A number of different drugs block sodium channels, but their mechanism of block is unclear. Tikhonov and Zhorov combine homology modeling with ligand docking and propose a pharmacophore for sodium channel blockers involving cationic and aromatic moieties.
Abstract
Local anesthetics, antiarrhythmics, and anticonvulsants include both charged and electroneutral compounds that block voltage-gated sodium channels. Prior studies have revealed a common drug-binding region within the pore, but details about the binding sites and mechanism of block remain unclear. Here, we use the x-ray structure of a prokaryotic sodium channel, NavMs, to model a eukaryotic channel and dock representative ligands. These include lidocaine, QX-314, cocaine, quinidine, lamotrigine, carbamazepine (CMZ), phenytoin, lacosamide, sipatrigine, and bisphenol A. Preliminary calculations demonstrated that a sodium ion near the selectivity filter attracts electroneutral CMZ but repels cationic lidocaine. Therefore, we further docked electroneutral and cationic drugs with and without a sodium ion, respectively. In our models, all the drugs interact with a phenylalanine in helix IVS6. Electroneutral drugs trap a sodium ion in the proximity of the selectivity filter, and this same site attracts the charged group of cationic ligands. At this position, even small drugs can block the permeation pathway by an electrostatic or steric mechanism. Our study proposes a common pharmacophore for these diverse drugs. It includes a cationic moiety and an aromatic moiety, which are usually linked by four bonds.
Introduction
Sodium channels are targets for a great variety of modulators, which belong to different chemical classes, bind to distinct sites, and act by different mechanisms (Hille, 2001; Catterall and Swanson, 2015). Many medicinally used drugs are small-molecule blockers of the ion-conducting pore. These include local anesthetics (LAs), anticonvulsants, and antiarrhythmics, which have common modes of action on sodium channels (Catterall, 1987, 2012) and a common binding region in the channel pore (Ragsdale et al., 1996; Kuo, 1998). The pore of the sodium channels is also blocked by antidepressants (Pancrazio et al., 1998; Dick et al., 2007), neuroprotective drugs (Hebert et al., 1994), and other medications. Numerous studies in academia and industry are aimed to understand binding sites, mechanisms of action, and structure–function relations of sodium channel drugs.
Sodium channel blockers have highly diverse chemical structures. Most LAs are flexible molecules that contain a protonatable amino group at one end, an aromatic moiety at the opposite end, and polar groups in the middle. Classical examples are lidocaine, bupivacaine, and tetracaine. Typical anticonvulsants such as phenytoin, lamotrigine, and carbamazepine (CMZ) are electroneutral molecules. Unlike LAs, they contain nonionizable polar groups at one end of the molecule and an aromatic moiety at the other end. However, some anticonvulsants, such as sipatrigine, contain an ionizable amino group (Liu et al., 2003). Sodium channels are also blocked by long flexible molecules, such as ranolazine and haloperidol. Structurally, such molecules resemble two LAs bridged by a moiety with an ionizable amino group. Antidepressants, which block sodium channels, have an ionizable amino group and a polycyclic moiety that contains aromatic and saturated rings. Another group of blockers contains bulky rigid moieties. Examples are cocaine, memantine, and quinidine.
Many sodium channel blockers are well studied electrophysiologically. The mechanism of block is rather complex and includes three typical phenomena observed in different experimental protocols. In case of channel activation by infrequent membrane depolarizations, so-called closed (resting) channel block is observed. Increase of the stimulation frequency increases the blocking effect. This phenomenon is called the frequency-dependent block. The maximal blocking effect is observed after long preconditioning membrane depolarization, which induces steady-state inactivation of the channels.
A generally accepted rationale for these phenomena is described by the so-called modulated receptor hypothesis (Hille, 1977) according to which blockers have different affinities to different functional states of the channel. The affinity to the closed state, which prevails at infrequent stimulations, is the lowest one. The affinity to the open state, which prevails at frequent stimulations, is bigger, and the inactivated states of the channel have the maximal affinity. This general phenomenon of the use dependence may significantly vary for different compounds. For example, benzocaine does not demonstrate frequency-dependent block likely because of very fast kinetics (Quan et al., 1996). In contrast, lacosamide demonstrates minor closed-channel block likely because of very slow kinetics (Wang and Wang, 2014).
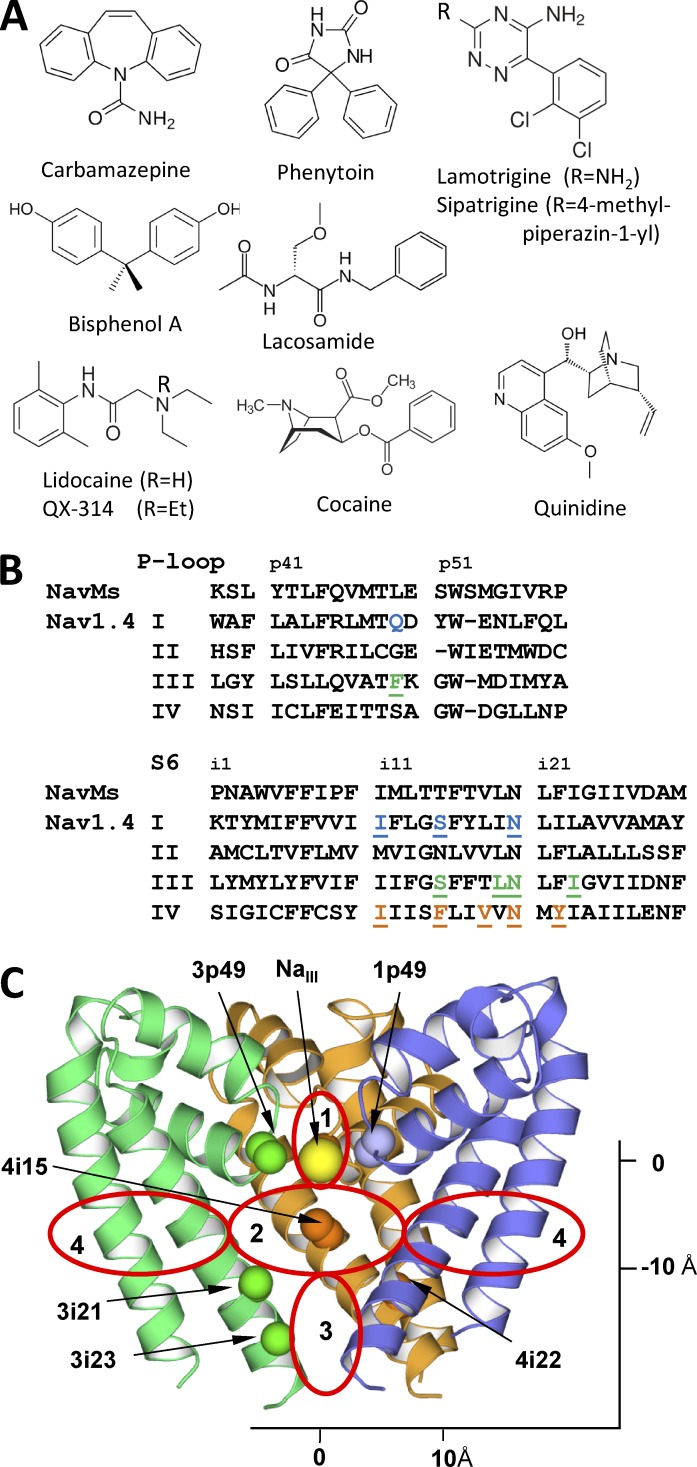
Mutational studies revealed that these structurally diverse drugs bind in the inner-pore region, which is lined by the S6 transmembrane helices and P-loop turns. Mutations in many positions affect action of the pore blockers. To our knowledge, mutations of a phenylalanine residue in the IVS6 helix affect action of all tested ligands. In the sequence alignment this is position 4i15, and the phenylalanine is labeled F4i15 (Fig. 1). Mutations of F4i15 affect the resting, frequency-dependent, and inactivated channel block. Systematic mutations in this position strongly suggest direct and specific involvement of F4i15 in binding of various ligands (Ragsdale et al., 1994; Yarov-Yarovoy et al., 2002; Ahern et al., 2008). Mutations of other residues in IS6, IIIS6, and IVS6 also affect binding of various pore blockers (Yarov-Yarovoy et al., 2001; Mike and Lukacs, 2010). However, a consensus conclusion on exact roles of these residues is hardly possible. For example, mutations of Y4i22 strongly affect the use-dependent but not the resting block (Wright et al., 1998; Li et al., 1999). Mutations in the selectivity filter (the DEKA locus) also affect the action, strongly suggesting electrostatic interactions with the selectivity filter (Sunami et al., 1997). More ligand-sensing residues are found at the P-loops (Tsang et al., 2005; Yamagishi et al., 2009), but the data on roles of these residues are fragmental. Thus, mutational studies unambiguously show the ligand-binding region and suggest candidate residues that contribute to receptors of various ligands, but they do not provide a complete picture of the channel interactions with the ligands.
Figure 1.
Ligand-sensing residues in the inner-pore region of Nav1.x channels. (A) Ligands considered in this study. (B) Sequences alignment of NavMs and Nav1.4. Key positions where mutations affect action of inner pore blockers (Mike and Lukacs, 2010) are highlighted. (C) The NavMs structure. Shown are Cα-Cβ bonds in key ligand-sensing residues and NaIII. Repeats I, III, and IV are blue, green, and orange, respectively. Repeat II is not depicted for clarity. Red ovals mark approximate borders of the selectivity filter region (1), the inner pore region (2), the activation gate region (3), and two of the four repeat interfaces (4).
Without experimental atomic-scale structures of eukaryotic sodium channels, attempts to rationalize a large body of experimental data in structural terms necessarily involve homology modeling using structures of similar channels (Lipkind and Fozzard, 2005, 2010; Scheib et al., 2006; Tikhonov and Zhorov, 2007; Bruhova et al., 2008; Browne et al., 2009; O’Reilly et al., 2012; Yang et al., 2012). The major challenge of the modeling is to provide structural rationale for the complex mechanism of action of structurally distinct ligands.
Generally, the modeling results are consistent with the schemes of ligand binding developed to rationalize mutational experiments (Ragsdale et al., 1994; Yarov-Yarovoy et al., 2002). In this consensus binding mode, the ligand’s charged moiety directs to the selectivity filter, and an aromatic moiety interacts with the hydrophobic environment in the lower levels of the inner pore. Attempts to apply this scheme to greatly diverse ligands, which according to mutational analysis bind in the same region and demonstrate generally similar mechanism of block, revealed serious contradictions. For example, the electrostatic mechanism of block was proposed for LAs because some of these molecules are small, bind tightly to IVS6, and do not interact significantly with IIS6 and therefore cannot sterically occlude the pore (Lipkind and Fozzard, 2005; Tikhonov et al., 2006). Because this mechanism is obviously not applicable to neutral drugs, a model was proposed in which phenytoin, CMZ, and lamotrigine sterically occlude the pore (Lipkind and Fozzard, 2010). A small neutral benzocaine and its analogues, which do not fit any of these models, are proposed to block the channel by stabilizing the closed state (Wang and Wang, 1994; Wang et al., 1997). Such a diversity of mechanisms for the ligands that target the same region and display similar electrophysiological characteristics seems excessive.
Another challenge is to explain the state-dependent action of blockers. Lipkind and Fozzard noted that lidocaine-like molecules are too big to fit into the KcsA-based closed-channel model in the same way as they fit into the MthK-based open-channel model (Lipkind and Fozzard, 2005). Later, a model was elaborated in which LAs extend along the pore axis in the open channel, as proposed before (Tikhonov and Zhorov, 2007), but adopt a horizontal orientation in the closed channel with the aromatic moiety protruding into the III/IV repeat interface (Bruhova et al., 2008). The latter is proposed to serve as a “sidewalk” for some blockers between the membrane and the channel pore (Tikhonov et al., 2006), the long-predicted hydrophobic access pathway (Hille, 1977), which is now seen as wide fenestrations in the x-ray structures of bacterial sodium channels (Payandeh et al., 2011). The only currently available structure of a complex of LA-like compound bound to a bacterial sodium channel (Bagnéris et al., 2014) indicates that the ligand binds in a horizontal orientation and protrudes from the sidewalk to the pore.
Thus, there is still no consensus view on the binding mode and molecular mechanism of action of structurally different small-molecule blockers of the inner pore of sodium channels. Impressive progress in structural studies of bacterial sodium channels provides more realistic templates for homology modeling and thus allows us to readdress the aforementioned problems. The x-ray structures of a bacterial sodium channel NavRh (Zhang et al., 2012) and an engineered bacterial calcium channel CavAb (Tang et al., 2014) contain calcium ions at the selectivity-filter region. In the NavMs bacterial sodium channel, a completely hydrated sodium ion NaIII is seen at the level of four backbone carbonyls of Tp48 residues, which are located at the turn region between the P1 helix and the selectivity filter (Naylor et al., 2016). This hydrated ion does not form any bonds with the channel protein and seems accessible for interactions with electronegative groups of the inner-pore–targeting ligands. Interactions with permeant ions have been predicted to stabilize binding of electroneutral ligands in sodium, calcium, and potassium channels (Zhorov and Tikhonov, 2013).
In this study, we have built a NavMs-based model of eukaryotic sodium channel Nav1.4, selected five cationic and five electroneutral compounds (Fig. 1), which represent a great variety of ligands targeting the inner pore of sodium channels, and used Monte Carlo (MC) energy minimizations to dock the ligands into the channel. We found that a sodium ion, which is attracted to an electroneutral ligand, or the ammonium group of a cationic ligand occurs at the channel narrowing close to the NaIII site, whereas the hydrophobic parts of the ligands typically extend into the inner pore. In such binding modes, even relatively small molecules would block the permeation.
Materials and methods
Our methodology of homology modeling and ligand docking is described in many studies (e.g., Garden and Zhorov, 2010; Tikhonov and Zhorov, 2012). In brief, we use the ZMM program that minimizes energy in the space of internal (generalized) coordinates, the MC energy minimization method (Li and Scheraga, 1987), and the AMBER force field (Weiner et al., 1984, 1986). Electrostatic interactions were calculated with the distance- and environment-dependent dielectric function (Garden and Zhorov, 2010). Atomic charges at ligands were calculated by MOPAC (Dewar et al., 1985). No distance cutoff was used to calculate electrostatic interactions involving ionized groups. For other interactions, we used the distance cutoff of 9 Å and a shifting function (Brooks et al., 1985).
Because we did not consider a possibility of the protein refolding, the MC minimization (MCM) sampling protocol randomized the channel side chains (but not backbone) torsions, as well as generalized coordinates that govern position, orientation, and conformation of the ligand. However, during energy minimizations, all generalized coordinates (both backbone and side-chain torsions of the channel, torsion angles of the ligand, and its bond angles) were treated as flexible. Because of limited precision of the homology modeling approach, we have used “pin” constraints to ensure similarity of the backbone conformation in the model and the template. A pin constraint is a flat-bottom parabolic energy function that imposes an energy penalty if an α carbon in the model deviates from the template position by more than 1 Å. For all constraints, the energy penalty was calculated using the force constant of 10 kcal/mol1/Å2.
We used several protocols for ligand docking. For the initial broad search of lidocaine and CMZ binding modes in the presence or absence NaIII, we seeded 10,000 random positions and orientations with the ligand mass center in the sphere of radius 8 Å. The sphere center was in the inner pore, at the cross section of the pore axis and level i15, which approximately corresponds to potassium binding site 5 in potassium channels. Each stating point was shortly MC minimized (100 energy minimizations). At the second stage, 100 energetically best complexes were further optimized in long MCM trajectories, each of which was terminated when 1,000 consecutive minimizations did not improve the energy. Intensive searches for low-energy binding modes were performed by submitting 100 long MCM trajectories from the starting points, which were generated from the best energy complexes obtained at the previous step. In the beginning of each trajectory, the ligand was randomly turned from the previous orientation by 0–30° and randomly shifted by 0–2 Å.
For each ligand–channel complex, structures with energy <7 kcal/mol from the apparent global minimum were accumulated and clustered in a stack whose size was limited to 200 records. During MC-energy minimizations, the stack was updated as described previously (Garden and Zhorov, 2010). Each record in the stack represented a family of complexes with similar positions, orientations, and conformations of the ligand and similar conformations of the channel side chains. To compare predicted ligand–channel interactions with data of mutational studies, we analyzed ligand–channel contacts. A ligand and a channel residue were considered to form a contact if their interaction energy was negative and they had at least one pair of heavy atoms separated by <5 Å.
Limitations of our modeling approach should be mentioned. Ligand docking in homology models is not expected to be as precise as docking in x-ray structures. Furthermore, repeat domains in heterotetrameric eukaryotic channels have asymmetric 3D structures as seen in the cryo-electron microscopy structure of the Cav1.1 calcium channel (Wu et al., 2016), and such asymmetry is hardly predictable when symmetric x-ray structures of homotetrameric prokaryotic channels are used as templates. Another source of uncertainly is approximate character of calculations that ignore entropy, do not use explicit water molecules, and use rather simple treatment of electrostatic interactions. Because of these limitations, we did not attempt to predict affinity of ligands, stability of their complexes with sodium ions, and considered ensembles of low-energy structures rather than single conformations that correspond to apparent global minima.
Online supplemental material
Table S1, available as a PDF, shows aligned sequences of specific segments (L45, S5, P-loop, and S6) in several potassium, sodium, and calcium channels. Dataset S1, included as a zipped file, contains coordinates of the sodium channel complexes with ligands available as the following files: Bisphenol_A.pdb, Carbamazepine.pdb, Cocaine.pdb, Lacosamide.pdb, Lamotrigine.pdb, Lidocaine.pdb, Phenotoyn.pdb, Quinidine.pdb, and Sipatrigine.pdb.
Results
Building the homology model
Previously, LAs and other ligands were docked in homology models, which were built using as templates the x-ray structures of KcsA (Lipkind and Fozzard, 2000; Bruhova et al., 2008), MthK (Lipkind and Fozzard, 2005), KvAP (Tikhonov et al., 2006), and NavAb (Tikhonov and Zhorov, 2012). Sequences of eukaryotic sodium channels are obviously much more similar to prokaryotic sodium channels than to potassium channels, and their alignment is much less ambiguous (Table S1). The sequences of bacterial sodium channels with available structures are rather similar, and their comparison does not allow us to select a preferable template. Among these structures, the NavMs bacterial channel (Naylor et al., 2016) presents a template that has three advantages for modeling eukaryotic channels with the inner-pore–bound ligands. First, this sodium channel is crystallized in the apparently open state. Second, this structure shows sodium ions in the selectivity-filter region. Third, the NavMs channel is also cocrystallized with a LA-like ligand (Bagnéris et al., 2014). Thus, using the NavMs template seems promising to advance our understanding of ligand interactions with eukaryotic sodium channels.
We have built the Nav1.4 channel model using the NavMs channel (PDB accession number 5BZB) as the template. The selectivity filter, which is different in bacterial and eukaryotic sodium channels channel, was modeled as in our previous studies (Tikhonov and Zhorov, 2012). A sodium ion NaIII was added to the model near the entrance to the inner pore as seen in the NavMs structure (Naylor et al., 2016). Position of NaIII is also close to site 4 for potassium ions in potassium channels. Other ions, which are seen in the x-ray structure (Naylor et al., 2016), were not added because according to our recent calculations, their electrostatic interactions with the inner-pore ligands are weak (Korkosh et al., 2016). Furthermore, the number of ions in the DEKA ring of eukaryotic channels may differ from that in the EEEE ring of NavMs. The sequence alignment, ligand-sensing residues found by mutational analysis and their positions in the NavMs structure are given in Fig. 1 (B and C). The model was optimized by a two-stage MCM protocol. At the first stage, the backbone torsions were kept rigid. At the second stage, both side-chain and backbone angles were varied and similarity between the template and the model was maintained by “pin” constraints imposed to α carbons (see Materials and methods). According to the MolProbity server (Chen et al., 2010), the quality (overall score) of the model (1.06) is similar to that of the NavMs template (1.27). The only region, where the backbone in our model differs from that of the template is short loops between P1 and P2 helices, in which insertions/deletions were introduced to accommodate experimental data on binding of tetrodotoxin (Tikhonov and Zhorov, 2012).
Hands-free docking of lidocaine and CMZ in the models with and without NaIII
For the initial hands-free docking we selected a protonated LA (lidocaine) and a neutral anticonvulsant (CMZ). Theoretical proposals and experimental data suggest significant interactions between blocking molecules and current-carrying ions (Zhorov and Tikhonov, 2013). Therefore, we docked each ligand in two channel models: with and without a sodium ion at site NaIII (Fig. 2). In agreement with previous studies (Sunami et al., 1997; Tikhonov and Zhorov, 2007), we observed repulsion between the charged lidocaine and the sodium ion NaIII. Positions of the lidocaine amino group in different binding modes were scattered over a wide region in the inner pore (Fig. 2 A). Contrary, in the absence of NaIII, the binding modes were found with the amino group clustered at the cation-attractive region in the pore, including site NaIII (Fig. 2 B).
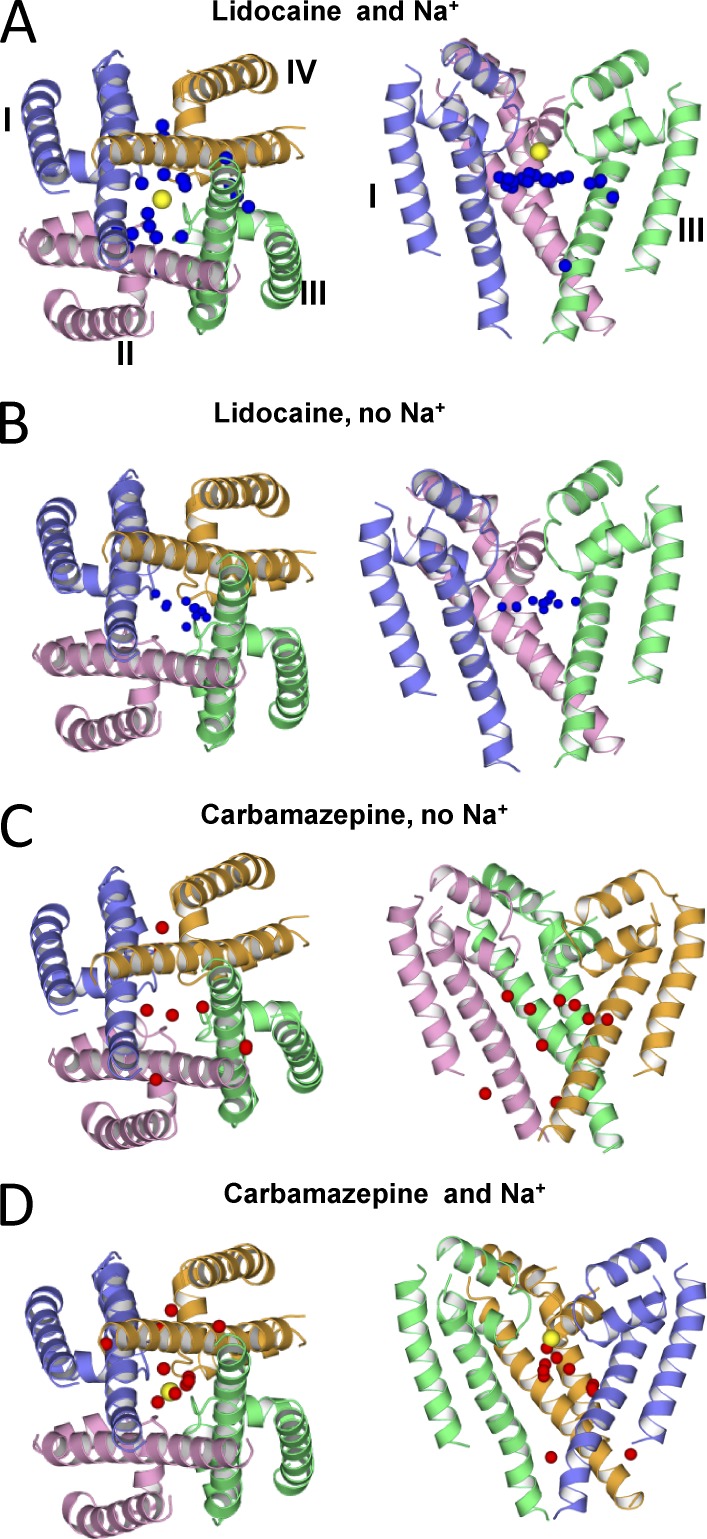
Figure 2.
Hands-free docking of lidocaine and CMZ in the Nav1.4 model with and without NaIII (yellow spheres). The oxygen atom of CMZ and the protonated nitrogen of lidocaine are shown as red and blue spheres, respectively. Distribution of ligands, as indicated by their key atoms, strongly depends on the presence of NaIII. In the presence of NaIII, low-energy positions of lidocaine and CMZ are scattered and clustered, respectively, and in the absence or presence of NaIII, the opposite distribution is observed. These results suggest that the NaIII site is a hotspot for ligands.
The hands-free docking of CMZ yielded opposite results. In the model without NaIII, CMZ binding modes were scattered in the pore and interfaces between helices (Fig. 2 C). This result is reminiscent to molecular dynamics (MD) simulations of the NavAb sodium channel with benzocaine and phenytoin, where many possible positions of the ligands were found (Boiteux et al., 2014). In the model with NaIII, low-energy binding modes of CMZ are clustered in the inner pore with the CMZ carbonyl oxygen often bound to NaIII (Fig. 2 D). These calculations suggest that the channel with NaIII favors binding of neutral ligands, whereas the channel without NaIII favors binding of charged ligands. Thus, the NaIII site appears to be the hotspot for binding of both charged and neutral ligands.
Docking lidocaine in the channel model without NaIII
We systematically explored possible binding modes of lidocaine in the model without NaIII (see Materials and methods). These computations yielded an ensemble of 63 low-energy ligand–channel complexes. Let us define axis Z, which coincides with the pore axis, originates at NIII and directs extracellularly. We characterized each binding mode of lidocaine by the Z-coordinate of the ammonium nitrogen, ZN+, its deviation from the pore axis, DN+, and respective coordinates of the aromatic carbon Cp in the para position to the amide nitrogen, ZCp and DCp. Characteristics of the ensemble are given in Fig. 3. Several pore-domain regions accessible for ligands and ions can be defined (Fig. 1 C). In the selectivity filter region (1), −2 Å < Z <5 Å and deviations from the pore axis are <2 Å. The inner-pore region (2) occupies the wide space between the P-loop turns and positions i21 with −10 Å < Z < −2 Å and deviations from the pore axis up to 8 Å. We define the activation gate region (3) as that where S6 helices differ significantly between the closed sodium channel NavAb and apparently open channel NavMs. This region is located below the pore region and has the coordinates Z < −10 Å, and deviations from the pore axis are <5 Å. The four interface regions (4) correspond to four fenestrations between the inner helices. Their main feature is large (>8 Å) deviations from the pore axis.
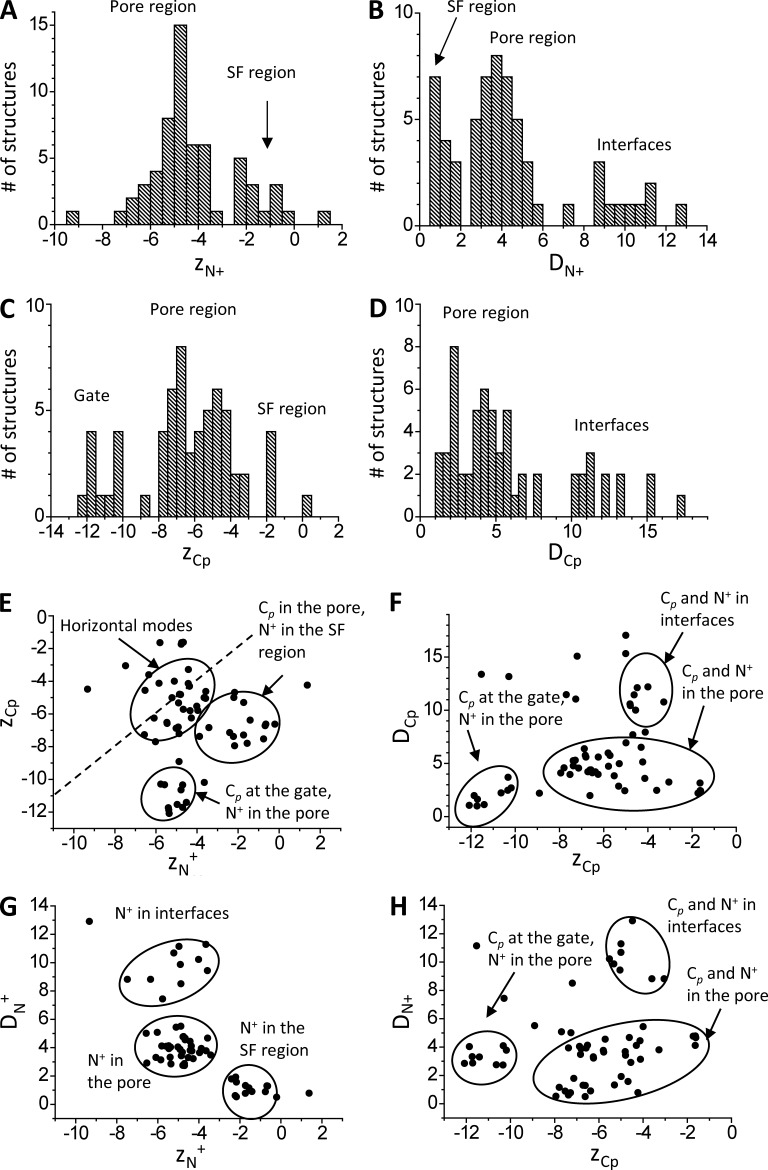
Figure 3.
Statistics of lidocaine-binding modes in the channel model without NaIII. Each binding mode is characterized by the Z-coordinates of the ammonium nitrogen, ZN+, and the carbon atom in para-position of the aromatic ring, ZCp, and deviations of these atoms from the Z-axis, DN+ and DCp. (A–D) Distributions of the characteristics of the ensemble of low-energy binding modes. (E–H) Correlation fields between the characteristics. Dots in E, which are close to the dashed line (ZCp = ZN), correspond to horizontal binding modes of lidocaine. All coordinates and distances are given if angstroms.
Distribution of ZN+ has peaks at level i15 (the inner-pore region) and immediately below site NaIII (Fig. 3 A). The ammonium group can approach the pore axis, or S6s, and also can be found in the interfaces or even at the pore domain outer surface (Fig. 3 B). Atom Cp occurs rarely at the NaIII level, most frequently in the pore, and also at the activation gate (Fig. 3 C), where N+ is practically not seen (Fig. 3 A). The aromatic ring of lidocaine can bind in the pore or in repeat interfaces (Fig. 3 D).
We further plotted four correlation fields and circled clusters with characteristic locations of N+ or/and Cp. In Fig. 3 E, points close to or far from the dashed line (ZCp = ZN+) represent, respectively, approximately horizontal or vertical lidocaine orientations. Among vertical orientations, the cluster dominates where N+ and Cp are, respectively, at site NaIII and in the pore. In the second cluster of vertical orientations, N+ and Cp are, respectively, in the pore and at the activation gate. In Fig. 3 F, the leftmost cluster represents binding modes with Cp at the activation gate, close to the pore axis because of rather narrow gate aperture. In the largest cluster, Cp is in the inner pore. In the topmost cluster, Cp is in interfaces and may significantly deviate from the pore axis. The lower, middle, and upper clusters in Fig. 3 G represent, respectively, structures where N+ is close to site NaIII, approaches S6s from inside the pore, and occurs in interfaces. In Fig. 3 H, binding modes are clustered as in Fig. 3 F. The leftmost cluster represents lidocaine-binding modes where location of Cp at the gate dictates vertical orientation of the ligand with small deviations of N+ from the pore axis. In the largest cluster, ZCp varies significantly, whereas N+ is close to the pore axis because of restriction imposed by S6s. In the topmost cluster, both N+ and Cp are in interfaces. The interface and inner-pore clusters are separated because the ammonium group avoids the narrowest hydrophobic part of the fenestrations; such separation is not seen in the ZCp/DCp correlation field (Fig. 3 F).
This analysis revealed three distinct binding modes with similar characteristics within each one. Two of these binding models (Fig. 4, B and D) correspond to previous results where lidocaine is seen in horizontal and vertical binding orientations (Tikhonov and Zhorov, 2007; Bruhova et al., 2008). In one of the vertical binding modes (Fig. 4 D), the ammonium group occurred at the cation-attractive region at the focus of P1 helices. This is a putative position NaIV, which is analogous to site 5 where a potassium ion is seen in the KcsA x-ray structure (Zhou et al., 2001). The ammonium group also approached F4i15 to form cation-pi contacts. The aromatic group of lidocaine occurred at the activation-gate level and contacted Y4i22 and I3i23, the experimentally known LA-sensing residues (Yarov-Yarovoy et al., 2002). In the horizontal binding mode, the ammonium group is approximately at the same level, but the aromatic moiety extends to repeat interfaces (Fig. 4 B). Different positions of lidocaine in the horizontal mode likely reflect its hydrophobic access route through the sidewalk between repeats III and IV.
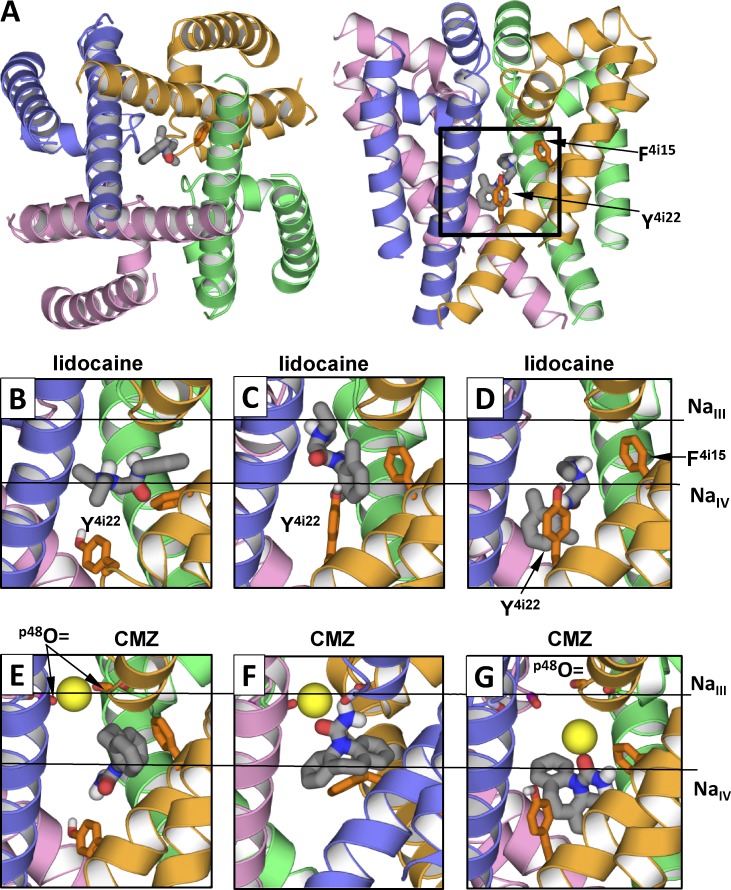
Figure 4.
Examples of low-energy binding modes of lidocaine and CMZ. Side chains of F4i15 and Y4i22 are shown as sticks. Horizontal lines in B–G show levels of site NaIII and putative site NaIV. (A) Location of ligand-binding region. (B–G) Representative structures from the ensembles of low-energy binding modes. (B) Horizontal binding mode of lidocaine. The aromatic group protrudes into the III/IV interface. (C and D) Vertical binding modes with the ammonium group at the levels of NaIII site and NaIV site, respectively. (E) CMZ without tight contacts with NaIII. (F and G) CMZ bound to NaIII. The sodium ion is bound at its innate site seen in the NavMs structure (F) or shifted toward putative site NaIV (G). Note that interactions of lidocaine and CMZ with F4i15 and Y4i22 depend on the ligand-binding modes.
A novel finding is the diversity of lidocaine vertical binding modes, which was not revealed in the sodium channel models based on potassium channel templates. Besides the aforementioned binding mode (Fig. 4 D), we found a new mode in which the cationic group approached the selectivity filter (Fig. 4 C) and occurred at the level where NaIII is seen in the NavMs structure (ZN+ is close to zero). The ammonium group formed favorable contacts with the backbone carbonyls in positions p48 and with side chains in position p49. In this mode, the aromatic moiety of lidocaine contacted F4i15 and Y4i22. The aromatic rings of the ligand and F4i15 formed face-to-face or edge-to-face contacts. Such a binding mode is not proposed in previous models, which are based on potassium channel x-ray structures, because the ligand ammonium group would not fit in the narrow level corresponding to site 4 in potassium channels.
Docking CMZ in the channel model with NaIII
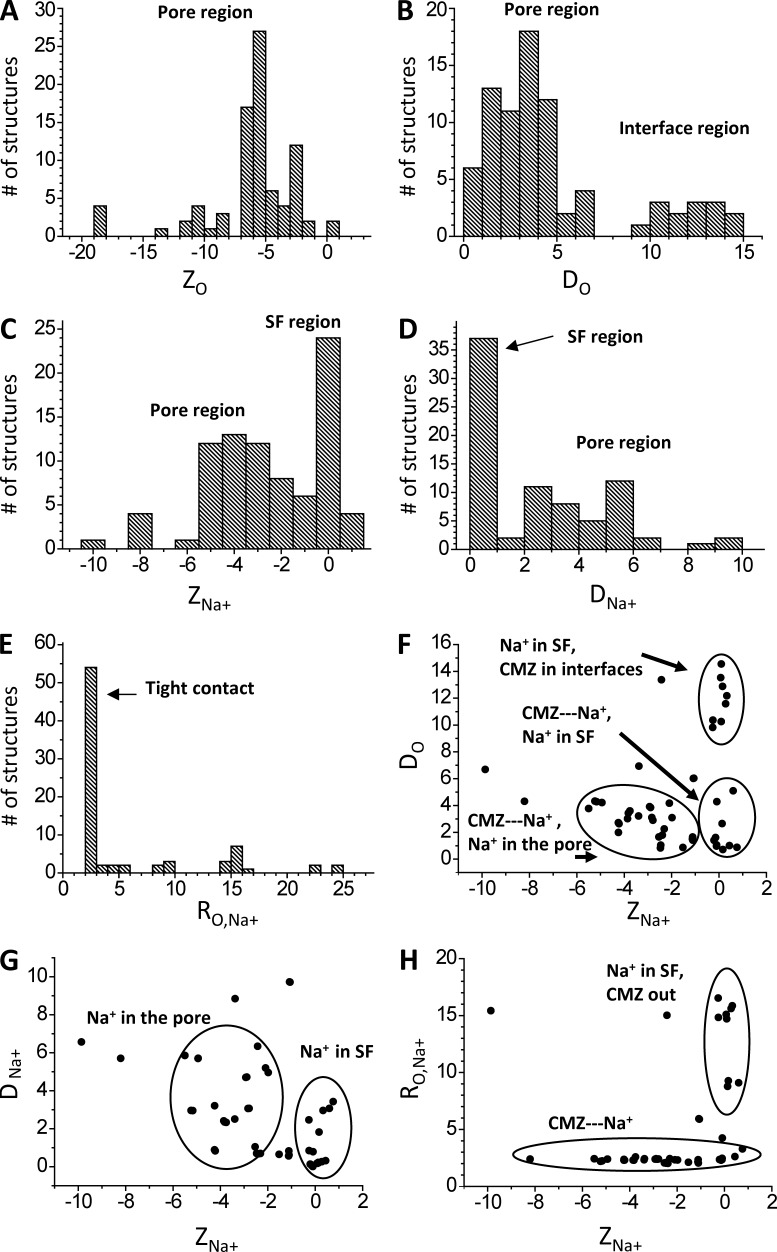
Intensive docking of CMZ in the channel with a sodium ion predicted the ensemble of 80 diverse low-energy structures. In these computations, the sodium ion was initially placed at site NaIII, but its position was randomized. We characterized mutual disposition of CMZ and Na+ by the Z-coordinates of the CMZ carbonyl oxygen (ZO) and Na+ (ZNa), deviations of these atoms from the pore axis (DO and DNa), and the distance between the CMZ oxygen and Na+, RO,Na+. The CMZ oxygen is seen in the inner pore and immediately below site NaIII (Fig. 5 A). It occurs most frequently in the pore but is also found in interfaces (Fig. 5 B). Na+ occurs frequently at site NaIII and is also found inside the pore, at the level corresponding to putative site NaIV or site 5 in potassium channels (Fig. 5 C). The sodium ion most frequently approached the pore axis but can also deviate from it (Fig. 5 D). The RO,Na+ distance distribution (Fig. 5 E) shows that in the vast majority of the structures, Na+ is coordinated by the CMZ oxygen. In the correlation field ZNa/DO (Fig. 5 F), the rightmost clusters represent structures with Na+ at site NaIII, either bound to or unbound from CMZ. The left cluster combines structures where the CMZ-bound Na+ is inside the pore. Correlation field ZNa/DNa (Fig. 5 G) shows groups with Na+ at site NaIII or inside the pore. Correlation field ZNa/RO,Na+ (Fig. 5 H) has two clusters. In the largest one, Na+ and CMZ are coordinated, but Na+ is found either at site NaIII or below it. In the smaller cluster, Na+ at site NaIII is not bound to the CMZ oxygen. In the modes where CMZ did not directly interact with NaIII, its aromatic groups interacted with different pore-facing S6 residues, including F4i15 (Fig. 4 E). When the CMZ-bound Na+ was at site NaIII, the CMZ tricyclic moiety contacted F4i15 (Fig. 4 F) When the CMZ-bound Na+ was between levels NaIII and NaIV, the CMZ amino group contacted F4i15, whereas its tricyclic moiety reached the activation gate level and contacted residues at positions i22 and i23 (Fig. 4 G).
Figure 5.
Statistics of CMZ and Na+ binding in the channel model. Each binding mode is characterized by the Z-coordinates of carbonyl oxygen, ZO, and sodium ion, ZNa, as well as deviations of these atoms from the Z-axis, DO and DNa. (A–E) Distributions of the characteristics of the ensemble of low-energy binding modes. (F–H) Correlation fields between the characteristics. All coordinates and distances are given in angstroms.
Similarity of lidocaine and sodium-bound CMZ binding modes
Comparison of low-energy binding modes of lidocaine and sodium-bound CMZ shows their intriguing similarity. First, the amino group of lidocaine and CMZ-bound sodium ion most frequently occurred in very similar positions, between the levels of NaIII and NaIV. Second, the aromatic moieties of both ligands interacted with the pore-facing S6 residues in similar ways. This similarity is caused by the fact that mutual disposition of aromatic and cationic moieties in lidocaine matches that in sodium-bound CMZ.
This finding allowed us to formulate the following working hypothesis. Both charged and neutral ligands bind to the channel in similar ways. The sodium ion bound to a neutral ligand occurs in the same position as the cationic group in a charged ligand and plays an analogous role by electrostatically preventing permeation. In later sections, we elaborate this hypothesis by considering representative examples of charged and neutral ligands that, according to experimental data, target the inner pore of sodium channels. We docked these ligands, obtained ensembles of low-energy structures, and subdivided the ensembles into binding modes with similar characteristics of ligand–channel interactions. Below, we describe these binding modes.
Charged ligands
We did not consider multiple LAs that are structurally similar to lidocaine because we did not expect that their docking would produce principally new results. We have chosen QX-314, which blocks the inner pore (Qu et al., 1995), but it was unclear if its triethylammonium group would fit the rather narrow pore at the level of the NaIII site. A similar motivation underlined our choice of even bulkier cocaine and quinidine, for which mutational data suggest binding in the inner pore (Ragsdale et al., 1996; O’Leary and Chahine, 2002). We have also chosen sipatrigine (Liu et al., 2003), an unusually long and rigid derivative of the well-studied anticonvulsant lamotrigine.
QX-314
In our model, QX-314 bound to the same site and in the same binding mode as lidocaine did. The triethylammonium group fits the NaIII site (Fig. 6 A) without forming repulsive contacts. The positive charge of QX-314, which is distributed over alkyl groups, is electrostatically attracted to the backbone carbonyls in position p48. Besides well-known LA-sensing residues in helices S6, our model predicted that the bulky ammonium group can also contact sidechains in positions p49. Mutations Q1p49C and F3p49C have been demonstrated to decrease action of QX-314 (Yamagishi et al., 2009). The authors of this study suggested an allosteric mechanism but did not rule out a possibility of direct interaction of residues p49 with QX-314. Our model is consistent with the latter possibility. Furthermore, F3p49 was predicted to interact with the pore-bound batrachotoxin (Tikhonov and Zhorov, 2005), and intensive mutational study confirmed this prediction (Wang et al., 2006).
Figure 6.
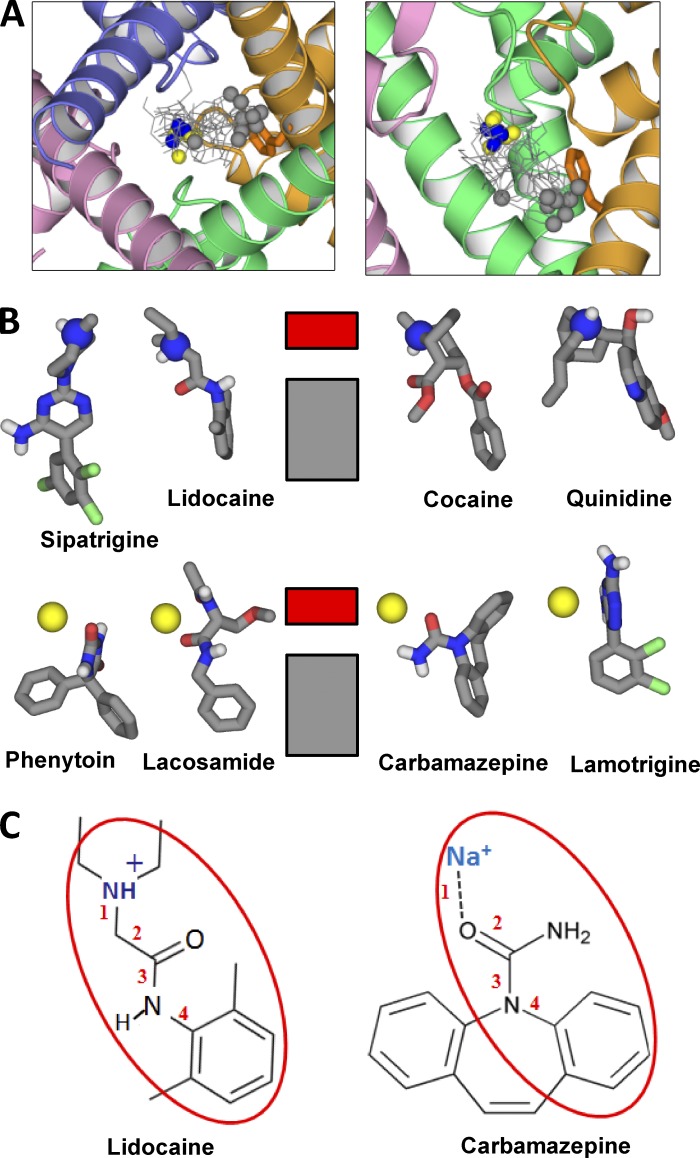
Cationic and sodium-bound electroneutral ligands in the channel. (A) The triethylammonium group of QX-314 is close to the NaIII site, the aromatic ring binds between F4i15 and Y4i22, and the ligand also interacts with Q1p49 and F3p49 in agreement with mutational data (Yamagishi et al., 2009). (B) The bulky moiety of quinidine fits the hotspot at the NaIII site, whereas the hydroxyl group donates an H bond to 3p48O=C. (C and D) Two binding modes of cocaine. In one binding mode (C), the cocaine amino group binds at the NaIII site. In another binding mode (D), the amino group binds close to the putative NaIV site and forms cation-π contacts with F4i15, whereas the opposite end of cocaine reaches I3i23. (E and F) The long sipatrigine molecule can bind in the horizontal (E) and vertical (F) modes, forming many contacts with the channel. In the vertical mode, sipatrigine extends from the NaIII site to the hydrophobic region at levels i22-i23. (G and H) Side and extracellular views of lamotrigine. In the side view (G), repeat II is removed for clarity. In the extracellular view (H), lamotrigine and backbone carbonyls p48O=C are space filled. Lamotrigine binds to NaIII, which is coordinated by 3p48O=C and 4p48O=C, donates an H bond to 2p48O=C, interacts with 1p48O=C, π-stacks with F4i15, and approaches Y4i22. (I and J) Side and extracellular views at two similar binding modes of bisphenol A. Both aromatic rings are involved in π-cation interactions with NaIII, two hydroxyls donate H bonds to carbonyls p48O=C, and methyl groups bind between F4i15 and Y4i22. (K and L) Side views of lacosamide and phenytoin. Both ligands interact with NaIII and form H bonds with Q1p49, whereas their aromatic rings bind between F4i15 and Y4i22.
Cocaine and quinidine
Unlike flexible alkylammonium groups of lidocaine and QX-313, the ionizable nitrogen in cocaine and quinidine is incorporated into bulky rigid bicyclic moieties. These structural peculiarities did not disfavor binding modes with the ammonium nitrogen at site NaIII (Fig. 6, B–D). Quinidine has a hydroxyl group, which may interact with the polar groups in positions p48 and p49 (Fig. 6 B). The aromatic groups of these ligands bound between F4i15 and Y4i22. For both ligands, the binding modes were also possible with the ammonium group at level i15 (Fig. 6 D).
Sipatrigine
Because of structural peculiarities of sipatrigine, its contacts with the channel markedly differ from those predicted for other cationic ligands. In the vertical binding mode, sipatrigine extended form the level of NaIII (where its ammonium group bound) to the level of i23 in the activation-gate region (Fig. 6 F). The long and rigid sipatrigine molecule stretched parallel to the pore axis and did not bind tightly against IVS6, as other cationic drugs typically bind. In the horizontal binding mode, sipatrigine stretched from the inner pore deeply into the interface between helices IIIP, IIIS6, and IVS6 (Fig. 6 E). Unlike other cationic ligands, the long sipatrigine molecule established multiple simultaneous contacts with residues both in the inner pore and in the interface.
Electroneutral ligands
Among many ligands, we have chosen those that have essentially different chemical structures (Fig. 1 A). Phenytoin has two amide groups in the five-membered ring. Lamotrigine has a triazine ring with two attached amino groups and lacks oxygen atoms. CMZ, which binds in the common site with lamotrigine and phenytoin, has an amide group attached to the bulky tricyclic moiety. Unlike the three semi-rigid ligands, lacosamide is a very flexible compound. We also considered bisphenol A, a pollutant that has two distant hydroxyls and, unlike other compounds, lacks nitrogen atoms. Experimental data strongly suggest binding of these ligands in the inner pore (Ragsdale et al., 1996; Kuo, 1998; Liu et al., 2003; O’Reilly et al., 2012; Wang and Wang, 2014). Despite the great structural diversity, docking of all these compounds revealed binding modes reminiscent to those of CMZ. First, all these compounds interacted with a sodium ion at site NaIII. Second, all these compounds donated H bonds to backbone carbonyls in position p48, thus substituting water molecules, which bridge NaIII to the backbone carbonyls in the ligand-free NavMs (Naylor et al., 2016). Third, nonpolar moieties of the drugs extended into the inner pore between F4i15 and Y4i22 in a manner similar to those found for the charged ligands.
Lamotrigine
In the predicted complexes, the sodium ion was bound to the triazine-ring plane. The amino groups donated H bonds to the backbone carbonyls p48 (Fig. 6, G and H). The dichloro-substituted aromatic ring bound between F4i15 and Y4i22 and also contacted V4i18. Alanine substitutions of these residues are known to affect action of lamotrigine (Liu et al., 2003).
Phenytoin
In the predicted low-energy complexes, the sodium ion was bound to the ligand’s carbonyl oxygen, which is located between two NH groups, and approached an aromatic ring. In this binding mode, the NH groups can donate H bonds to the backbone oxygen atoms in positions p48 and p49. For example, in the structure shown in Fig. 6 L, one NH group donated an H bond to 3p48O=C and another NH group donated an H bond to the sidechain of Q1p49.
Lacosamide
Batrachotoxin-activated Nav1.5 channels and the Nav1.5 mutant F4i15K are completely resistant to the lacosamide block, indicating that the lacosamide receptor overlaps with those for batrachotoxin and LAs (Wang and Wang, 2014). Only (R)-isomer of lacosamide is active (Porter et al., 2012). A very flexible lacosamide has three oxygen atoms and an aromatic ring capable of interacting with Na+. Therefore, the number of possible conformations in which lacosamide can interact with a sodium ion is unusually large. In the very diverse ensemble of lacosamide-binding modes, we found those that resemble more rigid ligands. In one of these modes, lacosamide chelated NaIII by two carbonyl oxygens, its aromatic ring bound between F4i15 and Y4i22, and the NH groups approached oxygen atoms in positions p48 and p49 (Fig. 6 K).
Bisphenol A
In the obtained low-energy structures, sodium ion in position NaIII bound between two aromatic rings, enjoying cation-π interactions with both of them. The two hydroxyl groups anchored this complex to two backbone carbonyls p48 (Fig. 6, I and J). Despite in this binding mode, bisphenol A did not protrude an aromatic group in the inner pore, but it did contact F4i15. This result is consistent with the data that mutation F4i15A affects action of bisphenol A in the Nav1.5 channel (O’Reilly et al., 2012). Interestingly, bisphenol A, phenytoin, and lamotrigine have a common feature: a rigid or semi-rigid structure incorporating chemical groups that can donate protons to the backbone carbonyls at the opposing sides of the p48 ring.
Common and specific features of ligand-binding modes
Our calculations demonstrated that low-energy binding modes of cationic ligands in the NavMs-based model of Nav1.4 do not contradict the earlier predicted binding modes where the ligands’ cationic groups occur at the focus of P1 helices, close to putative site NaIV (Tikhonov and Zhorov, 2007; Bruhova et al., 2008). In addition, the NavMs-based model allowed us to find that different cationic ligands adopted a previously unknown binding mode in which the cationic group binds at site NaIII. When this site is occupied by a sodium ion, it attracts various electroneutral ligands, which also form multiple contacts with the channel protein. Within the groups of cationic and electroneutral ligands, the contacts with the channel are rather similar. Importantly, such contacts are also similar between the two groups. Fig. 7 shows superposition of the considered ligands. The cationic nitrogen or the sodium ion at one end of the ligand is highlighted blue or yellow, respectively, and the most remote atom in the aromatic ring at another end of the ligand is highlighted gray. Positions of these atoms, which largely determine the binding mode, are conserved with two exceptions. One is bisphenol A, which lacks an aromatic group protruding in the inner pore. The second exception is sipatrigine, an unusually long ligand. Besides these exceptions, the principal features of the ligand–channel complexes (Fig. 7 B) are consistent with the classical pharmacophore model for LAs, which is proposed by Khodorov (1981). The pharmacophore includes a cation (the ammonium group or ligand-bound sodium ion) and an aromatic moiety, which are usually linked by four bonds (Fig. 7 C). We expect that multiple compounds that are structurally related to the considered ones should also have analogous binding modes.
Figure 7.
Similarity of binding modes of charged and neutral blockers. (A) Cytoplasmic and side views at superposition of the channel-bound ligands. The charged nitrogens and ligand-bound sodium ions are blue and yellow, respectively. The carbon atoms at the opposite end of the ligands are gray. Note the clustering of blue/yellow atoms at the NaIII site and gray atoms at F4i15. The gray outlier belongs to bisphenol A, which lacks an aromatic ring protruding into the pore. (B) Scheme of ligand–channel interactions. The charged nitrogen or ligand-bound sodium ion interacts with the ring of carbonyls p48O=C (NaIII site) indicated by the red rectangle. The aromatic groups interact with hydrophobic residues in the inner pore (gray rectangle), among which F4i15 plays the major role. (C) Lidocaine and sodium-bound CMZ have a common pharmacophore.
Discussion
Mechanism of the channel block
According to a large body of experimental data, structurally vastly different ligands bind in the same region of the inner pore and exhibit similar properties in electrophysiological studies (Catterall, 1987; Ragsdale et al., 1996). The common binding mode found in this study, which fits the classical pharmacophore model for LAs, allows us to propose that the mechanisms of the channel block by the charged and neural ligands have common features. The charged ligands block the channels by occupying sites NaIII or NaIV for permeant ions with their own charged group. Even if the permeation pathway is not completely occluded, the positive charge placed at the innate sites for the permeant ion would provide the block. We suggest that the neutral ligands clamp the permeant ion at site NaIII or NaIV, and the clamped ion would preclude permeation of other ions (i.e., it would block the channel by the electrostatic mechanism). According to MD simulations based on x-ray structures, the fast ion permeation through the selectivity filter of highly ion-selective P-loop channels is caused by the fine balance of interactions involving permeant ions, water molecules, and precisely positioned specific groups of the channel (Shrivastava and Sansom, 2000; Bernèche and Roux, 2001). In the presence of a ligand, this balance would be disrupted. First, immobile cation-attractive groups of the bound ligand would replace mobile water molecules from the ion hydration shell. Second, the hydrophobic and electropositive groups of the ligand, which occur in immediate proximity of the permeant ion in its innate binding sites, would also reduce the ion mobility. Because of these factors, even ligands that do not demonstrate high affinity to cations in the bulk solution would reduce the permeation rate and thus produce the blocking effect. This hypothesis provides a general explanation for the channel block by ligands of vastly different chemical structures considered in this study. We did not consider benzocaine in this study because it is unclear how many small neutral molecules can bind simultaneously (Boiteux et al., 2014). A mechanism of the channel block by several benzocaine molecules coordinating a sodium ion, which was proposed before (Tikhonov et al., 2006), generally agrees with present hypothesis. This hypothesis requires testing by further studies, including intensive MD simulations.
Our computations revealed the binding region for the ligand ammonium group or ligand-bound metal ion that spreads from site NaIII to putative site NaIV at level i15. Ligand–channel interactions in these binding modes are different. In particular, the cationic group at level i15 directly interacts with F4i15, but in position NaIII, the major determinants are backbone carbonyls in position p48. Our homology model is not precise enough to discriminate between these two binding modes basing on energetics only. However, in view of the mechanism of the block, binding in position NaIII, at the channel bottleneck, appears more likely because there is no room for the permeating ion to bypass the charged moiety of the blocker or the ligand-trapped sodium ion. Block at level i15 seems less likely because at this level, sodium channels are wider than potassium channels, and this level also involves pore-lining cation-attractive residues in positions i15. The electrostatic block by the ammonium group located at wide levels of inner pore would be inefficient. Indeed, engineered lysines in the inner-pore–lining positions, which are used to explore action of various drugs, do not block the current (e.g., Wang et al., 2000), although they reduce the channel conductance (McNulty et al., 2007). On the other hand, mexiletine, a lidocaine derivative with unsubstituted amino group, which is analogous to that of lysine, does block the sodium channels (Wang et al., 2004). In view of our model, the ammonium group of mexiletine would bind at the NaIII site and efficiently block the permeation by the electrostatic mechanism. Furthermore, sodium channel agonists, which are predicted to bind inside the pore (Tikhonov and Zhorov, 2005) at the site that largely overlaps with that of LAs (Wang et al., 2000, 2001), do not block the permeation, even if they carry a permanent positive change (Catterall, 1980).
The computational study focused on the NaIII site has become possible only after x-ray crystallography revealed a metal-binding site between the backbone carbonyls in positions p48 (Zhang et al., 2012; Tang et al., 2014; Naylor et al., 2016). Previous models of sodium channels, which are based on potassium channel templates, proposed that the ligands’ charged moieties bind in the inner pore, at the focus of P1 helices. Possibly, that is why the common pharmacophore for cationic and neutral ligands was not proposed before.
Mutational analysis
Multiple mutational studies using different ligands show involvement of F4i15 in ligand binding (for a review, see Mike and Lukacs, 2010). In the proposed here binding modes all the ligands do interact with this residue. Most of the ligands can establish both face-to-face and face-to-edge interactions of their aromatic ring with F4i15. In our models, Y4i22, another well-known determinant of the sodium channel block, also approaches the aromatic group of the bound ligands. It is also known that mutations of highly conserved asparagines in positions i20 (Fig. 1 B) affect the inner pore block. However, in our current and earlier (Tikhonov and Zhorov, 2007; Bruhova et al., 2008) models, as well as in available x-ray structures, these residues do not face the pore and cannot interact with the pore-bound ligands. The allosteric mechanism via stabilizing the open-pore geometry has been proposed to explain effects of such mutations (Tikhonov et al., 2015). Another residue whose mutations affect the channel block is I3i23 (Yarov-Yarovoy et al., 2002). In our models, a ligand with the cationic group in site NaIII is too far from the pore-facing position 3i23. However, in the binding modes with the cationic group at level i15, this group interacted with F4i15, whereas the aromatic group readily reached position 3i23. Possible rationale for the effect of mutations in position 3i23 involves indirect effects of mutations (as in the case of conserved asparagines) or the ligand fluctuation in the pore with interim interactions with I3i23. The possibility of fluctuations within the binding region is further supported by the fact that the action of ligands with a common pharmacophore is differently sensitive to the same mutations (Pless et al., 2011). This also seems consistent with rather low affinities of the inner pore blockers of sodium channels.
Ligands lacking a LA pharmacophore
Here, we have proposed a common pharmacophore for structurally very different blockers that have the common binding site and common mechanism of action. Obviously, there are blockers that do not fit this pharmacophore. One example is inflexible sipatrigine. According to our calculations, sipatrigine can bind in the vertical and horizontal modes, but the patterns of interactions for this unusually long ligand differ from those of other blockers. Mutational analysis supports the horizontal mode because alanine substitutions of the pore-facing residues V4i18 and Y4i22, which are obviously reachable by the vertically bound long ligand, have a much smaller impact on the action of sipatrigine than on the action of short ligands (Liu et al., 2003). The horizontal binding mode of sipatrigine resembles that of PL1 in the bacterial sodium channel NavMs (Bagnéris et al., 2014; Korkosh et al., 2016). There are many other blockers of sodium channels that have little in common with LAs, such as pancuronium, veratramine, lappaconitine, and methylstrychnine.
On the structure–activity relations
A recent cryo-electron microscopy structure of the eukaryotic calcium channel Cav1.1 (Wu et al., 2015) shows essential asymmetry in the inner pore region; certain asymmetry is also expected for other pseudo-heterotetrameric P-loop channels. Obviously, our homology model of the pseudo-heterotetrameric sodium channel, which is based on the homotetrameric template NavMs, is not expected to be precise enough to predict ligand affinities and quantitatively analyze structure–activity relations of drugs. Nevertheless, the proposed models allow us to consider qualitatively some aspects of structure–activity relations. The binding modes proposed by us agree with the classical LA pharmacophore (Khodorov, 1981) and thus should agree with numerous data on structure–activity relations fitting the pharmacophore. In addition, some other structure–activity features can be mentioned.
The antiseizure activity of lacosamide is caused by its (R)-enantiomer, whereas (S)-enantiomer is completely inactive (Porter et al., 2012). This fact is not easy to understand given that the chiral atom of lacosamide is located in the middle of the flexible chain, which in the wide inner pore would adapt its conformation to provide energetically favorable contacts with complementary protein moieties. In our model, lacosamide chelates a sodium ion by at least two groups. This would dramatically reduce lacosamide flexibility and provide nonadjustable mutual orientation of the aromatic and terminal polar moieties, which are connected to the chiral atom.
A chemical fingerprint of anticonvulsants is the conserved mutual disposition of electronegative atoms and polar hydrogens in the O=C-NH or =N-C-NH moieties of CMZ, phenytoin, lamotrigine, and similar electroneutral ligands (Unverferth et al., 1998; Anger et al., 2001). In view of our models, the electronegative atom binds to NaIII, whereas a polar hydrogen would be H-bonded to the oxygen atoms in positions p48/p49. Respective moieties of the ligand would substitute a rather mobile water molecule in the hydration shell of NaIII and bridge the latter to the channel protein, thus strengthening the channel block.
Conclusion
In conclusion, the NavMs-based model of the eukaryotic sodium channel accommodates a hydrated sodium ion at the NaIII site, which is reachable by ligands bound in the inner pore. Our calculations yielded models in which the ammonium group of cationic ligands or a sodium ion bound to electroneutral ligands occupies the innate NaIII site and can effectively block permeation by the combination of electrostatic and steric mechanisms. Our study for the first time proposes a common pharmacophore and atomic mechanism of action for drugs of highly diverse chemical structures.
Supplementary Material
Acknowledgments
Computations were made possible by the facilities of the Shared Hierarchical Academic Research Computing Network.
This work was supported by grants to B.S. Zhorov from the Natural Sciences and Engineering Research Council of Canada and Russian Foundation of Basic Research grants. D.B. Tikhonov acknowledges a Russian Academy of Sciences program “Molecular and Cell Biology” grant.
The authors declare no competing financial interests.
José D. Faraldo-Gómez served as editor.
Footnotes
Abbreviations used:
- CMZ
- carbamazepine
- LA
- local anesthetic
- MC
- Monte Carlo
- MCM
- MC minimization
- MD
- molecular dynamics
References
- Ahern C.A., Eastwood A.L., Dougherty D.A., and Horn R.. 2008. Electrostatic contributions of aromatic residues in the local anesthetic receptor of voltage-gated sodium channels. Circ. Res. 102:86–94. 10.1161/CIRCRESAHA.107.160663 [DOI] [PubMed] [Google Scholar]
- Anger T., Madge D.J., Mulla M., and Riddall D.. 2001. Medicinal chemistry of neuronal voltage-gated sodium channel blockers. J. Med. Chem. 44:115–137. 10.1021/jm000155h [DOI] [PubMed] [Google Scholar]
- Bagnéris C., DeCaen P.G., Naylor C.E., Pryde D.C., Nobeli I., Clapham D.E., and Wallace B.A.. 2014. Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism. Proc. Natl. Acad. Sci. USA. 111:8428–8433. 10.1073/pnas.1406855111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernèche S., and Roux B.. 2001. Energetics of ion conduction through the K+ channel. Nature. 414:73–77. 10.1038/35102067 [DOI] [PubMed] [Google Scholar]
- Boiteux C., Vorobyov I., French R.J., French C., Yarov-Yarovoy V., and Allen T.W.. 2014. Local anesthetic and antiepileptic drug access and binding to a bacterial voltage-gated sodium channel. Proc. Natl. Acad. Sci. USA. 111:13057–13062. 10.1073/pnas.1408710111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C.L., Pettitt B.M., and Karplus M.. 1985. Structural and energetic effects of truncating long ranged interactions in ionic and polar fluids. J. Chem. Phys. 83:5897–5908. 10.1063/1.449621 [DOI] [Google Scholar]
- Browne L.E., Blaney F.E., Yusaf S.P., Clare J.J., and Wray D.. 2009. Structural determinants of drugs acting on the Nav1.8 channel. J. Biol. Chem. 284:10523–10536. 10.1074/jbc.M807569200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhova I., Tikhonov D.B., and Zhorov B.S.. 2008. Access and binding of local anesthetics in the closed sodium channel. Mol. Pharmacol. 74:1033–1045. 10.1124/mol.108.049759 [DOI] [PubMed] [Google Scholar]
- Catterall W.A. 1980. Neurotoxins that act on voltage-sensitive sodium channels in excitable membranes. Annu. Rev. Pharmacol. Toxicol. 20:15–43. 10.1146/annurev.pa.20.040180.000311 [DOI] [PubMed] [Google Scholar]
- Catterall W.A. 1987. Common modes of drug action on Na+ channels: local anesthetics, antiarrhythmics and anticonvulsants. Trends Pharmacol. Sci. 8:57–65. 10.1016/0165-6147(87)90011-3 [DOI] [Google Scholar]
- Catterall W.A. 2012. Voltage-gated sodium channels at 60: Structure, function and pathophysiology. J. Physiol. 590:2577–2589. 10.1113/jphysiol.2011.224204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall W.A., and Swanson T.M.. 2015. Structural basis for pharmacology of voltage-gated sodium and calcium channels. Mol. Pharmacol. 88:141–150. 10.1124/mol.114.097659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B. III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., and Richardson D.C.. 2010. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66:12–21. 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar M.J.S., Zoebisch E.G., Healy E.F., and Stewart J.J.P.. 1985. Development and use of quantum mechanical molecular models. 76. AM1: A new general purpose quantum mechanical model. J. Am. Chem. Soc. 107:3902–3909. 10.1021/ja00299a024 [DOI] [Google Scholar]
- Dick I.E., Brochu R.M., Purohit Y., Kaczorowski G.J., Martin W.J., and Priest B.T.. 2007. Sodium channel blockade may contribute to the analgesic efficacy of antidepressants. J. Pain. 8:315–324. 10.1016/j.jpain.2006.10.001 [DOI] [PubMed] [Google Scholar]
- Garden D.P., and Zhorov B.S.. 2010. Docking flexible ligands in proteins with a solvent exposure- and distance-dependent dielectric function. J. Comput. Aided Mol. Des. 24:91–105. 10.1007/s10822-009-9317-9 [DOI] [PubMed] [Google Scholar]
- Hebert T., Drapeau P., Pradier L., and Dunn R.J.. 1994. Block of the rat brain IIA sodium channel alpha subunit by the neuroprotective drug riluzole. Mol. Pharmacol. 45:1055–1060. [PubMed] [Google Scholar]
- Hille B. 1977. Local anesthetics: Hydrophilic and hydrophobic pathways for the drug-receptor reaction. J. Gen. Physiol. 69:497–515. 10.1085/jgp.69.4.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. 2001. Ion Channels of Excitable Membranes. Third edition Sinauer Associates, Inc., Sunderland, MA. 814 pp. [Google Scholar]
- Khodorov B.I. 1981. Sodium inactivation and drug-induced immobilization of the gating charge in nerve membrane. Prog. Biophys. Mol. Biol. 37:49–89. 10.1016/0079-6107(82)90020-7 [DOI] [PubMed] [Google Scholar]
- Korkosh V.S., Zhorov B.S., and Tikhonov D.B.. 2016. Modeling interactions between blocking and permeant cations in the NavMs channel. Eur. J. Pharmacol. 780:188–193. 10.1016/j.ejphar.2016.03.048 [DOI] [PubMed] [Google Scholar]
- Kuo C.C. 1998. A common anticonvulsant binding site for phenytoin, carbamazepine, and lamotrigine in neuronal Na+ channels. Mol. Pharmacol. 54:712–721. [PubMed] [Google Scholar]
- Li Z., and Scheraga H.A.. 1987. Monte Carlo-minimization approach to the multiple-minima problem in protein folding. Proc. Natl. Acad. Sci. USA. 84:6611–6615. 10.1073/pnas.84.19.6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.L., Galue A., Meadows L., and Ragsdale D.S.. 1999. A molecular basis for the different local anesthetic affinities of resting versus open and inactivated states of the sodium channel. Mol. Pharmacol. 55:134–141. [DOI] [PubMed] [Google Scholar]
- Lipkind G.M., and Fozzard H.A.. 2000. KcsA crystal structure as framework for a molecular model of the Na(+) channel pore. Biochemistry. 39:8161–8170. 10.1021/bi000486w [DOI] [PubMed] [Google Scholar]
- Lipkind G.M., and Fozzard H.A.. 2005. Molecular modeling of local anesthetic drug binding by voltage-gated sodium channels. Mol. Pharmacol. 68:1611–1622. 10.1124/mol.105.014803 [DOI] [PubMed] [Google Scholar]
- Lipkind G.M., and Fozzard H.A.. 2010. Molecular model of anticonvulsant drug binding to the voltage-gated sodium channel inner pore. Mol. Pharmacol. 78:631–638. 10.1124/mol.110.064683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Yarov-Yarovoy V., Nobbs M., Clare J.J., Scheuer T., and Catterall W.A.. 2003. Differential interactions of lamotrigine and related drugs with transmembrane segment IVS6 of voltage-gated sodium channels. Neuropharmacology. 44:413–422. 10.1016/S0028-3908(02)00400-8 [DOI] [PubMed] [Google Scholar]
- McNulty M.M., Edgerton G.B., Shah R.D., Hanck D.A., Fozzard H.A., and Lipkind G.M.. 2007. Charge at the lidocaine binding site residue Phe-1759 affects permeation in human cardiac voltage-gated sodium channels. J. Physiol. 581:741–755. 10.1113/jphysiol.2007.130161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mike A., and Lukacs P.. 2010. The enigmatic drug binding site for sodium channel inhibitors. Curr. Mol. Pharmacol. 3:129–144. 10.2174/1874467211003030129 [DOI] [PubMed] [Google Scholar]
- Naylor C.E., Bagnéris C., DeCaen P.G., Sula A., Scaglione A., Clapham D.E., and Wallace B.A.. 2016. Molecular basis of ion permeability in a voltage-gated sodium channel. EMBO J. 35:820–830. 10.15252/embj.201593285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary M.E., and Chahine M.. 2002. Cocaine binds to a common site on open and inactivated human heart (Na(v)1.5) sodium channels. J. Physiol. 541:701–716. 10.1113/jphysiol.2001.016139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly A.O., Eberhardt E., Weidner C., Alzheimer C., Wallace B.A., and Lampert A.. 2012. Bisphenol A binds to the local anesthetic receptor site to block the human cardiac sodium channel. PLoS One. 7:e41667 10.1371/journal.pone.0041667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancrazio J.J., Kamatchi G.L., Roscoe A.K., and Lynch C. III. 1998. Inhibition of neuronal Na+ channels by antidepressant drugs. J. Pharmacol. Exp. Ther. 284:208–214. [PubMed] [Google Scholar]
- Payandeh J., Scheuer T., Zheng N., and Catterall W.A.. 2011. The crystal structure of a voltage-gated sodium channel. Nature. 475:353–358. 10.1038/nature10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless S.A., Galpin J.D., Frankel A., and Ahern C.A.. 2011. Molecular basis for class Ib anti-arrhythmic inhibition of cardiac sodium channels. Nat. Commun. 2:351 10.1038/ncomms1351 [DOI] [PubMed] [Google Scholar]
- Porter R.J., Dhir A., Macdonald R.L., and Rogawski M.A.. 2012. Mechanisms of action of antiseizure drugs. In Handbook of Clinical Neurology. Epilepsy, Part II. Stefan H. and Theodore W.H., editors. Elsevier, Amsterdam: 663–681. [DOI] [PubMed] [Google Scholar]
- Qu Y., Rogers J., Tanada T., Scheuer T., and Catterall W.A.. 1995. Molecular determinants of drug access to the receptor site for antiarrhythmic drugs in the cardiac Na+ channel. Proc. Natl. Acad. Sci. USA. 92:11839–11843. 10.1073/pnas.92.25.11839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan C., Mok W.M., and Wang G.K.. 1996. Use-dependent inhibition of Na+ currents by benzocaine homologs. Biophys. J. 70:194–201. 10.1016/S0006-3495(96)79563-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale D.S., McPhee J.C., Scheuer T., and Catterall W.A.. 1994. Molecular determinants of state-dependent block of Na+ channels by local anesthetics. Science. 265:1724–1728. 10.1126/science.8085162 [DOI] [PubMed] [Google Scholar]
- Ragsdale D.S., McPhee J.C., Scheuer T., and Catterall W.A.. 1996. Common molecular determinants of local anesthetic, antiarrhythmic, and anticonvulsant block of voltage-gated Na+ channels. Proc. Natl. Acad. Sci. USA. 93:9270–9275. 10.1073/pnas.93.17.9270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheib H., McLay I., Guex N., Clare J.J., Blaney F.E., Dale T.J., Tate S.N., and Robertson G.M.. 2006. Modeling the pore structure of voltage-gated sodium channels in closed, open, and fast-inactivated conformation reveals details of site 1 toxin and local anesthetic binding. J. Mol. Model. 12:813–822. 10.1007/s00894-005-0066-y [DOI] [PubMed] [Google Scholar]
- Shrivastava I.H., and Sansom M.S.. 2000. Simulations of ion permeation through a potassium channel: Molecular dynamics of KcsA in a phospholipid bilayer. Biophys. J. 78:557–570. 10.1016/S0006-3495(00)76616-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunami A., Dudley S.C. Jr., and Fozzard H.A.. 1997. Sodium channel selectivity filter regulates antiarrhythmic drug binding. Proc. Natl. Acad. Sci. USA. 94:14126–14131. 10.1073/pnas.94.25.14126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Gamal El-Din T.M., Payandeh J., Martinez G.Q., Heard T.M., Scheuer T., Zheng N., and Catterall W.A.. 2014. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature. 505:56–61. 10.1038/nature12775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov D.B., and Zhorov B.S.. 2005. Sodium channel activators: Model of binding inside the pore and a possible mechanism of action. FEBS Lett. 579:4207–4212. 10.1016/j.febslet.2005.07.017 [DOI] [PubMed] [Google Scholar]
- Tikhonov D.B., and Zhorov B.S.. 2007. Sodium channels: Ionic model of slow inactivation and state-dependent drug binding. Biophys. J. 93:1557–1570. 10.1529/biophysj.106.100248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov D.B., and Zhorov B.S.. 2012. Architecture and pore block of eukaryotic voltage-gated sodium channels in view of NavAb bacterial sodium channel structure. Mol. Pharmacol. 82:97–104. 10.1124/mol.112.078212 [DOI] [PubMed] [Google Scholar]
- Tikhonov D.B., Bruhova I., and Zhorov B.S.. 2006. Atomic determinants of state-dependent block of sodium channels by charged local anesthetics and benzocaine. FEBS Lett. 580:6027–6032. 10.1016/j.febslet.2006.10.035 [DOI] [PubMed] [Google Scholar]
- Tikhonov D.B., Bruhova I., Garden D.P., and Zhorov B.S.. 2015. State-dependent inter-repeat contacts of exceptionally conserved asparagines in the inner helices of sodium and calcium channels. Pflugers Arch. 467:253–266. 10.1007/s00424-014-1508-0 [DOI] [PubMed] [Google Scholar]
- Tsang S.Y., Tsushima R.G., Tomaselli G.F., Li R.A., and Backx P.H.. 2005. A multifunctional aromatic residue in the external pore vestibule of Na+ channels contributes to the local anesthetic receptor. Mol. Pharmacol. 67:424–434. 10.1124/mol.67.2. [DOI] [PubMed] [Google Scholar]
- Unverferth K., Engel J., Höfgen N., Rostock A., Günther R., Lankau H.J., Menzer M., Rolfs A., Liebscher J., Müller B., and Hofmann H.J.. 1998. Synthesis, anticonvulsant activity, and structure-activity relationships of sodium channel blocking 3-aminopyrroles. J. Med. Chem. 41:63–73. 10.1021/jm970327j [DOI] [PubMed] [Google Scholar]
- Wang G.K., and Wang S.Y.. 1994. Binding of benzocaine in batrachotoxin-modified Na+ channels. State-dependent interactions. J. Gen. Physiol. 103:501–518. 10.1085/jgp.103.3.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.K., and Wang S.Y.. 2014. Block of human cardiac sodium channels by lacosamide: Evidence for slow drug binding along the activation pathway. Mol. Pharmacol. 85:692–702. 10.1124/mol.113.091173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.K., Quan C., and Wang S.. 1997. A common local anesthetic receptor for benzocaine and etidocaine in voltage-gated mu1 Na+ channels. Pflugers Arch. 435:293–302. 10.1007/s004240050515 [DOI] [PubMed] [Google Scholar]
- Wang G.K., Russell C., and Wang S.Y.. 2004. Mexiletine block of wild-type and inactivation-deficient human skeletal muscle hNav1.4 Na+ channels. J. Physiol. 554:621–633. 10.1113/jphysiol.2003.054973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.Y., Nau C., and Wang G.K.. 2000. Residues in Na(+) channel D3-S6 segment modulate both batrachotoxin and local anesthetic affinities. Biophys. J. 79:1379–1387. 10.1016/S0006-3495(00)76390-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.Y., Barile M., and Wang G.K.. 2001. Disparate role of Na(+) channel D2-S6 residues in batrachotoxin and local anesthetic action. Mol. Pharmacol. 59:1100–1107. [DOI] [PubMed] [Google Scholar]
- Wang S.Y., Mitchell J., Tikhonov D.B., Zhorov B.S., and Wang G.K.. 2006. How batrachotoxin modifies the sodium channel permeation pathway: Computer modeling and site-directed mutagenesis. Mol. Pharmacol. 69:788–795. 10.1124/mol.105.018200 [DOI] [PubMed] [Google Scholar]
- Weiner S.J., Kollman P.A., Case D.A., Singh U.C., Ghio C., Alagona G., Profeta S., and Weiner P.. 1984. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 106:765–784. 10.1021/ja00315a051 [DOI] [Google Scholar]
- Weiner S.J., Kollman P.A., Nguyen D.T., and Case D.A.. 1986. An all atom force field for simulations of proteins and nucleic acids. J. Comput. Chem. 7:230–252. 10.1002/jcc.540070216 [DOI] [PubMed] [Google Scholar]
- Wright S.N., Wang S.Y., and Wang G.K.. 1998. Lysine point mutations in Na+ channel D4-S6 reduce inactivated channel block by local anesthetics. Mol. Pharmacol. 54:733–739. [PubMed] [Google Scholar]
- Wu J., Yan Z., Li Z., Yan C., Lu S., Dong M., and Yan N.. 2015. Structure of the voltage-gated calcium channel Cav1.1 complex. Science. 350:aad2395 10.1126/science.aad2395 [DOI] [PubMed] [Google Scholar]
- Wu J., Yan Z., Li Z., Qian X., Lu S., Dong M., Zhou Q., and Yan N.. 2016. Structure of the voltage-gated calcium channel Cav1.1 at 3.6 Å resolution. Nature. 537:191–196. 10.1038/nature19321 [DOI] [PubMed] [Google Scholar]
- Yamagishi T., Xiong W., Kondratiev A., Vélez P., Méndez-Fitzwilliam A., Balser J.R., Marbán E., and Tomaselli G.F.. 2009. Novel molecular determinants in the pore region of sodium channels regulate local anesthetic binding. Mol. Pharmacol. 76:861–871. 10.1124/mol.109.055863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Dib-Hajj S.D., Zhang J., Zhang Y., Tyrrell L., Estacion M., and Waxman S.G.. 2012. Structural modelling and mutant cycle analysis predict pharmacoresponsiveness of a Na(V)1.7 mutant channel. Nat. Commun. 3:1186 10.1038/ncomms2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarov-Yarovoy V., Brown J., Sharp E.M., Clare J.J., Scheuer T., and Catterall W.A.. 2001. Molecular determinants of voltage-dependent gating and binding of pore-blocking drugs in transmembrane segment IIIS6 of the Na(+) channel alpha subunit. J. Biol. Chem. 276:20–27. 10.1074/jbc.M006992200 [DOI] [PubMed] [Google Scholar]
- Yarov-Yarovoy V., McPhee J.C., Idsvoog D., Pate C., Scheuer T., and Catterall W.A.. 2002. Role of amino acid residues in transmembrane segments IS6 and IIS6 of the Na+ channel alpha subunit in voltage-dependent gating and drug block. J. Biol. Chem. 277:35393–35401. 10.1074/jbc.M206126200 [DOI] [PubMed] [Google Scholar]
- Zhang X., Ren W., DeCaen P., Yan C., Tao X., Tang L., Wang J., Hasegawa K., Kumasaka T., He J., et al. 2012. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 486:130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhorov B.S., and Tikhonov D.B.. 2013. Ligand action on sodium, potassium, and calcium channels: Role of permeant ions. Trends Pharmacol. Sci. 34:154–161. 10.1016/j.tips.2013.01.002 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Morais-Cabral J.H., Kaufman A., and MacKinnon R.. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 414:43–48. 10.1038/35102009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.