Abstract
Background
Colorectal cancers (CRCs) that lack DNA mismatch repair function exhibit the microsatellite unstable (MSI) phenotype and are characterized by the accumulation of frameshift mutations at short repetitive DNA sequences (microsatellites). These tumors recurrently show inactivating frameshift mutations in the tumor suppressor Transforming Growth Factor Beta Receptor Type 2 (TGFBR2) thereby abrogating downstream signaling. How altered TGFBR2 signaling affects exosome-mediated communication between MSI tumor cells and their environment has not been resolved. Here, we report on molecular alterations of exosomes shed by MSI cells and the biological response evoked in recipient cells.
Methods
Exosomes were isolated and characterized by electron microscopy, nanoparticle tracking, and western blot analysis. TGFBR2-dependent effects on the cargo and functions of exosomes were studied in a MSI CRC model cell line enabling reconstituted and inducible TGFBR2 expression and signaling. Microsatellite frameshift mutations in exosomal and cellular DNA were examined by PCR-based DNA fragment analysis and exosomal protein profiles were identified by mass spectrometry. Uptake of fluorescent-labeled exosomes by hepatoma recipient cells was monitored by confocal microscopy. TGFBR2-dependent exosomal effects on secreted cytokine levels of recipient cells were analyzed by Luminex technology and ELISA.
Results
Frameshift mutation patterns in microsatellite stretches of TGFBR2 and other MSI target genes were found to be reflected in the cargo of MSI CRC-derived exosomes. At the proteome level, reconstituted TGFBR2 expression and signaling uncovered two protein subsets exclusively occurring in exosomes derived from TGFBR2-deficient (14 proteins) or TGFBR2-proficient (five proteins) MSI donor cells. Uptake of these exosomes by recipient cells caused increased secretion (2–6 fold) of specific cytokines (Interleukin-4, Stem Cell Factor, Platelet-derived Growth Factor-B), depending on the TGFBR2 expression status of the tumor cell.
Conclusion
Our results indicate that the coding MSI phenotype of DNA mismatch repair-deficient CRC cells is maintained in their exosomal DNA. Moreover, we uncovered that a recurrent MSI tumor driver mutation like TGFBR2 can reprogram the protein content of MSI cell-derived exosomes and in turn modulate the cytokine secretion profile of recipient cells. Apart from its diagnostic potential, these TGFBR2-dependent exosomal molecular and proteomic signatures might help to understand the signaling routes used by MSI tumors.
Graphical Abstract
Fricke et al. uncovered coding microsatellite instability-associated mutations of colorectal tumor driver genes like TGFBR2 in MSI tumor cellderived exosomes. Depending on the TGFBR2 expression status of their donor cells, shed exosomes show distinct proteomic signatures and promote altered cytokine secretion profiles in recipient cells.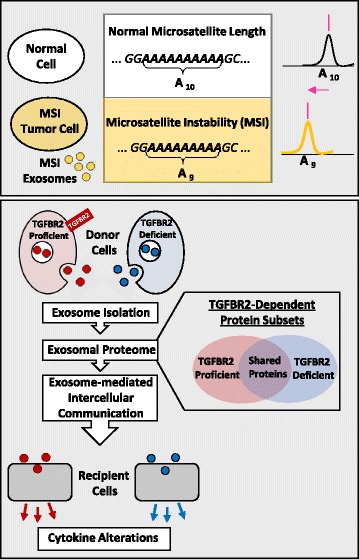
Electronic supplementary material
The online version of this article (doi:10.1186/s12964-017-0169-y) contains supplementary material, which is available to authorized users.
Keywords: Exosomes, Intercellular communication, Proteomics, Transforming Growth Factor Beta Receptor Type 2, DNA mismatch repair deficiency, Microsatellite instability, Colorectal cancer
Plain English summary
A subset of colorectal cancers show the so-called microsatellite instability (MSI) phenotype that is characterized by the accumulation of tumor cell-specific genetic alterations at repetitive DNA sequences. Like many other normal and malignant cells, these MSI tumor cells release 30–150 nm sized vesicles, termed exosomes, to communicate with other cells locally and at distant sites. How MSI tumor cell-specific genetic alterations in the parental cells impact the biology and function of shedded exosomes and in turn affect responder cells remains unresolved.
To address this question, we have isolated and characterized exosomes from MSI colorectal cancer cell lines. We uncovered that the MSI phenotype of the parental cells is shared by their secreted exosomes. It also was observed that the expression status of a tumor driver gene in MSI tumor cells led to specific alterations in the protein content of the released exosomes which in turn elicit a biological response in recipient cells by changing their cytokine secretion profile. Hence, the described molecular and proteomic signatures transmitted by MSI tumor cell-specific exosomes provide novel insights into the biological messages sent by their donor cells and might facilitate the development of new diagnostic and therapeutic approaches.
Background
Adaptation of tumor cells to their continuously changing microenvironment is reflected by genetic and epigenetic alterations and usually accompanied by significant changes in their cellular phenotype. Among several other mechanisms, extracellular vesicle shedding and release of their cargo to target cells can account for these effects [1, 2]. Exosomes constitute a distinct subset of extracellular vesicles about 30–150 nm in diameter and are released by all normal and neoplastic cells. They can accommodate proteins, nucleic acids, lipids, and metabolites [3] and are involved in different biological functions like intercellular communication, coagulation, and immune response modulation. Accumulating evidence suggests that exosomes play a key role in cancer [4]. It has been demonstrated that cancer cell-derived exosomes can transfer oncogenic proteins and nucleic acids to recipient cells, thereby promoting tumor growth, metastasis, and drug resistance [5–7]. For example, exosomes derived from breast cancer cells but not normal cells contain the complete pre-miRNA processing machinery which enables them to alter the transcriptome of target cells in a Dicer-dependent manner, thereby stimulating non-tumorigenic epithelial cells to form tumors [8]. Likewise, colon cancer cell-derived exosomes have been found to be enriched in ΔNp73 mRNA and the proliferation potential of target cells is greatly enhanced by incubation with ΔNp73-containing exosomes [9]. Thus, most of the genetic alterations that exist in the DNA, RNA or protein of the donor cells also occur in the exosomes derived thereof and contribute to the pathogenesis of cancer.
The pathogenesis of DNA mismatch repair-deficient (dMMR) colorectal cancers (CRC) has gained increasing attention, because of distinct clinico-histopathological and molecular features. When compared to their MMR-proficient counterparts they have a more favorable prognosis [10], show an altered chemo-responsiveness [11, 12], have a lower propensity to form distant metastases [13, 14], and exhibit an intense inflammatory response with tumor-infiltrating lymphocytes [15, 16]. As a molecular hallmark, these dMMR tumor cells accumulate numerous somatic small insertion/deletion mutations predominantly at short, repetitive DNA sequences (microsatellites), thereby manifesting the microsatellite instability (MSI) phenotype [17, 18].
When this instability occurs at coding mononucleotide repeats (cMNR), affected genes will generate transcripts with premature termination codons (PTC) that are usually recognized and degraded by the cellular nonsense-mediated RNA decay system [19, 20], but upon translation can give rise of truncated proteins with highly immunogenic frameshift peptide tails [21]. Several studies have identified a large number of cMNR harboring genes frequently affected by frameshift mutations in MSI colorectal tumors [22, 23], but only a limited set of recurrently mutated genes are considered to drive MSI tumorigenesis.
One of these prime MSI targets is the gene encoding the Transforming Growth Factor Beta Receptor Type 2 (TGFBR2), which is part of a key signaling pathway in colon epithelial cells. In canonical TGFBR2-mediated signaling, binding of the TGF-ß1 ligand to this transmembrane receptor causes hetero-tetramerization with type 1 (TGFBR1) receptors, that upon phosphorylation triggers SMAD-mediated signal propagation and execution of cell context-dependent expression programs [24, 25]. Altered TGFBR2 signaling in tumor cells can modulate a wide range of processes like epithelial-to-mesenchymal transition (EMT), migration and invasion, angiogenesis, immunomodulation, and cytokine secretion. Biallelic cMNR frameshift mutations within the TGFBR2 gene arise recurrently in most MSI colorectal tumors and are considered to drive MSI tumorigenesis [26]. In the present study, we explored whether the cellular MSI phenotype is maintained in exosomes and how MSI driver mutations in a major signaling pathway, as exemplified by the TGFBR2 tumor suppressor, can alter the exosomal content of MSI tumor cells and in turn elicit a biological response in specific target cells. It turned out, that the MSI status and the cMNR frameshift mutation allele patterns of MSI colorectal cancer cells is reflected by their shed exosomes. Moreover, using our MSI colorectal cancer cell line model system (HCT116-TGFBR2) that enables the analysis of TGFBR2-dependent cellular alterations in an isogenic background [27] we uncovered distinct differences in exosomal protein signatures depending on the TGFBR2 expression status of their donor cells. Similarly, these exosomes cause significant alterations in the cytokine secretion profile of HepG2 recipient cells in a TGFBR2-dependent manner with PDGF-B exhibiting the most prominent increase in protein expression levels. These results provide strong evidence for TGFBR2 being a potent modulator of exosomal protein content and a modulator of cytokine response in specific target cells.
Methods
Cell culture
dMMR CRC cell lines (HCT116, RKO, LoVo) and the MMR-proficient CRC cell line (SW948) were obtained from ATCC. The generation of the doxycycline-inducible cell line model system HCT116-TGFBR2 was reported previously [27]. KM12 cells were kindly provided by I.J. Fidler and HepG2 by K. Breuhahn. Cells were grown in RPMI 1640 (LoVo, KM12, RKO, HepG2) or DMEM (HCT116, HCT116-TGFBR2) medium supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin (Thermo Fisher Scientific Inc., USA) using standard conditions.
Isolation of exosomes
dMMR CRC cell lines were plated on T175 flasks and grown in complete medium as described above until they reached approximately 80–90% confluency. Cells were washed twice with phosphate-buffered saline (PBS) and cultured for 16 h in minimal volumes (17 ml/T175 flask) of serum-free medium. To investigate TGFBR2-dependent exosomal alterations, HCT116-TGFBR2 cells were cultured in the presence of TGF-ß1 (10 ng/ml) with or without doxycycline (Dox, 0.5 μg/ml). Cell culture media were collected and subjected to sequential centrifugations to remove floating cells (480 × g, 4 °C, 10 min) and cellular debris (2000 × g, 4 °C, 10 min). Supernatants were then passed through a 0.22 μm filter to reduce microparticle contamination and filtrates were concentrated to a final volume of 1 ml by 10,000 molecular weight-cutoff Vivaspin 20 centrifugal concentrators (4000 × g, 4 °C, 30 min; Sartorius, Germany). After addition of 500 μl Total Exosome Isolation Reagent (Thermo Fisher Scientific Inc., USA), samples were incubated overnight at 4 °C on a rotating wheel and then centrifuged at 10,000 x g and 4 °C for 1 h. Exosomal pellets were resuspended either (i) in 100 μl PBS for transmission electron microscopy, (ii) in 200 μl PBS for DNA isolation and fragment analysis, (iii) in 100 μl PBS containing 0.5% bovine serum albumin (BSA) for CFSE-labeling/uptake experiments, or (iv) in 60 μl RIPA buffer in the presence of protease inhibitors for Western blot and mass spectrometry analyses. Isolated extracellular vesicles were stored at −80 °C.
Transmission electron microscopy
Drops of thawed exosome suspensions were left to settle on 100 mesh formvar-coated copper grids (Plano GmbH, Germany), contrasted with 3% aqueous uranyl acetate (negative stain), air dried and visualized using a JEM-1400 transmission microscope (JEOL GmbH, USA) at 80 KV, equipped with a Tietz 2 K digital camera (TVIPS, Germany).
Western blotting
Western blot analysis was performed as described previously [27]. Briefly, 30 μg protein was separated on 4–12% SDS-PAGE gels (Thermo Fisher Scientific Inc., USA) and electro-blotted onto a nitrocellulose membrane. The following primary antibodies were used: mouse monoclonal CD63 antibody (1:500, 4 °C, overnight; Abcam, UK), mouse monoclonal CD9 antibody (1:200, 4 °C, overnight; Santa Cruz, Germany), and mouse anti-ß-Actin (1:1000, RT, 30 min; MP Biomedicals, USA). Subsequently, blots were incubated for 1 h at RT with a sheep anti-mouse-IgG HRP secondary antibody (1:5000; GE-Healthcare, UK). Signals were detected using Western Lightning Plus ECL (Perkin Elmer, USA).
Nanoparticle tracking analysis
Size profiling of isolated vesicles was performed in a dilution of 1:5000 by nanoparticle tracking analysis (NTA) using Nanosight LM10 equipped with a 405 nm laser (Malvern Instruments, UK). The analysis was conducted according to the manufacturer’s instructions by analyzing 30 s-measurements with a slider shutter of 1000 at five different positions per sample.
PCR-based DNA fragment analysis
Total cellular and exosomal DNA was isolated using QIAamp DNA Mini Kit (Qiagen, Germany). After precipitation by ethanol, the cellular and exosomal DNA was used for PCR-based frameshift analysis. PCR amplification of cMNR sequences was performed using specific primers (Additional file 1) designed with primer3 software (http://primer3.ut.ee/) and the following cycler program: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min and final extension for 7 min at 72 °C. Fragment analysis was carried out on an ABI 3130xl Genetic Analyzer (Applied Biosystems, Germany) using the Genescan Analysis Software (Applied Biosystems, Germany).
Mass spectrometry
Sample preparation
Total protein RIPA lysates of exosomes were precipitated using a methanol-chloroform-water mixture [28] followed by in-solution tryptic digestion. Precipitated exosomal proteins were redissolved in 10 μl 40 mM NH4HCO3 and treated with 2 μl 10 mM dithiothreitol (DTT) solution in 40 mM NH4HCO3 at 45 °C for 1 h to completely reduce disulfide bonds. Afterwards, the thiol groups were alkylated by addition of 1 μl 55 mM iodoacetamide solution in 40 mM NH4HCO3 and 30 min incubation in the dark at 25 °C. After adding 2.5 μl DTT solution, the mixture was incubated for 15 min at 37 °C to let all iodoacetamide react with a thiol group. Digestion was performed with 100 ng trypsin (Promega, USA) in 40 mM NH4HCO3 solution overnight at 37 °C. To stop the tryptic digestion, 7.5 μl of 0.1% TFA were added to the sample. In total, 1/5 (i.e. 5 μl) of the sample was subjected to the nano-Liquid Chromatography (LC) electrospray ionization MS/MS analysis. For the analysis, four biological replicates of each sample (JK2358_1–4: exosomes derived from dTGFBR2 cells; JK2358_5–8: exosomes derived from pTGFBR2) have been prepared.
Electrospray ionization MS/MS analysis
Tryptic peptide mixtures were separated by a nanoAcquity ultra-high-performance LC system. Peptides were trapped on a nanoAcquity C18 column (180 μm × 20 mm, 5-μm particle size). The liquid chromatography separation was performed on a C18 column (BEH 130 C18, 100 μm × 100 mm, 1.7-μm particle size) with a flow rate of 400 nl/min. For all samples, the chromatography was carried out using a 3 h gradient of solvent A (98.9% water, 1% acetonitrile, 0.1% formic acid) and solvent B (99.9% acetonitrile and 0.1% formic acid) in the sequence: from 0 to 4% B in 1 min, from 4 to 30% B in 140 min, from 30 to 45% B in 15 min, from 45 to 90% B in 5 min, 10 min at 90% B, from 90 to 0% B in 0.1 min, and 9.9 min at 0% B. The ultra-high-performance LC system was connected online to an LTQ Orbitrap XL mass spectrometer (Thermo Scientific, Germany). The mass spectrometer was operated in the sensitive mode with the adjusted parameters: capillary voltage 2400 V; capillary temperature 200 °C; normalized collision energy 35 V; activation time 30,000 ms. Data were obtained in scan cycles of one Fourier transform MS scan with a resolution of 60,000 at m/z 400 and a range from 370 to 2000 m/z in parallel with six MS/MS scans in the ion trap of the most frequent precursor ions.
Database search and evaluation
The MS files generated by Xcalibur software (version 2.0.6) were used for database searches with the MASCOT search engine (version 2.4; Matrix Science) against the Swiss-Prot database (SwissProt version 2013_02 (539165 sequences; 191456931 residues)) [29]. The taxonomy was set to “human”. The peptide mass tolerance was set to 5 ppm and the fragment mass tolerance was adjusted at 0.4 Dalton. Carbamidomethylation of cystein (C) was set as a fixed modification. Variable modifications included oxidation of methionine (M) and deamidation of asparagine (N) and glutamine (Q). One missed cleavage site in the case of partial trypsin hydrolysis was accepted. The false discovery rates (FDRs) at the protein and peptide level were set to 1%. Proteins were considered as identified, if more than one unique peptide had an individual ion score exceeding the MASCOT identity threshold (ion score cut-off 20). Identification under the performed search parameters refers to a match probability of p < 0.01, where p is the probability that the observed match is a random event. Candidate proteins were classified as differentially expressed (exclusively in exosomes derived from dTGFBR2 or pTGFBR2 cells), if detected in at least three of four biological replicates. The raw data of MS proteomics are deposited to the ProteomeXchange Consortium via the PRIDE partner repository [30] with the project accession number: PXD005620 and project DOI: 10.6019/PXD005620 [31].
Fluorescent labeling of isolated exosomes
Exosomes were isolated from HCT116-TGFBR2 cells as described above. To label intra-exosomal proteins, isolated exosomes were incubated in 5 μM 5(6)-carboxyfluorescein diacetate N-succinimidyl ester (CFSE; Sigma-Aldrich, Germany) at 37 °C for 30 min in the dark. The reaction was stopped by adding two volumes of RPMI medium. After ultracentrifugation (120,000 × g, 4 °C, 2 h; rotor: 100.2; UZ: TLA-100.2, Beckman Coulter, Germany), exosomes were resuspended in 50 μl cold PBS and used directly for uptake experiments.
Tracking of exosomes uptake by confocal microscopy
HepG2 cells were seeded in 0.5 ml of exosome-depleted medium onto a glass bottom dish (35 × 10 mm; Greiner Bio-One International GmbH, Austria) and treated with 50 μl of CFSE-labeled exosomes overnight in the dark at 37 °C in 5% CO2-atm. Next day, the medium was discarded and the cells were washed with 1 ml PBS. Uptake of exosomes by HepG2 cells was analyzed using a Zeiss LSM 710 ConfoCor3 confocal microscope (Carl Zeiss, Germany) equipped with an argon laser and a Plan-Apochromat 63x/1.40 Oil DIC objective (Carl Zeiss, Germany). Excitation was performed at 488 nm and detection was executed in a filter range of 493 to 630 nm. Images were analyzed using IMAGE J-Fiji software [32].
Luminex-based cytokine profiling of HepG2 cells
HepG2 cells (8.5 × 104 cells/cm2) were cultured in 6-well plates for 24 h. Cells were washed with PBS and grown in exosome-depleted 1% FBS-containing RPMI medium in the presence of HCT116-TGFBR2-derived exosomes (20 μg/ml of exosomal protein determined by Bradford Red assay). After 24 h of exosomal exposure, cell culture supernatants were collected and centrifuged (1000 × g, 4 °C, 15 min) and cell numbers in each well were counted. Supernatants were analyzed for 50 secreted cytokines and chemokines using pre-designed Bio-Plex panels (Bio-Plex Pro Human Cytokine 21-, 27-Plex Panel and ICAM-1-, VCAM-1 Assay; Bio-Rad Laboratories GmbH, Germany) according to the manufacturer’s protocol. The standard curve was prepared as a System; Bio-Rad Laboratories GmbH, Germany) with a 532 nm reporter laser and 635 nm classification laser. Cytokine concentrations [pg/ml] were calculated with Bio-Plex Manager software (version 4.0; Bio-Rad Laboratories GmbH, Germany) by optimizing the standard curves for each cytokine. Cytokine concentrations were normalized to corresponding cell numbers and fold changes (pTGFBR2/dTGFBR2) calculated from normalized values.
PDGF-B enzyme-linked immunosorbent assay (ELISA)
HepG2 cells were seeded as biological duplicates in 12-well plates at densities of 8.5 × 104 cells/cm2. Cells were grown in the presence of exosomes as described for the cytokine profiling. Supernatants were collected, centrifuged (1000 × g, 4 °C, 15 min) and the cell numbers of each well were counted. Supernatants were analyzed using the human PDGF-B ELISA (BlueGene Biotech CO., China) according to the manufacturer’s protocol. The OD of each sample was normalized to the corresponding cell number. After subtracting blank value (medium without cells), log10OD values were calculated. For analyzing the concentration [pg/ml] of each sample in accordance with the standard curve, the log10OD-values of the biological replicates were averaged arithmetically. Finally, fold changes were determined by calculating a concentration ratio (pTGFBR2/dTGFBR2).
Results
Characterization of extracellular vesicles Isolated from dMMR colorectal cancer cell lines
To determine the identity of isolated nanovesicles, three different approaches were pursued. First, transmission electron microscopy (TEM) was used for examining the structure of the vesicles. As indicated in Fig. 1a, representative exosomes consisted of cup-shaped closed vesicles that ranged in diameter from 30 to 120 nm and lacked apoptotic bodies and cellular debris. The exosomal appearance was observed in preparations from all dMMR cell lines (HCT116, LoVo, KM12, RKO, HCT116-TGFBR2). Second, Western blot analysis revealed expression of the exosomal marker proteins CD63 and CD9 in protein lysates of exosomes but not in whole cell lysates of HCT116-TGFBR2 donor cells (Fig. 1b). Third, nanoparticle tracking analysis (NTA) confirmed the size distribution of isolated nanovesicles. As shown in Fig. 1c, the major proportion of isolated vesicles ranged in a mono-peak fashion from 50 to 150 nm in diameter. These results indicate the isolation of bona fide exosomes from different dMMR donor cells and these exosomes were used for further investigations.
Fig. 1.
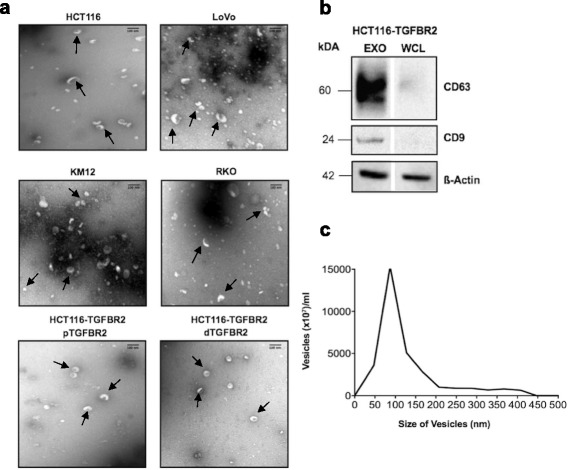
Characterization of isolated exosomes. a Transmission electron microscopy (TEM) illustrates the size and shape of exosomes (indicated by arrows) isolated from different MSI colorectal cancer cell lines (HCT116, LoVo, KM12, RKO) and from the model cell line HCT116-TGFBR2 (pTGFBR2: TGFBR2-proficient, dTGFBR2: TGFBR2-deficient). Smaller particles represent vesicle fragments resulting from the isolation procedure. Scale bar = 100 nm. b Western blot analysis shows tetraspanin (CD63, CD9) marker expression for whole cell lysates (WCL) of HCT116-TGFBR2 donor cells and lysates of derived exosomes (EXO). ß-actin served as a loading control. c Nanoparticle tracking analysis (NTA) indicates size distribution for vesicles isolated from HCT116-TGFBR2 cells (representative of six independent experiments)
Exosomes from different dMMR CRC cell lines accumulate frameshift mutations in coding mononucleotide repeats
In dMMR cells, numerous cMNR-harboring genes are affected by insertion/deletion mutations that constitute the MSI phenotype. Whether genomic DNA harboring such frameshift mutations are encased and detectable in exosomes secreted by dMMR cells is unresolved. Therefore, we isolated exosomal as well as cellular DNA from four dMMR CRC cell lines with known cMNR mutation status (Additional file 2) and examined the cMNR frameshift mutation pattern of three representative MSI target genes (TGFBR2 [A10], LMAN1 [A9], MARCKS [A11]). The allele profile of the CRC cell line SW948 served as control because these MMR-proficient (pMMR) and microsatellite stable cells lack cMNR mutations (Fig. 2). Using PCR-based DNA fragment analysis, allelic length shifts (-1/-2) affecting three representative cMNR gene sequences (MARCKS, TGFBR2, LMAN1) were observed in exosomes of four different dMMR cell lines. In terms of TGFBR2 and LMAN1, we found almost identical mutant and/or wildtype alleles in MSI cell lines and their derived exosomes. However, specific differences between the exosomal and cellular mutant allele pattern became apparent and are highlighted by circled areas in Fig. 2. For example, only mutant alleles (-1 deletions) affecting the A11-cMNR of the MARCKS gene were detected in the exosomes of 3/4 cell lines (HCT116, LoVo, RKO), whereas cellular DNA showed either wildtype alleles (HCT116, KM12, RKO) or a mixed normal and mutant allele pattern (LoVo). Similarly, exosomes of 3/4 cell lines (HCT116, LoVo, RKO) only contained mutant (-1) LMAN1 alleles, while identical mutant or wildtype LMAN1 allele pattern were observed among exosomal and cellular DNA in 3/4 cell lines (HCT116, KM12, LoVo). Interestingly, the most consistent exosomal and cellular cMNR pattern was identified for the TGFBR2 gene. All dMMR cell lines exclusively exhibited TGFBR2 frameshift mutations (−1, −2) and each cell line maintained identical allele pattern intracellularly and within exosomes. These data indicate that the genomic MSI phenotype of dMMR CRC cell lines principally is preserved in the exosomes released by these cells.
Fig. 2.
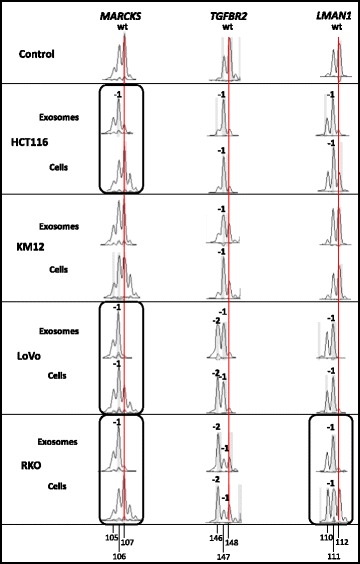
Gene-specific cMNR frameshift mutations in exosomal and cellular DNA of different MSI CRC cell lines. Frameshift mutations are recognized as shifts in allele length as determined by DNA fragment analysis. In the reference cMNR peak pattern of the microsatellite stable cell line SW948 (control) the highest peak refers to the normal wildtype (wt) allele length (red vertical line; reference mark), whereas additional peaks represent PCR-associated artefact peaks. Allele length shifts (-1, -2) were scored if novel peaks were obtained in cell lines compared to the microsatellite stable control SW948 cell line, or if the ratio of peak areas of corresponding peaks in cell lines and normal control revealed values ≤0.5 or ≥2. Differences between exosomal and cellular mutant allele pattern (circled) are indicated. Allele sizes are given by numbers
Expression status of the MSI target gene TGFBR2 modulates the exosomal proteome of dMMR HCT116 CRC cells
Since TGFBR2 cMNR frameshift mutations are considered drivers of dMMR tumors and HCT116 cells and derived exosomes both express only frameshift-mutated and no wildtype TGBFR2 gene pattern, we investigated whether the TGFBR2 expression status might affect the exosomal proteome composition of HCT116 cells. To address this question, we used our previously established model system of HCT116-TGFBR2 cells that have been genetically modified to regulate TGFBR2 expression and signaling in a doxycycline-dependent manner [27]. Exosomes were isolated from the supernatant of HCT116-TGFBR2 cells grown in the absence (dTGFBR2) or presence (pTGFBR2) of doxycycline and total exosomal protein lysates were analyzed by mass spectrometry. In total, 1453 different exosomal proteins have been identified. Among these, 1089 overlapping proteins were detected in exosomes derived from both pTGFBR2 and dTGFBR2 HCT116 cells and hence were considered unaffected by TGFBR2 expression of the dMMR donor cell. This shared exosomal proteome encompassed the top 25 exosomal cargo proteins that often have been identified in previous studies (www.exocarta.org). Furthermore, classical exosomal marker proteins like CD63, CD81, and TSG101 have been detected under both conditions thereby confirming the validity of our experimental approach.
In contrast to this shared proteome, a subset of 364 exosomal proteins was found to be associated with the TGFBR2 expression status of HCT116-TGFBR2 donor cells. In particular, 167 exosomal proteins were identified to be exclusively expressed in exosomes derived from dTGFBR2 cells, whereas 197 proteins remained restricted to exosomal vesicles shed by pTGFBR2 donor cells (Fig. 3). Applying further specific selection criteria revealed a small subset of exosomal proteins (n = 19) that presented consistent (3/4 biological replicates) TGFBR2-dependency and hence marked the exclusive TGFBR2-dependent exosomal proteome (Fig. 3, Table 1). The majority of these proteins were found to be expressed solely in dTGFBR2 exosomes (14/19) and thus might have potential impact on the pathogenesis of most MSI tumors that lack normal TGFBR2 function. Identified proteins include factors involved in RNA binding/processing, and transcriptional regulation (U2AF1, PCBP3), migration, extracellular matrix remodeling (COL3A1), epithelial-to-mesenchymal transition (FAMC3), chromatin structure (HIST2H2AB), cell junction structure (TJP1), regulators of cytoskeletal organization (LRRFIP1, INVS), nuclear/mitochondrial import (LRRC59, TOMM70A), DNA replication (RPA2), and vesicular trafficking (SYT5) as well as enzymes for purine nucleotide synthesis (GMPR2, PRPSAP1).
Fig. 3.
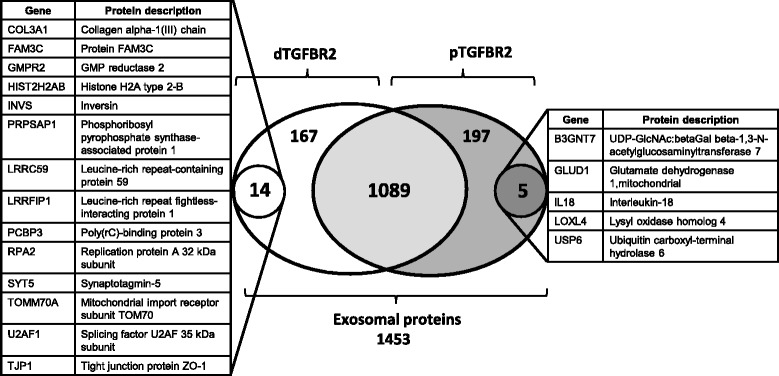
TGFBR2-dependent exosomal proteome profile. Numbers refer to exosomal proteins that comprise the total proteome (n = 1453) or define distinct protein subsets, whose expression is either shared by (n = 1089) or restricted to exosomes released by TGFBR2-deficient (n = 167) or TGFBR2-proficient (n = 197) donor cells. From these two latter proteomes two highly specific protein subsets emerged based on more stringent expression criteria (expression in at least three of four biological replicates, see Methods). Individual protein descriptions and gene names are listed
Table 1.
TGFBR2-dependent exosomal proteome of HCT116 MSI tumor cells
| Uni_Prot | Gene | Protein description | Scores | Mass [Da] | Sign Prot matches | Sign Prot sequences | Coverage [%] |
|---|---|---|---|---|---|---|---|
| dTGFBR2 | |||||||
| CO3A1_HUMAN | COL3A1 | Collagen alpha-1(III) chain | 34 47 42 | 139733 | 1 0 1 | 1 0 1 | 0.8 1.4 0.8 |
| FAM3C_HUMAN | FAM3C | Protein FAM3C | 47 33 52 | 24950 | 1 1 1 | 1 1 1 | 5.7 5.7 5.7 |
| GMPR2_HUMAN | GMPR2 | GMP reductase 2 | 49 37 53 | 38363 | 0 1 0 | 0 1 0 | 8.3 3.4 8.3 |
| H2A2B_HUMAN | HIST2H2AB | Histone H2A type 2-B | 245 261 293 291 | 13987 | 8 7 8 9 | 4 4 4 4 | 52.3 52.3 52.3 52.3 |
| INVS_HUMAN | INVS | Inversin | 31 36 42 | 118837 | 0 2 2 | 0 1 1 | 0.6 0.6 0.6 |
| KPRA_HUMAN | PRPSAP1 | Phosphoribosyl pyrophosphate synthase-associated protein 1 | 39 60 64 | 39654 | 1 1 1 | 1 1 1 | 6.5 10.4 6.5 |
| LRC59_HUMAN | LRRC59 | Leucine-rich repeat-containing protein 59 | 32 52 79 | 35308 | 1 1 1 | 1 1 1 | 3.3 7.2 9.1 |
| LRRF1_HUMAN | LRRFIP1 | Leucine-rich repeat flightless-interacting protein 1 | 55 37 75 | 89826 | 1 1 1 | 1 1 1 | 1.5 1.5 3.1 |
| PCBP3_HUMAN | PCBP3 | Poly(rC)-binding protein 3 | 209 232 212 | 39725 | 5 7 7 | 3 4 4 | 13.7 17.5 15.4 |
| RFA2_HUMAN | RPA2 | Replication protein A 32 kDa subunit | 58 45 94 | 29342 | 1 1 2 | 1 1 2 | 4.4 4.4 12.6 |
| SYT5_HUMAN | SYT5 | Synaptotagmin-5 | 50 54 43 | 43216 | 1 1 1 | 1 1 1 | 1.6 1.6 1.6 |
| TOM70_HUMAN | TOMM70A | Mitochondrial import receptor subunit TOM70 | 48 83 53 | 68096 | 1 1 1 | 1 1 1 | 3 3 3 |
| U2AF1_HUMAN | U2AF1 | Splicing factor U2AF 35 kDa subunit | 40 35 51 | 28368 | 0 1 1 | 0 1 1 | 11.2 3.8 11.2 |
| ZO1_HUMAN | TJP1 | Tight junction protein ZO-1 | 40 61 83 | 195682 | 1 1 1 | 1 1 1 | 0.7 1.4 1.4 |
| pTGFBR2 | |||||||
| B3GN7_HUMAN | B3GNT7 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 | 30 44 57 | 46471 | 0 1 1 | 0 1 1 | 4.7 4.7 4.7 |
| DHE3_HUMAN | GLUD1 | Glutamate dehydrogenase 1, mitochondrial | 91 60 49 | 61701 | 2 1 0 | 2 1 0 | 5.9 3.4 3.9 |
| IL18_HUMAN | IL-18 | Interleukin-18 | 42 44 36 | 22597 | 1 1 1 | 1 1 1 | 13.5 13.5 13.5 |
| LOXL4_HUMAN | LOXL4 | Lysyl oxidase homolog 4 | 110 221 169 | 86425 | 2 3 3 | 2 3 3 | 4.8 9.1 10.3 |
| UBP6_HUMAN | USP6 | Ubiquitin carboxyl-terminal hydrolase 6 | 34 34 31 | 160781 | 1 2 1 | 1 1 1 | 0.4 0.4 0.4 |
Different numbers in each column (scores, sign. prot. matches, sign. prot. sequences, coverage) refer to values for three or four biological replicates
In contrast, a much smaller number of exosomal proteins (5/19) were detected exclusively in vesicles secreted by pTGFBR2 HCT116 cells. Among these proteins were the pro-inflammatory cytokine interleukin 18 (IL-18) as well as four different enzymes like the beta 1,3-N-acetylglucosaminyltransferase 7 (B3GNT7), the mitochondrial glutamate dehydrogenase 1 (GLUD1), the lysyl oxidase-like 4 protein (LOXL4), and the ubiquitin-specific protease 6 (USP6). These observations suggest that the TGFBR2 expression status of MSI tumor cells determines the expression of specific protein subsets in derived exosomes that are expected to elicit TGFBR2-dependent responses in recipient cells.
Exosomes of MSI tumor cells cause alterations in the cytokine profile of HepG2 target cells
Since mutation in the TGF-ß pathway are among the most frequent genetic alterations found in CRCs and these tumors frequently metastasize to the liver, we examined the ability of HCT116-TGFBR2-derived exosomes to induce a biological response in HepG2 recipient cells. First, we analyzed the uptake of exosomes by these target cells using CFSE-labeled exosomes derived from HCT116-TGFBR2 cells. CFSE is a membrane permeable dye, which allows exosomal esterases to hydrolyze the dye by removing diacetate residues intra-exosomally. This reaction activates the green fluorescence of CFSE that is coupled to the amino ends of exosomal proteins. The appearance of green-fluorescent exosome-derived proteins inside of HepG2 cells was monitored by confocal microscopy (Fig. 4, Additional file 3).
Fig. 4.
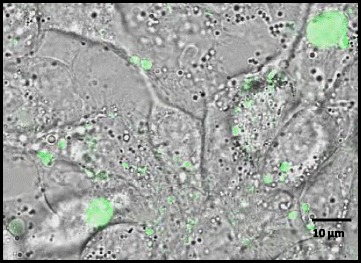
Uptake of exosomes by HepG2 recipient cells. HCT116-TGFBR2 derived exosomes were labeled with CFSE (green-fluorescent dye) and their uptake by HepG2 recipient cells was monitored by confocal microscopy. Scale bar = 10 μm
Additional file 3: Time lapse analysis of CFSE-labeled exosome uptake. Confocal laser scanning microscopy in the 3D-scanning modus was applied to continuously monitor CFSE-labeled exosomes by HepG2 recipient cells. The green fluorescence refers to intracellular signal accumulation of exosomes. Scale bar = 10 μm. (MOV 7290 kb)
These experiments clearly demonstrate the transmission of exosomal cargo proteins to HepG2 cells. This exosomal delivery of bioactive molecules is powerful to modulate various processes in recipient cells. One pronounced effect is the ability of exosomes to modify the spectrum of secreted cytokines upon cargo transmission. For example, it is known that exosomes derived from lung cancer cells can stimulate pro-inflammatory cytokine production in mesenchymal cells and hence favoring a tumor supportive milieu [33]. Since exosomes are known to modulate the spectrum of secreted cytokines in recipient cells, HepG2 cells were exposed to exosomes and any changes in the protein levels of 50 different secreted cytokines and chemokines were determined by Luminex technology.
As shown in Table 2, several cytokines exhibited significantly decreased or increased expression levels (cut off >1.5 fold) in a TGFBR2-dependent manner. Apart from IL-4 (2-fold) and SCF (2.5-fold), the secretion of Platelet-derived Growth Factor-B (PDGF-B) showed the most prominent TGFBR2-dependent response (6-fold) after exosomal exposure. Using a PDGF-B specific ELISA as an alternative method, confirmed the results obtained by Luminex assay. However, the PDGF-B concentration in the supernatant of HepG2 cells exposed to exosomes from dTGFBR2 cells was twice as high in the ELISA assay compared to the Luminex assay (87.61 pg/ml and 40.47 pg/ml, respectively) resulting in a decreased observed ratio of 3-fold (Table 3).
Table 2.
Luminex assay-based analysis of cytokine and chemokine secretion levels in response to exosomal exposure
| Cytokine/Chemokine concentration [pg/ml] | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IL-4 | IL-9 | IL-10 | IL-17 | G-CSF | IP-10 | PDGF-B | TNF-a | MIF | SCF | |
| pTGFBR2 | 0.92 | 6.21 | 47.03 | 23.98 | 18.90 | 6.64 | 255.93 | 13.62 | 589.72 | 85.33 |
| dTGFBR2 | 0.46 | 3.73 | 31.42 | 15.25 | 10.06 | 3.83 | 40.47 | 8.53 | 333.29 | 33.80 |
| Fold change (pTGFBR2/dTGFBR2) | 2.00 | 1.66 | 1.50 | 1.57 | 1.88 | 1.73 | 6.32 | 1.60 | 1.77 | 2.52 |
Fold change values indicate the ratio of secreted cytokine levels by HepG2 recipient cells upon exposure to exosomes derived from pTGFBR2 HCT116 cells in comparison to exosomes isolated from dTGFBR2 donor cells. G-SCF: Granulocyte Colony Stimulating Growth Factor; IP-10: Interferon-Gamma-Inducible Protein 10; PDGF-B: Platelet-derived Growth Factor-B; MIF: Macrophage Migration Inhibitory Factor; SCF: Stem Cell Factor
Table 3.
ELISA-based validation of PDGF-B secretion
| PDGF-B concentration [pg/ml] | |
|---|---|
| pTGFBR2 | 263.28 |
| dTGFBR2 | 87.61 |
| Fold change (pTGFBR2/dTGFBR2) | 3.01 |
Validation of PDGF-B secretion by HepG2 cells in response to exosomal treatment in a TGFBR2-dependent manner
In summary, these results demonstrate that the donor cell-dependent TGFBR2 expression status determines the cargo composition of secreted exosomes and that these exosomes are capable to modulate the biological/cytokine local environment of recipient cells.
Discussion
In this study, we show for the first time that exosomes harbor the same gene-specific cMNR frameshift mutations that have been identified in their cells of origin. One prominent example is the TGFBR2 gene, a recurrently mutated driver of MSI tumorigenesis. Although detectable at the DNA level, we did not observe any frameshift proteins in the exosomal proteome, which is well in line with previous studies from us and others that failed to detect such truncated proteins in total cell lysates from various dMMR MSI tumor cell lines [34]. The genetic alterations, i.e. coding microsatellite mutations, that we have identified in the exosomes of dMMR tumor cells are MSI tumor-specific and reflect the mutation profile of the donor cells. Preliminary experiments suggest that this also applies to diagnostic microsatellite markers routinely used for MSI classification of tumor specimens (Fricke et al., unpublished data). Evidently, this indicates the potential clinical and diagnostic utility of MSI-tumor cell derived exosomes. So far, our findings only apply to established MSI colorectal cancer cell lines and certainly need to be confirmed in exosomes isolated from liquid biopsies of MSI tumor patients. Building up on our findings, exosomes from cultured MSI cell lines might prove useful to define assay thresholds for the detection of cMNR mutations in specific MSI driver genes. Moreover, monitoring of exosomal MSI profiles might facilitate the development of strategies for enrichment of MSI-tumor specific exosomes.
Apart from this MSI-specific molecular fingerprint of exosomes, our results also provide experimental evidence for alterations at the proteome level. This was observed by focusing on the TGFBR2 signal transducer, a frequent mutation target and driver of MSI colorectal tumorigenesis [26, 35]. The results obtained in this study, clearly shows that the TGFBR2 expression/signaling status in the parental cells determined the protein profile of exosomes secreted by these cells. Although the results of our mass spectrometry data revealed no quantitative information, four enzymes (B3GNT7, GLUD1, LOXL4, USP6) and one pro-inflammatory cytokine (IL-18) were exclusively identified in exosomes released from pTGFBR2 cells. The cytokine IL-18 is a mediator of inflammation and an interferon-gamma-inducing factor that is involved in various processes of epithelial repair and collagen production that is emphasized by the prevention of TGF-ß-induced collagen gene expression due to IL-18 activity [36]. Moreover, IL-18 is a mediator of immune responses to eliminate cancer cells effectively [37]. However, IL-18 can also promote tumorigenesis by inducing angiogenesis, migration, metastasis, proliferation, and immune-escape [38, 39].
Apart from IL-18, the enzyme beta 1,3-N-acetylglucosaminyltransferase 7 (B3GNT7) was detected specifically in exosomes derived from pTGFBR2 HCT116-TGFBR2 cells. It has been reported that B3GNT7 is abundantly expressed in normal colon cells and significantly suppressed in colon cancer tissues by epigenetic alterations with effects on the metastatic spread potential [40]. Migratory and metastatic behavior of cancer cells can be further influenced by the lysyl oxidase-like 4 protein (LOXL4), an enzyme that is highly involved in the biogenesis of connective tissue and matrix re-modelling [41] and which has been identified as a protein in TGFBR2 expressing cell-derived exosomes. Another enzyme that is solely expressed in a pTGFBR2 manner is mitochondrial glutamate dehydrogenase 1 (GLUD1). This enzyme is involved in various metabolic processes and associated with poor prognosis in CRC [42]. Aberrant energy metabolism is a hallmark of many cancer cells [43] and plays a role in colorectal tumorigenesis. Recently, it was shown that over-expression of glutamate dehydrogenase (GDH) is associated with CRC metastasis and poor prognosis [44]. Finally, the expression of the enzyme ubiquitin-specific protease 6 (USP6) was exclusively identified in a pTGFBR2 fashion. USP6 can exert different functions by acting as an oncogene, promoting WNT signaling or modulating migration and cytokinesis [45, 46].
A larger subset of 14 proteins has been detected only in exosomes derived from dTGFBR2 cells. This protein subset is of particular clinical interest because it should reflect more accurately the situation existing in most primary MSI colorectal tumors with recurrent loss of TGFBR2 function. The majority of these candidate proteins are involved in migratory processes including EMT. For example, the candidate protein Leucine-rich repeat fightless-interacting protein 1 (LRRFIP1) is known to play a pivotal role in cytoskeletal organization. Using Vesiclepdia and Exocarta Databases [47, 48], this protein has already been identified in the cargo of CRC-derived exosomes [49]. The function of LRRFIP1 in cellular polarity/organisation is exemplified by a recent study that demonstrated LRRFIP1-stimulated CRC metastasis and invasion of hepatocytes through integrin-dependent RhoA activation [50]. Interestingly, RhoA can also be activated by Collagen alpha-1(III) chain (COL3A1) [51], another candidate exclusively identified from the cargo of exosomes derived from dTGFBR2 cells. The COL3A1 protein is upregulated in advanced ovarian carcinoma [52] and a known stimulator of growth acceleration in human osteoblastic cells [53]. The small GTPase RhoA is involved in various cellular processes and a prominent regulatory factor of cytoskeletal dynamics by the induction of stress fibers [54]. Furthermore, RhoA is considered as an essential driver of TGF-ß induced EMT [55].
Further candidates include the FAM3C protein, a promoter of EMT and metastatic progression [56, 57] that has previously been detected in exosomes from other CRC cell lines [49]. Studies have shown that cytoplasmic FAM3C expression might serve as a prognostic factor in colorectal malignancies [58]. Moreover, it has been proposed that elevated concentrations of FAM3C in the secretome of highly autophagic melanoma cell lines could serve as candidate autophagy biomarker [59]. The expression and metastatic proficiency of FAM3C is further regulated by TGF-ß at a post-transcriptional level [57, 60].
Another candidate protein linked to cellular polarity and cell-to-cell contacts is the tight junction protein ZO-1 (TJP1) that is also exclusively expressed in the dTGFBR2 exosomal proteome. Recently, it was shown that TGF-ß increases the expression of TJP1 and enhances cell motility of lung cancer cells [61]. Expression of the Inversin (INVS) protein also was restricted to exosomes derived from dTGFBR2 cells. Concerning the role of INVS in the organization of the cytoskeleton, it has been suggested that INVS modulates cellular polarity through positioning the mitotic spindle and at least partially by transcriptional regulation of genes involved in WNT signaling and pathways associated with the maintenance of the actin cytoskeleton/network and the migratory cellular potential [62, 63]. In another study, the migratory and metastatic potential of breast cancer is also correlated with another candidate protein, Leucine-rich-repeat-containing protein 59 (LRRC59), uncovered specially in the exosomal cargo derived from dTGFBR2 cells [64]. Although not yet confirmed by functional studies, our identified candidates suggest that exosomes derived from dTGFBR2 MSI CRC cells might modulate the migratory potential and/or cytokine profile of target cells.
Target cell changes due to exosomal exposure have been well described. For example, HepG2 cells can serve as target cells for exosomes derived from CRC cells [65]. Moreover, it was shown that exosomes derived from SW480 CRC cells are capable to stimulate the migratory behavior and thus driving cellular alterations of HepG2 cells [66]. When we examined HepG2 target cells that have been exposed to HCT116-TGFBR2 derived exosomes no obvious morphological changes were observed. Instead, our experiments revealed major differences in the cytokine profile in response to exosomal exposure. Depending on the TGFBR2 expression/signaling status of HCT116-TGFBR2 donor cells, exosomes-treated HepG2 target cells exhibited significantly increased secretion of interleukin-4 (2-fold, IL-4), stem cell factor (2.5-fold, SCF) and Platelet-derived Growth Factor-B (6-fold, PDGF-B). Although previous research could not observe any IL-4-specific cytokine concentration in liver tissue or liver metastasis [67], our results indicate that transmission of exosomes derived from HCT116-TGFBR2 CRC cells impact IL-4 secretion of recipient HepG2 cells in a TGFBR2-dependent fashion. For PDGF-B, the biological response was being validated by ELISA assay (3-fold). The observed difference in the ratio between Luminex and ELISA methodology might relate to different assay reagents such as the capture and reporter antibodies [68].
The exact mechanism how TGFBR2 reconstitution in the donor cells elicits exosome-linked PDGF-B release in HepG2 target cells is still unclear. From our previous work, we know that reconstituted TGFBR2 as well as ACVR2 signaling can cause an upregulation of PDGF-B at the transcriptional level in the HCT116 cell line model system [69]. Also, preliminary evidence suggests that PDGF-B transcripts are more abundant in exosomes of pTGFBR2 compared to dTGFBR2 HCT116 cells. However, it is not demonstrated by our experiments if these transcripts account for the high levels of PDGF-B secretion from HepG2 target cells. Metabolic labeling experiments could resolve whether this is attributable to exosomal cargo protein delivery to or de novo protein synthesis in recipient cells. Furthermore, the results obtained from HepG2 cells need to be validated in primary hepatocytes.
Several studies have shown that PDGF signaling can contribute to hepatocellular and colorectal tumor biology. For example, PDGF plays a fundamental role in the initiation of TGF-ß-mediated hepatocellular EMT [70]. Also, in colorectal tumors, expression of PDGF receptors is associated with a high metastatic spread potential [71] and expression of the PDGF-B receptor might contribute to aggressive phenotypes of colorectal tumors with mesenchymal characteristics and enhanced metastatic properties [72]. Moreover, increased levels of PDGF-B have been detected in the plasma of colorectal tumor patients [73]. Clearly, the in vitro model system used in the present study is limited to uncover the biological consequences associated with increased levels of cytokines and growth factors like PDGF-B and how exosomal cargo might shape local and distal environments for the benefit of primary MSI tumors. However, our data strongly suggest that TGFBR2-dependent reprogramming of exosomal cargo can convey MSI tumor cell-specific biological properties to specific target cells.
Conclusion
In conclusion, our data demonstrate that the coding MSI phenotype of dMMR cells is maintained in their secreted exosomes and a recurrent MSI driver mutation not only determines the protein content of MSI exosomes but also alters the cytokine profile of HepG2 recipient cells. Hence, MSI exosomes most likely elicit similar alterations in other potential target cells of endothelial, mesenchymal or hematopoietic origin which, altogether, might provide some mechanistic insights into the specific clinic-histopathological features of these MSI malignancies. Finally, the MSI- and/or TGFBR2-dependent proteome of serum exosomes of MSI tumor patients could serve as a novel source of MSI-specific diagnostic markers.
Additional files
Primers used for cMNR fragment analysis of exosomal and cellular DNA. (DOCX 46 kb)
Gene-specific cMNR frameshift mutant (-1/-2/+1) and wildtype (wt) alleles. (DOCX 46 kb)
Acknowledgments
The expert technical assistance of Marcel Karl, Sigrun Himmelsbach, Vera Bangert and Ulrike Ganserer is gratefully appreciated. Furthermore, we thank Dr. Thomas Worst for profound support. Moreover, we thank Dr. Felix Bestvater and Manuela Brom for the proficient technical assistance of confocal microscopy. Additionally, we are very thankful to Dr. Irina Nazarenko for helpful discussions. The encouragement of the Claussen-Simon-Stiftung is gratefully acknowledged. Finally, we acknowledge financial support by Deutsche Forschungsgemeinschaft and Ruprecht-Karls-Universität Heidelberg within the funding program Open Access Publishing.
Funding
This work was supported by intramural funding from the University Hospital Heidelberg to JG and JK. F.F is a PhD fellow of the Helmholtz International Graduate School for Cancer Research at DKFZ.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors’ contributions
JG, JK and FF planned the experiments. FF, UW, IH and MSC performed the experiments. FF, JG, JK, MS, JL, MM, IH and NH analyzed the data. FF, JG and JK wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- cMNR
Coding mononucleotide repeat
- CRC
Colorectal cancer
- dMMR
DNA mismatch repair deficiency
- dTGFBR2
TGFBR2-deficient
- MSI
Microsatellite instability
- pTGFBR2
TGFBR2-proficient
Contributor Information
Fabia Fricke, Email: f.fricke@dkfz-heidelberg.de.
Jennifer Lee, Email: jennylee@tissuegenesis.com.
Malwina Michalak, Email: malwina.michalak@med.uni-heidelberg.de.
Uwe Warnken, Email: u.warnken@dkfz-heidelberg.de.
Ingrid Hausser, Email: ingrid.hausser-siller@med.uni-heidelberg.de.
Meggy Suarez-Carmona, Email: meggy.suarez-carmona@nct-heidelberg.de.
Niels Halama, Email: niels.halama@med.uni-heidelberg.de.
Martina Schnölzer, Email: m.schnoelzer@dkfz-heidelberg.de.
Jürgen Kopitz, Email: juergen.kopitz@med.uni-heidleberg.de.
Johannes Gebert, Phone: +49-6221-56 4223, Email: johannes.gebert@med.uni-heidelberg.de.
References
- 1.Maas SL, Breakefield XO, Weaver AM. Extracellular Vesicles. Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2016;10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed]
- 2.Yamada N, Kuranaga Y, Kumazaki M, Shinohara H, Taniguchi K, Akao Y. Colorectal cancer cell-derived extracellular vesicles induce phenotypic alteration of T cells into tumor-growth supporting cells with transforming growth factor-beta1-mediated suppression. Oncotarget. 2016;7:27033–43. doi: 10.18632/oncotarget.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13:1554–71. doi: 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- 4.Tkach M, Thery C. Communication by extracellular vesicles: Where we are and where we need to go. Cell. 2016;164:1226–32. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 5.Federici C, Petrucci F, Caimi S, Cesolini A, Logozzi M, Borghi M, D’Ilio S, Lugini L, Violante N, Azzarito T, et al. Exosome release and low pH belong to a framework of resistance of human melanoma cells to cisplatin. PLoS One. 2014;9:e88193. doi: 10.1371/journal.pone.0088193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Wang XF. A niche role for cancer exosomes in metastasis. Nat Cell Biol. 2015;17:709–11. doi: 10.1038/ncb3181. [DOI] [PubMed] [Google Scholar]
- 7.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–48. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–21. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soldevilla B, Rodriguez M, San Millan C, Garcia V, Fernandez-Perianez R, Gil-Calderon B, Martin P, Garcia-Grande A, Silva J, Bonilla F, Dominguez G. Tumor-derived exosomes are enriched in DeltaNp73, which promotes oncogenic potential in acceptor cells and correlates with patient survival. Hum Mol Genet. 2014;23:467–78. doi: 10.1093/hmg/ddt437. [DOI] [PubMed] [Google Scholar]
- 10.Halama N, Michel S, Kloor M, Zoernig I, Pommerencke T, Schirmacher P, von Knebel-Doberitz M, Weitz J, Grabe N, Jager D. Immune infiltrates in liver metastases of colorectal cancer and response to chemotherapy. J Clin Oncol. 2009;27:e15069. [PMC free article] [PubMed] [Google Scholar]
- 11.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–26. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devaud N, Gallinger S. Chemotherapy of MMR-deficient colorectal cancer. Fam Cancer. 2013;12:301–6. doi: 10.1007/s10689-013-9633-z. [DOI] [PubMed] [Google Scholar]
- 13.Buckowitz A, Knaebel HP, Benner A, Blaker H, Gebert J, Kienle P, von Knebel DM, Kloor M. Microsatellite instability in colorectal cancer is associated with local lymphocyte infiltration and low frequency of distant metastases. Br J Cancer. 2005;92:1746–53. doi: 10.1038/sj.bjc.6602534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malesci A, Laghi L, Bianchi P, Delconte G, Randolph A, Torri V, Carnaghi C, Doci R, Rosati R, Montorsi M, et al. Reduced likelihood of metastases in patients with microsatellite-unstable colorectal cancer. Clin Cancer Res. 2007;13:3831–9. doi: 10.1158/1078-0432.CCR-07-0366. [DOI] [PubMed] [Google Scholar]
- 15.Dolcetti R, Viel A, Doglioni C, Russo A, Guidoboni M, Capozzi E, Vecchiato N, Macri E, Fornasarig M, Boiocchi M. High prevalence of activated intraepithelial cytotoxic T lymphocytes and increased neoplastic cell apoptosis in colorectal carcinomas with microsatellite instability. Am J Pathol. 1999;154:1805–13. doi: 10.1016/S0002-9440(10)65436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozek LS, Schmit SL, Greenson JK, Tomsho LP, Rennert HS, Rennert G, Gruber SB. Tumor-Infiltrating Lymphocytes, Crohn’s-Like Lymphoid Reaction, and Survival From Colorectal Cancer. J Natl Cancer Inst. 2016;108:1–8. doi: 10.1093/jnci/djw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–87. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woerner SM, Kloor M, von Knebel DM, Gebert JF. Microsatellite instability in the development of DNA mismatch repair deficient tumors. Cancer Biomark. 2006;2:69–86. doi: 10.3233/CBM-2006-21-208. [DOI] [PubMed] [Google Scholar]
- 19.Ionov Y, Nowak N, Perucho M, Markowitz S, Cowell JK. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer Cells with microsatellite instability. Oncogene. 2004;23:639–45. doi: 10.1038/sj.onc.1207178. [DOI] [PubMed] [Google Scholar]
- 20.El-Bchiri J, Buhard O, Penard-Lacronique V, Thomas G, Hamelin R, Duval A. Differential nonsense mediated decay of mutated mRNAs in mismatch repair deficient colorectal cancers. Hum Mol Genet. 2005;14:2435–42. doi: 10.1093/hmg/ddi245. [DOI] [PubMed] [Google Scholar]
- 21.Bauer K, Nelius N, Reuschenbach M, Koch M, Weitz J, Steinert G, Kopitz J, Beckhove P, Tariverdian M, von Knebel DM, Kloor M. T cell responses against microsatellite instability-induced frameshift peptides and influence of regulatory T cells in colorectal cancer. Cancer Immunol Immunother. 2013;62:27–37. doi: 10.1007/s00262-012-1303-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duval A, Hamelin R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res. 2002;62:2447–54. [PubMed] [Google Scholar]
- 23.Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013;155:858–68. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–84. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 25.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–30. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–8. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Ballikaya S, Schonig K, Ball CR, Glimm H, Kopitz J, Gebert J. Transforming growth factor beta receptor 2 (TGFBR2) changes sialylation in the microsatellite unstable (MSI) Colorectal cancer cell line HCT116. PLoS One. 2013;8:e57074. doi: 10.1371/journal.pone.0057074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–3. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 29.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 30.Vizcaino JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:11033. doi: 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Squizzato S, Park YM, Buso N, Gur T, Cowley A, Li W, Uludag M, Pundir S, Cham JA, McWilliam H, Lopez R. The EBI Search engine: providing search and retrieval functionality for biological data from EMBL-EBI. Nucleic Acids Res. 2015;43:585–8. doi: 10.1093/nar/gkv316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Wang S, Zhu R, Li H, Han Q, Zhao RC. Lung tumor exosomes induce a pro-inflammatory phenotype in mesenchymal stem cells via NFkappaB-TLR signaling pathway. J Hematol Oncol. 2016;9:42. doi: 10.1186/s13045-016-0269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.You KT, Li LS, Kim NG, Kang HJ, Koh KH, Chwae YJ, Kim KM, Kim YK, Park SM, Jang SK, Kim H. Selective translational repression of truncated proteins from frameshift mutation-derived mRNAs in tumors. PLoS Biol. 2007;5:e109. doi: 10.1371/journal.pbio.0050109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548–50. [PubMed] [Google Scholar]
- 36.Kim HJ, Song SB, Choi JM, Kim KM, Cho BK, Cho DH, Park HJ. IL-18 downregulates collagen production in human dermal fibroblasts via the ERK pathway. J Invest Dermatol. 2010;130:706–15. doi: 10.1038/jid.2009.302. [DOI] [PubMed] [Google Scholar]
- 37.Dinarello CA. Interleukin-18. Methods. 1999;19:121–32. doi: 10.1006/meth.1999.0837. [DOI] [PubMed] [Google Scholar]
- 38.Park S, Cheon S, Cho D. The dual effects of interleukin-18 in tumor progression. Cell Mol Immunol. 2007;4:329–35. [PubMed] [Google Scholar]
- 39.Palma G, Barbieri A, Bimonte S, Palla M, Zappavigna S, Caraglia M, Ascierto PA, Ciliberto G, Arra C. Interleukin 18: friend or foe in cancer. Biochim Biophys Acta. 1836;2013:296–303. doi: 10.1016/j.bbcan.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Lu CH, Wu WY, Lai YJ, Yang CM, Yu LC. Suppression of B3GNT7 gene expression in colon adenocarcinoma and its potential effect in the metastasis of colon cancer cells. Glycobiology. 2014;24:359–67. doi: 10.1093/glycob/cwu002. [DOI] [PubMed] [Google Scholar]
- 41.Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, Chi JT, Jeffrey SS, Giaccia AJ. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–6. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 42.Miyo M, Konno M, Nishida N, Sueda T, Noguchi K, Matsui H, Colvin H, Kawamoto K, Koseki J, Haraguchi N, et al. Metabolic adaptation to nutritional stress in human colorectal cancer. Sci Rep. 2016;6:38415. doi: 10.1038/srep38415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 44.Liu G, Zhu J, Yu M, Cai C, Zhou Y, Yu M, Fu Z, Gong Y, Yang B, Li Y, et al. Glutamate dehydrogenase is a novel prognostic marker and predicts metastases in colorectal cancer patients. J Transl Med. 2015;13:144. doi: 10.1186/s12967-015-0500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rueckert C, Haucke V. The oncogenic TBC domain protein USP6/TRE17 regulates cell migration and cytokinesis. Biol Cell. 2012;104:22–33. doi: 10.1111/boc.201100108. [DOI] [PubMed] [Google Scholar]
- 46.Madan B, Walker MP, Young R, Quick L, Orgel KA, Ryan M, Gupta P, Henrich IC, Ferrer M, Marine S, et al. USP6 oncogene promotes Wnt signaling by deubiquitylating Frizzleds. Proc Natl Acad Sci U S A. 2016;113:2945–54. doi: 10.1073/pnas.1605691113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borras FE, Breakefield X, Budnik V, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simpson RJ, Kalra H, Mathivanan S. ExoCarta as a resource for exosomal research. J Extracell Vesicles. 2012;doi:10.3402/jev.v1i0.18374. [DOI] [PMC free article] [PubMed]
- 49.Demory Beckler M, Higginbotham JN, Franklin JL, Ham AJ, Halvey PJ, Imasuen IE, Whitwell C, Li M, Liebler DC, Coffey RJ. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12:343–55. doi: 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ariake K, Ohtsuka H, Motoi F, Douchi D, Oikawa M, Rikiyama T, Fukase K, Katayose Y, Egawa S, Unno M. GCF2/LRRFIP1 promotes colorectal cancer metastasis and liver invasion through integrin-dependent RhoA activation. Cancer Lett. 2012;325:99–107. doi: 10.1016/j.canlet.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 51.Luo R, Jeong SJ, Jin Z, Strokes N, Li S, Piao X. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc Natl Acad Sci U S A. 2011;108:12925–30. doi: 10.1073/pnas.1104821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tapper J, Kettunen E, El-Rifai W, Seppala M, Andersson LC, Knuutila S. Changes in gene expression during progression of ovarian carcinoma. Cancer Genet Cytogenet. 2001;128:1–6. doi: 10.1016/S0165-4608(01)00386-7. [DOI] [PubMed] [Google Scholar]
- 53.Maehata Y, Takamizawa S, Ozawa S, Izukuri K, Kato Y, Sato S, Lee MC, Kimura A, Hata R. Type III collagen is essential for growth acceleration of human osteoblastic cells by ascorbic acid 2-phosphate, a long-acting vitamin C derivative. Matrix Biol. 2007;26:371–81. doi: 10.1016/j.matbio.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 54.Ridley AJ. The GTP-binding protein Rho. Int J Biochem Cell Biol. 1997;29:1225–29. doi: 10.1016/S1357-2725(97)00052-6. [DOI] [PubMed] [Google Scholar]
- 55.Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell. 2001;12:27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waerner T, Alacakaptan M, Tamir I, Oberauer R, Gal A, Brabletz T, Schreiber M, Jechlinger M, Beug H. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell. 2006;10:227–39. doi: 10.1016/j.ccr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 57.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–93. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao ZH, Lu C, Wang ZN, Song YX, Zhu JL, Gao P, Sun JX, Chen XW, Wang MX, Dong YL, Xu HM. ILEI: a novel marker for epithelial-mesenchymal transition and poor prognosis in colorectal cancer. Histopathology. 2014;65:527–38. doi: 10.1111/his.12435. [DOI] [PubMed] [Google Scholar]
- 59.Kraya AA, Piao S, Xu X, Zhang G, Herlyn M, Gimotty P, Levine B, Amaravadi RK, Speicher DW. Identification of secreted proteins that reflect autophagy dynamics within tumor cells. Autophagy. 2015;11:60–74. doi: 10.4161/15548627.2014.984273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song Q, Sheng W, Zhang X, Jiao S, Li F. ILEI drives epithelial to mesenchymal transition and metastatic progression in the lung cancer cell line A549. Tumour Biol. 2014;35:1377–82. doi: 10.1007/s13277-013-1188-y. [DOI] [PubMed] [Google Scholar]
- 61.Lee SH, Paek AR, Yoon K, Kim SH, Lee SY, You HJ. Tight junction protein 1 is regulated by transforming growth factor-beta and contributes to cell motility in NSCLC cells. BMB Rep. 2015;48:115–20. doi: 10.5483/BMBRep.2015.48.2.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caplan MJ. An inversin convergence: focus on “inversin modulates the cortical actin network during mitosis”. Am J Physiol Cell Physiol. 2013;305:22–23. doi: 10.1152/ajpcell.00126.2013. [DOI] [PubMed] [Google Scholar]
- 63.Veland IR, Montjean R, Eley L, Pedersen LB, Schwab A, Goodship J, Kristiansen K, Pedersen SF, Saunier S, Christensen ST. Inversin/Nephrocystin-2 is required for fibroblast polarity and directional cell migration. PLoS One. 2013;8:e60193. doi: 10.1371/journal.pone.0060193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Terp MG, Lund RR, Jensen ON, Leth-Larsen R, Ditzel HJ. Identification of markers associated with highly aggressive metastatic phenotypes using quantitative comparative proteomics. Cancer Genomics Proteomics. 2012;9:265–73. [PubMed] [Google Scholar]
- 65.Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep. 2012;28:1551–8. doi: 10.3892/or.2012.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chiba M, Watanabe N, Watanabe M, Sakamoto M, Sato A, Fujisaki M, Kubota S, Monzen S, Maruyama A, Nanashima N, et al. Exosomes derived from SW480 colorectal cancer cells promote cell migration in HepG2 hepatocellular cancer cells via the mitogen-activated protein kinase pathway. Int J Oncol. 2016;48:305–12. doi: 10.3892/ijo.2015.3255. [DOI] [PubMed] [Google Scholar]
- 67.Halama N, Zoernig I, Berthel A, Kahlert C, Klupp F, Suarez-Carmona M, Suetterlin T, Brand K, Krauss J, Lasitschka F, et al. Tumoral immune cell exploitation in colorectal cancer metastases can be targeted effectively by anti-CCR5 therapy in cancer patients. Cancer Cell. 2016;29:587–601. doi: 10.1016/j.ccell.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 68.Elshal MF, McCoy JP. Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA. Methods. 2006;38:317–23. doi: 10.1016/j.ymeth.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee J, Katzenmaier EM, Kopitz J, Gebert J. Reconstitution of TGFBR2 in HCT116 colorectal cancer cells causes increased LFNG expression and enhanced N-acetyl-d-glucosamine incorporation into Notch1. Cell Signal. 2016;28:1105–13. doi: 10.1016/j.cellsig.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 70.Gotzmann J, Fischer AN, Zojer M, Mikula M, Proell V, Huber H, Jechlinger M, Waerner T, Weith A, Beug H, Mikulits W. A crucial function of PDGF in TGF-beta-mediated cancer progression of hepatocytes. Oncogene. 2006;25:3170–85. doi: 10.1038/sj.onc.1209083. [DOI] [PubMed] [Google Scholar]
- 71.Kitadai Y, Sasaki T, Kuwai T, Nakamura T, Bucana CD, Hamilton SR, Fidler IJ. Expression of activated platelet-derived growth factor receptor in stromal cells of human colon carcinomas is associated with metastatic potential. Int J Cancer. 2006;119:2567–74. doi: 10.1002/ijc.22229. [DOI] [PubMed] [Google Scholar]
- 72.Steller EJ, Raats DA, Koster J, Rutten B, Govaert KM, Emmink BL, Snoeren N, van Hooff SR, Holstege FC, Maas C, et al. PDGFRB promotes liver metastasis formation of mesenchymal-like colorectal tumor cells. Neoplasia. 2013;15:204–17. doi: 10.1593/neo.121726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakamura Y, Tanaka F, Yoshikawa Y, Mimori K, Inoue H, Yanaga K, Mori M. PDGF-BB is a novel prognostic factor in colorectal cancer. Ann Surg Oncol. 2008;15:2129–36. doi: 10.1245/s10434-008-9943-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for cMNR fragment analysis of exosomal and cellular DNA. (DOCX 46 kb)
Gene-specific cMNR frameshift mutant (-1/-2/+1) and wildtype (wt) alleles. (DOCX 46 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


