Abstract
Background:
Next-generation sequencing (NGS) with a multi-gene panel is now available for patients with lung adenocarcinoma, but the performance characteristics and clinical utility of this testing are not well-described. We present the results of an extended 467 gene panel in a series of advanced, highly selected nonsmall cell lung cancer (NSCLC) patients using a range of specimens, including predominantly small biopsy and cytology specimens.
Materials and Methods:
A retrospective review of 22 NSCLC biopsies sent for NGS using an extended gene panel from January 2014 to July 2015. The customized NGS panel sequences 467 cancer-associated genes with exonic and intronic sequences obtained from purified tumor DNA. Genomic alterations, patient characteristics, and success of testing were determined.
Results:
The majority of samples tested were metastatic lung adenocarcinoma on final pathology. Of the 22 specimens tested, 5 (22.7%) were surgical resections and 17 (77.3%) were small biopsy and cytology specimens. Twenty-one (95%) of the specimens were adequate for full sequencing and yielded a total of 204 genomic alterations (average 8.9 per tumor), of which 17 (average 0.81 per tumor) were actionable and/or clinically relevant. Genomic alterations were found most commonly in the TP53, EGFR, EPHB1, MLL3, APC, SETD2, KRAS, DNMT3A, RB1, CDKN2A, ARID1A, EP300, KDM6B, RAD50, STK11, and BRCA2 genes.
Conclusions:
NGS using a comprehensive gene panel was performed successfully in 95% of all NSCLC cases in this series, including 94% small biopsy and cytology specimens and 100% surgical resections. This custom assay was performed on a range of tumor specimens and demonstrates that small specimens are able to provide a similar depth of information as larger ones. As many patients present at an advanced stage and only small specimens are obtained, the information these provide has the potential for guiding treatment in highly selected patients with advanced lung adenocarcinoma.
Keywords: Cytology, next-generation sequencing, nonsmall cell lung cancer
INTRODUCTION
Lung cancer remains the leading cause of cancer mortality, with the number of estimated deaths in the US exceeding 158,000 in 2015, representing nearly 27% of all cancer deaths.[1] Approximately, 85% of these malignancies will be non-small cell lung cancer (NSCLC), with adenocarcinoma accounting for over 50%.[2] The majority of patients have advanced disease at the time of their diagnosis. In adenocarcinomas, driver mutations in tumor cells have been identified which have potential prognostic significance and present potential therapeutic targets.[3] EGFR mutations, KRAS mutations, and translocations in ALK were among the earliest important genomic alterations identified in lung adenocarcinoma, but the list has grown significantly in the last 5 years.[4,5,6]
Consideration of genomic testing and genomic alteration-based targeted therapy is now the standard of care for some patients with lung adenocarcinoma, namely patients with metastatic and locally advanced disease.[7,8] The 2013 College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Guideline endorsed by the American Society of Clinical Oncology recommends EGFR and ALK testing for all patients with advanced stage lung adenocarcinoma.[7] Furthermore, the current National Comprehensive Cancer Network (NCCN) guidelines also include testing for ROS1, RET, BRAF, ERBB2, and MET in this population.[8] This testing sets the stage for genotype-driven decision-making for a significant number of patients with lung adenocarcinoma and has the potential to significantly impact cancer mortality.[9,10]
Many patients are now being diagnosed and staged with small biopsy and cytology samples such as endobronchial ultrasound (EBUS)-guided transbronchial needle aspiration.[11,12] However, it can be challenging to perform multiple single-gene molecular assays on small biopsy and cytology specimens for the seven cancer-associated genes recommended in the NCCN guidelines without depleting available tissue.
To maximize the available information from a single biopsy, multi-gene platforms have been developed. This testing offers an efficient, fast, and accurate way to identify a select panel of clinically relevant genomic alterations and can be integrated into routine clinical practice.[13,14] This testing modality, which has been referred to as targeted next-generation sequencing (NGS), can be used in small cytologic specimens, as well as small biopsy specimens, but the yield in these specimens is not well described.
Efforts by pathologists and cytopathologists to maximize the amount of tumor DNA for NGS are an evolving area of practice that also has yet to be standardized.[13] Furthermore, the ideal panel of genes targeted is constantly expanding. We describe our experience with targeted NGS using a customized 467 gene panel performed on small biopsy and cytology specimens in a series of 22 highly selected, advanced stage NSCLC patients to clarify expectations for its clinical utility.
MATERIALS AND METHODS
We retrospectively reviewed the records of 22 consecutive patients with NSCLC for whom tumor samples underwent NGS using the Columbia Combined Cancer Panel (CCCP) between January 2014 and July 2015. This customized, more comprehensive genomic profiling was specifically requested by the treating oncologist in metastatic or advanced NSCLC cases. The requests, as the data show, were mostly on small biopsy and cytology specimens, but resection specimens in this consecutive series are included to compare feasibility between the two. After the oncologist's request, the existing biopsy sample was assessed for adequacy by a pathologist. It is also our standard practice to microdissect specimens to enrich for tumor before attempting NGS. Tumors approved for the assay had at least 20% tumor present in the sample. There are no additional DNA or tissue quantity criteria required. Using these criteria, more than 90% of disease-associated variants in the region of interest are detected at a high specificity. This test was developed and its performance characteristics determined by the Molecular Pathology Laboratory of Columbia University. It has not been cleared or approved by the US Food and Drug Administration (FDA). This test was originally performed pursuant to waiver provided by the New York State Department of Health (NYSDOH), without a review by the Clinical Laboratory Review Program. It was subsequently fully approved by the NYSDOH after the study period ended. The customized NGS panel sequences 467 cancer-associated genes with exonic and intronic sequences obtained from DNA purified from tumor (with or without normal DNA). Custom Agilent SureSelect capture and Illumina HiSeq2500® sequencing is used. Resulting fastq files underwent alignment and variant calling by NextGENe software (SoftGenetics, LLC, State College, PA, USA). Reads with a Q-score of >20 and base matching of >90% were accepted. Variant calling was performed in regions with at least 15× coverage. The variant must have a frequency of 10% and be detected in at least three reads. Indels of up to 30 bp occurring in at least 5% of reads are reported. Samples have an average coverage of at least 500-fold, and at least 10-fold coverage of >98% of coding sequences in the region of interest.
Activating mutations detected were categorized into tiers depending on the known pathogenesis of the tumor type (lung cancer versus other malignancies) and whether or not the mutation was actionable. Actionable mutations were defined as those whose effect is potentially targetable using an FDA-approved and commercially available anticancer drugs. For the purposes of NSCLC, tier 1 variants are actionable driver mutations known in the pathogenesis of NSCLC. Tier 2 variants are either actionable driver mutations known in the pathogenesis of other tumor types or alterations in well-established cancer genes. Tier 3 variants are nonactionable alterations in other cancer-associated genes, and tier 4 variants are genetic alterations of uncertain significance. Actionable variants are confirmed by the use of an orthologous method.
The majority of samples were tested independently for individual genomic alterations as a part of a reflex testing protocol for lung adenocarcinoma in place at our institution. This protocol includes ALK break apart fluorescence in situ hybridization (FISH) testing (Abbott Molecular), direct di-deoxyterminator (Sanger) sequencing for EGFR exons 18-–21 and KRAS codons 12 and 13, and real-time polymerase chain reaction (RT-PCR) to detect the BRAF V600E variant. Sanger sequencing requires at least 50% tumor purity with a sensitivity of 25% for EGFR or 40% tumor for KRAS. RT-PCR requires 10% heterozygous mutant DNA or approximately 5% mutant allele (equivalent to approximately 10% heterozygous mutant tumor cells in a sample).
All testing performed on fine-needle aspiration (FNA) specimens was done using formalin-fixed cell blocks as previously described.[15,16,17] We report the concordance and discordance of individual gene testing with the results of the CCCP NGS.
Co-primary outcome measures include the feasibility of testing and genomic alterations detected. Secondary outcomes of biopsy method, the use of rapid on-site evaluation (ROSE) during specimen procurement, and concordance with single-gene, hotspot testing were recorded. ROSE was performed for FNAs by smears and core needle biopsies (CNBs) by touch imprints.
This research, including a waiver of consent, was approved by the institution review board.
RESULTS
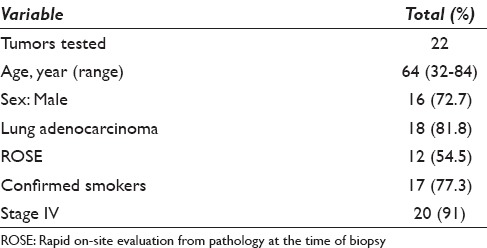
Baseline characteristics of the patients with available data are given in Table 1. There were 6 (27%) female and 16 (73%) male patients included in this study with a mean age of 64 years (range: 32–84), with the vast majority being metastatic lung adenocarcinoma on final pathology. Other final pathology included poorly differentiated adenocarcinoma and unclassifiable nonsmall cell carcinoma of the lung (4, 18%). Two patients had stage III adenocarcinoma.
Table 1.
Patient characteristics
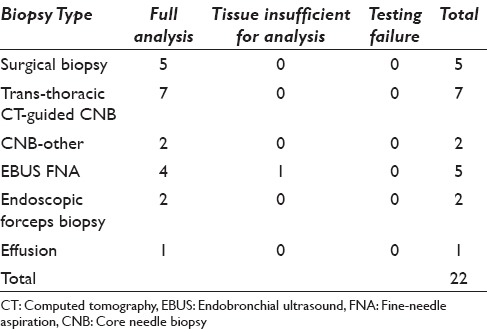
The success of NGS was categorized as either yielding a full, unqualified report (full analysis) or the inability to sequence the tumor secondary to insufficient DNA (tissue insufficient for analysis) or the inability to sequence the tumor secondary to test failure (testing failure). Table 2 shows the success of NGS according to the biopsy method. Twenty-one of the tumors tested (95%) yielded full reports. NGS was successfully performed on all five surgical biopsy specimens, which included three pleural biopsies, one lung wedge resection, and one brain tumor biopsy. Four of the five EBUS FNA samples were adequate for testing. The remaining EBUS FNA resulted in tissue insufficient for analysis after DNA isolation. One pleural effusion cytology sample, one endobronchial forceps biopsy, and one transbronchial forceps biopsy were tested and yielded full analyses. Seven transthoracic CNBs were fully analyzed along with one CNB of an abdominal wall mass, and one CNB of the sacrum. There were no samples with testing failure. ROSE by cytopathology was employed during tumor sampling in all five EBUS-FNA samples and seven of the nine CNBs.
Table 2.
Success of next-generation sequencing according to specimen type
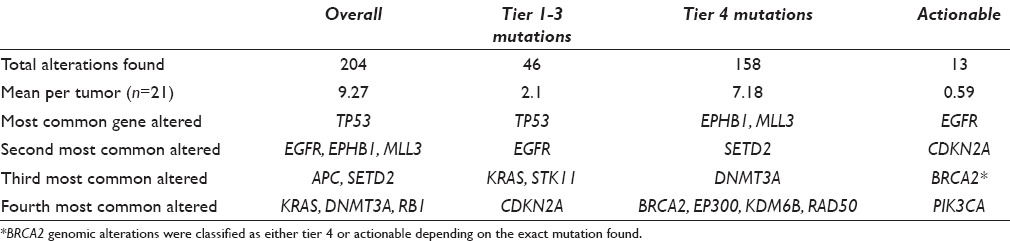
There were a total of 204 genomic alterations (range: 4–24, mean per tumor 8.9). The most common genes containing alterations are shown in Figure 1. The most frequent cancer-associated genomic alterations identified (tier 1–3 mutations) were in the TP53 (14/22 tumors), EGFR (7 mutations in 6/22 tumors), KRAS (4/22 tumors), and CDKN2A genes (3/22 tumors). A total of 158 mutations of uncertain significance (tier 4 mutations) were found (range: 2–20, mean per tumor 6.9). The most common mutations of uncertain significance were in the MLL3, APC, RB1, SETD2, and EPHB1 genes [Table 3].
Figure 1.
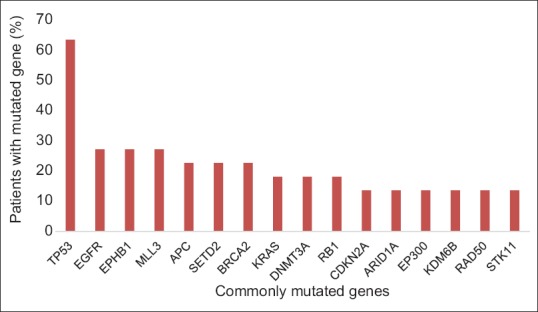
Most common genes containing alterations by next-generation sequencing. Mutations in these 16 genes represent 75 (37%) of the total alterations found. Percentage of patients with each mutated gene represented on the y-axis
Table 3.
Genetic alteration found in 21 fully sequenced nonsmall cell lung cancer tumors
Fifteen of the 22 tumors sent for testing had at least one tier 1 or tier 2 mutation. This included nine individual patients (41%) with 13 actionable mutations. The actionable mutations found were seven EGFR (eligible for tyrosine kinase inhibitors), three CDKNA2 (eligible for cyclin-dependent kinase 4 inhibitors), two BRCA2 (eligible for poly ADP ribose polymerase inhibitors), and one PIK3CA (eligible for mechanistic target of rapamycin inhibitor) mutations.
In addition to NGS, 20 of the 22 patients also had tumor specimens sent for traditional molecular testing using FDA approved single gene-targeted molecular assays or hotspot assays. Six of the seven EGFR mutations found by NGS were demonstrated on single-gene testing sent on the same tumor. One tumor had two separate EGFR mutations (EGFR L858R and T790M), of which only the L858R mutation was found on single-gene PCR testing. Our NGS assay detected both mutations using a separate biopsy 8 months after the original single-gene PCR test was sent. Notably, after this tumor originally tested positive for EGFR L858R by Sanger sequencing, the patient was placed on erlotinib therapy. Subsequent NGS testing from the new biopsy 8 months later revealed both the EGFR L858R and T790M mutations at 45% and 24% variant frequency, respectively. This new EGFR T790M mutation likely represents an acquired resistance mutation in the setting of EGFR TKI therapy; however, an underlying mutation present at a variant frequency below the sensitivity of the initial single-gene PCR cannot be completely excluded without additional testing. Two of the five patients with KRAS mutations on NGS had several separate tumor specimens sent for the single-gene KRAS assays. In one case, a metastatic bone lesion was KRAS mutated by NGS, while the primary lung tumor was KRAS wild-type on single-gene PCR assay. The second case was a lung adenocarcinoma with two distinct recurrences over a 9 years period after a primary tumor was biopsied. The most recent recurrence had a KRAS c. 34G>T mutation (G12C) on NGS, while the primary tumor 9 years earlier had a c. 35G>Y mutation (G12V) on single-gene testing. Meanwhile, a biopsy of the tumor's first recurrence demonstrated a wild-type KRAS on the single-gene testing. The one patient with insufficient DNA for NGS from an EBUS FNA had successful hotspot assays performed on another EBUS FNA collected at an earlier date. This patient had an EML4-ALK fusion gene detected by FISH.
DISCUSSION
NGS was successfully performed on 21 of 22 NSCLC specimens including an exfoliative cytology specimen, FNAs, forceps biopsies, CNBs, and small surgical biopsies. In this sample, 22 consecutive NSCLC cases underwent a customized, 467 gene panel after available tumor specimen was examined and approved by our molecular pathology laboratory.
These findings add to previous literature published by Kanagal-Shamanna et al. demonstrating that FNA specimens (smears and cell blocks) successfully underwent NGS of a smaller panel of 46 genes when pathologists selected 31 of 61 samples with adequate tumor cellularity.[18] However, our assay compares favorably to a larger series described by Hagemann et al. in which only 64% of pathology laboratory-approved samples referred for a 23 gene NGS panel were successfully sequenced.[19] The likelihood of successful sequencing was much lower in cytology and core needle specimens compared to excisional specimens in this series with yields of 33%, 40%, and 95%, respectively.[19] Similarly, König et al. reported a series of over 2100 lung adenocarcinoma patients in an NGS validation cohort using a 12 gene panel with < 70% of samples successfully tested.[20] As above, our samples were microdissected before attempting NGS. This may have contributed to a successful test on a sample with low tumor cellularity. Proper triage of specimens by tumor cellularity, avoidance of redundant testing, the role of microdissection, and cell block technique for FNA samples will be important subjects for future studies. Our series adds to prior cytopathology literature regarding the general importance of proper handling of small biopsy specimens, and that formalin-fixed cell blocks provide DNA of sufficient quality for NGS testing.[15,17,18,21]
ROSE was frequently employed to guide CNB and EBUS FNA. All 12 biopsies that utilized ROSE had an on-site evaluation of adequacy categorized as satisfactory or adequate. Eleven of the 12 biopsies that utilized ROSE were sequenced successfully. This may have also increased the yield of testing by ensuring high tumor cellularity at the time of biopsy.
A great number of potential driver mutations were found when tumors were sequenced using the CCCP assay. Most patients had several mutation types, across several different genes. The actionable mutations were mainly found in the EGFR gene in this selected set of patients. Several cases revealed hypermutated tumors with dozens of potential driver mutations. These hypermutated tumors may have a more favorable response to immune therapy.[22]
NGS correlated with single-gene molecular testing when the same tumor specimens were tested by both assays. This is consistent with a recent case series comparing multiple NGS platforms with dedicated hotspot assays for EGFR, KRAS, and BRAF mutations using formalin-fixed, paraffin-embedded tumor.[23,24] Evolving tumors can be tested in the same patient using single-gene assays after NGS has been employed. Comparing the genetic mutations in the new lesions to the known NGS results could be a method to further adjudicate tumor recurrence versus a second primary cancer.
NGS offers fast and more complete mutation analysis in lung adenocarcinoma, and the information has the potential to improve the outcome in individual patients and development of additional targeted therapies. This information can also present a major challenge to oncologists trying to understand the practical implications of numerous, concurrent, and novel mutations within a single patient's tumor. This is best demonstrated in the mutations of uncertain significance that represented 77% of the genomic alterations found.
There are several important limitations to our analysis. Notably, our sample size was small with genetic information for only 21 tumors. The retrospective nature of the data collection also limited our ability to collect important information surrounding the biopsy procedures. Another significant limitation is that the specimens were prescreened for suitability for NGS. Prescreening of specimens for suitability for NGS likely influenced a decision to pursue testing to an unknown degree. If this happened frequently, our results would be biased to reflect more successful assay. There may have also been several instances where a repeat biopsy was specifically requested after a review of available tumor samples. Biopsies solely dedicated to NGS are more likely to be successful as DNA is not being used for other testing prior to performing the assay.
Furthermore, the frequency of reported mutations in our cohort does not represent a random sampling, but it does highlight the broad diversity of genomic abnormalities that exist in lung adenocarcinomas beyond EGFR, ALK, and KRAS. Finally, our study provides no information on the impact of NGS on outcomes such as treatment selection, morbidity, quality of life, disease-free survival, and overall mortality. We lack the long-term follow-up in this study to comment on these outcomes.
NGS with a custom, 467 gene panel was highly successful in our sample of 22 NSCLC tumors. We demonstrate that targeted sequencing of an extensive gene panel is feasible in a range of small pathologic and cytologic specimens. This study shows that even the smallest of biopsy specimens at least has the potential to provide this information when close attention is paid to specimen collection and handling.
CONCLUSIONS
NGS using a large targeted gene panel was performed successfully in 94% of small biopsy and cytology samples compared to 100% of resections. Additional information gathered through large NGS panels has the potential to guide therapy in certain patients with NSCLC.
COMPETING INTERESTS STATEMENT BY ALL AUTHORS
The authors declare that they have no competing interests.
AUTHORSHIP STATEMENT BY ALL AUTHORS
All authors of this article declare that we qualify for authorship as defined by ICMJE http://www.icmje.org/#author.
Each author has participated sufficiently in the work and takes public responsibility for appropriate portions of the content of this article.
DD participated in the design of the study, data collection, and drafting of the manuscript. DR participated in data collection and editing of the manuscript. JH participated in editing the manuscript and interpretation of the data. CP participated in editing the manuscript and interpretation of the data. AS participated in the design of the study, study coordination, and editing of the manuscript. WB participated in the design of the study, study coordination, and editing of the manuscript. All authors read and approved the final manuscript.
ETHICS STATEMENT BY ALL AUTHORS
This study was conducted with approval from the Institutional Review Board of Columbia University Medical Center.
Authors take responsibility to maintain relevant documentation in this respect.
LIST OF ABBREVIATIONS (In alphabetic order)
CCCP - Columbia Combined Cancer Panel
CNB - Core needle biopsy
CT - Computed tomography
EBUS TBNA - Endobronchial ultrasound-guided transbronchial needle aspiration
FISH - Fluorescence in situ hybridization
FNA - Fine-needle aspirates
N/A - Not applicable
NCCN - National Comprehensive Cancer Network
NGS – Next-generation sequencing
NSCLC - Nonsmall cell lung cancer
PCR - Polymerase chain reaction
ROSE - Rapid on-site evaluation.
EDITORIAL/PEER-REVIEW STATEMENT
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through automatic online system.
Contributor Information
David M. DiBardino, Email: david.dibardino@uphs.upenn.edu.
David W. Rawson, Email: david.w.rawson@gmail.com.
Anjali Saqi, Email: aas177@cumc.columbia.edu.
Jonas J. Heymann, Email: jjh2110@cumc.columbia.edu.
Carlos A. Pagan, Email: cap2237@cumc.columbia.edu.
William A. Bulman, Email: wab10@cumc.columbia.edu.
REFERENCES
- 1.American Cancer Society. Cancer Facts & Figures. Atlanta: American Cancer Society; 2015. [Last accessed on 2014 Nov 15]. Available from: http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-031941.pdf . [Google Scholar]
- 2.Sharma SV, Settleman J. ErbBs in lung cancer. Exp Cell Res. 2009;315:557–71. doi: 10.1016/j.yexcr.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 3.Langer CJ, Besse B, Gualberto A, Brambilla E, Soria JC. The evolving role of histology in the management of advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:5311–20. doi: 10.1200/JCO.2010.28.8126. [DOI] [PubMed] [Google Scholar]
- 4.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuurbiers OC, Looijen-Salamon MG, Ligtenberg MJ, van der Heijden HF. A brief retrospective report on the feasibility of epidermal growth factor receptor and KRAS mutation analysis in transesophageal ultrasound- and endobronchial ultrasound-guided fine needle cytological aspirates. J Thorac Oncol. 2010;5:1664–7. doi: 10.1097/JTO.0b013e3181f0bd93. [DOI] [PubMed] [Google Scholar]
- 6.Keedy VL, Temin S, Somerfield MR, Beasley MB, Johnson DH, McShane LM, et al. American Society of Clinical Oncology provisional clinical opinion: Epidermal growth factor receptor (EGFR) Mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol. 2011;29:2121–7. doi: 10.1200/JCO.2010.31.8923. [DOI] [PubMed] [Google Scholar]
- 7.Leighl NB, Rekhtman N, Biermann WA, Huang J, Mino-Kenudson M, Ramalingam SS, et al. Molecular testing for selection of patients with lung cancer for epidermal growth factor receptor and anaplastic lymphoma kinase tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the study of lung cancer/association for molecular pathology guideline. J Clin Oncol. 2014;32:3673–9. doi: 10.1200/JCO.2014.57.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR, et al. Non-Small Cell Lung Cancer, Version 6.2015. J Natl Compr Canc Netw. 2015;13(5):515–24. doi: 10.6004/jnccn.2015.0071. [DOI] [PubMed] [Google Scholar]
- 9.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CK, Brown C, Gralla RJ, Hirsh V, Thongprasert S, Tsai CM, et al. Impact of EGFR inhibitor in non-small cell lung cancer on progression-free and overall survival: A meta-analysis. J Natl Cancer Inst. 2013;105:595–605. doi: 10.1093/jnci/djt072. [DOI] [PubMed] [Google Scholar]
- 11.Bulman W, Saqi A, Powell CA. Acquisition and processing of endobronchial ultrasound-guided transbronchial needle aspiration specimens in the era of targeted lung cancer chemotherapy. Am J Respir Crit Care Med. 2012;185:606–11. doi: 10.1164/rccm.201107-1199CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jurado J, Saqi A, Maxfield R, Newmark A, Lavelle M, Bacchetta M, et al. The efficacy of EBUS-guided transbronchial needle aspiration for molecular testing in lung adenocarcinoma. Ann Thorac Surg. 2013;96:1196–202. doi: 10.1016/j.athoracsur.2013.05.066. [DOI] [PubMed] [Google Scholar]
- 13.Sequist LV, Heist RS, Shaw AT, Fidias P, Rosovsky R, Temel JS, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol. 2011;22:2616–24. doi: 10.1093/annonc/mdr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Kung HJ, Mack PC, Gandara DR. Genotyping and genomic profiling of non-small-cell lung cancer: Implications for current and future therapies. J Clin Oncol. 2013;31:1039–49. doi: 10.1200/JCO.2012.45.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heymann JJ, Bulman WA, Maxfield RA, Powell CA, Halmos B, Sonett J, et al. Molecular testing guidelines for lung adenocarcinoma: Utility of cell blocks and concordance between fine-needle aspiration cytology and histology samples. Cytojournal. 2014;11:12. doi: 10.4103/1742-6413.132989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pareja F, Crapanzano JP, Mansukhani MM, Bulman WA, Saqi A. Cytomorphological features of ALK-positive lung adenocarcinomas: Psammoma bodies and signet ring cells. Cancer Cytopathol. 2015;123:162–70. doi: 10.1002/cncy.21507. [DOI] [PubMed] [Google Scholar]
- 17.Coley SM, Crapanzano JP, Saqi A. FNA, core biopsy, or both for the diagnosis of lung carcinoma: Obtaining sufficient tissue for a specific diagnosis and molecular testing. Cancer Cytopathol. 2015;123:318–26. doi: 10.1002/cncy.21527. [DOI] [PubMed] [Google Scholar]
- 18.Kanagal-Shamanna R, Portier BP, Singh RR, Routbort MJ, Aldape KD, Handal BA, et al. Next-generation sequencing-based multi-gene mutation profiling of solid tumors using fine needle aspiration samples: Promises and challenges for routine clinical diagnostics. Mod Pathol. 2014;27:314–27. doi: 10.1038/modpathol.2013.122. [DOI] [PubMed] [Google Scholar]
- 19.Hagemann IS, Devarakonda S, Lockwood CM, Spencer DH, Guebert K, Bredemeyer AJ, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer. 2015;121:631–9. doi: 10.1002/cncr.29089. [DOI] [PubMed] [Google Scholar]
- 20.König K, Peifer M, Fassunke J, Ihle MA, Künstlinger H, Heydt C, et al. Implementation of Amplicon parallel sequencing leads to improvement of diagnosis and therapy of lung cancer patients. J Thorac Oncol. 2015;10:1049–57. doi: 10.1097/JTO.0000000000000570. [DOI] [PubMed] [Google Scholar]
- 21.Yung RC, Otell S, Illei P, Clark DP, Feller-Kopman D, Yarmus L, et al. Improvement of cellularity on cell block preparations using the so-called tissue coagulum clot method during endobronchial ultrasound-guided transbronchial fine-needle aspiration. Cancer Cytopathol. 2012;120:185–95. doi: 10.1002/cncy.20199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinrichs JW, van Blokland WT, Moons MJ, Radersma RD, Radersma-van Loon JH, de Voijs CM, et al. Comparison of next-generation sequencing and mutation-specific platforms in clinical practice. Am J Clin Pathol. 2015;143:573–8. doi: 10.1309/AJCP40XETVYAMJPY. [DOI] [PubMed] [Google Scholar]
- 24.Tuononen K, Mäki-Nevala S, Sarhadi VK, Wirtanen A, Rönty M, Salmenkivi K, et al. Comparison of targeted next-generation sequencing (NGS) and real-time PCR in the detection of EGFR, KRAS, and BRAF mutations on formalin-fixed, paraffin-embedded tumor material of non-small cell lung carcinoma-superiority of NGS. Genes Chromosomes Cancer. 2013;52:503–11. doi: 10.1002/gcc.22047. [DOI] [PubMed] [Google Scholar]


