Abstract
Bioremediation as a method for removing polycyclic aromatic hydrocarbons (PAHs) from contaminated environments has been criticized for poor removal of potentially carcinogenic but less bioavailable high-molecular-weight (HMW) compounds. As a partial remedy to this constraint, we studied surfactant addition at sub-micellar concentrations to contaminated soil to enhance the biodegradation of PAHs remaining after conventional aerobic bioremediation. We demonstrated increased removal of 4- and 5-ring PAHs using two nonionic surfactants, polyoxyethylene(4)lauryl ether (Brij 30) and polyoxyethylene sorbitol hexaoleate (POESH), and analyzed bacterial community shifts associated with those conditions. Eight groups of abundant bacteria were implicated as potentially being involved in increased HMW PAH removal. A group of unclassified Alphaproteobacteria and members of the Phenylobacterium genus in particular showed significantly increased relative abundance in the two conditions exhibiting increased PAH removal. Other implicated groups included members of the Sediminibacterium, Terrimonas, Acidovorax, and Luteimonas genera, as well as uncharacterized organisms within the families Chitinophagaceae and Bradyrhizobiaceae. Targeted isolation identified a subset of the community likely using the surfactants as a growth substrate but few of the isolates exhibited PAH-degradation capability. Isolates recovered from the Acidovorax and uncharacterized Bradyrhizobiaceae groups suggest the abundance of those groups may have been attributable to growth on surfactants. Understanding the specific bacteria responsible for HMW PAH removal in natural and engineered systems and their response to stimuli such as surfactant amendment may improve bioremediation efficacy during treatment of contaminated environmental media.
Keywords: polycyclic aromatic hydrocarbons, surfactants, bacterial communities
Introduction
Bioremediation is among several options for the removal of polycyclic aromatic hydrocarbons (PAHs) from contaminated environments (Gan et al. 2009; U. S. Environmental Protection Agency 2007). Biological treatment of PAH-contaminated sites is dependent on the presence and activity of microorganisms capable of transforming the compounds of concern. While bioremediation is often successful in the removal of low-molecular-weight (LMW) PAHs such as naphthalene or phenanthrene, high-molecular-weight (HMW) PAHs of 4 or more rings, and particularly those of 5 or more rings, typically prove more resistant to microbial attack (Cerniglia 1993). Sixteen PAHs are designated priority pollutants by the U.S. Environmental Protection Agency (EPA) and seven of them, all HMW PAHs, are additionally considered probable human carcinogens (Agency for Toxic Substances and Disease Registry 2015; National Toxicology Program 2014). The removal of these larger, potentially more toxic compounds can thus be a determining factor when considering cleanup strategies at contaminated sites.
The physical properties of PAHs are at least partially to blame for the lesser removal of HMW PAHs during bioremediation. The larger PAHs are generally less soluble in water than their already hydrophobic LMW counterparts, with up to orders of magnitude differences in aqueous solubility observed for each additional ring (May et al. 1978). Given their hydrophobicity, PAHs can strongly sorb to organic components of soils commonly found at contaminated sites, thus limiting their bioavailability. Surfactant-amended bioremediation has been proposed as a means of increasing the availability of matrix-bound PAHs to microorganisms (Li and Chen 2009; Makkar and Rockne 2003; Yeom et al. 1996; Zhu and Aitken 2010). Surfactants can increase the rate of PAH desorption through two mechanisms: by maximizing the concentration gradient at the soil-water interface through micellar solubilization of PAH and by direct modification of the contaminant matrix. While micellar solubilization occurs at aqueous-phase surfactant concentrations above the critical micelle concentration (CMC), surfactants can also increase PAH desorption from field-contaminated soil at concentrations below the CMC (Adrion et al. 2016; Elliot et al. 2010; Frutos et al. 2011; Yeom et al. 1996; Zhu and Aitken 2010). Enhanced desorption at doses corresponding to aqueous-phase surfactant concentrations below the CMC in a soil-slurry system (sub-micellar or sub-CMC doses) has been attributed to increased PAH diffusivity within the contaminant matrix (Yeom et al. 1996) or to increased interfacial surface area caused by wetting (Dong et al. 2004) and dispersion of non-polar matrices (Churchill et al. 1995; Kile and Chiou 1989; Zhang and Miller 1992).
In addition to mobilizing matrix-sorbed hydrophobic compounds, surfactants can also impact the PAH-degrading activity of the endogenous microbial community in a contaminated environment (Makkar and Rockne 2003; Volkering et al. 1997). Some surfactants can be utilized as preferred carbon and energy sources for growth by microorganisms with a corresponding decrease in metabolism of the aromatic contaminants (Tiehm 1994). Growth on a high concentration of readily available, surfactant-derived carbon can also result in anoxic conditions in static systems, further limiting PAH degradation (Tiehm et al. 1997). Increased concentrations of surfactants, particularly ionic surfactants, have also been shown to be toxic to some PAH-degrading bacteria (Chen et al. 2000; Jin et al. 2007; Tiehm 1994). Conversely, surfactant-induced transport of PAHs from the environmental matrix into the aqueous phase can also result in increased bioavailability to surfactant-tolerant PAH-degrading microbes with a corresponding increase in transformation. Positive effects of surfactants on the biodegradation of PAHs is generally associated with situations under which PAH bioavailability is limited prior to surfactant addition (Adrion et al. 2016).
As a follow-up to previous work in which sub-CMC doses of the nonionic surfactant Brij 30 were shown to enhance PAH biodegradation (Zhu and Aitken 2010), we screened five nonionic surfactants of similar hydrophobicity for their ability to improve the removal of PAHs remaining in contaminated soil after aerobic biological treatment in a lab-scale, slurry-phase bioreactor (Adrion et al. 2016). Surprisingly, one of the surfactants, Brij 30, led to substantially greater removal of HMW PAHs at the lower of the two sub-CMC doses evaluated. In this study, we focused on the two most effective surfactants, Brij 30 and polyoxyethylene sorbitol hexaoleate (POESH), to determine their effects on the soil microbial communities using community fingerprinting, high-throughput sequencing, and targeted isolation. As differences in HMW PAH removal between treatment conditions likely resulted from differences in the microbial communities selected under each of the conditions, we hypothesized that some HMW PAH-degrading bacterial groups might be putatively identified by analysis of the whole community for members that maintained a high relative abundance, or increased relative abundance, in conditions exhibiting enhanced compound removal, and furthermore were not among those likely to be growing on an amended surfactant.
Methods and Materials
Chemicals
Brij 30 (polyoxyethylene (4) lauryl ether) and polyoxyethylene sorbitol hexaoleate (POESH) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Phenanthrene (98%) and chrysene (98%) were also purchased from Sigma-Aldrich. Pyrene (98%) was purchased from Acros Organics (Thermo Fisher Scientific, Waltham, MA, USA).
Soil and bioreactor treatment
Weathered, PAH-contaminated soil used in this experiment was collected from the site of a former manufactured gas plant in Salisbury, North Carolina, USA that was processed and characterized as described elsewhere (Hu et al. 2012; Richardson and Aitken 2011). Soil was treated in a lab-scale (2 L working volume), semi-continuous, slurry-phase, aerobic bioreactor as previously described (Singleton et al. 2011), except that the soil was slurried in a buffer containing 5 mM total phosphate and 2.5 mM NH4NO3 at a pH of 7.5 (“reactor buffer”). Twenty percent of the bioreactor contents was removed weekly and replaced with freshly slurried, untreated soil. Bioreactor-treated soil that had been centrifuged at 2130 x g for 20 minutes in a Sorvall RC5C plus centrifuge (Thermo Fisher Scientific) with a fixed-angle GSA rotor was used to set up incubations for this study. Soil moisture content was determined in triplicate by heating 1 g (wet weight) of centrifuged bioreactor-treated soil in pre-weighed ceramic crucibles over an open flame until a stable value of dry mass was obtained.
Soil slurry incubations with surfactants
To prepare incubations of bioreactor-treated soil with surfactants, 6 g (wet weight) aliquots of soil from centrifuged bioreactor slurry were added to 125-mL glass, screw-top Erlenmeyer flasks with PTFE-lined septa caps. Triplicate incubations in fresh reactor buffer were prepared for each of four post-bioreactor conditions: 4 mg/g Brij 30 (mg surfactant/g dry weight soil; designated as “BrijLow” samples), 12 mg/g Brij 30 (“BrijHigh”), 24 mg/g POESH (“POESH”), and no-surfactant controls (“NoSurf”). The surfactants were added to the soil slurries (15% w/w final solids content) in the incubation flasks to the desired dose. These doses corresponded to equilibrium aqueous-phase surfactant concentrations below the CMC in the soil-slurry system as determined in parallel work (Adrion et al. 2016). The headspaces of the flasks were then purged with nitrogen and put on a rotary shaker in darkness at 225 rpm for 48 hours to allow sorption of surfactant to the soil while minimizing aerobic biodegradation of the surfactants. After 48 hours, the flasks were uncapped daily for 5 minutes for a period of 14 days. Daily headspace exchange with air would have supplied over 100 mg O2 per g dry soil in each flask over the 14-day incubation. Even if a very high concentration of residual biodegradable contaminants in the soil removed from the bioreactor is assumed, and assuming a significant fraction of surfactant was aerobically biodegraded during the incubation, the O2 demand in the flask would have been less than 25 mg/g dry soil. Therefore, daily replacement of the headspace in each flask provided more than enough O2 to ensure aerobic conditions during the incubation period. On day 14 the flasks were sacrificed for extraction of DNA and determination of PAH concentrations as described below. Triplicate flasks of the initial centrifuged bioreactor soil used as inoculum (designated as “Bioreactor” samples) were prepared in the same manner as the experimental flasks and were processed immediately after preparation for DNA and PAH extractions.
Soil DNA extraction
To recover soil from incubations, 1450 μL of slurry sample from each flask (approximately 0.25 g soil dry weight) was centrifuged for 3 minutes at maximum speed in a benchtop microcentrifuge and the supernatant discarded. The pellet was used for DNA extraction using the FastDNA Spin Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) following the manufacturer’s directions except that cell lysis was achieved by vortexing horizontally secured tubes at maximum speed for 6 minutes. A portion of the recovered DNA was then run on a 0.8% agarose gel and HMW DNA (between 9–23 kbp) was purified using a Qiagen QIAEX II Gel Extraction Kit (Qiagen, Valencia, CA, USA). DNA quality was examined and concentrations estimated by running a portion of the purified DNA on a 1% agarose gel. Each of the triplicate samples of bioreactor slurry and each of the triplicate samples in the four treatment conditions were extracted and analyzed individually.
PAH extraction and quantification
The slurry remaining in each flask after the removal of samples for DNA extraction was used to quantify the concentrations of 14 PAHs. The slurry was transferred to 30-mL glass centrifuge tubes with PTFE-lined septa caps using the aqueous phase of the slurry to wash all soil mass into the centrifuge tubes. The slurry was then centrifuged at 2700 x g for 20 minutes in a Sorvall ST-16 benchtop centrifuge with swinging-bucket rotor. The supernatant was discarded (preliminary analyses indicated all individual PAH concentrations in supernatant were less than the limits of quantification, corresponding to no more than 5% of the initial mass of any individual PAH). Soil pellets were spiked with 200 μL of anthracene-D10 (98 mg·L−1) to serve as an internal measure of PAH recovery. The soil pellets were extracted overnight twice with 10-g anhydrous sodium sulfate and a mixture of 10-ml acetone/10-mL dichloromethane and the solvent extracts analyzed by high-pressure liquid chromatography with fluorescence detection to quantify PAH concentration using the mass of the soil added to each flask minus the amount removed for DNA extraction as previously described (Richardson et al. 2011). The recovery of the anthracene-D10 standard ranged from 91–100 ± 3–7%. The triplicate concentration values of each PAH for each of the four treatment conditions were compared using an ANOVA and Tukey’s HSD test using SAS Enterprise Guide 6.1 (SAS Institute, Cary, NC). Treatment conditions were assigned to significantly different (α=0.05) groups according to the results of those analyses. Percent disappearance was calculated using the means and standard deviations of values from the initial bioreactor samples and from the relevant experimental samples, accounting for propagation of error.
Community analyses
Broad differences among bacterial communities between treatments and the reproducibility of established communities within the treatments were examined using denaturing gradient gel electrophoresis (DGGE). DGGE-PCR was performed using primers 341FGC and 517R and a PCR program consisting of: 94°C for 5 minutes; 10 cycles of 94°C for 1 minute, 65°C for 1 minute (decreasing by 1°C per cycle), and 72°C for 3 minutes; 15 cycles of 94°C for 1 minute, 55°C for 1 minute, and 72°C for 3 minutes; with a final extension at 72°C for 7 minutes before cooling to 4°C. Products were run on a DCode™ Universal Mutation Detection system (Bio-Rad, Hercules, CA, USA) using a 10% acrylamide gel containing a denaturant range of 30–60% and a non-denaturing stacking gel. The gels were run for 16 hours at 60V prior to staining with ethidium bromide. Acquired images were cropped of regions not containing DNA bands, the image was negatively converted, and levels adjusted for the entire image using the GNU Image Manipulation Program v.2.8.
Sequencing of partial bacterial 16S rRNA genes in DNA extracted from soil was performed by MR DNA (Shallowater, TX, USA) on an Illumina MiSeq following the manufacturer’s guidelines. Briefly, variable region V4 was amplified using barcoded forward primer 515F, reverse primer 806R, and the HotStarTaq Plus Master Mix Kit (Qiagen). The PCR program consisted of an initial dwell of 94°C for 3 minutes, followed by 28 cycles of 94°C for 30 seconds, 53°C for 40 seconds and 72°C for 1 minute, after which a final elongation step at 72°C for 5 minutes was performed. PCR products were checked on a 2% agarose gel. Multiple samples were pooled in equal proportions based on their molecular weight and DNA concentrations, and purified using the Agencourt AMPure XP purification system (Beckman Coulter, Inc., Brea, CA, USA). The purified PCR product was used to prepare a library using the Illumina TruSeq DNA library preparation protocol.
The raw R1/R2 paired-end sequence libraries of partial 16S rRNA gene sequences were acquired from the sequencing facility and the make.contigs command of mothur v.1.36.1 (Schloss et al. 2009) was used to assemble the sequences. No sequences were obtained for the submitted negative control sample (DNA suspension buffer only), due to either the lack of PCR product or recovered sequences (Scot Dowd, MR DNA, personal communication). Analyses generally followed the Schloss MiSeq SOP (http://www.mothur.org/wiki/MiSeq_SOP; last accessed Feb. 2016) and followed an operational taxonomic unit- (OTU-) based analysis at a similarity of 97% (Kozich et al. 2013). Libraries were subsampled to match the smallest library for the purposes of diversity analyses. The implementation of get.oturep within mothur was used to determine representative sequences for each OTU. Principal coordinate analyses (PCoA) and non-metric multidimensional scaling (NMDS) analyses were carried out through their implementations in mothur. The relative abundance of each OTU within the sequence libraries was determined considering all sequences, not subsampled libraries. The phylogenetic classification of sequences was performed using the classify.seqs Bayesian classifier implementation of mothur with sequence and taxonomic data from training set 14 provided by the Ribosomal Database Project (RDP) (Wang et al. 2007).
Specific OTUs with increased representation in libraries exhibiting increased HMW PAH removal were identified using the metastats (White et al. 2009) implementation of mothur with the criteria of significant difference in representation between target and non-target libraries (p < 0.05), average relative abundance in target libraries ≥ 0.1%, and increased representation in target libraries compared to non-target libraries. Significant differences in the relative abundances of specific OTUs between experimental conditions were determined using ANOVA and Tukey’s HSD tests in R (version 3.1.3; R Foundation for Statistical Computing, Vienna, Austria).
Isolation of surfactant-degrading bacteria
Bacteria capable of using either Brij 30 or POESH as a sole source of energy and carbon were isolated from surfactant-amended soil slurries. Duplicate flasks with conditions identical to those used for PAH determination and DNA extractions, including an initial 2-day anoxic incubation period, were incubated for 15 days in the dark at room temperature (~23°C) with shaking at 225 rpm. Serial dilutions of slurry from each flask (10−4 to 10−9) were then prepared in phosphate-buffered saline (PBS; pH 7.5) and plated on sRB solid media with Brij 30 or POESH. The sRB medium was a modification of one used for the cultivation of Sulfuritalea hydrogenivorans and the composition of solutions can be found in the relevant citation and references within (Kojima and Fukui 2011). The sRB medium consisted of (per L): 993.3 mL of a 5 mM sodium-potassium phosphate buffer (pH 7.0) containing 5 mM NH4NO3, 100 μl of 1M MgSO4·7 H2O, 100 μl of 1M CaCl2·2 H2O, 1 mL of a trace element solution, 1 mL of a Se-W solution, and 1.5% agar added prior to autoclaving at 121°C for 20 minutes. After the media had cooled were added filtered-sterilized (0.2 μm pore-size) 1 mL of a vitamin solution, 1 mL of a thiamine solution, 1 mL of a vitamin B12 solution, and 1.5 mL of a thiosulfate solution (stored under N2). Surfactants were added to a concentration approximately equal to the expected sub-CMC concentrations in the aqueous-phase of the enrichment cultures after the 48h anoxic mixing period (BrijLow – 0.07%; BrijHigh – 0.2%; POESH – 0.4%). Plates were incubated inverted at room temperature. Moisture collected in lids of the inverted plates during incubation, likely due to the surfactants in the medium, and care was taken to ensure that free liquid did not disrupt colonies on the plate surfaces. Starting after a minimum of 15 days of incubation, well-isolated and morphologically distinct colonies were selected from the highest dilution plates that showed growth and struck out on sRB plates containing surfactant for isolation. Colonies recovered from those plates were maintained on R2A agar (Difco, BD, Franklin Lakes, NJ, USA) or in R-2A broth (Himedia, Mumbai, India). For long-term storage, bacterial isolates grown in R-2A broth were stored in 15% glycerol at −80°C.
PCR was conducted with cells suspended in water as template using general bacterial 16S rRNA gene primers 8F and 1492R, and DNA sequencing with the same primers (Eton Biosciences; Research Triangle Park, NC, USA) was used to obtain partial 16S rRNA gene sequences for isolate identification. Partial 16S rRNA gene sequences of at least 1275 bases for each isolate were assembled using Sequencher (version 5.0; Gene Codes, Ann Arbor, MI, USA). Blastn searches of the NCBI 16S ribosomal RNA sequences database were used to determine the closest described relatives (Altschul et al. 1990). Local blast+ searches of a database comprising reference sequences from OTUs in the amplicon libraries were used to associate isolates with OTUs using 97% sequence identity (Camacho et al. 2009). The largest OTU matching isolate sequences using this criterion was reported.
Transformation of the PAHs phenanthrene, pyrene, or chrysene by isolates was tested using a spray-plate method. Isolates were grown in R-2A broth to turbidity and 15 μL of that culture was spotted onto a region of an R2A plate; separate plates were inoculated for each tested PAH. The spotted culture was incubated at room temperature overnight to allow for initiation of growth as well as for liquid to absorb into the solid medium. The plates were then separately sprayed with one of the three PAHs (delivered as a solution of ~2% in acetone) with a thin-layer chromatography plate sprayer until a thin, crystalline film covered the entire surface of the plate. The plates were sealed with parafilm and incubated inverted at either 23°C or 30°C for one week. Cultures were examined at regular intervals for zones of clearing and/or color changes in the crystalline PAH layer and scored positively or negatively for such by visual examination. R2A plates were used due to generally strong growth of the isolates on that medium and to provide carbon sources for isolates either incapable of growth on the sprayed PAHs or requiring an alternate carbon source during transformation of those PAHs.
Sequence deposition
16S rRNA gene amplicon libraries derived from bioreactor and surfactant-amended samples were deposited in the NCBI Sequence Read Archive (SRA) under the accession number SRP070169. Partial 16S rRNA gene sequences of representative isolates were deposited in GenBank under the accession numbers KT888011 – KT888025.
Results
PAH removal
The concentrations of 14 of the 16 EPA priority pollutant PAHs were quantified in the bioreactor-treated soil slurry without additional incubation (“Bioreactor” samples), bioreactor-treated slurry incubated an additional 14 days with either of two sub-CMC doses of Brij 30 (“BrijLow” or “BrijHigh” samples) or a sub-CMC dose of POESH (“POESH” samples), and bioreactor-treated slurry without surfactant incubated an additional 14 days (no-surfactant control; “NoSurf” samples). Two weeks of additional incubation in a fresh buffer after bioreactor treatment with or without surfactant generally resulted in the increased disappearance of total PAHs (Figure 1, Supplementary Table S1). In surfactant-amended samples there was generally either no or modest impact on the further removal of LMW (2- or 3-ring) PAHs. Differences in the removal of HMW PAHs (4+ rings) in surfactant-amended incubations were more pronounced, with fluoranthene (4 rings), pyrene (4 rings), benz[a]anthracene (4 rings), chrysene (4 rings), and benzo[k]fluoranthene (5 rings) in particular showing large decreases, particularly in the BrijLow and/or POESH conditions. Consistent with previous observations (Adrion et al. 2016), incubations containing a higher concentration of Brij 30 (BrijHigh) were not significantly better than the no-surfactant control for removing HMW PAHs except for fluoranthene. The remaining 5- and 6-ring PAHs generally displayed similar removal in surfactant-amended samples and the no-surfactant control, or little variation from the bioreactor slurry.
Figure 1.
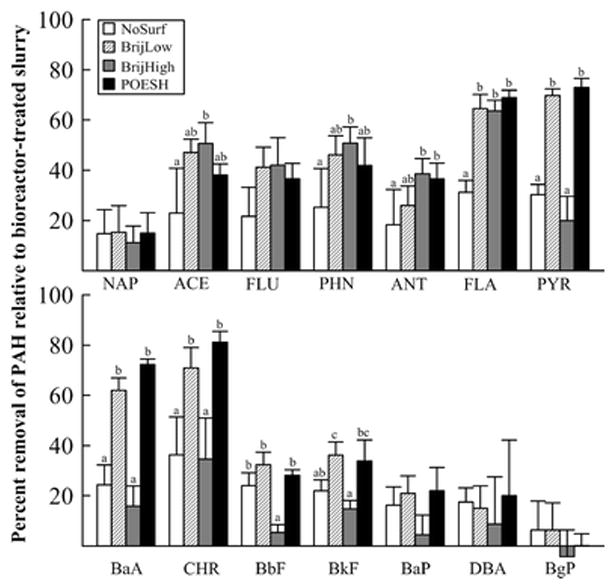
Removal (mean and standard deviation) of individual PAHs from bioreactor-treated soil after further incubation for 14 days under the experimental conditions. Identical letters above error bars indicate that those conditions were not significantly different (Tukey’s HSD; p < 0.05). PAHs with no letters above the error bars indicate that no significant difference was observed for any condition. Abbreviations: NAP – naphthalene, ACE – acenaphthene, FLU – fluorene, PHN – phenanthrene, ANT – anthracene, FLA – fluoranthene, PYR – pyrene, BaA – benz[a]anthracene, CHR – chrysene, BbF – benzo[b]fluoranthene, BkF – benzo[k]fluoranthene, BaP – benzo[a]pyrene, DBA – dibenz[a,h]anthracene, BgP – benzo[g,h,i]perylene.
Community analyses
Total bacterial communities in each of the treatments were analyzed by DGGE and barcoded amplicon high-throughput sequencing. DGGE was first used to examine the intratreatment reproducibility among the triplicate samples within each condition, as well as to examine the profiles for obvious intertreatment differences between conditions (Figure 2). The profiles of the initial bioreactor slurry and no-surfactant controls displayed consistent and highly similar banding patterns, with only minor differences between the two conditions. In contrast, surfactant-amended treatments displayed DGGE banding patterns dissimilar to both the unamended samples and each other. The profiles of BrijLow and POESH samples were internally consistent among the triplicates; however, one of the BrijHigh samples displayed a slightly variable pattern compared to the other two within that treatment. Major differences in the banding patterns between samples in the BrijLow and BrijHigh treatments also suggested that the Brij 30 concentration significantly impacted the apex bacterial communities.
Figure 2.
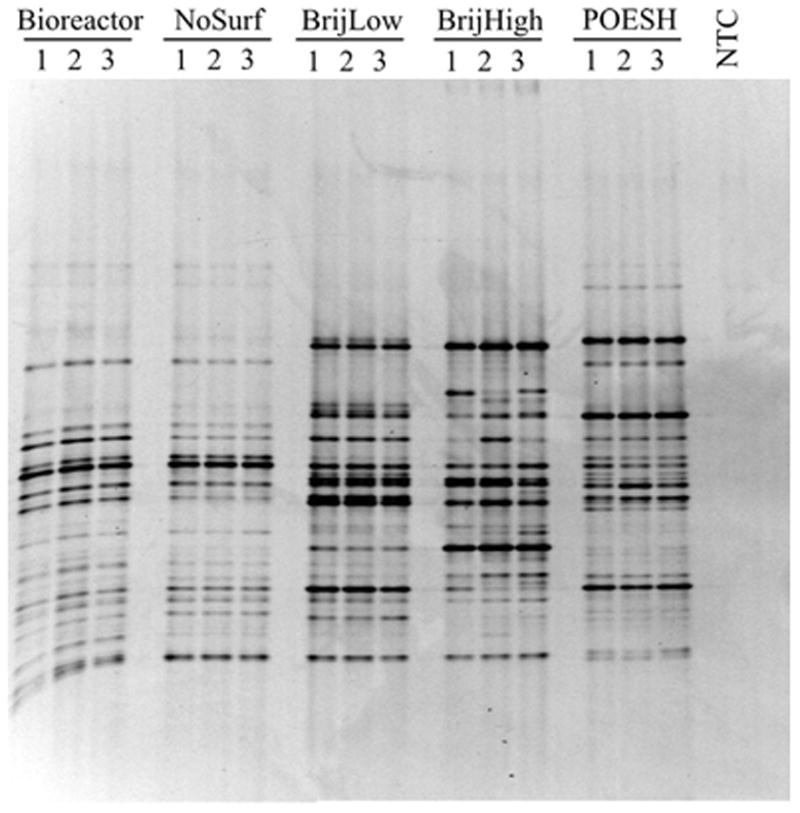
Denaturing gradient gel electrophoresis (DGGE) image showing community profiles from each incubation flask. NTC – No-template control (no DNA, PCR negative control).
Specific differences in the bacterial communities caused by the different treatments were further analyzed through 16S rRNA gene barcoded high-throughput sequencing. After processing a total of 1,079,787 partial 16S rRNA gene reads among all of the triplicate samples for the initial bioreactor slurry and each of the four treatment conditions for quality, length, and chimeras, the 15 finalized sequence libraries contained a total of 191,700 sequences clustered into 2016 OTUs (Table 1). The number of sequences in each library ranged between 8,194 and 24,162 sequences, with an average library size of 12,780 sequences. Estimates of OTU richness (chao1) and diversity (inverse Simpson diversity estimator, non-parametric estimate of the Shannon diversity index) were highest in the samples derived from the initial bioreactor slurry, and generally lower in all of the other samples incubated an additional 14 days (with or without surfactant amendment). Furthermore, the addition of the surfactants generally resulted in reduced diversity compared to the no-surfactant controls.
Table 1.
16S rRNA gene amplicon library statistics.
| Treatment Condition | No. Seqs per library | OTUsa | Coveragea | H′ab | Inv. Simp.a | Chao1a |
|---|---|---|---|---|---|---|
| Bioreactor | 11,956; 14,558; 24,162 | 417 (8) | 0.976 (0.002) | 4.29 (0.05) | 28.8 (2.2) | 865 (91) |
| NoSurf | 13,699; 13,992; 12,396 | 328 (14) | 0.983 (0.002) | 3.91 (0.02) | 21.3 (0.4) | 548 (80) |
| BrijLow | 11,433; 10,620; 11,968 | 223 (12) | 0.986 (0.002) | 3.34 (0.03) | 16.4 (0.6) | 510 (45) |
| BrijHigh | 11,334; 9,989; 8,194 | 207 (35) | 0.987 (0.003) | 3.00 (0.16) | 10.2 (0.8) | 527 (165) |
| POESH | 14,208; 12,767; 10,424 | 262 (10) | 0.983 (0.001) | 3.30 (0.12) | 12.2 (1.4) | 546 (47) |
based on a sub-sample of 8,194 sequences per library and an OTU definition of 97% similarity; values are means with standard deviation in parentheses (n=3 libraries per treatment)
nonparametric Shannon diversity index
Both NMDS (Figure 3) and PCoA (Supplementary Figure S1) demonstrated the reproducibility of 16S rRNA gene libraries derived from triplicate incubations and DNA extracts within each treatment condition. In both ordination methods the samples without amended surfactant (Bioreactor, NoSurf) were most similar to one another while the community libraries from surfactant-amended treatments each resulted in well-separated clusters. The outlier sample in the BrijHigh treatment indicated by DGGE (Figure 2) did not impact the clustering of that library with others within that treatment.
Figure 3.
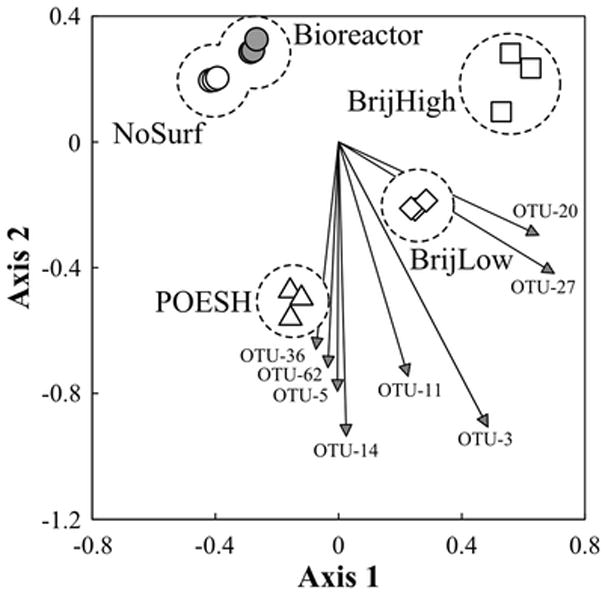
Non-metric multidimensional scaling (NMDS) plot of 16S rRNA gene amplicon libraries mapped to two dimensions. Symbols represent separate libraries derived from individual incubations; clusters of libraries are circled and labeled. Arrows indicate the influence of significant and abundant OTUs on the positioning of libraries. Labels indicate the designation of the specific OTU associated with each arrow.
OTUs enriched in BrijLow and POESH libraries
The phylogenetic information and relative abundance of all identified OTUs in the respective treatments was determined (Supplementary Table S2), with the most abundant OTUs summarized in Table 2. Using a non-parametric t-test built into the sequence analysis software, specific abundant OTUs with significantly increased representation in BrijLow and POESH libraries (p < 0.05) were determined and their influence on NMDS clustering plotted (Figure 3). Eight groups of sequences (OTUs 3, 5, 11, 14, 20, 27, 36, and 62) were identified in this manner. Of particular interest, two groups of sequences (OTUs 5 and 11) represented phylogenetic groups demonstrating significantly increased relative abundance in both BrijLow and POESH treatment libraries, the two conditions exhibiting increased HMW PAH removal, but not in libraries from other treatment conditions (Table 2). OTUs 5 and 11 were phylogenetically associated with uncharacterized Alphaproteobacteria and members of the Phenylobacterium genus, respectively. While sequences from the alphaproteobacterial group (OTU-5) possessed some similarity to members of the order Sphingomonadales, they could not be classified to that level with confidence. In addition to a number of environmental sequences from various habitats, the representative sequence of OTU-5 was identical to three 16S rRNA gene sequences from isolates in public databases including the recently described Sphingoaurantiacus polygranulatus MC 3718 isolated from Arctic soil (Kim et al. 2016), strain R-36935 recovered from Antarctica (Peeters et al. 2011), and an organism designated Sphingosinicella sp. ID0708 (GenBank accession KP326333) with no associated publication. The representative sequence of OTU-5 also possessed 99% sequence similarity to two isolates from a study of Brazilian soils (O'Neill et al. 2009), but no explicit connection to PAH degradation was apparent for any of these organisms. The representative sequence of the Phenylobacterium group (OTU-11) was similarly closely related to a variety of soil-derived environmental sequences, and possessed up to 99% 16S rRNA gene sequence similarity to a number of characterized Phenylobacterium strains.
Table 2.
Relative abundance of sequences from select, abundant OTUs in sequence libraries
| Average relative abundance (standard deviation)
|
||||||
|---|---|---|---|---|---|---|
| OTU | Classifier phylogeny | Bioreactor | NoSurf | BrijLow | BrijHigh | POESH |
| 1 | Uncl. Bacteroidetes | 10.4 (0.9) | 5.5 (0.9) | 10.8 (0.7) | 4.5 (7.5) | 5.2 (0.6) |
| 2 | Uncl. Gammaproteobacteria | 11.0 (0.6)b | 13.7 (0.6)a | 3.8 (0.5)c | 1.0 (0.5)d | 4.3 (1.0)c |
| (3) | Sediminibacterium | 0.6 (0.1)c | 1.0 (0.1)c | 7.3 (0.5)b | 5.7 (1.2)b | 21.3 (0.8)a |
| 4 | Pseudomonas | 0.4 (0.1)c | 0.3 (0.0)c | 8.8 (1.9)b | 21.1 (0.8)a | 0.3 (0.1)c |
| (5) | Uncl. Alphaproteobacteria | 2.1 (0.2)b | 3.7 (0.2)b | 8.4 (1.6)a | 1.7 (0.7)b | 8.3 (2.8)a |
| 6 | Uncl. Bacteroidetes | 5.5 (0.4)b | 10.2 (0.3)a | 0.6 (0.5)cd | 0.1 (0.0)d | 1.0 (0.2)c |
| 7 | Uncl. Moraxellaceae | 5.2 (0.3)ab | 3.0 (0.3)bc | 1.9 (0.2)bc | 0.4 (0.1)c | 6.8 (2.8)a |
| 8 | Uncl. Gammaproteobacteria | 2.7 (0.2)b | 6.9 (0.4)a | 3.8 (0.2)b | 3.7 (0.8)b | 1.1 (0.3)c |
| 9 | Caulobacter | 0.2 (0.0)b | 0.1 (0.0)b | 0.9 (0.2)b | 14.5 (2.2)a | 0.3 (0.0)b |
| 10 | Opitutus | 2.1 (0.3)b | 2.2 (0.3)b | 0.7 (0.1)c | 0.5 (0.1)c | 6.3 (0.7)a |
| (11) | Phenylobacterium | 0.2 (0.0)c | 0.2 (0.0)c | 10.6 (1.2)a | 0.2 (0.1)c | 2.1 (0.1)b |
| 12 | Uncl. Bacteria | 3.1 (0.4)ab | 3.8 (0.4)a | 1.7 (0.0)c | 0.2 (0.1)d | 2.2 (0.9)bc |
| 13 | Pseudoxanthomonas | 0.2 (0.0)c | 0.1 (0.0)c | 5.4 (0.2)b | 8.5 (1.6)a | 0.2 (0.0)c |
| (14) | Terrimonas | 0.3 (0.0)b | 0.8 (0.1)b | 1.5 (0.5)b | 0.5 (0.5)b | 8.9 (1.8)a |
| 15 | Lysobacter | 0.1 (0.0)b | 0.1 (0.0)b | 4.9 (0.3)a | 4.8 (2.2)a | 1.8 (0.2)b |
| 16 | Uncl. Chitinophagaceae | 2.9 (0.1)b | 4.0 (0.2)a | 0.2 (0.0)d | 0.2 (0.0)d | 1.7 (0.1)c |
| 17 | Novosphingobium | 0.5 (0.1)b | 0.5 (0.1)b | 0.6 (0.4)b | 0.3 (0.1)b | 7.5 (5.0)a |
| 19 | Rhizobium | 0.7 (0.1)bc | 0.3 (0.0)c | 1.3 (0.1)bc | 4.1 (1.3)a | 2.5 (1.1)ab |
| (20) | Uncl. Bradyrhizobiaceae | 0.1 (0.0)b | 0.1 (0.0)b | 8.1 (0.6)a | 0.5 (0.4)b | 0.1 (0.0)b |
| 25 | Uncl. Comamonadaceae | 1.5 (0.1)a | 0.9 (0.1)b | 1.5 (0.2)a | 0.1 (0.1)c | 0.5 (0.1)b |
| 26 | Sediminibacterium | 0.1 (0.0) | 0.1 (0.0) | 0.1 (0.0) | 4.6 (7.5) | 0.2 (0.0) |
| (27) | Acidovorax | 0.3 (0.0)ab | 0.3 (0.0)b | 2.0 (0.4)a | 1.2 (1.2)ab | 1.0 (0.6)ab |
| 28 | Sphingobium | 0.2 (0.0)b | 0.1 (0.0)b | 0.1 (0.0)b | 3.7 (0.9)a | 0.7 (0.2)b |
| 35 | Phenylobacterium | 0.2 (0.1)c | 0.1 (0.0)c | 0.7 (0.1)b | 2.6 (0.2)a | 0.1 (0.0)c |
| (36) | Uncl. Chitinophagaceae | 0.1 (0.0)bc | 0.2 (0.1)bc | 2.1 (0.3)a | 0.0 (0.0)c | 0.4 (0.1)b |
| (62) | Luteimonas | 0.1 (0.0)b | 0.1 (0.0)b | 0.1 (0.1)b | 0.1 (0.0)b | 0.9 (0.1)a |
OTUs in parentheses are those indicated as significantly associated with conditions exhibiting increased HMW PAH removal. Bold and italics indicate significantly greater and lower values, respectively, for surfactant-amended samples compared to NoSurf libraries (p < 0.05; Tukey’s HSD). Identical superscripted letters indicate no significant difference between values for that OTU. No superscripted letters among all conditions within a row indicate that the ANOVA for all samples was not significant (p < 0.05). Uncl – Unclassified.
Other OTUs indicated as significantly influencing the grouping of BrijLow and POESH libraries tended to be more strongly associated with just one of the treatment conditions. OTUs 20, 27, and 36 were most highly represented in libraries from the BrijLow condition and were associated with uncharacterized Bradyrhizobiaceae (8% relative abundance), Acidovorax (2%), and uncharacterized Chitinophagaceae (2%) sequences, respectively (Table 2). The representative partial gene sequence from OTU-20 was identical to that region of the 16S rRNA gene of Tardiphaga robiniae and additionally possessed 98% sequence identity to a number of Rhodopseudomonas species. The representative sequence from OTU-27 was identical to numerous Acidovorax strains, including one previously linked to phenanthrene degradation in our bioreactor treating a different PAH-contaminated soil (Singleton et al. 2009). OTU-36 sequences were identical to a large number of environmental sequences and were highly similar to multiple Flavihumibacter species (98 - 99% sequence identity). OTUs 14 and 62 were most prominent in libraries derived from incubations with POESH (9% and 1%, respectively).
OTU-14 was associated with the Terrimonas genus, with the representative sequence possessing 99% sequence identity to Terrimonas lutea. The OTU-62 representative sequence possessed 100% sequence similarity to a number of environmental sequences and was also identical to the isolates Luteimonas aquatica, Lysobacter brunescens, and Parnibacillus sanguinis, although the classifier implementation of mothur characterized the entire grouping of sequences within OTU-62 as likely belonging to the Luteimonas genus. While sequences from OTU-3, affiliated with the Sediminibacterium genus, were strongly represented in all three treatment conditions containing surfactants, they were significantly more abundant in the POESH libraries (21%) than the BrijLow (7%) and BrijHigh libraries (6%).
Additional OTUs not indicated statistically as significantly impacting the clustering of both BrijLow and POESH libraries under the criteria used nevertheless may have been involved in enhanced PAH removal. In particular, several OTUs were more abundant in POESH libraries than other libraries representing treatment conditions with post-bioreactor incubation, including OTUs 7, 10, and 17 (7%, 6%, and 8%, respectively). These groups represented sequences associated with uncharacterized Moraxellaceae, and the genera Opitutus and Novosphingobium. A group of uncharacterized sequences derived from the family Comamonadaceae (OTU-25) was slightly enriched in BrijLow libraries (2%), and the representative sequence for the group possessed 100% sequence similarity over the region sequenced to various strains within the Xylophilus, Ramlibacter, Acidovorax, and Variovorax genera. To facilitate comparisons with sequences and isolates from other studies, the partial 16S rRNA gene representative sequence from each OTU presented in Table 2 is available in Supplemental Material (Table S3).
Response of previously identified HMW PAH-degraders to surfactant amendment
Bacterial 16S rRNA gene sequences previously linked by stable-isotope probing to the removal of the HMW PAHs fluoranthene (FLA), benz[a]anthracene (BAA), and pyrene (PYR) in the untreated soil prior to bioreactor treatment were identified as members of the genera Sphingobium (FLA), Sphingomonas (FLA), Rhodobacter (BAA), Variovorax (BAA), and Rhizobium (BAA), as well as the uncharacterized “Pyrene Group 2” [PG2] (PYR, FLA, BAA) (Jones et al. 2011). In this experiment, the largest groups likely to include those previously determined sequences were OTUs 109, 104, 248, 27, 19, and 2, respectively. The identified Sphingobium, Sphingomonas, and Rhodobacter OTUs comprised < 0.1% relative abundance of any library (data not shown) and were unlikely to have significantly contributed to increased HMW PAH removal. Due to high similarity over the gene region sequenced, SIP-derived Variovorax sequences would likely have grouped with an OTU identified as Acidovorax (OTU-27), with significantly increased representation in BrijLow libraries compared to unamended samples (Table 2). Rhizobium sequences (OTU-19) displayed significantly increased representation in BrijHigh and POESH libraries (Table 2). PG2 sequences (OTU-2) were well represented in most libraries in this experiment, but with significantly lower relative abundance in conditions containing surfactants (Table 2).
Isolation of surfactant-degrading bacteria
In order to determine which abundant groups of organisms in amplicon libraries might have been influenced by amended carbon from surfactants, heterotrophic bacteria capable of growing on solid media containing each of the amended surfactants as a source of carbon and energy were isolated from surfactant-enriched, bioreactor-treated soil. A total of 28 distinct colonies representing 12 phylogenetic groups were isolated from surfactant-amended enrichments (Table 3); 8, 6, and 14 strains were isolated from surfactant-containing plates from BrijLow, BrijHigh, and POESH dilution series, respectively. Many of the isolates were closely related to amplicon library OTUs that displayed a marked increase in relative abundance in surfactant-amended samples (Tables 2 and 3), suggesting the potential for growth on the surfactant in those incubations.
Table 3.
Identification and PAH-transformation capabilities of surfactant-degrading isolates.
| Isolate(s) | OTUa | OTU classificationa | Closest relative by 16S rRNA gene (GenBank accession; % identity) | PHNb | PYRb | CHRb |
|---|---|---|---|---|---|---|
| BRJL1b
POES4 |
19 | Rhizobium |
Rhizobium selenitrireducens B1 (NR_044216; 100%) |
− | − | − |
| BRJL2 BRJH5 |
142 | Bosea |
Bosea robiniae R-46070 (NR_108516; 99.6%) |
− | − | − |
| BRJL4 | 22 | Alphaproteobacteria |
Enhydrobacter aerosaccus PAGU 1624 (NR_113385; 97.7%) |
− | − | − |
| BRJL7 BRJL11 |
44 | Bradyrhizobium |
Alfpia broomeae F186 (NR_029200; 99.9%) |
− | − | − |
| BRJL9 BRJL10 |
20 | Bradyrhizobiaceae |
Tardiphaga robiniae R-45977 (NR_117178; 99.6%) |
− | − | − |
| BRJL12 BRJH6 |
22 | Alphaproteobacteria |
Reyranella massiliensis 521 (NR_116005; 98.5%) |
− | − | − |
| BRJH1 | 4 | Pseudomonas |
Pseudomonas chlororaphis subsp.aurantiaca NCIB 10068 (NR_043925; 99.8%) |
− | − | − |
| BRJH2 BRJH7 |
4 | Pseudomonas |
Pseudomonas koreensis Ps 9–14 (NR_025228; 99.9%) |
− | − | − |
| BRJH4 | 19 | Rhizobium |
Beijerinckia fluminensis UQM 1685 (NR_116306; 99.8%)c |
− | − | − |
| POES1 POES3 POES10.2 |
17 | Novosphingobium |
Novosphingobium aromaticivorans
DSM 12444 (NR_074261; 98.1%)d |
+ | − | − |
| POES2 | 132 | Pseudomonas |
Pseudomonas chloritidismutans AW-1 (NR_115115; 99.9%)d |
+ | cc | − |
| POES5 | 25 | Comamonadaceae |
Ramlibacter henchirensis TMB834 (NR_025203; 96.7%) |
+ | − | − |
| POES6 POES7 POES10.1 POES11 POES12 |
27 | Acidovorax |
Acidovorax radicis N35 (NR_117776; 99.3%) |
+ | cc | − |
| POES8 | 500 | Sphingomonadales |
Altererythrobacter dongtanensis JM27 (NR_108695; 97.0%) |
− | − | − |
| POES9 | 7 | Moraxellaceae |
Perlucidibaca piscinae NBRC 102354 (NR_114062; 94%)d |
− | − | − |
OTU from amplicon libraries that would likely include the isolate(s) and mothur-derived phylogenetic affiliation.
PAH abbreviations as in Figure 1. BRJL : isolate from BrijLow condition, BRJH : isolate from BrijHigh condition, POES : isolate from POESH condition, “−“ : no transformation, “+” : zone of clearing, “cc” : color change of PAH
Misclassified genus – more likely a strain of Rhizobium radiobacter (Oggerin et al. 2009).
Identical similarity to multiple species or strains within the genus. For clarity, only one is shown.
All strains were also tested for the ability to transform each of three PAHs (phenanthrene, pyrene, or chrysene) on a solid medium. No isolates obtained from incubations with Brij 30 (either concentration) proved capable of transforming any of the tested PAHs. Ten of 13 strains isolated on POESH, representing four phylogenetic groups (Novosphingobium, Pseudomonas, Acidovorax, and uncharacterized Comamonadaceae), metabolized phenanthrene with a distinct zone of clearing around the inoculated culture. Seven of those 10 phenanthrene-transforming strains (representing isolates from the Acidovorax and Pseudomonas genera) additionally induced a color change in pyrene crystals in contact with the organisms (white to dark brown), although no clearing of the pyrene was apparent. No surfactant-degrading isolates transformed chrysene under the conditions tested. No isolates were associated with either of the amplicon library OTUs hypothesized to be the most relevant for the degradation of HMW PAHs in both BrijLow and POESH conditions (OTUs 5 and 11).
Discussion
In this study there were two conditions (BrijLow and POESH) under which the addition of sub-CMC doses of a surfactant resulted in significantly increased removal of HMW PAHs from contaminated soil that had already been subjected to conventional aerobic biological treatment. 16S rRNA gene sequences from bacteria in those conditions with significant or increased relative abundance compared to no-surfactant controls and a surfactant-amended condition (BrijHigh) not displaying the same levels of PAH removal were used to identify groups of organisms potentially contributing to the removal of those HMW PAHs.
The two groups whose 16S rRNA gene sequences increased significantly in relative abundance under both conditions displaying enhanced HMW PAH removal were OTUs 5 and 11, neither of which were represented by isolates from surfactant-containing media. Interestingly, a prior study to identify degraders of the oxy-PAH 9,10-anthraquinone using SIP in our group implicated members of the Phenylobacterium genus with sequences highly similar to OTU-11 (Rodgers-Vieira et al. 2015). Recent targeted isolation efforts for anthraquinone-degrading bacteria in our lab have produced pure strains from the Phenylobacterium genus capable of degrading not only anthraquinone, but more relevant to this work, some HMW PAHs as well (J. Vila, unpublished data). The capabilities of those isolates are currently being evaluated and will be the subject of a future publication. Sequences associated with the Phenylobacterium genus have also been observed responding positively to phenanthrene amendment in other contaminated soils (Ding et al. 2012; Thomas and Cébron 2016). Along with the Phenylobacterium group, uncharacterized members of the Alphaproteobacteria in OTU-5 were most highly represented in the sequence libraries derived from the two surfactant conditions correlated with HMW PAH removal (Table 2). Sequences highly similar to those in OTU-5 (99% 16S rRNA gene sequence identity) have been found in studies investigating soil community response to anthracene amendment (Núñez et al. 2012) and the influence of the rhizosphere on PAH removal (Tejeda-Agredano et al. 2013). The increased representation of these groups in the conditions demonstrating HMW PAH removal in this study warrant further investigation of the organisms represented by the two OTUs and their potential involvement in PAH degradation.
There were several other OTUs that increased significantly in relative abundance in sequence libraries derived from one of the two conditions that led to enhanced HMW PAH removal, but not both conditions. Notably, several recent studies have linked sequences or organisms from some of these groups to the transformation of HMW PAHs. For example, bacteria within the genus Terrimonas (OTU-14) have been linked to anthracene degradation (Zhang et al. 2011), but of more relevance to this work sequences from that genus were also recently linked to metabolism of the 5-ring HMW PAH benzo[a]pyrene in forest soil (Song et al. 2015). Sequences from the Terrimonas genus also comprised a significant percentage of a bacterial consortium degrading HMW PAHs derived from creosote that had been depleted of LMW-PAHs (Tauler et al. 2016). Sequences from the Luteimonas genus (OTU-62) were linked to the co-metabolism of benzo[a]pyrene in prior work in our group (Jones et al. 2014) and have been identified in a bacterial consortium that degraded pyrene in sediments (Bacosa and Inoue 2015). One sequence with high similarity to the uncharacterized Chitinophagaceae (OTU-36) representative sequence was an excised DGGE band derived from bacteria in heavy-oil-contaminated soil (Lladó et al. 2012) and notably, uncharacterized bacteria within the family Chitinophagaceae were also identified as putative HMW-PAH degraders in community analyses of a separate, creosote-contaminated soil (Lladó et al. 2015).
Two groups implicated as potentially contributing to HMW PAH removal represented organisms within the Acidovorax genus (OTU-27) and family Bradyrhizobiaceae (OTU-20), and each OTU was also represented in this study by multiple isolates from media containing a surfactant as a major carbon source. Targeted isolation in this study was not intended to cultivate HMW PAH-degrading organisms; in fact, we did not anticipate that isolation efforts with that goal would be successful. Bacteria growing on HMW PAHs tend to be more difficult to cultivate, and some of the OTUs of interest to this study represented bacterial groups with no prior history of successful isolation. Additionally, there are no published reports of bacterial isolates capable of growing solely on PAHs containing 5 or more rings; transformation of those PAHs is generally suspected to involve co-metabolism using some other growth substrate. Instead, we used isolation in this study as a tool to identify groups of organisms likely growing on the surfactants, which might in turn account for increased representation of those sequences in amplicon libraries. With some groups of sequences, for example OTU-4 (representing members of the Pseudomonas genus), growth on Brij 30 appeared highly likely with significant representation only in libraries derived from Brij 30-amended soil and even greater representation at the higher concentration of Brij 30 (Table 2), as well as multiple isolates recovered from Brij 30 enrichments (Table 3). Although some organisms isolated on surfactant-amended plates were also capable of transforming PAHs – for example, isolates from the Acidovorax genus (OTU-27) transformed phenanthrene and have been directly linked to LMW PAH degradation in our lab (Jones et al. 2011; Singleton et al. 2005; Singleton et al. 2009) – their increased representation in amplicon libraries in this instance might also be explained by growth on the surfactant rather than on PAHs or other compounds in the soil, particularly as most bioavailable LMW PAHs were removed during treatment in the primary bioreactor prior to surfactant amendment. Based on the recovery of isolates, growth on surfactant-derived carbon was also a possible explanation for the increased abundance of some Rhizobium sequences in this study (OTU-19), a group previously linked to the degradation of benz[a]anthracene in untreated soil (Jones et al. 2011). However, we cannot completely exclude organisms from this genus as potentially contributing to HMW PAH removal as a recent study demonstrated transformation of phenanthrene and benzo[a]pyrene by a Rhizobium tropici strain when grown in the presence of other nutrients (Yessica et al. 2013).
Members of the Sediminibacterium genus (OTU-3) have rarely been linked to PAH degradation, although they have historically been significant members of the bioreactor community (Singleton et al. 2011). Their high sequence abundance in libraries from all surfactant-amended conditions in this study may imply growth on the surfactants, despite a lack of recovered isolates. Thus their role in PAH removal, if any, remains unknown.
We also cannot exclude the additional possibility that less obvious or less abundant groups of sequences represented members primarily responsible for the increased HMW PAH removal. There may be functional redundancy in the community such that even though similar PAH removal was observed for two distinct surfactant-amended conditions, different groups of microorganisms within each condition may have been responsible for that activity. As support for this hypothesis, there exist several groups of sequences enriched in either the BrijLow library (e.g., OTU-25) or POESH library (e.g., OTUs 7, 10, and 17), but not both, that did not influence either community to the same extent as other OTUs. For two of these groups (OTU-17, Novosphingobium; OTU-25, uncharacterized Comamonadaceae), phenanthrene degradation by isolates capable of growth on surfactant-amended media was demonstrated in this study and it is conceivable that they may transform other PAHs (although their representation among the isolates may also indicate that their abundance was instead attributable to growth on surfactants as well). Members of the uncharacterized HMW PAH-degrading “Pyrene Group 2” (OTU-2) (Jones et al. 2011; Jones et al. 2008; Singleton et al. 2007; Singleton et al. 2006) appeared negatively impacted by surfactant amendment in this study, similar to what was documented in prior research (Zhu et al. 2010), but sequences from that OTU remained a significant percentage of the total communities in both BrijLow and POESH conditions; therefore, the associated organisms may have been active in PAH removal.
While other studies of the effects of sub-CMC concentrations of surfactants on LMW PAH degradation exist (Chang et al. 2014; Chen et al. 2000; Jahan et al. 1997; Noordman et al. 2000; Stelmack et al. 1999), less attention has been paid to the influence of surfactants at sub-CMC concentrations on the removal of HMW PAHs from contaminated soil. Most directly relevant to this study, we previously found that sub-CMC doses of Brij 30 enhanced both PAH desorption and PAH biodegradation (including HMW PAHs) from a different contaminated soil that had been treated in the same bioreactor used in the present study (Zhu and Aitken 2010). In that work we also reported that the addition of Brij 30 to the bioreactor-treated soil affected the populations of known PAH-degrading bacteria, positively in some cases and negatively for others (Zhu et al. 2010). In an additional series of experiments designed to screen five different nonionic surfactants, including Brij 30 and POESH (Adrion et al. 2016), the pattern of increased HMW PAH removal was consistent with that observed in the present study with the exception of benzo[a]pyrene (BaP) and benzo[b]fluoranthene (BbF); in the present study the removal of BaP or BbF in surfactant-amended conditions beyond that achieved in the no-surfactant control was not statistically significant (Figure 1), whereas in the screening study both of these carcinogenic PAHs were removed to a greater extent under the conditions corresponding to the BrijLow or POESH incubations (Adrion et al. 2016).
The effects of various surfactants on microbial cells and activity during PAH degradation have been studied for various permutations of surfactants and organisms, but total environmental communities are rarely investigated. Most studies are designed to test only one or a few (generally LMW) PAHs (Ahn et al. 2005; Liu et al. 1995), a limited number of specific PAH-degrading isolates or consortia (Boonchan et al. 1998; Doong and Lei 2003), or a combination of the two (Guha and Jaffé 1996; Pantsyrnaya et al. 2011; Yuan et al. 2000; Zhao et al. 2009). To the best of our knowledge only one other study has used high-throughput sequencing to evaluate the effects of a nonionic surfactant on the total microbial communities of a field-contaminated soil (Lladó et al. 2015). In that study, Brij 30 was added to creosote-contaminated soil that had been subjected to biopile treatment; the surfactant was added at a concentration of 4.5% (w/w), higher than either condition utilized in the present study. Amendment of Brij 30 at that concentration was concluded to potentially reduce degradation of HMW PAHs through inhibition of the specific degraders of those compounds (Lladó et al. 2013). Other studies using pure or mixed cultures have found that Brij 30 can reduce microbial PAH-degradation activity at concentrations above the CMC (Pantsyrnaya et al. 2012; Yuan et al. 2000), although amendments of Brij 30 with other organisms have shown no deleterious effects (Pantsyrnaya et al. 2011). In our earlier work, we also observed that Brij 30 inhibited HMW PAH degradation in bioreactor-treated soil when added at a dose corresponding to an aqueous-phase concentration above the CMC (Zhu and Aitken 2010). Interestingly, we found decreased HMW PAH removal in the present study when a higher but still sub-CMC level of Brij 30 was added to the bioreactor-treated soil. Deleterious effects on the HMW PAH-degrading microbial community due to the concentration of Brij 30 at the higher dose may help explain the differences in HMW PAH removal. This hypothesis is supported by the many significant differences in the microbial communities between the two Brij 30 doses in this study, and it is likely that both positive and negative effects of the surfactant on various organisms helped to shape the apex bacterial community, including PAH-degraders, which in turn affected the ability of the community to transform HMW PAHs.
This study begins to examine the underlying factors impacting the degradation of HMW PAHs in sub-CMC surfactant-amended soils. While the employed methodology by its nature results in correlative conclusions rather than causative, there is strong evidence for a biological explanation for the observed difference in PAH removal with several groups of bacteria being identified for future, targeted research. Aside from the obvious potential financial benefit of using lower concentrations of surfactant in future large-scale bioremediation schemes, the results of this study also suggest that sub-CMC concentrations of surfactant may have fewer negative impacts on indigenous HMW PAH-degrading bacteria while enhancing PAH availability and increasing overall PAH removal.
Supplementary Material
Figure S1. PCoA plot based on triplicate 16S rRNA gene amplicon libraries for each condition. Symbols for some Bioreactor and NoSurf replicates are obscured by other libraries within those conditions.
Table S1. Average (and standard deviation) concentrations of PAHsin bioreactor slurry and in treatments after 14 days of post-bioreactor incubation (μg/g dry soil).
Table S2. Classifierphylogeny and relative abundance of all OTUs with an average representation of at least 0.1% in at least one condition.
Table S3. Partial 16S rRNA gene representative sequences of all OTUs presented in Table 2.
Acknowledgments
Funding: This work was funded by the U.S. National Institute of Environmental Health Sciences (NIEHS) Superfund Research Program (grant P42 ES005948).
Footnotes
Compliance with Ethical Standards
Conflict of Interest: David R. Singleton, Alden C. Adrion, and Michael D. Aitken all declare that they have no conflict of interest. .
Ethical Approval: This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Adrion AC, Nakamura J, Shea D, Aitken MD. Screening nonionic surfactants for enhanced biodegradation of polycyclic aromatic hydrocarbons remaining in soil after conventional biological treatment. Environ Sci Technol. 2016;50:3838–45. doi: 10.1021/acs.est.5b05243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agency for Toxic Substances and Disease Registry. Priority List of Hazardous Substances. ATSDR; 2015. [Accessed 4/21/16]. [Google Scholar]
- Ahn TS, Lee GH, Song HG. Biodegradation of phenanthrene by psychrotrophic bacteria from Lake Baikal. J Microbiol Biotechnol. 2005;15:1135–1139. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bacosa HP, Inoue C. Polycyclic aromatic hydrocarbons (PAHs) biodegradation potential and diversity of microbial consortia enriched from tsunami sediments in Miyagi, Japan. J Hazard Mater. 2015;283:689–697. doi: 10.1016/j.jhazmat.2014.09.068. [DOI] [PubMed] [Google Scholar]
- Boonchan S, Britz ML, Stanley GA. Surfactant-enhanced biodegradation of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia. Biotechnol Bioeng. 1998;59:482–94. doi: 10.1002/(sici)1097-0290(19980820)59:4<482::aid-bit11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST plus : architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerniglia CE. Biodegradation of polycyclic aromatic hydrocarbons. Curr Opin Biotechnol. 1993;4:331–338. [Google Scholar]
- Chang Y-T, Hung C-H, Chou H-L. Effects of polyethoxylate lauryl ether (Brij 35) addition on phenanthrene biodegradation in a soil/water system. J Environ Sci Health, Part A. 2014;49:1672–1684. doi: 10.1080/10934529.2014.951228. [DOI] [PubMed] [Google Scholar]
- Chen P, Pickard M, Gray M. Surfactant inhibition of bacterial growth on solid anthracene. Biodegradation. 2000;11:341–347. doi: 10.1023/a:1011160004678. [DOI] [PubMed] [Google Scholar]
- Churchill SA, Griffin RA, Jones LP, Churchill PF. Biodegradation rate enhancement of hydrocarbons by an oleophilic fertilizer and a rhamnolipid biosurfactant. J Environ Qual. 1995;24:19–28. [Google Scholar]
- Ding GC, Heuer H, Smalla K. Dynamics of bacterial communities in two unpolluted soils after spiking with phenanthrene: soil type specific and common responders. Front Microbiol. 2012;3:290. doi: 10.3389/fmicb.2012.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Chowdhry B, Leharne S. Investigation of the wetting behavior of coal tar in three phase systems and its modification by poloxamine block copolymeric surfactants. Environ Sci Technol. 2004;38:594–602. doi: 10.1021/es026426q. [DOI] [PubMed] [Google Scholar]
- Doong RA, Lei WG. Solubilization and mineralization of polycyclic aromatic hydrocarbons by Pseudomonas putida in the presence of surfactant. J Hazard Mater. 2003;96:15–27. doi: 10.1016/s0304-3894(02)00167-x. [DOI] [PubMed] [Google Scholar]
- Elliot R, Singhal N, Swift S. Surfactants and bacterial bioremediation of polycyclic aromatic hydrocarbon contaminated soil—unlocking the targets. Crit Rev Environ Sci Technol. 2010;41:78–124. [Google Scholar]
- Frutos FJG, Escolano O, García S, Ivey GA. Mobilization assessment and possibility of increased availability of PAHs in contaminated soil using column tests. Soil Sediment Contam. 2011;20:581–591. [Google Scholar]
- Gan S, Lau EV, Ng HK. Remediation of soils contaminated with polycyclic aromatic hydrocarbons (PAHs) J Hazard Mater. 2009;172:532–549. doi: 10.1016/j.jhazmat.2009.07.118. [DOI] [PubMed] [Google Scholar]
- Guha S, Jaffé PR. Biodegradation kinetics of phenanthrene partitioned into the micellar phase of nonionic surfactants. Environ Sci Technol. 1996;30:605–611. [Google Scholar]
- Hu J, Nakamura J, Richardson SD, Aitken MD. Evaluating the effects of bioremediation on genotoxicity of polycyclic aromatic hydrocarbon-contaminated soil using genetically engineered, higher eukaryotic cell lines. Environ Sci Technol. 2012;46:4607–13. doi: 10.1021/es300020e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan K, Ahmed T, Maier WJ. Factors affecting the nonionic surfactant-enhanced biodegradation of phenanthrene. Water Environ Res. 1997;69:317–325. [Google Scholar]
- Jin D, Jiang X, Jing X, Ou Z. Effects of concentration, head group, and structure of surfactants on the degradation of phenanthrene. J Hazard Mater. 2007;144:215–221. doi: 10.1016/j.jhazmat.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Jones MD, Crandell DW, Singleton DR, Aitken MD. Stable-isotope probing of the polycyclic aromatic hydrocarbon-degrading bacterial guild in a contaminated soil. Environ Microbiol. 2011;13:2623–2632. doi: 10.1111/j.1462-2920.2011.02501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MD, Rodgers-Vieira EA, Hu J, Aitken MD. Association of growth substrates and bacterial genera with benzo[a]pyrene mineralization in contaminated soil. Environ Eng Sci. 2014;31:689–697. doi: 10.1089/ees.2014.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MD, Singleton DR, Carstensen DP, Powell SN, Swanson JS, Pfaender FK, Aitken MD. Effect of incubation conditions on the enrichment of pyrene-degrading bacteria identified by stable-isotope probing in an aged, PAH-contaminated soil. Microb Ecol. 2008;56:341–349. doi: 10.1007/s00248-007-9352-9. [DOI] [PubMed] [Google Scholar]
- Kile DE, Chiou CT. Water solubility enhancements of DDT and trichlorobenzene by some surfactants below and above the critical micelle concentration. Environ Sci Technol. 1989;23:832–838. [Google Scholar]
- Kim M, Kang O, Zhang Y, Ren L, Chang X, Jiang F, Fang C, Zheng C, Peng F. Sphingoaurantiacus polygranulatus gen. nov., sp. nov., isolated from high-Arctic tundra soil, and emended descriptions of the genera Sandarakinorhabdus, Polymorphobacter and Rhizorhabdus and the species Sandarakinorhabdus limnophila, Rhizorhabdus argentea and Sphingomonas wittichii. Int J Syst Evol Microbiol. 2016;66:91–100. doi: 10.1099/ijsem.0.000677. [DOI] [PubMed] [Google Scholar]
- Kojima H, Fukui M. Sulfuritalea hydrogenivorans gen. nov., sp. nov., a facultative autotroph isolated from a freshwater lake. Int J Syst Evol Microbiol. 2011;61:1651–5. doi: 10.1099/ijs.0.024968-0. [DOI] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013 doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-L, Chen B-H. Surfactant-mediated biodegradation of polycyclic aromatic hydrocarbons. Materials. 2009;2:76–94. [Google Scholar]
- Liu ZB, Jacobson AM, Luthy RG. Biodegradation of naphthalene in aqueous nonionic surfactant systems. Appl Environ Microbiol. 1995;61:145–151. doi: 10.1128/aem.61.1.145-151.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lladó S, Covino S, Solanas AM, Petruccioli M, D’annibale A, Viñas M. Pyrosequencing reveals the effect of mobilizing agents and lignocellulosic substrate amendment on microbial community composition in a real industrial PAH-polluted soil. J Hazard Mater. 2015;283:35–43. doi: 10.1016/j.jhazmat.2014.08.065. [DOI] [PubMed] [Google Scholar]
- Lladó S, Covino S, Solanas AM, Vinas M, Petruccioli M, D'Annibale A. Comparative assessment of bioremediation approaches to highly recalcitrant PAH degradation in a real industrial polluted soil. J Hazard Mater. 2013;248–249:407–14. doi: 10.1016/j.jhazmat.2013.01.020. [DOI] [PubMed] [Google Scholar]
- Lladó S, Solanas AM, de Lapuente J, Borràs M, Viñas M. A diversified approach to evaluate biostimulation and bioaugmentation strategies for heavy-oil-contaminated soil. Sci Total Environ. 2012;435–436:262–269. doi: 10.1016/j.scitotenv.2012.07.032. [DOI] [PubMed] [Google Scholar]
- Makkar RS, Rockne KJ. Comparison of synthetic surfactants and biosurfactants in enhancing biodegradation of polycyclic aromatic hydrocarbons. Environ Toxicol Chem. 2003;22:2280–92. doi: 10.1897/02-472. [DOI] [PubMed] [Google Scholar]
- May WE, Wasik SP, Freeman DH. Determination of the solubility behavior of some polycyclic aromatic hydrocarbons in water. Anal Chem. 1978;50:997–1000. [Google Scholar]
- National Toxicology Program. Report on carcinogens, thirteenth edition. U.S. Department of Health and Human Services Public Health Service; Research Triangle Park, NC: 2014. [Google Scholar]
- Noordman WH, Bruining J-W, Wietzes P, Janssen DB. Facilitated transport of a PAH mixture by a rhamnolipid biosurfactant in porous silica matrices. J Contam Hydrol. 2000;44:119–140. [Google Scholar]
- Núñez EV, Valenzuela-Encinas C, Alcántara-Hernández RJ, Navarro-Noya YE, Luna-Guido M, Marsch R, Dendooven L. Modifications of bacterial populations in anthracene contaminated soil. Appl Soil Ecol. 2012;61:113–126. [Google Scholar]
- O'Neill B, Grossman J, Tsai MT, Gomes JE, Lehmann J, Peterson J, Neves E, Thies JE. Bacterial community composition in Brazilian Anthrosols and adjacent soils characterized using culturing and molecular identification. Microb Ecol. 2009;58:23–35. doi: 10.1007/s00248-009-9515-y. [DOI] [PubMed] [Google Scholar]
- Oggerin M, Arahal DR, Rubio V, Marin I. Identification of Beijerinckia fluminensis strains CIP 106281T and UQM 1685T as Rhizobium radiobacter strains, and proposal of Beijerinckia doebereinerae sp. nov. to accommodate Beijerinckia fluminensis LMG 2819. Int J Syst Evol Microbiol. 2009;59:2323–8. doi: 10.1099/ijs.0.006593-0. [DOI] [PubMed] [Google Scholar]
- Pantsyrnaya T, Blanchard F, Delaunay S, Goergen JL, Guedon E, Guseva E, Boudrant J. Effect of surfactants, dispersion and temperature on solubility and biodegradation of phenanthrene in aqueous media. Chemosphere. 2011;83:29–33. doi: 10.1016/j.chemosphere.2011.01.024. [DOI] [PubMed] [Google Scholar]
- Pantsyrnaya T, Delaunay S, Goergen JL, Guedon E, Paris C, Poupin P, Guseva E, Boudrant J. Biodegradation of phenanthrene by Pseudomonas putida and a bacterial consortium in the presence and in the absence of a surfactant. Indian J Microbiol. 2012;52:420–6. doi: 10.1007/s12088-012-0265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters K, Hodgson DA, Convey P, Willems A. Culturable diversity of heterotrophic bacteria in Forlidas Pond (Pensacola Mountains) and Lundstrom Lake (Shackleton Range), Antarctica. Microb Ecol. 2011;62:399–413. doi: 10.1007/s00248-011-9842-7. [DOI] [PubMed] [Google Scholar]
- Richardson SD, Aitken MD. Desorption and bioavailability of polycyclic aromatic hydrocarbons in contaminated soil subjected to long-term in situ biostimulation. Environ Toxicol Chem. 2011;30:2674–81. doi: 10.1002/etc.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SD, Lebron B, Miller CT, Aitken MD. Recovery of phenanthrene-degrading bacteria after simulated in situ persulfate oxidation in contaminated soil. Environ Sci Technol. 2011;45:719–725. doi: 10.1021/es102420r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Vieira EA, Zhang Z, Adrion AC, Gold A, Aitken MD. Identification of anthraquinone-degrading bacteria in soil contaminated with polycyclic aromatic hydrocarbons. Appl Environ Microbiol. 2015;81:3775–3781. doi: 10.1128/AEM.00033-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Hunt M, Powell SN, Frontera-Suau R, Aitken MD. Stable-isotope probing with multiple growth substrates to determine substrate specificity of uncultivated bacteria. J Microbiol Methods. 2007;69:180–187. doi: 10.1016/j.mimet.2006.12.019. [DOI] [PubMed] [Google Scholar]
- Singleton DR, Powell SN, Sangaiah R, Gold A, Ball LM, Aitken MD. Stable-isotope probing of bacteria capable of degrading salicylate, naphthalene, or phenanthrene in a bioreactor treating contaminated soil. Appl Environ Microbiol. 2005;71:1202–1209. doi: 10.1128/AEM.71.3.1202-1209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Ramirez LG, Aitken MD. Characterization of a polycyclic aromatic hydrocarbon degradation gene cluster in a phenanthrene-degrading Acidovorax strain. Appl Environ Microbiol. 2009;75:2613–2620. doi: 10.1128/AEM.01955-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Richardson SD, Aitken MD. Pyrosequence analysis of bacterial communities in aerobic bioreactors treating polycyclic aromatic hydrocarbon-contamined soil. Biodegradation. 2011;22:1061–1073. doi: 10.1007/s10532-011-9463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Sangaiah R, Gold A, Ball LM, Aitken MD. Identification and quantification of uncultivated Proteobacteria associated with pyrene degradation in a bioreactor treating PAH-contaminated soil. Environ Microbiol. 2006;10:1736–1745. doi: 10.1111/j.1462-2920.2006.01112.x. [DOI] [PubMed] [Google Scholar]
- Song M, Luo C, Jiang L, Zhang D, Wang Y, Zhang G. Identification of benzo[a]pyrene-metabolizing bacteria in forest soils by using DNA-based stable-isotope probing. Appl Environ Microbiol. 2015;81:7368–7376. doi: 10.1128/AEM.01983-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelmack PL, Gray MR, Pickard MA. Bacterial adhesion to soil contaminants in the presence of surfactants. Appl Environ Microbiol. 1999;65:163–168. doi: 10.1128/aem.65.1.163-168.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauler M, Vila J, Nieto JM, Grifoll M. Key high molecular weight PAH-degrading bacteria in a soil consortium enriched using a sand-in-liquid microcosm system. Appl Microbiol Biotechnol. 2016;100:3321–3336. doi: 10.1007/s00253-015-7195-8. [DOI] [PubMed] [Google Scholar]
- Tejeda-Agredano MC, Gallego S, Vila J, Grifoll M, Ortega-Calvo JJ, Cantos M. Influence of the sunflower rhizosphere on the biodegradation of PAHs in soil. Soil Biol Biochem. 2013;57:830–840. [Google Scholar]
- Thomas F, Cébron A. Short-term rhizosphere effect on available carbon sources, phenanthrene degradation and active microbiome in an aged-contaminated industrial soil. Frontiers in Microbiology. 2016:7. doi: 10.3389/fmicb.2016.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiehm A. Degradation of polycyclic aromatic hydrocarbons in the presence of synthetic surfactants. Appl Environ Microbiol. 1994;60:258–263. doi: 10.1128/aem.60.1.258-263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiehm A, Stieber M, Werner P, Frimmel FH. Surfactant-enhanced mobilization and biodegradation of polycyclic aromatic hydrocarbons in manufactured gas plant soil. Environ Sci Technol. 1997;31:2570–2576. [Google Scholar]
- U. S. Environmental Protection Agency. Treatment technologies for site cleanup: annual status report. Washington, DC: 2007. [Google Scholar]
- Volkering F, Breure AM, Rulkens WH. Microbiological aspects of surfactant use for biological soil remediation. Biodegradation. 1997;8:401–417. doi: 10.1023/a:1008291130109. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comp Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom IT, Ghosh MM, Cox CD. Kinetic aspects of surfactant solubilization of soil-bound polycyclic aromatic hydrocarbons. Environ Sci Technol. 1996;30:1589–1595. [Google Scholar]
- Yessica G-P, Alejandro A, Ronald F-C, José AJ, Esperanza M-R, Samuel C-SJ, Remedios M-LM, Ormeño-Orrillo E. Tolerance, growth and degradation of phenanthrene and benzo[a]pyrene by Rhizobium tropici CIAT 899 in liquid culture medium. Appl Soil Ecol. 2013;63:105–111. [Google Scholar]
- Yuan SY, Wei SH, Chang BV. Biodegradation of polycyclic aromatic hydrocarbons by a mixed culture. Chemosphere. 2000;41:1463–1468. doi: 10.1016/s0045-6535(99)00522-6. [DOI] [PubMed] [Google Scholar]
- Zhang S-Y, Wang Q-F, Wan R, Xie S-G. Changes in bacterial community of anthracene bioremediation in municipal solid waste composting soil. J Zhejiang Univ Sci B. 2011;12:760–768. doi: 10.1631/jzus.B1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Miller RM. Enhanced octadecane dispersion and biodegradation by a Pseudomonas rhamnolipid surfactant (biosurfactant) Appl Environ Microbiol. 1992;58:3276–3282. doi: 10.1128/aem.58.10.3276-3282.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HP, Wu QS, Wang L, Zhao XT, Gao HW. Degradation of phenanthrene by bacterial strain isolated from soil in oil refinery fields in Shanghai China. J Hazard Mater. 2009;164:863–869. doi: 10.1016/j.jhazmat.2008.08.098. [DOI] [PubMed] [Google Scholar]
- Zhu H, Aitken MD. Surfactant-enhanced desorption and biodegradation of polycyclic aromatic hydrocarbons in contaminated soil. Environ Sci Technol. 2010;44:7260–7265. doi: 10.1021/es100112a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Singleton D, Aitken M. Effects of nonionic surfactant addition on populations of polycyclic aromatic hydrocarbon-degrading bacteria in contaminated soil. Environ Sci Technol. 2010;19:7266–7271. doi: 10.1021/es100114g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. PCoA plot based on triplicate 16S rRNA gene amplicon libraries for each condition. Symbols for some Bioreactor and NoSurf replicates are obscured by other libraries within those conditions.
Table S1. Average (and standard deviation) concentrations of PAHsin bioreactor slurry and in treatments after 14 days of post-bioreactor incubation (μg/g dry soil).
Table S2. Classifierphylogeny and relative abundance of all OTUs with an average representation of at least 0.1% in at least one condition.
Table S3. Partial 16S rRNA gene representative sequences of all OTUs presented in Table 2.


