The dynein motor is recruited to the kinetochore to capture spindle microtubules and control the spindle assembly checkpoint. Gama et al. reveal the molecular mechanism of how the Rod–Zw10–Zwilch complex and Spindly mediate dynein recruitment in Caenorhabditis elegans and human cells.
Abstract
The molecular motor dynein concentrates at the kinetochore region of mitotic chromosomes in animals to accelerate spindle microtubule capture and to control spindle checkpoint signaling. In this study, we describe the molecular mechanism used by the Rod–Zw10–Zwilch complex and the adaptor Spindly to recruit dynein to kinetochores in Caenorhabditis elegans embryos and human cells. We show that Rod’s N-terminal β-propeller and the associated Zwilch subunit bind Spindly’s C-terminal domain, and we identify a specific Zwilch mutant that abrogates Spindly and dynein recruitment in vivo and Spindly binding to a Rod β-propeller–Zwilch complex in vitro. Spindly’s N-terminal coiled-coil uses distinct motifs to bind dynein light intermediate chain and the pointed-end complex of dynactin. Mutations in these motifs inhibit assembly of a dynein–dynactin–Spindly complex, and a null mutant of the dynactin pointed-end subunit p27 prevents kinetochore recruitment of dynein–dynactin without affecting other mitotic functions of the motor. Conservation of Spindly-like motifs in adaptors involved in intracellular transport suggests a common mechanism for linking dynein to cargo.
Introduction
Chromosome segregation during cell division requires attachments between spindle microtubules and kinetochores, multiprotein complexes that assemble on each sister chromatid. The microtubule-based motor cytoplasmic dynein 1 (dynein) localizes to the outermost layer of the kinetochore, the fibrous corona, which expands in early prometaphase when kinetochores are not yet occupied by microtubules (Hoffman et al., 2001; Wynne and Funabiki, 2016). Corona expansion enlarges the kinetochore surface available for initial lateral microtubule capture by dynein, which drives transient poleward motion of chromosomes and accelerates the formation of stable end-coupled attachments by the Ndc80 complex (Rieder and Alexander, 1990; Yang et al., 2007; Vorozhko et al., 2008; Barisic et al., 2014; Cheerambathur and Desai, 2014; Magidson et al., 2015; Wynne and Funabiki, 2015). Corona expansion also facilitates amplification of the diffusible “wait anaphase” signal by the spindle assembly checkpoint (SAC) that prevents premature sister chromatid separation (Musacchio, 2015; Wynne and Funabiki, 2015). After kinetochore microtubule attachment, dynein contributes to corona disassembly and SAC silencing by transporting SAC components from kinetochores to spindle poles as part of a motor–cargo complex (Howell et al., 2001; Wojcik et al., 2001).
Dynein recruitment to kinetochores is dependent on the three-subunit Rod–Zw10–Zwilch (RZZ) complex and the coiled-coil protein Spindly (Starr et al., 1998; Griffis et al., 2007; Gassmann et al., 2008; Yamamoto et al., 2008; Chan et al., 2009; Barisic et al., 2010), but the molecular mechanism remains elusive. RZZ is also required for recruitment of Mad1/Mad2, whose presence at kinetochores is essential for SAC activation (Buffin et al., 2005; Karess, 2005; Kops et al., 2005; Essex et al., 2009). RZZ subunits are interdependent for kinetochore localization, consistent with the observation that their inhibition causes identical defects (Smith et al., 1985; Karess and Glover, 1989; Williams and Goldberg, 1994; Starr et al., 1997; Scaërou et al., 1999, 2001; Chan et al., 2000; Williams et al., 2003; Gassmann et al., 2008; Wainman et al., 2012). Anaphase chromosome bridges are a hallmark of RZZ inhibition in Drosophila melanogaster and Caenorhabditis elegans, and the resulting aneuploidy is lethal, underscoring the importance of RZZ for the fidelity of chromosome segregation.
Spindly acts downstream of RZZ as a kinetochore-specific adaptor for dynein, and in C. elegans it is also required for kinetochore recruitment of Mad1/Mad2 (Gassmann et al., 2008; Yamamoto et al., 2008). Whether RZZ directly binds to Spindly is unclear, nor is it known which RZZ subunits mediate Spindly recruitment. Like other adaptor proteins, Spindly forms a ternary complex with dynein and its cofactor dynactin, which facilitates processive movement of the motor in vitro (Splinter et al., 2012; McKenney et al., 2014). Spindly proteins contain a highly conserved motif in their N-terminal coiled-coil region, point mutations in which abrogate dynein–dynactin recruitment in vivo (Gassmann et al., 2010; Cheerambathur et al., 2013). The molecular basis of Spindly’s interaction with dynein–dynactin and the role of the Spindly motif are not known.
In this study, we describe protein–protein interactions that link RZZ to Spindly and Spindly to dynein and dynactin. We demonstrate the functional relevance of these interactions with engineered mutants biochemically and in vivo using the C. elegans early embryo and cultured human cells, thus providing a molecular view of how dynein is recruited to kinetochores. Our results suggest that the mechanism used by Spindly to engage dynein and dynactin is also relevant for how the motor interacts with cargo adaptors in the context of intracellular transport.
Results
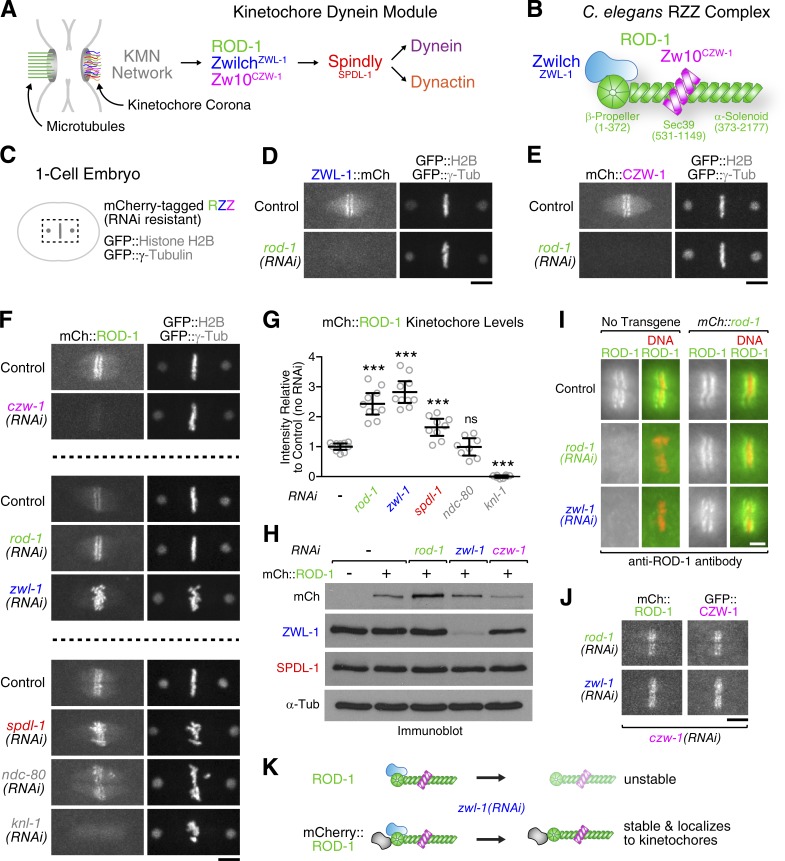
ROD-1 and Zw10CZW-1 can target to kinetochores without ZwilchZWL-1
To gain insight into the mechanisms of kinetochore dynein recruitment by the RZZ complex and Spindly (the kinetochore dynein module; Fig. 1 A), we asked how the three subunits of RZZ contribute to the function of the complex. We used the C. elegans early embryo, in which previous work had delineated the contribution of the kinetochore dynein module to chromosome segregation (Gassmann et al., 2008; Cheerambathur et al., 2013). We first defined the basic architecture of C. elegans RZZ using yeast two-hybrid mapping (Fig. S1, A–C). The predicted N-terminal β-propeller domain of ROD-1 (residues 1–373) was necessary and sufficient for the interaction with ZwilchZWL-1, and the Zw10CZW-1 binding region of ROD-1 mapped to residues 500–1,203 within ROD-1’s α-solenoid region, which corresponds to the Sec39 domain. Accordingly, residues 1–1,203 interacted with both ZwilchZWL-1 and Zw10CZW-1. No interaction was detected between ZwilchZWL-1 and Zw10CZW-1. Thus, ZwilchZWL-1 and Zw10CZW-1 bind independently to distinct domains within ROD-1’s N-terminal half (Fig. 1 B).
Figure 1.
ROD-1 and Zw10CZW-1 can target to kinetochores independently of ZwilchZWL-1. (A) Cartoon depicting localization dependencies of kinetochore dynein module components. (B) Summary of interactions between C. elegans RZZ subunits, based on yeast two-hybrid mapping (Fig. S1, A–C). Note that the intact RZZ complex likely exists as a dimer of the ROD-1–Zw10CZW-1–ZwilchZWL-1 trimer, based on hydrodynamic analysis of the human and D. melanogaster complexes (Williams et al., 2003; Çivril et al., 2010). (C) Cartoon of the one-cell embryo spindle region shown in subsequent panels. (D–F) Localization dependency analysis of mCherry-tagged RZZ subunits. Still images are from time-lapse sequences. (G) Quantification of mCherry::ROD-1 levels at kinetochores in the conditions shown in D–F. Circles correspond to measurements in individual one-cell embryos in metaphase. Error bars represent the SEM with a 95% confidence interval. The t test was used to determine statistical significance (***, P < 0.0001 compared with no RNAi control; ns, P > 0.05). (H) Immunoblots showing that depletion of Zw10CZW-1 but not ZwilchZWL-1 lowers mCherry::ROD-1 levels. α-Tubulin served as the loading control. (I) Immunofluorescence images of metaphase kinetochores stained for ROD-1. Endogenous ROD-1 is missing from kinetochores in ZwilchZWL-1-depleted embryos, whereas mCherry::ROD-1 localizes under the same conditions. (J) Stills from a time-lapse sequence showing that mCherry::ROD-1 supports kinetochore localization of GFP::Zw10CZW-1 in the absence of ZwilchZWL-1. Bars: (D–F and J) 5 µm; (I) 2µm. (K) Cartoon summary of the data shown in C–J.
Next, we generated animals expressing mCherry-tagged ROD-1, ZwilchZWL-1, and Zw10CZW-1 to examine the localization dependency of RZZ subunits (Fig. 1 C). The transgenes were made resistant to RNAi by codon reshuffling and integrated in single copy at a defined chromosomal locus (Fig. S1 D; Frøkjær-Jensen et al., 2012). Consistent with prior work (Gassmann et al., 2008), kinetochore localization of ZwilchZWL-1::mCherry and mCherry::Zw10CZW-1 was lost after depletion of ROD-1 (Fig. 1, D and E), and kinetochore localization of mCherry::ROD-1 depended on Zw10CZW-1 (Fig. 1 F). Immunoblotting showed that mCherry::ROD-1 loss from kinetochores after Zw10CZW-1 depletion could be attributed to destabilization of mCherry::ROD-1 (Fig. 1 H). In contrast, depletion of ZwilchZWL-1 did not destabilize mCherry::ROD-1, facilitating localization dependency analysis. Depletion of endogenous ROD-1 increased kinetochore mCherry::ROD-1 levels 2.4-fold (Fig. 1 G), presumably because mCherry::ROD-1 did not have to compete with endogenous ROD-1 for kinetochore targeting. Remarkably, and contrary to expectations, we observed a similar increase in kinetochore mCherry::ROD-1 levels after depletion of ZwilchZWL-1 (2.8-fold; Fig. 1, F and G). This suggested that endogenous ROD-1, but not mCherry::ROD-1, was destabilized in the absence of ZwilchZWL-1. Immunofluorescence with antibodies against ROD-1 confirmed that depletion of ZwilchZWL-1 abolished endogenous ROD-1 localization to kinetochores (Fig. 1 I), whereas robust kinetochore ROD-1 signal was detected in embryos expressing mCherry::ROD-1. Importantly, kinetochore-localized mCherry::ROD-1 recruited GFP::Zw10CZW-1 in ZwilchZWL-1-depleted embryos (Fig. 1 J), consistent with the independent binding of Zw10CZW-1 and ZwilchZWL-1 to ROD-1 observed in the yeast two-hybrid assay. These results indicate that ZwilchZWL-1 does not make a direct contribution to kinetochore targeting of ROD-1 and Zw10CZW-1. Instead, the displacement of ROD-1 and Zw10CZW-1 from kinetochores in ZwilchZWL-1-depleted embryos is likely an indirect consequence of ROD-1 destabilization. Fusing mCherry to ROD-1’s β-propeller domain, where ZwilchZWL-1 binds (Fig. S1, A–C), may stabilize ROD-1 when ZwilchZWL-1 is absent, thus facilitating mCherry::ROD-1 targeting to kinetochores along with Zw10CZW-1 (Fig. 1 K).
ZwilchZWL-1 is required for kinetochore recruitment of SpindlySPDL-1
We next asked whether the kinetochore-localized mCherry::ROD-1–Zw10CZW-1 subcomplex was sufficient to support the recruitment of SpindlySPDL-1. Depletion of endogenous ROD-1 confirmed that mCherry::ROD-1 could recruit GFP::SpindlySPDL-1 to kinetochores (Fig. 2 A). In contrast, depletion of ZwilchZWL-1 displaced GFP::SpindlySPDL-1 from kinetochores despite kinetochore-localized mCherry::ROD-1. Importantly, immunoblotting demonstrated that depletion of ZwilchZWL-1 did not affect total SpindlySPDL-1 levels (Fig. 1 H). These results directly implicated ZwilchZWL-1 in the kinetochore targeting of SpindlySPDL-1.
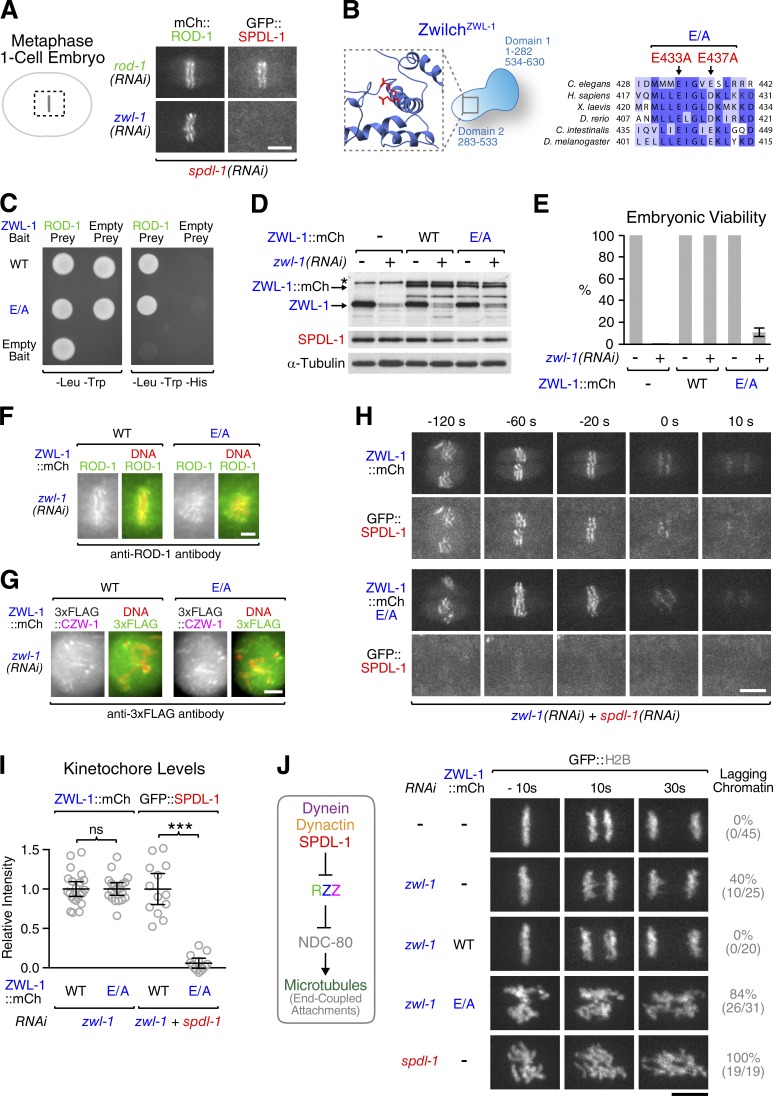
Figure 2.
ZwilchZWL-1 residues required for SpindlySPDL-1 recruitment to kinetochores. (A) Stills from time-lapse sequences showing that kinetochore-localized mCherry::ROD-1 does not support GFP::SpindlySPDL-1 recruitment in the absence of ZwilchZWL-1. (B) Cartoon of domain architecture and sequence alignment of Zwilch homologues. Residues E433 and E437 were both mutated to alanine (E/A). (C) Yeast two-hybrid assay showing that the ZwilchZWL-1 E/A mutant interacts with full-length ROD-1. Cells containing bait and prey plasmids grow on -Leu/-Trp plates, whereas -Leu/-Trp/-His plates select for bait–prey interaction. (D) Immunoblots of C. elegans adult worms expressing transgene-encoded ZwilchZWL-1::mCherry wild-type (WT) or the E/A mutant. The asterisk denotes a cross-reacting protein band of similar size as ZwilchZWL-1::mCherry. α-Tubulin served as the loading control. (E) Embryonic viability assay. More than 300 embryos from 12 or more mothers were counted for each condition. (F and G) Still images from a time-lapse sequence showing robust kinetochore localization of ROD-1 and 3×FLAG::Zw10CZW-1 in the ZwilchZWL-1 E/A mutant. (H) Selected frames from a time-lapse sequence showing that ZwilchZWL-1::mCherry E/A fails to recruit GFP::SpindlySPDL-1 (see Video 1). (I) Quantification of ZwilchZWL-1::mCherry and GFP::SpindlySPDL-1 levels on metaphase kinetochores in the conditions shown in H. Circles correspond to measurements in individual embryos. Error bars represent the SEM with a 95% confidence interval. The t test was used to determine statistical significance (***, P < 0.0001; ns, P > 0.05). (J, left) Cartoon summarizing the inhibitory cross-talk between the kinetochore dynein module and NDC-80. (Right) Selected frames from time-lapse sequences of the first embryonic division, demonstrating that the chromosome segregation defects of ZwilchZWL-1::mCherry E/A resemble those of SpindlySPDL-1 depletion (see Video 2). The frequencies of anaphase chromatin bridges are indicated in the percentages and absolute numbers of embryos in parentheses. Time is relative to the onset of sister chromatid separation. Bars: (A, H, and J) 5 µm; (F and G) 2 µm.
Identification of conserved ZwilchZWL-1 residues required for SpindlySPDL-1 recruitment to kinetochores
We sought to identify structural features in ZwilchZWL-1 required for kinetochore targeting of SpindlySPDL-1. Sequence alignments showed that E433 and E437 of ZwilchZWL-1 occupied the same positions in the αJ helix of domain 2 as human E422 and D426, which had previously been predicted to participate in a protein–protein interaction based on their conservation and surface-exposed position in the structure (Fig. 2 B; Çivril et al., 2010). Mutating both E433 and E437 to alanine (E/A) did not affect the yeast two-hybrid interaction between ZwilchZWL-1 and full-length ROD-1 (Fig. 2 C). Furthermore, when expressed recombinantly in insect cells, both wild-type ZwilchZWL-1 and the E/A mutant copurified with 6×His-tagged ROD-1 β-propeller (residues 1–372), and the complexes had identical elution profiles in size exclusion chromatography (Fig. S2, A and B). Thus, E433 and E437 of ZwilchZWL-1 are unlikely to be involved in RZZ complex formation. We generated animals expressing RNAi-resistant ZwilchZWL-1(E/A)::mCherry from the same chromosomal locus as our animals expressing the wild-type transgene. In contrast to wild-type ZwilchZWL-1::mCherry, the E/A mutant caused high embryonic lethality when endogenous ZwilchZWL-1 was depleted, despite identical expression levels (Fig. 2, D and E). No decrease in embryonic viability was observed in the presence of endogenous ZwilchZWL-1, demonstrating that the E/A mutant did not act as a dominant negative. ZwilchZWL-1(E/A)::mCherry supported kinetochore localization of endogenous ROD-1 and Zw10CZW-1 (Fig. 2, F and G), and both wild-type ZwilchZWL-1::mCherry and the E/A mutant localized equally well to kinetochores (Fig. 2, H and I). Wild-type ZwilchZWL-1::mCherry also recruited GFP::SpindlySPDL-1 to kinetochores (Fig. 2, H and I; and Video 1). In contrast, GFP::SpindlySPDL-1 was delocalized from kinetochores harboring the Zwilch E/A mutant. The Zwilch E/A mutant also delocalized dynein, dynactin, and Mad2MDF-2 from kinetochores and failed to activate the SAC, as predicted by the absence of SpindlySPDL-1 from kinetochores (Fig. S2, C–F). Depletion of SpindlySPDL-1 resulted in severe chromosome segregation defects because without dynein at kinetochores, RZZ inhibits the microtubule-binding activity of NDC-80 (Fig. 2 J; Gassmann et al., 2008; Cheerambathur et al., 2013). In contrast, depletion of RZZ subunits resulted in milder defects that reflect a loss of kinetochore dynein without NDC-80 inhibition. Visualization of chromosome dynamics showed that the ZwilchZWL-1 E/A mutant caused defects in chromosome segregation that were significantly more severe than depleting ZwilchZWL-1 on its own and resembled those of SpindlySPDL-1 depletions (Fig. 2 J and Video 2). This was also evident by quantifying the kinetics of spindle pole separation, which is a readout for the formation of end-coupled kinetochore–microtubule attachments capable of resisting tension (Fig. S2, G and H). Collectively, these results demonstrate that mutating residues E433 and E437 of ZwilchZWL-1 to alanine selectively displaces SpindlySPDL-1 from kinetochores without perturbing RZZ localization.
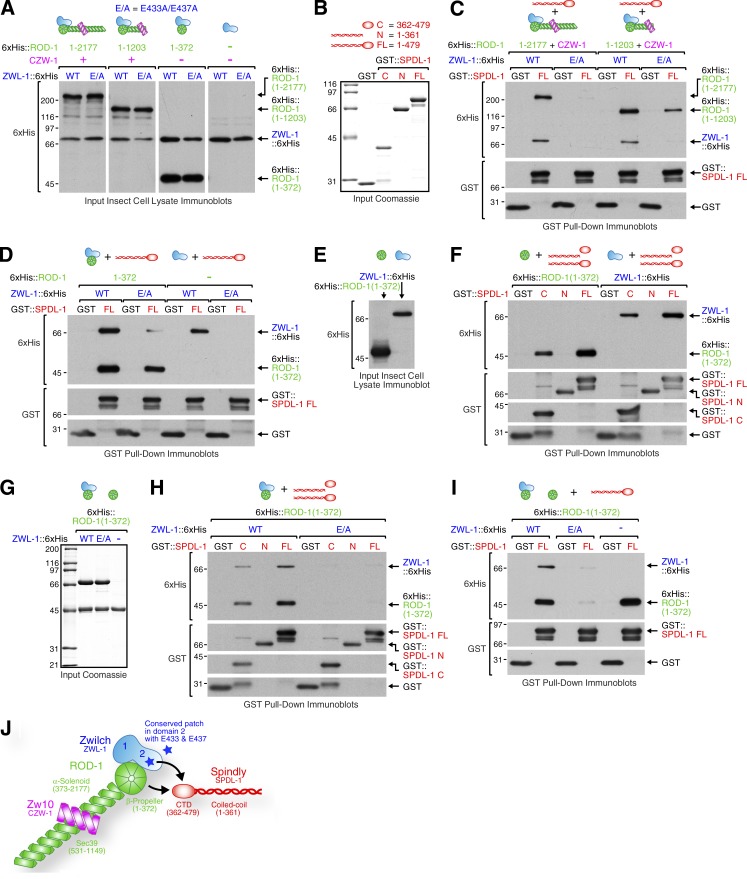
The C-terminal domain of SpindlySPDL-1 binds to ZwilchZWL-1 and the ROD-1 β-propeller
Next, we asked whether RZZ binds directly to SpindlySPDL-1. Initial attempts failed to reveal an interaction between ZwilchZWL-1 and SpindlySPDL-1 when both components were purified (unpublished data). We therefore performed pull-downs from insect cell lysates containing different recombinant versions of C. elegans RZZ (Fig. 3 A). Purified GST::SpindlySPDL-1 (Fig. 3 B) pulled down full-length RZZ, ROD-11–1,203ZZ, ROD-11–372–ZwilchZWL-1, and ZwilchZWL-1 by itself from corresponding lysates, and replacing wild-type ZwilchZWL-1 with the E/A mutant diminished these interactions (Fig. 3, C and D; and Fig. S3). We conclude that SpindlySPDL-1 interacts with RZZ through the ZwilchZWL-1 subunit and that the interaction requires residues E433 and/or E437 in ZwilchZWL-1. Interestingly, GST::SpindlySPDL-1 also pulled down ROD-11–372 by itself from lysate (Fig. 3, E and F), suggesting that SpindlySPDL-1 interacts with RZZ through both ZwilchZWL-1 and the ROD-1 β-propeller. To narrow down the binding region in SpindlySPDL-1, we purified its N-terminal coiled-coil region (residues 1–361) and the C-terminal domain (residues 362–479; Fig. 3 B). GST pull-downs from lysates revealed that the SpindlySPDL-1 C-terminal domain contains the binding sites for both ZwilchZWL-1 and ROD-11–372 (Fig. 3 F).
Figure 3.
SpindlySPDL-1 binds ZwilchZWL-1 and the ROD-1 β-propeller. (A) Lysates of insect Sf21 cells coinfected with viruses to express full-length (FL) ROD-11–2,177ZZ, ROD-11–1,203ZZ, ROD-11–372–ZwilchZWL-1, or ZwilchZWL-1 alone. ROD-1 and ZwilchZWL-1 were tagged with 6×His for detection on immunoblots. (B) Coomassie-stained protein gels showing purified recombinant GST::SpindlySPDL-1 used in pull-downs. (C and D) GST pull-downs from the lysates in A with purified GST::SpindlySPDL-1 from B. Protein fractions bound to beads were analyzed by immunoblotting using anti-6×His antibody. The same membranes were then reprobed with anti-GST antibody. (E) Lysates of insect Sf21 cells infected with viruses encoding for the ROD-1 β-propeller (residues 1–372) or ZwilchZWL-1, both tagged with 6×His. (F) Immunoblots of GST pull-downs from the lysates in E with purified GST::SpindlySPDL-1 (see A). (G) Coomassie-stained protein gels showing purified ROD-1 β-propeller alone and in complex with wild-type (WT) ZwilchZWL-1 or the E/A mutant. (H and I) Immunoblots of GST pull-downs using the purified proteins in B and G. (J) Cartoon showing the bipartite interaction between RZZ and the C-terminal domain (CTD) of SpindlySPDL-1. Molecular mass is indicated in kilodaltons.
Although GST::SpindlySPDL-1 pulled down ROD-11–372 by itself from lysates (Fig. 3 F), GST::SpindlySPDL-1 did not pull down full-length ROD-1 from lysates when ZwilchZWL-1 E/A was present (Figs. 3 C and S3 A). A possible explanation is that free ROD-1 β-propeller has higher affinity for SpindlySPDL-1 than ROD-1 β-propeller bound to ZwilchZWL-1, such that the SpindlySPDL-1–ZwilchZWL-1 interaction becomes essential for the binding of SpindlySPDL-1 to RZZ. The residual amounts of ROD-11–1,203 and ROD-11–372 pulled down by GST::SpindlySPDL-1 in the presence of ZwilchZWL-1 E/A (Fig. 3, C and D) may therefore represent a pool of free ROD-1 that is not bound to ZwilchZWL-1 E/A in the lysate (note that our in vivo data in Fig. 1 suggest that full-length ROD-1 is unstable when not bound to ZwilchZWL-1). To explore this idea further, we repeated the pull-downs after purifying ROD-11–372 on its own and in complex with ZwilchZWL-1 (Fig. 3 G). GST::SpindlySPDL-1 pulled down free ROD-11–372 and ROD-11–372 bound to wild-type ZwilchZWL-1, but not ROD-11–372 bound to the ZwilchZWL-1 E/A mutant (Fig. 3, H and I). Thus, although SpindlySPDL-1 binds free ROD-11–372, the SpindlySPDL-1–ROD-11–372 interaction is not sufficient to mediate binding of SpindlySPDL-1 to the ROD-11–372–ZwilchZWL-1 E/A complex. This result agrees with the observation that the ZwilchZWL-1 E/A mutant abrogates kinetochore recruitment of SpindlySPDL-1 despite kinetochore-localized ROD-1 (Fig. 2, F and H).
Collectively, biochemical and in vivo analysis suggests that RZZ recruits SpindlySPDL-1 to kinetochores through a bipartite interaction between its ZwilchZWL-1–ROD-1 β-propeller module and the SpindlySPDL-1 C-terminal domain (Fig. 3 J).
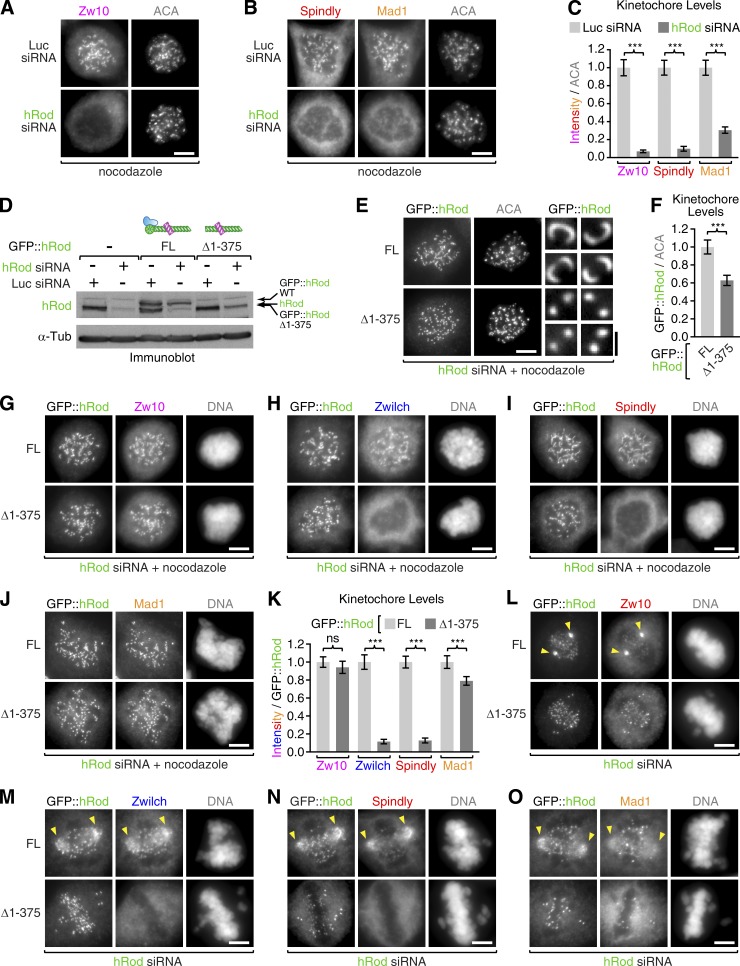
Human Rod without its β-propeller targets Zw10 to kinetochores but fails to recruit Zwilch and Spindly
To assess whether the function of the Zwilch–Rod β-propeller module is conserved in human cells, we characterized human Rod lacking the β-propeller domain (Δ1–375). We integrated expression constructs for full-length GFP::hRod and the Δ1–375 mutant into a single genomic locus in HeLa cells. Transgenes were resistant to an siRNA oligonucleotide that efficiently depleted endogenous hRod on immunoblots and delocalized Spindly, Mad1, and Zw10 from kinetochores by immunofluorescence (Fig. 4, A–D). Full-length GFP::hRod localized to kinetochores in cells treated with hRod siRNA and nocodazole (to avoid effects on kinetochore levels caused by microtubule binding) and supported kinetochore recruitment of Zwilch, Zw10, Spindly, and Mad1 (Fig. 4, E–K). GFP::hRod(Δ1–375) localized robustly to kinetochores in hRod-depleted cells (Fig. 4 E). However, the crescent morphology typically adopted by kinetochores after nocodazole treatment was distinctly missing (Fig. 4 E, blowups). This suggests that GFP::hRod(Δ1–375)-containing kinetochores were unable to undergo corona expansion in response to microtubule depolymerization. Accordingly, kinetochore levels of GFP::hRod(Δ1–375) in nocodazole-treated cells were reduced by 37% relative to full-length GFP::hRod (Fig. 4 F). Despite this difference, GFP::hRod(Δ1–375) was as efficient in recruiting Zw10 to kinetochores as full-length GFP::hRod (Fig. 4, G and K). In contrast, GFP::hRod(Δ1–375) failed to recruit Zwilch or Spindly to kinetochores (Fig. 4, H, I, and K). We also observed a modest reduction of Mad1 levels in GFP::hRod(Δ1–375) relative to full-length GFP::hRod (Fig. 4, J and K). In prometaphase cells without nocodazole treatment, full-length GFP::hRod was enriched at spindle poles along with Zw10, Zwilch, Spindly, and Mad1 (Fig. 4, L–O). Spindle pole enrichment was missing in cells expressing GFP::hRod(Δ1–375). This is consistent with the loss of Spindly from kinetochores, which abrogates dynein-mediated poleward transport of corona components (Gassmann et al., 2010). We conclude that the role of the Rod β-propeller–Zwilch module in Spindly recruitment is conserved in human cells. Furthermore, removing the β-propeller from hRod impairs corona expansion in response to microtubule depolymerization.
Figure 4.
Human Rod lacking its β-propeller localizes to kinetochores with Zw10 but fails to recruit Zwilch or Spindly. (A and B) HeLa cells coimmunostained with ACAs and Spindly/Mad1 (A) or Zw10 (B) after transfection with an siRNA oligonucleotide against hRod or luciferase (Luc) as a control. Cells were incubated in 1 µM nocodazole for 4 h before fixation. (C) Quantification of kinetochore levels of Spindly, Mad1, and Zw10 using immunofluorescence intensity measurements normalized to ACA signal. Each condition represents ≥50 kinetochore measurements from 10 or more different cells. Error bars represent the SEM with a 95% confidence interval. The t test was used to determine statistical significance (***, P < 0.0001). (D) Immunoblot of HeLa Flp-In T-REx cells with an antibody against human Rod (hRod), showing expression levels of endogenous hRod and RNAi-resistant full-length GFP::hRod (FL) or GFP::hRod lacking the β-propeller domain (Δ1–375). Luciferase siRNA was used as a control in RNAi experiments, and α-tubulin served as the loading control. Note that GFP::hRod(Δ1–375) migrates at the same size as endogenous hRod. (E) HeLa Flp-In T-REx cells expressing GFP::hRod or GFP::hRod(Δ1–375), depleted of endogenous hRod and immunostained with anti-GFP and ACAs. Blow-ups show examples of individual sister kinetochore pairs. (F) Quantification of full-length and Δ1–375 GFP::hRod levels at kinetochores as in C. (G–J) HeLa Flp-In T-REx cells immunostained for GFP and Zw10 (G), Zwilch (H), Spindly (I), or Mad1 (J) in cells expressing full-length and Δ1–375 GFP::hRod after depletion of endogenous hRod. (K) Quantification of kinetochore levels for the components in G–J as described in C using immunofluorescence intensity measurements normalized to GFP::hRod signal. (L–O) HeLa Flp-In T-REx cells in prometaphase immunostained as in G–J but without nocodazole treatment. Arrowheads point to the accumulation of GFP::hRod at spindle poles along with Zw10, Zwilch, Spindly, and Mad1. Polar accumulation is missing in cells expressing GFP::hRod(Δ1–375). Bars: (A, B, E [main images], G–J, and L–O) 5 µm; (E, zoom) 1 µm.
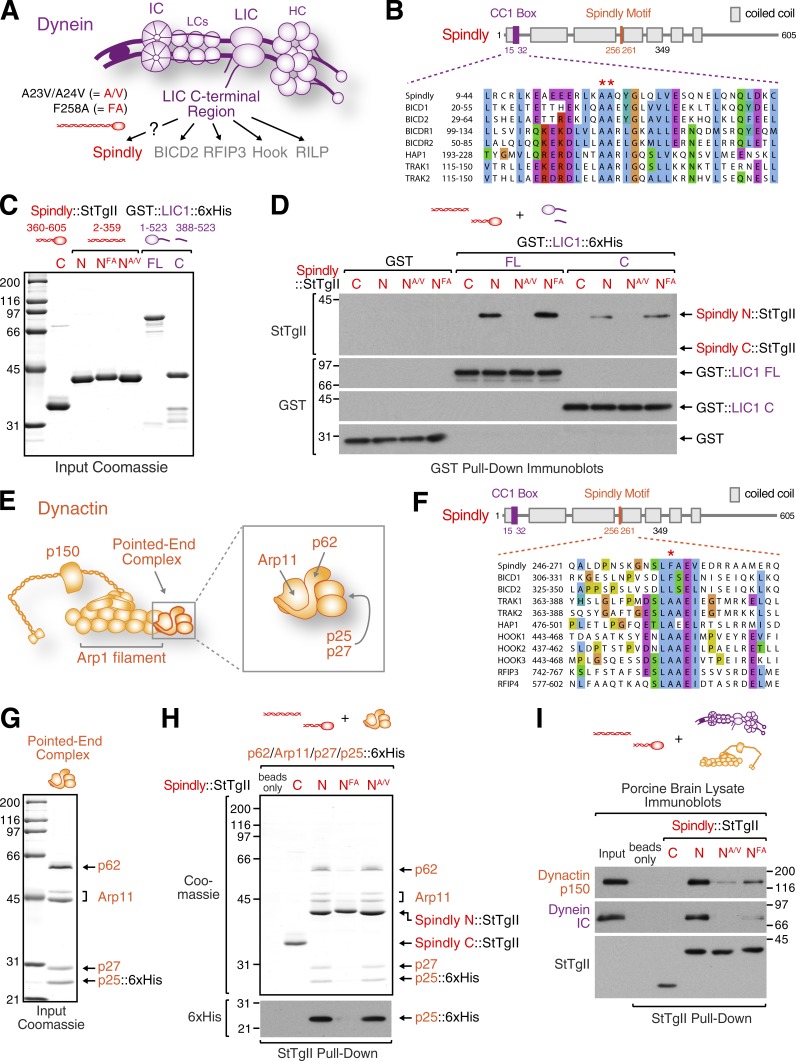
Spindly binds dynein light intermediate chain (LIC) and dynactin’s pointed-end complex through two conserved motifs
We next addressed how Spindly recruits dynein and dynactin. Spindly forms a ternary complex with dynein–dynactin in vitro, but molecular details are not known (McKenney et al., 2014). The C-terminal region of dynein LIC interacts with multiple adaptors for membrane cargo (Fig. 5 A; Schroeder et al., 2014; Schroeder and Vale, 2016). We therefore asked whether LIC also bound Spindly using purified human proteins. A GST fusion of full-length LIC1 (residues 1–523) or its C-terminal region (residues 388–523) pulled down Spindly’s N-terminal coiled-coil region (residues 2–359) but not its C-terminal region (residues 360–605; Fig. 5, C and D). Sequence alignments revealed a region in Spindly’s first coiled-coil segment that is conserved in multiple dynein adaptors, including BICD2 (Figs. 5 B and S4 A; Schlager et al., 2014; Hoogenraad and Akhmanova, 2016). Mutating the region’s two conserved alanines to valines in BICD2 inhibits complex formation with dynein–dynactin (Schlager et al., 2014), and the mutation A40V in D. melanogaster BICD causes a hypomorphic loss-of-function phenotype (Oh et al., 2000). We introduced the two alanine-to-valine (A/V) mutations into Spindly2–359 and BICD22–422 and found that both mutant proteins failed to bind LIC1 in GST pull-downs (Fig. 5, C and D; and Fig. S4, B–D). Furthermore, BICD22–422 displaced Spindly2–359 from GST::LIC1 in titration experiments (Fig. S4 E). We conclude that Spindly and BICD2 engage with the C-terminal region of LIC1 through a similar mechanism that involves a conserved region in their N-terminal coiled-coil segment, hereafter referred to as the CC1 box.
Figure 5.
Spindly binds dynein LIC and the dynactin pointed-end complex. (A) Cartoon of dynein, composed of two heavy chains (HCs), two ICs, two LICs, and six light chains (LCs). The C-terminal region of LIC binds dynein adaptors involved in membrane transport. The two point mutants of human Spindly used throughout this figure are indicated. (B) Sequence alignment showing a conserved region in the first coiled-coil segment of Spindly and other dynein adaptors (CC1 box). The two alanines mutated to valine in Spindly (A23/A24) and BICD2 (A43/A44) are marked with asterisks. (C) Coomassie-stained gels of purified Spindly fragments fused to Strep-tag II (StTgII) as well as GST-tagged full-length (FL) and C-terminal LIC1. (D) Immunoblots of GST pull-downs using the proteins in C. Protein fractions bound to beads were detected on blots with Strep-Tactin. The same membrane was then reprobed with anti-GST antibody. (E) Cartoon of dynactin with the pointed-end complex highlighted. (F) Sequence alignment showing that several dynein adaptors possess a motif that in Spindly is implicated in dynein–dynactin recruitment to kinetochores. The phenylalanine mutated to alanine in Spindly (F258) is marked with an asterisk. (G) Coomassie-stained gels of the purified recombinant dynactin pointed-end complex. The p25 subunit is tagged with 6×His. Arp11 appears as two distinct bands. We verified that both bands correspond to Arp11 by expressing Arp11::6×His (unpublished data). (H) Coomassie-stained gel and immunoblot of Strep-tag II pull-downs using the purified proteins in C and G. (I) Immunoblots of Strep-tag II pull-downs from porcine brain lysate using the purified proteins in C. The same membrane was probed for dynactin p150, dynein IC, and Strep-tagged Spindly. Molecular mass is indicated in kilodaltons.
We previously showed that point mutations in the Spindly motif, which is located more C-terminally to the CC1 box in Spindly’s coiled-coil region (Figs. 5 F and S4 A), abrogate dynein and dynactin recruitment to kinetochores in human cells (S256A or F258A) and C. elegans (F199A; Gassmann et al., 2010; Cheerambathur et al., 2013). We found that binding of Spindly to LIC1 was not affected by the F258A mutation (Fig. 5, C and D), implying that the Spindly motif mediated an additional distinct interaction with either dynein or dynactin. Structural work on the dynein–dynactin–BICD2 complex showed that the coiled-coil formed by BICD2’s N-terminal 270 residues binds along dynactin’s Arp1 filament, with the C-terminal end of the coiled-coil exiting at the filament’s pointed end (Urnavicius et al., 2015). If Spindly’s coiled-coil region also binds along the Arp1 filament, the Spindly motif around residue 258 would be predicted to reside near the filament’s pointed end, which is capped by a complex consisting of Arp11, p62, p27, and p25 (Fig. 5 E). We found that Strep-tagged Spindly2–359 pulled down purified recombinant pointed-end complex and that the association was diminished in the Spindly F258A mutant but not in the A/V mutant (Fig. 5, G and H). Thus, Spindly binds the pointed-end complex of dynactin in vitro, and this interaction involves a motif required for dynein–dynactin recruitment in vivo.
We next tested whether the mutations in the CC1 box and the Spindly motif affected binding of Spindly to intact dynein and dynactin in porcine brain lysate. Strep-tagged Spindly2–359 pulled down dynein and dynactin from lysate, as revealed by immunoblotting for dynactin p150 and dynein intermediate chain (IC; Fig. 5 I). In contrast, both Spindly2–359 A/V and F258A pulled down significantly less p150 and IC. Thus, binding of Spindly to LIC1 and the dynactin pointed end is required for the assembly of a stable ternary complex between dynein, dynactin, and Spindly.
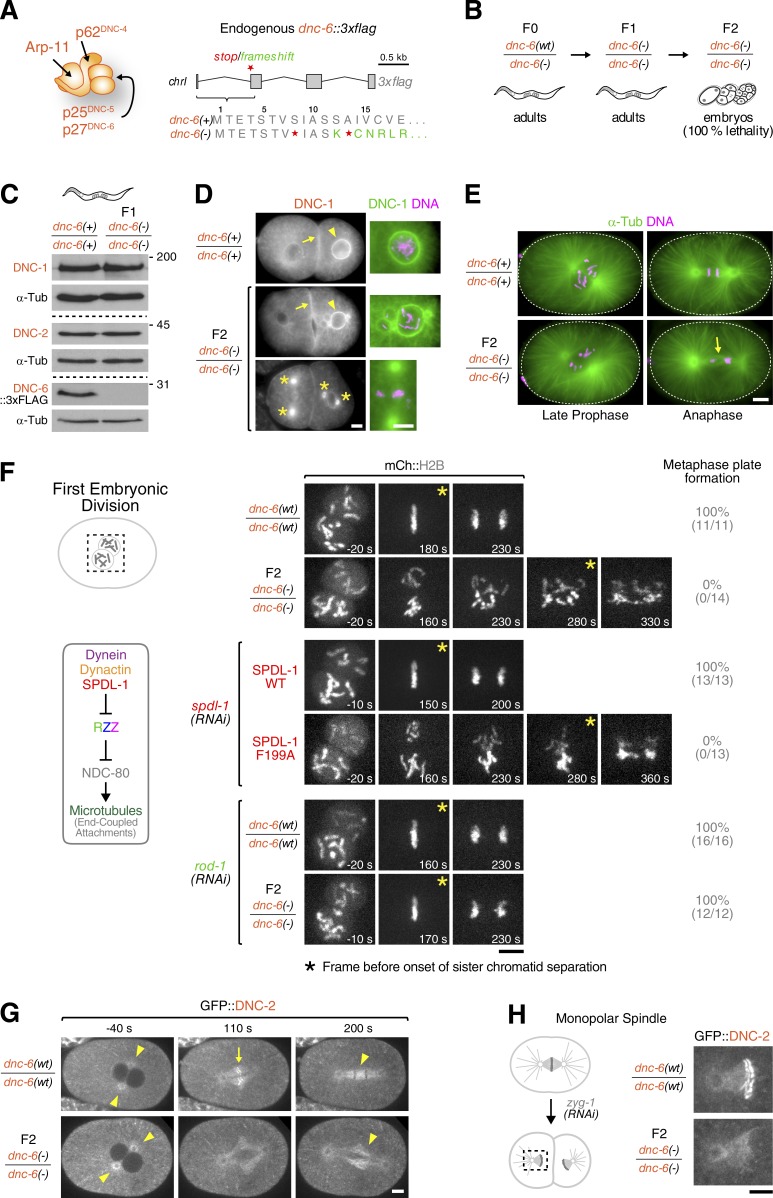
A null allele of p27dnc-6 prevents dynein–dynactin recruitment to kinetochores
Our results suggested that dynactin’s pointed-end complex has a key role in recruiting dynein–dynactin to kinetochores through Spindly. To directly test this prediction, we analyzed the loss-of-function phenotype of C. elegans pointed-end complex subunits in the mitotic one-cell embryo. RNAi-mediated depletion of Arp-11 and p62DNC-4 mimicked the phenotype of p150DNC-1 depletion, characterized by failure of centrosome separation and defective pronuclear migration (Fig. S5 A). This “dynactin-null” phenotype is consistent with a role for Arp-11 and p62DNC-4 in Arp-1 filament stabilization, as described for the mammalian proteins (Yeh et al., 2012). Similar, if less severe, defects in pronuclear migration were also evident after depletion of p25DNC-5. In contrast, depletion of p27DNC-6 had no adverse effects on centrosome separation, pronuclear migration, or spindle positioning (Fig. S5 A). Instead, we observed a specific defect in chromosome segregation (Fig. S5 B). To examine this phenotype without relying on RNAi knockdowns, we tagged endogenous p27dnc-6 with 3×flag and subsequently generated a genetic null allele, p27dnc-6(−) by introducing a stop/frameshift immediately after the p27dnc-6 start codon (Fig. 6 A). The p27dnc-6(−) allele was maintained by introduction of a balancer chromosome containing the wild-type locus, p27dnc-6(wt). F1 p27dnc-6(−/−) progeny, which receive a maternal supply of p27DNC-6, survived to adulthood and produced F2 p27dnc-6(−/−) progeny (Fig. 6 B). This permitted phenotypic analysis in a penetrant p27dnc-6-null background. Immunoblotting of F1 p27dnc-6(−/−) adults showed that p27DNC-6 was not required for stability of the p150DNC-1 and p50DNC-2 subunits (Fig. 6 C), and immunofluorescence of F2 p27dnc-6(−/−) embryos revealed normal p150DNC-1 localization to the nuclear envelope, the cell cortex, and centrosomes (Fig. 6 D). This suggested that the dynactin complex retained its structural integrity in the absence of p27DNC-6. Live imaging of F2 p27dnc-6(−/−) embryos using differential interference contrast confirmed that p27DNC-6 is dispensable for dynactin-dependent positioning of pronuclei and centrosomes in the first mitotic division (Video 3). However, despite successful bipolar spindle assembly (Fig. 6 E), F2 p27dnc-6(−/−) embryos became aneuploid because of chromosome missegregation (Fig. 6, D and E; and Video 3), providing an explanation for the penetrant lethality observed in F2 p27dnc-6(−/−) embryos. Visualization of chromosome dynamics with mCherry::histone H2B revealed severe defects in chromosome congression and segregation (Fig. 6 F and Video 4). The defects resembled those of the SpindlySPDL-1 motif mutant F199A, which delocalizes dynein–dynactin from kinetochores (Cheerambathur et al., 2013). Because RZZ inhibits the formation of stable kinetochore–microtubule attachments when kinetochores lack dynein–dynactin, the chromosome segregation defects of SpindlySPDL-1 F199A can be ameliorated by codepleting RZZ subunits. We observed a striking rescue of chromosome congression in p27dnc-6(−/−) embryos after rod-1(RNAi), suggesting that dynein–dynactin were delocalized from kinetochores in the absence of p27DNC-6 (Fig. 6 F). To test this directly, we crossed the p27dnc-6(−) allele with a gfp::p50dnc-2 knock-in allele. GFP::p50DNC-2 localized to centrosomes and the mitotic spindle in F2 p27dnc-6(−/−) embryos but was not detected at kinetochores (Fig. 6 G). We then triggered maximal kinetochore accumulation of GFP::p50DNC-2 by generating monopolar spindles. GFP::p50DNC-2 prominently accumulated at microtubule-unattached kinetochores of monopolar spindles in control embryos (Fig. 6 H). In contrast, GFP::p50DNC-2 was absent from kinetochores in F2 p27dnc-6(−/−) embryos, despite localizing to the monopolar spindle itself.
Figure 6.
Dynactin p27DNC-6 is required for dynein–dynactin recruitment to kinetochores. (A, left) Cartoon of the C. elegans dynactin pointed-end complex. (Right) Schematic of the endogenous p27dnc-6 locus modified with a C-terminal 3×flag tag. Stop codons (red stars) and a frameshift mutation (green lettering) were subsequently introduced to create a null allele, dnc-6(−). (B) Genetics of the p27dnc-6-null allele: F0 mothers heterozygous for dnc-6(−) contain a balancer chromosome with a wild-type copy, dnc-6(wt). F1 progeny homozygous for dnc-6(−) that survive to adulthood on maternally provided p27DNC-6 produce F2 dnc-6(−/−) embryos that completely lack p27DNC-6. (C) Immunoblots of F1 adults homozygous for dnc-6(−), demonstrating that levels of dynactin p150DNC-1 and p50DNC-2 are unaffected by the absence of p27DNC-6. Molecular mass is indicated in kilodaltons. (D) Immunofluorescence images of F2 dnc-6(−/−) embryos at the two-cell stage stained with anti-p150DNC-1 antibody, showing that dynactin without p27DNC-6 is recruited to the nuclear envelope (arrowheads), the cell cortex (arrows), and centrosomes (asterisks). Blowups show evidence of chromosome missegregation and aneuploidy in F2 dnc-6(−/−) embryos. (E) Immunofluorescence images showing successful bipolar spindle assembly in F2 dnc-6(−/−) embryos. The arrow indicates chromosome missegregation. (F) Selected time-lapse sequence frames of the first mitotic division in embryos expressing mCherry::histone H2B. F2 dnc-6(−/−) embryos exhibit severe chromosome segregation defects that are similar to those observed for the SpindlySPDL-1 F199A mutant. The defects are significantly ameliorated after depletion of ROD-1, as predicted by the inhibitory cross-talk between the kinetochore dynein module and NDC-80 (summarized in schematic on the left). The frequencies of embryos in which chromosomes congress to metaphase plates are indicated in the percentages and absolute numbers of embryos in parentheses. Time is relative to nuclear envelope breakdown. (G) Selected frames from a time-lapse sequence in F2 dnc-6(−/−) embryos expressing GFP::p50DNC-2, demonstrating that dynactin without p27DNC-6 localizes normally to centrosomes and the mitotic spindle (arrowheads) but is absent from kinetochores (arrow). (H, left) Experimental strategy to generate monopolar spindles in F2 dnc-6(−/−) embryos expressing GFP::p50DNC-2 by depleting the centriole duplication kinase ZYG-1. (Right) Stills from a time-lapse sequence showing that dynactin failed to accumulate on unattached kinetochores of monopolar spindles in the absence of p27DNC-6. Bars, 5 µm.
Thus, our analysis of an engineered p27dnc-6-null allele demonstrates that the dynactin pointed-end complex is essential for dynein–dynactin recruitment to kinetochores, consistent with our biochemical data and prior in vivo characterization of Spindly motif mutants in C. elegans and human cells.
Spindly-like motifs implicated in dynactin pointed-end binding are present in multiple dynein adaptors
Primary sequence analysis identified Spindly-like motifs in dynein adaptors involved in intracellular transport (Fig. 5 F). In all cases, the motif is preceded by a similarly sized coiled-coil region (260 residues on average), whose N-terminal segment also often harbors the LIC-binding CC1 box (Fig. S4 A). We found that Strep-tagged BICD22–422, which contains a Spindly-like motif at residues 336–340, pulled down the purified pointed-end complex in vitro (Fig. S4, B, C, and F). Mutating E339 to alanine (EA) weakened BICD22–422 binding to the pointed-end complex, and introducing four additional alanine mutations (5A) further diminished the interaction (Fig. S4, C and F). In contrast, BICD22–422 binding to LIC1 was not affected by the 5A mutant (Fig. S4 D). We conclude that binding of Spindly and BICD2 to dynactin’s pointed-end complex requires a motif shared by several functionally distinct dynein adaptors.
Discussion
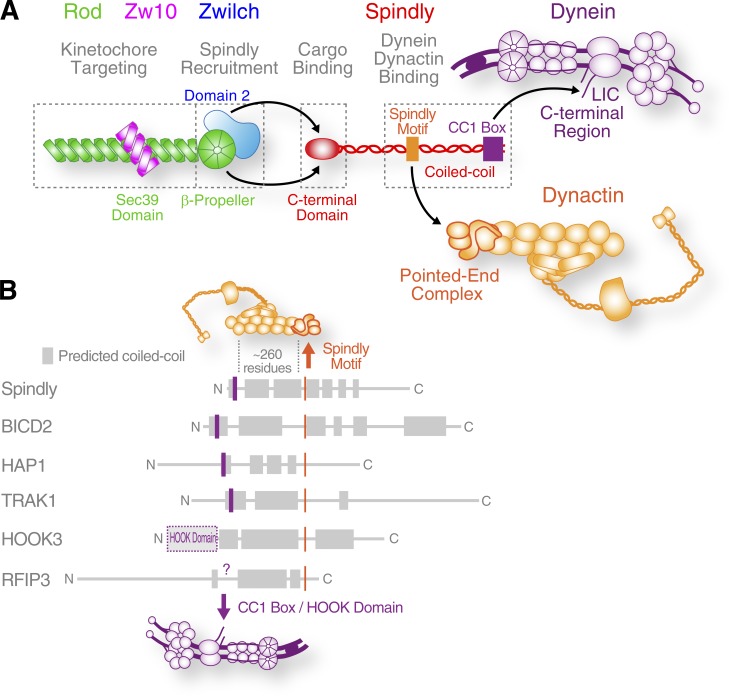
The molecular motor dynein is used in virtually all cellular processes that require microtubule minus end–directed motility. Dynein’s functional diversity requires that the motor associate with cofactors and cargo-specific adaptors, but how dynein is recruited and locally activated at subcellular structures remains poorly understood. In this paper, we provide a detailed view of how dynein is targeted to the mitotic kinetochore (Fig. 7 A). Our results suggest that the mechanism used by the kinetochore to engage dynein is also relevant for how dynein interacts with cargo in the context of intracellular transport (Fig. 7 B).
Figure 7.
Molecular interactions implicated in dynein recruitment to kinetochores. (A) Graphic summary of the physical interactions that link the RZZ complex to dynein and dynactin via the adaptor Spindly to recruit the motor to kinetochores. Note that the intact RZZ complex is dimeric but shown here as a monomer for simplicity. (B) Spindly and other dynein adaptors may use a similar mechanism to engage with dynein and dynactin (see Fig. S4 A). Common features include a Spindly-like motif implicated in pointed-end complex binding and an N-terminally located CC1 box for LIC binding. The intervening coiled-coil region likely binds along the Arp1 filament (Urnavicius et al., 2015).
Our analysis of C. elegans and human RZZ identifies a conserved module, consisting of Rod’s N-terminal β-propeller in complex with the Zwilch subunit, as the kinetochore receptor of the dynein adaptor Spindly. We also show that the Rod β-propeller–Zwilch module is dispensable for RZZ targeting to kinetochores and that this function instead resides in Rod’s α-solenoid, whose Sec39 domain binds the Zw10 subunit. Zw10 is known to interact with the outer kinetochore component Zwint (Starr et al., 2000; Vos et al., 2011), which in turn associates with the C-terminal domain of Knl1 (Petrovic et al., 2010). However, Zw10 mutants that cannot bind Zwint still localize to kinetochores (Famulski et al., 2008), and depletion of the putative C. elegans Zwint homologue has no effect on RZZ kinetochore levels (Varma et al., 2013). It is therefore likely that additional contacts exist between the Rod α-solenoid and Knl1 and/or Bub1 (Caldas et al., 2015; Silió et al., 2015; Zhang et al., 2015). In support of this idea, a recently described point mutation in D. melanogaster directly implicates Rod’s C-terminal region in kinetochore targeting of RZZ (Défachelles et al., 2015).
Using the C. elegans proteins, we show that the SpindlySPDL-1 C-terminal domain functions as the cargo-binding region of the adaptor that directly binds to both the ROD-1 β-propeller and ZwilchZWL-1. Analysis of the ZwilchZWL-1 E433A/E437A mutant demonstrates that the SpindlySPDL-1–ZwilchZWL-1 interaction is required for SpindlySPDL-1 kinetochore recruitment and for SpindlySPDL-1 binding to a ROD-1 β-propeller–ZwilchZWL-1 complex in vitro. The function of the ROD-1 β-propeller–SpindlySPDL-1 interaction remains to be determined. Interestingly, ROD-1’s β-propeller not only binds SpindlySPDL-1 but also interacts with the N-terminal tail of NDC-80 to inhibit microtubule binding of the NDC-80 complex (Cheerambathur et al., 2013). RZZ-mediated inhibition of NDC-80 is proposed to occur transiently during initial microtubule capture, when microtubules are laterally attached to kinetochores through dynein. Phenotypic analysis of the SpindlySPDL-1 F199A mutant (Cheerambathur et al., 2013) and the p27dnc-6-null allele as described in this study suggests that the presence of dynein at kinetochores is required to turn off RZZ-mediated inhibition of NDC-80, but the mechanism remains unclear. One attractive possibility is that the ROD-1 β-propeller–SpindlySPDL-1-dynein linkage we uncovered is used to directly modulate ROD-1 β-propeller binding to the NDC-80 tail.
Our analysis of human Rod lacking its β-propeller shows that the role of the Rod β-propeller–Zwilch module in Spindly recruitment is conserved. There is, however, a significant difference between C. elegans and human Spindly: the C-terminal cysteine in human Spindly is farnesylated, and this lipid modification is essential for Spindly targeting to kinetochores (Holland et al., 2015; Moudgil et al., 2015). As Spindly binding to Zwilch would be predicted to involve the two conserved acidic residues that we identified in C. elegans ZwilchZWL-1, which are unlikely to participate in a hydrophobic interaction, the binding site for Spindly’s farnesylated C terminus likely resides in the Rod β-propeller.
Intriguingly, the hRod Δ1–375 mutant, although localizing robustly to kinetochores, did not support formation of the crescent and ring shapes associated with corona expansion, which requires copolymerization of corona components (Hoffman et al., 2001; Wynne and Funabiki, 2015). We recently described a similar effect with farnesyltransferase inhibitors, which delocalize Spindly from kinetochores (Holland et al., 2015). We speculate that Spindly binding to the Rod β-propeller–Zwilch module may promote an oligomeric state of RZZ that contributes to corona expansion.
Recent work has shown that dynein associates with its essential cofactor dynactin in a ternary complex with adaptor proteins (Splinter et al., 2012; McKenney et al., 2014; Urnavicius et al., 2015; Carter et al., 2016). Using specific point mutants in Spindly’s N-terminal coiled-coil, we show that assembly of the dynein–dynactin–Spindly complex requires the CC1 box and the Spindly motif, which mediate binding to the LIC1 C-terminal region and the dynactin pointed-end complex, respectively.
The short Spindly motif defines the Spindly family because of its exceptional conservation against a near-complete divergence of the rest of the protein sequence. Single point mutations in the Spindly motif (S256A or F258A in humans; F199A in C. elegans) abrogate dynein–dynactin recruitment to kinetochores (Gassmann et al., 2010; Cheerambathur et al., 2013). Our results suggest that the molecular cause underlying this defect is the inability of Spindly F258A to bind dynactin’s pointed-end complex. Remarkably, a null allele of the C. elegans pointed-end subunit p27dnc-6 mimics the severe chromosome segregation defect of the SpindlySPDL-1 mutant F199A. This underscores the importance of the Spindly–dynactin pointed-end interaction for kinetochore dynein recruitment. Our results in C. elegans are in apparent contrast with work in mammalian cells that reported no reduction of kinetochore dynein–dynactin levels when p27 was depleted using RNAi (Raaijmakers et al., 2013; Yeh et al., 2013). One potential explanation for this difference is that the interaction with Spindly may involve different pointed-end subunits in mammals. Importantly, whether the Arp11 and p62 subunits play a direct role in kinetochore recruitment of dynein–dynactin cannot be assessed with inhibition experiments because both subunits are required for the stability of dynactin’s Arp1 filament (Raaijmakers et al., 2013; Yeh et al., 2013).
An important unresolved question is whether functionally diverse dynein adaptors use similar mechanisms to interact with dynein and dynactin. We find that Spindly shares the CC1 box and Spindly motif with adaptors involved in intracellular transport. Previous characterization of the A43V/A44V mutant in BICD2, which was generated based on a D. melanogaster mutant (Oh et al., 2000), implicated its CC1 box in dynein–dynactin binding (Schlager et al., 2014; Hoogenraad and Akhmanova, 2016). We show that mutating the two conserved alanines in the CC1 box to valine abrogates binding of both Spindly2–359 and BICD22–422 to LIC1 and that Spindly2–359 and BICD22–422 compete for LIC1 binding. BICD22–422 also binds to dynactin’s pointed-end complex, and mutations in BICD22–422’s Spindly-like motif weaken this interaction. Given the similarities between Spindly and BICD2, we hypothesize that other adaptors may also use their CC1 box and Spindly-like motif to interact with the LIC and dynactin’s pointed-end complex, respectively. In Spindly, BICD2, HAP1, TRAK1, and Hook3, the CC1 box (or the Hook domain, which also binds to LIC1; Schroeder and Vale, 2016) is separated from the Spindly-like motif by a coiled-coil segment predicted to bind along the groove of the Arp1 filament, as demonstrated for BICD2 (Chowdhury et al., 2015; Urnavicius et al., 2015). This configuration places the CC1 box/Hook domain at the barbed end and the Spindly-like motif near the pointed end of the Arp1 filament (Fig. 7 B). Interestingly, a recent study found that residues 402–552 of Hook3 are required for robust motility of a dynein–dynactin–Hook3 complex in vitro (Schroeder and Vale, 2016). This region contains the Spindly-like motif (residues 452–458; Fig. 5 F), raising the possibility that Hook3 binding to the pointed-end complex may contribute to dynein activation. In addition to its role in recruiting dynein to kinetochores that we describe here, dynactin’s pointed-end complex is implicated in linking dynein to a subset of membrane cargo through the p25 and p27 subunits (Zhang et al., 2011, 2014; Yeh et al., 2012). It is likely that the importance of the pointed-end complex for dynactin function has so far been underestimated, because Arp11 and p62’s requirement for dynactin integrity will have masked additional roles of these subunits. Further molecular and functional characterization of the interactions between dynein adaptors and the dynactin pointed-end promises to advance our understanding of how dynein is recruited and activated in different cellular contexts.
Materials and methods
Worm strains
Worm strains used in this study are listed in Table S1. Worm strains were maintained at 20°C on standard nematode growth media plates seeded with OP50 bacteria. A Mos1 transposon-based strategy (MosSCI) was used to generate strains stably expressing GFP or mCherry fusions of rod-1, zwl-1, czw-1, and spdl-1 (Frøkjær-Jensen et al., 2008, 2012). To make transgenes RNAi resistant, a region of several hundred bases was reencoded by codon shuffling. Transgenes were cloned into pCFJ151 for insertion on chromosome II (chrII; ttTi5605) or into pCFJ178 for insertion on chromosome IV (chrIV; cxTi10882). Single copy transgene insertions were generated by injecting a mixture of target plasmid, transposase plasmid pCFJ601, and selection markers into strains EG6429 (ttTi5605, chrII) or EG6700 (cxTi10882, chrIV), as described previously (Frøkjær-Jensen et al., 2012). Transgene integration was confirmed by PCR spanning both homology regions. The 3×flag::Zw10czw-1 knock-in allele, gfp::p50dnc-2 knock-in allele, p27dnc6::3×flag allele, and p27dnc-6-null allele were generated using a CRISPR-Cas9 method described previously (Arribere et al., 2014). Strains were outcrossed six times with the wild-type N2 strain, and the modifications were confirmed by sequencing. Other fluorescent markers were subsequently introduced by mating. For the experiments in Fig. 6, the p27dnc-6-null allele was maintained using a GFP-marked balancer. F1 p27dnc-6(−/−) progeny from balanced heterozygous mothers were identified by the lack of GFP fluorescence. GFP-negative F1 p27dnc-6(−/−) adults made up 14% of the total F1 adult population. Considering that progeny homozygous for the balancer is not viable, this indicates that maternally provided p27DNC-6 allows ∼50% of F1 p27dnc-6(−/−) progeny to survive to adulthood. All F2 p27dnc-6(−/−) progeny died during embryogenesis.
Antibodies
Affinity-purified rabbit polyclonal antibodies against a C-terminal fragment of human Rod/KNTC1 (residues 1,956–2,130), the N-terminal region of C. elegans ROD-1 (residues 1–350), a C-terminal region of DNC-1 (residues 1,037–1,351), and full-length DNC-2 were generated as described previously (Desai et al., 2003). In brief, GST fusions were expressed in Escherichia coli, purified, and injected into rabbits (Covance). Serum was affinity purified on HiTrap N-hydroxysuccinimide columns (GE Healthcare) against covalently coupled human Rod1,956–2,130, Sumo::ROD-11–350, and full-length DNC-2 or sequentially against coupled GST followed by coupled GST::DNC-11,037–1,351. The latter purification also yielded the anti-GST antibodies used in this paper.
Cell lines
Stable isogenic HeLa Flp-In T-REx cell lines expressing human Rod/KNTC1 were generated by FRT/Flp-mediated recombination as described previously (Tighe et al., 2004, 2008). Residues 1–2,209 of Rod (full-length) and a Rod fragment corresponding to residues 376–2,209 (Δ1–375) were cloned into a pcDNA5/FRT/TO-based vector (Invitrogen) modified to contain an N-terminal Myc-LAP epitope tag. The LAP tag consisted of GFP–tobacco etch virus (TEV)–S-peptide (Cheeseman and Desai, 2005).
Indirect immunofluorescence
For immunofluorescence experiments in tissue culture cells, HeLa cells were fixed immediately after aspiration of the medium with 4% formaldehyde in Phem buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH 6.9) for 5 min at room temperature, then permeabilized for 2 min with 0.1% Triton X-100 in Phem buffer, and rinsed three times in Phem buffer. For stainings with anti-Zw10 antibody, cells were fixed in methanol at −20°C for 45 min, followed by rehydration for 2 × 5 min in PBS/0.5% Triton X-100 and PBS/0.1% Triton X-100. Cells were blocked for 30 min in AbDil solution (4% IgG-free BSA [Jackson ImmunoResearch Laboratories, Inc.] and 0.1% Triton X-100) and incubated with primary antibody for 1 h at room temperature (or overnight at 4°C for anti-Mad1), diluted in AbDil (human anticentromere antibodies [ACAs], 1:5,000; rabbit anti-Zwilch, 1:900; rabbit anti-Zw10, 1:1,000; mouse anti-Mad1, 1:10; rabbit anti-Spindly OD174, 1:5,000; and goat anti-GFP, 1:15,000). After washing for 3 × 5 min in PBS/0.1% Triton X-100, cells were incubated with secondary antibodies conjugated with fluorescent dyes (Alexa Fluor 488, 594, and 647; Jackson ImmunoResearch Laboratories, Inc.). Cells were washed again for 3 × 5 min and mounted in ProLong Gold with DAPI stain (Invitrogen).
Immunofluorescence in C. elegans embryos was performed as described previously (Oegema et al., 2001) using the following primary antibodies: rabbit anti–ROD-1, 1 µg/ml; rabbit anti–DNC-1 GC2, 1:500; mouse anti–α-tubulin DM1α (Sigma-Aldrich), 1:1,000; and mouse anti-FLAG M2, 1:1,000 (Sigma-Aldrich).
Images were recorded on an Axio Observer microscope (ZEISS) at 1 × 1 binning with a 100×, 1.46 NA Plan-Apochromat objective and an Orca Flash 4.0 camera (Hamamatsu Photonics). Image files were imported into Fiji (ImageJ; National Institutes of Health) for further processing.
Image analysis
For image analysis in C. elegans embryos, intensity measurements were performed on selected frames from time-lapse series acquired by confocal microscopy. Kinetochore fluorescence intensities were determined as described previously (Moyle et al., 2014). In brief, a region was drawn around chromosomes (marked by GFP or mCherry::histone H2b), and the integrated fluorescence intensity was measured for the kinetochore component (mCherry::ROD-1, ZwilchZWL-1::mCherry, or GFP::SpindlySPDL-1). The chromosomal region was then expanded by 1–2 pixels on all sides, and the difference in integrated intensity between the expanded and chromosomal region was used to define background intensity. The final integrated intensity for the chromosomal region was calculated by subtracting background intensity.
Immunofluorescence kinetochore signals in human cells were quantified as described previously (Hoffman et al., 2001; Gassmann et al., 2010).
Statistical analysis
Statistical significance was evaluated with the t test using Prism 7 software (GraphPad Software).
RNAi in tissue culture cells
HeLa cells were maintained at 37°C in a 5% CO2 atmosphere in DMEM (Gibco) supplemented with 10% tetracycline-free FBS (Takara Bio Inc.), 100 u/ml penicillin, 100 u/ml streptomycin, and 2 mM l-glutamine. For immunofluorescence, cells were seeded on 12-mm poly-l-lysine–coated coverslips in 12-well plates 24 h before transfection with siRNAs. Cells were transfected with siRNA (On-Target Plus; GE Healthcare) targeting human Rod/KNTC1 (target sequence, 5′-GUAAAUAACUUGCGAGAGU-3′) as described previously (Gassmann et al., 2010). An siRNA against luciferase (5′-CGUACGCGGAAUACUUCGA-3′) was used in control experiments. Transgene expression was induced with 0.2 µg/ml tetracycline 24 h after transfection, and cells were fixed 20–24 h later. To depolymerize microtubules before immunofluorescence, cells were treated with 1 µM nocodazole for 4 h.
RNAi in C. elegans
For production of double-stranded RNA (dsRNA), oligonucleotides with tails containing T3 and T7 promoters were used to amplify regions from genomic N2 DNA or cDNA. The oligonucleotides are listed in Table S2. PCR reactions were cleaned and used as templates for T3 and T7 transcription reactions (MEGAscript; Invitrogen). Transcription reactions were cleaned (MEGAclear; Invitrogen) and mixed with 3× soaking buffer (32.7 mM Na2HPO4, 16.5 mM KH2PO4, 6.3 mM NaCl, and 14.1 mM NH4Cl). dsRNA was delivered by injecting L4 hermaphrodites, and animals were processed for live-imaging, immunofluorescence, or immunoblotting after incubation at 20°C for 45–50 h. For the experiment in Fig. 1 J, a partial knockdown of CZW-1 in strain GCP261 was performed (full depletion resulted in sterility) by incubating injected animals at 16°C for 40 h. Partial knockdown of CZW-1 prevented silencing of RNAi-resistant gfp::czw-1 transgene expression. Strains GCP171/GCP172 (Fig. 2 H) and GCP173 (Fig. 2 A) were propagated on HT115 bacteria expressing spdl-1–specific dsRNA to prevent silencing of the RNAi-resistant gfp::spdl-1 transgene.
Yeast two-hybrid analysis
Yeast two-hybrid analysis was performed according to the manufacturer’s guidelines (Matchmaker; Invitrogen). Yeast-containing bait (pGBKT7) and prey (pGADT7) vectors with the cDNAs of interest were plated on -Leu/-Trp/-His plates to select for interactions. Screening was performed using cDNA for zwl-1, czw-1, and the 15 rod-1 fragments listed in Fig. S1 B.
Immunoblotting
Samples were resolved by 10 or 12% SDS-PAGE and transferred to 0.2-µm nitrocellulose membranes (GE Healthcare). Membranes were blocked with 5% nonfat dry milk in TBST (20 mM Tris, 140 mM NaCl, and 0.1% Tween, pH 7.6) and probed at 4°C overnight with the following primary antibodies: rabbit anti-KNTC1 GC7, 1:3,000; mouse anti–α-tubulin B512 (Sigma-Aldrich), 1:5,000; rabbit anti-mCherry OD78, 1:17,000; rabbit anti–ZWL-1 OD89, 1:1,000; rabbit anti–SPDL-1 OD164, 1:1,000; rabbit anti-GST GC3, 1:7,500; mouse anti–6×His His.H8 (EMD Millipore), 1:2,500; mouse anti-p150 (BD), 1:500; mouse anti-dynein IC 70.1 (Sigma-Aldrich), 1:500; rabbit anti–DNC-1 GC2, 1:500; and rabbit anti–DNC-2 GC5, 1:5,000. Membranes were washed three times with TBST, incubated with goat secondary antibodies coupled to HRP (Jackson ImmunoResearch Laboratories, Inc.; 1:10,000) for 1 h at room temperature, and washed again three times with TBST. Blots were visualized by chemiluminescence using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) and x-ray film (Fujifilm). For Strep-Tactin detection, membranes were blocked with 3% BSA in PBS/0.5% Tween 20, washed three times with PBS/0.1% Tween 20, and probed at 4°C overnight with Strep-Tactin HRP conjugate.
Biochemistry
Expression constructs
The cDNAs of ZwilchZWL-1 (wild type and E433A/E437A), Zw10CZW-1, ROD-1 (residues 1–2,177, 1–1,203, and 1–372), DCTN4, DCTN5, DCTN6, and ARP11 were cloned into the pACEbac1 expression vector. ZwilchZWL-1 and DCTN5 were tagged C-terminally with 6×His, and the ROD-1 constructs were tagged N-terminally with 6×His followed by a linker containing a cleavage site for TEV protease. Bacmid recombination and virus production were performed as described previously (Bieniossek et al., 2008).
The cDNA of SpindlySPDL-1 (residues 1–479, 1–361, and 362–479) and DYNC1LI1 (residues 1–523 and 388–523) were cloned into pGEX-6P-1. DYNC1LI1 was also tagged C-terminally with 6×His. The cDNAs of human Spindly (residues 2–359 and 360–605) and human BICD2 (residues 2–422) were cloned into a 2CT expression vector containing an N-terminal 6×His::maltose-binding protein (MBP) fusion followed by a linker with a TEV protease cleavage site and a C-terminal linker followed by the Strep-tag II.
Purification of ROD-11–372–ZwilchZWL-1, ROD-11–372, and dynactin pointed-end complex from insect cells
ROD-11–372 and ZwilchZWL-1 (R1–372Z) were coexpressed in 500-ml cultures (SFM4 medium; Hyclone) of Sf21 cells (0.8 × 106 cells/ml) coinfected with corresponding viruses. For the purifications shown in Fig. S2 (A and B), only ROD-11–372 was tagged with 6×His. For the pull-downs in Fig. 3 (G–I), both ROD-11–372 and ZwilchZWL-1 were tagged with 6×His. Cells were harvested by centrifugation at 800 g for 5 min. Pellets were resuspended in lysis buffer A (50 mM Hepes and 200 mM NaCl, pH 8.0) supplemented with EDTA-free cOmplete Protease Inhibitor Cocktail (Roche), sonicated, and cleared by centrifugation at 34,000 g for 40 min. R1–372Z was purified by batch affinity chromatography using HIS-Select Nickel Affinity Gel beads (Sigma-Aldrich). Beads were washed with wash buffer A (50 mM Hepes, 200 mM NaCl, and 10 mM imidazole, pH 8.0), and R1–372Z was eluted on a gravity column with elution buffer A (50 mM Hepes, 200 mM NaCl, and 250 mM imidazole, pH 8.0). R1–372Z was further purified by size-exclusion chromatography using a Superose 6 10/300 column (GE HealthCare) equilibrated with 25 mM Hepes and 150 mM NaCl, pH 7.5. Fractions containing R1–372Z were pooled, and glycerol and DTT were added to a final concentration of 10% (vol/vol) and 1 mM, respectively. Aliquots were flash-frozen in liquid nitrogen and stored at −80°C. The same protocol was used for purification of 6×His::ROD-11–372 on its own and for purification of the dynactin pointed-end complex (p62–Arp11–p27–p25::6×His).
Protein purification from bacteria
All bacterial expression constructs were transformed into the E. coli strain BL21, and expression was induced with 0.1 mM IPTG at 18°C overnight at an OD600 of 0.5. Cells were harvested by centrifugation for 20 min at 4,000 g. For GST::SpindlySPDL-1, bacterial pellets were resuspended in lysis buffer B (50 mM Hepes, 500 mM NaCl, 0.1% Tween 20, 10 mM EDTA, 10 mM EGTA, 1 mM DTT, 1 mM PMSF, 2 mM benzamidine-HCl, and 1 mg/ml lysozyme, pH 8.0), disrupted by sonication, and cleared by centrifugation at 34,000 g for 40 min. GST::SpindlySPDL-1 was purified by batch affinity chromatography using Glutathione Agarose beads (Thermo Fisher Scientific). Beads were washed with wash buffer B (25 mM Hepes, 400 mM NaCl, 1 mM DTT, and 2 mM benzamidine-HCl, pH 8.0), and GST::SpindlySPDL-1 was eluted on a gravity column with elution buffer B (25 mM Hepes, 150 mM NaCl, and 10 mM reduced l-glutathione, pH 8.0). GST::SpindlySPDL-1 was further purified by size-exclusion chromatography using a Superose 6 10/300 column equilibrated with 25 mM Hepes and 150 mM NaCl, pH 7.5. Fractions containing GST::SpindlySPDL-1 were pooled, and glycerol and DTT were added to final concentrations of 10% (vol/vol) and 1 mM, respectively. Aliquots were flash-frozen in liquid nitrogen and stored at −80°C.
For human Spindly and BICD2 proteins, bacterial pellets were resuspended in lysis buffer C (50 mM Hepes, 250 mM NaCl, 10 mM imidazole, 0.1% Tween 20, 1 mM DTT, 1 mM PMSF, 2 mM benzamidine-HCl, and 1 mg/ml lysozyme, pH 8.0), disrupted by sonication, and cleared by centrifugation at 34,000 g for 40 min. Spindly and BICD2 were purified by tandem affinity chromatography using HIS-Select Nickel Affinity Gel beads (Sigma-Aldrich) followed by Strep-Tactin Sepharose (IBA). HIS-Select Nickel Affinity Gel beads were incubated in batch with the cleared lysates and then washed with wash buffer C (25 mM Hepes, 250 mM NaCl, 20 mM imidazole, 0.1% Tween 20, 1 mM DTT, and 2 mM benzamidine-HCl, pH 8.0), and proteins were eluted on a gravity column with elution buffer C (50 mM Hepes, 150 mM NaCl, 250 mM imidazole, 1 mM DTT, and 2 mM benzamidine-HCl, pH 8.0). Fractions containing the recombinant proteins were pooled, incubated overnight with TEV protease to cleave off the 6×His::MBP tag, incubated in batch with Strep-Tactin Sepharose beads, and washed with wash buffer D (25 mM Hepes, 250 mM NaCl, 0.1% Tween 20, 1 mM DTT, and 2 mM benzamidine-HCl, pH 8.0). Proteins were eluted on a gravity column with elution buffer D (50 mM Hepes, 150 mM NaCl, and 2.5 mM desthiobiotin, pH 8.0). Fractions containing the recombinant proteins were pooled and dialyzed against storage buffer (25 mM Hepes and 150 mM NaCl, pH 7.5), and glycerol and DTT were added to final concentrations of 10% (vol/vol) and 1 mM, respectively. Aliquots were flash-frozen in liquid nitrogen and stored at −80°C.
For GST::LIC1::6×His, bacterial pellets were resuspended in lysis buffer E (50 mM Hepes, 250 mM NaCl, 0.1% Tween 20, 10 mM EDTA, 10 mM EGTA, 1 mM DTT, 1 mM PMSF, 2 mM benzamidine-HCl, and 1 mg/ml lysozyme, pH 8.0). GST::LIC1::6×His was purified by tandem affinity chromatography using Glutathione Agarose beads followed by HIS-Select Nickel Affinity Gel beads. Glutathione Agarose beads were incubated in batch with the cleared lysates, then were washed with wash buffer E (25 mM Hepes, 250 mM NaCl, 0.1% Tween 20, 1 mM DTT, and 2 mM benzamidine-HCl, pH 8.0), and proteins were eluted on a gravity column with elution buffer E (50 mM Hepes, 150 mM NaCl, 10 mM reduced l-glutathione, 1 mM DTT, and 2 mM benzamidine-HCl, pH 8.0). Fractions containing the recombinant proteins were pooled, incubated in batch with Nickel Affinity Gel beads, and washed with wash buffer C (25 mM Hepes, 250 mM NaCl, 20 mM imidazole, 0.1% Tween 20, 1 mM DTT, and 2 mM benzamidine-HCl, pH 8.0). Proteins were eluted on a gravity column with elution buffer C (50 mM Hepes, 150 mM NaCl, 250 mM imidazole, 1 mM DTT, and 2 mM benzamidine-HCl, pH 8.0). Fractions containing the recombinant proteins were pooled and dialyzed against storage buffer (25 mM Hepes and 150 mM NaCl, pH 7.5), and glycerol and DTT were added to final concentrations of 10% (vol/vol) and 1 mM, respectively. Aliquots were flash frozen in liquid nitrogen and stored at −80°C.
Pull-downs from insect cell lysates
Insect cell lysates containing RZZ subunits (Fig. 3, A and E) were prepared from 50 ml of a 0.8 × 106 cell/ml suspension. Cells were harvested by centrifugation at 800 g for 5 min. Pellets were resuspended in 2 ml of pull-down buffer (50 mM Hepes, 50 mM NaCl, and 5 mM DTT, pH 7.5) supplemented with EDTA-free cOmplete Protease Inhibitor Cocktail, disrupted by sonication, and cleared by centrifugation at 20,000 g for 30 min. Cleared lysates were aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C. To decrease interference of endogenous glutathione-binding proteins present in insect cells, thawed lysate aliquots were first predepleted with Glutathione Agarose beads. Then, 15 µl of Glutathione Agarose beads coated with 225 pmol of the GST::SpindlySPDL-1 constructs or GST only were incubated with 250 µl of insect cell lysate for 1 h at 4°C. Beads were washed with 3 × 500 µl of pull-down buffer, and proteins were eluted in pull-down buffer supplemented with 15 mM reduced l-glutathione for 15 min at room temperature.
Pull-downs with purified ROD-11–372–ZwilchZWL-1 complex or ROD-11–372
50 pmol purified ROD-11–372–ZwilchZWL-1 complex or ROD-11–372 was incubated with 50 pmol GST::SpindlySPDL-1 for 1 h at 4°C in 150 µl pull-down buffer (50 mM Hepes, 50 mM NaCl, and 5 mM DTT, pH 7.5) supplemented with 15 µl of Glutathione Agarose beads. Beads were washed with 3 × 500 µl pull-down buffer, and proteins were eluted with pull-down buffer containing 15 mM reduced l-glutathione.
Pull-downs with purified LIC1
50 pmol of GST::LIC1::6×His were incubated with 50 pmol BICD2::Strep-tag II or 250 pmol Spindly::Strep-tag II for 1 h at 4°C in 150 µl pull-down buffer (50 mM Hepes, 50 mM NaCl, and 5 mM DTT, pH 7.5) supplemented with 15 µl of Glutathione Agarose beads. Beads were washed with 3 × 500 µl pull-down buffer, and proteins were eluted with pull-down buffer containing 15 mM reduced l-glutathione. For the competition experiment in Fig. S4 E, 50 pmol GST::LIC1::6×His was mixed with 250 pmol Spindly::Strep-tag II and 0, 50, 100, or 200 pmol BICD2::Strep-tag II.
Pull-downs with purified dynactin pointed-end complex
50 pmol Spindly::Strep-tag II or BICD2::Strep-tag II was incubated with 250 pmol dynactin pointed-end complex for 1 h at 4°C in 150 µl pull-down buffer (50 mM Hepes, 100 mM NaCl, 5 mM DTT, and 0.1% Tween 20, pH 7.5) supplemented with 15 µl of Strep-Tactin Sepharose beads. Beads were washed with 3 × 500 µl pull-down buffer, and proteins were eluted with buffer E (100 mM Tris/HCl, 150 mM NaCl, 1 mM EDTA, and 2.5 mM desthiobiotin, pH 8.0; IBA).
Pull-downs from porcine brain lysate
Porcine brain lysate was prepared as described previously (McKenney et al., 2014). In brief, fresh brains were broken into small chunks, flash-frozen in liquid nitrogen, and stored at −80°C. Frozen brain chunks were homogenized in equal weight/volume of buffer (50 mM Hepes, 50 mM Pipes, 1 mM EDTA, and 2 mM MgSO4, pH 7.0) using a waring blender, followed by glass pestle grinding. After clarification at 34,000 g for 45 min, the crude homogenate was flash frozen in 1-ml aliquots and stored at −80°C. For the pull-downs, 100 µl of brain lysate was diluted to 500 µl final volume with buffer A (30 mM Hepes, 50 mM K-acetate, 2 mM Mg-acetate, 1 mM EGTA, 10% glycerol, 1 mM PMSF, 5 mM DTT, and 0.1% NP-40, pH 7.4), and 50 pmol Spindly::Strep-tag II or BICD2::Strep-tag II were added together with 15 µl of Strep-Tactin Sepharose beads. After incubation for 1 h at 4°C, beads were washed with 3 × 500 µl buffer A, and proteins were eluted with elution buffer E (100 mM Tris/HCl, 150 mM NaCl, 1 mM EDTA, and 2.5 mM desthiobiotin, pH 8.0; IBA).
Online supplemental material
Fig. S1 summarizes two-hybrid experiments between RZZ subunits and describes RZZ subunit transgenes used for C. elegans strain generation. Fig. S2 extends the results in Fig. 2 with additional characterization of the ZwilchZWL-1 mutant E433A/E437A, including size-exclusion chromatography experiments with purified recombinant ZwilchZWL-1 bound to the ROD-1 β-propeller and phenotypic analysis of the mutant in the early embryo. Fig. S3 shows Coomassie-stained gels corresponding to the pull-downs presented in Fig. 3 (C and D). Fig. S4 shows the similarities in domain architecture between Spindly, BICD2, and additional dynein adaptors that contain a CC1 box and/or a Spindly-like motif and demonstrates that BICD2 interacts with LIC1 and dynactin’s pointed-end complex in vitro. Fig. S5 relates to Fig. 6 and describes the consequences of depleting dynactin pointed-end complex subunits in the C. elegans one-cell embryo by RNAi. Video 1 shows how the ZwilchZWL-1 E433A/E437A mutant displaces SpindlySPDL-1 from kinetochores. Video 2 shows how the ZwilchZWL-1 E433A/E437A mutant causes chromosome segregation defects that resemble those of SpindlySPDL-1 depletions. Video 3 shows the first embryonic divisions in the absence of p27DNC-6 imaged using differential interference contrast, showing normal pronuclear migration and spindle positioning but defective chromosome segregation leading to multinucleate cells. Video 4 show the first embryonic division in the absence of p27DNC-6 with mCherry-labeled histone H2B to visualize chromosome segregation. Table S1 is a list of worm strains used in this study. Table S2 is a list of oligonucleotides used in this study for dsRNA production. Tables S1 and S2 are included as Excel files.
Supplementary Material
Acknowledgments
We wish to thank Andrea Musacchio for antibodies, Kevin Corbett for the 6×His::MBP expression vector, and Arshad Desai for generous support during the early stages of this project.
This work was supported by a European Research Council Starting Grant (Dyneinome 338410) and a European Molecular Biology Organization Installation Grant to R. Gassmann. This work was also supported by funding from the Fundação para a Ciência e a Tecnologia to R. Gassmann (IF/01015/2013/CP1157/CT0006), C. Pereira (SFRH_BPD_95648_2013), and D.J. Barbosa (SFRH_BPD_101898_2014). Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440).
The authors declare no competing financial interests.
Author contributions: R. Gassmann, J.B. Gama, C. Pereira, P.A. Simões, and A.X. Carvalho conceived and designed experiments. J.B. Gama performed the protein biochemistry. C. Pereira and P.A. Simões performed the C. elegans work. R. Celestino constructed the dnc-6::3×flag and dnc-6–null alleles and contributed to protein purification with C. Carvalho. R.M. Reis performed the tissue culture work. D.J. Barbosa generated the GFP::DNC-2 strain and the anti–DNC-2 antibody, H.R. Pires generated the anti–DNC-1 antibody, J. Amorim performed the yeast two-hybrid experiments, and D.K. Cheerambathur generated the mCherry::ROD-1 strain and the anti–ROD-1 antibody. R. Gassmann, J.B. Gama, C. Pereira, and P.A. Simões prepared the figures and wrote the paper with advice from D.K. Cheerambathur and A.X. Carvalho.
Footnotes
Abbreviations used:
- ACA
- anticentromere antibody
- dsRNA
- double-stranded RNA
- IC
- intermediate chain
- LIC
- light intermediate chain
- MBP
- maltose-binding protein
- RZZ
- Rod–Zw10–Zwilch
- SAC
- spindle assembly checkpoint
- TEV
- tobacco etch virus
References
- Arribere J.A., Bell R.T., Fu B.X.H., Artiles K.L., Hartman P.S., and Fire A.Z.. 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 198:837–846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic M., Sohm B., Mikolcevic P., Wandke C., Rauch V., Ringer T., Hess M., Bonn G., and Geley S.. 2010. Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol. Biol. Cell. 21:1968–1981. 10.1091/mbc.E09-04-0356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic M., Aguiar P., Geley S., and Maiato H.. 2014. Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat. Cell Biol. 16:1249–1256. 10.1038/ncb3060 [DOI] [PubMed] [Google Scholar]
- Bieniossek C., Richmond T.J., and Berger I.. 2008. MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr. Protoc. Protein Sci. 5:Unit 5.20. [DOI] [PubMed] [Google Scholar]
- Buffin E., Lefebvre C., Huang J., Gagou M.E., and Karess R.E.. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 15:856–861. 10.1016/j.cub.2005.03.052 [DOI] [PubMed] [Google Scholar]
- Caldas G.V., Lynch T.R., Anderson R., Afreen S., Varma D., and DeLuca J.G.. 2015. The RZZ complex requires the N-terminus of KNL1 to mediate optimal Mad1 kinetochore localization in human cells. Open Biol. 5:150160 10.1098/rsob.150160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P., Diamant A.G., and Urnavicius L.. 2016. How dynein and dynactin transport cargos: a structural perspective. Curr. Opin. Struct. Biol. 37:62–70. 10.1016/j.sbi.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Chan G.K., Jablonski S.A., Starr D.A., Goldberg M.L., and Yen T.J.. 2000. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2:944–947. 10.1038/35046598 [DOI] [PubMed] [Google Scholar]
- Chan Y.W., Fava L.L., Uldschmid A., Schmitz M.H.A., Gerlich D.W., Nigg E.A., and Santamaria A.. 2009. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J. Cell Biol. 185:859–874. 10.1083/jcb.200812167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur D.K., and Desai A.. 2014. Linked in: formation and regulation of microtubule attachments during chromosome segregation. Curr. Opin. Cell Biol. 26:113–122. 10.1016/j.ceb.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur D.K., Gassmann R., Cook B., Oegema K., and Desai A.. 2013. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science. 342:1239–1242. 10.1126/science.1246232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005:pl1. [DOI] [PubMed] [Google Scholar]
- Chowdhury S., Ketcham S.A., Schroer T.A., and Lander G.C.. 2015. Structural organization of the dynein–dynactin complex bound to microtubules. Nat. Struct. Mol. Biol. 22:345–347. 10.1038/nsmb.2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çivril F., Wehenkel A., Giorgi F.M., Santaguida S., Di Fonzo A., Grigorean G., Ciccarelli F.D., and Musacchio A.. 2010. Structural analysis of the RZZ complex reveals common ancestry with multisubunit vesicle tethering machinery. Structure. 18:616–626. 10.1016/j.str.2010.02.014 [DOI] [PubMed] [Google Scholar]
- Défachelles L., Hainline S.G., Menant A., Lee L.A., and Karess R.E.. 2015. A maternal effect rough deal mutation suggests that multiple pathways regulate Drosophila RZZ kinetochore recruitment. J. Cell Sci. 128:1204–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., and Oegema K.. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435. 10.1101/gad.1126303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex A., Dammermann A., Lewellyn L., Oegema K., and Desai A.. 2009. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol. Biol. Cell. 20:1252–1267. 10.1091/mbc.E08-10-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulski J.K., Vos L., Sun X., and Chan G.. 2008. Stable hZW10 kinetochore residency, mediated by hZwint-1 interaction, is essential for the mitotic checkpoint. J. Cell Biol. 180:507–520. 10.1083/jcb.200708021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.-P., Grunnet M., and Jorgensen E.M.. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40:1375–1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M.W., Ailion M., and Jorgensen E.M.. 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods. 9:117–118. 10.1038/nmeth.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Essex A., Hu J.-S., Maddox P.S., Motegi F., Sugimoto A., O’Rourke S.M., Bowerman B., McLeod I., Yates J.R. III, et al. 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 22:2385–2399. 10.1101/gad.1687508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Holland A.J., Varma D., Wan X., Civril F., Cleveland D.W., Oegema K., Salmon E.D., and Desai A.. 2010. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 24:957–971. 10.1101/gad.1886810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E.R., Stuurman N., and Vale R.D.. 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 177:1005–1015. 10.1083/jcb.200702062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., and Salmon E.D.. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009. 10.1091/mbc.12.7.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Reis R.M., Niessen S., Pereira C., Andres D.A., Spielmann H.P., Cleveland D.W., Desai A., and Gassmann R.. 2015. Preventing farnesylation of the dynein adaptor Spindly contributes to the mitotic defects caused by farnesyltransferase inhibitors. Mol. Biol. Cell. 26:1845–1856. 10.1091/mbc.E14-11-1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad C.C., and Akhmanova A.. 2016. Bicaudal D family of motor adaptors: linking dynein motility to cargo binding. Trends Cell Biol. 26:327–340. 10.1016/j.tcb.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., and Salmon E.D.. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172. 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R. 2005. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 15:386–392. 10.1016/j.tcb.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Karess R.E., and Glover D.M.. 1989. rough deal: a gene required for proper mitotic segregation in Drosophila. J. Cell Biol. 109:2951–2961. 10.1083/jcb.109.6.2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G.J.P.L., Kim Y., Weaver B.A.A., Mao Y., McLeod I., Yates J.R. III, Tagaya M., and Cleveland D.W.. 2005. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169:49–60. 10.1083/jcb.200411118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V., Paul R., Yang N., Ault J.G., O’Connell C.B., Tikhonenko I., McEwen B.F., Mogilner A., and Khodjakov A.. 2015. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat. Cell Biol. 17:1134–1144. 10.1038/ncb3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney R.J., Huynh W., Tanenbaum M.E., Bhabha G., and Vale R.D.. 2014. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 345:337–341. 10.1126/science.1254198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudgil D.K., Westcott N., Famulski J.K., Patel K., Macdonald D., Hang H., and Chan G.K.T.. 2015. A novel role of farnesylation in targeting a mitotic checkpoint protein, human Spindly, to kinetochores. J. Cell Biol. 208:881–896. 10.1083/jcb.201412085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle M.W., Kim T., Hattersley N., Espeut J., Cheerambathur D.K., Oegema K., and Desai A.. 2014. A Bub1–Mad1 interaction targets the Mad1–Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J. Cell Biol. 204:647–657. 10.1083/jcb.201311015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A. 2015. The molecular biology of Spindle assembly checkpoint signaling dynamics. Curr. Biol. 25:R1002–R1018 (published erratum appears in Curr. Biol. 2015. 25:3017). 10.1016/j.cub.2015.08.051 [DOI] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., and Hyman A.A.. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. 10.1083/jcb.153.6.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Baksa K., and Steward R.. 2000. Functional domains of the Drosophila bicaudal-D protein. Genetics. 154:713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A., Pasqualato S., Dube P., Krenn V., Santaguida S., Cittaro D., Monzani S., Massimiliano L., Keller J., Tarricone A., et al. 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190:835–852. 10.1083/jcb.201002070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers J.A., Tanenbaum M.E., and Medema R.H.. 2013. Systematic dissection of dynein regulators in mitosis. J. Cell Biol. 201:201–215. 10.1083/jcb.201208098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., and Alexander S.P.. 1990. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J. Cell Biol. 110:81–95. 10.1083/jcb.110.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaërou F., Aguilera I., Saunders R., Kane N., Blottière L., and Karess R.. 1999. The rough deal protein is a new kinetochore component required for accurate chromosome segregation in Drosophila. J. Cell Sci. 112:3757–3768. [DOI] [PubMed] [Google Scholar]
- Scaërou F., Starr D.A., Piano F., Papoulas O., Karess R.E., and Goldberg M.L.. 2001. The ZW10 and Rough Deal checkpoint proteins function together in a large, evolutionarily conserved complex targeted to the kinetochore. J. Cell Sci. 114:3103–3114. [DOI] [PubMed] [Google Scholar]
- Schlager M.A., Serra-Marques A., Grigoriev I., Gumy L.F., Esteves da Silva M., Wulf P.S., Akhmanova A., and Hoogenraad C.C.. 2014. Bicaudal D family adaptor proteins control the velocity of Dynein-based movements. Cell Reports. 8:1248–1256. 10.1016/j.celrep.2014.07.052 [DOI] [PubMed] [Google Scholar]
- Schroeder C.M., and Vale R.D.. 2016. Assembly and activation of dynein–dynactin by the cargo adaptor protein Hook3. J. Cell Biol. 214:309–318. 10.1083/jcb.201604002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder C.M., Ostrem J.M., Hertz N.T., and Vale R.D.. 2014. A Ras-like domain in the light intermediate chain bridges the dynein motor to a cargo-binding region. eLife. 3:e03351 10.7554/eLife.03351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silió V., McAinsh A.D., and Millar J.B.. 2015. KNL1-Bubs and RZZ provide two separable pathways for Checkpoint activation at human kinetochores. Dev. Cell. 35:600–613. 10.1016/j.devcel.2015.11.012 [DOI] [PubMed] [Google Scholar]
- Smith D.A., Baker B.S., and Gatti M.. 1985. Mutations in genes encoding essential mitotic functions in Drosophila melanogaster. Genetics. 110:647–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D., Razafsky D.S., Schlager M.A., Serra-Marques A., Grigoriev I., Demmers J., Keijzer N., Jiang K., Poser I., Hyman A.A., et al. 2012. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol. Biol. Cell. 23:4226–4241. 10.1091/mbc.E12-03-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Williams B.C., Li Z., Etemad-Moghadam B., Dawe R.K., and Goldberg M.L.. 1997. Conservation of the centromere/kinetochore protein ZW10. J. Cell Biol. 138:1289–1301. 10.1083/jcb.138.6.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Williams B.C., Hays T.S., and Goldberg M.L.. 1998. ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 142:763–774. 10.1083/jcb.142.3.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Saffery R., Li Z., Simpson A.E., Choo K.H., Yen T.J., and Goldberg M.L.. 2000. HZwint-1, a novel human kinetochore component that interacts with HZW10. J. Cell Sci. 113:1939–1950. [DOI] [PubMed] [Google Scholar]
- Tighe A., Johnson V.L., and Taylor S.S.. 2004. Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J. Cell Sci. 117:6339–6353. 10.1242/jcs.01556 [DOI] [PubMed] [Google Scholar]
- Tighe A., Staples O., and Taylor S.. 2008. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J. Cell Biol. 181:893–901. 10.1083/jcb.200712028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnavicius L., Zhang K., Diamant A.G., Motz C., Schlager M.A., Yu M., Patel N.A., Robinson C.V., and Carter A.P.. 2015. The structure of the dynactin complex and its interaction with dynein. Science. 347:1441–1446. 10.1126/science.aaa4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D., Wan X., Cheerambathur D., Gassmann R., Suzuki A., Lawrimore J., Desai A., and Salmon E.D.. 2013. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J. Cell Biol. 202:735–746. 10.1083/jcb.201304197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorozhko V.V., Emanuele M.J., Kallio M.J., Stukenberg P.T., and Gorbsky G.J.. 2008. Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin. Chromosoma. 117:169–179. 10.1007/s00412-007-0135-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos L.J., Famulski J.K., and Chan G.K.. 2011. hZwint-1 bridges the inner and outer kinetochore: identification of the kinetochore localization domain and the hZw10-interaction domain. Biochem. J. 436:157–168. 10.1042/BJ20110137 [DOI] [PubMed] [Google Scholar]
- Wainman A., Giansanti M.G., Goldberg M.L., and Gatti M.. 2012. The Drosophila RZZ complex – roles in membrane trafficking and cytokinesis. J. Cell Sci. 125:4014–4025. 10.1242/jcs.099820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.C., and Goldberg M.L.. 1994. Determinants of Drosophila zw10 protein localization and function. J. Cell Sci. 107:785–798. [DOI] [PubMed] [Google Scholar]
- Williams B.C., Li Z., Liu S., Williams E.V., Leung G., Yen T.J., and Goldberg M.L.. 2003. Zwilch, a new component of the ZW10/ROD complex required for kinetochore functions. Mol. Biol. Cell. 14:1379–1391. 10.1091/mbc.E02-09-0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik E., Basto R., Serr M., Scaërou F., Karess R., and Hays T.. 2001. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 3:1001–1007. 10.1038/ncb1101-1001 [DOI] [PubMed] [Google Scholar]
- Wynne D.J., and Funabiki H.. 2015. Kinetochore function is controlled by a phospho-dependent coexpansion of inner and outer components. J. Cell Biol. 210:899–916. 10.1083/jcb.201506020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne D.J., and Funabiki H.. 2016. Heterogeneous architecture of vertebrate kinetochores revealed by three-dimensional superresolution fluorescence microscopy. Mol. Biol. Cell. 27:3395–3404. 10.1091/mbc.E16-02-0130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T.G., Watanabe S., Essex A., and Kitagawa R.. 2008. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J. Cell Biol. 183:187–194. 10.1083/jcb.200805185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Tulu U.S., Wadsworth P., and Rieder C.L.. 2007. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol. 17:973–980. 10.1016/j.cub.2007.04.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh T.-Y., Quintyne N.J., Scipioni B.R., Eckley D.M., and Schroer T.A.. 2012. Dynactin’s pointed-end complex is a cargo-targeting module. Mol. Biol. Cell. 23:3827–3837. 10.1091/mbc.E12-07-0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh T.-Y., Kowalska A.K., Scipioni B.R., Cheong F.K.Y., Zheng M., Derewenda U., Derewenda Z.S., and Schroer T.A.. 2013. Dynactin helps target Polo-like kinase 1 to kinetochores via its left-handed beta-helical p27 subunit. EMBO J. 32:1023–1035. 10.1038/emboj.2013.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Lischetti T., Hayward D.G., and Nilsson J.. 2015. Distinct domains in Bub1 localize RZZ and BubR1 to kinetochores to regulate the checkpoint. Nat. Commun. 6:7162 10.1038/ncomms8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yao X., Fischer L., Abenza J.F., Peñalva M.A., and Xiang X.. 2011. The p25 subunit of the dynactin complex is required for dynein-early endosome interaction. J. Cell Biol. 193:1245–1255. 10.1083/jcb.201011022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Qiu R., Arst H.N. Jr., Peñalva M.A., and Xiang X.. 2014. HookA is a novel dynein-early endosome linker critical for cargo movement in vivo. J. Cell Biol. 204:1009–1026. 10.1083/jcb.201308009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.