Quach and Quintana-Murci review the genetic and evolutionary history of our species, including how natural selection has shaped human genome diversity, and discuss the added value of population and functional genomic approaches in settings relevant to immunity and infection.
Abstract
More than a decade after the sequencing of the human genome, a deluge of genome-wide population data are generating a portrait of human genetic diversity at an unprecedented level of resolution. Genomic studies have provided new insight into the demographic and adaptive history of our species, Homo sapiens, including its interbreeding with other hominins, such as Neanderthals, and the ways in which natural selection, in its various guises, has shaped genome diversity. These studies, combined with functional genomic approaches, such as the mapping of expression quantitative trait loci, have helped to identify genes, functions, and mechanisms of prime importance for host survival and involved in phenotypic variation and differences in disease risk. This review summarizes new findings in this rapidly developing field, focusing on the human immune response. We discuss the importance of defining the genetic and evolutionary determinants driving immune response variation, and highlight the added value of population genomic approaches in settings relevant to immunity and infection.
Introduction
Studies of the history of the genus Homo have entered a golden age with the advent of genome-wide, deep-sequencing approaches. Surveys of genetic variation, such as the Exome Variant Server (Fu et al., 2013), the Exome Aggregation Consortium (Lek et al., 2016), the 1,000 Genomes Project (Auton et al., 2015), and the Simons Genome Diversity Project (Mallick et al., 2016), provide invaluable resources for the clinical interpretation of genetic variants identified in disease patients, and for dissections of the genetic architecture of morphological and physiological traits (Fig. 1, A and B). Genomic studies have also increased our understanding of the demographic history of our species (Veeramah and Hammer, 2014; Nielsen et al., 2017), including the contribution of interbreeding with archaic hominins, such as Neanderthals and Denisovans (Racimo et al., 2015), and the extent to which natural selection has acted on the human genome (Fan et al., 2016). This work has, in turn, provided insight into the mechanisms by which deleterious variants are removed from the population (Lohmueller, 2014; Henn et al., 2015) and the potential of humans to adapt to the broad range of environments they occupy (Jeong and Di Rienzo, 2014; Fan et al., 2016).
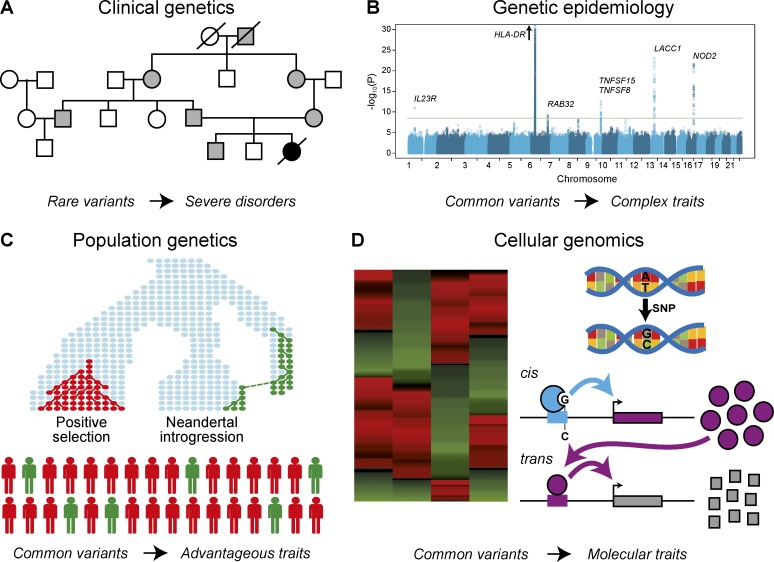
Figure 1.
Approaches to dissect the genetic basis of phenotypic traits and diseases. (A) Clinical genetics aims to identify rare mutations associated with severe clinical phenotypes. The genealogy illustrates the Mendelian inheritance of an autosomal recessive disorder, where one copy of the segregating disease mutation has no phenotype (gray), whereas two copies lead to the manifestation of the severe disorder (black). (B) Genetic epidemiology aims to dissect the genetic and environmental factors contributing to complex disease phenotypes by using population-based approaches. The Manhattan plot shows six loci associated with susceptibility to leprosy by a GWAS in the Chinese population (Liu et al., 2015). (C) Population genetics explores how past human demography, including admixture with archaic hominins, and natural selection have affected the genetic diversity of human populations. It represents a powerful approach to identify common variants associated with phenotypic traits that have played a major role in human survival and adaptation, including resistance to infectious diseases. (D) Cellular genomics allows the investigation of the genetic basis of intermediate molecular phenotypes, such as gene expression, which can be linked to ultimate organismal traits. The identification of genetic variants affecting gene expression in cis or trans, through the mapping of eQTL, helps to pinpoint cellular and molecular mechanisms involved in complex phenotypic traits, including immunity to infection.
Over the last decade, genomic scans of natural selection in the human genome have consistently identified genes and functions relating to immunity and host defense as recurrent targets of selection (Barreiro and Quintana-Murci, 2010; Quintana-Murci and Clark, 2013; Fumagalli and Sironi, 2014; Karlsson et al., 2014). There is also growing evidence to suggest that admixture with ancient hominins introduced advantageous immune variants into the modern human population (Racimo et al., 2015). Dissection of the action of selection on immune genes in settings relevant to immunity and infection, and investigations of the effects of genetic variants on responses to immune activation, through the mapping of expression quantitative trait loci (eQTL), are the key to identifying evolutionarily important determinants of host immune responsiveness and of the mechanisms underlying the diversity of immune phenotypes and disease risk (Fig. 1, C and D; Casanova et al., 2013; Fairfax and Knight, 2014).
This review focuses on the crucial contribution of genome-wide approaches to the understanding of human phenotypic diversity, by improving our knowledge of the demographic and adaptive history of our species, particularly as concerns pathogen pressures, and the admixture of Homo sapiens with archaic, now-extinct hominins, leading to the acquisition of advantageous variants. We will specially highlight recent advances in studies of the genetic, nongenetic, environmental, and evolutionary factors driving the inter-individual and inter-population phenotypic variability of immune responses, providing insight into the mechanisms underlying host immunity to infection and susceptibility to immune-related diseases.
Reconstructing the genomic history of Homo sapiens
Genomic surveys support an origin of modern humans in sub-Saharan Africa ∼200,000 yr ago, with subsequent dispersal out of Africa ∼40,000–80,000 yr ago, followed by the rapid colonization of South Asia, Australia, Europe, and East Asia (Novembre and Ramachandran, 2011; Veeramah and Hammer, 2014; Nielsen et al., 2017). Humans ultimately reached distant locations, such as the Americas, ∼15,000–35,000 yr ago, and the remote islands of Oceania, which were settled as recently as ∼1,000–4,000 yr ago (Fig. 2). African populations present the highest levels of diversity worldwide, and the diversity of non-African populations decreases with increasing distance from Africa (Abecasis et al., 2010, 2012; Campbell et al., 2014), attesting to the occurrence of bottlenecks and founder events during their migrations across the globe (Jakobsson et al., 2008; Henn et al., 2012). Recent studies based on whole-genome sequencing have shown that all contemporary non-Africans are descended from a single population that left Africa less than 60,000 yr ago (Malaspinas et al., 2016; Mallick et al., 2016). Modern Papua New Guineans present genomic traces of an ancient dispersal out of Africa ∼120,000 yr ago (Pagani et al., 2016), a dispersal that remains to be confirmed, but which clearly made little contribution to modern Eurasians.
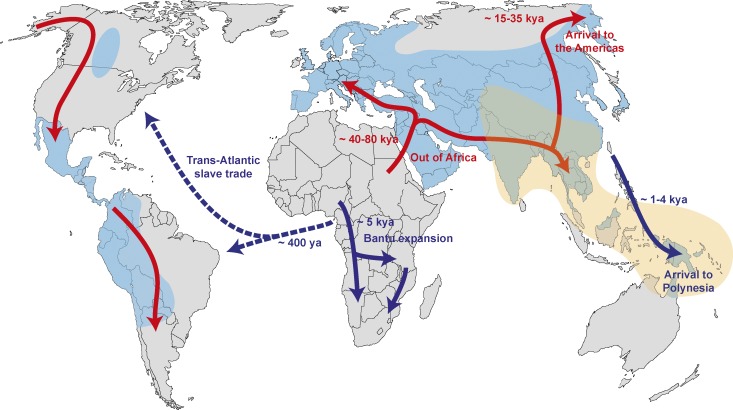
Figure 2.
Major human migrations across the globe and presence of archaic ancestry in the genomes of modern human populations. Red arrows indicate the major migrations of Homo sapiens to colonize the world after the out of Africa exodus. Blue arrows indicate some more recent migratory events (<5,000 yr ago), and dashed arrows represent the historical migrations related to the Trans-Atlantic slave trade. Approximate geographic areas of modern human populations presenting Neanderthal or Denisovan ancestry are shaded in blue and orange, respectively, according to the Simons Genome Diversity Project (Mallick et al., 2016). This map does not present a comprehensive evaluation of the populations presenting archaic ancestry, as only populations for which genetic profiles are available are plotted. The Neanderthal ancestry observed in American populations does not reflect in situ admixture with Neanderthals but, instead, their varying levels of European ancestry.
Genetic studies have also revealed substantial population structure (i.e., subdivision) within continents. In sub-Saharan Africa, population structure is correlated with geography, language family, and mode of subsistence (Henn et al., 2012; Campbell et al., 2014; Patin et al., 2014), whereas, in Europe, genetic diversity is strongly driven by geography, and population structure is detectable within even very small geographic areas (Novembre et al., 2008). The quantification of population structure has important implications for studies of disease, as it must be carefully controlled for in genome-wide association studies (GWAS). Genomic studies can also quantify major admixture events between populations, such as those between the ancestors of African-Americans (Bryc et al., 2010; Montinaro et al., 2015). Decomposing the genomes of admixed populations into segments of different ancestry facilitates the identification of variants associated with disease risk, particularly for traits with different prevalence in the parental populations of admixed groups (Smith and O’Brien, 2005).
Natural selection has driven human adaptation to environmental cues
During their spread across the globe, humans encountered a highly diverse set of climatic, nutritional, and pathogenic conditions, to which they had to adapt. Phenotypic traits increasing their chances to survive and reproduce in such environments are due, at least in part, to genetic variation and thus, transmitted across generations. The dissection of the legacy of past selection in the human genome has proved crucial for identification of genes underlying the broad morphological and physiological diversity observed across human populations, and for increasing our understanding of the genetic architecture of adaptive phenotypes (Vitti et al., 2013; Jeong and Di Rienzo, 2014; Key et al., 2014b; Fan et al., 2016).
Forms of natural selection
Although natural selection acts on phenotypes, it is primarily the genetics underlying the phenotype that is inherited, so genotypes will be acted on by selection. Selection can be classified according to whether an allele is disfavored (purifying selection) or favored (positive selection). Purifying selection removes deleterious alleles and is the most pervasive form of selection (Eyre-Walker and Keightley, 1999; Kryukov et al., 2007). Conversely, positive selection acts upon advantageous mutations after different evolutionary trends (Pritchard et al., 2010). According to the classic sweep model, positive selection acts upon a newly arising advantageous mutation that will increase in frequency, whereas selection on standing variation involves the selection of an allele that is already segregating in the population, which therefore increases in frequency, and polygenic adaptation involves the simultaneous selection of variants at many loci, each of which makes a small contribution to fitness. Genetic adaptation may also occur through balancing selection, which preserves functional diversity through heterozygote advantage, frequency-dependent selection, or pleiotropy (Charlesworth, 2006; Key et al., 2014b). Each selection form leaves specific molecular signatures around the selected region (Fig. 3 A), which are detected by an ever-increasing number of statistical methods that, depending on the nature of genetic data analyzed, can inform the time range at which selection occurred (Box 1).
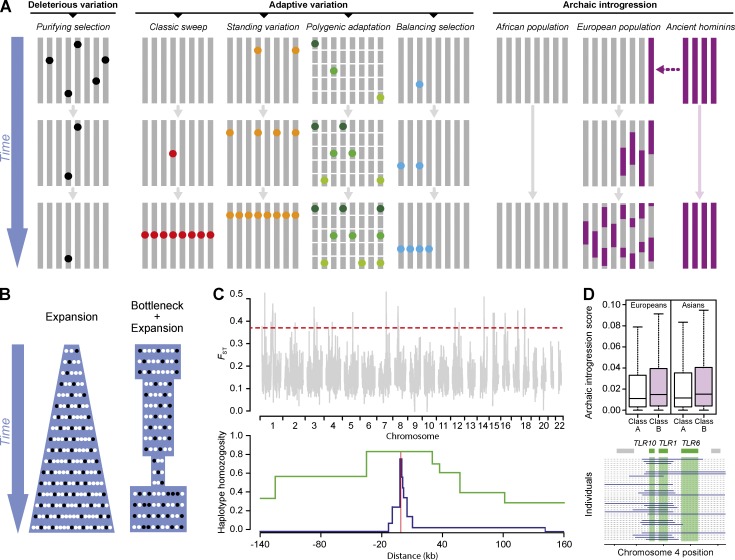
Figure 3.
Natural selection and archaic introgression affect human genome diversity. (A) Schematic representation of how different selective regimes and archaic introgression affect population genetic diversity. Purifying selection removes deleterious alleles (black dots) from the population. In turn, genetic variants conferring a selective advantage (e.g., disease resistance) increase in frequency, or are maintained, in the population though positive and balancing selection, respectively. Different models of positive selection are represented: the classic sweep, where a new mutation rapidly increases in frequency until fixation (red dots); selection on standing variation, where selection acts on a preexisting genetic variant in the population (orange dots); and polygenic adaptation, where multiple genetic variants located in different genomic regions simultaneously increase in frequency (green dots). Balancing selection is represented here by heterozygote advantage (blue dots). Archaic introgression illustrates ancient gene flow between archaic hominins (purple segments) and, in this case, the ancestors of modern Europeans. The traces of this archaic introgression are today detectable in the genomes of Europeans. (B) Genetic diversity and proportion of deleterious alleles in populations with different demographic regimes. A population that underwent a past expansion (left) presents higher levels of genetic diversity, i.e., higher absolute numbers of benign (white dots) and deleterious (black dots) variants, than a population that experienced a bottleneck followed by an expansion (right). However, the proportion of deleterious variants, with respect to those that are benign is lower in the expanding population due to the increased efficiency of purifying selection in removing deleterious alleles. (C) Some molecular signatures of positive selection. The FST statistic (top) measures genetic distances between populations; positive selection, which acts locally, tends to increase the genetic differentiation of particular loci, resulting in high FST values. The distribution of FST values, calculated for genome-wide SNPs across all chromosomes, is presented here. The dashed red line corresponds to the 99th percentile of the distribution, and loci above this line should be considered as strong candidates for positive selection. The degree of haplotype homozygosity (bottom) is another feature commonly used to detect advantageous variants. Positive selection can lead to an excess of homozygosity (green line) of the haplotypes carrying the positively selected allele (red line), with respect to the other haplotypes (blue line). This pattern is the result of the rapid increase in frequency of the advantageous allele, with recombination not having enough time to break down the haplotype on which the selected mutation arose. (D) Comparison of archaic introgression scores between two classes of genes (top); class B genes (e.g., innate immunity genes) present higher archaic ancestry than class A genes (e.g., the rest of the genome). The TLR1-6-10 gene cluster presents a high degree of archaic ancestry (bottom); archaic haplotypes (blue lines) surrounding this genomic region are represented here in European individuals (adapted from Deschamps et al., 2016).
Box 1. Methods to detect natural selection at different time scales
An increasing number of statistical tests are available to detect the occurrence, intensity, and time frame of selection acting on genomes. Frequency-based methods measure nucleotide diversity at a given locus, as well as the deviations of the distribution of its allele frequency spectrum from standard neutral expectations. These methods, including Tajima’s D and derivatives, generally detect selection events that occurred in the past ∼250,000 years: negative values of these tests indicate an excess of rare alleles, compatible with negative or positive selection, whereas positive values reflect an excess of intermediate frequency alleles, a pattern compatible with balancing selection (Kreitman, 2000; Nielsen, 2005).
Methods based on population genetic differences, such as FST and its derivatives, allow the identification of variants that have been favored locally (i.e., frequency increase of the advantageous allele in a population-specific manner) in the past ∼75,000 years by pinpointing marked population differences in frequency of these alleles (Holsinger and Weir, 2009). In turn, methods based on linkage disequilibrium generally measure the level of haplotype homozygosity associated with the selected allele, with respect to that associated with the corresponding neutral allele at a specific site. These include the long-range haplotype (LRH), the integrated haplotype score (iHS), and the cross population extended haplotype homozygosity (XP-EHH) tests; significant values of these tests highlight the rapid increase in frequency of the causal advantageous mutation, and their associated variants, supporting an event of positive selection occurring in the past ∼25,000 years (Sabeti et al., 2002; Voight et al., 2006; Wang et al., 2006). In the context of recent selection, a novel method, the singleton density score, allows the detection of adaptive events within the last ∼2,000 years, based on the low number of rare mutations present on the haplotype carrying the advantageous allele (Field et al., 2016).
Due to the moderate to high false discovery rate of some of these methods, together with the difficulty in identifying the causal selected variants, composite methods that combine multiple statistics into a single score, such as the composite likelihood ratio (CLR) and the composite of multiple signals (CMS), have been developed to increase power and minimize the detection of false-positive signals (Nielsen et al., 2005; Grossman et al., 2010, 2013).
More generally, the performance of methods to detect selection depends on several parameters, such as the age of the advantageous mutation, its selection coefficient, and the demographic history of the population studied, which affect allele frequencies and other aspects of the genetic data to different extents, thus impacting the interpretation of the results and the significance of the statistical tests. The extent to which currently used methods to detect selection are empowered and robust to some of these factors remains an active field of research in population genetics. Further details of these intraspecies methods, together with methods analyzing interspecies divergence to detect negative and positive selection, such as the ratio of substitution rates at non-synonymous and synonymous sites (dN/dS), Hudson-Kreitman-Aguadé (HKA), and McDonald-Kreitman tests, have been extensively reviewed elsewhere (Kreitman, 2000; Nielsen, 2005; Sabeti et al., 2006; Nielsen et al., 2007; Vitti et al., 2013).
Removal of deleterious mutations by purifying selection
Recent studies have shed new light on the ways in which population history can modify the mode in which selection removes deleterious variants (Lohmueller, 2014; Henn et al., 2015; Simons and Sella, 2016). Non-African populations have a higher proportion of deleterious variants of essential genes than Africans, a pattern consistent with less efficient purifying selection to purge deleterious alleles from small populations (Lohmueller et al., 2008; Gutenkunst et al., 2009; Tennessen et al., 2012). Population history appears to have a negligible impact on the mean burden of individual deleterious variants (Fu et al., 2014; Simons et al., 2014; Do et al., 2015), but it has been consistently shown that genetic drift affects the frequency of weakly deleterious mutations more strongly in bottlenecked than in large populations (Fig. 3 B). Indeed, the number of homozygous deleterious genotypes carried by individuals increases with distance from Africa (Henn et al., 2016), and founder populations, such as French-Canadians, and bottlenecked populations, such as the Finns, have larger proportions of deleterious variants, including loss-of-function variants and complete gene knockouts, than expanding populations, such as French or other European populations (Casals et al., 2013; Lim et al., 2014).
Population genetic adaptation to local environments
Genome-wide approaches have identified several hundreds of genomic regions as candidates for positive selection, based on molecular signatures such as the degree of population differentiation or haplotype homozygosity (Fig. 3 C; Voight et al., 2006; Sabeti et al., 2007; Barreiro et al., 2008; Grossman et al., 2013; Scheinfeldt and Tishkoff, 2013; Vitti et al., 2013). Iconic cases of positive selection include the lactase (LCT) gene, responsible for the persistence of lactose tolerance in adults (Bersaglieri et al., 2004; Tishkoff et al., 2007; Itan et al., 2009); genes involved in the variation of skin pigmentation and hair morphology (Lamason et al., 2005; Miller et al., 2007; Norton et al., 2007; Beleza et al., 2013); genes involved in adaptation to diverse habitats, from the rainforest to hypoxic conditions at high altitude (Bigham et al., 2010; Simonson et al., 2010; Yi et al., 2010; Jarvis et al., 2012); and genes related to infectious disease resistance and the immune response (Barreiro and Quintana-Murci, 2010; Quintana-Murci and Barreiro, 2010; Quintana-Murci and Clark, 2013; Fumagalli and Sironi, 2014; Karlsson et al., 2014; Siddle and Quintana-Murci, 2014). Table 1 presents a subset of strongly supported cases of positive selection occurring at different time scales, ranging from 45,000 to as recent as 1,500 yr, of an ever-growing number of genes and functions having participated in human adaptation to environmental cues.
Table 1. Some iconic cases of positive selection in the human genome.
| Gene | Function | Associated phenotype | Main selection method | Population | Date | Putative selective factor | References |
|---|---|---|---|---|---|---|---|
| yr | |||||||
| LCT | Glycoside hydrolase | Lactose tolerance | Haplotype homozygosity | Europeans | ∼7,000–11,000 | Dairy farming | Enattah et al., 2002; Bersaglieri et al., 2004; Tishkoff et al., 2007; Itan et al., 2009; Ranciaro et al., 2014; Mathieson et al., 2015 |
| Africans | ∼3,000–7,000 | ||||||
| EDAR | Cell surface receptor for ectodysplasin A | Hair straightness, changes of epidermal appendages | Haplotype homozygosity | East Asians | ∼10,000 | Thermo-regulation | Sabeti et al., 2007; Barreiro et al., 2008; Bryk et al., 2008; Tan et al., 2013 |
| SLC24A5 | Potassium-dependent sodium/calcium exchanger | Light skin color | Haplotype homozygosity | Europeans | ∼18,700 | Low environmental UV radiation exposure | Lamason et al., 2005; Barreiro et al., 2008; Beleza et al., 2013 |
| EPAS1 | Hypoxia-inducible factor | Response to hypoxic challenge | Haplotype homozygosity | Tibetans | ∼18,200 | Altitude-induced hypoxia | Simonson et al., 2010; Yi et al., 2010; Peng et al., 2011 |
| G6PD | Pentose monophosphate shunt of glycolysis | Resistance to malaria | Haplotype homozygosity | Africans | ∼3,800–11,700 | Plasmodium falciparum/vivax | Ruwende et al., 1995; Tishkoff et al., 2001; Saunders et al., 2002; Louicharoen et al., 2009 |
| Southeast Asians | ∼1,500 | ||||||
| DARC | Duffy antigen receptor for chemokines | Resistance to malaria | Population differentiation | Africans | ∼33,000 | Plasmodium vivax | Hamblin and Di Rienzo, 2000; Hamblin et al., 2002 |
| CD36 | Membrane protein | Resistance to malaria | Composite statistics; population differentiation | Africans | ∼3,900 | Plasmodium falciparum | Newbold et al., 1999; Ayodo et al., 2007; Fry et al., 2009; Bhatia et al., 2011; Deschamps et al., 2016 |
| TLR1 | Pathogen recognition receptor | Lower inflammatory response | Composite statistics; haplotype homozygosity | Europeans | ∼11,000 | ? | Barreiro et al., 2009; Deschamps et al., 2016 |
Genomic traces of admixture between archaic and modern humans
Genomic studies of ancient DNA have revealed that our species, Homo sapiens, interbred with other human forms, such as Neanderthals and Denisovans, present in Eurasia from ∼30,000–50,000 yr ago. This interbreeding resulted in a phenomenon known as archaic introgression; the legacy of this ancient admixture is observed in the genomes of modern human populations (Figs. 2 and 3 A; Kelso and Prüfer, 2014; Vattathil and Akey, 2015). Whole-genome sequences from ancient specimens have revealed the presence of DNA of Neanderthal ancestry accounting for ∼2% of the genome in Europeans to ∼4% in Asians (Green et al., 2010; Prüfer et al., 2014). DNA introgressed into modern humans from Denisovans is found mostly in Australo-Melanesians, in whom it may account for up to 6% of their genomes, and, to a lesser extent, in South East Asians (Reich et al., 2010, 2011; Meyer et al., 2012). These estimates are averages across the modern human genome, and specific regions of the genome may have degrees of Neanderthal ancestry as high as 64% in Europeans and 62% in Asians (Sankararaman et al., 2014).
However, there has been strong selection against archaic introgression, particularly among protein-coding genes, probably caused by its deleterious effects in modern humans (Sankararaman et al., 2014; Vernot and Akey, 2014; Fu et al., 2016). Identifying regions in the human genome strongly depleted of archaic ancestry can thus identify functions that may have contributed to the uniqueness of some modern human traits (Vernot et al., 2016). For example, a region devoid of Neanderthal ancestry has been identified around the forkhead box protein P2 (FOXP2) gene, mutations of which are associated with language disorders (Konopka and Roberts, 2016). The AMY1 gene, encoding an amylase enzyme responsible for starch digestion, also lies in a Neanderthal desert. Unlike modern humans, who carry multiple copies of AMY1, Neanderthals and Denisovans had only one copy of this gene, suggesting that the production of larger amounts of salivary amylase for starch digestion has been of great benefit to modern humans (Perry et al., 2015).
Despite the widespread signature of purifying selection against archaic alleles, modern humans have also acquired advantageous alleles via admixture with ancient hominins, through adaptive introgression (Kelso and Prüfer, 2014; Racimo et al., 2015; Vattathil and Akey, 2015). Genes involved in functions relating to keratin filaments, sugar metabolism, muscle contraction, and oocyte meiosis have been targeted by adaptive introgression from Neanderthals (Sankararaman et al., 2014). For example, EPAS1, associated with hemoglobin concentration and response to hypoxia, displays a high degree of Denisovan ancestry in Tibetans, suggesting that this population acquired advantageous alleles for life at high altitude through ancient admixture (Huerta-Sánchez et al., 2014). Whatever the potential benefits of archaic introgression in the past, alleles of Neanderthal origin have been shown to be associated with several neurological, dermatological, and immunological phenotypes, indicating an influence of ancient admixture on current disease risk in humans (Simonti et al., 2016). These studies show how the detection of introgression from archaic hominins can shed light on the impact of ancient hybridization on the morphological and physiological variability of modern human populations.
Evolutionary genetic dissection of human immunity
Population genetic studies have clearly indicated that immunity and host defense functions are among the functions most frequently subject to natural selection, whether purifying, positive or balancing (Bustamante et al., 2005; Voight et al., 2006; Sabeti et al., 2007; Barreiro et al., 2008; Grossman et al., 2013; Leffler et al., 2013). Identifying the causal agents of selection remains a daunting task, with the most supported cases of selection being primarily restricted to Plasmodium, the causative agent of malaria, which appears to be the true selective factor driving the variability of genes such as HBB, DARC, G6PD, or CD36 (Kwiatkowski, 2005; Siddle and Quintana-Murci, 2014). More generally, although pathogens appear to be the underlying pressure leading to selection signatures at immune defense genes (Fumagalli et al., 2011), it is difficult to rule out the contribution of nonpathogenic microbes, such as those present in the microbiota, and of other biological processes exerting pressure of host genes and pathways. Regardless of the true selective agent, the dissection of the form of selection acting on immunity genes allows to determine the biological relevance of these genes in natura—essentiality, redundancy or adaptability—and their potential contribution to variable immune traits and diseases (Quintana-Murci et al., 2007; Alcaïs et al., 2010; Casanova et al., 2013; Quintana-Murci and Clark, 2013).
Purifying selection and essentiality for immune responses
Genes evolving under purifying selection, and thus under strong constraints, are generally involved in mechanisms essential for host defense, and the variation of these genes can lead to severe disorders. Microbial sensors, such as endosomal TLRs and many NLRs, adaptors, such as MYD88 and TRIF, and effectors, such as some type-I IFNs and IFN-γ, have been shown to evolve under strong purifying selection, attesting to the unique, essential nature of the mechanisms in which they are involved (Quintana-Murci and Clark, 2013). Consequently, rare mutations of highly constrained genes, such as TLR3, TRIF, MYD88, STAT1, TRAF3 and the genes of the IFN-γ pathway, are associated with life-threatening diseases, such as HSV-1 encephalitis, pyogenic bacterial infections, and Mendelian susceptibility to mycobacterial disease (Pérez de Diego et al., 2010; Boisson-Dupuis et al., 2012; Casanova et al., 2013). More generally, a recent genome-wide study has shown that innate immunity genes have evolved under stronger purifying selection than the rest of the genome, with those associated with autosomal-dominant forms of primary immunodeficiencies subject to the strongest degree of purifying selection (Deschamps et al., 2016). These findings support the hypothesis that constrained genes are of major biological relevance in host survival and should be given priority in genetic studies aiming to identify new genetic etiologies of severe, infectious disease phenotypes.
Genetic adaptation and immune diversity
Genes undergoing adaptive evolution through positive selection provide evidence of the functional variability that has been beneficial to specific human populations. The list of immune system genes known to be subject to positive selection is continually growing (Barreiro and Quintana-Murci, 2010; Quintana-Murci and Clark, 2013; Brinkworth and Barreiro, 2014; Fumagalli and Sironi, 2014; Karlsson et al., 2014). Relevant cases of host defense and immune genes harboring advantageous variants include HBB, CD36, CR1, G6PD, DARC, and CD40LG, which are involved in malaria resistance in Africans; TLR1 and TLR5, which are involved in decreases in NF-κB signaling in Europeans and Africans, respectively (Barreiro et al., 2009; Grossman et al., 2013; Deschamps et al., 2016); type III IFN genes, implicated in the higher levels of spontaneous viral clearance in Europeans and Asians (Manry et al., 2011; Key et al., 2014a); and LARGE and IL21, which have been implicated in Lassa fever immunity in West Africans (Andersen et al., 2012). The evidence for balancing selection in humans is largely circumstantial, but host defense genes, such as those encoding several membrane glycoproteins, the KIR regions that coevolve with HLA ligands, and other genes encoding proteins involved in cell migration or innate immunity, are subject to this otherwise rare selective regimen (Norman et al., 2007; Single et al., 2007; Andrés et al., 2009; Leffler et al., 2013; DeGiorgio et al., 2014).
A recent study has estimated the time frame of selection targeting innate immunity genes, and found that most selective events occurred ∼6,000–13,000 yr ago (Deschamps et al., 2016), corresponding to the period at which human societies adopted agriculture, a major transition in human lifestyle that likely modified human exposure to pathogens and led to genetic adaptation. The detection of immune genes targeted by local adaptation can increase our understanding of the mechanisms involved in host resistance to infectious challenges at different time scales, the nature of which may also differ between human populations.
Present deleterious effects of past selection events
Environmental factors may change over time, and genetic variants inherited from our ancestors and that have been favored by past selection can become detrimental in modern societies, increasing the risk of inflammation or autoimmunity (Barreiro and Quintana-Murci, 2010; Sironi and Clerici, 2010; Brinkworth and Barreiro, 2014). For example, the prevalence of celiac disease, an autoimmune disorder caused by gluten intolerance, differs considerably between human populations, with individuals of European ancestry particularly affected (Kang et al., 2013). Population genetic analyses have shown that the high frequency of several risk alleles of genes associated with celiac disease, such as IL12A, IL18RAP, and SH2B3 (Hunt et al., 2008), in Europeans results from past positive selection events (Barreiro and Quintana-Murci, 2010; Zhernakova et al., 2010). Functional analyses of the SH2B3 risk variant have suggested that this otherwise deleterious variant was subject to positive selection in the past because it confers greater protection against infection (Zhernakova et al., 2010). However, the pleiotropic nature of SH2B3 (i.e., regulating the structural organization and development of platelets and endothelial cells), like that of many immune genes, suggests that alleles that currently increase the risk of inflammatory or autoimmune disorders have in the past conferred greater reproductive success via a broader range of beneficial phenotypes (Brinkworth and Barreiro, 2014). The case of celiac disease neatly illustrates the tradeoff between past selection and current maladaptation, and highlights the value of population genetics for elucidating the mechanisms contributing to pathological inflammation and its heterogeneous distribution across populations.
The immune legacy of archaic hominins in our genomes
There is increasing evidence to suggest that immune genes have not only been an important substrate of selection, but also that their diversity in modern humans has been affected by archaic introgression (Racimo et al., 2015). Innate immunity genes generally display a higher degree of Neanderthal ancestry than the rest of the genome, and genes encoding sensors, such the OAS and the TLR6-1-10 clusters, transcription factors, such as IRF6, and effector molecules, such as the restriction factors IFITM1-3 and the cytokines IL17A and IL17F, present the highest Neanderthal introgression scores at the genome-wide level (Fig. 3 D; Deschamps et al., 2016). In some cases, there is also evidence for selection acting on the archaic introgressed segments (i.e., adaptive introgression). For example, the HLA region, which evolves under balancing selection in humans, harbors functional variants that were probably introgressed from Neanderthals and Denisovans (Abi-Rached et al., 2011). Other candidates for adaptive introgression include STAT2, which is involved in the interferon response to viral infection; OAS1, which has been implicated in innate immune responses to viruses; and the TLR6-1-10 cluster, encoding proteins responsible for the sensing of microbial components on the cell surface (Mendez et al., 2012, 2013; Sankararaman et al., 2014; Dannemann et al., 2016; Deschamps et al., 2016; Sams et al., 2016).
The signals of adaptive introgression detected suggest an advantage for modern human survival, but recent findings have suggested that there may be a tradeoff for archaic introgression. For example, a nonsynonymous variant of the ZNF365D gene present in ∼26% of Europeans and absent from Africans was inherited from Neanderthals and is associated with a higher risk of Crohn’s disease (Sankararaman et al., 2014). Likewise, variants of TLR6-1-10 inherited from Neanderthals and Denisovans and present in Europeans and Asians have been associated with greater susceptibility to allergies (Dannemann et al., 2016). These studies collectively demonstrate that admixture with archaic hominins introduced variants into the gene pool of modern Eurasians some 30,000–50,000 yr ago, increasing the diversity of the modern human immune repertoire. These variants were initially neutral or advantageous for modern humans, but some are today associated with disease phenotypes.
Understanding immune response phenotypes through regulatory variation
Studies of the effects of selection on diversity at immune gene loci increase our understanding of the biological relevance of the functions concerned, complementing immunological, clinical, and epidemiological genetic studies (Casanova et al., 2013). Nevertheless, little is known about the relationship between genetic variation and immune phenotype diversity, and the nature of the immunological mechanisms under selection remains largely unexplored. The contribution of host genetic variants to variation in immune phenotypes, including those related to disease, is increasingly being documented by GWAS (Vannberg et al., 2011; Parkes et al., 2013; Abel et al., 2014). However, the multiple variants associated with immune phenotypes tend to have small individual effect sizes, and the identification of causal functional variants remains challenging (Manolio et al., 2009). In recent years, analyses of regulatory variants of gene expression (eQTL) have proved to be of considerable biomedical value (Montgomery and Dermitzakis, 2011), establishing links between intermediate phenotypes, such as gene expression, and organism traits, such as immunity to infection (Fig. 4 A; Fairfax and Knight, 2014). Furthermore, GWAS have shown that susceptibility to common immune diseases is primarily controlled by noncoding, probably regulatory, variants (Hindorff et al., 2009; Nicolae et al., 2010; Schaub et al., 2012; Fraser, 2013; Pickrell, 2014). However, eQTL studies have also shown that the genetic control of gene expression varies considerably across tissues and cell types (Dimas et al., 2009; Price et al., 2011; Powell et al., 2012), reinforcing the need to consider the appropriate cell context specificity of eQTL in studies aiming to increase knowledge on the genetic bases of phenotypic traits or diseases.
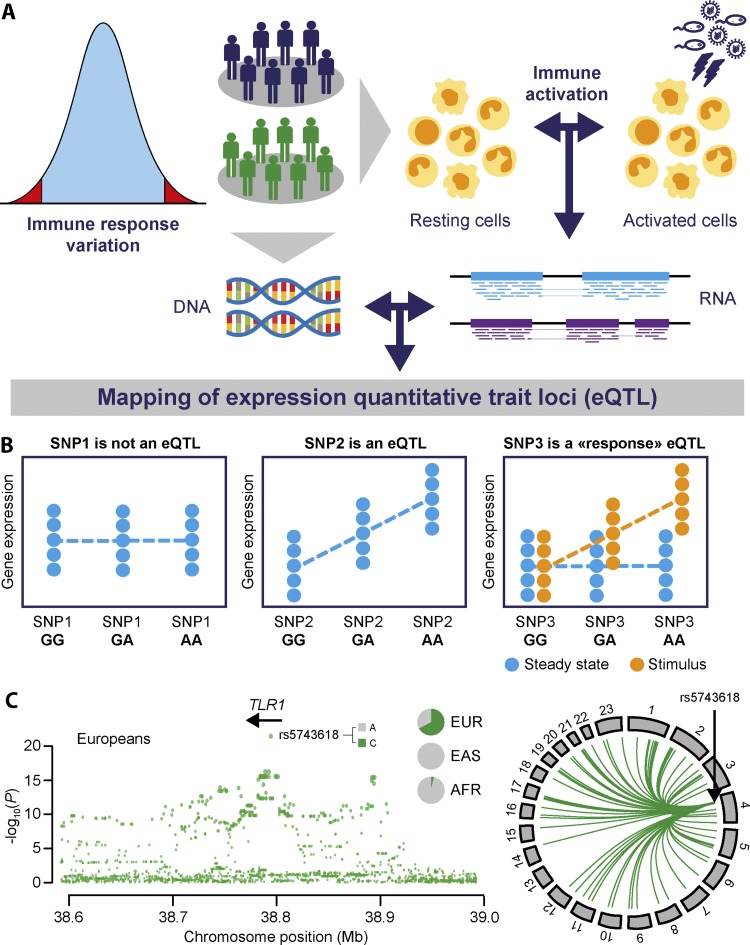
Figure 4.
Principles of eQTL mapping for investigating immune response variation. (A) General workflow to map the genetic basis of immune response variation in human populations; transcriptional responses, to various infections or immune stimulations, of primary immune cells from healthy donors are defined to subsequently assess the correlation between genotype variation and gene expression phenotypes, through eQTL mapping. (B) Plots representing eQTL; in contrast with SNP1, which does not behave as an eQTL (left), SNP2 is detected as an eQTL because its genotypes correlate with gene expression variation (middle). SNP3 behaves as a response eQTL; genotypes are correlated with gene expression variation only in stimulated conditions, indicating gene–environment interactions (right). (C) The fine mapping of the TLR1 genomic region (left) detected the SNP rs5743618 as the best trans-eQTL in Europeans (Quach et al., 2016), whose derived C allele is associated with the expression patterns of multiple genes upon Pam3CSK4 monocyte stimulation (right). The C allele presents signatures of local adaptation in Europeans (EUR), where it is present at very high frequency, whereas it is virtually absent in African (AFR) and East-Asian (EAS) populations.
Mapping eQTL for the immune response
Several studies have mapped eQTL in cell types relevant to immunity, such as primary monocytes and T lymphocytes, focusing on gene expression measurements at the basal state (Dimas et al., 2009; Fairfax et al., 2012; Raj et al., 2014), while others have more recently explored the genetic determinants of gene expression in the presence of external, immune stimuli (i.e., response eQTL), thereby increasing our understanding of gene–environment interactions in the context of infection (Fig. 4 B; Barreiro et al., 2012; Fairfax et al., 2014; Lee et al., 2014; Çalışkan et al., 2015; Nédélec et al., 2016; Quach et al., 2016). For example, a pioneering study, in which human dendritic cells from healthy individuals were infected with Mycobacterium tuberculosis (MTB), showed that immune response variation to MTB was often associated with genetic factors (Barreiro et al., 2012). Some of the regulatory variants detected overlapped genes already shown to be associated with susceptibility to pulmonary tuberculosis, including the MAPK phosphatase gene DUSP14 and the gene encoding the RIPK2 adapter of the NOD2-signaling pathway. A subsequent study in this experimental setting identified two loci, miR-326 and miR-1260, as controlling miRNA expression in response to MTB infection, providing the first assessment of the impact of genotype–infection interactions on miRNA molecular phenotypes (Siddle et al., 2014).
Two subsequent seminal eQTL studies provided mechanistic insights into the genetic basis of immune variation (Fairfax et al., 2014; Lee et al., 2014). The expression of hundreds of genes after the stimulation of monocytes from individuals of European descent with LPS or IFN-γ was shown to be under genetic control. The genes concerned include nodal genes and effector molecules involved in pathways such as those involving the products of the TLR4-related TIRAP, TRAF6, FADD, and MAP kinase genes; the inflammasome-related CASP1 and PYCARD genes; genes encoding downstream cytokines, such as IL6, CXCL9, and IFNB1; and many IFN-γ pathway genes (Fairfax et al., 2014). The other study focused on responses to LPS, influenza, or IFN-β in dendritic cells from individuals of various ethnic backgrounds (Lee et al., 2014). The authors identified a large number of response eQTL, and showed that some of the regulatory variants involved altered the binding of transcription factors released only upon immune activation, such as STAT1 and STAT2, which are released upon IFN-β treatment, providing some insight into the mechanisms underlying the gene–environment interactions detected. In some cases, response eQTL overlap variants associated with susceptibility to infectious and autoimmune diseases, as illustrated by CARD9 and Crohn’s disease, NOD2 and leprosy, or IRF7 and systemic lupus erythematosus (Fairfax et al., 2014; Lee et al., 2014). These findings support the notion that disease-risk alleles can display activity only after some sort of immune activation (Gaulton et al., 2010; Gregory et al., 2012), and highlight the value of response eQTL mapping for investigations of the mechanisms underlying complex disease phenotypes.
Genetic determinants of population variation in immune responses
Analyses of eQTL on lymphoblastoid cell lines from various human populations worldwide provided proof of concept that genetic variation accounts for differences in gene expression among ethnic groups (Spielman et al., 2007; Stranger et al., 2012). Using primary monocytes and T lymphocytes, it has been shown that cis-eQTL are largely shared across populations, with only a small number of them being population specific (Raj et al., 2014). Although this study focused on gene expression at the steady state, two recent studies have determined the degree, and underlying genetic control, of population differences in the response to immune stimulation (Nédélec et al., 2016; Quach et al., 2016). They defined transcriptional responses to immune challenges through RNA sequencing, and mapped eQTL in macrophages from African-Americans and European-Americans exposed to Listeria monocytogenes and Salmonella Typhimurium (Nédélec et al., 2016), and in monocytes from Africans and Europeans exposed to TLR ligands (activating TLR4, TLR1/2, and TLR7/8) and influenza A virus (Quach et al., 2016). Despite differences in the experimental settings, both these studies found that the genes for which the response to immune stimulation differed strongly between populations were enriched in genetic control, with the regulatory variants concerned presenting different allele frequencies in populations of different ancestries.
Individuals of African descent generally display a stronger response to bacterial infection, particularly for inflammatory response genes, than individuals of European descent (Nédélec et al., 2016). A large fraction of European individuals has a mutation at the TLR1 locus that impairs NK-κB activity (Barreiro et al., 2009) and acts as a population-specific trans-eQTL (Fig. 4 C). This TLR1 mutation is associated with a large network of genes displaying decreased levels of expression in response to immune activation only in Europeans (Quach et al., 2016). Altogether, these studies identified a large number of genes displaying considerable differences between populations, especially in the context of their responses to immune activation, revealing mechanisms that might account for differences in the clinical manifestations of immune-related diseases between individuals or ethnic groups.
Selection and archaic admixture have shaped immune response regulation
There is growing evidence to suggest that regulatory variants play a major role in population adaptation (Schaub et al., 2012; Fraser, 2013; Pickrell, 2014). The enrichment of immune-responsive regulatory variants in population-specific signals of positive selection thus facilitates the identification of genes and phenotypes of evolutionary importance that have contributed to human survival. Examples of population differences in immune responses resulting from local adaptation driven by regulatory variants include the expression of HLA-DQA1, associated with susceptibility to celiac disease; ERAP2, involved in susceptibility to Crohn’s disease; CCR1, limiting leukocyte recruitment and preventing inflammatory responses; and TLR1, associated with markedly lower levels of inflammatory response gene expression (Nédélec et al., 2016; Quach et al., 2016). The positive selection signature detected at the TLR1 trans-eQTL is consistent with attenuated TLR1-mediated signaling being advantageous to Europeans, highlighting the benefits of avoiding strong inflammatory responses that may be harmful to the host (Quach et al., 2016).
Introgression from Neanderthals also contributed to the diversification of transcriptional responses to infection in human populations. The genetic segments introgressed appear to have preferentially introduced regulatory variants into European genomes, with effects on steady-state expression and responses to TLR7/8 stimulation and influenza virus (Quach et al., 2016). Furthermore, several eQTL have been identified as potential candidates for adaptive introgression, in which the archaic variants confer greater adaptation through the regulation of gene expression. Examples include genes such as DARS, which is associated with neuroinflammatory and white matter disorders (Nédélec et al., 2016); the OAS locus, at which archaic variants appear to be associated with diverse flavivirus resistance phenotypes (Sams et al., 2016); and PNMA1, which harbors a response eQTL for influenza virus and encodes a protein that interacts with the viral protein PB2 and stimulate interferon production (Quach et al., 2016).
All these studies support the notion that variation in gene expression has been an important vehicle for human adaptation (Fraser, 2013), and clearly demonstrate that selection and archaic admixture have had a significant impact on present-day inter-population differences in immune responses, at least in terms of transcriptional variability. Only a partial understanding of the factors driving immune response variability can be gleaned from analyses based solely on eQTL, as these studies provide no information about the effects of genetic variants on other molecular phenotypes, such as protein production (protein quantitative trait loci, pQTL). Indeed, it has been shown that while a substantial fraction of genetic variants influence gene expression at all levels, from mRNA to steady-state protein abundance, the effects of eQTL are attenuated at the protein level, and some pQTL have little or no effect at the mRNA level suggesting that they affect posttranscriptional gene regulation (Battle et al., 2015). Likewise, one recent study identified genetic variants regulating cytokine production in response to various microbial stimuli, and showed that some of these variants co-localize with regions presenting signatures of positive selection (Li et al., 2016). Further investigations are required, but, by and large, the studies published to date have provided invaluable resources, by identifying genetic variants affecting intermediate molecular phenotypes in response to immune challenges in different cell types. They have also increased our understanding of the molecular and cellular mechanisms underlying host immunity to infection and susceptibility to disease.
Nongenetic and environmental factors driving immune response variation
Exploration of the epigenetic and environmental factors affecting the diversity of immune responses, across individuals and populations, is currently a major challenge, as a large fraction of the variation in immune responses cannot be attributed to genetic factors (Barreiro et al., 2012; Fairfax et al., 2014; Lee et al., 2014; Çalışkan et al., 2015; Nédélec et al., 2016; Quach et al., 2016). In this context, a recent study has shown a major impact of host nongenetic factors, such as age and gender, and environmental variables, such as annual seasonality, on the production of inflammatory cytokines in healthy donors upon cellular activation by 19 (non-) microbial stimuli; for example, the production of IFN-γ and IL-22 was significantly lower in elderly individuals (Ter Horst et al., 2016). Likewise, it has been shown that the composition and function of gut microbial communities also contribute to interindividual variation in cytokine responses to immune stimulation, providing a set of microbial-derived mediators that influence immune phenotypes in healthy individuals (Schirmer et al., 2016). Furthermore, there is growing evidence to suggest that other factors, such as social environment, can also affect immune response variation and, therefore, individual physiology and disease risk (Tung et al., 2012). In this context, a recent study has sought to decipher the influence of social status on immune system functions, using female rhesus macaques as a model (Snyder-Mackler et al., 2016). They showed that social rank affects immune cell proportions, gene expression levels and responses to immune stimuli, with a stronger proinflammatory and antibacterial response in low-status individuals. Together, these studies highlight the need to further investigate, using integrative approaches, the respective effects of host genetic and nongenetic factors, together with environmental parameters and lifestyle variables, on immune response variation, to improve our understanding of the different factors contributing to health disparities between individuals and populations.
Conclusion and perspectives
The collection of sequencing data for thousands of individuals from different populations worldwide, including some archaic hominins, has provided new insight into the demographic and selection history of our species, revealing the admixture of early Eurasians with other types of humans, such as Neanderthals and Denisovans, from whom they acquired advantageous alleles. Interestingly, genomic traces of archaic introgression have also been detected in Africans (Hammer et al., 2011; Lachance et al., 2012), raising the exciting possibility that other unknown archaic groups may have contributed to human genetic diversity. Sequencing additional samples from ancient hominins and from multiple human populations, and the development of robust statistical models and computational methods for detecting selection, will further deepen our knowledge of the contribution of archaic hominins to the diversity of human traits and complex diseases and will help to identify the functions and mechanisms that have contributed to human adaptation and survival over time.
The study of genetic variants with regulatory effects on gene expression (eQTL) has also provided insight into the genetic and evolutionary determinants of population phenotypic diversity, particularly for immune-related diversity (Fairfax and Knight, 2014). However, because the regulation of gene expression is highly dependent on cellular context (Dimas et al., 2009; Nica et al., 2011; Fairfax et al., 2012), it is essential to map eQTL, under basal conditions or after challenge, in a vast array of innate and adaptive cell types, to obtain a complete portrait of the genetic control of immune responses. In this context, the GTEx project explores the landscape of gene expression across 54 different tissues, providing the richest catalog of tissue-specific and shared eQTL (GTEx Consortium, 2015). The extension of this across-tissue rationale to multiple populations from different ethnic backgrounds will, together with population genetic analyses, help to provide a comprehensive picture of the immunological mechanisms underlying host adaptation to pathogen pressure and the maintenance of homeostasis.
An expansion of current efforts to map the genetics of gene expression onto other immune phenotypes, such as protein production, metabolite levels, and heterogeneity in immune cell populations, together with the influence of the microbiome, is increasing our understanding of the genetic, epigenetic, and environmental determinants of variation in immune responses and immunity to infection (Liston et al., 2016; Brodin and Davis, 2017). For example, studies analyzing twin cohorts, as well as a population from Sardinia, have dissected the heritable and nonheritable status of immune phenotypes, such as cell population frequencies and protein expression levels (Orrù et al., 2013; Brodin et al., 2015; Roederer et al., 2015), and found, for example, that age increases interindividual variation of these parameters, owing to increased exposure to pathogens, nonpathogenic microbes, or vaccination history (Brodin et al., 2015). Likewise, a recent study has characterized the genetic, epigenetic, and transcriptomic landscape of several major immune cell types and found that transcriptional variation at the majority of genes is a result of the presence of cis-regulatory genetic variants, but epigenetic influences were also observed for a small subset of biologically relevant genes (Chen et al., 2016). In this line, ongoing efforts, such as those of the Immune Variation (ImmVar) Project, the Human Functional Genomics Project, and the Milieu Intérieur Consortium, are focusing on the dissection of how interactions between host genetic and epigenetic variation and a large number of variables, including age, sex, commensal microbiota, nutrition, latent infections, and history of vaccination, drive the variation of human immune responses. Collectively, these studies constitute useful, comprehensive resources to investigate the interplay between genetic and nongenetic factors in modulating the plasticity of the immune system (Liston et al., 2016; Brodin and Davis, 2017).
In the context of how evolution has acted on immune phenotypes, it is important to keep in mind that potentially advantageous immune traits can be transmitted across generations not only through genetics but also via some environmental and cultural factors. These include, for example, nutritional and physical activity behaviors, smoking, mate choice, and access to medical care. Thus, another way to explore the evolution of human populations and the influence of selection is to use phenotypic approaches in the natural setting (Linnen and Hoekstra, 2009; Stearns et al., 2010). Focusing on phenotypic data collection, it has been reported that traits related to morphology (e.g., weight and height), physiology (e.g., cholesterol and systolic blood pressure), and life history (e.g., age at first and last births) have been targets of selection (Stearns et al., 2010). Future studies should develop comprehensive phenotypic databases as a complement to genetic studies to assess how the environmental and cultural changes operated by human populations can affect the evolution of our species at the genetic and phenotypic levels over time.
In conclusion, the integration of all of these datasets into a clinical, epidemiological, and population genetics framework will provide new insight, and probably a few surprises, concerning the history and immune responses of the genus Homo, and the ways in which our genetic and nongenetic makeup, together with changes in our environment and cultural behaviors, influence phenotypic variation in both health and disease.
Acknowledgments
We wish to thank the members of the Human Evolutionary Genetics Laboratory for discussions and critical reading of the manuscript.
Research in the Quintana-Murci laboratory was supported by the Institut Pasteur, the Centre National de la Recherche Scientifique (CNRS), the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases (grant no. ANR-10-LABX-62-IBEID), the Agence Nationale de la Recherche (ANR) grants IEIHSEER (ANR-14-CE14-0008-02) and TBPATHGEN (ANR-14-CE14-0007-02), and the European Research Council under the European Union’s Seventh Framework Program (FP/2007–2013)/ERC Grant Agreement No. 281297.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- eQTL
- expression quantitative trait loci
- GWAS
- genome-wide association study
- miRNA
- microRNA
- MTB
- Mycobacterium tuberculosis
- pQTL
- protein quantitative trait loci
References
- Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., and McVean G.A.. 1000 Genomes Project Consortium . 2010. A map of human genome variation from population-scale sequencing. Nature. 467:1061–1073. 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., and McVean G.A.. 1000 Genomes Project Consortium . 2012. An integrated map of genetic variation from 1,092 human genomes. Nature. 491:56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel L., Alcaïs A., and Schurr E.. 2014. The dissection of complex susceptibility to infectious disease: bacterial, viral and parasitic infections. Curr. Opin. Immunol. 30:72–78. 10.1016/j.coi.2014.07.002 [DOI] [PubMed] [Google Scholar]
- Abi-Rached L., Jobin M.J., Kulkarni S., McWhinnie A., Dalva K., Gragert L., Babrzadeh F., Gharizadeh B., Luo M., Plummer F.A., et al. 2011. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 334:89–94. 10.1126/science.1209202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaïs A., Quintana-Murci L., Thaler D.S., Schurr E., Abel L., and Casanova J.L.. 2010. Life-threatening infectious diseases of childhood: single-gene inborn errors of immunity? Ann. N. Y. Acad. Sci. 1214:18–33. 10.1111/j.1749-6632.2010.05834.x [DOI] [PubMed] [Google Scholar]
- Andersen K.G., Shylakhter I., Tabrizi S., Grossman S.R., Happi C.T., and Sabeti P.C.. 2012. Genome-wide scans provide evidence for positive selection of genes implicated in Lassa fever. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:868–877. 10.1098/rstb.2011.0299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés A.M., Hubisz M.J., Indap A., Torgerson D.G., Degenhardt J.D., Boyko A.R., Gutenkunst R.N., White T.J., Green E.D., Bustamante C.D., et al. 2009. Targets of balancing selection in the human genome. Mol. Biol. Evol. 26:2755–2764. 10.1093/molbev/msp190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., and Abecasis G.R.. 1000 Genomes Project Consortium . 2015. A global reference for human genetic variation. Nature. 526:68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayodo G., Price A.L., Keinan A., Ajwang A., Otieno M.F., Orago A.S., Patterson N., and Reich D.. 2007. Combining evidence of natural selection with association analysis increases power to detect malaria-resistance variants. Am. J. Hum. Genet. 81:234–242. 10.1086/519221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro L.B., and Quintana-Murci L.. 2010. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat. Rev. Genet. 11:17–30. 10.1038/nrg2698 [DOI] [PubMed] [Google Scholar]
- Barreiro L.B., Laval G., Quach H., Patin E., and Quintana-Murci L.. 2008. Natural selection has driven population differentiation in modern humans. Nat. Genet. 40:340–345. 10.1038/ng.78 [DOI] [PubMed] [Google Scholar]
- Barreiro L.B., Ben-Ali M., Quach H., Laval G., Patin E., Pickrell J.K., Bouchier C., Tichit M., Neyrolles O., Gicquel B., et al. 2009. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 5:e1000562 10.1371/journal.pgen.1000562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro L.B., Tailleux L., Pai A.A., Gicquel B., Marioni J.C., and Gilad Y.. 2012. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc. Natl. Acad. Sci. USA. 109:1204–1209. 10.1073/pnas.1115761109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle A., Khan Z., Wang S.H., Mitrano A., Ford M.J., Pritchard J.K., and Gilad Y.. 2015. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 347:664–667. 10.1126/science.1260793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beleza S., Santos A.M., McEvoy B., Alves I., Martinho C., Cameron E., Shriver M.D., Parra E.J., and Rocha J.. 2013. The timing of pigmentation lightening in Europeans. Mol. Biol. Evol. 30:24–35. 10.1093/molbev/mss207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri T., Sabeti P.C., Patterson N., Vanderploeg T., Schaffner S.F., Drake J.A., Rhodes M., Reich D.E., and Hirschhorn J.N.. 2004. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 74:1111–1120. 10.1086/421051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia G., Patterson N., Pasaniuc B., Zaitlen N., Genovese G., Pollack S., Mallick S., Myers S., Tandon A., Spencer C., et al. 2011. Genome-wide comparison of African-ancestry populations from CARe and other cohorts reveals signals of natural selection. Am. J. Hum. Genet. 89:368–381. 10.1016/j.ajhg.2011.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham A., Bauchet M., Pinto D., Mao X., Akey J.M., Mei R., Scherer S.W., Julian C.G., Wilson M.J., López Herráez D., et al. 2010. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 6:e1001116 10.1371/journal.pgen.1001116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dupuis S., Kong X.F., Okada S., Cypowyj S., Puel A., Abel L., and Casanova J.L.. 2012. Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes. Curr. Opin. Immunol. 24:364–378. 10.1016/j.coi.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkworth J.F., and Barreiro L.B.. 2014. The contribution of natural selection to present-day susceptibility to chronic inflammatory and autoimmune disease. Curr. Opin. Immunol. 31:66–78. 10.1016/j.coi.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P., and Davis M.M.. 2017. Human immune system variation. Nat. Rev. Immunol. 17:21–29. 10.1038/nri.2016.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P., Jojic V., Gao T., Bhattacharya S., Angel C.J., Furman D., Shen-Orr S., Dekker C.L., Swan G.E., Butte A.J., et al. 2015. Variation in the human immune system is largely driven by non-heritable influences. Cell. 160:37–47. 10.1016/j.cell.2014.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryc K., Auton A., Nelson M.R., Oksenberg J.R., Hauser S.L., Williams S., Froment A., Bodo J.M., Wambebe C., Tishkoff S.A., and Bustamante C.D.. 2010. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc. Natl. Acad. Sci. USA. 107:786–791. 10.1073/pnas.0909559107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk J., Hardouin E., Pugach I., Hughes D., Strotmann R., Stoneking M., and Myles S.. 2008. Positive selection in East Asians for an EDAR allele that enhances NF-kappaB activation. PLoS One. 3:e2209 10.1371/journal.pone.0002209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C.D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M.T., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., et al. 2005. Natural selection on protein-coding genes in the human genome. Nature. 437:1153–1157. 10.1038/nature04240 [DOI] [PubMed] [Google Scholar]
- Çalışkan M., Baker S.W., Gilad Y., and Ober C.. 2015. Host genetic variation influences gene expression response to rhinovirus infection. PLoS Genet. 11:e1005111 10.1371/journal.pgen.1005111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M.C., Hirbo J.B., Townsend J.P., and Tishkoff S.A.. 2014. The peopling of the African continent and the diaspora into the new world. Curr. Opin. Genet. Dev. 29:120–132. 10.1016/j.gde.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals F., Hodgkinson A., Hussin J., Idaghdour Y., Bruat V., de Maillard T., Grenier J.C., Gbeha E., Hamdan F.F., Girard S., et al. 2013. Whole-exome sequencing reveals a rapid change in the frequency of rare functional variants in a founding population of humans. PLoS Genet. 9:e1003815 10.1371/journal.pgen.1003815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.L., Abel L., and Quintana-Murci L.. 2013. Immunology taught by human genetics. Cold Spring Harb. Symp. Quant. Biol. 78:157–172. 10.1101/sqb.2013.78.019968 [DOI] [PubMed] [Google Scholar]
- Charlesworth D. 2006. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2:e64 10.1371/journal.pgen.0020064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yan Y., Kundu K., Ecker S., et al. 2016. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell. 167:1398–1414.e24. 10.1016/j.cell.2016.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemann M., Andrés A.M., and Kelso J.. 2016. Introgression of Neandertal- and Denisovan-like haplotypes contributes to adaptive variation in human Toll-like receptors. Am. J. Hum. Genet. 98:22–33. 10.1016/j.ajhg.2015.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGiorgio M., Lohmueller K.E., and Nielsen R.. 2014. A model-based approach for identifying signatures of ancient balancing selection in genetic data. PLoS Genet. 10:e1004561 10.1371/journal.pgen.1004561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps M., Laval G., Fagny M., Itan Y., Abel L., Casanova J.L., Patin E., and Quintana-Murci L.. 2016. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am. J. Hum. Genet. 98:5–21. 10.1016/j.ajhg.2015.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M., et al. 2009. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 325:1246–1250. 10.1126/science.1174148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do R., Balick D., Li H., Adzhubei I., Sunyaev S., and Reich D.. 2015. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat. Genet. 47:126–131. 10.1038/ng.3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enattah N.S., Sahi T., Savilahti E., Terwilliger J.D., Peltonen L., and Järvelä I.. 2002. Identification of a variant associated with adult-type hypolactasia. Nat. Genet. 30:233–237. 10.1038/ng826 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., and Keightley P.D.. 1999. High genomic deleterious mutation rates in hominids. Nature. 397:344–347. 10.1038/16915 [DOI] [PubMed] [Google Scholar]
- Fairfax B.P., and Knight J.C.. 2014. Genetics of gene expression in immunity to infection. Curr. Opin. Immunol. 30:63–71. 10.1016/j.coi.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfax B.P., Makino S., Radhakrishnan J., Plant K., Leslie S., Dilthey A., Ellis P., Langford C., Vannberg F.O., and Knight J.C.. 2012. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat. Genet. 44:502–510. 10.1038/ng.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfax B.P., Humburg P., Makino S., Naranbhai V., Wong D., Lau E., Jostins L., Plant K., Andrews R., McGee C., and Knight J.C.. 2014. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 343:1246949 10.1126/science.1246949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S., Hansen M.E., Lo Y., and Tishkoff S.A.. 2016. Going global by adapting local: A review of recent human adaptation. Science. 354:54–59. 10.1126/science.aaf5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y., Boyle E.A., Telis N., Gao Z., Gaulton K.J., Golan D., Yengo L., Rocheleau G., Froguel P., McCarthy M.I., and Pritchard J.K.. 2016. Detection of human adaptation during the past 2000 years. Science. 354:760–764. 10.1126/science.aag0776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H.B. 2013. Gene expression drives local adaptation in humans. Genome Res. 23:1089–1096. 10.1101/gr.152710.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.E., Ghansa A., Small K.S., Palma A., Auburn S., Diakite M., Green A., Campino S., Teo Y.Y., Clark T.G., et al. 2009. Positive selection of a CD36 nonsense variant in sub-Saharan Africa, but no association with severe malaria phenotypes. Hum. Mol. Genet. 18:2683–2692. 10.1093/hmg/ddp192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q., Posth C., Hajdinjak M., Petr M., Mallick S., Fernandes D., Furtwängler A., Haak W., Meyer M., Mittnik A., et al. 2016. The genetic history of Ice Age Europe. Nature. 534:200–205. 10.1038/nature17993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W., O’Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Rieder M.J., Altshuler D., Shendure J., et al. NHLBI Exome Sequencing Project . 2013. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 493:216–220. 10.1038/nature11690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W., Gittelman R.M., Bamshad M.J., and Akey J.M.. 2014. Characteristics of neutral and deleterious protein-coding variation among individuals and populations. Am. J. Hum. Genet. 95:421–436. 10.1016/j.ajhg.2014.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M., and Sironi M.. 2014. Human genome variability, natural selection and infectious diseases. Curr. Opin. Immunol. 30:9–16. 10.1016/j.coi.2014.05.001 [DOI] [PubMed] [Google Scholar]
- Fumagalli M., Sironi M., Pozzoli U., Ferrer-Admetlla A., Pattini L., and Nielsen R.. 2011. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 7:e1002355 10.1371/journal.pgen.1002355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton K.J., Nammo T., Pasquali L., Simon J.M., Giresi P.G., Fogarty M.P., Panhuis T.M., Mieczkowski P., Secchi A., Bosco D., et al. 2010. A map of open chromatin in human pancreatic islets. Nat. Genet. 42:255–259. 10.1038/ng.530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M., Patterson N., Li H., Zhai W., Fritz M.H., et al. 2010. A draft sequence of the Neandertal genome. Science. 328:710–722. 10.1126/science.1188021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory A.P., Dendrou C.A., Attfield K.E., Haghikia A., Xifara D.K., Butter F., Poschmann G., Kaur G., Lambert L., Leach O.A., et al. 2012. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 488:508–511. 10.1038/nature11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman S.R., Shlyakhter I., Karlsson E.K., Byrne E.H., Morales S., Frieden G., Hostetter E., Angelino E., Garber M., Zuk O., et al. 2010. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 327:883–886. 10.1126/science.1183863 [DOI] [PubMed] [Google Scholar]
- Grossman S.R., Andersen K.G., Shlyakhter I., Tabrizi S., Winnicki S., Yen A., Park D.J., Griesemer D., Karlsson E.K., Wong S.H., et al. 1000 Genomes Project . 2013. Identifying recent adaptations in large-scale genomic data. Cell. 152:703–713. 10.1016/j.cell.2013.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium 2015. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 348:648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst R.N., Hernandez R.D., Williamson S.H., and Bustamante C.D.. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5:e1000695 10.1371/journal.pgen.1000695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin M.T., and Di Rienzo A.. 2000. Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am. J. Hum. Genet. 66:1669–1679. 10.1086/302879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin M.T., Thompson E.E., and Di Rienzo A.. 2002. Complex signatures of natural selection at the Duffy blood group locus. Am. J. Hum. Genet. 70:369–383. 10.1086/338628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M.F., Woerner A.E., Mendez F.L., Watkins J.C., and Wall J.D.. 2011. Genetic evidence for archaic admixture in Africa. Proc. Natl. Acad. Sci. USA. 108:15123–15128. 10.1073/pnas.1109300108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn B.M., Cavalli-Sforza L.L., and Feldman M.W.. 2012. The great human expansion. Proc. Natl. Acad. Sci. USA. 109:17758–17764. 10.1073/pnas.1212380109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn B.M., Botigué L.R., Bustamante C.D., Clark A.G., and Gravel S.. 2015. Estimating the mutation load in human genomes. Nat. Rev. Genet. 16:333–343. 10.1038/nrg3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn B.M., Botigué L.R., Peischl S., Dupanloup I., Lipatov M., Maples B.K., Martin A.R., Musharoff S., Cann H., Snyder M.P., et al. 2016. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc. Natl. Acad. Sci. USA. 113:E440–E449. 10.1073/pnas.1510805112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., and Manolio T.A.. 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 106:9362–9367. 10.1073/pnas.0903103106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger K.E., and Weir B.S.. 2009. Genetics in geographically structured populations: defining, estimating and interpreting F(ST). Nat. Rev. Genet. 10:639–650. 10.1038/nrg2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Sánchez E., Jin X., Asan Z., Bianba Z., Peter B.M., Vinckenbosch N., Liang Y., Yi X., He M., Somel M., et al. 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 512:194–197. 10.1038/nature13408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt K.A., Zhernakova A., Turner G., Heap G.A., Franke L., Bruinenberg M., Romanos J., Dinesen L.C., Ryan A.W., Panesar D., et al. 2008. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 40:395–402. 10.1038/ng.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itan Y., Powell A., Beaumont M.A., Burger J., and Thomas M.G.. 2009. The origins of lactase persistence in Europe. PLOS Comput. Biol. 5:e1000491 10.1371/journal.pcbi.1000491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M., Scholz S.W., Scheet P., Gibbs J.R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., et al. 2008. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 451:998–1003. 10.1038/nature06742 [DOI] [PubMed] [Google Scholar]
- Jarvis J.P., Scheinfeldt L.B., Soi S., Lambert C., Omberg L., Ferwerda B., Froment A., Bodo J.M., Beggs W., Hoffman G., et al. 2012. Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African pygmies. PLoS Genet. 8:e1002641 10.1371/journal.pgen.1002641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C., and Di Rienzo A.. 2014. Adaptations to local environments in modern human populations. Curr. Opin. Genet. Dev. 29:1–8. 10.1016/j.gde.2014.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.Y., Kang A.H., Green A., Gwee K.A., and Ho K.Y.. 2013. Systematic review: worldwide variation in the frequency of coeliac disease and changes over time. Aliment. Pharmacol. Ther. 38:226–245. 10.1111/apt.12373 [DOI] [PubMed] [Google Scholar]
- Karlsson E.K., Kwiatkowski D.P., and Sabeti P.C.. 2014. Natural selection and infectious disease in human populations. Nat. Rev. Genet. 15:379–393. 10.1038/nrg3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelso J., and Prüfer K.. 2014. Ancient humans and the origin of modern humans. Curr. Opin. Genet. Dev. 29:133–138. 10.1016/j.gde.2014.09.004 [DOI] [PubMed] [Google Scholar]
- Key F.M., Peter B., Dennis M.Y., Huerta-Sánchez E., Tang W., Prokunina-Olsson L., Nielsen R., and Andrés A.M.. 2014a Selection on a variant associated with improved viral clearance drives local, adaptive pseudogenization of interferon lambda 4 (IFNL4). PLoS Genet. 10:e1004681 10.1371/journal.pgen.1004681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key F.M., Teixeira J.C., de Filippo C., and Andrés A.M.. 2014b Advantageous diversity maintained by balancing selection in humans. Curr. Opin. Genet. Dev. 29:45–51. 10.1016/j.gde.2014.08.001 [DOI] [PubMed] [Google Scholar]
- Konopka G., and Roberts T.F.. 2016. Insights into the neural and genetic basis of vocal communication. Cell. 164:1269–1276. 10.1016/j.cell.2016.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M. 2000. Methods to detect selection in populations with applications to the human. Annu. Rev. Genomics Hum. Genet. 1:539–559. 10.1146/annurev.genom.1.1.539 [DOI] [PubMed] [Google Scholar]
- Kryukov G.V., Pennacchio L.A., and Sunyaev S.R.. 2007. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am. J. Hum. Genet. 80:727–739. 10.1086/513473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski D.P. 2005. How malaria has affected the human genome and what human genetics can teach us about malaria. Am. J. Hum. Genet. 77:171–192. 10.1086/432519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance J., Vernot B., Elbers C.C., Ferwerda B., Froment A., Bodo J.M., Lema G., Fu W., Nyambo T.B., Rebbeck T.R., et al. 2012. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell. 150:457–469. 10.1016/j.cell.2012.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason R.L., Mohideen M.A., Mest J.R., Wong A.C., Norton H.L., Aros M.C., Jurynec M.J., Mao X., Humphreville V.R., Humbert J.E., et al. 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 310:1782–1786. 10.1126/science.1116238 [DOI] [PubMed] [Google Scholar]
- Lee M.N., Ye C., Villani A.C., Raj T., Li W., Eisenhaure T.M., Imboywa S.H., Chipendo P.I., Ran F.A., Slowikowski K., et al. 2014. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 343:1246980 10.1126/science.1246980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler E.M., Gao Z., Pfeifer S., Ségurel L., Auton A., Venn O., Bowden R., Bontrop R., Wall J.D., Sella G., et al. 2013. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science. 339:1578–1582. 10.1126/science.1234070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Exome Aggregation Consortium . 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536:285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Oosting M., Smeekens S.P., Jaeger M., Aguirre-Gamboa R., Le K.T., Deelen P., Ricaño-Ponce I., Schoffelen T., Jansen A.F., et al. 2016. A functional genomics approach to understand variation in cytokine production in humans. Cell. 167:1099–1110.e14. 10.1016/j.cell.2016.10.017 [DOI] [PubMed] [Google Scholar]
- Lim E.T., Würtz P., Havulinna A.S., Palta P., Tukiainen T., Rehnström K., Esko T., Mägi R., Inouye M., Lappalainen T., et al. Sequencing Initiative Suomi (SISu) Project . 2014. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 10:e1004494 10.1371/journal.pgen.1004494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnen C.R., and Hoekstra H.E.. 2009. Measuring natural selection on genotypes and phenotypes in the wild. Cold Spring Harb. Symp. Quant. Biol. 74:155–168. 10.1101/sqb.2009.74.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A., Carr E.J., and Linterman M.A.. 2016. Shaping variation in the human immune system. Trends Immunol. 37:637–646. 10.1016/j.it.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Liu H., Irwanto A., Fu X., Yu G., Yu Y., Sun Y., Wang C., Wang Z., Okada Y., Low H., et al. 2015. Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nat. Genet. 47:267–271. 10.1038/ng.3212 [DOI] [PubMed] [Google Scholar]
- Lohmueller K.E. 2014. The distribution of deleterious genetic variation in human populations. Curr. Opin. Genet. Dev. 29:139–146. 10.1016/j.gde.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Lohmueller K.E., Indap A.R., Schmidt S., Boyko A.R., Hernandez R.D., Hubisz M.J., Sninsky J.J., White T.J., Sunyaev S.R., Nielsen R., et al. 2008. Proportionally more deleterious genetic variation in European than in African populations. Nature. 451:994–997. 10.1038/nature06611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louicharoen C., Patin E., Paul R., Nuchprayoon I., Witoonpanich B., Peerapittayamongkol C., Casademont I., Sura T., Laird N.M., Singhasivanon P., et al. 2009. Positively selected G6PD-Mahidol mutation reduces Plasmodium vivax density in Southeast Asians. Science. 326:1546–1549. 10.1126/science.1178849 [DOI] [PubMed] [Google Scholar]
- Malaspinas A.S., Westaway M.C., Muller C., Sousa V.C., Lao O., Alves I., Bergström A., Athanasiadis G., Cheng J.Y., Crawford J.E., et al. 2016. A genomic history of Aboriginal Australia. Nature. 538:207–214. 10.1038/nature18299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., Zhao M., Chennagiri N., Nordenfelt S., Tandon A., et al. 2016. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 538:201–206. 10.1038/nature18964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., et al. 2009. Finding the missing heritability of complex diseases. Nature. 461:747–753. 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manry J., Laval G., Patin E., Fornarino S., Itan Y., Fumagalli M., Sironi M., Tichit M., Bouchier C., Casanova J.L., et al. 2011. Evolutionary genetic dissection of human interferons. J. Exp. Med. 208:2747–2759. 10.1084/jem.20111680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I., Lazaridis I., Rohland N., Mallick S., Patterson N., Roodenberg S.A., Harney E., Stewardson K., Fernandes D., Novak M., et al. 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 528:499–503. 10.1038/nature16152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez F.L., Watkins J.C., and Hammer M.F.. 2012. A haplotype at STAT2 Introgressed from neanderthals and serves as a candidate of positive selection in Papua New Guinea. Am. J. Hum. Genet. 91:265–274. 10.1016/j.ajhg.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez F.L., Watkins J.C., and Hammer M.F.. 2013. Neandertal origin of genetic variation at the cluster of OAS immunity genes. Mol. Biol. Evol. 30:798–801. 10.1093/molbev/mst004 [DOI] [PubMed] [Google Scholar]
- Meyer M., Kircher M., Gansauge M.T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prüfer K., de Filippo C., et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science. 338:222–226. 10.1126/science.1224344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.T., Beleza S., Pollen A.A., Schluter D., Kittles R.A., Shriver M.D., and Kingsley D.M.. 2007. cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 131:1179–1189. 10.1016/j.cell.2007.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery S.B., and Dermitzakis E.T.. 2011. From expression QTLs to personalized transcriptomics. Nat. Rev. Genet. 12:277–282. 10.1038/nrg2969 [DOI] [PubMed] [Google Scholar]
- Montinaro F., Busby G.B., Pascali V.L., Myers S., Hellenthal G., and Capelli C.. 2015. Unravelling the hidden ancestry of American admixed populations. Nat. Commun. 6:6596 10.1038/ncomms7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nédélec Y., Sanz J., Baharian G., Szpiech Z.A., Pacis A., Dumaine A., Grenier J.C., Freiman A., Sams A.J., Hebert S., et al. 2016. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell. 167:657–669.e21. 10.1016/j.cell.2016.09.025 [DOI] [PubMed] [Google Scholar]
- Newbold C., Craig A., Kyes S., Rowe A., Fernandez-Reyes D., and Fagan T.. 1999. Cytoadherence, pathogenesis and the infected red cell surface in Plasmodium falciparum. Int. J. Parasitol. 29:927–937. 10.1016/S0020-7519(99)00049-1 [DOI] [PubMed] [Google Scholar]
- Nica A.C., Parts L., Glass D., Nisbet J., Barrett A., Sekowska M., Travers M., Potter S., Grundberg E., Small K., et al. MuTHER Consortium . 2011. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 7:e1002003 10.1371/journal.pgen.1002003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., and Cox N.J.. 2010. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6:e1000888 10.1371/journal.pgen.1000888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. 2005. Molecular signatures of natural selection. Annu. Rev. Genet. 39:197–218. 10.1146/annurev.genet.39.073003.112420 [DOI] [PubMed] [Google Scholar]
- Nielsen R., Williamson S., Kim Y., Hubisz M.J., Clark A.G., and Bustamante C.. 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15:1566–1575. 10.1101/gr.4252305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Hellmann I., Hubisz M., Bustamante C., and Clark A.G.. 2007. Recent and ongoing selection in the human genome. Nat. Rev. Genet. 8:857–868. 10.1038/nrg2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Akey J.M., Jakobsson M., Pritchard J.K., Tishkoff S., and Willerslev E.. 2017. Tracing the peopling of the world through genomics. Nature. 541:302–310. 10.1038/nature21347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman P.J., Abi-Rached L., Gendzekhadze K., Korbel D., Gleimer M., Rowley D., Bruno D., Carrington C.V., Chandanayingyong D., Chang Y.H., et al. 2007. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat. Genet. 39:1092–1099. 10.1038/ng2111 [DOI] [PubMed] [Google Scholar]
- Norton H.L., Kittles R.A., Parra E., McKeigue P., Mao X., Cheng K., Canfield V.A., Bradley D.G., McEvoy B., and Shriver M.D.. 2007. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol. Biol. Evol. 24:710–722. 10.1093/molbev/msl203 [DOI] [PubMed] [Google Scholar]
- Novembre J., and Ramachandran S.. 2011. Perspectives on human population structure at the cusp of the sequencing era. Annu. Rev. Genomics Hum. Genet. 12:245–274. 10.1146/annurev-genom-090810-183123 [DOI] [PubMed] [Google Scholar]
- Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R., et al. 2008. Genes mirror geography within Europe. Nature. 456:98–101. 10.1038/nature07331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrù V., Steri M., Sole G., Sidore C., Virdis F., Dei M., Lai S., Zoledziewska M., Busonero F., Mulas A., et al. 2013. Genetic variants regulating immune cell levels in health and disease. Cell. 155:242–256. 10.1016/j.cell.2013.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani L., Lawson D.J., Jagoda E., Mörseburg A., Eriksson A., Mitt M., Clemente F., Hudjashov G., DeGiorgio M., Saag L., et al. 2016. Genomic analyses inform on migration events during the peopling of Eurasia. Nature. 538:238–242. 10.1038/nature19792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes M., Cortes A., van Heel D.A., and Brown M.A.. 2013. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 14:661–673. 10.1038/nrg3502 [DOI] [PubMed] [Google Scholar]
- Patin E., Siddle K.J., Laval G., Quach H., Harmant C., Becker N., Froment A., Régnault B., Lemée L., Gravel S., et al. 2014. The impact of agricultural emergence on the genetic history of African rainforest hunter-gatherers and agriculturalists. Nat. Commun. 5:3163 10.1038/ncomms4163 [DOI] [PubMed] [Google Scholar]
- Peng Y., Yang Z., Zhang H., Cui C., Qi X., Luo X., Tao X., Wu T., Ouzhuluobu, Basang, et al. 2011. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol. Biol. Evol. 28:1075–1081. 10.1093/molbev/msq290 [DOI] [PubMed] [Google Scholar]
- Pérez de Diego R., Sancho-Shimizu V., Lorenzo L., Puel A., Plancoulaine S., Picard C., Herman M., Cardon A., Durandy A., Bustamante J., et al. 2010. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 33:400–411. 10.1016/j.immuni.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G.H., Kistler L., Kelaita M.A., and Sams A.J.. 2015. Insights into hominin phenotypic and dietary evolution from ancient DNA sequence data. J. Hum. Evol. 79:55–63. 10.1016/j.jhevol.2014.10.018 [DOI] [PubMed] [Google Scholar]
- Pickrell J.K. 2014. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 94:559–573. 10.1016/j.ajhg.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J.E., Henders A.K., McRae A.F., Wright M.J., Martin N.G., Dermitzakis E.T., Montgomery G.W., and Visscher P.M.. 2012. Genetic control of gene expression in whole blood and lymphoblastoid cell lines is largely independent. Genome Res. 22:456–466. 10.1101/gr.126540.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Helgason A., Thorleifsson G., McCarroll S.A., Kong A., and Stefansson K.. 2011. Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals. PLoS Genet. 7:e1001317 10.1371/journal.pgen.1001317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J.K., Pickrell J.K., and Coop G.. 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20:R208–R215. 10.1016/j.cub.2009.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C., et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 505:43–49. 10.1038/nature12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach H., Rotival M., Pothlichet J., Loh Y.E., Dannemann M., Zidane N., Laval G., Patin E., Harmant C., Lopez M., et al. 2016. Genetic Adaptation and Neandertal admixture shaped the immune system of human populations. Cell. 167:643–656.e17. 10.1016/j.cell.2016.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L., and Barreiro L.B.. 2010. The role played by natural selection on Mendelian traits in humans. Ann. N. Y. Acad. Sci. 1214:1–17. 10.1111/j.1749-6632.2010.05856.x [DOI] [PubMed] [Google Scholar]
- Quintana-Murci L., and Clark A.G.. 2013. Population genetic tools for dissecting innate immunity in humans. Nat. Rev. Immunol. 13:280–293. 10.1038/nri3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L., Alcaïs A., Abel L., and Casanova J.L.. 2007. Immunology in natura: clinical, epidemiological and evolutionary genetics of infectious diseases. Nat. Immunol. 8:1165–1171. 10.1038/ni1535 [DOI] [PubMed] [Google Scholar]
- Racimo F., Sankararaman S., Nielsen R., and Huerta-Sánchez E.. 2015. Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 16:359–371. 10.1038/nrg3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj T., Rothamel K., Mostafavi S., Ye C., Lee M.N., Replogle J.M., Feng T., Lee M., Asinovski N., Frohlich I., et al. 2014. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 344:519–523. 10.1126/science.1249547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranciaro A., Campbell M.C., Hirbo J.B., Ko W.Y., Froment A., Anagnostou P., Kotze M.J., Ibrahim M., Nyambo T., Omar S.A., and Tishkoff S.A.. 2014. Genetic origins of lactase persistence and the spread of pastoralism in Africa. Am. J. Hum. Genet. 94:496–510. 10.1016/j.ajhg.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D., Green R.E., Kircher M., Krause J., Patterson N., Durand E.Y., Viola B., Briggs A.W., Stenzel U., Johnson P.L., et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 468:1053–1060. 10.1038/nature09710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D., Patterson N., Kircher M., Delfin F., Nandineni M.R., Pugach I., Ko A.M., Ko Y.C., Jinam T.A., Phipps M.E., et al. 2011. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am. J. Hum. Genet. 89:516–528. 10.1016/j.ajhg.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M., Quaye L., Mangino M., Beddall M.H., Mahnke Y., Chattopadhyay P., Tosi I., Napolitano L., Terranova Barberio M., Menni C., et al. 2015. The genetic architecture of the human immune system: a bioresource for autoimmunity and disease pathogenesis. Cell. 161:387–403. 10.1016/j.cell.2015.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwende C., Khoo S.C., Snow R.W., Yates S.N., Kwiatkowski D., Gupta S., Warn P., Allsopp C.E., Gilbert S.C., Peschu N., et al. 1995. Natural selection of hemi- and heterozygotes for G6PD deficiency in Africa by resistance to severe malaria. Nature. 376:246–249. 10.1038/376246a0 [DOI] [PubMed] [Google Scholar]
- Sabeti P.C., Reich D.E., Higgins J.M., Levine H.Z., Richter D.J., Schaffner S.F., Gabriel S.B., Platko J.V., Patterson N.J., McDonald G.J., et al. 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature. 419:832–837. 10.1038/nature01140 [DOI] [PubMed] [Google Scholar]
- Sabeti P.C., Schaffner S.F., Fry B., Lohmueller J., Varilly P., Shamovsky O., Palma A., Mikkelsen T.S., Altshuler D., and Lander E.S.. 2006. Positive natural selection in the human lineage. Science. 312:1614–1620. 10.1126/science.1124309 [DOI] [PubMed] [Google Scholar]
- Sabeti P.C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E.H., McCarroll S.A., Gaudet R., et al. International HapMap Consortium . 2007. Genome-wide detection and characterization of positive selection in human populations. Nature. 449:913–918. 10.1038/nature06250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sams A.J., Dumaine A., Nédélec Y., Yotova V., Alfieri C., Tanner J.E., Messer P.W., and Barreiro L.B.. 2016. Adaptively introgressed Neandertal haplotype at the OAS locus functionally impacts innate immune responses in humans. Genome Biol. 17:246 10.1186/s13059-016-1098-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S., Mallick S., Dannemann M., Prüfer K., Kelso J., Pääbo S., Patterson N., and Reich D.. 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 507:354–357. 10.1038/nature12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders M.A., Hammer M.F., and Nachman M.W.. 2002. Nucleotide variability at G6pd and the signature of malarial selection in humans. Genetics. 162:1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub M.A., Boyle A.P., Kundaje A., Batzoglou S., and Snyder M.. 2012. Linking disease associations with regulatory information in the human genome. Genome Res. 22:1748–1759. 10.1101/gr.136127.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeldt L.B., and Tishkoff S.A.. 2013. Recent human adaptation: genomic approaches, interpretation and insights. Nat. Rev. Genet. 14:692–702. 10.1038/nrg3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M., Smeekens S.P., Vlamakis H., Jaeger M., Oosting M., Franzosa E.A., Jansen T., Jacobs L., Bonder M.J., Kurilshikov A., et al. 2016. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 167:1125–1136.e8. 10.1016/j.cell.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddle K.J., and Quintana-Murci L.. 2014. The Red Queen’s long race: human adaptation to pathogen pressure. Curr. Opin. Genet. Dev. 29:31–38. 10.1016/j.gde.2014.07.004 [DOI] [PubMed] [Google Scholar]
- Siddle K.J., Deschamps M., Tailleux L., Nédélec Y., Pothlichet J., Lugo-Villarino G., Libri V., Gicquel B., Neyrolles O., Laval G., et al. 2014. A genomic portrait of the genetic architecture and regulatory impact of microRNA expression in response to infection. Genome Res. 24:850–859. 10.1101/gr.161471.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons Y.B., and Sella G.. 2016. The impact of recent population history on the deleterious mutation load in humans and close evolutionary relatives. Curr. Opin. Genet. Dev. 41:150–158. 10.1016/j.gde.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons Y.B., Turchin M.C., Pritchard J.K., and Sella G.. 2014. The deleterious mutation load is insensitive to recent population history. Nat. Genet. 46:220–224. 10.1038/ng.2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson T.S., Yang Y., Huff C.D., Yun H., Qin G., Witherspoon D.J., Bai Z., Lorenzo F.R., Xing J., Jorde L.B., et al. 2010. Genetic evidence for high-altitude adaptation in Tibet. Science. 329:72–75. 10.1126/science.1189406 [DOI] [PubMed] [Google Scholar]
- Simonti C.N., Vernot B., Bastarache L., Bottinger E., Carrell D.S., Chisholm R.L., Crosslin D.R., Hebbring S.J., Jarvik G.P., Kullo I.J., et al. 2016. The phenotypic legacy of admixture between modern humans and Neandertals. Science. 351:737–741. 10.1126/science.aad2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Single R.M., Martin M.P., Gao X., Meyer D., Yeager M., Kidd J.R., Kidd K.K., and Carrington M.. 2007. Global diversity and evidence for coevolution of KIR and HLA. Nat. Genet. 39:1114–1119. 10.1038/ng2077 [DOI] [PubMed] [Google Scholar]
- Sironi M., and Clerici M.. 2010. The hygiene hypothesis: an evolutionary perspective. Microbes Infect. 12:421–427. 10.1016/j.micinf.2010.02.002 [DOI] [PubMed] [Google Scholar]
- Smith M.W., and O’Brien S.J.. 2005. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat. Rev. Genet. 6:623–632. 10.1038/nrg1657 [DOI] [PubMed] [Google Scholar]
- Snyder-Mackler N., Sanz J., Kohn J.N., Brinkworth J.F., Morrow S., Shaver A.O., Grenier J.C., Pique-Regi R., Johnson Z.P., Wilson M.E., et al. 2016. Social status alters immune regulation and response to infection in macaques. Science. 354:1041–1045. 10.1126/science.aah3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman R.S., Bastone L.A., Burdick J.T., Morley M., Ewens W.J., and Cheung V.G.. 2007. Common genetic variants account for differences in gene expression among ethnic groups. Nat. Genet. 39:226–231. 10.1038/ng1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns S.C., Byars S.G., Govindaraju D.R., and Ewbank D.. 2010. Measuring selection in contemporary human populations. Nat. Rev. Genet. 11:611–622. 10.1038/nrg2831 [DOI] [PubMed] [Google Scholar]
- Stranger B.E., Montgomery S.B., Dimas A.S., Parts L., Stegle O., Ingle C.E., Sekowska M., Smith G.D., Evans D., Gutierrez-Arcelus M., et al. 2012. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 8:e1002639 10.1371/journal.pgen.1002639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Yang Y., Tang K., Sabeti P.C., Jin L., and Wang S.. 2013. The adaptive variant EDARV370A is associated with straight hair in East Asians. Hum. Genet. 132:1187–1191. 10.1007/s00439-013-1324-1 [DOI] [PubMed] [Google Scholar]
- Tennessen J.A., Bigham A.W., O’Connor T.D., Fu W., Kenny E.E., Gravel S., McGee S., Do R., Liu X., Jun G., et al. NHLBI Exome Sequencing Project . 2012. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 337:64–69. 10.1126/science.1219240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Horst R., Jaeger M., Smeekens S.P., Oosting M., Swertz M.A., Li Y., Kumar V., Diavatopoulos D.A., Jansen A.F., Lemmers H., et al. 2016. Host and environmental factors influencing individual human cytokine responses. Cell. 167:1111–1124.e13. 10.1016/j.cell.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff S.A., Varkonyi R., Cahinhinan N., Abbes S., Argyropoulos G., Destro-Bisol G., Drousiotou A., Dangerfield B., Lefranc G., Loiselet J., et al. 2001. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science. 293:455–462. 10.1126/science.1061573 [DOI] [PubMed] [Google Scholar]
- Tishkoff S.A., Reed F.A., Ranciaro A., Voight B.F., Babbitt C.C., Silverman J.S., Powell K., Mortensen H.M., Hirbo J.B., Osman M., et al. 2007. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39:31–40. 10.1038/ng1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J., Barreiro L.B., Johnson Z.P., Hansen K.D., Michopoulos V., Toufexis D., Michelini K., Wilson M.E., and Gilad Y.. 2012. Social environment is associated with gene regulatory variation in the rhesus macaque immune system. Proc. Natl. Acad. Sci. USA. 109:6490–6495. 10.1073/pnas.1202734109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannberg F.O., Chapman S.J., and Hill A.V.. 2011. Human genetic susceptibility to intracellular pathogens. Immunol. Rev. 240:105–116. 10.1111/j.1600-065X.2010.00996.x [DOI] [PubMed] [Google Scholar]
- Vattathil S., and Akey J.M.. 2015. Small amounts of archaic admixture provide big insights into human history. Cell. 163:281–284. 10.1016/j.cell.2015.09.042 [DOI] [PubMed] [Google Scholar]
- Veeramah K.R., and Hammer M.F.. 2014. The impact of whole-genome sequencing on the reconstruction of human population history. Nat. Rev. Genet. 15:149–162. 10.1038/nrg3625 [DOI] [PubMed] [Google Scholar]
- Vernot B., and Akey J.M.. 2014. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 343:1017–1021. 10.1126/science.1245938 [DOI] [PubMed] [Google Scholar]
- Vernot B., Tucci S., Kelso J., Schraiber J.G., Wolf A.B., Gittelman R.M., Dannemann M., Grote S., McCoy R.C., Norton H., et al. 2016. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 352:235–239. 10.1126/science.aad9416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitti J.J., Grossman S.R., and Sabeti P.C.. 2013. Detecting natural selection in genomic data. Annu. Rev. Genet. 47:97–120. 10.1146/annurev-genet-111212-133526 [DOI] [PubMed] [Google Scholar]
- Voight B.F., Kudaravalli S., Wen X., and Pritchard J.K.. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4:e72 10.1371/journal.pbio.0040072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E.T., Kodama G., Baldi P., and Moyzis R.K.. 2006. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc. Natl. Acad. Sci. USA. 103:135–140. 10.1073/pnas.0509691102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z.X., Pool J.E., Xu X., Jiang H., Vinckenbosch N., Korneliussen T.S., et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 329:75–78. 10.1126/science.1190371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A., Elbers C.C., Ferwerda B., Romanos J., Trynka G., Dubois P.C., de Kovel C.G., Franke L., Oosting M., Barisani D., et al. Finnish Celiac Disease Study Group . 2010. Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am. J. Hum. Genet. 86:970–977. 10.1016/j.ajhg.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]