Significance
Mosquitoes require a blood meal to reproduce and by blood feeding transmit some of the most dangerous human diseases. Female mosquitoes have extremely high metabolism, and the elucidation of regulatory pathways coordinating reproductive and metabolic events is essential. RNAi of the juvenile hormone receptor Methoprene-tolerant (Met) promoted triacylglycerol (TAG) catabolism and β-oxidation and diminished TAG levels, whereas ecdysone receptor (EcR) RNAi had an opposite effect. Hepatocyte nuclear factor 4 (HNF4) directly regulated TAG catabolism and β-oxidation genes, and its RNAi silencing phenocopied EcR RNAi. The expression of the HNF4 gene was downregulated by Met and activated by EcR and Target of rapamycin. Thus, HNF4 mediates hormonal and nutritional signaling of lipid metabolism regulation in reproducing female Aedes aegypti mosquitoes.
Keywords: juvenile hormone, 20-hydroxyecdysone, lipid, hepatocyte nuclear factor 4, beta-oxidation
Abstract
Mosquitoes transmit devastating human diseases because they need vertebrate blood for egg development. Metabolism in female mosquitoes is tightly coupled with blood meal-mediated reproduction, which requires an extremely high level of energy consumption. Functional analysis has shown that major genes encoding for enzymes involved in lipid metabolism (LM) in the mosquito fat bodies are down-regulated at the end of the juvenile hormone (JH)-controlled posteclosion (PE) phase but exhibit significant elevation in their transcript levels during the post-blood meal phase (PBM), which is regulated mainly by 20-hydroxyecdysone (20E). Reductions in the transcript levels of genes encoding triacylglycerol (TAG) catabolism and β-oxidation enzymes were observed to correlate with a dramatic accumulation of lipids in the PE phase; in contrast, these transcripts were elevated significantly and lipid stores were diminished during the PBM phase. The RNAi depletion of Methoprene-tolerant (Met) and ecdysone receptor (EcR), receptors for JH and 20E, respectively, reversed the LM gene expression and the levels of lipid stores and metabolites, demonstrating the critical roles of these hormones in LM regulation. Hepatocyte nuclear factor 4 (HNF4) RNAi-silenced mosquitoes exhibited down-regulation of the gene transcripts encoding TAG catabolism and β-oxidation enzymes and an inability to use lipids effectively, as manifested by TAG accumulation. The luciferase reporter assay showed direct regulation of LM-related genes by HNF4. Moreover, HNF4 gene expression was down-regulated by Met and activated by EcR and Target of rapamycin, providing a link between nutritional and hormonal regulation of LM in female mosquitoes.
Because of their unique biology as blood-feeding organisms, mosquitoes impose an enormous burden on human health and well-being, transmitting numerous devastating diseases (1–4). In female mosquitoes, every gonadotrophic cycle occurs in two phases. During the first, the posteclosion (PE) phase, the female mosquito’s development is controlled by a sesquiterpenoid juvenile hormone (JH), required for blood feeding and egg maturation. During this phase, female mosquitoes are actively involved in host seeking. The post blood meal (PBM) phase that commences with blood ingestion and is mainly regulated by an insect steroid hormone, 20-hydroxyecdysone (20E), results in rapid egg development supported by digestion of a large blood meal. These physiological functions of reproducing female mosquitoes require an unprecedented energy level supported by extremely high metabolism. Thus, a thorough understanding of metabolic processes at the molecular level will provide important insights into the reproductive biology of these disease vectors and may lead to the identification of previously unexplored targets for their control.
Lipid metabolism (LM) is essential for energy homeostasis of any organism. In mosquitoes, it is tightly coupled with a large array of physiological processes. It has been shown that diapausing mosquitoes, such as Culex pipiens and Aedes albopictus, use lipids as predominant energy stores (5, 6). LM plays an important role in mosquito immune responses to pathogens, such as Plasmodium and the Dengue virus (7–10). Moreover, lipids provide a potent source of energy for oocyte maturation. β-Oxidation is essential for mouse oocyte developmental competence (11), and pharmacological inhibition of this process impairs oocyte maturation and embryo development (12). In the mosquito Culex quinquefasciatus, ∼90% of the energy used by the developing embryo originates from lipids (13); in Aedes aegypti, 80% of lipids found in eggs of the first reproductive cycle originate from sugars taken up before a blood meal (14). However, although tightly regulated LM is crucial for maintaining normal functions in reproducing female mosquitoes, little is known about the molecular mechanisms that regulate mosquito LM during the reproductive cycle.
Our previous studies showed that carbohydrate metabolism plays an important role in supporting the drastic physiological changes in reproducing A. aegypti female mosquitoes (15). Moreover, they implicated JH and 20E in controlling carbohydrate metabolic pathways during the PE and PBM phases, respectively (15). Here we show that these hormones and their respective receptors, Methoprene-tolerant (Met) and Ecdysone receptor (EcR), play key roles in regulating mosquito LM. Our study also has linked the nuclear receptor (NR) Hepatocyte nuclear factor 4 (HNF4) with the control of triacylglycerol (TAG) catabolism and β-oxidation pathways in this mosquito. Furthermore, it has revealed that the expression of the HNF4 gene is down-regulated by JH/Met and is activated by 20E/EcR and Target of rapamycin (TOR), indicating the pivotal role of this NR in mediating hormonal and nutritional signaling of LM regulation in female A. aegypti mosquitoes.
Results
Lipid Metabolism in Female A. aegypti Mosquitoes Throughout the Reproductive Cycle.
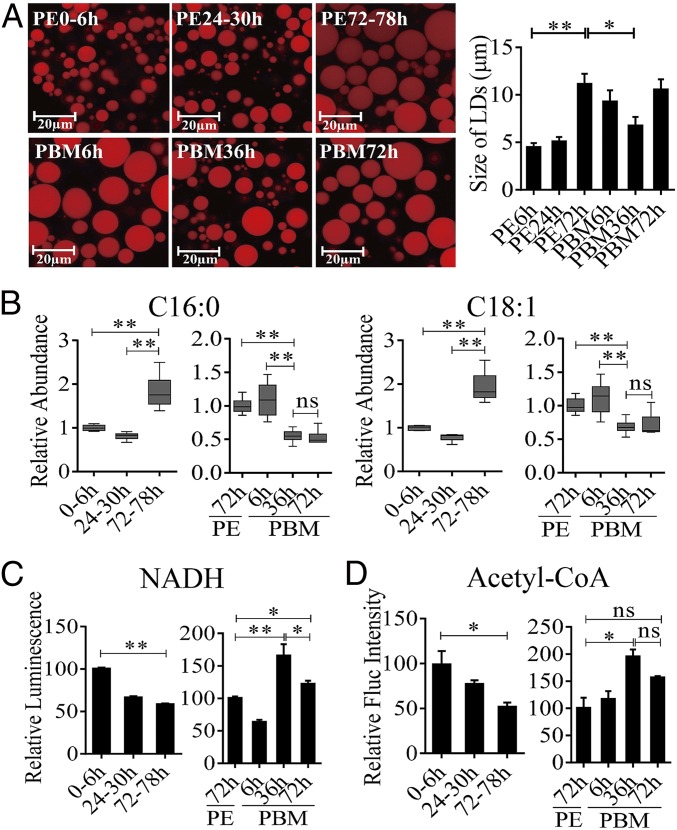
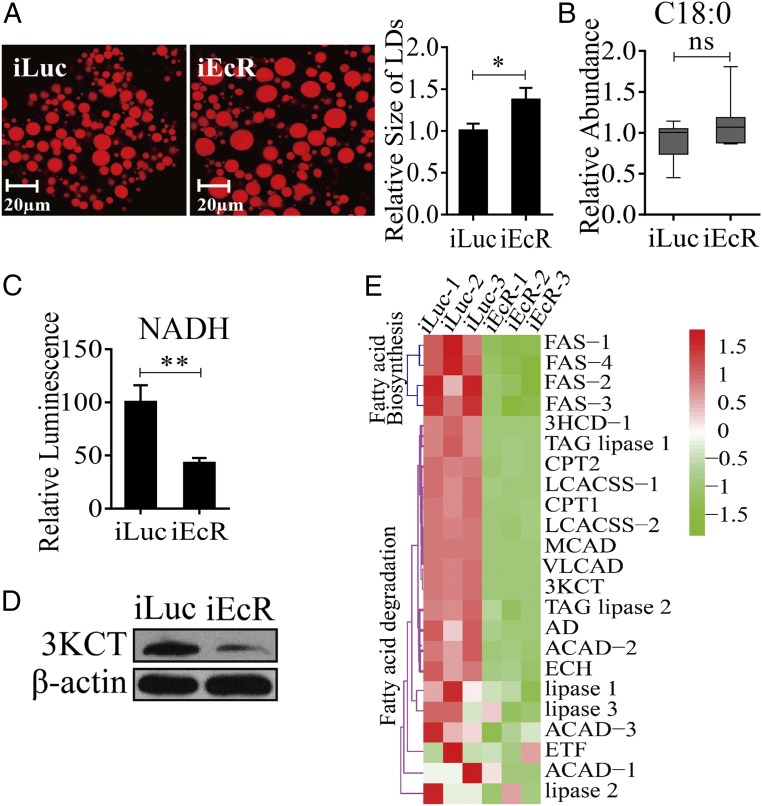
In organisms as different as flies and humans, body fat reserves are stored primarily as TAGs in lipid droplets (LDs), predominantly found intracellularly within specialized storage tissues such as the insect fat body (FB) or mammalian adipose tissue (16, 17). Our previous study has shown that in female A. aegypti mosquitoes, levels of TAGs exhibit biphasic fluctuations, being elevated by the end of the PE phase (72 h PE) and reduced after a blood feeding (15). Because TAGs are stored predominantly in LDs of the FB, we performed their staining using Nile Red and evaluated the FB lipid content by measuring LD abundance and relative size in this tissue. Both LD size and density increased dramatically at the late PE phase (72 h PE), indicating a shift in lipid homeostasis toward storage of energy reserves (Fig. 1A). At 6 h PBM, these LD parameters were moderately reduced, likely through the activation of lipases in the FB by blood meal-induced signals. LDs significantly diminished by 36 h PBM, suggesting an FB TAG mobilization to support oocyte development. By 72 h PBM, after the cessation of vitellogenesis and formation of the first batch of eggs, the size of FB LDs increased again (Fig. 1A).
Fig. 1.
LM changes in the FBs of A. aegypti females throughout the reproductive cycle. (A) LDs in the FBs of A. aegypti during the PE and PBM phases were detected using Nile red staining. (B) GC-MS was performed to analyze the relative levels of saturated (C16:0) and unsaturated (C18:1) FFAs throughout the reproductive cycle of mosquitoes. The relative abundances at PE and PBM time points were normalized to 0–6 h PE and 72 h PE, respectively. At least six independently collected samples with 15 mosquito FBs for each time point were analyzed. (C and D) The relative luminescence of NADH (C) and the fluorescence (Fluc) of acetyl-CoA (D) were measured during the PE and PBM phases. The relative abundance at PE and PBM time points was normalized to 0–6 h PE and 72 h PE, respectively. Amounts of NADH and acetyl-CoA were normalized to total protein levels. At least three independently collected samples with six mosquitoes for each time point were analyzed. Error bars represent the SEM; *P < 0.05, **P < 0.01.
To evaluate further the LM dynamics in female mosquitoes during the reproductive cycle, quantities of upstream metabolites, such as free fatty acids (FFAs), were determined by means of GC-MS. Relative quantities of both saturated and unsaturated FFAs were lower at 6 h and 24 h PE than at 72 h PE (Fig. 1B and Fig. S1). A blood meal triggers a significant increase in their levels at 6 h PBM relative to that at 72 h PE. By 36 h PBM, however, these levels decrease and remain stable until 72 h PBM (Fig. 1B and Fig. S1). We also evaluated changes in the β-oxidation status of these mosquitoes by measuring relative amounts of two key β-oxidation metabolites, NADH and acetyl-CoA (the end-product of β-oxidation). Both were higher at 6 h PE than at 72 h PE, suggesting a reduction in β-oxidation activity by the end of the PE phase. In contrast, blood feeding resulted in a significant increase in the amounts of these two β-oxidation metabolites by 36 h PBM (Fig. 1 C and D). Taken together, these results show that LM in female mosquitoes is biphasic, with contrasting dynamics during the PE and PBM phases of the reproductive cycle.
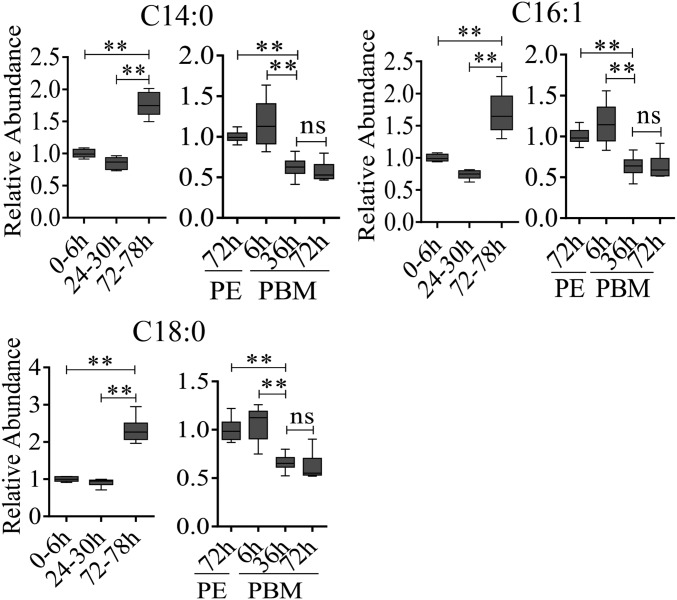
Fig. S1.
Dynamic changes of LM in the FB of A. aegypti female mosquitoes throughout the reproductive cycle. GC-MS was performed to analyze relative levels of saturated (C14:0, C18:0) and unsaturated (C16:1) FFAs during the mosquito reproductive cycle. Relative abundances at PE and PBM time points were normalized to 0–6 h PE and 72 h PE, respectively. At least six independently collected samples with 15 mosquito FBs for each time point were analyzed. **P < 0.01; ns, not significant.
Expression Kinetics of FB Genes Encoding LM Throughout the Reproductive Cycle of the Female Mosquito.
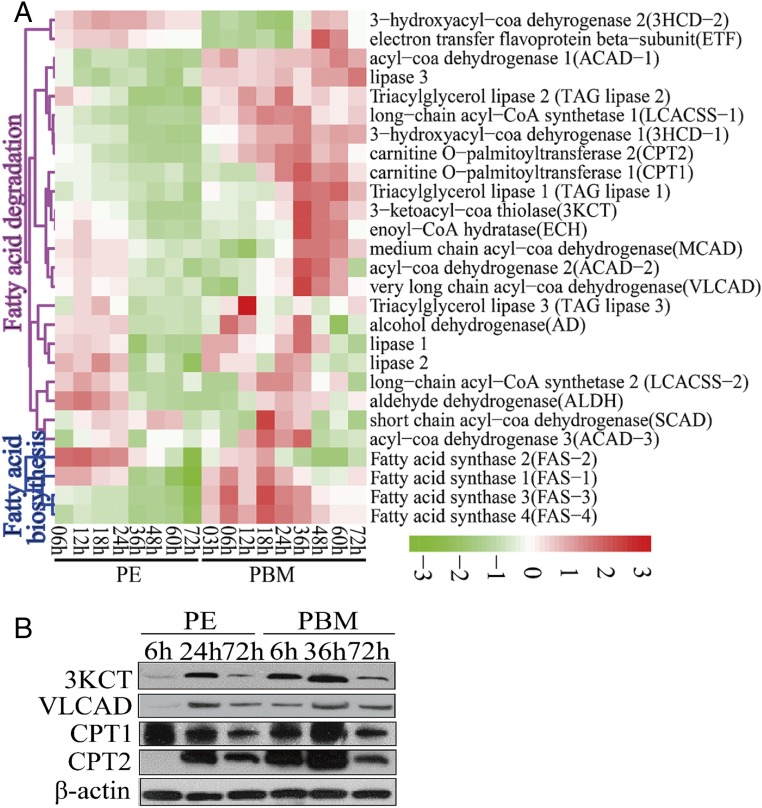
To understand further the role of the FB in lipid homeostasis, we investigated the overall expression kinetics of LM genes using two previously reported microarray transcriptomes of the mosquito FB covering the entire first reproductive cycle, one containing eight time points during the PE phase (6–72 h PE) and the other nine time points during the PBM phase (3–72 h PBM) (15, 18, 19). Transcripts of genes encoding fatty acid synthases (FASs) were at higher levels during the first 24 h PE and declined thereafter. Transcripts of six genes involved in TAG catabolism and of 17 genes associated with the β-oxidation pathway were also abundant during the first 24 h but declined by three- to fourfold at 72 h PE compared with their levels at 6 h PE (Fig. 2A and Dataset S1).
Fig. 2.
Temporal expression profiles of mRNA and protein levels involved in the LM pathway in the FBs of A. aegypti female mosquitoes throughout the reproductive cycle. (A) The heat map of expression patterns of LM genes during the PE and PBM phases. Only genes with a fold change >1.75 (0.8 in log2 scale) and a false-discovery rate of P < 0.01 are shown. (B) Protein levels of key LM enzymes (3KCT, VLCAD, CPT1, and CPT2) determined using Western blots. For each indicated time point, total protein extracts were prepared from eight female mosquito FBs. β-Actin was used as a control.
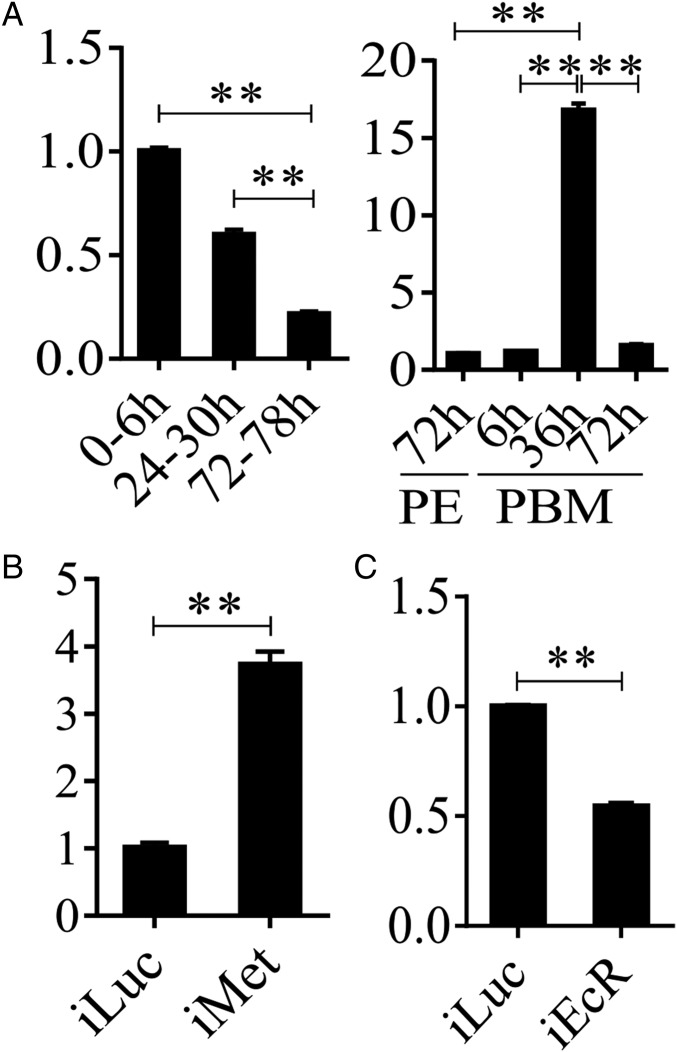
During the PBM phase, there was a stepwise sequential increase in transcript levels of LM genes (Fig. 2A). Transcripts of genes encoding FASs were elevated between 3 and 6 h PBM, reaching maximal levels at 18 h PBM, suggesting initial activation of lipid biosynthesis by blood meal-induced signals. At the same time, transcripts of genes encoding several lipases, including TAG lipases, were also increased, suggesting the onset of TAG degradation and an increase in FFA levels. Diminished levels of TAG and LD sizes point out that TAG catabolism predominates over biosynthesis at this time. Transcripts of genes encoding two FFA transport enzymes, carnitine O-acetyltransferase 1 (CPT1) and carnitine O-acetyltransferase 2 (CPT2), increased in abundance at 18–36 h PBM. Transcripts of genes encoding the key enzymes of the β-oxidation pathway, such as enoyl-CoA hydratase (ECH), 3-hydroxyacyl-coa dehydrogenase-1 (3HCD-1), 3-ketoacyl-CoA thiolase (3KCT), and very long chain acyl-CoA dehydrogenase (VLCAD), exhibited a significant simultaneous elevation 12 h later, at 36 h PBM, indicating a dramatic increase in lipid catabolism. Most genes of the LM pathways declined gradually after 48 h PBM. Their reduced mRNA levels at 72 h PBM may reflect the onset of preparation of female mosquitoes for a second gonadotrophic cycle. The quantitative real-time PCR (qRT-PCR) analysis of selected LM gene transcripts at three PE time points (6, 24, and 72 h PE) and three PBM time points (6, 36, and 72 h PBM) has validated these transcriptome data (Fig. S2).
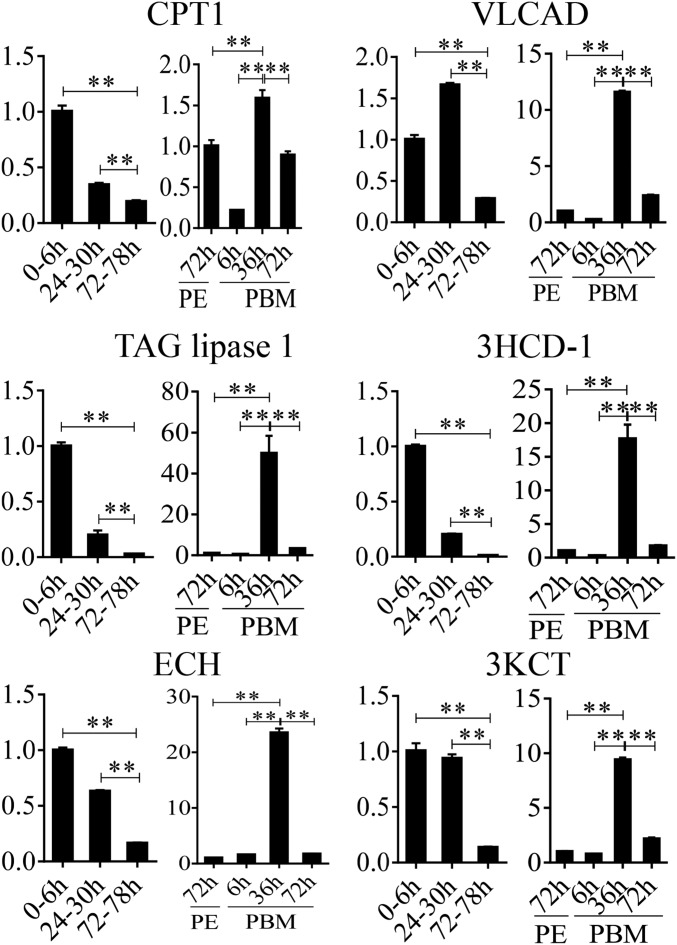
Fig. S2.
Expression profiles of FB LM genes throughout the reproductive cycle. qRT-PCR results exhibiting the relative mRNA levels of CPT1, VLCAD, TAG lipase 1, 3HCD-1, ECH, and 3KCT during the PE and PBM phases. Relative fold changes were normalized to 0–6 h PE and 72 h PE, respectively, and are indicated on the y axis of each graph. All experiments were performed at least three times. Error bars represent the SEM; **P < 0.01.
Western blot analysis of FB samples from the same six time points was performed using antibodies that recognize respective A. aegypti LM enzymes (two β-oxidation enzymes—3KCT and VLCAD—and two fatty acid transport enzymes—CPT1 and CPT2). This analysis has shown that the protein levels of these enzymes are coordinated with the expression of their respective genes (Fig. 2B).
The Role of JH and Its Receptor Met in LM Regulation.
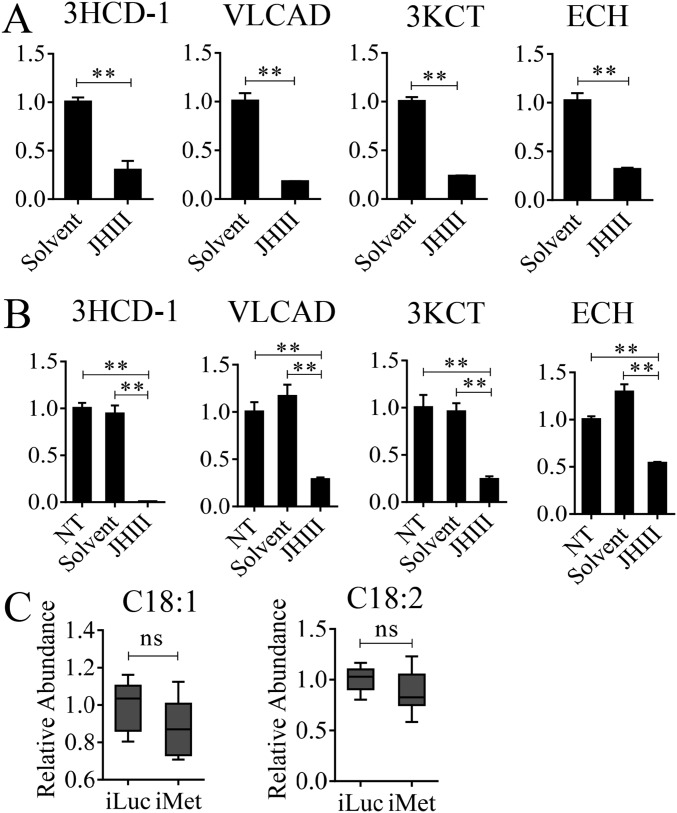
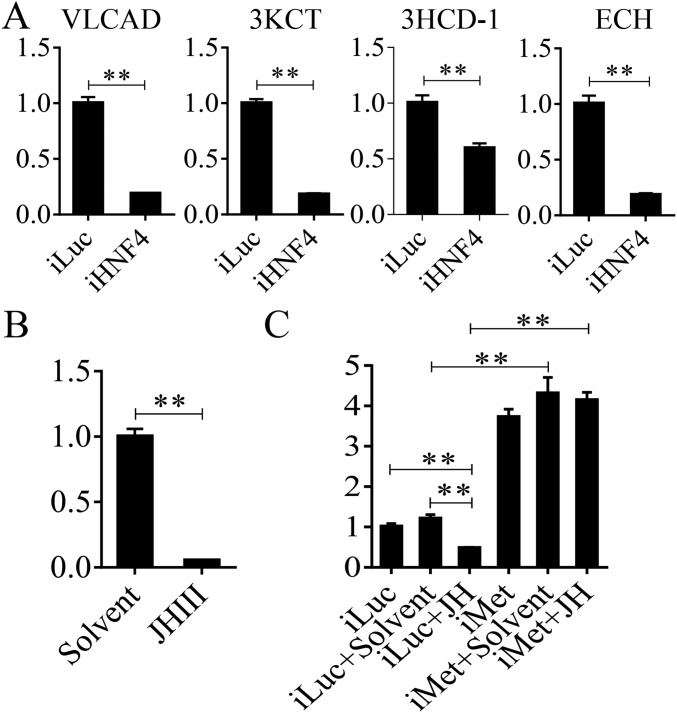
In A. aegypti female mosquitoes, JH III controls PE development and reproductive transitions (19). The JH III titer is low at eclosion, elevated by 12 h PE, reaches its maximal levels around 48–56 h PE, and maintained its high level until blood feeding (20). Thus, the JH titer negatively correlates with the pattern of LM gene transcript abundance during the PE phase, suggesting that JH may be responsible for the down-regulation of LM genes. To investigate this hypothesis, we first topically applied 10 μM JH III onto the abdomens of newly emerged female mosquitoes (3 h PE), and extracted total RNA 24 h after hormonal treatment. mRNA levels of selected genes (ECH, 3HCD-1, VLCAD, and 3KCT) were measured using qRT-PCR. The JH III application resulted in sharply reduced levels of these gene transcripts compared with those in mosquitoes treated with solvent (acetone) only (Fig. S3A). Next, we conducted an in vitro study, in which FB preparations, isolated from mosquitoes 10 h PE, were incubated in the culture medium containing 10 μM JH III in acetone or acetone alone for 8 h, as previously described (21). mRNA levels significantly decreased in FBs incubated in the presence of JH III compared with those from control FBs incubated in the presence of acetone or only the culture medium (Fig. S3B). Taken together, these results indicate that JH III is responsible for the down-regulation of expression of LM catabolic and β-oxidation genes during the PE phase.
Fig. S3.
The influence of JH and Met on representative LM genes and metabolites. (A) The in vivo effect of topical application of JH on LM gene expression. The fold change of LM genes in JH-treated mosquitoes was normalized to those treated with solvent (acetone) only and is indicated on the y axis of each graph. (B) The mRNA levels of LM genes after JH or solvent (acetone) was added to the medium of the in vitro FB culture. Relative fold changes with JH and solvent treatment were normalized to nontreated (NT) samples and are indicated on the y axis of each graph. Error bars represent the SEM; **P < 0.01 in A and B. (C) GC-MS results showing the relative level of C18:1 and C18:2 after Met RNAi knockdown. Error bars represent the SEM; ns, not significant.
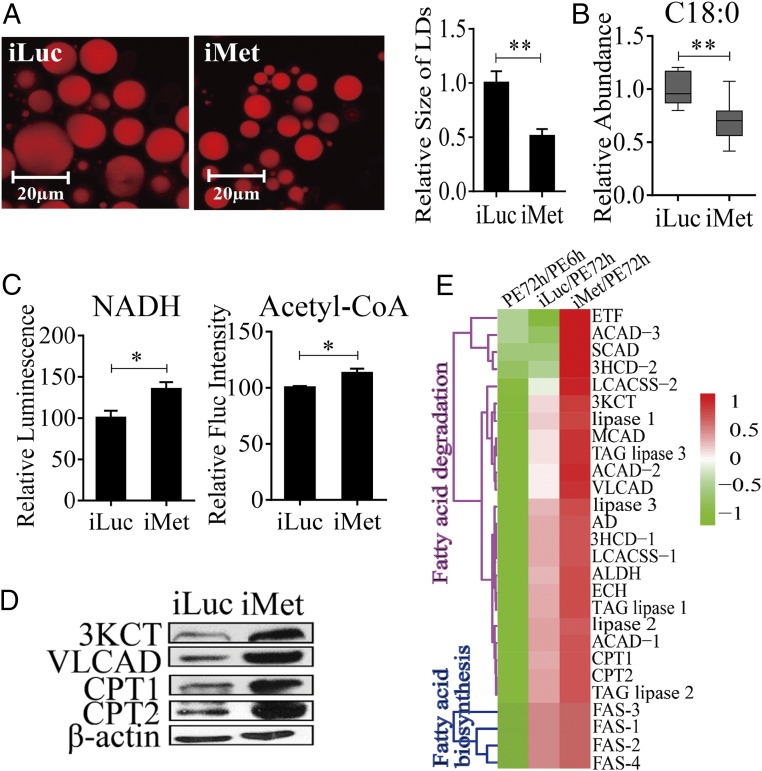
Our previous studies have shown that the JH receptor Met plays a central role in regulating FB gene expression in the PE female mosquitoes (15, 19, 22). To evaluate the role of Met in LM control, we conducted Met RNAi. Female mosquitoes 12 h PE were injected with either dsMet or dsLuc as a control. LM phenotypic manifestations of Met RNAi depletion were examined 4 d postinjection. LDs were significantly reduced in the FB, suggesting enhanced mobilization of TAG reserves in these mosquitoes (Fig. 3A). This result is in agreement with the previous observation of reduced total TAG levels in Met-depleted female PE mosquitoes (15). FFA levels were considerably lower in Met-depleted mosquitoes than in luciferase RNAi (iLuc) controls (Fig. 3B and Fig. S3C). Met depletion also resulted in the elevation of the β-oxidation metabolites NADH and acetyl-CoA, pointing to increased lipid consumption in Met RNAi-depleted mosquitoes (Fig. 3C). Moreover, Western blot analysis of FB samples from mosquitoes with depleted Met showed a dramatic elevation in the protein levels of two major β-oxidation enzymes—3KCT and VLCAD—and two fatty acid transport enzymes—CPT1 and CPT2 (Fig. 3D)—further indicating an increase in β-oxidation in Met-depleted mosquitoes.
Fig. 3.
Effect of Met RNAi knockdown (iMet) on LM during the PE phase in A. aegypti female mosquitoes. (A) Nile red staining of iMet and iLuc controls in female mosquito FBs. The relative LD sizes were decreased significantly after iMet treatment. (B) GC-MS analysis showing reduced FFA levels (C18:0) in iMet mosquitoes. RNAi knockdown of Met treatments was normalized to iLuc. At least six independently collected samples were used for each treatment. (C) NADH and acetyl-CoA levels in iMet and iLuc mosquitoes. Measurements were normalized to the total protein in each treatment. iMet results were further normalized to iLuc. At least three independently collected samples with six mosquitoes were used for each treatment. (D) Western blot showing increased levels of catabolic LM enzymes in the FBs of iMet mosquitoes. β-Actin was used as a loading control. (E) Heat map representing transcripts of LM-related genes in iMet mosquitoes. Error bars in A–C represent the SEM; *P < 0.05, **P < 0.01.
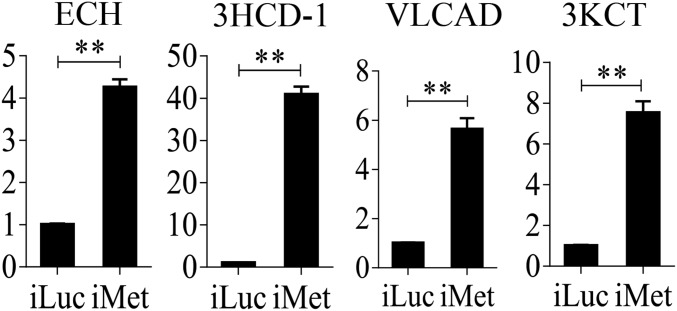
We used previously published FB transcriptome data obtained from Met RNAi-depleted female mosquitoes to analyze LM-related genes (19). This analysis showed that transcripts of TAG catabolism and β-oxidation genes were dramatically elevated (P = 1.192e-07, Wilcoxon signed rank test) (Fig. 3E and Dataset S2). In contrast, FAS genes were only marginally affected by Met RNAi (P = 0.0625, Wilcoxon signed rank test) (Fig. 3E and Dataset S2). We confirmed these results using qRT-PCR analysis through Met RNAi in female mosquitoes. Transcripts of genes encoding LM catabolic enzymes—ECH, VLCAD, 3KCT, and 3HCD-1—were significantly up-regulated in FBs of Met RNAi-depleted mosquitoes (Fig. S4).
Fig. S4.
qRT-PCR analysis of relative mRNA abundance of representative LM genes (ECH, 3HCD-1, VLCAD, and 3KCT) after Met RNAi knockdown. Relative fold changes were normalized to 0–6 h PE and 72 h PE, respectively, and are indicated on the y axis of each graph. Error bars represent the SEM; **P < 0.01.
The Role of 20E and EcR in LM Regulation.
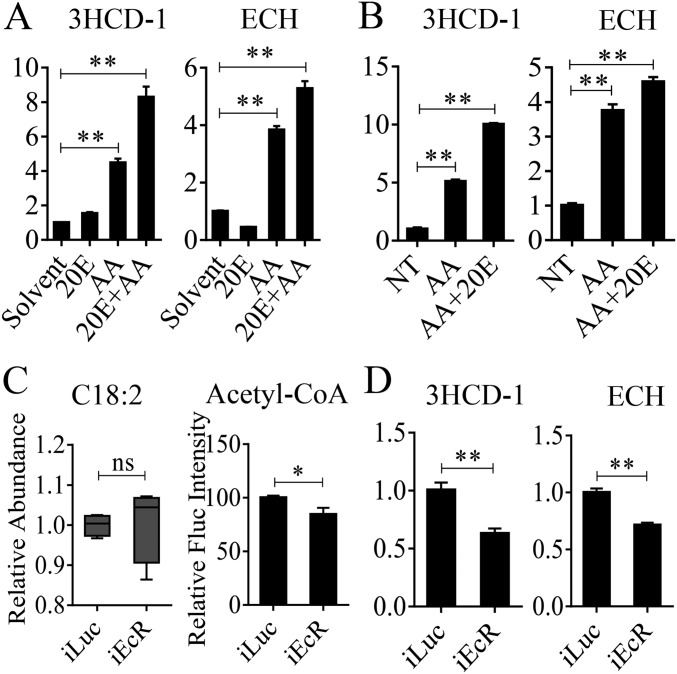
In mosquitoes, 20E plays a critical role in controlling egg maturation (23). To examine whether 20E is involved in the regulation of LM, we first conducted an in vivo experiment. Previtellogenic female mosquitoes, which at 72 h PE have a low level of endogenous 20E, were injected with 0.5 μL 10−6 M 20E alone or with addition of a mixture of amino acids. Solvent (ethanol) was used as a control. FBs were dissected 24 h postinjection, and the transcript abundance of two genes encoding β-oxidation enzymes (ECH and 3HCD-1) was evaluated using qRT-PCR. A combination of amino acids and 20E was observed to elicit a significant increase in these gene transcripts (Fig. S5A). Then, we conducted an in vitro FB culture experiment, as previously described (18). FBs were dissected from female mosquitoes at 72 h PE and were cultured in medium containing either amino acids or amino acids and 20E. 20E was added into the culture medium in two steps to mimic a natural rise in the 20E titer (18). FBs were incubated first in the presence of 5 × 10−8 M 20E for 4 h and then with 10−6 M 20E for an additional 4 h. Controls were incubated for 8 h. qRT-PCR analyses showed a significant up-regulation of ECH and 3HCD-1 after incubation with amino acids and 20E (Fig. S5B). Thus, both amino acids and 20E are implicated as positive regulators of β-oxidation.
Fig. S5.
Effect of 20E, amino acids, and EcR on LM gene expression and metabolite level. (A) The expression patterns of 3HCD-1 and ECH after the injection of amino acids and/or 20E into mosquitoes. Relative fold changes of amino acid- and/or 20E-treated mosquitoes were normalized to solvent (ethanol)-treated mosquitoes. (B) The mRNA levels of 3HCD-1 and ECH after amino acids and/or 20E were added to the medium of the in vitro FB culture. Relative fold changes of amino acid- and/or 20E-treated samples were normalized to nontreated (NT) samples. (C) EcR-depleted mosquitoes displayed a reduced rate of β-oxidation. FFAs (C18:2) increased, and the β-oxidation product acetyl-CoA decreased. (D) Relative mRNA expression levels of LM genes (ECH and 3HCD-1) were significantly reduced after EcR knockdown. Relative fold changes are indicated on the y axis of each graph in A, B, and D. Error bars represent the SEM; *P < 0.05, **P < 0.01; ns, not significant.
The action of 20E is mediated by the EcR (24). To gain insight into the involvement of this receptor in the regulation of LM, we conducted EcR RNAi-mediated knockdown. Twelve-hour-old female mosquitoes were injected with either dsEcR or dsLuc, were fed blood 4–5 d later, and were examined 36 h PBM for LM phenotypic manifestations. Overall, this analysis demonstrated that EcR RNAi profoundly affects LM by enhancing lipid storage and reducing β-oxidation and TAG catabolism. FB LDs increased in size in EcR-depleted female mosquitoes (Fig. 4A), a finding that is in agreement with previous data showing a total TAG increase in such mosquitoes (15). Furthermore, EcR-depleted mosquitoes exhibited a tendency towards increased FFA levels and reduced NADH and acetyl-CoA levels (Fig. 4 B and C and Fig. S5C). Significantly, the protein level of a representative β-oxidation enzyme (3KCT) was low relative to the iLuc controls, further indicating a decline in β-oxidation in these mosquitoes (Fig. 4D).
Fig. 4.
Effect of EcR RNAi knockdown (iEcR) on LM during the PBM phase in female A. aegypti mosquitoes. (A) Nile red staining of FBs was performed on iEcR mosquitoes and compared with iLuc control samples. The relative LD sizes increased significantly with iEcR treatment. (B) The GC-MS analysis of an FFA representative (C18:0) in iEcR mosquitoes. At least six independently collected samples were analyzed for each treatment. (C) Reduced levels of NADH in iEcR mosquitoes. iEcR results were further normalized to iLuc, which were normalized to the total protein levels for each treatment. At least three independently collected samples with six mosquitoes were analyzed for each treatment. (D) Western blot shows a decreased protein level of the enzyme 3KCT in iEcR mosquitoes. β-Actin was used as a loading control. (E) Heat map representing the transcripts of LM-related genes in FBs of iEcR mosquitoes. Error bars in A–C represent the SEM; *P < 0.05, **P < 0.01.
To understand whether EcR affects the LM gene expression, we performed an RNAi-based transcriptomic screen for A. aegypti EcR. Illumina RNA-sequencing (RNA-seq) technology was used to generate the FB transcriptional profiles of genes affected by EcR RNAi-mediated depletion in A. aegypti female mosquitoes; iLuc served as a control. Three biological replicates of RNA-seq libraries were constructed for each RNAi treatment. Each RNA-seq library contained 61–77 million clean reads (read length, 150 nt for the three replicates) (Dataset S3). These clean reads were obtained by adaptor removal and filtering of low-quality reads. Then the reads were mapped to the genome of A. aegypti (https://www.vectorbase.org/organisms/aedes-aegypti) using the TopHat2 program, and the Cufflinks program was used to estimate expression levels of all genes (25, 26). The resulting count table was transformed into fragments per kilobase of transcript per million fragments mapped (FPKM) values. Relative mRNA abundance in the iEcR and iLuc samples was compared according to FPKM values. From these RNA-seq data, 11,430 transcripts with FPKM >0 within at least two replicates were selected for further analysis. To gain insight into the differentially expressed genes (DEGs) in the iEcR and iLuc groups, we performed differential analysis using the DEGseq package in R environment (27). A P value less than 0.05 indicated that genes were differentially expressed. In total, we obtained 1,355 DEGs, of which 513 were down-regulated and 842 were up-regulated after EcR RNAi knockdown (Dataset S4). To analyze the gene-expression dynamics of LM genes after EcR knockdown, we performed hierarchical clustering of the LM genes identified during the previous step. Transcript levels of genes belonging to the β-oxidation pathway and TAG catabolism process significantly decreased (P = 1.907e-06, Wilcoxon signed rank test) in the dsEcR-treated mosquitoes (Fig. 4E and Dataset S5). FAS genes were less affected by EcR RNAi (P = 0.0625, Wilcoxon signed rank test) (Fig. 4E and Dataset S5). qRT-PCR analysis of selected LM genes (ECH and 3HCD-1) also showed a significant reduction in their transcript levels in EcR RNAi-depleted mosquitoes (Fig. S5D).
The Role of HNF4 in Regulation of LM Throughout the Female Mosquito Reproductive Cycle.
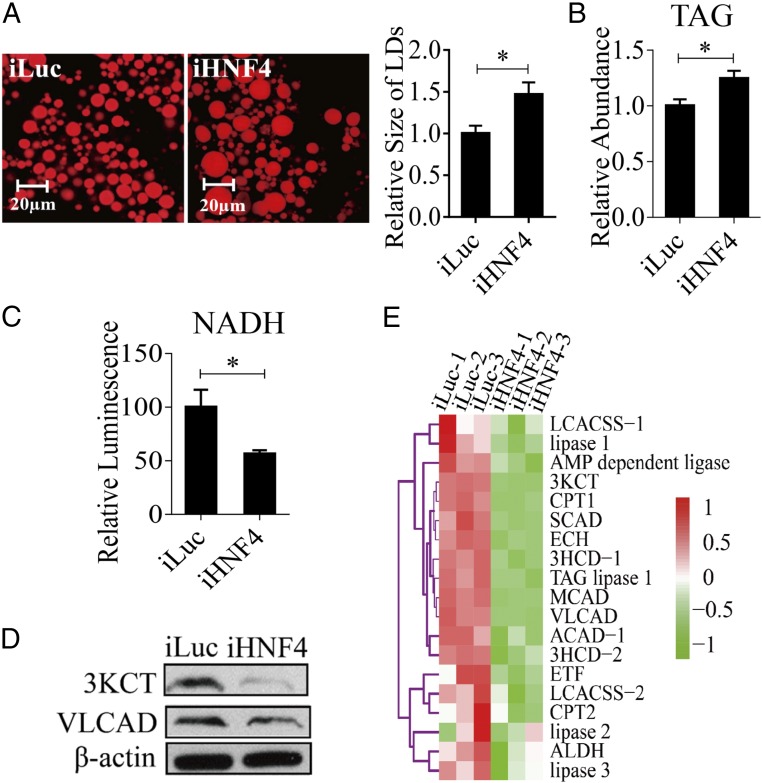
HNF4 has been reported to play an important role in regulating development and energy metabolism. Moreover, it has been found that these HNF4 actions result in part from its control of the β-oxidation pathway and TAG catabolism (28). To understand a possible role of HNF4 in female mosquitoes during reproduction, we first evaluated phenotypic manifestations of HNF4 RNAi. Mosquitoes were injected with HNF4 dsRNA at 12 h PE and were examined at 36 h PBM. Overall, the response to HNF4 RNAi depletion was highly similar to that of EcR RNAi knockdown. There was a significant increase in FB LD size as well as the elevation of TAG levels (Fig. 5 A and B). However, NADH levels were reduced in these mosquitoes (Fig. 5C). Protein levels of representative β-oxidation enzymes (3KCT and VLCAD) were low relative to the iLuc control, further indicating a decline in β-oxidation in these mosquitoes (Fig. 5D).
Fig. 5.
LM changes in the HNF4-depleted (iHNF4) female mosquitoes at 36 h PBM. (A) Nile red staining of FBs was performed on iHNF4 mosquitoes and compared with iLuc control samples. The LD sizes increased significantly in iHNF4 treatments. (B) TAG was measured in iHNF4 and iLuc mosquitoes. iHNF4 treatments were normalized to iLuc. At least three independently collected samples for each treatment were analyzed. (C) Measurements of NADH in iHNF4 and iLuc mosquitoes, which were normalized to the total protein levels for each treatment. The iHNF4 results were further normalized to iLuc. Measurements were performed using at least three independently collected samples with six mosquitoes for each treatment. (D) Western blot showing decreased protein levels of 3KCT and VLCAD enzymes in iHNF4 mosquitoes. β-Actin was used as a loading control. (E) Heat map representing transcripts of TAG degradation and β-oxidation genes in iHNF4 mosquito FBs. Error bars in A–C represent the SEM; *P < 0.05.
To investigate further the HNF4 effect on LM, an RNAi-based transcriptomic screen for this NR was performed. Transcriptional profiles of genes affected by HNF4 RNAi-mediated depletions in the FBs of A. aegypti female mosquitoes were generated by Illumina RNA-seq technology. iLuc served as a control. Three biological replicates of RNA-seq libraries were constructed with each library containing 49–93 million raw reads (read length, 150 nt for each replicate) (Dataset S6). After the adaptor was removed and low-quality reads were filtered, each library generated 47–89 million clean reads that were used for further analysis. These clean reads were mapped to the genome of A. aegypti (https://www.vectorbase.org/organisms/aedes-aegypti) using the TopHat2 program, and the Cufflinks program was used to estimate the expression level of all of the genes (25, 26). The resulting count table was transformed into FPKM values. The relative mRNA abundance in iHNF4 and iLuc samples was compared on the basis of FPKM values. From the RNA-seq data, 14,695 transcripts with FPKM >0 within at least two replicates were selected for further analysis. We then performed the analysis of these 14,695 transcripts using the DEGseq package in R environment (27). In total, we obtained 1,971 DEGs (P < 0.05), of which 1,004 genes were down-regulated and 967 were up-regulated after the HNF4 RNAi knockdown (Dataset S7). Transcript levels of genes belonging to the β-oxidation pathway as well as TAG catabolism were significantly decreased (P = 1.907e-06, Wilcoxon signed rank test) in the dsHNF4-treated mosquitoes (Fig. 5E and Dataset S8). The qRT-PCR analysis of selected β-oxidation genes (VLCAD, 3KCT, ECH, and 3HCD-1) also resulted in a significant reduction in their transcript levels in HNF4 RNAi-depleted mosquitoes (Fig. S6A).
Fig. S6.
Expression profiles of LM genes in HNF4- and Met RNAi-depleted mosquitoes and the effect of JH application on HNF4 expression level. (A) qRT-PCR shows the relative expression levels of LM genes (VLCAD, 3KCT, 3HCD-1, and ECH) after HNF4 RNAi knockdown. HNF4 dsRNA-treated mosquitoes were collected at 36 h PBM. (B) The expression patterns of HNF4 genes after topical application of JH on the abdomens of female A. aegypti mosquitoes. (C) The mRNA levels of HNF4 after topical application of JH or solvent (acetone) on Luc- and Met dsRNA-treated female mosquitoes. All experiments were repeated at least three times. Relative fold changes are indicated on the y axis of each graph. Error bars represent the SEM; **P < 0.01.
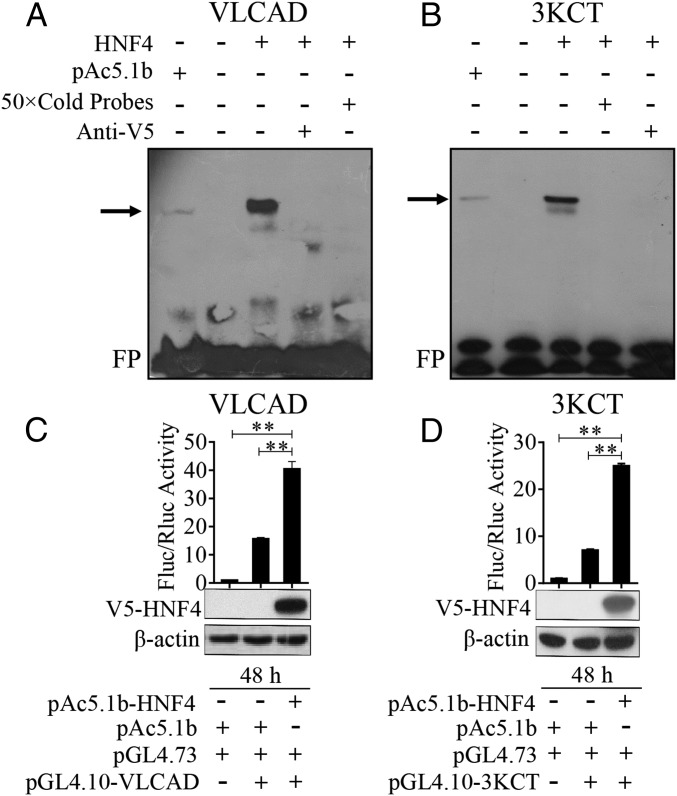
To understand the mode of HNF4 control of genes belonging to the β-oxidation pathway and TAG catabolism, we examined whether this NR interacts with promoters of these genes. We selected two genes (VLCAD and 3KCT) for this analysis. Promoters of VLCAD and 3KCT contain the putative HNF4-binding motifs with the consensus CAAAAGG/TT/CC/T predicted by the JASPAR server (jaspar.genereg.net/). Subsequently, an EMSA was performed using nuclear extracts from S2 cells transfected with either pAc5.1b/HNF4 or pAc5.1b empty vector. Single bands were visualized using the biotin-conjugated 25-bp VLCAD probe or biotin-conjugated 24-bp 3KCT probe (Fig. 6 A and B and Dataset S9). The bands disappeared after the addition of a 50× molar excess of unlabeled VLCAD or 3KCT probes to the respective binding reaction (Fig. 6 A and B). Preincubation of the nuclear extract with anti-V5 antibody resulted in the loss of mobility bands (Fig. 6 A and B). These results indicate that HNF4 binds specifically to the promoter regions of VLCAD and 3KCT and point to the direct control of these genes via binding to HNF4. It is noteworthy that, when HNF4 was replaced with the empty vector in the binding reaction, a vague band in VLCAD and 3KCT probes could also be seen (Fig. 6 A and B), possibly showing weak binding by endogenous HNF4 in S2 cells. To confirm further whether HNF4 directly regulates the transcription of key LM genes, upstream promoter sequences of VLCAD (nucleotides −2759 to +23) and 3KCT (nucleotides −781 to +24) were subcloned into the pGL4.10 vector (SI Results) to perform luciferase assays. The HNF4 cDNA (nucleotides 1–1695) was subcloned into a pAc5.1b/V5 vector to obtain the V5-HNF41–565 fusion protein in Drosophila S2 cells. Then, the constructs pGL4.10-VLCAD−2759 to +23 or pGL4.10-3KCT −781 to +24 were cotransfected along with pAc5.1b-HNF41−565 or empty vector pAc5.1b into Drosophila S2 cells to perform the dual luciferase reporter assays, and empty vector pAc5.1b was used as the control. Obviously, the luciferase activity in experimental groups significantly increased compared with controls (Fig. 6 C and D). Moreover, Western blot analysis using anti-V5 antibodies confirmed that the V5-HNF41–565 fusion protein was overexpressed in S2 cells (Fig. 6 C and D). Taken together, these data demonstrate that direct binding to promoters of VLCAD and 3KCT by HNF4 is required for the transcriptional activation of these genes.
Fig. 6.
(A and B) EMSA results showing specific binding between VLCAD (A) and 3KCT (B) probes and S2 cell nuclear extracts with expressed V5-HNF4 protein. The DNA probes containing HNF4 putative binding motifs (VLCAD: 5′-ATAGTGGGGCAAAAGTTCGAATCAG-3′; 3KCT: 5′-GAAGTTAATCAAAAGTCTCTAAAA-3′) and their flanking regions from the promoter regions of VLCAD and 3KCT were selected as probes. Unlabeled specific probes (50× cold probes) identified the specificity of the binding, and anti-V5 antibody confirmed the presence of HNF4 protein. The arrow indicates the specific complex. FP marks a free eluted probe. (C and D) Dual luciferase reporter assays, in which pGL4.10/VLCAD −2759 to +23 (C) or pGL4.10/3KCT −781 to +24 (D) were cotransfected into S2 cells with either the pAc5.1b-HNF4 or the pAc5.1b empty vector. pGL4.73 vector containing the hRluc reporter gene and an SV40 early promoter was used as an expression control in the luciferase assay. The relative luciferase activity was normalized to empty cell groups. Fluc/Rluc represents the ratio of Firefly to Renilla luciferase activity. Western blots show the V5-HNF4 protein input in S2 cells. The protein extracts from empty S2 cells and empty vector pAc5.1b were used as negative controls, and β-actin was used as a loading control. Error bars represent the SEM; **P < 0.01.
Hormonal and Nutritional Control of the HNF4 Gene Expression.
We used the qRT-PCR analysis to investigate the expression profile of the HNF4 gene throughout the reproductive cycle of A. aegypti female mosquitoes. This analysis revealed that the transcript level of the HNF4 gene was high at the beginning of the PE phase but reduced dramatically by 72 h PE (Fig. 7A). After a blood meal, HNF4 mRNA abundance was up-regulated, reaching its peak at 36 h PBM and dropping to its late PE stage level by 72 h PBM (Fig. 7A). This biphasic HNF4 expression profile suggests that HNF4 gene expression is differentially regulated during the PE and PMB phases. To understand the role of JH in regulation of the HNF4 gene expression, we performed in vivo and in vitro experiments as described above for LM genes (Fig. S6 B and C). The expression of HNF4 was dramatically decreased when JH was topically applied to 24-h-old mosquitoes (Fig. S6B). To confirm further the involvement of the JH receptor Met, we conducted its RNAi silencing and monitored the response of the HNF4 transcript using qRT-PCR analysis. In an in vitro test, Met RNAi rendered the HNF4 gene unresponsive to JH III (Fig. S6C). Moreover, RNAi Met depletion in PE female mosquitoes resulted in a significant elevation of the HNF4 transcript compared with iLuc control (Fig. 7B). RNAi EcR depletion was performed as described above. It showed a significant down-regulation of the HNF4 transcript compared with iLuc control, indicating involvement of EcR in the up-regulation of this gene expression (Fig. 7C). Taken together, these data clearly show that major hormones regulating reproductive events in female mosquitoes, JH and 20E, are also involved in the control of HNF4 gene expression, with Met downregulating this gene at the PE phase and EcR up-regulating it at the PBM phase.
Fig. 7.
Met and EcR regulate HNF4 gene expression. (A) Relative HNF4 mRNA levels throughout the A. aegypti female reproductive cycle. (B) Met RNAi up-regulates HNF4 gene expression at the PE phase. (C) EcR RNAi represses the HNF4 gene expression at the PBM phase. Metrics in the y axis in all figures represent relative fold changes. Error bars represent the SEM; **P < 0.01.
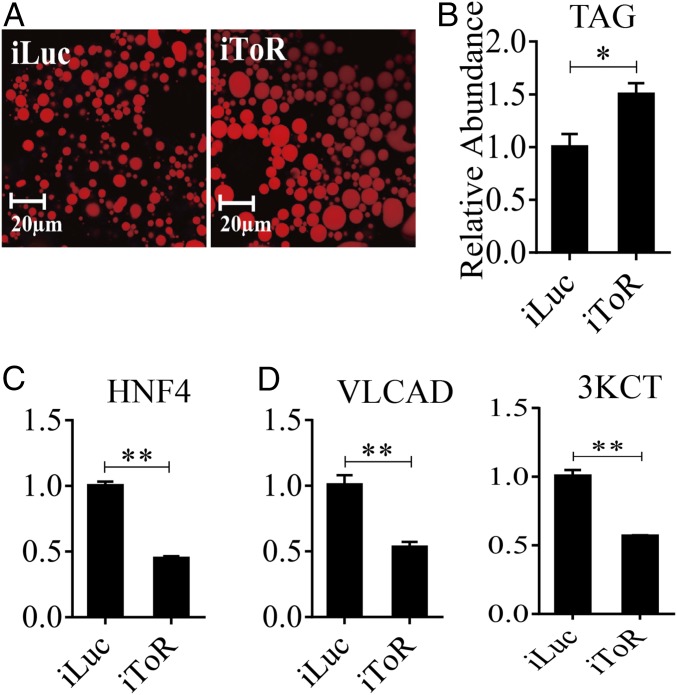
To assess further the involvement of nutritional factors in regulation of LM, we used RNAi for the TOR kinase C1, which is known to mediate amino acid signaling (29–34). TOR RNAi silencing was performed as described above for EcR and HNF4. It led to enlargement of LDs and an increase in the TAG content in the PBM mosquito FB, mimicking phenotypes after RNAi silencing of EcR and HNF4 (Fig. 8 A and B). Moreover, TOR RNAi resulted in down-regulation of the expression of genes encoding HNF4 and selected LM genes (VLCAD and 3KCT), indicating the involvement of the TOR pathway in regulation of the HNF4 expression (Fig. 8 C and D).
Fig. 8.
Effect of TOR on LM during PBM period. (A) Nile red staining of FBs was performed on iTOR female mosquitoes and compared with iLuc control samples. LD size increased in iTOR mosquitoes. (B) TAG was measured in iTOR and iLuc mosquitoes. dsTOR treatments were normalized to iLuc. At least three independently collected samples for each treatment were analyzed. (C and D) qRT-PCR results showing significantly reduced mRNA levels of HNF4 (C) and LM (D) genes (VLCAD and 3KCT) in iTOR mosquitoes. Metrics in the y axis in C and D represent relative fold changes. Error bars represent the SEM; *P < 0.05, **P < 0.01.
Discussion
To support their hematophagic lifestyle that involves intense host seeking, the utilization of a very large blood meal, and rapid egg development, female mosquitoes require tremendous energy expenditure. Accordingly, a high level of metabolism is tightly coordinated with the blood-mediated reproduction of these insects. Our present study has revealed that LM undergoes biphasic changes with opposite trends at the PE and PBM phases of the gonadotrophic cycle. Moreover, LM is synchronized at all levels: at gene expression; the translation of LM enzymes; levels of FFAs and TAG stores; and the final β-oxidation products, NADH and acetyl-CoA. Our transcriptomic analysis showed that LM genes in the mosquito FB are down-regulated by the end of the PE phase, but their transcript levels are significantly increased during the PBM phase. Reduction in the transcript levels of genes encoding TAG catabolism and β-oxidation enzymes correlates with a dramatic accumulation of lipids in the PE phase. In contrast, the elevation of LM gene transcripts is in accord with diminishing lipid stores during the PBM phase. Thus, taken together these observations suggest that LM, at least in part, is regulated at the transcriptional level.
Multiple physiological functions that are vital for female mosquitoes during reproduction are governed by alternating peaks of JH and 20E (23, 35). These hormones have been implicated in temporal coordination of mosquito carbohydrate metabolism, with JH/Met downregulating carbohydrate metabolic pathways at the PE phase and 20E/EcR up-regulating them at the PBM phase (15). Our present study has demonstrated that these hormones and their receptors play critical roles in controlling LM during female mosquito reproduction. Met RNAi silencing resulted in a reversal of the LM phenotype in PE female mosquitoes, manifested by the dramatic increase of LM gene transcripts, β-oxidation enzymes, and metabolites and by the decrease in levels of lipid stores and FFAs. The response to EcR RNAi silencing was the opposite, reversing all LM phenotypic signs associated with accelerated TAG mobilization and β-oxidation in PBM female mosquitoes to the state of lipid accumulation reminiscent of that of the PE phase.
Transcriptomic analysis coupled with Met RNAi silencing has confirmed the conclusion that Met is responsible for the down-regulation of LM genes. Met is a transcription factor belonging to the basic helix–loop–helix (bHLH)–Per-Amt-Sim (PAS) family that requires a heterodimeric partner to mediate the JH response (36, 37). Met interacts with a bHLH-PAS domain containing steroid receptor coactivator (SRC), also known as “Taiman” or “β-FTZ-F1 (fushi tarazu binding factor 1) interacting steroid receptor coactivator (FISC),” binds to E-box motifs in the regulatory regions of JH-controlled genes, and affects gene expression in a JH-dependent manner (22, 37–40). It appears that JH-mediated gene activation occurs via direct binding of Met/SRC, whereas gene repression is indirect and requires additional intermediate factors (19). Recently, it was shown that a long-distance repressor, Hairy, and its corepressor, Groucho, mediate the Met action in the repression of some JH-regulated genes (21). However, only 15% of Met-repressed genes have been shown to be controlled by Hairy/Groucho. Thus, further studies should uncover other mechanisms of Met gene repression.
The NR superfamily includes ligand-activated and orphan transcription factors that regulate gene expression by interacting with specific DNA sequences in regulatory regions of their target genes. NRs play essential roles in controlling development, reproduction, and metabolism and are important targets for therapy of metabolic diseases (41–43). Because of their ability to interact with fatty acids and other lipids, many NRs, including HNF4, are important regulators of LM (44–46). There is a remarkable evolutionary conservation of these lipophilic factors and mechanisms of their action between mammals and invertebrates (47). However, in contrast to mammals and nematodes, in which there are multiple NHF4 members, insects appear to have a single HNF4 ortholog that is highly similar to the mammalian HNF4α (28). HNF4 has been implicated in the regulation of lipid catabolism and β-oxidation during larval development of the model insect Drosophila melanogaster (28). Here, we reported a key role of HNF4 in controlling such functions in reproducing female mosquitoes. HNF4 RNAi-silenced mosquitoes exhibited an adverse LM phenotype similar to that of EcR RNAi mosquitoes. LM phenotypic abnormalities caused by HNF4 RNAi are manifested in the inability to use lipids and correlated with a dramatic down-regulation of gene transcripts encoding TAG catabolism and β-oxidation enzymes. We have demonstrated that, as in D. melanogaster (28), the mosquito HNF4 is involved in the transactivation of TAG catabolism and β-oxidation genes. However, we have additionally shown its direct action on gene representatives encoding β-oxidation enzymes. Our EMSA and the luciferase reporter assay have clearly shown that HNF4 controls the activation of representative β-oxidation genes (3KCT and VLCAD) via direct binding of their promoters to its cognate binding site with the consensus CAAAAGG/TT/CC/T. Interestingly, this action of the mosquito HNF4 is reminiscent of that of the mammalian HNF4α, which binds to the H4-specific CAAAGTCCA site and up-regulates its target LM genes (48).
The pattern of the HNF4 gene expression is synchronized with that of genes encoding TAG degradation and β-oxidation enzymes during the mosquito gonadotrophic cycle. This synchronization suggests that similar regulatory networks could be involved in its control. Indeed, HNF4 gene expression is repressed by JH and Met but is activated by EcR. Although the precise mechanisms of the respective repressive and activating actions of Met and EcR in controlling the HNF4 gene expression remain to be elucidated, our study has revealed a link between these major regulators of developmental timing governing mosquito reproduction and the HNF4 gene transcriptional regulation. Therefore, HNF4 likely acts as a mediator of JH and 20E signaling in setting the stage for temporal coordination of TAG degradation and β-oxidation with the energy demands of a reproducing female mosquito.
Clearly, regulation of metabolism is extremely complex and occurs at many physiological levels. Sensing the nutritional status is a critical adaptation for organism survival. The amino acid/TOR pathway is essential for the female mosquitoes; the amino acid pulse that follows early blood digestion signals to TOR that in turn acts synergistically with EcR to activate vitellogenic events (18, 29–34). Our data also demonstrate the importance of this pathway in LM. In vitro experiments showed that amino acids are required for 20E activity in up-regulating the expression of β-oxidation genes, and TOR RNAi depletion resulted in the down-regulation of these genes as well as HNF4. TOR also is required for TAG mobilization, similar to that of EcR and HNF4. Thus, TOR appears to be involved in maintaining the HNF4 expression level either directly or by affecting EcR. Another link to the nutritional status is provided by FFAs. In transgenic D. melanogaster, the key FFAs, such as palmitic acid (16:0) and oleic acid (18:1), are important as ligands for HNF4 transactivating activity (28). It is likely that the elevated FFA levels observed in female mosquitoes play similar roles. However, to perform its role in gene activation, HNF4 needs to be expressed and is controlled predominantly by EcR at the PBM phase.
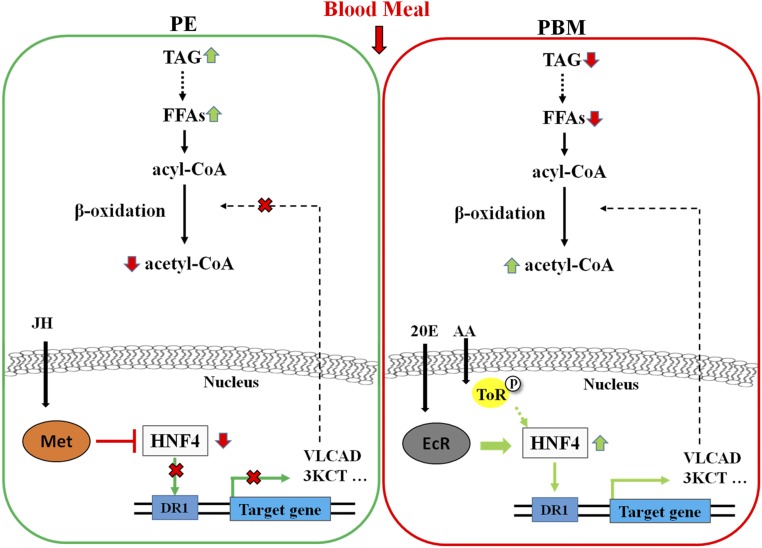
In summary, we have performed a comprehensive investigation of LM throughout the reproductive cycle of the A. aegypti female mosquito. Our transcriptome and metabolic data have revealed that transcript levels of genes encoding enzymes of the LM pathway and the LM intermediate metabolites exhibit biphasic dynamic changes during this cycle, with opposite trends during the PE and PBM phases of the cycle. We have identified that LM is controlled, at least in part, at the transcriptional level. HNF4 appears to be a direct regulator of the expression of genes encoding TAG catabolism and β-oxidation enzymes. The developmental hormones JH and 20E and their receptors set the stage for the temporal coordination of LM, mainly by controlling the HNF4 gene expression, with Met repressing this gene by the end of the PE phase and EcR up-regulating it during the PBM phase. In addition, the nutritional signaling exemplified by the amino acid/TOR pathway is involved synergistically with EcR in the control of HNF4 gene expression at the PBM phase of the mosquito reproductive cycle (Fig. S7). This work provides insight into the regulatory pathways controlling the temporal coordination of LM in female mosquitoes throughout their reproductive cycles.
Fig. S7.
A proposed scheme of LM regulation during the reproductive cycle of female A. aegypti mosquitoes. The expression of LM genes was repressed by the HNF4-mediated JH/Met pathway during the PE phase and was activated through the HNF4-mediated 20E/EcR pathway as well as by amino acid/TOR signaling pathway during the PBM phase.
Experimental Procedures
Animals.
The A. aegypti mosquitoes were raised as described previously (15). Adult mosquitoes were maintained continuously on water and 10% (wt/vol) sucrose solution. All procedures for vertebrate animal use were approved by the Animal Care and Use Committees of the Chinese Academy of Sciences Institute of Zoology and the University of California Riverside.
Bioinformatics Analysis.
Bioinformatics analysis was performed as previously described (49). DEG datasets from PE and PBM time-course microarrays were used to construct expression profiles of LM genes. Complete linkage hierarchical clustering was performed using the hclust function in R (19). Orthologous groups and pathway information, based on the KEGG, were downloaded from the database (50) and used in this study. Genes with a fold change greater than 1.75 (0.8 in log2 scale) and a false-discovery rate (P value) less than 0.01 were selected. RNA-seq analysis was carried out as described previously (51), and about 1.5 μg total RNA from each sample was used to enrich poly(A) mRNA using Oligo(dT) magnetic beads (Invitrogen). In the next step, paired-end 150 RNA-seq libraries were prepared following the Illumina’s library construction protocol. The libraries were sequenced using the Illumina HiSeq. 4000 platform (Illumina) at Novogene. Three replicates of each sample were pooled for analysis. Raw reads generated from the sequencing were processed by adaptor removal and filtering of low-quality reads. Afterward, these clean reads were mapped to the genome of A. aegypti (https://www.vectorbase.org/organisms/aedes-aegypti) using TopHat2, and Cufflinks was used to estimate the expression level of all genes (25, 26).
RNA Extraction and qRT-PCR.
Dissected FB tissues from 10–15 individual mosquitoes were homogenized using a motor-driven pellet pestle mixer and were lysed by TRIzol reagent (Invitrogen). Total RNA was extracted according to the protocol described previously (15). cDNA was synthesized using the FastQuant RT Kit (Tiangen). qRT-PCR was performed using SYBR Green SuperReal PreMix (Tiangen) in quadruplicate on the MX3005P system (Stratagene). Template concentrations were normalized to endogenous reference ribosomal protein S7 (rps7). Primers for qRT-PCR are shown in Dataset S9.
Cell Culture and Western Blot Analysis.
S2 cells were cultured in Schneider's insect medium (Sigma), and plasmids were transfected into the S2 cells using Lipofectamine 3000 (Invitrogen). Harvested cells were lysed in CelLytic M Cell Lysis Reagent (Sigma). The dissected FBs were lysed in breaking buffer [50 mM Tris (pH 7.4), 1% IGEPAL, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor mixture, 1× phosphatase inhibitor mixture] (Roche). Both protein extracts were used for Western blots, which were performed according to protocols described previously (15). Primary antibodies used in this study are shown in Dataset S10.
Determination of FFA Levels.
Fifteen mosquito FBs per time point were frozen in liquid nitrogen and ground in 1 mL of a 2% H2SO4/98% methanol solution. Samples were sealed with a cap and incubated at 80 °C for 1 h. After the addition of 0.3 mL of hexane and 1.5 mL of H2O, the fatty acid methyl esters were extracted into the hexane layer by shaking and then were centrifuged at 5,000 × g for 10 min. Samples of the organic phase were carried out by means of GC-MS on an Agilent Technologies 6890N GC-5973N mass selective detector. The GC was equipped with a HP-5MS column [60 mm × 0.25 mm (i.d.); film thickness 0.25 μm] (J&W Scientific). One-microliter samples were injected at a temperature of 280 °C; the GC-MS transfer line temperature was 280 °C, ion source 230 °C, and quadrupole 150 °C. All compounds were analyzed with 70 eV nominal electron energy and a scan range of 35–400 atomic mass units, with a solvent delay of 3 min. After injection, the column temperature was held at 50 °C for 0.5 min and then was increased to 200 °C at 5 °C/min, followed by an increase to 240 °C at 2 °C/min and then an increase to 250 °C at 5 °C/min and held for 10 min. Subsequently, the temperature was increased further to 280 °C at 3 °C/min and held for 3 min. Compounds were identified by comparing their retention time with those of authentic reference compounds and comparing the spectra with that of mass spectral library NIST02 (Rev. D.04.00; Agilent Technologies).
Lipid Droplet Staining.
Lipid was visualized by staining female mosquito FBs with Nile Red. Mosquitoes were dissected and washed two or three times with 1× PBS and then were incubated for 2 h in Nile Red mounting medium [20% (vol/vol) glycerol in PBS, with a 1:10,000 dilution of 10% (vol/vol) Nile Red in DMSO] at room temperature. After two or three washings with 1× PBS, FBs were mounted on glass slides and examined under a Zeiss LSM 710 confocal microscope at 63× magnification with an excitation wavelength of 543 nm and an emission wavelength of 626 nm. The size of LDs was measured using Zeiss ZEN software.
TAG, NADH, and Acetyl-CoA Measurements.
TAG was measured as described previously (15). Six mosquitoes were homogenized in 100 μL PBS containing 0.5% Tween-20 and were incubated at 70 °C for 5 min. Then, the samples were incubated with triglyceride reagent (Sigma) for 30 min at 37 °C. Following centrifugation, samples were transferred into 96-well plates, incubated with free glycerol reagent (Sigma) for 5 min at 37 °C, and then assayed using SpectraMax Plus384 with a wavelength of 540 nm.
For NADH measurements, six mosquitoes per sample were frozen in liquid nitrogen and ground in 5 mL PBS/bicarbonate/0.5% DTAB buffer; then 50 µL of each sample was transferred into 1.5-mL centrifuge tubes and heated in a water bath at 60–65 °C for 15 min. After cooling at room temperature for 10 min, 50 µL of a 1:1 mixture of 0.4 N HCl with 0.5 M Trizma base solution was added. Finally, 50 µL of the mixture was transferred into 96-well white assay plates to measure NADH with NAD/NADH-Glo luciferin detection reagent (Promega) using SpectraMax Plus 384. Determination of acetyl-CoA levels was conducted using the acetyl-coA assay kit (Sigma). Female mosquito samples were homogenized in ice-cold perchloric acid. After concentration, the supernatant was neutralized with 3 M potassium bicarbonate solution. After the pellet of potassium bicarbonate was removed, the remaining samples were brought to a final volume of 50 μL with acetyl-CoA assay buffer. To correct for background created by free CoA and succinyl-CoA, 10 μL of the acetyl-CoA quencher was used for each sample by incubation at room temperature for 5 min. After the quencher was removed with 2 μL quench remover, 50 μL of the acetyl-CoA assay reaction mix was added to each sample. Resulting samples were mixed either by using a horizontal shaker or by pipetting, and the reaction was incubated in darkness for 10 min at 37 °C. To detect acetyl-CoA, fluorescence intensity was measured using SpectraMax Plus384 with an excitation wavelength of 535 nm and an emission of 587 nm.
EMSA.
Drosophila S2 cells were transfected with pAc5.1b and pAc5.1b/HNF41-565 (V5-HNF4) using Lipofectamine 3000 (Invitrogen). At 48 h posttransfection, S2 cell nuclear protein extracts containing V5-HNF4 and empty vector were isolated using the NE-PER Nuclear and Cytoplasmic Extraction Reagents Kit (Thermo Scientific). The DNA probes (VLCAD: 5′-ATAGTGGGGCAAAAGTTCGAATCAG-3′; 3KCT: 5′-GAAGTTAATCAAAAGTCTCTAAAA-3′) were end-labeled with biotin and were purified by Sangon Biotech. EMSA was carried out using the Pierce LightShift Chemiluminescent EMSA Kit (Thermo Scientific). Nuclear protein extracts were incubated with biotin-labeled probes in binding buffer provided by the kit at room temperature for 30 min. For competition assays, 50× molar excess of unlabeled probes (50× cold probes) were added to the binding reagent. For supershift studies, the nuclear extracts were preincubated with anti-V5 (Invitrogen) antibody at 4 °C for 1 h before the addition of the labeled probes. The reaction mixtures were applied onto 6% (vol/vol) native polyacrylamide gels in 0.5× Tris-borate/EDTA buffer. The bands were detected according to the instructions provided with the EMSA kit.
Dual Luciferase Reporter Assay.
The sequence corresponding to the HNF4 coding region (amino acids1–565) was subcloned into the pAc5.1b/V5/His expression vector (Invitrogen); those corresponding to the promoter regions of VLCAD (−2759 to +23) and 3KCT (−781 to +24) were subcloned into the pGL4.10 vector (Promega) (Dataset S9). Afterward, pGL4.10-VLCAD−2759 to +23 and pGL4.10-3KCT−781 to +24 were cotransfected with pAc5.1b-HNF41–565 into Drosophila S2 cells and were kept at 27 °C in Schneider Drosophila medium supplemented with 10% (vol/vol) FBS. Cell transfections were carried out using Lipofectamine 3000 (Invitrogen). Luciferase activity was determined using the Dual-Luciferase Reporter Assay System and a GloMax 96 Microplate Luminometer (Promega) at 48 h posttransfection. The promoter sequences of VLCAD and 3KCT are shown in SI Results.
RNAi Experiments.
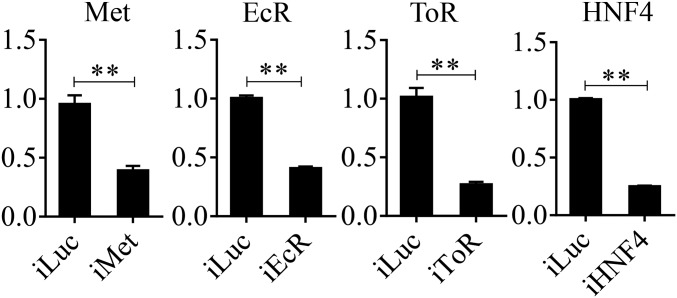
dsRNA was prepared using the T7 RiboMAX Express RNAi system (Promega) following the manufacturer’s instructions, as described previously (51). The bacterial luciferase gene was used to generate control Luc dsRNA. The corresponding dsRNAs were injected into the thorax of ice-anesthetized female mosquitoes within 12 h after emergence using a Nanoliter 2000 injector (World Precision Instruments). For Met RNAi, samples were collected 4 d after injection, and samples for EcR, HNF4, and TOR RNAi were collected at 36 h PBM. The efficiency of Met, EcR, HNF4, and TOR RNAi knockdown is shown in Fig. S8. All primers used for making dsRNA are listed in Dataset S9.
Fig. S8.
qRT-PCR determination of the RNAi efficiency of Met, EcR, HNF4, and TOR. Relative mRNA abundances of Met, EcR, HNF4, and TOR were measured in dsMet-, dsEcR-, dsHNF4-, and dsTOR-injected mosquitoes, respectively, and are represented on y axis of each figure. Error bars represent the SEM; **P < 0.01.
In Vivo Hormone Application and in Vitro FB Culture Experiments.
JH (10 μM) (Sigma) in acetone as solvent or acetone alone was topically applied to the abdomens of newly eclosed female mosquitoes (3 h PE) in the in vivo hormone application assays. 20E (Sigma), dissolved in ethanol and used at a final concentration of 1 ×10−6 M, was injected with or without amino acids into female mosquitoes at 72 h PE. Control samples were treated with the same volume of acetone and ethanol, respectively, instead of hormone. Amino acids used in this study were a mixture solution. All samples were collected at 24 h after treatment.
In vitro FB culture experiments and hormone applications were performed as described previously (15, 18). To test the effect of JH in vitro, FBs isolated from mosquitoes at 10 h PE were incubated in the medium containing 10 μM JH dissolved in solvent (acetone) or acetone alone for 8 h. To determine the effect of 20E in vitro, FBs were dissected from female mosquitoes at 72 h PE and were cultured in medium containing either amino acids or amino acids and 20E. 20E was added to culture medium in two steps to mimic a natural rise in the 20E titer: FBs were incubated first in the presence of 5 × 10−8 M 20E for 4 h and then in the presence of 10−6 M 20E for an additional 4 h. Controls were incubated for 8 h. Experiments were performed in triplicate under the same conditions.
Statistical Analysis.
The significance of RNA-seq data was calculated using a Wilcoxon signed rank test in R environment. In all other experiments, the mathematical significance of differences between samples was analyzed using the Student’s t test (GraphPad 6.0) and was expressed as a P value less than 0.05. All quantitative data are reported as mean ± SEM.
SI Results
The nucleotide promoter sequences of two selected LM genes, 3KCT and VLCAD, are shown. The 3KCT and VLCAD promoter sequences were cloned to the pGL4.10 vector to conduct the dual luciferase reporter assay. Probes used in the EMSA experiments are shown in bold. The core consensus HNF4-binding motifs are italicized.
>3KCT-Promoter (−781 to +24).
CAAAGTGGAATGAAGTCGATGTAGGAACTAAAACCATACTGTAGCCGATGAACACGATGAACTGCACAGTTGAGGAGGCAAGCCACATGCATACGAGATAGATTTATAACTACACTTACACGTAGGGGAGAAAACAGAGGCGAACTCACCTGGCCGGTATCTGTACCATACAGAAAGGTGTTGTGAAGAACATCGCAGAGAAAAAACGAGCATGGTAATGCTAAGTGACATACCAGTTTGTTATAACTTTTAACTTTTAAGTTCAAAAGTGTAGATACTAAAGAGTGTTTTTAAGGGCTTTCCTTTTAAAATCTTCCTAAAGAACGTGCAAAATCTCACGAACTTCAACCCTCAAAAGAAGGAAGAGTTATCTCTCGAGAATTGGCGTAGGAACAGGGGGGGCAGGGGGGCCTGGCCCCCTCCAGAGTCATTCATTCCGCGAAGTTAATCAAAAGTCTCTAAAAAATGTTGTCCTCAATATGGCTAACAAAAATAATTCTAAAGATAAGCCAAGGACTTTATACATTTTGATCTTCTCGGTGCAATTTTGATACAGATGTGTTATTAAATTGATAAGTTCCAACAACAAGGCAAATATTTGCTTATTTCACATGCTTGAAGTAAGTTTTCCAGAAATCATTTACTTACTTTACAATACTTGGTCCAGTACGTCTAAACACCGACTTCTTGATTTGAATTTCTCTATAGGAGCAATTCTCGCTGAAACCAGGCCGCCATCGGCACCCATCGTTAGAATTCCAATTTTATGTCACTGATCGCTAGTTTTCGATAAAACTTAAGGGTG.
>VLCAD-Promoter (−2759 to +23).
TATACCTGGAGCGTACGAGATAAACTAAAATTTGCAAATGTTGTTTCGTCCTTTGTCTAGAATATTATTTCAATTTTTCTACTTTGATTCTGGTTCTATCTTTCAAAACCCTCCCCCCCAACCCTAGATCCAAATGCTGGTTGCGCTACTGTGTGGTAATATCTGTAGTTTTGACGTAAAACATTGCCATCAGTACATTAACTTAAAATAGAAATCTTGAGTTTTTGAGCAAAACAAATCCTTACTTTTATGAGCCATAAAGTATCGCTTTTACTACCGCCCAACATACATATGAAAAATAGCGAAATTGAAAGATGAGAAGCACGCTTTTTTCTTATATGGACGTAATGCCGAAACGCAGGTGTATCATTTTGAGGTTGAAAAATCTGGTGGCCATCTTGGATTTCGACGCCATGCCATGTTTTTTTCGCAAACTGTATATCTAGGGGTCCACCAATTTGAGAAACCCGAAAGGACCACCCTTAAGTTTTATCGAAAATAGCGATCAATCAATAAAATAATTGTAATTCTAACGATGGGTGTTGATGGCGGGCTGGTTTCAGCGAGAATTGCTCAGTTCTTCTTCTCTTCTTCTTTCTGGCGTTACGTCCCAACTGAGACGAAGCCTGCTTCTCAGATTAATGTTCTTATGAGCACTTCCACAGTAATTAACTGAGAGCTTTCTTTGCCGATTGACCATTTTTGCATGTGTATATCGTGTGGCAGGTACGAAGCTACTCTATGCCCTGGGAATCGAGAAAATTTCCTTTACAAAAAGATCCTCGACCAGCGGGATTCGAACCCACGACCCTCAGCATGATCATGCTAAATAGCTGCGCGTTTACCACTACGGCTATCCGGGCCCAGTTATGGATCAAAAATAACTCGATTCCGAAAGAAACGCCGAAACGTATGCTTTCATTAACACACCATTCGTTTACCACGCAGATAAATTGTTGTTACAGTGTTTTGATCCATGATGTAAGTCGATTTAATCCAGAAAAAGAACCCTCCCCTCCCACTGATCCATATCTGTGGGATGTACAACGTTTGAAATTTCTATCGAAATGACCAATTTTTCATAAATAACTGCAAATTTTGAGGTGTACTCCCGAAAACAATTATAGTCTACACTGCCTTGGAGATATTTTTGTTTTGTACAATTTCACCATATGCGCTAGCTTCCAACATGACGGGAAGTCGCCGACGGTCATCTAGTTCCCGTTGAATAGCTGCACCACAACGGCTATTTGGGACTCAACATTTTATTAGACCCAATTTTCATTTCCGCACATGATGTTTTAAGGACCTACATTCCATTAAGTTTTTAAAATCAAATACAAGTAGCGGTATTGGAAAACGAAAATATTGCCTTCAAATTCGATGCACACAGCGCGGATAGTTCGGCACTTTTTGCTCTGATTTTTTTTTTTTTTTTTGCATAAGCAAAGATTTTCAAAGCTTCATACTTGTAGATCTGTAGAGTAAAGTGGGACAAAAGTTCGAGTGGAGCAAGAGTTTCTTTTTAAGATTTCTAGCTCATTTTAAAACAAAACTAATAAATGTCATGGTGGTTCGAGTGCTAGTCAAGTAAGAGACTTTCACTCCAAATCTCATAAAATTCAATTGAGATTTGGAAAAGTTATGGCTGTTTGTTGTTTTTTTTTATGTGAAAGTTGTAATGTTTGGTCGAACTTTTATTGCATGGAACCAAATAAACATACAATCTTTTTCGTTTTTTTTTGTTATGGCGTTTCTAGGCCTATCATAGAGATGCTTTGCCGTGTATTAGTTTTTTCATAAATGCTTGGAAACAAATTTTAGCCCATAGTGGGGCAAAAGTTCGAATCAGCGTGGCAAAAACTCATGGAAAAATTATCAAATTGCTTAAAATCCTAATATAATCTTTAAATTCAGCGAAATTTGCCTGATCGTGCGAAAAAAGTCACCAAAATTTCGAATTTTCATTTTGTTTGGGCAAAAGTTCGTTTAAGACAATCAATTTTAAAACTGTTATAACAAAAAATGGGTAATTATTTGGACAAAAGTTTGTTAAGCAAAATAAAGCCAGTATGTTGAAGATTCATTTTGTGGTATTTATTTCTGTTCAAATGCTGCTGTATTTCTGGAAATGTTGTTAAGATCACTAAGGTCGAACTTTTGCCCCACCTTACTCTATACTATATTCTTAGTCCTCTGGAGTTCATGGTAGGCACGACCAAGAATATGCATTACTTAAGATTAAACATGTCTAACTGACAAAGTAAAAGACCAAAACAAACCACATCATTTTGCGAAAATTATCTTTCCAATACACTATTTTCAGTTGAATCATCCTCACTAAATTTACGAGCTTCGTAAATTTGATTGACACAAAAAGGCTTTGTAAAACGAGATTTTAATGTATTTGGGAATCTGTTATTTTTTAATCATATTGGAATCCCTCTTCCTCTCCACTGATAATTTCCATCCTAAAATTTAACCTCTCTTATCTATAACTAATTGGAATAATCATGAAGCTTTGAACGACGTTCATAATAACCCAATTGGAATTCCTTCAACAGCCCAATCAAACAAAGAGCCAATTCATCGAGTTCACCACACAGGACGATAACGCACATACGGACAGAGATGAACCAGACGTTGCTTCTACGAGGGGGCCATGGATGTGTAATGGCTTTCGTATTGGATTGTTAGTTTTCTCTTTTGCGAAAAGCTTTAGCTTCCATAAAACCTTTACTATTACAAGCCGATCCGTCATTTGTCAACGAGCTCGTACAGT.
Supplementary Material
Acknowledgments
This work was supported by Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB11030600, National Basic Research Program of China Grant 2014CB138405, National Science Foundation of China Grants 31672291 and L1524009 (to Z.Z.), and National Institutes of Health/National Institute of Allergy and Infectious Diseases Award 5R01 AI036959 (to A.S.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619326114/-/DCSupplemental.
References
- 1.Weaver SC, et al. Zika virus: History, emergence, biology, and prospects for control. Antiviral Res. 2016;130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsetsarkin KA, Chen R, Weaver SC. Interspecies transmission and chikungunya virus emergence. Curr Opin Virol. 2016;16:143–150. doi: 10.1016/j.coviro.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fauci AS, Morens DM. Zika virus in the Americas--Yet another arbovirus threat. N Engl J Med. 2016;374(7):601–604. doi: 10.1056/NEJMp1600297. [DOI] [PubMed] [Google Scholar]
- 4.Barrett AD, Higgs S. Yellow fever: A disease that has yet to be conquered. Annu Rev Entomol. 2007;52:209–229. doi: 10.1146/annurev.ento.52.110405.091454. [DOI] [PubMed] [Google Scholar]
- 5.Reynolds JA, Poelchau MF, Rahman Z, Armbruster PA, Denlinger DL. Transcript profiling reveals mechanisms for lipid conservation during diapause in the mosquito, Aedes albopictus. J Insect Physiol. 2012;58(7):966–973. doi: 10.1016/j.jinsphys.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou G, Miesfeld RL. Energy metabolism during diapause in Culex pipiens mosquitoes. J Insect Physiol. 2009;55(1):40–46. doi: 10.1016/j.jinsphys.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheon HM, Shin SW, Bian G, Park JH, Raikhel AS. Regulation of lipid metabolism genes, lipid carrier protein lipophorin, and its receptor during immune challenge in the mosquito Aedes aegypti. J Biol Chem. 2006;281(13):8426–8435. doi: 10.1074/jbc.M510957200. [DOI] [PubMed] [Google Scholar]
- 8.Rono MK, Whitten MM, Oulad-Abdelghani M, Levashina EA, Marois E. The major yolk protein vitellogenin interferes with the anti-plasmodium response in the malaria mosquito Anopheles gambiae. PLoS Biol. 2010;8(7):e1000434. doi: 10.1371/journal.pbio.1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta L, et al. Apolipophorin-III mediates antiplasmodial epithelial responses in Anopheles gambiae (G3) mosquitoes. PLoS One. 2010;5(11):e15410. doi: 10.1371/journal.pone.0015410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perera R, et al. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 2012;8(3):e1002584. doi: 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunning KR, et al. Beta-oxidation is essential for mouse oocyte developmental competence and early embryo development. Biol Reprod. 2010;83(6):909–918. doi: 10.1095/biolreprod.110.084145. [DOI] [PubMed] [Google Scholar]
- 12.Dunning KR, Russell DL, Robker RL. Lipids and oocyte developmental competence: The role of fatty acids and β-oxidation. Reproduction. 2014;148(1):R15–R27. doi: 10.1530/REP-13-0251. [DOI] [PubMed] [Google Scholar]
- 13.Van Handel E. Fuel metabolism of the mosquito (Culex quinquefasciatus) embryo. J Insect Physiol. 1993;39(10):831–833. [Google Scholar]
- 14.Ziegler R, Ibrahim MM. Formation of lipid reserves in fat body and eggs of the yellow fever mosquito, Aedes aegypti. J Insect Physiol. 2001;47(6):623–627. doi: 10.1016/s0022-1910(00)00158-x. [DOI] [PubMed] [Google Scholar]
- 15.Hou Y, et al. Temporal coordination of carbohydrate metabolism during mosquito reproduction. PLoS Genet. 2015;11(7):e1005309. doi: 10.1371/journal.pgen.1005309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown DA. Lipid droplets: Proteins floating on a pool of fat. Curr Biol. 2001;11(11):R446–R449. doi: 10.1016/s0960-9822(01)00257-3. [DOI] [PubMed] [Google Scholar]
- 17.Martin S, Parton RG. Lipid droplets: A unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7(5):373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 18.Roy S, et al. Regulation of gene expression patterns in mosquito reproduction. PLoS Genet. 2015;11(8):e1005450. doi: 10.1371/journal.pgen.1005450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou Z, et al. Juvenile hormone and its receptor, methoprene-tolerant, control the dynamics of mosquito gene expression. Proc Natl Acad Sci USA. 2013;110(24):E2173–E2181. doi: 10.1073/pnas.1305293110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao B, et al. Determination of juvenile hormone titers by means of LC-MS/MS/MS and a juvenile hormone-responsive Gal4/UAS system in Aedes aegypti mosquitoes. Insect Biochem Mol Biol. 2016;77:69–77. doi: 10.1016/j.ibmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saha TT, et al. Hairy and Groucho mediate the action of juvenile hormone receptor Methoprene-tolerant in gene repression. Proc Natl Acad Sci USA. 2016;113(6):E735–E743. doi: 10.1073/pnas.1523838113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Mead EA, Zhu J. Heterodimer of two bHLH-PAS proteins mediates juvenile hormone-induced gene expression. Proc Natl Acad Sci USA. 2011;108(2):638–643. doi: 10.1073/pnas.1013914108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy S, et al. Regulation of reproductive processes in female mosquitoes. In: Alexander SR, editor. Advances in Insect Physiology. Vol 51. Academic; 2016. pp. 115–144. [Google Scholar]
- 24.Koelle MR, et al. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell. 1991;67(1):59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- 25.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 28.Palanker L, Tennessen JM, Lam G, Thummel CS. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab. 2009;9(3):228–239. doi: 10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen IA, Attardo GM, Park JH, Peng Q, Raikhel AS. Target of rapamycin-mediated amino acid signaling in mosquito anautogeny. Proc Natl Acad Sci USA. 2004;101(29):10626–10631. doi: 10.1073/pnas.0403460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Attardo GM, Hansen IA, Raikhel AS. Nutritional regulation of vitellogenesis in mosquitoes: Implications for anautogeny. Insect Biochem Mol Biol. 2005;35(7):661–675. doi: 10.1016/j.ibmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Park JH, Attardo GM, Hansen IA, Raikhel AS. GATA factor translation is the final downstream step in the amino acid/target-of-rapamycin-mediated vitellogenin gene expression in the anautogenous mosquito Aedes aegypti. J Biol Chem. 2006;281(16):11167–11176. doi: 10.1074/jbc.M601517200. [DOI] [PubMed] [Google Scholar]
- 32.Roy SG, Raikhel AS. The small GTPase Rheb is a key component linking amino acid signaling and TOR in the nutritional pathway that controls mosquito egg development. Insect Biochem Mol Biol. 2011;41(1):62–69. doi: 10.1016/j.ibmb.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy SG, Raikhel AS. Nutritional and hormonal regulation of the TOR effector 4E-binding protein (4E-BP) in the mosquito Aedes aegypti. FASEB J. 2012;26(3):1334–1342. doi: 10.1096/fj.11-189969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Attardo GM, Hansen IA, Shiao SH, Raikhel AS. Identification of two cationic amino acid transporters required for nutritional signaling during mosquito reproduction. J Exp Biol. 2006;209(Pt 16):3071–3078. doi: 10.1242/jeb.02349. [DOI] [PubMed] [Google Scholar]
- 35.Zhu J, Noriega FG. The role of juvenile hormone in mosquito development and reproduction. In: Alexander SR, editor. Advances in Insect Physiology. Vol 51. Academic; 2016. pp. 93–113. [Google Scholar]
- 36.Ashok M, Turner C, Wilson TG. Insect juvenile hormone resistance gene homology with the bHLH-PAS family of transcriptional regulators. Proc Natl Acad Sci USA. 1998;95(6):2761–2766. doi: 10.1073/pnas.95.6.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Xu J, Sheng Z, Sui Y, Palli SR. Steroid receptor co-activator is required for juvenile hormone signal transduction through a bHLH-PAS transcription factor, methoprene tolerant. J Biol Chem. 2011;286(10):8437–8447. doi: 10.1074/jbc.M110.191684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui Y, Sui Y, Xu J, Zhu F, Palli SR. Juvenile hormone regulates Aedes aegypti Krüppel homolog 1 through a conserved E box motif. Insect Biochem Mol Biol. 2014;52:23–32. doi: 10.1016/j.ibmb.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin SW, Zou Z, Saha TT, Raikhel AS. bHLH-PAS heterodimer of methoprene-tolerant and Cycle mediates circadian expression of juvenile hormone-induced mosquito genes. Proc Natl Acad Sci USA. 2012;109(41):16576–16581. doi: 10.1073/pnas.1214209109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charles JP, et al. Ligand-binding properties of a juvenile hormone receptor, Methoprene-tolerant. Proc Natl Acad Sci USA. 2011;108(52):21128–21133. doi: 10.1073/pnas.1116123109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulman IG. Nuclear receptors as drug targets for metabolic disease. Adv Drug Deliv Rev. 2010;62(13):1307–1315. doi: 10.1016/j.addr.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonoda J, Pei L, Evans RM. Nuclear receptors: Decoding metabolic disease. FEBS Lett. 2008;582(1):2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagy L, Schwabe JW. Mechanism of the nuclear receptor molecular switch. Trends Biochem Sci. 2004;29(6):317–324. doi: 10.1016/j.tibs.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 44.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: Opening the X-files. Science. 2001;294(5548):1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 45.Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med. 2004;10(4):355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 46.Repa JJ, Mangelsdorf DJ. The liver X receptor gene team: Potential new players in atherosclerosis. Nat Med. 2002;8(11):1243–1248. doi: 10.1038/nm1102-1243. [DOI] [PubMed] [Google Scholar]
- 47.Grönke S, et al. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1(5):323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 48.Fang B, Mane-Padros D, Bolotin E, Jiang T, Sladek FM. Identification of a binding motif specific to HNF4 by comparative analysis of multiple nuclear receptors. Nucleic Acids Res. 2012;40(12):5343–5356. doi: 10.1093/nar/gks190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou Z, et al. Transcriptome analysis of Aedes aegypti transgenic mosquitoes with altered immunity. PLoS Pathog. 2011;7(11):e1002394. doi: 10.1371/journal.ppat.1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanehisa M, et al. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34(Database issue):D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang YH, et al. A critical role for CLSP2 in the modulation of antifungal immune response in mosquitoes. PLoS Pathog. 2015;11(6):e1004931. doi: 10.1371/journal.ppat.1004931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.