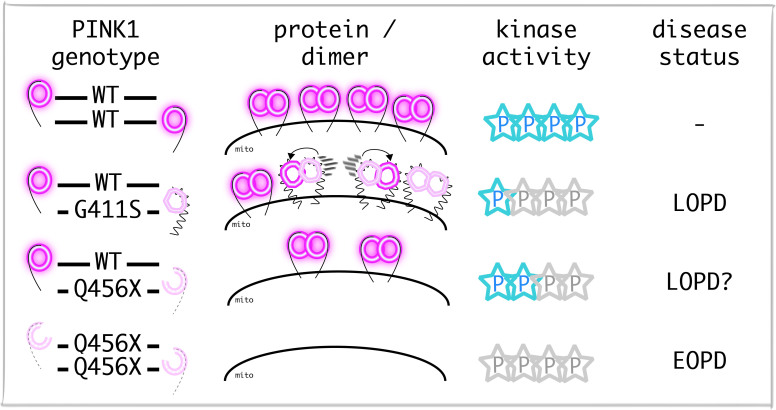
See Gandhi and Plun-Favreau (doi:10.1093/aww320) for a scientific commentary on this article.
Heterozygous mutations in recessive Parkinson’s disease genes have been postulated to increase disease risk. Puschmann et al. report a genetic association between heterozygous PINK1 p.G411S and Parkinson’s disease. They provide structural and functional explanations for a partial dominant-negative effect of the mutant protein, which impairs wild-type PINK1 activity through hetero-dimerization.
Keywords: Parkinson’s disease, PINK1, heterozygous mutation, ubiquitin, mitophagy
See Gandhi and Plun-Favreau (doi:10.1093/aww320) for a scientific commentary on this article.
Heterozygous mutations in recessive Parkinson’s disease genes have been postulated to increase disease risk. Puschmann et al. report a genetic association between heterozygous PINK1 p.G411S and Parkinson’s disease. They provide structural and functional explanations for a partial dominant-negative effect of the mutant protein, which impairs wild-type PINK1 activity through hetero-dimerization.
Abstract
See Gandhi and Plun-Favreau (doi:10.1093/aww320) for a scientific commentary on this article.
It has been postulated that heterozygous mutations in recessive Parkinson’s genes may increase the risk of developing the disease. In particular, the PTEN-induced putative kinase 1 (PINK1) p.G411S (c.1231G>A, rs45478900) mutation has been reported in families with dominant inheritance patterns of Parkinson’s disease, suggesting that it might confer a sizeable disease risk when present on only one allele. We examined families with PINK1 p.G411S and conducted a genetic association study with 2560 patients with Parkinson’s disease and 2145 control subjects. Heterozygous PINK1 p.G411S mutations markedly increased Parkinson’s disease risk (odds ratio = 2.92, P = 0.032); significance remained when supplementing with results from previous studies on 4437 additional subjects (odds ratio = 2.89, P = 0.027). We analysed primary human skin fibroblasts and induced neurons from heterozygous PINK1 p.G411S carriers compared to PINK1 p.Q456X heterozygotes and PINK1 wild-type controls under endogenous conditions. While cells from PINK1 p.Q456X heterozygotes showed reduced levels of PINK1 protein and decreased initial kinase activity upon mitochondrial damage, stress-response was largely unaffected over time, as expected for a recessive loss-of-function mutation. By contrast, PINK1 p.G411S heterozygotes showed no decrease of PINK1 protein levels but a sustained, significant reduction in kinase activity. Molecular modelling and dynamics simulations as well as multiple functional assays revealed that the p.G411S mutation interferes with ubiquitin phosphorylation by wild-type PINK1 in a heterodimeric complex. This impairs the protective functions of the PINK1/parkin-mediated mitochondrial quality control. Based on genetic and clinical evaluation as well as functional and structural characterization, we established p.G411S as a rare genetic risk factor with a relatively large effect size conferred by a partial dominant-negative function phenotype.
Introduction
Despite rapid progress in recent years, the genetic contribution to common disorders such as Parkinson’s disease has not been fully elucidated. The genetic factors that have been identified in Parkinson’s disease so far include clearly pathogenic mutations in rare familial forms and common genomic variants that confer a small increase or decrease in Parkinson’s disease risk (Ross et al., 2011; Puschmann, 2013; Mullin and Schapira, 2015). However, the heritability of Parkinson’s disease cannot be fully explained in terms of monogenic forms and the effects of common variants (Mullin and Schapira, 2015). Thus, it has been postulated that there may be a number of rare variants in the genome, which lead to a substantial increase in disease risk, without being pathogenic in all carriers. Only a few examples of such variants with intermediate effect size on the risk for Parkinson’s disease have been discovered, namely in the GBA and LRRK2 genes (Sidransky et al., 2009; Ross et al., 2011).
Homozygous or compound heterozygous mutations in PARK2/parkin, PINK1/PARK6 and PARK7/DJ1 are well-established causes of recessive, early-onset Parkinson’s disease (Puschmann, 2013). Careful analysis of families with mutations in recessive Parkinson’s disease genes revealed that heterozygous mutation carriers may also develop mild parkinsonism (Criscuolo et al., 2006; Klein et al., 2007; Eggers et al., 2010; Ricciardi et al., 2014). In a few instances, a seemingly autosomal dominant mode of inheritance was found in heterozygous carriers of mutations in recessive Parkinson’s disease genes (Klein et al., 2007). In particular, apparently autosomal dominant inheritance with complete or incomplete penetrance has been reported from families carrying the PINK1 c.1231G>A (rs45478900; p.G411S) variant (Abou-Sleiman et al., 2006; Toft et al., 2007; Mellick et al., 2009).
The proteins encoded by the PINK1 and PARK2 genes, PINK1, a mitochondrial kinase, and parkin, a cytosolic E3 ubiquitin ligase, together mediate a protective pathway that selectively eliminates damaged mitochondria from cells (Geisler et al., 2010a, b). Under physiological conditions, PINK1 protein is imported into the inner mitochondrial membrane (Jin et al., 2010; Shi et al., 2011; Yamano and Youle, 2013). Mitochondrial damage directs PINK1 to the outer mitochondrial membrane, where it rapidly accumulates, is auto-phosphorylated (Okatsu et al., 2012; Aerts et al., 2015) and dimerizes into a supermolecular protein complex (Liu et al., 2009; Okatsu et al., 2013). This activates PINK1's kinase activity, and activated PINK1 then phosphorylates ubiquitin (Kane et al., 2014; Kazlauskaite et al., 2014; Koyano et al., 2014; Shiba-Fukushima et al., 2014) and parkin (Kondapalli et al., 2012; Shiba-Fukushima et al., 2012) at a conserved residue (Ser65). Parkin is activated by phosphorylation and directed to damaged mitochondria, where it cooperates with PINK1 to amplify the formation of phosphorylated poly-ubiquitin chains (Ordureau et al., 2014; Okatsu et al., 2015), tagging damaged mitochondria for proteosomal and autophagic/lysosomal degradation (mitophagy) (Heo et al., 2015; Lazarou et al., 2015). Levels of phosphorylated p-Ser65-Ub are disease-relevant, mitochondrial stress-specific markers and reporters for PINK1 and parkin enzymatic activities (Fiesel et al., 2015a; Fiesel and Springer, 2015). Pathogenic mutations in either PINK1 or parkin disrupt mitochondrial quality control, albeit through different mechanisms and at distinct steps of a sequential process (Fiesel et al., 2015b).
Here, we studied co-segregation of PINK1 p.G411S with Parkinson’s disease in previously reported (Toft et al., 2007; Mellick et al., 2009) and newly identified families, and screened five independent Parkinson’s disease case-control series for this variant. In functional cell-based assays, we established a partial dominant-negative effect of p.G411S heterozygotes. Molecular modelling and dynamics simulations of wild-type PINK1 and mutant protein were used to provide a structural explanation. Altogether our data identify a novel mechanism by which p.G411S mutant protein interferes with wild-type PINK1 function through dimerization in heterozygotes, providing a link between clinically observed genetic risk and cellular phenotype.
Materials and methods
Case-control series
Parkinson’s disease patients and control subjects were enrolled in studies at Mayo Clinic Florida (USA; 748 Parkinson’s disease patients, 749 controls), in Katowice and Warsaw (Poland; 725 Parkinson’s disease patients, 238 controls), Trondheim (Norway; 418 Parkinson’s disease patients, 520 controls), Dublin (Ireland; 357 Parkinson’s disease patients, 361 controls), and Lund (Sweden; 312 Parkinson’s disease patients, 277 controls). All subjects were unrelated to each other and of European ancestry. A diagnosis of Parkinson’s disease was made by experienced neurologists and movement disorder specialists at local sites and according to established criteria (Ward and Gibb, 1990; Hughes et al., 1992). Clinical data were compiled; characteristics are provided in Supplementary Table 1. The study was not limited to early-onset cases and patients were included irrespective of their age at onset of Parkinson’s disease. Spouses, caregivers and other unrelated individuals served as control subjects. Blood was drawn and genomic DNA extracted according to standard protocols. PINK1 p.G411S genotype was assessed using TaqMan® single nucleotide polymorphism genotyping assays (Applied Biosystems). Direct sequencing was performed when results in TaqMan® cluster analyses of allele discrimination were positive or ambiguous. A two-sided Cochran-Mantel-Haenszel exact test, conditioning on the five different series, was used to compare the frequency of the p.G411S mutation between patients with Parkinson’s disease and controls. A two-sided Wilcoxon rank sum test was used to compare age at disease onset (which was available for ∼85% of patients) between carriers and non-carriers of p.G411S.
Meta-analysis
Previous studies that identified or excluded PINK1 p.G411S mutations in both Parkinson’s disease cases and controls were retrieved through a systematic search of the literature. Studies that were published by June 2016 and reported sequencing or high performance liquid chromatography of PINK1 exon 6 and/or PINK1 p.G411S in Parkinson’s disease patients and controls were searched in PubMed, OMIM, www.pdgene.org, and from other publications’ reference lists. In cases where one research group published several reports on the same population, we included the most recent publication to exclude sample overlap. Studies of single cases or families were not included. When supplementing these data with that of our aforementioned case-control study, the association between PINK1 p.G411S and Parkinson’s disease was evaluated using a two-sided Cochran-Mantel-Haenszel exact test, conditioning on each series.
Pedigree analysis
DNA and clinical information were available from affected and unaffected family members of Scandinavian PINK1 p.G411S patients, including Norwegian families previously published (Toft et al., 2007). Pedigrees were drawn according to information from the families where there was more than one affected family member. A previously published Australian family with this mutation (Mellick et al., 2009) was expanded and re-evaluated, and additional members studied clinically and genetically, and PINK1 p.G411S mutations were identified in two Ukrainian siblings (Fig. 1B). Results from additional genetic tests performed non-systematically for clinical purposes or within research were available for Scandinavian patients and were compiled. At a minimum, all patients with early-onset (before 50 years) disease were tested genetically for mutations in PARK2, PINK1 and PARK7/DJ1 by sequence analysis and for copy number variations.
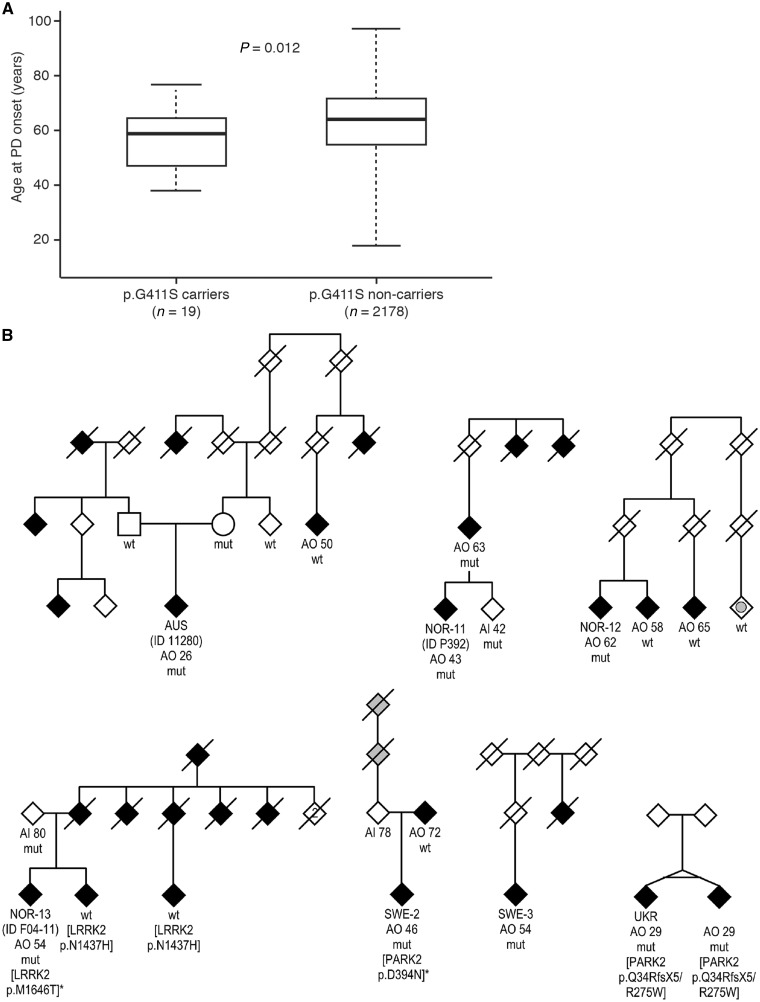
Figure 1.
Age at Parkinson’s disease onset of PINK1 p.G411S mutation carriers and pedigrees of the families with PINK1 p.G411S mutations. (A) Graph showing the distribution of the age at onset of the Parkinson’s disease (PD) patients with and without a PINK1 p.G411S mutation from the case-control series analysed. Median age at onset was 59 years for the patients carrying PINK1 p.G411S, which is higher than for patients homozygous for mutations in PINK1 but significantly lower than for the patients not carrying the mutation in our case-control series. (B) Black symbols indicate Parkinson’s disease; solid grey symbols, dementia; grey circle, tremor without parkinsonism. Diagonal lines denote the person was deceased. The index patient (proband, case) of each family is identified by AUS (Australian family), NOR (Norwegian), SWE (Swedish) and UKR (Ukrainian). ID indicates the individual identifier as used in previous publications (Toft et al., 2007; Mellick et al., 2009; Aasly et al., 2010). AO = age at onset; AI = age at inclusion in study. mut = PINK1 p.G411S mutation; wt = PINK1 p.G411 wild-type. In some families, sibling order has been changed to increase readability or to ensure confidentiality. Gender is disguised (diamonds) with the exception of the proband’s parents in the Australian family to enable comparison with its previous publication (Mellick et al., 2009). Additional mutations in genes for Parkinson’s disease are shown in square brackets. Besides carrying PINK1 p.G411S, both Ukrainian patients were compound heterozygous for PARK2 p.Q34RfsX5/p.R275W mutations that are considered pathogenic, Patient SWE-2 carried a PARK2 p.D394N substitution, for which there is no evidence for an association with Parkinson’s disease, and Patient NOR-13 carried a LRRK2 p.M1646T substitution, which has been shown to increase Parkinson’s disease risk (Wider et al., 2010; Ross et al., 2011). Affected relatives of Patient NOR-13 had an LRRK2 p.N1437H mutation but these individuals did not carry PINK1 p.G411S (Aasly et al., 2010). In three of the families, a proband’s first degree relative was affected and also carried the PINK1 p.G411S mutation, however in one family (Family UKR), this was the index patient’s monozygotic twin brother, and in another family (Family AUS), the first-degree relative did not have the full clinical picture of Parkinson’s disease but only resting tremor. In three families, PINK1 p.G411S was excluded in affected first-degree relatives.
Ethics
This study was approved by the Institutional Review Boards of the participating institutions. Written informed consent was obtained from all participants.
Cell culture
All cells were maintained at 37°C and 5% CO2 under humidified conditions. Control human dermal fibroblasts were received from Cell Applications. Other fibroblasts were collected under approved Mayo Clinic and Region Skåne (Sweden) ethical review board protocols (Supplementary Table 2). Parental human cervical carcinoma cells (HeLa) were obtained from ATCC (American Type Culture Collection) and clonal cells stably expressing EGFP-PARKIN were generated earlier (Fiesel et al., 2014). HeLa cells were cultured in high glucose Dulbecco’s modified Eagle medium (Invitrogen) supplemented with 10% foetal bovine serum (Biowest). Fibroblasts were supplemented with additional 1% non-essential amino acids and 1% penicillin-streptomycin (both Invitrogen). Induced neurons were generated from fibroblasts and cultivated as described previously (Fiesel et al., 2015a).
Chemical treatment and DNA, siRNA transfection of cells
To measure the induction of PINK1 and p-Ser65-Ub, cells were treated with 1 µM valinomycin (Axxora) or 10-15 µM carbonyl cyanide m-chlorophenyl hydrazine (CCCP; Sigma-Aldrich) for the indicated times. HeLa cells were transiently transfected with V5 and/or mCherry C-terminally tagged PINK1 variants (DNA ratio 1:1) and/or with 20 nM control (AllStars negative control) or PINK1-specific siRNA (5’-GACGCTGTTCCTCGTTATGAA-3’, both from Qiagen) using Lipofectamine® 2000 according to manufacturer’s protocol. Cell media was replaced 5 h after transfection for high content imaging and 24 h after transfection for immunoprecipitation and western blot analysis.
Quantitative reverse transcription-polymerase chain reaction
RNA was isolated using the RNeasy® mini kit (Qiagen) and 500 ng were reverse transcribed (Transcriptor High Fidelity cDNA synthesis kit, Roche) using oligo-dT primers. Two microlitres of 1/50 cDNA dilutions were mixed with 1/20 diluted primers (TaqMan® probes for PINK1: qHsaCEP0053094 and RPL27: qHsaCEP0051648, Bio-Rad) and 2.5 μl of 2× Universal Probe Supermix (Bio-Rad) in a 5 µl reaction. The PCR was executed using a 384-well block on a LightCycler 480 system (Roche). Absolute transcript levels for PINK1 and RPL27 were obtained by the second derivative method. Relative transcript levels were calculated as PINK1/RPL27 ratio and normalized to the relative expression level of the wild-type control.
Antibodies
The following primary antibodies were used for western blot (WB), immunofluorescence (IF) or meso scale discovery (MSD) assay: rabbit anti-PINK1 (WB, 1:2500; IF, 1:1000, #6946, D8G3), mouse anti-parkin (WB, 1:1000, #4211, Prk8, Cell Signaling Technology), mouse anti-Ub (WB, 1:2000, MSD, 1:500, MAB1510, ubi-1, Millipore), mouse anti-Mitofusin 2 (WB, 1:5000, ab56889, Abcam), mouse anti-TOM20 (IF, 1:100, sc-17764, Santa Cruz Biotechnology), rabbit anti-V5 (WB, 1:5000, ab9116, Abcam), rabbit anti-mCherry (WB, 1:2000, 5993-100, BioVision), rabbit anti-TUJ1 (WB, 1:1000, b3-tubulin, D71G9, Cell Signaling Technology), chicken anti-TUJ1 (IF, 1:500, β3-tubulin, AB9354, Millipore), mouse anti-GAPDH (WB, 1:150 000, H86504M, Meridian Life science), mouse anti-vinculin (WB, 1:250 000, V9131, Sigma-Aldrich). Rabbit anti-p-Ser65-Ub (1:5000 for WB, 1:250–500 for IF, and 1:250 for MSD) has been described recently (Fiesel et al., 2015a).
Western blots
Cells were harvested in 1× RIPA [50 mM Tris pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulphate (SDS)] or NP40 buffer (50 mM Tris pH 7.6, 150 mM NaCl, and 0.5% NP-40) containing protease and phosphatase inhibitors (Roche Applied Science). Protein concentration was determined by bicinchoninic acid (Pierce Biotechnology). Proteins were loaded onto 8–16% or 16% Tris-Glycine gels (Invitrogen) and transferred onto polyvinylidene fluoride membranes (Millipore), blocked and incubated with primary antibodies overnight at 4°C followed by horseradish peroxidase (HRP)-conjugated secondary antibodies (1:10 000; Jackson Immunoresearch). Bands were visualized with Immobilon Western Chemiluminescent HRP Substrate (Millipore) on Blue Devil Lite X-ray films (Genesee Scientific) or on a Fuji LAS-3000 (Fujifilm Life Science USA) system.
Immunofluorescence staining and high content imaging
Fibroblasts were seeded onto glass coverslips coated with poly-d-lysine (P0899, Sigma-Aldrich). On 96-well imaging plates (BD Biosciences), fibroblasts and induced neurons were grown on 0.01% poly-l-ornithine/3.3 µg/ml laminin (both Sigma-Aldrich) double-coated wells. Cells were fixed with 4% (w/v) paraformaldehyde, permeabilized with 1% Triton™ X-100, and incubated with primary antibodies followed by incubation with secondary antibodies (anti-rabbit Alexa Fluor® 488, anti-mouse IgG Alexa Fluor® 568, and/or anti-chicken Alexa Fluor® 647, 1:1000, Invitrogen). Nuclei were stained with Hoechst 33342 (1:5000, Invitrogen). For staining of PINK1 tyramide signal amplification (T20922, Invitrogen) was used. Coverslips were mounted onto microscope slides using fluorescent mounting medium (Dako). High content imaging plates were imaged on a BD Pathway 855 (BD Biosciences) and analysed as described (Fiesel et al., 2014). To collect signal only from cells transfected with siRNA and various PINK1-V5 constructs, co-expressed mCherry fluorescent intensity was used as a restriction parameter. High resolution confocal fluorescent images were taken with an AxioObserver microscope equipped with an ApoTome Imaging System (Zeiss).
Electrochemiluminescence assay
Ninety-six-well Meso Scale Discovery (MSD) assay plates were coated with 25 μl of p-Ser65-Ub capturing antibody with a final concentration of 3.08 µg/ml in carbonate-bicarbonate coating buffer (15 mM Na2CO3/35 mM NaHCO3, pH 9.4) overnight at 4°C. Plates were washed three times with 0.2% Tween-20 in Tris-buffered saline (TBST), blocked in 5% bovine serum albumin (BSA)/TBST for 3 h at room temperature and washed again. Protein lysates were diluted in 1% BSA/TBST to 0.16 µg/µl and 50 µl incubated overnight on the plate at 4°C. Plates were washed and incubated with total ubiquitin detection antibody for 1 h, followed by incubation with goat anti-mouse SULFO TAG secondary antibody (1:500 in 1% BSA/TBST, MSD) for 1 h at room temperature. Plates were washed and signal measured after addition of 150 μl/well 2× read buffer T with surfactant (MSD) on a SECTOR Imager 2400 using the MSD workbench Software.
(Co)-Immunoprecipitation
For PINK1-V5 immunoprecipitation (IP), HeLa cells treated with 15 µM CCCP were lysed in co-immunoprecipitation buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 0.5 mM EGTA, 10% glycerol and 0.2% NP-40) supplemented with protease and phosphatase inhibitors (Roche Applied Science) and briefly sonicated. Total cell lysate was incubated overnight at 4°C with mouse anti-V5-agarose beads (A7345, Sigma-Aldrich). Formed immuno-complex was spun down, washed three times with co-immunoprecipitation buffer and eluted from beads with 50 µl 2× SDS sample buffer and boiled at 95°C. Input cell lysates (10 µg) and 10 µl of immunoprecipitates were analysed by western blot.
In vitro kinase assay
For kinase assays, formed immuno-complexes were spun down, washed 2× with co-immunoprecipitation buffer and once with in vitro phosphorylation buffer (20 mM HEPES pH 7.4, 50 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA) with addition of 0.01% Triton™ X-100. Buffer was completely removed and replaced with 100 µl of in vitro reaction mixture containing phosphorylation buffer supplemented with 1 µg of N-terminally biotinylated ubiquitin (Boston Biochem), 1 mM TCEP (Gold Bio), 2 mM ATP trisodium salt (Sigma-Aldrich) and 0.01% Triton™ X-100. Reactions were carried out at 37°C under constant shaking for 20–24 h, stopped by addition of 6× SDS sample buffer, heated to 56°C for 15 min and analysed by western blot. Detection of biotinylated ubiquitin with streptavidin-HRP (Pierce) served as loading control.
Molecular modelling of PINK1, dynamics simulations and protein-protein docking
The full-length protein sequence of human PINK1, NP_115785.1 (581 amino acids) was retrieved from the NCBI Reference Sequence NM_032409.2. Each domain was modelled as a separate unit and built into a composite full-length structure. The hybrid model is derived from consensus between the programs PRIME (Prime v3.0, Schrödinger, LLC, New York, NY) (Jacobson et al., 2002; Krieger et al., 2009), YASARA SSP/Homology/PSSM Method (Hooft et al., 1996a, b; King and Sternberg, 1996; Altschul and Koonin, 1998; Jacobson et al., 2002; Qiu and Elber, 2006; Krieger et al., 2009, 2012), DISTILL (Porter) and TASSER (Zhou et al., 2007, 2009; Zhou and Skolnick, 2007, 2009, 2010, 2012) and combines homology, threading, ab initio, and compositing techniques. For details on the procedures for modelling of PINK1 structures, post-translational modifications, mutations, MDS and PINK1-ubiquitin docking see the online Supplementary material for the PINK1 model.
Results
Case-control series and meta-analysis for PINK1 p.G411S
In a case-control study on five different Caucasian series with a total of 2560 Parkinson’s disease patients and 2145 control subjects, PINK1 p.G411S substitution was present in 19 cases (0.74%) and five control subjects [0.23%; odds ratio (OR) 2.92, P = 0.032; Table 1], revealing a significant association with intermediate effect size of heterozygous PINK1 p.G411S carrier status with Parkinson’s disease. Median age at disease onset in the 19 cases carrying PINK1 p.G411S was 59 years (range: 38–77 years), and this was significantly lower than that of non-carrier cases (median: 64 years, range: 18–97 years, P = 0.012, Fig. 1A). p.G411S was most common in the Norwegian (1.44%), Polish (0.97%), and Swedish (0.96%) patients with Parkinson’s disease, with very low frequencies in controls for the Norwegian and Swedish series. A systematic search for previous publications examining this variant in both Parkinson’s disease patients and controls identified 14 additional studies (Supplementary Table 3), the majority included only patients with early-onset Parkinson’s disease. For the six studies involving Caucasian subjects, the PINK1 p.G411S mutation was found in three Parkinson’s disease patients (0.11%) compared to one control (0.06%). When supplementing the data from these six studies with that of our study, the increased risk of Parkinson’s disease for subjects with the p.G411S mutation remains evident (OR: 2.89, P = 0.027). Of note, results from prediction tools were rather ambiguous regarding the pathogenic nature of the mutation (Supplementary Table 4).
Table 1.
Association of PINK1 p.G411S heterozygotes with Parkinson’s disease
| Origin | Cases | Controls | |||
|---|---|---|---|---|---|
| p.G411S (HET) | p.G411 (WT) | p.G411S (HET) | p.G411(WT) | ||
| Norway | 6 (1.44%) | 412 (98.56%) | 1 (0.19%) | 519 (99.81%) | |
| Sweden | 3 (0.96%) | 309 (99.04%) | 0 (0%) | 277 (100%) | |
| Ireland | 0 (0%) | 357 (100%) | 0 (0%) | 361 (100%) | |
| Poland | 7 (0.97%) | 718 (99.03%) | 2 (0.84%) | 236 (99.16%) | |
| USA | 3 (0.40%) | 745 (99.60%) | 2 (0.27%) | 747 (99.73%) | |
| Total | 19 (0.74%) | 2541 (99.26%) | 5 (0.23%) | 2140 (99.77%) |
|
HET = heterozygous; WT = wild-type; CI = confidence interval.
Pedigree analysis of PINK1 p.G411S heterozygote carriers
We had access to relatives of seven patients with heterozygous PINK1 p.G411S mutation with at least one additional affected family member (Fig. 1B). These included the Norwegian (Families NOR9 and NOR11) and Australian (Family AUS) families where autosomal dominance previously had been suggested (Toft et al., 2007; Mellick et al., 2009), additional Scandinavian families (Families NOR12-14 and SWE2-3) from our case-control study, and a newly identified pair of monozygotic twins with this mutation from the Ukraine (Family UKR). PINK1 p.G411S did not co-segregate with Parkinson’s disease within families. We compared the clinical phenotype of the patients with heterozygous PINK1 p.G411S mutations and did not identify a clear pattern of clinical characteristics (Supplementary Table 5). All patients had all cardinal signs of Parkinson’s disease and all but one had tremor. Psychosis, hallucination and disorientation was noted in patients and first-degree relatives and might partly be explained by medication side effects; none of the patients had any obvious cognitive decline at the time of inclusion (Aasly et al., 2010; Wider et al., 2010).
p.G411S does not affect endogenous PINK1 protein levels, but impairs kinase activity in patients’ fibroblasts
We analysed primary skin fibroblasts from two unrelated patients with Parkinson’s disease heterozygous for PINK1 p.G411S to assay the increased disease risk from a functional, cell biological perspective. For comparison we used two cells per genotype from healthy control subjects, from Parkinson’s disease patients homozygous for PINK1 p.Q456X mutation, and from clinically unaffected family members heterozygous for PINK1 p.Q456X. First, we performed reverse transcription polymerase chain reaction (RT-PCR) and confirmed similar mRNA levels for wild-type PINK1 and p.G411S heterozygotes as well as a 50% reduction in p.Q456X heterozygotes (Supplementary Fig. 1). Transcripts with this mutation are known to be unstable due to nonsense-mediated mRNA decay resulting in loss of PINK1 full-length protein and kinase activity (Siuda et al., 2014). In p.G411S heterozygotes and in control cells with two wild-type alleles, PINK1 protein was almost undetectable under physiological conditions but increased swiftly upon mitochondrial depolarization with valinomycin as detected by western blot (Fig. 1A) and immunofluorescence (Supplementary Fig. 2). Levels of PINK1 protein were similar in p.G411S heterozygous cells compared to wild-type controls, but levels of p-Ser65-Ub persistently remained lower (Supplementary Fig. 3) and were significantly reduced at later time points (Fig. 2A and B). By contrast, in cells from patients with early-onset Parkinson’s disease homozygous for PINK1 p.Q456X, PINK1 protein levels were undetectable and phosphorylation of ubiquitin was absent. Consistently, cells from p.Q456X heterozygotes presented with lower PINK1 levels that initially also affected phosphorylation of ubiquitin at Ser65, however these differences did not remain significant over time (Fig. 2A and B and Supplementary Fig. 2 and 3). Parkin levels were comparable in controls or cells from all heterozygous PINK1 mutation carriers, but were strongly enhanced upon homozygous loss of PINK1 (Fig. 2A and C). While activation of parkin (as seen by reduction of the signal using an antibody that detects only inactive parkin) and the consequent ubiquitination of MFN2 was absent in p.Q456X homozygotes, effects on parkin and substrate levels in heterozygous cells were less clear, at least in western blot analyses.
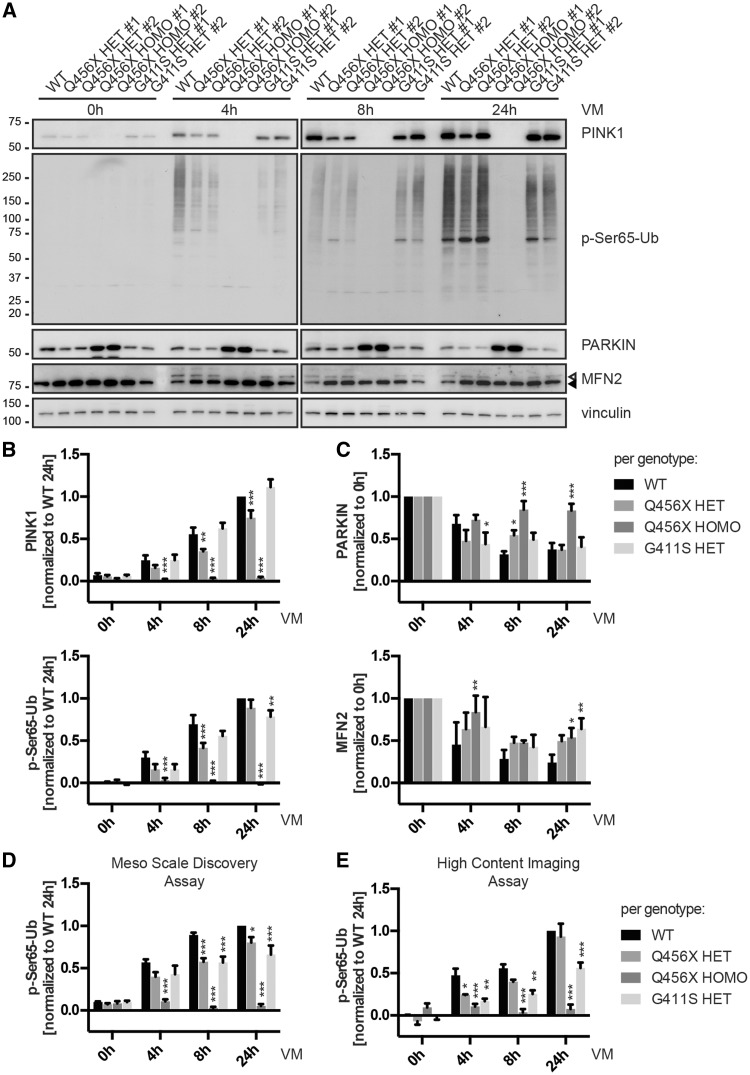
Figure 2.
p.G411S, but not p.Q456X heterozygotes show persistently impaired PINK1 kinase activity under endogenous conditions over time. Primary human skin fibroblasts were left untreated or stressed with 1 µM of the potassium ionophore valinomycin (VM) for the indicated times to depolarize mitochondrial membranes. Two cell lines per genotype were analysed. PINK1 genotypes of individuals are stated as wild-type (WT), heterozygous (HET) or homozygous (HOMO) along with the respective PINK1 mutation. (A) Representative western blots with total cell lysates were analysed for levels of PINK1, phosphorylated ubiquitin (p-Ser65-Ub), parkin, and its substrate mitofusin 2 (MFN2). Closed and open triangles indicate unmodified MFN2 and its ubiquitinated forms, respectively. Anti-vinculin was used as loading control. (B and C) Quantification of protein levels from western blots of six independent experiments. Shown is the mean per genotype ± SEM. Statistical significance was assessed with two-way ANOVA, Tukey’s post hoc test (*P < 0.05; **P < 0.005; ***P < 0.0005). (B) Shown are levels of PINK1 and p-Ser65-Ub normalized to wild-type at 24 h. Levels of total, unmodified ubiquitin were comparable from all cells and at all times (data not shown). p.Q456X heterozygotes showed significantly reduced levels of PINK1, concomitant with initially decreased levels of p-Ser65-Ub. However, p.G411S showed PINK1 levels similar to wild-type, but significantly reduced levels of p-Ser65-Ub over time. (C) Shown are relative levels of parkin and MFN2 normalized to 0 h valinomycin treatment. Absolute levels of parkin were similar in wild-type and all heterozygous PINK1 mutant cells, but were significantly elevated in p.Q456X homozygotes; however, no difference was seen for MFN2 (data not shown). (D) Quantitative MSD assay in 96-well plate format of p-Ser65-Ub levels from cell lysates. Cells were treated with 1 µM valinomycin (VM) for the indicated times, lysed and p-S65-Ub was captured with phospho-specific ubiquitin antibodies and detected with total ubiquitin antibodies. Shown is the mean per genotype ± SEM normalized to wild-type at 24 h from three independent experiments. Statistical significance was assessed with two-way ANOVA, Tukey’s post-hoc test (*P < 0.05; *** P < 0.0005). (E) Quantitative high content imaging in 96-well plate format of p-Ser65-Ub intensity in primary human fibroblasts before and after valinomycin (VM) treatment. Shown is the mean per genotype ± SEM normalized to wild-type at 0 h and 24 h from three independent experiments. Statistical significance was assessed with two-way ANOVA, Tukey’s post hoc test (*P < 0.05; **P < 0.005; ***P < 0.0005).
To corroborate genotype-dependent difference in p-Ser65-Ub response over time by alternative, quantitative methods, we developed an ELISA-like MSD assay using anti-phospho ubiquitin to capture and anti-total ubiquitin antibodies to detect p-Ser65-Ub in cell lysates. Using fibroblasts lysates, we could confirm a significant difference for both heterozygous genotypes at earlier time points compared to wild-type (Fig. 2D). However, p-Ser65-Ub signals remained reduced and highly significant over longer times of valinomycin treatment only in p.G411S heterozygotes. To ascertain these findings, we used automated high content imaging. This additional unbiased readout confirmed a persistent reduction of p-Ser65-Ub signal at longer times of valinomycin treatment only in p.G411S, but not in p.Q456X heterozygotes (Fig. 2E). p.Q456X homozygotes served as controls and accordingly showed no p-Ser65-Ub signals in MSD or high content imaging assay. Altogether, differences in pSer65-Ub levels, which were consistently observed across multiple, independent methods, suggest a pathomechanism for PINK1 p.G411S heterozygotes that is distinct from a simple loss-of-function such as p.Q456X.
Induced neurons recapitulate persistently reduced kinase activity in p.G411S heterozygotes
To corroborate findings from primary skin fibroblasts under endogenous conditions also in neuronal cells, we generated induced neurons by direct conversion via reprogramming of cellular miRNA pathways (Xue et al., 2013). Previously, we had already shown that the PINK1/parkin pathway is active and that p-Ser65-Ub levels are induced by mitochondrial stress in neurons (Fiesel et al., 2015a). Fibroblast were infected with a lentiviral polypyrimidine tract binding protein (PTB) short hairpin RNA and the resulting induced neurons were differentiated, treated with valinomycin and analysed by western blot. Similar to fibroblasts, induced neurons showed comparable levels of PINK1 in wild-type cells and p.G411S heterozygotes (Fig. 3A). PINK1 levels appeared slightly reduced in p.Q456X heterozygotes, albeit not as pronounced as in fibroblasts, and were absent from homozygotes. Consistently, p-Ser65-Ub levels were also reduced in induced neurons from both p.G411S and p.Q456X heterozygotes and completely absent in homozygotes while parkin levels were strongly elevated.
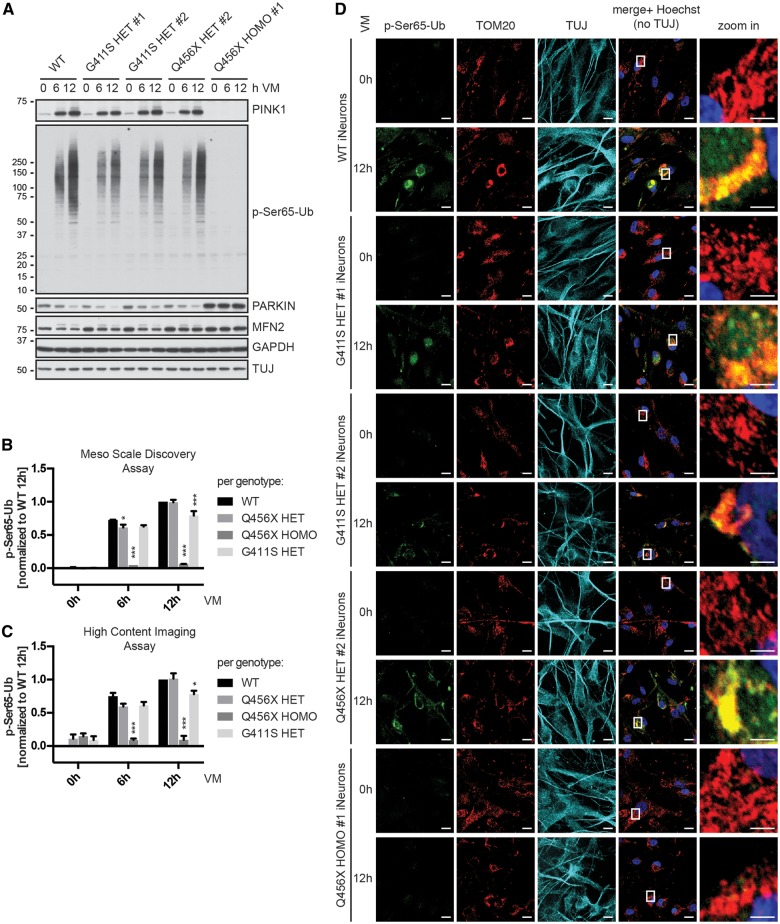
Figure 3.
p.G411S heterozygotes induced neurons show persistently reduced p-Ser65-Ub levels. Primary human fibroblasts were directly converted into induced neurons and treated with 1 µM valinomycin (VM) for the indicated times. Induced neurons (iNeurons) were analysed by independent methods that confirm a significant reduction of p-Ser65-Ub levels in p.G411S heterozygotes. (A) Western blot analysis of PINK1 stabilization and p-Ser65-Ub induction as well as parkin and MFN2 protein levels. Shown are representative images from three independent experiments. Anti-GAPDH served as a loading control and anti-TUJ as control for successful conversion to neurons. (B) Quantification of p-Ser65-Ub levels in lysates from induced neurons treated with valinomycin by MSD corroborates time- and genotype-dependent differences. Shown are mean values ± SEM per genotype from three independent experiments normalized to values of wild-type at 12 h. Statistical significance was assessed by two-way ANOVA with Tukey’s post hoc test (*P < 0.05; ***P < 0.0005). (C) Quantification of p-Ser65-Ub levels in induced neurons treated with valinomycin by high content imaging confirms time- and genotype-dependent differences. Shown are mean values ± SEM per genotype from three independent experiments normalized to wild-type at 12 h. Statistical significance was assessed by two-way ANOVA with Tukey’s post hoc test (*P < 0.05; ***P < 0.0005). (D) Representative images of induced neurons that were left untreated (0 h) or treated for 12 h with 1 µM valinomycin (VM) from three independent experiments. Cells were fixed and stained with anti-p-Ser65-Ub (green), anti-TOM20 (mitochondria, red), anti-TUJ1 (neuronal marker, cyan), and Hoechst (nuclei, blue). Shown is the merge of p-Ser65-Ub, TOM20, and Hoechst. Scale bars = 20 µm. Images to the right represent enlarged views of the boxed areas. Scale bars = 5 µm.
For quantification of p-Ser65-Ub in induced neurons, we used MSD and high content imaging assays (Fig. 3B and C). Consistent with data from fibroblasts, levels of p-Ser65-Ub though initially reduced in p.Q456X heterozygotes were not different from wild-type at longer times of valinomycin treatment. Yet, levels of p-Ser65-Ub remained reduced over time only in carriers of the p.G411S mutation. Conventional immunofluorescence corroborated the reduced p-Ser65-Ub signal measured by high content imaging at longer times valinomycin treatment for p.G411S heterozygotes (Fig. 3D). In summary, PINK1 genotype-specific responses were also present in disease-relevant induced neurons, where p-Ser65-Ub levels remained significantly reduced over time only in p.G411S, but not p.Q456X heterozygotes.
Molecular modelling of human full-length wild-type PINK1 and the p.G411S mutant
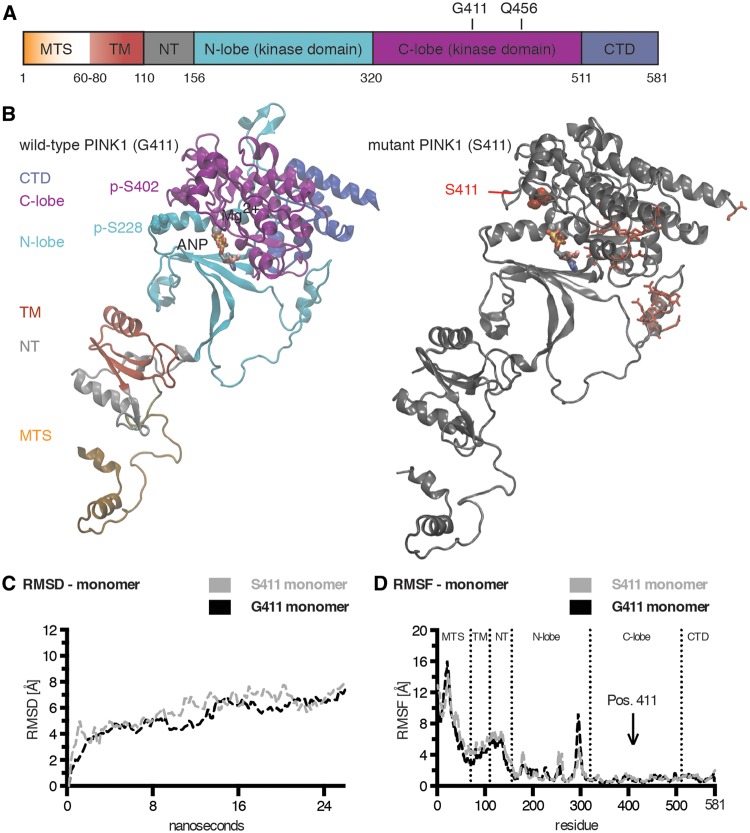
We used a structural computational approach to model wild-type and p.G411S mutated PINK1. As crystallographic data for PINK1 were not available, we performed molecular modelling with a combination of methods previously described (Caulfield et al., 2011; Caulfield and Devkota, 2012; Zhang et al., 2013). Individual (sub-)domains of PINK1 (Fig. 4A) were generated as separate units and built into a composite full-length protein model of human PINK1 at an all-atom resolution (Fig. 4B). The kinase domain of the obtained PINK1 structure was in good agreement with previous models (Beilina et al., 2005; Mills et al., 2008; Cardona et al., 2011; Sim et al., 2012; Trempe and Fon, 2013). In addition, our model also contained the N-terminal region that regulates import, sub-mitochondrial localization, and turnover of PINK1. A comparison of the primary/secondary/tertiary structure for this region between our model and predictions is given in Supplementary Fig. 4. To model an active kinase, PINK1 was phosphorylated in silico at Ser228 and Ser402. These two auto-phosphorylation sites gate the entry to the active site formed between the two lobes of the kinase domain (Fig. 4B, left), and are required for full kinase function (Okatsu et al., 2012, 2013; Aerts et al., 2015).
Figure 4.
Structural models for wild-type and p.G411S mutant forms of human PINK1. (A) Schematic representation of the protein domain composition of full-length PINK1. Individual regions are colour-coded from the N- to the C-terminus as follows: mitochondrial targeting sequence (MTS) in orange with gradient (residues 1–60); transmembrane region (TM) in red (residues 60–110); N-terminal regulatory region (NT) in grey (residues 110–155); N- and C-lobes of the kinase domain in cyan (residues 156–320) and magenta (residues 320–511), respectively; and the C-terminal domain in blue (residues 511–581). The locations of the residues Gly411 (G411) and Gln456 (Q456) in the activation loop within the C-lobe are highlighted. (B) Molecular modelling of autophosphorylated, full-length PINK1 wild-type (Gly411, left) and mutant (Ser411, right). The phosphorylated serine residues p-Ser228 and p-Ser402 are highlighted in Van der Waals presentation. The ATP analogue (ANP) is coloured by atom type and shown together with Mg2+ (Van der Waals sphere) in the active site of PINK1. The p.G411S mutation is depicted in red Van der Waals style. Red sticks highlight additional residues in distant regions with conformational shifts in the Ser411 mutant monomer compared to wild-type. (C) Shown is a comparison of the root mean square deviations (RMSD) of wild-type (G411, black) and mutant (S411, grey) PINK1 monomers over 24 ns MDS across the entire structures. (D) Shown is a comparison of the root mean square fluctuations (RMSF) of wild-type (G411, black) and mutant (S411, grey) PINK1 monomer structures over 24 ns MDS as a time-averaged sum for each residue. Dashed lines annotate domain regions of PINK1 and an arrow indicates the position of residue 411.
We generated corresponding models for wild-type PINK1 protein (Gly411) and p.G411S-mutated PINK1 (Ser411). The residue Gly411 is located in the activation loop within the C-lobe of the kinase domain close to the active and adjacent autophosphorylation sites. Exchange to Ser411 resulted in only minor local structural changes, but caused a few alterations more distant in the N-lobe of the kinase domain and at the very C-terminus of PINK1 without affecting overall conformation of PINK1 (Fig. 4B, right). Using free (unbiased) molecular dynamics simulation (MDS), we analysed structural motions of wild-type PINK1 and p.G411S mutant (Supplementary Videos 1 and 2, respectively). Focusing on the higher confidence prediction of the kinase domain, the N-lobes were more dynamic with several flexible loops compared to the C-lobes that had the most rigidity due to tightly packed alpha-helical motifs. The superposition overlay of Gly411 and Ser411 structures of the kinase domains showed only slight deviation in the initial conformation, but showed a clear conformational drift on MDS (Supplementary Fig. 5A). Notable changes also occurred in the active site region of Ser411 that may potentially reduce kinase activity.
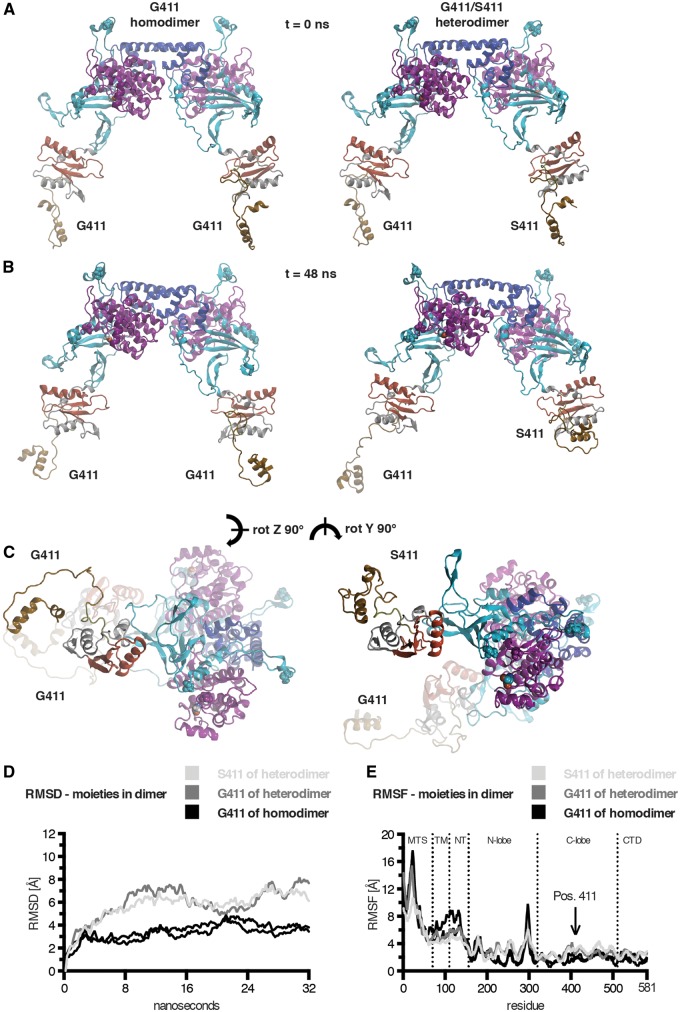
Structural alterations of the p.G411S propagate into wild-type in a PINK1 heterodimer
Next, we modelled dimeric forms of PINK1 kinase (wild-type G411/G411 homodimer and mutant G411/S411 heterodimer). The homodimeric Gly411 model of PINK1 showed a symmetric structure with interacting residues in the C-lobe of the kinase domain and residues 522–581 from the CTD (Fig. 5A, left). The heterodimeric G411/S411 complex only slightly deviated from the structure of the G411/G411 homodimer (Fig. 5A, right), consistent with only subtle changes in the Ser411 monomer. Upon free MDS (Fig. 5B and C), we found a significant retraction of the N-terminal domains of PINK1 in the Ser411 mutant subunit of the heterodimer (Supplementary Videos 3 and 4, respectively). Of note within the kinase domain, a beta-sheet around residue Thr257, which has been reported as a phosphorylation site (Kondapalli et al., 2012; Aerts et al., 2015), conformationally transitioned into an alpha-helical motif in the mutant subunit of the Gly411/Ser411 heterodimer (Supplementary Videos 5 and 6). Pronounced local differences were observed in a region 6 Å around the ATP analogue of the monomers; however, in a dimeric configuration each subunit showed more stabilization in the active site (Supplementary Fig. 5B), consistent with the idea that the PINK1 dimer is the active kinase.
Figure 5.
Structural models of PINK1 dimers and propagation of conformational effects from the mutant to the wild-type subunit in a PINK1 heterodimer. The PINK1 wild-type homodimer model is shown to the left and the Gly411/Ser411 heterodimer is shown to the right. Domains and residues of all PINK1 dimer subunits are colour-coded and depicted as in Fig. 4. Gly411 and Ser411 are highlighted in Van der Waals coloured by atom type. In addition to the two autophosphorylation sites p-Ser228 and p-Ser402, a third phosphorylation site in PINK1, Thr257, is also highlighted in Van der Waals coloured according to the domain colour. Both dimers are shown side-by-side before and after MDS in two orientations. (A) Similar conformations of the initial dimer models before MDS (time = 0 ns). (B) The PINK1 dimers are shown after 48 ns of simulation in large solvent box (>3.35 × 105 atoms) and reveal striking changes induced by the p.G411S mutation. (C) The same conformations are shown from a different angle as indicated (z-, y-rotated 90°). (D) Global comparisons of the individual moieties from the dimeric structures over time are shown. The root means square deviation (RMSD) was calculated and supports the propagation of mutational effects from the mutant (S411, light grey) into the wild-type subunit (G411, dark grey) of a PINK1 heterodimer in comparison to subunits of a wild-type homodimer (black). (D) The root means square fluctuation (RMSF) is shown for all subunits of wild-type and mutant heterodimer structures over the simulation as a time-averaged sum per residue. Dashed lines annotate domain regions of PINK1 and an arrow indicates the position of residue 411.
Interestingly, structural changes induced by the p.G411S mutation also resulted in motions at the dimerization interface (Supplementary Table 6) that further translated into considerable changes in the G411 subunit of a heterodimer. We next calculated the root mean square deviation of wild-type and mutant PINK1 dimers during free MDS. While both monomers had only minor global conformational changes (Fig. 4C), we noticed that the Ser411 mutation affected the motions of the wild-type moiety within a heterodimeric complex compared to either subunit of a wild-type PINK1 homodimer (Fig. 5D). We then compared fluctuations per residue as a time-averaged sum over the course of the individual simulations (root mean square fluctuations) to highlight regions with more mobility or greater stability. Monomers showed few greater peak changes in the N-lobe of the kinase domain (see red highlighted residues in Fig. 4B) along with several smaller ones in the entire N-terminal part (Fig. 4D). However, in the heterodimeric situation, substantial peak changes of both subunits were distributed across the entire molecules including the otherwise more rigid C-terminal parts as compared to both wild-type moieties of the homodimer (Fig. 5E).
Ubiquitin docking reveals reduced catalytic efficiencies for both PINK1 subunits of a heterodimer
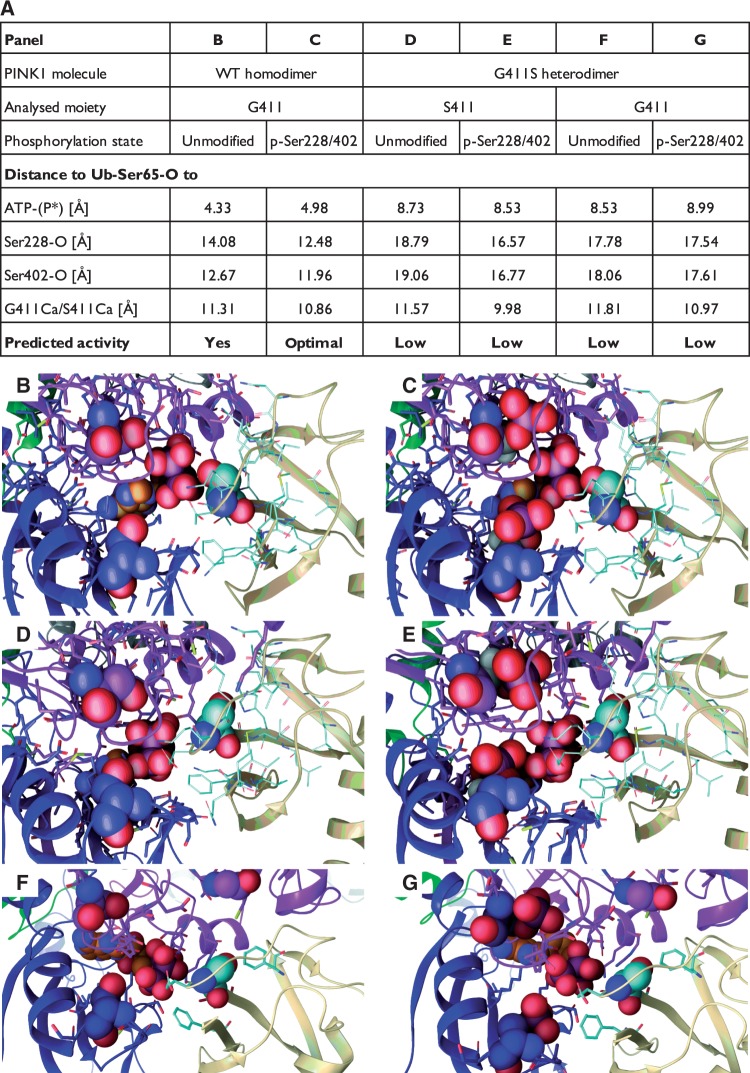
Despite the observed changes, the p.G411S mutation did not substantially affect accessibility to the catalytic site of PINK1 as measured by spacing between the C-alpha atoms of the gating residues p-Ser228 and p-Ser402 as well as distances to Gly411 or Ser411 (data not shown). However, in order to elucidate how structural changes could affect phosphorylation of the substrate, we docked ubiquitin to PINK1 homo- and heterodimers in complex with an ATP molecule. In the wild-type homodimer, we observed that interactions of His68 from ubiquitin stabilize the substrate in the kinase domain in a way that would facilitate transfer of the terminal phosphate from ATP to the oxygen of Ser65 (Fig. 6 and Supplementary Fig. 6). Notably, we detected that the auto-phosphorylation of PINK1 at Ser228 and Ser402 increased ubiquitin interactions via Lys63 and Lys6 (cf.Fig. 6B and C). These interactions not only stabilized the entire complex further, but resulted in a decrease of the atomic distances between kinase and substrate, which would positively influence phosphorylation of ubiquitin (Fig. 6 and Supplementary Fig. 7).
Figure 6.
Ubiquitin binding to PINK1 and prediction of phosphorylation efficiencies. Ubiquitin was docked as a substrate to each half of the PINK1 kinase wild-type homo- (G411/G411) and mutant (G411/S411) heterodimers. Given is a tabular summary of distances [Å] between the Ser65 oxygen of ubiquitin (Ub-Ser65-O) and the terminal phosphate of the bound ATP molecule as well as key PINK1 atoms (A). Relevant PINK1 atoms include the oxygens of Ser228 and Ser402 (Ser228/402-O) as well as the alpha carbons of either G411 or S411 (G411/S411-Cα). Both unmodified and auto-phosphorylated (p-Ser228/402) forms of PINK1 were analysed and compared for each subunit from the respective dimers. Greater distances between Ub-Ser65 and ATP as well as Ser228 and Ser402 of PINK1 in both wild-type and mutant subunits of the heterodimer, compared to wild-type homodimer, likely result in less efficient phosphorylation of the substrate. Interestingly, phosphorylation of PINK1 Ser228 and Ser402 in the wild-type homodimer facilitates an optimal alignment of ATP and Ub-Ser65 in the active site of PINK1. (B–G) The corresponding magnifications of the kinase domain (between the N- and C-lobes) of the PINK1 molecules with ubiquitin docked near the active site. Ser228 and Ser402 (or p-Ser228 and p-Ser402) are shown in Van der Waals spheres with the carbons coloured to match the domain colours. ATP is shown with orange carbons and structure in Van der Waals. The ubiquitin molecule is shown in beige ribbons with cyan carbon and the Ser65 residue in Van der Waals. G411 or S411 of PINK1 is shown above the site. Corresponding full-length PINK1 dimer structures in complex with ubiquitin can be found in Supplementary Fig. 6. A schematic representation of the analysed distances between the respective atoms is depicted in Supplementary Fig. 7.
In the p.G411S heterodimer, we observed that both subunits lost the interaction with His68 of ubiquitin. The entire ubiquitin structure was significantly pushed out by the conformational changes occurring in the S411 subunit (Fig. 6D and E). This was evidenced by distances between key atoms increasing by as much as 4–5 Å, placing the critical Ser65 oxygen too far from the ATP molecule for efficient catalysis. In addition, stabilizing effects of p-Ser228 and p-Ser402 were less capable of obtaining the interactions with lysine and histidine residues of ubiquitin, thus frustrating the enzymatic process. Importantly, we also found a deviation in positioning of the entire ubiquitin molecule to the G411 subunit of a PINK1 heterodimer (Fig. 6F and G). As a result of the structural defects that propagate from the S411 mutant to the G411 subunit, ubiquitin binding was decreased and the oxygen of Ser65 was placed outside desired distances to the terminal phosphate of the ATP. Taken together, these data support the idea that the p.G411S mutation transduces structural defects to the wild-type protein that adopts similar fold and motions in a PINK1 heterodimer and negatively affects positioning of the ubiquitin substrate and thus kinase activity.
PINK1 p.G411S exerts a dominant-negative mechanism
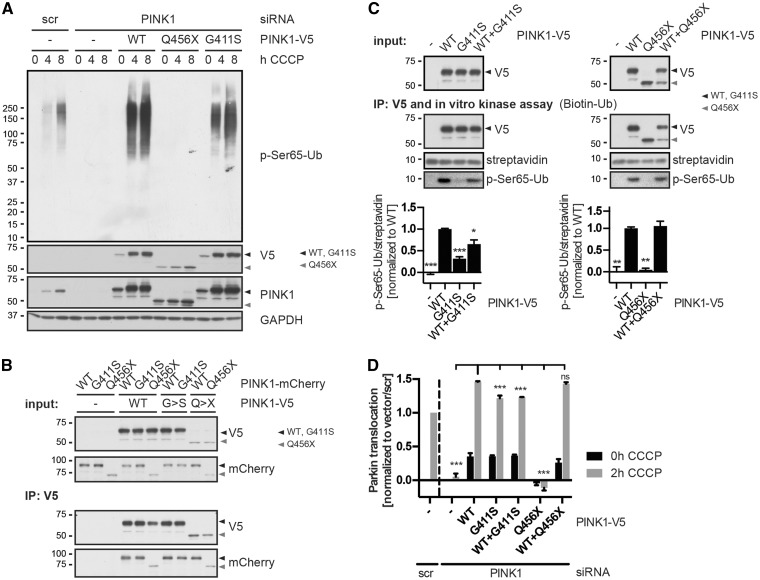
To provide further evidence for a dominant-negative effect of the p.G411S mutation on wild-type PINK1, we used an established HeLa cell model. We analysed overexpression of PINK1 wild-type, p.G411S and p.Q456X cDNAs in cells that had been depleted of endogenous PINK1 by RNAi (Fig. 7A). Consistent with the idea of a less efficient kinase activity, p.G411S overexpressing cells showed reduced p-Ser65-Ub levels compared to PINK1 wild-type, albeit expression levels and CCCP-dependent stabilization of both were similar. Expression of p.Q456X showed no p-Ser65-Ub signal, confirming the kinase deficiency of this mutant. To prove the predicted heterodimerization of PINK1 wild-type and p.G411S mutant, we expressed differentially tagged PINK1 variants and performed immunoprecipitations (Fig. 7B). Both PINK1 wild-type and p.G411S interacted with themselves and with each other to a similar extent. Also p.Q456X interacted with wild-type though to a lesser degree than full-length PINK1, in line with lack of some of the interaction surfaces in this truncated variant.
Figure 7.
The PINK1 p.G411S mutation exerts a dominant-negative mechanism. HeLa cells were used to confirm a dominant-negative effect of the p.G411S mutation on kinase activity of PINK1 wild-type that translated into reduced activation of parkin downstream. (A) HeLa cells were simultaneously transfected with specific PINK1 siRNA and siRNA-resistant PINK1-V5 wild-type or mutants (p.Q456X or p.G411S). Control cells were transfected with the corresponding empty vector (-) and with scrambled (scr) or PINK1 siRNA. Cells were treated with 15 µM CCCP for the indicated times and levels of phosphorylated ubiquitin were assessed by anti-p-Ser65-Ub. Endogenous and overexpressed PINK1 levels were monitored by anti-PINK1 and anti-V5 antibodies, respectively. Anti-GAPDH served as loading control. (B) HeLa cells were co-transfected with the indicated combinations of PINK1-V5 and PINK1-mCherry constructs or respective empty vector controls (-) and treated with 15 µM CCCP for 3 h. PINK1-V5 was immunoprecipitated (IP: V5) and the interaction between wild-type and mutant PINK1 was analysed by western blot. PINK1 wild-type and p.G411S strongly interacted with themselves and with each other to a similar extent. Black and grey triangles indicate full-length (wild-type and p.G411S) and truncated (p.Q456X) PINK1 protein, respectively. (C) In vitro ubiquitin phosphorylation assay confirms reduced kinase activity of p.G411S mutant and partial dominant-negative effects on PINK1 wild-type. HeLa cells were transfected with V5-tagged PINK1 wild-type, p.G411S, p.Q456X or a combination of wild-type plus p.G411S or p.Q456X. Cells were then treated with 15 µM CCCP for 3 h and PINK1 was immunoprecipitated with anti-V5. V5 immunoprecipitates were incubated with biotinylated ubiquitin in kinase reaction buffer. Anti-V5 antibody was used to show equal PINK1 levels in the IP. Black and grey triangles indicate full-length (wild-type and p.G411S) and truncated (p.Q456X) PINK1 protein, respectively. Phosphorylation of ubiquitin was determined by anti-p-Ser65-Ub antibody and total ubiquitin was detected by streptavidin-HRP that served as a loading control. Quantification of the p-Ser65-Ub/streptavidin ratio from three independent experiments is provided below. Values represent mean ± SEM, normalized to the average of wild-type values. Statistical significance was assessed by one-way ANOVA with Tukey’s post hoc test (*P < 0.05; **P < 0.005; ***P < 0.0005). (D) High content imaging of HeLa cells stably expressing EGFP–parkin was used to quantify parkin activation and translocation to damaged mitochondria upon overexpression of PINK1 variants. While co-expression of p.G411S with PINK1 wild-type significantly reduced parkin activation, p.Q456X showed no dominant-negative effect. HeLa cells were simultaneously transfected with specific PINK1 siRNA and siRNA-resistant PINK1-V5 wild-type or mutants (p.Q456X or p.G411S) or their combinations as indicated. mCherry was used as a transfection control. Control cells were transfected with empty vector with scrambled (scr) or PINK1 siRNA. Cells were left untreated (0 h) or treated with 10 µM CCCP for 2 h. Parkin translocation to mitochondria was measured in transfected, mCherry co-expressing cells only. Values represents mean ± SEM, n = 3. Statistical significance was assessed by two-way ANOVA with Tukey’s post hoc test (***P < 0.0005; ns = not significant).
Next, we aimed to mimic the heterozygous conditions by co-expression of both. Wild-type PINK1 and mutant p.G411S (or p.Q456X) were transfected either alone or in combination with wild-type, immunoprecipitated and subjected to an in vitro kinase assay using recombinant ubiquitin as a substrate (Fig. 7C). As expected, p.G411S showed strongly reduced kinase activity towards ubiquitin compared to PINK1 wild-type. Strikingly, co-expression of p.G411S along with PINK1 wild-type, significantly impaired ubiquitin phosphorylation. Importantly, ubiquitin phosphorylation by wild-type PINK1 was not affected in the presence of the p.Q456X mutant despite its lack of kinase activity. To further demonstrate translation into functional downstream defects, we monitored parkin translocation to damaged mitochondria by high content imaging (Fig. 7D). While overexpression of either wild-type PINK1 or the mutant p.G411S, but not p.Q456X, effectively activated parkin in this paradigm, the p.G411S mutant showed a small, but significant reduction compared to wild-type. Of note and in line with in vitro kinase assays, co-expression of p.G411S, but not the p.Q456X mutant, along with wild-type PINK1 also significantly reduced activation of parkin. Together, these data confirm the interaction of wild-type PINK1 and p.G411S mutant in a heterodimer as well as the reduction of kinase activity induced by the mutant in cis and in trans.
Discussion
Based on previous reports suggesting that heterozygous mutations in recessive Parkinson’s disease genes in general (Criscuolo et al., 2006; Klein et al., 2007; Eggers et al., 2010; Ricciardi et al., 2014) and PINK1 p.G411S in particular (Abou-Sleiman et al., 2006; Toft et al., 2007; Mellick et al., 2009) may increase the risk for Parkinson’s disease, we examined possible disease-association and pathogenic mechanisms of this variant. Genetic analyses as well as functional, cell-based and structural, computational characterization for the first time provided evidence for a partial dominant-negative function of the heterozygous PINK1 p.G411S mutation that confers a markedly increased risk for Parkinson’s disease.
Our pedigree analyses do not support co-segregation of PINK1 p.G411S with Parkinson’s disease in seven multi-incident families. In one of the Norwegian families that was reanalysed here, p.G411S was found in the proband (Subject NOR-11) and in the father. Two paternal uncles of the proband had Parkinson’s disease but were deceased and not tested genetically (Toft et al., 2007). A similar dominant mode of inheritance had previously been reported from an Australian family (Family AUS) (Mellick et al., 2009), but here expansion, clinical follow-up, and testing of additional family members has shown no co-segregation of the mutation with Parkinson’s disease in that family. This reduced penetrance may be due to multiple factors, for example, differences in allelic expression levels of the wild-type PINK1 allele relative to the p.G411S mutant in heterozygous individuals has not been analysed. In addition to transcriptional regulators at the PINK1 locus, we recently identified that expression is also regulated on the translational level, both of which could well contribute to this phenomenon (Kim et al., 2016).
In our case-control study, we found a significant association of heterozygote PINK1 p.G411S carrier status with Parkinson’s disease, and this association remained significant in a meta-analysis supplementing our results with data from previous publications. Heterozygous PINK1 p.G411S increased Parkinson’s disease risk with an effect size (OR 2.89), which is larger than that of known disease-associated common variants. The low frequency of PINK1 p.G411S, especially in non-Scandinavian populations, would explain why the PINK1 locus has not been identified in genome-wide association studies (IPDG Consortium, 2011). It has previously been reported from early-onset Parkinson’s disease case-control studies that more patients than controls subjects carry heterozygous mutations in PINK1 or PARK2/parkin, but without statistical significance. Differences in pathobiological effects among individual PINK1 mutations may be the reason why combined analysis of different PINK1 variants failed to show an association with Parkinson’s disease (Marongiu et al., 2008).
Incomplete penetrance of a mutation in PINK1 is also fully compatible with its biological role during stress-activated mitochondrial quality control. Levels of PINK1 engagement and thus robustness of the phenotype will be defined by levels of mitochondrial damage in the respective individual that result from a concert of various factors including age, environment and other genomic or somatic genetic variants (Coxhead et al., 2016). Incomplete penetrance is well-established for other pathogenic mutations in Parkinson’s disease genes, such as the common LRRK2 p.G2019S mutation or the disease-associated mutations in GBA (Sidransky et al., 2009; Ross et al., 2011). Interestingly, no homozygous carrier with PINK1 p.G411S mutation, nor compound heterozygosity has been described in studies of Parkinson’s disease (Kilarski et al., 2012); only one homozygous subject is present out of 60 266 subjects across all populations in the ExAC database (Cambridge, MA. http://exac. broadinstitute.org; accessed February 2016). The low frequency of homozygote p.G411S carriers may be due to the rarity of the mutation or could indicate it is particularly damaging or clinically manifests with an alternate phenotypic presentation. Replication of our genetic association in other case-control series and further clinical studies are now warranted.
Functional, cell-based assays with primary skin fibroblasts and induced neurons from human mutation carriers and controls were performed to determine the pathogenic mechanism of the PINK1 p.G411S mutation. Using an acute mitochondrial stress paradigm we focused on protein levels and kinase activity of PINK1. Western blots and conventional immunocytochemistry together with MSD and high content imaging consistently showed that, with protein levels similar to wild-type PINK1, total cellular PINK1 kinase activity remained significantly reduced over time in cells from p.G411S heterozygotes compared to wild-type controls and to p.Q456X heterozygotes. Findings of p.Q456X homo- and heterozygotes were compatible with a classical recessive loss-of-function mode, but our observations from p.G411S cells indicated a different mechanism for this particular heterozygous mutant. Our data demonstrate that p.G411S mutant PINK1 protein decreased the kinase activity of the wild-type protein in a dominant-negative mode, but inhibition was only partial. Yet, as PINK1 is rate-limiting for effective mitochondrial quality control, persistently reduced kinase activity further translated into compromised mitochondrial quality control that might well contribute to a significantly increased disease risk.
The p.G411S variant had previously been described as non-pathogenic based on prediction tools and experiments with cellular overexpression (Narendra et al., 2010). Overexpression of PINK1 p.G411S rescued parkin translocation to damaged mitochondria in mouse embryonic fibroblasts from PINK1 knockouts. However, here we thoroughly quantified effects and showed that the p.G411S mutation has significantly reduced, but not abrogated kinase activity towards the substrate ubiquitin. In addition to a reduced activity in cis (partial loss-of-function), p.G411S also reduced kinase activity of wild-type PINK1 in trans in a heterodimeric complex under endogenous conditions, on overexpression, and in vitro. Indeed p.G411S interacted with wild-type PINK1 and the partial dominant-negative function of p.G411S was dependent on presence of the mutated protein.
We used molecular modelling, dynamic simulations, and analyses of PINK1-ATP-ubiquitin complexes to provide a detailed structural explanation for the particular effect of mutant PINK1 p.G411S. Although our study is limited by the lack of X-ray structures and the lack of membrane insertion during modelling, we present a first time view of human full-length PINK1 at an all-atom resolution. To model PINK1 protein, its dynamics and enzymatic activity, we used a combination of methods that have been applied before to predict mutational effects and activation conformations of human parkin (Caulfield et al., 2014, 2015; Fiesel et al., 2015b), which were in good agreement with the latest structures of an activated form of the E3 ubiquitin ligase (Kumar et al., 2015; Sauve et al., 2015; Wauer et al., 2015). The C-terminus of our PINK1 model is very similar to structures of homologous kinases and existing models of the PINK1 kinase domain (Beilina et al., 2005; Mills et al., 2008; Cardona et al., 2011; Sim et al., 2012; Trempe and Fon, 2013).
For comparison of wild-type PINK1 and the p.G411S mutant, we focused mainly on the prediction of the catalytic kinase domain of PINK1. We modelled autophosphorylation at Ser228 and Ser402 as well as PINK1 dimerization, both of which correlate well with its enzymatic activity (Liu et al., 2009; Okatsu et al., 2012, 2013; Aerts et al., 2015). Although mutant PINK1 p.G411S showed some differences to wild-type in the monomers, effects of the mutation were more pronounced in a dimer. Fluctuations in the ATP binding sites as seen in the monomers were less prominent in all dimer subunits, suggesting that the dimeric configuration might indeed represent the active form of PINK1 kinase. Moreover, our models support the importance of the two PINK1 autophosphorylation sites Ser228 and Ser402 that gate the entry to the active site. Phosphorylation of both facilitated interaction of PINK1 and ubiquitin, stabilized the entire complex and ensured proximity to the bound ATP molecule. However, in contrast to wild-type PINK1, critical kinase-substrate interactions were absent in the p.G411S mutant, resulting in mis-positioning of ubiquitin in the active site, which significantly increased the distance between Ser65 and ATP. Strikingly, in a PINK1 heterodimer, effects of mutant p.G411S propagated into the wild-type subunit that adopted similar overall fold and motions including a misalignment of ubiquitin and ATP in the active site that likely frustrates the enzymatic process. While our models certainly need to be validated by structural data from crystals, our calculations together with functional cell-based data suggest the scenario of a mutant PINK1 p.G411S protein that heterodimerizes and interferes with wild-type PINK1 protein in heterozygotes.
It is conceivable that even simple reduction of PINK1 protein level would negatively affect the protective capacities of mitochondrial quality control during ageing. In three Italian families with the PINK1 p.W437X mutation, several heterozygotes had mild signs or Parkinson’s disease symptoms in a pattern of dominant inheritance (Criscuolo et al., 2006; Ricciardi et al., 2014). In a large German family, 8 of 10 PINK1 p.Q456X heterozygous relatives were found to have mild signs of parkinsonism, and a mean 20% reduction of striatal 18F-fluorodopa uptake in PET, as opposed to 60% in their homozygous relatives with early-onset Parkinson’s disease (Eggers et al., 2010). Although primary cells from p.Q456X heterozygotes were not obviously different from wild-type PINK1 with respect to overall cellular kinase activity over time, these acute conditions do not exclude a chronic haploinsufficiency that could increase the risk for Parkinson’s disease in vivo. Also, it is possible that heterozygous mutations in other recessive genes such as PARK2/parkin and PARK7/DJ1 similarly increase the risk for late-onset Parkinson’s disease over time and that disease penetrance is dependent on levels of mitochondrial stress. Future studies should focus on multi-genic combinations of variants to help provide a more complete picture on the genetic and environmental contributions and thus a better understanding of disease risk and pathogenic mechanisms.
Supplementary Material
Acknowledgements
We thank all patients and control persons who participated in our research studies. In Lund, Sweden, biobanking services were performed at Biobank, Labmedicin Skåne, University and Regional Laboratories Region Skåne, Sweden. We thank Håkan Widner, Per Odin and MultiPark – a strategic research environment at Lund University, Sweden, for providing DNA samples from MultiPark biobank. We thank the Mayo Clinic Center for Regenerative Medicine (CRM), Guojun Bu and the Neuroregeneration Lab for biobanking of patients cells at Mayo Clinic Florida. Results from the analysis of Scandinavian patients and from a systematic review of previous studies examining PINK1 have partially been included in a doctoral thesis: A.P. Heredity in Parkinson’s disease. From rare variants to common genetic risk factors. Lund University, Faculty of Medicine Doctoral Dissertation Series 2011:95.p1-174 (supervisor Christer Nilsson, Lund University, Sweden).
Funding
A.P. is partially supported by the Swedish Parkinson Academy, The Swedish Parkinson Foundation (Parkinsonfonden), governmental funding for clinical research within the Swedish National Health Services (ALF), MultiPark, Lund University, Sweden, and Bundy Academy, Lund, Sweden. W.S. is partially supported by the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS) [R01 #NS085070], the Michael J. Fox Foundation for Parkinson's Research and the Foundation for Mitochondrial Medicine, the Mayo Clinic Center for Regenerative Medicine (CRM), Center for Individualized Medicine (CIM), and Center for Biomedical Discovery (CBD), the Marriott Family Foundation, and a Gerstner Family Career Development Award. F.C.F. is the recipient of a fellowship from the American Parkinson Disease Association (APDA). M.A. acknowledges support from the Uehara Memorial Foundation. Mayo Clinic Florida is a Morris K. Udall Parkinson's Disease Research Center of Excellence [NIH/NINDS P50 #NS072187 to Z.K.W. and O.A.R.]. O.A.R. is supported by NIH/NINDS [R01 #NS078086]. Z.K.W. and O.A.R are supported by the Mayo Clinic Center for Individualized Medicine (CIM) and the Mayo Clinic Center for Regenerative Medicine (CRM). Z.K.W., O.A.R., and W.S. are recipients of Mayo Clinic Florida Neuroscience Focused Research Team Awards. O.H. is supported by European Research Council, Swedish Research Council, Swedish Brain Power and ALF. G.M. and M.F. acknowledge NHMRC project APP1084560. I.J-F. is supported by the CONCYT PhD program. M.S. is supported by MultiPark, Lund University, Sweden. The MultiPark sample collection was funded by MultiPark, Lund University, Sweden. V.L.D is supported by grants from the NIH/NINDS NS38377, MDSCRF 2007-MSCRFI-0420-00, 2009-MSCRFII-0125-00, MDSCRF 2012-MSCRFII-0268-00, MDSCRF 2013-MSCRFII-0105-00. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases, and is supported by grants from the NIH/NINDS NS38377, MDSCRF 2007-MSCRFI-0420-00, 2009-MSCRFII-0125-00, MDSCRF 2012-MSCRFII-0268-00, MDSCRF 2013-MSCRFII-0105-00, and the JPB Foundation. T.M.D. and V.L.D. acknowledge the joint participation by the Adrienne Helis Malvin Medical Research Foundation through their direct engagement in the continuous active conduct of medical research in conjunction with the Johns Hopkins Hospital and the Johns Hopkins University School of Medicine and the Foundation's Parkinson's Disease Program M-1.
Supplementary material
Supplementary material is available at Brain online.
Glossary
Abbreviations
- MDS
molecular dynamics simulation
- MSD
Meso Scale Discovery
- Ub
ubiquitin
References
- Aasly JO, Vilarino-Guell C, Dachsel JC, Webber PJ, West AB, Haugarvoll K, et al. Novel pathogenic LRRK2 p.Asn1437His substitution in familial Parkinson's disease. Mov Disord 2010; 25: 2156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou-Sleiman PM, Muqit MM, McDonald NQ, Yang YX, Gandhi S, Healy DG, et al. A heterozygous effect for PINK1 mutations in Parkinson's disease? Ann Neurol 2006; 60: 414–9. [DOI] [PubMed] [Google Scholar]
- Aerts L, Craessaerts K, De Strooper B, Morais VA. PINK1 kinase catalytic activity is regulated by phosphorylation on serines 228 and 402. J Biol Chem 2015; 290: 2798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Koonin EV. Iterated profile searches with PSI-BLAST–a tool for discovery in protein databases. Trends Biochem Sci 1998; 23: 444–7. [DOI] [PubMed] [Google Scholar]
- Beilina A, Van Der Brug M, Ahmad R, Kesavapany S, Miller DW, Petsko GA, et al. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc Natl Acad Sci USA 2005; 102: 5703–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona F, Sanchez-Mut JV, Dopazo H, Perez-Tur J. Phylogenetic and in silico structural analysis of the Parkinson disease-related kinase PINK1. Hum Mutat 2011; 32: 369–78. [DOI] [PubMed] [Google Scholar]
- Caulfield T, Devkota B. Motion of transfer RNA from the A/T state into the A-site using docking and simulations. Proteins 2012; 80: 2489–500. [DOI] [PubMed] [Google Scholar]
- Caulfield TR, Devkota B, Rollins GC. Examinations of tRNA range of motion using simulations of Cryo-EM microscopy and X-Ray data. J Biophys 2011; 2011: 219515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulfield TR, Fiesel FC, Moussaud-Lamodiere EL, Dourado DF, Flores SC, Springer W. Phosphorylation by PINK1 releases the UBL domain and initializes the conformational opening of the E3 ubiquitin ligase Parkin. PLoS Comput Biol 2014; 10: e1003935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulfield TR, Fiesel FC, Springer W. Activation of the E3 ubiquitin ligase Parkin. Biochem Soc Trans 2015; 43: 269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coxhead J, Kurzawa-Akanbi M, Hussain R, Pyle A, Chinnery P, Hudson G. Somatic mtDNA variation is an important component of Parkinson's disease. Neurobiol Aging 2016; 38: 217, e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo C, Volpe G, De Rosa A, Varrone A, Marongiu R, Mancini P, et al. PINK1 homozygous W437X mutation in a patient with apparent dominant transmission of parkinsonism. Mov Disord 2006; 21: 1265–7. [DOI] [PubMed] [Google Scholar]
- Eggers C, Schmidt A, Hagenah J, Bruggemann N, Klein JC, Tadic V, et al. Progression of subtle motor signs in PINK1 mutation carriers with mild dopaminergic deficit. Neurology 2010; 74: 1798–805. [DOI] [PubMed] [Google Scholar]
- Fiesel FC, Ando M, Hudec R, Hill AR, Castanedes-Casey M, Caulfield TR, et al. (Patho-)physiological relevance of PINK1-dependent ubiquitin phosphorylation. EMBO Rep 2015a; 16: 1114–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiesel FC, Caulfield TR, Moussaud-Lamodiere EL, Ogaki K, Dourado DF, Flores SC, et al. Structural and functional impact of Parkinson disease-associated mutations in the E3 ubiquitin ligase parkin. Hum Mutat 2015b; 36: 774–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiesel FC, Moussaud-Lamodiere EL, Ando M, Springer W. A specific subset of E2 ubiquitin-conjugating enzymes regulate Parkin activation and mitophagy differently. J Cell Sci 2014; 127 (Pt 16): 3488–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiesel FC, Springer W. Disease relevance of phosphorylated ubiquitin (p-S65-ubiquitin). Autophagy 2015; 11: 2125–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 2010a; 12: 119–31. [DOI] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Treis A, Skujat D, Weber SS, Fiesel FC, et al. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy 2010b; 6: 871–8. [DOI] [PubMed] [Google Scholar]
- Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN mitochondrial Ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell 2015; 60: 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft RW, Sander C, Scharf M, Vriend G. The PDBFINDER database: a summary of PDB, DSSP and HSSP information with added value. Comput Appl Biosci 1996a; 12: 525–9. [DOI] [PubMed] [Google Scholar]
- Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature 1996b; 381: 272. [DOI] [PubMed] [Google Scholar]
- Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson's disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry 1992; 55: 181–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPDG Consortium. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet 2011; 377: 641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MP, Friesner RA, Xiang Z, Honig B. On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol 2002; 320: 597–608. [DOI] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 2010; 191: 933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanes LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol 2014; 205: 143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J 2014; 460: 127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilarski LL, Pearson JP, Newsway V, Majounie E, Knipe MD, Misbahuddin A, et al. Systematic review and UK-based study of PARK2 (parkin), PINK1, PARK7 (DJ-1) and LRRK2 in early-onset Parkinson's disease. Mov Disord 2012; 27: 1522–9. [DOI] [PubMed] [Google Scholar]
- Kim J, Fiesel FC, Belmonte KC, Hudec R, Wang WX, Kim C, et al. miR-27a and miR-27b regulate autophagic clearance of damaged mitochondria by targeting PTEN-induced putative kinase 1 (PINK1). Mol Neurodegener. 2016; 11: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RD, Sternberg MJ. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci 1996; 5: 2298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Lohmann-Hedrich K, Rogaeva E, Schlossmacher MG, Lang AE. Deciphering the role of heterozygous mutations in genes associated with parkinsonism. Lancet Neurol 2007; 6: 652–62. [DOI] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol 2012; 2: 120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 2014; 510: 162–6. [DOI] [PubMed] [Google Scholar]
- Krieger E, Dunbrack RL Jr Hooft RW, Krieger B. Assignment of protonation states in proteins and ligands: combining pKa prediction with hydrogen bonding network optimization. Methods Mol Biol 2012; 819: 405–21. [DOI] [PubMed] [Google Scholar]
- Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 2009; 77 (Suppl 9): 114–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Aguirre JD, Condos TE, Martinez-Torres RJ, Chaugule VK, Toth R, et al. Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis. EMBO J 2015; 34: 2506–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015; 524: 309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Vives-Bauza C, Acin-Perez R, Yamamoto A, Tan Y, Li Y, et al. PINK1 defect causes mitochondrial dysfunction, proteasomal deficit and alpha-synuclein aggregation in cell culture models of Parkinson's disease. PLoS One 2009; 4: e4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marongiu R, Ferraris A, Ialongo T, Michiorri S, Soleti F, Ferrari F, et al. PINK1 heterozygous rare variants: prevalence, significance and phenotypic spectrum. Hum Mutat 2008; 29: 565. [DOI] [PubMed] [Google Scholar]
- Mellick GD, Siebert GA, Funayama M, Buchanan DD, Li Y, Imamichi Y, et al. Screening PARK genes for mutations in early-onset Parkinson's disease patients from Queensland, Australia. Parkinsonism Relat Disord 2009; 15: 105–9. [DOI] [PubMed] [Google Scholar]
- Mills RD, Sim CH, Mok SS, Mulhern TD, Culvenor JG, Cheng HC. Biochemical aspects of the neuroprotective mechanism of PTEN-induced kinase-1 (PINK1). J Neurochem 2008; 105: 18–33. [DOI] [PubMed] [Google Scholar]
- Mullin S, Schapira A. The genetics of Parkinson's disease. Br Med Bull 2015; 114: 39–52. [DOI] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 2010; 8: e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K, Koyano F, Kimura M, Kosako H, Saeki Y, Tanaka K, et al. Phosphorylated ubiquitin chain is the genuine Parkin receptor. J Cell Biol 2015; 209: 111–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K, Oka T, Iguchi M, Imamura K, Kosako H, Tani N, et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat Commun 2012; 3: 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K, Uno M, Koyano F, Go E, Kimura M, Oka T, et al. A dimeric PINK1-containing complex on depolarized mitochondria stimulates Parkin recruitment. J Biol Chem 2013; 288: 36372–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A, Sarraf SA, Duda DM, Heo JM, Jedrychowski MP, Sviderskiy VO, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell 2014; 56: 360–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschmann A. Monogenic Parkinson's disease and parkinsonism: clinical phenotypes and frequencies of known mutations. Parkinsonism Relat Disord 2013; 19: 407–15. [DOI] [PubMed] [Google Scholar]
- Qiu J, Elber R. SSALN: an alignment algorithm using structure-dependent substitution matrices and gap penalties learned from structurally aligned protein pairs. Proteins 2006; 62: 881–91. [DOI] [PubMed] [Google Scholar]
- Ricciardi L, Petrucci S, Guidubaldi A, Ialongo T, Serra L, Ferraris A, et al. Phenotypic variability of PINK1 expression: 12 years' clinical follow-up of two Italian families. Mov Disord 2014; 29: 1561–6. [DOI] [PubMed] [Google Scholar]
- Ross OA, Soto-Ortolaza AI, Heckman MG, Aasly JO, Abahuni N, Annesi G, et al. Association of LRRK2 exonic variants with susceptibility to Parkinson's disease: a case-control study. Lancet Neurol 2011; 10: 898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve V, Lilov A, Seirafi M, Vranas M, Rasool S, Kozlov G, et al. A Ubl/ubiquitin switch in the activation of Parkin. EMBO J 2015; 34: 2492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, Lee JR, Grimes DA, Racacho L, Ye D, Yang H, et al. Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson's disease. Hum Mol Genet 2011; 20: 1966–74. [DOI] [PubMed] [Google Scholar]
- Shiba-Fukushima K, Arano T, Matsumoto G, Inoshita T, Yoshida S, Ishihama Y, et al. Phosphorylation of mitochondrial polyubiquitin by PINK1 promotes Parkin mitochondrial tethering. PLoS Genet 2014; 10: e1004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, Sato S, et al. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep 2012; 2: 1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N Engl J Med 2009; 361: 1651–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim CH, Gabriel K, Mills RD, Culvenor JG, Cheng HC. Analysis of the regulatory and catalytic domains of PTEN-induced kinase-1 (PINK1). Hum Mutat 2012; 33: 1408–22. [DOI] [PubMed] [Google Scholar]
- Siuda J, Jasinska-Myga B, Boczarska-Jedynak M, Opala G, Fiesel FC, Moussaud-Lamodiere EL, et al. Early-onset Parkinson's disease due to PINK1 p.Q456X mutation–clinical and functional study. Parkinsonism Relat Disord 2014; 20: 1274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft M, Myhre R, Pielsticker L, White LR, Aasly JO, Farrer MJ. PINK1 mutation heterozygosity and the risk of Parkinson's disease. J Neurol Neurosurg Psychiatry 2007; 78: 82–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Fon EA. Structure and function of Parkin, PINK1, and DJ-1, the three musketeers of neuroprotection. Front Neurol 2013; 4: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CD, Gibb WR. Research diagnostic criteria for Parkinson's disease. Adv Neurol 1990; 53: 245–9. [PubMed] [Google Scholar]
- Wauer T, Simicek M, Schubert A, Komander D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature 2015; 524: 370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wider C, Ross OA, Wszolek ZK. Genetics of Parkinson disease and essential tremor. Curr Opin Neurol 2010; 23: 388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 2013; 152: 82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Youle RJ. PINK1 is degraded through the N-end rule pathway. Autophagy 2013; 9: 1758–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Caulfield T, Xu YF, Gendron TF, Hubbard J, Stetler C, et al. The dual functions of the extreme N-terminus of TDP-43 in regulating its biological activity and inclusion formation. Hum Mol Genet 2013; 22: 3112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Pandit SB, Lee SY, Borreguero J, Chen H, Wroblewska L, et al. Analysis of TASSER-based CASP7 protein structure prediction results. Proteins 2007; 69 (Suppl) 8: 90–7. [DOI] [PubMed] [Google Scholar]
- Zhou H, Pandit SB, Skolnick J. Performance of the Pro-sp3-TASSER server in CASP8. Proteins 2009; 77 (Suppl 9): 123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Skolnick J. Ab initio protein structure prediction using chunk-TASSER. Biophys J 2007; 93: 1510–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Skolnick J. Protein structure prediction by pro-Sp3-TASSER. Biophys J 2009; 96: 2119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Skolnick J. Improving threading algorithms for remote homology modeling by combining fragment and template comparisons. Proteins 2010; 78: 2041–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Skolnick J. Template-based protein structure modeling using TASSER(VMT). Proteins 2012; 80: 352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.