Abstract
Bovine leukemia virus (BLV) is the causative agent of enzootic bovine leukosis, a malignant B cell lymphoma that has spread worldwide and causes serious problems for the cattle industry. The BLV proviral load, which represents the BLV genome integrated into host genome, is a useful index for estimating disease progression and transmission risk. Here, we conducted a genome-wide association study to identify single nucleotide polymorphisms (SNPs) associated with BLV proviral load in Japanese Black cattle. The study examined 93 cattle with a high proviral load and 266 with a low proviral load. Three SNPs showed a significant association with proviral load. One SNP was detected in the CNTN3 gene on chromosome 22, and two (which were not in linkage disequilibrium) were detected in the bovine major histocompatibility complex region on chromosome 23. These results suggest that polymorphisms in the major histocompatibility complex region affect proviral load. This is the first report to detect SNPs associated with BLV proviral load in Japanese Black cattle using whole genome association study, and understanding host factors may provide important clues for controlling the spread of BLV in Japanese Black cattle.
Electronic supplementary material
The online version of this article (doi:10.1186/s12977-017-0348-3) contains supplementary material, which is available to authorized users.
Keywords: Bovine leukemia virus, Whole genome association study, Major histocompatibility complex
Main text
Bovine leukemia virus (BLV), which infects cattle worldwide [1–6], belongs to the family Retroviridae (genus Deltaretrovirus), together with human T cell leukemia virus types 1 and 2 (HTLV-1 and -2) [7]. Historically, the economic losses caused by BLV infection were thought to be related only to the onset of bovine leukosis, which occurs in only 1–5% of BLV-infected cows within 5 years post-infection [7]. However, recent reports show that BLV infection also reduces milk production [6, 8–11] and causes a high incidence of infectious disease [12] and reproductive inefficiency, resulting in high culling rates [13]; thus BLV eradication is of utmost importance.
Previous studies show that the proviral load is an important index for estimating the stage of BLV infection because it is associated with disease progression [14–16], lymphocyte count [17], viral biokinetics [18], and virus shedding into saliva and nasal secretions [19]. Indeed, one study shows that cattle with a low proviral load are not a source of BLV transmission [20]. Therefore, determining host factors associated with an increased proviral load is important if we are to develop eradication programs for BLV.
Studies of BLV-associated host factors identified polymorphisms within the bovine major histocompatibility complex (MHC) (BoLA) [21–29]. Recently, Miyasaka et al. revealed that polymorphisms within BoLA class II haplotypes were strongly associated with BLV proviral load in Japanese Black cattle, the main breed of beef cattle in Japan, but less so with that in European breeds [22]. However, no group has undertaken a genome-wide association study (GWAS) to identify such host factors.
Therefore, to identify proviral load-associated polymorphisms, we performed a GWAS using DNA samples from 676 Japanese Black cattle [30]. Genomic DNA was isolated from peripheral blood, and the BLV proviral load was measured using the BLV-CoCoMo-qPCR-2 method [31]. BLV provirus was detected in samples from 444 animals (range, 1 copy/105 cells to 132,230 copies/105 cells; median value, 5498 copies/105 cells) (Fig. 1a). We then compared the proviral load in animals used for the GWAS with that in Japanese Black cattle selected randomly from whole areas of Japan. We found no significant difference in the proviral load between animals used for GWAS and the randomly selected group (Fig. 1b). In most cases, the animals in both groups showed a proviral load of <10,000 copies/105 cells. A proviral load >100,000 copies/105 cells was rare.
Fig. 1.
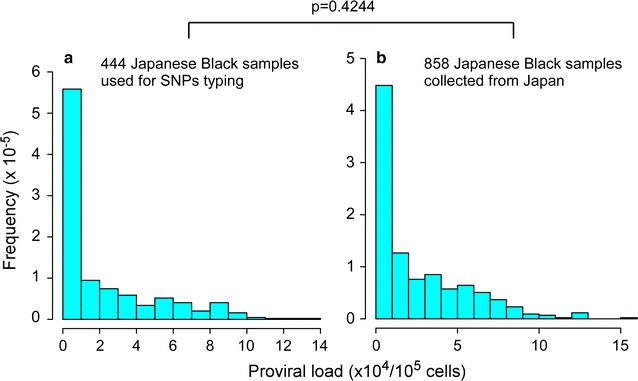
Proviral load estimated from SNP typing of DNA samples from 444 BLV-infected Japanese Black cattle (a) and 858 samples from Japanese Black cattle located in 22 prefectures of Japan (b) [17]. The proviral load in the 444 test samples was representative of the proviral load in Japanese Black cattle nationwide (p value, p = 0.4244; F test). Blood (collected in EDTA-2Na) was obtained from 444 Japanese black cows (aged >4 years), and genomic DNA was extracted from whole blood using the QIAsymphony kit (QIAGEN K.K., Tokyo, Japan). The BLV-CoCoMo-qPCR-2 method (RIKEN genesis, Kanagawa, Japan) was used to measure the BLV proviral load in 676 cattle at a single time-point; of these, 444 were positive for BLV and entered into the association study. Briefly, the BLV long terminal repeat region was amplified using a degenerate primer pair (CoCoMo-FRW and CoCoMo-REV) and an FAM-BLV probe. The BoLA-DRA gene (internal control) was amplified using the primer pair DRA-F and DRA-R and the FAM-DRA probe [31]
We categorized the 444 BLV-infected cows into four groups according to proviral load: Low (0 < provirus load ≤ 13,819, 266 heads), Medium (14,237 < provirus load ≤ 40,698, 85 heads), High (42,605 < provirus load ≤ 73,145, 60 heads), and Very High (76,397 < provirus load ≤ 132,230, 33 heads). We then performed a GWAS using these traits as a binary variable, as is done in 93 case (High + Very High group) − 266 control (Low group) studies. The 359 animals were genotyped using a SNP50 K BeadChip comprising probes targeting 54,001 single nucleotide polymorphisms (SNPs). In all, 32,919 autosomal SNPs met the quality control criteria (call rate >99%; minor allele frequency >0.01; Hardy–Weinberg equilibrium, p > 0.001). Analyses were then performed using GEMMA software [32], which uses a linear-mixed model approach based on a genetic-relationship matrix estimated from SNP genotypes to model correlations between the phenotypes of sample subjects. The genomic-inflation factor (λGC) for this analysis was 1.021, indicating that a sample was appropriate for inclusion in an association study. The quantile–quantile (Q–Q) plot showed that three SNPs showed a significant deviation from the null hypothesis (Fig. 2b, Bonferroni-corrected threshold for genome-wide significance (p < 1.5 × 10−6) add threshold line in A). Three significant genome-wide associations were detected: rs29026690 (p = 1.91 × 10−7, odds ratio = 2.745) and rs17872126 (p = 1.91 × 10−7, odds ratio = 0.414) on bovine chromosome 23 (BTA23) and rs110616206 (p = 5.37 × 10−7, odds ratio = 6.589) on BTA22 (Fig. 2b; Table 1). The two SNPs on BTA23 were found within an 800 Kb window located at 27,421,348–28,223,274 bp; these two SNPs did not show linkage disequilibrium (LD) (r 2 = 0.117), indicating that BTA23 harbored two independent quantitative trait loci (QTL)s (Figs. 2b, 3; Table 1).
Fig. 2.
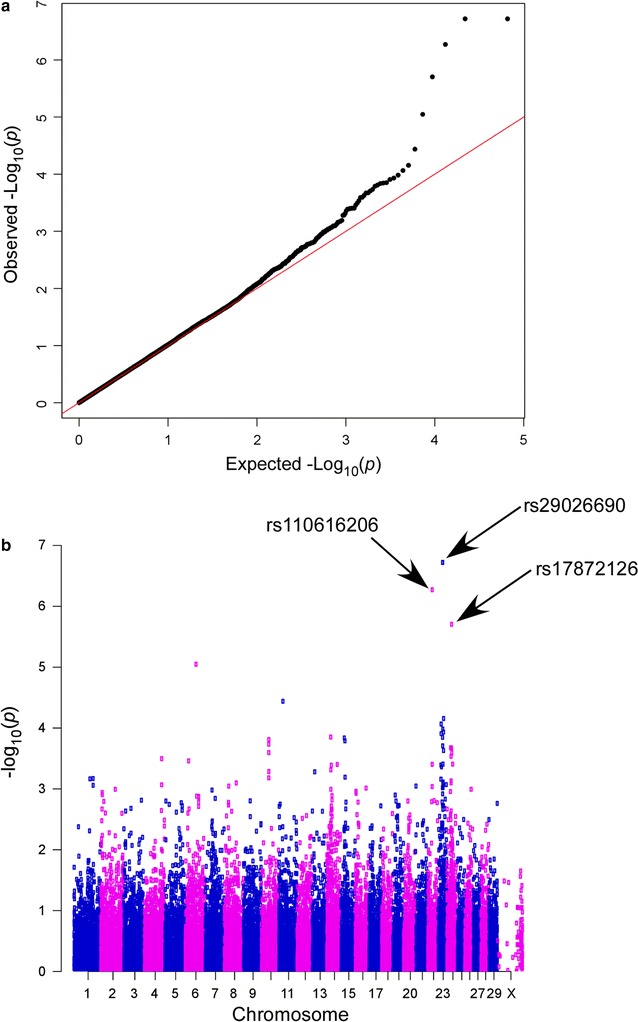
Three-hundred and fifty-nine BLV-infected cows were genotyped using a BovineSNP50 DNA Analysis BeadChip (Illumina Inc., San Diego, CA), and SNPs associated with the BLV proviral load were examined. a Quantile–quantile (Q–Q) plot. The observed distribution of the −log10 nominal p values (y-axis) demonstrates a significant departure from the null hypothesis (expected values are shown on the x-axis) (λGC = 1.021). Red line represents the line as y = x. b Manhattan plot showing the association between 33,006 SNPs (BovineSNP BeadChip) and the BLV proviral load in DNA samples from 359 Japanese Black cattle. The chromosomes are denoted by different colors (blue odd numbers; orange even numbers). The chromosome number is indicated on the x-axis. The blue line represents the Bonferroni-corrected threshold for genome-wide significance (−log10(p) = 5.82)
Table 1.
SNPs showing a significant association with BLV proviral load
| Chromosome | Ilumina_IDa | Reference cluster IDb | Positionc | p | Minor alleled | Minor alleled (case) | Minor alleled (control) | Major allelee | Odds ratiof |
|---|---|---|---|---|---|---|---|---|---|
| 23 | Hapmap57616-rs29026690 | rs29026690 | 27421348 | 1.91 × 10−7 | A | 0.2903 | 0.1297 | G | 2.745 |
| 23 | ARS-BFGL-NGS-113235 | rs17872126 | 28223274 | 1.91 × 10−7 | G | 0.2849 | 0.4906 | A | 0.414 |
| 22 | Hapmap33580-BTA-136506 | rs110616206 | 27280154 | 5.37 × 10−7 | A | 0.0914 | 0.01504 | G | 6.589 |
aSNP ID assigned by Illumina, Inc
bReference SNP (refSNP) ID assigned in the single nucleotide polymorphism database (dbSNP)
cPositions are based on the bovine genome, assembled in UMD3.1
dMinor allele is minor frequency allele determined in this study
eMajor allele is major frequency allele determined in this study
fOdds ratio is the effective value for estimating how strongly the SNPs associated to the proviral load, using following formula
Fig. 3.
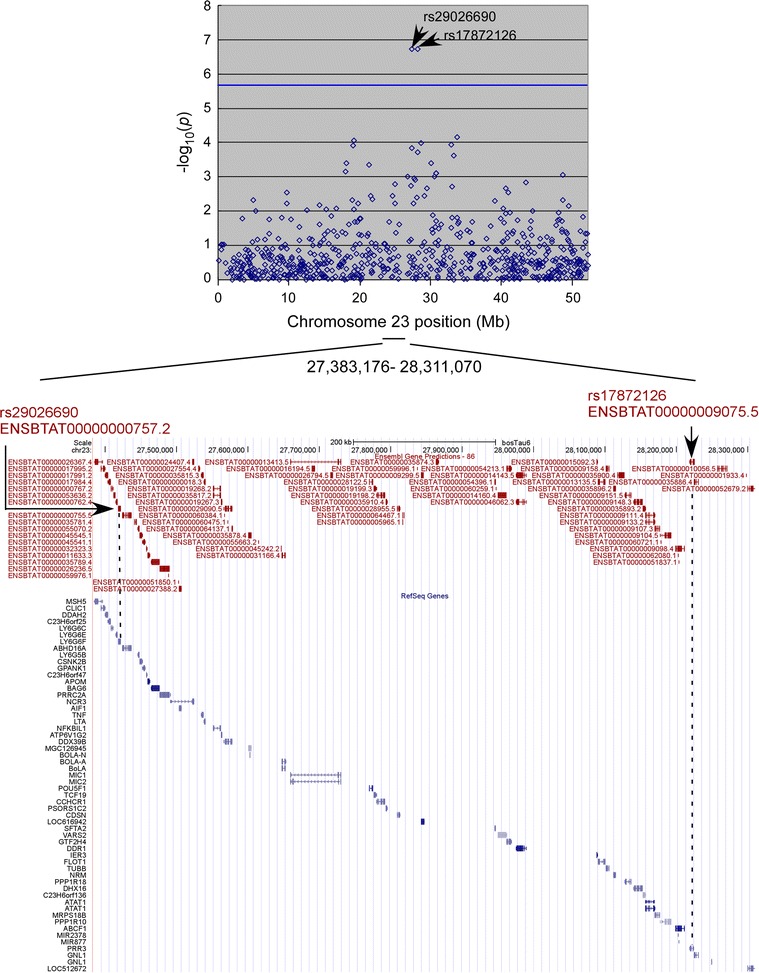
Regional Manhattan plot of the locus on chromosome 23 that harbors SNPs associated with BLV proviral load. The imputed SNPs are shown by arrows. Genes (Chr23:27,116,737 to Chr23:28,311,070) are listed, and the positions of SNPs associated with the BLV proviral load are indicated by arrows. The horizontal blue lines represent the Bonferroni-corrected thresholds for genome-wide significance (−log10(p) = 5.82). The indicated positions are based on the bovine genome (assembled in UMD3.1)
Genes within or near these regions were then analyzed using the UMD3.1 genome assembly tool. Hapmap57616-rs29026690 (27,421,348 bp on BTA23) was located between ENSBTAG00000000580 and ABHD16A (ENSBTAG00000000578) (Additional file 1: Table S1), whereas ARS-BFGL-NGS-113235 (28,223,274 bp on BTA23) was located between the 4th and 5th exons of PRR3 (ENSBTAG00000006914). These SNPs reside within the BoLA class III and class I regions, respectively (Figs. 2b, 3; Additional file 1: Table S1). Therefore, the gene density was much higher than that in other areas of the genome, and a number of candidate genes that could be used to estimate proviral load were present around the detected SNPs [33]. Hapmap33580-BTA-136506 was located on the centromeric side of BTA22, at a distance of 6.5 kb from the CONTACTIN3 (CNTN3) gene (Table 1; Additional file 1: Table S1, Additional file 2: Fig. S1).
To the best of our knowledge, this is the first report to detect SNPs associated with BLV proviral load in Japanese Black cattle using GWAS. Two of the identified SNPs were located in the BoLA region. We found it interesting that these two SNPs were located within the class III and class I regions because a previous study reported involvement of only class II genes [22]. The genome reference sequences for the BoLA region have many gaps, mainly because class I genes were difficult to genotype, making associations with class I genes difficult to determine. Target resequencing of high density SNPs across the MHC region using a next generation sequencer should be undertaken to confirm which genes are truly responsible for regulating the proviral load. Our result showed that the MHC polymorphism is important factor for proviral load. The reason why MHC polymorphisms were associated with proviral load is the polymorphism of classical MHC directly associate with antigen presentation and the difference of antigen presentation in each allele leads to the immunological difference in each host.
Taken together, the results described herein show that MHC genotyping of class III and class I alleles can identify cows with a low proviral load. In the farm with high infection rate, eliminating high proviral load cow is an effective way for eradicating BLV because proviral load is major risk factor for transmitting BLV to other host [20]. Therefore, farmer should frequently check the proviral load because the proviral load is variable, although it is not cost-effective. Taken together with the information of our finding 3 SNPs and our previously report about resistant BoLA class II allele [22], we can identify the BLV resistant cow. It will be helpful to develop a low cost method of eradicating BLV from farms because we can reduce the frequently measurement of proviral load.
Additional files
Additional file 1: Table S1. Genes around rs110616206 SNP (Tab "CNTN3") and rs29026690 and rs17872126 SNPs (Tab "BTA23").
Additional file 2: Figure S1. Regional Manhattan plot showing the association of 33,006 SNPs (BovineSNP BeadChip) with BLV proviral load in 359 Japanese Black cattle. Regional plot of the locus on chromosome 22 that harbors SNPs associated with BLV proviral load. The imputed SNPs are indicated by arrows. The horizontal blue lines represent the Bonferroni-corrected thresholds for genome-wide significance (−log10(p) = 5.82). The indicated positions are based on the bovine genome (assembled in UMD3.1).
Authors’ contributions
Study conception and design: YA. Data acquisition, analysis, and interpretation: SS, MP and ST. Contribution of reagents/materials/analysis tools: YA and YS. Drafting and revising the manuscript: ST, YA, and SS. Approval of the final version: ST, SS, MP, YS, and YA. All authors read and approved the final manuscript.
Acknowledgements
We thank the Support Unit at the Biomaterial Analysis, RIKEN BSI Research Resources Center, for help with sequence analysis.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets analyzed herein are available from the corresponding author upon request.
Ethics approval and consent to participate
All protocols for the collection of blood samples were reviewed and approved by the Shirakawa Institute of Animal Genetics Committee on Animal Research (H21-2).
Funding
The study was supported by Grants-in-Aid for Scientific Research (A and C) from the Japan Society for the Promotion of Science (JSPS) (Grant Nos. 18255013, 25450405), by a grant from the Integration Research for Agriculture and Interdisciplinary Fields in Japan (Grant No. 14538311). This research was also supported by grants from the Project of the NARO Bio-oriented Technology Research Advancement Institution (Grant No. 16817983) (the special scheme project on regional developing strategy).
Contributor Information
Shin-nosuke Takeshima, Email: takesima@riken.jp.
Shinji Sasaki, Email: sasakis@siag.or.jp.
Polat Meripet, Email: palati.mairepati@riken.jp.
Yoshikazu Sugimoto, Email: kazusugi@siag.or.jp.
Yoko Aida, Email: aida@riken.jp.
References
- 1.Erskine RJ, Bartlett PC, Byrem TM, Render CL, Febvay C, Houseman JT. Using a herd profile to determine age-specific prevalence of bovine leukemia virus in Michigan dairy herds. Vet Med Int. 2012;2012:350374. doi: 10.1155/2012/350374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polat M, Ohno A, Takeshima SN, Kim J, Kikuya M, Matsumoto Y, Mingala CN, Onuma M, Aida Y. Detection and molecular characterization of bovine leukemia virus in Philippine cattle. Arch Virol. 2015;160:285–296. doi: 10.1007/s00705-014-2280-3. [DOI] [PubMed] [Google Scholar]
- 3.Polat M, Takeshima SN, Hosomichi K, Kim J, Miyasaka T, Yamada K, Arainga M, Murakami T, Matsumoto Y, de la Barra Diaz V, et al. A new genotype of bovine leukemia virus in South America identified by NGS-based whole genome sequencing and molecular evolutionary genetic analysis. Retrovirology. 2016;13:4. doi: 10.1186/s12977-016-0239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polat M, Moe HH, Shimogiri T, Moe KK, Takeshima SN, Aida Y. The molecular epidemiological study of bovine leukemia virus infection in Myanmar cattle. Arch Virol. 2017;162:425–37. [DOI] [PubMed]
- 5.Zhang R, Jiang J, Sun W, Zhang J, Huang K, Gu X, Yang Y, Xu X, Shi Y, Wang C. Lack of association between bovine leukemia virus and breast cancer in Chinese patients. Breast Cancer Res. 2016;18:101. doi: 10.1186/s13058-016-0763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y, Fan W, Mao Y, Yang Z, Lu G, Zhang R, Zhang H, Szeto C, Wang C. Bovine leukemia virus infection in cattle of China: association with reduced milk production and increased somatic cell score. J Dairy Sci. 2016;99:3688–3697. doi: 10.3168/jds.2015-10580. [DOI] [PubMed] [Google Scholar]
- 7.Aida Y, Murakami H, Takahashi M, Takeshima SN. Mechanisms of pathogenesis induced by bovine leukemia virus as a model for human T-cell leukemia virus. Front Microbiol. 2013;4:328. doi: 10.3389/fmicb.2013.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emanuelson U, Scherling K, Pettersson H. Relationships between herd bovine leukemia-virus infection status and reproduction, disease incidence, and productivity in Swedish dairy herds. Prev Vet Med. 1992;12:121–131. doi: 10.1016/0167-5877(92)90075-Q. [DOI] [Google Scholar]
- 9.Da Y, Shanks RD, Stewart JA, Lewin HA. Milk and fat yields decline in bovine leukemia virus-infected Holstein cattle with persistent lymphocytosis. Proc Natl Acad Sci USA. 1993;90:6538–6541. doi: 10.1073/pnas.90.14.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norby B, Bartlett PC, Byrem TM, Erskine RJ. Effect of infection with bovine leukemia virus on milk production in Michigan dairy cows. J Dairy Sci. 2016;99:2043–2052. doi: 10.3168/jds.2015-10089. [DOI] [PubMed] [Google Scholar]
- 11.Nekouei O, VanLeeuwen J, Stryhn H, Kelton D, Keefe G. Lifetime effects of infection with bovine leukemia virus on longevity and milk production of dairy cows. Prev Vet Med. 2016;133:1–9. doi: 10.1016/j.prevetmed.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Sandev N, Koleva M, Binev R, Ilieva D. Influence of enzootic bovine leukosis virus upon the incidence of subclinical mastitis in cows at a different stage of infection. Vet Arh. 2004;74:411–416. [Google Scholar]
- 13.Bartlett PC, Norby B, Byrem TM, Parmelee A, Ledergerber JT, Erskine RJ. Bovine leukemia virus and cow longevity in Michigan dairy herds. J Dairy Sci. 2013;96:1591–1597. doi: 10.3168/jds.2012-5930. [DOI] [PubMed] [Google Scholar]
- 14.Jimba M, Takeshima SN, Matoba K, Endoh D, Aida Y. BLV-CoCoMo-qPCR: quantitation of bovine leukemia virus proviral load using the CoCoMo algorithm. Retrovirology. 2010;7:91. doi: 10.1186/1742-4690-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jimba M, Takeshima SN, Murakami H, Kohara J, Kobayashi N, Matsuhashi T, Ohmori T, Nunoya T, Aida Y. BLV-CoCoMo-qPCR: a useful tool for evaluating bovine leukemia virus infection status. BMC Vet Res. 2012;8:167. doi: 10.1186/1746-6148-8-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Somura Y, Sugiyama E, Fujikawa H, Murakami K. Comparison of the copy numbers of bovine leukemia virus in the lymph nodes of cattle with enzootic bovine leukosis and cattle with latent infection. Arch Virol. 2014;159:2693–2697. doi: 10.1007/s00705-014-2137-9. [DOI] [PubMed] [Google Scholar]
- 17.Ohno A, Takeshima SN, Matsumoto Y, Aida Y. Risk factors associated with increased bovine leukemia virus proviral load in infected cattle in Japan from 2012 to 2014. Virus Res. 2015;210:283–290. doi: 10.1016/j.virusres.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 18.Panei CJ, Takeshima SN, Omori T, Nunoya T, Davis WC, Ishizaki H, Matoba K, Aida Y. Estimation of bovine leukemia virus (BLV) proviral load harbored by lymphocyte subpopulations in BLV-infected cattle at the subclinical stage of enzootic bovine leucosis using BLV-CoCoMo-qPCR. BMC Vet Res. 2013;9:95. doi: 10.1186/1746-6148-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Y, Kitamura-Muramatsu Y, Saito S, Ishizaki H, Nakano M, Haga S, Matoba K, Ohno A, Murakami H, Takeshima SN, Aida Y. Detection of the BLV provirus from nasal secretion and saliva samples using BLV-CoCoMo-qPCR-2: comparison with blood samples from the same cattle. Virus Res. 2015;210:248–254. doi: 10.1016/j.virusres.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Juliarena MA, Barrios CN, Ceriani MC, Esteban EN. Hot topic: bovine leukemia virus (BLV)-infected cows with low proviral load are not a source of infection for BLV-free cattle. J Dairy Sci. 2016;99:4586–4589. doi: 10.3168/jds.2015-10480. [DOI] [PubMed] [Google Scholar]
- 21.Nikbakht Brujeni G, Ghorbanpour R, Esmailnejad A. Association of BoLA-DRB3.2 alleles with BLV infection profiles (persistent lymphocytosis/lymphosarcoma) and lymphocyte subsets in Iranian Holstein cattle. Biochem Genet. 2016;54:194–207. doi: 10.1007/s10528-016-9712-6. [DOI] [PubMed] [Google Scholar]
- 22.Miyasaka T, Takeshima SN, Jimba M, Matsumoto Y, Kobayashi N, Matsuhashi T, Sentsui H, Aida Y. Identification of bovine leukocyte antigen class II haplotypes associated with variations in bovine leukemia virus proviral load in Japanese Black cattle. Tissue Antigens. 2013;81:72–82. doi: 10.1111/tan.12041. [DOI] [PubMed] [Google Scholar]
- 23.Juliarena MA, Poli M, Sala L, Ceriani C, Gutierrez S, Dolcini G, Rodriguez EM, Marino B, Rodriguez-Dubra C, Esteban EN. Association of BLV infection profiles with alleles of the BoLA-DRB3.2 gene. Anim Genet. 2008;39:432–438. doi: 10.1111/j.1365-2052.2008.01750.x. [DOI] [PubMed] [Google Scholar]
- 24.Aida Y, Nishino Y, Amanuma H, Murakami K, Okada K, Ikawa Y. The role of tumor-associated antigen in bovine leukemia virus-induced lymphosarcoma. Leukemia. 1997;11(Suppl 3):216–218. [PubMed] [Google Scholar]
- 25.Zanotti M, Poli G, Ponti W, Polli M, Rocchi M, Bolzani E, Longeri M, Russo S, Lewin HA, van Eijk MJ. Association of BoLA class II haplotypes with subclinical progression of bovine leukaemia virus infection in Holstein–Friesian cattle. Anim Genet. 1996;27:337–341. [PubMed] [Google Scholar]
- 26.Xu A, van Eijk MJ, Park C, Lewin HA. Polymorphism in BoLA-DRB3 exon 2 correlates with resistance to persistent lymphocytosis caused by bovine leukemia virus. J Immunol. 1993;151:6977–6985. [PubMed] [Google Scholar]
- 27.Stear MJ, Dimmock CK, Newman MJ, Nicholas FW. BoLA antigens are associated with increased frequency of persistent lymphocytosis in bovine leukaemia virus infected cattle and with increased incidence of antibodies to bovine leukaemia virus. Anim Genet. 1988;19:151–158. doi: 10.1111/j.1365-2052.1988.tb00800.x. [DOI] [PubMed] [Google Scholar]
- 28.Lewin HA, Wu MC, Stewart JA, Nolan TJ. Association between BoLA and subclinical bovine leukemia virus infection in a herd of Holstein–Friesian cows. Immunogenetics. 1988;27:338–344. doi: 10.1007/BF00395129. [DOI] [PubMed] [Google Scholar]
- 29.Lewin HA, Bernoco D. Evidence for BoLA-linked resistance and susceptibility to subclinical progression of bovine leukaemia virus infection. Anim Genet. 1986;17:197–207. doi: 10.1111/j.1365-2052.1986.tb03191.x. [DOI] [PubMed] [Google Scholar]
- 30.Sasaki S, Ibi T, Watanabe T, Matsuhashi T, Ikeda S, Sugimoto Y. Variants in the 3′ UTR of general transcription factor IIF, polypeptide 2 affect female calving efficiency in Japanese Black cattle. BMC Genet. 2013;14:41. doi: 10.1186/1471-2156-14-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeshima SN, Kitamura-Muramatsu Y, Yuan Y, Polat M, Saito S, Aida Y. BLV-CoCoMo-qPCR-2: improvements to the BLV-CoCoMo-qPCR assay for bovine leukemia virus by reducing primer degeneracy and constructing an optimal standard curve. Arch Virol. 2015;160:1325–1332. doi: 10.1007/s00705-015-2377-3. [DOI] [PubMed] [Google Scholar]
- 32.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821-U136. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Consortium TMs Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Genes around rs110616206 SNP (Tab "CNTN3") and rs29026690 and rs17872126 SNPs (Tab "BTA23").
Additional file 2: Figure S1. Regional Manhattan plot showing the association of 33,006 SNPs (BovineSNP BeadChip) with BLV proviral load in 359 Japanese Black cattle. Regional plot of the locus on chromosome 22 that harbors SNPs associated with BLV proviral load. The imputed SNPs are indicated by arrows. The horizontal blue lines represent the Bonferroni-corrected thresholds for genome-wide significance (−log10(p) = 5.82). The indicated positions are based on the bovine genome (assembled in UMD3.1).
Data Availability Statement
The datasets analyzed herein are available from the corresponding author upon request.


