acuminata | E. guineensis | G. max | P. vulgaris | C. cajan | L. japonicus | M. truncatula | C. arietinum | T. cacao | S. lycopersicum | S. tuberosum | C. canephora |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nictaba | 4 | 3 | 1 | 7 | 13 | 5 | 3 | 1 | 9 | 12 | 2 | 8 | 14 | 19 | 8 |
| Nictaba/Nictaba | 1 | 1 | 1 | ||||||||||||
| F-box/Nictaba | 13 | 12 | 14 | 16 | 12 | 25 | 14 | 12 | 12 | 31 | 11 | 13 | 16 | 20 | 10 |
| F-box/Nictaba/Nictaba | 2 | 1 | 1 | ||||||||||||
| F-box/Nictaba/ζ toxin | 1 | ||||||||||||||
| Protein kinase/Nictaba | 1 | ||||||||||||||
| C1(6x or 10x)/Nictaba | 3 | ||||||||||||||
| MT/C1/Nictaba | 1 | ||||||||||||||
| MT/NB-ARC/C1/Nictaba | 1 | ||||||||||||||
| NB-ARC/LRR/LRR/ LRR/LRR/Nictaba | 1 | 1 | |||||||||||||
| TIR/Nictaba | 1 | 3 |
MT, methyltransferase
The domain organization of the NLL proteins can also be linked to their phylogenetic relationships. The domain sequences from clade IV are all part of proteins containing one or two Nictaba domains, while in the other clades Nictaba domain sequences were found originating from combinations of the Nictaba domain and the F-box domain (Figure 1). Considering the NLL proteins from soybean, the single-domain proteins containing the Nictaba domain and the amino acid sequence containing two tandem-arrayed Nictaba domains cluster in clades F and G of the phylogenetic tree, while the Nictaba domains that originate from F-box Nictaba proteins are found in clades A–E (Fig. S2). This is remarkable since the maximum likelihood tree was built using the amino acid sequences from the Nictaba domains alone. Similar observations were made with respect to the maximum likelihood tree of all 360 Nictaba domain sequences. Except for the domain architectures that include the C1 domain (which can all be found in clade II), Nictaba domain sequences from all other rare architectures (TIR, ζ, NB-ARC, LRR, protein kinase) are part of clade I.
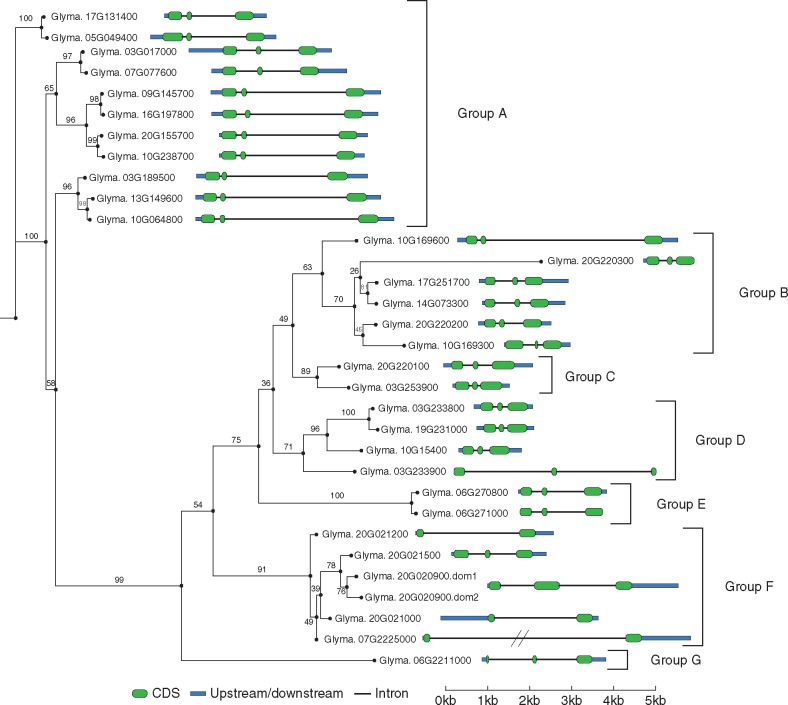
Analysis of the intron/exon gene structure demonstrated that most genes of the GmNLL family contain a conserved gene structure with three exons and two introns (Fig. 3, Table S1). However, some genes in clade F, which groups proteins with only the Nictaba domain, consist of two exons and one larger intron. Generally, closely related NLL genes show highly similar intron/exon gene structures. The intron size of the genes designated as belonging to clade A, for example, differs greatly from those in the other clades. This again demonstrates the stronger evolutionary relationship of genes within the same clade.
Fig. 3.
Maximum likelihood phylogenetic tree of Nictaba domains in soybean with the corresponding exon/intron structures for these NLL genes. Nictaba domain sequences were used to build the phylogenetic tree with RAxML. Gene intron/exon structures were generated using GSDS 2·0 (Hu et al., 2015). The coding sequences are visualized in green and the 5′ and 3′ untranslated regions are represented in blue.
Tandem and segmental duplications contributed to the expansion of NLL genes in all crops
Expansion of the NLL genes was investigated by identification of tandem duplication clusters and demonstrated that tandemly duplicated genes are present in all crop species (Table 3). In barrel clover, potato, oil palm, cacao and coffee, >50 % of the NLL genes were identified in tandem duplication clusters. For the tandemly duplicated soybean NLL genes, most genes originating from the same tandem duplication cluster group together in the same clade of the phylogenetic tree (Fig. S2).
Table 3.
Tandem duplications contributed to gene expansion
| Species | Number of NLL genes | Number of tandem duplication clusters | Total number of genes involved | Percentage |
|---|---|---|---|---|
| O. sativa | 20 | 2 | 7 | 35·0 |
| Z. mays | 16 | 3 | 6 | 37·5 |
| S. bicolor | 17 | 1 | 8 | 47·1 |
| M. acuminata | 23 | 1 | 3 | 13·0 |
| E. guineensis | 25 | 5 | 15 | 60·0 |
| G. max | 31 | 5 | 13 | 41·9 |
| P. vulgaris | 17 | 2 | 4 | 23·5 |
| C. cajan | 13 | 2 | 5 | 38·5 |
| L. japonicus | 21 | 3 | 6 | 28·6 |
| M. truncatula | 44 | 5 | 24 | 54·5 |
| C. arietinum | 13 | 1 | 3 | 23·1 |
| T. cacao | 27 | 5 | 15 | 55·6 |
| S. lycopersicum | 31 | 6 | 14 | 45·2 |
| S. tuberosum | 43 | 10 | 29 | 67·4 |
| C. canephora | 19 | 5 | 13 | 68·4 |
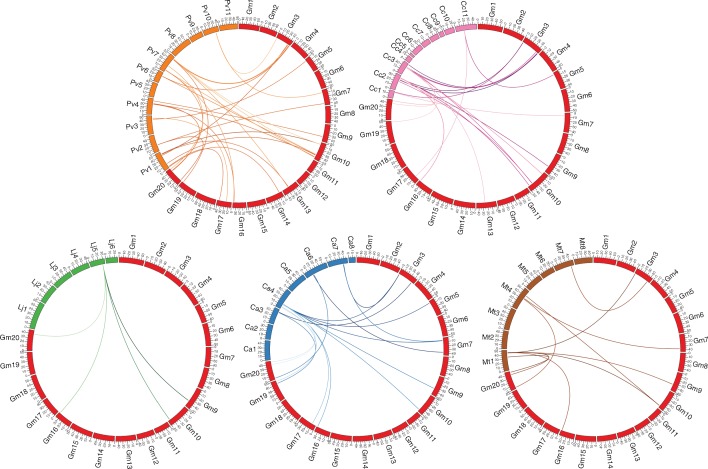
Additionally, the Plant Genome Duplication Database was used to explore the presence of orthologous NLL genes in the legume family. Segmental duplications of NLL genes between soybean and the other legumes are represented in Fig. 4. The NLL genes of G. max and P. vulgaris show the highest number of orthologous genes, while the lowest number of orthologues is found between G. max and L. japonicus. Strikingly, some of the soybean NLL genes have the same orthologues in all legumes (except for L. japonicus). These are mainly the NLL genes that are located on chromosomes 3, 5, 7, 9, 10, 16 and 20 of soybean.
Fig. 4.
Comparative analysis of orthologues of NLL genes between soybean and five legumes. Coloured lines represent orthologues between legume genomes. Bars in different colours represent the chromosomes of the different legumes in a circular way: Gm, G. max; Pv, P. vulgaris; Cc, C. cajan; Lj, L. japonicus; Ca, C. arietinum; Mt, M. truncatula.
Nictaba-related genes from soybean display great variability in their expression pattern
To get additional insight into the regulation of NLL genes, the expression pattern of soybean NLL genes was investigated using the available microarray database (Libault et al., 2010). Transcription profiles of the GmNLL genes in seven different tissues were collected and analysed. Expression data were unavailable for the following NLL genes: Glyma.20G220100, Glyma.20G220200, Glyma.07G222500, Glyma.03G233900 and Glyma.06G271000. Judging from the heat map shown in Fig. 5, the soybean NLL genes show a dynamic expression pattern. Not all genes belonging to the same phylogenetic clade show similar transcription profiles. While some genes are highly expressed in all examined tissues (Glyma.03G189500 and Glyma.10G169600), others show much lower expression or are hardly detectable (Glyma.20G220300, Glyma.06G270800).
Fig. 5.
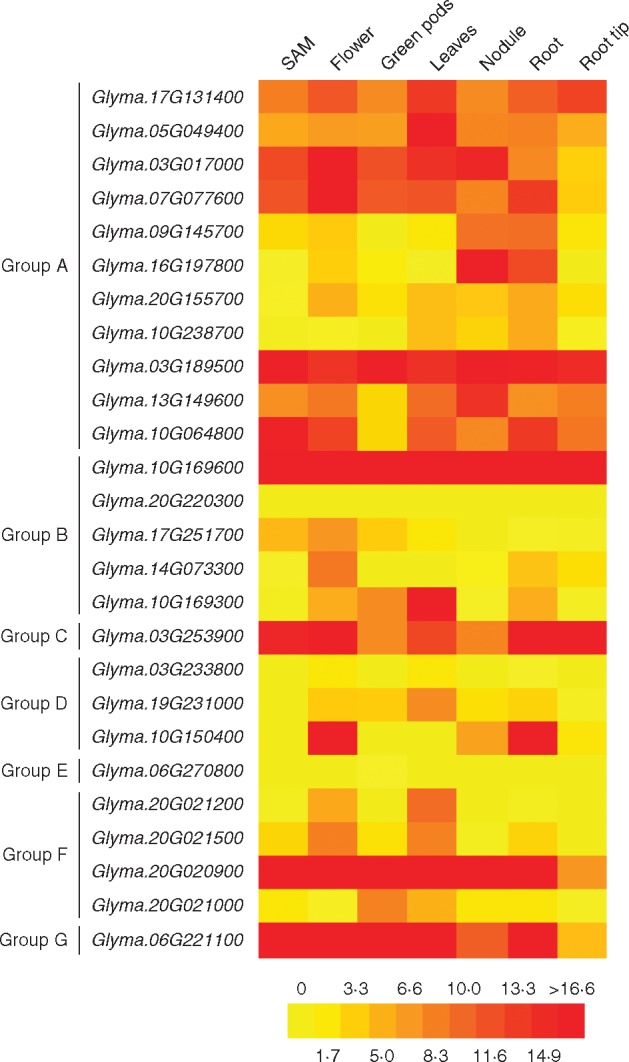
Expression levels of NLL genes in different tissues of soybean inferred from microarray data. Log2-transformed microarray data (Libault et al., 2010) were visualized in a heat map using the BAR HeatMapper Plus Tool (http://bar.utoronto.ca/ntools/cgi-bin/ntools_heatmapper_plus.cgi). SAM, shoot apical meristem.
Molecular modelling of an NLL protein from soybean reveals structural resemblance to the tobacco lectin
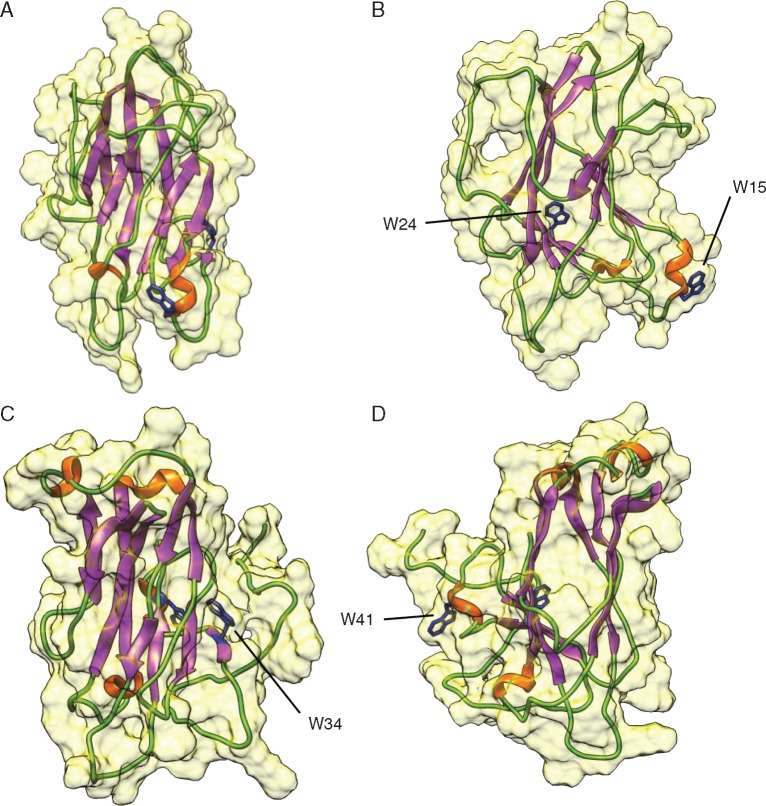
Despite the lack of a three-dimensional structure of the tobacco lectin, the availability of a structure model of Nictaba accommodated new insights in some amino acid residues that are important for the carbohydrate-binding activity of the lectin (Schouppe et al., 2010). Molecular models built for Nictaba and a selected soybean Nictaba homologue (GmNLL1) (encoded by Glyma.06G221100) revealed that both Nictaba and GmNLL1 exhibit the canonical β-sandwich core structure of the carbohydrate-binding module of glycoside hydrolase family 10 enzymes (Fig. 6). However, they differ in the size and the shape of the loops connecting the strands of β-sheets.
Fig. 6.
Molecular modelling of Nictaba and an NLL protein from soybean. Ribbon diagrams show front (A, C) and side (C, D) view of Nictaba and GmNLL1, respectively. The molecular surface, α-helices, β-sheets and loop/turns are coloured yellow, orange, purple and green, respectively. The conserved tryptophan residues important for the carbohydrate-binding activity of Nictaba (w15, w24, w34, w41) are indicated.
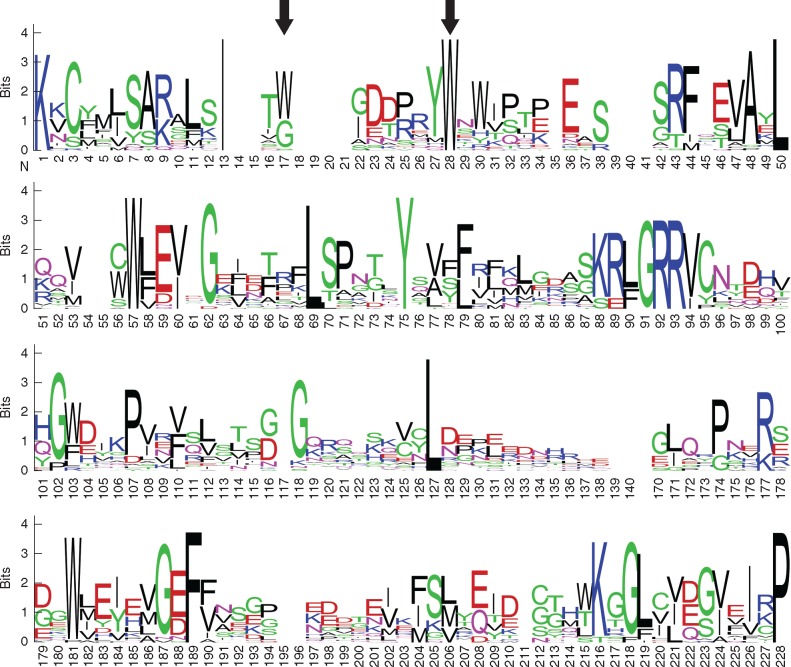
To gain insight into the conserved residues in the Nictaba domain sequences from soybean, sequence logos were created using WebLogo3 (Crooks et al., 2004). Several highly conserved amino acid residues are present in all GmNLL sequences, as depicted in Fig. 7. Interestingly, the two tryptophan residues that are necessary for the carbohydrate-binding activity of the tobacco lectin (Schouppe et al., 2010) are strongly conserved in the soybean NLL sequences (Fig. 7, positions 17 and 28). It is clear from the sequence logo that amino acid residues in other regions displayed varying levels of sequence conservation.
Fig. 7.
Logo of NLL amino acid sequences from soybean. The logo was created with WebLogo3 (Crooks et al., 2004) and consists of stacks of amino acids for each position. Sequence conservation at each position is indicated by the overall height of the stack while the height of an amino acid within a stack indicates the relative frequency of that amino acid. Positions 141–169 were deleted since these positions contained no information. The conserved tryptophan residues important for the carbohydrate-binding activity of Nictaba are marked with arrows.
DISCUSSION
A growing body of evidence has pointed to the involvement of NLL genes in plant stress responses. Transcript levels for the tobacco lectin are upregulated after jasmonate treatment and insect herbivory (Chen et al., 2002; Vandenborre et al., 2009). Presumably, Nictaba acts as a signalling molecule in response to stress, resulting in altered gene expression, caused by the interaction with O-GlcNAc (O-linked β-D-N-acetylglucosamine)-modified histones (Delporte et al., 2015). Recently, an F-box Nictaba protein from Arabidopsisthaliana was also linked to the plant stress response, since overexpression of this gene in A. thaliana showed reduced disease symptoms upon infection with Pseudomonas syringae (Stefanowicz et al., 2016). This study provides a comprehensive overview of the NLL gene family in 15 crop species across different lineages of vascular plants, with a special focus on soybean.
A total of 360 putative Nictaba lectin genes were identified with variable gene numbers (ranging from 11 to 44) for each species, which is in agreement with previous reports (Delporte et al., 2015; Van Holle and Van Damme, 2015). The discrepancy across species could not be explained by genome size or chromosome number. Furthermore, the NLL genes were randomly distributed over the chromosomes. For soybean, the high number of NLL genes spread over the different chromosomes can be attributed to the highly duplicated genome, in which 75 % of the genes are present in multiple copies, and where the duplication events were followed by many chromosome rearrangements (Schmutz et al., 2010). Analysis of the domain architectures indicated that the single-domain Nictaba protein and the multi-domain F-box Nictaba architectures are ubiquitous among all analysed species, consistent with the results of earlier studies (Lannoo et al., 2008; Delporte et al., 2015). Other architectures were found to be specific to a certain plant family. For example, the TIR Nictaba-encoding genes were restricted to the Solanaceae, and the combination of the NB-ARC domain, the Nictaba domain and LRRs was only identified in monocots. These different protein domains are known to be involved in disease resistance (van Loon et al., 2006). For example, the NB-ARC and LRR protein domain combination is typically encoded by R genes, key components of the plant immune system. The interaction between R proteins and specific pathogenic effectors leads to effector-triggered immunity (ETI), a mechanism that establishes protection of the plant against pathogens (Dangl and Jones, 2001; Jones and Dangl, 2006). As repeatedly discussed, formation of multi-domain proteins through domain combination is an important process that gives rise to proteins with new functions (Björklund et al., 2005; Kummerfeld and Teichmann, 2005; Vogel et al., 2005; Bashton and Chothia, 2007). Domain combination and convergence and divergence of protein domains could be driven by sub- and/or neofunctionalization of duplicated genes upon gene or genome duplications (Lynch and Conery, 2000; Taylor and Raes, 2004; Gough, 2005; Vogel and Morea, 2006). LRR domain-containing proteins, for example, are thought to be associated with the plant’s response to stress adaptation and tolerance (Schaper and Anisimova, 2015; Sharma and Pandey, 2016). Genes encoding proteins with a double Nictaba domain have been identified in three non-related species (S.bicolor, G. max and S.lycopersicum), but this could be the result of independent domain reorganizations within the different linages. Analysis of all currently known protein sequences indicated that repeats of the same domain in multi-domain architecture families is a very common phenomenon (Björklund et al., 2006; Levitt, 2009). What is more, new single-domain architecture families are emerging slowly while formation of multi-domain architecture families is growing exponentially by rearrangement and/or combination of existing domains (Moore et al., 2008; Levitt, 2009). The combination of the C1 domain with a lectin domain, as identified in T. cacao, is not unique. In cucumber, several proteins were identified in which the jacalin lectin domain is linked to multiple C1 domains (Dang and Van Damme, 2016). Concurrently with our data, it was shown that single-domain families are mostly shared by large groups of species, whereas multi-domain architectures are much more specific and account for species diversity (Levitt, 2009).
Gene family expansion is governed by tandem duplication, segmental duplication and gene transposition events (Ohno, 1970; Zhang, 2003; Cannon et al., 2004). Of all identified NLL genes, a significant share was shown to be involved in tandem duplications, explaining the greater expansion of NLL genes in some species. Furthermore, when interspecies gene orthologues were compared, several Nictaba-related genes from soybean showed extensive conservation with M. truncatula, P. vulgaris, C. arietinum and C. cajan. These results demonstrated the shared evolutionary relationship of some NLL genes from the investigated legumes, and are consistent with the documented WGD events in the legume family (Shoemaker et al., 2006; Schmutz et al., 2014). A large number of orthologues between the soybean genome and other legume genomes has previously also been reported for the heat-shock transcription factor gene family, the auxin gene family and the alcohol dehydrogenase gene family (Fukuda et al., 2005; Lin et al., 2014; Singh and Jain, 2015). The variability between the different species indicates lineage-specific gene gain or loss over time. These findings were further supported by the phylogenetic analysis revealing four clades in which all NLL genes could be classified. All soybean NLL genes from clades IV have a single-domain architecture, while domain architectures are diverse in the other clades, assuming that these genes are descendants of a shared ancestral NLL gene only containing the Nictaba domain. The soybean genes encoding F-box Nictaba proteins and the Nictaba (with one or two domains) proteins were found in distinct groups of the phylogenetic tree (Fig. S2). A similar tree was obtained using an alignment of the F-box domain sequences, demonstrating that the F-box and the Nictaba protein domains evolved together, and that the genes encoding F-box Nictaba proteins did not arise by re-shuffling of the individual protein domains. The data suggest that Nictaba-encoding genes are widespread throughout the plant kingdom, and the maintenance of these genes in all genomes during multiple rounds of genome duplications, gene loss and gene rearrangements points to a selective pressure on these genes.
Microarray data revealed that the NLL genes showed a diverse expression pattern between the different tissues of soybean, suggesting they play roles in multiple developmental stages. In addition, genes within the same phylogenetic clade did not show similar transcription profiles. Most of the genes showed moderate or high expression in one or more of the analysed tissues; however, two genes (Glyma.20G220300 and Glyma.06G270800) showed relatively low expression in all tissues. For five other NLL genes, no expression data were available. These findings could indicate that some NLL genes from soybean might only be expressed under stress conditions, similar to the NLL gene from tobacco (Chen et al., 2002). Additionally, the diverse expression pattern of the soybean NLL genes might be the result of sub- or neofunctionalization of duplicated genes and could explain the large number of NLL genes in soybean and why they were retained in the genome upon different WGD events (Van de Peer et al., 2009). To determine whether this divergence resulted in distinct functions of the soybean Nictaba homologues, functional analyses will have to be performed in the future. Recently, these microarray results have been validated via qRT-PCR (quantitative reverse transcription) for some of the GmNLLs. Soybean NLL genes displayed differential expression upon exposure to a diverse range of stress conditions and plants overexpressing these genes protected the plant against P. syringae infections, Aphis glycines infestation and salinity (Van Holle et al., 2016), making it tempting to speculate that GmNLL lectin genes might be involved in plant stress signalling.
Three-dimensional protein models for Nictaba and one of the soybean NLL proteins were made based on the structural homology with the carbohydrate-binding modules of some glycoside hydrolase family 10 proteins. Analysis revealed that, like Nictaba (Schouppe et al., 2010), the soybean homologue also consists of β-sheets. Structurally related proteins often share similar molecular functions (Brylinski and Skolnick, 2008; Drew et al., 2011; Rentzsch and Orengo, 2013). This is supported by two tryptophan resisdues, which were shown to be indispensable for lectin activity of Nictaba, are conserved in most of the GmNictaba domain sequences (Schouppe et al., 2010). These Nictaba homologues most likely represent functional carbohydrate-binding proteins. However, the sugar-binding specificity will probably not be conserved since multiple homologous lectin domains were shown to exhibit unique carbohydrate-binding specificities (Fouquaert and Van Damme, 2012; Stefanowicz et al., 2012). Further studies are necessary to elucidate the carbohydrate-binding specificities of NLL proteins in other species.
This research focused on the dynamic evolution of Nictaba-related genes in 15 crop species and revealed great divergence. A complex interplay of WGD and tandem and segmental duplication events probably resulted in the different domain architectures. Given the large number of identified Nictaba homologues, these are expected to play diverse roles in plant development and defence. We believe that these sub- or neofunctionalized genes were preserved in the different species as these new genes could help plants to adapt to a broader range of environmental conditions. More detailed analysis of Nictaba homologues in multiple species will facilitate insights related to their function in plant development and stress responses.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: maximum likelihood tree constructed with RAxML and based on all Nictaba domain sequences retrieved from the 15 genomes under study. Figure S2: phylogenetic relationships of soybean Nictaba-like domains. Table S1: characteristics of NLL lectin sequences in soybean. Table S2: overview of the number Nictaba domain-containing sequences in the 15 species, grouped according to clades of the phylogenetic tree shown in Fig. 1.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Research Council of Ghent University (project 01G00515).
LITERATURE CITED
- Arnold K, Bordoli L, Kopp J, Schwede T.. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- Bashton M, Chothia C.. 2007. The generation of new protein functions by the combination of domains. Structure 15: 85–99. [DOI] [PubMed] [Google Scholar]
- Benkert P, Biasini M, Schwede T.. 2011. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27: 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund ÅK, Ekman D, Light S, Frey-Skött J, Elofsson A.. 2005. Domain rearrangements in protein evolution. Journal of Molecular Biology 353: 911–923. [DOI] [PubMed] [Google Scholar]
- Björklund ÅK, Ekman D, Elofsson A.. 2006. Expansion of protein domain repeats. PLoS Computational Biology 2: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brylinski M, Skolnick J.. 2008. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proceedings of the National Academy of Sciences of the USA 105: 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G.. 2004. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biology 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, McKain MR, Harkess A, et al. 2015. Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Molecular Biology and Evolution 32: 193–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Peumans WJ, Hause B, et al. 2002. Jasmonate methyl ester induces the synthesis of a cytoplasmic/nuclear chitooligosaccharide-binding lectin in tobacco leaves. FASEB Journal 16: 905–907. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE.. 2004. WebLogo: a sequence logo generator. Genome Research 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJM, Lannoo N, Peumans WJ.. 2008. Plant lectins. Advances in Botanical Research 48: 107–209. [Google Scholar]
- Dang L, Van Damme EJM.. 2016. Genome-wide identification and domain organization of lectin domains in cucumber. Plant Physiology and Biochemistry 108: 165–176. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG.. 2001. Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Delporte A, De Vos WH, Van Damme EJM.. 2014. In vivo interaction between the tobacco lectin and the core histone proteins. Journal of Plant Physiology 171: 1149–1156. [DOI] [PubMed] [Google Scholar]
- Delporte A, Van Holle S, Lannoo N, Van Damme EJM.. 2015. The tobacco lectin, prototype of the family of Nictaba-related proteins. Current Protein and Peptide Science 16: 5–16. [DOI] [PubMed] [Google Scholar]
- Drew K, Winters P, Butterfoss GL, et al. 2011. The Proteome Folding Project: proteome-scale prediction of structure and function. Genome Research 21: 1981–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y.. 2009. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proceedings of the National Academy of Sciences of the USA 106: 5737–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research 44: 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquaert E, Van Damme EJM.. 2012. Promiscuity of the Euonymus carbohydrate-binding domain. Biomolecules 2: 415–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. 2009. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annual Review of Plant Biology 60: 433–453. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Yokoyama J, Nakamura T, et al. 2005. Molecular phylogeny and evolution of alcohol dehydrogenase (Adh) genes in legumes. BMC Plant Biology 5: 6:1–6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40: 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. 2005. Convergent evolution of domain architectures (is rare). Bioinformatics 21: 1464–1471. [DOI] [PubMed] [Google Scholar]
- Van Holle S, Van Damme EJM.. 2015. Distribution and evolution of the lectin family in soybean (Glycine max). Molecules 20: 2868–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holle S, Smagghe G, Van Damme EJM.. 2016. Overexpression of Nictaba-like lectin genes from Glycine max confers tolerance toward Pseudomonas syringae infection, aphid infestation and salt stress in transgenic Arabidopsis plants. Frontiers in Plant Science 7: 1590. doi:10.3389/fpls.2016.01590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G.. 2015. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31: 1296–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Misra G, Patel RK, et al. 2013. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant Journal 74: 715–729. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL.. 2006. The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kang YJ, Kim SK, Kim MY, et al. 2014. Genome sequence of mungbean and insights into evolution within Vigna species. Nature Communications 5: 5443. doi:10.1038/ncomms6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, Koraimann G, Vriend G.. 2002. Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins 47: 393–402. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Kummerfeld SK, Teichmann SA.. 2005. Relative rates of gene fusion and fission in multi-domain proteins. Trends in Genetics 21: 25–30. [DOI] [PubMed] [Google Scholar]
- Lannoo N, Peumans WJ, Van Damme EJM.. 2008. Do F-box proteins with a C-terminal domain homologous with the tobacco lectin play a role in protein degradation in plants? Biochemical Society Transactions 36: 843–847. [DOI] [PubMed] [Google Scholar]
- Lannoo N, Van Damme EJM.. 2014. Lectin domains at the frontiers of plant defense. Frontiers in Plant Science 5: 397. doi:10.3389/fpls.2014.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM.. 1993. PROCHECK: a program to check the steroechemical quality of protein structures. Journal of Applied Crystallography 26: 283–291. [Google Scholar]
- Leister D. 2004. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends in Genetics 20: 116–122. [DOI] [PubMed] [Google Scholar]
- Levitt M. 2009. Nature of the protein universe. Proceedings of the National Academy of Sciences of the USA 106: 11079–11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, et al. 2010. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant Journal 63: 86–99. [DOI] [PubMed] [Google Scholar]
- Lin Y, Cheng Y, Jin J, et al. 2014. Genome duplication and gene loss affect the evolution of heat shock transcription factor genes in legumes. PLoS One 9: e102825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CMJ.. 2006. Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology 44: 135–162. [DOI] [PubMed] [Google Scholar]
- Lynch M. 2013. Evolutionary diversification of the multimeric states of proteins. Proceedings of the National Academy of Sciences of the USA 110: 2821–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Melo F, Feytmans E.. 1998. Assessing protein structures with a non-local atomic interaction energy. Journal of Molecular Biology 277: 1141–1152. [DOI] [PubMed] [Google Scholar]
- Mitchell AL, Chang HY, Daugherty L, et al. 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Research 43: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AD, Bornberg-Bauer E.. 2012. The dynamics and evolutionary potential of domain loss and emergence. Molecular Biology and Evolution 29: 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AD, Björklund A, Ekman D, Bornberg-Bauer E, Elofsson A.. 2008. Arrangements in the modular evolution of proteins. Trends in Biochemical Sciences 33: 444–451. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1970. Why gene duplication? In: Evolution by gene duplication. Berlin: Springer, 59–88. [Google Scholar]
- Otto SP, Whitton J.. 2000. Polyploid incidence and evolution. Annual Review of Genetics 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A.. 2009. The evolutionary significance of ancient genome duplications. Nature Reviews Genetics 10: 725–732. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. Journal of Computational Chemistry 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Rentzsch R, Orengo CA.. 2013. Protein function prediction using domain families. BMC Bioinformatics 14 (Suppl): S5:1–S5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin A, Auer PL, Libault M, et al. 2013. The fate of duplicated genes in a polyploid plant genome. The Plant Journal 73: 143–153. [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, et al. 2008. Genome structure of the legume, Lotus japonicus. DNA Research 15: 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaper E, Anisimova M.. 2015. The evolution and function of protein tandem repeats in plants. New Phytologist 206: 397–410. [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, et al. 2010. Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Schmutz J, McClean PE, Mamidi S, et al. 2014. A reference genome for common bean and genome-wide analysis of dual domestications. Nature Genetics 46: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouppe D, Rougé P, Lasanajak Y, et al. 2010. Mutational analysis of the carbohydrate binding activity of the tobacco lectin. Glycoconjugate Journal 27: 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin AJ, Cannon SB, Graham MM, Grant D, Shoemaker RC.. 2011. Changes in twelve homoeologous genomic regions in soybean following three rounds of polyploidy. The Plant Cell 23: 3129–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Pandey GK.. 2016. Expansion and function of repeat domain proteins during stress and development in plants. Frontiers in Plant Science 6: 1218. doi:10.3389/fpls.2015.01218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RC, Schlueter J, Doyle JJ.. 2006. Paleopolyploidy and gene duplication in soybean and other legumes. Current Opinion in Plant Biology 9: 104–109. [DOI] [PubMed] [Google Scholar]
- Simpson PJ, Jamieson SJ, Abou-Hachem M, et al. 2002. The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase. Biochemistry 41: 5712–5719. [DOI] [PubMed] [Google Scholar]
- Singh VK, Jain M.. 2015. Genome-wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean. Frontiers in Plant Science 6: 918. doi:10.3389/fpls.2015.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanowicz K, Lannoo N, Proost P, Van Damme EJM.. 2012. Arabidopsis F-box protein containing a Nictaba-related lectin domain interacts with N-acetyllactosamine structures. FEBS Open Bio 2: 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanowicz K, Lannoo N, Zhao Y, et al. 2016. Glycan-binding F-box protein from Arabidopsis thaliana protects plants from Pseudomonas syringae infection. BMC Plant Biology 16: 213. doi:10.1186/s12870-016-0905-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Raes J.. 2004. Duplication and divergence: the evolution of new genes and old ideas. Annual Review of Genetics 38: 615–643. [DOI] [PubMed] [Google Scholar]
- Vandenborre G, Miersch O, Hause B, Smagghe G, Wasternack C, Van Damme EJM.. 2009. Spodoptera littoralis-induced lectin expression in tobacco. Plant and Cell Physiology 50: 1142–1155. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Chen W, Li Y, et al. 2011. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nature Biotechnology 30: 83–89. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Song C, Saxena RK, et al. 2013. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nature Biotechnology 31: 240–246. [DOI] [PubMed] [Google Scholar]
- Vogel C, Morea V.. 2006. Duplication, divergence and formation of novel protein topologies. BioEssays 28: 973–978. [DOI] [PubMed] [Google Scholar]
- Vogel C, Teichmann SA, Pereira-Leal J.. 2005. The relationship between domain duplication and recombination. Journal of Molecular Biology 346: 355–365. [DOI] [PubMed] [Google Scholar]
- Weiner J, Beaussart F, Bornberg-Bauer E.. 2006. Domain deletions and substitutions in the modular protein evolution. FEBS Journal 273: 2037–2047. [DOI] [PubMed] [Google Scholar]
- Wendel JF. 2000. Genome evolution in polyploids. Plant Molecular Biology 42: 225–249. [PubMed] [Google Scholar]
- Yang S, Bourne PE.. 2009. The evolutionary history of protein domains viewed by species phylogeny. PLoS One 4: e8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Tao Y, Zheng Z, et al. 2013. Draft genome sequence, and a sequence-defined genetic linkage map of the legume crop species Lupinus angustifolius L. PLoS One 8: e64799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GED, et al. 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2003. Evolution by gene duplication: an update. Trends in Ecology and Evolution 18: 292–298. [Google Scholar]
- Zhang M, Chekan JR, Dodd D, et al. 2014. Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes. Proceedings of the National Academy of Sciences of the USA 111: E3708–E3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.