Significance
The onset of androgen resistance is a major impediment in treating prostate cancer (PCa). However, the underlying molecular mechanisms are not fully understood. Here, we integrate multiple biophysical approaches that report conformational preferences of the intrinsically disordered protein (IDP) Prostate-Associated Gene 4 (PAGE4) with human cancer biology and nonlinear dynamics. Based on our biophysical, biochemical, and cellular data, a mathematical model is presented that suggests a mechanism by which phosphorylation-induced conformational changes in PAGE4 can switch from an androgen-dependent phenotype to an androgen-independent phenotype in PCa cells. The study underscores how IDPs that engage in multiple “promiscuous” interactions due to their conformational dynamics when overexpressed or aberrantly expressed can stochastically orchestrate phenotypic heterogeneity in PCa.
Keywords: intrinsic disorder, androgen resistance, prostate cancer, PAGE-4, phenotypic heterogeneity
Abstract
Intrinsically disordered proteins (IDPs) that lack a unique 3D structure and comprise a large fraction of the human proteome play important roles in numerous cellular functions. Prostate-Associated Gene 4 (PAGE4) is an IDP that acts as a potentiator of the Activator Protein-1 (AP-1) transcription factor. Homeodomain-Interacting Protein Kinase 1 (HIPK1) phosphorylates PAGE4 at S9 and T51, but only T51 is critical for its activity. Here, we identify a second kinase, CDC-Like Kinase 2 (CLK2), which acts on PAGE4 and hyperphosphorylates it at multiple S/T residues, including S9 and T51. We demonstrate that HIPK1 is expressed in both androgen-dependent and androgen-independent prostate cancer (PCa) cells, whereas CLK2 and PAGE4 are expressed only in androgen-dependent cells. Cell-based studies indicate that PAGE4 interaction with the two kinases leads to opposing functions. HIPK1-phosphorylated PAGE4 (HIPK1-PAGE4) potentiates c-Jun, whereas CLK2-phosphorylated PAGE4 (CLK2-PAGE4) attenuates c-Jun activity. Consistent with the cellular data, biophysical measurements (small-angle X-ray scattering, single-molecule fluorescence resonance energy transfer, and NMR) indicate that HIPK1-PAGE4 exhibits a relatively compact conformational ensemble that binds AP-1, whereas CLK2-PAGE4 is more expanded and resembles a random coil with diminished affinity for AP-1. Taken together, the results suggest that the phosphorylation-induced conformational dynamics of PAGE4 may play a role in modulating changes between PCa cell phenotypes. A mathematical model based on our experimental data demonstrates how differential phosphorylation of PAGE4 can lead to transitions between androgen-dependent and androgen-independent phenotypes by altering the AP-1/androgen receptor regulatory circuit in PCa cells.
Contrary to conventional wisdom that structure defines protein function (1), it is now increasingly evident that a large fraction of the human proteome is composed of intrinsically disordered proteins (IDPs) lacking rigid 3D structure (2–4). IDPs exist as conformational ensembles that are highly malleable, facilitating their interactions with multiple partners. These interactions are “wired” to form scale-free networks (5, 6) that represent the main conduit of information flow in the cell. Furthermore, the organization and properties of such protein interaction networks are evolutionarily conserved, underscoring their functional significance (7).
By occupying hub positions in such networks, IDPs play critical roles in many biological processes, such as transcription, translation, and signaling (8, 9). IDPs also participate in higher order phenomena, such as regulation of the cell division cycle (10–12), circadian rhythmicity (13, 14), and phenotypic plasticity (15, 16). Recent evidence suggests that several IDPs can act in a prion-like manner to create protein-based molecular memories that drive the emergence and inheritance of biological traits (17), further emphasizing their importance in state (or phenotype) switching. Moreover, if overexpressed, IDPs have the potential to engage in multiple “promiscuous” interactions with other proteins, which can lead to changes in phenotype and pathological states (18, 19). Therefore, there is considerable interest in understanding how IDPs interact with their biological targets.
The most prevalent view is that IDPs can undergo transitions from disordered conformational ensembles to folded structures upon binding to their cognate targets, a phenomenon referred to as “coupled folding and binding” (20). Two conceptually opposing ideas have been advanced to explain this phenomenon (21). Whereas the “induced fit” mechanism envisages folding of the IDP after association with the target, the “conformational selection” mechanism suggests that all potential conformations of the ensemble exist a priori and the ligand selects the most favored prefolded state from these conformations. However, as observed in the case of pKID/KIX and KIX/Myb interactions (22) and those interactions involving the C-terminal domain of the measles virus nucleoprotein (23), some combination of these two extremes may also be possible, suggesting that the intrinsic secondary structure propensities of the IDPs determine their binding mechanisms. Furthermore, some IDPs can shift the overall conformation of their ensembles while remaining disordered (24). Thus, some IDPs may be only marginally unstable and can be tipped to populate preferred conformations to become functionally active. Such changes in the structural ensemble sampled by the IDPs are similar conceptually to the conformational (fold) switching events seen in some marginally stable (“metamorphic”) folded proteins in response to mutation or environmental triggers that result in new functions (25). On the other hand, some IDPs, such as the nematode anhydrin (26), an N-terminal region of the human c-Myc (27), and the human Prostate-Associated Gene 4 (PAGE4) (28), that have important and pleotropic functions have been shown to remain largely disordered even while interacting with their biological targets.
PAGE4 is a cancer/testis antigen that is intrinsically disordered (>95%) (29) and is remarkably prostate-specific in the human male (30). It has the hallmarks of a protooncogene, with important roles in development and differentiation. Thus, the protein is highly expressed in the human fetal prostate, trophoblasts, and placenta (31, 32), as well as in reprogrammed induced pluripotent stem cells, but it is undetectable in the parent somatic cells used for reprogramming (reviewed in ref. 33). However, PAGE4 protein expression is elevated in prostate cancer (PCa) (31, 34–36), where it is up-regulated in response to a variety of stress factors (29, 31). PAGE4 is phosphorylated at S9 and T51 by Homeodomain-Interacting Protein Kinase 1 (HIPK1), a component of the cellular stress–response pathway, and functions as a strong potentiator of the oncoprotein c-Jun (32, 37). c-Jun is also a component of the stress–response pathway, and forms a heterodimer with members of the Fos family to produce Activator Protein-1 (AP-1). This group of early response transcription factors represents a paradigm for signal-responsive factors with important roles in the control of cell growth, apoptosis, and stress response (38). Phosphorylation at T51 is necessary because mutation of this residue to an alanine abolishes its activity by >90% (37).
By NMR spectroscopy, we had observed that although PAGE4 occupies a very heterogeneous conformational space, the polypeptide has local and long-range conformational preferences that are perturbed by phosphorylation, resulting in population shifts and altered intramolecular conformational dynamics (28). Thus, the population of transient turn-like structures increases upon phosphorylation in an ∼20-residue acidic region centered on T51. Further, consistent with our single-molecule fluorescence resonance energy transfer (smFRET) microscopy results (37), this central region becomes more compact and more negatively charged upon phosphorylation with increasing intramolecular contacts to basic sequence motifs near the N and C termini. Furthermore, we observed that although flexibility is decreased in the central region of the phosphorylated ensemble, the polypeptide chain remains disordered and highly dynamic overall. PAGE4 uses a transient helical structure adjacent to the central acidic region to bind c-Jun in vitro.
In this paper, we report that PAGE4 is also phosphorylated by CDC-Like Kinase 2 (CLK2), a dual-specificity kinase that phosphorylates serine-rich (SR) proteins of the spliceosome complex (39, 40). We show that in contrast to HIPK1, CLK2 phosphorylates PAGE4 at multiple S/T residues, including S9 and T51. Furthermore, we demonstrate that HIPK1 and CLK2 are differentially expressed in PCa cells that are either androgen-dependent or -independent, and that both kinases are up-regulated in PCa. Using multiple biophysical and biochemical techniques, we determined the conformational preferences of the differentially phosphorylated PAGE4 ensembles. Using a cell-based reporter assay, we demonstrate that phosphorylation of PAGE4 by HIPK1 and CLK2 has opposing functions with regard to c-Jun transactivation. Thus, in response to differential phosphorylation by HIPK1 and CLK2, PAGE4 is remodeled to populate distinct conformational ensembles with separate cellular functions. Overall, our results suggest that the phosphorylation-induced conformational dynamics of PAGE4 may play a role in modulating phenotypic plasticity in PCa (Fig. 1). Using mathematical modeling based on our experimental data, we demonstrate how differential phosphorylation of PAGE4 can lead to transitions between androgen-dependent and androgen-independent phenotypes by altering the AP-1/androgen receptor (AR) regulatory circuit in PCa cells.
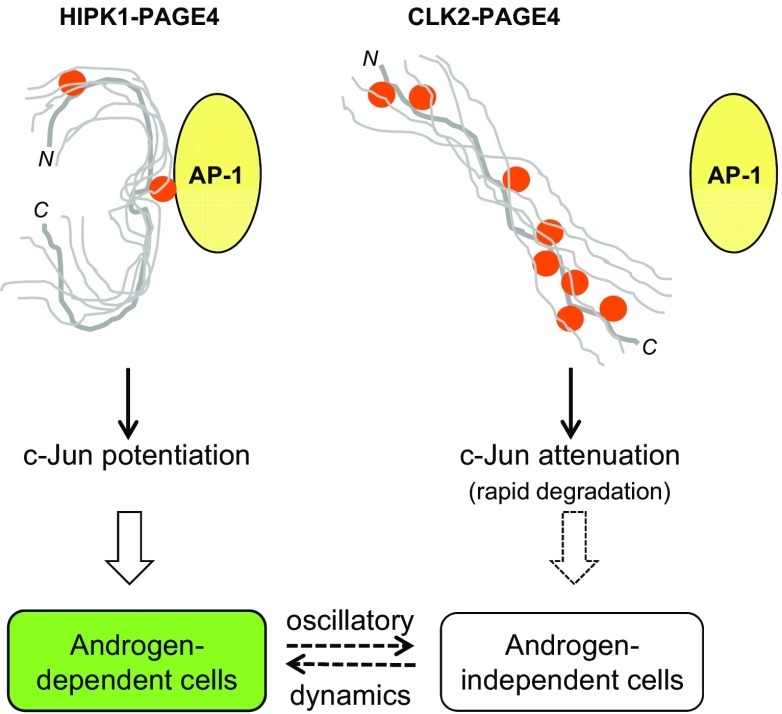
Fig. 1.
PAGE4 conformational dynamics and phenotypic heterogeneity in PCa cells. The stress–response kinase HIPK1 phosphorylates PAGE4 at S9 and T51, resulting in a relatively compact PAGE4 ensemble that can potentiate c-Jun (AP-1) in androgen-dependent PCa cells such as LNCaP. In contrast, the dual-specificity kinase CLK2 hyperphosphorylates PAGE4 at eight different S/T residues, including S9 and S51, leading to a more random-like PAGE4 ensemble that attenuates c-Jun transactivation and is likely to be degraded rapidly (9, 37, 77). Differential phosphorylation of PAGE4 by HIPK1 and CLK2 results in oscillations of the levels of HIPK1-PAGE4, CLK2-PAGE4, and CLK2 that can be modeled mathematically to correlate with the experimentally observed heterogeneity in a population of isogenic PCa cells (details are provided in the main text). Phosphorylated residues are indicated as solid orange circles.
Results
Identification of a Kinase CLK2 Acting on PAGE4.
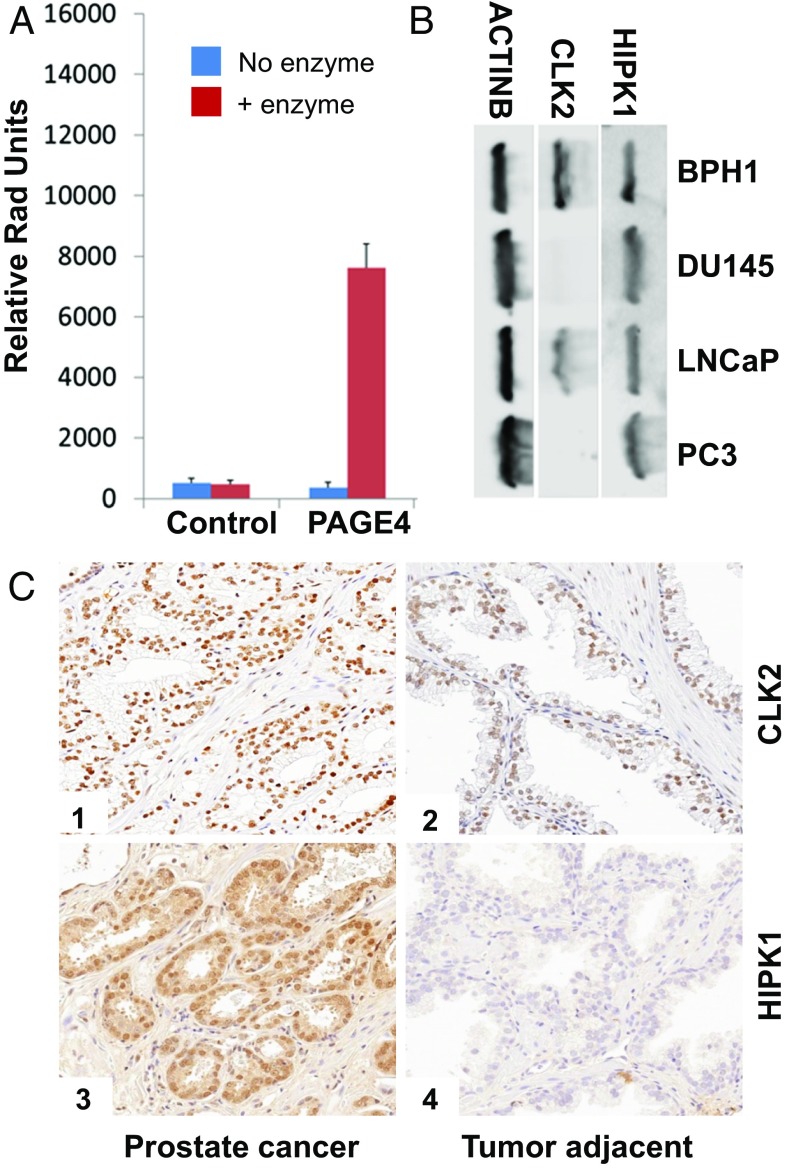
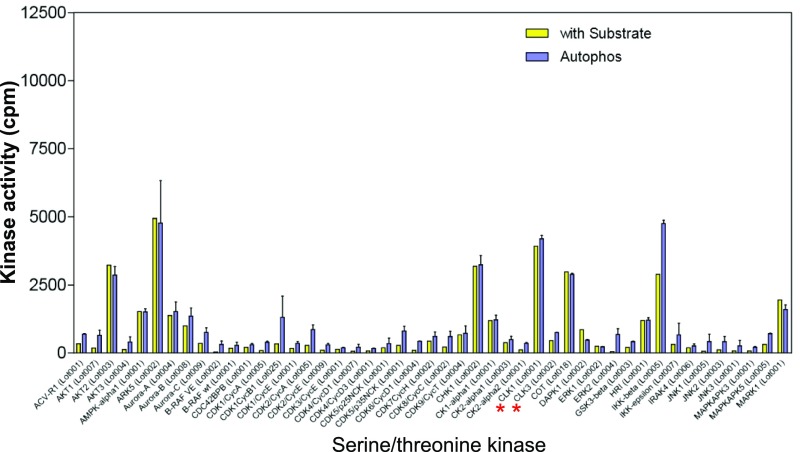
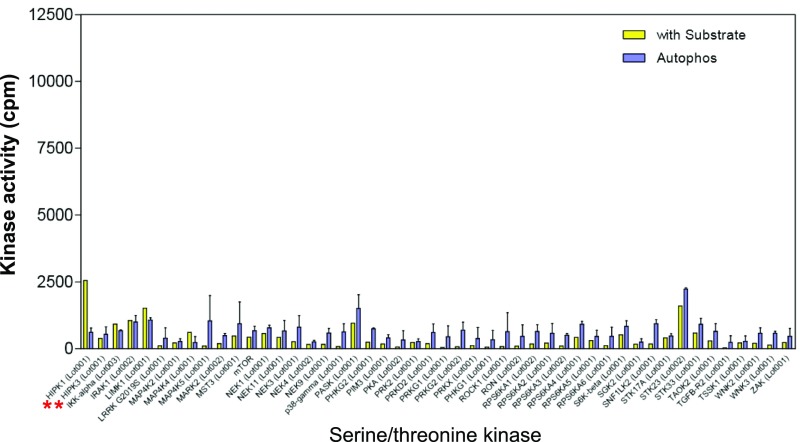
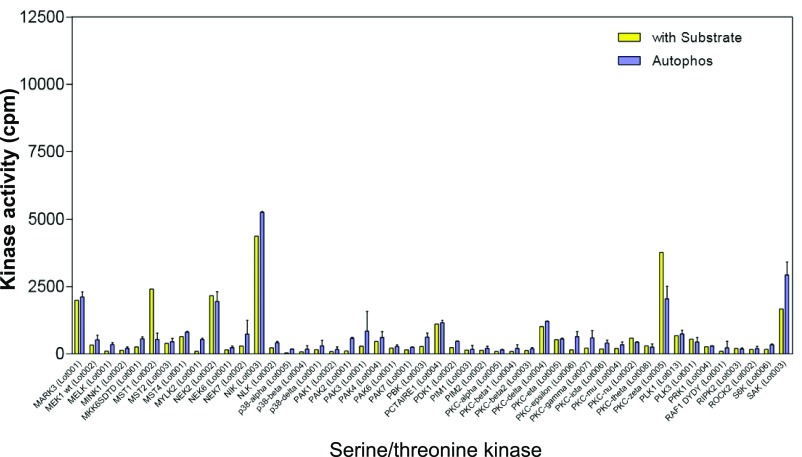
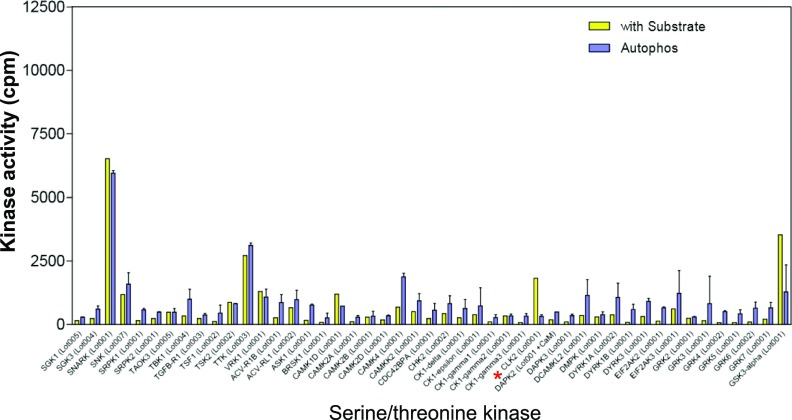
From a panel of 190 S/T kinases (Dataset S1), we identified CLK2 as a second kinase that phosphorylates PAGE4. CLK2 is a dual-specificity kinase known to phosphorylate SR proteins that are components of spliceosomes (40). As shown in Fig. 2A, phosphorylation was only observed in the presence of recombinant PAGE4 as a substrate and not when it was omitted from the in vitro reaction. The data also ruled out the possibility that the increase in phosphorylation is due to autophosphorylation of CLK2. Of note, the related kinases HIPK3, CLK1, and CLK3 that were part of the kinase panel do not phosphorylate PAGE4, highlighting the specificity of this posttranslational modification (Figs. S1–S4).
Fig. 2.
CLK2 activity and expression profile compared with HIPK1. (A) CLK2 phosphorylates PAGE4 in vitro. No enzyme control (blue) and the presence of enzyme (red) are shown. (B) Immunoblotting using specific antibodies against HIPK1, CLK2, and beta-actin as described in Materials and Methods. (C) Detection of CLK2 and HIPK1 protein expression in prostate tissue by qIHC in 80 paired cases of PCa and benign adjacent tissue obtained from radical prostatectomies. (Magnification: 20×, 0.5 μm/pixel resolution.)
Fig. S1.
Serine/threonine kinase screening. The 190 serine/threonine kinases screened are listed in Dataset S1. Enzyme assays using recombinant PAGE4 protein as the substrate and radioactive ATP were performed as described in Materials and Methods. The red asterisks indicate CLK1 and CLK3 in the panel.
Fig. S4.
Serine/threonine kinase screening. The 190 serine/threonine kinases screened are listed in Dataset S1. Enzyme assays using recombinant PAGE4 protein as the substrate and radioactive ATP were performed as described in Materials and Methods. The red asterisks indicate HIPK1 and HIPK3 in the panel.
Fig. S2.
Serine/threonine kinase screening. The 190 serine/threonine kinases screened are listed in Dataset S1. Enzyme assays using recombinant PAGE4 protein as the substrate and radioactive ATP were performed as described in Materials and Methods.
Fig. S3.
Serine/threonine kinase screening. The 190 serine/threonine kinases screened are listed in Dataset S1. Enzyme assays using recombinant PAGE4 protein as the substrate and radioactive ATP were performed as described in Materials and Methods. The red asterisk indicates CLK2 in the panel.
HIPK1 and CLK2 Are Differentially Expressed in Androgen-Dependent and Androgen-Independent PCa Cells.
PAGE4 is up-regulated in primary (organ-confined or localized), androgen-dependent PCa but not in advanced (metastatic), androgen-independent disease both at the mRNA level and at the protein level (31, 35, 36). Therefore, to discern the relevance of the posttranslational modification of PAGE4 by the two kinases in PCa, we determined their expression in both PCa cell lines and a benign prostatic hyperplasia (BPH) cell line. Three different PCa cell lines (LNCaP, DU145, and PC3) that are either dependent or independent on androgen and have different propensities for aggressiveness were examined. In addition, BPH-1, a nontumorigenic cell line established from a patient with BPH, was interrogated. When whole-cell lysates from these cell lines were probed with specific antibodies by immunoblotting (Fig. 2B), HIPK1 protein was detected at almost the same level in BPH-1 cells and in the LNCaP cells that are androgen-dependent and nonaggressive, as well as in the androgen-independent and aggressive DU145 and PC3 cells. In contrast, CLK2 was expressed only in BPH-1 cells and in the androgen-dependent LNCaP cells, which parallels the expression pattern of PAGE4 (35).
HIPK1 and CLK2 Are Up-Regulated in Primary, Androgen-Dependent PCa.
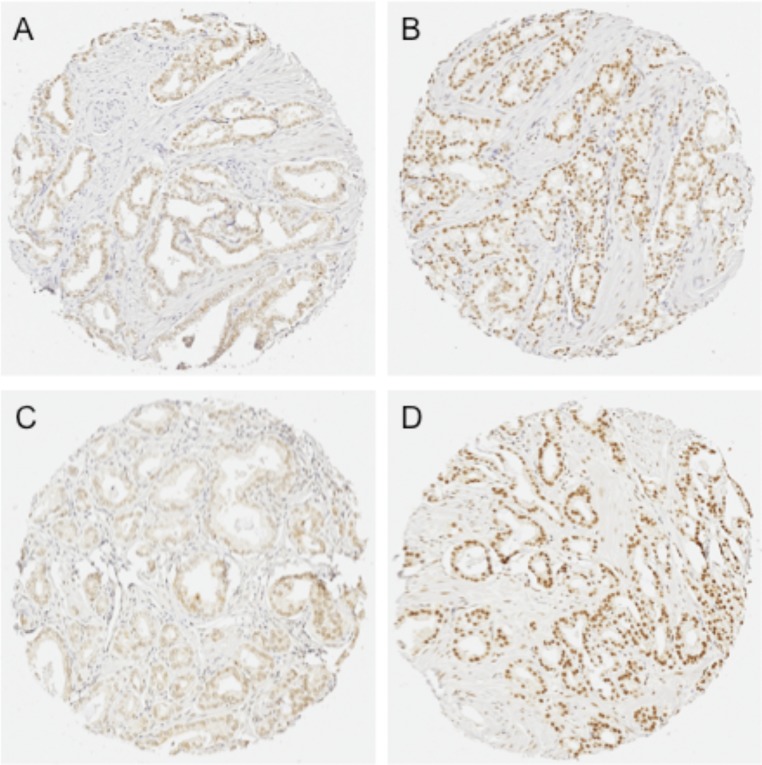
To corroborate the cell line expression patterns in human prostate tissue, quantitative immunohistochemistry (qIHC) was performed with tissue microarrays (TMAs) that were constructed using 80 paired cases of localized PCa and adjacent normal tissue obtained from radical prostatectomies. The same antibodies that were used for immunoblotting were used to stain the TMAs. As can been seen from Fig. 2, both CLK2 (Fig. 2 C, 1 and 2) and HIPK1 (Fig. 2 C, 3 and 4) were up-regulated in primary PCa compared with the adjacent normal prostate tissue. CLK2 showed strong positive nuclear staining, whereas HIPK1 localization was both cytoplasmic and nuclear. Both proteins were restricted to the epithelial cells, the main cell type afflicted by PCa, and no staining was noted in basal and/or tumor stromal cells.
Fig. 3.
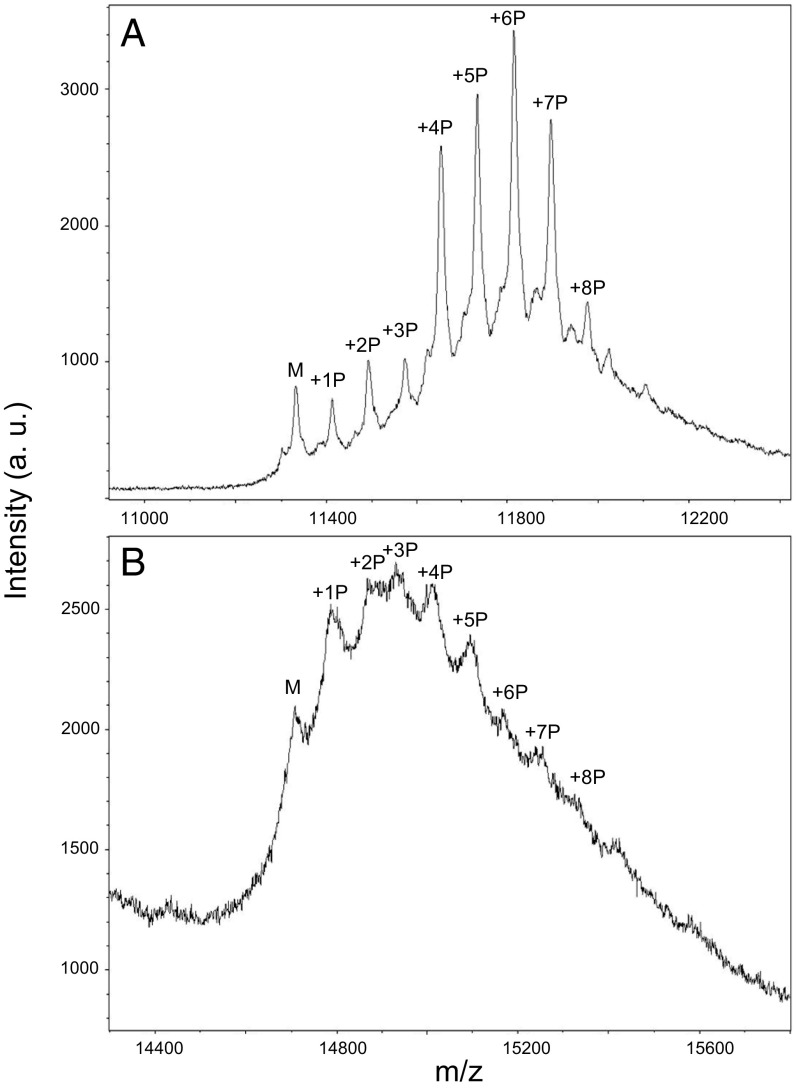
Characterization of CLK2-phosphorylated PAGE4 by MALDI mass spectrometry. (A) Mass spectrum of CLK2-phosphorylated PAGE4 obtained by coexpression in E. coli BL21DE3 cells. (B) Mass spectrum of Myc-DDK–tagged PAGE4 isolated by immunoprecipitation from LNCaP cells. a.u., arbitrary units.
Fig. 4.
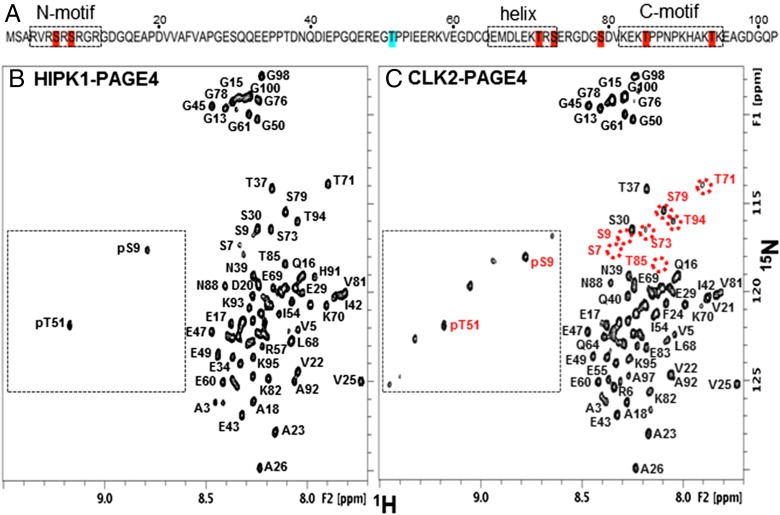
PAGE4 phosphorylation sites from NMR. (A) PAGE4 amino acid sequence highlighting the primary HIPK1 phosphorylation site (blue) and CLK2 phosphorylation sites (blue and red). Boxed regions denote key structural features. (B) Two-dimensional 1H-15N HSQC spectrum of HIPK1-PAGE4 with assignments for backbone amides. Assignments in crowded regions are omitted for clarity. (C) Two-dimensional 1H-15N HSQC spectrum of CLK2-PAGE4 with tentative assignments for likely CLK2 phosphorylation sites (red). Circled positions highlight where there is a significant loss of peak intensity relative to HIPK1-PAGE4, indicating CLK2 phosphorylation at that residue.
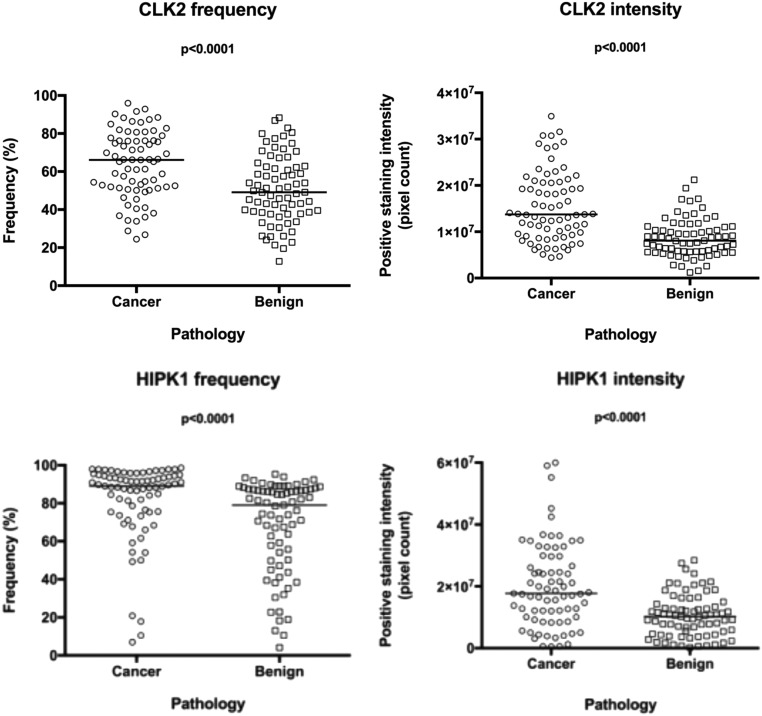
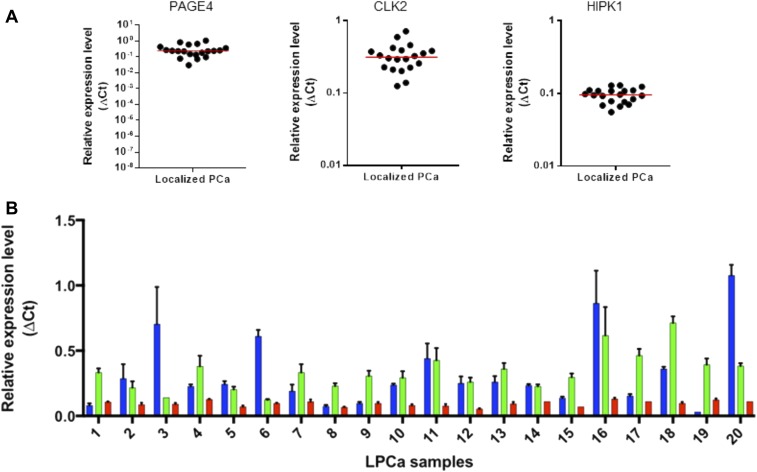
More importantly, however, we observed significant heterogeneity in the staining pattern and quantified these differences. In many cases, we observed stronger intensity in some regions (cores in the TMA) than in other regions of the same tumor, but we also detected different levels of protein expression in different samples. Such differences in intensity and frequency of positive staining were observed over a wide range both between the two groups of samples (cancer vs. benign) as well as within the same group (either cancer or benign) (Fig. S5). In addition, CLK2 stained differentially in different regions of the tumor from the same patient (Fig. S6). This heterogeneity in CLK2 (and HIPK1) expression in terms of both intensity and frequency is also captured in the same patient (Dataset S2). Consistent with the immunohistochemistry data, heterogeneity in the levels of HIPK1, CLK2, and PAGE4 was also confirmed at the mRNA level by quantitative RT-PCR (qRT-PCR) in 20 independent clinical samples of organ-confined PCa (Fig. S7). Taken together, the data underscore the differential expression, as well as up-regulation, of the two kinases in PCa. Furthermore, the scatter plots and the individual measurements for each core from the samples included in the TMA clearly demonstrate that CLK2 and HIPK1 expression is very heterogeneous and that the differences are significant, especially when evaluating different regions (cores) from the sample tumor.
Fig. S5.
Scatter plots representing frequency and intensity of the immunohistochemistry for protein detection. CLK2 and HIPK1 positivity was quantified, and the values are expressed in frequency (%) of positive cells and intensity (pixel count) of the positive signal. CLK2 and HIPK1 are significantly more expressed in the cancer areas than in the benign adjacent prostate tissue. Lines representing the median and P values were obtained by using the Wilcoxon nonparametric test for paired samples.
Fig. S6.
CLK2 protein expression in distinct regions from the same PCa sample. The upper (A and B) and lower (C and D) pairs of pictures represent two patients. In some of the cancer samples evaluated, it was possible to observe that different cores (distinct regions of the same tumor) presented differences in CLK2 levels as revealed by the intensity of staining. (Magnification: 20×, 0.5 μm/pixel resolution.)
Fig. S7.
Relative expression of PAGE4, CLK2, and HIPK1 mRNA in organ-confined PCa determined by qRT-PCR. (A) Scatter plots represent relative gene expression in samples from men with organ-confined (localized) prostate cancer (LPCa). (B) Histogram represents PAGE4 (blue), CLK2 (green), and HIPK1 (red) expression. Error bars represent SD, and qRT-PCR assays were performed in triplicate. ΔCt is the target gene expression (CLK2, HIPK1, and PAGE4) relative to GAPDH expression, where ΔCt = (Ct target gene – Ct GAPDH). Ct is the PCR cycle at which the amplification reaches the established threshold.
CLK2 Hyperphosphorylates PAGE4.
Having determined that PAGE4 is a substrate for CLK2 kinase and that CLK2 mirrors PAGE4 expression in PCa cells, we set out to elucidate the pattern of CLK2-mediated phosphorylation in PAGE4. As in our previous work on phosphorylation of PAGE4 with HIPK1 (HIPK1-PAGE4) (28, 37), we found that coexpression of the CLK2 kinase domain with PAGE4 in Escherichia coli BL21DE3 cells provided the most efficient route to obtaining CLK2-phosphorylated PAGE4 (CLK2-PAGE4). Mass spectrometric analysis of the resulting phosphorylated material by MALDI showed that CLK2 phosphorylates PAGE4 at a minimum of eight distinct sites (Fig. 3A). This finding is in contrast to HIPK1, which phosphorylates only at the S9 and T51 positions in PAGE4. The phosphorylation of PAGE4 in PCa cells was also investigated. Myc-DDK–tagged PAGE4 was overexpressed in LNCaP cells that express CLK2 endogenously (Fig. 2B), and the resulting modified PAGE4 was then isolated and purified by immunoprecipitation. The mass spectrum of the tagged PAGE4 from LNCaP cells indicated that multiple sites are also phosphorylated in vivo (Fig. 3B), although the distribution of these phosphorylated species is somewhat different from the distribution in the E. coli experiment.
The specific sites for CLK2 phosphorylation (Fig. 4A) were readily determined by comparing the 2D 1H-15N heteronuclear single-quantum coherence spectroscopy (HSQC) spectrum of 15N-labeled CLK2-PAGE4 with the previously assigned spectrum of HIPK1-PAGE4 (Fig. 4 B and C). Most CLK2-PAGE4 resonances could be assigned using this approach. The downfield-shifted peaks in these spectra (Fig. 4, boxed regions) are due to main chain amides for phosphorylated S and T residues. In CLK2-PAGE4, there are at least eight peaks in this region, consistent with observations from the MALDI spectrum. Relative to the HIPK1-PAGE4 spectrum, the loss of peak intensity for specific serine and threonine residues at their nonphosphorylated chemical shifts identifies which residues are phosphorylated by CLK2. The results show that CLK2-PAGE4 is hyperphosphorylated, with phosphorylation at S7 (>95%), S9 (>95%), T51 (>95%), T71 (∼50%), S73 (∼75%), S79 (∼50%), T85 (>95%), and T94 (∼60%).
CLK2-PAGE4 and HIPK1-PAGE4 Have Opposing Functions.
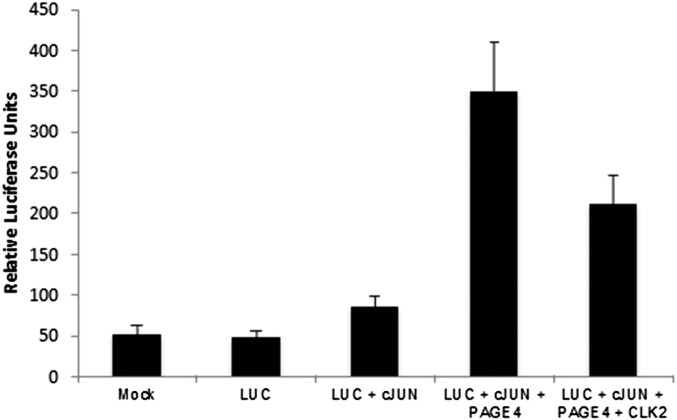
We had previously observed that PAGE4 phosphorylated by HIPK1 prefers a more compact conformation relative to the WT PAGE4 ensemble (37) and potentiates transactivation of c-Jun in a cell-based transactivation assay using a luciferase reporter (32). Therefore, we determined the effect of hyperphosphorylation of PAGE4 on c-Jun potentiation using the same luciferase reporter assay in PC3 cells that were cotransfected with full-length human CLK2 cDNA. However, as shown in Fig. 5, in contrast to HIPK1-PAGE, CLK2-PAGE4 attenuated potentiation by ∼50%, suggesting that hyperphosphorylation by CLK2 remodels the ensemble to populate a different conformation that hinders its interaction with AP-1.
Fig. 5.
CLK2-PAGE4 attenuates c-Jun transactivation. GAL4-cJun (aa 1–223) was cotransfected with either PAGE4 cDNA alone or with PAGE4- and CLK2-expressing cDNAs into PC3 (human prostate cancer cell line 3) cells. Luciferase (LUC) activity from a GAL4-binding site driving reporter construct was measured as described in the main text. Mock control (lane 1) represents background luminescence, and Luc (lane 2) represents background due to luciferase reporter. Potentiation of c-Jun transactivation in the absence of PAGE4 (lane 3) is compared with potentiation in the presence (lane 4) of PAGE4, and the effect of PAGE4 hyperphosphorylation by CLK2 (CLK2-PAGE4) on c-Jun potentiation is shown in lane 5.
Hyperphosphorylation of PAGE4 Shifts the Conformational Ensemble.
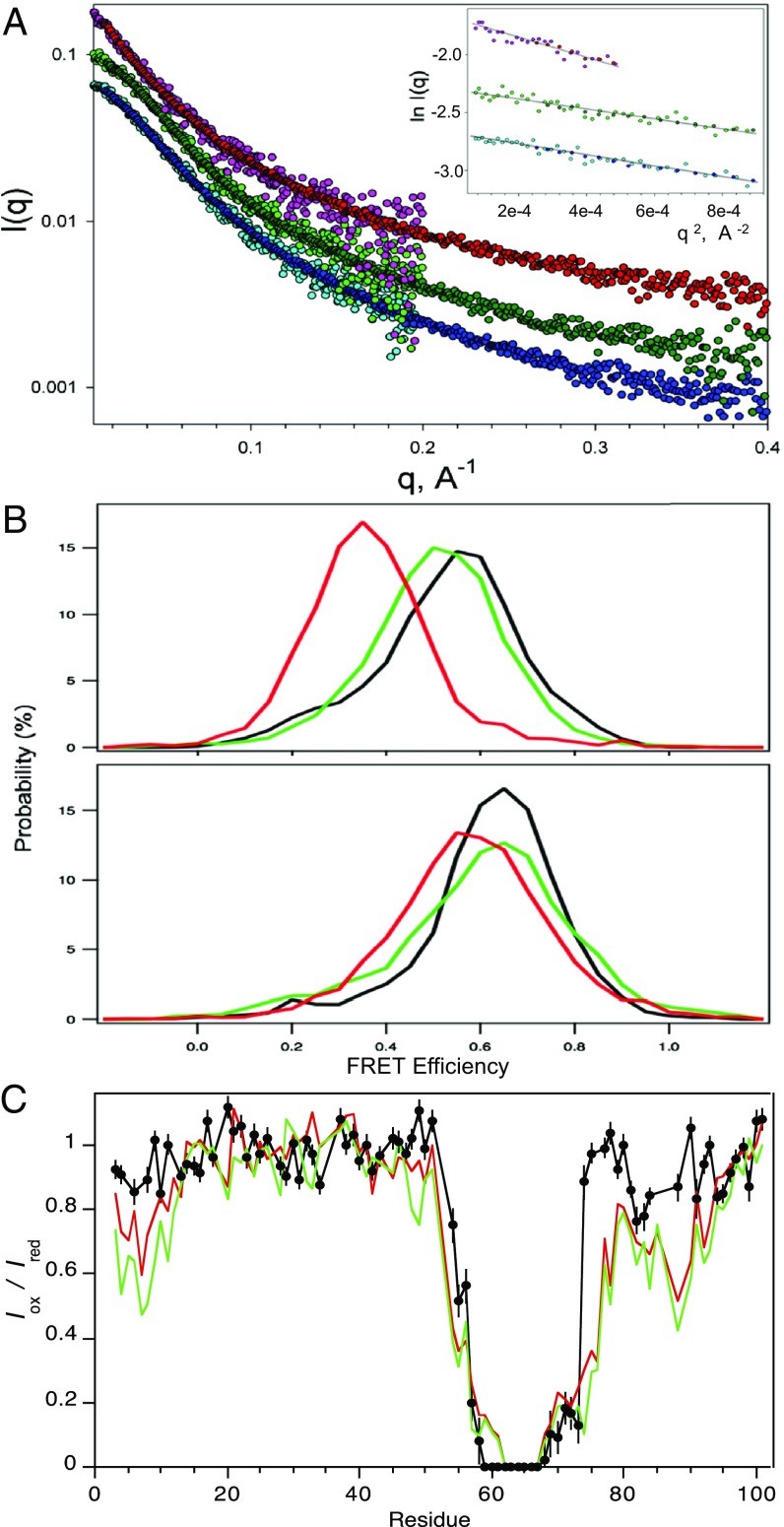
Given that CLK2-phosphorylated PAGE4 and HIPK1-PAGE4 have opposing regulatory functions, we wanted to understand the biophysical basis for these differences. Our earlier NMR results showed that although PAGE4 is an IDP without a unique 3D structure, its conformational ensemble has distinct local and long-range preferences. The longer range preferences in WT-PAGE4 and HIPK1-PAGE4 appear to be driven by favorable electrostatic interactions between a central acidic region and N- and C-terminal basic motifs (28). Therefore, to assess the effects of CLK2-phosphorylation on PAGE4 conformation, small-angle X-ray scattering (SAXS), smFRET, and NMR paramagnetic relaxation enhancement (PRE) experiments were used. Radius of gyration (Rgyr) values extracted from the SAXS data (Fig. 6A) demonstrate that WT-PAGE4 and HIPK1-PAGE4 both adopt relatively compact conformational ensembles (Rgyr: 36.2 ± 1.1 Å and 34.7 ± 1.2 Å, respectively), consistent with previous smFRET (37) and NMR (28) studies. However, CLK2-PAGE4 is significantly less compact and more like a random coil (Rgyr: 49.8 ± 1.9 Å).
Fig. 6.
Conformational expansion of PAGE4 upon hyperphosphorylation by CLK2. (A) Experimental X-ray scattering data for the WT-PAGE4 (bottom curve, cyan/blue), HIPK1-PAGE4 (middle curve, light green/dark green), and CLK2-PAGE4 (top curve, pink/red). For each of the variants, the two colors denote independent data collections probing lower-q and medium-q ranges of the scattering data. The curves are offset for clarity. (Inset) Guinier fits of the lowest q data that yield model-free estimates of the ensemble-averaged radii of gyration for the three variants. (B) smFRET measurements. (Upper) Distributions of smFRET efficiency measurements for PAGE4 with donor and acceptor sites at positions 18 and 63 WT-PAGE4 (black), HIPK1-PAGE4 (green), and CLK2-PAGE4 (red). (Lower) Donor and acceptor sites are at positions 63 and 102 for WT-PAGE4 (black), HIPK1-PAGE4 (green), and CLK2-PAGE4 (red). (C) PRE data for CLK2-PAGE4 (black) with an MTSL spin label at C63. Results are compared with earlier observations (28) for WT-PAGE4 (red) and HIPK1-PAGE4 (green).
To validate the results obtained with SAXS, an ensemble-averaged technique, we used smFRET microscopy, a complementary technique that reports on the conformation of individual molecules. We previously have applied smFRET to PAGE4 fluorescently labeled at cysteine mutations at either residue A18 or residue P102 to pair with the native cysteine at C63, resulting in two constructs with a FRET pair spanning either the N-terminal (A18:C63) (Fig. 6B, Upper) or C-terminal (C63:P102) (Fig. 6B, Lower) region. FRET measurements repeated here had FRET efficiency of 0.55 (A18:C63) and 0.65 (C63:P102) for WT-PAGE4 and FRET efficiency of 0.52 (A18:C63) and 0.65 (C63:P102) for HIPK1-PAGE4. These smFRET values were consistent with our previous studies (32, 37). Using the same labeling sites with CLK2-PAGE4, we found FRET efficiency of 0.35 (A18:C63) and 0.58 (C63:P102). The decreased FRET efficiency indicates a larger size for CLK2-PAGE4 compared with HIPK1-PAGE4 and WT-PAGE4, and confirms that hyperphosphorylation by CLK2 expands PAGE4.
We used these FRET measurements to estimate the Rgyr for PAGE4 treated with CLK2 or HIPK1. Similar to our previous work (32), we assumed a Gaussian probability distribution for a random fluctuating chain without self-avoidance to convert the FRET efficiencies to rms fluorophore separation distances, and then to the Rgyr for the entire protein (details are provided in Materials and Methods). This analysis resulted in smFRET-derived estimates for the PAGE4 gyration radius as Rgyr = 34 Å (WT-PAGE4), Rgyr = 35 Å (HIPK1-PAGE4), and Rgyr = 43 Å (CLK2-PAGE4). These values compare well with the SAXS results above. In particular, smFRET experiments agree with SAXS observations that CLK2-PAGE4 is substantially expanded compared with WT and HIPK1-PAGE4.
PRE measurements also support a more random-like ensemble of conformers for CLK2-PAGE4 than for WT-PAGE4 and HIPK1-PAGE4 (Fig. 6C). In these experiments, a stable nitroxide spin label [in this case, 1-oxy-2,2,5,5-tetramethylpyrroline-3-methyl)methane-thio-sulfonate (MTSL)] was covalently attached to the lone naturally occurring cysteine in PAGE4 at C63. Peak intensities were then determined for amide resonances in the paramagnetic species (Iox) and in the reduced diamagnetic species (Ired). The PRE results for CLK2-PAGE4 were compared with the PRE results obtained previously for WT-PAGE4 and HIPK1-PAGE4 (28). They show that the long-range interactions detected between C63-MTSL and the N- and C-terminal basic motifs of WT-PAGE4 and HIPK1-PAGE4 are substantially attenuated in the corresponding CLK2-phosphorylated protein. Most likely, this attenuation is because the negatively charged phosphoryl groups at S7 and S9 in the N-terminal motif and the negatively charged phosphoryl groups at T85 and T94 in the C-terminal motif serve to weaken any electrostatic attraction with the central acidic region adjacent to C63. Thus, even though WT-PAGE4 and HIPK1-PAGE4 are flexibly disordered, they are relatively compact, and CLK2 hyperphosphorylation remodels the conformational ensemble to a more open state.
PAGE4 Binding to c-Jun/c-Fos Depends on Its Phosphorylation State.
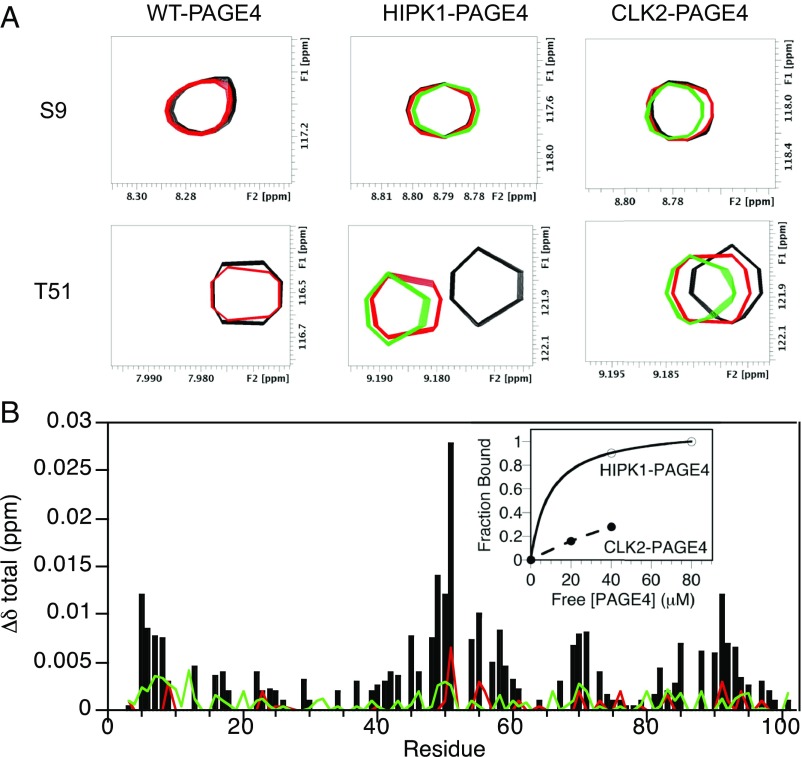
NMR binding experiments between differentially phosphorylated PAGE4 and the c-Jun/c-Fos heterodimer (AP-1) were carried out to gain insight into the underlying reasons for the observed differences in transactivation function. Truncated constructs were used for c-Jun (residues 150–331) and c-Fos (residues 1–220), which encompassed the coiled-coil and basic regions of each component of the heterodimer. Our previous studies (28) indicated that residues 1–150 of c-Jun were not required for potentiation by PAGE4. NMR spectroscopy offers advantages over other methods in that it can detect interactions with a wide range of binding affinities. A control 2D 1H-15N HSQC spectrum was recorded for 15N-labeled WT-PAGE4, HIPK1-PAGE4, or CLK2-PAGE4. Unlabeled c-Jun/c-Fos was then added, HSQC spectra were collected after each addition (Fig. 7A), and the chemical shift perturbations (CSPs) were plotted for each residue (Fig. 7B). The NMR data indicate that WT-PAGE4 and CLK2-PAGE4 have little or no interaction with AP-1. However, HIPK1-PAGE4 shows a series of CSPs centered around pT51 that are experimentally significant. Based on the NMR titration for HIPK1-PAGE4 with AP-1, we estimate a Kd of ∼10 μM (Fig. 7B, Inset). Therefore, the data indicate that although HIPK1-PAGE4 binds to AP-1, hyperphosphorylation with CLK2 weakens this binding (Kd > 150 μM), even though both HIPK1-PAGE4 and CLK2-PAGE4 have the T51 site phosphorylated. This finding suggests that the N- and C-terminal basic motifs also play a role in the interaction with the AP-1 complex, and that phosphorylation in these regions regulates the extent of the binding interaction. This idea is also supported by small but experimentally significant CSPs in the N- and C-terminal motifs for HIPK1-PAGE4, but not for CLK2-PAGE4 (Fig. 7B). Overall, the results from the binding experiments parallel the observations from the c-Jun transactivation assay. Whereas HIPK1-PAGE4 binds to AP-1 and stimulates c-Jun transactivation, CLK2 has weakened binding to AP-1 and attenuates activity.
Fig. 7.
PAGE4 binding to c-Jun/c-Fos monitored by NMR spectroscopy. (A) Regions from 2D 1H-15N HSQC spectra for 15N-labeled WT-PAGE4, HIPK1-PAGE4, and CLK2-PAGE4 as a function of unlabeled Jun/Fos concentration. Ratios of WT-PAGE4 to c-Jun/c-Fos are 1:0 (black) and 1:12.4 (red). Ratios of HIPK1-PAGE4 or CLK2-PAGE4 to c-Jun/c-Fos are 1:0 (black), 1:4 (red), and 1:8 (green). (B) Plot of CSP, Δδtotal, versus residue for c-Jun/c-Fos titrations with HIPK1-PAGE4 (black), CLK2-PAGE4 (red), and WT-PAGE4 (green). (Inset) Binding curves for HIPK1-PAGE4 (○) and CLK2-PAGE4 (●).
Mathematical Modeling Supports the Role of PAGE4 in Generating Phenotypic Heterogeneity in PCa Cells.
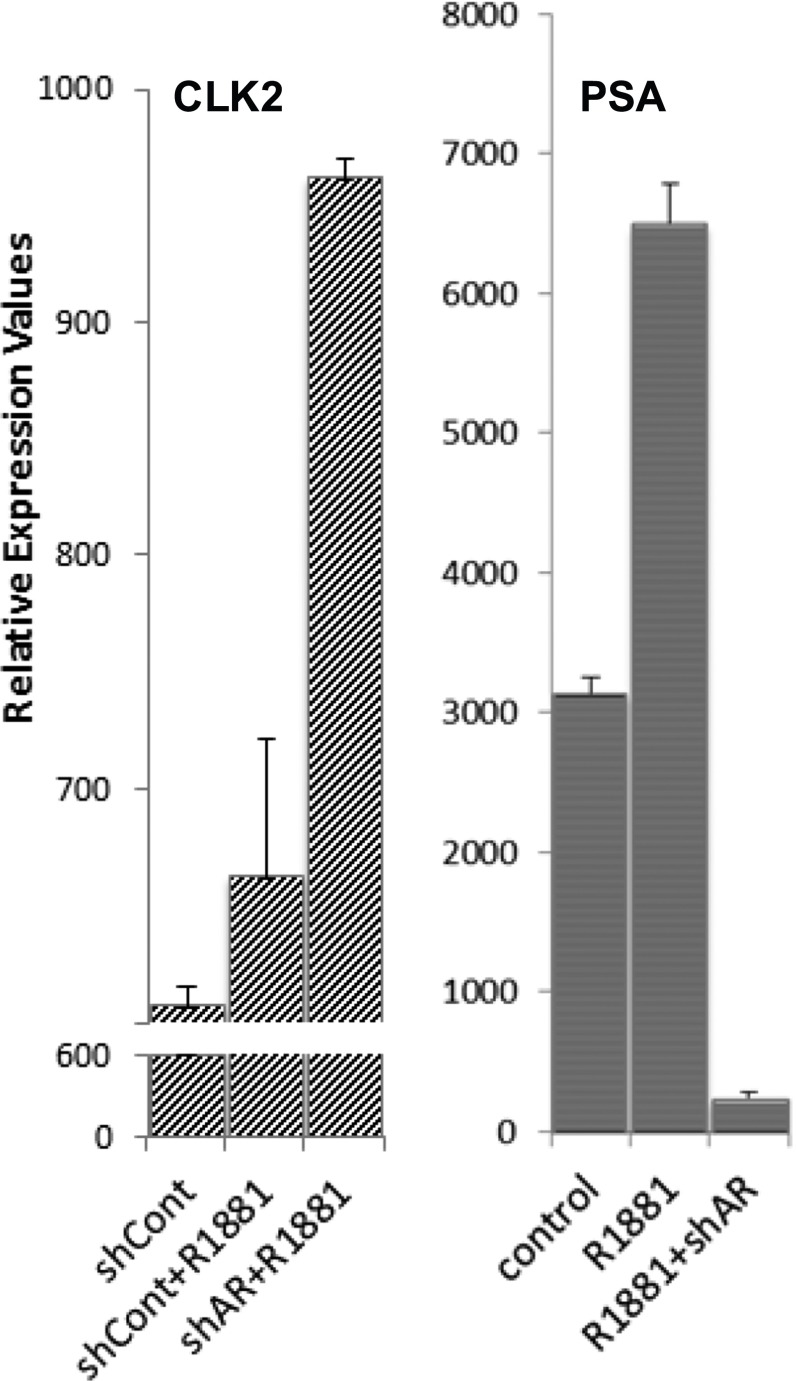
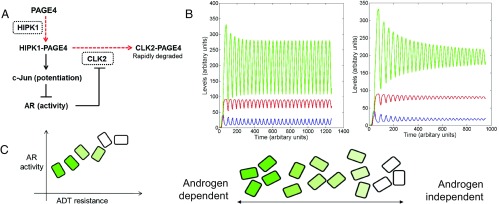
Previous studies have shown that AP-1 can negatively regulate AR activity (41, 42), and gene expression microarray data revealed that AR can transcriptionally inhibit CLK2 expression (Fig. S8). Furthermore, cells resistant to androgen deprivation therapy (ADT) often have enhanced AR activity (AR protein expression can increase >25 fold), suggesting a positive correlation between ADT resistance and AR activity (43, reviewed in ref. 44). Incorporating these interactions with the results presented here, we constructed a circuit representing the PAGE4/AP-1/AR interactions that drive nongenetic phenotypic heterogeneity in PCa cells (Fig. 8A) and developed a mathematical model to represent the dynamics of this circuit. The model predicts that this circuit can display sustained or damped oscillations (i.e., androgen dependence of a cell need not be a fixed state, but can vary temporally). Thus, cells can enter and exit the androgen-independent state or phenotype continually (Fig. 8B and Fig. S9). Even in the case of damped oscillations that eventually settle to one state, as seen in the deterministic model presented here (Fig. 8B, Lower), the system can display sustained oscillations under the effect of biological “noise.” Such noise can originate from multiple sources, such as (i) limited quantities of PAGE4, HIPK1, or CLK2, and (ii) conformational dynamics of PAGE4. Noise has been shown previously to increase the fitness of bacterial populations by expanding the range of stress levels to which bacteria can respond dynamically (45, 46). Similarly, noise can be expected to increase the fitness of the PCa population treated with ADT by driving phenotypic heterogeneity between androgen-dependent and androgen-independent states.
Fig. S8.
AR negatively regulates CLK2 expression in androgen-dependent LNCaP PCa cells. Gene expression microarray data were extracted from the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geoprofiles; GDS4113/203229_s_at) and analyzed. AR expression was knocked down using a specific shRNA, and changes in gene expression were interrogated using Affymetrix protocols and hybridized to the HG U133 Plus 2.0 array set. As can be seen, although AR-specific shRNA derepressed CLK2 mRNA, the control shRNA (shCont) did not. In contrast, knocking down the AR with the specific shRNA repressed prostate-specific antigen (PSA) expression, a direct target of the AR in LNCaP cells. However, the control shRNA did not repress PSA expression. R1881, synthetic androgen used to stimulate AR.
Fig. 8.
Modeling the PAGE4/AP-1/AR/CLK2 regulatory circuit. (A) Regulatory circuit for PAGE4/AP-1/AR/CLK2 interactions. Dashed red lines denote enzymatic reactions, and solid black lines denote nonenzymatic reactions. CLK2 and HIPK1, the two enzymes involved, are shown in dotted rectangles. (B) Dynamics of the circuit showing sustained and damped oscillations for HIPK1-PAGE4 (PAGE4M, shown in blue), CLK2-PAGE4 (PAGE4H, shown in red), and CLK2 (shown in green). Parameters are given in Fig. S9. (C) Distribution of androgen dependence for an isogenic population over a spectrum, as indicated by the shade of green. Dark green boxes denote highly androgen-dependent (i.e., ADT-sensitive) cells, and white boxes denote androgen-independent cells.
Fig. S9.
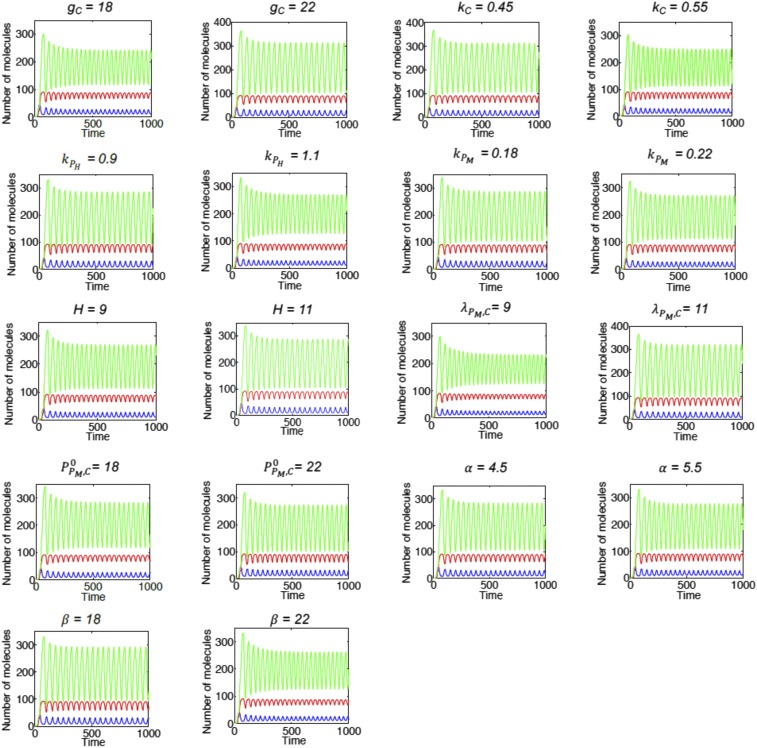
Sensitivity analysis. Dynamics of the circuit when the parameters of the model are varied by ±10% individually.
The intracellular oscillations of the levels of CLK2, PAGE4 monophosphorylated by HIPK1 (PAGE4M, also referred to as HIPK1-PAGE4), and PAGE4 hyperphosphorylated by CLK2 (PAGE4H, also referred to as CLK2-PAGE4) need not be synchronized across cells. Therefore, it is expected that individual cells in an isogenic population would have different levels of androgen dependence or independence at a given time point, consequently giving rise to nongenetic phenotypic heterogeneity or cellular diversity in PCa cells. In other words, androgen dependence is a trait whose values can display a broad distribution across the population of PCa cells (Fig. 8C). Indeed, this predicted heterogeneity in the levels of HIPK1, CLK2, and PAGE4 is corroborated by qIHC and qRT-PCR data. Of note, for modeling purposes, we have referred to the HIPK1-PAGE4 as “monophosphorylated.” The fact that T51 phosphorylation is critical for the transcriptional activity of PAGE4 further justifies this terminology.
The model (SI Text) is based on the following assumptions: (i) Initially (i.e., before the effect of HIPK1 begins), all of the available PAGE4 is unphosphorylated; (ii) total levels of PAGE4 are in a quasi-steady state (i.e., phosphorylation dynamics are much faster compared with the production of PAGE4); and (iii) the production of CLK2 by HIPK1-PAGE4 includes a delay, due to intermediary transcriptional mechanisms. Thus, in androgen-dependent cells, when PAGE4 is phosphorylated by HIPK1, HIPK1-PAGE4 potentiates c-Jun and represses AR activity. Consequently, the repression on CLK2 is relieved and induced CLK2 can hyperphosphorylate HIPK1-PAGE4 to form CLK2-PAGE4, thereby reducing the levels of HIPK1-PAGE4 and leading to the onset of oscillations in HIPK1-PAGE4 levels (Fig. 8B). These oscillatory levels of HIPK1-PAGE4 control the levels of CLK2 and CLK2-PAGE4, setting them to oscillate as well. The oscillations of CLK2 and CLK2-PAGE4 are in phase with one another, and out of phase with HIPK1-PAGE4, because CLK2, an indirect target of HIPK1-PAGE4, converts HIPK1-PAGE4 to CLK2-PAGE4.
SI Text
Mathematical Modeling of HIPK1/PAGE4/CLK2 Circuit.
In the model, there are three major components: monophosphorylated PAGE4 or HIPK1-PAGE4 (), hyperphosphorylated PAGE4 or CLK2-PAGE4 (), and CLK2 (). The temporal dynamics for each component are represented by rate in Eqs. S1–S3:
| [S1] |
| [S2] |
| [S3] |
In Eqs. S1–S3, , , and denote the innate degradation rates of monophosphorylated PAGE4 or HIPK1-PAGE4 (), hyperphosphorylated PAGE4 or CLK2-PAGE4 (), and CLK2 (), respectively. The first term in Eq. S1 shows conversion of unphosphorylated PAGE4 () by HIPK1 (represented as H), and the first term in Eq. S2 represents the conversion of monophosphorylated PAGE4 () to hyperphosphorylated PAGE4 (). These terms are denoted as following Michelis–Menten kinetics due to their enzymatic nature. In Eq. S3, is the basal production rate of CLK2 and is a shifted Hill function representing the indirect activation of CLK2 by with a time delay of .
The functional form of shifted Hill functions is given by
The parameters used in this modeling are: = 10, = 5, = 20, = 20, = 0.5, = 0.2, = 1, = 10, = 2, = 20, and = 15. Total levels of PAGE4 (sum of , , and ) is considered to be constant at 100. For Fig. 8B (Lower), = 25. We varied every parameter except total levels of PAGE4 by ±10% and yet observed oscillatory dynamics, suggesting robust behavior of the model to parameter perturbation (Fig. S9).
Discussion
In this paper, we have interrogated the role of IDP conformational dynamics in driving phenotypic heterogeneity using PAGE4 and androgen sensitivity in PCa as the respective paradigms. Androgen, by acting via the AR, also an IDP (47, 48), is known to play important roles not only in the development and differentiation of the prostate gland but also in its disease pathology. Thus, a central issue in PCa is that although the disease appears to be androgen-dependent and is initially responsive to ADT, it loses responsiveness over time and eventually becomes refractory to androgen, which can prove fatal. However, the development of hormone resistance remains quite elusive (44, 49, 50). Therefore, elucidating the molecular mechanisms underlying the transition from androgen sensitivity to androgen resistance in PCa is paramount to developing effective medical modalities to treat and manage the disease.
Earlier attempts to understand the relationship between AP-1 and AR in PCa had revealed that AP-1 can negatively regulate AR activity (41, 42, 51), but the role of PAGE4, a remarkably prostate-specific protein expressed only in androgen-dependent cells, had not been suspected, much less considered. The present study has not only uncovered a previously unappreciated role of PAGE4 in driving phenotypic heterogeneity (i.e., responsiveness to androgen in PCa) but also underscores the importance of conformational dynamics of this IDP. Such heterogeneity might be better able to evade the effects of ADT, and therefore drive androgen resistance, compared with a homogeneous PCa population. Furthermore, contrary to a “binary” model that assumes PCa cells to have an androgen-dependent or -independent phenotype, subpopulations of partially androgen-dependent and androgen-independent cells can also drive tumor progression synergistically through cell-to-cell communication as observed in some cases. For example, two E. coli strains can protect each other in the presence of two antibiotics (52). Similarly, a cluster of circulating tumor cells (CTCs) with “partial” phenotypes forms 50-fold more secondary tumors compared with the same number of individual CTCs (53).
The results also shed light on an important question in the field that remains equivocal: Is androgen resistance a later step in the linear “progression” of PCa as is generally held, or do slow-growing cells with different extents of androgen independence preexist in a population of androgen-dependent cells as has been reported by Terada et al. (54)? Our mathematical model suggests that, similar to bacterial persisters that form stochastically due to phenotypic switching and are highly tolerant to antibiotics (55, 56), a subpopulation of androgen-independent cells can exist a priori in a population of androgen-dependent cells. As the levels of monophosphorylated and hyperphosphorylated PAGE4 vary, the cells go on excursions of phenotypes with varying androgen deprivation sensitivities (as described in the legend for Fig. 8B). Thus, phenotypic plasticity in PCa may be driven by the underlying oscillatory dynamics in the PAGE4/AP-1/AR regulatory circuit rather than by genetic changes alone, supporting the idea that IDP conformational dynamics may play a role in driving state switching (57). Consistent with this argument, a growing number of studies focusing on well-studied systems that have been analyzed using multiple quantitative measurement and perturbation approaches demonstrate that cells can send and receive information by controlling the temporal behavior (dynamics) of their signaling molecules, without involving genetic changes (58). Additional studies that are outside the scope of the present work are needed to validate our model experimentally in PCa cells.
Although our model predicts the existence of a subpopulation of “persister-like” cells, the size of this subpopulation relative to the total population is highly dynamic and would depend on several factors, both extrinsic and intrinsic. Furthermore, whether the oscillations can induce phenotypical changes en masse or not would depend on several factors. For example, the amplitude of these oscillations (i.e., the fold change in levels of CLK2) needs to be robust to alter AR activity in a significant manner. Second, a negative feedback loop that can drive oscillations often involves a delay. We have assumed a delay in transcriptional production of CLK2 by HIPK1-PAGE4, but the relative time scale still needs to be measured experimentally. The lack of PAGE4 in metastatic disease that is resistant to ADT is consistent with our model predictions, but, of course, attaining that state may involve additional genetic/epigenetic changes in a population of PCa cells.
Interestingly, a recent study reported that a situation paralleling the preexistence of hormone-resistant cells in PCa is also encountered in breast cancer (59). When CTCs from patients with estrogen receptor-positive/human epidermal growth factor receptor 2-negative (HER2−) breast cancer who developed metastatic multidrug-resistant disease were cultured in vitro, they maintained a discrete population of Her2+ and Her2− cells that interconvert spontaneously with significant consequences for disease progression and drug response. Although the underlying mechanisms driving spontaneity in state switching were not elucidated, the study by Jordan et al. (59) and several other studies in different types of cancer (60–63) suggest that the phenomenon of state switching driven by nongenetic variability need not be restricted to bacterial “persisters” and may be fairly common in cancer. For example, cancer cells in a hybrid epithelial/mesenchymal state, when cultured, can give rise to epithelial and mesenchymal cells predominantly (64). Mathematical models have proposed that multistability emerging from the underlying gene regulatory circuit of epithelial-to-mesenchymal transition can drive such state switching (65, 66).
Our model suggests that an isogenic population of PCa cells displays a continuum of phenotypes with varying androgen dependence. These cells can reversibly switch between androgen-dependent and androgen-independent states, without any specific genetic perturbation. The findings might help explain why a new paradigm for treating patients with PCa, referred to as bipolar ADT (67), in which patients undergoing chemotherapy cycle through ADT followed by a supraphysiological dose of androgen, proved more beneficial than ADT alone. Because the function of AR in DNA replication is to help repair double-strand breaks, it has been argued that binding of testosterone to AR under supraphysiological conditions could prevent its normal function of repairing double-strand breaks, which, in turn, can lead to cell cycle failure and programmed cell death (43, 68), as seen in patients who responded to bipolar ADT. However, it has also been reported by others (69) that supraphysiological levels of androgens can induce cell cycle arrest and up-regulate markers of cellular senescence in human PCa cells. Therefore, the growth inhibitory role of androgens could also be due to cells entering dormancy rather than due to programmed cell death alone.
Thus, according to our model, it is possible that there exists a subset of “dormant” cells in this population in which bipolar ADT could restore the normal functioning of AR and reinitiate the PAGE4/AP-1/AR regulatory circuit and consequent oscillations. Furthermore, such cells with restored AR activity can also lower drug resistance because androgen-dependent cells are also more sensitive to chemotherapy than are androgen-independent cells (70). A corollary to the bipolar ADT treatment (67) that stems from our model is that higher levels of PAGE4 would predict a better prognosis. Indeed, PAGE4 expression is significantly correlated with longer biochemical recurrence-free time (71). Moreover, it has been noted that in hormone-naive PCa, the median survival of patients with tumors expressing high PAGE4 levels was 8.2 y compared with 3.1 y for patients with tumors expressing negative/low levels of PAGE4 (36), adding further support to our model. The present study demonstrates a plausible link between IDP conformational dynamics and oscillatory dynamics, and consequent state switching, in cancer. Thus, PAGE4, its phosphorylation status, and the kinases that modulate its conformational dynamics represent unique therapeutic and/or prognostic targets for the treatment and management of PCa.
Materials and Methods
Kinase Assay.
To identify the kinase(s) that phosphorylates PAGE4, the ProQinase GmbH service was utilized to screen a panel of 190 S/T protein kinases (Dataset S1) that were expressed in Sf9 cells as human recombinant GST-fusion proteins or His-tagged proteins by means of the baculovirus expression system. The kinases were purified by affinity chromatography using either glutathione-agarose (Sigma) or nickel-agarose (Qiagen). The purity and identity of each kinase was verified by SDS/PAGE, followed by Coomassie staining and immunoblotting. A kinase assay using 33Pγ-ATP was used to assess the recombinant enzyme’s ability to phosphorylate bacterially expressed 6HIS-PAGE4.
Immunoblotting.
Total protein from whole-cell lysates was obtained using radioimmunoprecipitation assay (RIPA) buffer. Cells were collected from 75-cm2 flasks during the exponential growth phase (∼80% confluent). After washing with 1× PBS, 1 mL of RIPA buffer containing protease inhibitors was added and cells were incubated in ice for 1 h. Supernatant was collected after centrifugation to eliminate cell debris. Total protein was quantified by the bicinchoninic acid assay for protein estimation method according to the manufacturer’s instructions (Pierce, Thermo Fisher Scientific). For each cell line, 20 μg of total protein was used for SDS/PAGE in a 12% (wt/vol) polyacrylamide gel (12% Mini-PROTEAN TGX Precast Gels; Bio-Rad). Transfer to a nitrocellulose membrane was performed using a semidry system (Trans-Blot Turbo Transfer System; Bio-Rad). The membrane was blocked with 5% milk (5 mg of powder fat free milk/100 mL of PBS) for 1 h at room temperature. Primary antibody was diluted in 5% (wt/vol) milk containing 0.2% Tween 20 (1:100 HIPK1, 1:500 CLK2, 1:1,000 ActinB) and incubated overnight at 4 °C. After serial washes in 1× TBST (Tris-buffered saline, 0.1% Tween), secondary fluorescent antibody (1:10,000 dilution) was added and incubated for 1 h at room temperature. The membrane was washed several times in 1× TBST and scanned using the Li-Cor Odyssey Infrared System (Li-Cor Biotechnology).
Immunohistochemistry.
For immunohistochemistry, CLK2 and HIPK1 protein expression in prostate tissue was performed by qIHC in 80 paired cases of PCa and normal adjacent tissue obtained from radical prostatectomies. The TMA slides were deparaffinized using xylene, and tissues were rehydrated in decreasing concentrations of ethanol (100%, 75%, 50%, and 25%; all vol/vol). Antigen retrieval was performed at controlled pH values under heat, followed by endogenous peroxidase inhibition using 0.3% hydrogen peroxidase. An unspecific protein block used Protein Block Serum Free reagent (Dako) for 1 h at room temperature. Primary antibody incubation was performed at 4 °C overnight using the ideal dilution for each antibody. Primary antibody was washed with 1× PBS, and secondary antibody (1:200) was added to the slides and incubated for 1 h at room temperature. Antigen localization was developed using 3,3′-diaminobenzidine chromogen. Tissue samples were counterstained in hematoxylin and dehydrated in ethanol and xylene. Rabbit anti-CLK2 polyclonal antibody (ab188141; Abcam) dilution in all immunohistochemistry staining reactions was diluted at 1:100, and mouse anti-HIPK1 monoclonal antibody (ab58136; Abcam) was diluted at 1:10.
For qIHC analysis, slides were scanned using the Aperio Scanscope XT (Leica Biosystems) and the staining quantifications were performed using Aperio Imagescope v12.3 software (Leica Biosystems). Intensity and frequency of positive staining are determined by the pixel count of the delimited area selected for analysis. Intensity (different brown-staining shades) for a determined area is given as the total brown pixel count for that region. The frequency (area of positive staining) is given by the ratio of positive brown region and the total area selected for analysis (positive + negative area). CLK2 and HIPK1 protein expression differences were compared using the Wilcoxon matched-pairs test, because tumor and benign adjacent tissues were collected from the same patient. The average for all cores available from each patient for qIHC analysis was calculated, and the values were used to compare medians between the groups (tumor vs. benign). Protein expression (frequency or intensity) was considered significantly different for a P value ≤0.05.
RNA Isolation and Gene Expression Analysis by qRT-PCR.
Total RNA from 20 organ-confined (localized PCa) samples was obtained from the Department of Urology at The University of Washington. One microgram of total RNA was utilized for cDNA synthesis using the iScript cDNA Synthesis Kit (Bio-Rad). The PCR reactions were performed with 0.2 μL of cDNA template in 25 μL of reaction mixture containing 12.5 μL of iQ SYBR Green Supermix (Bio-Rad) and 0.25 μmol/L each primer. PCR reactions were subjected to a hot start at 95 °C for 3 min, followed by 45 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s using the CFX96 Real-Time PCR Detection System (Bio-Rad). Analysis and fold differences were determined using the comparative threshold cycle method. Beta-actin (ACTB) was the housekeeping gene used for normalization. All reactions were performed in triplicate.
Transactivation Assays.
Potentiation of c-Jun by PAGE4 was determined using the luciferase reporter assay as described previously (32). Briefly, PC3 cells (8,000 cells per well) were seeded in 100 μL of RPMI/10% (vol/vol) FBS/PS (penicillin, streptomycin mix) in a 96-well plate and incubated overnight at 37 °C in a humidified, 5% (vol/vol) CO2 incubator. On the next day, cells were washed with RPMI and incubated for 4 h at 37 °C in a humidified, 5% (vol/vol) CO2 incubator with 100 μL of Lipofectamine 2000-cDNA mixture in RPMI medium. The medium was replaced with 100 μL of fresh RPMI/10% (vol/vol) FBS/PS and incubated for 48 h at 37 °C in a humidified, 5% (vol/vol) CO2 incubator. Luciferase activity was measured using the BrightGlo Luciferase Assay System from Promega (Promega Corp.). One hundred microliters of assay reagent was added to the cells and incubated on a shaker at room temperature for 5 min, and luminescence was measured using a Microlumat Plus luminometer (Berthold Technologies).
Protein Sample Preparation.
The PAGE4 gene was cloned into an eXact pPAL8 vector, and expressed and purified as described previously (72). HIPK1-phosphorylated PAGE4 was obtained by coexpressing PAGE4 with a pET28a gene construct of the human HIPK1 kinase domain (Origene Technologies) as described previously. CLK2-phosphorylated PAGE4 was prepared in a similar way using a pET28a construct of the human CLK2 kinase domain (Origene Technologies). Gene constructs for c-Jun (residues 150–331) and c-Fos (residues 1–220) were cloned into an eXact pPAL8 vector, and expressed and purified as for PAGE4. In all cases, purity was confirmed by polyacrylamide gel electrophoresis and MALDI mass spectrometry. Protein concentrations were measured using a colorimetric assay (73). For mass spectrometry, Myc-DDK–tagged PAGE4 from LNCaP cells was isolated as follows. Six million LNCaP cells were plated (1.2 × 106 cells per 100-mm tissue culture plate) in RPMI growth medium containing 10% (vol/vol) FBS and penicillin/streptomycin. After overnight incubation at 37 °C in a 5% (vol/vol) CO2, humidified incubator, cells were transfected with 5 μg of Myc-DDK–tagged PAGE4 encoding cDNA (Origene Technologies) using Effectene Transfection Reagent (Qiagen). After 48 h of transfection, cells were washed with PBS and lysed in 1 mL of lysis buffer (Cell Signaling Technologies) containing protease inhibitors (Roche Diagnostics). Anti-DDK agarose beads (Origene Technologies) were washed with PBS and incubated for 4 h at room temperature while shaking with LNCaP cell lysate. Agarose beads were pelleted and washed with PBS.
Mass Spectrometry.
MALDI-TOF analysis was performed on a Microflex Bruker instrument using a sinapic acid matrix. Mass spectra were acquired in positive ion linear mode using standard settings.
NMR Spectroscopy.
NMR spectra were recorded on a Bruker Avance III 600 MHz spectrometer equipped with a cryoprobe. Spectra were acquired at 10 °C for all experiments, and were processed using NMRPipe (74) and analyzed with Sparky (75). NMR titration experiments were performed using 5 μM concentrations of 15N-labeled WT-PAGE4, HIPK1-PAGE4, or CLK2-PAGE4 and unlabeled c-Jun/c-Fos. Before titration, both the PAGE4 and c-Jun/c-Fos samples were buffer-matched by dialysis in 100 mM KPi, 10 mM sodium azide, and 0.5 mM DTT. Unlabeled c-Jun/c-Fos samples (typically 5 μM in 0.8–3.2 mL of buffer) were added to 400 μL of 5 μM labeled PAGE4. Samples were then concentrated back to 400 μL using Centricon membrane filtration. Two-dimensional 1H-15N HSQC spectra were recorded for phosphorylated PAGE4/c-Jun/c-Fos molar ratios of 1:0:0, 1:4:4, and 1:8:8. For WT-PAGE4/c-Jun/c-Fos, molar ratios of 1:0:0 and 1:12.4:12.4 were used. CSPs were determined from Δδtotal = [(WHΔδH)2 + (WNΔδN)2]1/2, where ΔδH and ΔδN represent 1H and 15N chemical shift differences, respectively, between free and Jun/Fos-bound states of PAGE4, with weighting factors of WH = 1 and WN = 0.2. PRE experiments were carried out on CLK2-PAGE4 using methods described previously (28). A 5 μM 15N-labeled CLK2-PAGE4 sample was spin-labeled at C63 with MTSL (Santa Cruz Biotechnology). Two-dimensional 1H-15N HSQC spectra were collected on MTSL-labeled samples and on samples treated with DTT (5 eq). Peak intensities were measured for the paramagnetic (Iox) and diamagnetic (Ired) states using SPARKY.
SAXS.
Solution X-ray scattering data for the three variants of PAGE4 were acquired using a custom-built SAXSLAB Ganesha instrument at the Institute for Bioscience and Biotechnology Research. X-ray radiation at the 1.54-Å Cu Kα wavelength was produced by the Rigaku MicroMax-007HF rotating anode generator operating at 40 kV and 30 mA output. The incident beam was collimated by the two sets of programmable pinholes equipped with scatterless slits. Incident beam intensity and sample and buffer transmission factors were monitored using a photodiode mounted on a motorized beam stop arm. Scattered radiation was recorded with a Dectris Pilatus 300K area detector mounted on a programmable positioning stage within the vacuum chamber. Sample-to-detector distance and the incident beam position within the detector plane were determined using the powder scattering profile from the standard silver behenate sample, corresponding to the accessible q-ranges from 0.007 Å−1 to 0.24 Å−1 and from 0.015 Å−1 to 0.45 Å−1 for the two configurations used during this data collection. Totals of 12 sequential frames of 15 min each were recorded for all samples and matching buffers for both of the detector configurations. To decrease the likelihood of radiation damage further, each of the 50-μL sample and buffer loads was oscillated by a syringe pump during the entire data collection. Sample buffer match quality was further verified for all three samples using 2-h wide-angle data collections covering the q-range from 0.035 Å−1 to 0.87 Å−1. In addition to the stock samples (0.4–1.5 mg/mL), SAXS data were collected at twofold dilutions to investigate the concentration dependence of the scattering signal. Individual data frames were masked, corrected for stray cosmic radiation and for pixel sensitivities as provided by the manufacturer, normalized for the solid angle per each pixel, and radially integrated, producing 1D scattering profiles. These profiles were normalized by individually measured transmission factors and superimposed. The data were then averaged over all frames, with the uncertainties calculated as SDs divided by the square root of the number of exposures. Scattering intensities from the buffers were then subtracted from the sample scattering profiles. PRIMUS software (76) was used for derivation of the radii of gyration, zero-angle scattering intensities, and the associated uncertainties via Guinier fits subject to the Rgyr * qmax < 1.1 cutoff (qmax is the maximum q value used for the Guinier fit), while monitoring the fits for the absence of systematic bias in the residuals. The buffer-subtracted scattering curves were analyzed for the effects of the interparticle repulsion and concentration-dependent oligomerization equilibria by comparing the Guinier-fitted gyration radius and extrapolated zero-angle scattering intensity [I(0)] values normalized by the sample concentrations. Because no such effects were found, scattering data at the highest (stock) concentrations were used for subsequent analysis. No evidence of radiation damage or bubble formation during X-ray exposure was found.
smFRET.
Point mutation A18C or P102C was introduced into PAGE4 in the eXact pPAL8 vector and expressed with or without kinase expression vectors and purified as described above. These purified proteins were labeled at cysteines with maleimide-reactive Alexa Fluor 555 and Alexa Fluor 647; encapsulated in liposomes that were tethered to a quartz surface; and measured in a prism-type, total internal reflection smFRET microscope using an emCCD (electron-multiplying charge-coupled device) for detection. Additional details of these experiments and data analysis are identical to our previous descriptions (32). During measurements, PAGE4 was in a buffer solution containing 20 mM Tris (pH 7.8), 100 mM NaOAc, 5 mM MgCl2, 2% (wt/vol) glucose, 0.1 mM cyclooctatetraene, and oxygen scavenger enzymes (glucose oxidase and catalase) to extend fluorophore lifetime. We confirmed that changing the buffer to 100 mM KPi, 10 mM NaN3, and 0.5 mM DTT (pH 7.0) (as used for SAXS studies) did not substantially change the FRET efficiency of CLK2-PAGE4 (A18C:C63) (FRET = 0.35 vs. 0.36 for the two buffers).
Following the procedures described in our previous studies (32), we converted measured FRET efficiency to rms distance and then to Rgyr using a Förster radius of 5.24 nm and gamma = 1 (as verified for these samples). We assumed a Gaussian probability distribution for a random fluctuating chain to convert the FRET efficiencies to rms fluorophore separation distances: WT-PAGE4, 56 Å (A18:C63) and 50 Å (C63:P102); HIPK1-PAGE4, 59 Å (A18:C63) and 50 Å (C63:P102); and CLK2-PAGE4, 75 Å (A18:C63) and 55 Å (C63:P102). We modeled the domain from aa 1–18 with the same polymer scaling as the region spanning A18:C63 to get an rms distance from aa 1–63 and then assumed random polymer behavior without self-avoidance to combine these N-terminal and C-terminal distances as the sum of squares to give the full rms extension of the polymer. Converting to gyration radius using Rgyr = (1/√6) * (rms end-to-end distance), we finally find the FRET measurements correspond to full protein gyration radii as Rgyr = 34 Å (WT-PAGE4), Rgyr = 35 Å (HIPK1-PAGE4), and Rgyr = 43 Å (CLK2-PAGE4).
Supplementary Material
Acknowledgments
P.K. thanks Dr. Robert Vessella (University of Washington) for the generous contribution of the RNA samples used in this study. We thank our referees for constructive comments that greatly improved the paper. Research in the P.K. and J.O. laboratories was supported by NIH Grant CA181730. Research in the J.O. laboratory was also partly supported by NIH Grant GM62154. H.L. was supported by National Science Foundation (NSF) Center for Theoretical Biological Physics (NSF Grant PHY-1427654) and by NSF Grant DMS-1361411. The NMR facility is jointly supported by the University of Maryland, the National Institutes of Standards and Technology, and a grant from the W. M. Keck Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700082114/-/DCSupplemental.
References
- 1.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 3.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 4.Peng Z, et al. Exceptionally abundant exceptions: Comprehensive characterization of intrinsic disorder in all domains of life. Cell Mol Life Sci. 2015;72(1):137–151. doi: 10.1007/s00018-014-1661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barabasi A-L. Scale-free networks: A decade and beyond. Science. 2009;325(5939):412–413. doi: 10.1126/science.1173299. [DOI] [PubMed] [Google Scholar]
- 6.Barabási A-L, Albert R. Emergence of scaling in random networks. Science. 1999;286(5439):509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 7.Rangarajan N, Kulkarni P, Hannenhalli S. Evolutionarily conserved network properties of intrinsically disordered proteins. PLoS One. 2015;10(5):e0126729. doi: 10.1371/journal.pone.0126729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uversky VN. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015;282(7):1182–1189. doi: 10.1111/febs.13202. [DOI] [PubMed] [Google Scholar]
- 9.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16(1):18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galea CA, Wang Y, Sivakolundu SG, Kriwacki RW. Regulation of cell division by intrinsically unstructured proteins: Intrinsic flexibility, modularity, and signaling conduits. Biochemistry. 2008;47(29):7598–7609. doi: 10.1021/bi8006803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon M-K, Mitrea DM, Ou L, Kriwacki RW. Cell cycle regulation by the intrinsically disordered proteins p21 and p27. Biochem Soc Trans. 2012;40(5):981–988. doi: 10.1042/BST20120092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buske PJ, Mittal A, Pappu RV, Levin PA. An intrinsically disordered linker plays a critical role in bacterial cell division. Semin Cell Dev Biol. 2015;37:3–10. doi: 10.1016/j.semcdb.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurley JM, Larrondo LF, Loros JJ, Dunlap JC. Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered neurospora clock protein FRQ. Mol Cell. 2013;52(6):832–843. doi: 10.1016/j.molcel.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong P, et al. A dynamic interaction process between KaiA and KaiC is critical to the cyanobacterial circadian oscillator. Sci Rep. 2016;6:25129. doi: 10.1038/srep25129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue B, Oldfield CJ, Van YY, Dunker AK, Uversky VN. Protein intrinsic disorder and induced pluripotent stem cells. Mol Biosyst. 2012;8(1):134–150. doi: 10.1039/c1mb05163f. [DOI] [PubMed] [Google Scholar]
- 16.Mooney SM, Jolly MK, Levine H, Kulkarni P. Phenotypic plasticity in prostate cancer: Role of intrinsically disordered proteins. Asian J Androl. 2016;18(5):704–710. doi: 10.4103/1008-682X.183570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakrabortee S, et al. Intrinsically disordered proteins drive emergence and inheritance of biological traits. Cell. 2016;167(2):369–381.e12. doi: 10.1016/j.cell.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138(1):198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 19.Marcotte EM, Tsechansky M. Disorder, promiscuity, and toxic partnerships. Cell. 2009;138(1):16–18. doi: 10.1016/j.cell.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12(1):54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 21.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5(11):789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arai M, Sugase K, Dyson HJ, Wright PE. Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding. Proc Natl Acad Sci USA. 2015;112(31):9614–9619. doi: 10.1073/pnas.1512799112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Multiscaled exploration of coupled folding and binding of an intrinsically disordered molecular recognition element in measles virus nucleoprotein. Proc Natl Acad Sci USA. 2013;110(40):E3743–E3752. doi: 10.1073/pnas.1308381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi UB, McCann JJ, Weninger KR, Bowen ME. Beyond the random coil: Stochastic conformational switching in intrinsically disordered proteins. Structure. 2011;19(4):566–576. doi: 10.1016/j.str.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryan PN, Orban J. Proteins that switch folds. Curr Opin Struct Biol. 2010;20(4):482–488. doi: 10.1016/j.sbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakrabortee S, et al. Catalytic and chaperone-like functions in an intrinsically disordered protein associated with desiccation tolerance. Proc Natl Acad Sci USA. 2010;107(37):16084–16089. doi: 10.1073/pnas.1006276107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andresen C, et al. Transient structure and dynamics in the disordered c-Myc transactivation domain affect Bin1 binding. Nucleic Acids Res. 2012;40(13):6353–6366. doi: 10.1093/nar/gks263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y, et al. Phosphorylation-induced conformational ensemble switching in an intrinsically disordered cancer/testis antigen. J Biol Chem. 2015;290(41):25090–25102. doi: 10.1074/jbc.M115.658583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng Y, et al. The cancer/testis antigen prostate-associated gene 4 (PAGE4) is a highly intrinsically disordered protein. J Biol Chem. 2011;286(16):13985–13994. doi: 10.1074/jbc.M110.210765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prakash K, et al. Symptomatic and asymptomatic benign prostatic hyperplasia: Molecular differentiation by using microarrays. Proc Natl Acad Sci USA. 2002;99(11):7598–7603. doi: 10.1073/pnas.112191399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng Y, et al. Prostate-associated gene 4 (PAGE4) protects cells against stress by elevating p21 and suppressing reactive oxygen species production. Am J Clin Exp Urol. 2013;1(1):39–52. [PMC free article] [PubMed] [Google Scholar]
- 32.Rajagopalan K, et al. The Stress-response protein prostate-associated gene 4, interacts with c-Jun and potentiates its transactivation. Biochim Biophys Acta. 2014;1842(2):154–163. doi: 10.1016/j.bbadis.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulkarni P, Dunker AK, Weninger K, Orban J. Prostate-associated gene 4 (PAGE4), an intrinsically disordered cancer/testis antigen, is a novel therapeutic target for prostate cancer. Asian J Androl. 2016;18(5):695–703. doi: 10.4103/1008-682X.181818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iavarone C, et al. PAGE4 is a cytoplasmic protein that is expressed in normal prostate and in prostate cancers. Mol Cancer Ther. 2002;1(5):329–335. [PubMed] [Google Scholar]
- 35.Suyama T, et al. Expression of cancer/testis antigens in prostate cancer is associated with disease progression. Prostate. 2010;70(16):1778–1787. doi: 10.1002/pros.21214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sampson N, Ruiz C, Zenzmaier C, Bubendorf L, Berger P. PAGE4 positivity is associated with attenuated AR signaling and predicts patient survival in hormone-naive prostate cancer. Am J Pathol. 2012;181(4):1443–1454. doi: 10.1016/j.ajpath.2012.06.040. [DOI] [PubMed] [Google Scholar]
- 37.Mooney SM, et al. Cancer/testis antigen PAGE4, a regulator of c-Jun transactivation, is phosphorylated by homeodomain-interacting protein kinase 1, a component of the stress-response pathway. Biochemistry. 2014;53(10):1670–1679. doi: 10.1021/bi500013w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leppä S, Bohmann D. Diverse functions of JNK signaling and c-Jun in stress response and apoptosis. Oncogene. 1999;18(45):6158–6162. doi: 10.1038/sj.onc.1203173. [DOI] [PubMed] [Google Scholar]
- 39.Nayler O, Stamm S, Ullrich A. Characterization and comparison of four serine- and arginine-rich (SR) protein kinases. Biochem J. 1997;326(Pt 3):693–700. doi: 10.1042/bj3260693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duncan PI, Stojdl DF, Marius RM, Scheit KH, Bell JC. The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp Cell Res. 1998;241(2):300–308. doi: 10.1006/excr.1998.4083. [DOI] [PubMed] [Google Scholar]
- 41.Sato N, et al. Androgenic induction of prostate-specific antigen gene is repressed by protein-protein interaction between the androgen receptor and AP-1/c-Jun in the human prostate cancer cell line LNCaP. J Biol Chem. 1997;272(28):17485–17494. doi: 10.1074/jbc.272.28.17485. [DOI] [PubMed] [Google Scholar]
- 42.Tillman K, Oberfield JL, Shen XQ, Bubulya A, Shemshedini L. c-Fos dimerization with c-Jun represses c-Jun enhancement of androgen receptor transactivation. Endocrine. 1998;9(2):193–200. doi: 10.1385/ENDO:9:2:193. [DOI] [PubMed] [Google Scholar]
- 43.Isaacs JT, et al. Adaptive auto-regulation of androgen receptor provides a paradigm shifting rationale for bipolar androgen therapy (BAT) for castrate resistant human prostate cancer. Prostate. 2012;72(14):1491–1505. doi: 10.1002/pros.22504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karantanos T, Corn PG, Thompson TC. Prostate cancer progression after androgen deprivation therapy: Mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013;32(49):5501–5511. doi: 10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balázsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: From microbes to mammals. Cell. 2011;144(6):910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lenz P, Søgaard-Andersen L. Temporal and spatial oscillations in bacteria. Nat Rev Microbiol. 2011;9(8):565–577. doi: 10.1038/nrmicro2612. [DOI] [PubMed] [Google Scholar]
- 47.Lavery DN, McEwan IJ. Structural characterization of the native NH2-terminal transactivation domain of the human androgen receptor: A collapsed disordered conformation underlies structural plasticity and protein-induced folding. Biochemistry. 2008;47(11):3360–3369. doi: 10.1021/bi702221e. [DOI] [PubMed] [Google Scholar]
- 48.McEwan IJ. Intrinsic disorder in the androgen receptor: Identification, characterisation and drugability. Mol Biosyst. 2012;8(1):82–90. doi: 10.1039/c1mb05249g. [DOI] [PubMed] [Google Scholar]
- 49.Chandrasekar T, Yang JC, Gao AC, Evans CP. Mechanisms of resistance in castration-resistant prostate cancer (CRPC) Transl Androl Urol. 2015;4(3):365–380. doi: 10.3978/j.issn.2223-4683.2015.05.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wadosky KM, Koochekpour S. Molecular mechanisms underlying resistance to androgen deprivation therapy in prostate cancer. Oncotarget. 2016;7(39):64447–64470. doi: 10.18632/oncotarget.10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan H, et al. Suppression of the androgen receptor function by quercetin through protein-protein interactions of Sp1, c-Jun, and the androgen receptor in human prostate cancer cells. Mol Cell Biochem. 2010;339(1-2):253–262. doi: 10.1007/s11010-010-0388-7. [DOI] [PubMed] [Google Scholar]
- 52.Yurtsev EA, Conwill A, Gore J. Oscillatory dynamics in a bacterial cross-protection mutualism. Proc Natl Acad Sci USA. 2016;113(22):6236–6241. doi: 10.1073/pnas.1523317113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jolly MK, et al. Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front Oncol. 2015;5:155. doi: 10.3389/fonc.2015.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terada N, et al. Correlation of Sprouty1 and Jagged1 with aggressive prostate cancer cells with different sensitivities to androgen deprivation. J Cell Biochem. 2014;115(9):1505–1515. doi: 10.1002/jcb.24805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 56.Balaban NQ. Persistence: Mechanisms for triggering and enhancing phenotypic variability. Curr Opin Genet Dev. 2011;21(6):768–775. doi: 10.1016/j.gde.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 57.Mahmoudabadi G, et al. Intrinsically disordered proteins and conformational noise: Implications in cancer. Cell Cycle. 2013;12(1):26–31. doi: 10.4161/cc.23178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152(5):945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jordan NV, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537(7618):102–106. doi: 10.1038/nature19328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roesch A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141(4):583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 63.Ramirez M, et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun. 2016;7:10690. doi: 10.1038/ncomms10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruscetti M, et al. HDAC inhibition impedes epithelial-mesenchymal plasticity and suppresses metastatic, castration-resistant prostate cancer. Oncogene. 2016;35(29):3781–3795. doi: 10.1038/onc.2015.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu M, Jolly MK, Levine H, Onuchic JN, Ben-Jacob E. MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination. Proc Natl Acad Sci USA. 2013;110(45):18144–18149. doi: 10.1073/pnas.1318192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jia D, et al. OVOL guides the epithelial-hybrid-mesenchymal transition. Oncotarget. 2015;6(17):15436–15448. doi: 10.18632/oncotarget.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schweizer MT, et al. Effect of bipolar androgen therapy for asymptomatic men with castration-resistant prostate cancer: Results from a pilot clinical study. Sci Transl Med. 2015;7(269):269ra2. doi: 10.1126/scitranslmed.3010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Denmeade SR, Isaacs JT. Bipolar androgen therapy: The rationale for rapid cycling of supraphysiologic androgen/ablation in men with castration resistant prostate cancer. Prostate. 2010;70(14):1600–1607. doi: 10.1002/pros.21196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roediger J, et al. Supraphysiological androgen levels induce cellular senescence in human prostate cancer cells through the Src-Akt pathway. Mol Cancer. 2014;13:214. doi: 10.1186/1476-4598-13-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu C, et al. Functional p53 determines docetaxel sensitivity in prostate cancer cells. Prostate. 2013;73(4):418–427. doi: 10.1002/pros.22583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shiraishi T, et al. Cancer/testis antigens as potential predictors of biochemical recurrence of prostate cancer following radical prostatectomy. J Transl Med. 2011;9(9):153. doi: 10.1186/1479-5876-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruan B, Fisher KE, Alexander PA, Doroshko V, Bryan PN. Engineering subtilisin into a fluoride-triggered processing protease useful for one-step protein purification. Biochemistry. 2004;43(46):14539–14546. doi: 10.1021/bi048177j. [DOI] [PubMed] [Google Scholar]
- 73.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1–2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 74.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 75.Goddard T, Kneller D. (2008) SPARKY 3 (University of California, San Francisco)
- 76.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36(5):1277–1282. [Google Scholar]
- 77.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: From transcript synthesis to protein degradation. Science. 2008;322(5906):1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.