Significance
Salinity stress is common in many environments and is predicted to intensify. Such stress increases the expression of numerous genes in fish, but the corresponding regulatory mechanisms are unknown. Our study provides a toolkit for discovering and functionally validating cis-regulatory elements (CREs) that control inducible gene expression in fish. This toolkit was used for experimental identification of the first osmotic/salinity-responsive CREs in fish (OSRE1). Our findings greatly empower novel approaches for deciphering fish osmosensory signaling and gene regulatory networks. Because sequence variation in inducible CREs is critical for the evolution of stress tolerance, knowledge of osmolality/salinity-responsive enhancers is critical for revealing the evolution and function of regulatory networks responsible for euryhalinity of fish.
Keywords: biochemical evolution, compatible osmolytes, osmotic stress signaling, enhancer, CRE
Abstract
Fish respond to salinity stress by transcriptional induction of many genes, but the mechanism of their osmotic regulation is unknown. We developed a reporter assay using cells derived from the brain of the tilapia Oreochromis mossambicus (OmB cells) to identify osmolality/salinity-responsive enhancers (OSREs) in the genes of O. mossambicus. Genomic DNA comprising the regulatory regions of two strongly salinity-induced genes, inositol monophosphatase 1 (IMPA1.1) and myo-inositol phosphate synthase (MIPS), was isolated and analyzed with dual luciferase enhancer trap reporter assays. We identified five sequences (two in IMPA1.1 and three in MIPS) that share a common consensus element (DDKGGAAWWDWWYDNRB), which we named “OSRE1.” Additional OSREs that were less effective in conferring salinity-induced trans-activation and do not match the OSRE1 consensus also were identified in both MIPS and IMPA1.1. Although OSRE1 shares homology with the mammalian osmotic-response element/tonicity-responsive enhancer (ORE/TonE) enhancer, the latter is insufficient to confer osmotic induction in fish. Like other enhancers, OSRE1 trans-activates genes independent of orientation. We conclude that OSRE1 is a cis-regulatory element (CRE) that enhances the hyperosmotic induction of osmoregulated genes in fish. Our study also shows that tailored reporter assays developed for OmB cells facilitate the identification of CREs in fish genomes. Knowledge of the OSRE1 motif allows affinity-purification of the corresponding transcription factor and computational approaches for enhancer screening of fish genomes. Moreover, our study enables targeted inactivation of OSRE1 enhancers, a method superior to gene knockout for functional characterization because it confines impairment of gene function to a specific context (salinity stress) and eliminates pitfalls of constitutive gene knockouts (embryonic lethality, developmental compensation).
A major challenge of biology is understanding the evolutionary/adaptive significance of genetic variation and the biochemical mechanisms that govern the phenotypic diversity of organisms. Microevolutionary and functional autecology studies aim to explain how organisms adapt to environmental change and stress, but currently they rely heavily on correlations of phenotypes with particular SNPs or other sequence variations (1). Moreover, holistic systems biology approaches aimed at explaining physiological plasticity and acclimatory responses to environmental change and stress often rely on comprehensive correlations between specific environmental conditions and changes in the abundance of particular mRNAs or proteins. There is great need to complement such approaches with studies that establish causal links between sequence variation, changes in gene expression, and environmental signals to understand the mechanistic consequences of global climate change on organismal form and function. Climate change accelerates the melting of polar icecaps, the salinization of coastal areas, and the decrease in average ocean salinity (2). Because salinity is a major abiotic factor that controls the activity and distribution of aquatic animals, such changes significantly impact fish biodiversity and distribution.
Most fish (>25,000 extant species) are teleosts and osmoregulators, meaning that they maintain their extracellular body fluids at a relatively constant osmolality of ∼300 mOsmol/kg (isosmotic to a salinity of 9 g/kg). Only a small minority of fish (such as marine hagfish and elasmobranchs) are osmoconformers (3). Altering habitat salinity causes stress and evokes compensatory osmoregulatory responses in fish (3). Fish species that tolerate only a narrow salinity range are known as “stenohaline species”; those that can tolerate a wide salinity range are referred to as “euryhaline species” (4). Euryhaline fish have evolved special biochemical and physiological mechanisms that allow them to perceive and compensate for changes in the salinity of their aquatic habitat. They can sense osmotic stress, leading to the activation of osmosensory signaling mechanisms that, in turn, control osmoregulatory effectors to alleviate osmotic stress (5). A large number of osmoregulatory effector genes and their protein products are regulated when euryhaline fish experience salinity stress (3, 6, 7). The osmosensory signaling networks that control these effector genes are most potent and apparent in euryhaline species with highly dynamic osmoregulatory ability (8). One such species is Oreochromis mossambicus, which has a very wide salinity tolerance range of 0–120 g/kg (9) and is a well-established model for studies of teleost osmoregulation (10–12).
Evolutionary differences in the environmental regulation of gene expression are often the result of altered cis-regulatory elements (CREs) (13, 14). In fact, genetic variation in CREs and/or trans-acting factors represents an efficient and common evolutionary strategy for changing environmentally modulated gene-expression patterns (15–18). Changed gene-expression patterns, in turn, lead to altered phenotypes to achieve biochemical adaptation and the evolution of populations and species (15–18). Such variation modifies the highly dynamic nature of gene-regulatory networks, which embody the interaction of a large number of CREs and trans factors (19–22). Therefore, the identification and experimental validation of environmentally regulated CREs in organisms that are uniquely adapted to particular habitats is a critical prerequisite for understanding biochemical evolution (23).
Transcriptional enhancers are CREs that were discovered several decades ago, but their identification and the characterization of their environmental, developmental, or tissue-specific activation is still challenging (23). A popular and powerful approach for genome-wide mapping of enhancers is ChiP (24). However, this approach requires knowledge of specific epigenetic chromatin states (e.g., a particular histone posttranslational modification) or enhancer-specific transcription factors and is contingent on the availability of the corresponding antibodies. Computational approaches for de novo enhancer prediction represent an alternative to ChiP, but they often suffer from low-confidence results, particularly when novel types of enhancers or “nonmodel” species are being studied (23, 25). A more powerful approach for de novo identification of enhancers is the combination of computational prediction and experimental validation (26, 27).
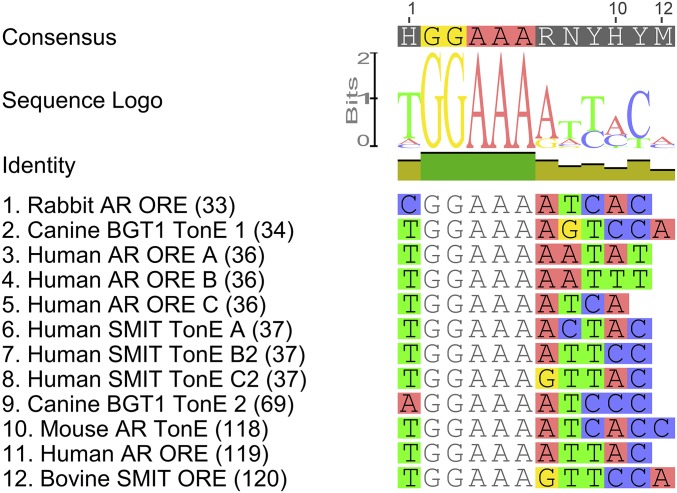
Unbiased experimental identification of novel enhancers is accomplished by enhancer trap reporter assays, e.g., dual luciferase assays, which are highly sensitive for quantifying gene expression over a wide dynamic range (28). These assays rely on established cell lines to permit the high-throughput screening of the enhancer activity of hundreds of different sequence fragments (29–32). Mammalian cell lines have been used to identify the first minimal tonicity-responsive enhancer (TonE), also known as an “osmotic-response element” (ORE) (33–35). Multiple copies of TonE/ORE are present in many mammalian genes that control the intracellular concentrations of compatible organic osmolytes, including betaine/γ-amino-butyric acid transporter (BGT1) (34), aldose reductase (AR) (33, 36), and sodium-myo-inositol transporter (SMIT) (37).
We recently established several immortalized O. mossambicus cell lines to render enhancer trap reporter assays feasible for euryhaline fish (38). Cultured cells are much more amenable to mechanistic and causal dissection of environmental stress responses than tissues of complex and long-lived organisms analyzed in situ (39, 40). Therefore, if the cellular and biochemical phenotypes observed in the tissues of whole organisms are reproducible in cell culture, their mechanistic basis can be revealed using cell lines as an alternative to animal models. For instance, the osmotic induction of the pathway for synthesis of the compatible organic osmolyte myo-inositol, which is evident in many O. mossambicus tissues, is fully reproducible in the OmB cell line derived from the brain of the tilapia O. mossambicus (38, 41–43).
Inositol monophosphatase 1 (IMPA1.1) and myo-inositol phosphate synthase (MIPS) are the two enzymes comprising the myo-inositol biosynthesis (MIB) pathway. This pathway plays a key physiological role in teleost osmoregulation, because it converts glucose-6-phosphate to the compatible organic osmolyte myo-inositol, which protects cells from salinity-induced damage (44). IMPA1.1 and MIPS mRNA, protein, and activity are all highly induced by salinity stress, resulting in elevated levels of the metabolite myo-inositol (41, 45). myo-Inositol is one of only a handful of compatible organic osmolytes that are universally used by all cells to protect macromolecular structure and function during osmotic stress (44, 46–48). Because the enzymes involved in compatible organic osmolyte synthesis and degradation are critical for osmoregulation, they represent excellent targets for elucidating the mechanisms by which osmosensory signaling networks control gene expression during salinity stress. In the current study we developed an enhancer trap reporter assay for the OmB cell line and used it to identify and functionally validate an osmotic/salinity-responsive CRE, OSRE1, that is present in multiple copies in the IMPA1.1 and MIPS genes of euryhaline fish.
Results
Development of the Luciferase Reporter Assay for Identifying CREs in Tilapia OmB Cells.
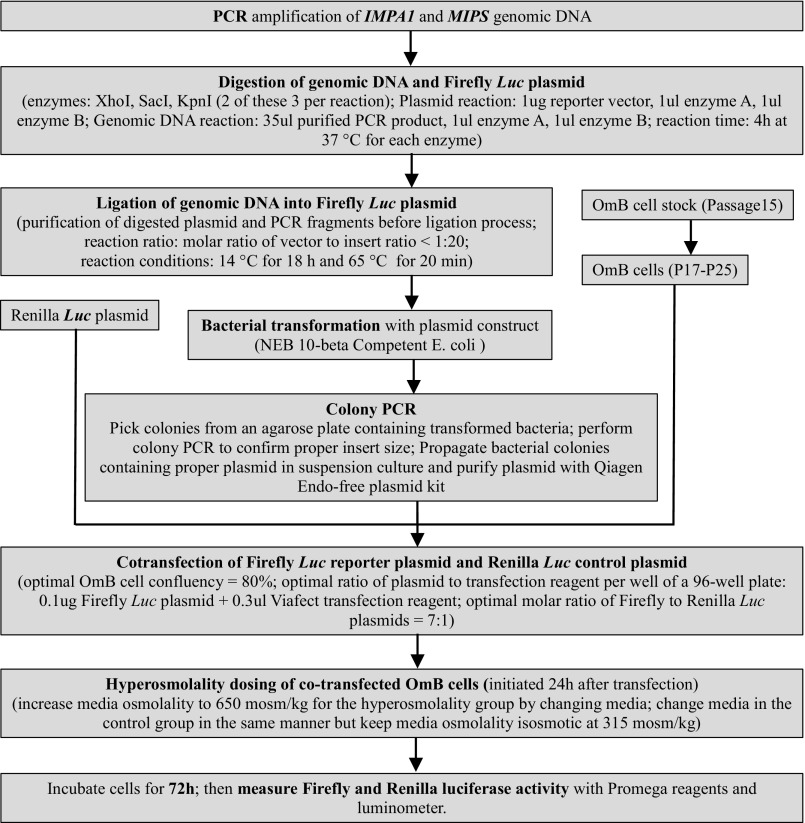
We developed an enhancer trap reporter assay for identifying OSREs by using firefly luciferase reporter plasmid, Renilla luciferase normalizer plasmid, and the OmB cell line. Transfection of OmB cells was most efficient at 80% cell confluency using ViaFect reagent (Fig. S1 A and B). The optimal ratio of Firefly to Renilla luciferase constructs transfected into OmB cells was 8:1. The highest dynamic range of reporter activity was accomplished when using a ratio of 1 µg DNA to 3 µL of transfection reagent. The optimal time of dosing cells in hyperosmotic medium was determined by transfecting OmB cells kept in isosmotic medium at different confluency (40–90%) with firefly luciferase reporter plasmid. Under these conditions luciferase activity represents the baseline, because reporter gene expression is driven only by the minimal promoter. These experiments established that reporter activity is highest at 80% cell confluency when measured 96 h after transfection (Fig. S1C). Therefore, all subsequent experiments were performed using cells at 80% confluency at the time of dosing. Dosing of cells in hyperosmotic medium and parallel isosmotic controls was performed 24 h after transfection, and luciferase reporter activity was measured 72 h after dosing to allow sufficient time for active luciferase enzyme to accumulate in the cells. This time course is supported by the observation that protein levels of IMPA1.1 and MIPS were much higher after 72-h than after 24-h exposure to hyperosmolality (Fig. 1). Therefore, an exposure of 72 h was chosen to maximize the signal-to-noise ratio and to increase the resolution for statistical analysis of reporter assays. The entire procedure and optimal conditions determined for each step are summarized in Fig. S2.
Fig. S1.
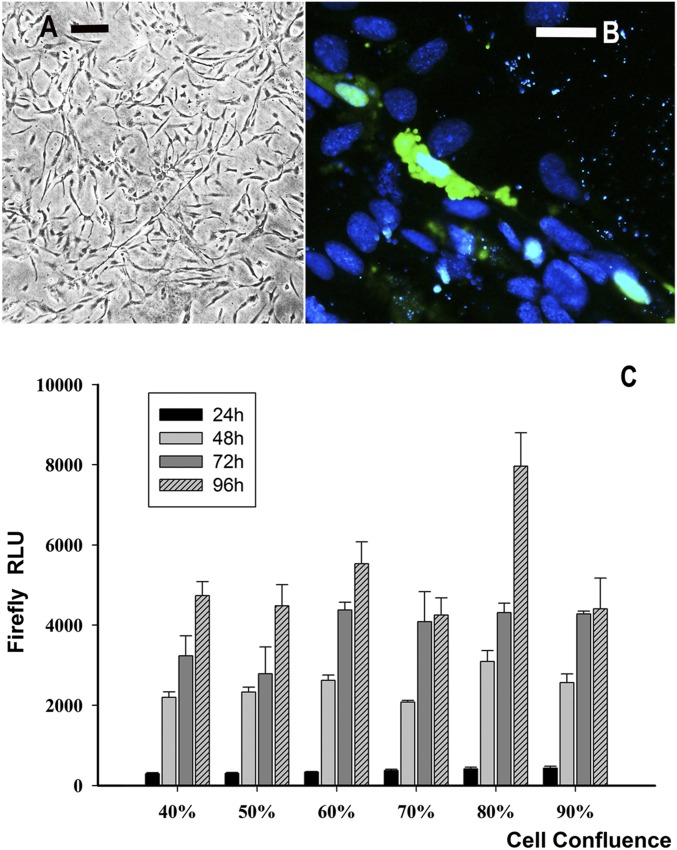
Optimization of transient transfection of tilapia OmB cells. (A) Phase-contrast micrograph showing OmB cells at 80% confluency. (Scale bar, 200 µM.) (B) Fluorescence micrograph showing OmB cells transiently transfected with eGFP plasmid (Addgene) and counterstained with NucBlue Live ReadyProbes Reagent (Thermo Fisher Scientific) (Scale bar, 25 µM.) (C) In addition to the eGFP reporter, luciferase reporter assays indicate that constitutive reporter gene activity (Firefly luciferase driven only by the minimal promoter) is highest when OmB cells are transfected at 80% confluency and luciferase activity is measured 96 h after transfection. All cells were cultured in isosmotic medium (315 mOsmol/kg) for these experiments; data represent means ± SD. RLU, relative luciferase activity units.
Fig. 1.
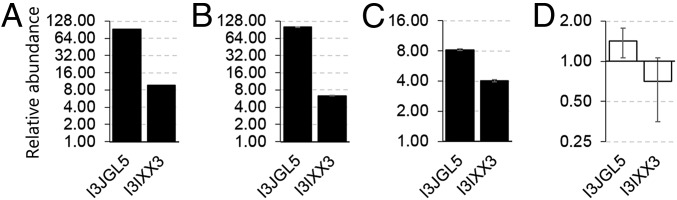
Targeted SWATH-MS/Skyline protein quantitation of IMPA1.1 and MIPS enzymes in cells grown in hyperosmotic (650 mOsmol/kg) medium relative to isosmotic (315 mOsmol/kg) controls, for which the abundance is 1. Proteomic analysis was performed on tryptic MIPS and IMPA1.1 peptides whose sequence is 100% conserved between O. mossambicus and O. niloticus using the O. niloticus proteome as a reference. Hence, accession numbers are I3JGL5 for IMPA1.1 and I3IXX3 for MIPS. Columns shown in black indicate a significant increase in protein abundance under hyperosmotic conditions (Benjamini–Hochberg adjusted P < 0.01). White columns indicate no significant effect of hyperosmolality (P > 0.05). (A and B) The increase in the abundance of IMPA1.1 and MIPS proteins after 72 h hyperosmolality did not differ in untransfected OmB cells (A) and transfected OmB cells (B). (C and D) In addition, both proteins were up-regulated after 24 h of hyperosmotic stress in the absence of actinomycin D (C), whereas the hyperosmotic up-regulation was completely inhibited by 10 µM actinomycin D (D). Data shown are means ± SEM, n = 5. Quantitative data and spectral libraries are accessible at Panorama public (https://panoramaweb.org/labkey/XW2016-1.url).
Fig. S2.
Optimal conditions for dual luciferase reporter assays and enhancer trapping in O. mossambicus OmB cells. A complete enhancer trap assay for evaluating a single sequence segment consists of cell culture; PCR and restriction digestion of genomic DNA and reporter plasmid; ligation of genomic DNA into reporter plasmid, bacterial propagation and plasmid purification; OmB cell transfection; hyperosmotic dosing; and measurement of dual luciferase activity.
Validation of Transcriptional Regulation as the Mechanism for Salinity-Induced Increases in MIPS and IMPA1.1.
To verify that the transfection procedure had no influence on the hyperosmotic up-regulation of MIPS and IMPA1.1, we compared the abundance of these proteins in transfected and nontransfected OmB cells grown for 72 h in isosmotic (315 mOsmol/kg) and hyperosmotic (650 mOsmol/kg) media. Targeted protein quantitation by sequential window acquisition of all theoretical fragment ion spectra (SWATH)-MS and Skyline analysis confirmed significant (P < 0.01) increases in IMPA1.1 and MIPS that were independent of the transfection procedure (Fig. 1 A and B). The mechanism of up-regulation of MIPS and IMPA1.1 was determined to be transcriptional using actinomycin D as a transcription inhibitor. For this purpose, cells were exposed to hyperosmolality (650 mOsmol/kg) for only 24 h because 48-h (and longer) exposure in the presence of actinomycin D was lethal for the majority of cells (Fig. S3). A significant (P < 0.01) increase in IMPA1.1 and MIPS protein abundance was also evident after 24-h exposure to 650 mOsmol/kg hyperosmolality, albeit at lesser magnitude than after 3 d (8.2 ± 0.2-fold for IMPA1.1 and 4.1 ± 0.1-fold for MIPS) (Fig. 1C). However, when transcription was inhibited by addition of actinomycin D, the hyperosmotic induction of both proteins was completely abolished (P > 0.05) (Fig. 1D). All data and metadata for SWATH-MS/Skyline targeted quantitation are available at Panorama Public (https://panoramaweb.org/labkey/XW2016-1.url).
Fig. S3.
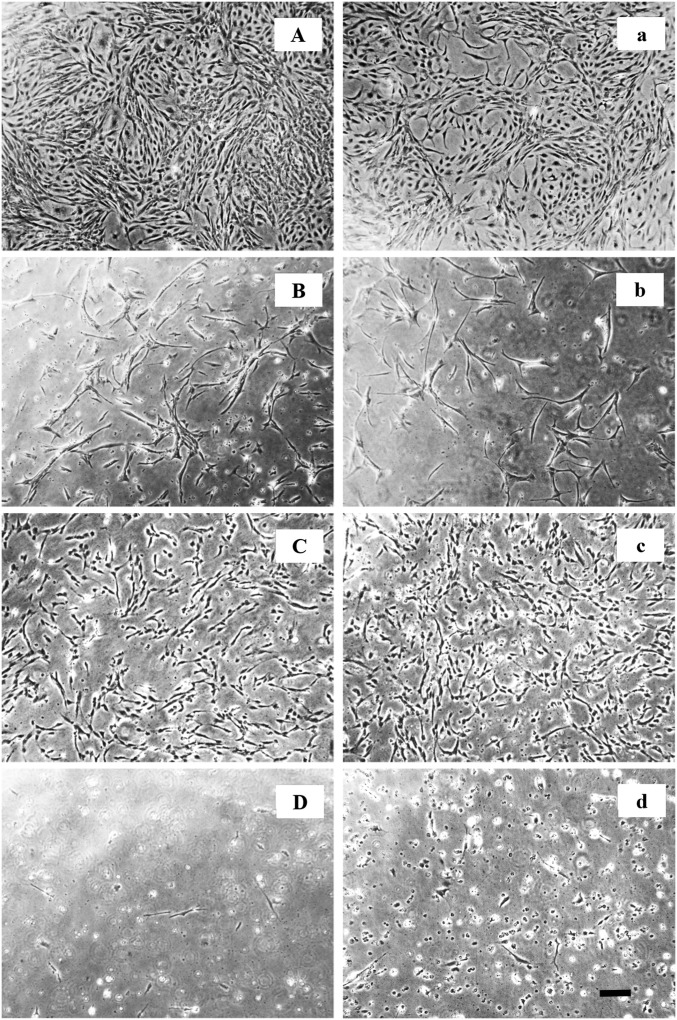
Morphology of OmB cells grown in isosmotic (Left) or hyperosmotic (Right) medium. (Scale bar, 200 µm.) (A and B) Transfection initially caused significant cell death and slowed growth rates at 24 h under isosmotic (B, Left) and hyperosmotic (B, Right) conditions relative to untransfected cells (A, Right and Left, respectively). Hyperosmolality (650 mOsmol/kg) slowed cell proliferation in untransfected (A, Right) and transfected (B, Right) cells relative to isosmotic conditions at 24 h (A, Left and B, Left), but the effect was small. (C) Adding 10 µM actinomycin D to isosmotic (Left) and hyperosmotic (Right) medium had some inhibitory effect on cell proliferation, but cell confluency is still high at 24 h. (D) In contrast, after 48-h exposure to 10 µM actinomycin D very few cells remain alive under both isosmotic (Left) and hyperosmotic (Right) conditions.
Cloning and Reporter Assay Screening of Large Fragments of IMPA1.1 and MIPS.
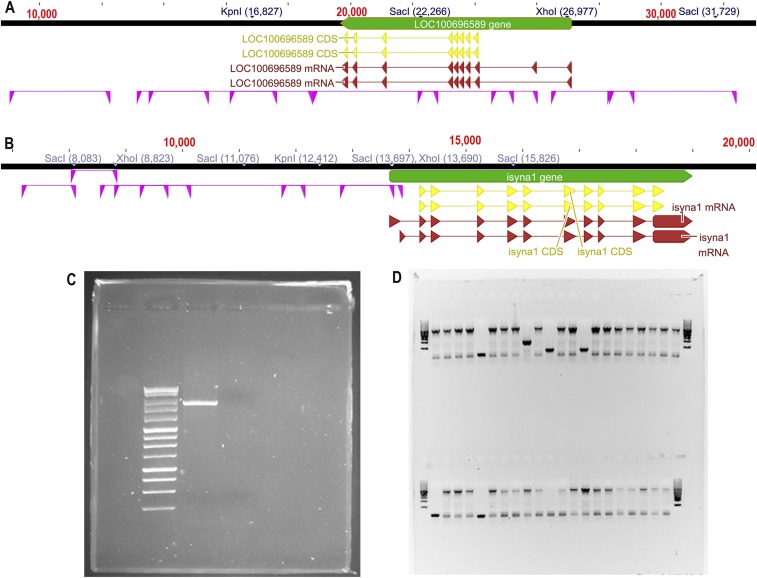
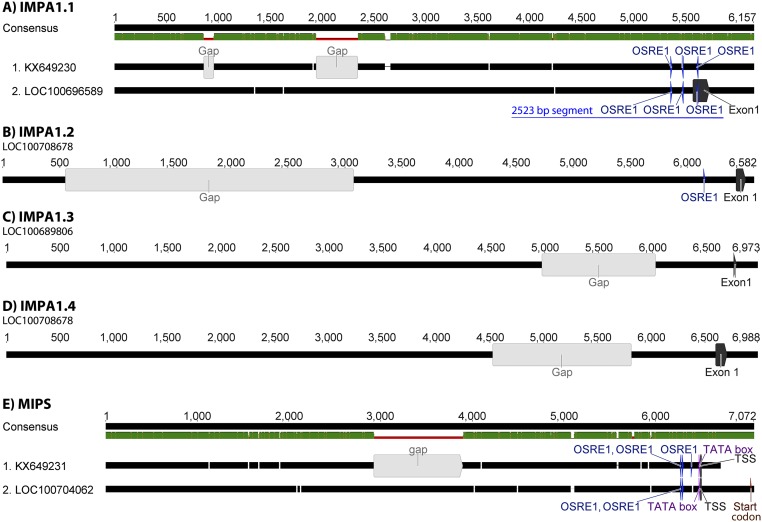
When cloning the 5′ regulatory sequence (RS), it is critical to consider the orientation of Oreochromis niloticus IMPA1.1 and MIPS genes. We used the National Center for Biotechnology Information (NCBI) database that shows that IMPA1.1 is encoded on the antisense strand and the MIPS gene is encoded on the sense strand (Fig. S4 A and B). The 5′ RS of O. mossambicus IMPA1.1 (8,031 bp preceding the start codon ATG) and large portions of the gene downstream of the 5′ RS (14,665 bp) were cloned in the form of 0.7- to 4.4-kb segments. Only one of those segments significantly induced reporter gene activity (26.9 ± 2.8-fold; P < 0.01) during hyperosmolality (Fig. 2A). This 2,523-bp segment spans −1941 to +582 bp relative to the transcription start site (TSS), which has been annotated by the NCBI for O. niloticus IMPA1.1 (https://www.ncbi.nlm.nih.gov/gene/100696589). The O. mossambicus IMPA1.1 5′ RS containing this segment was sequenced (GenBank accession no. KX649230) and aligned to the orthologous O. niloticus sequence. A single 7-bp deletion, a single 1-bp insertion, and 28 SNPs are present in the 2,523-bp 5′ RS segment of O. mossambicus (98.5% identity to O. niloticus) (Fig. S5A).
Fig. S4.
(A and B) Genomic structure of O. niloticus IMPA1.1 (A) and MIPS (B) genes serving as templates for cloning O. mossambicus orthologous sequences. Genes are indicated in green, mRNA in red, and the coding sequence (CDS) in yellow. Orthologous sequences cloned for O. mossambicus and assayed for hyperosmotic enhancer activity are indicated in purple. (C) Amplicon resulting from PCR of a genomic DNA fragment of O. mossambicus IMPA1.1. The first lane contains a 1-kb DNA ladder (Promega), and the second lane depicts a 6.5-kb amplicon used for enhancer trap reporter assays. (D) DNA agarose gel containing typical results of colony PCR. The four outermost lanes are loaded with 1-kb DNA ladder markers. It is evident that most (but not all) colonies harbor an insert of the expected size (6.5 kb). Bands at the bottom of each lane represent primer dimers.
Fig. 2.
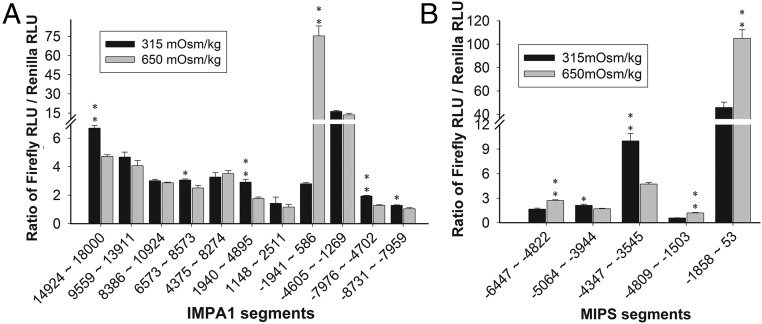
Dual-Glo luciferase reporter assays of long genomic portions of IMPA1.1 (A) and MIPS (B) genes. Black columns represent isosmotic controls (315 mOsmol/kg), and gray columns represent OmB cells dosed in hyperosmotic medium (650 mOsmol/kg). Numbers on the x axis denote the location of genomic sequence fragments relative to the TSS. Double asterisks indicate sequences in which the F/R ratio increased significantly during hyperosmotic stress (P < 0.01; F test followed by t test). One-way ANOVA combined with Tukey's test for assessing statistical significance yielded the same results. Data shown are means ± SD of four biological replicates; RLU, relative luciferase activity units.
Fig. S5.
Representation of tilapia IMPA1 and MIPS genomic DNA upstream of the translation start codon (ATG). All sequence fragments are depicted in the 5′→3′ direction. (A) Alignment of IMPA1.1 5′ RS from O. mossambicus (KX649230) with the orthologous O. niloticus sequence (LOC100696589). SNPs are indicated by orange lines within the green bar (green = 100% sequence identity). Deletions are indicated by white spaces in the black bar representing the O. mossambicus sequence, and insertions are indicated by white spaces in the black bar representing the O. niloticus sequence. OSRE1 motifs matching the DDKGGAAWWDWWYDNRB consensus are indicated in blue, and exon 1 containing the TSS is indicated in gray. (B–D) The corresponding 5′ RS of O. niloticus IMPA1.2, IMPA1.3, and IMPA1.4 paralogous genes with the OSRE1 motifs (if present) and exon 1 displayed as in A. (E) Alignment of MIPS 5′ RS from O. mossambicus (KX649231) with the orthologous O. niloticus sequence (LOC100704062). SNPs, OSRE1 motifs, and exon 1 are depicted as in A. The TATA box is shown in purple, and the start codon (ATG) is shown in ochre.
Enhancer trap reporter assays show that three fragments of the MIPS 5′ RS (−6447 to −4822, −4809 to −1503, and −1858 to +53) significantly increased the ratio of firefly to Renilla luciferase activity (the F/R ratio) during hyperosmotic stress (Fig. 2B). The region spanning these segments in the O. mossambicus MIPS gene was sequenced (GenBank accession no. KX649231) and compared with O. niloticus. Alignment of 5691bp of O. mossambicus MIPS 5′ RS with O. niloticus MIPS (https://www.ncbi.nlm.nih.gov/gene/100704062) revealed 77 SNPs, eight deletions (69 bp total), and seven insertions (51 bp total) in this region. Overall O. mossambicus MIPS was 96.5% identical with O. niloticus MIPS in the 5,691-bp 5′ RS region (Fig. S5B). The F/R ratio decreases in the −4347 to −3545 segment of MIPS and in several IMPA1.1 segments during hyperosmolality. Although the decrease is small (less than twofold in all cases), it is significant (P < 0.05 in all cases). We interpret this result as evidence for the presence of insulators or silencers in those regions (49).
Narrowing Sequences Containing Candidate OSREs to Shorter Fragments.
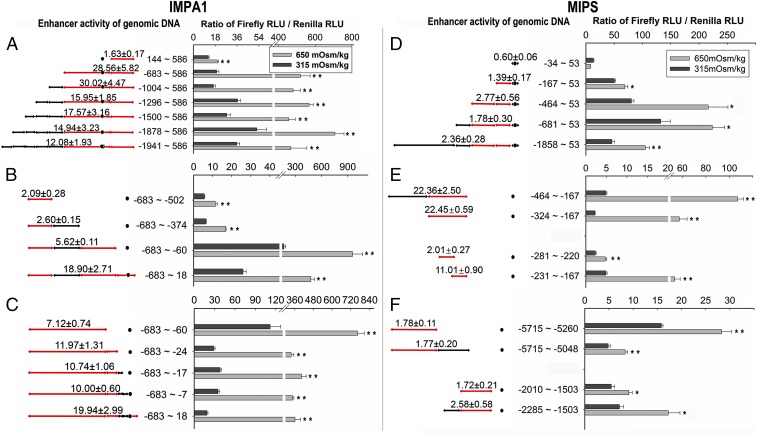
A large number of IMPA1.1 sequences representing sequentially shorter segments of the originally identified OSRE candidate sequence −1941 to +586 were cloned and sequenced. Sequences −683 to +586 and −1004 to +586 showed the highest degree of hyperosmotic trans-activation, whereas sequence +144 to +586 showed much lower hyperosmotic trans-activation (Fig. 3A). This result indicates that candidate IMPA1.1 OSRE(s) are present within an 828-bp sequence spanning −683 to +144 and possibly also within the +144 to +586 segment. The −683 to +144 region was narrowed further by cloning and screening sequentially shorter fragments that all start at position −683. Hyperosmolality enhanced reporter activity 19-fold for the fragment −683 to +18; a shorter fragment (−683 to −60) had three times lesser (sixfold) OSRE activity, and a longer fragment (−683 to 586) had 1.5× greater (29-fold) OSRE activity (Fig. 3 A–C). This result suggests that putative IMPA1.1 OSREs reside within the 78-bp region spanning −59 to +18, the 316-bp region spanning −375 to −60, the 182-bp region spanning −683 to −502, and the 568-bp region spanning 19–586. We were able to differentiate further two distinct sequence stretches (−6 to +18 and −24 to −59) within the 78-bp region (−59 to +18), each of which is capable of ca. twofold hyperosmotic trans-activation (Fig. 3C). Thus, for IMPA1.1, we identified five small regions that contain a candidate OSRE (region 1: −683 to −502; region 2: −375 to −60; region 3: −59 to −24; region 4: −6 to +18; and region 5: +19 to +586).
Fig. 3.
Dual-Glo luciferase reporter assays of candidate regions of the IMPA1.1 (A–C) and MIPS (D–F) genes. (Left) The sequence location, lengths, and fold change in reporter activity in hyperosmotic vs. isosmotic medium. The fold change represents the F/R ratio at hyperosmolality compared with the F/R ratio at isosmolality. Red lines indicate sequences containing one or more OSRE(s); a black dot indicates the TSS. (Right) The F/R ratio recorded for cells grown in isosmotic or hyperosmotic medium is plotted for each of the sequence fragments depicted at the left. Data represent means ± SD of four biological replicates. Asterisks indicate that the F/R ratio is significantly increased by hyperosmotic stress; **P < 0.01 and *P < 0.05 when using the F test followed by t test. One-way ANOVA combined with Tukey's test for assessing statistical significance yielded the same results, except that significance improved from P < 0.05 to P < 0.01 for MIPS segments −464 to +53 (D) and segments −2255 to +1503 (F). RLU, relative luciferase activity units.
The MIPS candidate sequence −1858 to +53 was sequentially shortened from the 5′ end while retaining the same 3′ end (position +53). Reporter activity of the resulting fragments indicates that three segments (−167 to −35, −464 to −167, and −1858 to −681) contain a candidate OSRE, but possible silencers or insulators also may be present in the −1858 to +53 region (Fig. 3D). We were able to isolate the −464 to −167 fragment and dissect it into shorter segments (Fig. 3E). The −324 to −167 segment showed 22-fold hyperosmotic trans-activation similar to that of the larger sequence spanning −464 to −167 (Fig. 3E). Two fragments within the −324 to −167 segment showed twofold (−281 to −220) and 11-fold (−231 to −167) hyperosmotic trans-activation (Fig. 3E). This result suggests that an OSRE is present between −281 and −220 and potentially multiple OSREs are present in the 66-bp sequence spanning −231 to −167. Sequentially shorter fragments also were generated for the other two large MIPS 5′ RS sequences (−6447 to −4822 and −4809 to −1503) harboring candidate OSREs. One putative OSRE was narrowed to within a 446-bp region (−5715 to −5260) displaying 1.8-fold hyperosmotic trans-activation (Fig. 3F). Another putative OSRE was narrowed to within a 783-bp region (−2285 to −1503) displaying 2.6-fold hyperosmotic trans-activation (Fig. 3F). Thus, for MIPS, we identified five regions containing OSREs (region 1: −5715 to −5260; region 2: −2285 to −1503; region 3: −280 to −220; region 4: −231 to −167; and region 5: −165 to −35).
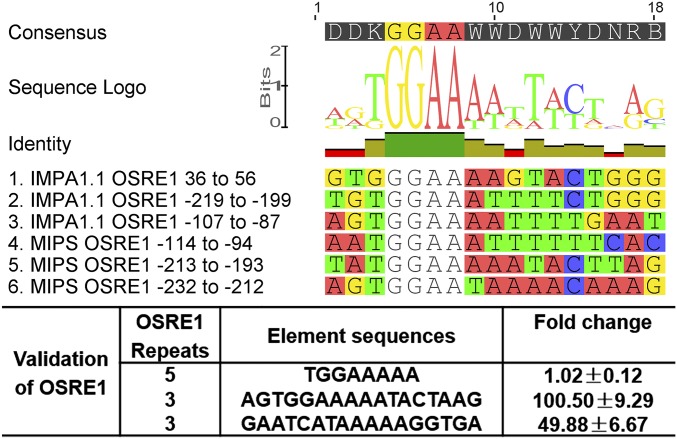
Prediction and Validation of OSRE1.
Manual screening of the minimal IMPA1.1 and MIPS regions that confer osmotic induction of reporter gene activity yielded 11 GGAA[N]A and one GGAAGGA candidate sequences that were present in three IMPA1.1 regions in either reverse or forward orientation (region 2: −372 to −368, −212 to −208, −143 to −138, and −100 to −95; region 4: −3 to +3; region 5: +43 to +48 and +572 to +577) and in all MIPS regions (region 1: −5578 to −5573 and −5367 to −5362; region 2: −2252 to −2248, −2025 to −2021, and −1698 to −1694; regions 3 and 4 (overlap): −225 to −220; region 4: −207 to −202 and −185 to −180; region 5: −108 to −104. The [N] in the GGAA[N]A motif designates an optional nucleotide. GGAA[N]A sequences are absent from the −683 to −502 and the −59 to −24 regions of IMPA1.1. Therefore, this motif cannot account for the hyperosmotic enhancer activity of these segments. To test the hypothesis that GGAA[N]A sequences of IMPA1.1 and MIPS confer hyperosmotic induction of those genes and to define a more specific consensus sequence, we tested them experimentally (Table 1). Synthetic constructs for 12 candidate GGAA[N]A-containing sequences were generated by self-annealing oligonucleotide primers that each contained two repeats of a single putative OSRE sequence. These constructs were tested separately for OSRE activity using dual luciferase reporter assays in OmB cells (Table 1).
Table 1.
Forward primer sequences containing two repeats of a single OSRE candidate sequence for self-annealing and amplification of the corresponding synthetic DNA
| Candidate no. | Primer sequence for self-annealing and amplification of synthetic DNA | Fold change |
| 1 | CTGTAAGTGGAAAATTTTGAATACTGTGTAAGTGGAAAATTTTGAATGGTAC | 29.67 ± 2.49 |
| 2 | CAATCAGAGGAAGAAACCATTCACTGAATCAGAGGAAGAAACCATTCGGTAC | 1.55 ± 0.16 |
| 3 | CCAAAGTGGGAAAAGTACTGGGACTGCAAAGTGGGAAAAGTACTGGGGGTAC | 16.31 ± 3.60 |
| 4 | CGATAGGTGGAAACAGCATGGAACTGGATAGGTGGAAACAGCATGGAGGTAC | 1.92 ± 0.19 |
| 5 | CCAATCAAGGAATAAAATTTCAACTGCAATCAAGGAATAAAATTTCAGAGCT | 2.07 ± 0.42 |
| 6 | CATATCAGGGAAGGAGAACTACACTGATATCAGGGAAGGAGAACTACGAGCT | 1.48 ± 0.09 |
| 7 | CATGTAGTGGAATAAAACAAAGACTGATGTAGTGGAATAAAACAAAGGAGCT | 6.40 ± 0.45 |
| 8 | CATGTTATGGAAAAATACTTAGACTGATGTTATGGAAAAATACTTAGGAGCT | 82.81 ± 4.10 |
| 9 | CCGCTGCAGGAAAATCGAGAAAACTGCGCTGCAGGAAAATCGAGAAAGAGCT | 1.48 ± 0.11 |
| 10 | CCCAGAATGGAAATTTTTTCACACTGCCAGAATGGAAATTTTTTCACGAGCT | 19.21 ± 1.54 |
| 11 | CCCCCCTGGGAAAATTGCACACACTGCCCCCTGGGAAAATTGCACACGAGCT | 1.44 ± 0.15 |
| 12 | CATGCACTGGAAAGACCGGGTCACTGATGCACTGGAAAGACCGGGTCGAGCT | 1.88 ± 0.30 |
Fold change represents the F/R ratio in 650 mOsmol/kg compared with the F/R ratio in 315 mOsmol/kg. Data shown are means ± SD. Italicized and underlined sequences represent putative OSREs.
Only five of the putative OSREs tested were highly potent in conferring hyperosmotic trans-activation of the reporter gene (more than sixfold to 83-fold) (Table 1). The remaining seven putative OSRE candidate tandem sequences were only marginally effective (up to twofold). Multiple sequence alignment of the five most highly effective OSREs yields the 17-bp consensus sequence DDKGGAAWWDWWYDNRB (Fig. 4), with the nomenclature according to ref. 50. We named this CRE “OSRE1.” Although not isolated for experimental validation (Table 1) the IMPA1.1 sequence −219 to −199 is included in the alignment (Fig. 4) because it is the only other candidate sequence matching the OSRE1 consensus. The most common variant of the OSRE1 consensus motif with regard to each position, AGTGGAAAAATACTAAG (Fig. 4), was synthesized de novo and evaluated for its potency to induce reporter activity during hyperosmolality. A triplet of this sequence enhanced reporter activity during hyperosmolality 100-fold when inserted in forward orientation and 50-fold when inserted in reverse orientation (Fig. 4). In contrast, a quintuplet of a shorter version of the OSRE1 consensus, TGGAAAAA, did not show any hyperosmotic enhancer activity (Fig. 4). This shorter version matches the mammalian TonE/ORE consensus, although the last residue of TonE/ORE is more commonly T than A (Fig. S6). This result demonstrates that the mammalian TonE/ORE and the less specific GGAA[N]A motifs are insufficient for hyperosmotic enhancer activity in fish.
Fig. 4.
(Upper) Multiple sequence alignment of six different regions in IMPA1.1 and MIPS genes that match a common consensus, which has OSRE activity. The CRE fitting this consensus motif was named OSRE1. (Lower) The table depicts the results of dual luciferase reporter assays for synthetic constructs representing a core within the OSRE1 consensus that is shared with mammalian TonE/ORE (Row 1) and the most common consensus of OSRE1 in sense (Row 2) and antisense (Row 3) orientation. The right-most column shows the fold change of reporter activity, i.e., the F/R ratio at hyperosmolality compared with the F/R ratio at isosmolality. Data are shown as means ± SD.
Fig. S6.
Alignment of mammalian TonE/ORE elements in AR, BGT1, and SMIT genes of different species depicting the consensus sequence for TonE/ORE. TonE/ORE motifs are labeled as in the original publications; reference numbers are given in parentheses following the sequence name.
Based on this knowledge of the OSRE1 consensus motif, we identified two additional putative OSREs in the IMPA1.1 region −59 to −24, GATGGTACATTCAC and TGCAACAA, (Table 2) that resemble (but do not fully match) the OSRE1 consensus. Synthetic DNA constructs were designed to test whether these sequences have OSRE activity. Reporter assays with these constructs show that neither of these sequences alone is sufficient to account for the enhancer activity of the −59 to −24 region (Table 2). Furthermore, because OSRE1 is not present in the IMPA1.1 −683 to −502 and MIPS −2285 to −1503 regions, another OSRE motif is responsible for the hyperosmotic enhancer activity of these regions.
Table 2.
OSRE candidate sequences in the −59 to −24 region of IMPA1.1
 |
Fold change represents the F/R ratio in 650 mOsmol/kg compared with the F/R ratio in 315 mOsmol/kg. Data shown are means ± SD. Bold italicized underlined sequences represent putative OSREs. The second repeat of synthetic triplet sequences is highlighted in gray.
Discussion
The IMPA1.1 and MIPS genes encode enzymes that comprise MIB pathway. This pathway converts glucose-6-phosphate to the compatible osmolyte myo-inositol and is highly salinity-induced in multiple tissues of O. mossambicus, including gill epithelium, brain, heart, larval epidermis, and OmB cells (38, 41–43, 51). Induction of these two genes is also evident in other euryhaline fish exposed to acute salinity stress. For instance, elevated salinity increases MIPS abundance in O. niloticus and Anguilla anguilla (52) and IMPA1.1 abundance in Gillichthys mirabilis and A. anguilla (53–56). The MIB pathway promotes the accumulation of high concentrations of the compatible osmolyte myo-inositol, which protects cells from salinity-induced damage (44, 48, 57). myo-Inositol and other compatible organic osmolytes are broadly relevant for the biology of all branches on the tree of life (44). In addition to their critical role in osmotic homeostasis, they have crucial functions as chemical chaperones for compensating changes in environmental parameters other than salinity (58, 59). Thus, efficient compatible osmolyte systems may explain why fish having a high salinity tolerance are often also highly tolerant of other types of environmental stress (60). The critical role of the MIB pathway for organismal salinity tolerance is illustrated by overexpression of recombinant MIPS from a highly salt-tolerant strain of cyanobacteria (Synechocystis sp. PCC6803), which significantly increases the salt tolerance of Escherichia coli and Schizosaccharomyces pombe (61). Furthermore, MIPS overexpression in several species of vascular plants significantly increases the salinity tolerance of these multicellular organisms, suggesting that the MIB pathway is a key biochemical determinant of euryhalinity across all major taxa (62–66). MIPS overexpression even increases pathogen resistance in sweet potato (67), as is consistent with the critical roles of compatible osmolytes as general cytoprotectants (58, 59).
An alternative pathway for myo-inositol accumulation is uptake via SMIT (37). Interestingly, intestinal SMIT mRNA increases threefold in euryhaline O. mossambicus but decreases fivefold in a less salinity-tolerant congener, O. niloticus during exposure to elevated salinity, suggesting genetic differences in myo-inositol control (68). Mammalian SMIT and the betaine transporter BGT1 were the first genes for which a TonE/ORE was identified (34, 37, 69). The TonE/ORE enhancer is also present in other mammalian genes that promote the accumulation of compatible organic osmolytes, including AR (33). Furthermore, salinity-responsive CREs have been identified in yeast (70) and in the quinoa plant (71). With the exception of TonE/ORE and osmotic-responsive element (OsmoE) (72) elements, no salinity-regulated CRE has been identified by experimental approaches in any animal, although computational algorithms (19) have been used to predict enhancers based on conserved sequence motifs. Using such algorithms, a TonE/ORE in the deiodinase 2 gene of Fundulus heteroclitus, which is induced by hypo- (rather than hyper-) osmotic stress but matches the mammalian TonE/ORE consensus, has been predicted (73). However, because of the short length of CRE sequences (generally 12–20 bp), their considerable degree of sequence variability, and the large number of possible false positives given the overall size of animal genomes, computational methods for predicting CREs are limited to “reduce the space of testable hypotheses and to drive experimental validation” (74). In addition, novel CREs cannot be discovered with computational methods, and functional motifs for CREs likely differ considerably across taxa (75). Therefore, we advanced an experimental approach for identifying OSRE CREs in O. mossambicus MIPS and IMPA1.1.
To enable enhancer trap reporter assays, we previously established immortalized O. mossambicus cell lines that are highly tolerant of acute hyperosmolality up to 700 mOsm/kg (38). Establishing cell lines was crucial because the isolation and identification of the specifies-specific transcriptional machinery and trans-acting factor that have coevolved with the salinity-induced CRE would be compromised in a heterologous background. Here we have optimized transient transfection of the OmB cell line with a reporter construct containing O. mossambicus genomic DNA positioned proximal to a firefly luciferase reporter gene. Constructs containing different parts of IMPA1.1 and MIPS genes were cotransfected with a Renilla luciferase plasmid serving as a transfection control followed by hyperosmotic induction of transfected cells (with corresponding isosmotic controls kept in parallel) and dual luciferase reporter assays. Before sequentially testing many different portions of MIPS and IMPA1.1 for the salinity-induced reporter (firefly/Renilla luciferase) activity, we showed that the increase in MIPS and IMPA1.1 mRNA and protein is the result of transcriptional regulation. This precaution was necessary because alternative (posttranscriptional) mechanisms are responsible for the increase in other mRNAs during hyperosmolality, e.g., TSC22D3 and osmotic stress transcription factor 1 (OSTF1) in O. mossambicus (76, 77), GADD45 and TonEBP in mammals (78, 79), and aquaporin in yeast (80). Actinomycin D, which is a well-characterized and widely used general transcriptional inhibitor (81), completely eliminated hyperosmotic induction of MIPS and IMPA1.1. The effect of actinomycin D was evident at the protein level. Therefore, posttranscriptional and posttranslational (e.g., mRNA and protein stabilization) mechanisms do not contribute to salinity-induced increases in MIPS and IMPA1.1.
Using dual luciferase reporter assays, we identified multiple OSRE CREs responsible for transcriptional induction of MIPS and IMPA1.1 during salinity stress. Five of these OSREs (two for IMPA1.1 and three for MIPS) share a common consensus motif (OSRE1). Alignment of 6 kbp of the IMPA1.1 5′ RS from O. mossambicus and O. niloticus shows that both contain three OSRE1 elements in close proximity to the TSS (Fig. S5A). The 5′ RS of O. niloticus IMPA1.2 contains only a single OSRE1 (Fig. S5B), and the other two IMPA1 paralogs of O. niloticus lack an OSRE1 in the corresponding entire 6,000-bp region (Fig. S5 C and D). These results are entirely consistent with our previous data showing that tilapia IMPA1.1 is highly induced, whereas IMPA1.2 is marginally induced in both OmB cells and brain of intact O. mossambicus exposed to salinity stress (38, 43). Moreover, in contrast to IMPA1.1 mRNA, the abundance of IMPA1.3 mRNA is not altered in any of the four O. niloticus tissues tested (gill, kidney, fin, and intestine) when salinity increases from fresh water (FW) to 50% or 100% seawater (SW) (53). O. niloticus IMPA1.4 mRNA even decreases under these conditions in gill, fin, and intestine, increases only slightly in kidneys of fish transferred from FW to 50% SW, and does not change in kidneys of fish transferred from FW to 100% SW (53). Alignment of the 6,000-bp MIPS 5′ RS from O. mossambicus and O. niloticus shows that OSRE1 elements are also located close to the TSS and that one OSRE1 is missing in O. niloticus compared with O. mossambicus because of a single-nucleotide deletion (Fig. S5E). This finding is consistent with the lower salinity tolerance of O. niloticus compared with O. mossambicus (68). However, a recently published article shows that MIPS is still potently induced by salinity stress in O. niloticus (52). Thus, the two remaining copies of OSRE1 and/or another OSRE may be sufficient for potent MIPS induction during salinity stress.
Altering the binding affinity for trans-acting factors by sequence variation represents one evolutionary mechanism for changing the environmental/developmental regulation of genes (82, 83). Our reporter assays demonstrate that even subtle sequence variation within the OSRE1 consensus itself alters enhancer activity, and such alterations could be important for tuning the degree of salinity induction of different genes. Another evolutionary mechanism for tuning environmental responsiveness of gene expression pertains to alteration of copy number for a particular CRE (84, 85). Thus, the difference in the number of OSRE1 motifs in the MIPS of O. mossambicus and O. niloticus (Fig. S5E) may be physiologically relevant. In mammals, strongly osmolality-induced genes also contain multiple copies of TonE/ORE. For instance, rabbit AR is osmotically induced via three TonE/OREs (86), dog BGT1 is controlled by two TonE/OREs that act synergistically to stimulate gene expression in response to hypertonicity (69), and human SMIT contains five TonE/OREs (37).
Teleost OSRE1 shares a high degree of sequence similarity with mammalian TonE/ORE. Both CREs have a common core sequence of TGGAAAA, which has been retained throughout vertebrate evolution. Our finding that this core sequence is common in mammals and fish facilitates computational approaches for predicting candidate salinity/osmolality-responsive CREs across vertebrate taxa. However, this core sequence on its own was completely ineffective in conferring salinity induction in our reporter assays (Fig. 4). Therefore, the TGGAAAA core motif is insufficient as a functional CRE, and additional residues that are more divergent between fish and mammals are necessary. For instance, extending the TGGAAAAA motif by two upstream and seven downstream residues to match the OSRE1 consensus (AGTGGAAAAATACTAAG) increases salinity induction from noninducible to 100-fold (Fig. 4). The six base-pair GGAAAA core of mammalian TonE/ORE alone also is not sufficient for conferring hyperosmotic responsiveness to gene expression in mammals (86). We conclude that the less conserved regions flanking the vertebrate GGAAWW core motif are inextricably linked to the functionality of these CREs and that coevolution of these flanking regions with the corresponding transcription factors may have occurred in vertebrates.
Bioinformatics identification of motifs that match the OSRE1 consensus in the −2500 to +50 region of MIPS from other fishes reveals that species closely related to O. niloticus (Astatotilapia burtoni, Pundamilia nyererei, and Maylandia zebra) also have two OSRE1 motifs in MIPS, one less than O. mossambicus. The salinity tolerance of these African cichlids is uncertain, but they are all of FW origin. If they are stenohaline, it is possible that silencer or insulator sequences in MIPS or the responsiveness of other essential osmoprotective genes is altered in these species. Of interest, some euryhaline Poecilia species also have two OSRE1 motifs, and several other euryhaline teleosts have one OSRE1 motif in this region of MIPS (Table S1). Although the comparison of stenohaline and euryhaline species in Table S1 is not comprehensive, it is intriguing that OSRE1 motifs were not found in any of the seven stenohaline species analyzed. Moreover, a MIPS gene is absent from the genome of stenohaline zebrafish (Danio rerio), suggesting lack of selective forces that favor its retention. However, OSRE1 motifs also are absent from MIPS of several euryhaline fishes (Table S1). Given the large phylogenetic distance between different orders of fish and the lack of strong conservation between mammalian TonE/ORE and tilapia OSRE1, it is likely that these CREs have coevolved with the corresponding transcription factor during the 500-My evolution of vertebrates.
Table S1.
The presence of the OSRE1 consensus motif DDKGGAAWWDWWYDNRB in the regulatory region −2500 to +50 of the MIPS gene for some other euryhaline and stenohaline fish
| Species | OSRE1 candidates | Position |
| Euryhaline species | ||
| Callorhinchus milii | None present | |
| Cynoglossus semilaevis | None present | |
| Poecilia reticulata | TTTGGAAAATTATAGGT | −2222 to −2206 |
| AATGGAATATTTCTGAC | −114 to −98 | |
| Poecilia latipinna | GTCAGAAATATTCCATT | −71 to −55 |
| Poecilia mexicana | GTCAGAAATATTCCATT | −68 to −52 |
| ACATATAATTTTCCAAA | −2219 to −2203 | |
| Poecilia formosa | GTCAGAAATATTCCATT | −119 to −103 |
| Xiphophorus maculatus | AATGGAATATTTCTGAC | −210 to −194 |
| Oryzias latipes | None present | |
| Cyprinodon variegatus | None present | |
| Austrofundulus limnaeus | None present | |
| Takifugu rubripes | None present | |
| Fundulus heteroclitus | None present | |
| Larimichthys crocea | None present | |
| Clupea harengus | ATCTGAAATTTTCCATT | 19–35 |
| Kryptolebias marmoratus | None present | |
| Stenohaline species | ||
| Esox lucius | None present | |
| Notothenia coriiceps | None present | |
| Astyanax mexicanus | None present | |
| Ictalurus punctatus | None present | |
| Danio rerio | MIPS gene is missing from genome | |
| Unknown salinity tolerance | ||
| Maylandia zebra | CTAAGTATTTTTCCATA | −221 to −205 |
| AGTGGAATAAAACAAAG | −244 to −228 | |
| Pundamilia nyererei | CTAAGTATTTTTCCATA | −246 to −230 |
| AGTGGAATAAAACAAAG | −269 to −253 | |
| Astatotilapia burtoni | CTTTGTTTTATTCCACT | −274 to −258 |
| TATGGAAAAATACTTGG | −251 to −235 | |
The transcription factor that binds to mammalian TonE/ORE has been identified and named “TonE-binding protein” (TonEBP); it is identical to a protein named “nuclear factor of activated T cells 5” (NFAT5) (69, 87). NFAT5 is a transcription factor of the Rel family, which includes NF-κB. Mammalian NFAT5 is regulated bidirectionally by osmolality: It is activated by hyperosmolality and inhibited by hypo-osmolality (88). Multiple mechanisms control NFAT5 activity during osmotic stress, including changes in phosphorylation (89, 90), nuclear localization (90, 91), trans-activation (92, 93), abundance (88), and sumoylation (94). In most vertebrates, including teleosts and mammals, NFAT5 is encoded by many paralogous isoforms. At least six NFAT5 isoforms are encoded in the O. niloticus genome (NCBI accession nos. XP_005467085, XP_005467086, XP_005467087, XP_005467088, XP_005467089, XP_005467090). Whether any of those fish NFAT5 proteins control transcription via OSRE1 binding remains to be investigated. Nevertheless, because of the GGAAWW core motif shared by OSRE1 and TonE/ORE, NFAT5 isoforms represent prime candidates for trans-acting factors that bind to OSRE1 and control its enhancer activity.
Another candidate protein for trans-activation of OSRE1 is OSTF1. OSTF1 was first identified as a salinity-induced protein in O. mossambicus gills and is orthologous to mammalian TSC22D3 (76, 77, 95). OSTF1 abundance also increases in other euryhaline teleosts during hyperosmotic stress (96–102). In addition to changes in mRNA abundance, phosphorylation (103) and alteration of translational preference via microRNA miR-429 (104) have been identified as mechanisms for osmotic regulation of OSTF1. Although OSRE1 clearly accounts for most of the salinity-induced trans-activation of MIPS and IMPA1.1, our results also suggest that more than one type of CRE is responsible for conferring hyperosmotic induction of the IMPA1.1 and MIPS genes. Both genes contain multiple OSRE1 motifs but also contain other sequences that do not match or even resemble the OSRE1 consensus but display robust OSRE activity (regions −59 to −24 and −683 to −502 in IMPA1.1 and −2285 to −1503 in MIPS). Thus, at least one other trans factor, in addition to the putative OSRE1-binding protein, is likely to contribute to the salinity-induced expression of MIB pathway genes in euryhaline fish. In fact, the presence of multiple different enhancers with additive effects on gene regulation represents another evolutionary mechanism by which the environmental control of gene expression can be tuned (105, 106). Such a mechanism has been demonstrated for the mammalian Na+/H+ exchanger (NHE2) gene, which is osmotically induced via OsmoE (GGGCCAGTTGGCGCTGGG) and TonE/ORE (GCTGGAAAACCGA) enhancers (72).
Tandem repeats of OSRE1 sequences identified in MIPS and IMPA1.1 were capable of very strong hyperosmotic reporter gene trans-activation (more than sixfold to 83-fold). In the reporter assays, some longer sequences showed lower hyperosmotic induction of trans-activation than the corresponding shorter sequences, although both contained the same number of OSRE1 elements. For instance, the −1004 to 586 fragment of IMPA1.1 showed 30-fold induction of trans-activation, whereas the larger −1296 to 586 fragment only showed 16-fold hyperosmotic induction of trans-activation. We interpret such unexpected differences as evidence for the presence of additional regulatory elements (e.g., silencers, repressors, or insulators) in the longer fragment (23, 49, 107). The statistical likelihood that such additional elements are present increases with the distance of OSRE elements from the TSS. The OSRE1 elements identified in MIPS and IMPA1.1 are all located in relatively close proximity to the TSS, and this location is consistent with the location of TonE/ORE elements in mammalian BGT1 and AR genes (69, 86). Of interest, one OSRE1 element of IMPA1.1 is located downstream of the TSS. However, this OSRE1 is still in very close proximity to the predicted TSS, and regions located 3′ of the TSS have been demonstrated to contain fully functional enhancers (108–110). The OSRE1 consensus sequence AGTGGAAAAATACTAAG is functional in both orientations (forward and reverse), as is consistent with the directional independence of other enhancers (111). However, the forward orientation is more effective (100-fold induction) than the reverse orientation (50-fold induction) of this OSRE1 sequence. The reason for this difference is currently unknown.
In summary, we have identified the main CRE, OSRE1, that enhances transcription of MIB pathway genes in euryhaline tilapia exposed to salinity stress. The 5′ RS of both genes (MIPS and IMPA1.1) also contains at least one additional type of salinity-inducible enhancer that is distinct from OSRE1. The identification of OSRE1 opens the way for future studies aimed at identifying the signaling mechanisms that confer salinity responsiveness to gene-regulatory networks in fish. For instance, ChIP-sequencing using NFAT5 and OSTF1 antibodies will allow testing of whether these transcription factors bind to OSRE1, and, if they do, will reveal genome-wide patterns of gene regulation via OSRE1. In addition to this candidate approach, unbiased approaches for identifying the putative OSRE1-binding protein are now feasible, e.g., pulldown assays using bead-immobilized OSRE1 sequences and identification of OSRE1-binding proteins by MS. Moreover, OSRE1-containing salinity-induced genes can now be rendered unresponsive to environmental salinity by genome editing of OSRE1 enhancers in specific target genes via CRISPR/Cas9 (112). Such an approach will allow studies of gene function in a specific environmental context (salinity stress) while retaining the constitutive expression of target genes under control conditions and eliminating concerns about embryonic lethality and developmental compensation, which are common pitfalls associated with constitutive gene knockouts in transgenic animals (113). In combination with revealing relevant insulators, repressors, and silencers in osmoregulated genes, future studies enabled by OSRE1 identification will propel our understanding of osmosensory signaling networks in euryhaline fish.
Materials and Methods
Cell Culture.
The OmB cell line was used in this study (38). Passage 11 (P11) OmB cells were thawed and maintained in L-15 medium containing 10% (vol/vol) FBS and 100 U/mL penicillin-streptomycin at 26 °C and 2% (vol/vol) CO2. Cells were passaged every 5–7 d using a 1:6 splitting ratio (38). A large supply of OmB cell superstock (P15) was generated and used for this study. All experiments were conducted on cells derived from this superstock at P17–P25. Hyperosmotic medium was prepared by adding an appropriate amount of NaCl to regular isosmotic medium, and osmolality was confirmed with a freezing point micro-osmometer (Advanced Instruments).
Cloning.
Total DNA was extracted from OmB cells using the PureLink Genomic DNA mini Kit (Invitrogen). PCR primers were designed using Geneious 7.1 (Biomatters) and PrimerQuest Tool (Integrated DNA Technologies) using the O. niloticus IMPA1.1 (XP_003439317) and MIPS (XP_003442861) genomic sequences as a reference (Fig. S4). The sequence CCCCC followed by a restriction enzyme site was added to the 5′ end of each primer. The restriction enzymes XhoI, SacI, and KpnI were chosen after screening IMPA1.1 and MIPS genes for restriction sites. PCR Master Mix (Promega) was used to amplify fragments <2.5 kb. Platinum PCR SuperMix (Thermo Fisher Scientific) was used to amplify fragments ranging from 2.5 to 8 kb. PCR was carried out as follows: initial denaturation at 94 °C for 3 min followed by 27–37 cycles of 94 °C for 30 s, 60–62 °C for 30 s, 72 °C for 1–5 min, and 72 °C for 15 min. Annealing and extension time and cycle number depended on the primers and amplicons. PCR products were checked by agarose gel electrophoresis (Fig. S4C) and were purified using the PureLink PCR Purification Kit (Thermo Fisher Scientific).
Amplified fragments of the MIPS 5′ RS, IMPA1.1 5′ RS, IMPA1.1 intragenic sequence, and IMPA1.1 3′ RS were cloned into the pGL4.23 vector (NCBI accession no. DQ904455.1; Promega) using XhoI, SacI, and KpnI enzymes (New England Biolabs). All amplicons were double-digested with two of these three enzymes. Restriction digests contained 35 µL purified PCR product, 4 µL buffer, and 1 µL of the first enzyme followed by 4- to 5-h incubation at 37 °C. Subsequently, an additional 4 µL of buffer and 1 µL of the second enzyme were added followed by another 4- to 5-h incubation at 37 °C. Finally, the enzymes were inactivated by 20-min incubation at 65 °C. The pGL4.23 vector was also double-digested using the same procedure except that 1 µg of vector (in MilliQ water) was used, and the final reaction volume was 20 µL for the first enzyme and 25 µL for the second enzyme. Digested inserts and vector were purified with the PureLink PCR Purification Kit (Thermo Fisher Scientific) and were ligated with T4 DNA ligase (Thermo Fisher Scientific) using 1 µL of vector (5 ng/µL), 14 µL of insert (<1:20 molar ratio), 4 µL of ligase buffer, and 1 µL of T4 ligase (1 U/µL) at 14 °C for 18 h and at 65 °C for 20 min.
The high-efficiency 10-beta–competent E. coli strain (New England Biolabs) was transformed with ligated plasmids. Interestingly, JM109-competent E. coli (Promega) was incompatible with tilapia genomic DNA. Transformation consisted of thawing competent bacteria on ice for 5 min, adding 5 µL ligation product, keeping the tube on ice for 30 min, applying heat shock (42 °C) for exactly 30 s, and placing the tube on ice for another 5 min. Transformed E. coli cells were propagated by the addition of 950 µL super optimal broth with catabolite repression (SOC medium, Thermo Fisher Scientific) and incubation at 500 × g and 37 °C for 90 min. Twenty microliters of this solution were spread onto prewarmed LB-ampicillin plates, and single colonies were picked for PCR to check ligation products. For colony PCR, samples were heated at 95 °C for 10 min and were quick-spun to collect any condensate. Forward and reverse primers were designed to bind to opposite ends of the multiple cloning site in pGL4.23 using Geneious 7.1 (Biomatters). Colony PCR was performed as follows: 94 °C for 3 min plus 27–37 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 1–5 min, and 72 °C for 15 min. Extension time and cycle number were adjusted to match amplicon size. PCR products were checked by agarose gel electrophoresis (Fig. S4D). One or two colonies that contained correctly sized fragments were chosen for plasmid purification. They were inoculated into liquid LB medium and were propagated overnight, and the corresponding plasmids were purified in sufficient quantity for transfection of OmB cells (Plasmid Mini Kit; Qiagen). Endotoxin removal buffer (Qiagen) was used after the sample neutralization step to minimize the toxicity of plasmid solutions during the transfection of OmB cells. DNA sequences of inserts were verified for all purified plasmids by Sanger sequencing at the University of California, Davis DNA Sequencing Facility.
Dual-Glo Luciferase Reporter Assay.
We developed an enhancer trap assay for O. mossambicus genomic DNA using conspecific OmB cells. This assay consists of a reporter vector pGL4.23 containing the genomic DNA prepared as described above and a control vector pGL4.73 (NCBI accession nos. DQ904455.1 and AY738229.1) (Promega). The reporter vector expresses firefly luciferase downstream of the multiple cloning site (MCS), and the control vector constitutively expresses Renilla luciferase. Both plasmids are cotransfected into OmB cells using conditions that were optimized using an eGFP expression plasmid (pMX229; Addgene) as an indicator of transient transfection efficiency (Fig. S1). Luciferase activity was measured in white 96-well plates with a SpectraFluor Plus luminometer (Tecan) and a GloMax luminometer (Promega). Development of this assay for fish cells included optimization of the transfection reagent, of the ratio of transfection reagent to reporter plasmids, of the time to assay following transfection, of the time of hyperosmolality dosing, and of instrument measurement time (Fig. S2). The conditions chosen represent a compromise between maximizing sustained robust induction of reporter activity during hyperosmolality and maintenance of high (>80%) OmB cell viability. Under these conditions technical error (noise vs. signal) is minimized, and the conditions reproduce the conditions under which IMPA1.1 and MIPS are highly induced by hyperosmolality (38). Four biological replicates (different batches of cells grown and treated in separate wells of a 96-well plate) were used for controls and hyperosmotic treatments to assess the effect of each sequence in enhancer trap reporter assays. Statistical analysis of the data was performed using the F test followed by a type-2 (homoscedasticity) or type-3 (heteroscedasticity) t test and by one-way ANOVA followed by Tukey's test (SPSS 19.0).
Measurement of MIPS and IMPA1.1 Protein Abundance Using SWATH-MS.
To confirm that the osmotically responsive enhancer(s) are activated in OmB cells under the conditions used for reporter assays, we quantified MIPS and IMPA1.1 levels. SWATH-MS was used to quantify protein abundances of MIPS and IMPA1.1 in isosmotic and hyperosmotic media. Twenty 60-mm dishes of OmB cells were seeded, grown to 90% confluency, and randomly divided into four groups of five dishes each. Two of these groups were cotransfected with firefly and Renilla luciferase plasmids, and the other two were not transfected (Fig. S3). One transfected and one untransfected group were exposed to hyperosmotic medium (650 mOsmol/kg) for 3 d; the other two groups were kept in isosmotic medium (315 mOsmol/kg) for the same duration. The medium was changed on the second day for all groups. At the end of the 3-d treatment period, cells were washed three times with PBS having the appropriate osmolality, and protein was extracted from cells. Cells then were dislodged, pipetted into a 1.5-mL MCF tube, and centrifuged for 5 s at 3,540 × g. Dishes were tilted, excess PBS was pipetted off, 200 µL of 10% (vol/vol) trichloroacetic acid/90% (vol/vol) acetone/0.2% DTT was added, and samples were incubated at −20 °C for 1 h. Protein extraction, protein assay, and in-solution trypsin digestion were performed as previously reported (114). The spectral library used for Skyline analysis of SWATH-MS data (115) and results are available in Panorama public (116) at https://panoramaweb.org/labkey/XW2016-1.url.
Confirmation of Transcriptional Induction of MIPS and IMPA1.1.
Actinomycin D, which is a well-known transcriptional inhibitor (76, 77, 117), was used to confirm that the mechanism causing an increase in abundance of MIPS and IMPA1.1 protein levels during hyperosmotic stress is transcriptional. Twenty 100-mm dishes containing 100% confluent OmB cells (Fig. S3) were randomly divided into four groups as follows: 315 mOsmol/kg without actinomycin D; 315 mOsmol/kg with 10 µM actinomycin D; 650 mOsmol/kg without actinomycin D; and 650 mOsmol/kg with 10 µM actinomycin-D. In this experiment cells were exposed to 650 mOsmol/kg hyperosmolality and isosmolality (315 mOsmol/kg, handling controls) for only 24 h. As a result, the extent of MIPS and IMPA1.1 increase is less than with 72-h exposure. However, this compromise was necessary to avoid killing cells by prolonged exposure to actinomycin D while retaining a significant hyperosmotic induction of MIPS and IMPA1.1 (Fig. S3). Following dosing in hyper- or isosmotic medium, samples were processed for SWATH-MS and Skyline analysis as described above. The resulting data and metadata are available at Panorama public, https://panoramaweb.org/labkey/XW2016-1.url.
Identification of OSREs for IMPA1.1 and MIPS.
Large portions of genomic DNA associated with IMPA1.1 (5′ RS, 3′ RS, and intragenic) and MIPS (5′ RS) were screened for enhancer activity (the F/R ratio at 650 mOsmol/kg compared with that ratio at 315 mOsmol/kg) using the dual luciferase reporter system. The screening of genomic DNA segments started with large (kilobase-sized) pieces, the length of which was decreased sequentially. If a segment displayed strong osmotically induced enhancer activity, it was divided into smaller segments, which then were assayed separately. Within the resulting smaller segments, segments with high osmotically induced enhancer activity were selected and divided further. The resulting short sequences then were evaluated for potential consensus motifs using Geneious 7.1 (Biomatters). Putative minimal osmotically responsive enhancer motifs predicted from these sequences were too short for cloning and PCR. Thus, they were synthesized de novo using the oligonucleotide annealing method (Integrated DNA Technologies) to assess their osmotically induced enhancer activity. Forward and reverse PCR primers used for this purpose served as the amplicon and contained KpnI and SacI recognition sites. Two complementary primers (forward and reverse) were dissolved in duplex buffer (Integrated DNA Technologies) and were added to the PCR mix in equimolar amounts followed by heating at 94 °C for 2 min and gradual cooling. PCR was performed, and the resulting amplicon was ligated into the reporter plasmid as described above. All synthetic sequences containing potential OSREs were evaluated with the dual luciferase reporter system in OmB cells as described above.
Acknowledgments
We thank Dr. Liqiao Chen and Dr. Erchao Li (East China Normal University) for their support and suggestions, and Toni N. Wiegers, Johnathon Li (University of California, Davis), and Dr. Sarah Teter (Promega) for helpful technical support. This project was supported by National Science Foundation Grant IOS-1355098 (to D.K.) and by China Scholarship Council Fellowship 201406140042 (to X.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KX649230 and KX649231) and are available at Panorama Public, https://panoramaweb.org/labkey/XW2016-1.url.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614712114/-/DCSupplemental.
References
- 1.Wray NR, et al. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14(7):507–515. doi: 10.1038/nrg3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Wijk EM, Rintoul SR. Freshening drives contraction of antarctic bottom water in the Australian Antarctic basin. Geophys Res Lett. 2014;41(5):1657–1664. [Google Scholar]
- 3.Kültz D. Physiological mechanisms used by fish to cope with salinity stress. J Exp Biol. 2015;218(Pt 12):1907–1914. doi: 10.1242/jeb.118695. [DOI] [PubMed] [Google Scholar]
- 4.Wurts WA, Stickney RR. Responses of red drum (Sciaenops ocellatus) to calcium and magnesium concentrations in fresh and salt water. Aquaculture. 1989;76(1-2):21–35. [Google Scholar]
- 5.Fiol DF, Kültz D. Osmotic stress sensing and signaling in fishes. FEBS J. 2007;274(22):5790–5798. doi: 10.1111/j.1742-4658.2007.06099.x. [DOI] [PubMed] [Google Scholar]
- 6.Kültz D. 2013. Osmosensing. Euryhaline Fishes. Fish Physiology, eds McCormick SD, Farrell AP, Brauner CJ (Academic Press, Oxford), Vol 32, pp 45–68.
- 7.Xu Z, et al. Transcriptome profiling and molecular pathway analysis of genes in association with salinity adaptation in Nile tilapia, Oreochromis niloticus. PLoS One. 2015;10(8):e0136506. doi: 10.1371/journal.pone.0136506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kültz D. The combinatorial nature of osmosensing in fishes. Physiology (Bethesda) 2012;27(4):259–275. doi: 10.1152/physiol.00014.2012. [DOI] [PubMed] [Google Scholar]
- 9.Stickney RR. Tilapia tolerance of saline waters: A review. Prog Fish-Cult. 1986;48(3):161–167. [Google Scholar]
- 10.Foskett JK, Bern HA, Machen TE, Conner M. Chloride cells and the hormonal control of teleost fish osmoregulation. J Exp Biol. 1983;106(Sep):255–281. doi: 10.1242/jeb.106.1.255. [DOI] [PubMed] [Google Scholar]
- 11.Kültz D, Bastrop R, Jürss K, Siebers D. Mitochondria-tich (MR) cells and the activities of the Na+/K+-ATPase and carbonic anhydrase in the gill and opercular epithelium of Oreochromis mossambicus adapted to various salinities. Comp Biochem Physiol B. 1992;102(2):293–301. [Google Scholar]
- 12.Kültz D, Jürss K, Jonas L. Cellular and epithelial adjustments to altered salinity in the gill and opercular epithelium of a cichlid fish (Oreochromis mossambicus) Cell Tissue Res. 1995;279(1):65–73. [Google Scholar]
- 13.Kratochwil CF, Meyer A. Closing the genotype-phenotype gap: Emerging technologies for evolutionary genetics in ecological model vertebrate systems. BioEssays. 2015;37(2):213–226. doi: 10.1002/bies.201400142. [DOI] [PubMed] [Google Scholar]
- 14.Cotney J, et al. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell. 2013;154(1):185–196. doi: 10.1016/j.cell.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheatle Jarvela AM, Hinman VF. Evolution of transcription factor function as a mechanism for changing metazoan developmental gene regulatory networks. Evodevo. 2015;6(1):3. doi: 10.1186/2041-9139-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villar D, Flicek P, Odom DT. Evolution of transcription factor binding in metazoans - mechanisms and functional implications. Nat Rev Genet. 2014;15(4):221–233. doi: 10.1038/nrg3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowell RD. Transcription factor binding variation in the evolution of gene regulation. Trends Genet. 2010;26(11):468–475. doi: 10.1016/j.tig.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Wagner GP, Lynch VJ. The gene regulatory logic of transcription factor evolution. Trends Ecol Evol. 2008;23(7):377–385. doi: 10.1016/j.tree.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Narlikar L, Ovcharenko I. Identifying regulatory elements in eukaryotic genomes. Brief Funct Genomics Proteomics. 2009;8(4):215–230. doi: 10.1093/bfgp/elp014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balleza E, et al. Regulation by transcription factors in bacteria: Beyond description. FEMS Microbiol Rev. 2009;33(1):133–151. doi: 10.1111/j.1574-6976.2008.00145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: The evolution of ribosomal regulation in yeast. Proc Natl Acad Sci USA. 2005;102(20):7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yaragatti M, Basilico C, Dailey L. Identification of active transcriptional regulatory modules by the functional assay of DNA from nucleosome-free regions. Genome Res. 2008;18(6):930–938. doi: 10.1101/gr.073460.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shlyueva D, et al. Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Mol Cell. 2014;54(1):180–192. doi: 10.1016/j.molcel.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 24.Levitsky VG, et al. Application of experimentally verified transcription factor binding sites models for computational analysis of ChIP-Seq data. BMC Genomics. 2014;15:80. doi: 10.1186/1471-2164-15-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: Five essential questions. Nat Rev Genet. 2013;14(4):288–295. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 27.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457(7231):854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allard STM, Kopish K. Luciferase reporter assays: Powerful, adaptable tools for cell biology research. Cell Notes. 2008;21:23–26. [Google Scholar]
- 29.Andruska N, Mao C, Cherian M, Zhang C, Shapiro DJ. Evaluation of a luciferase-based reporter assay as a screen for inhibitors of estrogen-ERα-induced proliferation of breast cancer cells. J Biomol Screen. 2012;17(7):921–932. doi: 10.1177/1087057112442960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clément T, Salone V, Rederstorff M. Dual luciferase gene reporter assays to study miRNA function. Methods Mol Biol. 2015;1296:187–198. doi: 10.1007/978-1-4939-2547-6_17. [DOI] [PubMed] [Google Scholar]
- 31.Rong J, et al. Cell-based high-throughput luciferase reporter gene assays for identifying and profiling chemical modulators of endoplasmic reticulum signaling protein IRE1. J Biomol Screen. 2015;20(10):1232–1245. doi: 10.1177/1087057115600414. [DOI] [PubMed] [Google Scholar]
- 32.Zhao SS, et al. Analyzing the promoters of two CYP9A genes in the silkworm Bombyx mori by dual-luciferase reporter assay. Mol Biol Rep. 2013;40(2):1701–1710. doi: 10.1007/s11033-012-2221-8. [DOI] [PubMed] [Google Scholar]
- 33.Ferraris JD, et al. ORE, a eukaryotic minimal essential osmotic response element: The aldose reductase gene in hyperosmotic stress. J Biol Chem. 1996;271(31):18318–18321. doi: 10.1074/jbc.271.31.18318. [DOI] [PubMed] [Google Scholar]
- 34.Takenaka M, Preston AS, Kwon HM, Handler JS. The tonicity-sensitive element that mediates increased transcription of the betaine transporter gene in response to hypertonic stress. J Biol Chem. 1994;269(47):29379–29381. [PubMed] [Google Scholar]
- 35.Burg MB, Kwon ED, Kültz D. Regulation of gene expression by hypertonicity. Annu Rev Physiol. 1997;59:437–455. doi: 10.1146/annurev.physiol.59.1.437. [DOI] [PubMed] [Google Scholar]
- 36.Ko BCB, Ruepp B, Bohren KM, Gabbay KH, Chung SSM. Identification and characterization of multiple osmotic response sequences in the human aldose reductase gene. J Biol Chem. 1997;272(26):16431–16437. doi: 10.1074/jbc.272.26.16431. [DOI] [PubMed] [Google Scholar]
- 37.Rim JS, et al. Transcription of the sodium/myo-inositol cotransporter gene is regulated by multiple tonicity-responsive enhancers spread over 50 kilobase pairs in the 5′-flanking region. J Biol Chem. 1998;273(32):20615–20621. doi: 10.1074/jbc.273.32.20615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardell AM, Qin Q, Rice RH, Li J, Kültz D. Derivation and osmotolerance characterization of three immortalized tilapia (Oreochromis mossambicus) cell lines. PLoS One. 2014;9(5):e95919. doi: 10.1371/journal.pone.0095919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fent K. Fish cell lines as versatile tools in ecotoxicology: Assessment of cytotoxicity, cytochrome P4501A induction potential and estrogenic activity of chemicals and environmental samples. Toxicol In Vitro. 2001;15(4-5):477–488. doi: 10.1016/s0887-2333(01)00053-4. [DOI] [PubMed] [Google Scholar]
- 40.Mazon AdeF, Nolan DT, Lock RAC, Wendelaar Bonga SE, Fernandes MN. Opercular epithelial cells: A simple approach for in vitro studies of cellular responses in fish. Toxicology. 2007;230(1):53–63. doi: 10.1016/j.tox.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 41.Sacchi R, Gardell AM, Chang N, Kültz D. Osmotic regulation and tissue localization of the myo-inositol biosynthesis pathway in tilapia (Oreochromis mossambicus) larvae. J Exp Zool A Ecol Genet Physiol. 2014;321(8):457–466. doi: 10.1002/jez.1878. [DOI] [PubMed] [Google Scholar]
- 42.Sacchi R, Li J, Villarreal F, Gardell AM, Kültz D. Salinity-induced regulation of the myo-inositol biosynthesis pathway in tilapia gill epithelium. J Exp Biol. 2013;216(Pt 24):4626–4638. doi: 10.1242/jeb.093823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gardell AM, et al. Tilapia (Oreochromis mossambicus) brain cells respond to hyperosmotic challenge by inducing myo-inositol biosynthesis. J Exp Biol. 2013;216(Pt 24):4615–4625. doi: 10.1242/jeb.088906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: Evolution of osmolyte systems. Science. 1982;217(4566):1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 45.Villarreal FD, Kültz D. Direct ionic regulation of the activity of myo-inositol biosynthesis enzymes in Mozambique tilapia. PLoS One. 2015;10(6):e0123212. doi: 10.1371/journal.pone.0123212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arakawa T, Timasheff SN. The stabilization of proteins by osmolytes. Biophys J. 1985;47(3):411–414. doi: 10.1016/S0006-3495(85)83932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolen DW. Protein stabilization by naturally occurring osmolytes. Methods Mol Biol. 2001;168:17–36. doi: 10.1385/1-59259-193-0:017. [DOI] [PubMed] [Google Scholar]
- 48.Burg MB. Coordinate regulation of organic osmolytes in renal cells. Kidney Int. 1996;49(6):1684–1685. doi: 10.1038/ki.1996.247. [DOI] [PubMed] [Google Scholar]
- 49.Chetverina D, Aoki T, Erokhin M, Georgiev P, Schedl P. Making connections: Insulators organize eukaryotic chromosomes into independent cis-regulatory networks. BioEssays. 2014;36(2):163–172. doi: 10.1002/bies.201300125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nomenclature Committee of the International Union of Biochemistry (NC-IUB) Nomenclature for incompletely specified bases in nucleic acid sequences. Recommendations 1984. Eur J Biochem. 1985;150(1):1–5. doi: 10.1111/j.1432-1033.1985.tb08977.x. [DOI] [PubMed] [Google Scholar]
- 51.Fiol DF, Chan SY, Kültz D. Identification and pathway analysis of immediate hyperosmotic stress responsive molecular mechanisms in tilapia (Oreochromis mossambicus) gill. Comp Biochem Physiol Part D Genomics Proteomics. 2006;1(3):344–356. doi: 10.1016/j.cbd.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 52.Kalujnaia S, Hazon N, Cramb G. myo-Inositol phosphate synthase expression in the European eel (Anguilla anguilla) and Nile tilapia (Oreochromis niloticus): Effect of seawater acclimation. Am J Physiol Regul Integr Comp Physiol. 2016;311(2):287–298. doi: 10.1152/ajpregu.00056.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalujnaia S, et al. Seawater acclimation and inositol monophosphatase isoform expression in the European eel (Anguilla anguilla) and Nile tilapia (Orechromis niloticus) Am J Physiol Regul Integr Comp Physiol. 2013;305(4):R369–R384. doi: 10.1152/ajpregu.00044.2013. [DOI] [PubMed] [Google Scholar]
- 54.Evans TG, Somero GN. A microarray-based transcriptomic time-course of hyper- and hypo-osmotic stress signaling events in the euryhaline fish Gillichthys mirabilis: Osmosensors to effectors. J Exp Biol. 2008;211(Pt 22):3636–3649. doi: 10.1242/jeb.022160. [DOI] [PubMed] [Google Scholar]
- 55.Kalujnaia S, McVee J, Kasciukovic T, Stewart AJ, Cramb G. A role for inositol monophosphatase 1 (IMPA1) in salinity adaptation in the euryhaline eel (Anguilla anguilla) FASEB J. 2010;24(10):3981–3991. doi: 10.1096/fj.10-161000. [DOI] [PubMed] [Google Scholar]
- 56.Kalujnaia S, Cramb G. Regulation of expression of the myo-inositol monophosphatase 1 gene in osmoregulatory tissues of the European eel Anguilla anguilla after seawater acclimation. Ann N Y Acad Sci. 2009;1163:433–436. doi: 10.1111/j.1749-6632.2009.04457.x. [DOI] [PubMed] [Google Scholar]
- 57.Beck FX, Schmolke M, Guder WG. Osmolytes. Curr Opin Nephrol Hypertens. 1992;1(1):43–52. doi: 10.1097/00041552-199210000-00007. [DOI] [PubMed] [Google Scholar]
- 58.Yancey PH. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol. 2005;208(Pt 15):2819–2830. doi: 10.1242/jeb.01730. [DOI] [PubMed] [Google Scholar]
- 59.Somero GN. Protons, osmolytes, and fitness of internal milieu for protein function. Am J Physiol. 1986;251(2 Pt 2):R197–R213. doi: 10.1152/ajpregu.1986.251.2.R197. [DOI] [PubMed] [Google Scholar]
- 60.Kültz D. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol. 2005;67:225–257. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- 61.Chatterjee A, et al. sll1981, an acetolactate synthase homologue of Synechocystis sp. PCC6803, functions as L-myo-inositol 1-phosphate synthase. Planta. 2006;224(2):367–379. doi: 10.1007/s00425-006-0221-4. [DOI] [PubMed] [Google Scholar]
- 62.Alter S, et al. DroughtDB: An expert-curated compilation of plant drought stress genes and their homologs in nine species. Database (Oxford) 2015;2015:bav046. doi: 10.1093/database/bav046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kusuda H, et al. Ectopic expression of myo-inositol 3-phosphate synthase induces a wide range of metabolic changes and confers salt tolerance in rice. Plant Sci. 2015;232:49–56. doi: 10.1016/j.plantsci.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 64.Joshi R, Ramanarao MV, Baisakh N. Arabidopsis plants constitutively overexpressing a myo-inositol 1-phosphate synthase gene (SaINO1) from the halophyte smooth cordgrass exhibits enhanced level of tolerance to salt stress. Plant Physiol Biochem. 2013;65:61–66. doi: 10.1016/j.plaphy.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 65.Kaur H, et al. Ectopic expression of the ABA-inducible dehydration-responsive chickpea L-myo-inositol 1-phosphate synthase 2 (CaMIPS2) in Arabidopsis enhances tolerance to salinity and dehydration stress. Planta. 2013;237(1):321–335. doi: 10.1007/s00425-012-1781-0. [DOI] [PubMed] [Google Scholar]
- 66.Patra B, Ray S, Richter A, Majumder AL. Enhanced salt tolerance of transgenic tobacco plants by co-expression of PcINO1 and McIMT1 is accompanied by increased level of myo-inositol and methylated inositol. Protoplasma. 2010;245(1-4):143–152. doi: 10.1007/s00709-010-0163-3. [DOI] [PubMed] [Google Scholar]
- 67.Zhai H, et al. A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnol J. 2016;14(2):592–602. doi: 10.1111/pbi.12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ronkin D, Seroussi E, Nitzan T, Doron-Faigenboim A, Cnaani A. Intestinal transcriptome analysis revealed differential salinity adaptation between two tilapiine species. Comp Biochem Physiol Part D Genomics Proteomics. 2015;13:35–43. doi: 10.1016/j.cbd.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 69.Miyakawa H, Rim JS, Handler JS, Kwon HM. Identification of the second tonicity-responsive enhancer for the betaine transporter (BGT1) gene. Biochim Biophys Acta. 1999;1446(3):359–364. doi: 10.1016/s0167-4781(99)00122-0. [DOI] [PubMed] [Google Scholar]
- 70.Dolz-Edo L, Rienzo A, Poveda-Huertes D, Pascual-Ahuir A, Proft M. Deciphering dynamic dose responses of natural promoters and single cis elements upon osmotic and oxidative stress in yeast. Mol Cell Biol. 2013;33(11):2228–2240. doi: 10.1128/MCB.00240-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Orsini F, et al. Beyond the ionic and osmotic response to salinity in Chenopodium quinoa: Functional elements of successful halophytism. Funct Plant Biol. 2011;38(10):818–831. doi: 10.1071/FP11088. [DOI] [PubMed] [Google Scholar]
- 72.Bai L, et al. Characterization of cis-elements required for osmotic response of rat Na(+)/H(+) exchanger-2 (NHE-2) gene. Am J Physiol. 1999;277(4 Pt 2):R1112–R1119. doi: 10.1152/ajpregu.1999.277.4.R1112. [DOI] [PubMed] [Google Scholar]
- 73.López-Bojórquez L, Villalobos P, García-G C, Orozco A, Valverde-R C. Functional identification of an osmotic response element (ORE) in the promoter region of the killifish deiodinase 2 gene (FhDio2) J Exp Biol. 2007;210(Pt 17):3126–3132. doi: 10.1242/jeb.004150. [DOI] [PubMed] [Google Scholar]
- 74.Quattrone A, Dassi E. Introduction to bioinformatics resources for post-transcriptional regulation of gene expression. Methods Mol Biol. 2016;1358:3–28. doi: 10.1007/978-1-4939-3067-8_1. [DOI] [PubMed] [Google Scholar]
- 75.Diao LT, et al. Conservation and divergence of transcriptional coregulations between box C/D snoRNA and ribosomal protein genes in Ascomycota. RNA. 2014;20(9):1376–1385. doi: 10.1261/rna.042309.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fiol DF, Mak SK, Kültz D. Specific TSC22 domain transcripts are hypertonically induced and alternatively spliced to protect mouse kidney cells during osmotic stress. FEBS J. 2007;274(1):109–124. doi: 10.1111/j.1742-4658.2006.05569.x. [DOI] [PubMed] [Google Scholar]
- 77.Fiol DF, Chan SY, Kültz D. Regulation of osmotic stress transcription factor 1 (Ostf1) in tilapia (Oreochromis mossambicus) gill epithelium during salinity stress. J Exp Biol. 2006;209(Pt 16):3257–3265. doi: 10.1242/jeb.02352. [DOI] [PubMed] [Google Scholar]
- 78.Chakravarty D, et al. Three GADD45 isoforms contribute to hypertonic stress phenotype of murine renal inner medullary cells. Am J Physiol Renal Physiol. 2002;283(5):F1020–F1029. doi: 10.1152/ajprenal.00118.2002. [DOI] [PubMed] [Google Scholar]
- 79.Cai Q, Ferraris JD, Burg MB. High NaCl increases TonEBP/OREBP mRNA and protein by stabilizing its mRNA. Am J Physiol Renal Physiol. 2005;289(4):F803–F807. doi: 10.1152/ajprenal.00448.2004. [DOI] [PubMed] [Google Scholar]
- 80.Leitch V, Agre P, King LS. Altered ubiquitination and stability of aquaporin-1 in hypertonic stress. Proc Natl Acad Sci USA. 2001;98(5):2894–2898. doi: 10.1073/pnas.041616498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2011;2(3):103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tümpel S, Cambronero F, Wiedemann LM, Krumlauf R. Evolution of cis elements in the differential expression of two Hoxa2 coparalogous genes in pufferfish (Takifugu rubripes) Proc Natl Acad Sci USA. 2006;103(14):5419–5424. doi: 10.1073/pnas.0600993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mukherjee R, Evans P, Singh LN, Hannenhalli S. Correlated evolution of positions within mammalian cis elements. PLoS One. 2013;8(2):e55521. doi: 10.1371/journal.pone.0055521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Richards S, et al. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 2005;15(1):1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mehrotra R, Sethi S, Zutshi I, Bhalothia P, Mehrotra S. Patterns and evolution of ACGT repeat cis-element landscape across four plant genomes. BMC Genomics. 2013;14:203. doi: 10.1186/1471-2164-14-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ferraris JD, Garcia-Perez A. Osmotically responsive genes: The mammalian osmotic response element (ORE) Am Zool. 2001;41(4):734–742. [Google Scholar]
- 87.Woo SK, Nahm O, Kwon HM. How salt regulates genes: Function of a Rel-like transcription factor TonEBP. Biochem Biophys Res Commun. 2000;278(2):269–271. doi: 10.1006/bbrc.2000.3798. [DOI] [PubMed] [Google Scholar]
- 88.Woo SK, Dahl SC, Handler JS, Kwon HM. Bidirectional regulation of tonicity-responsive enhancer binding protein in response to changes in tonicity. Am J Physiol Renal Physiol. 2000;278(6):F1006–F1012. doi: 10.1152/ajprenal.2000.278.6.F1006. [DOI] [PubMed] [Google Scholar]
- 89.Dahl SC, Handler JS, Kwon HM. Hypertonicity-induced phosphorylation and nuclear localization of the transcription factor TonEBP. Am J Physiol Cell Physiol. 2001;280(2):C248–C253. doi: 10.1152/ajpcell.2001.280.2.C248. [DOI] [PubMed] [Google Scholar]
- 90.Xu S, et al. Phosphorylation by casein kinase 1 regulates tonicity-induced osmotic response element-binding protein/tonicity enhancer-binding protein nucleocytoplasmic trafficking. J Biol Chem. 2008;283(25):17624–17634. doi: 10.1074/jbc.M800281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kwon MS, et al. Novel nuclear localization signal regulated by ambient tonicity in vertebrates. J Biol Chem. 2008;283(33):22400–22409. doi: 10.1074/jbc.M710550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee SD, Colla E, Sheen MR, Na KY, Kwon HM. Multiple domains of TonEBP cooperate to stimulate transcription in response to hypertonicity. J Biol Chem. 2003;278(48):47571–47577. doi: 10.1074/jbc.M308795200. [DOI] [PubMed] [Google Scholar]
- 93.Ferraris JD, et al. Activity of the TonEBP/OREBP transactivation domain varies directly with extracellular NaCl concentration. Proc Natl Acad Sci USA. 2002;99(2):739–744. doi: 10.1073/pnas.241637298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim JA, et al. Modulation of TonEBP activity by SUMO modification in response to hypertonicity. Front Physiol. 2014;5:200. doi: 10.3389/fphys.2014.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fiol DF, Kültz D. Rapid hyperosmotic coinduction of two tilapia (Oreochromis mossambicus) transcription factors in gill cells. Proc Natl Acad Sci USA. 2005;102(3):927–932. doi: 10.1073/pnas.0408956102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wong MK, Ozaki H, Suzuki Y, Iwasaki W, Takei Y. Discovery of osmotic sensitive transcription factors in fish intestine via a transcriptomic approach. BMC Genomics. 2014;15:1134. doi: 10.1186/1471-2164-15-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tse WK. The role of osmotic stress transcription factor 1 in fishes. Front Zool. 2014;11(1):86. doi: 10.1186/s12983-014-0086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tse WK, Chow SC, Wong CK. Eel osmotic stress transcriptional factor 1 (Ostf1) is highly expressed in gill mitochondria-rich cells, where ERK phosphorylated. Front Zool. 2012;9(1):3. doi: 10.1186/1742-9994-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tse WK, Lai KP, Takei Y. Medaka osmotic stress transcription factor 1b (Ostf1b/TSC22D3-2) triggers hyperosmotic responses of different ion transporters in medaka gill and human embryonic kidney cells via the JNK signalling pathway. Int J Biochem Cell Biol. 2011;43(12):1764–1775. doi: 10.1016/j.biocel.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 100.Breves JP, et al. Acute salinity challenges in Mozambique and Nile tilapia: Differential responses of plasma prolactin, growth hormone and branchial expression of ion transporters. Gen Comp Endocrinol. 2010;167(1):135–142. doi: 10.1016/j.ygcen.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 101.McGuire A, et al. Hyperosmotic shock adaptation by cortisol involves upregulation of branchial osmotic stress transcription factor 1 gene expression in Mozambique Tilapia. Gen Comp Endocrinol. 2010;165(2):321–329. doi: 10.1016/j.ygcen.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 102.Tse WK, Chow SC, Wong CK. The cloning of eel osmotic stress transcription factor and the regulation of its expression in primary gill cell culture. J Exp Biol. 2008;211(Pt 12):1964–1968. doi: 10.1242/jeb.017368. [DOI] [PubMed] [Google Scholar]
- 103.Chow SC, Tse WK, Wong CK. Dexamethasone (DEX) induces Osmotic stress transcription factor 1 (Ostf1) through the Akt-GSK3β pathway in freshwater Japanese eel gill cell cultures. Biol Open. 2013;2(5):487–491. doi: 10.1242/bio.20134135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yan B, Zhao LH, Guo JT, Zhao JL. miR-429 regulation of osmotic stress transcription factor 1 (OSTF1) in tilapia during osmotic stress. Biochem Biophys Res Commun. 2012;426(3):294–298. doi: 10.1016/j.bbrc.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 105.Huang Y, Krein PM, Winston BW. Characterization of IFN-gamma regulation of the complement factor B gene in macrophages. Eur J Immunol. 2001;31(12):3676–3686. doi: 10.1002/1521-4141(200112)31:12<3676::aid-immu3676>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 106.Ujaoney AK, Potnis AA, Kane P, Mukhopadhyaya R, Apte SK. Radiation desiccation response motif-like sequences are involved in transcriptional activation of the Deinococcal ssb gene by ionizing radiation but not by desiccation. J Bacteriol. 2010;192(21):5637–5644. doi: 10.1128/JB.00752-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Riethoven JJ. Regulatory regions in DNA: Promoters, enhancers, silencers, and insulators. Methods Mol Biol. 2010;674:33–42. doi: 10.1007/978-1-60761-854-6_3. [DOI] [PubMed] [Google Scholar]
- 108.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Blattler A, et al. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol. 2014;15(9):469. doi: 10.1186/s13059-014-0469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ott CJ, et al. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc Natl Acad Sci USA. 2009;106(47):19934–19939. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bagchi DN, Iyer VR. The determinants of directionality in transcriptional initiation. Trends Genet. 2016;32(6):322–333. doi: 10.1016/j.tig.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li M, et al. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics. 2014;197(2):591–599. doi: 10.1534/genetics.114.163667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maddison K, Clarke AR. New approaches for modelling cancer mechanisms in the mouse. J Pathol. 2005;205(2):181–193. doi: 10.1002/path.1698. [DOI] [PubMed] [Google Scholar]
- 114.Kültz D, Li J, Gardell A, Sacchi R. Quantitative molecular phenotyping of gill remodeling in a cichlid fish responding to salinity stress. Mol Cell Proteomics. 2013;12(12):3962–3975. doi: 10.1074/mcp.M113.029827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schubert OT, et al. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat Protoc. 2015;10(3):426–441. doi: 10.1038/nprot.2015.015. [DOI] [PubMed] [Google Scholar]
- 116.Sharma V, et al. Panorama: A targeted proteomics knowledge base. J Proteome Res. 2014;13(9):4205–4210. doi: 10.1021/pr5006636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cassé C, Giannoni F, Nguyen VT, Dubois MF, Bensaude O. The transcriptional inhibitors, actinomycin D and alpha-amanitin, activate the HIV-1 promoter and favor phosphorylation of the RNA polymerase II C-terminal domain. J Biol Chem. 1999;274(23):16097–16106. doi: 10.1074/jbc.274.23.16097. [DOI] [PubMed] [Google Scholar]
- 118.Daoudal S, Tournaire C, Halere A, Veyssière G, Jean C. Isolation of the mouse aldose reductase promoter and identification of a tonicity-responsive element. J Biol Chem. 1997;272(5):2615–2619. doi: 10.1074/jbc.272.5.2615. [DOI] [PubMed] [Google Scholar]
- 119.Ruepp B, Bohren KM, Gabbay KH. Characterization of the osmotic response element of the human aldose reductase gene promoter. Proc Natl Acad Sci USA. 1996;93(16):8624–8629. doi: 10.1073/pnas.93.16.8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou C, Cammarata PR. Cloning the bovine Na+/myo-inositol cotransporter gene and characterization of an osmotic responsive promoter. Exp Eye Res. 1997;65(3):349–363. doi: 10.1006/exer.1997.0335. [DOI] [PubMed] [Google Scholar]