Significance
Vertebrate rhodopsin (Rh) has been a model system for many G protein-coupled receptors for over a decade. However, due to its thus-far limited repertoire of active ligands, its use in assisting the development of new therapeutic modalities and drugs has been limited. This study elucidates a photocyclic G protein activation by Rh bound with a six-carbon ring retinal (Rh6mr), and thus broadens the diversity of such Rh signaling modulators. Rh6mr does not release its chromophore after light activation, but instead the resulting photoproduct is thermally reisomerized back to its inactive state, abrogating the necessity for a complex retinoid cycle to renew its chromophore. This photocyclic behavior of Rh6mr opens up several avenues for using optogenetic tools based on vertebrate Rhs.
Keywords: rhodopsin, vision, GPCR, isomerization, chromophore
Abstract
Vertebrate rhodopsin (Rh) contains 11-cis-retinal as a chromophore to convert light energy into visual signals. On absorption of light, 11-cis-retinal is isomerized to all-trans-retinal, constituting a one-way reaction that activates transducin (Gt) followed by chromophore release. Here we report that bovine Rh, regenerated instead with a six-carbon-ring retinal chromophore featuring a C11=C12 double bond locked in its cis conformation (Rh6mr), employs an atypical isomerization mechanism by converting 11-cis to an 11,13-dicis configuration for prolonged Gt activation. Time-dependent UV-vis spectroscopy, HPLC, and molecular mechanics analyses revealed an atypical thermal reisomerization of the 11,13-dicis to the 11-cis configuration on a slow timescale, which enables Rh6mr to function in a photocyclic manner similar to that of microbial Rhs. With this photocyclic behavior, Rh6mr repeatedly recruits and activates Gt in response to light stimuli, making it an excellent candidate for optogenetic tools based on retinal analog-bound vertebrate Rhs. Overall, these comprehensive structure–function studies unveil a unique photocyclic mechanism of Rh activation by an 11-cis–to–11,13-dicis isomerization.
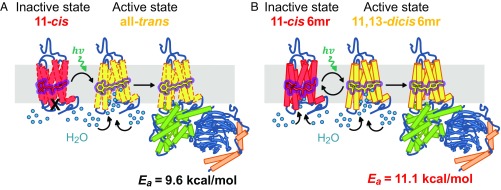
Rhodopsin (Rh) is the visual pigment found in rod outer segments of vertebrate and invertebrate photoreceptors that mediates the transformation of light into vision (1–5). By contrast, microbial Rhs mediate both the energy conversion and cell signaling required for cell survival (2, 6, 7). All classes of Rhs feature a heptahelical transmembrane structure that incorporates a covalently bound retinal chromophore, but they differ in their ability to interconvert between their two spectral absorption states driven by light exposure (2). Although vertebrate Rh undergoes a one-way photobleaching reaction of the retinal chromophore after light absorption (8), microbial Rhs exhibit an intrinsic photocyclic behavior with no chromophore release (2, 8). This makes vertebrate Rhs unsuitable for optogenetic applications that require reversible control over effector ligands. Nevertheless, all Rhs require a cis-trans/trans-cis isomerization of their chromophores to trigger a protein conformational change that mediates the downstream signaling cascade or energy conversion (2, 8). Previous studies on bovine Rh regenerated with six-carbon-ring retinal chromophores (Rh6mr) featuring a locked C11=C12 cis–trans isomerization (SI Appendix, Fig. S1) have implied their ability to activate the G protein transducin (Gt). These results suggest an alternative mechanism that can propagate the downstream visual cascades (9–14). In this study, we provide direct evidence that Rh6mr can activate Gt by an atypical cis-to-dicis isomerization that is also photocyclic, undergoing an 11,13-dicis–to–11-cis reisomerization. Even though it is believed that cis–trans isomerization is required to achieve the conformational change in opsin needed for Gt activation, our results generalize this prerequisite to any isomerization that can achieve the same conformational effect. This study demonstrates a complete transition from a one-way activation of a G protein-coupled receptor (GPCR) into a self-renewable cyclic activation by the mere addition of a cyclohexyl group in its inverse agonist. Additionally, this photocyclic behavior of Rh6mr opens up new avenues for using optogenetic tools based on retinal analog-bound vertebrate Rhs.
Results
Rh6mr Can Activate Gt Efficiently, Even Though It Cannot Undergo a cis–trans Isomerization at the C11=C12 Double Bond.
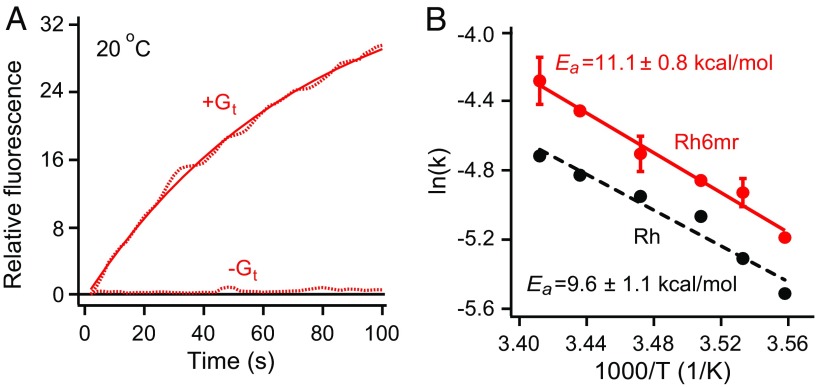
We evaluated the Gt activation ability of Rh6mr by monitoring the increase of intrinsic tryptophan fluorescence in the alpha subunit of Gt (Gtα) in the presence of light-activated Rh6mr at pH 7.0. Conditions for the assay were chosen such that the Gt activation rate was the same as that determined by GTPγS-induced complex dissociation (15). As expected, significant elevation of Gtα intrinsic fluorescence was observed upon GTPγS-induced complex dissociation with the light-activated Rh6mr (Fig. 1A). Similar Gt activation kinetics was observed with native bovine Rh, as shown in SI Appendix, Fig. S2A (16–20). Additionally, no significant Gtα fluorescence change was noted with Rh6mr in the absence of light stimulation (SI Appendix, Fig. S2B, black dashes). This indicates a light-induced change in Rh6mr similar to that seen in Rh activation.
Fig. 1.
Gt activation kinetics of Rh6mr. (A) The Gt activation ability of Rh6mr was monitored by an increase in the intrinsic tryptophan fluorescence of the Gt alpha subunit in the presence of light-activated Rh6mr at 20 °C (pH 7.0). (B) Arrhenius plot for Rh6mr Gt activation for a temperature range of 8 to 20 °C. Natural logarithms of the measured Gt activation rate constants were plotted against inverse temperatures. The slopes of Arrhenius lines (Rh6mr, solid red; Rh, black dashes) provide the energy required for the Rh6mr conformational change needed for Gt activation (Ea).
Gt activation rates of Rh6mr at different temperatures were then determined to obtain the temperature dependence of Gt activation kinetics associated with isomerization of the locked retinal chromophore. Gt activation rates at temperatures ranging from 8 to 20 °C fit the Arrhenius straight line, yielding the activation energy of 11.1 kcal/mol required for the conformational change of Rh6mr needed to activate Gt (Fig. 1B, red). The energy threshold for Gt activation by Rh6mr was similar to the Rh activation energy of 9.6 kcal/mol (Fig. 1B, black dashes) (20, 21). These data suggest a similar mechanism of Gt activation for both Rh6mr and Rh that involves opening of the opsin cytoplasmic side upon light illumination (22, 23) and subsequent binding to Gt. Even though the energy required for the conformational change necessary to activate Gt is similar for both Rh and Rh6mr, it is unknown whether their Gt activation efficiencies are equivalent. Overall, these results clearly show that Rh6mr can activate Gt, even though it cannot undergo a cis–trans isomerization at the C11=C12 double bond. Thus, another Rh6mr photoproduct formed upon light illumination could be responsible for activating Gt.
Rh6mr Has an Atypical Photocyclic Gt Activation Mechanism.
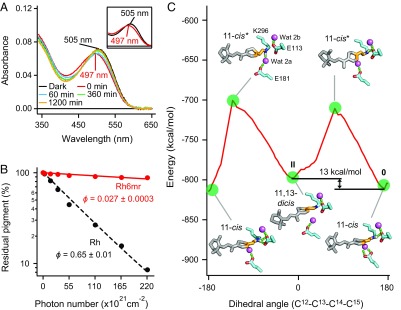
To investigate the formation of another Rh6mr photoproduct upon light illumination, we performed time-dependent UV-vis absorption spectral studies of Rh6mr upon light illumination at 20 °C and pH 7.0. As shown in Fig. 2A, the absorption maximum (λmax) of Rh6mr at 505 nm (black curve) exhibited an 8-nm blue shift (λmax, 497 nm; red curve) immediately (0 min) after illumination. This decrease in absorbance could be explained by a smaller extinction coefficient of the new Rh6mr photoproduct being formed. The same spectral shift also was previously reported, wherein λmax at 510 nm in the dark state was shifted to 494 nm after illumination (9). Our HPLC analysis, performed under conditions identical to the UV-vis spectroscopic measurements, revealed an increased intensity of peak 2 upon light illumination corresponding to the 11,13-dicis isoform of six-ring–locked retinal (6mr) (SI Appendix, Fig. S3). The identities of all HPLC peaks were determined by their corresponding 1H and NOESY NMR spectra (SI Appendix, Figs. S4–S7). Thus, a photoproduct of Rh6mr possessing the 11,13-dicis 6mr isomer as a chromophore can actively play a role similar to that of the Meta-II intermediate of Rh by activating Gt upon illumination (Fig. 1A and SI Appendix, Figs. S2 and S3). However, unlike Rh, Rh6mr requires longer illumination periods to reach its photostationary state, which clearly indicates its lower photosensitivity relative to that of Rh. Indeed, a photosensitivity analysis of Rh6mr accomplished by measuring the decrease in absorption at λmax under different light intensities (24–26) revealed a much lower photosensitivity compared with Rh (Fig. 2B). When the percentage of residual inactive pigment was plotted against the incident photon flux and fitted with an exponential function, the photosensitivity of Rh6mr was determined as only 0.042 ± 0.0004 relative to that of Rh. This value and the extinction coefficient of Rh6mr at 505 nm [41,200 M−1⋅cm−1 (10)] gave a quantum yield of 0.027 ± 0.0003, which was considerably lower than that of Rh [0.65 (24)]. It should be noted that the formation of the 11,13-dicis Rh6mr isoform is dependent on the incident photon flux and the pH of the system (SI Appendix, Fig. S3C). This suggests that the lower quantum yield of Rh6mr is unlikely to be the consequence of a second photon absorption by the 11,13-dicis conformer. The calculated quantum yield was also found to be in good agreement with the illumination periods required for Rh6mr to attain its Meta-II–like state during our Gt activation studies.
Fig. 2.
Photocyclic behavior and photosensitivity of Rh6mr. (A) Time-dependent UV-vis absorption spectra of Rh6mr in the dark and over a period of 20 h after a 1-min illumination at pH 7.0 at 20 °C. (A, Inset) Expanded absorption spectra of Rh6mr in the dark and immediately after a 1-min illumination. (B) The photosensitivity of Rh6mr and Rh. The percentage of residual pigment was plotted on a semilogarithmic scale against the incident photon flux and fitted with an exponential function (the Rh6mr photosensitivity plot is scaled down to show the comparison with Rh). The photosensitivity (ϕ) of Rh6mr (solid red), estimated by the slope of the fitting line, was 0.043 ± 0.0004 relative to that of Rh (black dashes). (C) Energy plot of Rh docked with the 11-cis 6mr isomer during the preferred anticlockwise rotation of the 6mr C12-C13-C14-C15 dihedral angle. The energy after a half-rotation (label II) is higher than the initial minimum at 170° (label 0), suggesting the plausibility of the 11,13-dicis–to–11-cis reisomerization. Waters 2a and 2b are shown as purple spheres, and hydrogen bonds are shown as dashed green sticks.
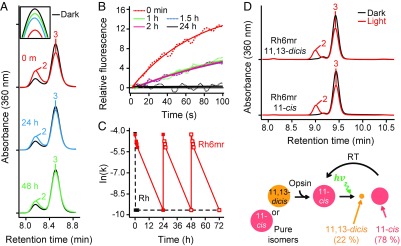
The Meta-II intermediate of Rh decays gradually by retinal Schiff base hydrolysis. In contrast, the Meta-II–like state of Rh6mr does not decay over time, but instead converts back to the Rh6mr inactive state, as noted by the gradual red shift of λmax from 497 nm back to 505 nm (Fig. 2A). This λmax backshift occurs over a period of hours and is reversible by additional illuminations. Our time-dependent HPLC analysis clearly shows a significant decrease in the intensity of peak 2 within 48 h compared with its intensity immediately after illumination (Fig. 3A). Additionally, the decrease in the intensity of peak 2 (11,13-dicis 6mr isomer) was accompanied by an increase in peak 3 intensity (11-cis 6mr isomer), suggesting reisomerization between the two isomers via a thermal isomerization around the C13=C14 double bond. Similar results were obtained with pure 6mr isomers, where the 11,13-dicis isomer converted to the 11-cis isomer after binding to opsin (Fig. 3D). These results suggest that Rh6mr can function in a photocyclic manner (Fig. 3D, Bottom). Our molecular mechanics (MM) analysis confirmed the plausibility of the 11,13-dicis–to–11-cis reisomerization or a 360° rotation of the C12-C13-C14-C15 dihedral angle. In particular, the energy minimum after half-rotation (−11°) is higher than the initial minimum at 170° (Fig. 2C and SI Appendix, Fig. S8, labels 0 and II, respectively). The ability of the 11,13-dicis isomer to reisomerize into the 11-cis isomer indicates the existence of a relatively small transition energy between the inactive and active forms of Rh6mr compared with Rh (27, 28). This small transition energy directly translates into a relatively small conformational change compared with Rh during light activation, and reiterates its striking similarity to microbial Rhs (29, 30).
Fig. 3.
Time-dependent HPLC analyses revealing prolonged Gt activation of Rh6mr. (A) Time-dependent HPLC analysis of photoactivated Rh6mr was performed over a 48-h period. Rh6mr was kept at 20 °C in the dark for either 24 or 48 h after a 1-min illumination with 480- to 520-nm light. Extraction of retinal-oximes from Rh6mr was performed as described in Materials and Methods. (A, Inset) Overlay of time-dependent HPLC chromatograms of peak 3 (11-cis 6mr isomer) showing reisomerization of the 11,13-dicis to the 11-cis form. (B) Comparison of Gt activation efficiencies of Rh6mr at different time points within a 24-h period. Samples were kept at 20 °C in the dark for 1, 1.5, 2.0, and 24 h after a 1-min illumination with 480- to 520-nm light at pH 7.0. Fluorescence intensities were monitored (dotted lines) and fitted by single-exponential functions (solid lines). (C) Prolonged and cyclic Gt activation of Rh6mr. The Gt activation rate constants of Rh6mr samples at time points 0, 1, 1.5, 2, and 24 h after illumination were plotted against time. Independent 24-h samples were subjected to an additional illumination, and the ensuing Gt activation rates were plotted similarly. Closed and open squares correspond to the experimental and extrapolated Gt activation rates, respectively (Rh6mr, red; Rh, black). (D) HPLC chromatograms of Rh6mr regenerated with pure 11,13-dicis and 11-cis 6mr isomers. (D, Top) HPLC analysis under dark (black) and light (red) conditions revealed that the 11,13-dicis isomer is converted exclusively to the 11-cis isomer after binding with the opsin protein moiety. (D, Bottom) Illustration showing the photocyclic behavior of Rh6mr regenerated with pure 11,13-dicis and 11-cis 6mr isomers. RT, room temperature.
A slow reisomerization from the 11,13-dicis back to the 11-cis form of Rh6mr also means a prolonged period of Gt activation. Indeed, Gt activation analysis of Rh6mr tested at different time points post illumination revealed Gt activation extending to over 2 h (Fig. 3B). Despite an initial drop in the Gt activation rate at 1 h, similar Gt activation kinetics were observed between 1 and 2 h post illumination (Fig. 3 B and C), again consistent with the slow photocyclic behavior of Rh6mr. Additionally, Rh6mr repeatedly activated Gt upon subsequent illuminations after 24-h dark recovery periods (Fig. 3C).
Previous computational modeling studies based on the Rh crystal structure showed that all four isomers of 6mr could fit into the opsin chromophore binding site, in good agreement with the accompanying spectral studies (11). However, our HPLC analysis identified the 11-cis 6mr isomer (peak 3) as the predominant form upon regeneration of opsin with the 6mr isomer mixture (Fig. 3A and SI Appendix, Fig. S3). This highlights the possibility that the chromophore binding site of Rh not only accommodates but drives the isomerization of all other 6mr isomers into the 11-cis form.
Further, we calculated energies of inactive Rh and the active Meta-II state docked with the 11-cis and 11,13-dicis 6mr isomers separately after 100-ns molecular dynamics (MD) simulations. The inactive Rh model bound to the 11-cis 6mr isomer (SI Appendix, Fig. S9A) displayed an energy minimum of −852 kcal/mol, significantly lower than that of inactive Rh docked with the 11,13-dicis 6mr isomer (−832 kcal/mol) (SI Appendix, Fig. S9B). Therefore, our MD analyses also suggest that the 11-cis isomer of 6mr fits energetically better to the inactive state of Rh. In contrast, the active Meta-II structure docked with the 11-cis 6mr isomer (SI Appendix, Fig. S9C) has an energy minimum of −838 kcal/mol, only 3 kcal/mol lower than that of the 11,13-dicis 6mr isomer-bound Meta-II (−835 kcal/mol) (SI Appendix, Fig. S9D). This difference in energy minima is significantly smaller than the 20 kcal/mol difference observed for inactive Rh. The energy gap of 3 kcal/mol can easily be overcome by small changes in the microenvironment of Rh mediated by water and lipid dynamics. These MD results further support the thermal reisomerization between the 11,13-dicis and 11-cis 6mr isomers.
11-cis 6mr Isomer Is the Preferred Configuration of Rh6mr.
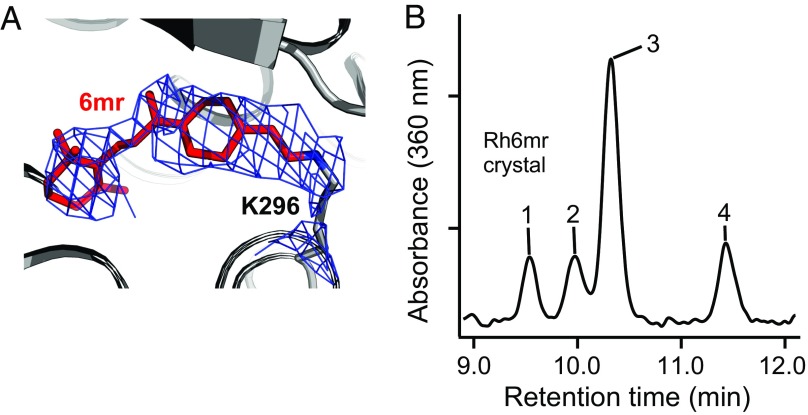
To visualize the configuration of the 6mr isomer that is preferentially accommodated in the Rh6mr-binding pocket, we crystallized Rh6mr in its inactive state. The crystal structure of Rh6mr bore a strong resemblance to the inactive Rh crystal structure. The Rh6mr structure refined at 4.01 Å shows the typical seven transmembrane helices connected by partially resolved extracellular (E1 to E3) and cytoplasmic (C1 to C3) loops and the cytoplasmic helix VIII, which runs along the membrane. Interestingly, Rh6mr crystallized in a unique P3121 space group (SI Appendix, Fig. S10), not observed in any Rh/opsin structure crystallized to date. It should be noted that the chromophore-binding pocket of Rh6mr revealed 6mr occupying the same binding pocket as 11-cis-retinal bound covalently to Lys296 on helix VII in Rh. The 2Fo-Fc omit map of the chromophore-binding pocket of Rh6mr fitted well with the 11-cis 6mr isomer (Fig. 4A). Thermal factor sharpening of 2Fo-Fc electron-density maps was performed at Bsharp below the negative Wilson B value of the diffraction data to upweight the higher-resolution terms (31–37). In addition to the Rh6mr crystal structure, bovine opsin lacking the 6mr chromophore was crystallized at 2.7 Å. Crystallization of bovine opsin was performed under the same conditions as Rh6mr except for the exclusion of 6mr. The opsin structure bears strong structural similarity to the previously reported active-like conformation (rmsd of 0.45 Å), wherein an n-octyl-β-d-glucopyranoside molecule was localized in the retinal-binding pocket (38). Initial refinement and comparison of the opsin crystal structure with Rh6mr clearly show the presence of a 6mr Fo-Fc density in the Rh6mr crystal structure (SI Appendix, Fig. S11). In contrast, the opsin crystal structure harbored an Fo-Fc density corresponding to an n-octyl-β-d-glucopyranoside molecule (SI Appendix, Fig. S11B) (38). HPLC analysis of washed Rh6mr crystals confirmed the presence of 6mr, with the 11-cis 6mr isomer being the dominant isoform (Fig. 4B). The dominance of the 11-cis 6mr isomer in Rh6mr crystals reinforces the conclusion that the opsin moiety itself promotes this favored isomerization state.
Fig. 4.
Preferential configuration of 6mr in Rh6mr crystals. (A) Crystal structure of Rh6mr. The retinal-binding pocket of Rh6mr shows a 2Fo-Fc density (blue mesh) corresponding to 6mr occupying the same binding pocket as 11-cis-retinal in Rh. The 2Fo-Fc density contoured at 1.1σ fits well with the 11-cis 6mr isomer covalently linked to Lys296. The 2Fo-Fc density map was B factor-sharpened (Bsharp = −178 Å2). (B) HPLC analysis of washed Rh6mr crystals showing the 11-cis 6mr isomer (peak 3) as the predominant configuration in the retinal-binding pocket. Rh6mr crystals were obtained by treating bleached ROS membranes with a 6mr isomer mixture. Five or six Rh6mr crystals (>100 μm in their longest dimension) were washed three times in 100 μL reservoir solution followed by extraction of 6mr-oximes and HPLC analysis.
Discussion
Although Rh6mr Gt activation capability has been discussed previously (9–14), its underlying molecular mechanism is poorly understood. In this study, we found that the 11,13-dicis 6mr isoform is generated as the photoproduct during illumination, and is exclusively responsible for the dynamic structural change in Rh6mr required to attain a Meta-II–like intermediate state that can activate Gt. This serves as one of the first atypical examples of photoactivation elicited by a non–cis–trans isomerization-based phenomenon. Furthermore, this 11,13-dicis–bound photoproduct is stable to Schiff base hydrolysis, and can thermally revert back to its inactive 11-cis form. Notably, the Rh6mr-specific photocycle is similar to those of microbial Rhs, such as bacterio-Rh, in which exposure to light isomerizes the all-trans-retinal chromophore to the 13-cis form that thermally reisomerizes back to the all-trans form (6–8). Thus, an isomerization around the C13=C14 double bond rather than the C11=C12 position plays a critical role in achieving an active Meta-II–like helical opening in Rh6mr.
Previous studies on Rh6mr also highlighted two potential reasons for its resistance to Schiff base hydrolysis (13), one being the nonaccessibility of bulk water to the retinal-binding pocket and the other the extremely low hydrolysis rate of the protonated Schiff base (PSB). Therefore, to obtain insight into the solvent accessibility of the Rh6mr chromophore-binding pocket, we assessed the effect of hydroxylamine (NH2OH) on the hydrolysis rate of the PSB with time-dependent UV-vis spectroscopy. As shown in SI Appendix, Fig. S12B, 200 mM NH2OH had a significant effect on the hydrolysis rate of light-illuminated Rh6mr. Interestingly, and unlike Rh, Rh6mr was susceptible to NH2OH in the dark with a T1/2 of 80 ± 3 min (SI Appendix, Fig. S12A). The sensitivity to NH2OH in the dark strongly suggests that 6mr binding to the opsin moiety renders the cytoplasmic side relatively open, allowing water and small molecules such as NH2OH to gain access to the retinal-binding pocket.
To confirm the existence of a relatively open cytoplasmic side in Rh6mr, we prepared proteoliposomes that contained Rh6mr preferentially oriented with their N termini located inside the liposomal lumen and C termini exposed to solvent (SI Appendix, Fig. S13 A and B) (39). As expected, extraction of retinal-oximes under nondenaturing conditions resulted in the quantitative liberation of 6mr-oximes from the proteoliposomes upon light illumination (SI Appendix, Fig. S13C). Interestingly, a similar release of 6mr-oximes was observed from Rh6mr proteoliposomes under dark conditions (SI Appendix, Fig. S13C). Considering the orientation of the Rh6mr molecules in the proteoliposomes, this further strengthens the supposition that small molecules enter the chromophore-binding pocket of Rh6mr from its relatively open cytoplasmic side compared with Rh. Quantitative comparison of the 6mr-oximes released from proteoliposomes under dark and illuminated conditions further supports an additional opening of the cytoplasmic side upon light stimulus, allowing Gt to bind the activated Rh6mr in an energetically favorable orientation (Fig. 5).
Fig. 5.
Proposed model for Gt activation by Rh6mr upon illumination. (A) In Rh, absorption of a photon leads to retinal cis–trans isomerization. This causes a conformational change in the opsin protein moiety, culminating in the formation of its activated Meta-II state. Upon Meta-II formation, the proton from the retinal Schiff base dissociates, disturbing the intramolecular hydrogen-bonding network in the retinal-binding pocket. This causes a conformational change, opening the cytoplasmic side and enabling Gt to interact with Meta-II. (B) Alternatively, in Rh6mr, the 11-cis 6mr isomer is isomerized to its 11,13-dicis form upon activation by a light stimulus, causing a helical structural change in the Rh6mr opsin moiety forming a Meta-II–like state. Unlike Rh, inactive Rh6mr has a relatively open cytoplasmic side, allowing bulk solvent and hydroxylamine to gain access to the protonated Schiff base. Upon light activation, the cytoplasmic side of Rh6mr undergoes additional opening that enables Gt to interact with the cytoplasmic surface of Rh6mr. Interestingly, activated Rh6mr can thermally revert back to its inactive form in a slow photocyclic manner.
In conclusion, we demonstrated the susceptibility of Rh6mr to bulk water/NH2OH even in the dark, indicating the accessibility of bulk solvent to the PSB. Thus, our results support the possibility of a low Schiff base hydrolysis rate as the reason for Rh6mr resistance to chromophore release. This could be caused by the Schiff base being protonated even in the active Meta-II–like state of Rh6mr. It is well-known that the active Meta-II state of Rh exhibits a 118-nm blue shift in the λmax that corresponds to the deprotonation of the Schiff base (2). However, the Rh intermediate state Meta-I exhibits a smaller blue shift (20 nm) and has the Schiff base protonated (1, 2, 4, 5). Therefore, the presence of the PSB also is consistent with a smaller 8-nm blue shift in λmax observed after light activation of Rh6mr (Fig. 2A). Previous FTIR studies show Glu113 to be protonated after light illumination of Rh6mr (14). Based on the combination of our spectral (Fig. 2A) and MM studies (SI Appendix, Fig. S9D), we propose that the Rh6mr Schiff base remains protonated after light activation. This PSB must be stabilized by a nearby nucleophile such as Glu181, a counterion switch in Rh Meta-II precursor states (40), to render Rh6mr resistant to PSB hydrolysis. Indeed, our MM analyses show a direct interaction between the PSB and Glu181 in the active Rh6mr bound with the 11,13-dicis 6mr isomer (SI Appendix, Fig. S9D), whereas Glu113 moves away from the PSB compared with the inactive Rh6mr (SI Appendix, Fig. S9A). This interaction could be enhanced by a retinal binding site rearrangement induced by the bulkier six-member ring culminating in an opsin conformational change required for Gt activation. Overall, these observations suggest an alternate pathway of Rh6mr activation that can attain a Meta-II–like state without deprotonation of the Schiff base.
In summary, this study broadens the diversity of Rh signaling modulators by showing a photocyclic G-protein activation by Rh bound to a noncanonical retinal. This demonstrates that Rh is more similar to other GPCRs in terms of its activation by chemically diverse ligands. We show that six-carbon-ring retinal chromophores can make Rh self-sustainable, thereby abrogating the necessity for a retinoid cycle to renew its chromophore. The comprehensive biophysical and structural analyses of Rh6mr reported here provide a basis for further medicinal chemistry efforts to improve 6mr for optogenetic and therapeutic applications. Although a new class of modified locked retinals with higher quantum yield and faster thermal recovery will be required, the current findings demonstrate the potential of vertebrate Rh to serve as a template for GPCR-based optogenetic applications. Additional structure–activity relationship-based chemical modifications in these synthetic chromophores could provide the characteristics required for specific applications.
Materials and Methods
Synthesis.
A mixture of (7E,9Z,11Z,13Z)-cyclohexyl-[1′,2′:10,13]-retinal (isomer 1; 9,11,13-tricis), (7E,9E,11Z,13Z)-cyclohexyl-[1′,2′:10,13]-retinal (isomer 2; 11,13-dicis), (7E,9E,11Z,13E)-cyclohexyl-[1′,2′:10,13]-retinal (isomer 3; 11-cis), and (7E,9Z,11Z,13E)-cyclohexyl-[1′,2′:10,13]-retinal (isomer 4; 9,11-dicis) was prepared by a method reported by Bhattacharya et al. (10). MS: m/z 297.3 (M+1). Pure 6mr isomers were separated by normal-phase HPLC on a preparative silica gel column (Phenomenex; Luna 10 μm silica, 250 × 21.2 mm) with a linear gradient of ethyl acetate (1 to 10%) in hexanes for 220 min at a flow rate of 5 mL/min. Peaks corresponding to the 9,11,13-tricis (peak 1), 11,13-dicis (peak 2), 11-cis (peak 3), and 9,11-dicis (peak 4) isomers appeared at 94, 100, 103, and 122 min, respectively.
MS analyses were performed with a single-quadrupole mass spectrometer with electrospray ionization. NMR spectra were recorded on a Bruker 500-MHz apparatus at either 10 or 25 °C, and referenced relative to the residual proton resonances of C6D6 (δ = 7.15 ppm). 1H NMR spectra of the 6mr isomers were collected with their corresponding 2D nuclear Overhauser effect spectroscopy, HNN-correlation spectroscopy, and total correlation spectroscopy spectra acquired at a concentration range of 1.9 to 2.4 mg for each isomer. Spectra were processed by MestReNova (Mestrelab Research). Proton chemical shifts of the purified 6mr isomers are provided in SI Appendix, Tables S1–S4.
Rh6mr and Rh Purification.
All experimental procedures were carried out in a darkroom under dim red light (>670 nm). Bovine rod outer segments (ROS) were prepared as described elsewhere (41, 42). ROS were washed with isotonic and hypotonic buffer to remove both soluble and membrane-associated ROS proteins (43). For Rh6mr preparation, purified ROS membranes (20 mg/mL Rh) were bleached with a 100-W white light for 40 min in the presence of 20 mM NH2OH. Bleached ROS/opsin membranes then were treated with a molar excess of six-carbon ring-locked retinal chromophore (6mr) for 24 h at 20 °C. Regenerated Rh6mr or native Rh membranes were solubilized by a zinc/alkyl-glucoside extraction method and centrifuged at 100,000 × g for 40 min to extract Rh6mr or Rh (44). Clear supernatants were loaded onto a 1D4-coupled CNBr-activated Sepharose 4B column and washed with buffer containing 10 mM MES (pH 6.4), 100 mM NaCl, and 0.02% n-dodecyl-β-d-maltoside (DDM) to either dispose of excess retinal or achieve further purification. Finally, purified Rh6mr or Rh was eluted with 0.5 mg/mL of the TETSQVAPA nanopeptide (from the Rh C-terminal sequence) (45).
Preparation of Rh6mr and Rh Proteoliposomes.
Rh6mr and Rh proteoliposomes were prepared as described previously (39). Briefly, a solution of commercially available soybean phospholipids (asolectin; Sigma-Aldrich) in buffer containing 10 mM MES (pH 5.9), 100 mM NaCl, and 1% n-nonyl-β-d-glucopyranoside (NG) was mixed with purified Rh6mr or Rh. The molar ratio of asolectin [assumed molecular mass, 760 (46)] to Rh6mr or Rh was 200:1 (39, 47). The protein–lipid suspension was kept at 20 °C for 2 h, followed by dialysis against buffer containing 10 mM MES (pH 5.9) and 100 mM NaCl (2,000-fold excess) at 4 °C in the dark for 48 h with four buffer changes. The proteoliposome suspension was pelleted at 100,000 × g at 4 °C for 1 h, and the pellet was washed and resuspended in 10 mM MES (pH 5.9) and 100 mM NaCl. Functional integrity of the preparation was examined spectrophotometrically by measuring the Rh6mr spectrum both in the dark and after a light stimulus. The orientation of Rh6mr or Rh in proteoliposomes was confirmed by Asp-N endoproteinase digestion (39, 47), which cleaves opsin specifically between Gly329 and Asp330 in the C-terminal tail (48).
Chromophore Extraction and Analysis.
Chromophores were extracted from irradiated or nonirradiated samples as described elsewhere (49). Briefly, samples were supplemented with 50 mM NH2OH followed by either light illumination with a 150-W fiber light (NCL-150; Volpi) delivered through a 480- to 520-nm band-pass filter (Chroma Technology) or kept in the dark depending on the type of experiment. NH2OH was added to the samples to achieve a final concentration of 100 mM. After a 10-min incubation on ice, samples were supplemented with methanol (250 μL) and mixed thoroughly to denature the protein. In experiments with proteoliposomes, samples were supplemented with 200 mM NH2OH followed by either light irradiation or kept in the dark and incubated for 2 h at 20 °C. Then retinoids were immediately extracted with 1 mL of hexanes. The organic phase was separated by centrifugation at 16,000 × g for 1 min. The hexane extraction was repeated twice and the retinoid composition was analyzed promptly by HPLC.
Retinoid composition was determined with an Agilent 1100 series HPLC. Retinoids extracted with hexanes were injected onto a normal-phase analytical HPLC column (Zorbax SIL 5 μm, 4.6 × 250 mm; Agilent Technology) equilibrated with 10% (vol/vol) ethyl acetate in hexanes. Retinal-oximes were separated by isocratic elution with the equilibration solvent at a flow rate of 1.4 mL/min.
Spectroscopy.
Absorption spectra of solubilized Rh6mr or Rh (10 mM MES, pH 6.4, 100 mM NaCl, 0.1% DDM) were measured with a Cary 50 UV-vis spectrophotometer (Varian) maintained at either 4 or 20 °C (TC 125 temperature controller; Quantum Northwest). Photobleaching experiments were carried out with a 150-W fiber light delivered through a 480- to 520-nm band-pass filter.
Gt Activation Assay.
Gt was extracted from frozen bovine ROS membranes as described elsewhere (50, 51). The intrinsic fluorescence increase from Gtα was measured with an L55 luminescence spectrophotometer (PerkinElmer Life Sciences) operating at excitation and emission wavelengths of 300 and 345 nm, respectively (15, 18, 19, 52). The ratio of Gt to either Rh6mr or Rh was 10:1, with Gt at a concentration of 1,500 nM and Rh6mr or Rh at 150 nM. This was followed by the addition of 300 µM GTPγS to determine the GTPγS-induced complex dissociation and fluorescence changes. Samples were bleached for 1 min with a fiber light delivered through a 480- to 520-nm long-pass wavelength filter before the fluorescence measurements. Gt activation rates were determined for the first 100 s of the Gt activation assay.
Photosensitivity Measurements of Rh6mr.
Photosensitivity measurements of Rh6mr and Rh were performed as described elsewhere (24–26, 49). Briefly, Rh6mr samples maintained at 20 °C were irradiated with light from a 150-W fiber light source delivered through a 500-nm band-pass interference filter. The cuvette (width, 4 mm; path length, 1 cm) was then inserted into a cell holder maintained at a desired temperature and the absorbance change was monitored with a UV-vis spectrophotometer. The light intensity was attenuated by a neutral-density filter (Thorlabs) with a maximum intensity set either to bleach <90% of Rh or deliver 500 μW to Rh6mr because of its low quantum yield compared with Rh. The amount of residual pigment after every irradiation was corrected with the dark spectrum of the pigment. The incident photon flux (s−1⋅cm−2) was calculated from the power (W) of the incident light measured by a power meter (Thorlabs). The intensity of the incident light was continuously monitored to correct for any fluctuations. The percentage of residual pigment was plotted on a semilogarithmic scale against the incident photon count and fitted with an exponential function wherein the slope of the fitting line corresponded to the relative photosensitivity of the pigment at the irradiating wavelength (24–26). Rh photosensitivity was used as a control for all measurements, and the photosensitivity of Rh6mr was determined as a value relative to that of Rh.
Molecular Mechanics Calculations.
All energy minimizations, equilibrations, and molecular dynamics simulations were carried out using NAMD version 2.10 (53) with the CHARMM36 force field (54). Structures of inactive Rh and the active Meta-II state of Rh [Protein Data Bank (PDB) ID codes 1U19 (55) and 3PQR (56), respectively] were used for the MM analysis. Internal water molecules present in both crystal structures were kept intact, and both key glutamic acid residues E113 and E181 were deprotonated. During the rotation around the C13=C14 double bond, a restraint was applied on the C20-C13-C14-C15 dihedral angle whereas all residues within a 6-Å vicinity of 6mr were allowed to move freely. Amino acid residues outside the chromophore-binding pocket were kept frozen but contributed to interactions with the freely moving area. A step size of 5° was used for the C20-C13-C14-C15 dihedral angle during both clockwise and anticlockwise rotations. The vicinity of retinal was optimized by using 10,000 steps of conjugation gradient energy minimization, followed by a 1-ns MD simulation at 27 °C and a final 10,000 steps of conjugation gradient energy minimization. The starting angle for the rotation scan was kept at 0° for the C20-C13-C14-C15 dihedral angle, which corresponds to ∼180° for the C12-C13-C14-C15 dihedral angle. The optimization procedure for every C20-C13-C14-C15 dihedral angle step was started from the optimized structure of the previous state to keep the changes continuous. The restraint applied on the C20-C13-C14-C15 dihedral angle rendered flexibility to the C12-C13-C14-C15 dihedral angle similar to that of native retinal flexibility during an 11-cis–to–all-trans isomerization in Rh. The MD simulations of both the inactive Rh and active Meta-II structures docked with the 11,13-dicis and 11-cis 6mr isomers were performed using the same optimization protocol, except for longer 100-ns MD simulations, and a lack of restraints applied on the dihedral angles.
Rh6mr Preparation and Crystallization.
Bovine Rh6mr and opsin membranes were prepared as described above (Rh6mr and Rh Purification). Rh6mr membranes (20 mg/mL opsin) were solubilized by the zinc/alkyl-glucoside extraction method as described (44), using 1% NG or 1% n-octyl-β-d-glucopyranoside (OG). The clear supernatant was purified further by size-exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare Life Sciences) equilibrated with 50 mM MES (pH 6.4) and either 1% NG or 1% OG. Fractions were concentrated and then used for crystallization. Alternatively, Rh6mr or opsin membranes were solubilized with 20 mM bis-Tris propane (pH 7.5) or 50 mM MES (pH 6.4), 130 mM NaCl, 1 mM MgCl2, 10% (wt/vol) sucrose, and 1% OG for 1 h at 20 °C. Insoluble material was removed by centrifugation at 16,110 × g for 5 min at 4 °C. Crystallization screens by the sparse matrix crystallization method (57) under conditions based upon previously published crystallization conditions for Rh and opsin (38, 56, 58–61) were carried out by both the hanging-drop and sitting-drop vapor-diffusion methods. Each hanging drop was prepared on a siliconized coverslip by mixing equal volumes of Rh6mr and reservoir solution. The reservoir solution contained 2.8 to 3.4 M ammonium sulfate in 0.05 to 0.1 M MES (pH 6.1 to 6.6) or 0.05 to 0.1 M NaAcO buffer (pH 5.2 to 5.6). Crystals appeared after 4 to 5 d at 4 °C and reached 50 to 200 μm in their longest dimension within 7 d. Crystals were transferred directly from the mother liquor into dual-thickness microloops (MiTeGen) and frozen by plunging them into liquid nitrogen.
Diffraction Data Collection and Structural Refinement.
X-ray data collection of Rh6mr and opsin crystals was performed at −173 °C with a cryoprotectant consisting of an 80% (vol/vol) reservoir solution and 20% (wt/vol) trehalose. Diffraction data were collected at the Northeastern Collaborative Access Team 24-ID-C beamline. Data were integrated with XDS and scaled using XSCALE (62). Initial phases for Rh6mr were obtained by molecular replacement using the 11-cis-retinal–removed bathoRh and inactive Rh structures as search models [PDB ID codes 2G87 (63) and 1U19 (55)] with the CCP4 program Phaser (64–66). Opsin initial phases were obtained by direct refinement using the ligand-removed opsin structure [PDB ID code 4J4Q (38)]. Initial models were improved by multiple rounds of REFMAC version 5.8 (64) refinement against Rh6mr or opsin datasets and manual model adjustments with Coot 0.8 (67). The final models had agreement factors Rfree and Rcryst of 34 and 30%, respectively, for Rh6mr and 25 and 24%, respectively, for opsin. The Rh6mr 2Fo-Fc density map was B factor-sharpened at −178 Å2. Because the B-sharpened map did not reveal any regions that were either degraded or noise-elevated relative to the nonsharpened maps, the B-sharpened map was maintained for model building. The stereochemical quality of the Rh6mr and opsin models was assessed with the MolProbity (68, 69) and wwPDB validation servers (70). Details of the diffraction data collection and structural refinement statistics are provided in SI Appendix, Table S5. Coordinates and structure factor amplitudes have been deposited in the PDB (PDB ID codes 5TE3 and 5TE5 for opsin and Rh6mr, respectively).
Supplementary Material
Acknowledgments
We thank Dr. Leslie T. Webster, Jr. for helpful comments regarding this manuscript. We thank Dr. Prashansa Agrawal for technical assistance with NMR. This work was supported by funding from National Institutes of Health Grants [EY021126, EY027283, and EY025214 (to K.P.); EY025214 (to B.J.); and CA157735 (to G.P.T.)], a JSPS Overseas Research Fellowship Award (to K.K.), Department of Veterans Affairs Award IK2BX002683 (to P.D.K.), and National Science Foundation Grant MCB-084480 (to G.P.T.). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (Grant P41 GM103403). The Pilatus 6M detector on the 24-ID-C beamline is funded by NIH-ORIP HEI Grant S10 RR029205) This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by the Argonne National Laboratory under Contract DE-AC02-06CH11357. K.P. is the John H. Hord Professor of Pharmacology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.D.O. is a Guest Editor invited by the Editorial Board.
Data deposition: The coordinates and structure factor amplitudes reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5TE3 and 5TE5 for opsin and Rh6mr, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617446114/-/DCSupplemental.
References
- 1.Palczewski K. Chemistry and biology of the initial steps in vision: The Friedenwald lecture. Invest Ophthalmol Vis Sci. 2014;55(10):6651–6672. doi: 10.1167/iovs.14-15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernst OP, et al. Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem Rev. 2014;114(1):126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubbard R, St. George RC. The rhodopsin system of the squid. J Gen Physiol. 1958;41(3):501–528. doi: 10.1085/jgp.41.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shichida Y, Imai H. Visual pigment: G-protein-coupled receptor for light signals. Cell Mol Life Sci. 1998;54(12):1299–1315. doi: 10.1007/s000180050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palczewski K. G protein-coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandori H. Ion-pumping microbial rhodopsins. Front Mol Biosci. 2015;2:52. doi: 10.3389/fmolb.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oesterhelt D. The structure and mechanism of the family of retinal proteins from halophilic archaea. Curr Opin Struct Biol. 1998;8(4):489–500. doi: 10.1016/s0959-440x(98)80128-0. [DOI] [PubMed] [Google Scholar]
- 8.Kiser PD, Golczak M, Palczewski K. Chemistry of the retinoid (visual) cycle. Chem Rev. 2014;114(1):194–232. doi: 10.1021/cr400107q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Grip WJ, et al. 10,20-Methanorhodopsins: (7E,9E,13E)-10,20-methanorhodopsin and (7E,9Z,13Z)-10,20-methanorhodopsin. 11-cis-locked rhodopsin analog pigments with unusual thermal and photo-stability. Eur J Biochem. 1990;191(1):211–220. doi: 10.1111/j.1432-1033.1990.tb19112.x. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharya S, Ridge KD, Knox BE, Khorana HG. Light-stable rhodopsin. I. A rhodopsin analog reconstituted with a nonisomerizable 11-cis retinal derivative. J Biol Chem. 1992;267(10):6763–6769. [PubMed] [Google Scholar]
- 11.Jang GF, et al. Mechanism of rhodopsin activation as examined with ring-constrained retinal analogs and the crystal structure of the ground state protein. J Biol Chem. 2001;276(28):26148–26153. doi: 10.1074/jbc.M102212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuksa V, et al. Biochemical and physiological properties of rhodopsin regenerated with 11-cis-6-ring- and 7-ring-retinals. J Biol Chem. 2002;277(44):42315–42324. doi: 10.1074/jbc.M206014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogel R, Fan GB, Ludeke S, Siebert F, Sheves M. A nonbleachable rhodopsin analogue with a slow photocycle. J Biol Chem. 2002;277(43):40222–40228. doi: 10.1074/jbc.M205032200. [DOI] [PubMed] [Google Scholar]
- 14.Fan G, Siebert F, Sheves M, Vogel R. Rhodopsin with 11-cis-locked chromophore is capable of forming an active state photoproduct. J Biol Chem. 2002;277(43):40229–40234. doi: 10.1074/jbc.M205033200. [DOI] [PubMed] [Google Scholar]
- 15.Fahmy K, Sakmar TP. Regulation of the rhodopsin-transducin interaction by a highly conserved carboxylic acid group. Biochemistry. 1993;32(28):7229–7236. doi: 10.1021/bi00079a020. [DOI] [PubMed] [Google Scholar]
- 16.Jastrzebska B, et al. Functional and structural characterization of rhodopsin oligomers. J Biol Chem. 2006;281(17):11917–11922. doi: 10.1074/jbc.M600422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahmy K, Sakmar TP. Light-dependent transducin activation by an ultraviolet-absorbing rhodopsin mutant. Biochemistry. 1993;32(35):9165–9171. doi: 10.1021/bi00086a023. [DOI] [PubMed] [Google Scholar]
- 18.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274(5288):768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 19.Heck M, Hofmann KP. Maximal rate and nucleotide dependence of rhodopsin-catalyzed transducin activation: Initial rate analysis based on a double displacement mechanism. J Biol Chem. 2001;276(13):10000–10009. doi: 10.1074/jbc.M009475200. [DOI] [PubMed] [Google Scholar]
- 20.Kojima K, Imamoto Y, Maeda R, Yamashita T, Shichida Y. Rod visual pigment optimizes active state to achieve efficient G protein activation as compared with cone visual pigments. J Biol Chem. 2014;289(8):5061–5073. doi: 10.1074/jbc.M113.508507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y, et al. Unusual kinetics of thermal decay of dim-light photoreceptors in vertebrate vision. Proc Natl Acad Sci USA. 2014;111(29):10438–10443. doi: 10.1073/pnas.1410826111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343(5):1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 23.Ahuja S, et al. Location of the retinal chromophore in the activated state of rhodopsin*. J Biol Chem. 2009;284(15):10190–10201. doi: 10.1074/jbc.M805725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JE, Tauber MJ, Mathies RA. Wavelength dependent cis-trans isomerization in vision. Biochemistry. 2001;40(46):13774–13778. doi: 10.1021/bi0116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dartnall HJ. The photosensitivities of visual pigments in the presence of hydroxylamine. Vision Res. 1968;8(4):339–358. doi: 10.1016/0042-6989(68)90104-1. [DOI] [PubMed] [Google Scholar]
- 26.Okano T, Fukada Y, Shichida Y, Yoshizawa T. Photosensitivities of iodopsin and rhodopsins. Photochem Photobiol. 1992;56(6):995–1001. doi: 10.1111/j.1751-1097.1992.tb09722.x. [DOI] [PubMed] [Google Scholar]
- 27.Koskelainen A, Ala-Laurila P, Fyhrquist N, Donner K. Measurement of thermal contribution to photoreceptor sensitivity. Nature. 2000;403(6766):220–223. doi: 10.1038/35003242. [DOI] [PubMed] [Google Scholar]
- 28.Birge RR, Barlow RB. On the molecular origins of thermal noise in vertebrate and invertebrate photoreceptors. Biophys Chem. 1995;55(1–2):115–126. doi: 10.1016/0301-4622(94)00145-a. [DOI] [PubMed] [Google Scholar]
- 29.Cooper A. Energy uptake in the first step of visual excitation. Nature. 1979;282(5738):531–533. doi: 10.1038/282531a0. [DOI] [PubMed] [Google Scholar]
- 30.Birge RR, Cooper TM. Energy storage in the primary step of the photocycle of bacteriorhodopsin. Biophys J. 1983;42(1):61–69. doi: 10.1016/S0006-3495(83)84369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schröder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464(7292):1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: Implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16(5):715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 33.Smith BJ, et al. Structural resolution of a tandem hormone-binding element in the insulin receptor and its implications for design of peptide agonists. Proc Natl Acad Sci USA. 2010;107(15):6771–6776. doi: 10.1073/pnas.1001813107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunger AT, DeLaBarre B, Davies JM, Weis WI. X-ray structure determination at low resolution. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 2):128–133. doi: 10.1107/S0907444908043795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brunger AT. Low-resolution crystallography is coming of age. Structure. 2005;13(2):171–172. doi: 10.1016/j.str.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Liu C, Xiong Y. Electron density sharpening as a general technique in crystallographic studies. J Mol Biol. 2014;426(4):980–993. doi: 10.1016/j.jmb.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Nicholls RA, Long F, Murshudov GN. Low-resolution refinement tools in REFMAC5. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):404–417. doi: 10.1107/S090744491105606X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JH, et al. Opsin, a structural model for olfactory receptors? Angew Chem Int Ed Engl. 2013;52(42):11021–11024. doi: 10.1002/anie.201302374. [DOI] [PubMed] [Google Scholar]
- 39.Jastrzebska B, Palczewski K, Golczak M. Role of bulk water in hydrolysis of the rhodopsin chromophore. J Biol Chem. 2011;286(21):18930–18937. doi: 10.1074/jbc.M111.234583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lüdeke S, et al. The role of Glu181 in the photoactivation of rhodopsin. J Mol Biol. 2005;353(2):345–356. doi: 10.1016/j.jmb.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 41.Papermaster DS. Preparation of retinal rod outer segments. Methods Enzymol. 1982;81:48–52. doi: 10.1016/s0076-6879(82)81010-0. [DOI] [PubMed] [Google Scholar]
- 42.Okada T, et al. Circular dichroism of metaiodopsin II and its binding to transducin: A comparative study between meta II intermediates of iodopsin and rhodopsin. Biochemistry. 1994;33(16):4940–4946. doi: 10.1021/bi00182a024. [DOI] [PubMed] [Google Scholar]
- 43.Baker BY, et al. Crystallization of proteins from crude bovine rod outer segments. Methods Enzymol. 2015;557:439–458. doi: 10.1016/bs.mie.2014.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okada T, Takeda K, Kouyama T. Highly selective separation of rhodopsin from bovine rod outer segment membranes using combination of divalent cation and alkyl(thio)glucoside. Photochem Photobiol. 1998;67(5):495–499. [PubMed] [Google Scholar]
- 45.Salom D, et al. Improvements in G protein-coupled receptor purification yield light stable rhodopsin crystals. J Struct Biol. 2006;156(3):497–504. doi: 10.1016/j.jsb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 46.O’Brien DF, Costa LF, Ott RA. Photochemical functionality of rhodopsin-phospholipid recombinant membranes. Biochemistry. 1977;16(7):1295–1303. doi: 10.1021/bi00626a009. [DOI] [PubMed] [Google Scholar]
- 47.Niu L, Kim JM, Khorana HG. Structure and function in rhodopsin: Asymmetric reconstitution of rhodopsin in liposomes. Proc Natl Acad Sci USA. 2002;99(21):13409–13412. doi: 10.1073/pnas.212518899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palczewski K, Buczyłko J, Imami NR, McDowell JH, Hargrave PA. Role of the carboxyl-terminal region of arrestin in binding to phosphorylated rhodopsin. J Biol Chem. 1991;266(23):15334–15339. [PubMed] [Google Scholar]
- 49.Tsutsui K, Imai H, Shichida Y. Photoisomerization efficiency in UV-absorbing visual pigments: Protein-directed isomerization of an unprotonated retinal Schiff base. Biochemistry. 2007;46(21):6437–6445. doi: 10.1021/bi7003763. [DOI] [PubMed] [Google Scholar]
- 50.Jastrzebska B, et al. Functional characterization of rhodopsin monomers and dimers in detergents. J Biol Chem. 2004;279(52):54663–54675. doi: 10.1074/jbc.M408691200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jastrzebska B, Orban T, Golczak M, Engel A, Palczewski K. Asymmetry of the rhodopsin dimer in complex with transducin. FASEB J. 2013;27(4):1572–1584. doi: 10.1096/fj.12-225383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park PS, et al. Modulation of molecular interactions and function by rhodopsin palmitylation. Biochemistry. 2009;48(20):4294–4304. doi: 10.1021/bi900417b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okada T, et al. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J Mol Biol. 2004;342(2):571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 56.Choe HW, et al. Crystal structure of metarhodopsin II. Nature. 2011;471(7340):651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 57.Lasala R, et al. Sparse and incomplete factorial matrices to screen membrane protein 2D crystallization. J Struct Biol. 2015;189(2):123–134. doi: 10.1016/j.jsb.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289(5480):739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 59.Salom D, Padayatti PS, Palczewski K. Crystallization of G protein-coupled receptors. Methods Cell Biol. 2013;117:451–468. doi: 10.1016/B978-0-12-408143-7.00024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lodowski DT, et al. Crystal packing analysis of rhodopsin crystals. J Struct Biol. 2007;158(3):455–462. doi: 10.1016/j.jsb.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454(7201):183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 62.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakamichi H, Okada T. Crystallographic analysis of primary visual photochemistry. Angew Chem Int Ed Engl. 2006;45(26):4270–4273. doi: 10.1002/anie.200600595. [DOI] [PubMed] [Google Scholar]
- 64.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 65.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 1):32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 68.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web Server issue):W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10(12):980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.