The Rod–Zw10–Zwilch (RZZ) complex assembles as a fibrous corona on kinetochores before microtubule attachment during mitotic spindle formation. Mosalaganti et al. provide new structural insight into the Spindly–RZZ complex that suggests that it resembles a dynein adaptor–cargo pair in the kinetochore corona.
Abstract
Kinetochores are macromolecular assemblies that connect chromosomes to spindle microtubules (MTs) during mitosis. The metazoan-specific ≈800-kD ROD–Zwilch–ZW10 (RZZ) complex builds a fibrous corona that assembles on mitotic kinetochores before MT attachment to promote chromosome alignment and robust spindle assembly checkpoint signaling. In this study, we combine biochemical reconstitutions, single-particle electron cryomicroscopy, cross-linking mass spectrometry, and structural modeling to build a complete model of human RZZ. We find that RZZ is structurally related to self-assembling cytosolic coat scaffolds that mediate membrane cargo trafficking, including Clathrin, Sec13–Sec31, and αβ’ε-COP. We show that Spindly, a dynein adaptor, is related to BicD2 and binds RZZ directly in a farnesylation-dependent but membrane-independent manner. Through a targeted chemical biology approach, we identify ROD as the Spindly farnesyl receptor. Our results suggest that RZZ is dynein’s cargo at human kinetochores.
Introduction
Molecular motors such as kinesin and cytoplasmic dynein 1 (hereafter referred to as dynein) hydrolyze ATP to move directionally on microtubules (MTs), actively transporting different cargos to shape the complex spatial organization of eukaryotic cells. Dynein, a member of the AAA family of ATPases, is the major retrograde (MT minus end–directed) motor in eukaryotic cells (Kardon and Vale, 2009; Cianfrocco et al., 2015; Carter et al., 2016). Virtually all functions of dynein also require an accessory subunit called dynactin, as well as a plethora of adaptor proteins that control the recruitment of dynein–dynactin to diversely localized cargos while also activating dynein’s motor activity (McKenney et al., 2014; Schlager et al., 2014). Understanding how adaptors bridge dynein–dynactin and cargo is a current challenge of great significance (Kardon and Vale, 2009; Cianfrocco et al., 2015; Carter et al., 2016).
During mitosis in metazoans, dynein is recruited to kinetochores, complex protein assemblies that link chromosomes to spindle MTs. Kinetochores are built on specialized chromosomal loci and consist of chromosome-proximal (inner) and -distal (outer) domains, respectively hosting the 16-subunit constitutive centromere-associated network, which binds directly to chromatin, and the 10-subunit KNL1, MIS12, NDC-80 (KMN) assembly, which binds directly to MTs (Pesenti et al., 2016). Dynein is recruited to an outermost poorly characterized domain of metazoan kinetochores named the fibrous corona, which is clearly visible only before MT attachment (Jokelainen, 1967; Rieder and Alexander, 1990; McEwen et al., 1993; Cooke et al., 1997; Yao et al., 1997; Hoffman et al., 2001; Magidson et al., 2015). Functional attributes of the corona are the expansion of the MT-binding surface of kinetochores into extended crescents, with the purpose of maximizing the likelihood of MT capture (Hoffman et al., 2001; Magidson et al., 2015; Wynne and Funabiki, 2015) and the promotion of spindle assembly checkpoint (SAC) signaling (Basto et al., 2000; Buffin et al., 2005; Kops et al., 2005), which protects genome integrity by restricting mitotic exit and sister chromatid separation (anaphase) to cells with bioriented sister chromatids (Musacchio, 2015).
Dynein may not be strictly required for either of these corona functions, but it is crucially involved in corona disassembly (hereafter referred to as “shedding”), which is required to silence the SAC and to promote conversion from lateral to end-on MT attachment. Corona shedding takes place when MTs bind kinetochores, in turn allowing dynein to transport corona and SAC components away from kinetochores (Williams et al., 1996; Howell et al., 2001; Wojcik et al., 2001; Basto et al., 2004; Mische et al., 2008; Varma et al., 2008; Sivaram et al., 2009).
The three-subunit ROD–Zwilch–ZW10 (RZZ) complex (named after the Drosophila melanogaster genes Rough Deal, Zwilch, and Zeste White 10; Fig. 1 A) is a primary corona component. Through a poorly understood mechanism, RZZ stabilizes kinetochore localization of the MAD1–MAD2 complex (Buffin et al., 2005; Kops et al., 2005; Matson and Stukenberg, 2014; Caldas et al., 2015; Silió et al., 2015; Zhang et al., 2015). This necessary function of RZZ in SAC signaling terminates with corona shedding, which depletes MAD1–MAD2 from kinetochores, silencing the SAC (Fig. 1 B; Karess, 2005). Importantly, RZZ is also required for kinetochore recruitment of dynein–dynactin, but this additionally requires Spindly, a dynein adaptor whose kinetochore localization also depends on RZZ (Starr et al., 1998; Griffis et al., 2007; Gassmann et al., 2008, 2010; Yamamoto et al., 2008; Chan et al., 2009; Barisic et al., 2010; Cheerambathur et al., 2013). Mutations within a conserved region of Spindly (the Spindly motif) abrogate kinetochore recruitment of dynein–dynactin, effectively blocking corona shedding and SAC silencing (Gassmann et al., 2010).
Figure 1.
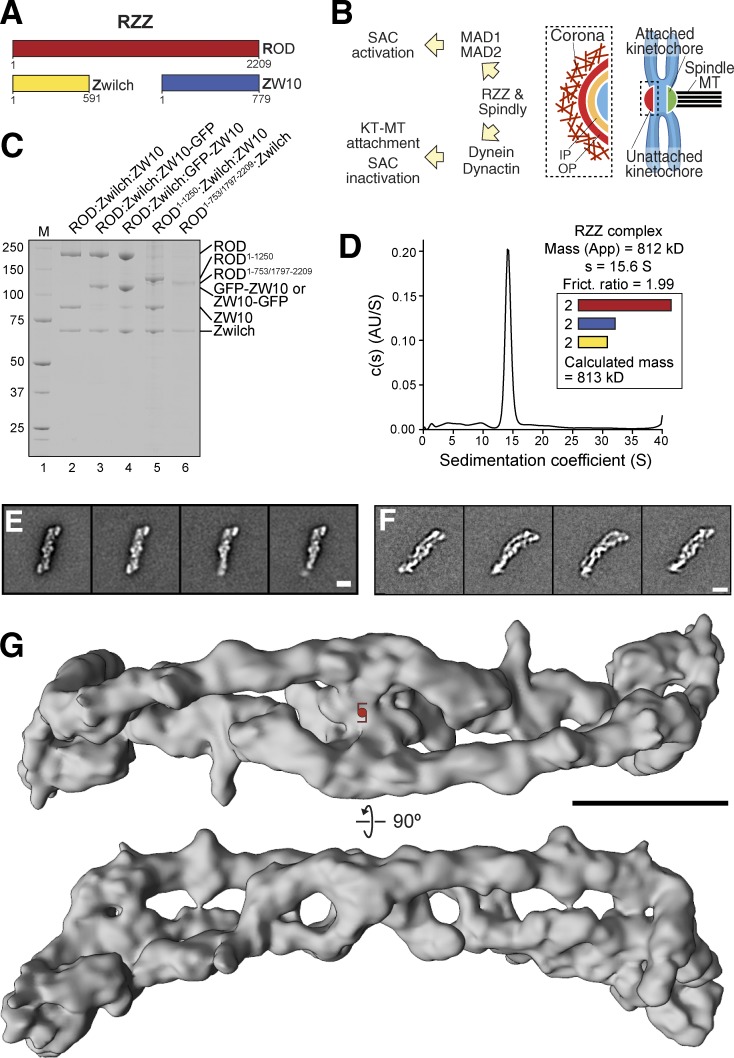
Reconstitution and structural analysis of the RZZ complex. (A) Schematic organization of the three RZZ subunits. (B) Biological functions of RZZ and Spindly. IP, inner plate of kinetochore; KT, kinetochore; OP, outer plate of kinetochore. (C) SDS-PAGE analysis of various RZZ complexes used in this paper. M, molecular weight marker. (D) Sedimentation velocity AUC profiles of the recombinant RZZ complex. “Mass (app)” denotes the apparent mass of RZZ derived from the velocity runs; “s” denotes the sedimentation coefficient; “Frict. ratio” is the frictional ratio of the RZZ particle. The calculated molecular mass for a complex containing two copies of each subunit is shown. The sedimentation velocity absorbance profiles and residuals of the fit showing the deviation of the c(S) model from the observed signals are shown, for this and other AUC profiles in this paper, in Fig. S1 E. AU, arbitrary units. (E) Typical class averages of negatively stained RZZ complexes. A clear twofold symmetry is evident. (F) Typical class averages of vitrified RZZ complexes. Bars, 10 nm. (G) Cryo-EM 3D reconstruction of the RZZ complex. The complex is ∼42 nm long and 10 nm wide. The position of a twofold symmetry axis is indicated in the top panel. The density maps of RZZ (here) and R1–1,250ZZ (Fig. 4 B) have been deposited into EMDataBank with the accession codes EMD-4103 and EMD-4104, respectively.
Spindly recruitment to kinetochores requires the posttranslational modification of Cys602 of Spindly with farnesyl, an isoprenoid lipid (Holland et al., 2015; Moudgil et al., 2015). Farnesylation is typically observed in small GTPases of the Ras family, regulators of many signaling and transport functions of eukaryotic cells, where it contributes to the association of GTPases with endomembranes (Wang and Casey, 2016). Precisely how Spindly farnesylation favors its kinetochore recruitment is currently unclear. Similarly to BicD2, another adaptor whose mode of dynein–dynactin binding is better characterized (Chowdhury et al., 2015; Urnavicius et al., 2015), Spindly activates dynein motility in vitro and is predicted to contain extensive coiled-coil segments (McKenney et al., 2014; Schlager et al., 2014; Cianfrocco et al., 2015; Hoogenraad and Akhmanova, 2016).
The structural organization of the RZZ complex and its influence on corona assembly are poorly understood. Previously, we determined the crystal structure of the smallest of the three subunits, Zwilch (Williams et al., 2003), and detected limited sequence similarity of ROD with the Clathrin heavy chain (Çivril et al., 2010). After reconstituting the RZZ complex (Altenfeld et al., 2015), we now report its overall structure by cryo-EM at an approximate resolution of 10 Å. By using the EM density and modeling, we generated a molecular model of the RZZ complex that satisfies several experimental spatial restraints. We now demonstrate a more general structural relatedness of ROD, the largest RZZ subunit, with proteins that oligomerize in proximity of biological membranes, including Clathrin, COPI, and COPII. We show that Spindly binds directly to the RZZ complex and that this requires an interaction of the C-terminal farnesyl moiety of Spindly with the ROD N-terminal region. Our results suggest a possible molecular basis for corona assembly and regulation at human kinetochores and implicate RZZ as dynein’s cargo at kinetochores.
Results
Reconstitution and structural analysis of the RZZ complex
We generated recombinant versions of the human RZZ complex and of several RZZ variants (including fluorescently tagged versions and deletion mutants) by baculovirus-driven insect cell expression (Fig. 1 C; see the Expression and purification of RZZ section of Materials and methods; Altenfeld et al., 2015). Sedimentation velocity analytical ultracentrifugation (AUC) of recombinant RZZ showed a single peak with a predicted molecular mass of 813 kD, very close to the theoretical mass of 812 kD calculated by assuming a 2:2:2 stoichiometry, confirming and extending less precise previous measurements (Scaërou et al., 2001; Çivril et al., 2010).
In negative-stain and cryo-EM, RZZ complexes appeared as elongated particles (≈42 nm long and ≈10 nm thick in the other two directions; (Fig. 1, E and F; and Fig. S1, A–D). In agreement with the 2:2:2 stoichiometry of the RZZ complex, a twofold symmetry axis, perpendicular to the long axis of the complex, is clearly discernible. We determined the 3D structure of the RZZ complex at a resolution of 10.4 Å using cryo-EM (Figs. 1 G and S1 C).
Molecular models of RZZ subunits
Probably because of its inherent flexibility (see Video 1), we could not obtain EM reconstructions of the RZZ complex at higher resolution. To facilitate our understanding of the molecular organization of the RZZ complex, we built molecular models of its subunits. The crystal structure of Zwilch has been determined previously (PDB ID 3IF8; Fig. 2 A; Çivril et al., 2010), but no direct structural information on ZW10 and ROD is available. Despite overall limited sequence identity to known structural scaffolds, the programs Phyre and HHpred (Söding et al., 2005; Kelley et al., 2015) generated high-confidence structural models for the majority of ZW10 and ROD (see Table S1 and Fig. 2 [B and C] for summaries). ZW10 is evolutionarily and structurally related to the S. cerevisiae proteins Dsl1 and Tip20 (Table S1), subunits of vesicle-tethering complexes consisting of two roughly equally sized helical domains (Tripathi et al., 2009). In line with homology modeling, negative-stain EM and single-particle analysis of ZW10 demonstrated that it consists of two approximately equally sized domains (Fig. 2 D) separated by a flexible linker, as was previously shown for Dsl1 and Tip20 (Tripathi et al., 2009).
Figure 2.
Summary of modeling. (A) Cartoon model of Zwilch (PDB ID: 3IF8). (B) Models of ROD were generated by a structure prediction program (I-TASSER). (C) Structures of the yeast homologue of ZW10 (Dsl1p), the N terminus (37–355; PDB ID: 3ETU), and the C terminus (333–684; PDB ID: 3K8P), were used for ZW10. NRH, NAG–ROD homology. (D) Typical negative-stain class averages of ZW10 obtained from ISAC. The averages highlight the inherent flexibility in the sample. Bar, 10 nm.
Phyre and HHpred identified Sec31, COP1, Sec39, Clathrin, and the nucleoporins Nup155 and Nup145 as high-confidence templates for structural modeling of ROD (Table S1 and Fig. 2 B). A common feature of these proteins is that they self-assemble in oligomeric/polymeric structures at or near cellular membranes. All these proteins share similar overall architectures, consisting of an N-terminal seven-bladed β-propeller domain and an extended α-helical domain built from tandem α-helical hairpins often separated by a structurally variable linker (ter Haar et al., 1998; Fotin et al., 2004; Stagg et al., 2006, 2007; Fath et al., 2007; Brohawn et al., 2008; Lee and Goldberg, 2010). In ROD, the predicted β-propeller domain occupies ≈350 residues from the N terminus and is followed by a predicted helical domain of ≈1,850 residues, in which a few subdomains are recognizable (Fig. 2 B). ROD was unstable in the absence of other RZZ subunits, and we could not yet address its structure in isolation. However, the results from homology modeling, and especially the observation that models based on tandem α-helical hairpin repeats are consistently predicted along the entire length of ROD after the N-terminal β-propeller, give confidence to the model and strongly suggest that ROD shares common ancestry with these proteins, despite limited overall sequence identity (Table S1).
Building a structural model of the RZZ complex
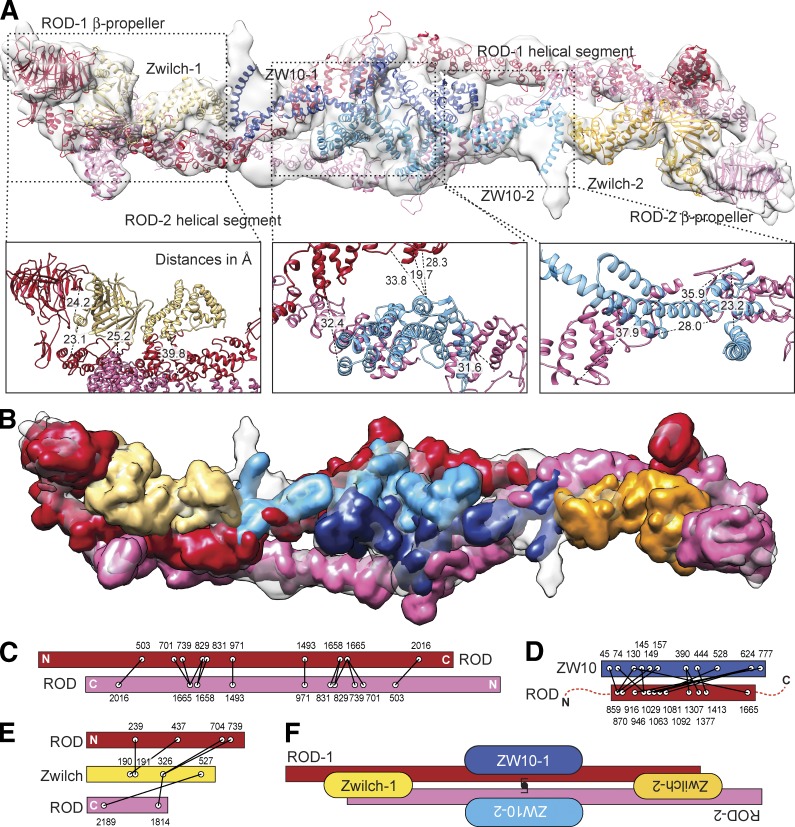
We used the Fit in Map rigid body fitting tool of program UCSF Chimera (Pettersen et al., 2004) to position experimental or homology models of Zwilch, ZW10, ROD396–1,191, and ROD1,319–1,797 (see Table S2 for fitting scores and an explanation of fitting procedures). We then manually positioned the homology models for ROD1–376 (β-propeller) and ROD1,798–2,189 in the residual density. Uninterrupted ribbons of density suggest that ROD spans the entire length of the complex, adopting an antiparallel configuration, with the ROD β-propellers at opposite ends (Fig. 3 A and Video 2). Zwilch fits snugly in the density from which it directly contacts the β-propeller and the C-terminal region of the first and second ROD protomers, respectively, in agreement with previous observations that the N-terminal region of ROD binds Zwilch (Çivril et al., 2010; Cheerambathur et al., 2013). ZW10 is positioned near the middle of the structure and of the twofold axis, where it interacts with the central domain of ROD in agreement with a previous yeast two-hybrid analysis (Scaërou et al., 2001).
Figure 3.
A molecular model of the RZZ complex. (A) The molecular model of the RZZ, consisting of a combination of experimental (for Zwilch) or homology models (for ROD and ZW10) of RZZ subunits was fitted in the cryo-EM density. (B) Map calculated from the molecular model at a resolution of 10.4 Å. (C–E) Summary of XL-MS data reporting various classes of intersubunit cross-links (listed in Table S3). (F) Schematic view of the organization of the RZZ complex.
Validation of the structural model
At an equivalent resolution, a map calculated from our molecular model of the RZZ complex displayed excellent correlation with the experimental map of the RZZ complex, testifying to the reliability of the model (Fig. 3 B and Table S2). As a further validation strategy, we used disuccinimidyl suberate, a bifunctional cross-linker, to cross-link the primary amines of neighboring lysines in the RZZ complex, and we identified cross-links by mass spectrometry (MS; cross-linking MS [XL-MS]; Herzog et al., 2012). The majority of observed cross-links (listed in Table S3 and displayed schematically in Fig. 3, C–E) were consistent with the organization and physical interactions of RZZ subunits predicted by molecular modeling, confirming the antiparallel organization of ROD as well as interactions of ZW10 with the central domain of ROD and of Zwilch with the N and C termini of ROD. Furthermore, a large majority of observed cross-links were compatible with lysine–lysine distances calculated from the molecular model of RZZ (Table S3), indicating that in spite of limited resolution of the EM map, the model is largely consistent with experimental spatial restraints.
The relative orientation of ZW10 and Zwilch in the model further suggests that they do not interact, or do so weakly. Size-exclusion chromatography (SEC) of individually expressed and purified ZW10 and Zwilch showed that they did not coelute, confirming the prediction (Fig. 4 A). In agreement with the cross-linking and EM data, a recombinant RZZ variant incorporating a ROD deletion mutant lacking the central domain (RODΔ754–1,796; Fig. 1 C, lane 6; shown schematically in Fig. S2 A) was sufficient to bind Zwilch but failed to interact with ZW10 in SEC experiments (Fig. S2 B). Conversely, a ROD deletion mutant consisting of the 1,250 N-terminal residues of ROD (ROD1–1,250, shown schematically in Fig. S2 A) was sufficient to bind both Zwilch and ZW10 (Fig. 1 C, lane 5), even if XL-MS predicted the existence of additional interactions of ZW10 with downstream segments of ROD (Fig. 3 D). A negative-stain EM 3D reconstruction of the ROD1–1,250–Zwilch–ZW10 (R1–1,250ZZ) complex revealed a structure with an overall extension of 26 nm, approximately half that of the full-length RZZ complex (Fig. 4 B, top; and Fig. S2, C and D). The reconstruction is shown together with the portion of the cryo-EM reconstruction of the complete RZZ that corresponds to an RZZ model in which ROD was truncated at residue 1,250 (Fig. 4 B, bottom).
Figure 4.
Validation of the RZZ model. (A) Elution profiles and SDS-PAGE of SEC experiments on Zwilch, ZW10, or their stoichiometric combination. Molecular mass is indicated in kilodaltons. AU, arbitrary units. (B) Top: 3D reconstruction of the negatively stained ROD1–1,250ZZ complex. Bottom: the density corresponding to a model of the ROD1–1,250ZZ complex was cut out of the complete RZZ complex for comparison. (C) NiNTA-gold labeling of the N-terminal histidine tag of ROD. The bottom-right panel represents part of a field in which labeling of ROD was performed after proteolytic removal of the His tag. (D) Micrographs of negatively stained GFP-RZZ or ROD–Zwilch–GFP-ZW10 complexes. Bars, 10 nm. (E) Summary of positions identified in experiments in C and D mapped onto the molecular model of RZZ.
Further validation of the RZZ model included visualization, by gold or antibody labeling, of the N terminus of ROD and of the N and C termini of ZW10 (Fig. 4, C–E), with results that were again in very good agreement with the proposed model (summarized in Fig. 3 F). Thus, we generated a molecular model of the RZZ complex that relies on experimental and high-confidence homology models and that accounts for various spatial restraints derived from EM and cross-linking experiments. Although the RZZ model we have described cannot yet provide atomic detail, it provides a first reference to understanding the topology and general organization of the RZZ complex (Fig. 3 F).
A direct RZZ–Spindly interaction
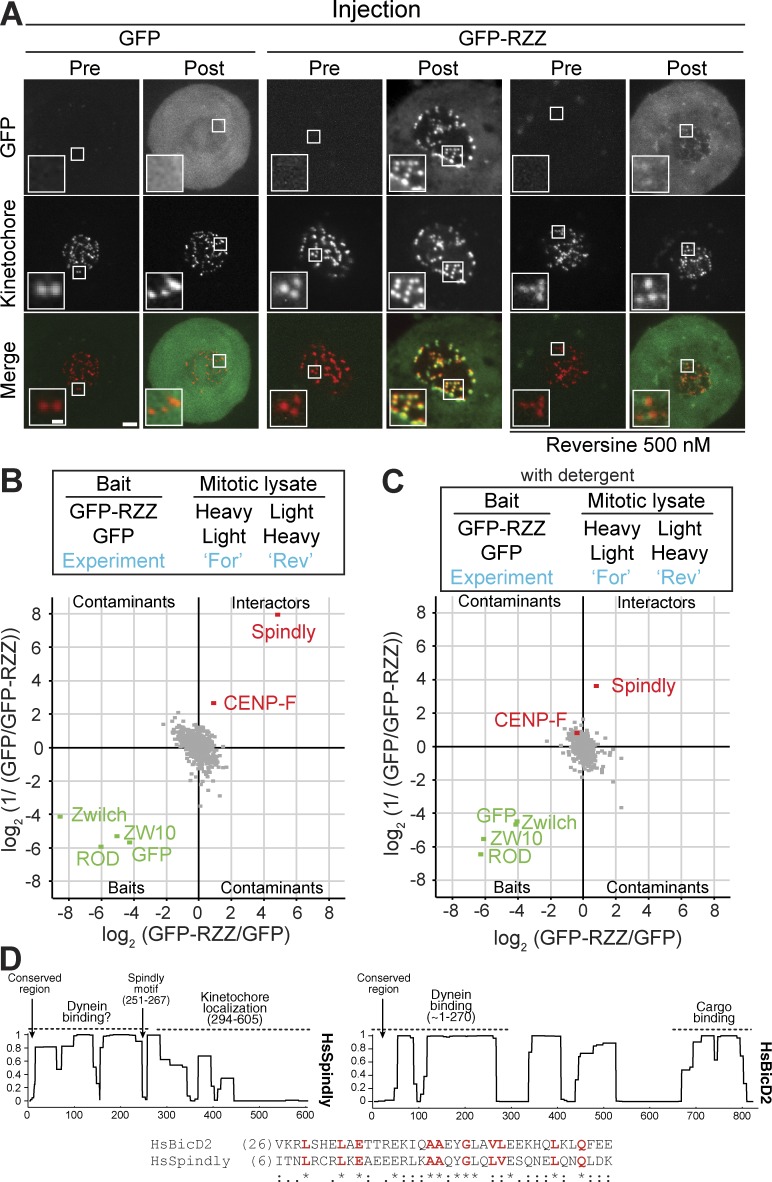
Recombinant GFP-RZZ (Fig. 1 C, lane 3) brightly decorated kinetochores after injection in HeLa cells, whereas Reversine, a small-molecule inhibitor of the SAC kinase MPS1 (Santaguida et al., 2010), suppressed recruitment (Fig. 5 A). These results further confirm that we have reconstituted a native or near-native recombinant version of RZZ, which can reach its natural destination in the cellular environment.
Figure 5.
RZZ interacts with Spindly. (A) Representative images of the localization of the recombinant GFP-RZZ complex injected in mitotic HeLa cells transiently expressing mCherry-CENPA (to visualize kinetochores). Cells were synchronized in G2 by treatment with the Cdk1 inhibitor RO3306 and released into mitosis in the presence of the indicated drugs. Mitotic cells were live imaged before (Pre) and after (Post) microinjection with recombinant GFP or GFP–RZZ. Bars: (main images) 2 µm; (insets) 0.5 µm. Numbers of cells injected: for GFP, n = 2; for GFP-RZZ n = 8; for GFP in Reversine-treated cells, n = 2 (not depicted); for GFP-RZZ in Reversine-treated cells, n = 8. (B and C) Forward (For) and reverse (Rev) SILAC experiments (see the SILAC immunoprecipitation experiments section in Materials and methods) were performed as indicated in the upper part of the figure. Spindly is the most prominent component of the “Interactors” quadrant, which identifies specific binders of the GFP-RZZ complex in comparison with GFP. The experiments were performed in the absence (B) or presence (C) of detergent in lysis buffer. Baits are shown in green, interaction partners in red, and all quantified background proteins in gray. (D) Coiled-coil content of human Spindly and BicD2 predictions obtained with the program COILS (Lupas et al., 1991). Sequence alignment of Spindly and BicD2 shows excellent conservation in Spindly of a highly conserved motif previously identified in BicD2 (Hoogenraad and Akhmanova, 2016). Residues shown in red indicate conservation of signatures within motifs, but sequence conservation is more extensive and extends to the entire dynein-binding domain (not depicted).
Because direct binding partners of the RZZ are currently unknown, we incubated mitotic lysates from SILAC (stable isotope labeling with amino acid in culture; Ong et al., 2002)-labeled HeLa cells with recombinant baits consisting of immobilized GFP (negative control) or GFP-RZZ. This identified Spindly as the most prominent binding partner of GFP-RZZ. The kinetochore protein CENP-F was the only other enriched binder identified (Fig. 5 B). Both interactions of GFP-RZZ were strongly reduced in the presence of detergents in the lysis buffer (Fig. 5 C).
Human Spindly is 605 residues and contains extensive coiled-coil segments. The “Spindly motif” (residues 251–267; Fig. 5 D) is required for the interaction with dynein–dynactin, and its mutation prevents the release of MAD1–MAD2 from kinetochores and SAC silencing (Gassmann et al., 2010). Other dynein adaptors, including members of the bicaudal D (BICD) and BICD-related (BICDR) families such as BicD2, also contain several predicted coiled-coil segments (Fig. 5 D). The BicD2 N-terminal region (∼270 residues) interacts directly with dynein–dynactin (Urnavicius et al., 2015), and a strongly conserved sequence signature is identified in this region (Hoogenraad and Akhmanova, 2016). Interestingly, we found that Spindly and BicD2 are clearly evolutionarily related, with extensive sequence similarity all along their sequences and a strictly conserved N-terminal signature (Fig. 5 D and Table S4). Thus, Spindly and BicD2 are likely to bind dynein in structurally similar ways, i.e., by engaging their N termini and using their C termini for interactions with cargo.
RZZ interaction with Spindly is direct and requires farnesylation
As Spindly is the most prominent binding partner of GFP-RZZ (Fig. 5, B and C), we asked whether stoichiometric amounts of RZZ and Spindly coeluted from an SEC column (Fig. 6 A). A partial shift in the elution volume of Spindly suggested a low-affinity substoichiometric interaction. Because recruitment of Spindly to kinetochores requires farnesylation (Holland et al., 2015; Moudgil et al., 2015), we subjected Spindly to farnesylation in vitro with farnesyl transferase (FTase) and farnesyl pyrophosphate (Fpp) as substrate (Nguyen et al., 2007). By MS, we found Spindly to be homogeneously modified at Cys602 in this reaction (Fig. S3, A and B). Prominent shifts in the SEC elution volumes of both farnesylated Spindly (SpindlyFarn) and RZZ demonstrated that SpindlyFarn and RZZ bind stoichiometrically, thus identifying Spindly farnesylation as a crucial determinant of the interaction (Fig. 6 A). The modest interaction of Spindly with RZZ observed in the absence of FTase and Fpp is probably caused by a low degree of farnesylation of Spindly upon its expression in insect cells (detected by MS; not depicted). Spindly and SpindlyFarn showed essentially identical elution volumes (Fig. 6 A), suggesting that prenylation does not grossly change the oligomerization or structural organization of Spindly.
Figure 6.
RZZ interaction with Spindly is direct and requires farnesylation. (A) Elution profiles and SDS-PAGE analysis of SEC experiments with the indicated proteins. The elution volume of the RZZ–Spindly complex after treatment with FTase and Fpp as substrate shifts significantly, whereas farnesylated Spindly (SpindlyFarn) elutes like unmodified Spindly. (B) Summary of XL-MS data reporting ROD–Spindly intersubunit cross-links. (C) Sedimentation velocity AUC profiles of recombinant mCherry-Spindly. The calculated molecular mass for a complex containing two copies of each subunit is shown. (D) Sedimentation velocity AUC profiles of the RZZ–SpindlyFarn complex. The calculated molecular mass for a complex containing two copies of each subunit is shown. (E–G) Sedimentation velocity AUC profiles of the RZZ–mCherry–SpindlyFarn at 5, 10, and 15 µM. “Mass (app)” denotes the apparent mass of RZZ derived from the velocity runs; “s” denotes the sedimentation coefficient; “Frict. ratio” is the frictional ratio of the Spindly particle. In E, the calculated molecular mass for a complex containing two copies of each subunit is shown. AU, arbitrary units.
XL-MS analysis of the SpindlyFarn–RZZ identified multiple contacts between SpindlyFarn and ROD but not other RZZ subunits (Fig. 6 B and Table S3, section C). Neither Zwilch nor ZW10 bound SpindlyFarn in isolation in SEC experiments (Fig. S2, G and H), whereas the RODΔ1,251-CZZ complex bound SpindlyFarn, confirming a prominent role for ROD and also suggesting that the C-terminal region of ROD is dispensable for SpindlyFarn binding (Fig. S2 I). The pattern of cross-links suggests that Spindly projects its C-terminal region (where the farnesyl acceptor residue Cys602 resides) in the direction of the ROD β-propeller. Residues 294–605 of Spindly have been previously identified as the minimal kinetochore recruitment domain (Moudgil et al., 2015), suggesting that farnesylation augments the binding affinity of the interaction but is not its sole determinant. Indeed, RZZ did not bind to a variety of short (<6 residues) farnesylated peptides (unpublished data).
Isolated nonfarnesylated mCherry–Spindly behaved as a monomer in sedimentation velocity AUC experiments (Fig. 6 C). Conversely, the RZZ–SpindlyFarn and RZZ–mCherry–SpindlyFarn complexes (at a concentration of 5 µM) showed monodisperse samples with very good agreement of the apparent molecular mass with that calculated assuming two copies of each subunit (Fig. 6, D and E). When bound to dynein–dynactin, BicD2 forms a dimeric, parallel coiled-coil (Urnavicius et al., 2015), and we speculate that dimerization on RZZ promotes a similar transition in the N-terminal region of Spindly. Both RZZ–SpindlyFarn and RZZ–mCherry–SpindlyFarn complexes had slightly increased frictional ratios (i.e., they behaved as more elongated particles) in comparison with the isolated RZZ complex (2.14 and 2.34 against 1.99; Fig. 6, D and E), which likely explains the reduced elution volume of RZZ–SpindlyFarn in comparison with RZZ in SEC experiments. Negative-stain EM analysis at low sample concentration did not reveal major structural changes of RZZ–SpindlyFarn in comparison with RZZ (Fig. S1 F), suggesting that the increased frictional ratio may be caused by regions of Spindly that do not occupy a fixed position relative to the core of the complex. At higher concentrations (10 and 15 µM), RZZ–mCherry–SpindlyFarn complexes sedimented in broader peaks (Fig. 6, F and G), suggesting initial aggregation- or concentration-driven high-order oligomerization.
Mapping the RZZ–Spindly interaction
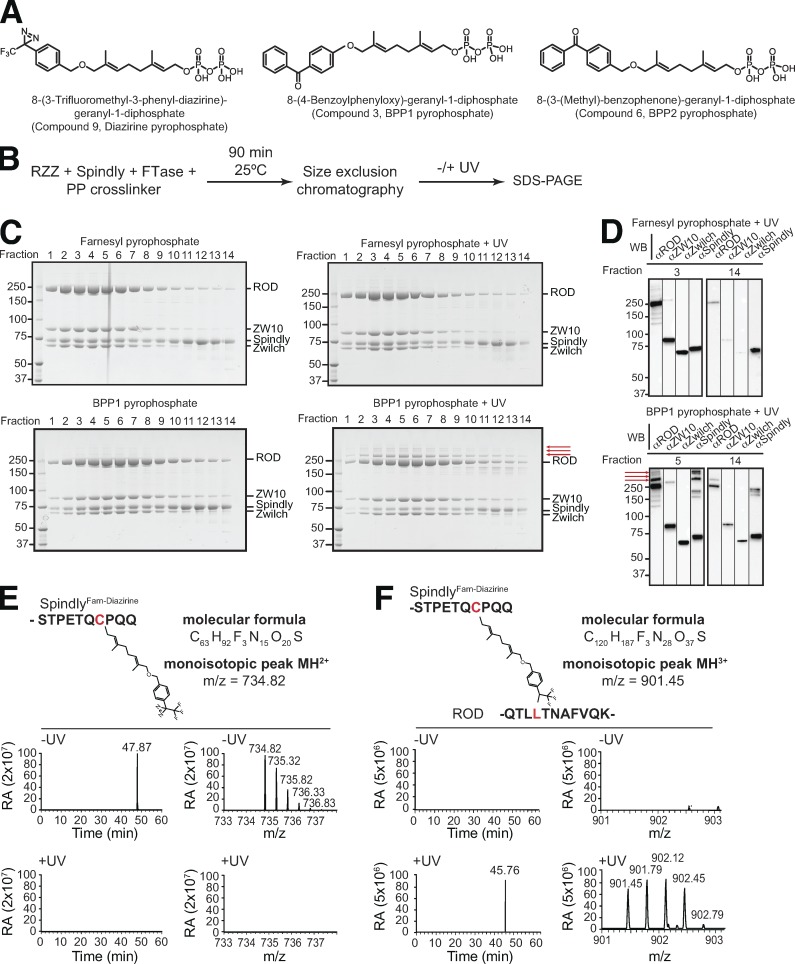
To begin to shed light on how prenylation contributes to the interaction of the RZZ complex with Spindly, we resorted to a chemical biology approach exploiting pyrophosphate precursors of farnesyl derivatives carrying UV-activatable cross-linking groups (benzophenone 1 [BPP1], benzophenone 2 [BBP2], and diazirine [DPP], shown in Fig. 7 A; Kuhlmann et al., 2002). After applying the first step of the protocol schematized in Fig. 7 B, we found by MS that FTase had efficiently incorporated all three farnesyl derivatives on Spindly (Fig. S3, C–E). The RZZ–SpindlyFarn mixes were separated by SEC, and each fraction was exposed to UV. RZZ–SpindlyFarn complexes harboring photoactivatable cross-linkers revealed slow-migrating bands in SDS-PAGE gels, suggestive of successful cross-linking. No equivalent bands were observed upon UV treatment of the unmodified RZZ–SpindlyFarn complex (Figs. 7 D and S4, A–H). By Western blotting, we determined that the slow-migrating bands contain ROD and Spindly (Figs. 7 D and S4, A–H), suggesting that ROD acts as the receptor for the Spindly farnesyl moiety.
Figure 7.
ROD binds the Spindly farnesyl group. (A) Chemical structure of Fpp analogues incorporating photoactivatable cross-linking groups. (B) Schematic of the UV-induced RZZ–SpindlyFarn cross-linking experiment. PP, pyrophosphate. (C) FTase was incubated with Spindly (also in presence of RZZ) to incorporate farnesyl or a modified farnesyl harboring the cross-linking group BPP1 in Spindly. After separation by SEC, elution fractions were subjected to UV irradiation and visualized by Coomassie staining of SDS-PAGE gels. UV-induced cross-linked bands (the most prominent of which are indicated by red arrows) appear in the sample containing BPP1. Equivalent experiments with BPP2 and diazirine are shown in Fig. S4 (A–H). (D) Western blotting (WB) analysis of the indicated selected fractions (from SDS-PAGE in C) with antibodies against ROD, ZW10, Zwilch, and Spindly. Molecular mass is indicated in kilodaltons. (E and F) MS identifies UV-induced cross-links of Spindly’s farnesyl diazirine with residues Leu120 of ROD (shown in red). For each panel, left traces show the chromatographic peak and right traces show the isotopic pattern of the corresponding peptide identified in the MS survey scan. Upper traces represent samples before UV treatment (−UV) and lower traces after UV treatment (+UV). Corresponding MS/MS spectra containing sequence information are shown in Fig. S3 E. Equivalent MS analyses with BPP1 and BPP2 are shown in Fig. S4 (I–L) and MS/MS spectra in Fig. S3 (C and D). RA, relative abundance.
We used MS to identify covalent adducts between Spindly and ROD. Peaks corresponding to (unmodified) photoactivatable cross-linkers conjugated to Cys602 were readily identified in the absence of UV but disappeared when UV was applied to the Spindly–RZZ samples (this is shown for diazirine in Fig. 7, E and F; and for BPP1 and BPP2 in Fig. S4, I–L; tandem MS [MS/MS] fragmentation spectra are shown in Fig. S3, C–H). Correspondingly, new peaks appearing after exposure to UV revealed that all three photoactivatable farnesyl derivatives had become cross-linked with Leu120 in the β-propeller of ROD. Thus, our analysis identifies the β-propeller of ROD as the farnesyl receptor of Spindly.
Discussion
In this study, we have taken advantage of a cryo-EM map, extensive homology modeling, and a comprehensive validation strategy to generate the first structural model of the RZZ complex. ROD, the largest RZZ subunit, is structurally related to proteins that toggle between monomeric and polymeric states, including Clathrin, the αβ’ε-COP (COPI) complex, and the Sec13–Sec31 complex (COPII; Fig. 8 A; Stagg et al., 2007). Underlying the polymerization behavior of these proteins is a common structural design of precursor cage proteins, consisting of an N-terminal β-propeller domain and an extended C-terminal helical domain. The organization of the COPII assembly unit, an inverted dimer of the Sec13–Sec31 complex, with β-propeller domains capping each end, is strongly reminiscent of the dimeric antiparallel arrangement of ROD in RZZ (Stagg et al., 2006, 2007; Fath et al., 2007). These observations make us speculate that RZZ, in analogy to these proteins, may act as the structural precursor of the corona and drive the expansion of the kinetochore into a crescent, as explained in the Introduction (Fig. 8 A). Importantly, there is no evidence at present that the corona exhibits a regular lattice, but this may not be a precondition for its establishment.
Figure 8.
Comparison of self-assembly properties of coat proteins and implications for RZZ–Spindly. (A) The units of self-assembly in COPI, COPII, and Clathrin cages are proteins consisting of an N-terminal β-propeller followed by an α-solenoid. These units often form oligomers (e.g., the inverted tetramers of Sec13–31), which further self-assemble to create finite structures like cages. Whether the kinetochore fibrous corona reflects an ordered assembly of RZZ–Spindly complexes is currently unclear. Kinetochores may act to nucleate self-assembly of cytosolic RZZ–Spindly units and to promote the formation of crescents. (B) Summary of interactions and pathways discussed in this paper.
If RZZ assembles in higher-order oligomers, is seems likely that kinetochores may be required to limit the process spatially. A speculative working model is that full display of high-order oligomerization by RZZ is only possible after kinetochore recruitment and local activation. Kinetochore-localized mitotic kinases have been implicated in kinetochore expansion and may trigger the self-assembly process to limit it to kinetochores (Wynne and Funabiki, 2015). In the process of cargo-stimulated assembly of coated vesicles, coat units are cytosolic and their self-assembly in cells is strictly controlled by interaction with specific cargo adaptors near membranes (Russell and Stagg, 2010; McMahon and Boucrot, 2011; Jackson, 2014). In view of the structural similarity of the RZZ with Clathrin and COPs, we speculate that kinetochore-localized assembly of the corona bears similarity with processes best known to happen near membranes. The exact mechanism of kinetochore recruitment of the RZZ has not yet been unveiled, but previous studies demonstrated a requirement for the outer kinetochore NDC-80 complex, the main MT receptor (Miller et al., 2008; Chan et al., 2009; Cheerambathur et al., 2013; Samejima et al., 2015). This, however, may also reflect an indirect requirement, as the NDC-80 complex acts as recruitment platform for the MPS1 kinase, whose activity is necessary for RZZ kinetochore recruitment (see, e.g., Martin-Lluesma et al. [2002], Santaguida et al. [2010], and this study). We speculate that once established, the corona, through the oligomeric structure of RZZ, facilitates the formation of multiple cooperative low-affinity interactions that “trap” additional corona proteins (most of which are themselves coiled-coil–rich oligomers, like the MAD1–MAD2 tetramer and the CENP-E and CENP-F dimers; Fig. 8 B). Because the fibrous network of the corona only exists at kinetochores and dissolves rapidly upon cell lysis, this might explain previous difficulties in precisely defining the kinetochore receptors of these proteins. Thus, an important goal of future studies will be the identification of binding partners and regulators of RZZ at kinetochores capable of unleashing its predicted high-order oligomerization potential. We aim to approach this problem by reconstituting corona assembly on artificial kinetochores (Weir et al., 2016).
Recent work reported that Spindly farnesylation is required for kinetochore recruitment and its interaction with the RZZ (Holland et al., 2015; Moudgil et al., 2015). Our results extend these previous findings by showing that the interaction of Spindly with RZZ is direct and requires Spindly farnesylation. Furthermore, by using a chemical biology approach, we mapped the binding site for the Spindly farnesyl group to the ROD β-propeller domain. Thus, ROD is a new example of a very small cohort of isoprenoid-binding proteins that also includes PDEδ, the guanine–nucleotide dissociation inhibitor (GDI) for the small GTPases Rab and Rho, and a 14-3-3 protein (Wang and Casey, 2016). PDEδ has high binding affinity for the isolated isoprenoid moiety (Ismail et al., 2011); RabGDI and RhoGDI require the GTPase domain in addition to the prenyl group for tight binding (Hoffman et al., 2000; Rak et al., 2003); finally, 14-3-3 recognizes a hybrid phosphorylated and prenylated motif in the target RND GTPase (Riou et al., 2013). Our results, combined with the previous observation that the minimal kinetochore-binding region of Spindly comprises the entire second half of Spindly (Moudgil et al., 2015), strongly suggest that the binding affinity of ROD for Spindly results from the combined recognition of the farnesyl group and of an extended interaction interface on Spindly. An interaction of Spindly with the kinetochore and dynein-binding protein CENP-F, FTase, and the MAD1–MAD2 spindle checkpoint complex in cellular lysates has been previously described (Holland et al., 2015). Like Spindly, CENP-F is also farnesylated (Ashar et al., 2000; Hussein and Taylor, 2002), and its identification as a binding partner of RZZ raises the possibility that RZZ acts as a more general receptor for farnesylated proteins.
The evolutionary relationship of Spindly with BicD2, which had escaped previous analyses, suggests that Spindly, like BicD2, binds dynein through its N-terminal domain while interacting with cargo via its C-terminal region (Fig. 8 B). This hypothesis is consistent with the sufficiency of the Spindly C-terminal region in kinetochore binding (Moudgil et al., 2015). In light of our result that Spindly binds RZZ directly and of previous evidence that RZZ is required for recruitment of Spindly to the kinetochore, it can be predicted that the C-terminal region of Spindly is sufficient for an interaction with RZZ. Our identification of ROD as the Spindly farnesyl–binding site is consistent with this prediction. This network of interactions suggests that the RZZ is a dynein cargo at kinetochores, an interpretation that raises the interesting question of whether RZZ augments the relatively modest ability of the Spindly adaptor to turn dynein into a processive motor (McKenney et al., 2014). Our future studies will address these important unresolved mechanistic questions.
Materials and methods
Expression and purification of mCherry–Spindly
Spindly was produced as an mCherry fusion using the biGBac system for baculovirus expression (Weissmann et al., 2016). Specifically, the coding sequence of Spindly was subcloned into the multiple cloning site of pLIB. The mCherry coding sequence was then inserted by PCR at the 5′ end of Spindly. Bacmid was produced from EMBacY cells and subsequently used to transfect Sf9 cells and produce baculovirus. The latter was amplified through three rounds of amplification and used to infect TnaO38 cells (Hashimoto et al., 2012). Cells infected with the mCherry–Spindly virus were cultured for 72 h before harvesting. Cells were resuspended in lysis buffer (50 mM Hepes, pH 8, 250 mM NaCl, 10% glycerol, 20 mM imidazole, and 2 mM tris(2-carboxyethyl)phosphine [TCEP]). Resuspended cells were lysed by sonication in the presence of the protease inhibitor mix HP Plus (Serva), DNaseI (Roche), and 1 mM PMSF before clearance at 100,000 g at 4°C for 45 min. The cleared lysate was applied to a 5-ml NiNTA Fast Flow column (GE Healthcare) preequilibrated in lysis buffer. The column was washed with 100 column volumes of lysis buffer, and the bound protein was eluted with lysis buffer supplemented with 300 mM imidazole. The eluate was diluted five times with dilution buffer (50 mM Hepes, pH 8, and 2 mM TCEP) to a final NaCl concentration of 50 mM and applied to a 6-ml Resource Q anion exchange column (GE Healthcare) preequilibrated in buffer A (50 mM Hepes, pH 8, 50 mM NaCl, and 2 mM TCEP). Elution of bound protein was achieved by a linear gradient from 50–400 mM NaCl in 20 column volumes. Relevant fractions were concentrated in 30-kD molecular mass cutoff Amicon concentrators (EMD Millipore) and applied to a Superose6 10/300 column (GE Healthcare) equilibrated in SEC buffer (50 mM Hepes, pH 8, 250 mM NaCl, and 2 mM TCEP). SEC was performed under isocratic conditions at a flow rate of 0.4 ml/min, and the relevant fractions were pooled, concentrated, flash frozen in liquid nitrogen, and stored at −80°C.
Expression and purification of Spindly
Expression of human Spindly was performed with the biGBac system in TnaO38 cells. Lysis of a pellet from 500 ml expression culture was performed by sonication in 100 ml lysis buffer (50 mM Hepes, pH 8.5, 200 mM NaCl, 10% glycerol, 20 mM imidazole, 5 mM β-mercaptoethanol [βMe], 1 mM PMSF, and 1 mM protease inhibitor cocktail [Serva]). The cleared lysate was loaded onto an equilibrated 5-ml HisTrap Fast Flow column (GE Healthcare) using a peristaltic pump (2 ml/min flow rate). The column was washed with 500 ml wash buffer (50 mM Hepes, pH 8.5, 200 mM NaCl, 10% glycerol, 20 mM imidazole, and 5 mM βMe). Elution was performed with wash buffer supplemented with 250 mM imidazole. The 2-ml fractions were analyzed by SDS-PAGE, and those containing Spindly were concentrated up to a volume of 10 ml. The protein solution was diluted five times with buffer A and subsequently purified on a Resource Q anion exchange column. Peak fractions were analyzed by SDS-PAGE, and those containing Spindly were concentrated up to 500 µl and applied to a Superose 6 10/300 column. The peak fractions were analyzed by SDS-PAGE, and those containing pure Spindly were concentrated up to 20 mg/ml, flash frozen in liquid nitrogen, and stored at −80°C.
Expression and purification of RZZ
A protocol for the expression of RZZ has been described previously (Altenfeld et al., 2015). For a pellet of 500 ml of expression culture, cell lysis was performed in 100 ml lysis buffer (50 mM Hepes, pH 8.5, 200 mM NaCl, 10% glycerol, 20 mM imidazole, 5 mM βMe, 1 mM PMSF, and 1 mM protease inhibitor cocktail) by sonication. The cleared lysate was loaded onto an equilibrated 5-ml HisTrap Fast Flow column using a peristaltic pump (2 ml/min flow rate). The column was washed with 500 ml wash buffer (50 mM Hepes, pH 8.5, 200 mM NaCl, 10% glycerol, 20 mM imidazole, and 5 mM βMe), and elution was generally performed by an overnight on-column cleavage with Prescission protease at a ratio of 1:10 (protease/target) and in a circulating buffer volume of 20 ml (15 h, flow rate = 1 ml/min). The eluate was analyzed by SDS-PAGE, concentrated up to a volume of 2 ml, and diluted 1:5 in buffer A (25 mM Hepes, pH 8.5, 25 mM NaCl, and 4 mM TCEP) before loading onto a Resource Q anion exchange column. The diluted sample was loaded onto the column at a flow rate of 1 ml/min and washed (3 column volumes) with buffer A. The buffer was adjusted to a concentration of 50% buffer B (25 mM Hepes, pH 8.5, 1 M NaCl, and 4 mM TCEP) in 20 column volumes. The resulting fractions were pooled and loaded on a Superose 6 10/300 SEC column in SEC buffer (25 mM Hepes, pH 8.5, 250 mM NaCl, and 4 mM TCEP). The eluted RZZ complex was concentrated up to 8–10 mg/ml, flash frozen in liquid nitrogen in aliquots of 20 µl, and then stored at −80°C.
Cell culture, plasmid transfection, microinjections, and imaging
HeLa cells were grown in DMEM (PAN Biotech) supplemented with 10% tetracycline-free FBS (PAN Biotech), penicillin/streptomycin (Gibco), and l-glutamine (PAN Biotech). Cells were grown at 37°C in the presence of 5% CO2. Cdk1 inhibiton, microinjections, and live imaging were performed in complemented CO2-independent media (Gibco) at 37°C. A cDNA segment encoding human CENP-A was cloned in a pcDNA5/FLP recombination target/tetracycline operator–internal ribosome entry site–mCherry (pcDNA5/FRT/TO-IRES-mCherry) vector, a modified version of pcDNA5/FRT/TO vector (Invitrogen) generated in house by M. Mattiuzzo and A. De Antoni as a C-terminal fusion to mCherry. Transient transfections of pcDNA5/FRT/TO-IRES-mCherry were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions, and the mCherry–CENP-A fusion was expressed by addition of 200 ng/ml doxycycline (Sigma-Aldrich) for 48 h.
Where indicated, nocodazole (Sigma-Aldrich) was used at 3.3 µM, RO-3306 (EMD Millipore) was used at 9 µM for 18 h, MG-132 (EMD Millipore) was used at 10 µM, and Reversine (Cayman Chemical) was used at 500 nM. Microinjections were performed using a combination of FemtoJet, InjectMan-NI2, and Femtotip-II, all purchased from Eppendorf. Recombinant GFP and GFP-RZZ complex were injected at concentrations of 4 µM and 10 µM, respectively. Live-cell images were taken before injection and 1–3 min after injection using the spinning-disk confocal microscopy of a Marianas system (Intelligent Imaging Innovations, Inc.) equipped with an Axio Observer Z1 microscope (ZEISS), a CSU-X1 confocal scanner unit (Yokogawa Electric Corporation), Plan-Apochromat 63× or 100×/1.4 NA objectives (ZEISS), and an Orca Flash 4.0 sCMOS Camera (Hamamatsu Photonics). Images were acquired as Z sections using Slidebook Software (5.5; Intelligent Imaging Innovations, Inc.) or LCS 3D software (Leica Microsystems) and converted into maximal intensity projection TIFF files for illustrative purposes.
FTase α/β
Escherichia coli BL21(DE3)-Codon-plus-RIPL cells containing the pGATEV plasmid encoding the GST fusion of the FTase mutant (W102T/Y154T) were grown in Luria–Bertani medium at 37°C to an OD600 of 1. Protein expression was induced by the addition of 0.1 mM IPTG, and cells were incubated at 37°C for 5 h before harvesting. Cells were resuspended in lysis buffer (50 mM Hepes, pH 7.5, 500 mM LiCl, and 2 mM TCEP). Resuspended cells were lysed by sonication in the presence of protease-inhibitor mix HP Plus and 2 mM of PMSF before clearance at 100,000 g at 4°C for 45 min. The cleared lysate was applied to a 20-ml GSH4 Fast Flow column (GE Healthcare) preequilibrated in lysis buffer. The column was washed with 20 column volumes of lysis buffer, and the bound protein was eluted with 30 ml lysis buffer supplemented with 20 mM glutathione. The GST tag was then cleaved by addition of tobacco etch virus protease to the eluate for 3 h at 4°C. The eluate concentrated in 10-kD molecular mass cutoff Amicon concentrators (EMD Millipore) and was applied to a Superdex200 16/60 column (GE Healthcare) equilibrated in SEC buffer (25 mM Hepes, pH 7.5, 150 mM NaCl, and 2 mM TCEP). SEC was performed under isocratic conditions at a flow rate of 1 ml/min, and the relevant fractions were pooled, concentrated, flash frozen in liquid nitrogen, and stored at −80°C.
In vitro farnesylation and cross-linking reactions
Spindly constructs were incubated with RZZ and FTase for 90 min at 25°C in the reaction buffer (50 mM Hepes, pH 8.0, 250 mM NaCl, 10 mM MgCl2, and 2 mM TCEP) containing Fpp, BPP1 pyrophosphate, BPP2 pyrophosphate, or DPP pyrophosphate. Samples were then loaded onto a Superose 6 5/150 column (GE Healthcare) preequilibrated in the reaction buffer. Eluted fractions containing proteins were then either directly analyzed on SDS-PAGE or exposed to UV (60 min for Fpp, BPP1, and BPP2 and 15 min for DPP) before SDS-PAGE and immunoblotting analysis.
SILAC immunoprecipitation experiments
For SILAC, light cells were grown under normal conditions in DMEM without lysine and arginine and supplemented with 10% FCS and 1% penicillin/streptomycin. FCS was dialyzed against a 10-kD cutoff filter to remove possible residual sources of amino acids and ensure efficient labeling. Heavy labeled cells were grown in medium supplemented with [15N2],[13C6]lysine and [15N4],[13C6]arginine. To ensure complete incorporation of labeled amino acids into cellular proteins, cells were passaged at least five times in the corresponding medium. To generate mitotic populations for immunoprecipitation experiments, cells were treated with 330 nM nocodazole for 16 h. Mitotic cells were then harvested by shakeoff and lysed by sonication in lysis buffer (150 mM KCl, 75 mM Hepes, pH 7.5, 1.5 mM EGTA, 1.5 mM MgCl2, 10% glycerol, and 0.075% NP-40) or in lysis buffer without detergents (20 mM Hepes-KOH, pH 7.5, 10 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 1 mM EDTA) supplemented with protease inhibitor cocktail and PhosSTOP phosphatase inhibitors (Roche). Extracts were precleared with a mixture of protein A–Sepharose (CL-4B; GE Healthcare) and protein G–Sepharose (rec-Protein G Sepharose 4B; Invitrogen) for 1 h at 4°C and incubated with GFP-trap beads (3 µl/mg of extract; ChromoTek) presenting GFP-RZZ (2 µg) or GFP (in equimolar amounts) for 15 h at 4°C. After three washing steps, the “light” immunoprecipitate of the reference (GFP alone) was mixed with the “heavy” immunoprecipitate of the bait (GFP-RZZ) in a 1:1 ratio (experiment 1; “For”). In a second experiment with swapped labels, the “heavy” immunoprecipitate of the reference (GFP alone) was mixed with the “light” immunoprecipitate of the bait (GFP-RZZ) in a 1:1 ratio (experiment 2; “Rev”). After two more washes, the respectively combined immunoprecipitates were prepared for analysis by liquid chromatography (LC)–MS/MS.
Sample preparation for MS
Samples of in vitro farnesylated Spindly alone or in complex with RZZ before and after UV treatment were adjusted to a final concentration of 6 M urea and treated with 10 mM DTT and 55 mM chloroacetamide to reduce disulfide bridges. Then, urea was lowered to 4 M, and LysC (129-02541; Wako Pure Chemical Industries) was added for 3 h (protein/enzyme ratio, 50:1). After dilution with 2 vol of 50 mM ammonium bicarbonate, pH 8.3, to a final concentration of 2.0 M urea, peptides were digested overnight at 25°C with sequencing-grade modified trypsin (03708985001; Roche) at a protein/enzyme ratio of 50:1. Resulting peptides were desalted on C18 stage tips (Rappsilber et al., 2007). Approximately 100–200 ng of peptides were injected for every MS run. For GFP/GFP-RZZ pull-downs, 8 M urea was directly applied to the beads, and samples were reduced and alkylated. Bound proteins were then digested in 4 M urea directly from the beads using LysC for 1 h. Subsequently, urea was further diluted to 2 M with 50 mM ammonium bicarbonate, pH 8.3, and proteins bound to the beads were additionally digested with trypsin for 1 h. Supernatants from LysC and trypsin were further digested for 15 h with trypsin. Samples were purified on C18 stage tips, and half of the sample was injected for every MS run.
LC-MS/MS
Peptides were separated on either an UltiMate 3000 Nano LC system or an EASY-nLC 1000 Nano LC System (Thermo Fisher Scientific). Columns (75-µm inner diameter and 25-cm length) were obtained from New Objective (FS36PF7508-250H363). Peptides were loaded in buffer A (double-distilled H2O and 0.1% formic acid) and separated with a gradient from 5–60% buffer B (100% acetonitrile [ACN] and 0.1% formic acid) within 50 min (in vitro farnesylation) or 120 min (for pull-downs) at 200 nl/min. The column temperature was set to 40°C. A quadrupole Orbitrap mass spectrometer (Q Exactive or Q Exactive HF; Thermo Fisher Scientific; Scheltema et al., 2014) was directly coupled to the LC via a nanoelectrospray source. The Q Exactive machines were operated in the data-dependent mode after choosing, from one survey scan, the most abundant precursors for sequencing. The survey scan range was set from 300 to 1,650 Th, with a resolution of 70,000 at m/z 200 for the Q Exactive and 60,000 at m/z 200 for the Q Exactive HF. The target value was set to 3 × 106 and maximum injection time to 20 ms. For sequencing, up to 10 (Q Exactive) or up to 15 (Q Exactive HF) of the most abundant isotope patterns with a charge ≥2 were subjected to higher-energy collisional dissociation with a target value of 1e5. Normalized collision energy was set to 25, and an isolation window of 3 Th for the Q Exactive and 1.4 Th for the Q Exactive HF was used. Resolution for higher-energy collisional dissociation spectra was set to 17,500 at m/z 200 with a maximum ion injection time of 120 ms on the Q Exactive and 15,000 at m/z 200 with maximum ion injection time of 25 ms on the Q Exactive HF. Dynamic exclusion of sequenced peptides was set to 20 s.
Data analysis
To process MS raw files, MaxQuant software (1.5.2.18) was used (Cox and Mann, 2008). We used the Andromeda search engine, which is integrated into MaxQuant, to search MS/MS spectra. For in vitro samples, we searched against a reduced database containing only Spindly and RZZ. Pull-downs were searched against the human UniProt FASTA database (version January 2015). Enzyme specificity was set to C-terminal lysine and arginine and up to two miscleavages were allowed. Peptides had to have a minimum length of seven amino acids to be considered hits. For in vitro samples, carbamidomethylation of cysteine, deamidation (NQ), and oxidation of methionine were always set as variable modifications. Farnesylation or farnesyl-like modifications containing the UV-active compounds (diazirine pyrophosphate, BPP1, and BPP2) were set as variable modification on cysteine according to the experiment. To identify UV-induced cross-links between Spindly and RZZ, the mass of the C-terminal peptide of Spindly containing the farnesyl-modified cysteine plus the mass of the different UV-active compounds were given as variable modifications for the database search. In addition, the y3 ion fragment (PQQ; 372.1878 Th) of Spindly was used as diagnostic marker to identify possible cross-links with RZZ subunits. Because the compounds react in a sequence-independent manner with the backbone of amino acids, searches for every amino acid were performed individually to avoid an exponential increase of the database. MS and MS/MS spectra of possible cross-links were carefully inspected manually, and precursors were required to have a charge state of >2. For pull-down experiments, carbamidomethylation of Cys was given as fixed modification. Deamidation (NQ), oxidation of Met, and N-terminal acetylation of proteins were given as variable modifications. A false discovery rate cutoff of 1% was applied at the peptide, protein levels, and the modified site fraction. An initial precursor mass deviation of up to 4.5 ppm and a fragment mass deviation up to 20 ppm were allowed. Precursor ion mass accuracy was improved by time-dependent recalibration algorithms in MaxQuant.
Negative-stain EM
4 µl of the sample was adsorbed for 1 min at 25°C onto freshly glow-discharged carbon-coated grids (G2400C; Agar Scientific). The sample was blotted (no. 4; Whatman), and the grids were subsequently washed with the SEC buffer (respective buffers in which each of the samples was prepared otherwise; Bröcker et al., 2012). All the images for negatively stained specimens were collected on an electron microscope (JEM1400; JEOL) equipped with a LaB6 cathode operated at 120 kV. All the images were collected under low-dose conditions (19 e/Å2) at a magnification of 67,200× on a 4,000 × 4,000 camera (F416; TVIPS).
Image processing of negative stained samples
All micrographs were recorded with a pixel size of 2.32 Å/pixel, unless stated otherwise. Particles were manually selected, aligned, and classified using reference-free alignment and K-means classification procedures implemented in SPARX (Hohn et al., 2007) and EMAN2 (Ludtke, 2010). For the ZW10 dataset, we used the iterative stable alignment and clustering (ISAC) procedure implemented in SPARX, as the K-means classification procedure failed to give well-defined 2D averages (Yang et al., 2012).
For the 3D reconstructions of the RZZ and the miniRZZ complexes, tilt pairs were collected at tilt angles of 55° and 0°, and particle pairs were selected respectively using the e2RCTboxer program in EMAN2. Random conical tilt reconstructions of the tilted particles were calculated from the respective best class average by back projection followed by back projection refinement (Radermacher et al., 1987). The resulting reconstructions were then submitted to several rounds of 3D multireference projection matching against the untilted datasets, respectively, using SPARX. The resolution of the final reconstructions was estimated by the Fourier shell correlation (FSC) criterion of 0.5. Table 1 lists the number of particles and other parameters for the various datasets.
Table 1. Number of particles and other parameters for the various datasets.
| Dataset | Particles | Defocus | Other details |
|---|---|---|---|
| µm | |||
| RZZ | 4,000 | −1.5 | |
| RZZ (tilt-pairs, RCT) | 1,700 | −1.8 and −1.5 | Tilted and untilted images were recorded at 55° and 0°, with a nominal negative defocus of 1.8 and 1.5, respectively. |
| ZW10 | 10,111 | −1.5 | |
| R1–1,250ZZ | 3,424 | −1.5 | |
| R1–1,250ZZ (tilt-pairs, RCT) | 1,200 | −1.8 and −1.5 | Tilted and untilted images were recorded at 55° and 0°, with a nominal negative defocus of 1.8 and 1.5, respectively. |
Fitting of homology models in EM densities
Homology models of ROD, ZW10, and the crystallographic structure of Zwilch (PDB ID: 3IF8) were automatically fitted in the density using UCSF Chimera Fit in Map tool. Individual rigid-body fittings were performed using the systematic global search option starting from 10,000 random initial placements of a simulated 10.5-Å map of the proteins within the EM map. The cross-correlation score between the simulated map and the EM map was used as a fitting metric. For each of the placements, local optimization was performed. The best-fitting structures had the best cross-correlation (Table S2) and upon visual examination showed excellent correspondence between the fitted model and the EM map.
Sample preparation for cryo-EM
Holey carbon grids (Quantifoils R2/1; Protochips) were freshly glow discharged (two times on the darker side and once on the lighter side of the grid), and 4 µl of the RZZ was applied. 0.02% (wt/vol) Triton was added to the sample just before its application onto the grid. After 30 s, the sample was manually blotted (no. 4; Whatman) followed by application of another 1.7 µl of the sample that was vitrified by plunging (1-s blotting) into liquid ethane using a Cryoplunge3 (Cp3; Gatan). The humidity was maintained to be ∼90%.
Image processing of the cryo-EM dataset
For optimizing the best conditions, images were collected on an electron microscope (3200FSC; JEOL) equipped with a field-emission gun at an operation voltage of 200 kV and an underfocus range of 1.5–3.5 µm. The omega in-column energy filter of the microscope was used to estimate the best ice conditions (∼40–60 nm thickness). A test dataset was collected where the images were recorded manually on an 8,000 × 8,000 CMOS camera (F816; TVIPS) at a pixel size of 1.25 Å/pixel using minimal dose conditions (∼20 e− Å−2).
The final dataset was collected on a spherical aberration–corrected transmission electron microscope (Titan Krios; FEI) equipped with an extra-high brightness field emission gun and operated at a voltage of 300 kV at the Netherlands Centre for Electron Nanoscopy (NeCEN), Leiden, Netherlands. Images were recorded on a back-thinned 4,000 × 4,000 direct detection camera (FalconII) under minimal dose conditions using the automatic data collection software EPU (FEI). The images were recorded at a magnification of 122,807×, corresponding to a pixel size of 1.14 Å. We collected seven consecutive frames during each exposure corresponding to a dose of 2.1 e− Å2 per frame. The frames were collected from 85–475 ms of exposure. A final integrated 1-s image with a total dose of ∼38.2 e− Å−2 was also recorded. A total number of 7,575 images with a defocus range of 1.5–3.0 µm were acquired automatically using the EPU software. The frames of the micrographs collected with the Falcon II detector were aligned to each other using the program MotionCorr to reduce beam-induced blurring of the images (Li et al., 2013). All seven frames were used for processing of the data. The defocus of the micrographs was determined using both CTFFIND4 and “sxcter” program (SPARX; Hohn et al., 2007). We excluded micrographs whose relative defocus error and astigmatism angular error as estimated by sxcter was larger than the defocus and astigmatism error percentage. The error is computed as (SD defocus)/defocus*100%. 49,718 particles were manually handpicked using e2boxer. Particle alignment, classification, and 3D reconstruction were performed in SPARX. The dataset was extensively cleaned up by 2D alignment and classification procedure. In brief, the electron density map from the negative-stain reconstruction was low-pass filtered to 70 Å and used as a starting reference. C2 symmetry was applied during refinement. Refinement of orientation parameters and reconstruction was performed where an FSC-based filter was applied to each volume (Scheres and Chen, 2012). The final reconstruction was calculated from 11,666 particles to a mean resolution of 10.4 Å (FSC0.5 Criterion).
Visualization and analysis of EM density
For analysis, visualization, and segmentation of densities, the program Chimera (Pettersen et al., 2004) was used.
Immunoblotting
Samples were diluted in Laemmli sample buffer, boiled at 95°C, resolved on SDS-PAGE with NuPAGE Bis-Tris 4–12% gradient gels (Invitrogen), and transferred onto nitrocellulose membranes (GE Healthcare). The following primary antibodies were used: anti-ROD (mouse monoclonal; clone CB22-1; 1:500), anti-Zwilch (mouse monoclonal; clone CE47-3; 1:500), anti-ZW10 (mouse monoclonal; clone CO-45-2; 1:500), and anti-Spindly (rabbit polyclonal; A301-354A; 1:5,000; Bethyl Laboratories, Inc.). Mice immunized with recombinant full-length RZZ complex were used to generate anti-ROD, anti-ZW10, and anti-Zwilch antibodies. Antibodies were then affinity purified from sera by using immobilized antigens. Antibody productions, purifications, and biochemistry were performed at the Istituto FIRC di Oncologia Molecolare–Instituto Europeo de Oncologia (IFOM-IEO) Campus Biochemistry Unit. Secondary antibodies were anti–mouse and anti–rabbit (working dilutions, 1:10,000; GE Healthcare). After incubation with the ECL Western blotting system (GE Healthcare), images were acquired with the ChemiDoc MP Imaging System (Bio-Rad Laboratories) in 8-bit Tiff format.
AUC
Sedimentation velocity experiments were performed in an analytical ultracentrifuge (Optima XL-A) with epon charcoal-filled double-sector quartz cells and an An-60 Ti rotor (Beckman Coulter). Samples were centrifuged at 203,000 g at 20°C, and 500 radial absorbance scans were collected with a time interval of 1 min. Data were analyzed using the SEDFIT software (Brown and Schuck, 2006) in terms of continuous distribution function of sedimentation coefficients (c(S)). The protein partial specific volume was estimated from the amino acid sequence using the program SEDNTERP. Data were plotted using the program GUSSI comprised in the SEDFIT software (Brown and Schuck, 2006). Analysis of Spindly constructs bound to RZZ was performed at 20°C in 50 mM Hepes, pH 8, 250 mM NaCl, and 2 mM TCEP, leading to values of buffer density of 1.01306 g/ml and viscosity of 1.002 cP (centipoise). The values of the partial specific volume of mCherry–Spindly–RZZ and Spindly–RZZ are 0.74268 and 0.74335 ml/g at 20°C, respectively (assuming 1:1 stoichiometry).
Expression and purification of ROD1–1,250ZZ and RODΔ754–1,796
The expression and purification of ROD1–1,250ZZ was performed almost identically to the RZZ complex by coexpression from individual viruses. Purification was performed within 1 d to limit the effects of reduced stability of the complex. Elution from the NiNTA column was performed by step elution using 250 mM imidazole, and no ion exchange chromatography was performed. To build RODΔ754–1,796, we fused ROD segments 1–753 and 1,797–2,209 with a (GGSG)4 linker (ROD1–753-(GGSG)4-1,797–2,209) and subcloned it in the His6-POPIN vector. To assemble a complex with Zwilch, we coinfected TnaO38 cells. Further coinfection with virus for expression of ZW10 did not lead to incorporation of ZW10 in the complex with ROD–Zwilch, indicative of a lack of interaction. For a 500-ml expression culture, cell lysis was performed in 100 ml lysis buffer (50 mM Hepes, pH 8.5, 200 mM NaCl, 10% glycerol, 20 mM imidazole, 5 mM βMe, 1 mM PMSF, and 1 mM protease inhibitor cocktail) by sonication. The cleared cell lysate was loaded on a HisTrap Fast Flow column previously equilibrated with wash buffer (50 mM Hepes, pH 8.5, 200 mM NaCl, 10% glycerol, 20 mM imidazole, and 5 mM βMe) using a peristaltic pump at 4°C and a flow rate of 2 ml/min, and the column was washed with 500 ml of wash buffer. Elution was performed with 250 mM imidazole added to wash buffer and collecting 1.5-ml fractions. The fractions were analyzed by SDS-PAGE, and those containing RODΔ754–1,796–Zwilch were concentrated up to 500 µl and applied to a Superose 6 10/300 column using SEC buffer (25 mM Hepes, pH 8.5, 250 mM NaCl, and 4 mM TCEP). Peak fractions were analyzed by SDS-PAGE, and those containing pure RODΔ754–1,796–Zwilch were concentrated up to 5 mg/ml and flash frozen in liquid nitrogen in aliquots of 20 µl and stored at −80°C.
Chemical cross-linking and MS of the RZZ–Spindly complex
Cross-linking of the RZZ–(farnesyl)Spindly complex was performed by mixing 60 µg of the complex (at 0.3 mg/ml) with 450 µM of an equimolar mixture of isotopically light (d0) and heavy (d6) labeled disuccinimidyl suberate (Creative Molecules) for 30 min at 37°C or at 4°C for 15 h. The reactions were quenched by adding a final concentration of 100 mM ammonium bicarbonate for 20 min at 37°C. Cross-linked proteins were enzymatically digested, and cross-linked peptides were identified by LC coupled to MS/MS as reported previously (Herzog et al., 2012). In brief, cross-linked proteins were denatured by adding two sample volumes of 8 M urea (Sigma Aldrich) and were reduced by incubating with 5 mM TCEP (Thermo Fisher Scientific) at 35°C for 15 min. Proteins were alkylated with 10 mM iodoacetamide (Sigma Aldrich) for 35 min at room temperature in the dark. Samples were proteolytically digested by incubating with lysyl endopeptidase (1:50, wt/wt; Wako Pure Chemical Industries) for 2 h at 35°C followed by adding trypsin (1:50, wt/wt; Promega) overnight at a final concentration of 1 M urea. Proteolysis was stopped by the addition of 1% (vol/vol) trifluoroacetic acid (Sigma Aldrich). Acidified peptides were purified by reversed-phase chromatography on C18 columns (Sep-Pak; Waters). Eluates were dried and reconstituted in 20 µl of mobile phase (water/ACN/trifluoroacetic acid, 75:25:0.1), and cross-linked peptides were enriched on a Superdex Peptide PC 3.2/30 column (GE Healthcare) at a flow rate of 50 µl/min. Fractions of the cross-linked peptides were analyzed by LC-MS/MS using an LTQ Orbitrap Elite (Thermo Fisher Scientific) instrument. Finally, the cross-link fragment ion spectra were searched, and the peptides were identified by the open source software xQuest (Walzthoeni et al., 2012). False discovery rates were calculated using xProphet, and results were filtered using the following parameters: delta score <0.85, MS1 tolerance window of −4 to 4 ppm and score >22.
Synthesis of photoactivatable compounds
All commercially available compounds were used as provided without further purifications. Chemicals and solvents were purchased from the companies Acros Organics, Alfa Aesar, Sigma-Aldrich, TCI Europe, and Thermo Fisher Scientific. Dry solvents (e.g., tetrahydrofuran [THF] and ACN) were used without further purifications. Dichloromethane was freshly distilled over calcium hydroxide. Analytical thin-layer chromatography was performed on silica gel aluminum plates (Merck) with F254 indicator (Sigma-Aldrich). Compounds were visualized by irradiation with UV light or potassium permanganate staining. Column chromatography was performed using silica gel Acros 60 A. [1H]NMR, [13C]NMR, and [31P]NMR were measured on a DRX-400 (400 MHz), DRW-500 (500 MHz; Bruker), or INOVA 600 (600 MHz; Agilent Technologies) using CDCl3 or 25 mM ND4OD as solvent and internal standard. The high-resolution mass spectra were recorded on an LTQ Orbitrap mass spectrometer. The preparative HPLC purifications were performed on an Agilent HPLC (1100 series; Agilent Technologies) using a reversed-phase C4 column (NUCLEODUR C4). Method: flowrate 6.0 ml/min from 10% to 100% ACN over 25 min, with 25 mM NH4OH water as cosolvent.
Synthesis of the photoactivatable probes
Synthesis of 8-hydroxy-geranyl-t-butyldimethylsilylether (1)
8-Hydroxy-geranyl-t-butyldimethylsilylether 1 was synthesized according to a previous study (Alexander et al., 2009). See Fig. S5 A.
Synthesis of 8-(4-benzoylphenyloxy)-geraniol (2)
8-(4-Benzoylphenyloxy)-geraniol 2 was synthesized according to a previous study (Alexander et al., 2009). See Fig. S5 B.
Synthesis of 8-(4-benzoylphenyloxy)-geranyl-1-diphosphate (3)
The compound 2 (104 mg and 0.3 mmol) and PPh3 (polymer-supported beads; 503.5 mg and 0.86 mmol) were dissolved in CH2Cl2 (5 ml) and stirred for 30 min to allow the beads to swell. Afterward, a solution of tetrabromomethane (170.3 mg and 0.5 mmol, in 2 ml CH2Cl2) was added, and the mixture was stirred overnight at room temperature. After filtration of the beads, the crude product was extracted with ethyl acetate. The organic phases were combined, and the solvent was removed under reduced pressure. Because of the instability of the allylic bromide, it was directly used without any purification step. See Fig. S5 C.
Therefore, the bromide was dissolved in 6 ml ACN, and (n-Bu4N)3HP2O5 (714 mg and 0.79 mmol) was added slowly. The reaction was allowed to stir for 3 h, and then the solvent was removed under reduced pressure. An ion-exchange column (AG 50W-X8; Bio-Rad Laboratories) was used to convert the product to its ammonium form. The resin was packed and washed using 3 column volumes of 25 vol% NH3 in water, followed by an equilibration step with 3 column volumes of 2 vol% propanol in aqueous 25 mM NH4HCO3 solution. The dark red residue was dissolved in a minimal volume of solvent and loaded on the column. 3 column volumes of the equilibration solution were applied to the column to convert the product in its ammonium form. Fractions containing the product were combined and lyophilized to dryness. The resulting white powder was purified using preparative HPLC.
Yield: 17%.
[1H]NMR: (600 MHz and 25 mM ND4OD): δ = 7.85–7.82 (m, 2H), 7.78–7.75 (m, 2H), 7.67–7.62 (m, 2H), 7.13 (m, 2H), 5.69 (t, J = 7.0 Hz, 1H), 5.55 (t, J = 6.5 Hz, 1H), 4.63 (s, 2H), 4.56 (t, J = 6.5 Hz, 2H), 2.30 (t, J = 7.3 Hz, 2H), 2.19 (t, J = 7.6 Hz, 2H), and 1.80 (s, 6H).
[13C]NMR: (150 MHz and 25 mM ND4OD): δ = 199.51, 162.81, 142.15, 137.36, 133.16, 133.09, 130.67, 130.01, 129.94, 129.42, 128.58, 120.35, 120.29, 114.98, 114.92, 74.31, 62.44, 62.41, 38.35, 25.57, 22.74, 15.74, and 13.15.
[31P]NMR: (243 MHz and 25 mM ND4OD): δ = −6.33 (d, J = 22.2 Hz) and −10.32 (d, J = 22.2 Hz).
HRMS: calculated exact mass for C23H29O9P2 m/z 511.1281 [M + H]+; found m/z 511.1288 [M + H]+.
Synthesis of 8-(3-(methyl)-benzophenone)-geranyl-t-butyldimethylsilylether (4)
The alcohol 1 (300 mg and 1.1 mmol) was slowly added under ice cooling to a solution of NaH (84.3 mg and 2.1 mmol) in THF (15 ml). The mixture was stirred for 30 min under ice cooling before 3-(bromomethyl) benzophenone (580.2 mg and 2.1 mmol) was slowly added. After 2 h at 40°C, the mixture was brought to room temperature and quenched with water (30 ml). The solution was extracted three times with diethyl ether (each 30 ml). Afterward, the combined organic phases were dried over MgSO4. The solvent was removed under reduced pressure and purified using flash chromatography on silica gel (cyclohexane/ethyl acetate). See Fig. S5 D.
Yield: 88%.
Rf: 0.16 (5:1, cyclohexane/ethyl acetate [vol/vol]).
[1H]NMR: (400 MHz and CDCl3): δ = 7.82–7.76 (m, 4H), 7.63–7.55 (m, 1H), 7.53–7.43 (m, 4H), 5.50–5.40 (m, 1H), 5.36–5.26 (m, 1H), 4.53 (d, J = 6.6 Hz, 2H), 4.19 (d, J = 6.3 Hz, 2H), 3.94 (s, 2H), 2.25–2.01 (m, 4H), 1.70 (s, 3H), 1.64 (s, 3H), 0.90 (s, 9H), and 0.07 (s, 6H).
Synthesis of 8-(3-(methyl)-benzophenone)-geraniol (5)
The silylether 4 (517 mg and 1.08 mmol) was dissolved in 5 ml THF. Tetrabutylammoniumfluoride (1.296 ml and 1.3 mmol, 1 M in THF) was added dropwise over 30 min under ice cooling. After 4 h at room temperature, the solution was quenched with brine (20 ml), and the crude product was extracted with diethyl ether (30 ml). The pooled organic phases were dried over MgSO4, and the solvent was removed under reduced pressure. The product was purified using flash chromatography on silica gel (cyclohexane/ethyl acetate). See Fig S5 E.
Yield: 95%.
Rf: 0.18 (2:1, cyclohexane/ethyl acetate [vol/vol]).
[1H]NMR: (500 MHz and CDCl3): δ = 7.82–7.77 (m, 4H), 7.61–7.57 (m, 1H), 7.51–7.43 (m, 4H), 5.45–5.40 (m, 2H), 4.80 (s, 1H), 4.53 (s, 2H), 4.15 (d, J = 6.9 Hz, 2H), 3.94 (s, 2H), 2.24–2.06 (m, 4H), 1.70 (s, 3H), and 1.69 (s, 3H).
[13C]NMR: (125 MHz and CDCl3): δ = 196.61, 143.63, 139.30, 137.85, 136.86, 132.49, 132.37, 130.54, 130.38, 130.15, 128.42, 128.40, 128.28, 127.33, 126.54, 123.90, 76.77, 71.00, 59.51, 39.20, 26.09, 16.39, and 14.14.
HRMS: calculated exact mass for C24H29O3 m/z 365.2111 (M + H)+; found m/z 365.2119 (M + H)+.
Synthesis of 8-(3-(methyl)-benzophenone)-geranyl-1-diphosphate (6)
The compound 5 (150 mg and 0.41 mmol) and PPh3 (polymer-supported beads; 484 mg and 0.82 mmol) were dissolved in CH2Cl2 (5 ml) and stirred for 30 min to allow the beads to swell. A solution of tetrabromomethane (164 mg and 0.5 mmol in 2 ml CH2Cl2) was added, and the mixture was allowed to stir overnight at room temperature. After filtration of the beads, the crude product was extracted three times with ethyl acetate (each 20 ml). The organic phases were combined and the solvent was removed under reduced pressure. Because of the instability of the allylic bromide, it was directly used without any purification step. See Fig S5 F.
The bromide was dissolved in 6 ml ACN, and then (n-Bu4N)3HP2O5 (680.73 mg and 0.75 mmol) was added slowly. The reaction was allowed to stir for 3 h, and afterward the solvent was removed under reduced pressure. An ion-exchange column (AG 50W-X8; Bio-Rad Laboratories) was used to convert the product to its ammonium form. The resin was packed and washed using 3 column volumes of 25 vol% NH3, followed by an equilibration step with 3 column volumes of 2 vol% propanol in aqueous 25 mM NH4HCO3. The dark red residue was solved in a minimal volume of solvent and loaded on the column. 3 column volumes of equilibration solvent were applied to the column to convert the product in its ammonium form. Fractions containing the product were pooled and lyophilized to dryness. The resulting white powder was purified using preparative HPLC.
Yield: 32%.
[1H]NMR: (600 MHz and 25 mM ND4OD): δ = 7.93–7.88 (m, 4H), 7.85–7.80 (m, 1H), 7.71–7.62 (m, 4H), 5.62 (t, J = 6.7 Hz, 1H), 5.58 (t, J = 6.7 Hz, 1H), 4.69 (s, 2H), 4.59–4.54 (m, 2H), 4.11 (s, 2H), 2.32 (t, J = 7.1 Hz, 2H), 2.22 (t, J = 7.4 Hz, 2H), 1.82 (s, 3H), and 1.78 (s, 3H).
[13C]NMR: (150 MHz and 25 mM ND4OD): δ = 200.67, 143.53, 142.27, 136.92, 136.36, 133.59, 131.72, 130.72, 130.34, 130.27, 128.65, 128.25, 120.34, 120.28, 76.57, 70.37, 62.39, 38.41, 25.55, 15.66, and 13.31.
[31P]NMR: (243 MHz and 25 mM ND4OD): δ = −6.35 (d, J = 22.3 Hz) and −10.38 (d, J = 22.4 Hz).
HRMS: calculated exact mass for C24H31O9P2 m/z 525.1438 (M + H)+; found m/z 525.1446 (M + H)+. Calculated exact mass for C24H30O9NaP2 m/z 547.1257 (M + Na)+; found m/z 547.1265 (M + Na)+.
Synthesis of 8-(3-trifluoromethyl-3-phenyl-diazirine)-geranyl-t-butyldimethylsilylether (7)
The alcohol 1 (300 mg and 1.05 mmol) was slowly added under ice cooling to a solution of NaH (51 mg and 2.1 mmol) in THF (7.5 ml). The mixture was stirred for 30 min under ice cooling before the 4-(3-(trifluoromethyl)-3H-diazirin-3-yl)benzyl bromide (589 mg and 2.1 mmol) was slowly added. After 4 h at room temperature, the mixture was brought to room temperature and quenched with water. The solution was extracted three times with diethyl ether (each 20 ml), and the combined organic phases were dried over MgSO4. The solvent was removed under reduced pressure and purified using flash chromatography on silica gel (cyclohexane/ethyl acetate). See Fig. S5 G.
Yield: 81%.
Rf: 0.20 (10:1, cyclohexane/ethyl acetate).
[1H]NMR: (500 MHz and CDCl3): δ = 7.37 (d, J = 8.7 Hz, 2H), 7.17 (d, J = 7.9 Hz, 2H), 5.40 (d, J = 7.0 Hz, 1H), 5.34–5.29 (m, 1H), 4.44 (s, 2H), 4.19 (dd, J = 6.3, 0.8 Hz, 2H), 3.88 (s, 2H), 2.22–2.14 (m, 2H), 2.08–2.02 (m, 2H), 1.67 (s, 3H), 1.63 (s, 3H), 0.90 (s, 9H), and 0.06 (s, 6H).
HRMS: calculated exact mass for C25H38F3N2O2Si m/z 483.2655 (M + H)+; found m/z 483.2649 (M + H)+.
Synthesis of 8-(3-Trifluoromethyl-3-phenyl-diazirine)-geraniol (8)
The silylether 7 (320 mg and 0.66 mmol) was dissolved in 6 ml THF. Tetrabutylammoniumfluoride (796 µl and 0.8 mmol, 1 M in THF) was added dropwise over 30 min under ice cooling. After 4 h at room temperature, the solution was quenched with brine (20 ml), and the crude product was extracted with diethyl ether (30 ml). The combined organic phases were dried over MgSO4, and the solvent was removed under reduced pressure. The product was purified using flash chromatography on silica gel (cyclohexane/ethyl acetate). See Fig. S5 H.
Yield: 91%.
Rf: 0.67 (2:1, cyclohexane/ethyl acetate).
[1H]NMR: (500 MHz and CDCl3): δ = 7.37 (d, J = 8.5 Hz, 2H), 7.17 (d, J = 7.8 Hz, 2H), 5.41 (d, J = 7.3 Hz, 2H), 4.44 (s, 2H), 4.15 (d, J = 6.7 Hz, 2H), 3.88 (s, 2H), 2.24–2.15 (m, 2H), 2.11–2.04 (m, 2H), 1.68 (d, J = 6.9 Hz, 3H), and 1.67 (s, 3H).
HRMS: calculated exact mass for C19H22F3N2O1 m/z 351.1679 ([M − H2O] + H)+; found m/z 351.1679 ([M − H2O] + H)+.
Synthesis of 8-(3-trifluoromethyl-3-phenyl-diazirine)-geranyl-1-diphosphate (9)
The compound 8 (50 mg and 0.14 mmol) and PPh3 (polymer-supported beads; 159.68 mg and 0.27 mmol) were dissolved in CH2Cl2 (5 ml) and stirred for 30 min to allow the beads to swell. A solution of tetrabromomethane (57 mg and 0.16 mmol in 2 ml CH2Cl2) was added, and the mixture was stirred overnight at room temperature. After filtration of the beads, the crude product was extracted with ethyl acetate. The organic phases were combined, and the solvent was removed under reduced pressure. Because of the instability of the allylic bromide, it was directly used without any purification step. See Fig. S5 I.
The bromide was dissolved in 6 ml ACN, and (n-Bu4N)3HP2O5 (332 mg and 0.37 mmol) was added slowly. The reaction was allowed to stir for 3 h, and afterward, the solvent was removed under reduced pressure. An ion-exchange column (AG 50W-X8; Bio-Rad Laboratories) was used to convert the product to its ammonium form. The resin was packed and washed using 3 column volumes of 25 vol% NH3, followed by an equilibration step with 3 column volumes of 2 vol% propanol in aqueous 25 mM NH4HCO3 solution. The dark red residue was solved in a minimal volume of solvent and loaded on the column. 3 column volumes of equilibration solvent were applied to the column to convert the product in its ammonium form. Fractions containing the product were pooled and lyophilized to dryness. The resulting white powder was purified using preparative HPLC.
Yield: 14%.
[1H]NMR: (600 MHz and 25 mM ND4OD): δ = 7.54 (s, 2H), 7.39 (s, 2H), 5.56 (s, 2H), 4.57 (s, 2H), 4.53 (s, 2H), 4.04 (s, 2H), 2.34–2.18 (m, 4H), 1.79 (s, 3H), and 1.73 (s, 3H).
[13C]NMR: (150 MHz and 25 mM ND4OD): δ = 167.02, 165.70, 142.31, 139.57, 131.64, 130.21, 128.94, 128.31, 126.75, 120.25, 76.31, 70.18, 64.25, 62.41, 38.40, 25.52, 23.78, 15.67, and 13.28.
[31P]NMR: (243 MHz and 25 mM ND4OD): δ = −6.37 (d, J = 21.9 Hz) and −10.37 (d, J = 22.3 Hz).
HRMS: calculated exact mass for C24H31O9P2 m/z 525.1438 (M + H)+; found m/z 525.1446 (M + H)+. Calculated exact mass for C19H26O8N2F3P2 m/z 529.1111 (M + H)+; found m/z 529.11155 (M + H)+.
Online supplemental material
Fig. S1 shows additional EM data and analyses of the RZZ and RZZ–Spindly complexes as well as sedimentation velocity absorbance profiles associated with experiments shown in Figs. 1 and 6. Fig. S2 shows additional EM and biochemical analyses of different RZZ deletion mutants and SEC analyses of RZZ intersubunit interactions. Fig. S3 shows MS/MS spectra demonstrating incorporation of UV photoactivatable cross-linker farnesyl derivatives on Cys602 as well as MS/MS spectra of their adducts with ROD. Fig. S4 shows additional biochemical and MS analyses of the UV cross-linking experiments. Fig. S5 displays sequences from the Synthesis of the photoactivatable probes section of Materials and methods. Video 1 demonstrates the considerable flexibility of the RZZ complex. Video 2 illustrates the fitting of molecular model into the EM density of the RZZ complex. Table S1 summarizes the strategy to create molecular models of the RZZ subunits. Table S2 summarizes the quality of model fitting experiments. Table S3 summarizes results with bifunctional cross-linkers and MS. Table S4 displays the multiple alignment using fast Fourier transform (MAFFT) of the indicated BicD2 and Spindly sequences.
Supplementary Material
Acknowledgments
We thank Marta Mattiuzzo and Anna De Antoni for sharing unpublished reagents, and we thank members of our laboratories for helpful discussions.
J. Keller acknowledges support by the European Molecular Biology Organization long-term fellowship ALTF 331-2010. A. Wehenkel acknowledges support by the European Molecular Biology Organization long-term fellowship ALTF 662-2008 and Marie Curie Intra-European Fellowship. A. Musacchio acknowledges funding by the European Research Council Advanced Grant RECEPIANCE (grant 669686) and the Deutsche Forschungsgemeinschaft Collaborative Research Centre (CRC) 1093. F. Herzog is supported by the European Research Council StG MolStruKT (grant 638218) and by the Deutsche Forschungsgemeinschaft (grant GRK1721). S. Raunser gratefully acknowledges the Max Planck Society and the European Council under the European Union’s Seventh Framework Programme (FP7/2007–2013; grant 615984).
The authors declare no competing financial interests.
Author contributions: S. Mosalaganti collected EM data and performed EM computational work, and M. Saur additionally contributed to this part of the work. P. Rombaut and F. Herzog performed XL-MS experiments and data analysis. A. Altenfeld, J. Keller, A. Wehenkel, and S. Wohlgemuth expressed and purified RZZ variants and/or Spindly for various experiments and set up the FFase binding assay. A. Petrovic and J. Keller performed and analyzed AUC experiments. M. Winzker synthesized the photoactivatable farnesyl derivatives under supervision of H. Waldmann; J. Keller and S. Wohlgemuth performed UV cross-linking assays with photoactivatable farnesyl derivatives. F. Müller and T. Bange performed MS/MS experiments to identify UV-activated cross-links. S. Maffini carried out RZZ microinjection experiments. J. Keller carried out all the modeling of the RZZ structure. A. Musacchio, S. Raunser, S. Mosalaganti, A. Altenfeld, and J. Keller designed the majority of experiments. S. Raunser coordinated the EM work and wrote sections of the manuscript. A. Musacchio coordinated the team and wrote most of the manuscript.
Footnotes
Abbreviations used:
- ACN
- acetonitrile
- AUC
- analytical ultracentrifugation
- Fpp
- farnesyl pyrophosphate
- FRT
- FLP recombination target
- FSC
- Fourier shell correlation
- FTase
- farnesyl transferase
- GDI
- guanine–nucleotide dissociation inhibitor
- ISAC
- iterative stable alignment and clustering
- LC
- liquid chromatography
- MS
- mass spectrometry
- MT
- microtubule
- RZZ
- ROD–Zwilch–ZW10
- SAC
- spindle assembly checkpoint
- SEC
- size-exclusion chromatography
- SILAC
- stable isotope labeling with amino acid in culture
- TCEP
- tris(2-carboxyethyl)phosphine
- THF
- tetrahydrofuran
- XL-MS
- cross-linking MS
References
- Alexander M., Gerauer M., Pechlivanis M., Popkirova B., Dvorsky R., Brunsveld L., Waldmann H., and Kuhlmann J.. 2009. Mapping the isoprenoid binding pocket of PDEδ by a semisynthetic, photoactivatable N-Ras lipoprotein. ChemBioChem. 10:98–108. 10.1002/cbic.200800275 [DOI] [PubMed] [Google Scholar]
- Altenfeld A., Wohlgemuth S., Wehenkel A., Vetter I.R., and Musacchio A.. 2015. Complex assembly, crystallization and preliminary X-ray crystallographic analysis of the human Rod-Zwilch-ZW10 (RZZ) complex. Acta Crystallogr. F Struct. Biol. Commun. 71:438–442. 10.1107/S2053230X15004343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashar H.R., James L., Gray K., Carr D., Black S., Armstrong L., Bishop W.R., and Kirschmeier P.. 2000. Farnesyl transferase inhibitors block the farnesylation of CENP-E and CENP-F and alter the association of CENP-E with the microtubules. J. Biol. Chem. 275:30451–30457. 10.1074/jbc.M003469200 [DOI] [PubMed] [Google Scholar]
- Barisic M., Sohm B., Mikolcevic P., Wandke C., Rauch V., Ringer T., Hess M., Bonn G., and Geley S.. 2010. Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol. Biol. Cell. 21:1968–1981. 10.1091/mbc.E09-04-0356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Gomes R., and Karess R.E.. 2000. Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila. Nat. Cell Biol. 2:939–943. 10.1038/35046592 [DOI] [PubMed] [Google Scholar]
- Basto R., Scaerou F., Mische S., Wojcik E., Lefebvre C., Gomes R., Hays T., and Karess R.. 2004. In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr. Biol. 14:56–61. 10.1016/j.cub.2003.12.025 [DOI] [PubMed] [Google Scholar]
- Bröcker C., Kuhlee A., Gatsogiannis C., Balderhaar H.J., Hönscher C., Engelbrecht-Vandré S., Ungermann C., and Raunser S.. 2012. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. USA. 109:1991–1996. 10.1073/pnas.1117797109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S.G., Leksa N.C., Spear E.D., Rajashankar K.R., and Schwartz T.U.. 2008. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 322:1369–1373. 10.1126/science.1165886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.H., and Schuck P.. 2006. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 90:4651–4661. 10.1529/biophysj.106.081372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffin E., Lefebvre C., Huang J., Gagou M.E., and Karess R.E.. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 15:856–861. 10.1016/j.cub.2005.03.052 [DOI] [PubMed] [Google Scholar]
- Caldas G.V., Lynch T.R., Anderson R., Afreen S., Varma D., and DeLuca J.G.. 2015. The RZZ complex requires the N-terminus of KNL1 to mediate optimal Mad1 kinetochore localization in human cells. Open Biol. 5:150160 10.1098/rsob.150160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P., Diamant A.G., and Urnavicius L.. 2016. How dynein and dynactin transport cargos: a structural perspective. Curr. Opin. Struct. Biol. 37:62–70. 10.1016/j.sbi.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Chan Y.W., Fava L.L., Uldschmid A., Schmitz M.H., Gerlich D.W., Nigg E.A., and Santamaria A.. 2009. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J. Cell Biol. 185:859–874. 10.1083/jcb.200812167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur D.K., Gassmann R., Cook B., Oegema K., and Desai A.. 2013. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science. 342:1239–1242. 10.1126/science.1246232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S., Ketcham S.A., Schroer T.A., and Lander G.C.. 2015. Structural organization of the dynein-dynactin complex bound to microtubules. Nat. Struct. Mol. Biol. 22:345–347. 10.1038/nsmb.2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfrocco M.A., DeSantis M.E., Leschziner A.E., and Reck-Peterson S.L.. 2015. Mechanism and regulation of cytoplasmic dynein. Annu. Rev. Cell Dev. Biol. 31:83–108. 10.1146/annurev-cellbio-100814-125438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çivril F., Wehenkel A., Giorgi F.M., Santaguida S., Di Fonzo A., Grigorean G., Ciccarelli F.D., and Musacchio A.. 2010. Structural analysis of the RZZ complex reveals common ancestry with multisubunit vesicle tethering machinery. Structure. 18:616–626. 10.1016/j.str.2010.02.014 [DOI] [PubMed] [Google Scholar]
- Cooke C.A., Schaar B., Yen T.J., and Earnshaw W.C.. 1997. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma. 106:446–455. 10.1007/s004120050266 [DOI] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Fath S., Mancias J.D., Bi X., and Goldberg J.. 2007. Structure and organization of coat proteins in the COPII cage. Cell. 129:1325–1336. 10.1016/j.cell.2007.05.036 [DOI] [PubMed] [Google Scholar]
- Fotin A., Cheng Y., Sliz P., Grigorieff N., Harrison S.C., Kirchhausen T., and Walz T.. 2004. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 432:573–579. 10.1038/nature03079 [DOI] [PubMed] [Google Scholar]
- Gassmann R., Essex A., Hu J.S., Maddox P.S., Motegi F., Sugimoto A., O’Rourke S.M., Bowerman B., McLeod I., Yates J.R. III, et al. 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 22:2385–2399. 10.1101/gad.1687508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Holland A.J., Varma D., Wan X., Çivril F., Cleveland D.W., Oegema K., Salmon E.D., and Desai A.. 2010. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 24:957–971. 10.1101/gad.1886810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E.R., Stuurman N., and Vale R.D.. 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 177:1005–1015. 10.1083/jcb.200702062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Zhang S., Zhang S., Chen Y.R., and Blissard G.W.. 2012. Correction: BTI-Tnao38, a new cell line derived from Trichoplusia ni, is permissive for AcMNPV infection and produces high levels of recombinant proteins. BMC Biotechnol. 12:12 10.1186/1472-6750-12-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F.U., Ban N., et al. 2012. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 337:1348–1352. 10.1126/science.1221483 [DOI] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., and Salmon E.D.. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009. 10.1091/mbc.12.7.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman G.R., Nassar N., and Cerione R.A.. 2000. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 100:345–356. 10.1016/S0092-8674(00)80670-4 [DOI] [PubMed] [Google Scholar]
- Hohn M., Tang G., Goodyear G., Baldwin P.R., Huang Z., Penczek P.A., Yang C., Glaeser R.M., Adams P.D., and Ludtke S.J.. 2007. SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157:47–55. 10.1016/j.jsb.2006.07.003 [DOI] [PubMed] [Google Scholar]
- Holland A.J., Reis R.M., Niessen S., Pereira C., Andres D.A., Spielmann H.P., Cleveland D.W., Desai A., and Gassmann R.. 2015. Preventing farnesylation of the dynein adaptor Spindly contributes to the mitotic defects caused by farnesyltransferase inhibitors. Mol. Biol. Cell. 26:1845–1856. 10.1091/mbc.E14-11-1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad C.C., and Akhmanova A.. 2016. Bicaudal D family of motor adaptors: linking dynein motility to cargo binding. Trends Cell Biol. 26:327–340. 10.1016/j.tcb.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., and Salmon E.D.. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172. 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein D., and Taylor S.S.. 2002. Farnesylation of Cenp-F is required for G2/M progression and degradation after mitosis. J. Cell Sci. 115:3403–3414. [DOI] [PubMed] [Google Scholar]
- Ismail S.A., Chen Y.X., Rusinova A., Chandra A., Bierbaum M., Gremer L., Triola G., Waldmann H., Bastiaens P.I., and Wittinghofer A.. 2011. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol. 7:942–949. 10.1038/nchembio.686 [DOI] [PubMed] [Google Scholar]
- Jackson L.P. 2014. Structure and mechanism of COPI vesicle biogenesis. Curr. Opin. Cell Biol. 29:67–73. 10.1016/j.ceb.2014.04.009 [DOI] [PubMed] [Google Scholar]
- Jokelainen P.T. 1967. The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J. Ultrastruct. Res. 19:19–44. 10.1016/S0022-5320(67)80058-3 [DOI] [PubMed] [Google Scholar]
- Kardon J.R., and Vale R.D.. 2009. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 10:854–865. 10.1038/nrm2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R. 2005. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 15:386–392. 10.1016/j.tcb.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., and Sternberg M.J.. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10:845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G.J., Kim Y., Weaver B.A., Mao Y., McLeod I., Yates J.R. III, Tagaya M., and Cleveland D.W.. 2005. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169:49–60. 10.1083/jcb.200411118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann J., Tebbe A., Völkert M., Wagner M., Uwai K., and Waldmann H.. 2002. Photoactivatable synthetic Ras proteins: “baits” for the identification of plasma-membrane-bound binding partners of Ras. Angew. Chem. Int. Ed. Engl. 41:2546–2550. [DOI] [PubMed] [Google Scholar]
- Lee C., and Goldberg J.. 2010. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell. 142:123–132. 10.1016/j.cell.2010.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Mooney P., Zheng S., Booth C.R., Braunfeld M.B., Gubbens S., Agard D.A., and Cheng Y.. 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 10:584–590. 10.1038/nmeth.2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke S.J. 2010. 3-D structures of macromolecules using single-particle analysis in EMAN. Methods Mol. Biol. 673:157–173. 10.1007/978-1-60761-842-3_9 [DOI] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., and Stock J.. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Magidson V., Paul R., Yang N., Ault J.G., O’Connell C.B., Tikhonenko I., McEwen B.F., Mogilner A., and Khodjakov A.. 2015. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat. Cell Biol. 17:1134–1144. 10.1038/ncb3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke V.M., and Nigg E.A.. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 297:2267–2270. 10.1126/science.1075596 [DOI] [PubMed] [Google Scholar]
- Matson D.R., and Stukenberg P.T.. 2014. CENP-I and Aurora B act as a molecular switch that ties RZZ/Mad1 recruitment to kinetochore attachment status. J. Cell Biol. 205:541–554. 10.1083/jcb.201307137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B.F., Arena J.T., Frank J., and Rieder C.L.. 1993. Structure of the colcemid-treated PtK1 kinetochore outer plate as determined by high voltage electron microscopic tomography. J. Cell Biol. 120:301–312. 10.1083/jcb.120.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney R.J., Huynh W., Tanenbaum M.E., Bhabha G., and Vale R.D.. 2014. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 345:337–341. 10.1126/science.1254198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H.T., and Boucrot E.. 2011. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12:517–533. 10.1038/nrm3151 [DOI] [PubMed] [Google Scholar]
- Miller S.A., Johnson M.L., and Stukenberg P.T.. 2008. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80Hec1. Curr. Biol. 18:1785–1791. 10.1016/j.cub.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mische S., He Y., Ma L., Li M., Serr M., and Hays T.S.. 2008. Dynein light intermediate chain: an essential subunit that contributes to spindle checkpoint inactivation. Mol. Biol. Cell. 19:4918–4929. 10.1091/mbc.E08-05-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudgil D.K., Westcott N., Famulski J.K., Patel K., Macdonald D., Hang H., and Chan G.K.. 2015. A novel role of farnesylation in targeting a mitotic checkpoint protein, human Spindly, to kinetochores. J. Cell Biol. 208:881–896. 10.1083/jcb.201412085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A. 2015. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 25:R1002–R1018 (published erratum appears in Curr. Biol. 2015. 25:3017). 10.1016/j.cub.2015.08.051 [DOI] [PubMed] [Google Scholar]
- Nguyen U.T., Cramer J., Gomis J., Reents R., Gutierrez-Rodriguez M., Goody R.S., Alexandrov K., and Waldmann H.. 2007. Exploiting the substrate tolerance of farnesyltransferase for site-selective protein derivatization. ChemBioChem. 8:408–423. 10.1002/cbic.200600440 [DOI] [PubMed] [Google Scholar]
- Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., and Mann M.. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 1:376–386. 10.1074/mcp.M200025-MCP200 [DOI] [PubMed] [Google Scholar]
- Pesenti M.E., Weir J.R., and Musacchio A.. 2016. Progress in the structural and functional characterization of kinetochores. Curr. Opin. Struct. Biol. 37:152–163. 10.1016/j.sbi.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Radermacher M., Wagenknecht T., Verschoor A., and Frank J.. 1987. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J. Microsc. 146:113–136. 10.1111/j.1365-2818.1987.tb01333.x [DOI] [PubMed] [Google Scholar]
- Rak A., Pylypenko O., Durek T., Watzke A., Kushnir S., Brunsveld L., Waldmann H., Goody R.S., and Alexandrov K.. 2003. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 302:646–650. 10.1126/science.1087761 [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., and Ishihama Y.. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Rieder C.L., and Alexander S.P.. 1990. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J. Cell Biol. 110:81–95. 10.1083/jcb.110.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou P., Kjær S., Garg R., Purkiss A., George R., Cain R.J., Bineva G., Reymond N., McColl B., Thompson A.J., et al. 2013. 14-3-3 proteins interact with a hybrid prenyl-phosphorylation motif to inhibit G proteins. Cell. 153:640–653 (published erratum appears in Cell 2013. 153:1164). 10.1016/j.cell.2013.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C., and Stagg S.M.. 2010. New insights into the structural mechanisms of the COPII coat. Traffic. 11:303–310. 10.1111/j.1600-0854.2009.01026.x [DOI] [PubMed] [Google Scholar]
- Samejima I., Spanos C., Alves F.L., Hori T., Perpelescu M., Zou J., Rappsilber J., Fukagawa T., and Earnshaw W.C.. 2015. Whole-proteome genetic analysis of dependencies in assembly of a vertebrate kinetochore. J. Cell Biol. 211:1141–1156. 10.1083/jcb.201508072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Tighe A., D’Alise A.M., Taylor S.S., and Musacchio A.. 2010. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190:73–87. 10.1083/jcb.201001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaërou F., Starr D.A., Piano F., Papoulas O., Karess R.E., and Goldberg M.L.. 2001. The ZW10 and Rough Deal checkpoint proteins function together in a large, evolutionarily conserved complex targeted to the kinetochore. J. Cell Sci. 114:3103–3114. [DOI] [PubMed] [Google Scholar]
- Scheltema R.A., Hauschild J.P., Lange O., Hornburg D., Denisov E., Damoc E., Kuehn A., Makarov A., and Mann M.. 2014. The Q Exactive HF, a Benchtop mass spectrometer with a pre-filter, high-performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol. Cell. Proteomics. 13:3698–3708. 10.1074/mcp.M114.043489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H., and Chen S.. 2012. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 9:853–854. 10.1038/nmeth.2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager M.A., Hoang H.T., Urnavicius L., Bullock S.L., and Carter A.P.. 2014. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 33:1855–1868. 10.15252/embj.201488792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silió V., McAinsh A.D., and Millar J.B.. 2015. KNL1-Bubs and RZZ Provide Two Separable Pathways for Checkpoint Activation at Human Kinetochores. Dev. Cell. 35:600–613. 10.1016/j.devcel.2015.11.012 [DOI] [PubMed] [Google Scholar]
- Sivaram M.V., Wadzinski T.L., Redick S.D., Manna T., and Doxsey S.J.. 2009. Dynein light intermediate chain 1 is required for progress through the spindle assembly checkpoint. EMBO J. 28:902–914. 10.1038/emboj.2009.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J., Biegert A., and Lupas A.N.. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248. 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg S.M., Gürkan C., Fowler D.M., LaPointe P., Foss T.R., Potter C.S., Carragher B., and Balch W.E.. 2006. Structure of the Sec13/31 COPII coat cage. Nature. 439:234–238. 10.1038/nature04339 [DOI] [PubMed] [Google Scholar]
- Stagg S.M., LaPointe P., and Balch W.E.. 2007. Structural design of cage and coat scaffolds that direct membrane traffic. Curr. Opin. Struct. Biol. 17:221–228. 10.1016/j.sbi.2007.03.010 [DOI] [PubMed] [Google Scholar]
- Starr D.A., Williams B.C., Hays T.S., and Goldberg M.L.. 1998. ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 142:763–774. 10.1083/jcb.142.3.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E., Musacchio A., Harrison S.C., and Kirchhausen T.. 1998. Atomic structure of clathrin: a β propeller terminal domain joins an α zigzag linker. Cell. 95:563–573. 10.1016/S0092-8674(00)81623-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A., Ren Y., Jeffrey P.D., and Hughson F.M.. 2009. Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex. Nat. Struct. Mol. Biol. 16:114–123. 10.1038/nsmb.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnavicius L., Zhang K., Diamant A.G., Motz C., Schlager M.A., Yu M., Patel N.A., Robinson C.V., and Carter A.P.. 2015. The structure of the dynactin complex and its interaction with dynein. Science. 347:1441–1446. 10.1126/science.aaa4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D., Monzo P., Stehman S.A., and Vallee R.B.. 2008. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. J. Cell Biol. 182:1045–1054. 10.1083/jcb.200710106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzthoeni T., Claassen M., Leitner A., Herzog F., Bohn S., Förster F., Beck M., and Aebersold R.. 2012. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat. Methods. 9:901–903. 10.1038/nmeth.2103 [DOI] [PubMed] [Google Scholar]
- Wang M., and Casey P.J.. 2016. Protein prenylation: unique fats make their mark on biology. Nat. Rev. Mol. Cell Biol. 17:110–122. 10.1038/nrm.2015.11 [DOI] [PubMed] [Google Scholar]
- Weir J.R., Faesen A.C., Klare K., Petrovic A., Basilico F., Fischböck J., Pentakota S., Keller J., Pesenti M.E., Pan D., et al. 2016. Insights from biochemical reconstitution into the architecture of human kinetochores. Nature. 537:249–253. 10.1038/nature19333 [DOI] [PubMed] [Google Scholar]
- Weissmann F., Petzold G., VanderLinden R., Huis in 't Veld P.J., Brown N., Lampert F., Westermann S., Stark H., Schulman B.A., and Peters J.-M.. 2016. biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc. Natl. Acad. Sci. USA. 113:E2564–E2569. 10.1073/pnas.1604935113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.C., Gatti M., and Goldberg M.L.. 1996. Bipolar spindle attachments affect redistributions of ZW10, a Drosophila centromere/kinetochore component required for accurate chromosome segregation. J. Cell Biol. 134:1127–1140. 10.1083/jcb.134.5.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.C., Li Z., Liu S., Williams E.V., Leung G., Yen T.J., and Goldberg M.L.. 2003. Zwilch, a new component of the ZW10/ROD complex required for kinetochore functions. Mol. Biol. Cell. 14:1379–1391. 10.1091/mbc.E02-09-0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik E., Basto R., Serr M., Scaërou F., Karess R., and Hays T.. 2001. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 3:1001–1007. 10.1038/ncb1101-1001 [DOI] [PubMed] [Google Scholar]
- Wynne D.J., and Funabiki H.. 2015. Kinetochore function is controlled by a phospho-dependent coexpansion of inner and outer components. J. Cell Biol. 210:899–916. 10.1083/jcb.201506020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T.G., Watanabe S., Essex A., and Kitagawa R.. 2008. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J. Cell Biol. 183:187–194. 10.1083/jcb.200805185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Fang J., Chittuluru J., Asturias F.J., and Penczek P.A.. 2012. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 20:237–247. 10.1016/j.str.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Anderson K.L., and Cleveland D.W.. 1997. The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 139:435–447. 10.1083/jcb.139.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Lischetti T., Hayward D.G., and Nilsson J.. 2015. Distinct domains in Bub1 localize RZZ and BubR1 to kinetochores to regulate the checkpoint. Nat. Commun. 6:7162 10.1038/ncomms8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.