Significance
Advanced spring flowering has been described as a fingerprint of climate change—a public, visible display of the detrimental effects of global warming. However, warming experiments fail to account for the full magnitude of observed changes in phenology, suggesting that other factors may play important roles. We show that peak flowering time shifts earlier for most species when we experimentally reduce plant diversity. Additionally, peak flowering times of plant species are more evenly distributed across the season in high-diversity plots. Overall, these results demonstrate the importance of biotic interactions in influencing flowering times and indicate that advancing phenology, one of the most well-described and well-publicized phenomena linking global warming to plant communities, may result equally from biodiversity declines.
Keywords: biodiversity–ecosystem functioning, nitrogen, soil moisture, soil temperature, species interactions
Abstract
Observational studies and experimental evidence agree that rising global temperatures have altered plant phenology—the timing of life events, such as flowering, germination, and leaf-out. Other large-scale global environmental changes, such as nitrogen deposition and altered precipitation regimes, have also been linked to changes in flowering times. Despite our increased understanding of how abiotic factors influence plant phenology, we know very little about how biotic interactions can affect flowering times, a significant knowledge gap given ongoing human-caused alteration of biodiversity and plant community structure at the global scale. We experimentally manipulated plant diversity in a California serpentine grassland and found that many plant species flowered earlier in response to reductions in diversity, with peak flowering date advancing an average of 0.6 days per species lost. These changes in phenology were mediated by the effects of plant diversity on soil surface temperature, available soil N, and soil moisture. Peak flowering dates were also more dispersed among species in high-diversity plots than expected based on monocultures. Our findings illustrate that shifts in plant species composition and diversity can alter the timing and distribution of flowering events, and that these changes to phenology are similar in magnitude to effects induced by climate change. Declining diversity could thus contribute to or exacerbate phenological changes attributed to rising global temperatures.
Evidence is mounting that anthropogenic effects are altering the phenology of many organisms. Several global metaanalyses (1–3) and many regional studies (e.g., refs. 4, 5) have shown that species phenology has shifted earlier by several days to weeks, generally in step with rising temperatures attributed to global warming. These observations have been supported by experimental studies linking warming to phenological shifts, and have sparked concern over the possibility of a global warming-induced “phenological mismatch” between mutualistic partners, such as flowering plants and their pollinators (6).
Despite linkages with rising temperature, plant phenology is not the product of a single environmental cue (7). In addition to temperature, phenology responds to a variety of environmental cues, including day length, rainfall, nutrient availability, and timing of snow melt (8). Many of these phenological cues are being altered by human activity. In addition to the influence of global warming on phenological shifts, nitrogen deposition and altered precipitation regimes have been linked to changes in flowering times (9, 10). Although phenological studies have tended to focus on abiotic drivers of change, feedbacks between biotic and abiotic processes suggest that changes in species composition, abundance, or diversity could also affect phenology. Because humans are having major impacts on the composition and diversity of plant communities worldwide (11, 12), understanding the role that biotic interactions have on phenology is critical to understanding the combined anthropogenic effects on leaf out, flowering timing, and other phenological events.
Altering plant community structure could influence phenology indirectly, via effects on abiotic processes and resource availability, or directly, via biotic interactions. Years of experimental manipulations of plant species diversity have demonstrated that diversity affects abiotic ecosystem processes, such as soil moisture and nutrient pools (13, 14). These effects mimic, on a smaller spatial scale, many of the main effects of anthropogenic nitrogen deposition and changing precipitation patterns, but it remains untested whether the magnitude of these diversity effects is sufficient to cause detectable shifts in phenology. Mechanisms driving the direct effects of biotic interactions on flowering phenology remain largely understudied, but plant density has been shown to affect flowering timing in monospecific assemblages (15) and co-occurring species may partition flowering phenology across time (16), reducing pollinator competition or interspecific pollen transfer. Although this partitioning often appears to be a product of trait displacement over evolutionary time rather than a plastic response to community changes (though see ref. 17), phenological plasticity in response to biotic interactions may be advantageous in highly variable communities, where the identity of co-occurring species shifts over small spatial or temporal scales.
In this study, we asked the following: (i) Do changes in plant diversity affect flowering timing; (ii) if so, how are these changes linked to the effects of diversity loss on abiotic variables; and (iii) do changes in plant diversity affect the dispersion of flowering times across the flowering season? We assessed the effect of plant community diversity on flowering phenology by experimentally manipulating plant species diversity in field plots from two to 16 species (18) in a serpentine grassland at Coyote Ridge near San Jose, CA. We measured peak flowering date for nine experimental species by monitoring the number of flowers per species per plot from February through May while simultaneously measuring diversity effects on several abiotic variables in each plot, including available soil N, soil moisture, and soil surface temperature.
Results
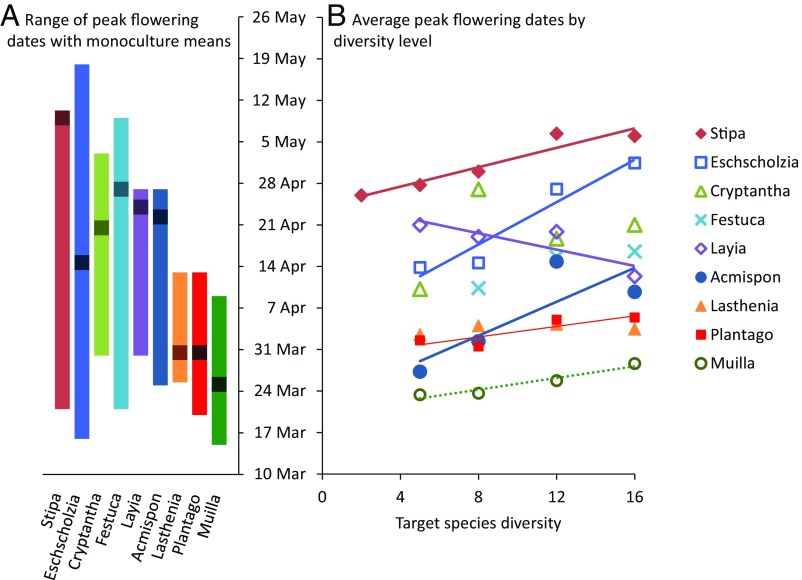
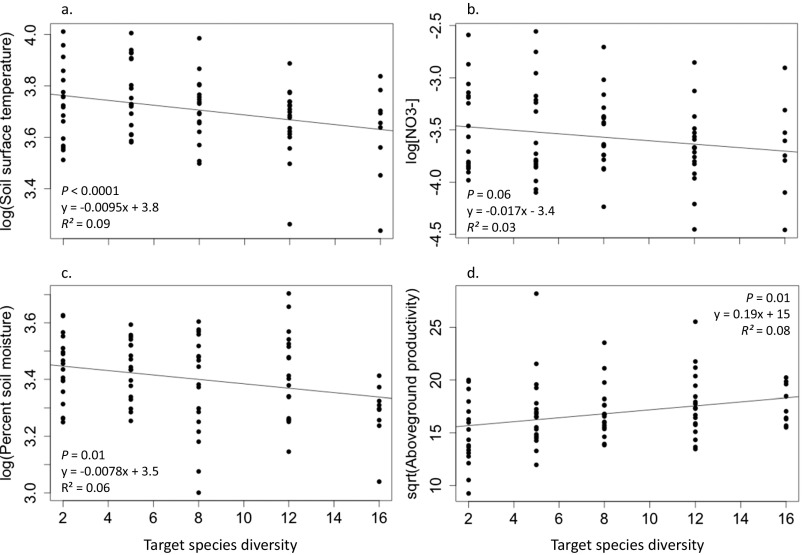
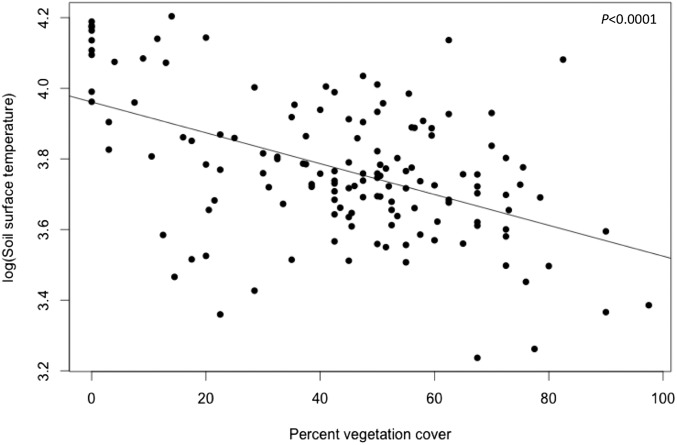
Reducing species diversity significantly advanced overall peak flowering date (P < 0.0001; Fig. 1 and Table 1), but the effects of diversity loss were not homogeneous. Five of the nine species examined flowered significantly earlier at lower diversity levels, whereas one flowered significantly later at lower diversity levels. Consistent with many previous biodiversity–ecosystem functioning experiments, declining diversity significantly affected the abiotic variables we measured (Fig. S1). Soil surface temperature, soil moisture, and soil available N all increased with declining diversity. Soil temperature was strongly correlated with percentage of vegetation cover (Fig. S2); vegetation cover has been found to decline with decreasing diversity in this experimental system (18).
Fig. 1.
Peak flowering dates for the nine species in this study. (A) Range of peak flowering dates for each species, with the mean of each species’ monoculture plots shown as a dark point on each line. (B) Mean peak flowering dates by species diversity. Error bars are omitted for clarity, and SE values are reported in Table 1. Regression lines are shown for species with a significant relationship between diversity and peak flowering time (P < 0.05); the dotted line denotes marginal significance (P < 0.1). Slopes and significance values are reported in Table 1.
Table 1.
Peak flowering date (Julian date ± SE) and, in parentheses, the number of plots used per species per diversity level
| Plot diversity | Stipa pulchra | Eschscholzia californica | Cryptantha flaccida | Festuca microstachys | Layia gaillardioides | Acmispon wranglianus | Lasthenia californica | Plantago erecta | Muilla maritima |
| 16 | 127.0 ± 1.7 (4) | 122.4 ± 2.4 (9) | 112.0 ± 3.4 (9) | 107.5 ± 4.7 (9) | 103.3 ± 2.6 (9) | 100.7 ± 3.1 (7) | 94.4 ± 1.5 (9) | 96.4 ± 2.3 (9) | 88.6 ± 3.3 (4) |
| 12 | 127.4 ± 2.0 (10) | 118.0 ± 3.1 (12) | 109.7 ± 5.9 (6) | 106.6 ± 2.6 (16) | 110.9 ± 2.8 (7) | 105.8 ± 4.0 (6) | 95.3 ± 1.3 (16) | 96.0 ± 1.5 (16) | 85.8 ± 2.1 (10) |
| 8 | 121.0 ± 4.7 (11) | 105.6 ± 8.4 (5) | 118.0 ± 7.1 (4) | 101.4 ± 3.1 (14) | 110.0 ± 4.7 (4) | 92.4 ± 3.0 (4) | 95.0 ± 3.0 (5) | 91.5 ± 1.6 (14) | 83.6 ± 1.4 (12) |
| 5 | 118.7 ± 4.7 (11) | 104.8 ± 10.0 (3) | 101.2 ± 1.4 (3) | 112.0 ± 4.5 (4) | 87.3 ± 2.8 (3) | 93.5 ± 3.3 (3) | 92.5 ± 1.3 (12) | 83.4 ± 1.0 (11) | |
| 2 | 117.0 ± 4.1 (10) | ||||||||
| Monoculture | 130.0 ± 0.0 (3) | 105.7 ± 9.3 (3) | 111.5 ± 1.5 (3) | 118.0 ± 0.0 (3) | 115.0 ± 3.0 (3) | 113.3 ± 4.7 (3) | 90.5 ± 0.0 (3) | 90.5 ± 0.0 (3) | 85.2 ± 3.2 (3) |
| Diversity effect size | 0.8177* | 1.7841** | 0.5715 | 0.7702 | −0.6885* | 1.4314* | 0.0744 | 0.4443* | 0.4922† |
The final row is the diversity effect size (the slope of the relationship between plot diversity and peak flowering date), showing the change in peak flowering date in days per species lost. Positive effect sizes indicate an advancement of peak flowering as diversity decreases; a negative effect size indicates a delay of peak flowering as diversity decreases. The significance of the diversity effect size is indicated: **P < 0.01; *P < 0.05; †P < 0.1 (marginally significant).
Fig. S1.
Influence of plot diversity on soil surface temperature (A, °C), available soil N (B, μg of N g−1⋅d−1), soil moisture (C, %), and above-ground primary productivity (D, g⋅m−2⋅y−1). Lines, linear equations, and R2 values in each panel are based on regressions between diversity and each presented metric. P values are the significance of the relationships via generalized linear model (GLM) with block as a random factor (n = 18 for diversity levels 2, 5, 8, and 12; n = 9 for 16-species plots). sqrt, square root.
Fig. S2.
Correlation between percentage of vegetation cover and soil surface temperature (°C) across all plots, including monocultures, polycultures, and bare plots (R2 = 0.25).
To distinguish between biotic and abiotic effects on the observed phenological response to diversity, we (i) examined pairwise relationships between abiotic response variables and phenological change and (ii) performed model averaging, including both biotic (target species diversity) and abiotic (soil surface temperature, soil moisture, and soil available N) factors applied to both the entire plant community phenology dataset and to each species individually (Table 2). Among pairwise relationships, only soil surface temperature was individually correlated with peak flowering date: As soil surface temperature increased, peak flowering date advanced (P = 0.002). When all variables were considered together, the best predictors of peak flowering times for all nine species included both biotic and abiotic variables based on model averaging results. Specifically, declining species diversity, declining soil moisture, and increasing soil surface temperature all contributed to peak flowering time shifting earlier. For individual species, both abiotic and biotic variables were often included in the top model set (Table 2), with the factors exerting the strongest influence dependent on species identity. In general, flowering time of individual species advanced in response to lower diversity and increasing soil surface temperature, but responses to soil moisture and N availability were more varied.
Table 2.
Model-averaged effects of biotic (species diversity) and abiotic variables on timing of peak flowering when all species are considered together (first row) or separately (subsequent rows)
| Biotic effect | Abiotic effects | |||||
| Species | Model parameters | Species diversity | Soil surface temperature | Soil moisture | Soil NO3− | (Intercept) |
| All species | Est | 0.154 ± 0.04 | −0.075 ± 0.04 | 0.071 ± 0.04 | 0.061 ± 0.04 | 0.014 ± 0.24 |
| I | 1 | 0.67 | 0.65 | 0.56 | ||
| P | 0.0005 | 0.06 | 0.06 | 0.1 | 0.95 | |
| Stipa pulchra | Est | 0.299 ± 0.15 | −0.145 ± 0.15 | 0.046 ± 0.15 | −0.025 ± 0.16 | 0 ± 0.14 |
| I | 0.73 | 0.24 | 0.11 | 0.15 | ||
| P | 0.04 | 0.34 | 0.76 | 0.88 | 1 | |
| Eschscholzia californica | Est | 0.474 ± 0.17 | −0.025 ± 0.18 | −0.038 ± 0.18 | 0.216 ± 0.17 | −0.003 ± 0.18 |
| I | 1 | 0.12 | 0.12 | 0.26 | ||
| P | 0.006 | 0.89 | 0.83 | 0.21 | 0.99 | |
| Cryptantha flaccida | Est | 0.122 ± 0.23 | −0.268 ± 0.22 | 0.159 ± 0.23 | 0.181 ± 0.22 | 0 ± 0.22 |
| I | 0.11 | 0.21 | 0.12 | 0.13 | ||
| P | 0.59 | 0.22 | 0.48 | 0.42 | 1 | |
| Layia gallardioides | Est | −0.439 ± 0.19 | −0.005 ± 0.19 | −0.069 ± 0.21 | 0.333 ± 0.19 | 0 ± 0.19 |
| I | 0.81 | 0.11 | 0.11 | 0.51 | ||
| P | 0.02 | 0.98 | 0.74 | 0.08 | 1 | |
| Festuca microstachys | Est | 0.297 ± 0.16 | −0.268 ± 0.15 | 0.428 ± 0.15 | 0.066 ± 0.15 | 0 ± 0.14 |
| I | 0.64 | 0.57 | 1 | 0.2 | ||
| P | 0.06 | 0.08 | 0.005 | 0.65 | 1 | |
| Acmispon wranglianus | Est | 0.447 ± 0.21 | 0.144 ± 0.23 | 0.388 ± 0.22 | −0.25 ± 0.19 | 0.024 ± 0.27 |
| I | 0.66 | 0.12 | 0.33 | 0.18 | ||
| P | 0.03 | 0.53 | 0.08 | 0.2 | 0.93 | |
| Lasthenia californica | Est | −0.074 ± 0.16 | −0.403 ± 0.16 | −0.035 ± 0.16 | 0.331 ± 0.16 | −0.003 ± 0.16 |
| I | 0.17 | 0.93 | 0.1 | 0.73 | ||
| P | 0.65 | 0.01 | 0.82 | 0.03 | 0.99 | |
| Plantago erecta | Est | 0.296 ± 0.15 | 0.046 ± 0.15 | −0.163 ± 0.14 | 0.137 ± 0.14 | 0 ± 0.14 |
| I | 0.81 | 0.17 | 0.35 | 0.25 | ||
| P | 0.04 | 0.76 | 0.25 | 0.33 | 1 | |
| Muilla maritima | Est | 0.287 ± 0.17 | −0.241 ± 0.18 | 0.191 ± 0.16 | −0.105 ± 0.17 | 0 ± 0.17 |
| I | 0.52 | 0.36 | 0.32 | 0.19 | ||
| P | 0.09 | 0.19 | 0.24 | 0.53 | 1 | |
All factors have been standardized. Entries in bold and italics indicate significant (P < 0.05) or marginally significant (P < 0.1) factors. Est, estimate; I, importance value; P, P value.
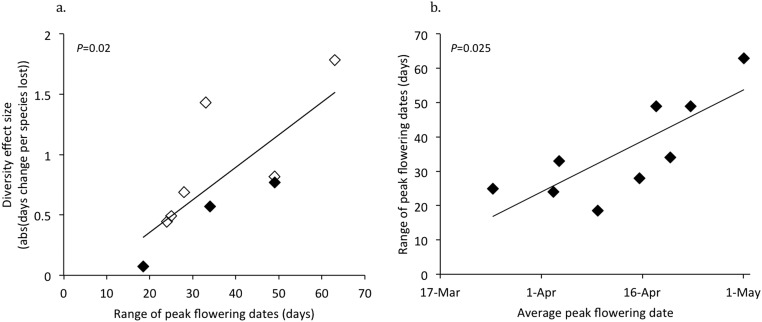
Diversity effect size (i.e., the slope of the line describing the relationship between diversity and peak flowering time) was positively correlated with the range in peak flowering time (P = 0.02; Fig. S3A), with species that flowered later in the season having broader ranges in peak flowering times (P = 0.03; Fig. S3B). However, we found no significant relationship between the mean date of peak flowering time and diversity effect size (P = 0.2, R2 = 0.2).
Fig. S3.
(A) Correlation between the range of peak flowering dates for a species (defined as the difference in dates between the plot with the latest peak flowering date for a species and the plot with the earliest peak flowering date for a species) and the absolute value of the diversity effect size (defined in main text) for a species (n = 9 species; R2 = 0.57). Open symbols (♢) represent species with a significant or marginally significant relationship between peak flowering date and diversity level. Closed symbols (♦) represent species with no significant relationship between peak flowering date and diversity level. (B) Correlation between average peak flowering date (averaged across all plots containing a species, including monoculture and polyculture plots) and the range of peak flowering dates (as defined for A) (n = 9 species; R2 = 0.62).
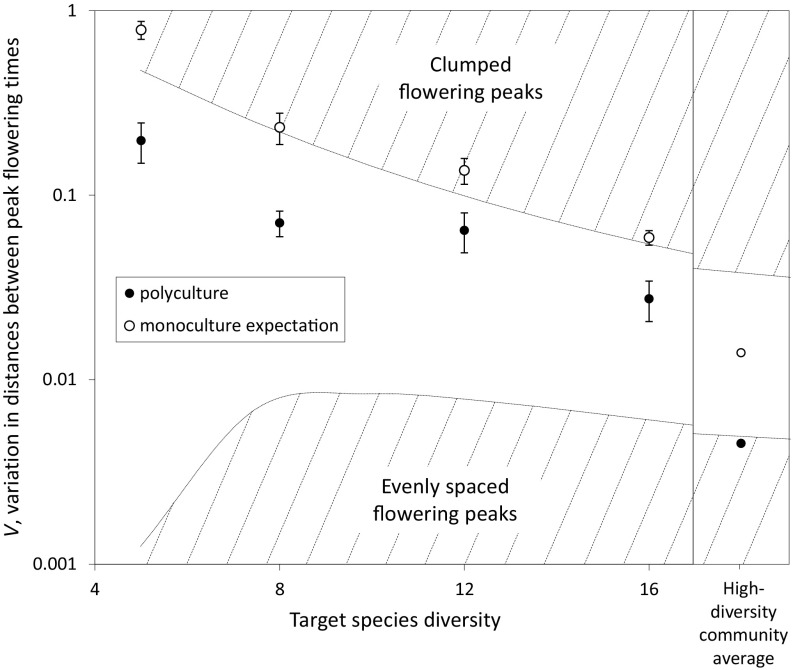
Changing species diversity also affected the distribution of flowering peaks across time. We calculated V, a metric of the variability in pairwise distances between flowering peaks (19, 20), for each individual plot as well as for the mean of high-diversity plots (Fig. 2). Across diversity levels, flowering peaks of species growing in polyculture were always less clumped than expected based on the flowering time of those same species grown separately in monocultures (P < 0.0001). Peak flowering times across the mean of all high-diversity plots were significantly overdispersed (P < 0.05); that is, the time between peak flowering dates of different species was more even than expected based on peak flowering times of species observed in monocultures (Fig. 2).
Fig. 2.
Values for V, the variation in the distances between peak flowering times across species, as observed in polyculture (●) and as expected based on monocultures (○), as a function of plant species diversity. Note the log scale on the y axis. Polyculture V by diversity level, on the left side of the graph, is calculated at the individual plot scale based on observed peak flowering times for each species within each plot, with monoculture expected V also calculated on the plot level. On the right side of the graph, V is calculated at the community level rather than the plot level. Polyculture V is calculated from the average peak flowering times for each species across the high-diversity (16-species) plots, whereas monoculture V is calculated from the average peak flowering times for each species grown in monoculture. Also shown at the boundaries of the hatched areas are isoclines for values of P > 0.975 and P < 0.025 across species diversity levels (20). Values of V that fall within these hatched regions represent significant deviation at α < 0.05 from the null hypothesis of random distribution of peak flowering times, with the alternative hypotheses being clumped flowering peaks at the top of the graph (high values of V) or evenly spaced flowering peaks at the bottom of the graph (low values of V).
Discussion
Experimentally reducing plant diversity in a California serpentine grassland resulted in earlier flowering of many species, with an average advancement in peak flowering date of 0.6 d per species lost. As detailed below, the magnitude of this change in flowering timing is similar to the magnitude of phenological change previously attributed to global warming. In addition to changes in the timing of flowering, we found changes in the dispersion of flowering times across species: Peak flowering dates between species were more dispersed in high-diversity plots than expected based on monocultures. Taken together, these findings demonstrate a strong role of biotic processes and plant–plant interactions on plant phenology that should be considered alongside the more commonly addressed effects of abiotic processes, such as rising global temperatures, N deposition, and altered precipitation regimes.
The effects of changing diversity on flowering phenology appear to be mediated by significant effects of diversity on abiotic properties, including soil surface temperature as well as nutrient and water availability. The positive correlation between increasing soil surface temperature and earlier flowering is consistent with the direction of phenological shifts attributed to global warming (2). Temperature effects on phenology due to global warming are largely attributed to direct effects, such as degree days and frost dates (9), but indirect effects of temperature on other phenological cues, such as soil moisture, are also possible (2). Although plant diversity loss and climate warming affect temperature on very different spatial scales, our results suggest that the phenological response of plants to changing temperature cues across scales is similar; shifts in plant community composition can alter local-scale abiotic processes sufficiently to cause detectable shifts in flowering phenology.
Despite the influence of abiotic factors, diversity remained an important factor in models where abiotic effects were considered alongside biotic effects (Table 2). Several indirect effects of diversity on phenology may explain why the abiotic variables did not account for a larger proportion of the variation in peak flowering times. First, although temperature, moisture, and nitrogen availability are the cues most often associated with phenological change due to human impacts, phenology may respond to other abiotic variables (e.g., light infiltration, other nutrient concentrations) that we did not measure. Second, phenology may be influenced by the abiotic variables we measured at finer spatial or temporal scales than our measurements allowed. In addition to possible indirect effects, a direct biotic effect of biodiversity on phenology is possible. Plants exchange chemical signals in response to environmental stimuli, such as jasmonic acid signaling in response to herbivory (21), but no mechanism is known for a chemical signal linked to plant community composition, diversity, or flowering.
Despite the differences in the drivers of phenological change, the magnitude of phenological change we observed due to diversity loss is similar to previously observed phenological responses to climate change. Phenological advancement in plants from long-term observational datasets is estimated at 1.1 d per decade for forbs and grasses (22) or ∼4.5 d per degree Celsius for woody and herbaceous plants (2). This phenological shift is largely attributed to global climate warming, which has proceeded at ∼0.12 °C per decade since 1951 (23). Peak flowering time in this study advanced by an average of 0.6 d per species lost, ranging from an advancement of 1.8 d per species lost to a delay of 0.7 d per species lost. Based on these data, the phenological change attributable to the loss of two species from our study approximates the average phenological change due to one decade of global warming. Previous work has demonstrated that warming experiments significantly underestimate observed phenological shifts (2). Our findings suggest that changes to community structure may account for some of this difference, because warming experiments that occur over short time scales are unlikely to capture the full effects of community shifts.
In addition to changes in diversity causing earlier flowering times, we found that peak flowering times of species growing in polyculture were always more dispersed than would be expected based on monocultures. Ecologically, this finding is intriguing because more dispersed peak flowering times generate phenological complementarity, which may allow a reduction in pollinator competition across the plant community. Because all plots were seeded from the same seed pool, the differences in flowering time between high- and low-diversity plots are due to phenological plasticity rather than underlying intraspecific genetic differences. Native serpentine grassland plants are often highly plastic (24), and the phenological plasticity demonstrated here may be adaptive, given the dramatic year-to-year variation in rainfall and concomitant changes in community species composition and abundance (25) in this ecosystem. Phenological partitioning has been observed in other ecosystems: Sympatric agaves partition flowering time (16), although in that case, the partitioning is thought to be an evolved response rather than a plastic response. Co-occurring acacias also partition flowering times at both daily and seasonal temporal scales (17), although it is unclear whether this response is plastic. To our knowledge, dispersion of flowering times of co-occurring species, especially across multiple genera and families, has not been previously demonstrated experimentally. Plastic phenological partitioning could be a product of indirect biotic interactions, such as soil resource partitioning or competition for soil resources. Alternatively, there may be more direct biotic interactions, such as plant-to-plant signaling, that lead to dispersion of flowering times; further work is needed to distinguish between these possible mechanisms.
Understanding which species will exhibit phenological shifts is a major goal of climate change research. The flowering times of all three perennial species in our dataset were affected by decreasing biodiversity, and three of the six annual species were also affected. Interestingly, intraspecific phenological variability was a good predictor of how species’ peak flowering times in this study were affected by changing diversity. In our study, species with the largest ranges in peak flowering times had the strongest responses to changing biodiversity. Conversely, the species with little underlying variation in peak flowering times responded only weakly to changing biodiversity. Although some studies note that earlier flowering species exhibit larger phenological shifts than later flowering species (8), we did not see that pattern. Other studies have found that phenotypic plasticity plays a large role in long-term phenological responses to climate change (26); the ability of plants to respond plastically to both biotic and abiotic phenological cues may underlie previous findings demonstrating that species unable to track climate change phenologically are at increased risk in a warming climate (27).
Although most of our target species showed earlier flowering times with decreasing diversity, one annual species, Layia gaillardioides, responded to decreasing diversity with a delayed mean flowering time. This unexpected response does not fit any patterns previously established in the literature; although some studies suggest that late-season plants may delay flowering times with warming temperatures (28), Layia gaillardioides flowers near the middle of the growing season. There is no indication from our models that Layia gaillardioides responds differently than other species to the abiotic variables we measured. Rather, Layia gaillardioides appears to respond very differently to species diversity alone. The lack of explanation for this pattern in Layia gaillardioides underlines the fact that species phenological responses, especially to biotic factors, are highly varied and complex, and may often be tied to traits beyond those traits that are typically measured (e.g., seasonality of flowering, growth form, life history).
We conducted this study over one growing season in one type of ecosystem. Our results demonstrate that the biotic community can have large effects on flowering phenology, although much work remains to be done before we fully understand year-to-year variation, differences across ecosystems, and the mechanisms driving the observed biotic effect. Understanding the mechanistic underpinnings of biotic effects on phenology will necessitate parsing the potential direct phenological effects of the biotic community (e.g., plant signaling pathways) from indirect effects (e.g., changes to phenology mediated through abiotic effects of changing plant communities). Our study suggests that smaller scale abiotic changes due to plant community change affect phenology in similar ways to larger scale abiotic changes; those larger scale abiotic changes, such as increasing temperatures due to climate change, generally have consistent effects across a range of communities, although there is significant variation in how individual species respond to phenological cues (8, 28). These results open several new areas of questioning. Are the phenological effects of climate change and declining diversity additive, synergistic, or mutually exclusive? Are biotic effects on phenology more or less important under stressful conditions, such as drought? How do invasive species change biotic effects on phenology? Which species exert the strongest effects on phenology, and which show the strongest response to biotic effects? Future research should address the variation of potential biotic effects on phenology, the interactions between biotic and abiotic effects, and the importance of biotic effects on phenology across a range of climatic conditions and global ecosystems.
Overall, our findings demonstrate that changing plant community composition and diversity can alter both flowering phenology and the dispersion of peak flowering across a plant community. In addition, the effect size of biodiversity loss on phenology is similar in magnitude to the effect of rising global temperatures. Although biodiversity loss is not ubiquitous (29, 30; though see ref. 31), it is a significant threat to many ecosystems worldwide (32), and shifts in species distributions (33) and abundances, including increases in the number of rare species (34), are also well documented. Our study indicates that changes in plant species composition that affect ecosystem functioning (e.g., percentage of vegetation cover, resource availability, surface temperature), including community homogenization, species invasions, and diversity loss, can affect phenology independent of the larger scale abiotic impacts of rising temperatures, changing precipitation patterns, or N deposition. Incorporating these community effects into our understanding of past and future phenological change is critical for fully understanding and predicting the effects of human activities on the timing of biological events, as well as for accurately attributing changes in phenology to anthropogenic causes.
Methods
Field Site.
Biodiversity manipulations occurred at Coyote Ridge near San Jose, CA, a serpentine grassland reserve with a Mediterranean climate. Biodiversity manipulations began in the 2007–2008 growing season; the data reported here were collected in the 2011–2012 growing season. Further details on the experimental design and results have been published previously (18, 35, 36).
Experimental Design.
Biodiversity was manipulated based on two loss orders: realistic or random. From a species pool of 16 species (Table S1), the realistic loss order was based on drought-induced diversity loss, whereas species were chosen at random to populate the random loss order. Each block (n = 9) contained a single replicate plot of each diversity level for each loss order; each plot was a 58-cm diameter circle. Individual plots consisted of two, five, eight, or 12 species (n = 18 for each diversity level across the two loss orders), and each block also contained one plot with the full complement of 16 species (n = 9). Additionally, each species was grown in monoculture in three plots and there were nine bare plots. As soon as seedlings could be identified to species, nontarget native species were removed from each plot on a monthly basis. Invasive species (Bromus hordeaceus and Festuca perennis) were allowed to remain in polyculture plots (plots with two or more target species); to maintain true monocultures, these invasive species were weeded out of all monoculture plots. Because of this difference between monoculture and polyculture plots, monocultures are not used in analyses examining the effects of diversity on experimental outcomes (18, 35).
Table S1.
List of species used in this experiment and their growth forms
| Species | Growth form |
| Stipa pulchra (formerly Nassella pulchra) | Perennial grass |
| Chlorogalum pomeridianum | Perennial forb |
| Calystegia subacaulis | Perennial forb |
| Muilla maritima | Perennial forb |
| Plantago erecta | Annual forb |
| Lessingia nemaclada | Annual forb |
| Festuca microstachys (formerly Vulpia microstachys) | Annual grass |
| Microseris douglasii | Annual forb |
| Lasthenia californica | Annual forb |
| Hesperevax sparsiflora | Annual forb |
| Eschscholzia californica | Perennial forb |
| Calandrinia ciliata | Annual forb |
| Layia gaillardioides | Annual forb |
| Acmispon wrangelianus (formerly Lotus wrangelianus) | Annual forb (N-fixing) |
| Cryptantha flaccida | Annual forb |
| Hemizonia congesta | Annual forb |
Species in bold are those species for which phenological data were recorded. Species names follow Baldwin et al. (42), with previous names shown in parentheses.
The abundance of each species was based on field observations of realistic species densities, with the total stem density of each plot set at 1,285 plants per square meter (18) (planting density of species at each diversity level is shown in Tables S2 and S3). These planting densities were based on observations from a 30-y dataset of species presence and abundance at this field site. Seeds for all species and perennating organs for perennials were collected from near the study site and dispersed into plots coinciding with the first soaking rain of each year, generally in late October to early November.
Table S2.
Stem targets for plots in the randomized diversity treatments
| Species richness | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 |
| Replicate no. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| Stipa pulchra | 367 | 257 | 130 | 121 | 63 | 139 | 80 | 8 | 13 | 12 | 8 | 8 | 9 | |||||||||||||||||||||||
| Chlorogalum pomeridianum | 343 | 56 | 18 | 27 | 92 | 15 | 8 | 22 | 14 | 13 | 29 | 29 | 17 | 26 | 17 | 18 | 20 | |||||||||||||||||||
| Calystegia subacaulis | 317 | 26 | 84 | 30 | 16 | 20 | 18 | 10 | 8 | 13 | 12 | 8 | 8 | S | ||||||||||||||||||||||
| Muilla maritima | 235 | 24 | 26 | 74 | 17 | 24 | 11 | 14 | 9 | 111 | 91 | 89 | 135 | 90 | 93 | 105 | ||||||||||||||||||||
| Plantago erecta | 208 | 115 | 178 | 164 | 116 | 147 | 194 | 129 | 120 | 9 | 7 | 12 | 12 | 7 | 11 | 7 | ||||||||||||||||||||
| Lessingia nemaclada | 221 | 35 | 100 | 22 | 28 | 22 | 13 | 17 | 68 | 113 | 67 | 101 | 68 | 69 | 79 | |||||||||||||||||||||
| Festuca microstachys | 316 | 172 | 187 | 77 | 218 | 144 | 86 | 80 | 25 | 21 | 35 | 21 | 31 | 21 | 24 | |||||||||||||||||||||
| Microseris douglasii | 151 | 137 | 35 | 26 | 113 | 56 | 29 | 55 | 37 | 30 | 75 | 61 | 102 | 101 | 60 | 61 | 63 | |||||||||||||||||||
| Lasthenia californica | 5 | 84 | 130 | 120 | 91 | 60 | 80 | 53 | 49 | 91 | 75 | 123 | 73 | 74 | 76 | 86 | ||||||||||||||||||||
| Hesperevax sparsiflora | 29 | 148 | 154 | 259 | 55 | 104 | 122 | 62 | 57 | 5 | 4 | 7 | 7 | 6 | 4 | 5 | ||||||||||||||||||||
| Eschscholzia californica | 55 | 9 | 13 | 28 | 19 | 12 | 15 | 12 | 7 | 4 | 4 | 6 | 6 | 4 | 5 | |||||||||||||||||||||
| Calandrinia ciliata | 86 | 28 | 78 | 34 | 9 | 6 | 7 | 4 | 15 | 12 | 20 | 20 | 12 | 12 | 12 | |||||||||||||||||||||
| Layia gaillardioides | 164 | 20 | 32 | 45 | 94 | 31 | 20 | 18 | 11 | 16 | 13 | 21 | 21 | 13 | 19 | 15 | ||||||||||||||||||||
| Acmispon wrangelianus | 298 | 14 | 24 | 27 | 31 | 24 | 15 | 13 | 13 | 12 | 8 | 8 | 9 | |||||||||||||||||||||||
| Cryptantha flaccida | 286 | 12 | 25 | 54 | 8 | 16 | 19 | 9 | 10 | 13 | 13 | 8 | 8 | 9 | ||||||||||||||||||||||
| Hemizonia congesta | 74 | 7 | 28 | 7 | 15 | 22 | 17 | 8 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | |||||||||||||||||||||
| Total stem target | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 | 372 |
Table S3.
Stem targets for plots in the realistic richness diversity treatment and 16 species plots (n = 9 plots per richness level)
| Species richness | 2 | 5 | 8 | 12 | 16 |
| Stipa pulchra | 324 | 158 | 72 | 47 | 45 |
| Chlorogalum pomeridianum | 48 | 24 | 9 | 7 | 7 |
| Calystegia subacaulis | 23 | 18 | 15 | 14 | |
| Muilla maritima | 20 | 10 | 7 | 8 | |
| Plantago erecta | 146 | 132 | 81 | 64 | |
| Lessingia nemaclada | 11 | 7 | 6 | ||
| Festuca microstachys | 87 | 61 | 42 | ||
| Microseris douglasii | 33 | 19 | 14 | ||
| Lasthenia californica | 55 | 67 | |||
| Hesperevax sparsiflora | 67 | 71 | |||
| Eschscholzia californica | 4 | 4 | |||
| Calandrinia ciliata | 3 | 2 | |||
| Layia gaillardioides | 8 | ||||
| Acmispon wrangelianus | 9 | ||||
| Cryptantha flaccida | 6 | ||||
| Hemizonia congesta | 7 | ||||
| Total stem target | 372 | 372 | 372 | 372 | 372 |
Soil moisture, defined as volumetric soil water content at a depth of ∼10 cm, was measured in each plot each month from February through June 2012. Available soil N was assessed with resin bags buried in the center of each plot that were replaced every other month (35). Soil surface temperature was measured in each plot with an infrared thermometer (Omega 05530HR; Omega Engineering) as the average of three readings per plot. Further details of these abiotic measurements can be found in SI Methods.
Phenological observations were made from late February to mid-May of 2012. Of the 16 species in our experiment (Table S1), we were able to collect phenological data for nine species. On each measurement date, we visited every plot and recorded the presence and abundance of fully opened flowers for every target species. Flower abundance per species on a given date was recorded in log-based bins: 0, no flowers; 1, one to two flowers; 2, three to five flowers; 3, six to 10 flowers; 4, 11–20 flowers; 5, 21–50 flowers; 6, 51–100 flowers; and 7, 100+ flowers. A biomass harvest of a portion of each plot was conducted on April 16–18, 2012; following this harvest, flower abundance was recorded as raw number of flowers per species, corrected for the area harvested, and then assigned to an abundance bin. We revisited the site throughout May and June of 2012 to ensure that our phenological measurements captured peak flowering of all nine species for which data are presented.
Because the abundance of each species was manipulated as part of the experiment, first flowering date, a commonly reported phenological metric, was not appropriate for this study (37). Instead, we used peak flowering date as our metric of flowering phenology because it is mathematically independent of species abundance (37). In our study, phenological measurements were taken at ∼9-d intervals; although first flowering date is highly sensitive to measurement interval, peak flowering date has been shown to be relatively unaffected by the interval between sampling dates (37). Miller-Rushing et al. (37) demonstrated that although the likelihood of detecting a significant change decreases with sampling interval, the date of peak flowering itself does not significantly change with decreasing sampling frequency. In addition, peak flowering date represents a population average rather than an individual extreme, and therefore may be a more relevant community-based metric than first flowering date (37). Peak flowering date was determined for each species for each plot, and was defined as the sampling date at which flower abundance was highest; if more than one date had an equal flower abundance, peak flowering date was taken as the midpoint between those dates. Because the two loss orders (random and realistic) in our experiment did not significantly vary in their effects on flowering phenology, plots from both species loss orders were combined based on diversity level.
Individual plots were used to represent a species only if peak flowering consisted of three or more flowers for a particular species; species diversity levels were only included for a species if that species was represented in at least three plots of a given diversity level. The number of plots fitting these criteria is given in Table 1, along with the average peak flowering date and SE per species per diversity level. The expected number of plots occupied by each species differs by species and diversity level because of differential representation of species in the two loss orders and due to decreasing representation of species as diversity decreases. Overall representation of species in plots into which they were seeded was excellent. At least one flowering individual of Cryptantha, Layia, Plantago, Lasthenia, and Festuca was observed in every plot in which they were expected. Flowering Eschscholzia individuals were missing from three expected plots, Acmispon from two plots, and Muilla from one plot. Flowers of Stipa, the most commonly missing species, were missing from 11 expected plots, although they were still represented in over 70% of plots into which Stipa was seeded. However, there were many nonflowering Stipa individuals, so lack of flowers does not equate with lack of species representation in those plots.
Data Analysis.
We first tested whether target species diversity was predictive of overall peak flowering time using the lme4 package (38) in R 3.1.2 (39). We tested species individually, with block included as a random factor, and with the full nine-species dataset, with block and species identity (ID) as random factors. We also tested soil surface temperature, soil N, and soil moisture as individual predictors of overall peak flowering time. Second, we fit linear mixed-effects models to assess the combined influences of both biotic (target species diversity) and abiotic (soil surface temperature, soil moisture, and available soil N) predictor variables on species-specific peak flowering time, as well as on the community overall. Again, block was included as a random factor in the individual-species models; in the overall model including all species, both block and species ID were included as random factors. We used a model-averaging approach to assess the relative importance of the abiotic and biotic factors (40). Before fitting models, all numeric variables were standardized using the scale function in R. We used the dredge function in the MuMIn package (41) of R to fit all possible models; we then selected only the models with a difference in Akaike information criterion (AIC) scores corrected for small sample size (ΔAICC) less than 4 to include for model averaging using the model.avg function in the MuMIn package. We estimated parameter values, SEs, and AICC-weighted importance values for each variable. In addition, we tested for correlations between our predictor variables (Table S4). Further details of data transformations and other analysis can be found in SI Methods.
Table S4.
Correlation matrix between variables used to predict peak flowering times
| Predictor variable | Species diversity | Soil moisture | Soil surface temperature | Available soil N |
| Species diversity | 1 | −0.24* | −0.30** | −0.18† |
| Soil moisture | 1 | 0.016 | 0.17 | |
| Soil surface temperature | 1 | −0.068 | ||
| Available soil N | 1 |
Species diversity was deliberately manipulated; soil moisture (%), soil surface temperature (°C), and available soil N (micrograms of N per gram of resin per day) were measured for each plot. Soil moisture, soil surface temperature, and available soil N were log-transformed for this and all other analyses. The significance of the correlations is indicated: **P < 0.01; *P < 0.05; †P < 0.1 (marginally significant).
We defined the diversity effect size as the species-specific slope of the regression between plot diversity and peak flowering date. To examine which species were most affected by changes in community diversity and composition, we looked at several possible explanatory factors for the magnitude of the diversity effect size, including plant life history (perennial vs. annual), average date of peak flowering, and the range of peak flowering dates (defined as the difference between the maximum and minimum peak flowering date for each species). The correlation between these factors and the diversity effect size was assessed by linear model.
The degree of randomness vs. dispersion of peak flowering times was calculated with the Var statistic (19) modified as V by Williams (20):
where SSdist is the sum of the squared distances between peak flowering dates of each species, and range is the distance between the earliest and latest measurements. This statistic was developed specifically for examining dispersion vs. randomness of flowering times, although it has also been used to detect character displacement. We calculated V at two different spatial scales. First, we calculated V at the plot scale separately for each plot (polyculture V). Only plots represented by at least three of our target species were included in this analysis, corresponding to an overall diversity level of at least five species. For each of these plots, we also calculated expected V based on the average monoculture data (monoculture expectation) for each target species in the plot. For example, if the target species in a plot were Stipa, Cryptantha, and Plantago, we used the average monoculture peak flowering date for those three species to calculate expected V. Differences between polyculture V and monoculture expectation by plot were assessed by a paired t test. Second, we calculated a community-based V using the mean peak flowering time of all species growing in the highest diversity plots, and compared that with expected V based on the mean peak flowering time of each of the species in monoculture. Critical values for the V statistic, as displayed in Fig. 2, are from Williams (20); note that n is determined by the number of species for which we have phenology data in each plot rather than by the target species diversity of a plot.
SI Methods
Abiotic Measurements.
Soil moisture, as volumetric soil water content, was measured in each plot using a time domain reflectrometry probe (Mesa Systems Co.) and polycarbonate access tubes permanently installed in the center of each plot. Three measurements were taken in each plot at each sampling time; these three measurements were then averaged.
Nylon bags were used as resin bags for soil N. They were filled with ∼6 g of mixed-bed ion exchange resin. To minimize disturbance from inserting and collecting the bags several times per growing season, as well as to minimize damage from insects and herbivores, bags were installed inside perforated PVC tubes that were closed on the top and open on the bottom. After collection, resin beads were extracted with 2 M KCl and analyzed for NO3− and NH4+ concentrations using a discrete analyzer (WestCo SmartChem 200; Unity Scientific).
To standardize soil temperature across plots, three measurements were taken from the same height at the same three compass directions from the center of each plot.
Data Transformations and Analysis.
Before analysis, all variables were tested for assumptions of normality; soil available N, soil moisture, and soil surface temperature were log-transformed to meet these assumptions. Because we expected species diversity to influence many of the abiotic factors in our models, we examined the correlation between our predictor variables (Table S2). Species diversity was significantly correlated with all three abiotic variables, although there were no significant correlations between the abiotic predictor variables. Calculating the variance inflation factor demonstrated that multicollinearity did not complicate our models.
Acknowledgments
C. Morozumi, E. Wells, K. Philpott, I. Holl, J. Villa, and S. Weiss provided help with field and laboratory work. J. Pasari and D. Hernández helped design and establish the experiment. Comments from T. Young and two anonymous reviewers improved the manuscript. Funding for this project came from National Science Foundation Grant DEB0918785.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608357114/-/DCSupplemental.
References
- 1.Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421(6918):37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- 2.Wolkovich EM, et al. Warming experiments underpredict plant phenological responses to climate change. Nature. 2012;485(7399):494–497. doi: 10.1038/nature11014. [DOI] [PubMed] [Google Scholar]
- 3.Root TL, et al. Fingerprints of global warming on wild animals and plants. Nature. 2003;421(6918):57–60. doi: 10.1038/nature01333. [DOI] [PubMed] [Google Scholar]
- 4.Forrest J, Inouye DW, Thomson JD. Flowering phenology in subalpine meadows: Does climate variation influence community co-flowering patterns? Ecology. 2010;91(2):431–440. doi: 10.1890/09-0099.1. [DOI] [PubMed] [Google Scholar]
- 5.Miller-Rushing AJ, Primack RB. Global warming and flowering times in Thoreau’s Concord: A community perspective. Ecology. 2008;89(2):332–341. doi: 10.1890/07-0068.1. [DOI] [PubMed] [Google Scholar]
- 6.Miller-Rushing AJ, Høye TT, Inouye DW, Post E. The effects of phenological mismatches on demography. Philos Trans R Soc Lond B Biol Sci. 2010;365(1555):3177–3186. doi: 10.1098/rstb.2010.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forrest J, Miller-Rushing AJ. Toward a synthetic understanding of the role of phenology in ecology and evolution. Philos Trans R Soc Lond B Biol Sci. 2010;365(1555):3101–3112. doi: 10.1098/rstb.2010.0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleland EE, Chuine I, Menzel A, Mooney HA, Schwartz MD. Shifting plant phenology in response to global change. Trends Ecol Evol. 2007;22(7):357–365. doi: 10.1016/j.tree.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Cleland EE, Chiariello NR, Loarie SR, Mooney HA, Field CB. Diverse responses of phenology to global changes in a grassland ecosystem. Proc Natl Acad Sci USA. 2006;103(37):13740–13744. doi: 10.1073/pnas.0600815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoover SER, et al. Warming, CO2, and nitrogen deposition interactively affect a plant-pollinator mutualism. Ecol Lett. 2012;15(3):227–234. doi: 10.1111/j.1461-0248.2011.01729.x. [DOI] [PubMed] [Google Scholar]
- 11.Tylianakis JM, Didham RK, Bascompte J, Wardle DA. Global change and species interactions in terrestrial ecosystems. Ecol Lett. 2008;11(12):1351–1363. doi: 10.1111/j.1461-0248.2008.01250.x. [DOI] [PubMed] [Google Scholar]
- 12.Tilman D, Lehman C. Human-caused environmental change: Impacts on plant diversity and evolution. Proc Natl Acad Sci USA. 2001;98(10):5433–5440. doi: 10.1073/pnas.091093198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardinale BJ, et al. The functional role of producer diversity in ecosystems. Am J Bot. 2011;98(3):572–592. doi: 10.3732/ajb.1000364. [DOI] [PubMed] [Google Scholar]
- 14.Hooper DU, et al. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol Monogr. 2005;75(1):3–35. [Google Scholar]
- 15.Antonovics J, Levin DA. The ecological and genetic consequences of density-dependent regulation in plants. Annu Rev Ecol Syst. 1980;11:411–452. [Google Scholar]
- 16.Rocha M, Valera A, Eguiarte LE. Reproductive ecology of five sympatric Agave Littaea (Agavaceae) species in central Mexico. Am J Bot. 2005;92(8):1330–1341. doi: 10.3732/ajb.92.8.1330. [DOI] [PubMed] [Google Scholar]
- 17.Stone GN, Willmer P, Rowe JA. Partitioning of pollinators during flowering in an African Acacia community. Ecology. 1998;79(8):2808–2827. [Google Scholar]
- 18.Selmants PC, Zavaleta ES, Pasari JR, Hernandez DL. Realistic plant species losses reduce invasion resistance in a California serpentine grassland. J Ecol. 2012;100(3):723–731. [Google Scholar]
- 19.Poole RW, Rathcke BJ. Regularity, randomness, and aggregation in flowering phenologies. Science. 1979;203(4379):470–471. doi: 10.1126/science.203.4379.470. [DOI] [PubMed] [Google Scholar]
- 20.Williams MR. Critical-values of a statistic to detect competitive displacement. Ecology. 1995;76(2):646–647. [Google Scholar]
- 21.Heil M, Silva Bueno JC. Within-plant signaling by volatiles leads to induction and priming of an indirect plant defense in nature. Proc Natl Acad Sci USA. 2007;104(13):5467–5472. doi: 10.1073/pnas.0610266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parmesan C. Influences of species, latitudes and methodologies on estimates of phenological response to global warming. Glob Change Biol. 2007;13(9):1860–1872. [Google Scholar]
- 23.Pachauri RK, et al. Climate Change 2014: Synthesis Report. IPCC; Geneva: 2014. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; p. 151. [Google Scholar]
- 24.Baythavong BS, Stanton ML. Characterizing selection on phenotypic plasticity in response to natural environmental heterogeneity. Evolution. 2010;64(10):2904–2920. doi: 10.1111/j.1558-5646.2010.01057.x. [DOI] [PubMed] [Google Scholar]
- 25.Hobbs RJ, Yates S, Mooney HA. Long-term data reveal complex dynamics in grassland in relation to climate and disturbance. Ecol Monogr. 2007;77(4):545–568. [Google Scholar]
- 26.Anderson JT, Inouye DW, McKinney AM, Colautti RI, Mitchell-Olds T. Phenotypic plasticity and adaptive evolution contribute to advancing flowering phenology in response to climate change. Proc Biol Sci. 2012;279(1743):3843–3852. doi: 10.1098/rspb.2012.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cleland EE, et al. Phenological tracking enables positive species responses to climate change. Ecology. 2012;93(8):1765–1771. doi: 10.1890/11-1912.1. [DOI] [PubMed] [Google Scholar]
- 28.Sherry RA, et al. Divergence of reproductive phenology under climate warming. Proc Natl Acad Sci USA. 2007;104(1):198–202. doi: 10.1073/pnas.0605642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vellend M, et al. Global meta-analysis reveals no net change in local-scale plant biodiversity over time. Proc Natl Acad Sci USA. 2013;110(48):19456–19459. doi: 10.1073/pnas.1312779110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dornelas M, et al. Assemblage time series reveal biodiversity change but not systematic loss. Science. 2014;344(6181):296–299. doi: 10.1126/science.1248484. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez A, et al. Estimating local biodiversity change: A critique of papers claiming no net loss of local diversity. Ecology. 2016;97(8):1949–1960. doi: 10.1890/15-1759.1. [DOI] [PubMed] [Google Scholar]
- 32.Urban MC. Climate change. Accelerating extinction risk from climate change. Science. 2015;348(6234):571–573. doi: 10.1126/science.aaa4984. [DOI] [PubMed] [Google Scholar]
- 33.Chen IC, Hill JK, Ohlemüller R, Roy DB, Thomas CD. Rapid range shifts of species associated with high levels of climate warming. Science. 2011;333(6045):1024–1026. doi: 10.1126/science.1206432. [DOI] [PubMed] [Google Scholar]
- 34.Hull PM, Darroch SA, Erwin DH. Rarity in mass extinctions and the future of ecosystems. Nature. 2015;528(7582):345–351. doi: 10.1038/nature16160. [DOI] [PubMed] [Google Scholar]
- 35.Selmants PC, Zavaleta ES, Wolf AA. Realistic diversity loss and variation in soil depth independently affect community-level plant nitrogen use. Ecology. 2014;95(1):88–97. doi: 10.1890/13-1192.1. [DOI] [PubMed] [Google Scholar]
- 36.Wolf AA, Zavaleta ES. Species traits outweigh nested structure in driving the effects of realistic biodiversity loss on productivity. Ecology. 2015;96(1):90–98. doi: 10.1890/14-0131.1. [DOI] [PubMed] [Google Scholar]
- 37.Miller-Rushing AJ, Inouye DW, Primack RB. How well do first flowering dates measure plant responses to climate change? The effects of population size and sampling frequency. J Ecol. 2008;96(6):1289–1296. [Google Scholar]
- 38.Bates D, Maechler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1):1–48. [Google Scholar]
- 39.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2014. [Google Scholar]
- 40.Grueber CE, Nakagawa S, Laws RJ, Jamieson IG. Multimodel inference in ecology and evolution: Challenges and solutions. J Evol Biol. 2011;24(4):699–711. doi: 10.1111/j.1420-9101.2010.02210.x. [DOI] [PubMed] [Google Scholar]
- 41.Bartoń K. 2016 MuMIn: Multi-Model Inference. R Package version 1.15.6. Available at cran.r-project.org/web/packages/MuMIn/index.html. Accessed June 1, 2016.
- 42.Baldwin BG, Goldman DH, Keil DJ, Patterson R, Rosatti TJ. The Jepson Manual: Vascular Plants of California. Univ of California Press; Berkeley, CA: 2012. [Google Scholar]