Significance
Clinical treatments for metabolic diseases rely on agents with high selectivity to specific targets often within a class of structurally and functionally related proteins. In this paper, we uncover physical and chemical features governing selective small-molecule binding to peroxisome proliferator-activated receptor (PPAR) δ concomitant with distinct conformational changes in the receptor, key to therapeutic modulation of lipid catabolism, transport, and storage. These studies reveal the subtle interplay between ligand configuration and chemistry coupled to modulation of PPARδ structural dynamics. This set of structure–activity relationships (SARs) guide synthetic ligand designs necessary to refine therapeutic leads for temporally and spatially regulating PPARδ during the course of metabolic disease onset and progression.
Keywords: nuclear receptors, drug discovery, structure-based design, peroxisome proliferator-activated receptor, cation–π interaction
Abstract
The peroxisome proliferator-activated receptor (PPAR) family comprises three subtypes: PPARα, PPARγ, and PPARδ. PPARδ transcriptionally modulates lipid metabolism and the control of energy homeostasis; therefore, PPARδ agonists are promising agents for treating a variety of metabolic disorders. In the present study, we develop a panel of rationally designed PPARδ agonists. The modular motif affords efficient syntheses using building blocks optimized for interactions with subtype-specific residues in the PPARδ ligand-binding domain (LBD). A combination of atomic-resolution protein X-ray crystallographic structures, ligand-dependent LBD stabilization assays, and cell-based transactivation measurements delineate structure–activity relationships (SARs) for PPARδ-selective targeting and structural modulation. We identify key ligand-induced conformational transitions of a conserved tryptophan side chain in the LBD that trigger reorganization of the H2′–H3 surface segment of PPARδ. The subtype-specific conservation of H2′–H3 sequences suggests that this architectural remodeling constitutes a previously unrecognized conformational switch accompanying ligand-dependent PPARδ transcriptional regulation.
When gated with lipid signals (1–3), the peroxisome proliferator-activated receptor (PPAR) family of nuclear receptors (NRs) induces the expression of genes involved in lipid biosynthesis, oxidation, storage, and transport (4). The three subtypes, PPARα, PPARγ, and PPARδ, serve as sensors for dietary and endogenous fats; and therefore, they coordinately regulate transcription of genes linked to glucose and lipid metabolism (5, 6). PPARα plays pivotal roles in hepatic lipid and cholesterol metabolism (7, 8), whereas PPARγ is considered the master regulator of adipogenesis (9–11). PPARδ is highly expressed in the liver, intestine, adipose tissue, and skeletal muscle, where it regulates lipid catabolism, transport, and storage, making PPARδ an attractive target for treating hyperlipidemia and insulin resistance associated with obesity, the so-called metabolic syndrome (12, 13). Nevertheless, the physiological ligands of PPARδ remain uncertain, slowing the clinical development of PPARδ leads. In contrast, synthetic small-molecule therapeutics such as fibrates and thiazolidinediones have been successfully advanced for PPARα and PPARγ, respectively. These PPARα and PPARγ agonists are currently prescribed as hypolipidemic agents and insulin sensitizers (14, 15).
In recent years, the critical role of PPARδ in energy expenditure through its regulation of lipid metabolism, particularly in skeletal muscle and brown fat (16–19), has increased interest in the δ-subtype as a bona fide drug target. Indeed, an early PPARδ agonist, GW501516, showed beneficial effects in primate models of metabolic disorders (20, 21). This observation, combined with side effects after long-term regiments of prescribed PPARα and PPARγ agonists (22, 23), has attracted considerable interest in next-generation drug discovery for PPARδ-specific therapeutic intervention, with improved isoform selectivity and reduced off-target side effects.
Activity-based assays identified a plethora of ligands capable of regulating PPARδ, including saturated fatty acids, unsaturated fatty acids, eicosanoids (24), and very low-density lipoprotein–derived fatty acids (25). Protein X-ray crystallographic structures of PPARδ bound to Z-octadec-11-enoic acid (26) and eicosapentaenoic acid (EPA) (27) identified a “Y”-shaped ligand-binding cavity of ∼1,300 Å3 formed by a sandwich of six α-helices and two β-strands. This three-pronged ligand-binding site accounts for PPARδ’s ability to accommodate structurally diverse chemicals. The binding of EPA, a weak ligand with 4 mM affinity, is especially interesting because it illustrates two alternative binding modes that define the “Y”-shaped, three-arm binding cavity (27). EPA’s carboxylate moiety anchors next to the activation factor 2 (AF-2) helix at the base of the “Y” (arm I), and the lipophilic tail binds in a “tail-up” (arm III) or a “tail-down” (arm II) orientation (SI Appendix, Fig. S1A). The cocrystal structures of synthetic ligands such as GW2433 (SI Appendix, Fig. S1B) (27) and GW2331 (SI Appendix, Fig. S1C) (28) confirm that small molecules exploit the alternative ligand-binding modes of PPARδ by simultaneously occupying arms II and III.
Results and Discussion
Structure-Based Design of PPARδ-Specific Agonists.
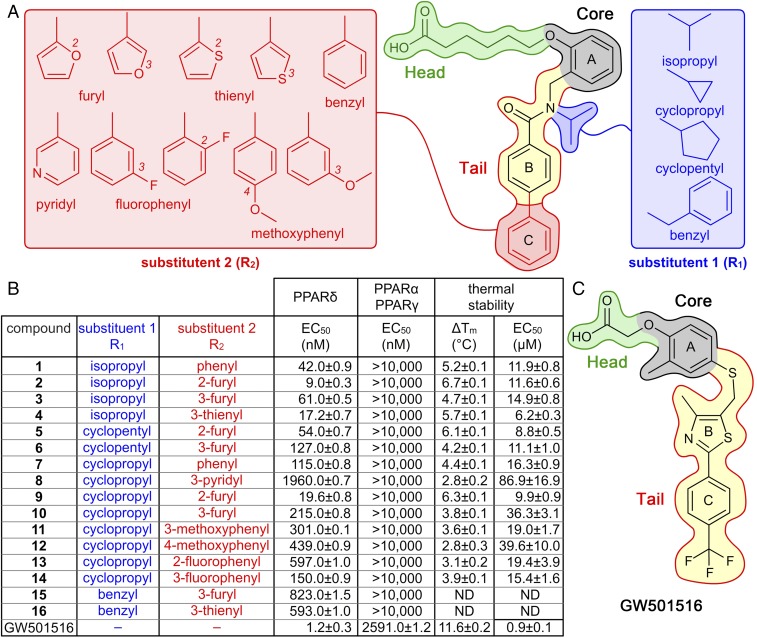
Using the hPPARδ ligand-binding domain (hPPARδ-LBD) as a target for rational ligand design, a synthetic motif was developed bearing a carboxylate head group to mimic fatty-acid ligands (green, Fig. 1A). The carboxyl moiety was attached to a central aromatic ring through a phenoxyl ether linkage (grey, ring A, Fig. 1A). A dialkylbenzamide core was coupled to the ortho-position of ring A (yellow, Fig. 1A). The core was appended with two substituents, R1 and R2. R1 probed amide nitrogen modifications proximal to ring B (blue, Fig. 1A). R2 explored the chemistry and accessible volume of ring B para-position moieties using a varied set of aromatic units (red, Fig. 1A). This scaffold incorporated three design principles. First, it placed a suitably spaced carboxylate (head) in the arm I cavity abutting the AF-2 helix, and secondary amides (tail) with lipophilic modules of varying volumes in the arm II and III cavities. Second, the dialkylbenzamide core constrained conformations of the synthetic ligands to facilitate optimal insertion into the arm II cavity adjacent to H3. Third, aryl units sat proximal to lipophilic residues on H6 and H7 (Fig. 1A and SI Appendix, Fig. S1).
Fig. 1.
Chemical structures of compounds 1–16, GW501516, and their hPPARδ-targeting activity. (A) Structure of compound 1 highlights scaffold design. Regions that occupy arm I (dark-green lines), core (black lines), and arm II (blue or red lines) in the hPPARδ ligand-binding cavity are highlighted. These sectors are accentuated by color fills: head group (light green), core (gray), fixed unit of the tail (yellow), substituent R1 (blue), and substituent R2 (red). (B) Functional groups and associated activities for compounds 1–16 and GW501516. hPPARδ transcriptional responses shown as EC50 values quantified by cell-based luciferase reporter assays. hPPARδ-LBD thermal stabilities shown as both maximal Tm values and EC50 values from fitted dose–response curves (SI Appendix, Fig. S6). ND indicates not determined. (C) Structure of GW501516 with comparable color-coded group designations as in A.
A panel of compounds prepared using this motif (Fig. 1A and SI Appendix, Schemes S1–S3) was screened using cell-based reporter assays that identified 16 unique molecules exhibiting quantifiable transactivation of hPPARδ (1–16, Fig. 1B) comparable to GW501516-mediated transactivation (Fig. 1 B and C). From this set, six compounds (1–5 and 9) with EC50 values <100 nM and eight compounds (6–8 and 10–14) with attenuated activities (EC50 values ∼ 100–2,000 nM) were chosen for further study. Compounds 1–7 and 9–14 were specific for hPPARδ compared with hPPARα and hPPARγ. Remarkably, each of these compounds offered ∼17-fold (compound 13) to ∼1,000-fold (compound 2) selectivity for hPPARδ (Fig. 1B). Furthermore, treatment of skeletal muscle cells with 9 induced the expression of PPARδ target genes involved in fatty-acid metabolism including angiopoietin-like protein 4 (Angptl4) and mitochondrial pyruvate dehydrogenase (acetyl-transferring) kinase isozyme 4 (Pdk4) genes to similar extents as GW501516, confirming PPARδ agonist activity (SI Appendix, Fig. S2 A and B). Moreover, differentiated C2C12 myotubes treated with 9 showed significant increases in fatty-acid oxidation without measurable effects on glucose oxidation as quantified using a Seahorse XF Analyzer (SI Appendix, section A.2 and Fig. S2C).
Structure–activity relationships (SARs) from these initial data correlated with SAR predictions. The hexanoic acid unit was optimal for the head motif. At R2, aromatic substituents were favored for in vivo potency: 2-furyl > 3-furyl ∼ phenyl ∼ 3-thienyl > Br >> 2-thienyl (data not shown). Small aliphatic isopropyl or cyclopropyl groups were optimal for the R1 substituents, as activity dropped considerably with increasing size of R1 as observed for 15 and 16 (Fig. 1B).
Structures of Ligand-Bound hPPARδ-LBDs.
Compounds 1–16 (Fig. 1 A and B) and GW501516 (Fig. 1C) were cocrystallized with recombinant hPPARδ-LBD comprising residues 170–441. Crystals were grown using similar conditions [40 mM Bis-Tris propane, pH 7.5–8.8, 200 mM KCl, 4–14% (wt/vol) PEG 8000, 2.5% (vol/vol) 1,2-propandiol, 0.5% (wt/vol) heptyl-β-d-glucopyranoside, 1 mM EDTA, and 10 mM DTT] (27). The structures were solved by molecular replacement, and refined to 1.5- to 2.1-Å resolutions, providing high-quality electron density maps of hPPARδ-LBDs and associated ligands with two polypeptide chains per asymmetric unit (SI Appendix, Tables S1 and S2).
Comparison with previously reported NR structures revealed that compounds 1–16 and GW501516 stabilized hPPARδ-LBDs in “canonical” active conformations of NRs with ordered AF-2 helical segments (29, 30). These well-resolved C-terminal AF-2 helical segments, H12s, abut the ligand-binding site and the carboxyl moiety of hPPARδ ligands (Fig. 2A). The helical structures of AF-2s are crucial for ligand-dependent transactivation activity of NRs by providing hydrophobic docking sites for NR coactivators (31). In our structures, these hydrophobic grooves are filled by two heptyl-β-d-glucopyranoside molecules. These two detergent molecules stack together and rest between the interfaces of the two hPPARδ-LBD polypeptide chains in each asymmetric unit (SI Appendix, Fig. S3).
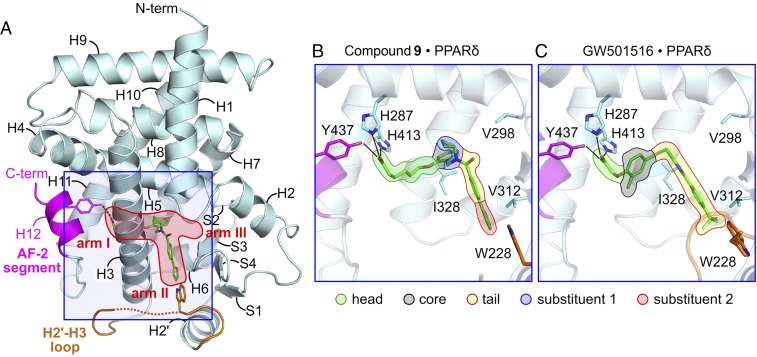
Fig. 2.
Active 3D structures of hPPARδ-LBD stabilized by compound 9 and GW501516. (A) Overall ribbon diagram of 9•hPPARδ-LBD. Bound compound 9 is shown in stick format atom-colored with carbon (green), nitrogen (blue), oxygen (red), and sulfur (orange). The H2′–H3 loop (in the flexible conformation) and H12 (AF-2 segment) are colored orange and magenta, respectively. Missing residues in the H2′–H3 loop are depicted as a dotted orange curve. (B) Close-up view of the ligand-binding site for 9•hPPARδ-LBD. (C) Close-up view of the ligand-binding site for GW501516•hPPARδ-LBD. For both B and C, the ligand is outlined to emphasize portions of the compounds occupying arm I (dark-green lines), core (black lines), and arm II (blue or red lines). Semitransparent color fills highlight head group (light green), core (gray), fixed unit of the tail (yellow), substituent R1 (blue), and substituent R2 (red).
Compounds 1–16 and GW501516 adopt similar binding modes compared with other PPARδ modulators (27, 31, 32). The ligand-binding cavity spans three extended arms, arms I, II, and III. The carboxylate head groups of compounds 1–16 (Fig. 2B) and GW501516 (Fig. 2C) reside in arm I, and the aromatic R2 tails occupy arm II, leaving arm III partially empty, as the R1 substituents do not fully access this subcavity. The carboxylate moieties of these ligands form hydrogen bonds with conserved hydrophilic side chains of H287, H413, and Y437 pointing into arm I. Direct bonding with Y437's phenolic hydroxyl group is crucial for stabilizing AF-2 in the active helical state, and intermolecular engagement of Y437 is key for potency of hPPAR agonists generally (30, 33).
Comparative Analyses of Bound Ligands.
The high-resolution structures were used to systematically interrogate the architectural interplay between hPPARδ-LBD and compounds 1–16 as well as GW501516. Structural bases for receptor selectivity of compounds 1–16 compared with PPARα and PPARγ cross-reactivity observed with GW501516, were also investigated (Fig. 1B). Shared ligand–protein interactions are seen with compounds 1–16 and GW501516 in arm I, including conserved hydrogen bonds with the side chains of H287, H413, and Y437, and van der Waals interactions with F246, C249, T253, I327, M417, and L433 (SI Appendix, Fig. S4). In contrast, compounds 1–16 also establish additional contacts in the hPPARδ ligand-binding cavities.
The phenoxy–ether linkages mediate packing interactions with the side chains of I328 and K331 in the central regions of the “Y-shaped” three-arm cavities, whereas the ligands’ aromatic tails tuck into arm II, where the distal C rings exhibit increased interplay with hPPARδ-LBDs than observed with the trifluoromethyl tail of GW501516 (Fig. 2C). These latter interactions are marked by additional contacts with W228, R248, and L219 at the end of arm II (SI Appendix, Fig. S4). In addition, the rigidity of these unique ligands result in repositioning of their aromatic R2 tails closer to the antiparallel β-strands forming one side of arm II (Fig. 3A). These positional shifts induce favorable interactions with small, hydrophobic residues lining the proximal β-sheet of PPARδ-LBD, including L303, V305, and V312 (SI Appendix, Fig. S4). The presence of larger side chains projecting into the center of arm II and the proximal β-sheets of hPPARα and hPPARγ serve as structural barriers mitigating binding of ligands such as 1–16 to PPARα and hPPARγ, thereby conferring PPARδ specificity (Fig. 3A).
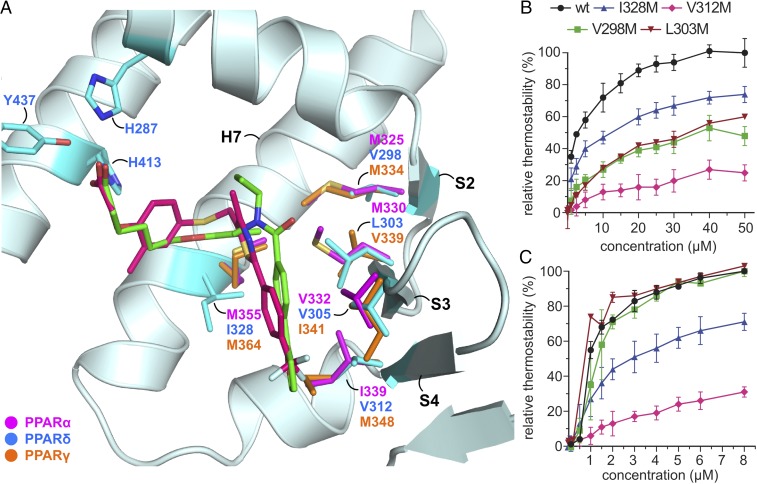
Fig. 3.
Side-chain variants in the ligand-binding cavities of hPPARα and hPPARγ act as structural barriers for high-affinity recognition of PPAR ligands. (A) Superimposition of GW501516•hPPARδ-LBD and 9•hPPARδ-LBD with agonist-bound hPPARα-LBD and agonist-bound hPPARγ-LBD. Select side chains for each shown in stick formats. Compound 9 and GW501516 are depicted as green and pink sticks for carbon, respectively, with nitrogen (blue), oxygen (red), and sulfur (orange). Protein side chains of 9•hPPARδ-LBD are cyan, whereas protein side chains of hPPARα (PDB ID code 2P54) (34) and hPPARγ (PDB ID code 2PRG) (32) are purple and orange, respectively. (B) Ligand-dependent thermal-shift assays of wild-type and mutated hPPARδ-LBDs dosed with compound 9. (C) Complementary assays of wild-type and mutated hPPARδ-LBDs dosed with GW501516. Data points are shown as mean ± SE, where n ≥ 3.
Functional Bases of PPARδ Targeting.
To support our hypothesis regarding PPARδ-specific targeting, V298, L303, V312, and I328 in hPPARδ-LBD were mutated individually to methionines to mimic the large side chains found in PPARα and PPARγ (Fig. 3A). Thermal-shift binding assays were used to quantify ligand recognition through stabilization of the LBD fold to thermal denaturation (35, 36). Wild-type and mutated hPPARδ-LBDs displayed comparable, synchronous melting curves (SI Appendix, Fig. S5A), confirming that the mutations did not introduce significant perturbations to hPPARδ-LBD thermal stability. Furthermore, the midpoint melting temperatures (Tm) of wild-type hPPARδ-LBD increased in ligand dose-dependent manners (SI Appendix, Fig. S5 B and C), confirming that bound ligands stabilized the protein folds (Fig. 1B and SI Appendix, Fig. S5).
Substitutions of methionines in hPPARδ-LBDs’ ligand-binding cavities via V312M and I328M mutations reduced binding of 9 and GW501516 by ∼70% and ∼30%, respectively (Fig. 3 B and C), and similarly impacted the binding of compounds 1–8 and 10–16 (SI Appendix, Fig. S7), consistent with previous reports (37). V312 resides at the end of arm II in hPPARδ, whereas hPPARα and hPPARγ have methionines at the equivalent positions. In these latter two cases, methionine residues substantially shorten arm II. Although the structural analyses suggest that I328M replacements in hPPARδ should abrogate recognition of compounds 1–16, thermal-shift assays reveal that I328M mutants only impair ligand bindings by ∼30%. Examination of ligand-bound hPPARα and hPPARγ structures reveals that the methionine side chains corresponding to I328 in hPPARδ (M355 and M364 in hPPARα and hPPARγ, respectively) adopt alternative conformations. The conformational flexibility of this region may be sufficient to accommodate bulkier ligands such as 1–16, explaining the modest impact of I328M replacements on the binding and efficacy of compounds 1–16 to hPPARδ.
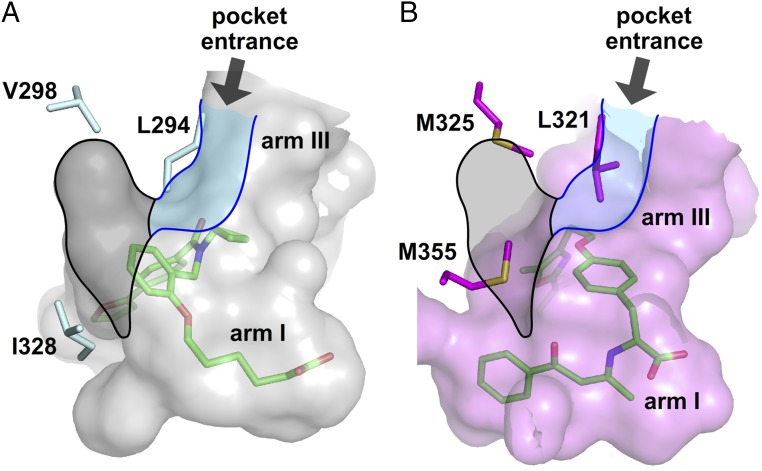
The L303M mutation, located on the β-sheet lining arm II, impaired the binding of 9 by ∼50% without affecting GW501516 binding (Fig. 3 B and C). This mutational result is consistent with our structural hypothesis suggesting that compounds 1–16 should be more sensitive to bulky side chains on the arm II β-sheet than GW501516. Unexpectedly, the V298M mutation profoundly impacted ligand binding, despite being relatively remote from the ligand-interaction surface in hPPARδ, with compounds 1–16 losing 50–70% of their potencies (Fig. 3B and SI Appendix, Fig. S7). Examination of the X-ray crystallographic structures revealed that the smaller R1 substituents (blue, Fig. 1A) reside next to the entrance of arm III (Fig. 4A). These functional groups appear to buttress the side chain of L294 between R1 substituents and ring A of 1–16 (Fig. 1 A and B). The bulkier V298M mutation introduces significant steric repulsion in this region, thus impairing binding of 1–16 to PPARδ. In contrast, GW501516 binding was minimally affected by the V298M mutation in hPPARδ (Fig. 3C). Comparison with the agonist-bound hPPARα structure revealed that the L321 and M325 side chains in hPPARα (L294 and V298 in hPPARδ, respectively) substantially narrow the entrance to arm III (Fig. 4B). This partial occlusion, unique to the hPPARα and γ subtypes, most likely prevents the binding of ligands with R1 substituents like those found in compounds 1–16 to PPARα and γ, but would have limited consequences for ligands such as GW501516. These studies identify the amino-acid residues at positions 298, 303, 312, and 328 (hPPARδ numbering) as key determinants in ligand recognition and specificity in the PPAR subfamily of NRs.
Fig. 4.
Side-chain variants at the hub of the three-arm ligand-binding sites of hPPARα-LBD and hPPARδ-LBD. (A) Accessible surface of the ligand-binding site of 9•hPPARδ-LBD. (B) Accessible surface of agonist-bound hPPARα-LBD (PDB ID code 1K7L) (31). For A and B, the two structures were superimposed to present the three-arm cavities of their ligand-binding sites in the same orientations. Surfaces rendered and colored transparent gray for hPPARδ and transparent purple for hPPARα. Representative protein side chains shown in stick format and labeled hPPARδ (cyan) and hPPARα (magenta) accordingly. Accessible volumes of the central region (black) and arm III (blue) in 9•hPPARδ-LBD are superimposed facilitating comparison with the same sectors in hPPARα. Bound ligands shown in stick format color-coded by atom type, carbon (green), nitrogen (blue), oxygen (red), and sulfur (orange).
SARs.
A combination of cell-based transactivation assays, PPARδ-LBD temperature-dependent stability measurements, and atomic-resolution protein X-ray crystallography revealed generalized rules governing PPARδ SARs for this unique class of compounds. A hexanoate head group offered optimal activity versus shorter fatty-acid chains (data not shown). Moving down the scaffold, we examined the functionality of the two tail substituents, R1 and R2 (Fig. 1 A and B). Substituent R1 explored functionality on the nitrogen atom proximal to the scaffold’s core. Although variants at this position (i.e., isopropyl, cyclopropyl, cyclopentyl, and benzyl groups) had modest effects on ligand potency (Fig. 1B), a cyclopentyl group was generally less effective in driving hPPARδ transactivity, suggesting that increased bulk at R1 impairs ligand binding. This reduced transactivation activity is supported by spotty electron density maps of the cyclopentane rings in hPPARδ-LBDs complexes with ligands and their higher B-factors compared with neighboring hPPARδ-LBD residues in the structures of 5•hPPARδ-LBD and 6•hPPARδ-LBD (SI Appendix, Fig. S8 A–C).
Unexpectedly, substitution of bulkier but rotationally flexible benzyl groups at R1 resulted in interpretable electron density maps, suggesting that repositioning of the benzyl moieties away from the ligand cores are tolerated in hPPARδ-LBD complexes. Accordingly, in the structures of 15•hPPARδ-LBD and 16•hPPARδ-LBD (SI Appendix, Fig. S8D), we observe the L294 side chains pointing toward arm III to accommodate the benzyl groups. In turn, this alternative rotameric conformation of L294 seals off arm III turning the typical Y-shaped, ligand-binding cavity into an L-shaped chamber (SI Appendix, Fig. S9 A and B). L294 acts as a gating residue providing PPARδ with the necessary flexibility to sense and respond to diverse ligands. This observation is also consistent with previously published PPARδ structures (38–41), wherein L294 adopts alternative conformations that substantially narrow arm III accessible volumes (SI Appendix, Fig. S9C).
The more distal tail substitutions (R2, Fig. 1) sit at the end of arm II. By fixing R1 and varying R2, we found the 2-furyl C ring enhanced hPPARδ transactivation potency. In the crystal structures, the 2-furyl–containing compounds adopt nearly coplanar conformations of their B and C rings (SI Appendix, Fig. S10A). This generally unfavorable biaryl conformation suggests that a planar orientation of the B–C biaryl system is compensated for by binding to hPPARδ. Replacement of ring C with groups possessing additional ortho-hydrogens (R2 = 3-furyl, 3-thienyl, or phenyl) further increases the energetic barriers for adopting planar biaryl systems. These energetic perturbations result in compounds possessing more twisted biaryl conformations with reduced B–C ring planarities. Indeed, compounds with the greatest potency possess largely coplanar conformations of their B and C rings, whereas those lacking biaryl planarity, with torsion angles, Φt, of −10° to −26° (SI Appendix, Fig. S10 B–D and Table S3), have reduced activity. For instance, compound 8, which has a low EC50 value (Fig. 1B) in transactivation assays, lacked B–C ring planarity (average Φt of −10.4°) of the two bound ligands per asymmetric unit due to mutual repulsions between their ortho-positions (SI Appendix, Fig. S10E). Methoxyphenyl or fluorophenyl substituents in 11–14 introduce even more repulsive forces, favoring greater out-of-plane biaryl conformations. Compounds 11–14 in the hPPARδ-LBD cocrystal structures have average Φts of 16.4°, −17.4°, −25.9°, and −14.8°, respectively. These disfavored binding conformations further reduce transactivation potencies with EC50 values >100 nM (Fig. 1B).
Ligand-Induced Conformational Switching of H2′–H3.
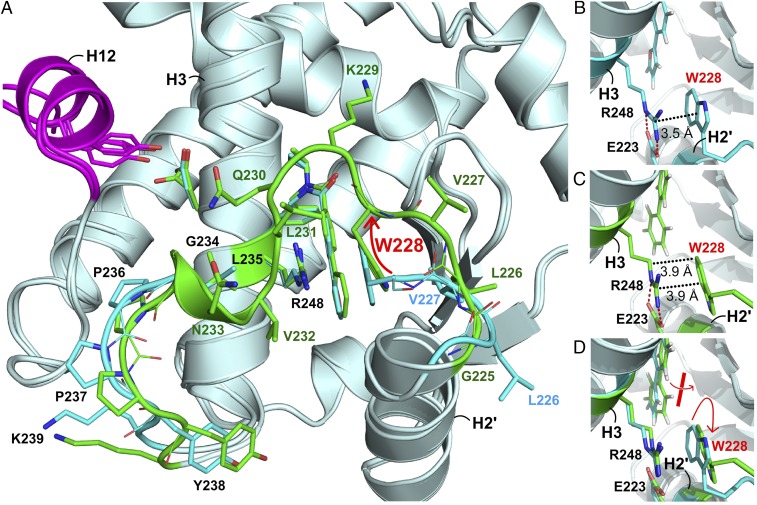
In addition to ligand selectivity and AF-2 stabilization, a newly characterized ligand-induced conformational switch in the polypeptide segment between H2′ and H3 (G225 to K239) was observed (Fig. 5A). The largest structural change occurred for G225–G234. For four of our solved structures, 2•hPPARδ-LBD, 5•hPPARδ-LBD, 9•hPPARδ-LBD, and 15•hPPARδ-LBD, these residues were not visible in electron density maps (SI Appendix, Fig. S11A). However, the structures complexed to ligands 1, 4, 7, 10–12, 14, 16, and GW501516 displayed unambiguous electron density maps for H2′–H3 (SI Appendix, Fig. S11B). Strikingly, the single ligand-contacting residue in H2′–H3, W228, undergoes a 180° indole ring flip to a preferred side-chain rotamer when presented with a subset of R2 substituents. This indole ring flip triggers the transition of H2′–H3 from a disordered to ordered conformation (SI Appendix, Fig. S11 C and D). W228’s side-chain rotamer repositions its backbone (red arrow, Fig. 5A), which, in turn, stabilizes L226, V227, and L229–N233 through newly formed backbone–backbone hydrogen bonds (SI Appendix, Fig. S11E).
Fig. 5.
Ligand-dependent conformational switching of hPPARδ-LBD H2′–H3. (A) Close-up superimposed views of H2′–H3 in 9•hPPARδ-LBD (cyan) and 1•hPPARδ-LBD (green). Secondary structure elements are shown as gray ribbons. W228 is labeled. Red arrow highlights main-chain movements triggered by alternative W228 rotamers. (B) Close-up view of 9•hPPARδ-LBD illustrating the W228–R248 cation–π interaction boarding 9’s 2-furyl C-ring residing in the crevice on the top of the cation–π motif. This set of local interactions correlates with flexible H2′–H3 conformations. (C) Close-up view of 1•hPPARδ-LBD emphasizing the 180° indole ring flip of W228 triggered by the out-of-plane conformation of 1’s B–C biaryl moiety. This ligand–W228 interaction, in turn, disrupts the energetically favorable spacing and orientation of the W228–R248 cation–π interaction, leading to stabilization of H2′–H3. Ligands and selected residues are shown in stick formats. For both panels, the distances between W228 and R248 side chains are indicated by dotted lines and labeled accordingly. The conserved salt bridges between R248 and E223 are shown as red dotted lines. (D) Superimposition of B and C. Torsion angle differences between the B and C rings of 9 and 1 are highlighted using a red bar and arrow. The directional reorientation of W228’s indole ring is also emphasized using a red arrow.
In the 2•hPPARδ-LBD, 5•hPPARδ-LBD, 9•hPPARδ-LBD, and 15•hPPARδ-LBD complexes, W228’s side chains assume alternative rotameric conformations (SI Appendix, Table S3). These less preferred rotamers are compensated for by W228’s indole ring forming favorable parallel stacking interactions with the plane of the positively charged guanidinium group of R248 extending from H3 (Fig. 5B). These nearly ideal cation–π interactions (42, 43) set up conformational changes that induce flexibility of H2′–H3 in PPARδ-LBD. Moreover, in these four structures, the aromatic biaryl motifs of the bound ligands are nearly coplanar (SI Appendix, Table S3), forming extended π systems, that favorably position the C rings within the W228–R248 cation–π systems (Fig. 5B). Deviations from planarity within the biaryl systems for compounds 1, 4, 7, 10–12, 14, and 16 reduce the extent of the resonance-stabilized π systems and induce steric repulsions with W228 indole rings directing W228 away from R248. The net effects are disruption of the cation–π interactions (Fig. 5 C and D), reorganization of W228 side-chain rotamers, and compensatory stabilization of H2′–H3 segments through hydrogen bonds between W228’s Nε1 and the main-chain oxygen of A222. Also, the L229–N233 polypeptide segments move inward toward the ligand-binding cavities establishing additional hydrogen bonds to stabilize the conformations of H2′–H3 (SI Appendix, Fig. S11E).
GW501516 also stabilizes this alternate W228 rotamer that disrupts the cation–π interaction leading to compensatory stabilization of H2′–H3 (Fig. 2C and SI Appendix, Table S3). Superimposing GW501516•hPPARδ-LBD with 9•hPPARδ-LBD demonstrates that the trifluoromethyl group of GW501516 clashes with the indole moiety of W228 in the flexible H2′–H3 conformation. Consistent with our observation, stabilization of H2′–H3 also occurs in two published hPPARδ-LBD structures, one bound to GW0742 (analog of GW501516) and another with a synthetic ligand possessing a terminal trifluoromethyl group (PDB ID codes 2XYX and 3TKM, respectively) (37, 44) (SI Appendix, Table S3). Together with this SAR study of hPPARδ-LBDs interacting with a unique class of synthetic ligands, these previous structural investigations support a model intimating that bulky groups at the tail end of hPPARδ ligands, much like the twisted biaryl B–C ring arrangements in a subset of our compounds, trigger the H2′–H3 conformational switch from a flexible to an ordered conformation. With some ligands possessing smaller deviations of B–C ring planarities, for instance in 2•hPPARδ-LBD, 3•hPPARδ-LBD, 6•hPPARδ-LBD, and 15•hPPARδ-LBD, we observe mixtures of H2′–H3 conformational states likely due to smaller repulsive forces between the ligands’ C rings and the R248–W228 cation–π interactions. Collectively, these structure–function studies suggest this unique set of synthetic hPPARδ ligands cannot only modulate PPAR selectivity in a subtype-specific manner but also tune the conformational states of PPARδ H2′–H3 polypeptide segments.
H2′ and the H2′–H3 segment are structural elements unique to the PPAR NR family, viewed as structurally flexible “lips” for LBD adaptation to chemically diverse ligands (45). However, the amino-acid sequences of these polypeptide segments are highly conserved in each subtype but distinct across the three PPARs (SI Appendix, Fig. S12). Our studies suggest that H2′ and H2′–H3 segments may have defined roles in mediating subtype-specific functions including ligand-dependent protein–protein interaction modules for each PPAR member and additional components of PPAR transcriptional regulation. Comparative structural analyses of compounds 1–16 bound to hPPARδ-LBD correlate the H2′–H3 3D conformation and dynamics to the chemistry of this unique set of PPARδ ligands. Notably, the observed ligand-triggered H2′–H3 conformational switch is set up by a network of energetically coupled interactions from ligand biaryl systems to W228 to the G225–G234 segments (Fig. 5A). G225 is absolutely conserved in PPARδ subtypes and the flexibility of this glycine plays crucial roles in the structural transitions described here. The N-Cα (Phi, φ) and Cα-C (Psi, ψ) torsion angles of G225 reside in the disallowed region of the Ramachandran plot for nonglycine residues (φ/ψ = 156°/−28°) when H2′–H3 adopt the flexible/disordered conformations as seen in the 9•hPPARδ-LBD structure. In the ordered H2′–H3 conformation, G225’s φ/ψ torsion angles reside in the allowed region of the Ramachandran plot (φ/ψ = −105°/−24°) as seen in the 1•hPPARδ-LBD structure. Importantly, replacement of the residue equivalent to G225 in hPPARδ by Cα-branched amino-acid residues such as threonine in PPARα and lysine in PPARγ would disfavor the φ/ψ torsion angles observed for G225 in PPARδ. G225, together with W228 and R248, are strictly conserved in PPARδ subtypes (SI Appendix, Fig. S12). This deep phylogenetic pattern indicates that these three residues serve as adaptive linchpins in an evolutionarily conserved energetic network that affords selective, ligand-induced conformational changes in H2′–H3 of PPARδ.
Conclusion
Protein X-ray crystallographic analyses of a unique set of highly specific PPARδ agonists cocrystallized with hPPARδ LBDs reveal the structural basis for PPARδ synthetic ligand specificity. Unexpectedly, this series of high-resolution X-ray crystallographic structures uncover a conformational switch in the H2′–H3 loop of PPARδ’s LBD upon ligand binding, a mechanism that may be shared across the superfamily of PPAR NRs. Studies of PPARγ have suggested that structural features of PPAR ligands may guide the conformations of H2′–H3 (46). As architectural changes and dynamics of H2′–H3 polypeptide segments induce significant differences in the surface features surrounding H2′–H3, it is likely that these ligand-mediated effects steer PPARδ interactions with coregulators. Supporting this hypothesis, the acetylation state of a highly conserved Lys residue located on the H2′–H3 loop of PPARγ (corresponding to K229 in hPPARδ) is crucial for the interplay of PPARγ with coregulators (47). In short, conformational coupling between NR ligands and the H2′–H3 loop stage additional ligand-dependent protein–protein interaction surfaces and posttranslational modifications affording further levels of PPAR-mediated transcriptional regulation.
These next-generation PPAR ligands also manifest enhanced PPARδ specificity. Selectivity arises from PPAR subtype-specific sequence variation within the larger family of NR LBDs matched to stereoelectronically tuned biaryl units embodied in the design and syntheses of these newly deployed PPARδ agonists. Nevertheless, GW501516 has 10-fold or greater potency against hPPARδ and other PPARs than these newly described ligands. This enhanced affinity may be partially attributed to the greater rigidity of GW501516s shortened carboxylate head group compared with the longer carboxylates of this newly designed set of compounds. The conservation of the intricate set of stabilizing hydrogen bonds between these carboxyl moieties and the conserved hydrophilic side chains of H287, H413, and Y437 across the PPAR subfamily of NRs are crucial for stabilizing AF-2s in their active helical states. Conformational stabilization of GW501516s shortened carboxylate head group may be key for enhanced potency of hPPAR agonists generally when presented near the AF-2 regions (30, 33). In contrast, although this next generation of PPAR ligands generally exhibit reduced potencies against hPPARδ, they convincingly demonstrate substantially increased specificity for PPARδ relative to other PPAR subtypes.
Accordingly, this study advances hPPARδ ligand design clinically as previously demonstrated for hPPARγ (48). Enhancing hPPARδ specificity in a predictable manner should greatly reduce adverse side effects through avoidance of full activation of hPPARγ-driven genes in the treatment of metabolic disorders (49). Moreover, having chemical tools in hand to specifically manipulate hPPAR agonism should facilitate resolution of the controversial role of PPARs in oncogenesis (50). Studies are now underway to determine whether the cell proliferation enhancement and cancer promoting activities of GW501516 (51–53) are related to hPPARδ targeting or arise due to unanticipated off-target effects.
Most importantly, this study extends our understanding of hPPARδ structure–function relationships at the atomic level. This predictive chemical template for modulation of hPPARδ structural dynamics, therefore, offers a vital next step toward clinically treating metabolic diseases using a repertoire of synthetically accessible small molecules (<900 Da) to fine-tune hPPARδ transcriptional responses.
Methods
Complete methods for sample preparation, cell-based assays, protein crystallization, structure determination, site-directed mutagenesis, thermal-shift binding assays, and general chemical procedures are described in SI Appendix.
Supplementary Material
Acknowledgments
This research was supported by National Science Foundation Grant EEC-0813570 (to J.P.N.) and NIH Grants DK057978, HL105278, DK090962, HL088093, ES010337, and CA014195, as well as the Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED-002) (to R.M.E.). R.M.E. holds a March of Dimes Chair in Molecular and Developmental Biology at the Salk Institute. J.P.N. holds the Arthur and Julie Woodrow Chair at the Salk Institute. R.M.E. and J.P.N. are investigators of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: J.P.N., T.J.B., M.D., and R.M.E are coinventors of PPARδ molecules/ligands and methods of use and may be entitled to royalties.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 5U3Q, 5U3R, 5U3S, 5U3T, 5U3U, 5U3V, 5U3W, 5U3X, 5U3Y, and 5U3Z (for compounds 1–10•hPPARδ-LBD complex structures, respectively), 5U40 and 5U41 (for 15 and 16•hPPARδ-LBD complex structures, respectively), and 5U42, 5U43, 5U44, 5U45, and 5U46 (for 11–14•hPPARδ-LBD and GW501516•hPPARδ-LBD complex structures, respectively)].
See Commentary on page 3284.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621513114/-/DCSupplemental.
References
- 1.Schupp M, Lazar MA. Endogenous ligands for nuclear receptors: Digging deeper. J Biol Chem. 2010;285(52):40409–40415. doi: 10.1074/jbc.R110.182451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kliewer SA, et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc Natl Acad Sci USA. 1997;94(9):4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forman BM, et al. 15-Deoxy-delta 12,14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 1995;83(5):803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 4.Poulsen Ll, Siersbæk M, Mandrup S. PPARs: Fatty acid sensors controlling metabolism. Semin Cell Dev Biol. 2012;23(6):631–639. doi: 10.1016/j.semcdb.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Monsalve FA, Pyarasani RD, Delgado-Lopez F, Moore-Carrasco R. Peroxisome proliferator-activated receptor targets for the treatment of metabolic diseases. Mediators Inflamm. 2013;2013:549627. doi: 10.1155/2013/549627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmadian M, et al. PPARγ signaling and metabolism: The good, the bad and the future. Nat Med. 2013;19(5):557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kersten S, et al. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103(11):1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costet P, et al. Peroxisome proliferator-activated receptor alpha-isoform deficiency leads to progressive dyslipidemia with sexually dimorphic obesity and steatosis. J Biol Chem. 1998;273(45):29577–29585. doi: 10.1074/jbc.273.45.29577. [DOI] [PubMed] [Google Scholar]
- 9.Vernochet C, et al. C/EBPalpha and the corepressors CtBP1 and CtBP2 regulate repression of select visceral white adipose genes during induction of the brown phenotype in white adipocytes by peroxisome proliferator-activated receptor gamma agonists. Mol Cell Biol. 2009;29(17):4714–4728. doi: 10.1128/MCB.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tontonoz P, Spiegelman BM. Fat and beyond: The diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 11.He W, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA. 2003;100(26):15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreyer C, et al. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68(5):879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 13.Kliewer SA, et al. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci USA. 1994;91(15):7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arner P. The adipocyte in insulin resistance: Key molecules and the impact of the thiazolidinediones. Trends Endocrinol Metab. 2003;14(3):137–145. doi: 10.1016/s1043-2760(03)00024-9. [DOI] [PubMed] [Google Scholar]
- 15.Kahn CR, Chen L, Cohen SE. Unraveling the mechanism of action of thiazolidinediones. J Clin Invest. 2000;106(11):1305–1307. doi: 10.1172/JCI11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gan Z, et al. The nuclear receptor PPARβ/δ programs muscle glucose metabolism in cooperation with AMPK and MEF2. Genes Dev. 2011;25(24):2619–2630. doi: 10.1101/gad.178434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang YX, et al. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell. 2003;113(2):159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 18.Wang YX, et al. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2(10):e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka T, et al. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc Natl Acad Sci USA. 2003;100(26):15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dressel U, et al. The peroxisome proliferator-activated receptor beta/delta agonist, GW501516, regulates the expression of genes involved in lipid catabolism and energy uncoupling in skeletal muscle cells. Mol Endocrinol. 2003;17(12):2477–2493. doi: 10.1210/me.2003-0151. [DOI] [PubMed] [Google Scholar]
- 21.Oliver WR, Jr, et al. A selective peroxisome proliferator-activated receptor delta agonist promotes reverse cholesterol transport. Proc Natl Acad Sci USA. 2001;98(9):5306–5311. doi: 10.1073/pnas.091021198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahn SE, et al. A Diabetes Outcome Progression Trial (ADOPT) Study Group Rosiglitazone-associated fractures in type 2 diabetes: An analysis from A Diabetes Outcome Progression Trial (ADOPT) Diabetes Care. 2008;31(5):845–851. doi: 10.2337/dc07-2270. [DOI] [PubMed] [Google Scholar]
- 23.Nesto RW, et al. Thiazolidinedione use, fluid retention, and congestive heart failure: A consensus statement from the American Heart Association and American Diabetes Association. Diabetes Care. 2004;27(1):256–263. doi: 10.2337/diacare.27.1.256. [DOI] [PubMed] [Google Scholar]
- 24.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci USA. 1997;94(9):4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chawla A, et al. PPARdelta is a very low-density lipoprotein sensor in macrophages. Proc Natl Acad Sci USA. 2003;100(3):1268–1273. doi: 10.1073/pnas.0337331100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fyffe SA, et al. Recombinant human PPAR-beta/delta ligand-binding domain is locked in an activated conformation by endogenous fatty acids. J Mol Biol. 2006;356(4):1005–1013. doi: 10.1016/j.jmb.2005.12.047. [DOI] [PubMed] [Google Scholar]
- 27.Xu HE, et al. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol Cell. 1999;3(3):397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 28.Takada I, et al. Alteration of a single amino acid in peroxisome proliferator-activated receptor-alpha (PPAR alpha) generates a PPAR delta phenotype. Mol Endocrinol. 2000;14(5):733–740. doi: 10.1210/mend.14.5.0456. [DOI] [PubMed] [Google Scholar]
- 29.Aagaard MM, Siersbæk R, Mandrup S. Molecular basis for gene-specific transactivation by nuclear receptors. Biochim Biophys Acta. 2011;1812(8):824–835. doi: 10.1016/j.bbadis.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 30.Delfosse V, Maire AL, Balaguer P, Bourguet W. A structural perspective on nuclear receptors as targets of environmental compounds. Acta Pharmacol Sin. 2014;35:88–101. doi: 10.1038/aps.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu HE, et al. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc Natl Acad Sci USA. 2001;98(24):13919–13924. doi: 10.1073/pnas.241410198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nolte RT, et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395(6698):137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 33.Xu HE, et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415(6873):813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- 34.Sierra ML, et al. Substituted 2-[(4-aminomethyl)phenoxy]-2-methylpropionic acid PPARalpha agonists. 1. Discovery of a novel series of potent HDLc raising agents. J Med Chem. 2007;50(4):685–695. doi: 10.1021/jm058056x. [DOI] [PubMed] [Google Scholar]
- 35.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2(9):2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 36.Ngaki MN, et al. Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature. 2012;485(7399):530–533. doi: 10.1038/nature11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batista FA, et al. Structural insights into human peroxisome proliferator activated receptor delta (PPAR-delta) selective ligand binding. PLoS One. 2012;7(5):e33643. doi: 10.1371/journal.pone.0033643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Epple R, et al. 3,4,5-Trisubstituted isoxazoles as novel PPARdelta agonists. Part 2. Bioorg Med Chem Lett. 2006;16(21):5488–5492. doi: 10.1016/j.bmcl.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 39.Oyama T, et al. Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 8):786–795. doi: 10.1107/S0907444909015935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Connors RV, et al. Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma. Bioorg Med Chem Lett. 2009;19(13):3550–3554. doi: 10.1016/j.bmcl.2009.04.151. [DOI] [PubMed] [Google Scholar]
- 41.Luckhurst CA, et al. Discovery of isoindoline and tetrahydroisoquinoline derivatives as potent, selective PPARδ agonists. Bioorg Med Chem Lett. 2011;21(1):492–496. doi: 10.1016/j.bmcl.2010.10.117. [DOI] [PubMed] [Google Scholar]
- 42.Crowley PB, Golovin A. Cation-pi interactions in protein-protein interfaces. Proteins. 2005;59(2):231–239. doi: 10.1002/prot.20417. [DOI] [PubMed] [Google Scholar]
- 43.Dougherty DA. Cation-pi interactions involving aromatic amino acids. J Nutr. 2007;137(6) Suppl 1:1504S–1508S, discussion 1516S–1517S. doi: 10.1093/jn/137.6.1504S. [DOI] [PubMed] [Google Scholar]
- 44.Keil S, et al. Sulfonylthiadiazoles with an unusual binding mode as partial dual peroxisome proliferator-activated receptor (PPAR) γ/δ agonists with high potency and in vivo efficacy. ChemMedChem. 2011;6(4):633–653. doi: 10.1002/cmdc.201100047. [DOI] [PubMed] [Google Scholar]
- 45.Zoete V, Grosdidier A, Michielin O. Peroxisome proliferator-activated receptor structures: Ligand specificity, molecular switch and interactions with regulators. Biochim Biophys Acta. 2007;1771(8):915–925. doi: 10.1016/j.bbalip.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 46.Waku T, et al. Structural insight into PPARgamma activation through covalent modification with endogenous fatty acids. J Mol Biol. 2009;385(1):188–199. doi: 10.1016/j.jmb.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 47.Qiang L, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell. 2012;150(3):620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi JH, et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature. 2010;466(7305):451–456. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi JH, et al. Antidiabetic actions of a non-agonist PPARγ ligand blocking Cdk5-mediated phosphorylation. Nature. 2011;477(7365):477–481. doi: 10.1038/nature10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peters JM, Shah YM, Gonzalez FJ. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat Rev Cancer. 2012;12(3):181–195. doi: 10.1038/nrc3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang D, et al. Crosstalk between peroxisome proliferator-activated receptor delta and VEGF stimulates cancer progression. Proc Natl Acad Sci USA. 2006;103(50):19069–19074. doi: 10.1073/pnas.0607948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stephen RL, et al. Activation of peroxisome proliferator-activated receptor delta stimulates the proliferation of human breast and prostate cancer cell lines. Cancer Res. 2004;64(9):3162–3170. doi: 10.1158/0008-5472.can-03-2760. [DOI] [PubMed] [Google Scholar]
- 53.Gupta RA, et al. Activation of nuclear hormone receptor peroxisome proliferator-activated receptor-delta accelerates intestinal adenoma growth. Nat Med. 2004;10(3):245–247. doi: 10.1038/nm993. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.