Inoue et al. show that Foxo1 controls not only GC polarization but also GC B cell proliferation, the latter of which is mediated by Foxo1-dependent BATF up-regulation.
Abstract
Germinal center (GC) B cells cycle between two states, the light zone (LZ) and the dark zone (DZ), and in the latter they proliferate and hypermutate their immunoglobulin genes. How this functional transition takes place is still controversial. In this study, we demonstrate that ablation of Foxo1 after GC development led to the loss of the DZ GC B cells and disruption of the GC architecture, which is consistent with recent studies. Mechanistically, even upon provision of adequate T cell help, Foxo1-deficient GC B cells showed less proliferative expansion than controls. Moreover, we found that the transcription factor BATF was transiently induced in LZ GC B cells in a Foxo1-dependent manner and that deletion of BATF similarly led to GC disruption. Thus, our results are consistent with a model where the switch from the LZ to the DZ is triggered after receipt of T cell help, and suggest that Foxo1-mediated BATF up-regulation is at least partly involved in this switch.
Introduction
After their initial encounter with T cell–dependent antigens, B cells migrate to the interface between the T and B cell zones in lymphoid organs, where they interact with cognate T cells to form antigen-specific cell clusters. After leaving these clusters, B cells undergo brisk proliferation before entering the GC reaction or developing into short-lived plasmablasts (herein called the preGC period; Victora and Mesin, 2014; De Silva and Klein, 2015).
Once a GC is established in the B cell follicle, the dark zone (DZ) and light zone (LZ) form and the GC B cells then cycle between them (Allen et al., 2007; Victora and Nussenzweig, 2012). B cells in these two zones can be identified based on expression levels of the signature surface proteins CXCR4, CD83, and CD86; DZ GC cells express higher levels of CXCR4, but lower levels of CD83 and CD86, whereas LZ cells are CXCR4low, CD83hi, and CD86hi. Proliferation and somatic hypermutation (SHM) occur in the DZ, and then the B cells shuttle to the LZ, where they exit the cell cycle. In the LZ, de novo mutated BCRs capture antigen and internalize it for MHC class II (MHC-II) presentation to follicular helper T (TFH) cells. According to the current model (Allen et al., 2007; Victora et al., 2010; Liu et al., 2015), GC B cells expressing high-affinity BCRs are selected in response to signals provided by cognate TFH cells in the LZ. Next, as cells transit from the LZ back to the DZ, proliferation is induced. Therefore, it has been argued that induction of proliferation after receipt of TFH cell help is well coupled to the LZ-to-DZ transition. Ultimately, after several such iterative cycles of proliferation, diversification, and selection, the GC generates high-affinity plasma cells and memory B cells.
In regard to the molecular processes for DZ cyclic reentry, it has been demonstrated that c-Myc plays an important role because it is expressed by a small fraction of LZ GC B cells that are enriched for high-affinity BCRs and have recently entered the S phase of the cell cycle (Calado et al., 2012; Dominguez-Sola et al., 2012; Gitlin et al., 2015). Transient c-Myc expression can be induced by forcing T–B cell interactions, leading to reentry into the DZ and stimulation of cell division.
Recently, the role of Foxo1 in the transition from the LZ-to-DZ program has been explored by two studies, both indicating that this transcription factor plays a regulatory role in the formation and maintenance of the GC DZ, as in its absence there was no DZ in the GC (Dominguez-Sola et al., 2015; Sander et al., 2015). Notably, in these studies the overall GC size was intact even in the absence of Foxo1, a finding somewhat at odds with the aforementioned coupling model between proliferation and the LZ-to-DZ transition. Because the chemokine receptor CXCR4 is one of the direct physiological Foxo1 targets (Dubrovska et al., 2012; Dominguez-Sola et al., 2015), the observed DZ defect in Foxo1-deficient GC B cells has been explained, at least in part, by down-regulation of CXCR4. However, functionally, the Foxo1-deficient GC B cells appear to be more severely affected than in the CXCR4 knockout (Bannard et al., 2013). For instance, down-regulation of CD86 occurred in both Cxcr4−/− and wild-type GC B cells, whereas this regulation was perturbed in Foxo1-deficient GC B cells. These data imply that the loss of CXCR4 expression is necessary but not sufficient to account for the defective DZ program resulting from Foxo1 ablation, prompting us to investigate how Foxo1 participates in the transition from the LZ to the DZ program.
Here, we report that, in addition to CXCR4, Foxo1 participates in up-regulation of the transcription factor BATF. BATF was transiently expressed in a small fraction of LZ GC cells, and depletion of BATF, like that of Foxo1, led to the GC disruption. Hence, our results suggest that Foxo1 controls not only GC polarization, but also GC proliferation, at least in part, through mediating BATF expression, together contributing to the transition from the LZ to DZ program.
Results
Hyperexpansion of preGC B cells after Foxo1 ablation
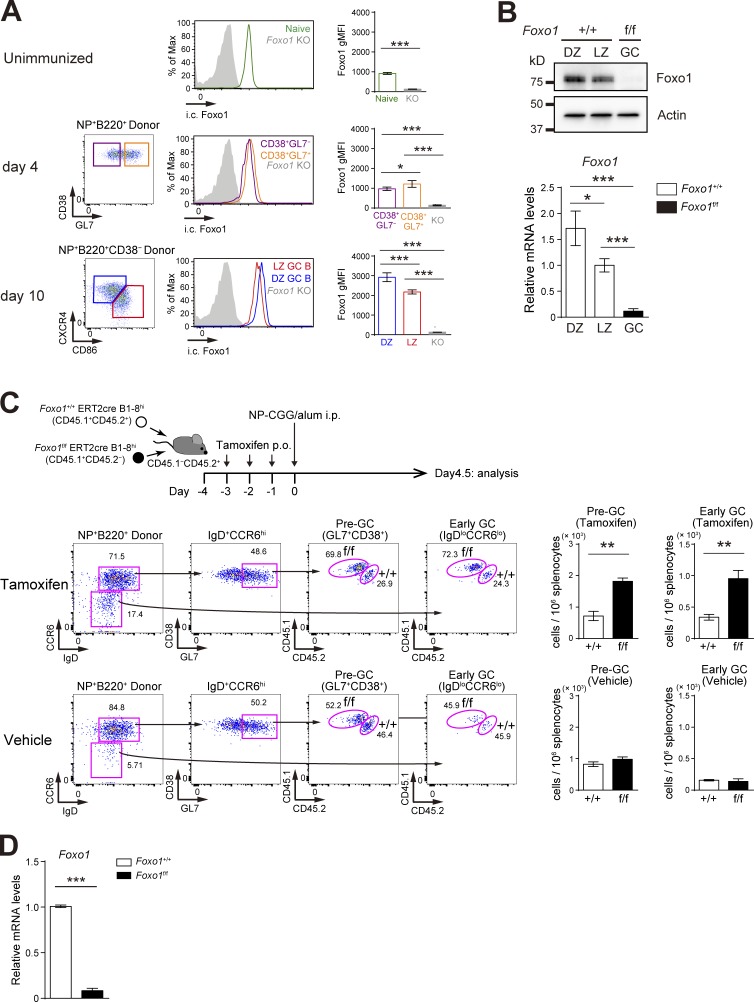
To examine the effects of Foxo1 ablation on antigen-driven clonal expansion and GC differentiation, we used adoptive transfer experiments using B1-8hi BCR heavy chain knock-in B cells, which are specific for the hapten 4-hydroxy-3-nitrophenylacetyl (NP) when combined with Igλ light chains. Before examining the B cell intrinsic biological roles of Foxo1, we analyzed Foxo1 expression during an immune response. B1-8hi B cells were transferred into congenically marked mice and immunized with NP-chicken γ globulin (CGG)/alum. Foxo1 protein expression level, assessed by intracellular flow cytometry, was similar or slightly higher in proliferative CD38+GL7− or CD38+GL7+ B cells than in their naive B cell counterparts (Fig. 1 A). Compared with parental naive B cells, the Foxo1 expression level in LZ GC B cells was higher and the expression level in the DZ GC B cells was higher than that in LZ GC B cells (Fig. 1 A). This differential Foxo1 expression between the DZ and LZ GC B cells was confirmed by Western blot and was also apparent in Foxo1 mRNA levels (Fig. 1 B).
Figure 1.
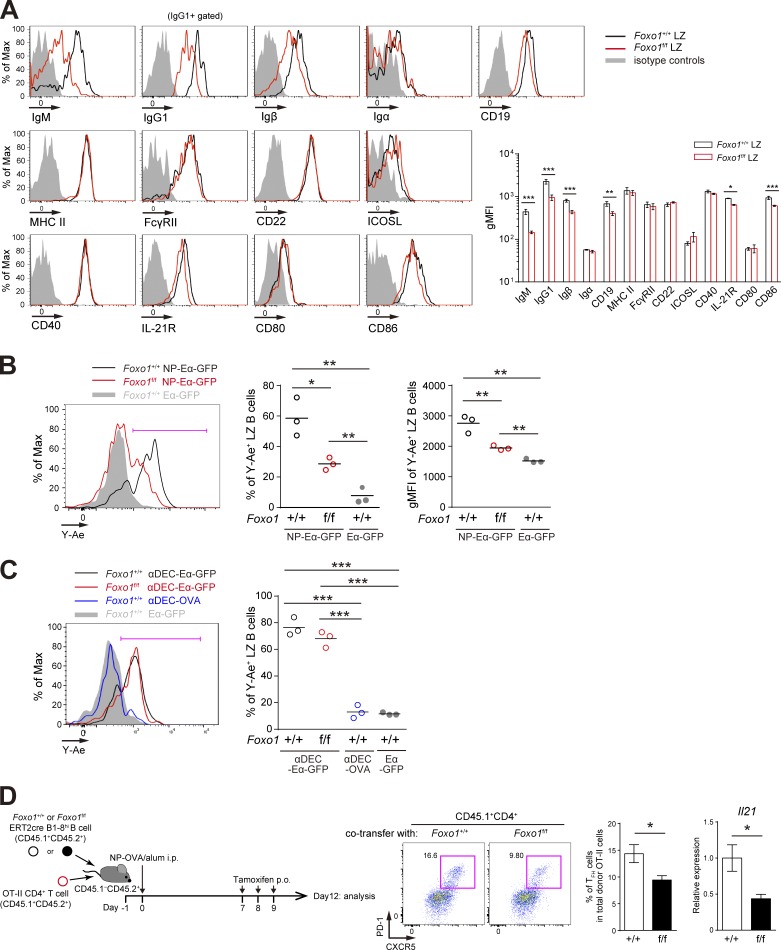
Hyperexpansion of preGC B cells with Foxo1 ablation. (A) Left, flow cytometry of intracellular Foxo1 protein expression in naive B cells at day 0 (CD45.1+B220+NP+CD38+), activated B cells on day 4 (CD45.1+B220+NP+CD38+GL7− or CD45.1+B220+NP+CD38+GL7+), LZ (CD45.1+B220+NP+CD38−CD86hiCXCR4lo), and DZ (CD45.1+B220+NP+CD38−CD86loCXCR4hi) GC B cells on day 10. Wild-type mice were transferred with B1-8hi CD45.1+ B cells and then immunized i.p. with NP-CGG/alum on day 0. Foxo1 KO staining controls (gray histograms) were prepared as previously described in Figs. 1 C and 2 A. (right) Histograms indicating the geometric mean fluorescence intensity (gMFI) of each population. n = 4 biological replicates. (B) Analysis of Foxo1 protein and mRNA expression in DZ and LZ GC B cells by Western blot (top) and real-time qPCR (bottom). Foxo1-proficient LZ, DZ, and Foxo1-deficient GC B cells were sorted from mice prepared as described in Fig. 2 A. Actin, loading control. n = 3 biological replicates. (C, top) Schematic illustration of the experimental protocol. (bottom left) Flow cytometry of NP-specific donor B cells (CD45.1+B220+NP+). (bottom right) Histograms representing the cell number of preGC (Donor B220+NP+GL7+CD38+IgD+CCR6hi) and early GC (Donor B220+NP+IgDloCCR6lo) B cells in 106 splenocytes. n = 3 biological replicates. (D) Real-time qPCR analysis of Foxo1 mRNA expression in Foxo1+/+ERT2cre B1-8hi and Foxo1f/f ERT2cre B1-8hi preGC B cells. Error bars represent SD. Data are representative of three (A) or two (B and C) independent experiments, and from one experiment with three biological replicates (D). *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test (A, B, and D) and paired Student’s t test (C).
To examine the role of Foxo1 in expansion of preGC B cells, we co-transferred equal numbers of Foxo1f/f ERT2cre B1-8hi and Foxo1+/+ ERT2cre B1-8hi B cells into congenically marked mice, which were then immunized with NP-CGG/alum after tamoxifen treatment (Fig. 1 C). In this protocol, deletion of Foxo1 was very efficient (Fig. 1 D), and did not result in any induction of Foxo3 or Foxo4 mRNA (not depicted). Expansion of preGC B1-8hi B cells (GL7+CD38+IgD+CCR6hi) and their subsequent differentiation toward early GC B cells (IgDloCCR6lo; Schwickert et al., 2011) were enhanced when Foxo1 was eliminated; control vehicle treatment had essentially no effects. These observations appear to be consistent with the conventional view that Foxo family transcription factors act as tumor suppressors (Dansen and Burgering, 2008; Hedrick, 2009).
GC maintenance requires Foxo1
The idea that Foxo1 might play a proliferative, rather than antiproliferative, role during the GC phase has been suggested by an earlier study showing that GC-derived lymphomas frequently carry mutations in Foxo1 that prevent its inactivation by Akt (Trinh et al., 2013). Hence, we wished to delete Foxo1 specifically during the GC stage. To do this, we used the same co-transfer experiments as Fig. 1 C, but used the time course depicted in Fig. 2 A, because almost all the transferred B1-8hi B cells in our experimental conditions were already differentiated into GC B cells by day 7. If tamoxifen was administered at earlier time points, for instance at day 4, deletion of Foxo1 would occur at both preGC and GC stages, possibly complicating interpretation of the data.
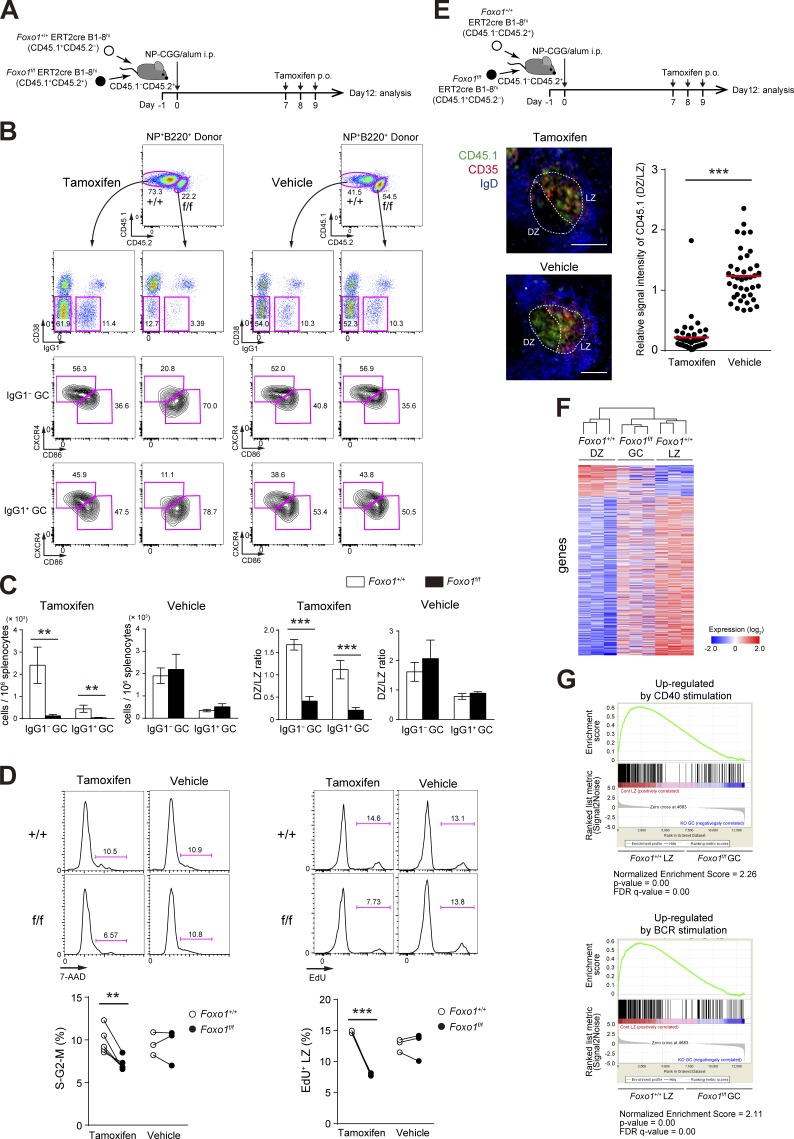
Figure 2.
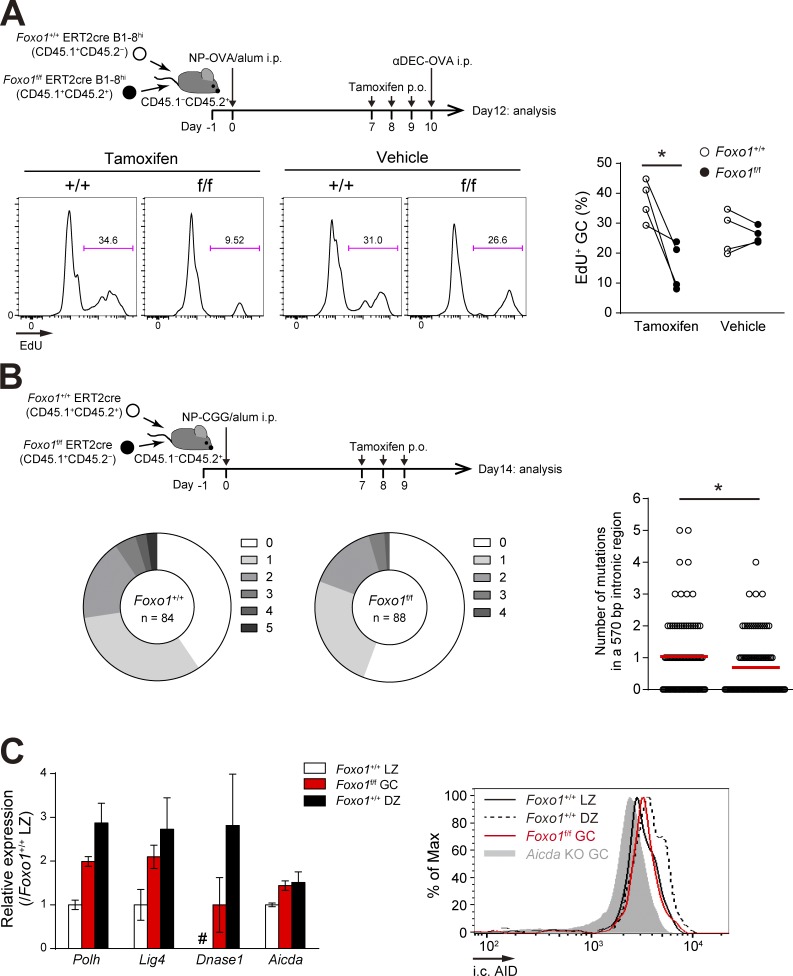
Foxo1 is required for GC maintenance. (A) Schematic illustration of the experimental protocol for B–D, F, and G. (B) Flow cytometry of NP-specific donor B cells (CD45.1+B220+NP+). (C) Histograms representing the number of donor IgG1− GC B cells (CD45.1+B220+NP+CD38−IgG1−) and IgG1+ GC B cells (CD45.1+B220+NP+CD38−IgG1+) in 106 splenocytes (left), and the ratio of DZ:LZ cells (right). n = 3 biological replicates. (D, left) DNA content measurement of Foxo1+/+ and Foxo1f/f LZ GC B cells assessed by 7-AAD staining. n = 5 and 3 biological replicates for tamoxifen and vehicle treatment, respectively. (right) Proliferation status of Foxo1+/+ and Foxo1f/f LZ GC B cells assessed by EdU incorporation 30 min after an EdU injection. n = 3 biological replicates. (E) Immunohistochemical analysis. (top) Schematic illustration of the experimental protocol. (bottom left) Representative images of immunofluorescence microscopy of spleen sections showing expression of CD45.1 (Foxo1f/f-derived donor cells), CD35 (FDC network), and IgD (follicular B cells). DZ and LZ defined by the presence of CD35+ FDCs are surrounded by dashed lines. Bars, 100 µm. (bottom right) Quantification of relative CD45.1 signal intensity in the DZ compared with that in the LZ. Each symbol represents a single GC, and red bars indicate the mean. n = 43 (tamoxifen) and 40 (vehicle) GC pooled from three animals. (F) Hierarchical clustering of the gene expression profiles of Foxo1+/+ DZ, Foxo1f/f GC, and Foxo1+/+ LZ B cells using genes differentially expressed (more than twofold) between Foxo1+/+ DZ and Foxo1+/+ LZ B cells (normalized log2 values based on RNA-seq analysis). n = 3 biological replicates. (G) Gene set enrichment analysis showing the enrichment for genes up-regulated after ligation of CD40 (top) and BCR (bottom) compared of Foxo1+/+ LZ B cells with Foxo1f/f GC B cells. Error bars represent SD. Data are representative of three (B and C) or two independent experiments (D and E) and from one experiment (F and G). **, P < 0.01; ***, P < 0.001; unpaired Student’s t test (E) and paired Student’s t test (C and D).
Tamoxifen injection resulted in selective diminution of Foxo1f/f ERT2cre B1-8hi GC B cells; both IgG1− and IgG1+ B1-8hi GC B cells were similarly affected by Foxo1 ablation, whereas control vehicle treatment had no effect (Fig. 2, B and C). In this protocol, Foxo1 protein and mRNA were efficiently depleted in GC B cells (labeled as Foxo1f/f in Fig. 1 B). The similar loss of IgG1− and IgG1+ GC B cells indicates that deletion of Foxo1 takes place after completion of class switch recombination (CSR) in our protocol, being different from the previous two studies by using a Cγ1-cre (Dominguez-Sola et al., 2015; Sander et al., 2015); indeed, previous studies demonstrate the involvement of Foxo1 in CSR processes.
To further examine whether Foxo1 ablation affects cell division, we compared the frequency of Foxo1-deficient and -proficient LZ GC B cells in the S–G2–M phase of the cell cycle. By DNA content measurement, Foxo1-deficient LZ GC B cells exhibited less proliferation status than control cells (Fig. 2 D, left), and a similar result was obtained when incorporation of the nucleotide analogue 5-ethynyl-2’-deoxyuridine (EdU) over a 0.5-h period was compared (Fig. 2 D, right). Hence, we conclude that Foxo1 ablation in GC cells decelerates cell cycle progression.
We then examined the effects of Foxo1 deletion on DZ/LZ compartmentalization. Consistent with previous studies (Dominguez-Sola et al., 2015; Sander et al., 2015), the majority of the Foxo1-deficient GC B cells were CXCR4lowCD86hi, reminiscent of LZ cells, although a DZ-like population (CXCR4int) was to some extent detectable, particularly among IgG1− cells (Fig. 2 B). The compartmentalization defect was also confirmed histologically. To do this, we also used co-transfer experiments (Fig. 2 E). In contrast to control vehicle treatment, localization of tamoxifen-treated Foxo1f/f ERT2cre B1-8hi GC cells was largely restricted to the LZ, which was defined by the presence of the CD35+ follicular dendritic cell (FDC) network. These results suggest that Foxo1-deficient cells are locked in the LZ even in a GC where overall DZ/LZ compartmentalization is preserved. Together, deletion of Foxo1 in the GC phase resulted in GC disruption and defective polarization.
The loss of the DZ compartment in Foxo1-deficient GC cells was further substantiated by gene expression profile analysis. We prepared mRNA from Foxo1+/+ ERT2cre B1-8hi DZ and LZ GC B cells and from Foxo1f/f ERT2cre B1-8hi total GC B cells after tamoxifen treatment at day 12 (Fig. 2 A), and performed transcriptome analysis by RNA-seq. A hierarchical clustering analysis indicated that the Foxo1-deficient GC B cells are more similar to normal LZ than DZ B cells; the mutant GC profile exhibited a significant absence of gene signatures associated with the DZ program (Fig. 2 F). Hence, our results demonstrate that Foxo1 directly or indirectly controls a major part of the program that distinguishes DZ from LZ GC B cells. However, when compared with normal LZ GC B cells, some differences were also noted in the Foxo1-deficient GC B cells. Indeed, gene set enrichment analysis (Foxo1-deficient GC B cells versus control LZ GC B cells) indicated that the Foxo1-deficient cells were more weakly imprinted with the signatures of CD40 and BCR signaling (Fig. 2 G).
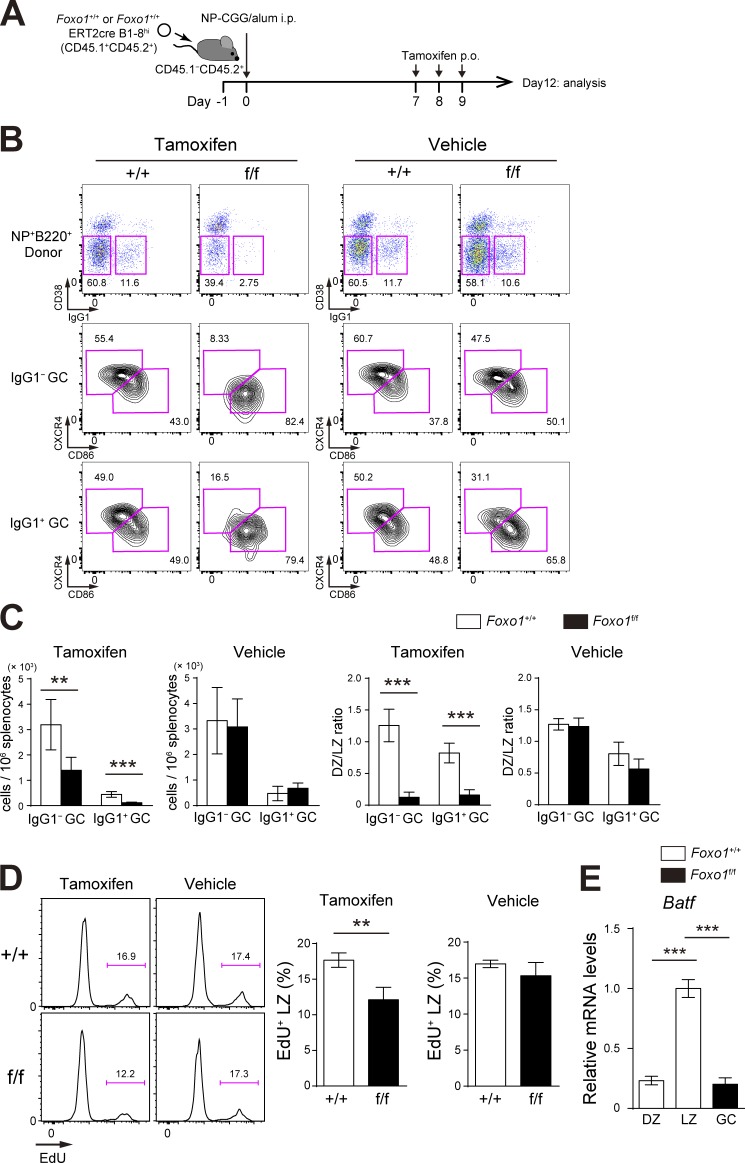
When Foxo1+/+ ERT2cre B1-8hi and Foxo1f/f ERT2cre B1-8hi B cells were individually transferred (Fig. 3 A), instead of co-transferred, the similar compartmentalization defect was observed (Fig. 3 B). The proliferation defect was also observed in the individual transfer experiments, but the defective extent in the absence of Foxo1 was relatively small, compared with co-transfer experiments (Fig. 3, C and D). Hence, we conclude that Foxo1 deletion directly causes the proliferative changes, although, in the aforementioned mixed B cell chimera experimental settings, cell competition takes place.
Figure 3.
Assessment of Foxo1 requirement for GC maintenance and proliferation in single transfer experiments. (A) Schematic illustration of the experimental protocol. (B) Flow cytometry of NP-specific donor B cells (CD45.1+B220+NP+). (C) Histograms representing the number of donor IgG1− GC B cells (CD45.1+B220+NP+CD38−IgG1−) and IgG1+ GC B cells (CD45.1+B220+NP+CD38−IgG1+) in 106 splenocytes (left), and the ratio of DZ:LZ cells (right). n = 5 biological replicates. (D) Proliferation status of Foxo1+/+ and Foxo1f/f LZ GC B cells assessed by EdU incorporation 30 min after an EdU injection. n = 3 biological replicates. (E) Analysis of Batf mRNA expression in Foxo1+/+ DZ, LZ, and Foxo1f/f GC B cells by real-time qPCR. n = 3 biological replicates. Error bars represent SD. Data are representative of three (B and C) or two independent experiments (D and E). **, P < 0.01; ***, P < 0.001; unpaired Student’s t test.
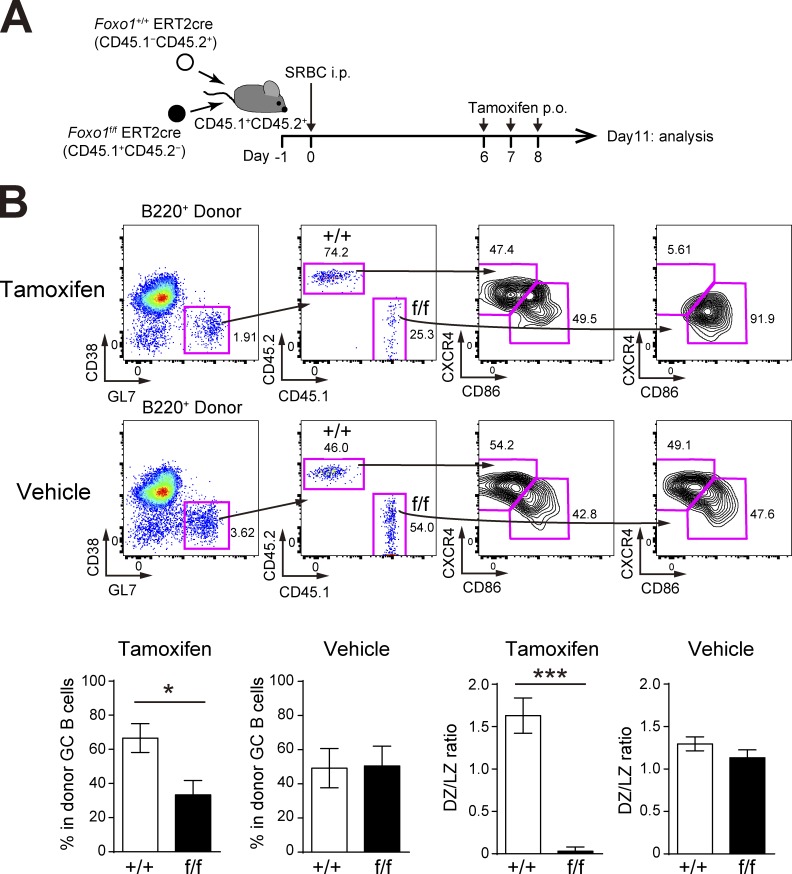
Our conclusion that the number of Foxo1-deficient GC B cells is decreased differs from previous studies demonstrating that the GC size is unchanged in the absence of Foxo1 (Dominguez-Sola et al., 2015; Sander et al., 2015). As these studies analyzed polyclonal GC responses, we reasoned that the polyclonal versus our monoclonal system might cause such differences. To test this possibility, we co-transferred equal numbers of Foxo1f/f ERT2cre and Foxo1+/+ ERT2cre donor B cells into congenically marked mice and immunized them with sheep RBCs (SRBCs). Foxo1 was then deleted as depicted in Fig. 4 A and, overall, both GC maintenance and GC compartmentalization were decreased by the loss of Foxo1 (Fig. 4 B). However, compared with the aforementioned results using B1-8hi B cells, the decrease in polyclonal Foxo1-deficient GC B cell numbers was relatively small. Given the hyperexpansion of B cells caused by deletion of Foxo1 at the preGC stage, one possibility is that, because of the asynchrony of the polyclonal immune responses, some activated B cells might still have been at the preGC stage when Foxo1 was deleted (Fig. 4 A), thereby diluting the effects by GC-specific deletion of Foxo1. It is also possible that monoclonal BCR transgenic B cells might enhance the activation kinetics, which in turn makes more differences particularly in the competitive settings.
Figure 4.
GC phenotype with Foxo1 ablation during a polyclonal immune response. (A) Schematic representation of the experimental protocol. Because entering the GC in the polyclonal stimulation was a little earlier than the monoclonal condition (Fig. 2 A), we treated mice with tamoxifen on day 6, 7, and 8, and analyzed on day 11. (B, top) Flow cytometry of donor B cells (B220+CD45.1−CD45.2+ and B220+CD45.1+CD45.2−). (bottom left) Percentage of each donor in total donor GC B cells (CD45.1−CD45.2+ or CD45.1+CD45.2−B220+GL7+CD38−). (bottom right) Ratio of DZ:LZ cells. n = 5 biological replicates. Error bars represent SD. *, P < 0.05; ***, P < 0.001; paired Student’s t test. Data are representative of two independent experiments.
Antigen presentation is inhibited
As GC B cell proliferation is only marginally affected in Cxcr4−/− mice (Bannard et al., 2013), the aforementioned findings suggest that, in addition to regulating access into the anatomical DZ, Foxo1 utilizes other mechanisms, thereby regulating the transition from the LZ-to-DZ program. According to the current model (Allen et al., 2007; Victora et al., 2010; Liu et al., 2015), the LZ-to-DZ transition is thought to be regulated by two consecutive processes; first, the BCR on LZ B cells functions as an endocytic receptor that shuttles antigen into the MHC-II pathway for processing and presentation to TFH cells; second, the antigen-specific TFH cells activated by cognate interaction with the LZ B cells in turn provide them with T cell help, driving their transition to the DZ program. Hence, we reasoned that Foxo1 might participate in these two processes.
Before addressing whether the first processes are affected by Foxo1 ablation, we analyzed receptors relevant to this process and found that expression of BCR and Igβ in Foxo1-deficient GC cells was lower compared with control cells (Fig. 5 A). Then, to examine the antigen presentation activity on GC B cells to TFH cells, we generated a chemical conjugate of NP and recombinant Eα-GFP, which contains the Eα peptide from the I-E molecule (pEα) fused GFP (Pape et al., 2007). This reagent allows for quantification of the amount of peptide MHC class II (pMHC-II) presentation. The pMHC-II can be monitored with the Y-Ae monoclonal antibody, which is specific for pEα–I-Ab complexes (Murphy et al., 1992).
Figure 5.
Decreased antigen presentation of Foxo1-deficient LZ GC B cells. (A, left) Flow cytometry of surface molecules on Foxo1+/+ and Foxo1f/f LZ GC B cells. Mice were immunized as described in Fig. 2 A, and LZ GC B cells were prepared on day 12. (right) Histograms indicating the gMFI of each population. n = 3 biological replicates. (B) In vitro antigen presentation assay. Mice were prepared as described in Fig. 3 A. Purified splenic B cells were incubated with NP-Eα-GFP or Eα-GFP for 1 h at 37°C. Antigen-presenting LZ GC B cells were detected by staining the LZ GC B cells (CD45.1+CD138−GL7+CD38−CD86hiCXCR4lo) with Y-Ae antibody. (left) Flow cytometry of LZ GC B cells; (middle) the percentage of Y-Ae+ (bracketed line in the left) LZ GC B cells; (right) the gMFI of Y-Ae of Y-Ae+ LZ GC B cells. n = 3 biological replicates. (C) In vitro antigen presentation of Foxo1-deficient LZ GC B cells using αDEC-Eα-GFP. Eα-GFP and αDEC-OVA were used as negative controls. Mice were prepared as described in Fig. 3 A. (left) Flow cytometry of LZ GC B cells; (right) the percentage of Y-Ae+ (bracketed line in the left) LZ GC B cells. n = 3 biological replicates. (D) Effects of Foxo1 ablation in LZ GC B cells on TFH cell number and cytokine production. (left) Schematic representation of the experimental protocol. Foxo1+/+ ERT2cre B1-8hi CD45.1+CD45.2+ or Foxo1f/f ERT2cre B1-8hi CD45.1+CD45.2+ B cells were co-transferred together with CD4+ T cells from OT-II CD45.1+CD45.2+ mice into wild-type mice (CD45.1−CD45.2+), which were immunized with NP-OVA/alum i.p. on day 0. Mice were administered tamoxifen p.o. on day 7, 8, and 9, and analyzed on day 12. (middle) Flow cytometry of OT-II CD4+ T cells (CD45.1+CD4+). (right) The percentage of TFH cells (CD45.1+CD4+CXCR5+PD-1+) among total donor OT-II cells, and the quantification of Il21 mRNA expression in sorted TFH cells by real-time qPCR. n = 8 biological replicates. Error bars represent SD. Data are representative of three (A) or two (B–D) independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test.
Although we detected antigen capture and pMHC-II on activated B1-8hi B cells at the preGC stage after NP-Eα-GFP/alum immunization, neither could be detected on GC B cells (unpublished data). The most likely explanation for this is that the antigen availability in the microenvironments of the GC is very limited, thereby making it undetectable in vivo by our experimental system. Therefore, in the case of GC B cells, we performed ex vivo experiments. As shown in Fig. 5 B, when antigen was provided, Foxo1-deficient B1-8hi LZ GC B cells were able to present pMHC-II lesser extent than control cells; both population of pMHC-II+ cells and their expression levels were low. Next, to ask the question of why mutant GC B cells possess the low antigen presentation activity, we delivered the Eα-GFP antigen to GC B cells in a surrogate manner. For this purpose, instead of targeting to the BCR, we used DEC205. DEC205 is a cell surface receptor, which was expressed to similar extents between wild-type and Foxo1-deficient LZ GC B cells (unpublished data). Eα-GFP was fused to an antibody specific to DEC205 (anti–DEC-Eα-GFP). Upon provision of anti–DEC-Eα-GFP, control and Foxo1-deficient B1-8hi LZ GC B cells showed similar presentation of the pMHC-II complex on the cell surface (Fig. 5 C), indicating that the antigen presentation machinery, itself, operates similarly in control and Foxo1-deficient GC B cells. Collectively, the defective antigen presentation activity in Foxo1-deficient GC B cells is most likely explained by the low level of BCR expression on them.
Because CD19 is known to act as a BCR co-receptor, we wondered whether the decreased CD19 expression (Fig. 5 A) might affect antigen presentation activity. To examine this possibility, we immunized Cd19cre/+ mice, because Cd19cre/+ mice retain only one functional Cd19 allele (Rickert et al., 1997). The ex vivo antigen presentation activity of the LZ GC B cells was unchanged by haploinsufficiency of CD19 (unpublished data).
Because activation of GC TFH cells requires pMHC-II on LZ GC B cells, the aforementioned observations predict that ablation of Foxo1 in GC B cells would render the GC TFH cells less active in vivo. To test this prediction, after transferring OVA-specific TCR transgenic (OT-II) CD4+ T cells together with Foxo1f/f ERT2cre B1-8hi or Foxo1+/+ ERT2cre B1-8hi B cells, we immunized the mice with NP-OVA and deleted Foxo1 as depicted in Fig. 5 D. 3 d after Foxo1 ablation in GC B cells, both the GC TFH cell numbers and production of IL-21 were decreased. These results suggest that Foxo1 contributes to antigen presentation by GC B cells and, subsequently, to activation of GC TFH cells.
In addition to this defective antigen presentation on Foxo1-deficient LZ GC B cells, the lower expression of CD86 in the mutant cells, albeit being a small change (Fig. 5 A), might also be involved in subsequent defective activation of GC TFH cells.
Upon access to T cell help, Foxo1-deficient GC cells have a proliferation defect
We then specifically examined the second process, namely Foxo1 involvement in the transition from the LZ to the DZ state after receipt of T cell help. To address this issue, we adopted an in vivo approach to enforce T cell–B cell interactions by using a T cell antigen (OVA) fused to anti-DEC205 antibody (anti–DEC-OVA; Bonifaz et al., 2002). As shown in Fig. 6 A, after co-transferring of Foxo1+/+ ERT2cre B1-8hi and Foxo1f/f ERT2cre B1-8hi B cells, the recipient mice were immunized with NP-OVA. When stimulated with anti-DEC-OVA after tamoxifen treatment, Foxo1+/+ but not Foxo1-deficient GC B cells showed a burst of proliferation; control vehicle treatment did not induce such differential responses. Thus, we conclude that, even after they receive sufficient T cell help, Foxo1-defcient GC B cells cannot induce the normal proliferation program.
Figure 6.
Foxo1-deficient GC B cells have defects in proliferation and SHM. (A, top) Schematic representation of the experimental protocol. Mice were injected i.p. with αDEC-OVA on day 10 and analyzed on day 12. Bottom, proliferation status of Foxo1+/+ and Foxo1f/f GC B cells assessed by EdU incorporation 30 min after an EdU injection. n = 4 biological replicates. (B) JH4 intron SHM analysis in control and Foxo1-deficient LZ GC B cells. (top) Schematic representation of the experimental protocol. Bottom left, the pie charts showing the relative frequency of sequences with 0–5 mutations from mice of each genotype. n = number of sequences analyzed. (bottom right) Mutation frequencies (number of mutations in a JH4 570 bp intronic region). Red bars indicate the mean. (C, left) Expression of selected SHM-related genes in Foxo1+/+ LZ, Foxo1f/f GC, and Foxo1+/+ DZ B cells based on RNA-seq analysis. Relative fragments per kilobase of exon per million reads (FPKM) values normalized with Foxo1+/+ LZ cells are shown. For Dnase1, the FPKM value of Foxo1f/f GC B cells was set as 1. #, undetected. n = 3 biological replicates. (right) Flow cytometry of intracellular AID protein expression in Foxo1+/+ LZ, Foxo1f/f GC, and Foxo1+/+ DZ GC B cells. Gray histogram, control signal in Aicda KO GC B cells, which were prepared from immunized Aicdaf/f ERT2cre mice treated with tamoxifen. Error bars represent SD. Data are representative of two independent experiments (A and C, right), pooled from two independent animals (B), and from one experiment with three biological replicates (C, left). *, P < 0.05; paired Student’s t test (A) and Mann-Whitney test (B).
Consistently, the mutation load in a 570 bp intronic region downstream of JH4 exon of the Igh locus was lower in the Foxo1-deficient LZ GC B cells compared with the controls (Fig. 6 B). Therefore, efficient SHM requires Foxo1. Polh, Lig4, Dnase1, and Aicda genes are involved in SHM and their transcripts are abundant in the DZ B cells (Victora et al., 2012). Compared with Foxo1-proficient LZ GC B cells, all had higher mRNA expression levels in Foxo1-deficient GC B cells, whereas their levels did not reach those in wild-type DZ GC B cells, except Aicda (Fig. 6 C, left). Despite the similar mRNA levels of Aicda between Foxo1-deficient GC B cells and Foxo1-proficient DZ GC B cells, its protein expression in the mutant cells did not reach that in wild-type DZ GC B cells (Fig. 6 C, right). Thus, key enzymes involved in SHM were up-regulated, to some extents, even in the absence of Foxo1, but not corresponding to the level in wild-type DZ GC cells.
T cell help triggers BATF in a Foxo1-dependent manner and is required for GC maintenance
Having demonstrated the importance of Foxo1 in regulating the GC B cell proliferation program upon receipt of T cell help, we wished to address how Foxo1 exerts this function. To do this, we first examined the expression of CD40 and IL-21R, critical receptors for receiving T cell help (Victora and Nussenzweig, 2012), on Foxo1-deficient B1-8hi LZ GC B cells. Expression of CD40 was similar, but that of IL-21R was somewhat lower in Foxo1-deficient LZ GC B cells compared with control cells (Fig. 5 A). To examine whether the decreased expression of IL-21R affects the proliferation program of GC B cells, we generated bone marrow chimeras that lack IL-21R on B cells by reconstituting mice with a mixture of bone marrow cells from μMT and Il21r+/− mice, and immunized them with NP-CGG/alum. Despite two-fold lower expression of IL-21R on Il21r+/− LZ GC B cells, the number of GC B cells was unaffected (unpublished data). Moreover, IL-21R expression by Foxo1-deficient LZ GC B cells was higher than that of IL-21R haploinsufficient cells (unpublished data). Thus, the expression level of key receptors for receiving T cell help appears not to be solely responsible for the defective proliferation in Foxo1-deficient GC B cells.
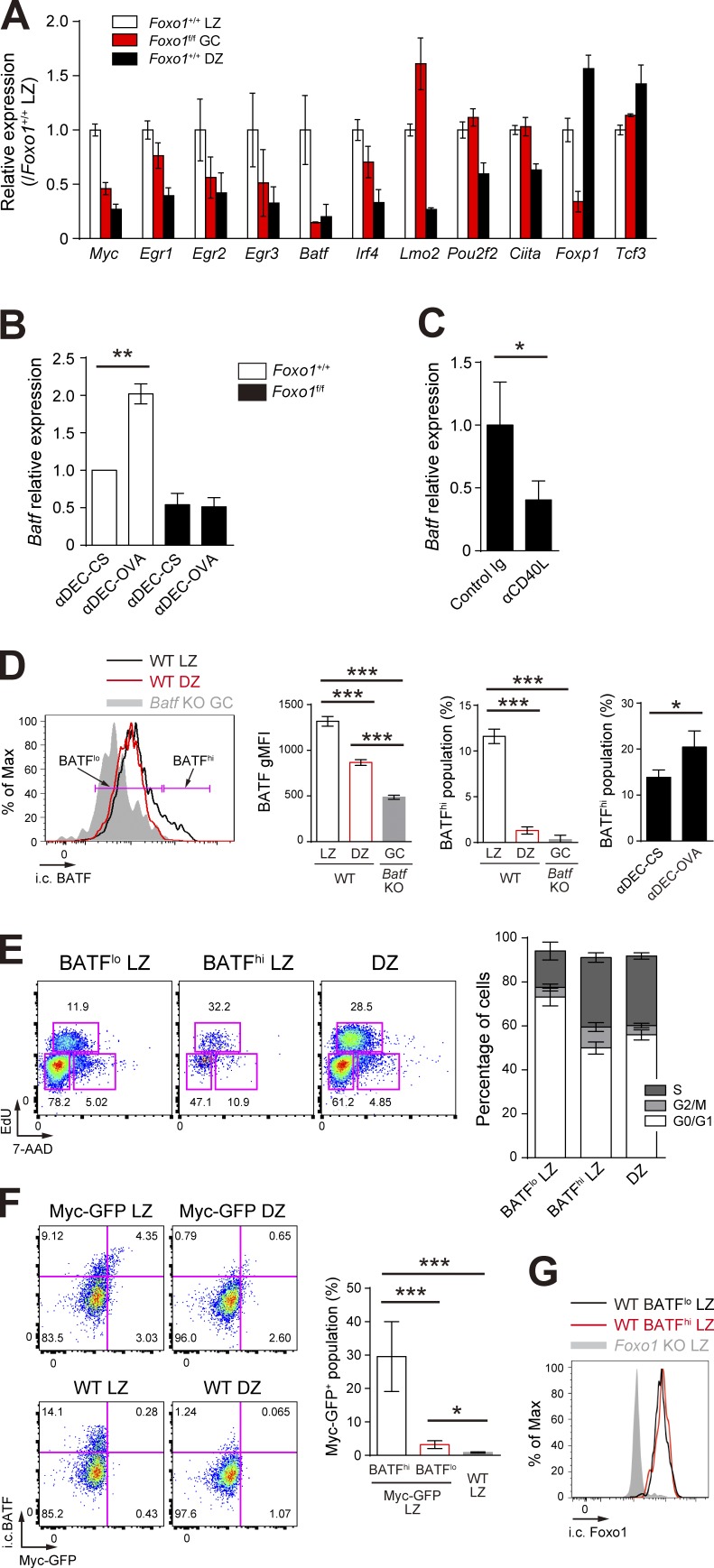
Previous studies have shown that transient c-Myc expression only in the LZ is induced by forcing T–B cell interactions and is required for stimulating cell division (Calado et al., 2012; Dominguez-Sola et al., 2012). We thus hypothesized that such cell division-related key transcription factors might exist in the LZ GC B cells, induced by T cell help in a Foxo1-dependent manner. Among transcription factors differentially expressed between LZ and DZ GC B cells (Victora et al., 2012), c-Myc, Egr2, Egr3, BATF, and Foxp1 were significantly down-regulated and Lmo2 was up-regulated in Foxo1-deficient B1-8hi GC B cells (Fig. 7 A). A previous study demonstrated that transgenic expression of FoxP1 leads to a significant decrease in GC B cells (Sagardoy et al., 2013). In addition, Egr2 and Egr3 have been shown to be negative regulators of T cell activation (Safford et al., 2005; Zheng et al., 2012), and Lmo2 has been suggested as a proto-oncogene in T cell acute lymphoblastic leukemia and in other type of cancers (Chambers and Rabbitts, 2015). Based on these observations, involvement of Egr2, Egr3, Foxp1, and Lmo2 in GC proliferation in a Foxo1-dependent manner appeared to be somewhat unlikely. Because c-Myc was already well characterized (Calado et al., 2012; Dominguez-Sola et al., 2012), we focused our analysis here on BATF.
Figure 7.
T cell help triggers BATF expression in a Foxo1-dependent manner. (A) Expression of selected transcription factors in Foxo1+/+ LZ, Foxo1f/f GC, and Foxo1+/+ DZ B cells based on RNA-seq analysis. Relative FPKM values normalized with Foxo1+/+ LZ cells are shown. (B) Real-time qPCR analysis of Batf mRNA in control and Foxo1-deficient LZ GC B cells after injection of anti–DEC-OVA. Data were normalized with control cells treated with anti–DEC-CS. n = 3 biological replicates. (C) Real-time qPCR analysis of Batf mRNA in wild-type LZ GC B cells after injection of anti-CD40L. n = 4 biological replicates. (D, left) Flow cytometry of intracellular BATF protein expression in LZ and DZ GC B cells. Gray histogram, control signal in Batf KO cells prepared as described in Fig. 8 A. Bracketed lines indicate the BATFlo and BATFhi populations. (middle left) BATF gMFI of each population. n = 3 biological replicates. (middle right) Percentage of BATFhi population. n = 3 biological replicates. (right) Percentage of BATFhi population in the LZ B cells. B1-8hi CD45.1+ B cells were transferred into wild-type mice (CD45.1−CD45.2+), which were immunized with NP-OVA/alum i.p. on day 0. Mice were injected i.p. with anti–DEC-OVA or anti–DEC-CS on day 10. CD45.1+B220+NP+ LZ B cells were analyzed 24 h after injection of each antibody. n = 3 biological replicates. (E, left) Cell cycle analysis of BATFlo LZ, BATFhi LZ, and DZ GC B cells. (right) The percentage of GC B cells at different stages of the cell cycle. S, EdU+ 7-AADint; G2/M, EdU− 7-AADhi; G0/G1, EdU− 7-AADlo. Wild-type mice were immunized with NP-CGG/alum i.p., followed by injection of EdU on day 10, 30 min before analysis of the LZ (B220+GL7+CD38−CD86hiCXCR4lo), and DZ (B220+GL7+CD38−CD86loCXCR4hi) GC B cells. n = 5 biological replicates. (F, left) Flow cytometry of intracellular BATF and c-Myc-GFP expression in GC B cells of c-Myc-GFP and wild-type mice. (right) Percentage of c-Myc-GFP+ populations in BATFhi and BATFlo LZ B cells of c-Myc-GFP mice and total LZ B cells of wild-type B cells. n = 3 biological replicates. (G) Flow cytometry of intracellular Foxo1 protein expression in BATFhi and BATFlo LZ GC B cells. Gray histogram, control signal in Foxo1 KO LZ B cells. Error bars represent SD. Data are from one experiment with three biological replicates (A), or representative of two independent experiments (B–G). *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test.
Indeed, provision of in vivo T cell help by injection of anti–DEC-OVA induced up-regulation of Batf mRNA in B1-8hi GC LZ B cells, and this was dependent on Foxo1 (Figs. 3 E and 7 B). Conversely, inhibition of the T-B interactions in vivo by CD40 blockade decreased Batf expression (Fig. 7 C). Next, we wondered whether BATF, like c-Myc, is dominantly expressed in a small fraction of LZ GC B cells and, if so, whether this small BATFhi LZ fraction is more actively cycling than the BATFlo LZ cells. After NP-CGG immunization of wild-type mice, BATFhi and BATFlo fractions were present among the LZ GC B cells, as assessed by intracellular cytometry analysis (Fig. 7 D), and the population of the BATFhi fraction was increased by anti–DEC-OVA injection (Fig. 7 D). Moreover, the BATFhi cells were more actively cycling (Fig. 7 E). To examine the relationship between BATFhi cells and c-Myc+ cells in the LZ, we used NP-CGG–immunized mice harboring c-Myc-GFP (Huang et al., 2008). According to these staining patterns, c-Myc+ cells were mainly included in the BATFhi LZ fraction; ∼30% among BATFhi cells coexpressed c-Myc (Fig. 7 F). BATFhi and BATFlo LZ fractions expressed similar levels of Foxo1, assessed by intracellular cytometric analysis (Fig. 7 G).
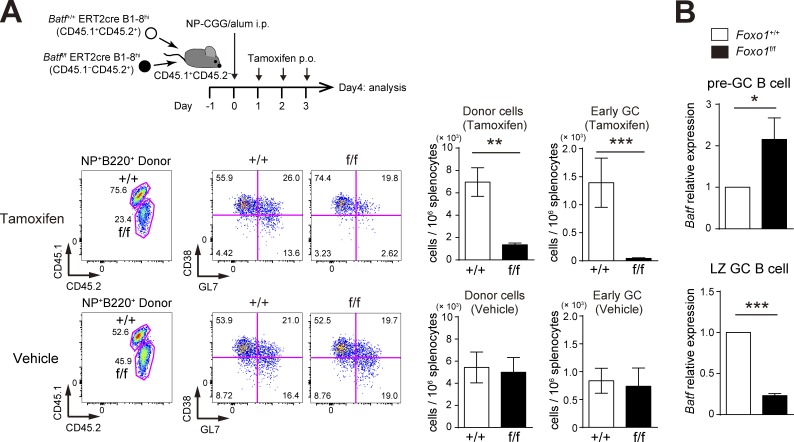
Next, to address the function of BATF in GC maintenance, we used the experimental protocol depicted in Fig. 8 A; both Batff/f ERT2cre B1-8hi and Batf+/+ ERT2cre B1-8hi B cells were co-transferred into congenically marked mice, which were immunized with NP-CGG, and then treated with tamoxifen to delete Batf in GC B cells (Fig. 8 D). As shown in Fig. 8 B, in contrast to control vehicle treatment, tamoxifen injection resulted in a selective decrease of Batff/f ERT2cre B1-8hi GC B cells; the decrease was more prominent in the DZ. Furthermore, when EdU was incorporated over 0.5 h, BATF-deficient LZ GC B cells showed less proliferation than control cells (Fig. 8 C). Thus, BATF is up-regulated by T cell help in a Foxo1-dependent manner, and is required for GC B cell proliferation.
Figure 8.
BATF is required for GC maintenance. (A) Schematic illustration of the experimental protocol for B–D. (B, top) Flow cytometry of NP-specific donor B cells (CD45.2+B220+NP+). (bottom left) Histograms showing the donor GC B cell (CD45.2+B220+NP+Fas+CD38−) number in 106 splenocytes. (bottom right) The ratio of DZ:LZ cells. n = 4 biological replicates. (C) Proliferation status of Batf+/+ and Batff/f LZ GC B cells assessed by EdU incorporation 30 min after an EdU injection. n = 4 biological replicates. (D) Real-time qPCR analysis of Batf mRNA expression in Batf+/+ERT2cre B1-8hi and Batff/f ERT2cre B1-8hi GC B cells. n = 4 biological replicates. (E) Schematic illustration of the experimental protocol for F. Note that viral infection efficiencies were comparable between samples as assessed by GFP+ population (∼15–20%) before transfer. (F, left) Flow cytometry of CD45.1+B220+ cells. (right) Histograms showing the number of GFP+ GC B cell (CD45.1+B220+GFP+CD38−) in 106 splenocytes. n = 8 (Foxo1+/+) and 10 (Foxo1f/f) biological replicates. Error bars represent SD. Data are representative of two independent experiments (B and C), from one experiments with three biological replicates (D), representative of four independent experiments (F, left), and are pooled from four independent experiments (F, right). *, P < 0.05; **, P < 0.01; ***, P < 0.001; paired Student’s t test (B and C) and unpaired Student’s t test (D and F).
The aforementioned findings raise the question of whether Foxo1 ablation-mediated anomalies could be explained by the defective expression of BATF. To explore this question, we used a retroviral transduction system, allowing for both inducible expression of BATF and inducible deletion of endogenous Foxo1 (Fig. 8 E). Retrovirus-infected donor B cells were transferred into congenically marked mice, which were immunized and then treated with tamoxifen. Transduction with BATF was monitored by GFP, and deletion of Foxo1 in Foxo1f/f donor B cells was confirmed by RT-qPCR (unpublished data). Foxo1-proficient cells transduced by BATF exhibited not significant expansion of GC B cells, suggesting that, in these proficient conditions, BATF is not a limiting factor. Foxo1-deficient cells transduced by empty retrovirus manifested the expansion defect, as expected, whereas this defect was restored by transduction of BATF, suggesting that BATF is critical for the effects of Foxo1 on GC proliferation (Fig. 8 F). Nevertheless, Foxo1 ablation-induced polarization defect could not be restored, consistent with the previous data that CXCR4 is a direct target of Foxo1 (Dubrovska et al., 2012; Dominguez-Sola et al., 2015). Thus, the LZ-dominant phenotype induced by BATF ablation (Fig. 8 B) is likely a result of secondary effects caused by proliferation anomalies, but not by direct down-regulation of CXCR4 as observed in Foxo1 ablation.
Foxo1 acts in a context-dependent manner
The aforementioned results raised the question of why Foxo1 ablation leads to opposite proliferation outcomes, depending on whether it occurs in the preGC or GC stage (Fig. 1 C and 2 B). To gain insight into this point, we performed BATF deletion experiments at the preGC stage. Expansion of BATF-deficient B1-8hi B cells was significantly decreased after tamoxifen treatment (Fig. 9 A), similar to what was observed with BATF deletion at the GC phase. However, effects of Foxo1 deletion on BATF expression differs between the preGC and GC stages. As shown in Fig. 9 B, in preGC B cells BATF was up-regulated in Foxo1-deficient B1-8hi B cells. A straightforward explanation of these results is that the mechanisms by which Foxo1 regulates BATF differ between preGC (inhibitory) and GC (stimulatory) phases, although the role of BATF in B cell proliferation is similar between these two phases. Hence, our data suggest that the regulatory network operating through Foxo1 is highly context-specific.
Figure 9.
BATF is required for preGC B cell expansion and is up-regulated in Foxo1-deficient preGC B cells. (A, top) Schematic illustration of the experimental protocol. Batf+/+ ERT2cre B1-8hi (CD45.1+CD45.2+) and Batff/f ERT2cre B1-8hi (CD45.1−CD45.2+) B cells were co-transferred into recipient mice (CD45.1+CD45.2−), which were immunized with NP-CGG/alum i.p. on day 0. Mice were administered tamoxifen or vehicle p.o. for 3 d and analyzed on day 4. (bottom left) Flow cytometry of NP-specific donor B cells (CD45.2+B220+NP+). (bottom right) Histograms showing the cell number of total donor cells (CD45.2+NP+B220+) and early GC B cells (CD45.2+NP+B220+GL7+CD38−) in 106 splenocytes. n = 4 biological replicates. (B) Real-time qPCR analysis of Batf mRNA expression in control and Foxo1-deficient B1-8hi preGC (donor B220+NP+CD38+GL7+) and LZ GC B cells (donor B220+NP+GL7+CD38−CD86hiCXCR4lo). preGC B cells and LZ GC B cells were prepared from mice as described in Fig. 1 C and Fig. 2 A, respectively. n = 3 biological replicates. Error bars represent SD. Data are representative of three (A) or two independent experiments (B). *, P < 0.05; **, P < 0.01; ***, P < 0.001; paired Student’s t test (A) and unpaired Student’s t test (B).
Discussion
By focusing on Foxo1 functions in GC B cells, we demonstrate that Foxo1 participates in two consecutive processes occurring in the LZ, antigen presentation to GC TFH cells and subsequent activation through T cell help, and that through these mechanisms Foxo1 regulates the GC B cell proliferation program. The latter conclusion is seemingly inconsistent with previous studies demonstrating apparently normal expansion of Foxo1-deficient GC B cells (Dominguez-Sola et al., 2015; Sander et al., 2015). By ablating Foxo1 before or after GC establishment, we found that, in contrast to ablation at the GC stage, hyper-expansion of preGC B cells was induced by Foxo1 deletion at the preGC stage. Therefore, considering that class-switching begins at the preGC stage (Toellner et al., 1998; Shlomchik and Weisel, 2012), it is likely that the previous studies using a Cγ1-cre have led to deletion of Foxo1 in preGC cells, at least to some extents. The observed phenotype would thus result from a combination of effects of Foxo1 deletion at preGC and GC cell stages.
GC B cells expressing higher amounts of surface pMHC-II have been thought to receive better quality/quantity of help from GC TFH cells, thereby contributing to their preferential proliferation. Indeed, it was shown that B cells deficient in the MHC family molecule H2-O, an inhibitor of peptide loading into MHC-II, outcompete H2-O–sufficient B cells in mixed knockout and wild-type GCs, even under conditions in which BCR affinity for NP was equalized (Draghi and Denzin, 2010). Conversely, Foxo1-deficient GC B cells with lower levels of surface pMHC-II were less capable of activating cognate GC TFH cells. This could be most likely caused by down expression of BCR in the absence of Foxo1. Supporting this possibility, we found that when a T cell antigen was targeted to DEC205, instead of BCR, on GC B cells, similar surface pMHC-II presentation was induced between mutant and control GC cells.
How does Foxo1 contribute to proper BCR expression by LZ GC B cells? Because IgH, and Igα, but not Igβ are expressed to the same extents at the protein level in Foxo1-deficient LZ GC B cells as in controls (unpublished data), the lower level of Igβ is most likely responsible for the lower BCR levels on the surface of mutant LZ GC B cells. Indeed, in developing B cells, the importance of Igβ for the assembly and the cell surface expression of the preBCR has been documented (Gong and Nussenzweig, 1996; Kurosaki, 2002). The protein, but not mRNA, level of Igβ was decreased in Foxo1-deficient GC cells (unpublished data). In this regard, Itch, one of the HECT family of E3 ligases, was reported to mediate the constitutive ubiquitinylation of Igβ, and that this was required for normal sorting through the endocytic pathway (Zhang et al., 2007). Hence, one possibility is that such ubiquitin-mediated mechanisms might control constitutive Igβ protein levels in a Foxo1-dependent manner.
By taking a strategy of providing T cell help by targeting a T cell antigen to GC B cells through DEC205, we could show that even after receipt of sufficient T cell help, Foxo1-deficient GC B cells cannot induce the normal proliferation program. Thus, in in vivo physiological settings, the malfunctioning DZ program in the Foxo1-deficient LZ GC cells is likely to result from two defects, the mutant B cells receive weaker help from GC TFH cells and are less capable of responding to it. Therefore, our data are consistent with a model in which T cell help plays a critical role in the LZ-to-DZ switch.
The DZ is believed to be tightly linked with the cell cycle; indeed, expression of cell cycle–related genes is increased in the DZ (Victora et al., 2012). Here, B cells in GCs devoid of the DZ as a result of the absence of Foxo1 manifest three features; 1) defective proliferation; 2) insufficient elevation of many cell cycle-related genes; and 3) defective induction of c-Myc and BATF, critical regulators for inducing the cell cycle progression. Because CXCR4 is a target of Foxo1, Foxo1 controls both access to the anatomical DZ and the proliferation program, possibly making it the key coupling molecule between these two DZ functions.
As expected because of defective GC B cell proliferation and defective expression of key enzymes involved in SHM, SHM is also perturbed in the absence of Foxo1. SHM and selection are thought to take place in distinct zones, the DZ and LZ, respectively. This spatial separation has also been suggested to be important for normal SHM, because GC B cells deficient in CXCR4 displayed defective SHM, despite apparently normal cell division. Hence, in the case of Foxo1-deficient GC B cells, in addition to the aforementioned defect, the inability to physically separate sites of SHM and selection might also contribute to defective SHM.
BATF belongs to the AP-1 family of transcription factors that include Fos, Jun, and Atf (Dorsey et al., 1995). A previous study demonstrates that BATF is required for GCs (defined by GL7+Fas+ B cells in this study) in a T cell– but not B cell–dependent manner (Ise et al., 2011). CD38, rather than Fas, is a better marker to delineate these cells. Indeed, both GL7+CD38+ (intermediate cells toward mature GL7+CD38− GCs) and GL7+CD38− cells express FAS to the same extents (Taylor et al., 2012). Here, by using the GL7+CD38− marker, we showed the requirement for B cell–dependent BATF in mature GCs; BATF-deficient B cells, when singly transferred, gave rise to GL7+CD38+, but not GL7+CD38− GCs (unpublished data). Thus, it is likely that most of the GL7+Fas+ B cells detected by the previous study could correspond to GL7+CD38+ cells.
As observed in the previous study (Dominguez-Sola et al., 2015), we also found that BATF expression is severely reduced in Foxo1-deficient LZ GC B cells. An additional three lines of evidence allow us to conclude that the GC proliferation defect seen in the absence of Foxo1 is due, at least in part, to defective up-regulation of BATF. First, BATF is up-regulated by enforced T cell help in a Foxo1-dependent manner. Second, ablation of BATF during the GC reaction, like Foxo1 ablation, led to proliferation defect. Finally, forced expression of BATF restores the defective GC expansion induced by Foxo1 ablation. Because BATF lacks a transcriptional activation domain, it acts through its interactions with other Jun or IRF family members (Murphy et al., 2013). Probably, not appreciably increase in the GC expansion by overexpression of BATF in Foxo1-proficient cells could be explained by requirement for other partners to exert the BATF action. Despite the importance of BATF, it is clearly only one of several critical factors underlying the defective proliferation induced by Foxo1 ablation. For instance, incomplete up-regulation of c-Myc in the absence of Foxo1 also appears to contribute to.
In regard to regulation of BATF by Foxo1, no significant binding of Foxo1 to Batf locus was observed in human GC B cells in a previous ChIP-seq analysis (Dominguez-Sola et al., 2015). Instead, our gene set enrichment analysis showed that Foxo1-deficient LZ GC B cells were more weakly imprinted by BCR and CD40 signature genes, simply suggesting the involvement of Foxo1 in BCR and CD40 signaling. Moreover, BATFhi expression is restricted to a small fraction of LZ GC B cells, which also express c-Myc. The c-Myc+ GC B cell fraction has been reported to be enriched for high affinity BCRs, which are required for strong T cell help. Taking these observations together, we speculate that, rather than direct transcriptional regulation of BATF by Foxo1, Foxo1 is required for generating strong BCR and CD40 signaling by modulating the expression of such receptors, associated signaling molecules, and/or transcriptional regulators, which in turn, leads to up-regulation of BATF.
Considering the previous conventional idea that Foxo1 acts as a tumor suppressor, it has been puzzling why GC-derived diffuse large B cell lymphomas frequently carry mutations in Foxo1 that prevent its inactivation by Akt (Trinh et al., 2013). A large number of Foxo1-bound sites was reported to co-localize with sites bound by Bcl6, suggesting that Foxo1 and Bcl6 cooperate in the transcriptional modulation of a subset of targets (Dominguez-Sola et al., 2015). Such modulation of Foxo1 activity by Bcl6 likely makes Foxo1 exert an unconventional proliferative role specifically in GC B cells, providing an explanation for the generation of GC-derived lymphomas by active Foxo1.
Materials and methods
Mice
Foxo1f/f (Paik et al., 2007), B1-8hi (Shih et al., 2002), c-Myc-GFP (Huang et al., 2008), and Cd19cre/+ (Rickert et al., 1997) mice were provided by R.A. DePinho (The University of Texas MD Anderson Cancer Center, Houston, TX), M.C. Nussenzweig (Tha Rockefeller University, New York, NY), B.P. Sleckman (Washington University School of Medicine, St. Louis, MO), and K. Rajewsky (Max Delbruck Center for Molecular Medicine in the Helmholtz Alliance, Berlin-Buch, Germany), respectively. Rosa26-ERT2cre mice were obtained from Taconic Farms. Il21+/− and OT-II mice were obtained from The Jackson Laboratory. Batff/f (Kuwahara et al., 2016) and μMT (Kitamura et al., 1991) mice were described previously. Aicdaf/f mice were generated by homologous recombination using Bruce4 ES cells so that exons 2 and 3 were flanked with two loxp sites. Positive ES clones were used for microinjection to obtain chimeric mice, which were then crossed with C57BL/6 mice to obtain germline transmitted animals. C57BL/6 mice were purchased from CLEA Japan. Sex-matched 8–15-wk-old mice were used for all the experiments. For the experiments with mixed bone marrow chimeras, C57BL/6 mice were lethally irradiated (8.5 Gy) and reconstituted with a mixed inoculum of 80% μMT and 20% Il21+/− bone marrow cells at least 8 wk before immunization. All mice were bred and maintained under specific pathogen–free conditions and all animal experiments were performed under the institutional guidelines of the Institute of Physical and Chemical Research Research Institute, Osaka University, and Washington University in St. Louis.
Immunization
Mice were immunized with 100 µg of NP-CGG or NP-OVA precipitated with Imject alum (Thermo Fisher Scientific), or with ∼5 × 108 SRBCs by i.p. injection. The deletion of the loxP-flanked allele of the target genes was induced by oral administration (p.o.) of 2 mg tamoxifen (Sigma-Aldrich) in sunflower oil (Sigma-Aldrich) once per day for 3 d.
Flow cytometry analysis
Single-cell suspensions of splenocytes were prepared and analyzed as previously described (Inoue et al., 2015). Anti-B220 (RA3-6B2), IgG1 (A85-1), CD138 (281–2), CD45.2 (104), IgM[a] (DS-1), PD-1 (J43), CXCR5 (2G8), CD19 (1D3), and Fas (Jo2) were purchased from BD. Anti-GL7 (GL7), CXCR4 (2B11), CD4 (GK1.5), Igα (HM47), MHC-II (M5/114.15.2), CD16/32 (93), ICOSL (HK5.3), CD80 (16-10A1), IgD (11-26c), and AID (mAID-2) were purchased from eBioscience. Anti-CD38 (90), CD86 (GL-1), CD45.1 (A20), Igβ (HM79-12), CD22 (OX-97), CD40 (3/23), CD83 (Michel-19), CCR6 (29-2L17), and Brilliant Violet 510 Streptavidin were purchased from BioLegend. For staining with an isotype control for CD86 (Fig. 4 C), we used anti-CD83 instead of anti-CD86 for gating LZ GC B cells. For intracellular staining, the cells were fixed and permeabilized using a Foxp3 staining kit (eBioscience) for anti-Foxo1 (C29H4; Cell Signaling Technology) staining, or a True-Nuclear Transcription Factor Buffer Set (BioLegend) for anti-BATF (D7C5; Cell Signaling Technology) staining. Cells were incubated with first antibody, and then incubated with Alexa Fluor 488– or Alexa Fluor 647–conjugated anti–rabbit antibody (Thermo Fisher Scientific). APC-conjugated NP was prepared as described previously (Shinnakasu et al., 2016). For EdU incorporation assays, mice were injected i.p. with 1 mg EdU (Thermo Fisher Scientific) in PBS and sacrificed 30 min later. Splenocytes were surface-stained, and then EdU-incorporated cells were identified using a Click-iT Plus EdU Flow Cytometry Assay kit (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Adoptive transfers
Adoptive transfer experiments were performed as described previously (Shinnakasu et al., 2016). In brief, splenic B cells were purified by magnetic cell depletion using anti-CD43 MicroBeads and the AutoMACS system (Miltenyi Biotec). For B1-8hi B cell transfer experiments, purified B1-8hi B cells containing 105 NP-binders (a 1:1 mixture of 5 × 104 NP-binders for co-transfer experiments) were transferred i.v. into recipient mice. For B cell co-transfer experiments in Figs. 4 and 6 B, 107 purified B cells (a 1:1 mixture of 5 × 106 B cells) were used for donor.
SHM sequence analysis of the JH4 intron
Mice were co-transferred with Foxo1+/+ ERT2cre (CD45.1+CD45.2–) and Foxo1f/f ERT2cre (CD45.1+CD45.2+) B cells, and immunized with NP-CGG/alum on day 0. On day 7, 8, and 9, mice were orally administered tamoxifen. On day 14, LZ GC B cells from each donor were sorted (∼103 cells) for genomic DNA purification. JH4 intron sequences were amplified using Ex Taq DNA polymerase (Takara Bio Inc.) with primers 5′-TCCTAGGAACCAACTTAAGAGT-3′ and 5′-TGGAGTTTTCTGAGCATTGCAG-3′ (Gitlin et al., 2014). PCR products were subcloned into the pGEM-T vector (Promega) and single clones (∼50 per mouse) were sequenced.
In vitro antigen presentation assay
Purified splenic B cells (∼2.5 × 106 cells) were incubated with 1 µg/ml NP-Eα-GFP, Eα-GFP (Itano et al., 2003; Pape et al., 2007), αDEC-OVA (Boscardin et al., 2006), or αDEC-Eα-GFP for 1 h at 37°C. Surface pMHC-II was detected by staining the NP-specific LZ GC B cells with biotin-conjugated Y-Ae antibody (eBioscience). For generation of αDEC-Eα-GFP, cDNA encoding Eα-GFP was inserted in frame with the carboxyl terminus of the heavy chain of mouse αDEC-205 (Boscardin et al., 2006), and the fusion antibody was expressed using Expi293 Expression System (Thermo Fisher Scientific) and purified on HiTrap Protein G HP column (GE Healthcare).
RNA-seq analysis
Wild-type mice were co-transferred with Foxo1+/+ ERT2cre B1-8hi (CD45.1+CD45.2−) and Foxo1f/f ERT2cre B1-8hi (CD45.1+CD45.2+) B cells, and immunized with NP-CGG precipitated in alum on day 0. On day 7, 8, and 9, mice were orally administered tamoxifen. On day 12, Foxo1+/+ LZ (CD45.1+CD45.2−B220+NP+GL7+CD38−CD86hiCXCR4lo) and DZ GC B cells (CD45.1+CD45.2–B220+NP+GL7+CD38−CD86loCXCR4hi), and Foxo1f/f GC B cells (CD45.1+CD45.2−B220+NP+GL7+CD38−) were sorted for RNA preparation. Three biological replicates were used in each genotype. Construction of DNA libraries for RNA-seq and sequencing were performed as described previously (Shinnakasu et al., 2016). In brief, the DNA library was constructed using an NEBNext Ultra RNA Library Prep kit for Illumina (NEB) from total RNA purified from ∼104 sorted cells. RNA-sequencing was performed on a HiSeq 1500 sequencer (Illumina) in a 49-bp single-end read mode. Gene set enrichment analysis was performed using GSEA software v2.2.2 (Subramanian et al., 2005). The lists of gene signature of up-regulated genes by CD40 and BCR stimulation for gene set enrichment analysis were previously described (Victora et al., 2010; Shinnakasu et al., 2016). The RNA-seq data are available at Gene Expression Omnibus database under accession no. GSE93554.
Anti–DEC-OVA and anti-CD40L treatment
For anti–DEC-OVA treatment, wild-type mice were co-transferred with Foxo1+/+ ERT2cre B1-8hi (CD45.1+CD45.2−) and Foxo1f/f ERT2cre B1-8hi (CD45.1+CD45.2+) B cells, and immunized with NP-OVA precipitated in alum on day 0. On day 7, 8, and 9, mice were orally administered tamoxifen. The mice were injected i.p. with 10 µg anti–DEC-OVA (Victora et al., 2010) on day 10, and sacrificed for analysis on day 12. As a control, we used a chimeric antibody in which anti-DEC205 was fused to an irrelevant antigen, Plasmodium falciparum circumsporozoite protein (anti–DEC-CS). For anti-CD40L treatment, wild-type mice were immunized i.p. with NP-CGG in alum on day 0. The mice were injected i.p. with 250 µg IgG1 anti-CD40L (MR1; Bio X Cell) or control hamster IgG (Innovative Research) on day 7 and sacrificed for analysis 12 h later.
Immunohistochemistry
Spleens were fixed in 4% paraformaldehyde for 2 h on ice, and then incubated in PBS overnight at 4°C. Next, the spleens were embedded in Tissue-Tek OCT Compound (Sakura Finetek) and frozen at –80°C. 8-µm-thick sections were mounted on glass slides and blocked with CAS-Block (Invitrogen) for 10 min at room temperature. The samples were first stained with biotin anti-CD45.1, followed by incubation with Alexa Fluor 546 streptavidin (Thermo Fisher Scientific), eFluor 450 anti-IgD (11-26c; eBioscience), and APC anti-CD21/35 (7G6; BD). Images were acquired using a Fluoview FV10i confocal microscope system (Olympus). CD45.1 signal intensity was quantified using ImageJ software (National Institutes of Health), by which relative mean intensity values of CD45.1 signals in the DZ compared with those in the LZ was calculated.
Western blot
Sample preparation and Western blot were performed as described previously (Inoue et al., 2015). Immunoblotting was performed using anti-Foxo1 (C29H4; Cell Signaling Technology) and anti–β-actin (C-11; Santa Cruz Biotechnology, Inc.).
qPCR analysis
Real-time qPCR was performed as described previously (Inoue et al., 2015). Actb mRNA levels were used for normalization. The following primers were used for qPCR analysis: Foxo1 forward, 5′-AAGAGCGTGCCCTACTTCAA-3′; Foxo1 reverse, 5′-CTGTTGTTGTCCATGGACGC-3′; Foxo3 forward, 5′-AGGATAAGGGCGACAGCAAC-3′; Foxo3 reverse, 5′-CCCGTGCCTTCATTCTGA-3′; Foxo4 forward, 5′-CTTCCTCGACCAGACCTCG-3′; Foxo4 reverse, 5′-ACAGGATCGGTTCGGAGTGT-3′; Il21 forward, 5′-TCAGCTCCACAAGATGTAAAGGG-3′; Il21 reverse, 5′-GGGCCACGAGGTCAATGAT-3′; Batf forward, 5′-GTTCTGTTTCTCCAGGTCC-3′; Batf reverse, 5′-GAAGAATCGCATCGCTGC-3′; Actb forward, 5′-CCGCCACCAGTTCGCCATG-3′; Actb reverse, 5′-TACAGCCCGGGGAGCATCGT-3′.
Inducible retroviral expression of BATF in vivo
To inducibly express BATF in Foxo1-deficient B cells in vivo, Foxo1+/+ ERT2cre B1-8hi (CD45.1+CD45.2+) or Foxo1f/f ERT2cre B1-8hi (CD45.1+CD45.2+) mice were immunized i.p. with 50 µg of NP-Ficoll in PBS. 6 h later, splenic B cells were purified from these mice and cultured with 2 µg/ml anti-CD40 for 18 h in vitro. Cells were then retrovirally transduced with an inducible BATF expression cassette by spin infection (800 g, 90 min, 25°C) with polybrene (8 µg/ml, Millipore) and virus supernatant produced in PLAT-E cells. The retroviral vector was constructed by inserting loxp-flanked human CD2, mouse BATF cDNA, and IRES-EGFP into pMYs vector (Fig. 8 E). 3 h after infection, 106 B cells were transferred i.v. into CD45.1−CD45.2+ wild-type mice, which were immunized i.p. with 100 µg of NP-CGG/alum on day 0, followed by tamoxifen injection p.o. on day 6, 7, and 8 to induce Foxo1 deletion and BATF expression, and analysis on day 15.
Statistical analysis
Statistical analyses were performed with a two-tailed unpaired Student’s t test, a two-tailed paired Student’s t test, or a Mann-Whitney test using GraphPad Prism software.
Acknowledgments
We thank R.A. DePinho for Foxo1f/f mice, M.C. Nussenzweig for B1-8hi mice, anti–DEC-OVA, and anti–DEC-CS, K. Rajewsky for Cd19cre/+ mice, B.P. Sleckman for c-Myc-GFP mice, G.D. Victora (The Rockefeller University, New York, NY) for gene sets for the gene set enrichment analysis, M.K. Jenkins (University of Minnesota Medical School, Minneapolis, MN) for Eα-GFP construct, Y. Baba, K. Kometani, M. Tochigi, N. Takahashi, and S. Moriyama for technical advice and assistance, and P.D. Burrows for critical reading of the manuscript.
This work was supported by grants to T. Inoue, R. Shinnakasu, W. Ise, and T. Kurosaki from the Ministry of Education, Culture, Sports, Science and Technology in Japan and to T. Egawa from the National Institutes of Health (AI097244-01A1 and AI114593-01A1).
The authors declare no competing financial interests.
Author contributions: T. Inoue, R. Shinnakasu, W. Ise, and T. Kurosaki designed the research; T. Inoue, R. Shinnakasu, and W. Ise performed the experiments with technical assistance of C. Kawai; T. Egawa provided Il21r+/− mice and technical advice; and T. Inoue and T. Kurosaki wrote the paper with the assistance of all other authors.
Footnotes
Abbreviations used:
- CGG
- chicken γ globulin
- DZ
- dark zone
- EdU
- 5-ethynyl-2’-deoxyuridine
- FDC
- follicular DC
- FPKM
- fragments per kilobase of exon per million reads
- GC
- germinal center
- gMFI
- geometric mean fluorescence intensity
- LZ
- light zone
- NP
- 4-hydroxy-3-nitrophenylacetyl
- SHM
- somatic hypermutation
- SRBC
- sheep red blood cell
- TFH
- follicular helper T
References
- Allen C.D., Okada T., Tang H.L., and Cyster J.G.. 2007. Imaging of germinal center selection events during affinity maturation. Science. 315:528–531. 10.1126/science.1136736 [DOI] [PubMed] [Google Scholar]
- Bannard O., Horton R.M., Allen C.D., An J., Nagasawa T., and Cyster J.G.. 2013. Germinal center centroblasts transition to a centrocyte phenotype according to a timed program and depend on the dark zone for effective selection. Immunity. 39:912–924. 10.1016/j.immuni.2013.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaz L., Bonnyay D., Mahnke K., Rivera M., Nussenzweig M.C., and Steinman R.M.. 2002. Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. J. Exp. Med. 196:1627–1638. 10.1084/jem.20021598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscardin S.B., Hafalla J.C., Masilamani R.F., Kamphorst A.O., Zebroski H.A., Rai U., Morrot A., Zavala F., Steinman R.M., Nussenzweig R.S., and Nussenzweig M.C.. 2006. Antigen targeting to dendritic cells elicits long-lived T cell help for antibody responses. J. Exp. Med. 203:599–606. 10.1084/jem.20051639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado D.P., Sasaki Y., Godinho S.A., Pellerin A., Köchert K., Sleckman B.P., de Alborán I.M., Janz M., Rodig S., and Rajewsky K.. 2012. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat. Immunol. 13:1092–1100. 10.1038/ni.2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers J., and Rabbitts T.H.. 2015. LMO2 at 25 years: a paradigm of chromosomal translocation proteins. Open Biol. 5:150062 10.1098/rsob.150062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansen T.B., and Burgering B.M.. 2008. Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 18:421–429. 10.1016/j.tcb.2008.07.004 [DOI] [PubMed] [Google Scholar]
- De Silva N.S., and Klein U.. 2015. Dynamics of B cells in germinal centres. Nat. Rev. Immunol. 15:137–148. 10.1038/nri3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D., Victora G.D., Ying C.Y., Phan R.T., Saito M., Nussenzweig M.C., and Dalla-Favera R.. 2012. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat. Immunol. 13:1083–1091. 10.1038/ni.2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D., Kung J., Holmes A.B., Wells V.A., Mo T., Basso K., and Dalla-Favera R.. 2015. The FOXO1 Transcription Factor Instructs the Germinal Center Dark Zone Program. Immunity. 43:1064–1074. 10.1016/j.immuni.2015.10.015 [DOI] [PubMed] [Google Scholar]
- Dorsey M.J., Tae H.J., Sollenberger K.G., Mascarenhas N.T., Johansen L.M., and Taparowsky E.J.. 1995. B-ATF: a novel human bZIP protein that associates with members of the AP-1 transcription factor family. Oncogene. 11:2255–2265. [PubMed] [Google Scholar]
- Draghi N.A., and Denzin L.K.. 2010. H2-O, a MHC class II-like protein, sets a threshold for B-cell entry into germinal centers. Proc. Natl. Acad. Sci. USA. 107:16607–16612. 10.1073/pnas.1004664107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovska A., Elliott J., Salamone R.J., Telegeev G.D., Stakhovsky A.E., Schepotin I.B., Yan F., Wang Y., Bouchez L.C., Kularatne S.A., et al. 2012. CXCR4 expression in prostate cancer progenitor cells. PLoS One. 7:e31226 10.1371/journal.pone.0031226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin A.D., Shulman Z., and Nussenzweig M.C.. 2014. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 509:637–640. 10.1038/nature13300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin A.D., Mayer C.T., Oliveira T.Y., Shulman Z., Jones M.J., Koren A., and Nussenzweig M.C.. 2015. HUMORAL IMMUNITY. T cell help controls the speed of the cell cycle in germinal center B cells. Science. 349:643–646. 10.1126/science.aac4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S., and Nussenzweig M.C.. 1996. Regulation of an early developmental checkpoint in the B cell pathway by Ig beta. Science. 272:411–414. 10.1126/science.272.5260.411 [DOI] [PubMed] [Google Scholar]
- Hedrick S.M. 2009. The cunning little vixen: Foxo and the cycle of life and death. Nat. Immunol. 10:1057–1063. 10.1038/ni.1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.Y., Bredemeyer A.L., Walker L.M., Bassing C.H., and Sleckman B.P.. 2008. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur. J. Immunol. 38:342–349. 10.1002/eji.200737972 [DOI] [PubMed] [Google Scholar]
- Inoue T., Morita M., Hijikata A., Fukuda-Yuzawa Y., Adachi S., Isono K., Ikawa T., Kawamoto H., Koseki H., Natsume T., et al. 2015. CNOT3 contributes to early B cell development by controlling Igh rearrangement and p53 mRNA stability. J. Exp. Med. 212:1465–1479. 10.1084/jem.20150384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise W., Kohyama M., Schraml B.U., Zhang T., Schwer B., Basu U., Alt F.W., Tang J., Oltz E.M., Murphy T.L., and Murphy K.M.. 2011. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol. 12:536–543. 10.1038/ni.2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itano A.A., McSorley S.J., Reinhardt R.L., Ehst B.D., Ingulli E., Rudensky A.Y., and Jenkins M.K.. 2003. Distinct dendritic cell populations sequentially present antigen to CD4 T cells and stimulate different aspects of cell-mediated immunity. Immunity. 19:47–57. 10.1016/S1074-7613(03)00175-4 [DOI] [PubMed] [Google Scholar]
- Kitamura D., Roes J., Kühn R., and Rajewsky K.. 1991. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 350:423–426. 10.1038/350423a0 [DOI] [PubMed] [Google Scholar]
- Kurosaki T. 2002. Regulation of B cell fates by BCR signaling components. Curr. Opin. Immunol. 14:341–347. 10.1016/S0952-7915(02)00344-8 [DOI] [PubMed] [Google Scholar]
- Kuwahara M., Ise W., Ochi M., Suzuki J., Kometani K., Maruyama S., Izumoto M., Matsumoto A., Takemori N., Takemori A., et al. 2016. Bach2-Batf interactions control Th2-type immune response by regulating the IL-4 amplification loop. Nat. Commun. 7:12596 10.1038/ncomms12596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Xu H., Shih C., Wan Z., Ma X., Ma W., Luo D., and Qi H.. 2015. T-B-cell entanglement and ICOSL-driven feed-forward regulation of germinal centre reaction. Nature. 517:214–218. 10.1038/nature13803 [DOI] [PubMed] [Google Scholar]
- Murphy D.B., Rath S., Pizzo E., Rudensky A.Y., George A., Larson J.K., and Janeway C.A. Jr. 1992. Monoclonal antibody detection of a major self peptide. MHC class II complex. J. Immunol. 148:3483–3491. [PubMed] [Google Scholar]
- Murphy T.L., Tussiwand R., and Murphy K.M.. 2013. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 13:499–509. 10.1038/nri3470 [DOI] [PubMed] [Google Scholar]
- Paik J.H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., et al. 2007. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 128:309–323. 10.1016/j.cell.2006.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape K.A., Catron D.M., Itano A.A., and Jenkins M.K.. 2007. The humoral immune response is initiated in lymph nodes by B cells that acquire soluble antigen directly in the follicles. Immunity. 26:491–502. 10.1016/j.immuni.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Rickert R.C., Roes J., and Rajewsky K.. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25:1317–1318. 10.1093/nar/25.6.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safford M., Collins S., Lutz M.A., Allen A., Huang C.T., Kowalski J., Blackford A., Horton M.R., Drake C., Schwartz R.H., and Powell J.D.. 2005. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat. Immunol. 6:472–480. 10.1038/ni1193 [DOI] [PubMed] [Google Scholar]
- Sagardoy A., Martinez-Ferrandis J.I., Roa S., Bunting K.L., Aznar M.A., Elemento O., Shaknovich R., Fontán L., Fresquet V., Perez-Roger I., et al. 2013. Downregulation of FOXP1 is required during germinal center B-cell function. Blood. 121:4311–4320. 10.1182/blood-2012-10-462846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander S., Chu V.T., Yasuda T., Franklin A., Graf R., Calado D.P., Li S., Imami K., Selbach M., Di Virgilio M., et al. 2015. PI3 Kinase and FOXO1 Transcription Factor Activity Differentially Control B Cells in the Germinal Center Light and Dark Zones. Immunity. 43:1075–1086. 10.1016/j.immuni.2015.10.021 [DOI] [PubMed] [Google Scholar]
- Schwickert T.A., Victora G.D., Fooksman D.R., Kamphorst A.O., Mugnier M.R., Gitlin A.D., Dustin M.L., and Nussenzweig M.C.. 2011. A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. J. Exp. Med. 208:1243–1252. 10.1084/jem.20102477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih T.A., Roederer M., and Nussenzweig M.C.. 2002. Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nat. Immunol. 3:399–406. 10.1038/ni776 [DOI] [PubMed] [Google Scholar]
- Shinnakasu R., Inoue T., Kometani K., Moriyama S., Adachi Y., Nakayama M., Takahashi Y., Fukuyama H., Okada T., and Kurosaki T.. 2016. Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol. 17:861–869. 10.1038/ni.3460 [DOI] [PubMed] [Google Scholar]
- Shlomchik M.J., and Weisel F.. 2012. Germinal center selection and the development of memory B and plasma cells. Immunol. Rev. 247:52–63. 10.1111/j.1600-065X.2012.01124.x [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.J., Pape K.A., and Jenkins M.K.. 2012. A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J. Exp. Med. 209:597–606. 10.1084/jem.20111696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toellner K.M., Luther S.A., Sze D.M., Choy R.K., Taylor D.R., MacLennan I.C., and Acha-Orbea H.. 1998. T helper 1 (Th1) and Th2 characteristics start to develop during T cell priming and are associated with an immediate ability to induce immunoglobulin class switching. J. Exp. Med. 187:1193–1204. 10.1084/jem.187.8.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh D.L., Scott D.W., Morin R.D., Mendez-Lago M., An J., Jones S.J., Mungall A.J., Zhao Y., Schein J., Steidl C., et al. 2013. Analysis of FOXO1 mutations in diffuse large B-cell lymphoma. Blood. 121:3666–3674. 10.1182/blood-2013-01-479865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., and Mesin L.. 2014. Clonal and cellular dynamics in germinal centers. Curr. Opin. Immunol. 28:90–96. 10.1016/j.coi.2014.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Victora G.D., Schwickert T.A., Fooksman D.R., Kamphorst A.O., Meyer-Hermann M., Dustin M.L., and Nussenzweig M.C.. 2010. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 143:592–605. 10.1016/j.cell.2010.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., Dominguez-Sola D., Holmes A.B., Deroubaix S., Dalla-Favera R., and Nussenzweig M.C.. 2012. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 120:2240–2248. 10.1182/blood-2012-03-415380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Veselits M., O’Neill S., Hou P., Reddi A.L., Berlin I., Ikeda M., Nash P.D., Longnecker R., Band H., and Clark M.R.. 2007. Ubiquitinylation of Ig beta dictates the endocytic fate of the B cell antigen receptor. J. Immunol. 179:4435–4443. 10.4049/jimmunol.179.7.4435 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Zha Y., Driessens G., Locke F., and Gajewski T.F.. 2012. Transcriptional regulator early growth response gene 2 (Egr2) is required for T cell anergy in vitro and in vivo. J. Exp. Med. 209:2157–2163. 10.1084/jem.20120342 [DOI] [PMC free article] [PubMed] [Google Scholar]