Significance
Our study provided a comprehensive view of the transcriptional landscape across the cell cycle. We revealed lag between transcription and steady-state RNA expression at the cell-cycle level and characterized a large amount of active transcription during early mitosis. In addition, our analysis identified thousands of enhancer RNAs and related transcription factors that are highly correlated with cell-cycle–regulated transcription but not with steady-state expression, thus highlighting the importance of transcriptional and epigenetic dynamics during cell-cycle progression.
Keywords: GRO-seq, nascent RNA, transcriptional regulation, epigenetics, cell cycle
Abstract
Steady-state gene expression across the cell cycle has been studied extensively. However, transcriptional gene regulation and the dynamics of histone modification at different cell-cycle stages are largely unknown. By applying a combination of global nuclear run-on sequencing (GRO-seq), RNA sequencing (RNA-seq), and histone-modification Chip sequencing (ChIP-seq), we depicted a comprehensive transcriptional landscape at the G0/G1, G1/S, and M phases of breast cancer MCF-7 cells. Importantly, GRO-seq and RNA-seq analysis identified different cell-cycle–regulated genes, suggesting a lag between transcription and steady-state expression during the cell cycle. Interestingly, we identified genes actively transcribed at early M phase that are longer in length and have low expression and are accompanied by a global increase in active histone 3 lysine 4 methylation (H3K4me2) and histone 3 lysine 27 acetylation (H3K27ac) modifications. In addition, we identified 2,440 cell-cycle–regulated enhancer RNAs (eRNAs) that are strongly associated with differential active transcription but not with stable expression levels across the cell cycle. Motif analysis of dynamic eRNAs predicted Kruppel-like factor 4 (KLF4) as a key regulator of G1/S transition, and this identification was validated experimentally. Taken together, our combined analysis characterized the transcriptional and histone-modification profile of the human cell cycle and identified dynamic transcriptional signatures across the cell cycle.
The process of cell division is vital to the growth and development of an organism as a single fertilized cell develops into a mature organism and organs undergo cellular renewal or repair (1–3). Tight control of molecular events during the cell cycle guarantees fidelity in preserving genetic information and the prevention of unwarranted cell division. The progression of the cell cycle involves well-orchestrated transcriptional and epigenetic controls (4–7). Dysregulation of this process can lead to various diseases, including cancer (8–10). Because the proportion of actively dividing cells is considerably higher in cancers than in normal tissues, targeting the cell cycle is an attractive therapeutic option for cancer treatment (11, 12).
Many proteins that carry out important functions during the cell cycle display a cyclic expression pattern that is often regulated on the transcriptional level (13, 14). Because it has been shown that cell-cycle gene expression serves as a tumor signature (15), extensive efforts have been devoted to identify periodically expressed genes across the cell cycle using microarray platforms (4, 16–20). In addition, identification of the cell-cycle–regulated genes and follow-up mechanistic studies of individual genes have greatly advanced our understanding of cell-cycle progression and the development of diseases. For instance, the transcriptional regulation of cell-cycle genes was found to be governed by the RB-E2F, DREAM, and MMB-FOXM1 complexes (21–23). However, analyses using microarray or RNA sequencing (RNA-seq) can identify only the accumulated, steady-state gene-expression level; the temporal regulation of transcriptional dynamics during the cell cycle remains elusive.
The nuclear run-on assay has long been used to investigate RNA polymerase activity and nascent RNA transcription for individual genes (24). The global nuclear run-on followed by RNA sequencing (GRO-seq) assay enables the investigation of temporal transcription at a genome-wide scale (25–28). In addition, GRO-seq also can be used to analyze transcription from active regulatory sequences such as enhancers (29). Recent studies have found that the loading of RNA polymerase II (Pol II) at enhancer regions can lead to widespread active transcription and production of enhancer RNAs (eRNAs) (30, 31). Since then, eRNAs have been demonstrated to play important roles in regulating enhancer–promoter interactions and target gene transcription, rather than merely being transcriptional noises from enhancers (32–35). GRO-seq has been applied widely to study transcriptional regulation in a variety of biological systems but has not been explored in the context of the cell cycle (26, 29, 36).
In this study, we used a combination of GRO-seq, RNA-seq, and histone-modification ChIP sequencing (ChIP-seq) to investigate transcriptional and epigenetic dynamics across the cell cycle. The multilevel data of nascent transcription, steady-state expression level, and chromatin status provide insights not only into genes periodically regulated during the cell cycle but also into the underlying regulatory mechanisms. Our data depict a comprehensive transcriptional and epigenetic landscape of the human cell cycle and will be a valuable resource for cell-cycle studies.
Results
Mapping the Transcriptional and Epigenetic Landscape Across the Cell Cycle.
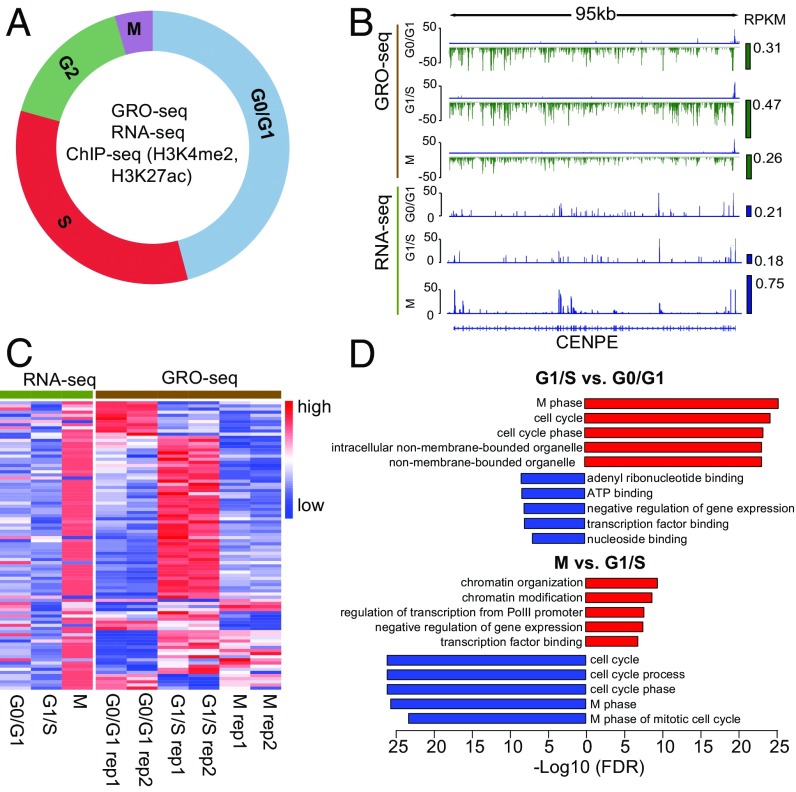
To investigate dynamic transcriptional and epigenetic gene regulation across the cell cycle, we performed GRO-seq (25), RNA-seq, and ChIP-seq of histone 3 lysine 27 acetylation (H3K27ac) and histone 3 lysine 4 methylation (H3K4me2), two histone modifications that mark promoters and enhancers, in the synchronized human breast cancer cell line MCF-7 (Fig. 1A). The cells were synchronized to G0/G1 with hormone starvation, to G1/S with double thymidine treatment, and to early M phase with thymidine-nocodazole treatment (Materials and Methods). The degree of synchrony at each cell-cycle stage was monitored by flow cytometry analysis of DNA content using propidium iodide staining (SI Appendix, Fig. S1) (37). Nuclei were isolated from two biological replicates of the synchronized cells and were subjected to the GRO-seq procedure (Materials and Methods) for nascent RNA analysis. Total RNA from the same batch of synchronized cells was subjected to RNA-seq analysis to investigate steady-state expression levels. Chromatin profiling was conducted using replicated H3K27ac and H3K4me2 ChIP-seq at all three cell-cycle stages.
Fig. 1.
GRO-seq and RNA-seq identify different cell-cycle–regulated genes. (A) Illustration of transcriptional dynamics analysis across the cell-cycle stages in MCF-7 cells. GRO-seq and ChIP-seq experiments were performed in two biological replicates, and RNA-seq was performed without replicates. (B) Transcription and expression of CENPE as measured by GRO-seq and RNA-seq at different cell-cycle stages. Green and blue bars on the right side of the signal tracks represent the CENPE transcription and expression levels as measured by reads per kilobase per million mapped reads (RPKM). (C) Transcription (GRO-seq) and expression (RNA-seq) of curated mitotic genes. The genes specifically up-regulated at G2/M were curated from published datasets (Materials and Methods). Read counts of each gene were normalized among the three cell-cycle stages so that their mean equals 0 and the SD equals 1, with red representing higher signal and blue representing lower signal. (D) GO analysis of cell-cycle-stage–specific genes identified by GRO-seq analysis. Bar length represents the −log10 FDR. Red bars indicate terms enriched for up-regulated genes; blue bars indicate terms enriched for down-regulated genes. The top five enriched terms are shown for each comparison.
Approximately 30–40 million reads were uniquely mapped to the human genome for each GRO-seq sample, and the biological replicates for each cell-cycle stage were highly correlated (SI Appendix, Table S1). Most reads (70%) align on the coding strand within the boundaries of annotated National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) genes, and the typical bidirectional transcription at the transcriptional start site (TSS) was observed (SI Appendix, Fig. S2 and Table S1) (37). Thirty million reads were uniquely mapped for each RNA-seq sample, more than 50% of which were mapped to the annotated RefSeq gene exons and UTRs (SI Appendix, Table S2). Fifteen to twenty million reads were uniquely mapped for each ChIP-seq sample, and the correlation between biological replicates was more than 0.96 (SI Appendix, Table S3).
GRO-Seq and RNA-Seq Identify Different Cell-Cycle–Regulated Genes.
To investigate the correlation between nascent transcription and steady-state expression levels, we compared the GRO-seq and RNA-seq read counts for all the RefSeq annotated genes (Materials and Methods). Although the correlations between GRO-seq samples at different cell-cycle stages ranged from 0.89–0.99, and those of the RNA-seq samples ranged from 0.78–0.90, the correlations between GRO-seq and RNA-seq data were below 0.5 (SI Appendix, Fig. S3), indicating significant difference between newly synthesized and accumulated RNA levels. As an example, we plotted the histogram of GRO-seq and RNA-seq read counts for Centromere-associated protein E precursor (CENPE) (Fig. 1B), a centrosome-associated mitotic kinesin (38). Although the RNA-seq signal for CENPE is three- to fourfold higher at M phase than at G0/G1 and G1/S, the GRO-seq signal plateaus at G1/S (Fig. 1B), suggesting a lag of RNA-seq in reflecting transcription at cell-cycle stages. This lagging effect was observed for most of the mitotic genes we curated from publicly available datasets profiling cell-cycle expression (Fig. 1C) (16–20).
To assess the dynamics of transcriptional regulation at different cell-cycle stages, we identified genes differentially transcribed across the cell cycle. GRO-seq read counts for RefSeq genes at different cell-cycle stages were normalized using spiked-ins as described previously (39), and the normalized read counts were subjected to DESeq2 analysis (SI Appendix, Fig. S4) (40). Gene ontology (GO) analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (41) was conducted on genes with significant [false-discovery rate (FDR) <0.01] differential transcription among cell-cycle stages (Fig. 1D, SI Appendix, Fig. S5, and Dataset S1). Genes with higher transcription in G1/S than in G0/G1 and M phases were enriched for the GO term “M phase,” suggesting that mitotic genes are actively transcribed in G1/S and accumulated at M phase. Importantly, most of the mitotic genes we curated (Materials and Methods) overlapped with genes in the M phase GO term and had the highest steady-state expression at M phase (Fig. 1C). In addition, genes highly transcribed at G0/G1 were enriched in GO terms of “ribonucleotide binding” (SI Appendix, Fig. S5), suggesting that genes required for DNA synthesis at S phase are, at least in part, actively transcribed at G0/G1. Finally, genes with decreased transcription at G0/G1 compared with the G1/S and M phases were enriched in GO terms of “cytosolic ribosomes” and “organelles” (Fig. 1D and SI Appendix, Fig. S5), and the duplication of organelles and cytosolic components are the major activities at the G1 phase. Taking these results together, we observed a prevailing lag of mRNA abundance compared with gene transcription, suggesting that the transcriptional activation of genes precedes the accumulation of their transcribed products at the cell-cycle level. These results demonstrate well-orchestrated transcriptional dynamics during the cell cycle.
Active Transcription at Early Mitosis.
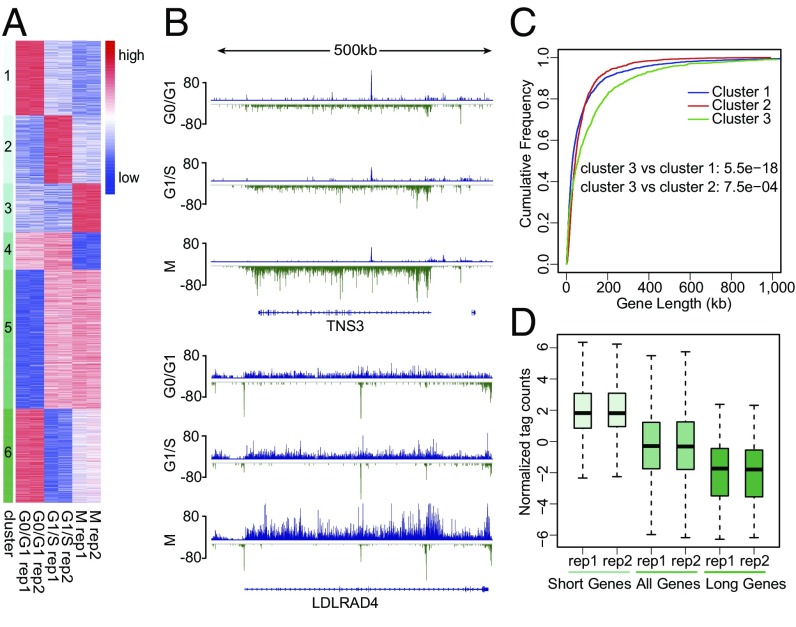
To investigate further the dynamic pattern of transcription at different cell-cycle stages, we performed unsupervised k-means clustering of all the differentially transcribed genes in GRO-seq samples (Fig. 2A). Among the six dynamic transcription patterns, cluster 3 contains genes with highest transcription at M phase (Fig. 2A). The transcription complexes are in general inactivated and disassembled from chromatin during chromosome condensation in the prophase, and the mitotic phase is known for silence of transcription (42). Although a fraction of transcription factors, including mixed-lineage leukemia (MLL), bromodomain containing 4 (BRD4), Forkhead box A1 (FOXA1), and GATA-binding protein 1 (GATA1), are retained on mitotic chromosomes, they are thought to facilitate rapid gene reactivation post mitosis (43–46). Our thymidine-nocodazole blocking followed by a shake-off method enriches cells at the early mitotic phase. The identification of genes with peak transcription at early M phase using GRO-seq was intriguing. Analysis of RNA-seq data revealed that this group of genes had the highest expression level at G0/G1 (SI Appendix, Figs. S6 and S7), consistent with a lag of RNA-seq in reflecting the transcription observed for the mitotic genes (Fig. 1C).
Fig. 2.
Active transcription at early M phase. (A) Clustering of differentially transcribed genes at different cell-cycle stages identified by GRO-seq analysis. Genes are classified into six clusters using unsupervised k-means clustering. (B) Representative long genes up-regulated at M phase. Blue and green bars represent the GRO-seq signal from the plus and minus strands, respectively. Signals were normalized to a total of 10 million reads, and replicates were combined. (C) Empirical cumulative distribution of gene length. Blue, red, and green traces represent the groups of genes with higher GRO-seq signal at G0/G1, G1/S, and M phases, respectively. (D) GRO-seq signal of short (top 25% shortest, two far-left boxes), all (middle two boxes), and long (top 25% longest, two far-right boxes) genes that are highly expressed at M phase (red trace in C).
Further inspection of the top differentially transcribed genes in cluster 3, such as TNS3 and LDLRAD4, found them to be of larger size (Fig. 2B). We thus analyzed the length distribution of the genes in the six clusters. The one-tailed Wilcox rank sum test between each cluster pair showed that cluster 3 is significantly enriched for longer genes (Fig. 2C and SI Appendix, Fig. S8A). We then binned all differentially transcribed genes into 10 length intervals (with interval 1 containing the shortest genes and interval 10 containing the longest genes) and plotted the proportion of genes in each interval for the six clusters. More than 20% of the cluster 3 genes were in interval 10, compared with 6–11% of the genes in the other clusters (SI Appendix, Fig. S8B). The longer genes had relatively lower transcription levels than shorter ones (Fig. 2D).
To compare the transcription pattern of these long genes during the cell cycle, we plotted the GRO-seq signal along the gene body for the cluster 3 genes in the longest gene length interval (SI Appendix, Fig. S8C). Genes were equally divided into 50 bins from the TSS to cleavage/polyadenylation (CPA) site, and the average GRO-seq signal within each bin was summarized. Many of these genes had a strong GRO-seq signal at the TSS and a weak signal in the gene body at G0/G1, indicating paused Pol II. Interestingly, as the cell cycle progressed the signal in the gene body became stronger, especially at M phase. To determine whether the strong GRO-seq signal observed at M phase is from paused or actively elongating Pol II, we reanalyzed publically available Pol II ChIP-seq data in mitotic HeLa cells treated with or without flavopiridol, which inhibits the transcription elongation factor P-TEFb and the release of promoter-proximal paused Pol II (47). Flavopiridol treatment resulted in a significantly stronger Pol II signal at the promoter regions (SI Appendix, Fig. S9), suggesting that these Pol II were actively released in early mitotic cells without flavopiridol treatment. Collectively, our analysis identified genes actively transcribed at early M phase, which tend to be longer genes with lower expression.
H3K27ac and H3K4me2 Signals Increase Globally at Mitosis.
Given the importance of changes in chromatin structure during the cell cycle (48–50), we performed histone-modification ChIP-seq of H3K4me2 and H3K27ac to investigate chromatin dynamics across the cell cycle. When normalized to same sequencing depth, the peak numbers at different cell-cycle stages were very similar (SI Appendix, Table S3). In addition, the correlations of the ChIP-seq signal among different cell-cycle stages were very high (SI Appendix, Figs. S10 and S11), indicating that the local histone-modification states were remarkably stable despite the dramatic changes in chromosome organization across the cell cycle (5).
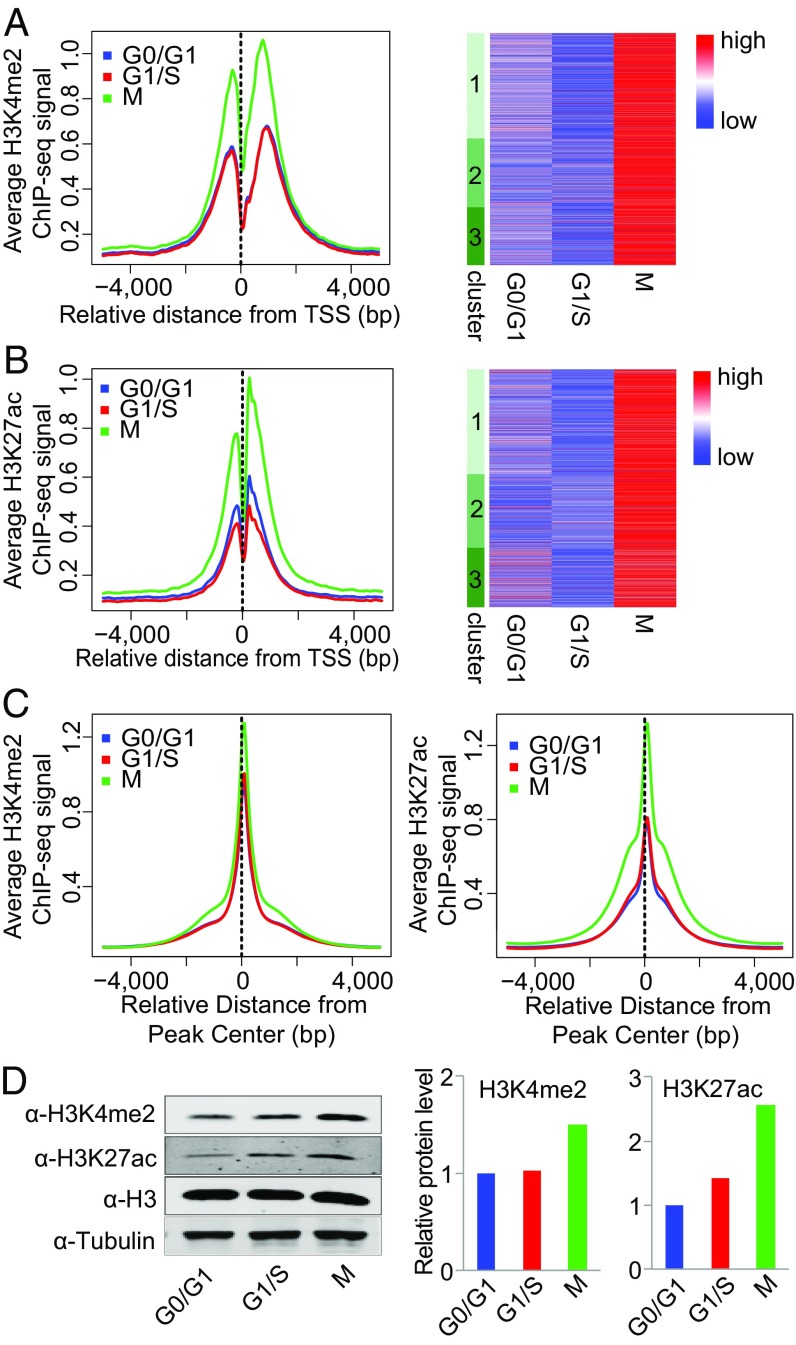
We then grouped genes into high, medium, and low categories based on the expression level calculated from RNA-seq data and plotted the normalized tag counts from the histone marks ChIP-seq data at promoter regions (TSS ± 1 kb). Consistent with a previous report (51), both H3K4me2 and H3K27ac were positively correlated with gene-expression level (SI Appendix, Fig. S12 A and B). A similar trend was observed when genes were grouped based on the transcription level calculated from GRO-seq data (SI Appendix, Fig. S12 A and B). We then sought to determine whether the differential transcription across the cell cycle was correlated with changes in local histone-modification states. Intriguingly, when normalized to the same sequencing depth, the H3K4me2 and H3K27ac signals were much stronger at M phase for all the differentially transcribed genes, regardless of their transcription patterns across the cell-cycle stages (Fig. 3 A and B and SI Appendix, Fig. S8 C and D).
Fig. 3.
Global increase in H3K4me2 and H3K27ac signals at early M phase. (A and B) H3K4me2 (A) and H3K27ac (B) ChIP-seq signal at promoter regions across different cell-cycle stages. (Left) Average H3K4me2 (A) and H3K27ac (B) signal (RPKM) of genes in clusters 1, 2, and 3. (Right) Heatmap of H3K4me2 (A) and H3K27ac (B) signals of genes in clusters 1, 2, and 3. (C, Left) Average H3K4me2 ChIP-seq signal at all H3K4me2 peak regions. (Right) Average H3K27ac ChIP-seq signal at all H3K27ac peak regions. (D, Left) Western blot of H3K4me2 and H3K27ac at different cell-cycle stages. H3 and tubulin were used as loading controls. (Right) Bar plots represent the quantitative value (normalized to H3) of H3K4me2 and H3K27ac protein levels from the Western blot.
To investigate whether this correlation results from the global increase of the histone modifications at M phase, we plotted the ChIP-seq signal at the union of peaks identified from all the samples and indeed found both the H3K4me2 and the H3K27ac signals were significantly higher at M phase (Fig. 3C). We then performed Western blot analysis of H3K4me2 and H3K27ac at different cell-cycle stages. When normalized to total H3 or tubulin, the protein levels of modified H3 were higher at M phase than at the G0/G1 and G1/S phases (Fig. 3D), as was consistent with the ChIP-seq signals.
Identification of Cell-Cycle–Regulated eRNAs.
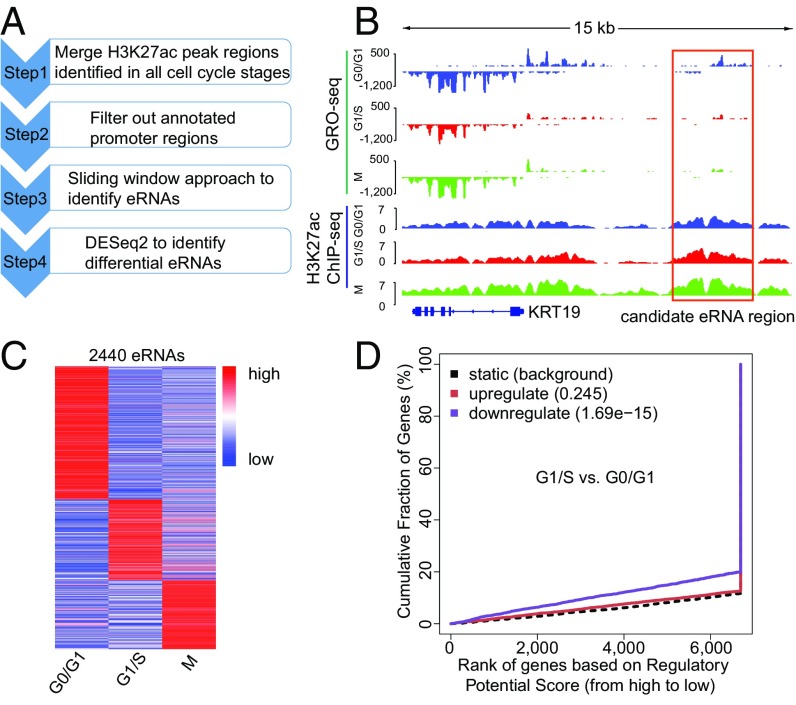
Pol II and many other transcription factors associate with a large number of enhancers marked by H3K4me1 and H3K27ac and produce noncoding eRNAs that have been demonstrated to play an important role in transcriptional regulation (30, 31). Therefore we sought to determine whether there were cell-cycle–regulated eRNAs and to identify their role in cell-cycle progression. To this end, we developed a computational pipeline to characterize cell-cycle–regulated eRNAs (Fig. 4A). H3K27ac ChIP-seq peaks at different cell-cycle stages were merged, and promoter regions were filtered out, leaving ∼50,000 peaks as potential active enhancers. Because eRNAs are often transcribed bidirectionally from enhancer regions (31), we applied a sliding window approach to identify enhancer regions with bidirectional transcription (Fig. 4B). This analysis identified a total of 4,922 eRNAs, 2,440 of which were differentially transcribed at different cell-cycle stages (Fig. 4C).
Fig. 4.
Identification of cell-cycle–regulated eRNAs. (A) Workflow for identification of cell-cycle-stage–specific eRNAs. (B) Example of a candidate eRNA near gene KRT19. (C) Heatmap of 2,440 differential eRNAs. Color shows the relative GRO-seq signal at each eRNA region at three different stages. (D) Correlation between cell-cycle–regulated eRNAs and differentially transcribed genes.
To investigate the role of eRNAs in transcriptional regulation and cell-cycle progression, we analyzed the correlation between differentially transcribed eRNAs and genes. Binding and Expression Target Analysis (BETA) software (52) was used to reveal a relationship between eRNAs and genes. Accordingly, a strong correlation was identified between eRNAs and differentially transcribed genes as determined by GRO-seq (Fig. 4D and SI Appendix, Fig. S13). Similar analysis performed with RNA-seq data found much weaker correlation between the eRNAs and differentially expressed genes (SI Appendix, Fig. S14), further emphasizing the advantage of analyzing temporal transcription regulation from GRO-seq rather than analyzing steady-state gene expression from RNA-seq.
Analysis of eRNAs Identifies Kruppel-Like Factor 4 KLF4 as a Key Regulator of G1/S Transition.
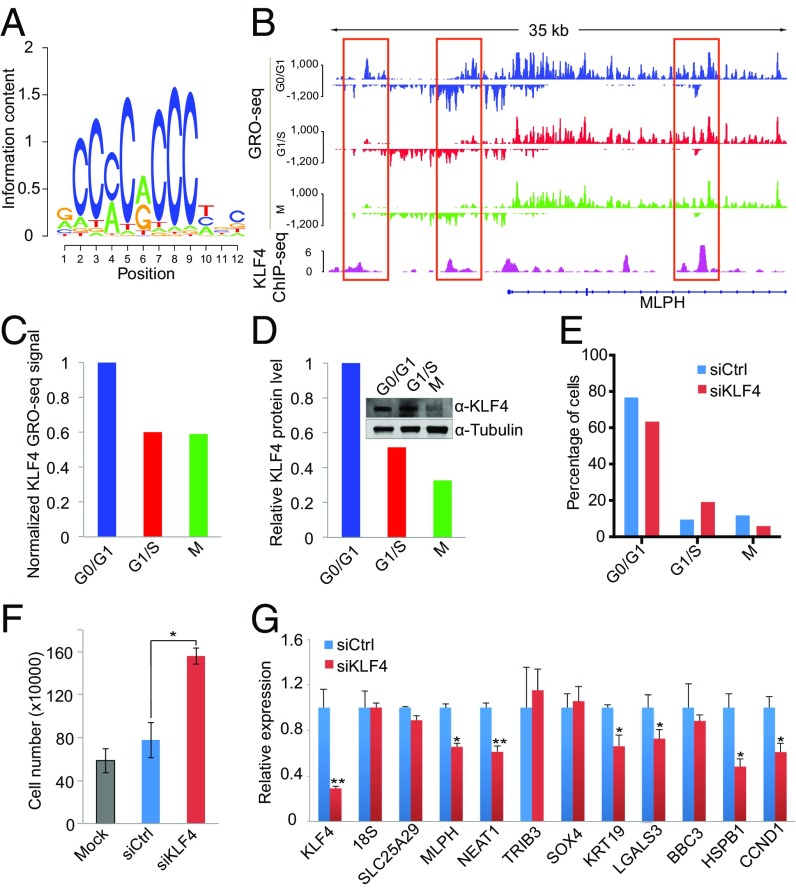
After confirming the correlation between cell-cycle–regulated eRNAs and genes, we sought to identify the transcription factors that regulate these eRNAs. Transcription factors usually bind to open chromatin regions. To improve the spatial resolution of the transcription factor-binding sites in eRNA regions, we performed DNase sequencing (DNase-seq) in MCF-7 cells to identify open chromatin regions that were hypersensitive to DNase I digestion. Subsequently, we performed motif analysis at DNase I hypersensitivity sites (DHS) in the eRNA regions (SI Appendix, Fig. S15). This analysis identified motifs of multiple transcription factors including Kruppel-like factor 4 (KLF4), which ranked top in the eRNA regions that are highly transcribed at G0/G1 (Fig. 5A and SI Appendix, Table S4). KLF4 is important in cell-cycle control, cellular differentiation, and carcinogenesis (53–55). We then compared published KLF4 ChIP-seq peaks (56) with eRNAs and found that KLF4 binding was enriched in eRNA regions with peaked transcription at G0/G1 (Fig. 5B and SI Appendix, Fig. S16).
Fig. 5.
KLF4 regulates eRNAs and target genes to control G1/S transition. (A) The KLF4 motif was identified as the most significant one enriched in G0/G1-specific eRNA regions. (B) Example of KLF4 eRNAs (marked with red boxes) near the target gene MLPH. (C and D) KLF4 RNA (C) and protein levels (D) across different cell-cycle stages. Tubulin was used as the loading control for Western blot analysis. (E) Flow cytometry cell-cycle analysis of MCF-7 cells in which KLF4 was silenced with siRNAs. (F) Cell growth analysis of MCF-7 cells upon silencing of KLF4. Mock, without transfection. (G) Validation of candidate KLF4 target genes identified through eRNA analysis. RT-PCR was performed for candidate KLF4 target genes upon silencing of KLF4. Error bars indicate the SEM; *P < 0.05; ** P < 0.01 (t test).
In addition, GRO-seq revealed that the KLF4 transcription level was high at G0/G1 and decreased at the G1/S and M phases (Fig. 5C). A more dramatic decrease was observed at the protein level through Western blotting (Fig. 5D). To evaluate the role of KLF4 in cell-cycle progression, we silenced KLF4 by siRNAs in MCF-7 cells. FACS analysis showed a significant increase in the proportion of cells at S phase (Fig. 5E). Moreover, cell-proliferation analysis showed that silencing KLF4 promotes MCF-7 cell growth (Fig. 5F). Together the results indicate that KLF4 has an important role in blocking G1/S transition. Because transcription factors function by regulating the transcription of specific gene targets, we aimed to identify a KLF4-dependent gene signature that modulates this G1/S transition through the regulation of eRNAs. BETA analysis identified 10 potential KLF4 direct target genes, six of which were significantly down-regulated upon silencing of KLF4 (Fig. 5G). Among the six genes down-regulated by KLF4 silencing, KRT19 has been reported to suppress cell proliferation, and silencing of KRT19 leads to an increased proportion of cells at S phase (57). On the other hand, CCND1, a gene with decreased expression at S phase, is a well-characterized cell-cycle regulator that promotes the G1/S transition (58), and NEAT1 and HSPB1 have been reported to promote the proliferation of breast cancer cells (59, 60). Thus the exact mechanism by which KLF4 controls G1/S transition is unclear and warrants further investigation.
Discussion
In this study, we systematically investigated transcriptional and epigenetic dynamics during the cell cycle by analyzing GRO-seq, RNA-seq, and histone marks ChIP-seq data at G0/G1, G1/S, and M phases in the MCF-7 breast cancer cell line. Our study revealed (i) a lag between transcription and steady-state RNA expression at the cell-cycle level; (ii) a large amount of active transcription during early mitosis; (iii) a global increase in active histone modifications at mitosis; (iv) thousands of cell-cycle–regulated eRNAs; and (v) dynamic eRNAs bound by transcription factors such as KLF4 that regulate cell-cycle progression.
Steady-state mRNA abundance is influenced by a few factors, including transcription, RNA processing, maturation, and degradation. Therefore, measuring steady-state mRNA levels by microarray or RNA-seq techniques may not accurately reflect active transcription. Indeed, GRO-seq and 4-thiouridine metabolic labeling followed by sequencing (4sU-seq) analyses that measure nascent transcription have revealed a broad inconsistency between transcription rate and mRNA levels (25, 28, 61, 62). Specifically, there is a delay in steady-state expression reflecting the transcription and mass production of rapidly degraded transcripts that are not detectable at the steady-state expression level. Most of the previous nascent transcription analyses were performed with unsynchronized cells or with synchronized cells within a short time window that was insufficient to cover multiple cell-cycle stages (26, 28, 29, 32, 35, 36, 62). Importantly, our GRO-seq and RNA-seq analysis at different cell-cycle stages revealed a lag between active transcription and steady-state expression during the cell cycle. The RNA degradation rate has been considered the most prominent measurable factor that contributes to the lag between transcription and accumulated RNA levels. Recent studies demonstrated that the half-lives of mammalian genes range from less than 1 min to more than 3 h (61, 62). In agreement with these observations, our data showed that mitotic genes are most highly transcribed at G1/S, and the genes most highly transcribed at M phase are more abundant at G0/G1, suggesting that these genes have an extremely long half-life.
Mitotic chromatin is transcriptionally inactive in general, and even ongoing transcriptions are aborted to ensure the integrity of the separating chromosomes (63). However, exceptions have been found in which the promoter of the cyclin B1 gene maintains an open chromatin configuration, and the gene is actively transcribed during mitosis (64). Recently, additional large-scale studies have revealed that part of the mitotic chromatin remains accessible to Pol II and transcription factors such as MLL, BRD4, GATA1, FOXA1, and AR (43–46, 65). Our GRO-seq data showed that although CCNB1 transcription peaks at G1/S, strong nascent transcription was observed at M phase. More interestingly, we identified a group of genes with a transcription peak at M phase. The observation that this group was enriched for unusually long genes made us hypothesize that the GRO-seq signal was from the incomplete transcription from previous stages (66). We therefore compared the GRO-seq signal along the gene body to identify the longest quarter of genes with the highest GRO-seq signal at M phase. If the hypothesis is correct, we should be able to observe a GRO-seq signal pattern shifted from the TSS toward the CPA site during the cell-cycle progression from G0/G1 to M phase. Our analysis revealed a uniform distribution of signal along the gene body for most genes. In addition, reanalysis of publically available Pol II ChIP-seq data in early mitotic cells pretreated with and without flavopiridol (47) confirmed that Pol II is actively engaged at the TSS of these genes. Together, the results suggested that the high GRO-seq signal of these genes arose from active transcription at early M phase rather than from incomplete transcription at the G0/G1 and G1/S phases. Importantly, Liang et al. (47) recently reported mitotic transcriptional activation as a mechanism to clear actively engaged Pol II from mitotic chromatin; this mechanism is consistent with our observation of active transcription at early mitotic cells.
In support of active transcription at M phase, we observed extremely stable chromatin states marked by active histone modifications H3K4me2 and H3K27ac across different cell-cycle stages. In addition, the total H3K4me2 and H3K27ac levels increased significantly at M phase. Previous studies have identified mitotic-specific H4K20 methylation and the dynamics of H3K36 and H3K27 methylations across the cell cycle (67–69). The functional role of posttranscriptional histone modifications in the cell cycle is still largely unknown and warrants further analysis. It is worth noting that these observations were made in cancer cells with uncontrolled cell division; these cells may differ from normal cells with more stringent cell-cycle regulation (70). Future studies are warranted to explore the mechanisms underlying the active transcription during mitosis in normal and cancer cells.
Taken together, our analyses identified thousands of eRNAs and related transcription factors that are highly correlated with cell-cycle–regulated transcription but not with steady-state expression, thus highlighting the importance of transcriptional and epigenetic dynamics during cell-cycle progression. Overall, our study provides a comprehensive view of transcriptional landscape across the cell cycle and deepens our understanding of transcriptional dynamics during cell cycle. Future studies combining transcription, expression, and proteomics data at more detailed time courses are warranted to provide a more comprehensive view of cell-cycle regulation.
Materials and Methods
Cell Culture and Synchronization.
The MCF-7 cells were obtained from ATCC and were cultured in DMEM medium supplemented with 10% (vol/vol) FBS, 1% penicillin-streptomycin, and 1% glutamine in a 5% (vol/vol) CO2 humidified incubator. Cells were synchronized to G0/G1, G1/S, and M phase with hormone starvation, thymidine double treatment, and thymidine-nocodazole treatment, respectively. For detailed operations, see SI Appendix, Supplemental Methods.
GRO-Seq Library Construction.
Nuclear run-on experiments (SI Appendix, Supplemental Methods) were performed as described previously (25). The resultant RNA was purified further with the TURBO DNA-free kit (AM1907; Life Technologies) to remove residue DNA contamination. Libraries then were constructed with the Encore Complete RNA-Seq DR Multiplex System 1–8 (0333-32; NuGEN Technologies, Inc.) and were sequenced to 50 bp with an Illumina HiSeq machine.
Additional experimental procedures and methods are described in SI Appendix, Supplemental Methods.
Availability of Data and Material.
MCF-7 GRO-seq, RNA-seq, ChIP-seq, and DNase-seq raw sequence tags and processed bed files have been submitted to the National Center for Biotechnology Gene Expression Omnibus (GEO) database under accession no GSE94479.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China Grant 31329003 (to X.S.L.), the Princess Margaret Cancer Foundation (H.H.H.), Canada Foundation for Innovation and Ontario Research Fund Grant CFI32372 (to H.H.H.), Natural Sciences and Engineering Research Council of Canada Discovery Grant 498706 (to H.H.H.), NIH Grant 1R01GM099409 (to X.S.L.), US Public Health Service Grants R01CA63113 and R01CA173023 (to J.A.D.), and a German National Academy of Sciences Leopoldina Fellowship (to M.F.). H.H.H. holds an Institute Community Support (ICS)-Institute of Genetics Maud Menten New Principal Investigator Prize (ICS-145381); an Office of Management, Information, and Research (OMIR) Early Researcher Award; a Terry Fox New Investigator Award; and a Canadian Institutes of Health Research (CIHR) New Investigator Salary Award.
Footnotes
The authors declare no conflict of interest.
Data deposition: The MCF-7 GRO-seq, RNA-seq, ChIP-seq, and DNase-seq raw sequence tags and processed bed files reported in this paper have been submitted to the National Center for Biotechnology Gene Expression Omnibus (GEO) database (accession no. GSE94479).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617636114/-/DCSupplemental.
References
- 1.Okayama H, et al. Cell cycle control in fission yeast and mammals: Identification of new regulatory mechanisms. Adv Cancer Res. 1996;69:17–62. doi: 10.1016/s0065-230x(08)60859-3. [DOI] [PubMed] [Google Scholar]
- 2.McGill CJ, Brooks G. Cell cycle control mechanisms and their role in cardiac growth. Cardiovasc Res. 1995;30(4):557–569. [PubMed] [Google Scholar]
- 3.Piatelli M, Tanguay D, Rothstein T, Chiles T. Cell cycle control mechanisms in B-1 and B-2 lymphoid subsets. Immunol Res. 2003;27(1):31–52. doi: 10.1385/IR:27:1:31. [DOI] [PubMed] [Google Scholar]
- 4.Fraser HB. Cell-cycle regulated transcription associates with DNA replication timing in yeast and human. Genome Biol. 2013;14(10):R111. doi: 10.1186/gb-2013-14-10-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schafer KA. The cell cycle: A review. Vet Pathol. 1998;35(6):461–478. doi: 10.1177/030098589803500601. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Ma H, Pugh BF. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res. 2011;21(6):875–884. doi: 10.1101/gr.117465.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramachandran S, Henikoff S. Transcriptional Regulators Compete with Nucleosomes Post-replication. Cell. 2016;165(3):580–592. doi: 10.1016/j.cell.2016.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bello MJ, Rey JA. The p53/Mdm2/p14ARF cell cycle control pathway genes may be inactivated by genetic and epigenetic mechanisms in gliomas. Cancer Genet Cytogenet. 2006;164(2):172–173. doi: 10.1016/j.cancergencyto.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Dash BC, El-Deiry WS. Cell cycle checkpoint control mechanisms that can be disrupted in cancer. Methods Mol Biol. 2004;280:99–161. doi: 10.1385/1-59259-788-2:099. [DOI] [PubMed] [Google Scholar]
- 10.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 11.Wäsch R. Targeting mitotic exit for cancer treatment. Expert Opin Ther Targets. 2011;15(7):785–788. doi: 10.1517/14728222.2011.577420. [DOI] [PubMed] [Google Scholar]
- 12.Dominguez-Brauer C, et al. Targeting mitosis in cancer: Emerging strategies. Mol Cell. 2015;60(4):524–536. doi: 10.1016/j.molcel.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Dynlacht BD. Regulation of transcription by proteins that control the cell cycle. Nature. 1997;389(6647):149–152. doi: 10.1038/38225. [DOI] [PubMed] [Google Scholar]
- 14.Brandeis M, Hunt T. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 1996;15(19):5280–5289. [PMC free article] [PubMed] [Google Scholar]
- 15.Nevins JR, Potti A. Mining gene expression profiles: Expression signatures as cancer phenotypes. Nat Rev Genet. 2007;8(8):601–609. doi: 10.1038/nrg2137. [DOI] [PubMed] [Google Scholar]
- 16.Whitfield ML, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13(6):1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bar-Joseph Z, et al. Genome-wide transcriptional analysis of the human cell cycle identifies genes differentially regulated in normal and cancer cells. Proc Natl Acad Sci USA. 2008;105(3):955–960. doi: 10.1073/pnas.0704723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peña-Diaz J, et al. Transcription profiling during the cell cycle shows that a subset of Polycomb-targeted genes is upregulated during DNA replication. Nucleic Acids Res. 2013;41(5):2846–2856. doi: 10.1093/nar/gks1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant GD, et al. Identification of cell cycle-regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol Biol Cell. 2013;24(23):3634–3650. doi: 10.1091/mbc.E13-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26(5):474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadasivam S, DeCaprio JA. The DREAM complex: Master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer. 2013;13(8):585–595. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertoli C, Klier S, McGowan C, Wittenberg C, de Bruin RA. Chk1 inhibits E2F6 repressor function in response to replication stress to maintain cell-cycle transcription. Curr Biol. 2013;23(17):1629–1637. doi: 10.1016/j.cub.2013.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer M, Grossmann P, Padi M, DeCaprio JA. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 2016;44(13):6070–6086. doi: 10.1093/nar/gkw523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gariglio P, Bellard M, Chambon P. Clustering of RNA polymerase B molecules in the 5′ moiety of the adult beta-globin gene of hen erythrocytes. Nucleic Acids Res. 1981;9(11):2589–2598. doi: 10.1093/nar/9.11.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Step SE, et al. Anti-diabetic rosiglitazone remodels the adipocyte transcriptome by redistributing transcription to PPARγ-driven enhancers. Genes Dev. 2014;28(9):1018–1028. doi: 10.1101/gad.237628.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hah N, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145(4):622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23(8):1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8(5):e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39(4):170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh CL, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci USA. 2014;111(20):7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, et al. Enhancer RNA-driven looping enhances the transcription of the long noncoding RNA DHRS4-AS1, a controller of the DHRS4 gene cluster. Sci Rep. 2016;6:20961. doi: 10.1038/srep20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498(7455):516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474(7351):390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Min IM, et al. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25(7):742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barisic M, et al. Mitosis. Microtubule detyrosination guides chromosomes during mitosis. Science. 2015;348(6236):799–803. doi: 10.1126/science.aaa5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lovén J, et al. Revisiting global gene expression analysis. Cell. 2012;151(3):476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 42.Segil N, Guermah M, Hoffmann A, Roeder RG, Heintz N. Mitotic regulation of TFIID: Inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10(19):2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 43.Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat Cell Biol. 2011;13(11):1295–1304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caravaca JM, et al. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 2013;27(3):251–260. doi: 10.1101/gad.206458.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kadauke S, et al. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell. 2012;150(4):725–737. doi: 10.1016/j.cell.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blobel GA, et al. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell. 2009;36(6):970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang K, et al. Mitotic transcriptional activation: Clearance of actively engaged Pol II via transcriptional elongation control in mitosis. Mol Cell. 2015;60(3):435–445. doi: 10.1016/j.molcel.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pederson T. Chromatin structure and the cell cycle. Proc Natl Acad Sci USA. 1972;69(8):2224–2228. doi: 10.1073/pnas.69.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nair VD, et al. Involvement of histone demethylase LSD1 in short-time-scale gene expression changes during cell cycle progression in embryonic stem cells. Mol Cell Biol. 2012;32(23):4861–4876. doi: 10.1128/MCB.00816-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh AM, et al. Cell-Cycle Control of Bivalent Epigenetic Domains Regulates the Exit from Pluripotency. Stem Cell Rep. 2015;5(3):323–336. doi: 10.1016/j.stemcr.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 52.Wang S, et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc. 2013;8(12):2502–2515. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu D, et al. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat Commun. 2015;6:8419. doi: 10.1038/ncomms9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu D, Zhou Z, Davidson NE, Huang Y, Wan Y. Novel insight into KLF4 proteolytic regulation in estrogen receptor signaling and breast carcinogenesis. J Biol Chem. 2012;287(17):13584–13597. doi: 10.1074/jbc.M112.343566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gamper AM, et al. Regulation of KLF4 turnover reveals an unexpected tissue-specific role of pVHL in tumorigenesis. Mol Cell. 2012;45(2):233–243. doi: 10.1016/j.molcel.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mohammed H, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Reports. 2013;3(2):342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ju JH, et al. Regulation of cell proliferation and migration by keratin19-induced nuclear import of early growth response-1 in breast cancer cells. Clin Cancer Res. 2013;19(16):4335–4346. doi: 10.1158/1078-0432.CCR-12-3295. [DOI] [PubMed] [Google Scholar]
- 58.Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459(7247):722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ke H, et al. NEAT1 is Required for Survival of Breast Cancer Cells Through FUS and miR-548. Gene Regul Syst Bio. 2016;10(Suppl 1):11–17. doi: 10.4137/GRSB.S29414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cayado-Gutiérrez N, et al. Downregulation of Hsp27 (HSPB1) in MCF-7 human breast cancer cells induces upregulation of PTEN. Cell Stress Chaperones. 2013;18(2):243–249. doi: 10.1007/s12192-012-0367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 62.Rabani M, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;29(5):436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shermoen AW, O’Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67(2):303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sciortino S, et al. The cyclin B1 gene is actively transcribed during mitosis in HeLa cells. EMBO Rep. 2001;2(11):1018–1023. doi: 10.1093/embo-reports/kve223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McNair C, et al. Cell cycle-coupled expansion of AR activity promotes cancer progression. Oncogene. 2016;10(1038):1–14. doi: 10.1038/onc.2016.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44(6):966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 67.Rice JC, et al. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 2002;16(17):2225–2230. doi: 10.1101/gad.1014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zee BM, Britton LM, Wolle D, Haberman DM, Garcia BA. Origins and formation of histone methylation across the human cell cycle. Mol Cell Biol. 2012;32(13):2503–2514. doi: 10.1128/MCB.06673-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma Y, Kanakousaki K, Buttitta L. How the cell cycle impacts chromatin architecture and influences cell fate. Front Genet. 2015;6:19. doi: 10.3389/fgene.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shah MA, Denton EL, Arrowsmith CH, Lupien M, Schapira M. A global assessment of cancer genomic alterations in epigenetic mechanisms. Epigenetics Chromatin. 2014;7(1):29. doi: 10.1186/1756-8935-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.