Significance
Rare recessive monogenic diseases are often found in isolated populations. In one such population, we identified a child carrying a homozygous nonsense mutation in an understudied smooth muscle-restricted gene called Leiomodin1 (LMOD1). Heterozygous parents showed no disease; however, the child died shortly after birth from a rare condition known as megacystis microcolon intestinal hypoperistalsis syndrome. A mouse model with a similar Lmod1 mutation, engineered with CRISPR-Cas9 genome editing, exhibited the same gastrointestinal and urinary bladder phenotypes as seen in the newborn child. Phenotyping revealed insights into the underlying cause of the disease. Results demonstrate the conserved function of LMOD1 in human and mice and the importance of this protein in the molecular regulation of contractility in visceral smooth muscle cells.
Keywords: CRISPR-Cas9, genetics, Leiomodin, myopathy, smooth muscle
Abstract
Megacystis microcolon intestinal hypoperistalsis syndrome (MMIHS) is a congenital visceral myopathy characterized by severe dilation of the urinary bladder and defective intestinal motility. The genetic basis of MMIHS has been ascribed to spontaneous and autosomal dominant mutations in actin gamma 2 (ACTG2), a smooth muscle contractile gene. However, evidence suggesting a recessive origin of the disease also exists. Using combined homozygosity mapping and whole exome sequencing, a genetically isolated family was found to carry a premature termination codon in Leiomodin1 (LMOD1), a gene preferentially expressed in vascular and visceral smooth muscle cells. Parents heterozygous for the mutation exhibited no abnormalities, but a child homozygous for the premature termination codon displayed symptoms consistent with MMIHS. We used CRISPR-Cas9 (CRISPR-associated protein) genome editing of Lmod1 to generate a similar premature termination codon. Mice homozygous for the mutation showed loss of LMOD1 protein and pathology consistent with MMIHS, including late gestation expansion of the bladder, hydronephrosis, and rapid demise after parturition. Loss of LMOD1 resulted in a reduction of filamentous actin, elongated cytoskeletal dense bodies, and impaired intestinal smooth muscle contractility. These results define LMOD1 as a disease gene for MMIHS and suggest its role in establishing normal smooth muscle cytoskeletal–contractile coupling.
Megacystis microcolon intestinal hypoperistalsis syndrome (MMIHS) (MIM155310) is a rare congenital defect of visceral smooth muscle, primarily affecting females who present at birth with functional obstruction of intestine, microcolon, dilation of bladder, and secondary hydronephrosis. More than 250 cases have been reported since the initial description of MMIHS in five young girls in 1976 (1–3). Total parenteral nutrition (TPN), adequate intermittent catheterization of bladder, and surgical corrections for intestinal malrotation are frequent modes of treatment for this disease without which rapid death ensues. In some instances, multivisceral organ transplantation has been indicated with some success. Despite these clinical interventions, MMIHS often leads to premature death due to complications of therapy (3, 4).
The majority of MMIHS cases are sporadic, stemming from de novo, heterozygous missense mutations in the smooth muscle-restricted ACTG2 gene (5, 6). These mutations are thought to render the ACTG2 protein defective for normal actin polymerization and contractile activity in visceral smooth muscle organs although formal in vivo proof for this ACTG2 loss of function is lacking (6, 7). Heterozygous mutations in ACTG2 are also observed in patients with autosomal dominant MMIHS (6, 8), but there is emerging evidence for a recessive mode of inheritance. For example, a homozygous loss-of-function variant in the myosin heavy chain 11 (MYH11) gene, which is another highly specific contractile gene for smooth muscle lineages (9), was reported in a patient with MMIHS from a consanguineous (i.e., genetically related) couple (10). To date, no other smooth muscle-restricted contractile genes have been linked to MMIHS.
In this report, a patient with MMIHS is described from a consanguineous couple in whom no variants of ACTG2 or MYH11 were found. Rather, an autosomal recessive inherited subtype of MMIHS has been discovered involving a homozygous nonsense mutation in Leiomodin1 (LMOD1), an understudied smooth muscle-restricted gene (11) that is a direct target of the serum response factor (SRF)/Myocardin transcriptional switch (12). CRISPR-Cas9 (CRISPR-associated protein) genome editing of the murine Lmod1 gene resulted in a similar nonsense mutation and a nearly identical MMIHS phenotype. Functional studies in both human and mouse support a previously unrecognized role for LMOD1 in visceral smooth muscle function. We conclude that LMOD1 is an MMIHS disease gene that expands the genetic heterogeneity of this devastating visceral myopathy.
Results
Clinical Case Study of a Neonate with MMIHS.
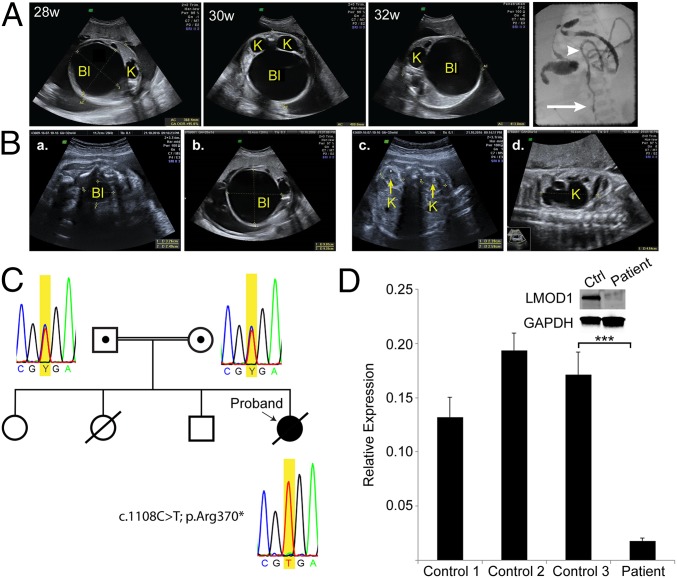
The affected proband was a female neonate from a consanguineous couple, originating from a genetic isolate in the Netherlands. Most of the inhabitants in this region are descendants of a small group of founders; a plague epidemic in the 17th century killed the majority of the population, except for 151 inhabitants that survived (13, 14). During a routine ultrasound sonography (USG) examination at 20 wk gestation, a distended bladder (megacystis) and bilateral hydronephrosis, a condition that typically occurs when the kidney swells due to the failure of normal drainage of urine from the kidney to the bladder, were detected (Fig. 1A); bladder distention and hydronephrosis are common features of MMIHS that are not evident in normal age-matched controls (Fig. 1B). Polyhydramnios, an excess of amniotic fluid in the amniotic sac, was later acknowledged, and delivery was medically induced at 34 wk gestation due to mechanical problems of the mother. Physical examination in the neonatal period revealed distended and lax abdominal wall, causing the intestine to be externally palpable. Further postnatal USG investigation did not reveal abnormalities of the heart or brain. Hydronephrosis, bilateral distention of the proximal ureter, and distended bladder were persistent during postnatal life of the patient. No peristaltic movement of the gastrointestinal tract was noted. Evidence of a microcolon was demonstrated through a barium enema test (Fig. 1A). Conventional treatments included TPN, stomach suction, and catheterization of the atonic bladder. Because the patient died at 5 d of age due to sepsis, there were no further clinical measures. Although the patient was the first child in the family diagnosed with MMIHS, the parents acknowledged a previous intrauterine fetal death in which routine 20 wk USG detected abdominal distention of the fetus. Two other siblings in this family are healthy and show no signs of bladder or intestinal abnormalities (Fig. 1C).
Fig. 1.
Loss-of-function mutation in LMOD1 gene of an MMIHS patient. (A) Timed ultrasonography at 28, 30, and 32 wk of gestation showing distended bladder (Bl) and hydronephrotic kidney (K). A postnatal barium enema confirmed MMIHS (Far Right); white arrow and arrowhead indicate the sigmoid colon and descending colon, respectively. (B) In utero ultrasonography of control (a and c) and MMIHS patient (b and d) demonstrating megacystis (Bl) and hydronephrosis (K) in the MMIHS patient. (C) Pedigree of the consanguineal family and results of Sanger sequencing. A heterozygous variant is identified in DNA of the parents and a homozygous variant generating a PTC (TGA) is detected in the proband. (D) RT-qPCR analysis of LMOD1 using RNA from control and patient-derived dermal fibroblasts. (Inset) Protein expression of LMOD1 in control (Ctrl) and patient dermal fibroblasts. Data shown are the mean expression of LMOD1 ± SD of three controls and the patient, performed in triplicate. ***P < 0.001 by unpaired t test.
Copy Number Variation Profiling and Homozygosity Mapping.
Seven large copy number variants (CNVs) were identified, all of which were common polymorphisms inherited from one of the unaffected parents. Runs of homozygosity (ROH) larger than 1 Mb within the neonate were considered to be inherited by descent because these regions were heterozygous in the parental samples and contained over 50 consecutive homozygous probes (SI Appendix, Table S1). These putative inherited-by-descent regions contained 19 recessive single nucleotide polymorphisms (SNPs) with a minor allele frequency below 1% (SI Appendix, Table S2). Only three of these variants were predicted to be deleterious. The degree of parental relatedness was determined by F statistics using all ROH larger than 1 Mb and at least 50 consecutive probes using the Genomic Oligoarray and SNP array evaluation tool v3.0. An F value ≥ 0.0156 indicates a progeny from a consanguineous marriage (15). Father (F = 0.015625 and 64 Mb of ROH), mother (F = 0.03125 and 86 Mb of ROH), and patient (F = 0.046875 and 115 Mb of ROH) had a relatively high F number (13, 14), suggesting that the patient was indeed a progeny from a consanguineous couple and that the parents originated from a genetically isolated population (13).
A Homozygous Nonsense Mutation of LMOD1 in the MMIHS Patient.
Targeted analysis of the exome data identified no deleterious variants in the known disease genes, ACTG2 and MYH11. In total, 526 variants had a minor allele frequency below 0.1%. Prioritization to yield the most likely deleterious variants resulted in the identification of 70 variants in total: 7 were nonsense variants, 10 were predicted to affect splicing, and 53 had a Combined Annotation Dependent Depletion (CADD) score of ≥20, which is indicative of possible pathogenicity. Only 3 out of 70 variants were homozygous. One was a missense variant in the Interleukin24 gene (NM_001185156.1:c.541T>G; p.Leu181Val). This variant was relatively frequent in the normal population (minor allele frequency of 0.4%), making it an unlikely candidate for the MMIHS phenotype. The two remaining recessive variants were rare: One resulted in an intronic SNP predicted to affect splicing of Cellular Repressor of E1A Stimulated Genes 2 (CREG2) (NM_153836.3: c.442-5G>A), and the other resulted in a premature termination codon (PTC) in exon two of Leiomodin1 (LMOD1) (NM_012134.2:c.1108C>T; p.Arg370*). These two recessive variants are each located within a large ROH (SI Appendix, Table S2). CREG2 is expressed primarily in brain (16) whereas the LMOD1 gene exhibits mRNA and protein expression in both bladder and intestine, and the encoded protein is part of the smooth muscle network of cytocontractile elements (11, 12). Further, we showed that the PTC in LMOD1 disrupted normal mRNA and protein expression in dermal fibroblasts derived from the proband compared with control fibroblasts (Fig. 1D). Sanger sequencing confirmed that both parents carried one copy of the PTC whereas the proband was homozygous, suggesting a recessive mode of inheritance (Fig. 1C). Of note, the parents exhibited no MMIHS-like symptoms. These findings prompted us to consider LMOD1 as the most likely candidate gene underlying MMIHS in the proband.
LMOD1 Expression in Human Intestinal and Bladder Smooth Muscle.
Evidence exists for LMOD1 protein expression in smooth muscle of the developing mouse embryo (12). To extend these findings to human, immunohistochemical studies were undertaken in the developing intestine and bladder. Intense staining of LMOD1 protein was observed in both visceral and vascular smooth muscle of the intestine and bladder over a number of developmental time points (SI Appendix, Fig. S1). Further evidence for LMOD1 protein in these tissues was demonstrated by Western blotting (SI Appendix, Fig. S1). These findings, coupled with the sharp reduction in LMOD1 mRNA and LMOD1 protein in dermal fibroblasts of the proband compared with control cells (Fig. 1D), support LMOD1 as a disease gene for MMIHS.
CRISPR-Cas9 Genome Editing of the Mouse Lmod1 Gene.
To provide more formal proof of an association between loss in LMOD1 and MMIHS, we turned to CRISPR-Cas9 genome editing in the mouse. Initial efforts to inactivate Lmod1 were based on transgenic studies showing the critical importance of two conserved SRF-binding CArG boxes for Lmod1 promoter activity in vivo (12) and a more recent report that revealed near abolishment of target gene expression upon CRISPR-Cas9 genome editing of a single CArG box (17). In contrast to the latter finding, there was little change in Lmod1 mRNA or LMOD1 protein in mice homozygous for three-component CRISPR genome editing (18) of both CArG boxes (SI Appendix, Fig. S2). Moreover, homozygous Lmod1 CArG-null mice were viable and fertile and showed no overt signs of pathology. These results suggest that the two proximal CArG boxes are dispensable for LMOD1 expression and normal smooth muscle cell (SMC) function in mice.
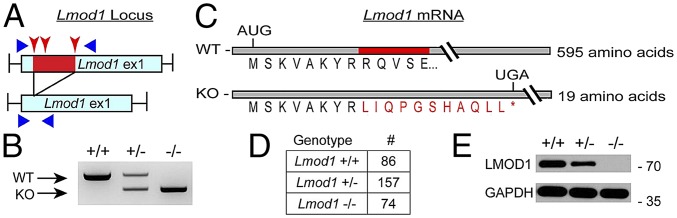
Parallel studies using two-component CRISPR genome editing (18) of the first exon of Lmod1 resulted in a 151-bp deletion (Fig. 2 A and B). Sequence analysis showed that the deletion preserved the first eight amino acids of LMOD1 but introduced an 11-aa frameshift followed by a PTC (Fig. 2C). Using CRISPResso (19), we did not detect any off-targeted events for any of the guide RNAs (SI Appendix, Table S3). Intercrossing heterozygous mice revealed an expected Mendelian ratio of live born pups (Fig. 2D). Western blotting showed a twofold decrease in LMOD1 protein in stomach of heterozygous mice whereas undetectable levels were observed in homozygous mice (hereafter referred to as Lmod1−/−) (Fig. 2E). Similar findings were noted in aorta and bladder (SI Appendix, Fig. S3 A and B). Compound heterozygous mice in which one Lmod1 allele had both CArG boxes mutated and the other allele carried the 151-bp deletion exhibited even lower levels of LMOD1 protein; however, such compound heterozygous mice were without noticeable defect, indicating a low-level threshold of LMOD1 protein exists for normal homeostasis (SI Appendix, Fig. S3B). Loss of LMOD1 expression had little to no effect on the expression of other SMC contractile proteins (SI Appendix, Fig. S3B). Taken together, these CRISPR-Cas9 genome editing results highlight salient differences in phenotypic outcome dependent upon the strategy employed.
Fig. 2.
Two-component CRISPR genome editing of mouse Lmod1. (A) Schematic of targeting strategy to disrupt exon one of Lmod1. Injection of Cas9 mRNA and three guide RNAs (red arrowheads) resulted in deletion of 151 bp (red) within exon one of the mouse Lmod1 gene. PCR primers (blue triangles) were used to confirm deletion and sequence changes. (B) PCR products generated with forward and reverse primers (shown in A) indicating WT (+/+), heterozygous (+/−), and homozygous (−/−) deletion of Lmod1. (C) Deletion of the 151-bp region resulted in a frameshift of the ORF and the introduction of a PTC within Lmod1. WT amino acid is in black, and the frame-shifted sequence is in red (asterisk denotes the UGA stop codon). (D) Number of live-born pups of each genotype from heterozygous intercross (P = 0.79 versus expected Mendelian numbers by χ2 test). (E) Western blotting of LMOD1 protein in stomach of different Lmod1 genotypes.
Lmod1−/− Mice Exhibit an MMIHS-Like Phenotype.
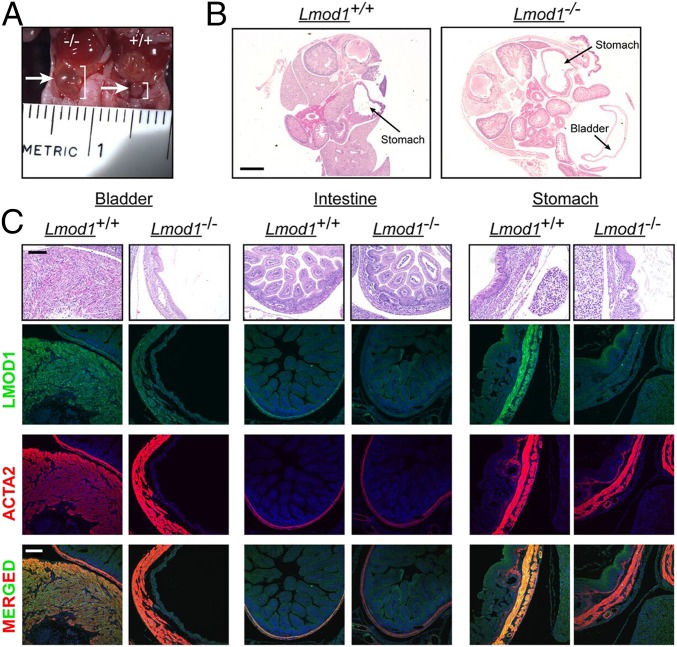
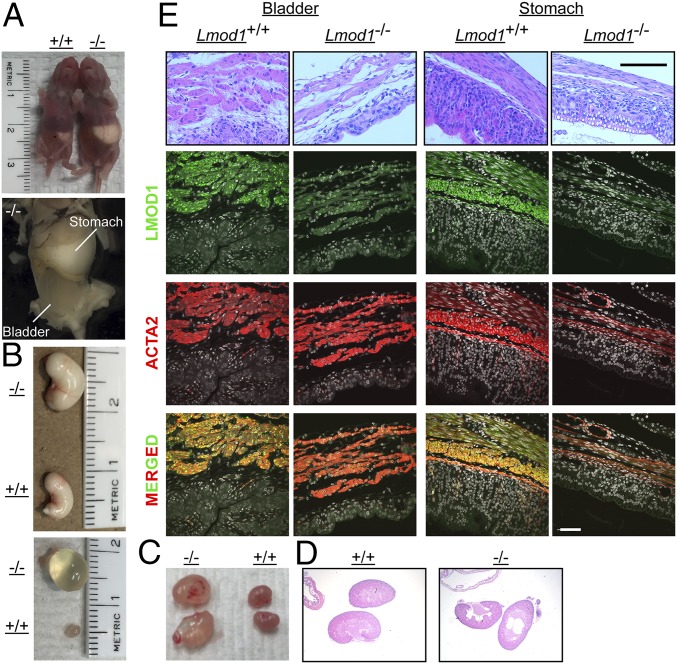
Consistent with the above homozygous LMOD1 neonatal child, Lmod1−/− mice displayed distention of the bladder beginning at 18.5 d of embryonic development (Fig. 3A). In some cases, the developing bladder expanded to such a great extent as to encroach upon the abdominal cavity (Fig. 3B). Further histological analysis revealed early onset thinning of detrusor muscle of the bladder in Lmod1−/− mice whereas fewer histological changes were seen in intestine and stomach (Fig. 3C). Expression of LMOD1 protein coincided with ACTA2 in bladder, intestine, and stomach of WT mice whereas mutant mice showed the expected loss in LMOD1 protein (Fig. 3C). After parturition and the first feeding, 100% of homozygous mutant mice showed enlargement of the stomach and urinary bladder (Fig. 4 A and B), the latter of which is consistent with clinical observations in human MMIHS patients. Hydronephrosis, due to the failure of urine emptying into the bladder, was noted in some mutant mice (Fig. 4 C and D). Histological analysis of postnatal mice demonstrated thinning and compaction of the visceral smooth muscle layers in both stomach and bladder, along with severe loss of the transitional epithelium of the bladder (Fig. 4E and SI Appendix, Fig. S4). Immunofluorescence microscopy of Lmod1−/− tissues of postnatal mice showed the absence of LMOD1 protein within smooth muscle layers of the stomach, bladder, and intestine; the apparent positive staining in some mutant tissues here and below is attributable to occasional nonspecific staining of the primary antibody (Fig. 4E and SI Appendix, Fig. S4). All segments of the small intestine exhibited loss of LMOD1 protein expression, with little evidence of overt histopathology (Fig. 5). There was a significant increase in Ki67 staining of SMCs in the outer longitudinal layer of the intestine (SI Appendix, Fig. S5 A and B), but no obvious signs of cell death as measured by TUNEL staining (SI Appendix, Fig. S5C). Interestingly, a significant decrease in intestinal length was noted in Lmod1−/− mice (SI Appendix, Fig. S6 A and B), but there was no evidence of a microcolon (SI Appendix, Fig. S6C). Further, there were no obvious signs of histopathology in Lmod1−/− aorta or esophagus, despite the absence of LMOD1 protein within SMCs of these tissues (SI Appendix, Fig. S6D). Comparative analysis of human and mouse LMOD1 protein revealed 82% primary amino acid homology, similar domain structure, and the relative positions of each PTC (SI Appendix, Fig. S7). Taken together, these findings demonstrate an analogous MMIHS-like phenotype in both human and mice lacking LMOD1.
Fig. 3.
MMIHS phenotype in Lmod1−/− mice at embryonic (e) day 18.5. (A) Gross pathology showing distended bladder in Lmod1−/− mice at e18.5 (white arrow and white bracket) compared with Lmod1+/+ littermate control. (B) H&E staining shows distended bladder and stomach in the same plane of section from an e18.5 Lmod1−/− mouse. (Scale bar: 1 mm.) (C) H&E staining and immunofluorescence staining of indicated SMC marker proteins in sections of bladder, intestine, and stomach from e18.5 Lmod1−/− versus Lmod1+/+ mice. (Scale bars: 100 μm.)
Fig. 4.
MMIHS phenotype in postnatal Lmod1−/− mice. (A) Gross image of Lmod1+/+ (leftmost) and Lmod1−/− (rightmost) pups showing abdominal distention in the mutant due to bladder and milk-filled stomach (Top). A higher magnified image of a mutant is shown at Bottom. (B) Dissected gross images of stomach and bladder from indicated Lmod1 genotypes. (C) Dissected kidneys and hydronephrosis in some postnatal day 5 Lmod1−/− mice. (D) H&E-stained sections of kidneys from each genotype. Note the large renal cysts in the Lmod1−/− section, representing evidence of previously retained water in the mutant kidneys. (E) Sections of Lmod1+/+ versus Lmod1−/− stomach and bladder stained with H&E (Top row) or antibodies to the indicated proteins. DAPI nuclear stain was pseudocolored white. (Scale bars: H&E sections, 100 μm; immunofluorescence sections, 50 μm.)
Fig. 5.
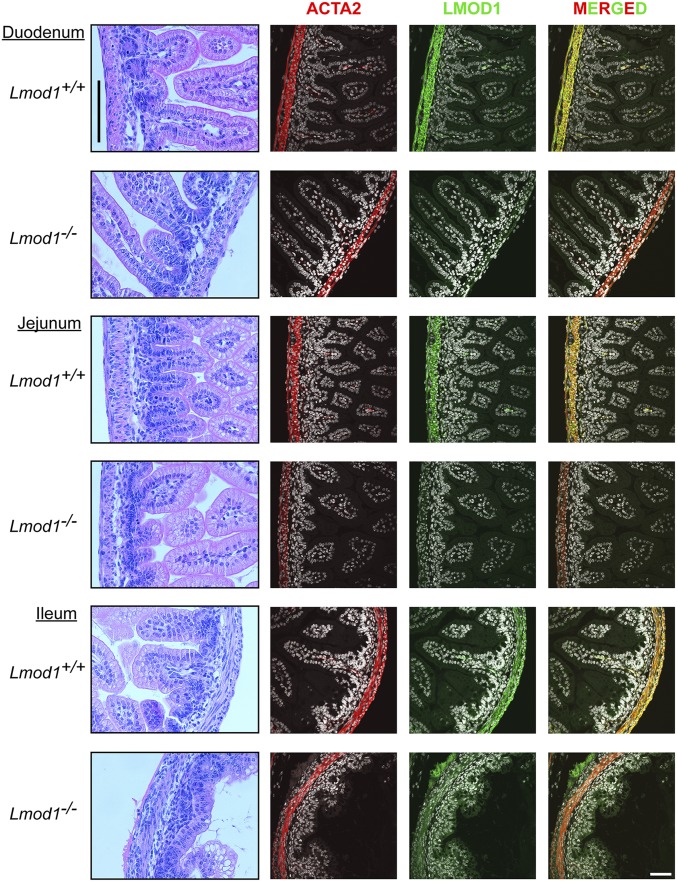
Histology and LMOD1 immunofluorescence in three segments of intestine. H&E-stained sections of duodenum, jejunum, and ileum (Left column) and immunofluorescence microscopy of ACTA2 and LMOD1 show absence of LMOD1 protein in visceral smooth muscle of intestinal segments of Lmod1−/− mice. [Scale bars: H&E sections, 100 μm (vertical line at Top Left); immunofluorescence sections, 50 μm (Bottom Right).] Note, the apparent faint immunostaining for LMOD1 in mutants is nonspecific (SI Appendix, Fig. S4).
Cytoskeletal–Contractile Defects with Deficiency of LMOD1.
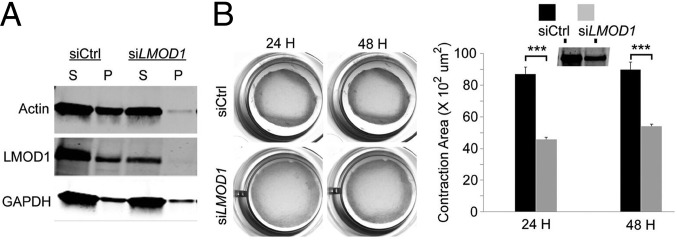
To begin gaining mechanistic insight into how loss of LMOD1 leads to the characteristic hypoperistalsis in MMIHS, we performed biochemical and functional studies in cultured human intestinal smooth muscle cells (hiSMCs). siRNA knockdown of LMOD1 in purified hiSMCs resulted in a reduction of filamentous actin pelleted by ultracentrifugation (Fig. 6A). To determine whether there was a functional loss of contractile competence in LMOD1-deficient hiSMCs, a collagen gel contraction assay was used. Results showed a 40% decrease in contractile activity in hiSMCs with reduced LMOD1 (Fig. 6B).
Fig. 6.
LMOD1 knockdown in human intestinal SMCs decreases actin filament formation and contractility. (A) In vitro G-actin:F-actin assay in cultured human intestinal smooth muscle cells (hiSMCs) with LMOD1 knockdown. S and P refer to supernatant (G-actin) and pellet (F-actin), respectively. (B) Collagen contractility assays (Left) in hiSMCs transfected with siCtrl or siLMOD1 for 24 or 48 hours (H). Quantitative data are shown at Right with Western blot of LMOD1 protein (Inset). Data shown are mean ± SD of three independent experiments, performed in triplicate. ***P < 0.001 by unpaired t test.
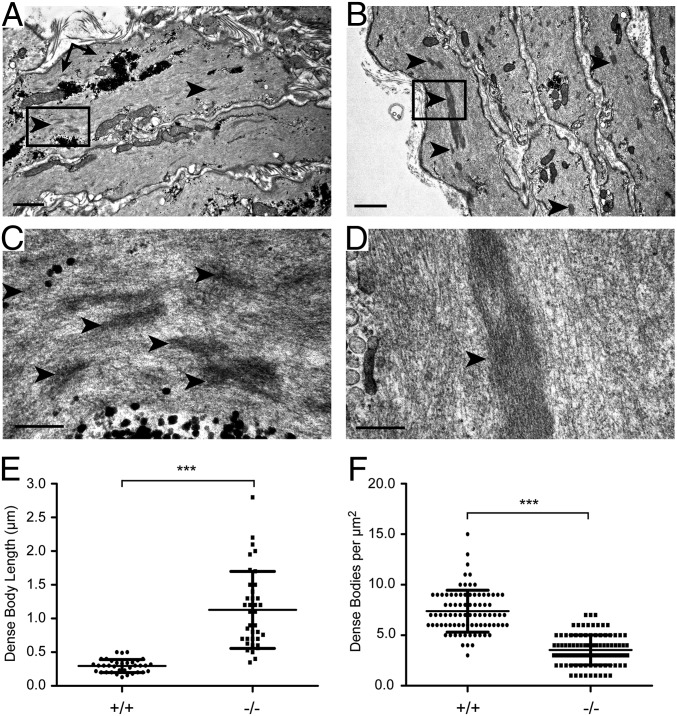
We next turned to the Lmod1−/− mouse model of MMIHS. Ultracentrifugation and phalloidin staining showed a reduction in filamentous actin in the Lmod1−/− mouse (SI Appendix, Figs. S8 and S9). Surprisingly, however, there was little evidence of ultrastructural actin filament perturbations in the bladder or intestine of Lmod1−/− mice. However, a notable defect was observed in dense bodies of bladder detrusor SMCs (Fig. 7 A–D and SI Appendix, Fig. S10) and, to a lesser extent, intestine (SI Appendix, Fig. S10). The changes in dense bodies were quite pervasive in two independent mutant animals, and these structures bore some resemblance to nemaline bodies that were recently described in humans carrying mutations in LMOD3 (20) although the structures reported here lacked the characteristic 145-Å periodicity of cross-striations (Fig. 7D). The length of dense bodies in longitudinally oriented SMCs of Lmod1−/− mice was nearly three times greater than WT controls (Fig. 7E). Further, there was a twofold decrease in the concentration of dense bodies in mutant bladder (Fig. 7F). Collectively, these results suggest ultrastructural changes in the normal cytoskeleton of visceral SMCs in Lmod1−/− mice.
Fig. 7.
Ultrastructure of bladder smooth muscle in postnatal Lmod1−/− mice. Lmod1+/+ WT (A and C) versus Lmod1−/− mutant (B and D) SMC dense bodies (arrowheads) that are much larger in mutants. Boxed regions in A and B are shown at higher magnifications in C and D, respectively. [Scale bars: A and B, 1 μm; C and D, 200 nm (black in Lower Left of each panel).] Arrows point to glycogen granules, which are more prevalent in WT SMCs. (E) Quantitative analysis of dense body length in WT (+/+) versus mutant (−/−) SMCs (n = 35 for WT and n = 37 for mutant). Scatterplot depicts the means ± SD (***P < 0.0001 by paired t test). (F) Quantitative analysis of dense body concentration in WT (+/+) versus mutant (−/−) SMCs (n = 86 for WT and n = 102 for mutant). Scatterplot depicts the mean ± SD (***P < 0.0001 by paired t test from two independent and blinded observers).
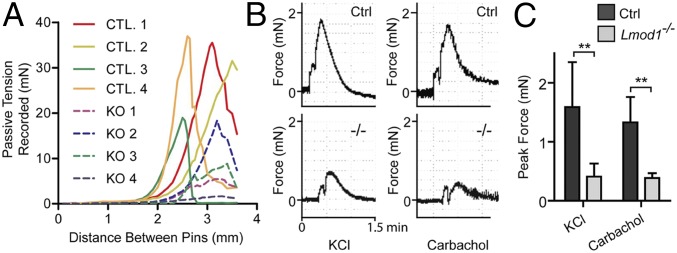
To assess the functional consequence of loss in LMOD1 in the intestinal tract, we performed physiological measures of passive tensile strength and contractile competence. Myography experiments revealed a notable lack of tensile strength in jejunum ring segments of Lmod1−/− mice as indicated by a lower level of passive tension in response to mechanical expansion (Fig. 8A and SI Appendix, Fig. S11A). Importantly, a more than threefold decrease in KCL- or carbachol-induced contractility was demonstrated in intestinal ring segments from Lmod1−/− mice (Fig. 8 B and C). These results suggest that loss in LMOD1 protein expression leads to compromised structural integrity and impaired contraction of visceral smooth muscle-containing tissues. The near identical phenotypes found in human and mouse support the notion that loss in LMOD1 leads to the functional obstruction of intestine and urinary bladder commonly seen in MMIHS.
Fig. 8.
Loss of LMOD1 results in reduced passive tensile strength and agonist-induced contractility in intestinal rings. (A) Lmod1−/− jejunum ring segments exhibit decreased tensile strength after dilation using wire myography. Ring segments were mounted and equilibrated before being dilated at set distances, and the passive tension of the ring segment was recorded. (B) Representative myography traces showing force of contraction versus time for control (Ctrl) and mutant (−/−) mouse jejunum ring segments treated with 60 mM KCl or 1 μM carbachol. (C) Quantitation of data from B. Shown are the means ± SD from n = 6 knockout or control groups receiving KCl treatment and n = 4 for carbachol treatment (**P < 0.01 for all graphs by paired t test analysis).
Discussion
Previous studies have documented spontaneous and autosomal dominant mutations in ACTG2 or a recessive mutation in MYH11 in patients diagnosed with MMIHS, a rare congenital disease in visceral smooth muscle of the intestine and urinary bladder (8, 10). However, there is emerging evidence for considerable heterogeneity in the disease, and additional recessive mutations are likely (4). In this report, we have identified an SNP resulting in a premature termination codon (PTC) within exon two of human LMOD1 in a family from a genetically isolated population (13). Although the heterozygous parents exhibited no discernable disease, the homozygous neonate displayed classic signs of MMIHS, both in utero and after parturition. The PTC severely reduced expression of LMOD1 in patient-derived dermal fibroblasts. The fact that mice homozygous for a similar PTC located in exon one of Lmod1 exhibited loss of LMOD1 protein and phenotypes analogous to those seen in the homozygous neonatal child strongly supports LMOD1 as a disease gene for MMIHS. Indeed, both the affected child and homozygous mutant mice displayed the highly penetrant megacystis (distended bladder) during prenatal and postnatal development. The distended bladder likely precipitated hydronephrosis, a trait also observed in patients diagnosed with MMIHS.
Another pathognomonic feature of human MMIHS is intestinal hypoperistalsis (2). In mice homozygous for the PTC in Lmod1, as well as cultured human intestinal SMCs deficient in LMOD1, a dramatic decrease in agonist-induced contractility was observed. Intestinal hypoperistalsis likely caused the gastroparesis observed in all mutant mice; it is noteworthy that failure to empty the stomach has also been reported in some cases of human MMIHS (4). We were unable to observe intestinal transit due to technical limitations inherent in gavage-feeding neonatal mice. One notable difference between the child and mice null for Lmod1 was the absence of a microcolon in mutant mice. It is unclear as to why this aspect of the disease was not manifest in the Lmod1-null mouse. Nevertheless, the weight of evidence supports an expansion of the heterogeneity of MMIHS by showing a direct link between nonsense mutations in LMOD1 and clinical and experimental cases of the disease.
LMOD1 was originally cloned from a human thyroid cDNA library (21) and subsequently was shown to be enriched in smooth muscle-containing tissues (22). An early report showed that the amino terminus of LMOD1 binds tropomyosins in an isoform-specific manner (23). We now recognize LMOD1 as one of three gene paralogs that function in the nucleation of actin monomers in association with tropomyosin (20, 24, 25). Recent gene knockout experiments have further illuminated the role of LMOD family members. For example, Lmod2 knockout mice exhibit shortened cardiac actin filaments and dilated cardiomyopathy although no known LMOD2 mutations have been uncovered in humans (26). On the other hand, human mutations in LMOD3 are linked to nemaline myopathy (20), a complex congenital skeletal muscle disease characterized by rod-like nemaline bodies that are thought to cause muscle weakness (27, 28). Similar to humans with mutations in LMOD3, mice in which Lmod3 has been genetically knocked out exhibit skeletal muscle weakness and the presence of nemaline bodies (29, 30). Notably, similar rod-like structures representing elongated dense bodies were frequently seen in visceral smooth muscle of Lmod1-null mice. Although they did not have the cross-striations of 145-Å periodicity seen in sarcomeric nemaline bodies, very similar structures were described in gastric smooth muscle of a patient diagnosed with nemaline myopathy (31). Of note, no such structures have yet been reported in patients with MMIHS. Smooth muscle dense bodies are thought to act as anchor points bridging the actin cytoskeleton with thin filaments (32, 33), suggesting that changes in dense body characteristics reported here uncouple the actin cytoskeleton from the contractile apparatus of visceral smooth muscle-containing organs such as stomach, intestine, and bladder of Lmod1−/− mice. Further confirmation and characterization of visceral smooth muscle nemaline-like bodies is needed in other animal models of MMIHS, as well as MMIHS patients, to establish a pathological role of these intriguing structures.
The cumulative mouse genetic data thus far would suggest an essential role for each LMOD family member in the fidelity of the actin cytoskeleton and normal contractile activity of smooth (LMOD1), cardiac (LMOD2), and skeletal (LMOD3) muscle. Because LMOD2 and LMOD3 are barely detectable in smooth muscle-containing tissues (11, 12) and were not up-regulated in tissues of Lmod1−/− mice (SI Appendix, Fig. S11B), no genetic compensation was evident upon loss of LMOD1 in visceral smooth muscle. Similarly, LMOD1 apparently cannot compensate for loss in either LMOD2 or LMOD3. Future genetic complementation studies will be needed to provide insight into the relative activities of each LMOD isoform in the complex setting of the mouse.
Several unexpected findings emerged from the CRISPR-mediated edits of Lmod1. First, the elimination of SRF-binding CArG boxes upstream of Lmod1 had little to no effect on baseline LMOD1 protein or mRNA expression. This finding should be contrasted with an earlier report showing near abolishment of CNN1, another SMC-restricted gene product (34), upon CRISPR-mediated editing of a single intronic CArG box (17). Similarly, a decrease in expression of smooth muscle myosin light chain kinase with attenuated intestinal smooth muscle contractility was noted after deletion of an upstream CArG box (35). The absence of change in LMOD1 expression with the double CArG box mutation suggests that, whereas proximal promoter CArG boxes are sufficient for Lmod1 promoter activity (12), they are not necessary in their native genomic context, which would imply that other, more distal CArG boxes coordinate SRF-dependent expression of LMOD1 in both human and mouse. Chromosome conformation capture assays may unveil these and other important regulatory elements for Lmod1 expression. A second unexpected finding was the profound attenuation in transitional epithelium of the bladder and, to a lesser extent, the stomach of Lmod1−/− mice, particularly in the postnatal period when these organs exhibited dramatic distention. In contrast, there was little evidence of epithelial loss in each segment of the intestine, perhaps due to the absence of mechanical stress as seen in stomach and bladder. Another unanticipated finding in Lmod1 mutant mice was the seemingly normal architecture and function of airways of the lung and vasculature, both of which have a rich supply of smooth muscle. Similarly, patients with MMIHS generally do not present with respiratory or cardiovascular pathology. It must be stressed, however, that any defects in these systems may be masked by the profound changes in gastrointestinal and urinary tracts. Future work should further assess the functional consequences of loss in LMOD1 in these organ systems using more restrictive means of inactivating the Lmod1 gene. Finally, it was somewhat surprising to have not observed an obvious defect in actin filaments at the ultrastructural level, despite clear evidence of reduced filamentous actin as measured by phalloidin staining of visceral organs of the mouse deficient in LMOD1. We cannot explain this apparent paradox although there could be subtle perturbations in filamentous actin or its association with, for example, tropomyosin that are not resolved by the electron microscope. It may prove informative to generate an allelic series of mice in which the actin-binding, tropomyosin, or Wiskott–Aldrich syndrome homology 2 domains of LMOD1 (SI Appendix, Fig. S7) are mutated to pinpoint the critical regions necessary for LMOD1 function in vivo.
It is interesting to contrast the findings reported here with several other smooth muscle-restricted actin-binding proteins that are dispensable for similar functional activity in these organs, including CNN1 (17, 36), TAGLN (37), CSRP1 (38), and CALD1 (39), which would suggest that compensatory pathways exist to preserve normal organ function in the absence of each of these actin-binding proteins. On the other hand, genetic inactivation of Myh11, encoding the major thick filament protein in smooth muscle, leads to megacystis, a decrease in intestinal motility, defective contraction of the bladder, and early postnatal death, all of which are reminiscent of the Lmod1−/− phenotype and human MMIHS (40). Further, smooth muscle-specific knockout of myosin light chain kinase results in early postnatal demise due to similar phenotypes observed in the Lmod1 and Myh11 knockouts (41). Although human mutations in the latter have not yet been documented, there is a report of a consanguineous family with a nonsense mutation in MYH11 associated with MMIHS (10). Inactivation of SmoothelinA, encoding a visceral smooth muscle restricted actin-binding protein (42), results in impaired intestinal contractility and 50% mortality in homozygous mutants by postnatal day 20 (43); no human mutations have yet been reported for SmoothelinA. Surprisingly, mice in which the smooth muscle alpha actin gene (Acta2) is inactivated survive with mild hypotension and reduced blood flow under thermal challenge; no gastrointestinal or urinary pathologies were reported (44). Compensatory increases in the skeletal alpha actin isoform (ACTA1), as well as the dominant expression of the gamma isoform of smooth muscle actin (ACTG2) in visceral smooth muscle (45), likely compensate for loss of ACTA2 in the gastrointestinal and urinary tracts. However, there are some de novo mutations in human ACTA2 (R179H and R179C) that associate with hypotonic bladder, malrotation, and intestinal hypoperistalsis (46, 47). Notably, the Actg2 knockout mouse has yet to be reported. In this context, levels of Actg2 mRNA and other smooth muscle markers were unchanged in Lmod1−/− mice, indicating that the MMIHS-like phenotype observed here is not attributable to a secondary effect on expression of other smooth muscle actin-binding proteins.
In summary, we have used human genetics coupled with CRISPR-Cas9 genome editing in mice to establish LMOD1 as a disease gene for MMIHS. Defining all genes linked to this devastating disorder will provide insight into potential treatment strategies involving either genome editing and/or viral-mediated therapies.
Methods
SNP-Array Analysis.
To determine chromosomal abnormalities in the patient and to identify the presence of runs of homozygosity (ROH), we processed 250 ng of DNA on the Illumina HumanOmniExpress BeadChip v1 array (Illumina, Inc.) using standard protocols. Normalized output was generated using Genome Studio version 2011.1 (Illumina). ROH and copy number variants (CNVs) were analyzed and visualized in Nexus CN8.0 (Biodiscovery Inc.). ROH regions had to contain at least 50 probes and be larger than 1 Mb in size in line with established criteria (48, 49). CNVs were profiled according to previously described prioritization methods (50). To determine the consanguinity coefficient (f) and the inbreeding coefficient (F), we used the genomic oligoarray and SNP-array evaluation tool v3.0 (51) and a 50 probes minimum for ROH region.
Whole Exome Sequencing.
Genomic DNA was fragmented using acoustic technologies (Covaris, Inc.). Exome capture was performed with the SureSelect Human All Exon 50 Mb targeted exome enrichment kit v4 (Agilent Technologies, Inc.), and the library was paired-end sequenced (2 × 101 bp) using Illumina TruSeq v4 (Illumina). More than 5 Gb of raw sequence data were processed using the NARWAHL pipeline and aligned to the hg19 reference genome with Burrows–Wheeler Aligner v0.6.2 (52). The Bayesian Genotyper, within the genome analyzer toolkit v1.2.9, was used to determine variants, which were subsequently uploaded to Cartagenia Bench NGS v4.1.8 (Cartagenia Inc.) for filtering and prioritization. CNVs and ROH regions were exported from Biodiscovery Nexus CN8.0 to Cartagenia Bench to inspect for overlap between identified ROH regions with rare variants.
We excluded variants with a high frequency in our in-house control cohort (n = 200, with similar capture, alignment, and variant calls) and with an allele frequency above 0.1% in public databases (ExAC release 0.3, ESP6500SI-v2; 1000 Genomes Phase 3 release v5.20130502; and GoNL SNPs and Indels release 5). Next, we annotated variants using the framework described by Kircher et al. (53) and retained all splicing, coding, noncoding, and nonsense variants with a CADD score of >20 representing the top 1% ranked variants in the framework. The higher this Phred base-calling score is, the more likely the variant has a deleterious effect (CADD v1.3; cadd.gs.washington.edu/home). Heterozygous variants with an allele frequency of ≤0.1% in the above public databases were considered of interest; for homozygous variants, a more lenient cutoff of 1% was used. The deleterious variant is described in the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) using the study description, “Whole exome sequencing studies in patients with megacystis microcolon intestinal hypoperistalsis syndrome (MMIHS)” under the following accession number: SCV000299346. Genes harboring candidate variants were inspected for their mRNA and protein expression patterns in the Human Integrated Protein Expression Database and the Genotype Tissue Expression Protein Database (54), incorporated in GeneCards (55), and overlapped with smooth muscle functioning in the Reactome Database (56).
Sanger Sequencing.
PCR amplification across exon two of LMOD1 was performed on genomic DNA isolated from peripheral blood lymphocytes to confirm the nonsense mutation using sets of primers (SI Appendix, Table S4). Sequencing reactions on purified PCR products were done on an ABI 3130XL genetic analyzer (Applied Biosystems), and sequencing reads were analyzed using SeqScape v2.5 software (Thermo Fisher Scientific).
CRISPR-Cas9 Genome Editing of Lmod1 in Mouse.
All mice were housed in a temperature- and humidity-controlled facility under the supervision of trained laboratory support personnel. Guide RNAs (SI Appendix, Table S4) targeted either to the paired CArG boxes or exon one of mouse Lmod1 were cloned into the BbsI site of pX330-U6 chimeric BB-CBh-hSpCas9 plasmid (42230; Addgene). The guide RNAs were in vitro transcribed using a HiScribe T7 mRNA synthesis kit (New England BioLabs) and purified before assessment on an Agilent Bioanalyzer as described (18). Mouse zygotes were microinjected into the cytoplasm with Cas9 mRNA (100 ng/μL) and each guide RNA (100 ng/μL each) as described previously (17, 57). After injection, zygotes were cultured in KSOM+AA medium for 1 h before transfer into the oviductal ampullae (10 to 15 zygotes per oviduct) of either CD1 (two-component CRISPR) or C57BL/6J (three-component CRISPR) female mice that had been mated, respectively, with vasectomized CD1 or C57BL/6J males the previous evening. Genomic DNA was isolated from tail-snips or toe-clips of founder mice by overnight proteinase K digestion. Primers flanking exon one were used in a PCR to detect the deletion using AccuStart II Supermix (Quanta). PCR conditions were as follows: 3 min incubation at 95 °C, followed by 35 cycles of 30 s at 95 °C, 30 s at 58 °C, and 30 s at 72 °C and a 5-min extension at 72 °C. PCR genotyping of the CArG mutant mice was done with a duplex PCR assay as previously described (17). All PCR products were resolved in a 2% agarose gel.
A guide RNA design tool (58) was used to calculate the 10 most highly ranked off-targets for each of the three guide RNAs. PCR primers flanking each potential off-target sequence were designed and are listed in SI Appendix, Table S4. Adaptor sequences were added to the 5′ end of each primer (5′-ACACTGACGACATGGTTCTACA-3′ for forward primers and 5′-TACGGTAGCAGAGACTTGGTCT-3′ for reverse primers). PCR was carried out for 28 cycles (sequences 95 °C for 30 s, 53 °C for 30 s, and 72 °C for 30 s) using AccuStart II Supermix (Quanta). All 30 PCR reactions were pooled and Sanger sequenced by a core facility (Research Resources Center at the University of Illinois at Chicago). Sequence analysis of FASTQ files was done using CRISPResso (19).
Quantitative Real-Time qPCR.
RNA extraction from patient and control fibroblasts was done using an RNA extraction kit (Qiagen) according to the manufacturer’s instructions. One microgram of total RNA was converted into cDNA using an iScript cDNA synthesis kit (Bio-Rad). A mixture of cDNA and primers (SI Appendix, Table S4) was used with a KAPA SYBR FAST qPCR Kit (Kapa Biosystems), and real-time (RT)-qPCR was performed in an IQ5 cycler machine (Bio-Rad). Expression levels were normalized with the expression of two housekeeping genes (COPS5 and CLK2) and then averaged and presented as fold changes. Total RNA was isolated from mouse tissues using an miRNeasy Mini Kit (Qiagen), and cDNA was synthesized with an iScript cDNA Synthesis Kit (Bio-Rad). RT-qPCR was performed using PerfeCTa SYBR Green SuperMix (Quanta) and PCR primers (SI Appendix, Table S4) in a CFX Connect Real-Time qPCR system (Bio-Rad).
Protein Isolation and Western Blotting.
Fibroblasts (dermal) and human intestinal SMCs were washed with PBS and incubated with standard lysis buffer and protease inhibitors (Roche) for 30 min on ice. Cell lysates were collected by scraping and cleared by centrifugation at 9,500 × g for 15 min at 4 °C. Protein was quantitated using bicinchoninic acid (Thermo Scientific), and 30 μg from each sample were loaded in a TGX precast 4% to 15% gel (Bio-Rad). Proteins were transferred to nitrocellulose membranes and incubated with primary antibodies to LMOD1 (1:200; Proteintech), alpha actin (1:250; Cytoskeleton), and GAPDH (1:1,000; Millipore). Secondary antibodies used were as follows: IRDye 800CW goat anti-mouse and IRDye 680RD goat anti-rabbit (Li-Cor). Whole mouse tissue protein was extracted in Cell Lysis Buffer (Cell Signaling) supplemented with complete mini EDTA-free protease inhibitor mixture tablets (Roche) by mechanical homogenization. Cleared lysates were resolved in 12% polyacrylamide gels, blotted to nitrocellulose, and incubated with primary antibodies to LMOD1 (15117-1-AP, 1:800; Proteintech), TAGLN (ab14106, 1:5,000; Abcam), MYH11 (BT-562, 1:500; Biomedical Technology), Pan-actin (AAN01, 1:1,000; Cytoskeleton), and TUBB (2128, 1:1,000; Cell Signaling).
Immunohistochemistry/Immunofluorescence Microscopy.
Formalin-fixed paraffin-embedded human small intestinal tissues from controls were obtained from the repository of the Department of Pathology, Erasmus University Medical Center. Immunostaining was performed using specific antibody directed against LMOD1 (1:1,000; Proteintech), followed by a biotinylated secondary antibody and then avidin–biotin complex with peroxidase. Sections were then incubated with 3′3′-diaminobenzidine chromagen (Ventana) and then counterstained with hematoxylin. All images were taken with a Nanozoomer 2.0-HT (Hamamatsu Photonics) and analyzed using the Nanozoomer Digital Pathology viewer software (Hamamatsu Photonics). Mouse tissues were fixed in methanol/H2O/acetic acid (60:30:10, vol/vol) overnight, embedded in paraffin, and sectioned at 5 μm thickness. After deparaffinization, slides were either stained with hematoxylin and eosin (H&E) or underwent antigen retrieval using a pressure cooker set at high pressure for 5 min in a pH 9 solution (Dako S2367) for primary antibody staining. For TUNEL and Ki67 staining, slides were similarly treated with high pressure steam in pH 6 solution (S1699; Dako) for 5 min and 10 min, respectively. Primary antibodies were LMOD1 (1:100), Cy3-conjugated ACTA2 (C6198; Sigma), MYH11 (BT-562, 1:100; Biomedical Technology), and Ki67 (652402, 1:100; BioLegend); slides were incubated with each primary antibody overnight at 4 °C. The next day, slides were washed in 1× PBS before incubation with secondary antibody (AF-488 anti-rabbit, A11008, Thermo Scientific), AF-488 anti-mouse (A11012; Thermo Scientific) for 1 h at room temperature. All images were taken with a confocal microscope (FV 1000; Olympus) with voltage adjusted to control (WT) slides and held constant for heterozygous and homozygous null sections.
Transmission Electron Microscopy.
Samples of jejunum intestine and bladder were obtained from Lmod1−/− and WT mice after perfusion fixation with 2.5% glutaraldehyde/4% paraformaldehyde in Millonig’s phosphate buffer. Tissues were postfixed in buffered 1% osmium tetroxide, dehydrated, and embedded into EPON/Araldite epoxy resin. Think sections (70 nm) on formvar/carbon slot grids were imaged as described previously (59). Random measurements of dense body length and concentration were made and analyzed in a blinded manner by two independent investigators.
Cell Culture and Transfections.
Primary human intestinal SMCs (hiSMCs) were obtained from Sciencell Research Laboratories and cultured in SMC medium mixed with 1% smooth muscle growth supplement and 1% penicillin/streptomycin (Sciencell). Cells (300,000 per 35-mm dish) were maintained at 37 °C and 5% CO2 24 h before transfection. Cells were transfected with either scrambled siRNA or siLMOD1 (100 nM per well; Ambion) using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific). After 6 h, the medium was refreshed with normal SMC medium, and cells were allowed to grow for 72 h before analysis.
G-Actin/F-Actin Analysis.
hiSMCs were transfected with scrambled siRNA or siLMOD1 cells and after 72 h cells were harvested and lysed in detergent-based lysis buffer that preserves monomeric actin (G-actin) and filamentous actin (F-actin). Further isolation to separate both forms of actin from cell debris was done according to the manufacturer’s instructions (BK037; Cytoskeleton). F-actin was pelleted by ultracentrifugation whereas G-actin remained in the supernatant. Samples of supernatant and pellet were mixed with 5× SDS sample buffer and loaded into an SDS/PAGE system for Western blotting using an actin antibody (Cytoskeleton), GAPDH (Millipore), and LMOD1 (Proteintech). Bladder tissue from WT and Lmod1 knockout mice was frozen in liquid nitrogen before being assayed in the same manner as hiSMCs.
Contractility Assays.
Twenty-four hours after transfection with scrambled siRNA or siLMOD1, hiSMCs were trypsinized and used for a collagen contractility assay according to the manufacturer’s instructions (Cell Biolabs). Jejunum from WT or Lmod1 knockout mice were rapidly dissected and placed in ice-cold PSS buffer (130 mM NaCl, 4.7 mM KCl, 1.18 mM KH2PO4, 1.17 mM MgSO4, 14.9 mM NaHCO3, 5.5 mM dextrose, 0.026 mM EDTA, and 2.5 mM CaCl2). Rings of jejunum were mounted on a 610M Myograph (DMT) in PSS buffer with constant oxygenation, and the temperature was maintained at 37 °C over a 30-min equilibration period. Passive tension was applied by mechanically expanding jejunum ring segments at 100- to 200-μm intervals, and buffer was exchanged before tissues were allowed to equilibrate for an additional 20 min. Tissues were treated with K-PSS buffer (PSS with 60 mM KCl, 74.7 mM NaCl) or 100 μM carbachol to induce contraction. For KCl treatment, the mean peak contractile force was taken for several independent rounds of contraction over a 30-min period.
Statistics.
Student’s unpaired t test was used to compare independent datasets. For all data, error bars represent SD. Analysis of Mendelian ratios was done using Chi Squared analysis to determine whether the difference between expected and observed ratios was significant. A P value less than 0.05 was considered statistically significant. All statistical analyses were performed using Prism 6.0 (GraphPad Software) or Microsoft Excel.
Study Approval.
Ethical approval of an MMIHS patient from consanguineous parents was granted by the Erasmus Medical Center Ethical Committee (Medisch Ethische Toetsings Commissie, Metc 2011/148, ABR form, NL35920.042.11). The family approved a written informed consent form before inclusion in the study. Mouse experiments were approved by the Institutional Animal Care and Use Committees of the University of Rochester School of Medicine and Dentistry and the University of Nevada, Reno.
Supplementary Material
Acknowledgments
We thank the family involved in this study. We thank the University of Rochester Mouse Genome Editing Resource for the microinjection experiments involving three-component CRISPR editing of the Lmod1 CArG boxes. We thank Gayle Schneider of the University of Rochester’s Electron Microscope Shared Resource Laboratory for thin sectioning of the transmission electron microscopy specimens. The two-component CRISPR knockout of Lmod1 was conducted in the Genome Editing Core of the University of Nevada, Reno School of Medicine, which is supported, in part, by a COBRE grant from the NIH (1P30GM110767). J.M.M. is supported by NIH Grants HL-117907 and HL-112793. R.M.W.H. is supported by a grant from the Vrienden van het Sophia.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620507114/-/DCSupplemental.
References
- 1.Berdon WE, et al. Megacystis-microcolon-intestinal hypoperistalsis syndrome: A new cause of intestinal obstruction in the newborn. Report of radiologic findings in five newborn girls. AJR Am J Roentgenol. 1976;126(5):957–964. doi: 10.2214/ajr.126.5.957. [DOI] [PubMed] [Google Scholar]
- 2.Mc Laughlin D, Puri P. Familial megacystis microcolon intestinal hypoperistalsis syndrome: A systematic review. Pediatr Surg Int. 2013;29(9):947–951. doi: 10.1007/s00383-013-3357-x. [DOI] [PubMed] [Google Scholar]
- 3.Wymer KM, Anderson BB, Wilkens AA, Gundeti MS. Megacystis microcolon intestinal hypoperistalsis syndrome: Case series and updated review of the literature with an emphasis on urologic management. J Pediatr Surg. 2016;51(9):1565–1573. doi: 10.1016/j.jpedsurg.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Moreno CA, et al. Visceral myopathy: Clinical and molecular survey of a cohort of seven new patients and state of the art of overlapping phenotypes. Am J Med Genet A. 2016;170(11):2965–2974. doi: 10.1002/ajmg.a.37857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wangler MF, Beaudet AL. ACTG2-related disorders. In: Pagon RA, et al., editors. GeneReviews. University of Washington; Seattle, WA: 1993. [Google Scholar]
- 6.Halim D, et al. ACTG2 variants impair actin polymerization in sporadic megacystis microcolon intestinal hypoperistalsis syndrome. Hum Mol Genet. 2016;25(3):571–583. doi: 10.1093/hmg/ddv497. [DOI] [PubMed] [Google Scholar]
- 7.Thorson W, et al. De novo ACTG2 mutations cause congenital distended bladder, microcolon, and intestinal hypoperistalsis. Hum Genet. 2014;133(6):737–742. doi: 10.1007/s00439-013-1406-0. [DOI] [PubMed] [Google Scholar]
- 8.Wangler MF, et al. Baylor-Hopkins Center for Mendelian Genomics Heterozygous de novo and inherited mutations in the smooth muscle actin (ACTG2) gene underlie megacystis-microcolon-intestinal hypoperistalsis syndrome. PLoS Genet. 2014;10(3):e1004258. doi: 10.1371/journal.pgen.1004258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miano JM, Cserjesi P, Ligon KL, Periasamy M, Olson EN. Smooth muscle myosin heavy chain exclusively marks the smooth muscle lineage during mouse embryogenesis. Circ Res. 1994;75(5):803–812. doi: 10.1161/01.res.75.5.803. [DOI] [PubMed] [Google Scholar]
- 10.Gauthier J, et al. A homozygous loss-of-function variant in MYH11 in a case with megacystis-microcolon-intestinal hypoperistalsis syndrome. Eur J Hum Genet. 2015;23(9):1266–1268. doi: 10.1038/ejhg.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conley CA, Fritz-Six KL, Almenar-Queralt A, Fowler VM. Leiomodins: Larger members of the tropomodulin (Tmod) gene family. Genomics. 2001;73(2):127–139. doi: 10.1006/geno.2000.6501. [DOI] [PubMed] [Google Scholar]
- 12.Nanda V, Miano JM. Leiomodin 1, a new serum response factor-dependent target gene expressed preferentially in differentiated smooth muscle cells. J Biol Chem. 2012;287(4):2459–2467. doi: 10.1074/jbc.M111.302224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Visser KL, et al. Familial aggregation between the 14th and 21st century and type 2 diabetes risk in an isolated Dutch population. PLoS One. 2015;10(7):e0132549. doi: 10.1371/journal.pone.0132549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeegers MP, van Poppel F, Vlietinck R, Spruijt L, Ostrer H. Founder mutations among the Dutch. Eur J Hum Genet. 2004;12(7):591–600. doi: 10.1038/sj.ejhg.5201151. [DOI] [PubMed] [Google Scholar]
- 15.Bittles A. Consanguinity and its relevance to clinical genetics. Clin Genet. 2001;60(2):89–98. doi: 10.1034/j.1399-0004.2001.600201.x. [DOI] [PubMed] [Google Scholar]
- 16.Kunita R, Otomo A, Ikeda JE. Identification and characterization of novel members of the CREG family, putative secreted glycoproteins expressed specifically in brain. Genomics. 2002;80(5):456–460. [PubMed] [Google Scholar]
- 17.Han Y, Slivano OJ, Christie CK, Cheng AW, Miano JM. CRISPR-Cas9 genome editing of a single regulatory element nearly abolishes target gene expression in mice: Brief report. Arterioscler Thromb Vasc Biol. 2015;35(2):312–315. doi: 10.1161/ATVBAHA.114.305017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miano JM, Zhu QM, Lowenstein CJ. A CRISPR path to engineering new genetic mouse models for cardiovascular research. Arterioscler Thromb Vasc Biol. 2016;36(6):1058–1075. doi: 10.1161/ATVBAHA.116.304790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinello L, et al. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat Biotechnol. 2016;34(7):695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuen M, et al. Leiomodin-3 dysfunction results in thin filament disorganization and nemaline myopathy. J Clin Invest. 2014;124(11):4693–4708. doi: 10.1172/JCI75199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong Q, Ludgate M, Vassart G. Cloning and sequencing of a novel 64-kDa autoantigen recognized by patients with autoimmune thyroid disease. J Clin Endocrinol Metab. 1991;72(6):1375–1381. doi: 10.1210/jcem-72-6-1375. [DOI] [PubMed] [Google Scholar]
- 22.Conley CA. Leiomodin and tropomodulin in smooth muscle. Am J Physiol Cell Physiol. 2001;280(6):C1645–C1656. doi: 10.1152/ajpcell.2001.280.6.C1645. [DOI] [PubMed] [Google Scholar]
- 23.Kostyukova AS. Leiomodin/tropomyosin interactions are isoform specific. Arch Biochem Biophys. 2007;465(1):227–230. doi: 10.1016/j.abb.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Boczkowska M, Rebowski G, Kremneva E, Lappalainen P, Dominguez R. How Leiomodin and Tropomodulin use a common fold for different actin assembly functions. Nat Commun. 2015;6:8314. doi: 10.1038/ncomms9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chereau D, et al. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320(5873):239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pappas CT, et al. Knockout of Lmod2 results in shorter thin filaments followed by dilated cardiomyopathy and juvenile lethality. Proc Natl Acad Sci USA. 2015;112(44):13573–13578. doi: 10.1073/pnas.1508273112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shy GM, Engel WK, Somers JE, Wanko T. Nemaline myopathy. A new congenital myopathy. Brain. 1963;86:793–810. doi: 10.1093/brain/86.4.793. [DOI] [PubMed] [Google Scholar]
- 28.Wallgren-Pettersson C, Sewry CA, Nowak KJ, Laing NG. Nemaline myopathies. Semin Pediatr Neurol. 2011;18(4):230–238. doi: 10.1016/j.spen.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Cenik BK, et al. Severe myopathy in mice lacking the MEF2/SRF-dependent gene leiomodin-3. J Clin Invest. 2015;125(4):1569–1578. doi: 10.1172/JCI80115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian L, et al. Leiomodin-3-deficient mice display nemaline myopathy with fast-myofiber atrophy. Dis Model Mech. 2015;8(6):635–641. doi: 10.1242/dmm.019430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shafiq SA, Dubowitz V, Peterson HdeC, Milhorat AT. Nemaline myopathy: Report of a fatal case, with histochemical and electron microscopic studies. Brain. 1967;90(4):817–828. doi: 10.1093/brain/90.4.817. [DOI] [PubMed] [Google Scholar]
- 32.Bond M, Somlyo AV. Dense bodies and actin polarity in vertebrate smooth muscle. J Cell Biol. 1982;95(2 Pt 1):403–413. doi: 10.1083/jcb.95.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Small JV, Gimona M. The cytoskeleton of the vertebrate smooth muscle cell. Acta Physiol Scand. 1998;164(4):341–348. doi: 10.1046/j.1365-201X.1998.00441.x. [DOI] [PubMed] [Google Scholar]
- 34.Miano JM, Olson EN. Expression of the smooth muscle cell calponin gene marks the early cardiac and smooth muscle cell lineages during mouse embryogenesis. J Biol Chem. 1996;271(12):7095–7103. doi: 10.1074/jbc.271.12.7095. [DOI] [PubMed] [Google Scholar]
- 35.Chen M, et al. Regulation of 130-kDa smooth muscle myosin light chain kinase expression by an intronic CArG element. J Biol Chem. 2013;288(48):34647–34657. doi: 10.1074/jbc.M113.510362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshikawa H, et al. Mice lacking smooth muscle calponin display increased bone formation that is associated with enhancement of bone morphogenetic protein responses. Genes Cells. 1998;3(10):685–695. doi: 10.1046/j.1365-2443.1998.00214.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang JCL, et al. Analysis of SM22alpha-deficient mice reveals unanticipated insights into smooth muscle cell differentiation and function. Mol Cell Biol. 2001;21(4):1336–1344. doi: 10.1128/MCB.2001.21.4.1336-1344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lilly B, et al. Loss of the serum response factor cofactor, cysteine-rich protein 1, attenuates neointima formation in the mouse. Arterioscler Thromb Vasc Biol. 2010;30(4):694–701. doi: 10.1161/ATVBAHA.109.200741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo H, et al. Ablation of smooth muscle caldesmon affects the relaxation kinetics of arterial muscle. Pflugers Arch. 2013;465(2):283–294. doi: 10.1007/s00424-012-1178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morano I, et al. Smooth-muscle contraction without smooth-muscle myosin. Nat Cell Biol. 2000;2(6):371–375. doi: 10.1038/35014065. [DOI] [PubMed] [Google Scholar]
- 41.He WQ, et al. Myosin light chain kinase is central to smooth muscle contraction and required for gastrointestinal motility in mice. Gastroenterology. 2008;135(2):610–620. doi: 10.1053/j.gastro.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rensen SSM, et al. Expression of the smoothelin gene is mediated by alternative promoters. Cardiovasc Res. 2002;55(4):850–863. doi: 10.1016/s0008-6363(02)00491-1. [DOI] [PubMed] [Google Scholar]
- 43.Niessen P, et al. Smoothelin-a is essential for functional intestinal smooth muscle contractility in mice. Gastroenterology. 2005;129(5):1592–1601. doi: 10.1053/j.gastro.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 44.Schildmeyer LA, et al. Impaired vascular contractility and blood pressure homeostasis in the smooth muscle alpha-actin null mouse. FASEB J. 2000;14(14):2213–2220. doi: 10.1096/fj.99-0927com. [DOI] [PubMed] [Google Scholar]
- 45.McHugh KM, Crawford K, Lessard JL. A comprehensive analysis of the developmental and tissue-specific expression of the isoactin multigene family in the rat. Dev Biol. 1991;148(2):442–458. doi: 10.1016/0012-1606(91)90263-3. [DOI] [PubMed] [Google Scholar]
- 46.Milewicz DM, et al. De novo ACTA2 mutation causes a novel syndrome of multisystemic smooth muscle dysfunction. Am J Med Genet A. 2010;152A(10):2437–2443. doi: 10.1002/ajmg.a.33657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meuwissen ME, et al. ACTA2 mutation with childhood cardiovascular, autonomic and brain anomalies and severe outcome. Am J Med Genet A. 2013;161A(6):1376–1380. doi: 10.1002/ajmg.a.35858. [DOI] [PubMed] [Google Scholar]
- 48.Auton A, et al. Global distribution of genomic diversity underscores rich complex history of continental human populations. Genome Res. 2009;19(5):795–803. doi: 10.1101/gr.088898.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu A, et al. Comparison of human genetic and sequence-based physical maps. Nature. 2001;409(6822):951–953. doi: 10.1038/35057185. [DOI] [PubMed] [Google Scholar]
- 50.Brosens E, et al. Copy number variations in 375 patients with oesophageal atresia and/or tracheoesophageal fistula. Eur J Hum Genet. 2016;24(12):1715–1723. doi: 10.1038/ejhg.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wierenga KJ, Jiang Z, Yang AC, Mulvihill JJ, Tsinoremas NF. A clinical evaluation tool for SNP arrays, especially for autosomal recessive conditions in offspring of consanguineous parents. Genet Med. 2013;15(5):354–360. doi: 10.1038/gim.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brouwer RW, van den Hout MC, Grosveld FG, van Ijcken WF. NARWHAL, a primary analysis pipeline for NGS data. Bioinformatics. 2012;28(2):284–285. doi: 10.1093/bioinformatics/btr613. [DOI] [PubMed] [Google Scholar]
- 53.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Consortium GT. GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fishilevich S, et al. 2016. Genic insights from integrated human proteomics in GeneCards. Database (Oxford) 2016:baw030.
- 56.Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(Database issue):D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oliver D, Yuan S, McSwiggin H, Yan W. Pervasive genotypic mosaicism in founder mice derived from genome editing through pronuclear injection. PLoS One. 2015;10(6):e0129457. doi: 10.1371/journal.pone.0129457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Albinsson S, et al. Smooth muscle miRNAs are critical for post-natal regulation of blood pressure and vascular function. PLoS One. 2011;6(4):e18869. doi: 10.1371/journal.pone.0018869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.