Chlamydiae are intracellular pathogens that depend on the host for their survival and development. Chowdhury et al. demonstrate that Chlamydia trachomatis infection can prevent mitochondrial fission in primary cells by reducing DRP1 abundance via miR-30c–dependent inhibition of p53.
Abstract
Obligate intracellular bacteria such as Chlamydia trachomatis depend on metabolites of the host cell and thus protect their sole replication niche by interfering with the host cells’ stress response. Here, we investigated the involvement of host microRNAs (miRNAs) in maintaining the viability of C. trachomatis–infected primary human cells. We identified miR-30c-5p as a prominently up-regulated miRNA required for the stable down-regulation of p53, a major suppressor of metabolite supply in C. trachomatis–infected cells. Loss of miR-30c-5p led to the up-regulation of Drp1, a mitochondrial fission regulator and a target gene of p53, which, in turn, severely affected chlamydial growth and had a marked effect on the mitochondrial network. Drp1-induced mitochondrial fragmentation prevented replication of C. trachomatis even in p53-deficient cells. Additionally, Chlamydia maintain mitochondrial integrity during reactive oxygen species–induced stress that occurs naturally during infection. We show that C. trachomatis require mitochondrial ATP for normal development and hence postulate that they preserve mitochondrial integrity through a miR-30c-5p–dependent inhibition of Drp1-mediated mitochondrial fission.
Introduction
The obligate intracellular bacterial pathogen Chlamydia trachomatis is the most prevalent bacterial cause of sexually transmitted diseases and the causative agent of preventable infectious blindness (Belland et al., 2004). C. trachomatis exhibits a biphasic developmental life cycle unique to the members of the phylum Chlamydiae. The small (∼0.3 µm) elementary body (EB) is the infectious form of the pathogen, which attaches to the host cell and undergoes endocytosis. After endocytosis, EBs dwell within a membrane-bound inclusion and eventually transit into the metabolically active reticulate bodies (RBs; Matsumoto, 1988; Moulder, 1991; Abdelrahman and Belland, 2005) The RBs replicate by binary fission and differentiate back into the EB form to bring the developmental cycle to fruition. At the end of the developmental cycle, the infected cells lyse and release infectious EBs that infect new cells (Todd and Caldwell, 1985; Hybiske and Stephens, 2007; Lutter et al., 2013).
C. trachomatis, an extremely sophisticated and adaptive pathogen, has a very small, albeit streamlined (∼1.04 Mb), genome (Stephens et al., 1998). Even though the chlamydial genome encodes several glucose-metabolizing enzymes, it also expresses an ATP/ADP transporter as an immediate early gene, highlighting the fact that the pathogen is not entirely independent of the host cell ATP production (Moulder, 1991; McClarty, 1994; Wylie et al., 1997; Omsland et al., 2012; Fisher et al., 2013). Indeed, cells afflicted with C. trachomatis infection exhibit elevated lipid biosynthesis and NADPH consumption (Fukuda et al., 2005; Szaszák et al., 2011). Thus, to ensure the supply of metabolites for chlamydial development and replication, the host cell is required to withstand and survive the enormous stress generated as a result of the infection.
C. trachomatis uses a multitude of strategies to inhibit host cell apoptosis (Fan et al., 1998; Rajalingam et al., 2008; Kun et al., 2013). Among other pathways, degradation of p53 is one of the key aspects of such resilience of the C. trachomatis–infected host cell to pro-apoptotic stimuli (González et al., 2014). We have recently shown that C. trachomatis infection is affected by the p53-mediated down-regulation of the pentose phosphate pathway (Siegl et al., 2014), which connects C. trachomatis–induced apoptotic resistance to the host cell bioenergetics.
Whereas the bioenergetics of C. trachomatis–infected cells has been the focus of several studies, very little is known about the effect of the infection on the mitochondria and associated dynamics of the mitochondrial network. Mitochondria play a significant role in apoptosis, and evidence shows that alterations in the mitochondrial architecture and dynamics contribute to and are governed by the general metabolic health of the cell (Campello et al., 2006; Labbé et al., 2014). Furthermore, mitochondrial fission and the regulator of mitochondrial fission Drp1 are influenced by increased metabolic demands and apoptotic stress (Cereghetti et al., 2010; Rambold et al., 2011). Recently, it has come to light that the regulation of mitochondrial dynamics is also governed by miRNAs (Wang et al., 2011a; Tak et al., 2014).
Small noncoding RNAs such as miRNAs have emerged as regulators of host cell apoptotic pathways by influencing gene expression of several key modulators, including p53 (Le et al., 2009; Park et al., 2009; Hu et al., 2010). miRNAs have also been implicated in the regulation of host cell bioenergetics and metabolism (Fang et al., 2012; Goedeke et al., 2013; Ramírez et al., 2013). Additionally, several studies show that bacterial infections directly and indirectly affect the host immuno-metabolic (Fassi Fehri et al., 2010; Eisenreich et al., 2013), apoptotic (Hatch et al., 1982; Stephens et al., 1998; Moffatt and Lamont, 2011), and autophagic (Wang et al., 2013) responses by modulation of miRNA and gene expression profiles.
In this study, we applied a high-throughput miRNA sequencing approach to study the effect of C. trachomatis infection on the miRNA expression profile of host cells. We show that the C. trachomatis–induced up-regulation of the host miRNA miR-30c-5p (hereafter, miR-30c) plays an important role in the p53-mediated down-regulation of Drp1, thereby inhibiting stress-induced mitochondrial fission and promoting cell survival during infection.
Results
C. trachomatis infection affects the miRNome of the host and down-regulates p53 in a miR-30c–dependent manner
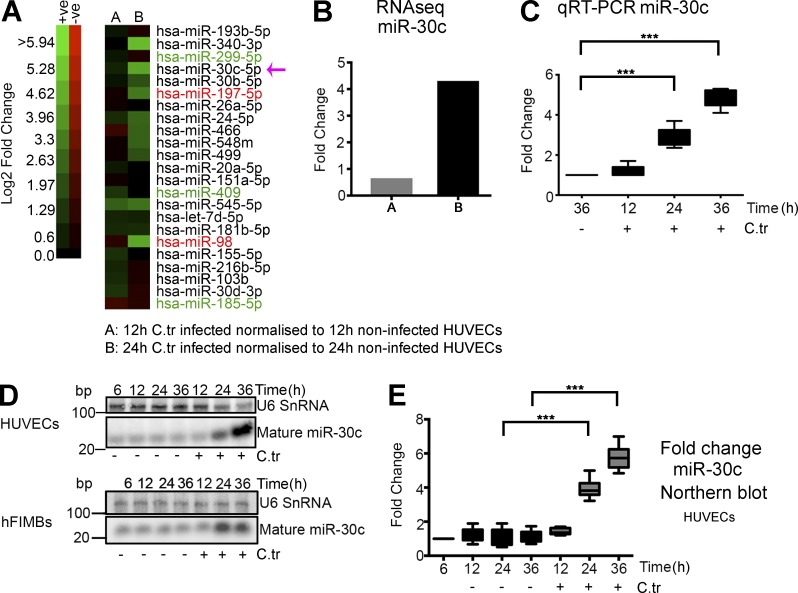
To identify differentially expressed miRNAs in C. trachomatis–infected primary human umbilical vein endothelial cells (HUVECs) versus noninfected samples, we used a high-throughput approach to sequence the miRNome of the samples. Analysis of miRNA deep sequencing data and validation of some of the candidate miRNAs revealed that C. trachomatis–infected HUVECs exhibit differential regulation of the selected miRNAs (Fig. 1 A and Fig. S1, A–D), some of which have been shown to be involved in cell death regulation and carcinogenesis (Wang et al., 2011b; Li et al., 2012; Fiori et al., 2014). One of the up-regulated miRNAs, miR-30c, has been shown to directly target the p53 mRNA (Li et al., 2010), which is concordant with the previous observations of p53 down-regulation in C. trachomatis–infected cells (González et al., 2014; Siegl et al., 2014). Therefore, we focused further research on miR-30c. After 24 h of C. trachomatis infection, up-regulation of miR-30c could be detected not only by miRNA sequencing in HUVECs (Fig. 1, A and B), but also by quantitative real-time PCR (qRT-PCR) in HUVECs (Fig. 1 C) and Northern blot in HUVECs, primary epithelial cells of the human fallopian tube fimbriae (hFIMB cells; Fig. 1, D and E), and primary human foreskin fibroblasts (HFF; Fig. S1 E). We then modulated the levels of miR-30c by transfecting mimics and inhibitors into HUVECs before infection. Transfection of miR-30c mimic promotes, whereas inhibition negatively affects, chlamydial growth in HUVECs (Fig. 2 A). Additionally, we used an inducible miR-30c sponge to create a miR-30c knockdown HeLa cell line. Anhydrous tetracycline (AHT)–induced expression of the sponge, determined by increase in p53, caspase 3, and DRP1 levels (Fig. S1, F and G) and GFP expression (Fig. S1, H and I), reduced the ability of C. trachomatis to grow and develop (Fig. 2, B–D) and produce infectious progeny (Fig. S1 J). The effect of AHT alone on C. trachomatis growth was insignificant (Fig. S1 K). At the same time, HeLa cells expressing the miR-30c sponge exhibited a marked decrease in mitochondrial fragment length and an increase in mitochondrial fragment count as observed by confocal microscopy (Fig. 2, E and F). A similar effect on mitochondria was observed when miR-30c was artificially modulated in HUVECs using mimics and inhibitors (Fig. S1, L–N).
Figure 1.
C. trachomatis infection increases miR-30c abundance in multiple cell types. (A) Heat map represents log2 fold changes of several miRNAs derived from RNA sequencing. miRNAs reported to be pro-apoptotic are labeled in green, and those reported to be anti-apoptotic are labeled in red. (B) Graph represents the log2 fold changes of miR-30c determined by miRNA deep sequencing of HUVECs after 12 and 24 h of C. trachomatis (C.tr) infection compared with noninfected samples. (C) Graph represents quantification of miR-30c up-regulation upon C. trachomatis infection by qRT-PCR. U6 snRNA was used as endogenous control for qRT-PCR. Cells were infected for 12, 24, and 36 h, and fold changes were normalized to miR-30c expression of noninfected cells at 36 h. Fold change with qRT-PCR for HUVECs (± SD) at 24 h, 2.96 ± 0.47; and 36 h, 4.87 ± 0.46 (n = 6). (D and E)Northern blots of HUVEC and hFIMB cell RNA, probed for U6 SnRNA, miR-30c, and the chlamydial small RNA, CtrR5. Fold change with Northern blot for HUVECs (± SD) at 24 h, 3.9 ± 0.58; and 36 h, 5.76 ± 0.72 (n = 5). Fold changes were normalized to miR-30c expression in noninfected cells at 12, 12, and 36 h, respectively (n = 6 experiments). All data represents mean ± SD. Asterisks denote significance by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant. See also Fig. S1.
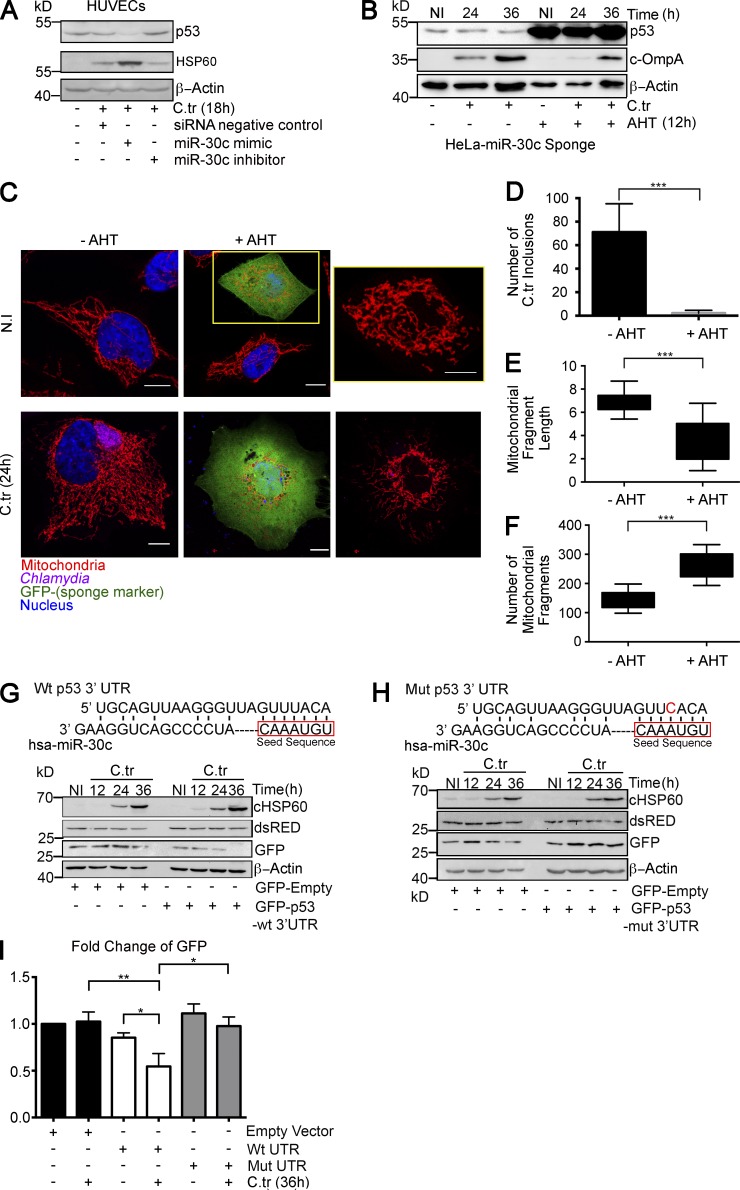
Figure 2.
C. trachomatis infection down-regulates p53 in a miR-30c–dependent manner. (A) Immunoblot of HUVECs transfected with siRNA negative control, miR-30c mimic, or inhibitor for 36 h followed by C. trachomatis (C.tr) infection for 12 h. The blots were probed with antibodies against the miR-30c target p53, chlamydial outer membrane protein A (cOmpA), and β-actin. (B) Immunoblot of HeLa cells expressing miR-30c sponge after AHT induction. Blots were probed with antibodies against miR-30c target p53, cOmpA, and β-actin. Induced and noninduced cells were infected with C. trachomatis for 24 and 36 h. (C) Sponge-mediated miR-30c depletion in HeLa results in mitochondrial fragmentation and affects chlamydial growth. Bars, 10 µm. Yellow box and inset point out the damaged mitochondrial architecture. (D) Quantification of chlamydial inclusion in induced and noninduced samples infected with C. trachomatis for 24 h. The samples were stained against cHSP60 (Cy3, 570 nm), and MitoTracker Deep Red FM was used to label the mitochondria. The number of inclusions was quantified using Object Count; ∼600 cells were analyzed from a random selection of six regions of interest (ROIs)/sample; n = 3. (E and F) Graphs represent the analysis of mitochondrial fragment length distribution and number of individual mitochondrial fragments in noninduced and AHT-induced miR-30c sponge cells. n = 6; ∼20 cells were analyzed from a random selection of ∼10 ROIs in each sample. Immunoblot of chimeric GFP protein under the control of the wild-type p53 3′ UTR (G) and mutant p53 3′ UTR (H). Blots were probed with antibodies against GFP, cHSP60, dsRed, and β-actin. (I) Quantification of chimeric GFP expression normalized to the expression of the dsRed transfection control (n = 3). All data represent mean ± SD. Asterisks denote significance by Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001. See also Fig. S1.
We have previously shown that C. trachomatis infection in HUVECs results in a dramatic down-regulation of the tumor suppressor p53 in a phosphoinositide 3 kinase–Akt–dependent manner via HDM2 regulation (Siegl et al., 2014). We further tested whether chlamydial infection can regulate p53 in a miR-30c–dependent manner. We created GFP-based reporter constructs that carry the wild type p53 3′ UTR or the p53 3′ UTR in which the miR-30c binding site was altered. C. trachomatis infection of HUVECs transfected with the reporter construct down-regulated the levels of GFP under the control of the wild-type p53 3′ UTR, whereas the levels of GFP under the control of the mutated 3′ UTR remained unchanged (Fig. 2, G–I). Our results show that C. trachomatis–induced down-regulation of the host p53 is not dependent solely on the E3 ligase activity of HDM2, but depends also on the regulation of p53 mRNA by miR-30c.
Down-regulation of p53 during C. trachomatis infection is a prerequisite for normal chlamydial development (Siegl et al., 2014). Therefore, it is very likely that the strong interference of miR-30c down-regulation on chlamydial development (Fig. 2, A–D) depends on the stabilization of p53 resulting from the reduction in miR-30c abundance (Fig. S1, G–I). Additionally, modulation of miR-30c had noticeable effects on the mitochondrial architecture of both HUVECs and HeLa cells.
C. trachomatis infection reduces Drp1 protein levels and inhibits accumulation of Drp1 aggregates on the mitochondrial surface
The cellular mitochondrial network is greatly influenced by the general metabolic health of the cell (Campello et al., 2006; Labbé et al., 2014). A cell stressed with increased energy demand will exhibit hyperfused mitochondria (Gomes et al., 2011; Rambold et al., 2011), whereas fission is necessary for quality control and degradation of damaged organelles (Twig et al., 2008). The delicate balance between fusion and fission of mitochondrial fragments is maintained by a dedicated group of cytoplasmic and mitochondrial proteins, which in turn are regulated by immune response pathways (Kang et al., 2015), cellular stress (Breckenridge et al., 2003; Karbowski et al., 2004), oxidative stress (Pletjushkina et al., 2006), and miRNAs (Wang et al., 2011a; Tak et al., 2014). Dynamin-related protein 1 (Drp1) is one such cytoplasmic GTPase protein that functions as a master regulator of mitochondrial fission (Smirnova et al., 2001). Several groups have studied the regulation of Drp1 in depth (Cribbs and Strack, 2007; Karbowski et al., 2007; Braschi et al., 2009). Recently, it has come to light that miRNAs also influence the fission-inducing activity of Drp1 via modulation of the tumor suppressor protein p53 (Li et al., 2010; Wang et al., 2011a). Both p53 and Drp1 have pivotal roles in mitochondrial regulation of apoptosis (Gomes et al., 2011), and Drp1-dependent mitochondrial remodeling is a crucial step for induction of apoptosis (Losón et al., 2013).
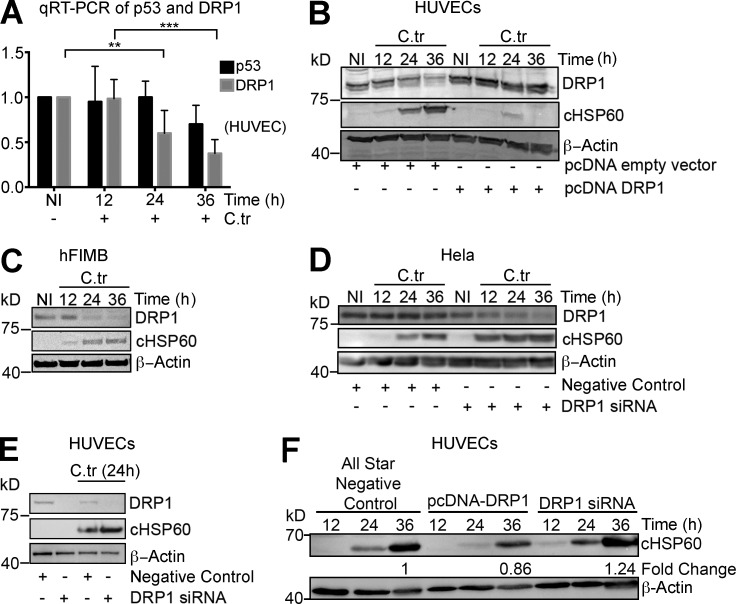
Overexpression of miR-30c and p53 suppression can reduce disease-associated mitochondrial fragmentation by inhibiting the transcription of Drp1 in specific cell types (Li et al., 2010). Additionally, p53 is reported to induce Drp1 translocation to the mitochondrial surface by suppression of miR-499 expression, which in turn regulates the expression of the Drp1 activator, calcineurin (Wang et al., 2011a). Thus, we determined whether C. trachomatis infection affects the levels of Drp1 in HUVECs. C. trachomatis–infected HUVECs exhibit decreased levels of Drp1 mRNA and protein 24 and 36 h after infection. Correspondingly, the amount of p53 mRNA changes as well (Fig. 3, A and B). Similar changes in the protein levels of Drp1 were also observed in hFIMB cells (Fig. 3 C). This observation corresponds to the C. trachomatis–induced up-regulation of miR-30c (Fig. 1, B–E) and down-regulation of p53 expression (Fig. 2 A). Overexpression of Drp1 almost completely abrogated C. trachomatis growth in HUVECs (Fig. 3 B). Consistent with the previous observations, targeted down-regulation of Drp1 in HeLa cells and HUVECs enhances chlamydial growth and production of infectious progeny (Fig. 3, D–F, and Fig. S2 A), Furthermore, Operetta-based high-throughput characterization of chlamydial inclusion in cells with overexpression or knockdown of Drp1 revealed significant differences in inclusion size. Although C. trachomatis was barely able to establish infection in cells expressing enhanced levels of DRP1, the inclusions in DRP1 siRNA-transfected cells were significantly bigger compared with control (Fig. S2, B and C)
Figure 3.
C. trachomatis infection reduces Drp1 protein levels upon infection and is affected by artificial modulation of Drp1 levels. (A) Quantification of qRT-PCR for p53 and Drp1 mRNA levels in noninfected and C. trachomatis (C.tr)–infected HUVECs for 12, 24, and 36 h. Fold change of Drp1 (± SD) at 24 h, 0.6 ± 0.30; and 36 h, 0.325 ± 0.10 (n = 6). (B) Immunoblot of HUVECs transfected with vector control or pcDNA-Drp1 overexpression vector and infected with C. trachomatis for 12, 24, and 36 h. Blots were probed with antibodies against Drp1, chlamydial heat shock protein 60 (cHSP60), and actin. (C) Immunoblot of hFIMB cells infected with C. trachomatis for 12, 24, and 36 h. Blots were probed with antibodies against Drp1, cHSP60, and actin. (D) Immunoblot of HeLa cells transfected with siRNA negative control and Drp1 siRNA infected with C. trachomatis for 12, 24, and 36 h. Blots were probed with antibodies against Drp1, cHSP60, and actin. (E) Immunoblot of HUVECs transfected with siRNA negative control or siRNA against Drp1 for 24 h and infected with C. trachomatis for 18 h. Blots were probed with antibodies against Drp1, cHSP60, and actin. (F) Immunoblot of infectivity assays of 36 h of primary C. trachomatis infection in HUVECs transfected with siRNA negative control, pcDNA-Drp1 overexpression vector, or Drp1 siRNA pool. All data represents mean ± SD. Asterisks denote significance by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant.
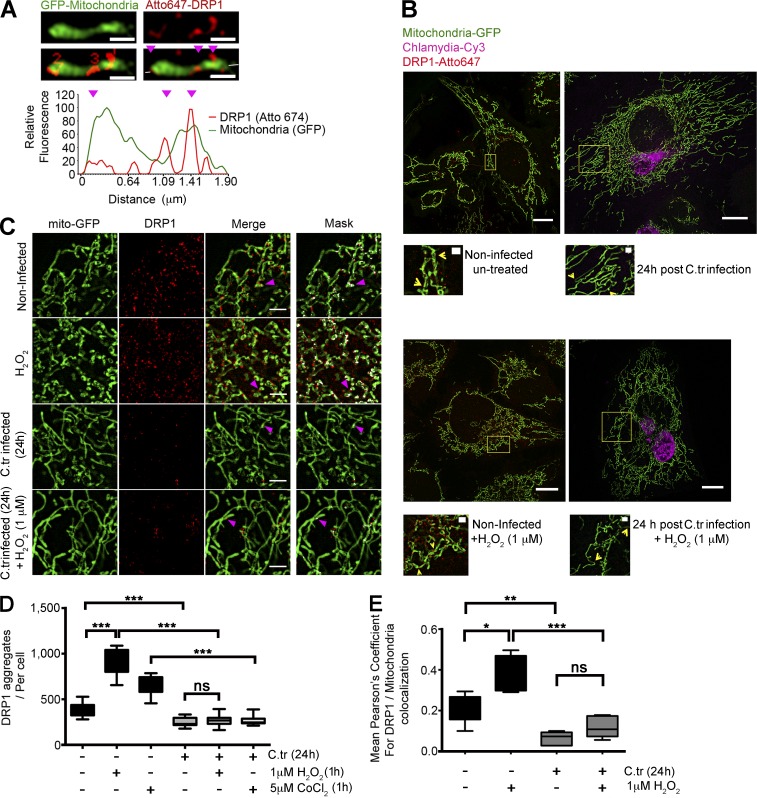
Drp1 undergoes extensive posttranslational modifications such as phosphorylation (Cribbs and Strack, 2007), S-nitrosylation (Haun et al., 2013), ubiquitination (Karbowski et al., 2007), and SUMOylation (Braschi et al., 2009), which govern its translocation to the mitochondrial surface and capacity to form ring-like aggregates. Using superresolution structured illumination microscopy (SR-SIM), we plotted the intensity profiles of mitochondrial fragments and Drp1 to identify mitochondrial fission sites. Using the intensity profiles, we determined the presence of Drp1 aggregates on regions of the mitochondria exhibiting constrictions (Fig. 4 A). Quantification of the number of Drp1 aggregates on the surface of the GFP-tagged mitochondria of uninfected and C. trachomatis–infected HUVECs (mito-HUVECs) revealed a reduction in the number of Drp1 aggregates in the infected cells (Fig. 4, B–D). Furthermore, quantification of constricted and dilated Drp1 rings in control and C. trachomatis–infected cells showed that infected cells had reduced instances of both (Fig. S2, D and E).
Figure 4.
C. trachomatis infection inhibits accumulation of Drp1 aggregates on the mitochondrial surface. (A) Graph represents fluorescence intensity profile plot of a mitochondrial fragment (green) with Drp1 aggregates (red) on constriction sites (magenta arrowheads). The number of Drp1 aggregates was quantified, and intensities in both channels were measured along the depicted axis (white line in the last panel through the fragment). Bars, 1 µm. (B) Structured illumination micrographs of mito-HUVECs infected with C. trachomatis (C.tr; stained against cHSP60; Cy3), treated with 1 µM H2O2 for 1 h, or treated with 1 µM H2O2 for 1 h after 24 h of C. trachomatis infection. The samples were probed against Drp1 and stained with Atto647N. Bars, 10 µm. Zoomed insets (yellow boxes) show DRP 1 aggregation at mitochondrial constriction sites. Bars, 1 µm. (C and D) Quantification of Drp1 aggregates (sizes 100 to 280 nm) in C. trachomatis–infected and uninfected HUVECs. Indicated samples were treated with 1 µM H2O2 for 1 h or treated with 1 µM H2O2 for 1 h after 24 h of C. trachomatis infection. Number of Drp1 aggregates per cell (± SD) for noninfected and infected cells was 377.2 ± 65.86 and 251.8 ± 46.44 (∼60 cells were analyzed from random selection of 20 regions of interest [ROIs] in each sample). 2 or 3 random 10 × 10-µm ROIs were chosen in each selected cell. Significance was determined by Mann–Whitney test; n = 10). (C and E) Structured illumination micrographs and graph represent colocalization analysis (Pearson’s coefficient) between Drp1 and mitochondrial GFP using the COLOC2 plugin; ∼20 cells were analyzed per sample. 2 or 3 random 10 × 10-µm ROIs were chosen in each selected cell. n = 10). Bars, 2 µm. All data represent mean ± SD. Asterisks denote significance by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant. See also Figs. S2 and S3.
Additionally, comparison of the degree of colocalization of the Drp1 aggregates with mitochondria between C. trachomatis–infected and noninfected HUVECs showed a marked decrease in the presence of Drp1 aggregates on the mitochondrial surface of C. trachomatis–infected cells (Fig. 4, D and E). Treatment with cobalt chloride (CoCl2) and hydrogen peroxide (H2O2) does not significantly increase the abundance of Drp1 aggregates or colocalization with mitochondria in C. trachomatis–infected cells (Fig. 4, C–E). CoCl2 acts by inducing a reactive oxygen species (ROS)-mediated up-regulation of p53 expression, whereas H2O2, in addition to p53 up-regulation, depletes the miR-30 family, leading to an eventual up-regulation of Drp1-mediated mitochondrial fission (An et al., 1998; Li et al., 2010). However, upon treatment with H2O2, the levels of miR-30c remained unchanged in C. trachomatis–infected HUVECs, and levels of p53 remained down-regulated (Fig. S2, F and G). This supports an essential role of maintaining high miR-30c levels in infected cells even under ROS stress.
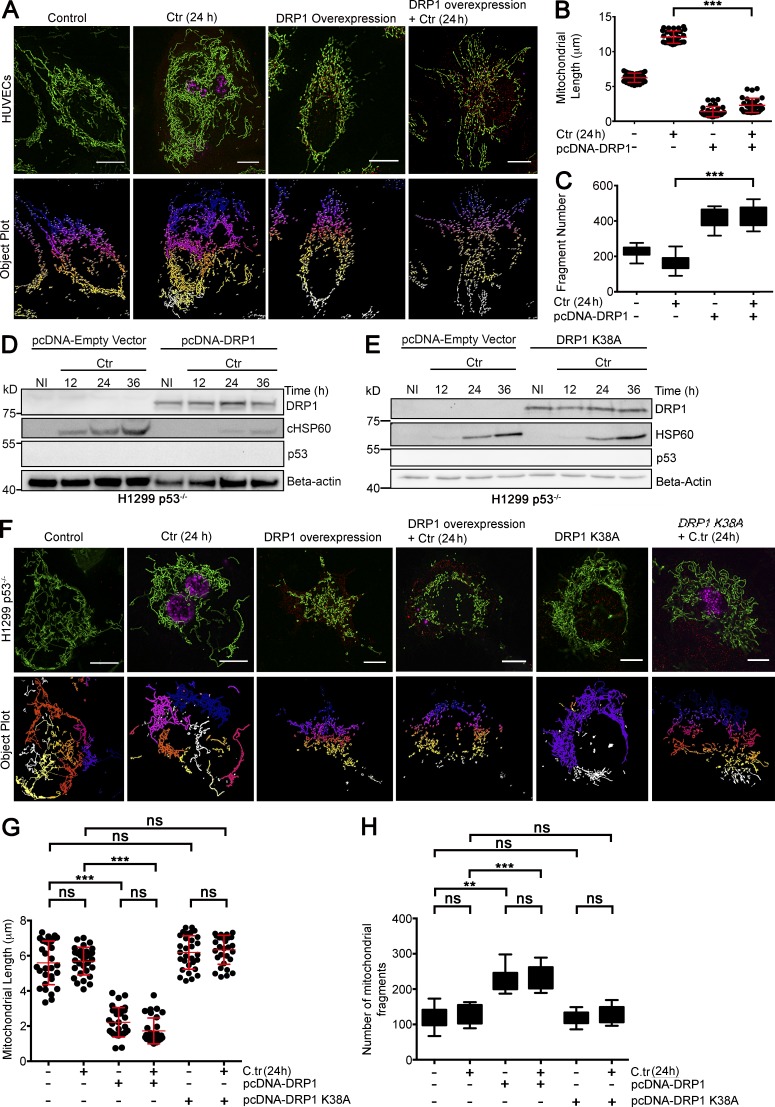
Drp1 has been shown to form ring-like aggregates on the mitochondrial surface, which cause outer mitochondrial membrane scission in a GTP-dependent conformational change of the ring diameter (Ingerman et al., 2005; Mears et al., 2011; Rosenbloom et al., 2014). Overexpression of Drp1 induced massive mitochondrial fragmentation and inhibited chlamydial infection not only in HUVECs (Fig. 3 B; and Fig. 5, A–C) but also in H1299 p53−/− cells (Fig. 5, D–H), demonstrating that Drp1 can induce fission and inhibit C. trachomatis even in the absence of p53. However, this inhibitory effect of DRP1 on the growth and development of C. trachomatis was not observed when H1299 p53−/− cells were transfected with a catalytically inactive and dominant negative variant of DRP1 (DRP1K38A; Smirnova et al., 1998), which fails to induce and inhibits successful mitochondrial fission events (Fig. 5, E–H). Interestingly, C. trachomatis infection had no significant effect on the mitochondrial length of H1299 cells, possibly because of the lack of p53.
Figure 5.
Functional Drp1 overexpression in a p53-null background fragments mitochondria and inhibits C. trachomatis infection. (A) Structured illumination micrograph of mito-HUVECs infected with C. trachomatis (C.tr) for 24 h, transfected with pcDNA-Drp1 overexpression vector, or both. Graphs represent quantification of mitochondrial length and the number of mitochondrial fragments in HUVECs upon C. trachomatis infection, overexpression of Drp1, or both. Bars, 10 µm. (B) Analysis of mitochondrial fragment length distribution in HUVECs infected with C. trachomatis and transfected with Drp1 overexpression vector. Dots represent the mean mitochondrial fragment length of ∼3 cells in a region of interest (ROI) chosen at random within samples. 30 such dots were plotted on the graph. Mean mitochondrial fragment length (μm; ± SD) in control, 6.25 ± 0.66; C. trachomatis (24 h), 12.08 ± 0.87; Drp1 overexpression, 1.39 ± 0.74; and Drp1 overexpression + C. trachomatis (24 h), 2.3 ± 1.02. (C) Analysis of mitochondrial fragment count in HUVECs infected with C. trachomatis and transfected with Drp1 overexpression vector. Mean mitochondrial fragment count (± SD) in control, 240 ± 30.08; C. trachomatis (24 h), 133 ± 44.31; Drp1 overexpression, 388 ± 51.69; and DRP1 overexpression + C. trachomatis (24 h), 399 ± 58.58 (n = 3 for B and C; ∼30 cells were analyzed from random selection of ∼10 ROI in each sample). (D and E) Immunoblots for different durations of C. trachomatis infection in control H1299 p53−/− cells and H1299 p53−/− cells transfected with pcDNA-Drp1 overexpression vector or dominant negative variant of Drp1-Drp1K38A (Smirnova et al., 2001). Blots were probed for Drp1, p53, cHSP60, and β-actin (n = 3). (F) Structured illumination micrograph of H1299 p53−/− cells expressing GFP targeted to mitochondria. Samples were transfected with pcDNA-Drp1 overexpression vector or DRP1K38A and infected with C. trachomatis for 24 h. Bars, 10 µm. (G) Graph represents quantification of the mitochondrial length distribution in H1299 p53−/− cells upon C. trachomatis infection and overexpression of Drp1 or Drp1K38A before infection. Dots represent the mean mitochondrial fragment length of ∼3 cells in an ROI chosen at random within samples. 30 such dots were plotted on the graph. Mean mitochondrial fragment length (μm; ± SD) in control, 5.59 ± 1.24; C. trachomatis, 5.69 ± 0.78; DRP1 overexpression, 2.21 ± 0.83; Drp1 overexpression + C. trachomatis (24 h), 1.72 ± 0.73; Drp1K38A overexpression, 6.18 ± 0.95; and DRP1 K38 overexpression + C. trachomatis (24 h), 6.33 ± 0.82. (H) Analysis of mitochondrial fragment count in H1299 p53−/− cells upon C. trachomatis infection, overexpression of Drp1 or DRP1K38A before infection. Mean mitochondrial fragment count (± SD) in control, 121.31 ± 27.56; C. trachomatis, 130.56 ± 25.56; DRP1 Overexpression = 225.87 ± 29.63; Drp1 overexpression + C. trachomatis (24 h), 233.43 ± 32.45; Drp1K38A overexpression, 119.12 ± 32.45; and Drp1K38A overexpression + C. trachomatis infection (24 h), 128 ± 22.96 (n = 3 for panels G and H; ∼30 cells were analyzed from random selection of ∼10 ROIs in each sample). All data represent mean ± SD. Asterisks denote significance by one-way analysis of variance followed by Tukey’s multiple comparisons test for panel C and panel D: **, P < 0.01; ***, P < 0.001; ns, nonsignificant.
Similarly to the effect of Drp1 overexpression on C. trachomatis in H1299 p53−/− cells, overexpression of p53 pre- and postinfection also has an inhibitory effect on C. trachomatis growth. Additionally, forced overexpression of p53 increases both Drp1 expression and its mitochondrial localization in H1299 p53−/− cells (Fig. S3, A–F).
Thus, we argue that C. trachomatis–induced up-regulation of miR-30c diminishes p53 expression and, as a result, reduces expression and accumulation of functional Drp1 on the mitochondrial surface. Defects in Drp1 have been shown to result in hyperfused mitochondria and resistance to mitochondrial fragmentation and apoptosis (Qian et al., 2012; Thomas and Jacobson, 2012). Collectively, our observations corroborate the previously known fact that C. trachomatis can inhibit apoptosis in HUVECs by down-regulating p53 (Siegl et al., 2014) and imply that C. trachomatis infection affects mitochondrial fission in the host via p53-mediated down-regulation of Drp1.
C. trachomatis infection modulates ROS production within the mitochondrial matrix
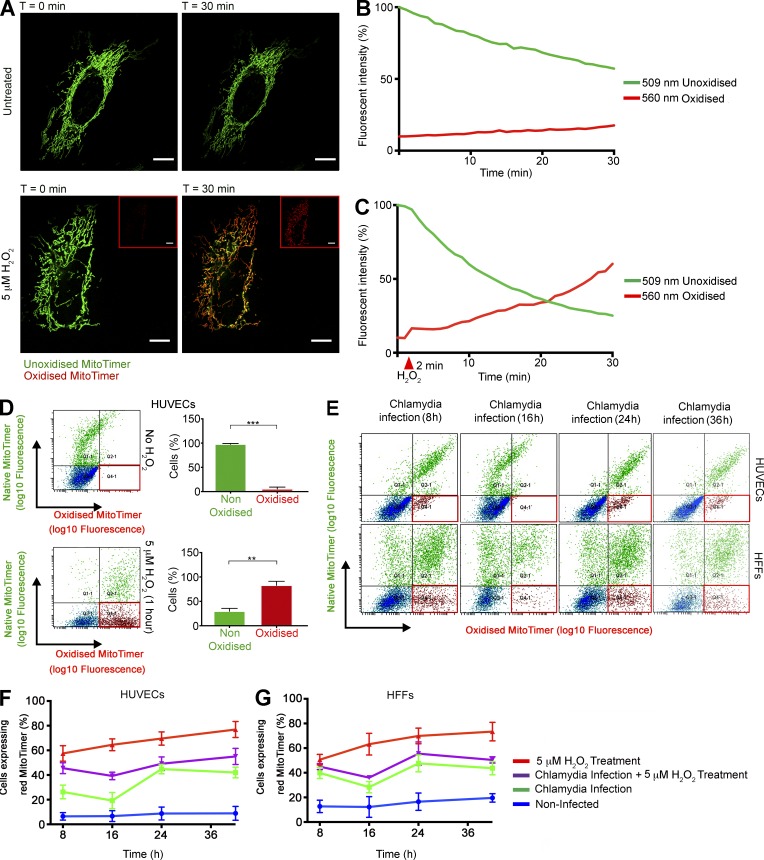
It has been reported that C. trachomatis induces ROS production upon infection (Boncompain et al., 2010), but the effect of such an event on mitochondria and the mitochondrial matrix has not been studied. We measured the effect of C. trachomatis infection on the mitochondrial matrix using a Timer protein targeted to mitochondria by fusing the mitochondrial targeting sequence of the human cytochrome c oxidase subunit VIII (Cox8) gene (MitoTimer; Laker et al., 2014). MitoTimer exhibits a shift in fluorescence spectrum (500 to 596 nm) upon encountering ROS. We transiently expressed the MitoTimer protein in HUVECs and HFF cells and confirmed the localization in the mitochondrial matrix (Fig. S4, A and B). The expression of MitoTimer is observable 8 h after transfection as a green fluorescence (unoxidized form). The protein eventually matures to red (oxidized form), which can be prevented to an extent by washing and exchanging medium every 4 h (Fig. 6 A). Although the MitoTimer protein has previously been used to exhibit protein turnover in mitochondria and equilibration of mitochondrial protein age (Ferree et al., 2013), other studies have shown the susceptibility of MitoTimer to oxidative stress (Laker et al., 2014). To confirm this observation, we performed live-cell imaging with MitoTimer-transfected HUVECs in the presence and absence of 5 µM H2O2. Upon H2O2 treatment, MitoTimer was found in its oxidized form (Fig. 6, A–D; and Videos 1 and 2), which is consistent with the effect of H2O2 to create oxidative stress within the mitochondrial matrix. HUVECs transfected with MitoTimer were infected with C. trachomatis, and the shift in fluorescence spectrum was measured by quantifying the percentage of cells expressing the red oxidized form of MitoTimer by flow cytometry. We observed that 8 h after infection, there is a significant increase in the population of cells exhibiting the red variant of MitoTimer within the mitochondrial matrix; 16 h after infection, however, this effect is reversed and the population of cells expressing the red variant of MitoTimer is reduced significantly. There is a second peak of fluorescence shift at 24 h after infection, during which ∼44% (± 3.8%; ± SD) of the cells begin to exhibit the red variant of MitoTimer. This population gradually reaches a plateau phase after 24 h. By that time, ∼10% of the noninfected HUVECs begin exhibiting red fluorescence within the mitochondrial matrix (Fig. 6, E and F) possibly because of the accumulation of metabolites. Similar effects were also observed in MitoTimer-transfected HFF cells (Fig. 6, E and G). Interestingly, treatment of HUVECs with 1 µM H2O2 30 min before C. trachomatis infection does not significantly alter the fluorescence shift pattern upon infection, whereas such treatment in uninfected cells results in a dramatic increase in cell populations expressing the red variant of Timer with time. C. trachomatis–infected cells can also resist the H2O2-induced down-regulation of miR-30c and up-regulation of p53 (Fig. S2, F and G).
Figure 6.
C. trachomatis infection modulates ROS production within the mitochondrial matrix. (A) Micrographs represent start (t = 0 min) and endpoints (t = 30 min) of Videos 1 and 2. pcDNA-MitoTimer was transfected into HUVECs, and the mitochondrial localization was ascertained by confocal laser scanning microscopy after 12 h. Bars, 10 µm. The samples were imaged simultaneously at 500 nm (green; native, unoxidized form) and 596 nm (red; oxidized form) to detect the fluorescence shift. 5 µM H2O2 was added into imaging media of indicated samples after 2 min of untreated imaging. Insets show the red channel images of the H2O2 -treated cells at t = 0 and 30 min. Images were captured every 60 s for 30 min. (B and C) Graphs indicate the change in fluorescent properties of the Timer protein in the absence and presence of H2O2 treatment. The arbitrary fluorescent intensity values were normalized to the starting fluorescent intensity value of the Timer protein in the green channel and depicted as percentage of that value. (D) Scatterplots indicate the change in fluorescent properties of the Timer protein in the absence and presence of H2O2 treatment as determined by FACS. Graphs indicate the percentages of cell population that exhibit fluorescence in the red channel (red box). (E) Scatter plots of MitoTimer transfected HUVEC and HFF cells after C. trachomatis (C.tr) infection for 8, 16, 24, and 36 h. A second group of cells were pretreated with 5 µM H2O2 for 30 min, washed, and infected with C. trachomatis for 8, 16, 24, and 36 h. Finally, as a control, HUVEC and HFF cells were treated with 5 µM H2O2 for 30 min, washed, and allowed to grow for 8, 16, 24, and 36 h. All samples were transfected 12 h before infection or treatment. The red box indicates the fraction of cell population expressing the red MitoTimer. (F and G) Graphs indicate the percentages of HUVEC and HFF cell populations that exhibit fluorescence in the red channel after treatments (n = 3). All data represent mean ± SD. Asterisks denote significance by Student’s t test. **, P < 0.01; ***, P < 0.001; ns, nonsignificant. See also Videos 1 and 2.
We next transfected MitoTimer-expressing HUVECs with p53 and DRP1 siRNAs (separately) and treated them with 5 µM H2O2. Although siRNA-mediated knockdown of p53 and DRP1 did not have significant effects on the percentage of cells expressing the red variant of MitoTimer, the samples of HUVECs transfected with both p53 and DRP1 siRNAs and treated with 5 µM H2O2 exhibited significantly lower percentages of cells expressing the oxidized variant of MitoTimer (Fig. S4, C and D).
In conclusion, our data show that C. trachomatis infection reversibly induces stress within the mitochondrial matrix and at the same time provides protection from external sources of oxidative stress. This protection could be partially attributed to the down-regulation of p53 and Drp1 upon infection.
C. trachomatis infection induces changes in the mitochondrial architecture by inhibiting fragmentation
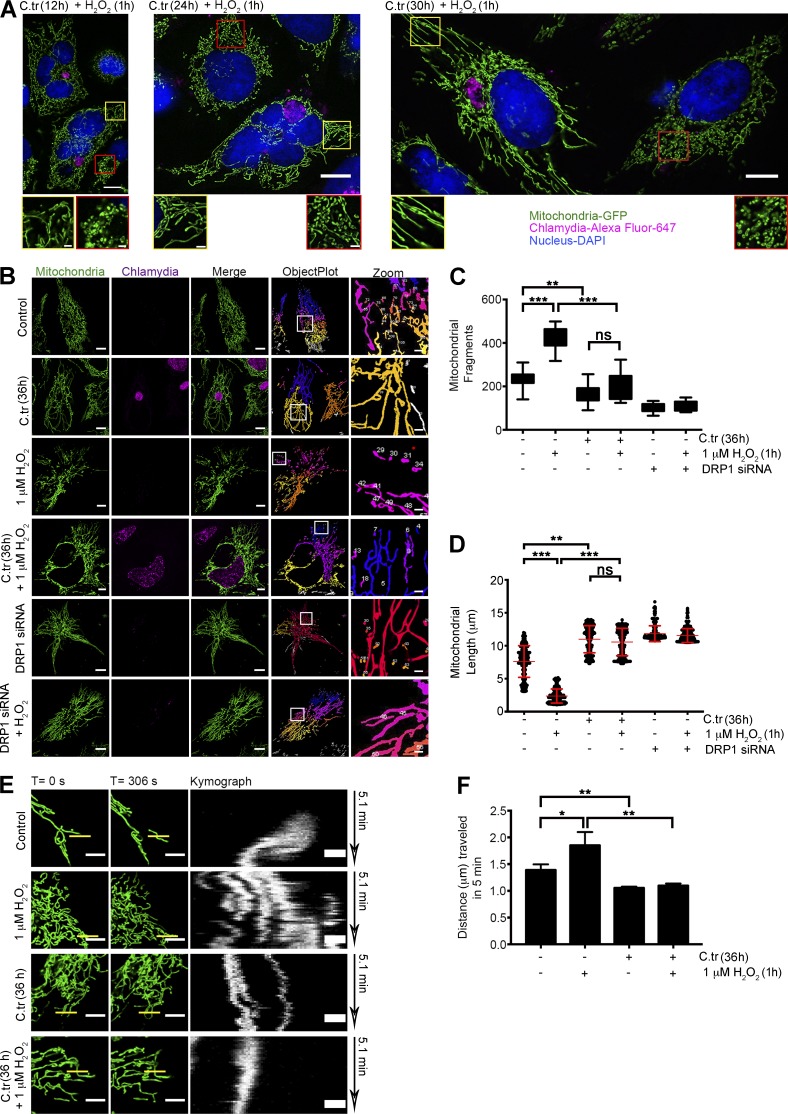
To gauge the extent to which C. trachomatis affects the mitochondrial morphology of the host, we studied the changes in mitochondrial morphology in C. trachomatis–infected mito-HUVECs after H2O2 treatment by SR-SIM (Fig. 7 A). Comparison of the mitochondrial fragment distribution and total mitochondrial area between noninfected and C. trachomatis–infected mito-HUVECs showed that infected cells exhibited mitochondrial phenotypes similar to those in Drp1 siRNA–transfected cells with elongated and hyperfused mitochondria (Fig. 7, B and C). C. trachomatis infection decreased the occurrence of isolated mitochondrial fragments, while simultaneously increasing the overall connectivity and length of the mitochondrial fragments. Whereas treatment with 1 µM H2O2 for 1 h increased fragmentation in uninfected cells, C. trachomatis–infected cells were resistant to such profragmentation stress (Fig. 7, A–D).
Figure 7.
C. trachomatis infection induces changes in the mitochondrial architecture by inhibiting fragmentation. (A) Structured Illumination micrographs of mito-HUVECs posttreated with 1 µM H2O2 for 1 h after 12, 24, and 30 h of C. trachomatis (C.tr) infection (C. trachomatis stained with Alexa Fluor 647 against cHSP60 and DAPI to mark the nucleus). Bar, 10 µm. Red-bordered insets focus on uninfected cells, and yellow-bordered insets focus on infected cells. Bar, 1.5 µm. (B) Micrographs represent data processing and mitochondrial segmentation by rolling ball thresholding and 3D object count. Bars, 10 µm. (C) Analysis of mitochondrial fragment length distribution in cells treated with 1 µM H2O2 and infected with C. trachomatis. Dots represent the mean mitochondrial fragment length of approximately three cells in a region of interest (ROI) chosen at random within samples. 200 such dots were plotted on the graph. Mean mitochondrial fragments length in control (± SD), 7.607 ± 2.39; H2O2, 2.36 ± 1.06; C. trachomatis, 10.98 ± 2.05; and C. trachomatis + H2O2, 9.90 ± 1.84 (n = 20; ∼30 cells were analyzed from random selection of ∼10 ROIs in each sample). (D) Comparison of fragment distribution between infected and noninfected cells. Treatment with 1 µM H2O2 is taken as positive control for fragmentation, and 36 h of siRNA-mediated Drp1 knockdown is used as negative control. Number of fragments in control (± SD), 235.4 ± 36.34; H2O2, 414.6 ± 50.39; C. trachomatis, 169.1 ± 41.12; and C. trachomatis + H2O2, 198 ± 61.52 (n = 20 for C and D; ∼30 cells were analyzed from random selection of ∼10 ROIs in each sample). (E) Freeze-frame of 20 × 20-µm sections of uninfected and C. trachomatis–infected mito-HUVECs from time-lapse video micrographs at t = 0 and 306 s (Videos 3–6). Indicated samples were treated with 1 µM H2O2. White bars represent 5 µm. Yellow bars (0.5 µm in thickness) represent arbitrary lines drawn to determine lateral displacement of mitochondrial fragments with time as represented by the Kymograph panels (unquantified). (F) Graph represents differences in mitochondrial mobility between HUVECs after 36 h of C. trachomatis infection, 1 µM H2O2 treatment for 1 h, or treatment with 1 µM H2O2 after 36 h of C. trachomatis infection. The distance moved by the mitochondrial fragments in 5 min was determined by analyzing Videos 3–6 with the mitoCRWLR macro (n = 5; three ROIs were analyzed from individual cells, with six cells being chosen at random from every sample). All data represent mean ± SD. Asterisks denote significance by one-way analysis of variance followed by Tukey’s multiple comparisons test for C and D: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant. See also Video 3–6.
We also measured the rates of mitochondrial mobility using the novel macro script, mitoCRWLR (see Online supplemental material), because the changes in the rates of mitochondrial movement are largely governed by alterations in the fusion/fission ratio of the mitochondrial network (Okamoto and Shaw, 2005). Although H2O2 treatment of noninfected cells enhances mitochondrial fragmentation and random movements, C. trachomatis–infected cells were resistant to these changes (Fig. 7, E and F; and Videos 3, 4, 5, and 6).
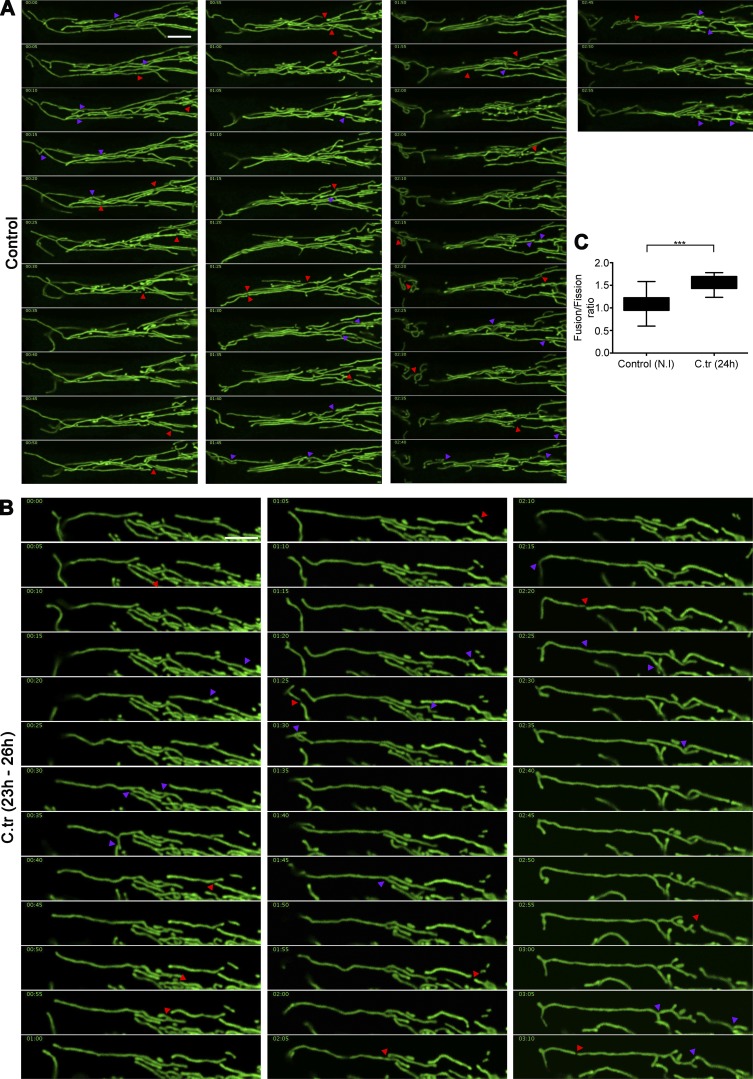
To understand if the mitochondrial elongations seen in the C. trachomatis–infected cells were a result of increased number of fusion events or decreased fission events, we manually tracked segments of mitochondria within infected and noninfected cells for ∼3 h (Fig. 8, A and B; and Videos 7 and 8). We observed that, whereas infected cells exhibited a slight decrease in the occurrence of mitochondrial fusion, fission events were found to be quite rare. This reduced occurrence of fission events results in a slightly higher fusion/fission ratio in C. trachomatis–infected cells compared with noninfected controls, which favors elongation of the mitochondrial fragments (Fig. 8 C).
Figure 8.
Changes in the mitochondrial fusion/fission ratio induce elongation of mitochondrial fragments in C. trachomatis–infected cells. Montage of time-lapse confocal micrography of noninfected (A) and 24-h C. trachomatis (C.tr)–infected (B) HUVEC mitochondria (Videos 7 and 8). The samples were imaged for 3 h with pictures taken every 5 min. The purple arrowheads indicate fusion events, and the red arrowheads indicate fission events. Bars, 10 µm. (C) Graph represents differences in mitochondrial fusion/fission ratio between noninfected and C. trachomatis–infected samples. Fusion/fission ratio in control (± SD), 1.09 ± 0.22; and C. trachomatis, 1.58 ± 0.17 (n = 5; two regions of interest were analyzed from individual cells, with three cells being chosen at random from every sample). Asterisks denote significance by Student’s t test. ***, P < 0.001. See also Video 7.
C. trachomatis infection is affected by regulation of ATP synthesis in the host mitochondria
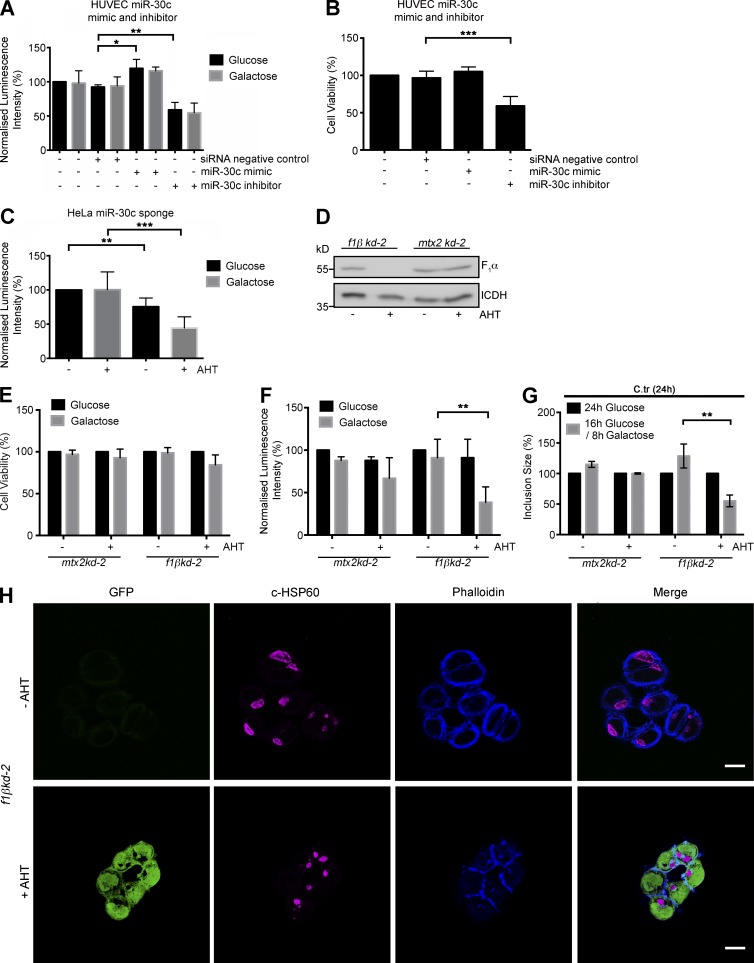
Changes in mitochondrial morphology caused by aberrant fission/fusion ratios are among the earliest observable hallmarks of cellular stress (van der Bliek et al., 2013). An increasingly tubular nature of the mitochondrial network indicates a heightened requirement for oxidative phosphorylation (Sansome et al., 2001). Intracellular C. trachomatis, particularly the RBs, takes up ATP from the host cell (Omsland et al., 2012). This results in a drain on the cellular resources of ATP, and thus preservation of the mitochondrial network is essential for C. trachomatis to keep the available mitochondrial capacity for ATP production. In line with this hypothesis, we observed that depleting miR-30c severely affected ATP production and cell viability in HUVECs (Fig. 9, A and B). A similar decrease in ATP levels was also observed in induced HeLa-miR-30c sponge cells upon diminishing ATP production via glycolysis by growing the cells in galactose-containing medium (Fig. 9 C). Interestingly, overexpression of Drp1 and p53 lowers the ATP levels of HeLa and H1299 p53−/− cells by a narrow margin (Fig. S4, E and F). Conversely, although siRNA-mediated knockdown of p53 and DRP1 did not have a major effect on the ATP levels of untreated HUVECs, the transfection of p53 or Drp1 siRNAs (separately) significantly ameliorated the effect of 1 µM H2O2 treatment on the ATP levels compared with that of control cells (Fig. S4 G). These results suggest that even though Drp1, independently of p53, affects cell physiology by regulating mitochondrial fragmentation, reduction of both p53 and Drp1 expression can lower the depletion of ATP within when the cell is faced with external oxidative stress. To further investigate the role of mitochondrial ATP in chlamydial survival we generated a stable HeLa cell with AHT-inducible knockdown of the F1β subunit of the mitochondrial F1FO-ATPase. As a control, we used a cell line with an inducible knockdown of metaxin 2, a mitochondrial outer membrane protein, which is known not to affect mitochondrial morphology and protein levels within the time frame of the experiment (Ott et al., 2012). Silencing the expression of the F1β subunit was efficient (Fig. 9 D) but had almost no effect on cell viability (Fig. 9 E), and only a mild effect was observed on cellular ATP levels, because tumor cells such as HeLa produce ATP mainly via glycolysis. However, inhibition of glycolysis by growing the cells in medium containing galactose (Rossignol et al., 2004) for 8 h caused a strong depletion of ATP (Fig. 9 F) without affecting cell survival. Under these conditions, C. trachomatis growth was severely affected (Fig. 9, G and H). At the same time, the knockdown of metaxin 2 had no effect on ATP production, cell viability, or C. trachomatis growth (Fig. 9, E–G). These results indicate that mitochondrial ATP is an essential metabolite for normal chlamydial development.
Figure 9.
C. trachomatis infection is affected by regulation of ATP synthesis in the host mitochondria. (A) Graph represents quantification of total endogenous ATP levels in HUVECs transfected with siRNA negative control miR-30c mimic and inhibitors. Indicated samples were incubated in glucose-free galactose-supplemented media and tested using a luminescent ATP detection kit according to the manufacturer’s protocol. ATP luminescence was normalized to ATP levels of untreated and nontransfected cells grown in glucose medium. (B) Cell viability measurements of HUVECs transfected with siRNA negative control, miR-30c mimic, and inhibitors and grown in glucose medium. (C) Graph represents quantification of total endogenous ATP levels in noninduced and induced HeLa-miR-30c sponge cells grown in glucose-free galactose-supplemented media. ATP luminescence was normalized to ATP levels of noninduced cells grown in glucose medium. (D) Immunoblot of isolated mitochondria from noninduced and induced (AHT) f1βkd-2 and mtx2kd-2 cells. Blots were probed with antibodies against F1α subunit, and isocitrate dehydrogenase (ICDH) was used as a loading control. (E) Cell viability measurements of noninduced and induced mtx2kd-2 and f1βkd-2 cells incubated in glucose-containing medium or for 16 h in glucose followed by 8 h in galactose-supplemented medium. (F) Graph represents ATP measurements of mtx2kd-2 and f1βkd-2 cells. The cells were incubated in glucose-containing medium or for 16 h in glucose followed by 8 h in galactose-supplemented medium. ATP luminescence was normalized to ATP levels of noninduced cells grown in glucose medium. (G) Graph represents quantification of chlamydial inclusion sizes in noninduced and induced mtx2kd-2 and f1βkd-2 cells infected with C. trachomatis for 24 h in glucose-containing medium or for 16 h in glucose followed by 8 h in galactose-supplemented medium (n = 3). (H) Representative epifluorescence micrographs of noninduced and induced f1βkd-2 cells infected with C. trachomatis for 24 h and treated as mentioned earlier and grown in glucose followed by 8 h in galactose-supplemented medium. Cells were stained against cHSP60 (magenta) and phalloidin-647 for actin (blue). GFP (green) indicates induction of shRNA. Bars, 10 µm. All data represent mean ± SD. Asterisks denote significance by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (n = 3 for all panels). See also Fig. S4.
Discussion
Obligate intracellular pathogens such as C. trachomatis necessitate the survival of the host to ensure the availability of the metabolites required for the completion of the developmental cycle. Noting the lack of several key metabolic pathway components in its genome, C. trachomatis is heavily influenced by host metabolic conditions and particularly sensitive to the perturbations that interfere with the production and availability of amino acids, nucleotides, and lipids (Saka and Valdivia, 2010). Previous studies show that C. trachomatis requires the production of ROS for normal development (Boncompain et al., 2010; Chumduri et al., 2013; Siegl et al., 2014). Our data further show that C. trachomatis infection results in reversible stress on the host mitochondrial network. Normally, the presence of such stress factors would induce an overall negative effect on host mitochondrial health, leading to rapid fragmentation and induction of apoptotic pathways. However, our study and previous evidence show that the mitochondrial damage and pro-apoptotic effects of oxidative damage can be ameliorated to a degree by suppressing the activity or expression of the p53-dependent mitochondrial fission regulator Drp1 (Cereghetti et al., 2010; Oettinghaus et al., 2016). Here we show that C. trachomatis infection, in addition to HDM2-induced degradation of p53, up-regulates miR-30c within infected cells to down-regulate p53 and suppress Drp1 expression.
Our data show that the down-regulation of Drp1 upon C. trachomatis has a significant effect on the host mitochondria as exhibited by the elongated and fused nature of the network, a hallmark of cells that are facing increased energy demand (Tondera et al., 2009). Furthermore, we show that enforced mitochondrial fission by Drp1 overexpression in the absence of p53 also prevents chlamydial development, highlighting the need for Drp1 suppression. Additionally, it has been shown that Drp1 stabilization negatively affects intracellular ATP levels as a result of excessive mitochondrial fragmentation (Roos and Kaina, 2006), which makes the reduction of Drp1 activity indispensable for C. trachomatis growth. Interestingly, new evidence has shown that IFNγ-induced autophagy inhibits C. trachomatis growth (Al-Zeer et al., 2013). In context of the increased demand for metabolites and amino acids during C. trachomatis growth and development, resulting in a crippling nutrient-deprived (amino acids, ATP, and glucose) state for the host cell, it is interesting to note that loss of DRP1 possibly prevents the necessary mitochondrial fission that precedes mitophagy (Toyama et al., 2016). Furthermore, mild oxidative stress, akin to the sort generated by C. trachomatis infection, although incapable of inducing cell death or nonselective autophagy, is capable of inducing mitochondrial fragmentation and mitophagy (Frank et al., 2012). Frank et al. (2012) also demonstrate that induction of mitophagy by such stress and by starvation can be inhibited by inhibition of DRP1 function. Additionally, it has been shown that elongated mitochondria are not only capable of maintaining ATP levels during nutrient deprivation but are also selectively spared from autophagic degradation (Gomes et al., 2011; Rambold et al., 2011). It is hard to tell at this stage, however, whether it is the loss of DRP1 that restricts the induction of autophagy during the C. trachomatis growth-induced nutrient stress.
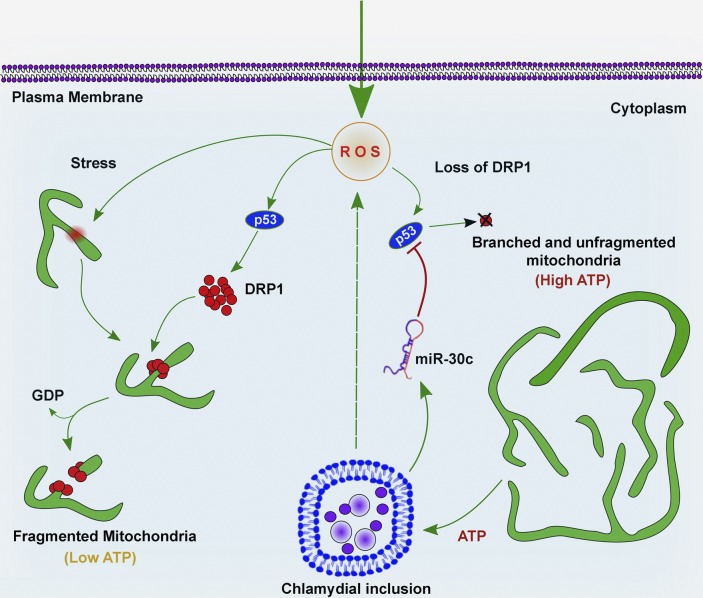
For intracellular bacteria such as C. trachomatis, survival of the host and maintenance of the metabolite supply is of paramount importance. Therefore, we suggest that the down-regulation of Drp1 in a miR-30c/p53–dependent manner serves to stabilize the mitochondrial network to maintain the metabolic integrity of the host cell in the presence of the severe stress caused by C. trachomatis infection (Fig. 10). This is a unique example of a mechanism by which an intracellular pathogen influences the host mitochondrial network by deregulation of a miRNA.
Figure 10.
Model of the effects of C. trachomatis on the mitochondrial network. Graphical representation of the effect of C. trachomatis infection on the mitochondrial network via miR-30c–mediated regulation of p53 and Drp1.
Materials and methods
Reagents and antibodies
Antibodies against cHSP60 (A57-B9), p53 (DO-1), GFP (B-2), and caspase-3 were purchased from Santa Cruz Biotechnology, Inc. Rabbit polyclonal anti-Drp1 was purchased from Genetex. Mouse monoclonal anti-Drp1 antibody (C-5) used for immunoblots was purchased from Santa Cruz Biotechnology, Inc. Equal loading was confirmed by using antibodies against β-actin (A5441; Sigma-Aldrich). Anti–mouse and anti–rabbit IgG Cy3TM- or Cy5TM-linked goat secondary antibodies were purchased from Dianova. All oligonucleotides used for real-time PCR, cloning and Northern blots were purchased from Sigma-Aldrich. FlexiTube GeneSolution (GS10059; QIAGEN) was used for siRNA knockdown of Drp1 and p53. Syn-hsa-miR-30c-5p miScript miRNA Mimic (MSY0000244), anti–hsa-miR-30c-5p miScript miRNA inhibitor (MIN0000244)and AllStars negative control siRNA were purchased from QIAGEN. MitoTracker Deep Red FM was purchased from Thermo Fisher Scientific. All buffers and reagents were purchased from Sigma-Aldrich unless otherwise mentioned.
Cell culture
HUVECs were cultured in Medium 200 supplemented with low serum growth supplement (Gibco) at 37°C in 5.0% CO2 unless otherwise mentioned. HeLa229, HeLa-miR-30c sponge cells, f1βkd-2, and mtx2kd-2 cells were cultured in RPMI-1640 medium supplemented with 10% FCS (Gibco) at 37°C in 5.0% CO2 unless otherwise mentioned. HFF cells were cultured in fibroblast basal medium (Gibco) supplemented as per manufacturer recommendation. hFIMB cells were prepared (Bono et al., 2012) and cultured in RPMI-1640 medium supplemented with 10% FCS (Gibco) at 37°C in 5.0% CO2. The use of the cells was in accordance with the guidelines imposed by the local ethics committee.
Propagation of C. trachomatis and infection of cells
HeLa cells infected with C. trachomatis LGV serovar L2 and C. trachomatis serovar D (ATCC) for 48 h were collected with a rubber policeman, transferred to a 50-ml tube, and lysed by vortexing with glass beads. To remove cell debris, the supernatant was centrifuged at 500 g at 4°C for 10 min (Hermle). To pellet down C. trachomatis, the supernatant was removed and centrifuged at 45,000 g for 45 min at 4°C in a SS34 rotor (Thermo Fisher Scientific). C. trachomatis was washed in SPG buffer (0.22 M sucrose, 10 mM Na2HPO4, 3.8 mM KH2PO4, and 5 mM glutamate, pH 7.4), aliquoted, and stored at −80°C.
For experiments, cells were infected with C. trachomatis with a multiplicity of infection of 1 in Medium 200 supplemented with low serum growth supplement (for HUVECs) and RPMI-1640 with 5% FCS (for HeLa) at 35°C for 2.5 h. The infected cells were incubated for necessary time periods. The cells were then processed for experiments as described. For Fig. 4 A, a multiplicity of infection of 0.6 was used to include noninfected cells in the imaging fields.
Infectivity assay
Infectivity assays for C. trachomatis were performed as described previously (Siegl et al., 2014). In brief, C. trachomatis–infected cells (36 hpi) and uninfected control cells were washed once with PBS and lysed manually by glass beads. A second set of cells was seeded the day before and infected by one-hundredth of the cell lysate containing infectious EBs, incubated for 12, 24, and 36 h at 35°C, and lysed for immunoblot analysis.
Transfection
All plasmid transfections in HUVECs and HFF cells were performed using Lipofectamine 2000 (Invitrogen) or X-tremeGENE HP DNA Transfection Reagent (Roche) according to the manufacturer’s protocol. miR-30c mimic, miR-30c inhibitor, and Drp1 siRNA transfection in HUVECs was performed with RNAiFect Transfection Reagent (QIAGEN) for 24 h before infection or processing. Plasmid DNA constructs were transfected into HeLa229 cells using polyethylenimine according to the protocol published by Longo et al. (2013).
RNA sequencing of small RNA fraction
Total RNA, including the small RNA fraction, was extracted from HUVECs after 12 and 24 h of C. trachomatis infection along with noninfected samples (0, 12, and 24 h) using miRNAeasy kit (QIAGEN) according to the manufacturer’s protocol. Vertis Biotechnology AG performed library preparation, and the RNA-sequencing was performed on an Illumina HiSeq 2000 sequencer at the Max Planck-Genome-centre Cologne, Cologne, Germany, as described in Maudet et al. (2014).
Computational analysis of RNA-seq data
RNA-seq analysis was performed using the READemption pipeline v0.3.3 (Förstner et al., 2014) with Segemehl v0.1.7 (Otto et al., 2014) for the mapping of transcripts against human mature miRNA sequences obtained from miRBase (Maudet et al., 2014), the human and C. trachomatis genomes (NCBI references NC_021049 and NC_021052). Parameter cutoffs of 14 nt as minimum read length and 95% as minimum similarity were used. Differential gene expression analysis was performed with DESeq 1.12.1 (Anders and Huber, 2010) comparing infected samples to noninfected samples collected at the same time point. Pathway-based analysis of selected miRNAs was done with the DIANA-miRPath v2.0 web server (Vlachos et al., 2012).
Designing of wild-type and mutant P53 3′ UTR
The human P53 full-length 3′ UTR was PCR-amplified from HUVECs using forward and reverse primers that introduced XhoI and NotI restriction sites at the 5′ and 3′ ends, respectively. The sequence of the primers used are as follows: forward, 5′-TAGAATTCTAGACCCTAGGGGTCTTGGCCATTC-3′, and reverse, 5′–TAGAATGCGGCCGC GCAGTCAGACAGCTTCTTTATTTGACT-3′; underlined sequences are the Xho1 and NotI sites, respectively. The 943-bp PCR product was ligated in the Empty GFP plasmid to create the GFP-p53-wt 3′ UTR vector. The vector carries an additional dsRed expression cassette independent of the GFP promoter, unaffected by any UTR, and serves as a normalization control. GFP-p53-mut 3′ UTR construct was prepared by performing site-directed mutagenesis on the wild-type construct. The sequences of the primers used to mutate the miR-30c binding sites are as follows: forward, 5′-TCTTGCAGTTAAGGGTTAGTTCACAATCAGCCACATTCTAGGT-3′, and reverse, 3′-ACCTAGAATGTGGCTGATTGTGAACTAACCCTTAACTGCAAGA-5′; underlined sequences denote the mutated miR-30c binding sites. Quick-change site-directed mutagenesis protocol was used to generate a point mutation in the miR-30c binding site. The oligonucleotides described carry a single nucleotide change compared with the wild-type binding site and were extended in a PCR reaction using Phusion polymerase (Thermo Fisher Scientific). The polymerase was inactivated by 5-min incubation at 80°C. The original template was digested with DpnI for 1 h at 37°C followed by an inactivation step at 80°C for 20 min. The presence of the desired mutation was verified by DNA sequencing (SeqLab, Gottingen, Germany). The mutated and wild-type vectors were transformed into and propagated in chemo-competent Escherichia coli XL1Blue.
Designing of presequence GFP, GFP-P53, and Drp1 overexpression construct
To construct a mitochondrially targeted GFP (mtGFP) expression plasmid, the egfp were amplified from the pEGFP-N1 vector (Takara Bio Inc.). This was cloned into a modified pcDNA3.1 vector downstream of the mitochondria targeting presequence to generate the necessary constructs. For the Drp1 overexpression vector, the Drp1 insert was amplified from HUVEC cDNA using the following primers: forward, 5′-TCAGGCGGCCGCAGCGCATGGCCTGCCGGGA-3′, and reverse, 5′-CTAGCTCGAGCTACTCTATACGGTTATGTTCCAAAG-3′. The product was ligated into an empty pcDNA3.1 vector.
To generate GFP-P53 fusion construct, P53 was amplified from the pCDNA3-HA-p53 (Marin et al., 2000) and cloned into the pCDNA3 vector already containing a GFP gene, so the product represented a p53 fused with its amino terminus to the GFP, including two linker amino acids between the proteins originating from the EcoRI restriction site. pcDNA3-Drp1K38A was a gift from A. van der Bliek (David Geffen School of Medicine at University of California, Los Angeles, South Los Angeles, CA) and R. Youle (National Institute of Neurological Disorders and Stroke, Porter Neuroscience Research Center, Bethesda, MD; plasmid 45161; Addgene). pMitoTimer was a gift from Z. Yan (University of Virginia, Charlottesville, VA; plasmid 52659; Addgene).
Designing of miR-30c sponge
For miR30c sponge preparation, mature miRNA sequence of miR-30c was obtained from miRBase. Oligo sequences carrying six miR30c target sites (italicized) each carrying a two-nucleotide bulge (underlined) and a central unrelated loop (marked in bold) were synthesized (Sigma-Aldrich), annealed, and cloned into the pLV-THM vector using MluI and ClaI sites downstream of H1 promoter sequences. Large-scale preparation of DNA required for lentivirus particle formation was performed using the endotoxin-free plasmid Midi prep system (MACHEREY-NAGEL). miR-30c-oligo, 5′-CGCGTCCCCCTGAGGATGTAGGATGTTTACATGACTGAGGATGTAGGATGTTTACAAGCGGCTGAGGATGTAGGATGTTTACACCCGGTGCATGACTAAGCTAGCCTGAGGATGTAGGATGTTTACACGACTGAGGATGTAGGATGTTTACAACGCGCTGAGGATGTAGGATGTTTACATTTTTGGAAAT-3′.
Generation of sponge, shRNA, and mitochondrial GFP cell lines
The miR-30c sponge and shRNA-expressing vectors were constructed by cloning the necessary miR-30c sponge oligo and shRNA sequence into the pLVTHM vector. The constructs were verified by sequencing. The sequence of the F1β shRNA was as follows: 5′-GCAGAATCATGAATGTCATTG-3′. The inducible miR-30c sponge and F1β KD (f1βkd-2) cell lines were constructed as described previously (Ott et al., 2012). In brief, the lentiviral construct, pLVTHM-tTR-KRAB-Red vector, was transduced into HeLa cells in presence of the cationic polymer, Polybrene (Sigma-Aldrich). Single-cell clones were sorted by selecting the cells for dsRed expression by flow cytometry using a BD FACS Aria III. One single-cell clone was used for infection with the miR-30c sponge and F1β KD shRNA-carrying virus. After allowing the cells to proliferate, single-cell clones were obtained by a second sorting step by selecting for expression of dsRed to avoid miR-30c depletion–induced cell defects or F1β deficiency–induced defects. For expression of the miR-30c sponge, the cells were cultivated in RPMI-1640 plus 10% FBS supplemented with 1 µg/ml AHT for 24 h. For expression of the F1β KD shRNA, the cells were cultivated in RPMI-1640 plus 10% FBS supplemented with 1 µg/ml AHT for 5 to 7 d. The sponge expression was confirmed by Western blot for miR-30c targets (Fig. S1, F and G). An inducible knockdown cell line for the mitochondrial outer membrane protein metaxin2 (Ott et al., 2012) was used as a control for all experiments involving F1β KD cells. To detect the effect of AHT on chlamydial growth, HeLa cells were treated with 0.5, 1, and 1.5 µg/ml AHT followed by C. trachomatis infection for 24 h. Chlamydial growth was monitored by immunoblot (Fig. S1 K). HUVECs stably expressing the mitochondrial targeted GFP were created by cloning the mitochondrial targeted GFP into a pLVTHM vector and transducing the cells with the lentivirus as described earlier. pLVTHM and pLV-tTRKRAB-red was a gift from Didier Trono (plasmid 12250; Addgene).
cDNA preparation and qRT-PCR
cDNA preparation and qRT-PCR for miRNA were performed using the miScript II RT kit and miScript SYBR Green PCR kit (QIAGEN) according to manufacturer’s protocols. Custom-made primers for all the miRNAs studied was also bought from QIAGEN. cDNA preparation and qRT-PCR for p53 and Drp1 were performed using the RevertAid RT kit (Thermo Fisher Scientific) and PerfeCTa SYBR Green FastMix (Quanta Biosciences). Primers for qRT-PCR were as follows: Drp1 forward, 5′-GCGCTGATCCCGCGTCAT-3′, and reverse, 5′-CCGCACCCACTGTGTTGA-3′; p53 forward, 5′-TCAACAAGATGTTTTGCCAACTG-3′, and reverse, 5′-ATGTGCTGTGACTGACTGACTGCTT-3′; and GAPDH forward, 5′-CGTCTTCACCACCATGGAGAAGGC-3′, and reverse, 5′-AAGGCCATGCCAGTGAGCTTCCC-3′.
All qRT-PCRs of miRNAs were performed using the miScript PCR System (QIAGEN) or miRCURY LNA universal RT miRNA PCR (Exiqon) with specific forward primers or LNA primer sets for individual miRNAs (QIAGEN or Exiqon) and endogenous control, U6 SnRNA (QIAGEN or Exiqon). All experiments were performed on a StepOnePlus real-time PCR platform (Applied Biosystems) according to the manufacturer’s protocol. Data were analyzed using StepOne Software v2.3 and GraphPad Prism v6.
Northern blot
Northern blots for miR-30c were performed using oligonucleotide probes for miR-30c (5-GCTGAGAGTGTAGGATGTTTACA-3′) and U6 snRNA (5-CACGAATTTGCGTGTCATCCTT-3′) according to the protocol described by the Narry Kim Lab (http://www.narrykim.org/en/protocols). U6 snRNA was used as loading control for all experiments done in HUVECs. The chlamydial small RNA described as CtrR5 (Albrecht et al., 2010) was used as a marker for C. trachomatis infection (5′-CAGCACCCCTCTGAGTTCTCCC-3′). Hybond-XL (GE Healthcare) nylon membranes were used for Northern blots. Decade Markers System from Thermo Fisher Scientific was used to radiolabel RNA ladder for Northern blots.
Immunoblotting
Infected or uninfected control cells were lysed in Laemmli sample buffer (100 mM Tris/HCl, pH 6.8, 4% SDS, 20% glycerin, 1.5% β-mercaptoethanol, and 0.2% bromophenol blue) and resolved by 12% SDS-PAGE. Proteins were transferred to PVDF membranes (EMD Millipore) and blocked with 5% milk/TBS-T. The membranes were probed with respective primary antibodies and subsequently with HRP-conjugated secondary antibodies. Proteins were detected by ECL (GE Healthcare) in the linear response range using an ECL Imager (Intas Science Imaging). β-Actin was used as loading control and for normalization in all cases unless stated otherwise.
Cell viability assay
Cells were plated in 96-well flat-bottom plates and transfected with the described constructs or siRNAs or treated with necessary reagents. At specific time points, 20 µl of 5 mg/ml MTT solution was added to the wells and incubated for 4 h. 170 µl DMSO was added after aspiration of media, and the absorbance (540 nm) was determined using an Infinite 200 PRO multimode reader (Tecan). Three technical replicate wells were used for each condition, and standard deviation was determined. Experiments were repeated three or more times, and statistical significance was calculated by unpaired t test.
Total ATP measurement
HUVECs, Hela-miR30c-sponge cells, or H1299 p53−/− cells were plated in white-bottom 96-well plates for 12–24 h. The cells were incubated in necessary medium (glucose-free or galactose-supplemented) for indicated time periods after transfection or H2O2 treatment. Total endogenous ATP levels were measured using the Luminescent ATP Detection Assay kit (Abcam) according to the manufacturer’s protocol. Three technical replicate wells were used for each condition, and standard deviation was determined. Experiments were repeated three or more times, and statistical significance was calculated by unpaired t test.
Flow cytometry
HUVECs or HFF cells were seeded in six-well plates for 12 h and transfected with pMitoTimer for 24 h before infection or H2O2 treatment. Cells were washed every 4–6 h to prevent metabolite accumulation. At the endpoint, cells were trypsinized and suspended in cell-specific medium. The fluorescence of MitoTimer was measured simultaneously in FITC (509-nm) and PE (560-nm) channels by flow cytometry using a BD FACS Aria III.
Immunostaining and fixed-cell microscopy
To visualize the mitochondrial division sites, HUVECs expressing GFP tagged with a mitochondrial presequence were stained with an anti–rabbit Atto-647N secondary antibody (Sigma-Aldrich) against the rabbit polyclonal anti-Drp1 antibody. Cells were seeded in 12-well plates on glass coverslips, transfected or treated with necessary constructs, siRNA, or reagents as required, and infected with C. trachomatis for the indicated times. Cells were washed in PBS and fixed with 4% PFA for 15 min. After washing, the cells were permeabilized using 0.2% Triton X-100 in PBS for 15 min and blocked with 2% FCS in PBS for 45 min. Cells were incubated for 1 h with the primary antibody in blocking buffer (2% FCS in PBS), washed three times in PBS, and stained with the corresponding secondary antibody. After three washes with PBS, a poststaining fixation process was performed for all samples imaged on the structured illumination platform. After a final wash with PBS, the samples were mounted onto slides using 2.5% Mowiol-DABCO (Carl Roth). Samples were imaged on a TCS SP5 confocal microscope (Leica Biosystems) using a 63× oil-immersion UV objective with a numerical aperture of 1.4. For superresolution imaging of Drp1 fission rings, images were acquired using a Zeiss ELYRA S.1 SR-SIM structured illumination platform using a Plan-Apochromat 63× oil-immersion objective with a numerical aperture of 1.4. The images were reconstructed using the ZEN 2012 image-processing platform with a SIM module.
Live-cell videomicroscopy
In brief, HUVECs or H1299 cells were seeded 2 d before infection in µ-Dish 35-mm, high glass bottom (81158; Ibidi) to reach appropriate confluence. These cells were then transfected or treated with necessary constructs, siRNA, or reagents as required and infected with C. trachomatis for the indicated times. 10 min before imaging, cells were washed in PBS and bathed in prewarmed RPMI-1640 containing 25 mM Hepes buffer (without phenol red and Ca2+/Mg2+). The samples were imaged on a TCS SP5 confocal microscope (Leica Biosystems) using a 63× oil-immersion UV objective with a numerical aperture of 1.4 and equipped with a live-cell microscopy incubation chamber (Life Imaging Systems), prewarmed to 37°C. Images were collected on live cells using the Leica Biosystems Application Suite software package. Unless otherwise noted, 1,024 × 1,024-pixel images were recorded in 8-bit mode and collected at necessary time intervals.
Image analysis
The surface area of inclusions relative to the cell surface for Fig. S2 (C and D) was determined via automated fluorescence microscopy as previously described (Reimer et al., 2015). All image-processing steps were performed using Fiji (Schindelin et al., 2012). The numbers of C. trachomatis inclusions and Drp1 fission rings were quantified by processing the confocal micrographs with the Fiji Object Counter plugin after appropriate thresholding. Background was subtracted using the rolling ball background subtraction model. A threshold for detecting Drp1 aggregates was applied to the original micrographs by measuring the mean pixel intensity of the control samples in the Drp1 channel and normalizing the pixel intensity of all other samples to this constant. Additionally, several hundred individual Drp1 rings were measured using the Profile function of the ZEN 2012 image-processing platform and profile plot plugin (Plot Profile) from Fiji to determine the maximum and minimum ring diameters of constricted and dilated Drp1 rings. Based on this step, the threshold of object detection by the Object Profile algorithm was set to include Drp1 particles that exhibited a diameter between 100 (constricted) and 360 nm (Ingerman et al., 2005; Mears et al., 2011; Rosenbloom et al., 2014; Ji et al., 2015). Within this range, Drp1 aggregates with a ring size of >180 nm were classified as dilated fission rings, and the aggregates with smaller diameter were considered to be of constricted form. Mitochondrial fission sites were defined by profiling regions of low mitochondrial intensity and high Drp1 signals along a linear path through mitochondrial fragments using the Plot Profile plugin from Fiji (Fig. 4 A).
The length of mitochondrial fragments was measured by a further modification of the Object Count plugin from Fiji. In brief, while keeping an equal threshold for all images, mitochondrial GFP fluorescence was converted to binary signals, and the algorithm was allowed to numerically categorize the mitochondria as a continuous network or individual fragment and finally determine the area covered by the mitochondrial fragments in micrometers squared. The area was divided by a factor of 0.39 nm, the mean width of HUVEC mitochondria determined by measuring ∼300 individual HUVEC mitochondrial fragments using the Profile function of ZEN 2012. Similarly, the length of H1299 p53−/− mitochondrial fragments was determined by dividing the area by a factor of 0.46 nm. The mitochondrial fragments with no visible connections with neighboring networks were considered to be individual fragments. Cells with >300 individual fragments were considered to be hyperfragmented, and ones with <160 fragments were considered hyperfused.
Drp1 colocalization with the mitochondria was determined using the COLOC2 plugin from Fiji (Schindelin et al., 2012; http://imagej.net/Coloc_2). The degree of colocalization was ascertained using Pearson’s colocalization coefficient. Analysis of mitochondrial mobility was performed using MitoCRWLR (see Online supplemental material). In brief, the GFP channels of the movies were opened with Fiji. We measured differences for each of the pairs of two consecutive time frames and analyzed the area of each difference image by the Analyze Particles function of Fiji. At the same time, the total area of fluorescently labeled mitochondria was recorded and used to normalize the differences. Normalized mitochondrial differences of treated, infected, or control cells were thus determined in triplicate and plotted to visualize the impact of various treatments and C. trachomatis infection on mitochondrial mobility. Statistics of the measurements were analyzed using GraphPad Prism version 6.
Statistical analysis
All statistical calculations were performed using GraphPad Prism 6.0. Error bars displayed on graphs represent the means ± SD of three or more independent replicates of an experiment. D’Agostino–Pearson normality test was used to determine whether datasets were normally distributed (Fig. 4 D; Fig. 5, B, C, G, and H; and Fig. 7, C and D). Statistical significance was calculated using Student’s t test or one-way analysis of variance followed by Tukey’s multiple comparisons test. For image analysis, six or more biological replicates per sample-condition were used to generate the representative data. Western blot, qRT-PCR, Northern blot, cell viability, and ATP assay data presented are representative of at least three independent experiments.
Online supplemental material
Fig. S1 shows validation and pathway analysis of the RNA sequencing data along with validation of miR-30c mimic, inhibitor, and sponges. We also demonstrate the effect of miRNA sponge on the infectivity of chlamydial progeny and mitochondrial integrity. Fig. S2 demonstrates the effect of Drp1 modulation on chlamydial inclusion sizes and the identification of Drp1 rings using SIM. Fig. S2 also illustrates the effect of peroxide exposure on miR-30c and p53 in infected and noninfected HUVECs. Fig. S3 details the effect of p53 overexpression on mitochondrial morphology, Drp1 expression, and chlamydial growth in H1299 p53−/− cells. Fig. S4 shows matrix localization of the MitoTimer to the mitochondrial matrix. Fig. S4 also demonstrates the effect of DRP1 and p53 modulation on MitoTimer oxidation and ATP levels in different cell types. Videos 1 and 2 show live-cell imaging of mito-HUVECs with and without H2O2 treatment. Videos 3–6 show the effects of C. trachomatis infection and H2O2 on HUVECs expressing mitochondrially targeted GFP. Videos 7 and 8 are time-lapse confocal micrography of the effects of C. trachomatis infection in mito-HUVECs. The mitoCRWLR MACRO script was used to analyze the mitochondrial mobility of the GFP-tagged mitochondrial particles within live cells.
Supplementary Material
Acknowledgments
We thank Markus Sauer for access to super resolution microscopy, Jörg Wischhusen for the FIMB tissue, and Stefan Gaubatz for the HA-p53 construct. We thank Franziska Hagen and Kathrin Stelzner for assistance with the FACS analysis. We also thank Sudip Das and Dominik Kiser for suggesting and critically examining the proper statistical methods used for data analysis.
This research was funded in part by a fellowship to S.R. Chowdhury from the Graduate School of Life Sciences and grants from the Deutsche Forschungsgemeinschaft (SPP1580 and Infect-ERA CINOCA) to T. Rudel.
The authors declare no competing financial interests.
Author contributions: S.R. Chowdhury, V. Kozjak-Pavlovic, and T. Rudel designed research; S.R. Chowdhury, A. Reimer, and K. Karunakaran performed research; S.R. Chowdhury, M. Sharan, V. Kozjak-Pavlovic, A. Eulalio, M. Fraunholz, and T. Rudel analyzed data; B.K. Prusty provided important reagents; S.R. Chowdhury, V. Kozjak-Pavlovic, and T. Rudel wrote the paper.
Footnotes
Abbreviations used:
- AHT
- anhydrous tetracycline
- EB
- elementary body
- HFF
- human foreskin fibroblast
- HUVEC
- human umbilical vein endothelial cell
- qRT-PCR
- quantitative real-time PCR
- RB
- reticulate body
- ROS
- reactive oxygen species
- SR-SIM
- superresolution structured illumination microscopy
References
- Abdelrahman Y.M., and Belland R.J.. 2005. The chlamydial developmental cycle. FEMS Microbiol. Rev. 29:949–959. 10.1016/j.femsre.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Albrecht M., Sharma C.M., Reinhardt R., Vogel J., and Rudel T.. 2010. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res. 38:868–877. 10.1093/nar/gkp1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Zeer M.A., Al-Younes H.M., Lauster D., Abu Lubad M., and Meyer T.F.. 2013. Autophagy restricts Chlamydia trachomatis growth in human macrophages via IFNG-inducible guanylate binding proteins. Autophagy. 9:50–62. 10.4161/auto.22482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W.G., Kanekal M., Simon M.C., Maltepe E., Blagosklonny M.V., and Neckers L.M.. 1998. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature. 392:405–408. 10.1038/32925 [DOI] [PubMed] [Google Scholar]
- Anders S., and Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland R., Ojcius D.M., and Byrne G.I.. 2004. Chlamydia. Nat. Rev. Microbiol. 2:530–531. 10.1038/nrmicro931 [DOI] [PubMed] [Google Scholar]
- Boncompain G., Schneider B., Delevoye C., Kellermann O., Dautry-Varsat A., and Subtil A.. 2010. Production of reactive oxygen species is turned on and rapidly shut down in epithelial cells infected with Chlamydia trachomatis. Infect. Immun. 78:80–87. 10.1128/IAI.00725-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono Y., Kyo S., Takakura M., Maida Y., Mizumoto Y., Nakamura M., Nomura K., Kiyono T., and Inoue M.. 2012. Creation of immortalised epithelial cells from ovarian endometrioma. Br. J. Cancer. 106:1205–1213. 10.1038/bjc.2012.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braschi E., Zunino R., and McBride H.M.. 2009. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 10:748–754. 10.1038/embor.2009.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge D.G., Stojanovic M., Marcellus R.C., and Shore G.C.. 2003. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J. Cell Biol. 160:1115–1127. 10.1083/jcb.200212059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campello S., Lacalle R.A., Bettella M., Mañes S., Scorrano L., and Viola A.. 2006. Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J. Exp. Med. 203:2879–2886. 10.1084/jem.20061877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghetti G.M., Costa V., and Scorrano L.. 2010. Inhibition of Drp1-dependent mitochondrial fragmentation and apoptosis by a polypeptide antagonist of calcineurin. Cell Death Differ. 17:1785–1794. 10.1038/cdd.2010.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumduri C., Gurumurthy R.K., Zadora P.K., Mi Y., and Meyer T.F.. 2013. Chlamydia infection promotes host DNA damage and proliferation but impairs the DNA damage response. Cell Host Microbe. 13:746–758. 10.1016/j.chom.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Cribbs J.T., and Strack S.. 2007. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 8:939–944. 10.1038/sj.embor.7401062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenreich W., Heesemann J., Rudel T., and Goebel W.. 2013. Metabolic host responses to infection by intracellular bacterial pathogens. Front. Cell. Infect. Microbiol. 3:24 10.3389/fcimb.2013.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T., Lu H., Hu H., Shi L., McClarty G.A., Nance D.M., Greenberg A.H., and Zhong G.. 1998. Inhibition of apoptosis in chlamydia-infected cells: Blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187:487–496. 10.1084/jem.187.4.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R., Xiao T., Fang Z., Sun Y., Li F., Gao Y., Feng Y., Li L., Wang Y., Liu X., et al. 2012. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J. Biol. Chem. 287:23227–23235. 10.1074/jbc.M112.373084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassi Fehri L., Koch M., Belogolova E., Khalil H., Bolz C., Kalali B., Mollenkopf H.J., Beigier-Bompadre M., Karlas A., Schneider T., et al. 2010. Helicobacter pylori induces miR-155 in T cells in a cAMP-Foxp3-dependent manner. PLoS One. 5:e9500 10.1371/journal.pone.0009500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree A.W., Trudeau K., Zik E., Benador I.Y., Twig G., Gottlieb R.A., and Shirihai O.S.. 2013. MitoTimer probe reveals the impact of autophagy, fusion, and motility on subcellular distribution of young and old mitochondrial protein and on relative mitochondrial protein age. Autophagy. 9:1887–1896. 10.4161/auto.26503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiori M.E., Barbini C., Haas T.L., Marroncelli N., Patrizii M., Biffoni M., and De Maria R.. 2014. Antitumor effect of miR-197 targeting in p53 wild-type lung cancer. Cell Death Differ. 21:774–782. 10.1038/cdd.2014.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D.J., Fernández R.E., and Maurelli A.T.. 2013. Chlamydia trachomatis transports NAD via the Npt1 ATP/ADP translocase. J. Bacteriol. 195:3381–3386. 10.1128/JB.00433-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstner K.U., Vogel J., and Sharma C.M.. 2014. READemption: A tool for the computational analysis of deep-sequencing-based transcriptome data. Bioinformatics. 30:3421–3423. 10.1093/bioinformatics/btu533 [DOI] [PubMed] [Google Scholar]
- Frank M., Duvezin-Caubet S., Koob S., Occhipinti A., Jagasia R., Petcherski A., Ruonala M.O., Priault M., Salin B., and Reichert A.S.. 2012. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim. Biophys. Acta. 1823:2297–2310. 10.1016/j.bbamcr.2012.08.007 [DOI] [PubMed] [Google Scholar]
- Fukuda E.Y., Lad S.P., Mikolon D.P., Iacobelli-Martinez M., and Li E.. 2005. Activation of lipid metabolism contributes to interleukin-8 production during Chlamydia trachomatis infection of cervical epithelial cells. Infect. Immun. 73:4017–4024. 10.1128/IAI.73.7.4017-4024.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedeke L., Vales-Lara F.M., Fenstermaker M., Cirera-Salinas D., Chamorro-Jorganes A., Ramírez C.M., Mattison J.A., de Cabo R., Suárez Y., and Fernández-Hernando C.. 2013. A regulatory role for microRNA 33* in controlling lipid metabolism gene expression. Mol. Cell. Biol. 33:2339–2352. 10.1128/MCB.01714-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes L.C., Di Benedetto G., and Scorrano L.. 2011. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13:589–598. 10.1038/ncb2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E., Rother M., Kerr M.C., Al-Zeer M.A., Abu-Lubad M., Kessler M., Brinkmann V., Loewer A., and Meyer T.F.. 2014. Chlamydia infection depends on a functional MDM2-p53 axis. Nat. Commun. 5:5201 10.1038/ncomms6201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch T.P., Al-Hossainy E., and Silverman J.A.. 1982. Adenine nucleotide and lysine transport in Chlamydia psittaci. J. Bacteriol. 150:662–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun F., Nakamura T., Shiu A.D., Cho D.H., Tsunemi T., Holland E.A., La Spada A.R., and Lipton S.A.. 2013. S-nitrosylation of dynamin-related protein 1 mediates mutant huntingtin-induced mitochondrial fragmentation and neuronal injury in Huntington’s disease. Antioxid. Redox Signal. 19:1173–1184. 10.1089/ars.2012.4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Chan C.S., Wu R., Zhang C., Sun Y., Song J.S., Tang L.H., Levine A.J., and Feng Z.. 2010. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol. Cell. 38:689–699. 10.1016/j.molcel.2010.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hybiske K., and Stephens R.S.. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. USA. 104:11430–11435. 10.1073/pnas.0703218104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerman E., Perkins E.M., Marino M., Mears J.A., McCaffery J.M., Hinshaw J.E., and Nunnari J.. 2005. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170:1021–1027. 10.1083/jcb.200506078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W.K., Hatch A.L., Merrill R.A., Strack S., and Higgs H.N.. 2015. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife. 4:e11553 10.7554/eLife.11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.J., Bang B.R., Han K.H., Hong L., Shim E.J., Ma J., Lerner R.A., and Otsuka M.. 2015. Regulation of NKT cell-mediated immune responses to tumours and liver inflammation by mitochondrial PGAM5-Drp1 signalling. Nat. Commun. 6:8371 10.1038/ncomms9371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., Arnoult D., Chen H., Chan D.C., Smith C.L., and Youle R.J.. 2004. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J. Cell Biol. 164:493–499. 10.1083/jcb.200309082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., Neutzner A., and Youle R.J.. 2007. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 178:71–84. 10.1083/jcb.200611064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kun D., Xiang-Lin C., Ming Z., and Qi L.. 2013. Chlamydia inhibit host cell apoptosis by inducing Bag-1 via the MAPK/ERK survival pathway. Apoptosis. 18:1083–1092. 10.1007/s10495-013-0865-z [DOI] [PubMed] [Google Scholar]
- Labbé K., Murley A., and Nunnari J.. 2014. Determinants and functions of mitochondrial behavior. Annu. Rev. Cell Dev. Biol. 30:357–391. 10.1146/annurev-cellbio-101011-155756 [DOI] [PubMed] [Google Scholar]
- Laker R.C., Xu P., Ryall K.A., Sujkowski A., Kenwood B.M., Chain K.H., Zhang M., Royal M.A., Hoehn K.L., Driscoll M., et al. 2014. A novel MitoTimer reporter gene for mitochondrial content, structure, stress, and damage in vivo. J. Biol. Chem. 289:12005–12015. 10.1074/jbc.M113.530527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le M.T., Teh C., Shyh-Chang N., Xie H., Zhou B., Korzh V., Lodish H.F., and Lim B.. 2009. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 23:862–876. 10.1101/gad.1767609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Nie H., Wang M., Su L., Li J., Yu B., Wei M., Ju J., Yu Y., Yan M., et al. 2012. MicroRNA-409-3p regulates cell proliferation and apoptosis by targeting PHF10 in gastric cancer. Cancer Lett. 320:189–197. 10.1016/j.canlet.2012.02.030 [DOI] [PubMed] [Google Scholar]
- Li J., Donath S., Li Y., Qin D., Prabhakar B.S., and Li P.. 2010. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 6:e1000795 10.1371/journal.pgen.1000795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo P.A., Kavran J.M., Kim M.S., and Leahy D.J.. 2013. Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol. 529:227–240. 10.1016/B978-0-12-418687-3.00018-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losón O.C., Song Z., Chen H., and Chan D.C.. 2013. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 24:659–667. 10.1091/mbc.E12-10-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter E.I., Barger A.C., Nair V., and Hackstadt T.. 2013. Chlamydia trachomatis inclusion membrane protein CT228 recruits elements of the myosin phosphatase pathway to regulate release mechanisms. Cell Reports. 3:1921–1931. 10.1016/j.celrep.2013.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin M.C., Jost C.A., Brooks L.A., Irwin M.S., O’Nions J., Tidy J.A., James N., McGregor J.M., Harwood C.A., Yulug I.G., et al. 2000. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat. Genet. 25:47–54. 10.1038/75586 [DOI] [PubMed] [Google Scholar]
- Matsumoto A. 1988. Structural characteristics of chlamydial bodies. CRC Press, Boca Raton, FL. [Google Scholar]
- Maudet C., Mano M., Sunkavalli U., Sharan M., Giacca M., Förstner K.U., and Eulalio A.. 2014. Functional high-throughput screening identifies the miR-15 microRNA family as cellular restriction factors for Salmonella infection. Nat. Commun. 5:4718 10.1038/ncomms5718 [DOI] [PubMed] [Google Scholar]
- McClarty G. 1994. Chlamydiae and the biochemistry of intracellular parasitism. Trends Microbiol. 2:157–164. 10.1016/0966-842X(94)90665-3 [DOI] [PubMed] [Google Scholar]
- Mears J.A., Lackner L.L., Fang S., Ingerman E., Nunnari J., and Hinshaw J.E.. 2011. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 18:20–26. 10.1038/nsmb.1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt C.E., and Lamont R.J.. 2011. Porphyromonas gingivalis induction of microRNA-203 expression controls suppressor of cytokine signaling 3 in gingival epithelial cells. Infect. Immun. 79:2632–2637. 10.1128/IAI.00082-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder J.W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettinghaus B., D’Alonzo D., Barbieri E., Restelli L.M., Savoia C., Licci M., Tolnay M., Frank S., and Scorrano L.. 2016. DRP1-dependent apoptotic mitochondrial fission occurs independently of BAX, BAK and APAF1 to amplify cell death by BID and oxidative stress. Biochim. Biophys. Acta. 1857:1267–1276. 10.1016/j.bbabio.2016.03.016 [DOI] [PubMed] [Google Scholar]
- Okamoto K., and Shaw J.M.. 2005. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 39:503–536. 10.1146/annurev.genet.38.072902.093019 [DOI] [PubMed] [Google Scholar]
- Omsland A., Sager J., Nair V., Sturdevant D.E., and Hackstadt T.. 2012. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc. Natl. Acad. Sci. USA. 109:19781–19785. 10.1073/pnas.1212831109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott C., Ross K., Straub S., Thiede B., Götz M., Goosmann C., Krischke M., Mueller M.J., Krohne G., Rudel T., and Kozjak-Pavlovic V.. 2012. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol. Cell. Biol. 32:1173–1188. 10.1128/MCB.06388-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto C., Stadler P.F., and Hoffmann S.. 2014. Lacking alignments? The next-generation sequencing mapper segemehl revisited. Bioinformatics. 30:1837–1843. 10.1093/bioinformatics/btu146 [DOI] [PubMed] [Google Scholar]
- Park S.Y., Lee J.H., Ha M., Nam J.W., and Kim V.N.. 2009. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat. Struct. Mol. Biol. 16:23–29. 10.1038/nsmb.1533 [DOI] [PubMed] [Google Scholar]
- Pletjushkina O.Y., Lyamzaev K.G., Popova E.N., Nepryakhina O.K., Ivanova O.Y., Domnina L.V., Chernyak B.V., and Skulachev V.P.. 2006. Effect of oxidative stress on dynamics of mitochondrial reticulum. Biochim. Biophys. Acta. 1757:518–524. 10.1016/j.bbabio.2006.03.018 [DOI] [PubMed] [Google Scholar]
- Qian W., Choi S., Gibson G.A., Watkins S.C., Bakkenist C.J., and Van Houten B.. 2012. Mitochondrial hyperfusion induced by loss of the fission protein Drp1 causes ATM-dependent G2/M arrest and aneuploidy through DNA replication stress. J. Cell Sci. 125:5745–5757. 10.1242/jcs.109769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalingam K., Sharma M., Lohmann C., Oswald M., Thieck O., Froelich C.J., and Rudel T.. 2008. Mcl-1 is a key regulator of apoptosis resistance in Chlamydia trachomatis-infected cells. PLoS One. 3:e3102 10.1371/journal.pone.0003102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold A.S., Kostelecky B., Elia N., and Lippincott-Schwartz J.. 2011. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA. 108:10190–10195. 10.1073/pnas.1107402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez C.M., Goedeke L., Rotllan N., Yoon J.H., Cirera-Salinas D., Mattison J.A., Suárez Y., de Cabo R., Gorospe M., and Fernández-Hernando C.. 2013. MicroRNA 33 regulates glucose metabolism. Mol. Cell. Biol. 33:2891–2902. 10.1128/MCB.00016-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer A., Blohm A., Quack T., Grevelding C.G., Kozjak-Pavlovic V., Rudel T., Hentschel U., and Abdelmohsen U.R.. 2015. Inhibitory activities of the marine streptomycete-derived compound SF2446A2 against Chlamydia trachomatis and Schistosoma mansoni. J. Antibiot. 68:674–679. 10.1038/ja.2015.54 [DOI] [PubMed] [Google Scholar]
- Roos W.P., and Kaina B.. 2006. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 12:440–450. 10.1016/j.molmed.2006.07.007 [DOI] [PubMed] [Google Scholar]
- Rosenbloom A.B., Lee S.H., To M., Lee A., Shin J.Y., and Bustamante C.. 2014. Optimized two-color super resolution imaging of Drp1 during mitochondrial fission with a slow-switching Dronpa variant. Proc. Natl. Acad. Sci. USA. 111:13093–13098. 10.1073/pnas.1320044111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol R., Gilkerson R., Aggeler R., Yamagata K., Remington S.J., and Capaldi R.A.. 2004. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 64:985–993. 10.1158/0008-5472.CAN-03-1101 [DOI] [PubMed] [Google Scholar]
- Saka H.A., and Valdivia R.H.. 2010. Acquisition of nutrients by Chlamydiae: Unique challenges of living in an intracellular compartment. Curr. Opin. Microbiol. 13:4–10. 10.1016/j.mib.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansome C., Zaika A., Marchenko N.D., and Moll U.M.. 2001. Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett. 488:110–115. 10.1016/S0014-5793(00)02368-1 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegl C., Prusty B.K., Karunakaran K., Wischhusen J., and Rudel T.. 2014. Tumor suppressor p53 alters host cell metabolism to limit Chlamydia trachomatis infection. Cell Reports. 9:918–929. 10.1016/j.celrep.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Smirnova E., Shurland D.L., Ryazantsev S.N., and van der Bliek A.M.. 1998. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 143:351–358. 10.1083/jcb.143.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E., Griparic L., Shurland D.L., and van der Bliek A.M.. 2001. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 12:2245–2256. 10.1091/mbc.12.8.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens R.S., Kalman S., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Tatusov R.L., Zhao Q., et al. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 282:754–759. 10.1126/science.282.5389.754 [DOI] [PubMed] [Google Scholar]
- Szaszák M., Steven P., Shima K., Orzekowsky-Schröder R., Hüttmann G., König I.R., Solbach W., and Rupp J.. 2011. Fluorescence lifetime imaging unravels C. trachomatis metabolism and its crosstalk with the host cell. PLoS Pathog. 7:e1002108 10.1371/journal.ppat.1002108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak H., Kim J., Jayabalan A.K., Lee H., Kang H., Cho D.H., Ohn T., Nam S.W., Kim W., and Lee E.K.. 2014. miR-27 regulates mitochondrial networks by directly targeting the mitochondrial fission factor. Exp. Mol. Med. 46:e123 10.1038/emm.2014.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K.J., and Jacobson M.R.. 2012. Defects in mitochondrial fission protein dynamin-related protein 1 are linked to apoptotic resistance and autophagy in a lung cancer model. PLoS One. 7:e45319 10.1371/journal.pone.0045319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd W.J., and Caldwell H.D.. 1985. The interaction of Chlamydia trachomatis with host cells: Ultrastructural studies of the mechanism of release of a biovar II strain from HeLa 229 cells. J. Infect. Dis. 151:1037–1044. 10.1093/infdis/151.6.1037 [DOI] [PubMed] [Google Scholar]
- Tondera D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S., Da Cruz S., Clerc P., Raschke I., Merkwirth C., et al. 2009. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 28:1589–1600. 10.1038/emboj.2009.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama E.Q., Herzig S., Courchet J., Lewis T.L. Jr., Losón O.C., Hellberg K., Young N.P., Chen H., Polleux F., Chan D.C., and Shaw R.J.. 2016. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 351:275–281. 10.1126/science.aab4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446. 10.1038/sj.emboj.7601963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek A.M., Shen Q., and Kawajiri S.. 2013. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 5:pii:a011072 10.1101/cshperspect.a011072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos I.S., Kostoulas N., Vergoulis T., Georgakilas G., Reczko M., Maragkakis M., Paraskevopoulou M.D., Prionidis K., Dalamagas T., and Hatzigeorgiou A.G.. 2012. DIANA miRPath v.2.0: Investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 40(W1):W498W–504 10.1093/nar/gks494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.X., Jiao J.Q., Li Q., Long B., Wang K., Liu J.P., Li Y.R., and Li P.F.. 2011a miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat. Med. 17:71–78. 10.1038/nm.2282 [DOI] [PubMed] [Google Scholar]
- Wang J., Yang K., Zhou L., Minhaowu Y., Wu Y., Zhu M., Lai X., Chen T., Feng L., Li M., et al. 2013. MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS Pathog. 9:e1003697 10.1371/journal.ppat.1003697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Tang Y., Cui H., Zhao X., Luo X., Pan W., Huang X., and Shen N.. 2011b Let-7/miR-98 regulate Fas and Fas-mediated apoptosis. Genes Immun. 12:149–154. 10.1038/gene.2010.53 [DOI] [PubMed] [Google Scholar]
- Wylie J.L., Hatch G.M., and McClarty G.. 1997. Host cell phospholipids are trafficked to and then modified by Chlamydia trachomatis. J. Bacteriol. 179:7233–7242. 10.1128/jb.179.23.7233-7242.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.