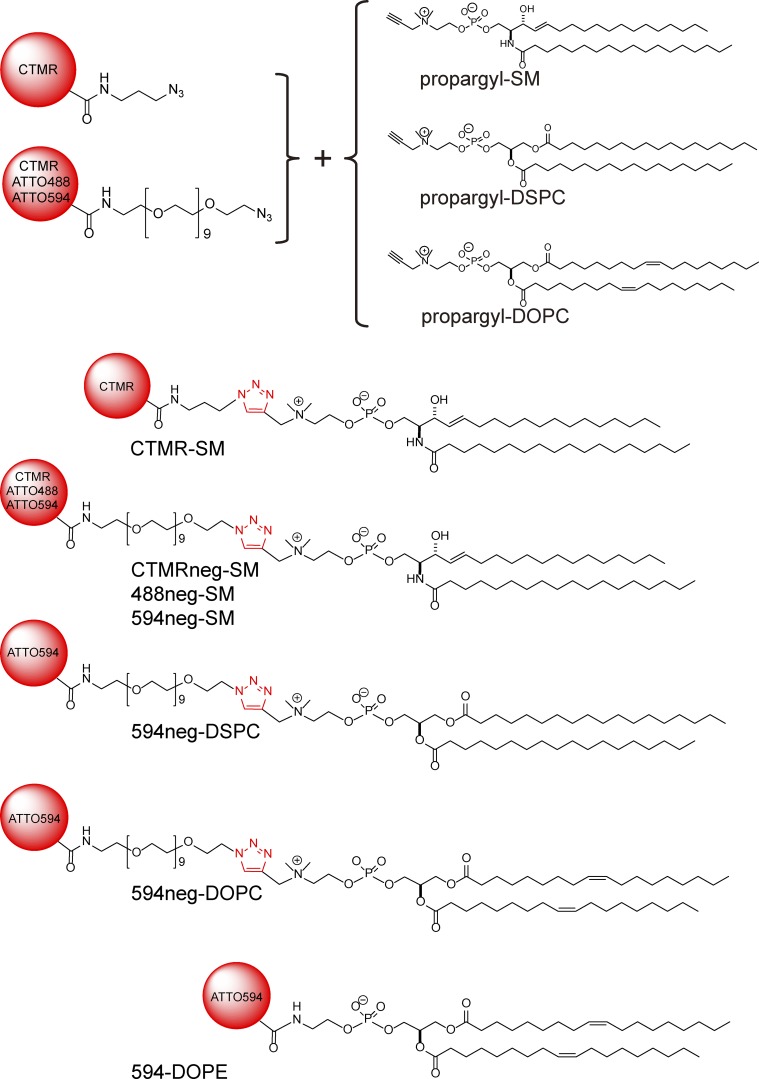
The behavior of sphingomyelin (SM) in the plasma membrane is unclear. Using new fluorescent SM analogs and single-molecule tracking, Kinoshita et al. find that SM transiently and continually interacts with the glycosylphosphatidylinositol (GPI)-anchored protein CD59 in oligomer size–, cholesterol-, and GPI anchoring–dependent manners.
Abstract
Sphingomyelin (SM) has been proposed to form cholesterol-dependent raft domains and sphingolipid domains in the plasma membrane (PM). How SM contributes to the formation and function of these domains remains unknown, primarily because of the scarcity of suitable fluorescent SM analogs. We developed new fluorescent SM analogs by conjugating a hydrophilic fluorophore to the SM choline headgroup without eliminating its positive charge, via a hydrophilic nonaethylene glycol linker. The new analogs behaved similarly to the native SM in terms of their partitioning behaviors in artificial liquid order-disorder phase-separated membranes and detergent-resistant PM preparations. Single fluorescent molecule tracking in the live-cell PM revealed that they indirectly interact with each other in cholesterol- and sphingosine backbone–dependent manners, and that, for ∼10–50 ms, they undergo transient colocalization-codiffusion with a glycosylphosphatidylinositol (GPI)-anchored protein, CD59 (in monomers, transient-dimer rafts, and clusters), in CD59-oligomer size–, cholesterol-, and GPI anchoring–dependent manners. These results suggest that SM continually and rapidly exchanges between CD59-associated raft domains and the bulk PM.
Introduction
Sphingomyelins (SMs) are phospholipids that are considered to reside in the outer leaflet of the plasma membrane (PM) and to be key molecules to generate cholesterol-dependent raft domains (Lingwood and Simons, 2010; van Meer and Hoetzl, 2010; Sáenz et al., 2012, 2015; Lin and London, 2015) and sphingolipid domains (Frisz et al., 2013; Abe and Kobayashi, 2014; Shen et al., 2014; Benda et al., 2015; Kishimoto et al., 2016). The term “raft domain” has not been solidly defined. Therefore, in this report, we define it as a domain or a group of molecules in the PM containing at least three molecules, formed by positive and/or negative interactions of acyl chains (saturated and unsaturated chains, respectively) and cholesterol, following Kusumi et al. (2004), which we think is a general, useful working definition. These domains are likely to perform critical functions as platforms for signal transduction in the PM. For example, SMs are essential for the raft-based formation of Fas-associated signaling clusters to induce apoptosis (Miyaji et al., 2005). However, the exact interactions of SMs with other raft-associated molecules for raft formation and function remain essentially unknown (Simons and Ikonen, 1997; Lin and London, 2015; Holowka and Baird, 2016). Fluorescence microscopy at high spatiotemporal resolutions (DeWitt and Dunn, 2015) would be suitable for addressing molecular behaviors in nano- to mesoscale domains by visualizing SM distributions, dynamics, and interactions with other molecules (Sezgin et al., 2012; Hori et al., 2013; Watanabe et al., 2014), but suitable fluorescent SM analogs have scarcely been available (Makino et al., 2015).
Native SMs primarily partition into cold detergent–resistant membranes (DRMs) prepared from the cell and into the liquid-ordered (Lo) domains, rather than the liquid-disordered (Ld) domains, in Lo-Ld phase–separated giant unilamellar vesicles (GUVs; Lingwood and Simons, 2010; Yasuda et al., 2015). However, the currently available fluorescent SM analogs, with the fluorescent compound bound to the second acyl chain or the headgroup, favored the Ld domains, rather than the Lo domains, in phase-separated GUVs (also see Fig. S1; Vicidomini et al., 2011; Sezgin et al., 2012; Klymchenko and Kreder, 2014), although native SMs (deuterated SM) prefer Lo domains (Beutel et al., 2014). A large fluorophore attached to the acyl chain might hamper the incorporation of the labeled SMs into the Lo domains, as found in the case of fluorescent ganglioside probes (Komura et al., 2016). A large hydrophobic fluorescent probe linked to the SM headgroup may intercalate into the hydrophobic interior of the membrane, and the bulkiness of the dye again might prevent the incorporation of the SM analogs into the Lo-like domain.
Another analog labeled with a polyene acyl chain partitioned into Lo domains, but the polyene gave a low fluorescence signal, rapidly photobleached, and required UV excitation (Kuerschner et al., 2005). Thus, the currently available fluorescent SM analogs are quite inadequate for probing the SM behaviors in the PMs.
To alleviate these problems and to understand how SMs participate in the formation and function of raft domains, we developed new fluorescent SM analogs that behave quite similarly to their native counterparts, in terms of partitioning into artificial raft-related membrane domains/preparations. Our strategy for their development was to attach more hydrophilic fluorescent compounds to the SM headgroup and to place it some distance away from the SM headgroup toward the bulk aqueous phase, while keeping a positive charge at the choline group. Previously, we attached the propargyl group to the choline residue in the SM headgroup, while retaining the positive charge at the choline group, and found that the propargyl-SM thus synthesized (Fig. 1) exhibited close similarity to the behavior of natural SMs in multilamellar membranes (Sandbhor et al., 2009; Goretta et al., 2012). Therefore, we first synthesized propargyl-SM, and then, using the carbon–carbon triple bond of the propargyl group for the Huisgen cycloaddition reaction, obtained fluorescent SM analogs. As good fluorescent SM analogs, we selected fluorescently conjugated SMs (18:0) that primarily partition into DRMs prepared from the cell and into the Lo domains in Lo-Ld phase–separated GUVs, just like native SMs.
Figure 1.
Schemes for preparing fluorescently labeled SMs and PCs synthesized in this study, using Huisgen cycloaddition. Chemical structures of fluorophores are omitted.
As control molecules for fluorescent SMs, we additionally synthesized fluorescently labeled phosphatidylcholine (PC). PC possesses the same phosphorylcholine headgroup as SM and can thus be fluorescently conjugated in the same manner as that for SM. Unlike SMs, which contain the sphingosine backbone, PC has the glycerol backbone. We used l-α-distearoyl-PC (di 18:0; DSPC) and l-α-dioleoyl-PC (di 18:1; DOPC).
We observed the dynamic behaviors of SMs in the PMs of living cells. We used advanced single–molecule imaging, at time resolutions down to 0.5 ms, the highest time resolution currently available for single fluorescent-molecule imaging, developed in our laboratory (Hiramoto-Yamaki et al., 2014), and simultaneous, two-color (two-molecular-species), single-molecule imaging. We investigated the interactions of fluorescent SM analogs with human CD59, a glycosylphosphatidylinositol (GPI)-anchored receptor, in its three assembly states in the PMs of live cells: CD59 monomers, transient CD59 homodimer rafts, and CD59-cluster rafts (containing a mean of five CD59 molecules), which trigger intracellular signals (Suzuki et al., 2007a,b, 2012).
Our analyses of single-molecule trajectories provided the first direct evidence that SMs and GPI-anchored receptors interact in cholesterol-dependent and GPI anchorage–dependent manners. The fluorescent SM analogs underwent transient colocalization-codiffusion with CD59 in cholesterol- and GPI anchorage–dependent manners, with mean lifetimes in the range of 12–50 ms. The colocalization lifetimes increased in the order of CD59 monomers, CD59 transient homodimer rafts in quiescent cells, and engaged CD59-cluster signaling rafts. These results unequivocally showed that SMs continually and very dynamically enter and exit from the CD59-associated raft domains and the local lipid environment generated by CD59 (in its various assembly states).
Results
Synthesis of fluorescent SM analogs and their partitioning into the Lo versus Ld domains in phase-separated GUV membranes
We first synthesized propargyl-SM (18:0) and then conjugated it with a fluorophore with an azide group by using the propargyl group in the Huisgen cycloaddition reaction (Fig. 1 and Online supplemental material). We conjugated carboxytetramethylrhodamine (CTMR) to propargyl-SM with a short alkyl linker (CTMR-SM; Fig. 1) and then examined the partitioning of CTMR-SM between the Lo and Ld domains in Lo-Ld phase–separated GUV membranes (SM[18:0]/DOPC/cholesterol [1:1:1 by moles]) at 28.5°C (see Materials and methods; all GUV observations were conducted under these conditions). This was because our previous solid-state deuterium nuclear magnetic resonance (NMR) analysis revealed that deuterated SM (18:0, without the fluorophore) in multilamellar vesicles, with the same lipid composition and at the same temperature, partitioned into the Lo domains 3.6-fold more than into the Ld domain, in terms of the number density of deuterated SM (Yasuda et al., 2015; see Materials and methods for calculation details).
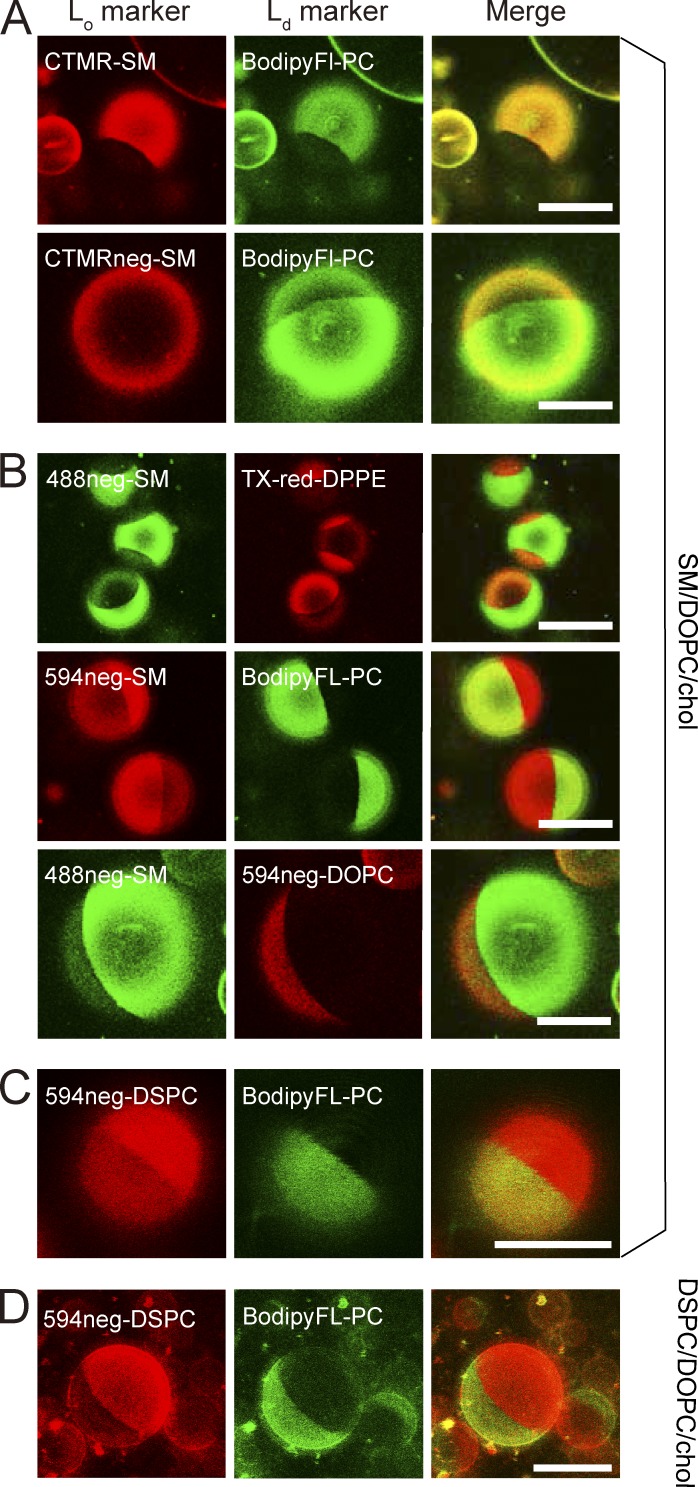
Unlike the deuterated SM (18:0), which is expected to exhibit properties quite similar to those of native SM (18:0), CTMR-SM primarily partitioned into the Ld domain in Lo-Ld phase–separated GUV membranes (Fig. 2 A). Such partitioning behavior was also detected with the commercially available TopFluor-SM and N-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl] (NBD)–SM (fluorescent groups attached to the acyl chain at C11 and C6, respectively), despite their previous uses as raft markers (Fig. S1). These results suggest that the CTMR moiety might interfere with its alkyl-chain interactions with other lipids, including cholesterol.
Figure 2.
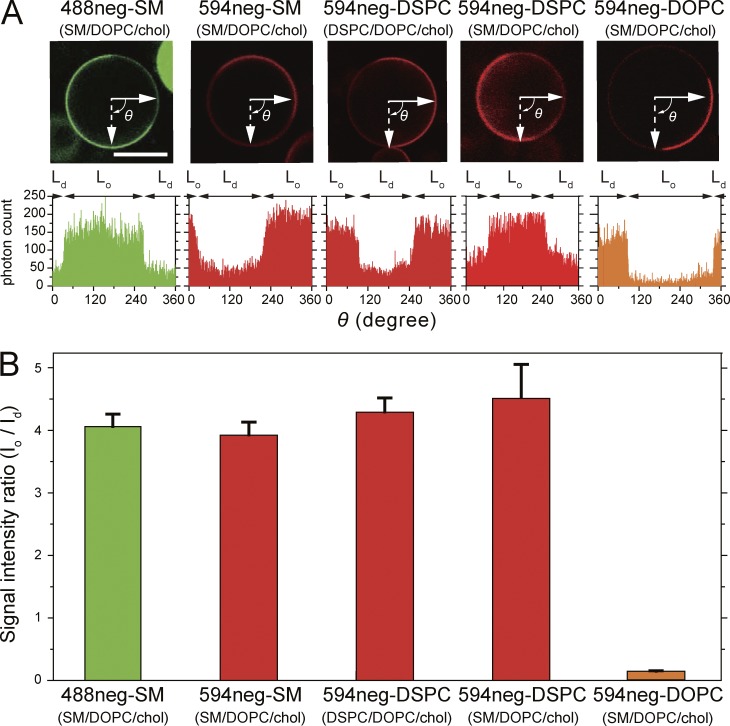
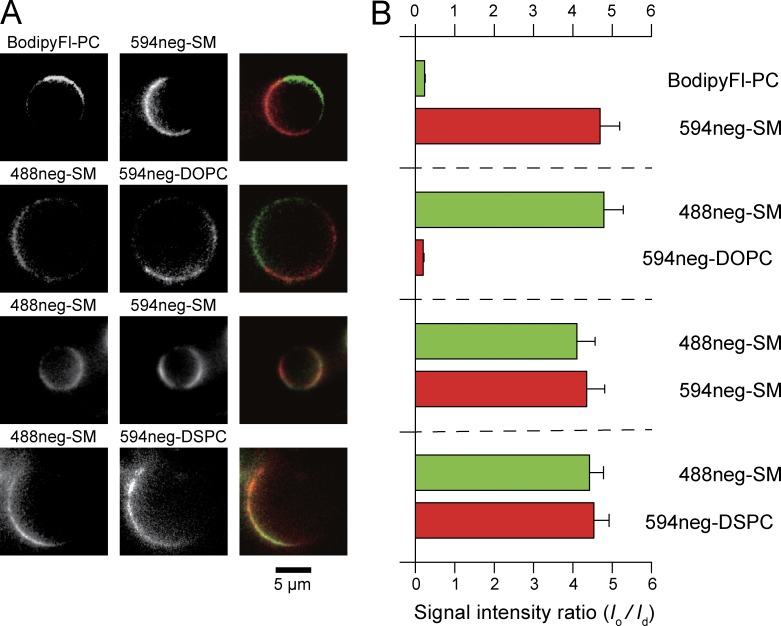
Partitioning of various fluorescent SM, DSPC, and DOPC analogs into Lo versus Ld domains in phase-separated GUV membranes at 28.5°C, observed by confocal fluorescence microscopy. Phase-separated GUVs consisting of ternary mixtures of SM(18:0)/DOPC/cholesterol and DSPC/DOPC/cholesterol (1:1:1 by moles) were used as host membranes. BodipyFL-PC (green) and Texas-red-DPPE (red) are typical Ld-domain markers. Bars, 20 µm. (A) CTMR-SM, in which CTMR was directly conjugated to the choline headgroup of SM(18:0), preferentially partitions into Lo domains, whereas CTMRneg-SM, in which CTMR was conjugated to the SM headgroup by way of the NEG moiety, partitions almost equally between Lo and Ld domains. The GUVs used consisted of SM/DOPC/cholesterol. (B) Both 488neg- and 594neg-SMs partitioned more into Lo domains than Ld domains, whereas 594neg-DOPC partitioned into Ld domains in phase-separated SM/DOPC/cholesterol-GUVs. (C) In GUVs consisting of SM/DOPC/cholesterol, 594neg-DSPC partitioned more into Lo domains than Ld domains. (D) In GUVs consisting of DSPC/DOPC/cholesterol, 594neg-DSPC preferentially partitioned into Lo domains. See Fig. 3 for the quantitative evaluation of the partitioning behavior.
To alleviate this possible problem, we next used a longer, hydrophilic linker, a nonaethylene glycol (NEG; n = 9) chain between the fluorophore and the SM choline headgroup, termed CTMRneg-SM (Fig. 1). It partitioned into the Ld and Lo domains equally (Fig. 2 A), suggesting quite improved partitioning behavior, although its Lo domain preference was much less than that of deuterated SM. However, this improvement suggested that placing the somewhat hydrophobic CTMR probe farther away from the membrane, by using the hydrophilic NEG linker, would be beneficial. Therefore, we replaced CTMR with more hydrophilic (i.e., more water soluble) dyes, ATTO488 and ATTO594, and kept the same NEG linker (488neg-SM and 594neg-SM, respectively; Fig. 1).
The 488neg- and 594neg-SMs preferentially partitioned into the Lo domain over the Ld domain by factors of 4.1 ± 0.19 and 3.9 ± 0.15 (mean ± standard error [SE], used throughout), respectively (Figs. 2 B and 3). This partitioning is similar to that of deuterated SM (18:0, without the fluorophore) in multilamellar vesicles with the same lipid composition and at the same temperature (Yasuda et al., 2015). This agreement suggests that these fluorescent SM analogs will faithfully mimic the native SM, in terms of partitioning into the Lo domain, and thus perhaps into various raft-related membranes/preparations. Therefore, in the next part of this article, we focus on the 488neg- and 594neg-SMs, rather than other fluorescently conjugated SMs.
Figure 3.
Quantitative analysis of confocal fluorescence microscopic images. In Lo-Ld phase–separated GUVs, 488neg-SM, 594neg-SM, and 594neg-DSPC preferentially partitioned into Lo domains at levels similar to that of deuterated SM (observed by solid-state NMR), whereas 594neg-DOPC preferentially partitioned into Ld domains. (A, top) Typical confocal fluorescent micrographs of the equatorial planes of Lo-Ld phase–separated GUVs (top). Bar, 20 µm. (bottom) Fluorescence signal intensity profiles along the equatorial circumferences of the GUVs are shown above. 594neg-DSPC was examined in Lo-Ld phase–separated GUVs consisting of SM(18:0)/DOPC/cholesterol and those made of DSPC/DOPC/cholesterol (1:1:1 by moles). (B) Ratio of the mean fluorescence signal intensity in the Lo domain versus that in the Ld domain (Io/Id), based on the signal intensity profiles shown in A (mean ± SE; n = 22, 15, 21, 22, and 21 GUVs from left to right).
Partitioning into DRMs prepared from cells
In the erythrocyte ghost membrane, 78% of the endogenous SMs detected by high-performance thin-layer chromatography remained in the DRMs, whereas 41% of the endogenous PC remained in the DRMs (Fig. S2 A). Meanwhile, epifluorescence microscopy revealed that 488neg-SM and 594neg-SM preincorporated in the PM entirely remained in the DRMs in the erythrocyte ghost as well as in CHO-K1 and T24 cells (Fig. S2, B–D). Therefore, in this prototypical biochemical assay, which suggests the raft association of molecules, both 488neg-SM and 594neg-SM behaved like typical raft-associated molecules.
Comparison with 594neg-DSPC and 594neg-DOPC
We next synthesized 594neg-DSPC (Fig. 1). DSPC has often been selected as a major phospholipid component to form Lo domains, together with cholesterol, in the Lo-Ld phase–separated GUVs (Zhao et al., 2007; Heberle et al., 2010). The headgroup of PC, phosphorylcholine, is the same as that of SM, and thus the headgroups of 594neg-DSPC and 594neg-SM are the same, containing the same charged groups and the same hydrophilic linker. Furthermore, the alkyl chains of 594neg-DSPC (18:0–18:0) and 594neg-SM (18:0 + the alkane 14-carbon chain of sphingosine) were quite similar. The major difference between them was the glycerol backbone of 594neg-DSPC and the sphingosine backbone of 594neg-SM (particularly the groups located close to the glycerol backbone in the membrane). Therefore, these two probes allowed us to examine whether this chemical structure difference affects their interactions with raft domains and other raft-associated molecules, a very interesting subject matter for understanding raft domain formation and dynamics. We found that 594neg-DSPC preferentially partitioned into the Lo domain over the Ld domain by a factor of 4.3 ± 0.24 (Figs. 1, 2 [C and D], and 3), which is comparable to 3.9 ± 0.15 for 594neg-SM.
Furthermore, in the partitioning assay using DRMs, 594neg-DSPC preincorporated in the PMs of the erythrocyte ghost, CHO-K1, and T24 cells almost entirely remained in the DRMs, just as with 488neg-SM and 594neg-SM, as determined by epifluorescence microscopy (Fig. S2, B–D). Collectively, 594neg-DSPC behaved like 594neg-SM (and 488neg-SM) in terms of the partitioning behaviors in both Lo-Ld phase–separated GUV membranes and DRMs. However, as will be described later, 594neg-SM and 594neg-DSPC exhibited very different behaviors in the PMs of live cells.
For comparison, we synthesized 594neg-DOPC and ATTO594-l-α-dioleoylphosphatidylethanolamine (594-DOPE; Fig. 1). DOPC and DOPE are typical nonraft molecules (Yasuda et al., 2015; Komura et al., 2016). 594neg-DOPC exhibited much less partitioning into the Lo domain than the Ld domain (by a factor of 0.14 ± 0.01; Figs. 2 B and 3), and the majorities (∼95%) of 594neg-DOPC and 594-DOPE were extracted by the cold-Triton treatment (Fig. S2, E and F).
Dynamics of SMs in the Lo and Ld domains of Lo-Ld phase–separated GUVs
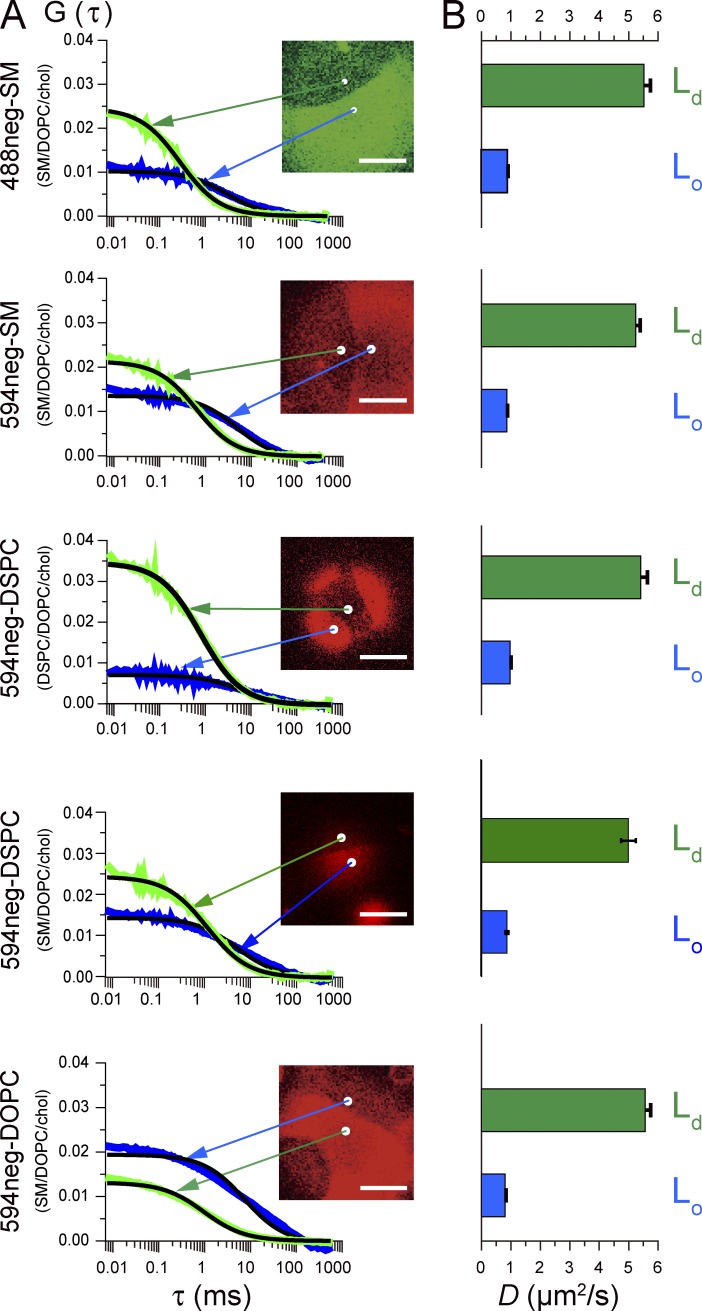
Our knowledge of diffusion coefficients of membrane molecules in the Lo and Ld domains in phase-separated GUV membranes is quite limited (for DiI-C18 data, see Kahya et al. [2003]). As shown in Figs. 2 and 3, because the fluorescent-NEG-conjugated SMs, DSPC, and DOPC partition into both the Lo and Ld domains in GUV membranes (although with quite different efficiencies), we measured their diffusion coefficient, D, in the Lo and Ld domains in the phase-separated GUVs using fluorescence correlation spectroscopy (FCS; Kahya et al., 2003; Fig. 4 A).
Figure 4.
Diffusion coefficients of 488neg-SM, 594neg-SM, 594neg-DSPC, and 594neg-DOPC in Lo and Ld domains in Lo-Ld phase–separated GUVs, determined by FCS. (A) Autocorrelation functions G(τ) values for the time-dependent changes of the signal intensities of the fluorescent probes observed at typical diffraction-limited areas (white spots) shown in the image on the right in Lo and Ld domains (the spots in the images are expanded 5× to show their locations clearly). The black solid smooth curves indicate the best-fit functions, providing the diffusion coefficients (D). Bars, 10 µm. 594neg-DSPC was examined in both Lo-Ld phase–separated GUVs consisting of SM(18:0)/DOPC/cholesterol and those made of DSPC/DOPC/cholesterol (1:1:1 by moles). (B) Mean ± SE values of D in the Ld and Lo domains (n = 24, 24, 31, 27, 24, 31, 25, 24, 25, and 25 GUVs from top to bottom). See also Figs. 2 and 3.
Mean ± SE values of D in the Lo and Ld domains are shown in Fig. 4 B. These values did not depend on the concentrations of the probes used in the present study. Notably, in each Lo and Ld domain, the diffusion coefficients of the Lo-preferring molecules (SM and DSPC analogs) and the Ld-preferring molecule DOPC were similar to each other. Their diffusion coefficients in the Ld domain were greater than those in the Lo domain by a factor of ∼5.9–7.1. The diffusion coefficients of phospholipid analogs evaluated here (28°C) were slightly greater than those of DiI-C18 (22°C) and were generally consistent with the pulsed field-gradient NMR data (Filippov et al., 2004).
Partitioning into the Lo-like versus Ld-like domains in phase-separated giant plasma membrane vesicles
In phase-separated giant plasma membrane vesicles (GPMVs) prepared from RBL-2H3 cells at 10°C, 488neg-SM, 594neg-SM, and 594neg-DSPC preferentially partitioned into the Lo-like domain over the Ld-like domain (here we used RBL-2H3 cells because the protocol for preparing GPMVs is well established [Baumgart et al., 2007]; Fig. 5). Meanwhile, 594neg-DOPC partitioned much less into the Lo-like domain than the Ld-like domain (Fig. 5). The preferential partitioning of 488neg- and 594neg-SMs and 594neg-DSPC into the raft-related membrane domains/preparations found in Lo-Ld phase–separated GUVs, DRMs, and phase-separated GPMVs suggests that these fluorescent phospholipid analogs might be useful for investigating SM involvement in raft formation and function.
Figure 5.
In phase-separated GPMVs from RBL-2H3 cells at 10°C, 488neg- and 594neg-SMs as well as 594neg-DSPC preferentially partitioned into Lo-like domains, whereas 594neg-DOPC partitioned into Ld-like domains. (A) Representative still images taken from simultaneous two-color video sequences. First and second rows, 594neg- and 488neg-SM exhibited partitioning complementary with Ld-domain marker molecules. Third and fourth rows, 488neg- and 594neg-SMs and 594neg-DSPC exhibited similar partitioning. (B) Ratio of the mean fluorescence signal intensity in the Lo-like domain versus that in the Ld-like domain (Io/Id; mean ± SE; n = 17, 22, 17, and 18 GPMVs from top to bottom). The same probe exhibited slightly different ratios because of variations in the leak signal from the other dye molecule in the pair. The mean ratios for each probe under various labeling conditions used here are as follows (mean ± SE, n = number of GPMVs examined): 488neg-SM, 4.5 ± 0.2 (n = 57); 594neg-SM, 4.5 ± 0.3 (n = 34); 594neg-DSPC, 4.5 ± 0.4 (n = 18); 594neg-DOPC 0.2 ± 0.02 (n = 22); and Bodipy FL-PC, 0.2 ± 0.02 (n = 17).
Distribution of 594neg-SM in the PMs of living cells
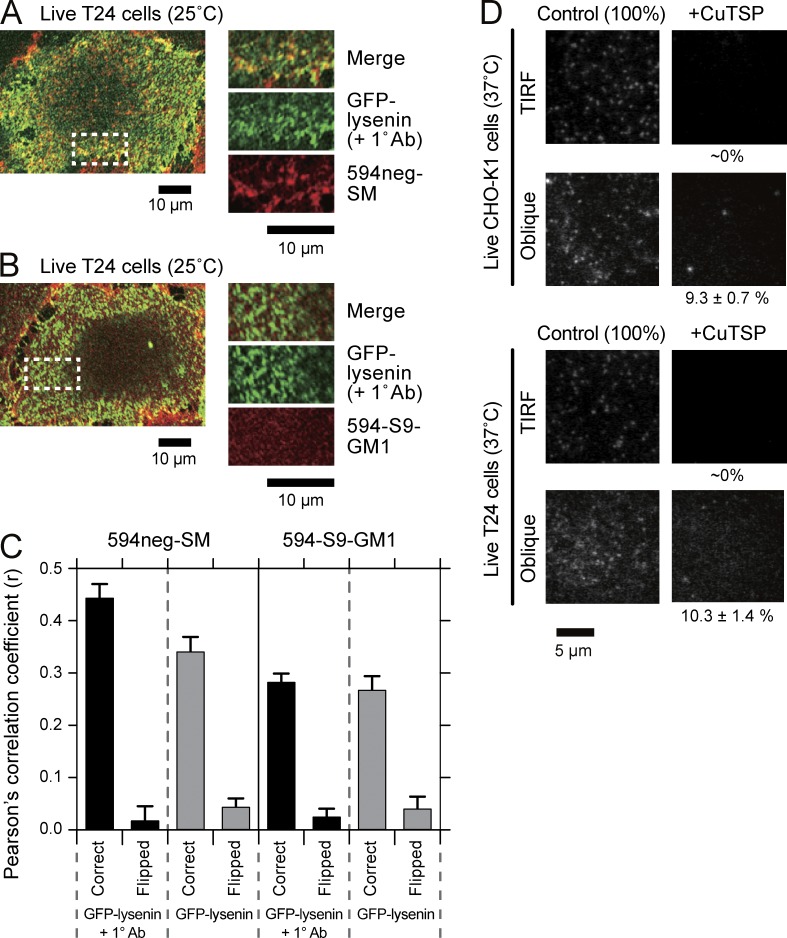
Lysenin is known to bind to SM, but because lysenin exhibited greatly enhanced affinities when SM is located in so-called raft domains (Kiyokawa et al., 2005; Abe and Kobayashi, 2014; Kishimoto et al., 2016), lysenin might not be an ideal probe for all the SM molecules distributed in the PM. However, because lysenin is still probably the best probe available for labeling endogenous SM, we observed whether 594neg-SM colocalized with exogenously added GFP-lysenin and GFP-lysenin clusters induced by the addition of polyclonal anti-GFP antibodies (Fig. 6 A; with a control using a fluorescent ganglioside analog 594-S9-GM1 [Fig. 6 B] described by Komura et al. [2016]; the density of 594neg-SM in the PM was ∼100 molecules/µm2). The Pearson statistical analysis (evaluated for the maximal rectangular regions that could be taken within each cell) showed a quite good correlation of 594neg-SM and GFP-lysenin (0.34 vs. the value for the negative control, 0.04; Fig. 6 C), although the colocalization of these two molecules was difficult to detect by visual examination (not depicted) because both GFP-lysenin and 594neg-SM were distributed rather homogeneously at the spatial resolution of confocal fluorescence microscopy.
Figure 6.
594neg-SM colocalizes with GFP-lysenin at multimolecular levels in PMs and resides in the PM outer leaflet; ∼10% is internalized 15 min after its application. (A and B) Representative confocal fluorescence images of GFP-lysenin and 594neg-SM (A) and 594-S9-GM1 (B) after the application of anti-GFP antibodies to T24 cells. All reactions before the observation were performed at 37°C, and the live cells were observed with a confocal fluorescence microscope at 25°C. (C) Pearson’s correlation coefficients (r) were evaluated for the maximal rectangular regions that could be taken within each cell. Correct, the images of GFP-lysenin and 594neg-SM or 594-S9-GM1 were compared spatially correctly. Flipped, one of the images was rotated by 180° and r was calculated. The difference in r before and after the addition of primary antibodies for 594neg-SM and that without the antibody addition between 594neg-SM and 594-S9-GM1 were statistically significant (P < 0.001). The numbers of images examined were 12 and 11 for 594neg-SM and 13 and 12 for 594-S9-GM1 before and after the addition of primary antibodies, respectively. Error bars indicate standard errors. (D) After incubating T24 and CHO-K1 cells with 594neg-SM for 15 min on the microscope stage, the cells were quickly observed by TIRF and oblique-angle illuminations with a focus near the ventral (basal) PM at single-molecule sensitivities. The membrane-impermeable quencher CuTSP was then added, followed by a second set of observations using TIRF and oblique-angle illuminations (37°C; same view field before and after CuTSP addition). For quantitative data for 594neg-SM, as well as 594neg-DOPC and 594-DOPE, and the method for evaluating percentage molecular fractions in the PM inner leaflet and the cytoplasm, see Table S1 and its notes as well as Materials and methods.
After the addition of polyclonal anti-GFP antibodies, which induce spatial fluorescent patterns (heterogeneous distributions) of GFP-lysenin, the colocalization of GFP-lysenin clusters with the 594neg-SM signals became visible, and Pearson’s correlation coefficient increased to 0.44 (Fig. 6 C).
This colocalization might simply be caused by the recruitment of any raft-associated molecules to the stabilized “rafts” formed by SMs cross-linked by GFP-lysenin and its antibodies. Therefore, as a control, the colocalization of GFP-lysenin and its clusters by another raft-associated molecule, 594-S9-GM1, was investigated. The results (Fig. 6, B and C) indicated colocalization, but the levels of colocalization were significantly smaller compared with those for 594neg-SM (P < 0.001; Fig. 6 C). Collectively, these results indicate that 594neg-SM colocalized with endogenous SM reasonably well in the live-cell PM.
A possible problem of this experiment is that substantial fractions of 594neg-SM might be bound by GFP-lysenin, and therefore, the colocalization might simply represent behaviors of 594neg-SM bound by GFP-lysenin. We examined whether GFP-lysenin binds 594neg-SM using an enzyme-linked immunosorbent assay (ELISA; Makino et al., 2015). The results indicated that, under the conditions where GFP-lysenin strongly binds brain SM but not egg PC (Kiyokawa et al., 2005), GFP-lysenin binds to 594neg-SM with an affinity of ∼60% of that of the GFP-lysenin to brain SM (the readouts of optical density at 490 nm were 0.47 ± 0.04, 0.038 ± 0.007, and 0.26 ± 0.02 for brain SM, egg PC, and 594neg-SM, respectively; mean ± SD, n = 4). These results clearly show that 594neg-SM is recognized by GFP-lysenin. This further supports the usefulness of 594neg-SM as a fluorescent SM analog.
Under the 594neg-SM incorporation conditions used for the multimolecular experiments shown in Fig. 6, the ratio of the molecular number of 594neg-SM versus that of endogenous SM in the PM outer leaflet is probably one to several hundred (for example, <1:400), assuming that the 594neg-SM number density is proportional to the added concentration of 594neg-SM and that ∼20% of the phospholipids in the PM outer leaflet are SM (and that all SM molecules reside in the outer leaflet). Therefore, the majority (>99% if the binding affinity of GFP-lysenin is the same for endogenous SM and 594neg-SM) of GFP-lysenin is bound to endogenous SM, and thus GFP-lysenin would represent the behavior of endogenous SM.
Meanwhile, from the fluorescence signal intensities of GFP-lysenin compared with those of 594neg-SM (see Fig. 6 A for typical images), GFP-lysenin concentration on the cell surface would be greater than 594neg-SM, but by less than a factor of 10. Therefore, 1 of several tens (say <1:40) of endogenous SM and 594neg-SM would be bound by GFP-lysenin, indicating that probably >97% of 594neg-SM molecules were not bound by GFP-lysenin.
Collectively, the 594neg-SM distributions on the cell surface shown in Fig. 6 are likely to represent behaviors of 594neg-SM that are mostly not bound by GFP-lysenin. Therefore, it is concluded that the spatial distribution of 594neg-SM in the PM is quite similar to that of endogenous SM.
594neg-SM, 594neg-DOPC, and 594-DOPE in the PM exclusively resided in the outer leaflet, whereas ∼10% of these molecules were internalized in 15 min
The lipid analogs were incorporated in the PM at densities of ∼0.5 molecules/µm2 for single-molecule observations. When a membrane-impermeable quencher, Cu(II)meso-tetra(4-sulfonatophenyl)porphine (CuTSP; Hiramoto-Yamaki et al., 2014), was added to the live T24 and CHO-K1 cells that had incorporated fluorescent lipid analogs, 594neg-SM, 594neg-DOPC, and 594-DOPE, for 15 min, their signals from the ventral PM by total internal reflection fluoroscopy (TIRF) illumination totally disappeared. This indicates that virtually all of the lipid analog molecules in the PM resided in the outer leaflet (Fig. 6 D and Table S1).
When the cells were observed with oblique-angle illuminations with a focus near/at the ventral PM after they had been incubated with the lipid analogs for 15 min, before and after the addition of CuTSP, ∼10% of the fluorescence signal intensity remained. When the microscope focus was shifted, no signal was detectable, indicating that no lipid analogs existed deeper in the cytoplasm. Therefore, these results indicate that ∼10% of the lipid analog molecules (all three species of lipid analogs) was internalized in 15 min (Fig. 6 D and Table S1). Because all our observations were finished within 15 min after the addition of the lipid analogs, <10% of the analog lipids would have existed in the cytoplasm during our observations. Furthermore, because these molecules in the cytoplasm were hardly visible by the TIRF illumination used in the present study, the results reported in this paper would not be affected by the internalized fluorescent lipid analogs.
Cholesterol-dependent homo-colocalization of SM, but not DSPC, in the live-cell PM
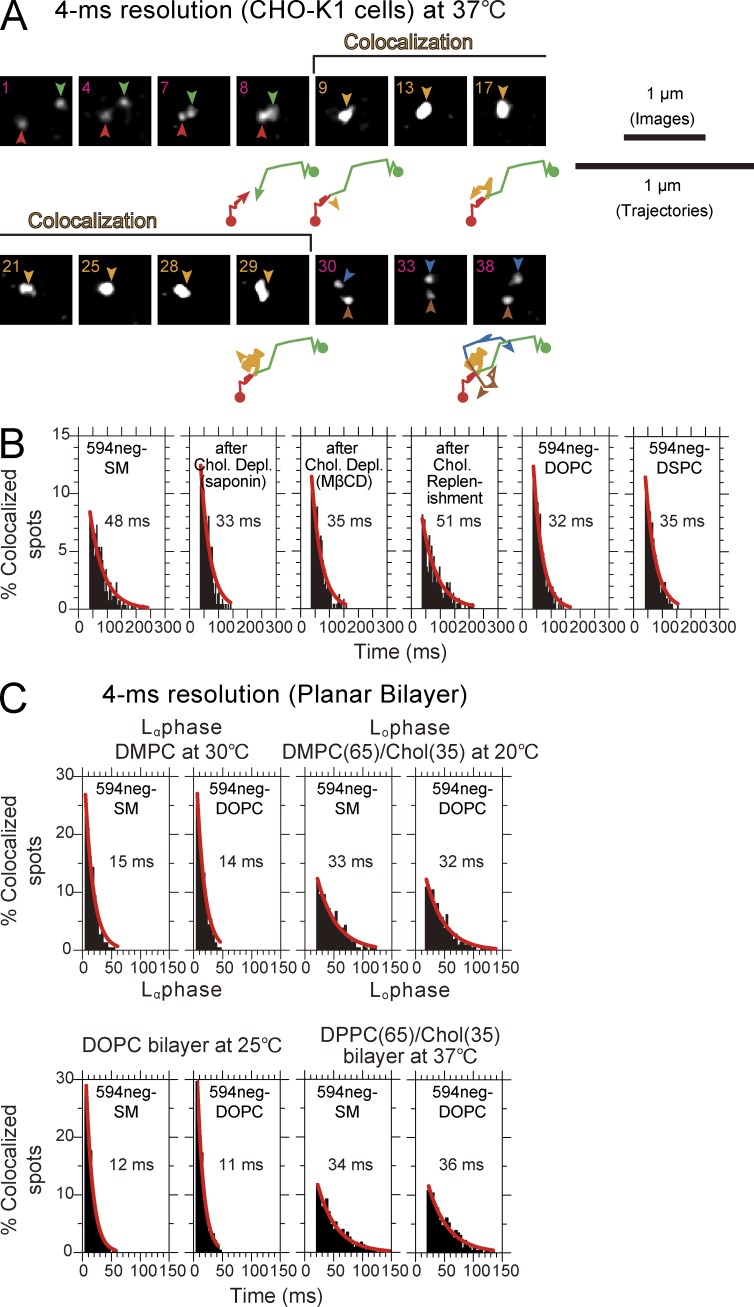
594neg-SM molecules were incorporated in the PM of live CHO-K1 cells, and their behaviors were observed at the level of single molecules at a time resolution of 4 ms. Single 594neg-SM molecules in the CHO-K1-cell PM exhibited frequent temporary colocalization (Fig. 7 A). (Colocalization of two fluorescent single molecules was defined as the event where two fluorescent spots become localized within 240 nm of each other, as described in Koyama-Honda et al. [2005], Kasai et al. [2011], and Suzuki et al. [2012]. Using this definition, the probability that two molecules located at exactly the same position are found to be colocalized by this method will be 99.7%.) Each time we found a colocalization event, we measured its duration, and after observing many colocalization events, we obtained the histogram showing the distribution of the colocalization durations (Fig. 7 B, left). The distribution could be fitted by a single exponential decay function, with a decay time constant of 48 ± 3 ms.
Figure 7.
Transient (∼15-ms) molecular-scale colocalization of two 594neg-SM molecules at raft-related structures in the CHO-K1-cell PM at 37°C, recorded at a 4-ms resolution, which was undetectable in artificial lipid bilayers. (A) Typical image sequence of two diffusing 594neg-SM molecules (red and green arrowheads before colocalization; blue and brown arrowheads after colocalization) undergoing transient colocalization and codiffusion (orange arrowheads). The number in the upper-left corner in each image indicates the number of frames. Trajectories are shown in a twofold magnified scale (displayed intermittently). The initial points of the trajectories are shown by closed circles. The end points of the trajectory shown by the arrowheads matched the fluorescent spots indicated by arrowheads in the image right above the trajectory. The green, red, orange, dark-blue, and brown colors indicate molecules (colors matched with those in the arrows in the images) before, during, and after colocalization, respectively. (B) Duration distributions for individual homo-colocalization events for single molecules of 594neg-SM (under various conditions), 594neg-DOPC, and 594neg-DSPC. All of the distributions could be fitted by single-exponential decay functions, but because the short molecular interaction lifetimes, the durations for incidental colocalization and those for colocalization including the molecular-scale interaction could not be separated. The decay time constants are shown in the boxes. The number of the observed colocalization events were 260, 223, 237, 220, 206, and 243 from left to right. (C) The duration distributions of individual homo-colocalization events for single 594neg-SM and 594neg-DOPC molecules in artificial planar bilayers in Lα or Lo phase observed at a 4-ms time resolution, showing that the homo-colocalization of 594neg-SM, which was prolonged from that of 594neg-DOPC, in the cellular PM (48 vs. 33 ms), did not occur in either Lo or Lα artificial planar lipid bilayers. For Lα membranes, DMPC bilayers (30°C) and DOPC bilayers (25°C) were used, whereas for Lo membranes, DMPC-cholesterol (65:35 mol/mol; 20°C) and DPPC-cholesterol (65:35 mol/mol; 37°C) were used. The numbers of the observed colocalization events were 223, 225, 206, and 210 from left to right in the top row and 221, 243, 227, and 231 from left to right in the bottom row.
When both 488neg- and 594neg-SM were incorporated in the PM and dual-color single fluorescent-molecule tracking was performed, 488neg- and 594neg-SM became colocalized, with a colocalization lifetime of 46 ± 3 ms (Fig. S3, A and B), which is comparable to the 594neg-SM’s homo-colocalization lifetime of 48 ms. Because these colocalization lifetimes were much less than the photobleaching lifetimes of ATTO488 and ATTO594 (0.49 s [123 frames] and 2.2 s [550 frames], respectively, at a frame rate of every 4 ms; Suzuki et al., 2012), no correction for photobleaching was conducted.
Partial cholesterol depletion from the CHO-K1-cell PM, using saponin or methyl-β-cyclodextrin (MβCD), reduced the homo-colocalization lifetime of 594neg-SM (single-color experiment) to 33 or 35 ms (Fig. 7 B), respectively, which was significantly shorter than that under control conditions (P < 0.001; log-rank test) and comparable to that for 594neg-DOPC (32 ms; P > 0.4; Fig. 7 B). Cholesterol replenishment after the MβCD treatment restored the SM colocalization lifetime found under control conditions (51 ms, P < 0.001). The lifetime of hetero-colocalization of 488neg-SM with 594neg-DOPC was 30 ms (Fig. S3 B), which was significantly shorter than the homo-colocalization lifetime of SM molecules (P < 0.001) and comparable to the colocalization lifetime of two 594neg-DOPC molecules as well as that of two 594neg-SM molecules after partial cholesterol depletion (P > 0.4).
These results suggest (a) that SM homo-colocalization is mediated by raft–lipid interactions involving cholesterol, and (b) that the ∼30–35-ms colocalization lifetime (a mean value of 33 ms will be used in this article) would represent the duration of incidental colocalization, in which two molecules incidentally encounter within a distance of 240 nm, the optical diffraction limit, without any molecular-level interactions, and thus would provide the baseline for the colocalization lifetimes. (The diffusion coefficients of 594neg-SM, 594neg-DOPC, and 594neg-DSPC, in CHO-K1 cells in the time window of 18 ms were practically the same [0.64 µm2/s for n ≥ 193], and thus the incidental colocalization time of 33 ms would be the same for all three of the lipid analogs. It might be ideal if the colocalization time could be predicted by a Monte Carlo simulation, but such a prediction would not be robust to the details of the simulation methods and parameters, particularly variations of diffusion coefficients for individual single molecules [perhaps because of spatial variations of the PM structure and dynamics], and thus it was not performed. See the following section.) Therefore, the actual duration for the cholesterol-mediated homo-interactions of the SM is considered to last for 15 ms (48 − 33 ms). These results further confirm that the transient SM colocalization was not driven by the interaction of the dyes conjugated to SM. Because the PM contains abundant native SMs, homo-colocalization of the fluorescent SM analogs found here suggests frequent and transient occurrences of cholesterol-dependent nano- to mesoscale (<240 nm in diameter) domains enriched in SMs. Meanwhile, other more complicated mechanisms that are affected by cholesterol depletion are possible, and this should always be kept in mind (Table S2, see note §).
Surprisingly, the colocalization lifetime for two single 594neg-DSPC molecules was only 35 ms, which is much shorter than that for 594neg-SMs (48 ms; P < 0.001), and comparable to that for 594neg-DOPC (32 ms; P > 0.4; Fig. 7 B). This suggests the possibility that the occurrence of prolonged colocalization of two SM molecules depends not only on raft-lipid interactions but also on SM structure–specific interactions, such as those involving the sphingosine backbone (rather than the glycerol backbone of glycerophospholipids).
SM homo-colocalization is not linked to SM immobilization or entrapment within nanodomains
Previously, a nonraft fluorescent SM analog was found to be trapped quite often (>50% of the time) in SM-specific stationary membrane domains of <20 nm in diameter in the PMs of PtK2 cells, at both 23°C and 37°C (Sahl et al., 2010; Honigmann et al., 2014). Therefore, we examined whether 594neg-SMs, 594neg-DSPC, and 594neg-DOPC undergo such confinement in the PM of the same cell line PtkK2 under the same conditions (as well as in the PM of T24 cells; it was difficult to incorporate 594neg-DSPC in the PM of CHO-K1 cells, and thus T24 cells were used here). The time resolution was also set at the same value (0.5 ms), but in contrast to the previous method where only a single molecule was tracked at a time (without imaging), we simultaneously visualized many single molecules in a ∼10 × 10-µm field at a time resolution of 0.5 ms, which is the fastest in single-molecule imaging.
No significant entrapment/immobilization longer than 5 ms within <20-nmφ domains was detectable with any of these lipid analogs (Table S3). Consistently, the ensemble-averaged behavior suggested simple Brownian diffusion for 594neg-SM, 594neg-DSPC, and 594neg-DOPC on the time scale between 1 and 25 ms (Fig. S4, A and B) with virtually the same mean diffusion coefficients at both 23°C and 37°C in both T24 and PtK2 cells (P > 0.4; Table S4 and Fig. S5 A, histograms). If 594neg-SM had been entrapped in special static nanoscale domains for significant time fractions, its mean diffusion coefficient would have been significantly smaller than that of 594neg-DOPC. In addition, we found that 594neg-SM and 594neg-DOPC diffused ∼1.8 times faster than 594-DOPE (P < 0.002; Fig. S5 B), in which ATTO594 was directly linked to the amine residue in the headgroup (Fig. 1) and thus changed the charge of the headgroup from neutral to −2 (ATTO594 has a negative charge). Although the reason for such a difference is unknown, this result suggests that 594neg-DOPC could be a better phospholipid probe than 594-DOPE.
Homo-colocalization of the fluorescent SM does not occur in artificial planar membranes, at variance with its behavior in the PM
We wondered whether the 594neg-SM homo-colocalization represents the formation of true 594neg-SM homodimers based on their direct molecular interaction (but that involving cholesterol) or simple colocalization within a nanodomain (with sizes smaller than the diffraction limit). To examine this issue, 594neg-SM was observed at the level of single molecules, in artificial homogeneous planar bilayers consisting of l-α-dimyristoylphosphatidylcholine (DMPC) or l-α-dipalmitoylphosphatidylcholine (DPPC) and cholesterol (65:35) in the Lo phase (20°C or 37°C, respectively) and those consisting of only DMPC or only DOPC in the Lα phase (30°C or 25°C, respectively). (The planar lipid bilayers rather than GUVs were used in this investigation because single-molecule colocalization observation was difficult to perform in GUVs.)
The homo-colocalization lifetimes of 594neg-SM were the same as those of 594neg-DOPC, a typical nonraft lipid, in all artificial bilayers (32–36 and 11–15 ms in Lo and Lα bilayers, respectively; P > 0.4 between 594neg-SM and 594neg-DOPC in both Lo and Lα membranes; Fig. 7 C). Namely, the prolonged homo-colocalization of 594neg-SM, compared with 594neg-DOPC, detected in the cellular PM (48 vs. 33 ms) did not occur in either Lo or Lα artificial planar lipid bilayers.
These results indicate that the homo-colocalization of SM molecules in the PM is prolonged, compared with that under the conditions of partial cholesterol depletion or that between DOPC molecules, by 15 ms (48 − 33 ms), but this is not induced by (cholesterol-induced) SM dimerization. Instead, the results suggest that two SM molecules were held together at distances less than 240 nm by interactions with nano- to mesoscale structures or molecular complexes containing cholesterol in the PM. Such structures/complexes might resemble raft domains, but perhaps a particular subset of raft domains includes at least one other molecular species that, together with cholesterol, induces cholesterol-dependent prolonged SM colocalization. Because the raft-induced elongation of the colocalization lifetime for two single 594neg-DSPC molecules did not occur even in the PM (Fig. 7 B), this third molecular species in the PM might work by interacting with the SM-specific structure, such as the sphingosine backbone (rather than the glycerol backbone of glycerophospholipids). We primarily explained these results based on particular raft–lipid interactions that are closely linked to SM-specific structures like the sphingosine backbone, but other more complicated mechanisms that are affected by cholesterol depletion might be possible, including cholesterol-dependent, but more direct, lipid–protein interactions and cholesterol-based membrane topological effects.
Recruitment of SM to monomers and transient homodimer rafts of CD59, a GPI-anchored protein
In the PM of quiescent cells before extracellular stimulation, all of the GPI-anchored proteins examined thus far were found to be mobile almost all the time. In addition, they continually formed transient (∼200-ms lifetime) homodimers through ectodomain protein interactions, stabilized by the presence of the GPI-anchoring chain and cholesterol (termed GPI-anchored protein’s homodimer rafts; GPI-anchored proteins are forming transient homodimer rafts and homodimers are dissociating to monomers all the time; Suzuki et al., 2012). Using CD59 as a representative GPI-anchored protein, we examined whether 594neg-SM molecules are recruited to CD59 monomers and transient homodimer rafts. We fused the cDNA encoding acyl-carrier protein (ACP) to the human CD59 cDNA (at its N terminus), expressed the fused protein ACP-CD59 in CHO-K1 cells, which lack endogenous CD59, and labeled ACP-CD59 with ATTO488. This way, ∼95% of the CD59 in the PM could be fluorescently labeled, making possible the clear observation of ACP-CD59 monomers and dimers. Then, we simultaneously observed single molecules of 594neg-SM and ATTO488-ACP-CD59 in the PM of live CHO-K1 cells.
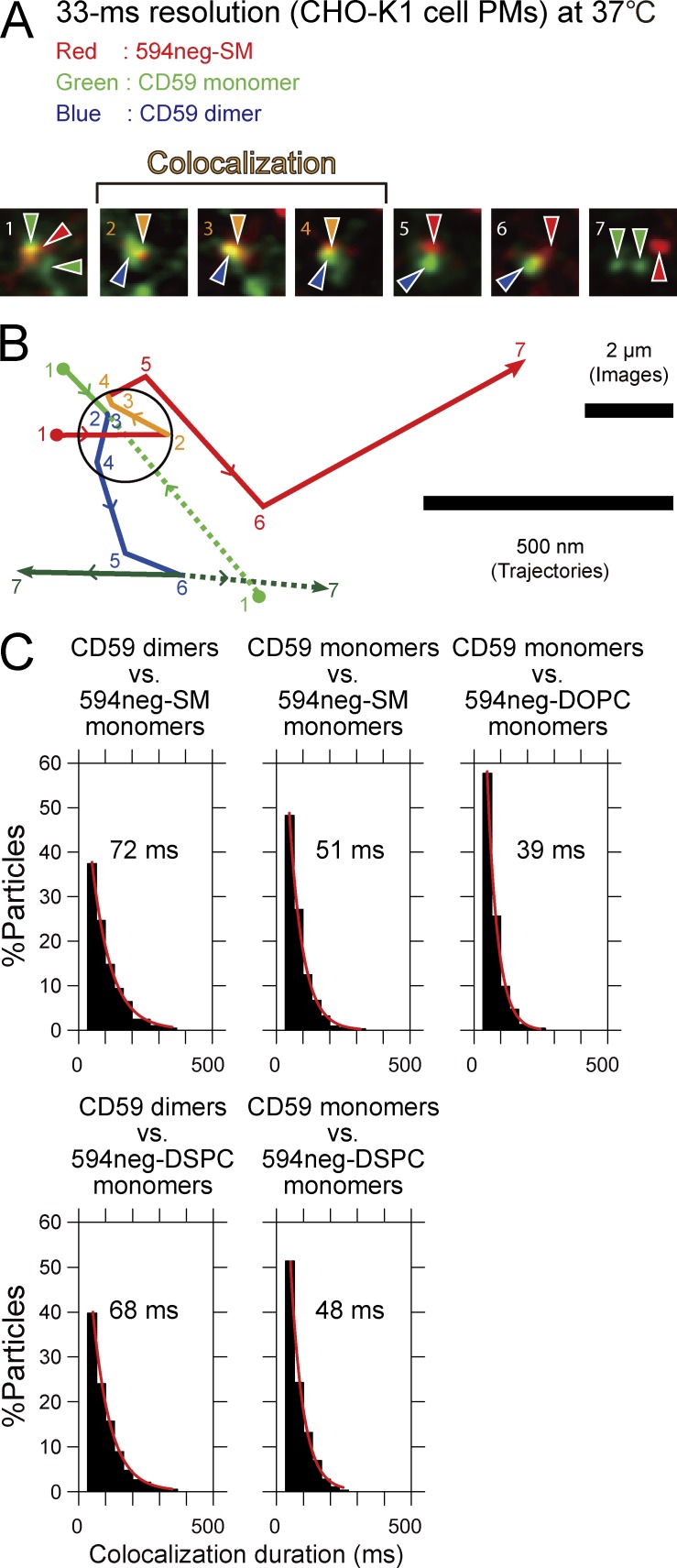
Single 594neg-SM molecules were found to become colocalized with CD59 monomers transiently and frequently (Fig. 8, A and B). The colocalization duration distribution could be fitted by an exponential function, with a decay lifetime of 51 ms (Fig. 8 C and Table S5). Meanwhile, 594neg-DOPC exhibited a colocalization lifetime of 39 ms (P = 0.005), which was considered to be the duration for an incidental encounter of the two molecules. Therefore, we concluded that 594neg-SM molecules interact with CD59 monomers, either directly or indirectly, with a lifetime of 12 ms (51 − 39 ms).
Figure 8.
Single molecules of 594neg-SM were frequently and transiently recruited to CD59 monomers and CD59-homodimer rafts in quiescent cells for 12 and 33 ms, respectively. (A) Typical superimposed videoframe sequences of a 594neg-SM molecule (red spot) and two CD59 molecules (green spots) in the CHO-K1-cell PM, observed at a 33-ms resolution. Two individual CD59 monomers formed a dimer and diffused together (distances ≤240 nm) for the period of four video frames (between frame 2 and frame 6), and simultaneously, these CD59 molecules became colocalized with a 594neg-SM molecule between frame 2 and frame 4. (B) Trajectories of the molecules shown in A. The positions of individual molecules in each image frame shown in A are indicated by the frame numbers. Color coding: 594neg-SM in red (orange during colocalization with CD59); CD59 monomers in green solid and dotted lines, and homo-colocalized CD59 molecules (CD59 homodimer raft) in blue. Colocalization of 594neg-SM and CD59 is shown in the circle, with a 240-nm diameter (see Online supplemental material). (C) Colocalization duration distributions of single molecules of 594neg-SM (+ 594neg-DSPC and 594neg-DOPC) and CD59 monomers or CD59 homodimer rafts. Single-exponential fitting of each histogram provided the colocalization lifetime, which is shown in each box. The numbers of the observed colocalization events were 203, 313, 234, 191, and 288 from left to right in the top row and then the bottom row.
Furthermore, 594neg-SM molecules were colocalized with CD59 homodimer rafts (72 ms) significantly longer than with CD59 monomers (51 ms; P < 0.001; Fig. 8, A–C; and Table S5). This result suggests that 594neg-SM molecules interact with CD59 homodimer rafts, either directly or indirectly, with a lifetime of 33 ms (72 − 39 ms). These interaction lifetimes (12 and 33 ms) are much shorter than the CD59 homodimer raft lifetime (160 ms; Suzuki et al., 2012) or the photobleaching times of ATTO488 and ATTO594 (0.49 and 2.2 s, respectively), and therefore, no corrections were applied.
Because many other raft-associated molecules must exist in the PM, the CD59 monomers might also be (transiently) associated with raft domains. Therefore, the 594neg-SM interaction with CD59 monomers, which is prolonged by 12 ms from the duration of the DOPC interaction with CD59 monomers, will probably be induced by the raft–lipid interaction. Meanwhile, a 594neg-SM interaction duration with CD59 homodimer rafts of 33 ms is approximately three times longer than that with CD59 monomers (or CD59-monomer containing rafts, 12 ms). Such prolongation, as well as the similar extent of prolongation for both SM and DSPC, suggests that the raft–lipid interaction in CD59 homodimer rafts enhanced by CD59’s protein–protein interactions (Suzuki et al., 2012) might have prolonged the SM residency time in CD59 homodimer rafts, compared with the SM residency time with CD59 monomers or that in the raft domains with which CD59 monomers associate.
The colocalization lifetimes of 594neg-DSPC with CD59 monomers and homodimer rafts were very similar to those of 594neg-SM (P > 0.3; Fig. 8 C and Table S5), clearly indicating that 594neg-DSPC behaves like 594neg-SM in terms of its interactions with CD59 monomers and homodimer rafts (as well as with CD59 clusters, as shown in the next section), and further supporting the proposal that the interactions observed for 594neg-SM with CD59 are “raft-based” and not dependent on the sphingosine backbone (also see Table S5, note ∞).
Recruitment of SM to engaged CD59-cluster signaling rafts
Upon stimulation, CD59 forms clusters containing four to five CD59 molecules in a cholesterol-dependent manner, and these CD59 clusters in turn transiently recruit trimeric G proteins, Src-family kinases, and PLCγ2 and trigger intracellular IP3-Ca2+ responses (Suzuki et al., 2007a,b). The dynamics of the ligand-induced CD59 clusters and their downstream signaling induction were mimicked very well by the CD59 clusters consisting of a mean of five CD59 molecules, generated by 40-nmφ fluorescent beads coated with anti-CD59 monoclonal antibody IgG (Suzuki et al., 2007a,b). Therefore, both of these CD59 clusters have been called “CD59-cluster signaling rafts.”
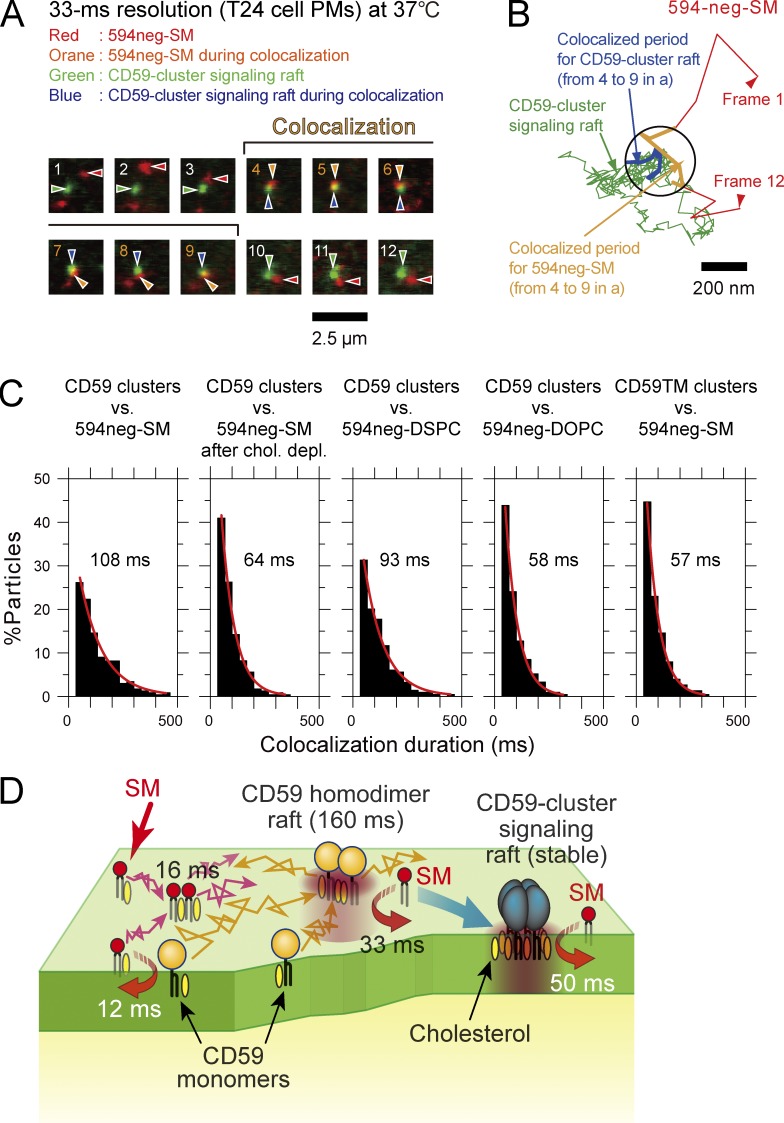
In the present study, we performed simultaneous two-color single-molecule imaging-tracking of 594neg-SM molecules and fluorescent bead–induced CD59-cluster signaling rafts and found that 594neg-SM molecules became colocalized with CD59-cluster rafts transiently but frequently (Fig. 9, A and B; and Table S5, note ¶). The histogram of the colocalization durations could be fitted by an exponential function, with a decay lifetime of 108 ms (Fig. 9 C and Table S5). The colocalization lifetime of 594neg-DOPC and CD59-cluster signaling rafts was significantly shorter (58 ms; P < 0.0001; Fig. 9 C). This lifetime is considered to be the duration of incidental colocalization. This colocalization lifetime of 58 ms was longer than the incidental colocalization lifetime between 594neg-DOPC molecules and CD59 monomers of 39 ms, probably because the diffusion of CD59-cluster signaling rafts was at least 10 times slower than that of CD59 monomers (Suzuki et al., 2007a,b). Therefore, the interaction lifetime of 594neg-SM and CD59-cluster signaling rafts was estimated to be 50 ms (108 − 58 ms).
Figure 9.
Single molecules of 594neg-SMs were frequently and transiently recruited to CD59-cluster signaling rafts for 50 ms. (A) Typical superimposed videoframe sequences of a CD59-cluster signaling raft and a single molecule of 594neg-SM, including a colocalization period (frames 4–9), observed at a 33-ms resolution. (B) Trajectories of the CD59-cluster raft and the single molecule of 594neg-SM shown in A. Colocalization within 240 nm (circle) is shown in the orange (594neg-SM) and blue (CD59-cluster raft) trajectories. (C) Duration distributions of colocalization of CD59-cluster-rafts (and CD59TM clusters as a control) by single molecules of 594neg-SM, 594neg-DSPC, and 594neg-DOPC. The exponential colocalization decay time is shown in each box. The numbers of the observed colocalization events were 233, 232, 214, 212, and 226 from left to right. (D) Schematic figure showing four types of transient interactions of SM with SM and CD59 in the PM. (1) Two SM molecules interact by way of a third molecule in a manner dependent on cholesterol and the sphingosine backbone (16-ms lifetime). SM molecules, in addition, transiently associate with CD59 monomers (2; monomer-associated rafts; 12 ms), CD59 transient homodimer rafts (3; whose lifetime is ∼160 ms; 33 ms), and CD59-cluster signaling rafts (4; whose lifetimes are on the order of several to several tens of minutes; 50 ms), in a cholesterol-dependent manner (in a manner dependent on the raft–lipid interaction).
After cholesterol depletion, the lifetime of colocalization between 594neg-SM and CD59-cluster signaling rafts was reduced to 64 ms (Fig. 9 C and Table S5), significantly shorter than that under control conditions (108 ms, P < 0.0001), suggesting that the raft–lipid interaction is important to induce prolonged colocalization between 594neg-SM and CD59-cluster signaling rafts. This was further confirmed by using CD59TM, in which the GPI-anchoring chain was replaced by the transmembrane (TM) domain of the LDL receptor, which is a prototypical nonraft molecule; i.e., we evaluated the colocalization lifetime between 594neg-SM and myc-CD59TM clustered by 40-nmφ beads coated with anti-myc monoclonal antibody IgG. The colocalization lifetime was decreased to 57 ms (Fig. 9 C), which is significantly shorter than that for the control pair of 594neg-SM and CD59-cluster rafts (P < 0.0001) and is comparable to that for the control pair after cholesterol depletion (P > 0.4). These results suggest that the raft-based lipid interaction is important for inducing 594neg-SM colocalization with CD59-cluster signaling rafts (as well as with other forms of CD59; see Fig. 9 D).
Is the sphingosine backbone involved in the interaction of 594neg-SM with CD59-cluster signaling rafts? The lifetime of colocalization of 594neg-DSPC with CD59-cluster signaling rafts was 93 ms, which was significantly longer than that of 594neg-DOPC (58 ms; P = 0.0002) and slightly shorter or comparable to that of 594neg-SM (108 ms; P = 0.05). Collectively, these results suggest that CD59-cluster signaling rafts recruit 594neg-SM largely through raft–lipid interactions, with the slight involvement of the sphingosine interaction. This mechanism is quite different from that of the SM homo-colocalization, which strongly depends on the sphingosine backbone interactions.
Recruitment of SM to engaged FcεRI clusters but not to FcεRI monomers in quiescent cells
We further examined the colocalization of 594neg-SM with a second PM protein, the high-affinity receptor for Fcε, FcεRI, before and after the multivalent antigen ligation, which induces receptor clustering (Fig. 10, A and B). This experiment is biologically interesting because the issue of whether FcεRI is associated with so-called raft domains has been quite controversial (Holowka et al., 2005; Baumgart et al., 2007).
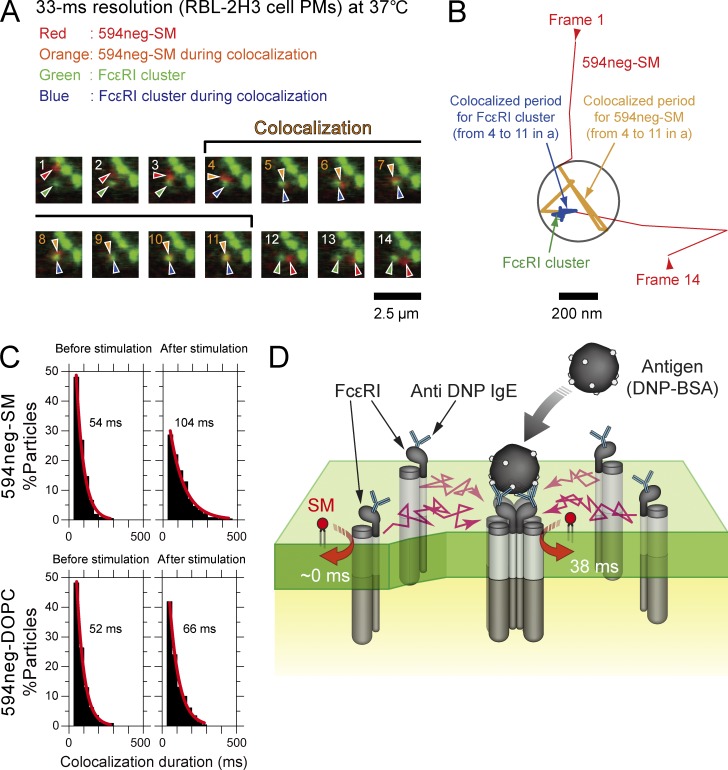
Figure 10.
Single molecules of 594neg-SM were not recruited to FcεRI monomers in quiescent cells, but were recruited to FcεRI clusters induced by multivalent antigens. (A) Typical superimposed videoframe sequences of FcεRI clusters induced by multivalent antigens and a single molecule of 594neg-SM. (B) Trajectories of the FcεRI cluster and the single molecule of 594neg-SM, pointed out by the arrows in A. Colocalization within 240 nm (circle) is shown in the orange (594neg-SM) and blue (FcεRI cluster) trajectories. (C) Duration distributions of the colocalizations of single molecules of 594neg-SM and 594neg-DOPC with FcεRI monomers (before multivalent antigen stimulation) or FcεRI clusters (after stimulation). The exponential colocalization decay time is shown in each box. The number of the observed colocalization events were 212, 241, 224, and 230 from left to right in the top row and then the bottom row. (D) Schematic figure showing transient interactions of SM with FcεRI clusters (after multivalent antigen stimulation) in the PM, but not with FcεRI monomers (before stimulation).
In quiescent mast cells (RBL-2H3 cells), the colocalization lifetime of 594neg-SM with FcεRI (monomers) was as short as that of 594neg-DOPC (54 and 52 ms, respectively; Fig. 10 C). However, after the addition of a multivalent antigen (DNP-conjugated BSA), which induced FcεRI oligomers (under the conditions used here, ∼50% and ∼25% of FcεRI molecules existed in clusters of sizes between dimers and tetramers and in those containing five or more molecules, respectively, whereas ∼25% remained as monomers, according to Andrews et al. [2009]), the colocalization lifetime of 594neg-SM with FcεRI oligomers was dramatically prolonged (104 ms; Fig. 10 C and Table S6). After subtracting incidental colocalization lifetimes, evaluated by using 594neg-DOPC (52 and 66 ms before and after the antigen addition), the interaction lifetimes of 594neg-SM with FcεRI monomers and ligated oligomers were estimated to be ∼0 and 38 ms, respectively (Fig. 10 D). These results suggest that FcεRI becomes capable of recruiting SMs for periods longer than simple collision only after ligand-induced clustering, perhaps because of raft–lipid interactions enhanced by the clustering of FcεRI molecules.
Discussion
Fluorescent SM analogs, in which rather hydrophilic fluorophores are linked to the SM headgroup choline group (without varying its positive charge) via a hydrophilic NEG linker, were developed. Single-molecule tracking of these probes revealed that SM interactions with raft domains and raft-associated molecules, including the raft domains containing SM molecules and CD59 monomers, CD59’s transient homodimer rafts in quiescent cells, and engaged CD59-cluster signaling rafts, occur dynamically on the order of 10 ms in cholesterol-dependent and GPI anchorage–dependent manners. Furthermore, it revealed that SM interacts with engaged FcεRI clusters, whereas it hardly does with nonengaged FcεRI. These results suggest that the fluorescent SM analogs developed here will be extremely useful for studying SM interactions with raft-associated molecules and raft domains, as well as for investigating membrane organization and transport (Diaz-Rohrer et al., 2014; Simons, 2016; Zurzolo and Simons, 2016). In particular, transient homo-colocalization of two SM analogs that occurred in manners dependent on both cholesterol and the sphingosine backbone (in the presence of abundant native nonfluorescent SMs) suggests that several SM molecules frequently and transiently assemble by sphingosine-based interaction with a third unknown molecules in the nano- to mesoscale (<240-nmφ) raft-like structures.
High-resolution secondary ion mass spectrometry analysis with spatial resolutions of 50–100 nm, after fixation with glutaraldehyde and osmium tetroxide, previously detected sphingolipid microdomains with mean diameters of ∼200 nm in the PM (Frisz et al., 2013; Wilson et al., 2015), and therefore, the SM homo-colocalization detected in this investigation might occur in some of these 200-nm sphingolipid microdomains. Frisz et al. (2013) further found that partial cholesterol depletion reduced the number of sphingolipid domains, which might be related to the shorter SM homo-colocalization lifetimes in cholesterol-depleted PMs.
General design strategies for fluorescent phospholipid analogs
We propose the following general design strategies for fluorescent SM and PC probes, which could be useful for synthesizing fluorescent analogs of other phospholipids: (a) attach a fluorophore to the phospholipid headgroup with the fewest modifications of the charged groups, certainly keeping the original ionic state; (b) use a hydrophilic linker with a proper length, such as NEG, between the fluorophore and the phospholipid headgroup; and (c) use fluorophores more hydrophilic than CTMR. All of these factors appear to contribute to keeping the fluorophore away from the hydrophobic region of the parent phospholipid to which the fluorophore is linked. This point, the absence of any disturbance of the hydrophobic domain, will be particularly important for phospholipids involved in raft domains. These three strategies will also be useful for generating phospholipids conjugated by biotin or photoaffinity probes.
Previously, Honigmann et al. (2013) used a much longer polyethylene glycol linker (n = 45) for the conjugation of a fluorophore to phosphatidylethanolamines (unsaturated, long-chain [18:1cis] DOPE and saturated, long-chain [18:0] DSPE). These probes lacked the native molecule’s positive charge at the ammonium group, unlike the method used here. However, the authors found that fluorescent DSPE preferentially partitioned into the Lo domain over the Ld domain by a factor >4, whereas fluorescent DOPE did so at levels of <0.33, indicating that these fluorescent PE probes largely preserved the partitioning behaviors of native PEs. These results further suggested that, to keep the relatively hydrophobic fluorescent probes away from the hydrophobic domain of the membrane, using a hydrophilic linker (between the lipid headgroup and the fluorescent probe) with the proper length might be the most important factor among the three strategies. Nevertheless, we raise the possibility that a polyethylene linker as long as n = 45 might create problems as a lipid reporter, because it might be susceptible to steric hindrance effects from various structures on the PM outer surface, or it might make the lipid probe too hydrophilic to detect changes in the hydrophobic PM domains.
Interaction of SM with CD59 monomers, homodimer rafts, and cluster signaling rafts
594neg-SM dynamically interacts with all three forms of CD59, monomers and homodimer rafts in quiescent cells, and CD59-cluster signaling rafts (containing approximately five CD59 molecules) after stimulation, with lifetimes of 12, 33, and 50 ms, respectively. In all cases, the interactions are raft-based. Unlike the SM–SM interactions (homo-colocalization), the involvement of the sphingosine backbone in the SM-CD59 colocalization (interaction) was limited.
The SM molecules might enter the nanoscale raft domains created by CD59 monomers, transient CD59 homodimers (homodimer rafts), and CD59-cluster signaling rafts as a result of raft-based lipid interactions, stay there for periods on the order of 10 ms, and then leave promptly (Fig. 9). The CD59-cluster signaling raft might persist for a long time (several minutes to several tens of minutes; Suzuki et al., 2012; Hiramoto-Yamaki et al., 2014), but its constituent SM molecules exchange continually with those in the bulk domain, with a residency time within the CD59-cluster raft of 50 ms (Fig. 9).
These results are in general agreement with recently reported data showing that GPI-anchored receptors moved in and out of the domain formed by monoclonal antibody–induced dimers of another GPI-anchored receptor species (small, but stabilized raft domains; Huang et al., 2015). More recently, the gangliosides GM1 and GM3 were found to transiently interact with all forms of CD59 in the PM, on time scales very similar to those found here (12, 40, and 48 ms with CD59 monomers, homodimer rafts, and cluster signaling rafts, respectively, vs. SM interaction durations of 12, 33, and 50 ms, respectively), as a result of raft–lipid interactions via their alkyl chains, without any specific interaction between the ganglioside glycochains and the ecto glycoprotein domain of CD59 (Komura et al., 2016). Because the SM results obtained in the present research are quantitatively similar to the results obtained previously with gangliosides, together these results suggest that the raft–lipid interactions involving cholesterol are more important for the interactions of SM or glycosphingolipids with CD59, rather than the interactions of the sphingosine backbone or the headgroups of the lipids (ganglioside glycochains and the SM’s phosphorylcholine) with CD59. This is in contrast with the (indirect and cholesterol-dependent) interactions of two SM molecules, where both the sphingosine backbone interaction and the raft–lipid interaction are important.
The results described here, as well as those reported by us previously (Kusumi et al., 2004; Suzuki et al., 2007a,b, 2012; Komura et al., 2016), indicate that all of the raft constituent molecules we examined thus far, including GPI-anchored proteins, gangliosides, and SM, move in and out of raft domains continuously, with residency times on the order of milliseconds to seconds. The next key question would be to reveal the lifetime of each raft domain, which might be longer than the residency times of constituent molecules there. Such dynamic molecular exchanges might be useful for accelerating molecular interactions and enhancing the variety of molecules that go into interaction.
Materials and methods
Materials
Porcine brain SM; synthetic DOPC, DSPC, DMPC, and DOPE; and commercially available fluorescent SMs were purchased from Avanti Polar Lipids, Inc. Cholesterol and NEG were obtained from Sigma-Aldrich. Stearoyl-SM was purified from the porcine brain SM by HPLC, and the purity was monitored by thin-layer chromatography. These samples were dissolved at 1 mg/ml in CHCl3/MeOH (4:1 vol/vol) and stored at −20°C until use. ATTO488 and ATTO594 were purchased from ATTO-TEC, Texas-red-DPPE and Bodipy-PC were from Invitrogen, and CTMR was from Thermo Fisher Scientific. These dye compounds were stored in the dark at −20°C until use. PBS was obtained from Nacalai Tesque, NaH2PO4 and Na2HPO4 were from Wako Pure Chemical Industries, HBSS was from Nissui, Pipes was from Dojindo, and Triton X-100 was from ICN Biomedicals. Other chemicals were purchased from Nacalai Tesque.
Statistical analysis methods
Unless otherwise stated, all the statistical analyses were performed using Mann–Whitney U test. All the histograms for colocalization durations could be fitted by single exponential functions, and therefore, the statistical analysis of these distributions was performed using the log-rank test (statistical survival analysis). Colocalization of 594neg-SM with GFP-lysenin on the cell surface using conventional confocal fluorescence microscopy was statistically evaluated by Pearson’s correlation coefficients (r) using ImageJ.
GUV preparation
GUVs of ternary mixtures of lipids were prepared by electroformation (Angelova and Dimitrov, 1986). In brief, 5–10 µl of a SM(18:0)/DOPC/cholesterol equimolar solution (1 mg/ml) for 488neg-SM, 594neg-SM, 594neg-DSPC, or 594neg-DOPC observations and a DSPC/DOPC/cholesterol equimolar solution (1 mg/ml) for 594neg-DSPC observations were spread on the electrode surface (platinum wires, Φ = 100 µm) in the presence of 0.2 mol% fluorescent probes, and the remaining solvent was removed under vacuum for 24 h. Two electrodes coated with the thin lipid film were attached on both sides of a square-shaped rubber seal (1 mm thick, with a square window of 15 × 15 mm, containing 0.4 ml Milli-Q water), and then were sandwiched by two cover glasses (24 × 60 mm, 0.12- to 0.17-mm thickness; Matsunami). This chamber was placed on a temperature-controlled sample stage (Tokai Hit) on an inverted confocal fluorescence microscope, and then a low-frequency (10 Hz) sinusoidal current (10 Vpp) was applied, using an Agilent Technologies function generator (model 33120A) at 55°C for 60 min (the temperature was calibrated by a K-type thermocouple sensor, AD-5602A; Sanyo Industries). The GUVs formed in this chamber were maintained at 25°C for 15 min, and then the Lo-Ld phase–separated GUVs attached to the coverslip were observed at 28.5°C (Farkas and Webb, 2010).
Confocal fluorescence observations
Confocal laser-scanning fluorescence microscopy was performed with an FV1000-D microscope (IX81; Olympus), using an objective lens with a long working distance (LUCPLFLN 60×, NA 0.7). The image was obtained by the acquisition software FV10-ASW4.2. The contrast and the brightness of the obtained images were adjusted with Adobe Photoshop.
Estimation of the number density of the deuterated SM in the Lo and Ld domains
The SM number densities in the Lo and Ld domains (ρo and ρd, respectively) were evaluated using the partition ratios of SM, cholesterol, and DOPC into the Lo versus Ld domains obtained by a solid-state NMR study of deuterated SM (18:0; nSM, nchol, and nDOPC, respectively; Yasuda et al., 2014), as well as other molecular parameters reported previously (Alwarawrah et al., 2010). ρo was calculated as
where and represent the number of SM molecules in the Lo domains and the total area of the Lo domains, respectively; NSM, Nchol, and NDOPC represent the total numbers of SM, cholesterol, and DOPC, respectively, in the entire ternary mixture; and , , and represent the lateral areas occupied by SM, cholesterol, and DOPC in the Lo domain (Alwarawrah et al., 2010; Yasuda et al., 2014). Because ρd can be calculated in the same manner, the relative number density of SM in the Lo to Ld domains (ρ) was calculated as
FCS measurements
FCS measurements of GUV surfaces were performed at 28°C with the same microscope used to obtain the confocal fluorescence images, with an oil-immersion apochromat objective lens (Olympus PLAPON60XO, 60×, NA 1.4), following the protocol published previously (Korlach et al., 1999; Kahya et al., 2003). The excitation laser light (488/594 nm) was focused on the flat top part of the GUV membrane (the membrane opposite from the coverslip side). The confocal geometry was ensured by obtaining the confocal images before FCS measurements.
The diffusion coefficient, D, was obtained by fitting the autocorrelation function of the time-dependent changes of the signal intensities of fluorescent probe molecules in diffraction-limited spots, G(τ), with the following equation for two-dimensional simple Brownian diffusion:
where N is the mean number of fluorescent particles in the detection area; W0 is the beam waist (radius) in the focal plane (W0 = 0.14 and 0.18 µm for excitation wavelengths of 488 and 594 nm, respectively); D is the diffusion coefficient; and τ is the delay time. The W0 value for the 543-nm laser line was calibrated using GUVs containing 0.002 mol% of DiIC18 (Kahya et al., 2003), and the W0 values for the 488- and 594-nm laser lines were calculated assuming that the W0 values are proportional to the wavelength for the same objective lens. When the FCS measurements were performed in the Lo and Ld domains in a single GUV, 0.008 mol% 488neg-SM, 594neg-SM, or 594neg-DSPC or 0.016 mol% 594neg-DOPC was added to the lipid mixture (Fig. 4). When the FCS measurements in the Lo and Ld domains were performed independently in different GUVs, 0.002 mol% 488neg-SM, 594neg-SM, or 594neg-DSPC or 0.016 mol% 594neg-DOPC was included in the lipid mixture for the observations of the Lo domain, whereas 0.008 mol% 488neg-SM, 594neg-SM, or 594neg-DSPC or 0.002 mol% 594neg-DOPC was added to the lipid mixture for the observations of the Ld domain. The results obtained by these methods gave virtually the same diffusion coefficients and thus were combined (Fig. 4 B).
Preparation of human erythrocyte ghosts, labeling with fluorescent lipid analogs, and cold-Triton extraction
Human erythrocyte ghost membranes were prepared essentially as previously reported (Dodge et al., 1963). In brief, erythrocytes obtained from one of the authors (M. Kinoshita) were pelleted by centrifugation, and after the removal of the buffy coat, 100 µl of the erythrocyte pellet was resuspended in 900 µl prechilled hypotonic 5 mM NaH2PO4-Na2HPO4 buffer (pH 8.0; 5P8) and incubated on ice for 20 min, to induce cell lysis. The cells were then washed four times with a 10× volume of prechilled 5P8 buffer, by resuspension/centrifugation (12,000 g, 10 min, 4°C).
The erythrocyte ghost membranes were labeled with fluorescent lipid analogs by incubation with 1 µM fluorescent lipid analog in PBS, pH 7.6, at 25°C for 7.5 min in the dark. They were then washed four times with a 10× volume of PBS by resuspension/centrifugation (12,000 g, 10 min, 4°C) and finally diluted in PBS at ∼10% (vol/vol). The ghost membrane suspension was placed on poly-l-lysine–coated glass-base dishes, and then the ghost membranes attached to the glass surface were observed by epifluorescence microscopy. For cold-Triton extraction, the ghost membranes bound to the glass surface were incubated with 1% cold Triton X-100 in PBS on ice water at 2.8°C for 20 min, and then washed three times with prechilled PBS.
Estimating the partitioning of endogenous SM into DRM and non-DRM fractions in ghost membranes
Ghost membranes (sedimented volume of 100 µl) were mixed with 200 µl of 1% Triton X-100 at 0°C. The mixture was incubated in ice water for 20 min and then centrifuged (2°C, 12,000 g, 5 min). The lipids in both the supernatant and pellet were extracted (Bligh and Dyer, 1959), and after the solvent was completely removed under vacuum for 24 h, the extracted lipids were dissolved in 100 µl CHCl3/CH3OH (2:1 vol/vol). A 2-µl portion of the lipid solution was applied to the TLC plate (2 × 5.5 cm, silica gel 60 F254; EMD Millipore), together with standard samples of SM (18:0) and DOPC, and the plate was developed with CHCl3/CH3OH/CH3COOH/HCOOH/H2O (35:15:6:2:1 by vol) until the solvent front ascended to 5 cm above the dotted line (Macala et al., 1983). The phospholipids on the TLC plate were quantitated by densitometry, after visualization with phosphomolybdic acid.
Cell culture, fluorescent phospholipid analog incorporation into the cellular PM, cold-Triton treatment, and evaluation of fluorescent analog partitioning into DRMs and non-DRMs
CHO-K1 and T24 cells were cultured in HAM’s F12 medium (Gibco) supplemented with 10% FBS. RBL-2H3 cells were maintained in Eagle’s minimal essential medium (Sigma-Aldrich), supplemented with 1 mM sodium pyruvate (Sigma-Aldrich), 0.1 mM MEM nonessential amino acids (Wako Pure Chemical Industries; 139-15651), and 15% FBS (Sigma-Aldrich). PtK2 cells were grown in MEM supplemented with 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, and 10% FBS. Fluorescent lipid analogs were incorporated in the PMs of the ghost, CHO-K1, and T24 cells by incubations with 1 µM (final concentration) fluorescent lipid analogs in HBSS (without phenol red and sodium bicarbonate) and buffered at pH 7.4 with 2 mM Pipes (P-HBSS), at 22°C for 15 min in the dark, followed by three washes with P-HBSS. The cells were further washed with prechilled P-HBSS three times and incubated with prechilled 1% (vol/vol) Triton X-100 in P-HBSS on ice water for 20 min, as described previously (Suzuki et al., 2012). They were washed three times with cold P-HBSS and twice with prechilled PBS, then fixed with 4% PFA in PBS for 90 min. The cold Triton–treated cells were observed with an IX70 epifluorescence microscope (Olympus), equipped with a cooled CCD camera (Photometrics Quantix). The fluorescence intensities in 10 × 10-µm areas in the PM were measured before and after the cold Triton treatment (n = 31–53 cells).
Formation and microscopic observation of GPMVs exhibiting separations into Lo-like and Ld-like domains
RBL-2H3 cells grown on a glass-base dish (Iwaki) were washed twice with 1 ml P-HBSS. Fluorescent lipid analogs were suspended in P-HBSS after vigorous vortexing for 1 min followed by sonication for 5 min (2 nmol probe in 200 µl P-HBSS). Subsequently, the cells on the glass were incubated in 0.1 ml of this probe suspension at 22°C for 12 min. After three washes with P-HBSS, membrane blebs were induced by incubating the cells in 25 mM formaldehyde and 2 mM DTT in P-HBSS at 37°C for 1 h (Baumgart et al., 2007). During this incubation, numerous blebs were generated and then detached from the cells to form GPMVs. The dish was moved onto the microscope stage of a home-built TIRF microscope, quietly left at 20°C for 15 min to allow the GPMVs to settle on the bottom glass, and then cooled by circulating a chilled water–ethanol solution (−5°C) so that the temperature of the top surface of the glass became stabilized at 10°C. Under these conditions, the majority (>90%) of the GPMVs exhibited two coexisting domains. The focus was adjusted to collect the fluorescence signal emitted near the equatorial plane of the blebs. Images were simultaneously recorded in green and red channels at a rate of 30 Hz. To estimate the partitioning coefficients of 488neg-SM, 594neg-SM, and 594neg-DOPC, only the first or second frame of the movie was used to minimize the effect of photobleaching.
All 8-bit images were analyzed with ImageJ. First, the background fluorescence in the image was estimated, using the rolling ball algorithm. The radius of the rolling ball was 50 pixels (many times wider than the bleb outline itself; for example, see Sternberg, 1983). This background image was subtracted from the original image, and all further operations were performed on the background-subtracted image. Next, using the freehand selection tool, the regions of interest were drawn around the domains marked by test and control lipid molecules. Then, the mean fluorescence intensity values in the regions of interest were obtained. The ratio of the mean intensity in the Lo-like domain versus that in the Ld-like domain was obtained as the partition coefficient.
Examination of colocalization of 594neg-SM with GFP-lysenin on the cell surface using conventional confocal fluorescence microscopy
T24 cells were seeded on glass-base dishes (4 × 103 cells on the 12-mmφ glass window in a 35-mmφ dish, 0.15-mm-thick glass; Iwaki) and grown for 2 d before observation. 594neg-SM or 594-S9-GM1 was incorporated in the PMs of the T24 cells by incubating the cells with 1 µM (final concentration) lipid analogs in HAM’s F21 medium containing 450 mM sucrose (S-HAM, to slow the internalization process) at 37°C for 10 min, followed by three washes with S-HAM. The cells were then incubated with 5 µg/ml (final concentration) GFP-lysenin (purified as described by Kiyokawa et al. [2005] and Abe and Kobayashi [2014]) in S-HAM at 37°C for 15 min, followed by washing with S-HAM three times. Some of the specimens were further incubated with rabbit anti-GFP antibody IgG (10 µg/ml; Medical & Biological Laboratories) in S-HAM for 15 min at 37°C and washed three times with P-HBSS containing 450 mM sucrose. The basal PMs of the labeled live cells were observed at RT (25°C) with an FV1000 confocal microscope (Olympus), using excitation wavelengths of 488 and 594 nm. To quantitatively evaluate colocalization, Pearson’s correlation coefficients (r) were obtained for the pair of images of the same view field within a single cell, using ImageJ. As a negative control, one of the images was rotated 180° (the flipped image) and Pearson’s correlation coefficient was evaluated.
Evaluating the amounts of fluorescent lipid analogs located in the PM outer and inner leaflets and in the cytoplasm using a membrane-impermeable fluorescence quencher
T24 or CHO-K1 cells cultured on a glass-base dish were washed with P-HBSS twice at 37°C, and then 2 nM (final concentration) of 594neg-SM, 594neg-DOPC or 594-DOPE in P-HBSS was added to the incubation-observation medium (P-HBSS). The dish was then immediately moved to the microscope stage and incubated at 37°C there. At 15 min after the addition of the lipid analog, the cells were quickly observed by TIRF and oblique-angle illuminations of the 594-nm laser at a single-molecule detection sensitivity (for molecules located in/near the PM and those located in/near the PM plus in the cytoplasm, respectively), and then 100 mM CuTSP in P-HBSS was further added to achieve a final concentration of 3.3 mM. The same cells were observed again using TIRF and oblique-angle illuminations. The total fluorescence intensity of ATTO594 was estimated by measuring the fluorescence signal intensities of individual spots of the lipid analog molecules in areas of 300 × 300 pixels (17.1 × 17.1 µm) and then summing the signal intensities of the spots located in this area. The fluorescence intensities before and after CuTSP addition were then compared. Virtually no signals of fluorescent lipid analogs were detected when the focus was shifted deeper in the cytoplasm, all the way to the top membrane, using the oblique-angle illumination. Therefore, the amounts of lipid in the cytoplasm were evaluated using the oblique-angle illumination with the focal plane on the basal PM (the same focal plane as that with TIRF illumination).
Cell preparation for single-molecule observations and cholesterol depletion and repletion
CHO-K1, T24, and PtK2 cells were sparsely seeded on glass-base dishes (4 × 103 cells on the glass window of 12 mmφ in a 35-mmφ dish, 0.15-mm-thick glass; Iwaki), and grown for 2 d before observation. Fluorescent lipid analogs, 594neg-SM, 594neg-DOPC, 594neg-DSPC, and 594-DOPE, were incorporated in the CHO-K1, T24, and PtK2 cell PMs by incubating the cells with 5 nM of these lipid analogs in 200 µl P-HBSS.
Partial cholesterol depletion was performed by incubating the cells in 200 µg/ml saponin (Sigma-Aldrich) in P-HBSS on ice for 15 min or in 4 mM methyl-β-cyclodextrin (MβCD, Sigma-Aldrich) in P-HBSS for 30 min at 37°C, as previously reported (Suzuki et al., 2012). Cholesterol was replenished by incubating the cholesterol-depleted cells in 10 mM MβCD–cholesterol complex (1:1) in P-HBSS for 30 min at 37°C (Shigematsu et al., 2003). The overall amounts of cholesterol per CHO-K1 cell after cholesterol depletion with MβCD and after the subsequent repletion were found to be 63 ± 4% and 129 ± 7% of the original amount, as determined with a Wako cholesterol E-test kit.
Single fluorescent-molecule tracking of fluorescent lipid analogs in the PMs of cultured cells and colocalization detection
Colocalizations of 594neg-SM, 594neg-DSPC, or 594neg-DOPC with CD59 monomers or homodimers (using ACP-CD59, covalently labeled with ATTO488-conjugated acetyl-CoA) were examined in CHO-K1 cells, which do not endogenously express CD59. CHO-K1 cells were transfected with cDNA encoding ACP-CD59 by using LipofectAMINE PLUS (Thermo Fisher Scientific), according to the manufacturer’s recommendations. cDNA encoding ACP-CD59 was prepared as previously reported (Suzuki et al., 2012). In brief, cDNA sequence of ACP-CD59 was placed in the Epstein-Barr virus–based episomal vector pOsTet15T3, which carries the tetracycline-regulated expression units, the transactivator (rtTA2-M2), and the TetO sequence (a Tet-on vector).
Single molecules of the fluorescent lipid analogs in the bottom PMs (the PMs facing the cover glass) of CHO-K1 cells were observed at 37°C at a 4-ms resolution, using a home-built, objective lens–type, TIRF microscope, based on an inverted microscope (Olympus IX-70, 100× 1.49-NA oil objective), equipped with a two-stage microchannel plate intensifier (C8600-03; Hamamatsu Photonics) coupled to an sCMOS camera (ORCA-Flash4.0; Hamamatsu Photonics), as described previously (Suzuki et al., 2012). All of the fluorescent spot identifications and colocalization determinations were performed by home-built computer software (machine vision) as described previously (Suzuki et al., 2012). The position determination precisions for single ATTO594 molecules immobilized on the glass surface were ±12 nm and ±17 nm for observations at video rate (33-ms resolution) and 4-ms resolution, respectively.
Colocalization of two fluorescent single molecules was defined as the event where two fluorescent spots become localized within 240 nm of each other, as described (Koyama-Honda et al., 2005; Kasai et al., 2011; Suzuki et al., 2012). Using this definition, the probability that two molecules located at exactly the same position are found to be colocalized by this method will be 99.7%.
Each time a colocalization event was detected, its duration was measured, and after observing many colocalization events, a histogram of the distribution of colocalization durations was obtained. All of these histograms could be fitted by single exponential decay functions, using nonlinear least-squares fitting by the Levenberg–Marquardt algorithm provided in the MicroCal Origin package, version 7.5. For each fitting of the exponential decay function, the 68.3% confidence limit was obtained as the fitting error for the decay time. The maximal error of the decay time throughout this article was ±8% of the best-fit value. Statistical analysis of these distributions was performed using the log-rank test (statistical survival analysis). The colocalized periods were then corrected for the photobleaching lifetime of the probes, based on the equations described previously (Kasai et al., 2011; Suzuki et al., 2012), using the equation , where τoff is the colocalization lifetime corrected for photobleaching lifetime, τapp is the apparent lifetime directly obtained from experiments using the histogram of colocalized durations, and τb1 and τb2 are exponential photobleaching lifetimes for the two dyes used in the experiments. In the case of single-color experiments, the equation was used.
Induction of CD59-cluster signaling rafts using 40-nm beads coated with anti-CD59 monoclonal antibody IgG
CD59-cluster signaling rafts were induced by applying 40-nmφ carboxylate-modified yellow-green beads conjugated with anti-CD59 monoclonal antibody IgG to the cells, as described previously (Suzuki et al., 2007a,b). In brief, carboxylate-modified yellow-green beads (1.4 × 1015 particles/ml; FluoSpheres; Invitrogen) were diluted 10-fold with 2 mM phosphate buffer, pH 7.0, and centrifuged at 20,000 g for 10 min to remove the clumps. A 6-µl portion of the supernatant was mixed with 1 µl (2.1 mg/ml) of the anti-CD59 IgG antibody (MEM43/5) or the anti–c-myc mouse monoclonal antibody (9E10.2), as well as 3 µl (1.25 mg/ml) of a nonspecific IgG antibody (Zymed), and then the total volume was adjusted to 200 µl with 2 mM phosphate buffer. The mixture was incubated on a slowly tumbling shaker at 4°C for 4 h. The beads were further stabilized with 0.02% Carbowax 20 M (Sigma-Aldrich). After three washes by sedimentation (100,000 g for 15 min) and resuspension in 0.02% Carbowax 20 M in 2 mM phosphate buffer, the beads were finally resuspended in P-HBSS containing 0.02% Carbowax 20 M and used within 8 h. After 10-fold dilution with P-HBSS, a 200-µl portion of the bead suspension was added to T24 cells, and single-molecule tracking of the beads was performed on the apical/dorsal membrane.
Binding of anti-DNP IgE and DNP-BSA (multivalent antigen) to FcεRI in RBL-2H3 cells
RBL-2H3 cells in culture medium were incubated with 1 ng/ml Alexa Fluor 488–labeled mouse anti-DNP monoclonal IgE (clone SPE-7; Sigma-Aldrich; final concentration) at 37°C under 5% CO2 for 15 min, followed by washing with the culture medium three times. After three further washes with P-HBSS, the fluorescent lipid analog, 594neg-SM or 594neg-DOPC, was incorporated in the RBL-2H3-cell PM by incubating the cells with 5 nM lipid analog in 200 µl P-HBSS. Single molecules of FcεRI bound by Alexa Fluor 488–labeled anti-DNP IgE (FcεRI stays as monomers) and the fluorescent lipid analog in the bottom PM (the PM facing the cover glass) of RBL-2H3 cells were simultaneously observed at 37°C at a 33-ms resolution, using the TIRF microscope.
For multivalent antigen stimulation, RBL-2H3 cells were first incubated with 1 µg/ml nonlabeled anti-DNP-IgE in the culture medium at 37°C for 15 min. After washing the cells with the cell culture medium three times and then with P-HBSS three times, the fluorescent lipid analog, 594neg-SM or 594neg-DOPC, was incorporated in the RBL-2H3-cell PM by incubating the cells with 5 nM lipid analog in 200 µl P-HBSS. The cells were washed with P-HBSS twice, and then the medium was changed to P-HBSS containing 1% BSA. The cells were then incubated with Alexa Fluor 488–labeled DNP-BSA (EMD Millipore) at a final concentration of 1 ng/ml at 37°C for 5 min, which cross-linked FcεRI (under these conditions, ∼50% and ∼25% of FcεRI molecules existed in clusters of sizes between dimers and tetramers and in those containing five or more molecules, respectively, whereas ∼25% remained as monomers [Andrews et al., 2009]), followed by washing with P-HBSS three times. Simultaneous dual-color (488 and 594 nm) single-molecule observations of cross-linked FcεRI and the fluorescent lipid analogs were performed at 37°C at a 33-ms resolution.
High-speed single fluorescent-molecule observations and detection of temporary confinement in nanoscale domains
594neg-SM and 594neg-DOPC were observed in the bottom PMs of T24 cells at 23°C (37°C was not used here because this study was conducted to compare the data with those reported by Eggeling et al. [2009] and Sahl et al. [2010] obtained at RT), using a home-built, objective lens–type, TIRF microscope (described in the previous section), but the images produced by a two-stage microchannel plate intensifier (C8600-03; Hamamatsu Photonics) were projected onto a specially designed CMOS sensor-based camera (Photron) by way of an optical-fiber bundle (Hiramoto-Yamaki et al., 2014). This system allowed imaging at frame rates up to 2,000 Hz (time resolutions down to 0.5 ms; Hiramoto-Yamaki et al., 2014).
Attempts to detect temporary confinement/binding events or TALL events were made using the method developed by Sahl et al. (2010), with our previous modifications (Komura et al., 2016). Trajectories longer than 50 frames were used for the analysis (>170 trajectories for both of the molecules, with a total number of frames for each molecule >29,000). The detection circle radius and the threshold residency time were set at 50 nm or smaller and 10 frames (5 ms), respectively, according to Sahl et al. (2010). ATTO594 fixed on a glass surface was used as the standard for immobile molecules. The results obtained by this method were comparable to those previously reported by Simson et al. (1995) and our laboratory (Suzuki et al., 2007a,b, 2012).
Preparation of planar lipid bilayers
Small unilamellar vesicles (∼50 nm in diameter), composed of DMPC-cholesterol (65:35 mol/mol), DMPC (100%), DPPC-cholesterol (65:35 mol/mol), or DOPC (100%) were prepared with a Lipofast extruder (Avestin). The vesicles were incubated for 10 min on a coverslip cleaned by the method of Yamada et al. (2013), which led to the formation of a large planar lipid bilayer on the coverslip. After washing the bilayers on the coverslip with Milli-Q water, a 500-µl portion of 2 nM 594neg-SM or 594neg-DOPC suspension in P-HBSS was added, and then single lipid probe molecules were observed at a 4-ms resolution at either 30°C (DMPC bilayer in the Lα phase), 20°C (DMPC-cholesterol bilayer in the Lo phase), 25°C (DOPC bilayer in the Lα phase), or 37°C (DPPC-cholesterol bilayer in the Lo phase).
ELISA measurement of GFP-lysenin binding to 594neg-SM
The binding of GFP-lysenin to 594neg-SM was measured by ELISA as described previously (Makino et al., 2015). In brief, 50 µl of lipid (10 µM) in ethanol was added to a well of a microtiter plate (Immulon 1; Dynatech Laboratories). After the solvent was evaporated at RT, 100 µl of 30 mg/ml BSA in Tris-buffered saline (10 mM Tris-HCl, pH 7.5, and 150 mM NaCl) was added to each well. After washing, the wells were incubated with 50 µl of GFP-lysenin in Tris-buffered saline containing 10 mg/ml BSA for 1 h at RT. The bound GFP-lysenin was detected by incubation with mouse anti-His antibody (1/1,000 dilution, RT, 1 h) followed by a second incubation with HRP-conjugated anti–mouse IgG antibodies (1/1,000 dilution, RT, 1 h). The intensity of the color developed after the addition of o-phenylenediamine and H2O2 was measured with an ELISA reader (SpectraMax M2; Molecular Devices), reading the absorption at 490 nm.
Organic synthesis of fluorescent analogs of SM (18:0) and PC (di-18:0, di-18:1)
In brief, an azide-derivatized dye compound was linked to NEG to form CTMRneg, 488neg, and 594neg, and then these compounds were conjugated to propargyl-SM, providing the CTMRneg-, 488neg-, and 594neg-SMs, respectively (Fig. 1 A).
In more detail, to a solution of the N-hydroxysuccinate ester of a fluorescent dye (CTMR, ATTO488, or ATTO594, 1.45 μmol) in dimethylformamide (450 µl) were added triethylamine (TEA, 3.71 µl, 18.3 µmol) and a dimethylformamide solution of NEG (2.18 µmol/1.5 eq) or 3-azido-1-propanamine (5.52 µmol/3.8 eq). After stirring at RT for 7 h, the solvent was removed by a Biotage V-10 solvent evaporation system. The residue was then dissolved in CHCl3/MeOH/H2O (13:9:2 by volume) and applied to a silica gel column equilibrated with the same solution, followed by gel permeation chromatography (JAIGEL-GS; MeOH), to obtain the purified azide derivative of the fluorescent dye. The azide derivative (1.0 μmol), CuSO4 (0.5 µmol), and sodium ascorbate (1.5 µmol) were dissolved in 400 µl t-BuOH/H2O (4:1) and mixed with a MeOH solution of propargyl-SM, DSPC, or DOPC (2.0 µmol/100 µl) prepared as previously reported (Sandbhor et al., 2009; Goretta et al., 2012). The solution was stirred at RT for 24 h, and the solvent was removed by a Biotage V-10 solvent evaporation system. The residue was dissolved in CHCl3, and subjected to preparative silica gel TLC (CHCl3/MeOH/H2O, 13:9:1 by volume) and then gel permeation chromatography (JAIGEL-GS; MeOH), to produce purified CTMR-SM, CTMRneg-SM, 488neg-SM, 594neg-SM, 594neg-DSPC, and 594neg-DOPC (∼20% yields; Fig. 1).
The results of the examination by proton NMR and high-resolution mass spectroscopy (HRMS) are described in the next subsections.
CTMR-SM
NMR (500 MHz, CD3OH) δ 0.88 (t, J = 6.9 Hz, 10H), 1.16–1.43 (m, 88H), 1.49–1.63 (m, 4H), 1.97–2.03 (m, 4H), 2.12–2.18 (m, 3H), 2.33 (t, J = 6.7 Hz, 2H), 3.21 (s, 6H), 3.48 (t, J = 6.6 Hz, 2H), 3.60–3.67 (m, 3H), 3.92–4.02 (m, 3H), 4.08–4.14 (m, 1H), 4.33–4.40 (m, 2H), 4.61 (t, J = 6.7 Hz, 2H), 5.32 (t, J = 5.5 Hz, 1H), 5.41 (dd, J = 15.6, 7.9 Hz, 1H), 5.62–5.69 (m, 1H), 6.92 (d, J = 2.4 Hz, 2H), 7.00 (dd, J = 9.5, 2.6 Hz, 2H), 7.22 (d, J = 9.5 Hz, 2H), 7.35 (d, J = 8.0 Hz, 1H), 8.05 (dd, J = 7.7, 1.8 Hz, 1H), 8.45 (s, 1H), and 8.51 (b, 3H).
HRMS calculated for C71H112N8O10P+ [M + H]+, 1,267.8234; found, 1,267.8571. The reason for this 0.0337 difference is unknown.
CTMRneg-SM
NMR (500 MHz, CD3OH) δ 0.90 (t, J = 6.8 Hz, 6H), 1.08–1.42 (m, 50H), 1.51–1.64 (m, 2H), 1.99–2.06 (m, 2H), 2.15–2.21 (m, 2H), 3.29 (s, 6H), 3.55–3.74 (m, 40H, PEG), 3.92–4.00 (m, 4H), 4.05 (t, J = 8.3 Hz, 1H), 4.10–4.15 (m, 1H) 4.34–4.39 (m, 2H), 4.65–4.67 (m, 1H), 5.44 (dd, J = 15.3, 7.8 Hz, 1H), 5.67–5.73 (m, 1H), 6.94 (t, J = 2.5 Hz, 2H), 7.03 (dt, J = 9.5, 2.6 Hz, 2H), 7.26 (dd, J = 9.4, 8.0 Hz, 2H), 7.37 (d, J = 7.9 Hz, 0.5H), 7.73 (d, J = 1.4 Hz, 0.5H), 8.07 (dd, J = 7.9, 1.7 Hz, 0.5H), 8.11 (dd, J = 8.2, 1.4 Hz, 0.25H), 8.15 (d, J = 8.2 Hz, 0.25H), 8.41 (d, J = 2.4 Hz, 1H), and 8.53 (d, J = 1.6 Hz, 1H).
HRMS calculated for C90H151N8O20P+ [M + 2H]2+, 847.5386; found, 847.5347.
488neg-SM
NMR (400 MHz, CD3OD) δ 0.07 (d, J = 15.1 Hz, 2H), 0.90 (t, J = 6.6 Hz, 6H), 1.07–1.50 (m, 50H), 1.52–1.69 (m, 2H), 1.89 (d, J = 7.8 Hz, 1H), 1.94–2.08 (m, 2H), 2.11–2.24 (m, 2H), 2.87 (s, 3H), 3.17 (s, 6H), 3.44–3.76 (m, 40H, PEG), 3.82–4.16 (m, 4H), 4.22–4.41 (m, 2H), 4.58–4.70 (m, 1H), 5.40–5.51 (m, 1H), 5.61–5.78 (m, 1H), 6.94–7.03 (m, 2H, aromatic), 7.23–7.28 (m, 2H, aromatic), 7.33–7.81 (m, 4H, aromatic), and 8.36–8.60 (m, 1H).
HRMS calculated for C90H151N9O25PS2+ [M]+, 1,852.9995; found, 1,852.9949.
594neg-SM
NMR (500 MHz, CD3OD) δ 0.10 (s, 1H), 0.90 (t, J = 6.7 Hz, 6H), 1.20–1.45 (m, 50H), 1.55 (d, J = 3.3Hz, 3H) 1.58–1.62 (m, 4H), 1.78 (t, J = 5.8 Hz, 1H), 1.97–2.09 (m, 2H), 2.13–2.23 (m, 2H), 2.64 (s, 1H), 2.72 (s, 1H), 3.19 (s, 6H), 3.43–3.52 (m, 2H), 3.54–3.70 (m, 40H, PEG), 3.70–3.78 (m, 2H), 3.85 (d, J = 15.8 Hz, 1H), 3.90–4.01 (m, 2H), 4.03–4.09 (m, 1H), 4.10–4.18 (m, 1H), 4.33–4.39 (m, 1H), 4.58 (s, 12H), 4.64–4.68 (m, 1H), 4.76 (s, 2H), 5.41–5.48 (m, 1H), 5.66–5.74 (m, 1H), 5.89 (s, 1H), 6.79 (s, 1H), 7.38 (d, J = 15.3 Hz, 1H), 7.46–7.53 (m, 1H), 7.57–7.64 (m, 1H), 7.68–7.76 (m, 2H), and 8.41 (s, 1H).
HRMS calculated for C106H173N9Na3O25PS22+ [M-2H+3Na]2+, 1,068.5719; found, 1,068.5717.
594neg-DSPC
NMR (400 MHz, CD3OD) δ 0.90 (t, J = 6.9 Hz, 6H), 1.25–1.40 (m, 56H), 1.56 (d, J = 3.3 Hz, 3H) 1.56–1.65 (m, 4H), 2.15 (t, 7.8 Hz, 1H), 2.33 (q, J = 8.1 Hz, 4H), 2.72 (s, 1H), 2.79 (s, 1H), 3.20 (s, 6H), 3.56–3.68 (m, 40H, PEG), 3.93 (t, J= 5.0 Hz, 2H), 4.02 (t, J = 6.0 Hz, 2H), 4.18 (dd, J = 12.1, 7.1 Hz, 1H), 4.32–4.39 (m, 2H), 4.45 (dd, J= 12.1, 3.0 Hz, 1H), 4.66 (t, J = 4.8 Hz, 2H), 4.77 (s, 1H), 5.21–5.28 (m, 1H), 5.89 (s, 1H), 6.79 (s, 1H), 7.38 (d, J = 12.8 Hz, 1H), 7.59–7.65 (m, 1H), 7.69–7.75 (m, 1H), 8.41 (s, 1H), and 8.55 (s, 1H).
HRMS calculated for C109H181N8O27PS22+ [M+H]2+, 1,065.1119; found, 1,065.1129.
594neg-DOPC
NMR (500 MHz, CD3OD) δ 0.10 (s, 1H), 0.90 (t, J = 6.6 Hz, 6H), 1.22–1.45 (m, 44H), 1.55 (d, J = 2.8Hz, 3H), 1.58–1.65 (m, 4H), 2.00–2.07 (m, 8H), 2.21–2.26 (m, 3H), 2.33 (q, J =10.3 Hz, 4H), 2.63 (s, 1H), 2.72 (s, 2H), 2.89 (s, 1H), 3.13 (s, 1H), 3.19 (s, 6H), 3.47–3.53 (m, 4H), 3.56–3.66 (m, 40H, PEG), 3.73 (q, J = 8.5 Hz, 5H), 3.84 (d, J = 14.2 Hz, 2H), 3.93 (t, J= 6.0 Hz, 2H), 4.02 (t, J = 7.5Hz, 2H), 4.17 (dd, J = 12.6 Hz, 1H), 4.32–4.38 (m, 2H), 4.44 (d, J= 9.2Hz, 1H), 4.58 (s, 1H), 4.66 (t, J = 5.5Hz, 2H), 4.67 (s, 1H), 4.78–4.84 (m, 1H), 5.23–5.27 (m, 1H), 5.34 (t, J= 6.0Hz, 4H), 5.89 (s, 3H), 6.79 (s, 3H), 7.38 (d, J = 13.3 Hz, 3H), 7.48–7.52 (m, 1H), 7.59–7.66 (m, 2H), 7.70–7.75 (m, 4H), 8.41 (s, 1H), and 8.47–8.57 (m, 8H).
HRMS calculated for C109H177N8O27PS22+ [M+H]2+, 1,063.0962; found, 1,063.0982.
Online supplemental material
Fig. S1 shows partitioning of commercially available SM analogs into Lo and Ld domains in Lo-Ld phase–separated GUVs. Fig. S2 shows cold-Triton (in)solubility of endogenous SM and PC in erythrocyte ghosts, and 488neg-SM, 594neg-SM, 594neg-DSPC, 594neg-DOPC, and 594-DOPE preincorporated in human erythrocyte ghosts and the PMs of CHO-K1 and T24 cells. Fig. S3 shows prolonged colocalization of SM probes in CHO-K1-cell PM as revealed by simultaneous two-color single-molecule tracking of 488neg-SM and 594neg-SM as well as single-color single molecule tracking of 594neg-SM. Fig. S4 shows that 594neg-SM and DOPC exhibited no signs of temporary immobilization in T24- and PtK2-cell PMs. Fig. S5 shows that diffusion coefficients of single molecules of 594neg-SM, 594neg-DOPC, and 594neg-DSPC observed at 0.5-ms resolution in T24- and PtK2-cell PMs were similar to each other. Table S1 shows the fractions of 594neg-SM, 594neg-DOPC, and 594-DOPE located in the PM inner leaflet. Table S2 shows the lifetimes of homo- and hetero-colocalization of lipid analogs in CHO-K1 cells at 37°C as well as in the Lα- and Lo-phase domains of planar lipid bilayers. Table S3 shows the time fractions of mobile, TALL, and immobile periods for 594neg-SM, 594neg-DOPC, and 594neg-DSPC trajectories obtained at a time resolution of 0.5 ms in the intact PMs of the T24 and PtK2 cell lines. Table S4 shows diffusion coefficients on the time scale of 2.3 ms for 594neg-SM, DOPC, and DSPC in the T24-cell PM and the PtK2-cell PM at 23°C and 37°C. Table S5 shows the colocalization lifetimes of CD59 clusters with 594neg-SM, 594neg-DOPC, and 594neg-DSPC (T24 cells) and those of CD59 transient homodimer rafts and monomers with 594neg-SM and 594neg-DOPC (CHO-K1 cells) at 37°C. Table S6 shows the colocalization lifetimes of FcεRI clusters with 594neg-SM and 594neg-DOPC (RBL-2H3 cells) at 37°C.
Supplementary Material
Acknowledgments
We thank Masahiro Sokabe (Nagoya University School of Medicine), Barbara Baird (Cornell University), and Vaclav Horejsi (Academy of Sciences of the Czech Republic) for their kind gifts of human epithelial T24 cells, RBL-2H3 cells, and mouse hybridoma cell line MEM43/5, which produces the anti-CD59 monoclonal antibody, respectively, as well as Rinshi S. Kasai (Kyoto University) for setting up the single-molecule tracking station used in this work and for valuable discussions.
This work was supported in part by Grants-in-Aid for scientific research from the Japan Society for the Promotion of Science (Kiban B to K.G.N. Suzuki [24370055 and 15H04351], T. Fujiwara [16H04775], N. Matsumori [15H03121], and K. Morigaki [26291031], Challenging Exploratory Research to K.G.N. Suzuki [15K14477], and Kiban A and S to A. Kusumi [24247029 and 16H06386, respectively]), Grants-in-Aid for Innovative Areas from the Japan Society for the Promotion of Science to K.G.N. Suzuki (23110001 and 16H01358), N. Matsumori (26102527 and 16H00773), and T. Fujiwara (26115707 and 15H01212), a grant from Core Research for Evolutional Science and Technology project “Creation of Fundamental Technologies for Understanding and Control of Biosystem Dynamics” of the Japan Science and Technology Agency to A. Kusumi, and the Lipid Active Structure Project supported by Exploratory Research for Advanced Technology Organization of the Japan Science and Technology Agency to M. Murata. Institute for Integrated Cell-Material Sciences of Kyoto University is supported by the World Premiere Research Center Initiative of the Ministry of Education, Culture, Sports, Science and Technology.
The authors declare no competing financial interests.
Author contributions: M. Kinoshita performed a large majority of observations using GUVs, and K.G.N. Suzuki performed a large majority of experiments using the PM and GPMVs. M. Kinoshita conducted FCS measurements, and K.G.N. Suzuki carried out single-molecule imaging-tracking. K. Morigaki and K.G.N. Suzuki performed experiments using planar lipid bilayers. M. Abe, A. Makino, and T. Kobayashi prepared GFP-lysenin and performed ELISA assay and, together with K.G.N. Suzuki, performed the cell staining experiments. K.M. Hirosawa and K.G.N. Suzuki performed the experiments monitoring the SM interaction with FcεRI monomers and engaged clusters. T. Fujiwara and A. Kusumi developed the single-molecule imaging camera system and set up the single-molecule instruments. M. Takada and H. Ano synthesized fluorescent SM and PC analogs under the guidance of N. Matsumori and M. Murata. N. Matsumori, A. Kusumi, and M. Murata conceived and formulated this project, and evaluated and discussed data. M. Kinoshita, K.G.N. Suzuki, and A. Kusumi wrote the manuscript, and all authors participated in revising the manuscript.
Footnotes
Abbreviations used:
- ACP
- acyl-carrier protein
- CTMR
- carboxytetramethylrhodamine
- CuTSP
- Cu(II)meso-tetra(4-sulfonatophenyl)porphine
- DMPC
- l-α-dimyristoylphosphatidylcholine
- DOPC
- l-α-dioleoylphosphatidylcholine
- DOPE
- l-α-dioleoylphosphatidylethanolamine
- DPPC
- l-α-dipalmitoylphosphatidylcholine
- DRM
- cold detergent–resistant membrane
- DSPC
- l-α-distearoylphosphatidylcholine
- ELISA
- enzyme-linked immunosorbent assay
- FCS
- fluorescence correlation spectroscopy
- GPI
- glycosylphosphatidylinositol
- GPMV
- giant plasma-membrane vesicle
- GUV
- giant unilamellar vesicle
- Ld
- liquid-disordered
- Lo
- liquid-ordered
- MβCD
- methyl-β-cyclodextrin
- NEG
- nonaethylene glycol
- NMR
- nuclear magnetic resonance
- PC
- phosphatidylcholine
- PM
- plasma membrane
- SE
- standard error
- SM
- sphingomyelin
- TIRF
- total internal reflection fluoroscopy
- TM
- transmembrane
References
- Abe M., and Kobayashi T.. 2014. Imaging local sphingomyelin-rich domains in the plasma membrane using specific probes and advanced microscopy. Biochim. Biophys. Acta. 1841:720–726. 10.1016/j.bbalip.2013.07.003 [DOI] [PubMed] [Google Scholar]
- Alwarawrah M., Dai J., and Huang J.. 2010. A molecular view of the cholesterol condensing effect in DOPC lipid bilayers. J. Phys. Chem. B. 114:7516–7523. 10.1021/jp101415g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N.L., Pfeiffer J.R., Martinez A.M., Haaland D.M., Davis R.W., Kawakami T., Oliver J.M., Wilson B.S., and Lidke D.S.. 2009. Small, mobile FcepsilonRI receptor aggregates are signaling competent. Immunity. 31:469–479. 10.1016/j.immuni.2009.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelova M.I., and Dimitrov D.S.. 1986. Liposome electroformation. Faraday Discuss. Chem. Soc. 81:303–311. 10.1039/dc9868100303 [DOI] [Google Scholar]
- Baumgart T., Hammond A.T., Sengupta P., Hess S.T., Holowka D.A., Baird B.A., and Webb W.W.. 2007. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. USA. 104:3165–3170. 10.1073/pnas.0611357104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benda A., Ma Y., and Gaus K.. 2015. Self-calibrated line-scan STED-FCS to quantify lipid dynamics in model and cell membranes. Biophys. J. 108:596–609. 10.1016/j.bpj.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutel O., Nikolaus J., Birkholz O., You C., Schmidt T., Herrmann A., and Piehler J.. 2014. High-fidelity protein targeting into membrane lipid microdomains in living cells. Angew. Chem. Int. Ed. Engl. 53:1311–1315. 10.1002/anie.201306328 [DOI] [PubMed] [Google Scholar]
- Bligh E.G., and Dyer W.J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- DeWitt B.N., and Dunn R.C.. 2015. Interaction of cholesterol in ternary lipid mixtures investigated using single-molecule fluorescence. Langmuir. 31:995–1004. 10.1021/la503797w [DOI] [PubMed] [Google Scholar]
- Diaz-Rohrer B.B., Levental K.R., Simons K., and Levental I.. 2014. Membrane raft association is a determinant of plasma membrane localization. Proc. Natl. Acad. Sci. USA. 111:8500–8505. 10.1073/pnas.1404582111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge J.T., Mitchell C., and Hanahan D.J.. 1963. The preparation and chemical characteristics of hemoglobin-free ghosts of human erythrocytes. Arch. Biochem. Biophys. 100:119–130. 10.1016/0003-9861(63)90042-0 [DOI] [PubMed] [Google Scholar]
- Eggeling C., Ringemann C., Medda R., Schwarzmann G., Sandhoff K., Polyakova S., Belov V.N., Hein B., von Middendorff C., Schönle A., and Hell S.W.. 2009. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 457:1159–1162. 10.1038/nature07596 [DOI] [PubMed] [Google Scholar]
- Farkas E.R., and Webb W.W.. 2010. Precise and millidegree stable temperature control for fluorescence imaging: Application to phase transitions in lipid membranes. Rev. Sci. Instrum. 81:093704–093708. 10.1063/1.3483263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippov A., Orädd G., and Lindblom G.. 2004. Lipid lateral diffusion in ordered and disordered phases in raft mixtures. Biophys. J. 86:891–896. 10.1016/S0006-3495(04)74164-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisz J.F., Lou K., Klitzing H.A., Hanafin W.P., Lizunov V., Wilson R.L., Carpenter K.J., Kim R., Hutcheon I.D., Zimmerberg J., et al. . 2013. Direct chemical evidence for sphingolipid domains in the plasma membranes of fibroblasts. Proc. Natl. Acad. Sci. USA. 110:E613–E622. 10.1073/pnas.1216585110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goretta S.A., Kinoshita M., Mori S., Tsuchikawa H., Matsumori N., and Murata M.. 2012. Effects of chemical modification of sphingomyelin ammonium group on formation of liquid-ordered phase. Bioorg. Med. Chem. 20:4012–4019. 10.1016/j.bmc.2012.05.015 [DOI] [PubMed] [Google Scholar]
- Heberle F.A., Wu J., Goh S.L., Petruzielo R.S., and Feigenson G.W.. 2010. Comparison of three ternary lipid bilayer mixtures: FRET and ESR reveal nanodomains. Biophys. J. 99:3309–3318. 10.1016/j.bpj.2010.09.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto-Yamaki N., Tanaka K.A.K., Suzuki K.G.N., Hirosawa K.M., Miyahara M.S., Kalay Z., Tanaka K., Kasai R.S., Kusumi A., and Fujiwara T.K.. 2014. Ultrafast diffusion of a fluorescent cholesterol analog in compartmentalized plasma membranes. Traffic. 15:583–612. 10.1111/tra.12163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowka D., and Baird B.. 2016. Roles for lipid heterogeneity in immunoreceptor signaling. Biochim. Biophys. Acta. 1861(8 Pt B, 8 Pt B):830–836. 10.1016/j.bbalip.2016.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowka D., Gosse J.A., Hammond A.T., Han X., Sengputa P., Smith N.L., Wagenknecht-Wiesner A., Wu M., Young R.M., and Baird B.. 2005. Lipid segregation and IgE receptor signaling: A decade of progress. Biochim. Biophys. Acta. 1746:252–259. 10.1016/j.bbamcr.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Honigmann A., Mueller V., Hell S.W., and Eggeling C.. 2013. STED microscopy detects and quantifies liquid phase separation in lipid membranes using a new far-red emitting fluorescent phosphoglycerolipid analogue. Faraday Discuss. 161:77–89. 10.1039/C2FD20107K [DOI] [PubMed] [Google Scholar]
- Honigmann A., Mueller V., Ta H., Schoenle A., Sezgin E., Hell S.W., and Eggeling C.. 2014. Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. Nat. Commun. 5:5412 10.1038/ncomms6412 [DOI] [PubMed] [Google Scholar]
- Hori Y., Norinobu T., Sato M., Arita K., Shirakawa M., and Kikuchi K.. 2013. Development of fluorogenic probes for quick no-wash live-cell imaging of intracellular proteins. J. Am. Chem. Soc. 135:12360–12365. 10.1021/ja405745v [DOI] [PubMed] [Google Scholar]
- Huang H., Simsek M.F., Jin W., and Pralle A.. 2015. Effect of receptor dimerization on membrane lipid raft structure continuously quantified on single cells by camera based fluorescence correlation spectroscopy. PLoS One. 10:e0121777 10.1371/journal.pone.0121777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahya N., Scherfeld D., Bacia K., Poolman B., and Schwille P.. 2003. Probing lipid mobility of raft-exhibiting model membranes by fluorescence correlation spectroscopy. J. Biol. Chem. 278:28109–28115. 10.1074/jbc.M302969200 [DOI] [PubMed] [Google Scholar]
- Kasai R.S., Suzuki K.G.N., Prossnitz E.R., Koyama-Honda I., Nakada C., Fujiwara T.K., and Kusumi A.. 2011. Full characterization of GPCR monomer-dimer dynamic equilibrium by single molecule imaging. J. Cell Biol. 192:463–480. 10.1083/jcb.201009128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto T., Ishitsuka R., and Kobayashi T.. 2016. Detectors for evaluating the cellular landscape of sphingomyelin- and cholesterol-rich membrane domains. Biochim. Biophys. Acta. 1861(8 Pt B, 8 Pt B):812–829. 10.1016/j.bbalip.2016.03.013 [DOI] [PubMed] [Google Scholar]
- Kiyokawa E., Baba T., Otsuka N., Makino A., Ohno S., and Kobayashi T.. 2005. Spatial and functional heterogeneity of sphingolipid-rich membrane domains. J. Biol. Chem. 280:24072–24084. 10.1074/jbc.M502244200 [DOI] [PubMed] [Google Scholar]
- Klymchenko A.S., and Kreder R.. 2014. Fluorescent probes for lipid rafts: From model membranes to living cells. Chem. Biol. 21:97–113. 10.1016/j.chembiol.2013.11.009 [DOI] [PubMed] [Google Scholar]
- Komura N., Suzuki K.G.N., Ando H., Konishi M., Koikeda M., Imamura A., Chadda R., Fujiwara T.K., Tsuboi H., Sheng R., et al. . 2016. Raft-based interactions of gangliosides with a GPI-anchored receptor. Nat. Chem. Biol. 12:402–410. 10.1038/nchembio.2059 [DOI] [PubMed] [Google Scholar]
- Korlach J., Schwille P., Webb W.W., and Feigenson G.W.. 1999. Characterization of lipid bilayer phases by confocal microscopy and fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. USA. 96:8461–8466. 10.1073/pnas.96.15.8461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama-Honda I., Ritchie K., Fujiwara T., Iino R., Murakoshi H., Kasai R.S., and Kusumi A.. 2005. Fluorescence imaging for monitoring the colocalization of two single molecules in living cells. Biophys. J. 88:2126–2136. 10.1529/biophysj.104.048967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuerschner L., Ejsing C.S., Ekroos K., Shevchenko A., Anderson K.I., and Thiele C.. 2005. Polyene-lipids: A new tool to image lipids. Nat. Methods. 2:39–45 http://dx.doi.org/. 10.1038/nmeth728 [DOI] [PubMed] [Google Scholar]
- Kusumi A., Koyama-Honda I., and Suzuki K.. 2004. Molecular dynamics and interactions for creation of stimulation-induced stabilized rafts from small unstable steady-state rafts. Traffic. 5:213–230. 10.1111/j.1600-0854.2004.0178.x [DOI] [PubMed] [Google Scholar]
- Lin Q., and London E.. 2015. Ordered raft domains induced by outer leaflet sphingomyelin in cholesterol-rich asymmetric vesicles. Biophys. J. 108:2212–2222. 10.1016/j.bpj.2015.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D., and Simons K.. 2010. Lipid rafts as a membrane-organizing principle. Science. 327:46–50. 10.1126/science.1174621 [DOI] [PubMed] [Google Scholar]
- Macala L.J., Yu R.K., and Ando S.. 1983. Analysis of brain lipids by high performance thin-layer chromatography and densitometry. J. Lipid Res. 24:1243–1250. [PubMed] [Google Scholar]
- Makino A., Abe M., Murate M., Inaba T., Yilmaz N., Hullin-Matsuda F., Kishimoto T., Schieber N.L., Taguchi T., Arai H., et al. . 2015. Visualization of the heterogeneous membrane distribution of sphingomyelin associated with cytokinesis, cell polarity, and sphingolipidosis. FASEB J. 29:477–493. 10.1096/fj.13-247585 [DOI] [PubMed] [Google Scholar]
- Miyaji M., Jin Z.X., Yamaoka S., Amakawa R., Fukuhara S., Sato S.B., Kobayashi T., Domae N., Mimori T., Bloom E.T., et al. . 2005. Role of membrane sphingomyelin and ceramide in platform formation for Fas-mediated apoptosis. J. Exp. Med. 202:249–259. 10.1084/jem.20041685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáenz J.P., Sezgin E., Schwille P., and Simons K.. 2012. Functional convergence of hopanoids and sterols in membrane ordering. Proc. Natl. Acad. Sci. USA. 109:14236–14240. 10.1073/pnas.1212141109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáenz J.P., Grosser D., Bradley A.S., Lagny T.J., Lavrynenko O., Broda M., and Simons K.. 2015. Hopanoids as functional analogues of cholesterol in bacterial membranes. Proc. Natl. Acad. Sci. USA. 112:11971–11976. 10.1073/pnas.1515607112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl S.J., Leutenegger M., Hilbert M., Hell S.W., and Eggeling C.. 2010. Fast molecular tracking maps nanoscale dynamics of plasma membrane lipids. Proc. Natl. Acad. Sci. USA. 107:6829–6834. 10.1073/pnas.0912894107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandbhor M.S., Key J.A., Strelkov I.S., and Cairo C.W.. 2009. A modular synthesis of alkynyl-phosphocholine headgroups for labeling sphingomyelin and phosphatidylcholine. J. Org. Chem. 74:8669–8674. 10.1021/jo901824h [DOI] [PubMed] [Google Scholar]
- Sezgin E., Levental I., Grzybek M., Schwarzmann G., Mueller V., Honingmann A., Belov V.N., Eggeling C., Coskun Ü., Simons K., and Schwille P.. 2012. Partitioning, diffusion, and ligand binding of raft lipid analogs in model and cellular plasma membranes. Biomchim. Biophys. Acta. 1818:1777–1784. 10.1016/j.bbamem.2012.03.007 [DOI] [PubMed] [Google Scholar]
- Shen H., Giordano F., Wu Y., Chan J., Zhu C., Milosevic I., Wu X., Yao K., Chen B., Baumgart T., et al. . 2014. Coupling between endocytosis and sphingosine kinase 1 recruitment. Nat. Cell Biol. 16:652–662. 10.1038/ncb2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu S., Watson R.T., Khan A.H., and Pessin J.E.. 2003. The adipocyte plasma membrane caveolin functional/structural organization is necessary for the efficient endocytosis of GLUT4. J. Biol. Chem. 278:10683–10690. 10.1074/jbc.M208563200 [DOI] [PubMed] [Google Scholar]
- Simons K. 2016. Cell membranes: A subjective perspective. Biochim. Biophys. Acta. 1858:2569–2572. 10.1016/j.bbamem.2016.01.023 [DOI] [PubMed] [Google Scholar]
- Simons K., and Ikonen E.. 1997. Functional rafts in cell membranes. Nature. 387:569–572. 10.1038/42408 [DOI] [PubMed] [Google Scholar]
- Simson R., Sheets E.D., and Jacobson K.. 1995. Detection of temporary lateral confinement of membrane proteins using single-particle tracking analysis. Biophys. J. 69:989–993 http://dx.doi.org/10.1016%2FS0006-3495(95)79972-6. 10.1016/S0006-3495(95)79972-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg R.S. 1983. Biomedical image processing. Computer 16:22–34. 10.1109/MC.1983.1654163 [DOI] [Google Scholar]
- Suzuki K.G.N., Fujiwara T.K., Sanematsu F., Iino R., Edidin M., and Kusumi A.. 2007a GPI-anchored receptor clusters transiently recruit Lyn and G α for temporary cluster immobilization and Lyn activation: Single-molecule tracking study 1. J. Cell Biol. 177:717–730. 10.1083/jcb.200609174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K.G.N., Fujiwara T.K., Edidin M., and Kusumi A.. 2007b Dynamic recruitment of phospholipase C γ at transiently immobilized GPI-anchored receptor clusters induces IP3-Ca2+ signaling: Single-molecule tracking study 2. J. Cell Biol. 177:731–742. 10.1083/jcb.200609175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K.G.N., Kasai R.S., Hirosawa K.M., Nemoto Y.L., Ishibashi M., Miwa Y., Fujiwara T.K., and Kusumi A.. 2012. Transient GPI-anchored protein homodimers are units for raft organization and function. Nat. Chem. Biol. 8:774–783. 10.1038/nchembio.1028 [DOI] [PubMed] [Google Scholar]
- van Meer G., and Hoetzl S.. 2010. Sphingolipid topology and the dynamic organization and function of membrane proteins. FEBS Lett. 584:1800–1805. 10.1016/j.febslet.2009.10.020 [DOI] [PubMed] [Google Scholar]
- Vicidomini G., Moneron G., Han K.Y., Westphal V., Ta H., Reuss M., Engelhardt J., Eggeling C., and Hell S.W.. 2011. Sharper low-power STED nanoscopy by time gating. Nat. Methods. 8:571–573. 10.1038/nmeth.1624 [DOI] [PubMed] [Google Scholar]
- Watanabe R., Soga N., Fujita D., Tabata K.V., Yamauchi L., Hyeon Kim S., Asanuma D., Kamiya M., Urano Y., Suga H., and Noji H.. 2014. Arrayed lipid bilayer chambers allow single-molecule analysis of membrane transporter activity. Nat. Commun. 5:4519 10.1038/ncomms5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.L., Frisz J.F., Klitzing H.A., Zimmerberg J., Weber P.K., and Kraft M.L.. 2015. Hemagglutinin clusters in the plasma membrane are not enriched with cholesterol and sphingolipids. Biophys. J. 108:1652–1659 10.1016/j.bpj.2015.02.026. 10.1016/j.bpj.2015.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Imaishi H., and Morigaki K.. 2013. Microarrays of phospholipid bilayers generated by inkjet printing. Langmuir. 29:6404–6408. 10.1021/la400570h [DOI] [PubMed] [Google Scholar]
- Yasuda T., Kinoshita M., Murata M., and Matsumori N.. 2014. Detailed comparison of deuterium quadrupole profiles between sphingomyelin and phosphatidylcholine bilayers. Biophys. J. 106:631–638. 10.1016/j.bpj.2013.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T., Tsuchikawa H., Murata M., and Matsumori N.. 2015. Deuterium NMR of raft model membranes reveals domain-specific order profiles and compositional distribution. Biophys. J. 108:2502–2506. 10.1016/j.bpj.2015.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Wu J., Heberle F.A., Mills T.T., Klawitter P., Huang G., Costanza G., and Feigenson G.W.. 2007. Phase studies of model biomembranes: Complex behavior of DSPC/DOPC/cholesterol. Biochim. Biophys. Acta. 1768:2764–2776. 10.1016/j.bbamem.2007.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurzolo C., and Simons K.. 2016. Glycosylphosphatidylinositol-anchored proteins: Membrane organization and transport. Biochim. Biophys. Acta. 1858:632–639. 10.1016/j.bbamem.2015.12.018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.