Significance
Oncogene MYC is up-regulated in up to 75% of all cancers. Suppression of MYC expression or inhibition of MYC function by mutant protein has been shown efficacious in a variety of cancer models. Even though selective MYC-targeting agents have been long sought after, no drugs are available yet. Herein, we demonstrate that a pharmacological agent, stauprimide, selectively suppresses MYC transcription in a variety of cancer cells in vitro and inhibits tumor growth in xenograft mouse models. This study provides not only a proof-of-concept for pharmacologically targeting MYC transcription as an effective strategy for cancer treatment, but also evidence to support further development of stauprimide as an anticancer drug candidate.
Keywords: MYC, stauprimide, NME2, nuclear localization, cancer
Abstract
Stauprimide is a staurosporine analog that promotes embryonic stem cell (ESC) differentiation by inhibiting nuclear localization of the MYC transcription factor NME2, which in turn results in down-regulation of MYC transcription. Given the critical role the oncogene MYC plays in tumor initiation and maintenance, we explored the potential of stauprimide as an anticancer agent. Here we report that stauprimide suppresses MYC transcription in cancer cell lines derived from distinct tissues. Using renal cancer cells, we confirmed that stauprimide inhibits NME2 nuclear localization. Gene expression analysis also confirmed the selective down-regulation of MYC target genes by stauprimide. Consistent with this activity, administration of stauprimide inhibited tumor growth in rodent xenograft models. Our study provides a unique strategy for selectively targeting MYC transcription by pharmacological means as a potential treatment for MYC-dependent tumors.
The transcription factor MYC participates in diverse cellular processes by regulating the transcription of a large number of genes involved in gene expression, cell division, apoptosis, cell adhesion, stem cell self-renewal and differentiation, and metabolism (1–3). MYC is an oncogene whose expression is elevated in up to 75% of all cancers with various tissue origins (4); MYC expression levels also correlate with prognostic outcomes in patients of various types of malignancies (5, 6). In addition to its well-established roles in tumor initiation and cancer cell survival and proliferation, MYC has been shown to be involved in tumor microenvironment remodeling, drug resistance, and cancer stem cell maintenance (7–9). Recent studies have also revealed a role for MYC in regulating the transcription of immune checkpoint genes, including CD47 and PD-L1 in cancer cells, suggesting that MYC is involved in cancer escape from immune surveillance (10).
Overexpression of MYC is able to induce malignant transformation in skin (11), lung (12), liver (13), breast (14, 15), and hematopoietic cells (16) in transgenic mouse models, and termination of MYC overexpression results in halted cancer cell proliferation, increased apoptosis and terminal differentiation, and tumor regression. Furthermore, inhibition of endogenous MYC by the inducible expression of a dominant-negative variant of MYC, Omomyc, is able to induce tumor regression in transgenic mouse models of lung cancer (17). More intriguingly, transient deactivation of MYC overexpression (18) and episodic expression of Omomyc (19) have been shown to induce irreversible tumor regression in osteosarcoma and lung cancer transgenic mouse models, suggesting the potential of MYC-targeted anticancer therapies.
Drug discovery efforts have attempted to directly and indirectly modulate MYC-dependent transcription. For example, compounds have been identified that disrupt the interaction between MYC and its transcriptional partner MAX to suppress downstream gene transcription (20–24). In addition, molecules have been developed that stabilize the DNA G-quadruplex secondary structure in the MYC promoter region to down-regulate its own transcription (25, 26). More recently, bromodomain and extraterminal (BET) inhibitors have been shown to effectively suppress MYC transcription in various types of cancer cells and inhibit tumor growth in animal models (27), which has led to the development of drug candidates currently in clinical trials. However, the selectivity of these epigenetic-based approaches for targeting MYC transcription is not yet fully established; indeed, in some cancers, the anticancer activity of BET inhibitors has been attributed to their inhibition of genes other than MYC (28–30).
In previous studies, we identified a small molecule, stauprimide, which promotes embryonic stem cell (ESC) differentiation by suppressing MYC transcription (31) through its effect on the nuclear localization of the transcription factor NME2. These data prompted us to investigate the potential of stauprimide as an anticancer agent. Herein, we show that stauprimide suppresses MYC transcription in cancer cells from different tissue origins in vitro, and inhibits tumor growth in xenograft mouse models using renal cancer cells. mRNA sequence-based (mRNA-seq) global gene expression and gene set enrichment analyses (GSEA) indicated that the effects of stauprimide on MYC transcription were direct and selective. Our study provides a unique approach for targeting NME2-mediated MYC transcription as an effective anticancer strategy.
Results
Stauprimide Suppresses MYC Transcription in Various Cancer Cell Lines of Different Tissue Origins.
To assess its effect on MYC transcription in malignant cells, we tested stauprimide in a panel of cancer cell lines representing different tissue origins (including breast cancer, melanoma, renal carcinoma, leukemia, prostate cancer, hepatoma, colorectal cancer, lung cancer, and pancreatic cancer). After 6 h, total RNA was isolated and MYC transcript levels were analyzed using quantitative RT-PCR (qRT-PCR). Consistent with its effects in ESCs (31), stauprimide suppresses MYC transcription in the majority of cell lines tested with EC50s ranging from 30 nM to 8 μM, and decreased MYC levels between 15% to over 90% at 10 μM (Table S1). The different responses to stauprimide in different cells may be attributable to the diversity of signaling cascades, transcriptional programs, and epigenetic modifications regulating MYC promotor activities in different cellular contexts (32). Furthermore, stauprimide suppresses MYC transcription in murine cancer cells, including the melanoma cell line B16 and lung cancer cell line MLE12 at concentrations and magnitudes comparable to those in their human counterparts, suggesting relatively conserved MYC transcription regulation across species (33, 34).
Table S1.
MYC transcription suppression by stauprimide in various cancer cell lines
| Cancer type | Cell line | EC50, nm | Maximum Myc suppression, % |
| Breast cancer | MCF7 | 337 | 40 |
| MDA-MB-231 | 1,177 | 86 | |
| MDA-MB-468 | 1,326 | 36 | |
| Lung cancer | A549 | 29 | 40 |
| H441 | 1,112 | 17 | |
| H358 | 1,455 | 72 | |
| Melanoma | A375 | 716 | 78 |
| A2058 | 1,116 | 31 | |
| LOX-IMVI | 280 | 59 | |
| MDA-MB-435 | 39 | 35 | |
| MEWO | 119 | 33 | |
| MALME-3M | 2,691 | 38 | |
| Osteosarcoma | SK-ES-1 | 1,858 | 32 |
| SJSA-1 | 1,978 | 100 | |
| 143B | 177 | 100 | |
| G292 | 2,884 | 65 | |
| HOS | 563 | 95 | |
| U2OS | 916 | 69 | |
| SAOS | 3,038 | 63 | |
| Renal cancer | RXF393 | 609 | 100 |
| SN12C | 527 | 60 | |
| ACHN | 5,000 | 80 | |
| UO31 | 1,272 | 97 | |
| 786-0 | 3,252 | 70 | |
| A498 | 270 | 73 | |
| CAKI-1 | 962 | 100 | |
| TK10 | 426 | 100 | |
| Hematopoietic | KG1A | 399 | 89 |
| RAMOS | — | — | |
| CA46 | — | — | |
| MEG01 | 634 | 70 | |
| MOLT4 | 715 | 65 | |
| Prostate | PC3 | 1,055 | 41 |
| DU145 | 364 | 15 | |
| Colon cancer | Colo 205 | 8,000 | 70 |
| HT29 | 1,070 | 53 | |
| Ovary cancer | OVCAR3 | 130 | 17 |
| OVCAR4 | 274 | 47 | |
| OVCAR5 | 762 | 83 | |
| SKOV3 | 1,378 | 100 | |
| Murine cancer | B16 | 3,366 | 82 |
| MLE12 | 1,993 | 96 |
Cells were treated with stauprimide at various concentrations for 6 h and mRNA was extracted, relative MYC mRNA levels were quantified by qRT-PCR using either GAPDH or β-actin for normalization.
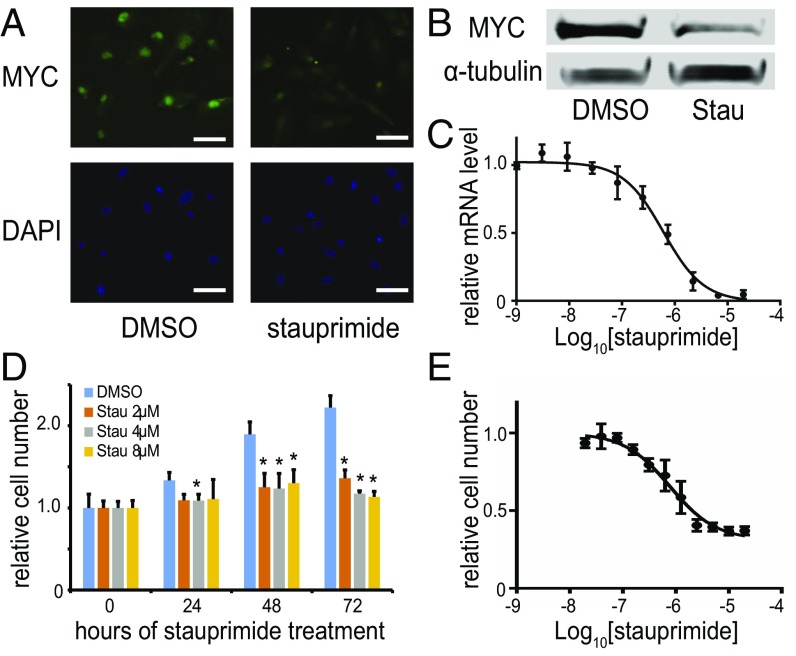
To further investigate the effects of stauprimide on MYC transcription in cell culture and in vivo, we chose to focus on the renal cancer cell line RXF 393 cells, based on the potency of stauprimide and near complete MYC transcription suppression in this cell line. Stauprimide treatment decreased MYC protein levels by more than 60% in RXF 393 cells as shown by immunofluorescent staining and Western blotting (Fig. 1 A and B). Moreover, when stauprimide was included in the cell culture media for 24–72 h, cell proliferation was almost completely inhibited by stauprimide at concentrations of 2–8 μM (Fig. 1D). The IC50 for inhibition of cell proliferation (780 ± 160 nM; Fig. 1E) was comparable to the IC50 of MYC transcription suppression (610 ± 90 nM; Fig. 1C). Similar effects on cell proliferation by stauprimide were observed in other cell lines, including CAKI-1 and TK-10 cells (renal cancer), as well as KG1A cells (leukemia). These results are in agreement with the well-established role of MYC in cell cycle progression.
Fig. 1.
Stauprimide suppresses MYC transcription in cancer cells. (A) Immunostaining of MYC in RXF 393 cells treated by stauprimide (5 μM) or DMSO for 24 h; nuclei were stained with DAPI. (Scale bars: 50 μm.) (B) Western blot of RXF 393 cell lysate for MYC upon stauprimide (5 μM) treatment for 24 h; α-tubulin was used as a loading control. (C) qRT-PCR for MYC mRNA in RXF 393 cells treated with stauprimide for 6 h in dose–response format. (D) Proliferation of RXF 393 cells upon stauprimide treatment at indicated concentrations assessed at various time points. Asterisk indicates statistically significant difference compared with DMSO-treated controls, P < 0.05. (E) Dose–response of RXF 393 cell proliferation to stauprimide treatment measured at 72 h. All data are presented as mean ± SD, n = 3.
Stauprimide Suppresses MYC Transcription by Inhibiting NME2 Nuclear Translocation.
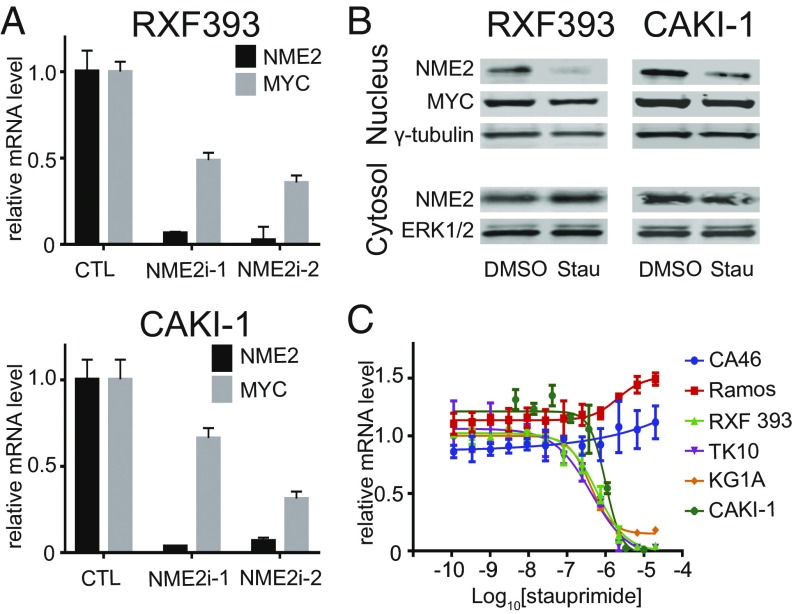
In previous studies, we demonstrated that stauprimide binds to NME2 and inhibits its nuclear localization, which leads to the decreased MYC promoter occupancy by NME2 (31). To confirm that this same mechanism operates in cancer cells, we studied the role of NME2 in regulating MYC transcription in RXF 393 and CAKI-1 cells. When NME2 was knocked down by NME2-targeting siRNAs, MYC transcription was suppressed to levels comparable to stauprimide treatment (Fig. 2A). This result is consistent with previous findings that NME2 is a MYC transcription factor. Next, we assessed the effect of stauprimide on NME2. As seen in ESCs, nuclear localized NME2 significantly decreased upon stauprimide treatment in both RXF 393 and CAKI-1 cells as early as at 3-h time point as shown by cell nuclear fractionation followed by Western blotting (Fig. 2B). Because the vast majority of NME2 is localized in the cytoplasm, the attempt to use immunostaining to quantify the altered NME2 nuclear localization by stauprimide treatment was unsuccessful. Furthermore, stauprimide did not affect the overall mRNA or protein levels of NME2, which is in line with the notion that stauprimide binds to and regulates NME2 subcellular distribution.
Fig. 2.
NME2-mediated MYC transcription is inhibited by stauprimide in cancer cells. (A) qRT-PCR analysis of NME2 and MYC mRNA levels in RXF 393 and CAKI-1 renal cancer cells following transfection of NME2 targeting siRNAs in the cells. (B) Western blotting of indicated proteins for nuclear and cytosolic fractions of RXF 393 and CAKI-1 cell lysates upon stauprimide treatment (5 μM) for 3 h. (C) qRT-PCR for MYC mRNA in indicated cell lines treated with stauprimide for 6 h in dose–response format.
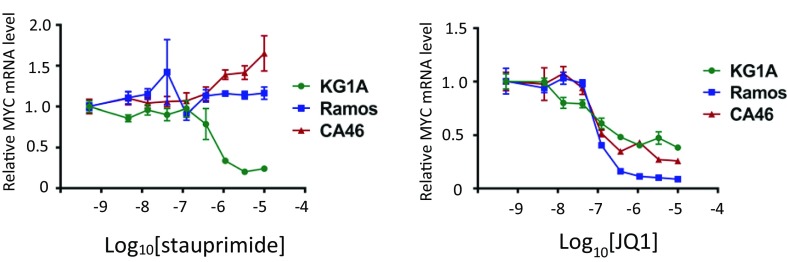
NME2 regulates MYC transcription by binding to the nuclease hypersensitive element III (NHE III) region of the MYC promoter close to the transcription start site (35–37). Given the mode of action of stauprimide, we hypothesized that cancer cells with different genomic regulatory sequences around the NHE III region may respond differently to stauprimide. To explore this notion, we studied the effects of stauprimide on three hematopoietic cancer cell lines with differing NHE III regions: a leukemia cell line KG1A with the wild-type genomic arrangement of MYC promoter; a Burkitt’s lymphoma cell line CA46 with a chromosomal translocation involving chromosome 8 and 14 that results in an insertion of Ig promoter between NHE III and transcription start site in the MYC promoter; and another Burkitt’s lymphoma cell line RAMOS RA1 with a chromosomal translocation in the MYC promoter upstream of the NHE III region. As seen in other cancer cells with the wild-type MYC promoter, stauprimide suppresses MYC transcription in KG1A cells efficiently with an EC50 of 400 ± 50 nM and a maximum suppression of ∼90%; in contrast, CA46 cells were resistant to stauprimide treatment (Fig. 2C), which supports our hypothesis that the regulation of MYC transcription by NME2 depends on the involvement of the NHE III sequence.
Unexpectedly, RAMOS RA1 cells also showed resistance to stauprimide treatment, even though the NHE III region is conserved in these cells. The NHE III region has high guanine content that may form secondary DNA structures (a G-quadruplex) that negatively affects the binding of the RNA polymerase complex to the MYC promoter. NME2 recognizes the G-quadruplex and releases the negative regulatory effect of the latter. We speculate that in RAMOS RA1 cells, even though the NHE III is conserved, the formation of G-quadruplex structure may be affected by upstream genomic sequences that make MYC transcription less sensitive to NME2 regulation. Moreover, the formation of a G-quadruplex and its recognition by NME2 may depend on the epigenetic status of the cell. The insertion of a transcriptionally active Ig promoter may affect the epigenetic status of the MYC locus and the arrangement of nucleosome, which renders these cells resistant to stauprimide. Indeed, a recent study by Yadav et al. (38) demonstrated a relationship between NME2 occupancy and nucleosome positioning in lung cancer cells. Distinct from stauprimide, the bromodomain inhibitor JQ1 suppressed MYC transcription in all three cell lines (Fig. S1), which is consistent with its mode of action of targeting epigenetically highly active transcriptional programs regardless of the different promoter arrangements in these cancer cell lines. The difference in MYC suppressing activities between stauprimide and JQ1 implies distinct modes of action between the two compounds, and suggests that stauprimide selectively suppresses MYC transcription driven by the wild-type promoter.
Fig. S1.
Dose–response of MYC mRNA levels upon stauprimide and JQ1 treatment in CA46, RAMOS, and KG1A cells. The cells were treated with compounds at indicated concentrations for 6 h. mRNA was extracted and analyzed by qRT-PCR for MYC; GAPDH was used as control for normalization. Data are presented as mean ± SD.
Stauprimide Selectively Suppresses MYC and Its Target Gene Transcription.
We assessed the effects of stauprimide on global gene expression in RXF 393 renal cancer cells. mRNA was collected from cells treated with either DMSO or stauprimide at 5 μM for 6, 12, and 24 h, and mRNA-seq was performed to quantify the levels of all transcripts in each sample. At as early as 6 h, MYC mRNA levels were significantly decreased in the presence of stauprimide. Other potential NME2 target genes, including CTGF, FRMD6, and MITF, which have previously been shown to be regulated by NME2 (39), also made the list of top 50 genes down-regulated by stauprimide (Table S2), which supports the notion that NME2 transcriptional programs are directly impacted by stauprimide treatment. At 12 and 24 h, the number of genes whose mRNA levels were affected by stauprimide treatment and the magnitude of these changes both increased, compared with those at 6-h time point (Fig. 3A).
Table S2.
mRNA-seq expression analysis top 50 down-regulated genes at 6 h post stauprimide treatment
| Rank | Gene symbol | Normalized abundance, log2 | Differential expression, log2 | |
| DMSO | Stauprimide | |||
| 1 | CTGF | 11.40 | 8.25 | −3.15 |
| 2 | ANKRD1 | 9.51 | 6.87 | −2.64 |
| 3 | CYR61 | 13.33 | 11.00 | −2.34 |
| 4 | EDN1 | 9.58 | 7.58 | −2.00 |
| 5 | THBS1 | 13.25 | 11.41 | −1.85 |
| 6 | SGK1 | 9.83 | 8.16 | −1.67 |
| 7 | PLK2 | 11.93 | 10.26 | −1.67 |
| 8 | TGFB2 | 9.66 | 8.04 | −1.62 |
| 9 | FLG | 9.88 | 8.59 | −1.29 |
| 10 | F3 | 12.98 | 11.74 | −1.24 |
| 11 | ADAMTS1 | 12.12 | 10.91 | −1.21 |
| 12 | NEXN | 10.07 | 8.91 | −1.16 |
| 13 | GDF6 | 6.46 | 5.31 | −1.15 |
| 14 | ITGB8 | 11.10 | 9.95 | −1.15 |
| 15 | TRPC4 | 7.05 | 5.96 | −1.10 |
| 16 | BDNF | 8.06 | 7.00 | −1.06 |
| 17 | CITED2 | 11.08 | 10.02 | −1.06 |
| 18 | LMCD1 | 9.39 | 8.35 | −1.04 |
| 19 | FILIP1L | 8.76 | 7.73 | −1.03 |
| 20 | FRMD6 | 11.28 | 10.25 | −1.03 |
| 21 | LBH | 11.02 | 9.99 | −1.03 |
| 22 | DLC1 | 9.86 | 8.84 | −1.02 |
| 23 | AMOTL2 | 11.63 | 10.62 | −1.01 |
| 24 | MAFF | 9.27 | 8.26 | −1.01 |
| 25 | HBEGF | 9.21 | 8.22 | −0.99 |
| 26 | AMIGO2 | 12.63 | 11.65 | −0.99 |
| 27 | ADAMTS5 | 9.05 | 8.08 | −0.97 |
| 28 | ENC1 | 10.64 | 9.69 | −0.95 |
| 28 | ARHGAP29 | 11.76 | 10.82 | −0.94 |
| 30 | CD274 | 8.92 | 8.01 | −0.91 |
| 31 | DDAH1 | 10.61 | 9.71 | −0.90 |
| 32 | SERTAD4 | 7.17 | 6.28 | −0.89 |
| 33 | IFFO2 | 10.10 | 9.22 | −0.87 |
| 34 | TUFT1 | 10.05 | 9.18 | −0.87 |
| 35 | HEG1 | 10.81 | 9.94 | −0.87 |
| 36 | DUSP8 | 7.17 | 6.31 | −0.86 |
| 37 | PHLDB2 | 10.43 | 9.56 | −0.86 |
| 38 | STARD13 | 9.34 | 8.49 | −0.85 |
| 39 | GPRC5A | 8.62 | 7.76 | −0.85 |
| 40 | MYC | 11.39 | 10.55 | −0.84 |
| 41 | ATP8B1 | 9.11 | 8.27 | −0.83 |
| 42 | ARID5B | 8.07 | 7.23 | −0.83 |
| 43 | DUSP1 | 11.15 | 10.32 | −0.83 |
| 44 | MITF | 6.90 | 6.07 | −0.83 |
| 45 | INHBB | 11.13 | 10.32 | −0.81 |
| 46 | RBM24 | 5.79 | 4.98 | −0.81 |
| 47 | MB21D2 | 7.85 | 7.05 | −0.80 |
| 48 | MBNL2 | 9.34 | 8.55 | −0.79 |
| 49 | PDCD1LG2 | 7.51 | 6.72 | −0.79 |
| 50 | DCDC2 | 9.40 | 8.62 | −0.78 |
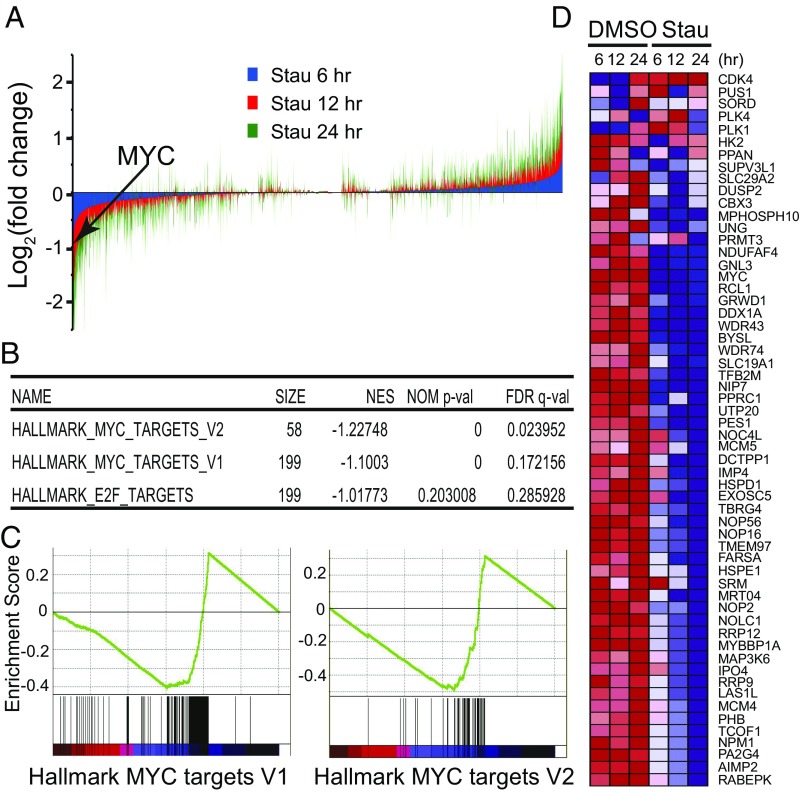
Fig. 3.
Stauprimide selectively suppresses MYC target gene transcription. (A) Genome-wide gene expression analysis by mRNA-seq upon stauprimide treatment (5 μM) in RXF 393 cells at 6, 12, and 24 h time points. The expression levels of each gene were normalized to the total mRNA abundance of each sample and compared with that of DMSO-treated controls. (B) The list of the top gene sets that are enriched over the time course of stauprimide treatment. GSEA analysis was performed using HALLMARK gene set database. (C) Enrichment plot of genes in HALLMARK_MYC_TARGETS_V1 and HALLMARK-MYC_TARGETS_V2 gene sets during the time course of stauprimide treatment. (D) Heat map of the mRNA levels of the genes listed in the HALLMARK_MYC_TARGETS_V2 gene set during the time course of stauprimide treatment.
Next, we carried out a GSEA (40) using the mRNA-seq expression data from the time course of stauprimide treatment. When analyzed using the hallmark gene sets that cover well-defined biological states and processes in the Molecular Signature Database (Broad Institute), the gene expression data rendered only two gene sets among a total of 50 gene sets with statistically significant enrichment between stauprimide and DMSO treatments: HALLMARK_MYC_TARGETS_V1 and HALLMARK_MYC_TARGETS_V2 [false discovery rate (FDR) q value threshold <0.25, nominal p value threshold <0.01], which consist of a host of well-characterized genes whose transcription is directly regulated by the transcription factor MYC (199 genes on the V1 list and 58 genes on the V2 list, respectively). Both gene sets were negatively correlated with the time course of stauprimide treatment (decreased gene expression with increased treatment duration), with a normalized enrichment score of −1.227 and −1.100, respectively (Fig. 3B). Furthermore, the majority (∼80%) of the members of both MYC target gene lists (166 of 199 genes of HALLMARK_MYC_TARGETS_V1 and 46 of 58 genes of HALLMARK_MYC_TARGETS_V2, respectively) were recognized as contributing to the enrichment score (Fig. 3C). The heat maps exhibited a consistent, time-dependent down-regulation of the majority of MYC target genes in both gene sets (Fig. 3D and Fig. S2). Interestingly, in contrast to the time-dependent trend of MYC target gene down-regulation, the magnitude of down-regulation of MYC’s own mRNA by stauprimide was rather unchanged over time (from 44% at 6 h to 39% at 12 h, and 38% at 24 h, respectively). These data suggest that the effects of stauprimide on MYC transcription are rapid and direct, whereas the suppression of MYC target genes, as a consequence of MYC down-regulation, becomes appreciable at later time points. The third gene set on the enrichment list following the two MYC target gene sets was HALLMARK_E2F_TARGETS; however, its enrichment did not meet the statistical significance threshold (FDR q value 0.286, nominal p value 0.203). Thus, the down-regulation of MYC and other NME2 target genes supports the notion that stauprimide inhibits the transcriptional activity of NME2, and the GSEA provides convincing evidence that inhibition of NME2 leads to a direct and selective suppression of MYC transcription.
Fig. S2.
Heat map of mRNA levels of genes in the HALLMARK_MYC_target_V1 gene set.
Stauprimide Inhibits Tumor Growth in Xenograft Mouse Models.
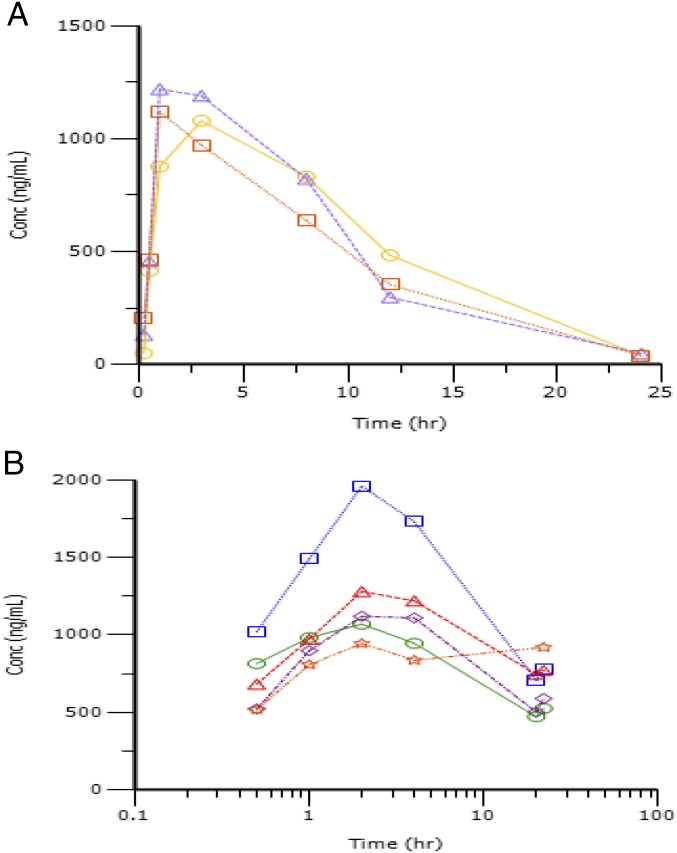
To assess the effects of stauprimide in vivo, we carried out pharmacokinetics (PK) and tolerability studies. Stauprimide exhibited favorable systemic exposure upon oral administration at 20 mg/kg with maximum plasma concentrations in a range of 1.85 to 2.09 μM, comparable to its in vitro cellular active concentrations, and a half-life of ∼4 h. Next, we explored the tolerability of stauprimide upon oral administration at 50 mg/kg once per day for 7 d. The regimen was well tolerated without any adverse effects observed on body weight, motor function, or plasma chemistry. In addition, we assessed the PK profile during the last dosing cycle. Stauprimide showed elevated plasma levels at all time points of sampling (Table S3 and Fig. S3), encouraging us to carry out subsequent efficacy studies.
Table S3.
Stauprimide pharmacokinetic profile
| Study | Initial PK | Tolerability |
| Dose, mg⋅kg | 20 | 50 |
| Administration | Single dose | Seven doses, SID |
| Cmax, ng⋅mL; nM | 1,140 ± 72.1; 1,951 ± 123 | 1,275 ± 359; 2,182 ± 614 |
| Tmax, h | 1.7 ± 1.15 | 2 ± 0 |
| T1/2, h | 3.86 ± 0.28 | 16.78 ± 4.73 |
| AUC0–24, h⋅ng⋅mL−1 | 12,050 ± 982 | 21,476 ± 4,087 |
| Cl, mL⋅min⋅kg−1 | 27.2 ± 2.24 | 22.83 ± 4.09 |
| Vd, L⋅kg | 9.11 ± 1.21 | 33.41 ± 8.2 |
| MRT, h | 9.09 ± 1.33 |
Fig. S3.
Plasma stauprimide levels upon oral administration in the initial PK study and the tolerability study. (A) Plasma stauprimide levels in the initial PK study. Stauprimide was administered as single dose at 20 mg/kg in 75% (vol/vol) PEG300; 25% (vol/vol) D5W. (B) Plasma stauprimide levels in the tolerability study. Samples were collected following the last dose in a 7-d, once-per-day dosing scheme. Stauprimide was orally administered at 50 mg/kg. Each line represents one animal in the dosing group.
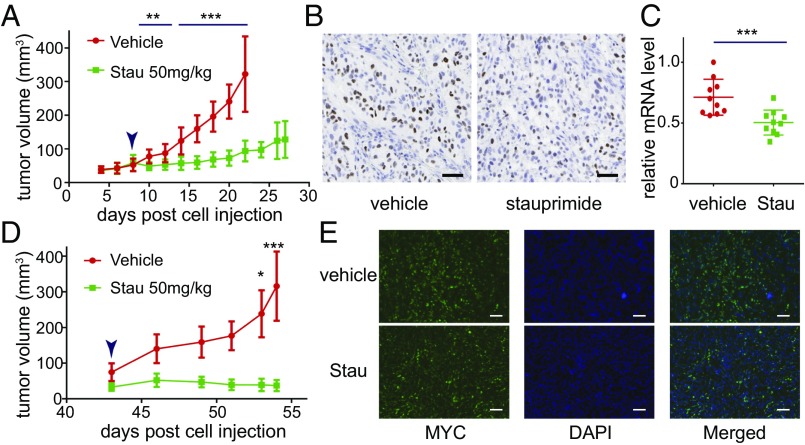
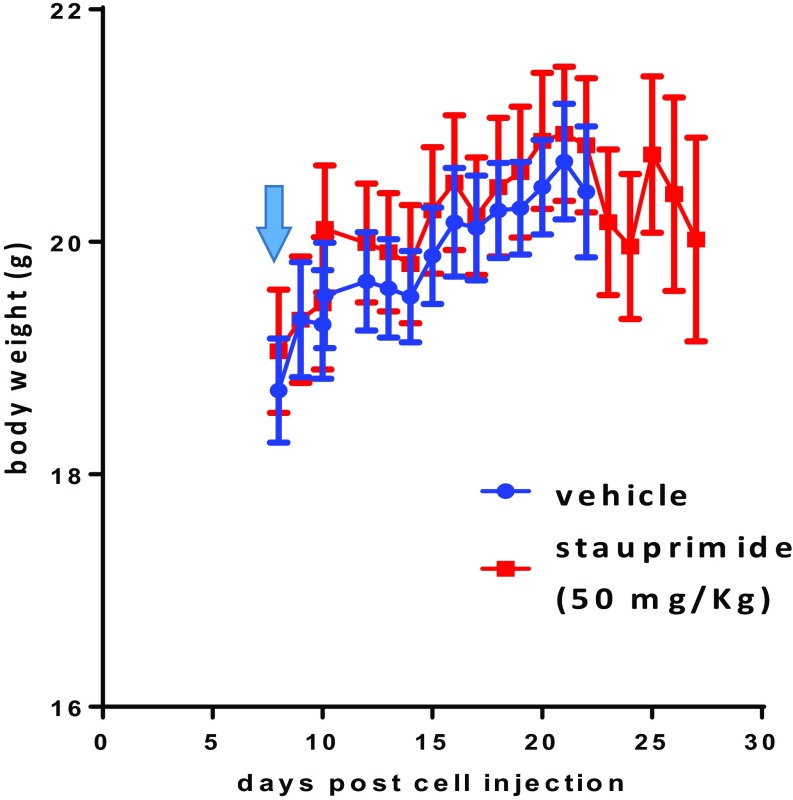
Based on the in vitro sensitivity of cancer cell lines to stauprimide, we chose renal cancer cell lines RXF 393 and CAKI-1 to carry out xenograft tumor models. The cancer cells (5 × 106 cells per injection) were injected into immunocompromised mice [nonobese diabetic/severe combined immunodeficiency (NOD/SCID)] subcutaneously. When tumors reached a size of 50–100 mm3, oral stauprimide administration was started once per day at 50 mg/kg. Stauprimide treatment almost completely blocked tumor growth in mice injected with either RXF 393 or CAKI-1 cells during the dosing periods (Fig. 4 A and D). No difference in body weight was observed between vehicle and stauprimide-treated groups (Fig. S4). Tumor samples were collected at the end of the studies and analyzed for stauprimide levels at 3, 10, and 24 h post last dose. Stauprimide was detectable at all time points with concentrations between 1.8 and 3.6 μM (Table S4; assuming tissue density is the same as water, 1.0 g/mL), higher than its in vitro EC50s in both cell lines (610 ± 90 nM in RXF 393 cells and 1,004 ± 142 nM in CAKI-1 cells, respectively). Due to the weak signal of NME2 immunostaining in the nucleus compared with that in the cytoplasm, we were unable to quantify the altered NME2 nuclear localization upon stauprimide treatment in tumor tissues. However, immunohistochemistry staining of MYC in the nuclei clearly showed that stauprimide reduced MYC protein levels in xenograft tumors (Fig. 4 B and E). In addition, qRT-PCR analysis confirmed the inhibition of MYC transcription in the RXF 393 tumor samples upon stauprimide treatment (Fig. 4C). These data confirm the effects of stauprimide on MYC transcription in vivo, consistent with its mode of action in the cellular assays.
Fig. 4.
Stauprimide inhibits tumor growth in xenograft tumor models. (A) Tumor volume measurement of RXF 393 cells injected s.c. into NOD/SCID mice and treated with stauprimide (50 mg/kg, once per day) or with vehicle [75% (vol/vol) PEG300; 25% (vol/vol) D5W], n = 10/group. (B) Immunohistochemistry staining of tumor samples using anti-MYC antibody at the end of the dosing period of A. (Scale bars: 100 μm.) (C) qRT-PCR analysis of human MYC mRNA in tumor samples at the end of the dosing period of A; mRNA abundance was normalized to human GAPDH. (D) Tumor volume measurement of CAKI-1 cells injected s.c. into NOD/SCID mice treated with stauprimide (50 mg/kg, once per day) or with vehicle [75% (vol/vol) PEG300; 25% (vol/vol) D5W], n = 5/group. (E) Fluorescent immunostaining of tumor samples using anti-MYC antibody at the end of dosing period of D. (Scale bars: 100 μm.) Arrowheads in A and D indicate the start of dosing. All data are presented as mean ± SD; statistical analyses were carried out using t test, *P < 0.05, **P < 0.01, ***P < 0.001.
Fig. S4.
Body weight of mice treated with stauprimide in the xenograft model with RXF 393 cell injection. Arrow indicates the time dosing was started. Data are presented as mean ± SD, n = 10.
Table S4.
Stauprimide exposure in xenograft tumor at the end of the in vivo efficacy study with RXF 393 cell injection
| Time post dosing, h | Concentration, nM |
| 3 | 3,557 ± 1,357 |
| 10 | 4,024 ± 884 |
| 24 | 1,807 ± 690 |
Discussion
MYC plays critical roles in almost all aspects of cancer biology, including cancer cell proliferation and survival, cancer stem cell self-renewal and differentiation, cancer cell interactions with extracellular matrix, and other tumor resident cells, including fibroblasts and immune cells, cancer cell drug resistance, and metastasis. The plurality of these activities makes MYC an attractive target for anticancer drug development; however, no approved MYC-targeting drugs are available to date. In the current study, we demonstrate that stauprimide selectively suppresses MYC transcription in a number of different cancer cell lines. The down-regulation of MYC by stauprimide leads to the inhibition of cell proliferation in vitro and halts tumor growth in rodent xenograft tumor models using renal cancer cells.
NME2 is a MYC transcription factor that binds the NHE III site of the MYC promoter and releases the negative regulatory effect by the G-quadruplex secondary DNA structure on MYC transcription. Stabilization of the G-quadruplex by small molecules has been shown effective in suppressing MYC transcription in vitro and in vivo. Previously, we demonstrated that stauprimide does not act as a broad spectrum kinase inhibitor like staurosporine, but rather binds to NME2 and blocks its nuclear localization in ESCs, which results in down-regulation of MYC transcription (31). This mechanism was confirmed in cancer cells by the suppression of MYC transcription upon NME2 knockdown by siRNAs and the blockade of nuclear localization of NME2 by stauprimide. Furthermore, MYC promoter-translocated cancer cells, including Burkitt’s lymphoma cell lines RAMOS RA1 and CA46, are resistant to stauprimide treatment, supporting the notion that NME2 regulates MYC transcription by recognizing the wild-type MYC promoter.
mRNA-seq–based global gene expression and gene set enrichment analyses provided further evidence that stauprimide directly and selectively inhibits NME2 transcriptional activity and subsequently suppresses MYC transcription. Stauprimide significantly reduced the transcript levels of a list of NME2 target genes, including MYC, CTGF, FRMD6, and MITF at a time point as early as 6 h; among all 50 hallmark gene sets representing a host of cellular processes, only MYC target gene sets showed statistically significant enrichment during the time course of stauprimide treatment. Moreover, stauprimide treatment affected the transcription of ∼80% of MYC target genes listed in both gene sets. Even though the regulation of MYC transcription by NME2 and its cognate G-quadruplex DNA secondary structure has been extensively studied, the mechanism by which NME2 shuffles between cytosol and nucleus, and how stauprimide interferes with this process, remains to be determined.
Stauprimide suppresses MYC transcription in a context-dependent manner: it acts with different EC50s and with different degrees of maximal MYC mRNA down-regulation in different cell lines. This observation is in line with previous findings that MYC is tightly regulated by upstream cell signaling and transcription programs depending on the particular cellular context (32). The cellular context, including genetic backgrounds (gene mutations, chromosomal translocations, and epigenetic status) and cellular signaling cascades may be responsible for the observed difference in sensitivity to stauprimide among difference cancer cells. In addition to its role in regulating cancer cell biology, MYC has recently been shown to up-regulate the expression of checkpoint genes, including PD-L1 and CD47, which may be responsible for cancer escape of immune surveillance.
Stauprimide suppresses MYC transcription in both human and murine cancer cells, which allows for further investigation of its cellular and therapeutic effects using additional, physiologically more relevant cancer models, especially transgenic mouse models with intact immune systems to explore the role MYC plays in cancer immune escape and the therapeutic potential of targeting MYC as a strategy to potentiate the endogenous immune response. In conclusion, the data presented in this study provide a proof-of-concept for selectively targeting NME2-mediated MYC transcription as a promising strategy of anticancer therapy, and support the further development of stauprimide through medicinal chemistry efforts to afford for more potent analogs as potential drug candidates for clinical applications.
Materials and Methods
Cell Culture.
Cancer cell lines were obtained either from the National Cancer Institute (NCI) or ATCC, cultured in RPMI medium containing 10% (vol/vol) FBS at 37 °C with 5% CO2. Cells were treated with vehicle (DMSO at 0.1%) or test compounds at the indicated concentrations and time periods and processed for downstream analysis.
Stauprimide Synthesis.
Stauprimide was synthesized following previously established procedures and the purity was >95% (31). In some of the experiments, stauprimide was purchased from LC Laboratories.
RNA Extraction and qRT-PCR.
RNA was extracted from cells using the RNeasy Kit (Qiagen) or TRIzol reagent (Life Technologies), and cDNA synthesized using the First Strand Reverse Transcription Kit (Life Technology) following manufacturers’ recommendations. Gene-specific TaqMan assays were purchased from Life Technologies; the relative quantities of the genes of interest were analyzed using the ΔΔCT relative quantification method following manufacturer’s recommendations. Either GAPDH or β-actin was used as internal control.
Cell Proliferation Assay.
Cells were plated in 384-well solid-bottom white microtiter plates at ∼50% confluency and let to attach overnight; test compounds were added to the culture media at the indicated concentrations. Relative cell numbers were determined using CellTiter Glo assay (detecting cellular ATP content; Promega) every 24 h.
Nuclear Fractionation and Western Blotting.
Nuclear and cytosolic content fractionation was performed following the protocol reported in previous studies (31, 41). Fractions were subjected to SDS/PAGE electrophoresis and Western blotting using primary antibodies recognizing target proteins, including NME2 (Abcam), MYC (Cell Signaling), ERK1/2 (Cell Signaling), α- and γ-tubulin (Sigma), and fluorophore-conjugated secondary antibodies (LI-COR) and scanned using an Odyssey CLx imager.
Fluorescent Immunostaining.
At the end of each experiment, cells were fixed with 10% (vol/vol) formalin solution at room temperature for 10 min, permeabilized with 0.5% Triton X-100 (Sigma) in PBS solution for 5 min, and then blocked with PBS containing 0.1% Triton X-100, 10% (vol/vol) horse serum, and 1% BSA at room temperature for 1 h. The cells were incubated with primary antibodies in PBS solution containing 0.1% Triton X-100 and 0.1% BSA at 4 °C overnight and then rinsed with PBS three times and incubated with fluorescently labeled secondary antibodies in PBS solution containing 0.1% Triton X-100, 0.1% BSA, and DAPI at room temperature for 1 h. The cells were rinsed with PBS containing 0.1% Triton X-100 at least three times and subjected to microscopic analysis.
siRNA Gene Silencing.
ON-TARGETplus siRNAs targeting NME2 and siGLO nontargeting control siRNA were purchased from Dharmacon. The cells were transfected with individual siRNAs using Lipofectamine 2000 (Life Technologies) following the manufacturer’s recommendation. The sequences of siRNA that effectively down-regulated NME2 are NME2i-1, GCGAGAUCAUCAAGCGCUU (catalog no. J-005102-07); and NME2i-2, CUGAAGAACACCUGAAGCA (catalog no. J-005102-10). Gene knockdown efficiency was assessed by qRT-PCR using mRNAs collected 24 and 48 h posttransfection.
High-Throughput Whole-Transcriptome Sequencing (mRNA-seq) and Gene Set Enrichment Analysis.
Total RNA was isolated using the RNeasy Kit (Qiagen) and digested with TURBO DNase (Life Technologies). Qubit (Invitrogen) was used to determine RNA concentration, and Agilent Tape Station was used to determine RNA integrity numbers before library preparation. mRNA-seq libraries were prepared using the TruSeq Stranded mRNA Library Prep Kit following the manufacturer’s instructions (Illumina). Briefly, RNA with poly-A tail was isolated using magnetic beads conjugated to poly-T oligos. mRNA was then fragmented and reverse-transcribed into cDNA. dUTPs were incorporated, followed by second-strand cDNA synthesis. The dUTP containing second strand was not amplified. cDNA was then end repaired, index adapter ligated, and PCR amplified. AMPure XP beads (Beckman Coulter) were used to purify nucleic acid after each step of the library preparation. All sequencing libraries were then quantified, pooled, and sequenced at single-end 50 bp using the Illumina HiSeq 2500 at the Salk Next Generation Sequencing (NGS) Core. Raw sequencing data were demultiplexed and converted into FASTQ files using CASAVA (v1.8.2). Libraries were sequenced at an average depth of 30 million reads per library. The relative abundances of the transcripts in each sample were used to perform GSEA (Broad Institute) in the time-course format. The analysis parameters were selected following the software instruction at the GSEA website.
Xenograft Tumor Mouse Model.
Animals for tumor engraftment studies were obtained from Jackson Laboratories and maintained in a 12-h light-cycle room with chow and water given ad libitum. The cancer cells (5 × 106 RXF 393 or CAKI-1 cells per injection) were suspended in 100 μL PBS and injected s.c. into the right flanks of 8- to 10-wk-old female NOD/SCID mice. Tumor growth was monitored externally up to three times per week with a caliper to measure length and width. Tumor volumes were calculated by L⋅W⋅W/2 according to NCI standards. The compound or vehicle [75% (vol/vol) PEG300; 25% (vol/vol) D5W] was administered orally at 10 mL/kg once per day starting at the indicated time following cell inoculation. At the end of the studies, tumors were harvested and processed for immunohistochemistry staining or RNA extraction and qRT-PCR analysis. All protocols were approved by the Institutional Animal Care and Use Committee at Calibr.
Statistical Analyses.
Statistical analyses were performed using Microsoft Excel. Student’s t test (two-tailed) was performed to determine significance when comparing data from different treatment groups. P values were calculated, and P < 0.05 was considered to represent significant difference.
Acknowledgments
We thank Drs. Heiko Wurdak, Luke Lairson, and Michael Bollong for constructive discussions and proofreading of the manuscript. The RNA-seq analysis was supported by the NGS Core Facility of the Salk Institute with funding from NIH-NCI Cancer Center Support Grants: P30 014195, the Chapman Foundation, and the Helmsley Charitable Trust.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702663114/-/DCSupplemental.
References
- 1.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soucek L, Evan GI. The ups and downs of Myc biology. Curr Opin Genet Dev. 2010;20(1):91–95. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez PC, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17(9):1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dang CV, O’donnell KA, Juopperi T. The great MYC escape in tumorigenesis. Cancer Cell. 2005;8(3):177–178. doi: 10.1016/j.ccr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Kim S, et al. MYC and BCL2 overexpression is associated with a higher class of Memorial Sloan-Kettering Cancer Center prognostic model and poor clinical outcome in primary diffuse large B-cell lymphoma of the central nervous system. BMC Cancer. 2016;16:363. doi: 10.1186/s12885-016-2397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kübler K, et al. c-Myc copy number gain is a powerful prognosticator of disease outcome in cervical dysplasia. Oncotarget. 2015;6(2):825–835. doi: 10.18632/oncotarget.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitfield JR, Soucek L. Tumor microenvironment: Becoming sick of Myc. Cell Mol Life Sci. 2012;69(6):931–934. doi: 10.1007/s00018-011-0860-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sodir NM, et al. Endogenous Myc maintains the tumor microenvironment. Genes Dev. 2011;25(9):907–916. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laurenti E, Wilson A, Trumpp A. Myc’s other life: Stem cells and beyond. Curr Opin Cell Biol. 2009;21(6):844–854. doi: 10.1016/j.ceb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Casey SC, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352(6282):227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: Induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3(5):565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 12.Ehrhardt A, et al. Development of pulmonary bronchiolo-alveolar adenocarcinomas in transgenic mice overexpressing murine c-myc and epidermal growth factor in alveolar type II pneumocytes. Br J Cancer. 2001;84(6):813–818. doi: 10.1054/bjoc.2000.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431(7012):1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 14.Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38(3):627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- 15.Leder A, Pattengale PK, Kuo A, Stewart TA, Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: Multiple neoplasms and normal development. Cell. 1986;45(4):485–495. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- 16.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4(2):199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 17.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455(7213):679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain M, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297(5578):102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 19.Soucek L, et al. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27(5):504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, et al. Disruption of Myc-Max heterodimerization with improved cell-penetrating analogs of the small molecule 10074-G5. Oncotarget. 2013;4(6):936–947. doi: 10.18632/oncotarget.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clausen DM, et al. In vitro cytotoxicity and in vivo efficacy, pharmacokinetics, and metabolism of 10074-G5, a novel small-molecule inhibitor of c-Myc/Max dimerization. J Pharmacol Exp Ther. 2010;335(3):715–727. doi: 10.1124/jpet.110.170555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong KC, et al. Intravesical instillation of c-MYC inhibitor KSI-3716 suppresses orthotopic bladder tumor growth. J Urol. 2014;191(2):510–518. doi: 10.1016/j.juro.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 23.Brown RV, Danford FL, Gokhale V, Hurley LH, Brooks TA. Demonstration that drug-targeted down-regulation of MYC in non-Hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J Biol Chem. 2011;286(47):41018–41027. doi: 10.1074/jbc.M111.274720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi J, Stover JS, Whitby LR, Vogt PK, Boger DL. Small molecule inhibitors of Myc/Max dimerization and Myc-induced cell transformation. Bioorg Med Chem Lett. 2009;19(21):6038–6041. doi: 10.1016/j.bmcl.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh S, Pradhan SK, Kar A, Chowdhury S, Dasgupta D. Molecular basis of recognition of quadruplexes human telomere and c-myc promoter by the putative anticancer agent sanguinarine. Biochim Biophys Acta. 2013;1830(8):4189–4201. doi: 10.1016/j.bbagen.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 26.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99(18):11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogg SJ, et al. BET-inhibition induces apoptosis in aggressive B-cell lymphoma via epigenetic regulation of BCL-2 family members. Mol Cancer Ther. 2016;15(9):2030–2041. doi: 10.1158/1535-7163.MCT-15-0924. [DOI] [PubMed] [Google Scholar]
- 29.da Motta LL, et al. The BET inhibitor JQ1 selectively impairs tumour response to hypoxia and downregulates CA9 and angiogenesis in triple negative breast cancer. Oncogene. 2017;36(1):122–132. doi: 10.1038/onc.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loganathan SN, et al. BET bromodomain inhibitors suppress EWS-FLI1-dependent transcription and the IGF1 autocrine mechanism in Ewing sarcoma. Oncotarget. 2016;7(28):43504–43517. doi: 10.18632/oncotarget.9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu S, et al. A small molecule primes embryonic stem cells for differentiation. Cell Stem Cell. 2009;4(5):416–426. doi: 10.1016/j.stem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Levens D. You don’t muck with MYC. Genes Cancer. 2010;1(6):547–554. doi: 10.1177/1947601910377492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnaud-Dabernat S, et al. Nm23-M2/NDP kinase B induces endogenous c-myc and nm23-M1/NDP kinase A overexpression in BAF3 cells. Both NDP kinases protect the cells from oxidative stress-induced death. Exp Cell Res. 2004;301(2):293–304. doi: 10.1016/j.yexcr.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 34.Reddoch JF, Miller DM. Inhibition of nuclear protein binding to two sites in the murine c-myc promoter by intermolecular triplex formation. Biochemistry. 1995;34(23):7659–7667. doi: 10.1021/bi00023a012. [DOI] [PubMed] [Google Scholar]
- 35.Dexheimer TS, et al. NM23-H2 may play an indirect role in transcriptional activation of c-myc gene expression but does not cleave the nuclease hypersensitive element III(1) Mol Cancer Ther. 2009;8(5):1363–1377. doi: 10.1158/1535-7163.MCT-08-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thakur RK, et al. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009;37(1):172–183. doi: 10.1093/nar/gkn919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang D, Hurley LH. Structure of the biologically relevant G-quadruplex in the c-MYC promoter. Nucleosides Nucleotides Nucleic Acids. 2006;25(8):951–968. doi: 10.1080/15257770600809913. [DOI] [PubMed] [Google Scholar]
- 38.Yadav VK, et al. Promoter-proximal transcription factor binding is transcriptionally active when coupled with nucleosome repositioning in immediate vicinity. Nucleic Acids Res. 2014;42(15):9602–9611. doi: 10.1093/nar/gku596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thakur RK, et al. Non-metastatic 2 (NME2)-mediated suppression of lung cancer metastasis involves transcriptional regulation of key cell adhesion factor vinculin. Nucleic Acids Res. 2014;42(18):11589–11600. doi: 10.1093/nar/gku860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson K, et al. A stem cell-based approach to cartilage repair. Science. 2012;336(6082):717–721. doi: 10.1126/science.1215157. [DOI] [PubMed] [Google Scholar]