Significance
Critical for regulating cell function, integral membrane proteins (MPs) are key engineering targets. MP engineering is limited because these proteins are difficult to express with proper plasma membrane localization in heterologous systems. We investigate the expression, localization, and light-induced behavior of the light-gated MP channel, channelrhodopsin (ChR), because of its utility in studying neuronal circuitry. We used structure-guided SCHEMA recombination to generate libraries of chimeric ChRs that are diverse in sequence yet still capable of efficient expression, localization, and useful light-induced functionality. The conservative nature of recombination generates unique protein sequences that tend to fold and function. Recombination is also innovative: chimeric ChRs can outperform their parents or even exhibit properties not known in natural ChRs.
Keywords: membrane proteins, channelrhodopsin, structure-guided recombination, chimeragenesis
Abstract
Integral membrane proteins (MPs) are key engineering targets due to their critical roles in regulating cell function. In engineering MPs, it can be extremely challenging to retain membrane localization capability while changing other desired properties. We have used structure-guided SCHEMA recombination to create a large set of functionally diverse chimeras from three sequence-diverse channelrhodopsins (ChRs). We chose 218 ChR chimeras from two SCHEMA libraries and assayed them for expression and plasma membrane localization in human embryonic kidney cells. The majority of the chimeras express, with 89% of the tested chimeras outperforming the lowest-expressing parent; 12% of the tested chimeras express at even higher levels than any of the parents. A significant fraction (23%) also localize to the membrane better than the lowest-performing parent ChR. Most (93%) of these well-localizing chimeras are also functional light-gated channels. Many chimeras have stronger light-activated inward currents than the three parents, and some have unique off-kinetics and spectral properties relative to the parents. An effective method for generating protein sequence and functional diversity, SCHEMA recombination can be used to gain insights into sequence–function relationships in MPs.
Integral membrane proteins (MPs) serve diverse and critical roles in controlling cell function. Their receptor, channel, and transporter functions make MPs common targets for pharmaceutical discovery and important tools for studying complex biological processes (1–4). Biochemical studies of MPs and their engineering for biotechnological applications are often limited by poor expression and membrane localization in heterologous systems (5, 6). Unlike soluble proteins, MPs must go through the additional steps of membrane targeting and insertion as well as rigorous posttranslational quality control (7, 8). Functional diversity depends on sequence diversity, but it is challenging to design highly diverse variants that retain membrane localization while at the same time revealing other useful functionality (9). To address this challenge, we demonstrate that structure-guided SCHEMA recombination (10) can create functional MP chimeras from related yet sequence-diverse channelrhodopsins (ChRs). The resulting chimeric ChRs retain their ability to localize to the plasma membrane of mammalian cells but exhibit diverse, potentially useful functional properties.
ChRs are light-gated ion channels with seven transmembrane α-helices. They were first identified in photosynthetic algae, where they serve as light sensors in phototaxic and photophobic responses (11, 12). ChR’s light sensitivity is imparted by a covalently linked retinal chromophore (13). With light activation, ChRs open and allow a flux of ions across the membrane and down the electrochemical gradient (14). When ChRs are expressed in neurons, their light-dependent activity can stimulate action potentials, allowing cell-specific control over neuronal activity (15, 16). This has led to extensive application of these proteins as tools in neuroscience (3). The functional limitations of available ChRs have led to efforts to engineer and/or discover unique ChRs, for example, ChRs activated by far-red light, ChRs with altered ion specificity, or ChRs with increased photocurrents with low light intensity (14). The utility of any ChR, however, depends on its ability to express in eukaryotic cells of interest and localize to the plasma membrane. Our goal is to generate sequence-diverse ChRs whose functional features are useful for neuroscience applications and have not been found in natural environments.
MP engineering is still in its infancy compared with soluble protein engineering. Significant progress in increasing microbial expression and stability of MPs has been made using high-throughput screening methods to identify variants with improved expression from large mutant libraries (6, 17–19). The main motivation was to generate MP mutants that are stable and produced in sufficient quantities for crystallographic and biochemical characterization. This pioneering work demonstrated that MP expression in Escherichia coli and yeast can be enhanced by directed evolution. Because there is not a good method for high-throughput screening of ChR function, however, we chose to focus on introduction of sequence diversity using structure-guided SCHEMA recombination.
SCHEMA recombination offers a systematic method for modular, rational diversity generation that conserves the protein’s native structure and function but allows for large changes in sequence (20–22). SCHEMA divides structurally similar parent proteins into blocks that, when recombined, minimize the library-average disruption of tertiary protein structure (10). Two different structure-guided recombination methods have been developed—one restricts blocks to be contiguous in the polypeptide sequence (10, 23), whereas the other allows for design of structural blocks that are noncontiguous in the polypeptide sequence but are contiguous in 3D space (24). SCHEMA has enabled successful recombination of parental sequences with as low as 34% identity (25), which is not possible using random DNA recombination methods such as DNA shuffling (26). SCHEMA recombination has been used to create a variety of functionally diverse soluble proteins (25, 27–30), but it has not yet been applied to MP engineering. Our goals in this study were to (i) test whether structure-guided recombination produces chimeric MPs that express and localize; (ii) measure the fraction of chimeric sequences in a SCHEMA library that express and localize; and (iii) assess the functional diversity of the MPs that successfully localize to the membrane.
We used SCHEMA to design two libraries of chimeric ChRs, using three parental ChRs having 45–55% amino acid sequence identity. The parent ChRs show different levels of expression and localization in mammalian cells, differences in channel current strength, and differences in the optimal wavelength for channel activation. The SCHEMA recombination libraries, one contiguous and the other noncontiguous, were designed with 10 blocks, yielding an overall library size of 2 × 310, or more than 118,000 possible sequences. On average, chimeras are 73 mutations from the closest parent. We chose and synthesized a set of 218 chimeric genes from these libraries and assayed the proteins for expression and membrane localization in mammalian cells. Our results offer insight into the sequence dependence of ChR expression and localization, and reveal unique functional variation in diverse, well-localizing ChR chimeras. We show that SCHEMA recombination can rapidly and efficiently generate functionally diverse MPs.
Results
Parents for ChR Chimera Library.
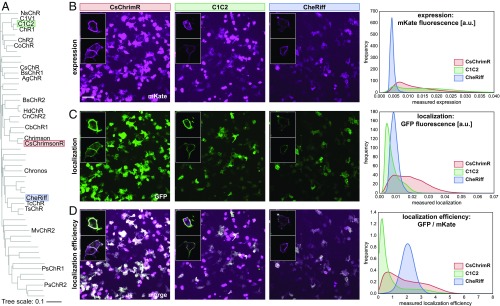
Since the initial discovery and characterization of channelrhodopsins ChR1 (31) and ChR2 (32) from the alga Chlamydomonas reinhardtii, a number of ChRs have been isolated and characterized, for example, VChR1 (33), VChR2 (34, 35), MvChR1 (36), CaChR1 (37), DChR (4), and PsChR (38). De novo transcriptome sequencing of 127 species of algae led to the discovery of 14 ChRs that express and function in mammalian neurons (39). To create unique ChRs by SCHEMA recombination, we chose CsChrimsonR (39), C1C2 (40), and CheRiff (41) as parents. These three ChRs are representative of the available sequence diversity and share 45–55% amino acid identity (Fig. 1A). CsChrimsonR (CsChrimR) is a fusion between the N terminus of CsChR from Chloromonas subdivisa and the C terminus of CnChR1 from Chlamydomonas noctigama and contains a single mutation (K176R) that improves the off-kinetics (the time it takes the channel to close after it is exposed to light) (39). C1C2 is a fusion between ChR1 (N-terminal) and ChR2 (C-terminal), both from Chlamydomonas reinhardtii (40). C1C2 is the only ChR with a solved crystal structure, making it a useful parent for structure-guided recombination. CheRiff is SdChR, from Scherffelia dubia with a single mutation (E154A) that speeds up the off-kinetics and provides a blue-shifted peak in the action spectrum (the current strength achieved by different wavelengths of light) (41). These three parental sequences are fully functional in mammalian cells and have distinct spectral properties. The peak activation wavelengths for CsChrimR, C1C2, and CheRiff are 590, 480, and 460 nm, respectively.
Fig. 1.
Parental ChRs and their properties. (A) Phylogenetic tree of published ChR sequences. Sequences with an alias (e.g., NsChR) have been characterized for expression and functionality in HEK cells and/or mammalian neurons. The three parental sequences (C1C2, CsChrimsonR, and CheRiff) are highlighted. (B–D) HEK cells were transfected with a parental ChR. Membrane-localized ChR was labeled using SpyCatcher-GFP assay, and ChR expression was measured using mKate. HEK cell populations were imaged and processed to measure expression [mean mKate fluorescence (in arbitrary units)], plasma membrane localization [mean GFP fluorescence (in arbitrary units)], and localization efficiency (mean GFP fluorescence/mean mKate fluorescence). Example images show population expression (B), localization (C), and localization efficiency (D) for each parental construct. For both CsChrimR and C1C2, there are cells with very high levels of mKate signal that do not perfectly colocalize with the GFP localization label. These cells express at high levels, much of which is not trafficked to the plasma membrane. (Scale bar: 100 μm.) Insets show confocal images for a few representative cells expressing each parental construct. HEK cell population images were segmented, and the ChR expression, localization, and localization efficiency were measured for each cell. The distribution of these properties for the population of transfected cells is plotted for each parent using kernel density estimation for smoothing.
Quantifying ChR Expression and Localization.
Fluorescent protein fusions have been used extensively as markers for ChR expression (42). To quantify ChR expression, we fused the red fluorescent protein, mKate2.5 (mKate) (43), to the C termini of the ChRs. To quantify membrane insertion and plasma membrane localization, we used the SpyTag/SpyCatcher labeling method (44). Briefly, SpyTag is a 13-aa tag that forms a covalent bond with its interaction partner, SpyCatcher (45). For each ChR, SpyTag was cloned after the native N-terminal signal sequence. This tag is displayed on the extracellular surface of the cell if the ChR is correctly localized to the plasma membrane. Surface-exposed SpyTag can be quantified using exogenously added SpyCatcher protein fused to GFP, which specifically and covalently binds to the SpyTag of correctly localized SpyTag-ChR. Using these methods, we assayed ChR expression (mKate fluorescence: Fig. 1B) and localization (GFP fluorescence: Fig. 1C) in human embryonic kidney (HEK) cells and measured the localization efficiency, or fraction of total protein localized, using the ratio of GFP fluorescence signal to mKate fluorescence signal (Fig. 1D).
HEK cells were transfected in a 96-well plate format, labeled with SpyCatcher-GFP, and imaged for mKate and GFP fluorescence as described in Materials and Methods. For the three parental ChRs, images have been processed by cell segmentation to show the distribution of protein expression and localization levels across the population of expressing cells. Alternative image processing, measuring the whole population intensity, was used to quantify the expression (mean mKate intensity), plasma membrane localization (mean GFP intensity), and localization efficiency (mean mKate intensity/mean GFP intensity) of each ChR construct (Materials and Methods). The whole-population intensity measurements provide a single intensity measurement for each property for a given population of expressing cells. There is significant cell-to-cell variability in transient transfections. To account for this, we measured the properties of each ChR in quadruplicate and calculated the deviation of single intensity measurements between these replicates.
Expression, Localization, and Localization Efficiency of the Three Parent ChRs.
Fig. 1 B–D shows the expression, localization, and localization efficiency of each parent protein in HEK cells. Each parent ChR has an easily distinguishable signature expression and localization profile that can be seen in example images and in the distributions of expression, localization, and localization efficiency for the three parents (Fig. 1 B–D). Both CsChrimR and C1C2 have very high expression levels with large cell-to-cell variation, whereas CheRiff expresses at a significantly lower yet consistent level (Fig. 1B). CsChrimR has the highest level of localization, whereas CheRiff and C1C2 have lower localization levels (Fig. 1C). Localization efficiency shows a different ranking among the parent proteins: CheRiff has the highest localization efficiency and C1C2 has the lowest (Fig. 1D). The wide range in parent ChR mean expression, localization, and localization efficiency should facilitate generation of chimeras with different levels of these properties.
SCHEMA Recombination Library Design.
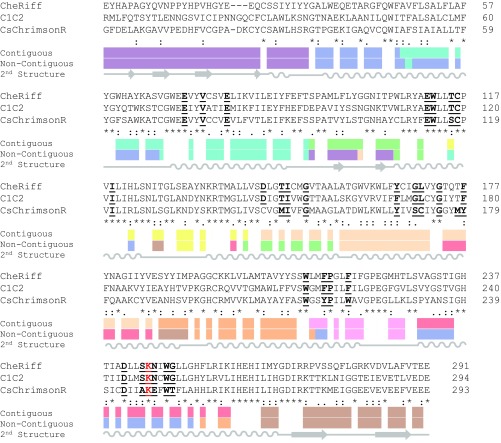
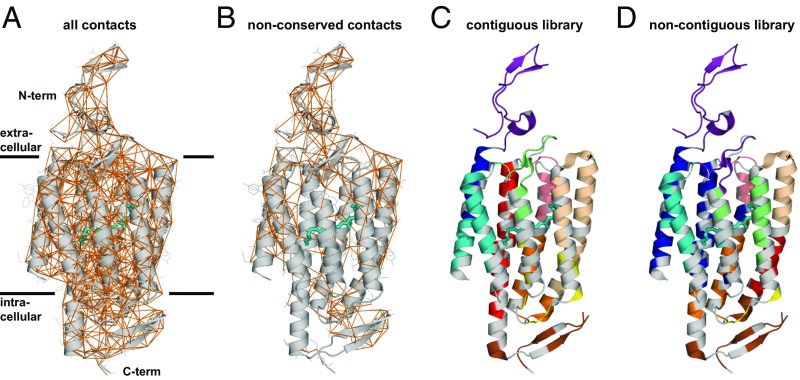
Using the three ChR parents, the known structure of C1C2, and the SCHEMA algorithm (10, 23), we designed two 10-block recombination libraries. SCHEMA is a scoring function that predicts block divisions that minimize the disruption of protein structure when swapping homologous sequence elements among parental proteins. SCHEMA works by defining pairs of residues that are in “contact” and identifying a block design (size and location of sequence blocks) that minimizes the average number of broken amino acid contacts in the resulting library. Two residues are defined to be in contact if they contain nonhydrogen atoms that are within 4.5 Å of each other. If a chimera inherits a contacting pair that is not present in a parent sequence, that contact is said to be broken. Contacts can only be identified in regions of the ChR protein with reliable structural information. The C1C2 structure provides such information for part of the N-terminal extracellular domain (residues 49–84), the seven-helix integral membrane domain (residues 85–312), and the intracellular C-terminal β-turn (residues 313–342) (40). A parental alignment was made for the structurally modeled residues of C1C2 (49–342) and homologous regions of CheRiff (23–313) and CsChrimR (48–340) (Fig. S1). The full contact map calculated from the C1C2 structure is shown in Fig. 2A. Only contacts between nonconserved residues are relevant for the library design (Fig. 2B), because only these can be broken upon recombination. Although contacts are distributed throughout the ChR structure, the nonconserved contacts are far denser at the termini and on the outer surface of the protein; these are the areas of the protein with most sequence diversity (Fig. 2).
Fig. S1.
Amino acid alignment of parental sequences and recombination block designs. Alignment showing the contiguous and noncontiguous block designs. Each color represents a different block, and white shows the conserved residues. Amino acids thought to be important for ChR spectral properties are bolded and underlined. The conserved lysine residue that participates in a Schiff base linkage with retinal is highlighted in red text. The secondary structure is shown below the alignment.
Fig. 2.
Structure-guided recombination library design. (A) Contact map highlighting all amino acids within 4.5 Å of each other (orange lines) in the ChR structure. (B) For library design, we only considered those contacts that can be broken when a different parent block is inserted. Contiguous and noncontiguous libraries were built using the three parental ChRs. The structural cartoon representation of the two libraries is shown for both the contiguous library (C) and noncontiguous library (D). Residues conserved among the parents are shown in gray, and the different sequence blocks are color coded. All-trans-retinal (ATR) is shown covalently linked to the protein by the conserved lysine residue using a teal-colored stick representation.
Two SCHEMA libraries were designed: contiguous (10, 23) and noncontiguous (24). Contiguous libraries are designed so that blocks are contiguous in the amino acid sequence, whereas noncontiguous libraries swap blocks in the 3D structure that are not necessarily contiguous in the primary structure. Using the parental alignment and the contact map, SCHEMA generates a list of possible library designs with a minimized library-average disruption score, the E value, that is, the average number of broken parental contacts per chimera in the library. A 10-block contiguous library was selected (Fig. 2C) with roughly even-length blocks (14–43 residues), a relatively low average E value (E = 25), and whose sequences have an average of 73 mutations from the nearest parent. The selected 10-block noncontiguous library has a low average E value (E = 23), block sizes comparable to the contiguous library, and an average of 71 mutations from the nearest parent (Fig. 2D). The noncontiguous library design also maintains the presumptive dimer interface. For these libraries, the “mutations” introduced into any one parent are limited to the nonconserved residues of the other two parents. Each of the 10-block, three-parent libraries gives 59,049 possible chimeras (310), for a total of 118,098 possible chimeras.
The two library designs both place block boundaries in positions that may not be obvious in the protein structure. For example, that several boundaries appear in the middle of α-helices indicates that naive chimeragenesis by simply swapping elements of secondary structure would be more disruptive than design based on conservation of native contacting residue pairs. To test this, we calculated the average E value for libraries with block boundaries within the loops between transmembrane α-helices such that the N-terminal domain, the C-terminal domain, and each helix form separate blocks for a total of nine blocks. Within the loops, there are multiple possible locations for block boundaries. We built 128 different designs with block boundaries within loops and calculated library average E values that range from 36 to 43. These values are significantly higher than those for the SCHEMA designs and indicate that naive helix swapping is more disruptive than SCHEMA recombination.
Production of Chimeras for Characterization.
We chose a set of 223 sequences from the recombination libraries for gene synthesis and characterization of expression and localization properties of the ChRs in mammalian cells. This set included all 120 proteins with single-block swaps from both libraries. These chimeras consist of nine blocks of one parent and a single block from one of the other two parents. An additional 103 sequences were designed to maximize mutual information (46) between chosen chimeras and the remainder of the chimeric library, using the rationale described by Romero et al. (29). Seventeen of these sequences were designed with a constraint on the number of mutations from the nearest parent (<40 mutations). This set, referenced as the “maximally informative with mutation cap,” provided chimeras composed of, on average, six blocks of one dominant parent and four blocks of a mix of the other two parents. The remaining 86 of the “maximally informative” sequences are highly diverse, consisting of blocks from all three parents and containing, on average, 84 mutations compared with the most sequence-related parent. This set of 223 genes was synthesized and cloned in a mammalian expression vector at Twist Bioscience. Two hundred and fifteen of the designed sequences were synthesized successfully and cloned into the expression vector; with the three parent sequences, this gave a total of 218 sequences for the library characterization studies.
Localization and Expression of ChR Chimeras.
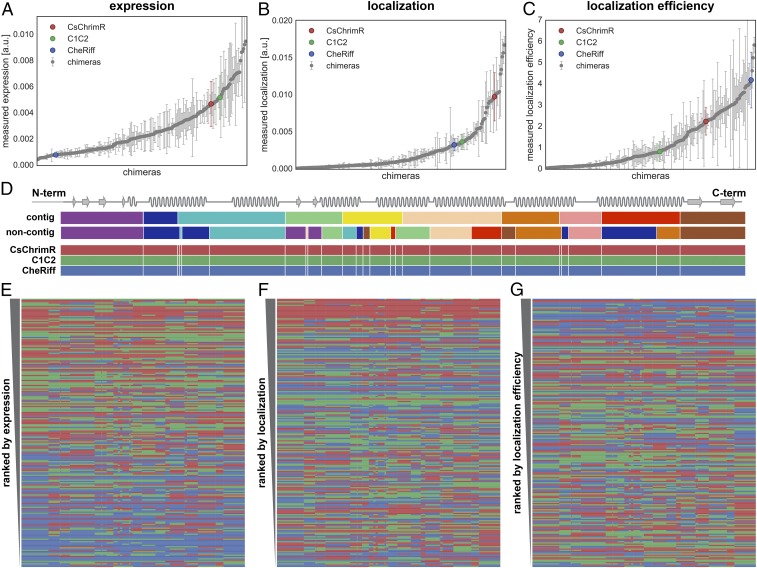
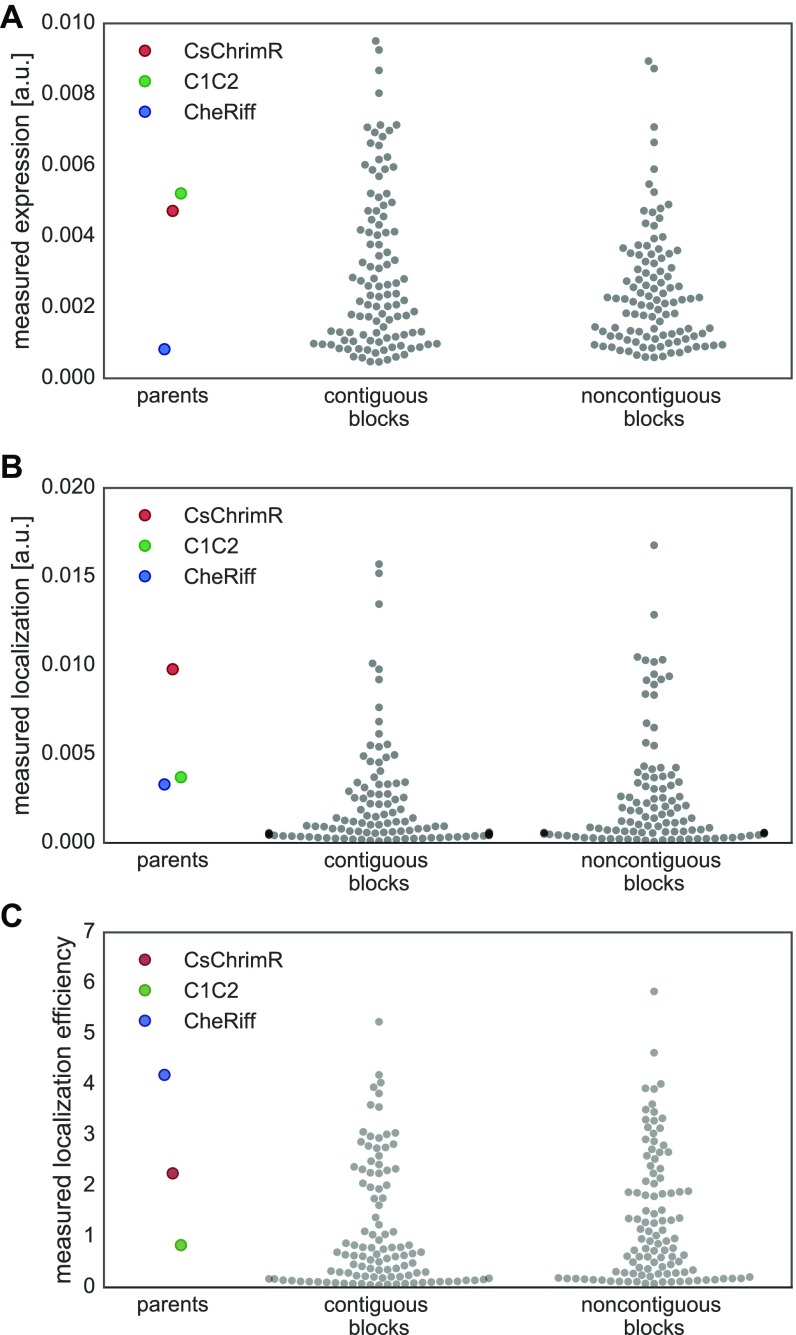
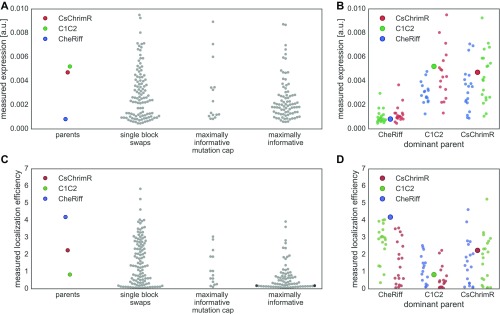
HEK cell expression and localization were measured for each chimera using at least 150 and up to 100,000 transfected cells from at least four replicate HEK cell transfections (Dataset S1). Chimeras were benchmarked to the lowest performing parent. CheRiff is the lowest performing parent for expression and localization, and C1C2 is the lowest performing parent for localization efficiency. The majority (89%) of the chimeras have higher expression levels than the lowest parent (Fig. 3A) whereas a lower number, amounting to 23%, have higher localization levels than the lowest parent (Fig. 3B). Forty-four percent of the chimeras have better localization efficiency than the lowest parent (Fig. 3C). The difference between the number of chimeras that express well and the number of chimeras that localize well suggests that the sequence demands for localization are more stringent.
Fig. 3.
Chimera expression, localization, and localization efficiency. A–C show the measured expression [mean mKate fluorescence (in arbitrary units)] (A), localization [mean GFP fluorescence (in arbitrary units)] (B), and localization efficiency (mean mKate/GFP fluorescence) (C), respectively, of all 218 chimeras with the properties of the three parental constructs highlighted in color. Error bars represent the SD of measurements from, at least, quadruplicate replicates with each replicate representing >150 transfected cells. Each chimera is ranked according to its performance for each property (expression, localization, and localization efficiency) in ascending order. D shows the contiguous (contig) and noncontiguous (noncontig) 10-block library designs with each block in a different color aligned with a schematic of the ChR secondary structure. The block coloring of the contig and noncontig block designs match Fig. 2 and Fig. S1, although, for clarity, the conserved locations are not shown in gray. Block boundaries (white lines) for the combined contiguous and noncontiguous library designs are shown on the three parents below the individual library designs. E–G show the block identity of the chimeras ranked according to their performance for each given property with the best-ranking chimera at the top of the list. Each row represents a chimera. The colors represent the parental origin of the block (red, CsChrimR; green, C1C2; and blue, CheRiff).
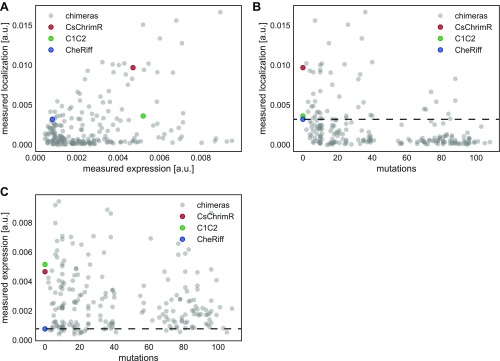
Measurements show no clear correlation between chimera expression and localization (Fig. S2A), and chimeras localize more frequently if they are only a single-block swap away from the nearest parent (<40 mutations) (Fig. S2B). On the other hand, most chimeras express, even with as many as 108 mutations from the nearest parent (Fig. S2C). Only 9% of the sequences in the maximally informative set localize as well as the lowest localizing parent, whereas 24% of the maximally informative mutation cap set localize as well as the lowest localizing parent, and 33% of the sequences with a single-block swap localize as well as the lowest parent (Fig. 4A). Thus, sequences from the maximally informative set are less likely to localize than the sequences with single-block swaps or sequences with a mutation cap. These results highlight the difficulty of finding highly mutated ChR sequences (>40 mutations from the nearest parent) that localize well. Nonetheless, we found 51 ChRs in this test set of 218 that localize to the plasma membrane at least as well as the worst parent, and 8 of those are more than 40 mutations away from the closest parent. Although less diverse than the maximally informative chimeras, the single-block swap chimeras still contain on average 15 mutations compared with the closest parent. This is a significant amount of diversity to introduce while still maintaining localization, given that even a single mutation can destroy a protein’s ability to fold or function (22).
Fig. S2.
Interdependencies of chimera properties. Chimera data are plotted as gray points, and parental data points are highlighted in color (red, CsChrimR; green, C1C2; and blue, CheRiff). (A) Plot of measured localization [mean GFP fluorescence (in arbitrary units)] vs. measured expression [mean mKate fluorescence (in arbitrary units)] shows no clear correlation. (B) Plot of measured localization vs. number of mutations from closest parent. (C) Plot of measured expression vs. number of mutations from closest parent. Dashed lines in B and C show the measured properties of the lowest-performing parent (CheRiff).
Fig. 4.
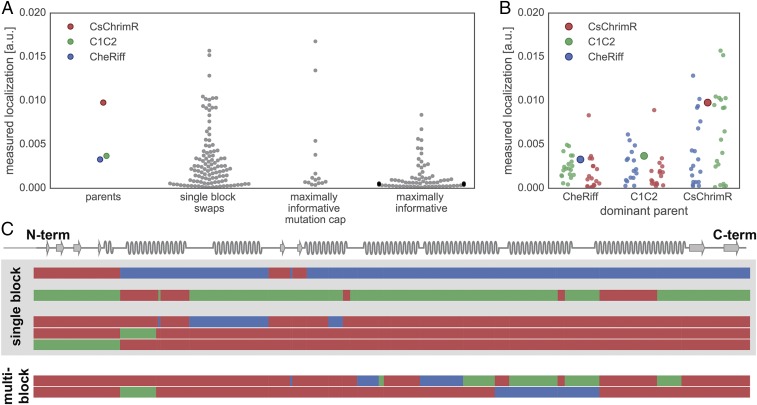
Comparison of membrane localization for different chimeras. (A) Swarm plots of measured localization [mean GFP fluorescence (in arbitrary units)] for the parent constructs and each chimera set: single-block swaps, maximally informative with mutation cap, and maximally informative. Chimera data are plotted as gray points; parental data are highlighted in color (red, CsChrimR; green, C1C2; and blue, CheRiff). (B) Comparison of measured localization [mean GFP fluorescence (in arbitrary units)] of single-block swap chimeras relative to their dominant parent. Each single-block swap chimera is grouped based on the dominant parent with data points colored according to the identity of the single block being swapped into the dominant parent (red, CsChrimR block; green, C1C2 block; and blue, CheRiff block). The large point in each group shows the performance of the dominant parent. (C) Shown is the block identity of selected single-block swap and multiblock swap chimeras aligned with the ChR secondary structure. The top two single-block swap chimeras are the top-performing chimeras for the CheRiff- and C1C2-dominant parents. The bottom three single-block swap chimeras are the top-performing single-block swaps in the CsChrimR-dominant parent. For the noncontiguous design, a single (structural) block may be disconnected along the primary sequence. Thus, single-block swap chimeras from the noncontiguous library may have swapped sequence elements in more than one location along the primary sequence. The two multiblock swap chimeras are the top two-performing chimeras in the maximally informative with mutation cap chimera set. Each row represents a chimera. The three different colors represent blocks from the three different parents (red, CsChrimR; green, C1C2; and blue, CheRiff).
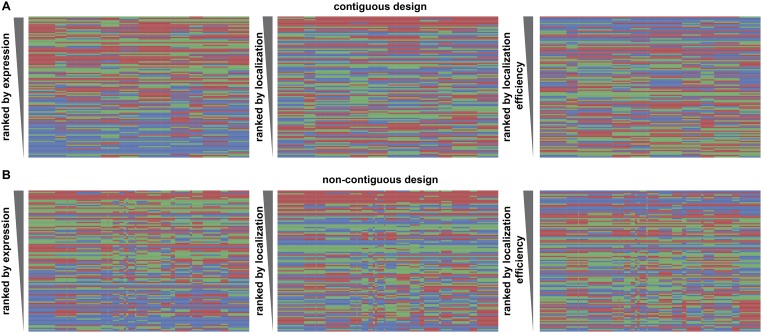
Performance ranking of chimera sequences for each property of interest (expression, localization, and localization efficiency) shows that sequences dominated by CheRiff generally rank low in expression but have the highest rankings for localization efficiency (Fig. 3 E and G), whereas sequences dominated by CsChrimR have the highest ranking for localization (Fig. 3F). These trends are seen for both the contiguous and noncontiguous libraries (Fig. S3). No clear patterns or specific blocks of sequence emerge from the data that determine chimera performance, suggesting that each sequence/structural block behaves differently in different contexts. However, the single-block–swapped chimeras offer insight into the sequence dependence of properties in the context of the parental ChRs.
Fig. S3.
Chimeras from the contiguous and noncontiguous libraries, ranked by expression, localization, and localization efficiency. Block identity of the chimeras ranked according to performance for each given property with the best-ranking chimera at the top of the list for the contiguous (A) and noncontiguous (B) library chimeras. Each row represents a chimera. The colors represent the parental origin of the block (red, CsChrimR; green, C1C2; and blue, CheRiff). The properties shown are measured expression [mean mKate fluorescence (in arbitrary units)], localization [mean GFP fluorescence (in arbitrary units)], and localization efficiency (mean mKate/GFP fluorescence).
We also wanted to compare the two library design strategies. Both the contiguous and noncontiguous SCHEMA recombination libraries have the same number of blocks, similar average disruption scores (E values) (25 and 23, respectively), similar average number of mutations (73 and 71, respectively), but different design strategies. We found that chimeras show similar ranges in measured properties whether they were designed to be contiguous in the primary or tertiary structure (Fig. S4). These results suggest that, for ChRs, library design is less important than the average disruption score and average number of mutations per chimera. For soluble proteins, the average disruption score and average number of mutations of SCHEMA libraries have been shown to correlate with the fraction of the recombination library that does not fold and function (25).
Fig. S4.
Comparison of chimeras from the contiguous and noncontiguous recombination libraries. Swarm plot showing each chimera’s expression [mean mKate fluorescence (in arbitrary units)] (A), localization [mean GFP fluorescence (in arbitrary units)] (B), and localization efficiency (mean mKate/GFP fluorescence) (C) for the contiguous and noncontiguous recombination libraries. Chimera data are plotted as gray points, and parental data points are highlighted in color (red, CsChrimR; green, C1C2; and blue, CheRiff).
Comparison of Chimeras with Good Localization.
Chimeras with single-block swaps indicate which individual blocks increase localization (Fig. 4B), expression (Fig. S5B), and localization efficiency (Fig. S5D). For both the CheRiff and C1C2 parents, there is a single-block swap from CsChrimR that results in a chimera with large improvements in localization (Fig. 4B). Interestingly, the block from CsChrimR that boosts CheRiff’s localization is different from the CsChrimR block that improves C1C2’s localization: the former contains the CsChrimR N terminus and an associated extracellular loop and the latter contains the first and (structurally adjacent) seventh CsChrimR helices. In fact, the CsChrimR block that causes a nearly twofold increase in C1C2’s localization causes a twofold decrease in CheRiff localization when chimeras are compared with their respective dominant parent. This result stresses again the importance of context when assessing the sequence dependence of a property as complex as localization.
Fig. S5.
Comparison of measured expression and membrane localization efficiency for each chimera set. Swarm plots of expression [mean mKate fluorescence (in arbitrary units)] (A) and localization efficiency (mean mKate/GFP fluorescence) (C) showing measurements for each dataset compared with parents: single-block swaps, maximally informative with mutation cap, and maximally informative. Chimera data are plotted as gray points, and parental data points are highlighted in color (red, CsChrimR; green, C1C2; and blue, CheRiff). Comparison of single-block swap chimeras measured expression (B) and localization efficiency (D) relative to the dominant parent. Each single-block swap chimera is grouped based on the dominant parent with data points colored based on the identity of the single block being swapped in (red, CsChrimR block; green, C1C2 block; and blue, CheRiff block). The large point in each group shows the performance of the dominant parent.
There are also single blocks from both the CheRiff and C1C2 parents that significantly increase localization of CsChrimR (Fig. 4B). This is interesting because both the CheRiff and C1C2 parents have lower localization levels than the CsChrimR parent. This result illustrates recombination’s ability to produce progeny that outperform all of the parental sequences. The three single-block swaps that produce chimeras that outperform CsChrimR are at the N terminus, first helix, and second helix (Fig. 4C). It is expected that swapping the N terminus of the protein could influence localization (47), but it is not clear why the first and second helix swaps are important for localization. Finally, there are two maximally informative mutation cap sequences that also outperform the top parent, CsChrimR (Fig. 4A). These chimeras have blocks from all three parents spread across the protein sequence (Fig. 4C).
Functional Characteristics of Chimeras That Localize.
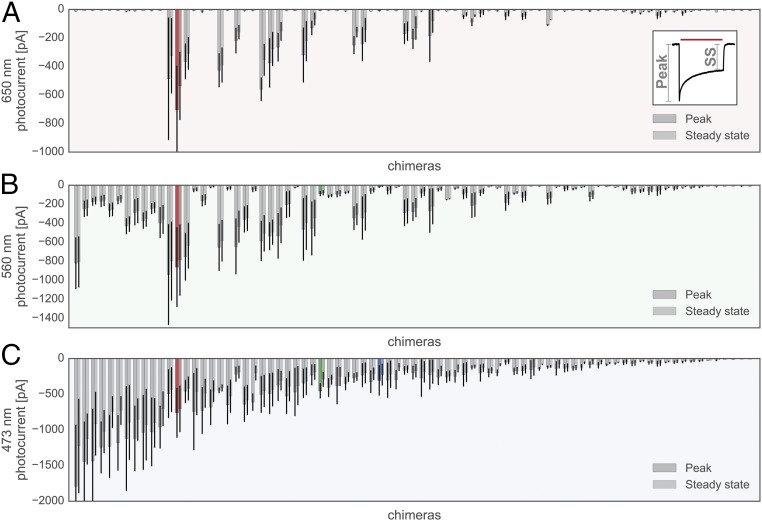
Seventy-five chimeras with localization levels above or within 1 SD of the CheRiff parent or localization efficiency above or within 1 SD of the C1C2 parent were analyzed for other functional characteristics (Dataset S2). Each chimera was expressed in HEK cells and its light-inducible currents were measured using patch-clamp electrophysiology in voltage-clamp mode upon sequential exposure to three different wavelengths of light (473, 560, and 650 nm). ChRs have a characteristic light-activated current trace with an initial peak in inward current occurring immediately after light exposure followed by a decay of inward current to a constant, or steady-state, current (Fig. 5, Inset). The majority of tested chimeras were functional, with only 5 of the 75 tested chimeras having light-activated steady-state inward currents less than 20 pA (Fig. 5). Different chimeras are optimally activated by different wavelengths. All 70 of the active chimeras are activated by 473-nm light, whereas only 18 chimeras show robust activation with 650-nm light (Fig. 5). When activated with 473-nm light, 10 chimeras have stronger peak and steady-state photocurrents than the parental protein with the strongest photocurrents (CsChrimR) (Fig. 5C), demonstrating again that recombination can generate MPs that outperform any of the parents.
Fig. 5.
Chimera photocurrents with 650-, 560-, and 473-nm light. Peak and steady-state photocurrents induced by a 1-s exposure to 650-nm (A, red shading), 560-nm (B, green shading), and 473-nm (C, blue shading) wavelength light for each chimera measured. Inset shows the canonical ChR peak vs. steady-state (SS) inward current observed when the channel is exposed to light. All chimera data are plotted as gray bars, and parental data are highlighted in color (red, CsChrimR; green, C1C2; and blue, CheRiff). Peak and SS current are measured for n = 4–10 cells for each chimera. Bars show the mean, and error bars represent SD of measured cells for both peak and SS current.
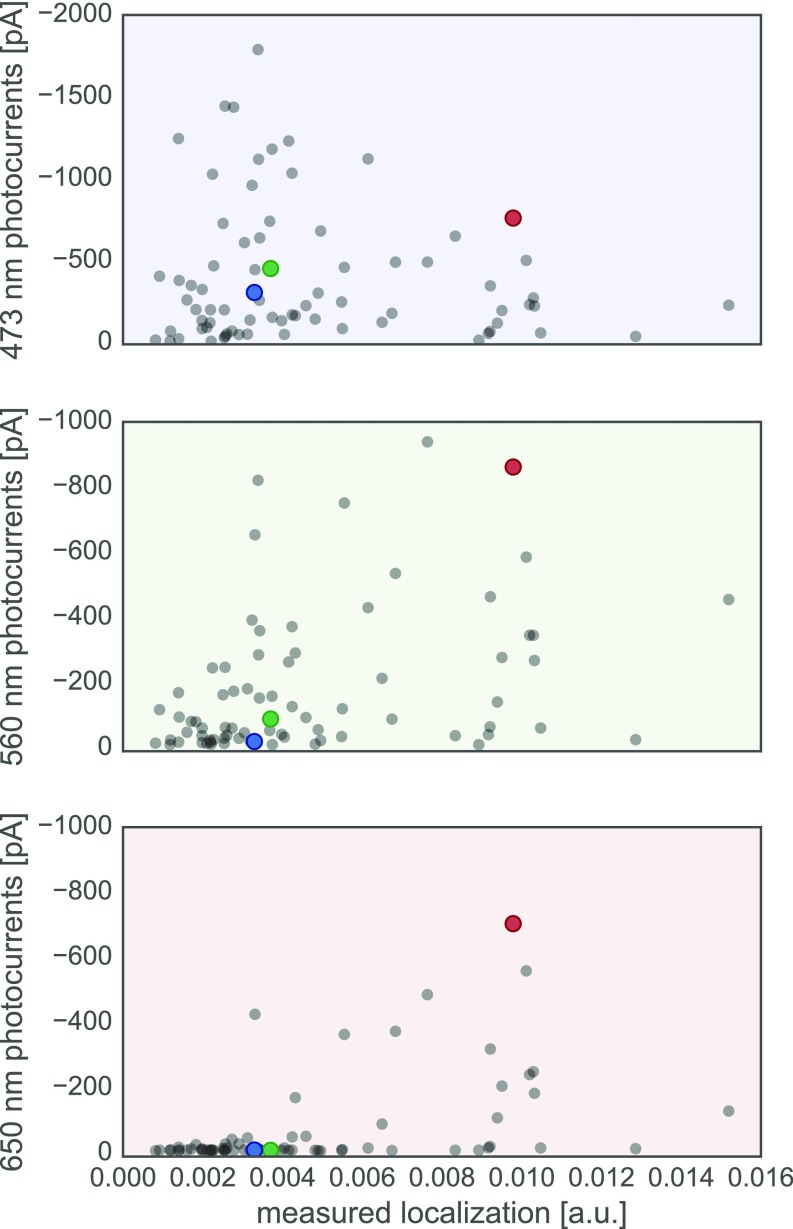
Although localization is a prerequisite for channel function, a chimera that localizes well does not necessarily provide stronger currents than a chimera that localizes less well. In addition to the amount of protein in the membrane, the channel’s conductance properties also affect current strength. The mutations in these ChR sequences could cause a change in channel conductance. To test whether changes in current strength are due to differences in localization or conductance, we compared the measured localization and peak current strength for each chimera (Fig. S6). That we did not find a strong positive correlation between these two measurements suggests that differences in chimera currents are dominated by changes in their conductance. That is, as long as an adequate fraction of a ChR is able to localize to the plasma membrane, the major factor determining current strength is the chimera’s specific conductance properties, which is sequence dependent and can be tuned by mutation.
Fig. S6.
Photocurrents vs. measured localization for all tested chimeras. Chimera data are plotted as gray points and parental data points are highlighted in color (red, CsChrimR; green, C1C2; and blue, CheRiff). Plot of measured photocurrents vs. measured localization [mean GFP fluorescence (in arbitrary units)] for three different wavelengths: 473 nm (Top, blue shading), 560 nm (Middle, green shading), and 650 nm (Bottom, red shading).
ChR Chimeras with Altered Photocurrent Properties.
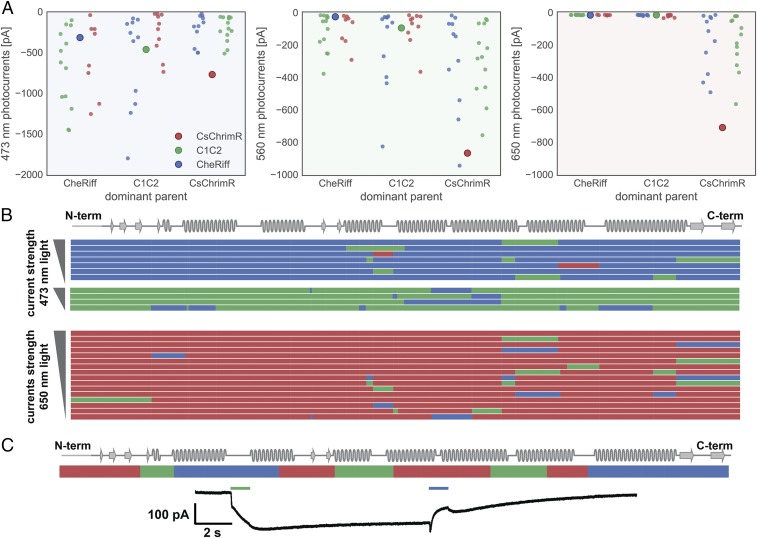
Analysis of the photocurrent properties of single-block swap chimeras activated with 473-nm light show that there are many single-block changes to both the CheRiff and C1C2 parent that cause large increases in current strength (Fig. 6A). The CheRiff parent shows large increases in current strength with single blocks from either C1C2 or CsChrimR, whereas C1C2 performs best with single blocks from CheRiff, even though CheRiff has the weakest currents of the three parents. Comparison of the sequences of these highly functional chimeras shows that single blocks swapped at many different positions in the ChR sequence can have a positive effect on current strength and that no single-block position alone accounts for the improved currents (Fig. 6B).
Fig. 6.
Comparison of chimeras with significantly altered photocurrent properties. (A) Peak photocurrent for each single-block swap chimera grouped based on the dominant parent with data points colored based on the identity of the single block being swapped in (red, CsChrimR block; green, C1C2 block; and blue, CheRiff block). The large point in each group shows the performance of the dominant parent. (B) Shown is the block identity of top-performing single-block swap chimeras aligned with the ChR secondary structure. Single-block swap chimeras that outperform CsChrimR with 473-nm light are shown (top six performing single-block swap chimeras with the CheRiff-dominant parent and the top four performing single-block swap chimeras with the C1C2-dominant parent). All chimeras that produce photocurrents >50 pA upon 650-nm light exposure are also shown. These single-block swap chimeras all have the CsChrimR-dominant parent. Chimeras are grouped based on the identity of the dominant parent and ranked based on photocurrent with either 473-nm light or 650-nm light. For the noncontiguous design, a single (structural) block may be disconnected along the primary sequence. Thus, single-block swap chimeras from the noncontiguous library may have swapped sequence elements in more than one location along the primary sequence. Each row represents a chimera. The colors represent the parental origin of the block (red, CsChrimR; green, C1C2; and blue, CheRiff). (C) One multiblock swap chimera (c96) has unique light activation properties relative to the parents. This ChR chimera is activated by 560-nm light and closes with 473-nm light. The chimera block identity is shown.
Significant effort has been taken to find ChR sequences with red-shifted properties (activation by ∼650-nm light), because red light has enhanced tissue penetration and decreased phototoxicity compared with higher energy blue light (33, 39). Three natural ChRs have been shown to be activated with red light: CsChR/Chrimson (39), VChR1 (33), and MChR1 (36). Here, we show that recombination generates many chimeras that are activated with 650-nm light and that have significant sequence diversity compared with their red-light–activated parent (a mean of 15 and as many as 70 mutations) (Figs. 5A and 6A). All of the single-block swap chimeras capable of producing photocurrents with 650-nm light have CsChrimR as the dominant parent (Fig. 6A). The CsChrimR parent can tolerate single-block swaps from either C1C2 or CheRiff at many positions in the ChR sequence and still retain strong currents activated by 650-nm light (>50-pA peak current) (Fig. 6B), showing that none of its single-block positions is necessary for CsChrimR’s red-light–activated current.
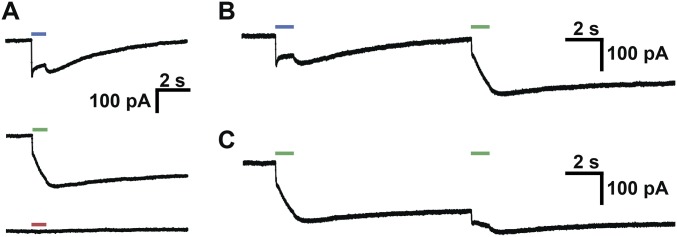
Some chimeras have unique spectral properties, exhibited by none of the three parent ChRs. One multiblock swap chimera from the maximally informative set, for example, shows strong activation with 560-nm light but atypical properties once the light is turned off (Fig. 6C). This chimera shows a gradual increase in inward current once the green light is turned off, followed by a very slow decrease in current. This inward current can be turned off with 473-nm light, causing a brief depolarization, then a decrease in inward current while the 473-nm light is on. Once the 473-nm light is turned off, there is a brief depolarization followed by a decrease in current to baseline levels. When activated by 473-nm light without preexposure to 560-nm light, this chimera produces inward currents with unusual light-off behavior (Fig. S7A). Sequential 1-s exposures to 560-nm light causes continued depolarization (Fig. S7 B and C). This type of bistable excitation, step function opsin (SFO), has been reported previously, in ChRs generated with site-directed mutagenesis at a single position (C128) in ChR2 (48). However, this SFO is activated by blue (470-nm) light and terminated by green (542-nm) light (48). The unusual light-off behavior, with inward currents that continue to increase ∼0.5 s after the light has been turned off, suggests an altered photocycle (48).
Fig. S7.
One multiblock swap chimera with unique properties. (A) Chimera photocurrents upon 1-s exposure to 473-nm (Top), 560-nm (Middle), and 650-nm (Bottom) light. (B) Sequential activation of chimera with 473-nm and then 560-nm light. (C) Sequential activation of chimera with 560-nm and then 560-nm light.
Discussion
SCHEMA uses structural information to guide the choice of block boundaries for creating libraries of chimeric proteins from homologous parents. Both conservative and innovative, recombination generates large changes in sequence without destroying the features required for proper folding, localization, and function. Recombination is conservative because the sequence diversity source has passed the bar set by natural selection for fold and function. Recombination thus introduces limited diversity and at positions that are tolerant to mutation, for example, at the protein termini or the surface interacting with the lipid bilayer. In contrast, conserved functional residues and those in the structural core experience little or no change upon recombination. The sequence changes that are made can nonetheless lead to functional properties that may not be selected for in nature.
In the largest screen of ChR sequences and properties to date, we found that a high proportion of chimeras made by recombining three parent integral membrane ChRs retain the ability to localize to the plasma membrane and exhibit high photocurrents despite having an average of 43 mutations with respect to the closest parent. In HEK cells, 89% of the 218 tested chimeras expressed at least as well as the lowest performing parent, and 23% localized better than the lowest performing parent. Moreover, 70 out of 75 well-localizing chimeras show light-activated inward currents. The innovative nature of SCHEMA recombination was observed in ChR expression, localization, and photocurrents under activation by 473-nm light, for which 5–15% of the tested chimeras outperformed the best-performing parent. In particular, six single-block swap chimeras showed between a 1.5- and 2-fold increase in photocurrent relative to the parent with the strongest photocurrents (CsChrimR) when activated by 473-nm light. From one of the heavily mutated chimeras, we also discovered that the photophysical properties of a ChR can be modified dramatically and unexpectedly. Recombination can create sequences with properties that may not be selected in nature. For example, red wavelengths do not penetrate to the water depths typically occupied by algae, and thus red-light–activated ChRs are rare in nature, with only three natural such ChRs discovered to date (33, 36, 39). We purposefully biased our recombination libraries by choosing a red-light–activated parent, CsChrimR, and found a number of sequence-diverse progeny that were also red-light activated. Although the retinal binding pockets of the two blue-shifted parents are nearly identical, almost one-half of the residues in the retinal-binding pocket of CsChrimR are different. Including CsChrimR as a parent thus allowed us to explore sequence diversity in this vital region of the protein and enrich for properties desirable for neuroscience applications but not necessarily favored in nature. This type of enrichment in recombination libraries depends on the choice and availability of parent proteins.
Two of the parent proteins for this study came from the 61 ChR homologs that were discovered from de novo transcriptome sequencing of 127 species of algae (39). Of the 50 of these ChR homologs assayed for expression and photocurrents in HEK cells, 25 produced photocurrents, whereas the other 25 did not. Fourteen of these sequences were then characterized and shown to retain function in mammalian neurons (39). Although interesting and useful genes can to be found in nature, it is not always clear where to look for them. SCHEMA recombination, on the other hand, offers a systematic, straightforward method for generating artificial diversity from a set of natural sequences. Furthermore, the type of systematic diversity in a recombination library is useful for analyzing how sequence features determine protein properties. Such analysis is greatly simplified by the greatly reduced sequence space (i.e., 10 blocks with only three possible sequences at each block).
This ChR chimera dataset offers insights into the robustness of ChR expression, localization, and function to changes in sequence. Although almost all of the chimeric sequences express, localization is more rare, indicating that the sequence and structural constraints on localization are greater than those on expression. Among sequences that successfully localize, most are functional light-activated channels, but there is significant sequence-based variability in activation wavelength and conductance. This suggests that membrane localization is a principal hurdle to engineering ChR sequences with unique functions. Simply extrapolating the fraction of well-localized chimeras in our 218-chimera sample set to the overall library, we could expect 10,000–27,000 of the 118,000 chimeras to localize to the membrane.
The ability to predict which sequences are likely to localize will remove a key roadblock to identifying unique, functional sequences. Changes throughout the ChR protein can enhance localization and photocurrents, and no single sequence block determines the observed improvements. This suggests that each sequence/structural block behaves differently in different contexts. For certain soluble protein properties (e.g., thermostability), it has been shown that block contributions are additive, that is, context independent, and that chimera stability can be predicted using linear regression (28, 29, 49, 50). Our data suggest that ChR localization and photocurrent properties, however, require a more complex model to account for the nonlinear dependence of function on block sequence. Our future work will explore the use of statistical models to provide sequence/structure insights into the features that determine localization and photocurrent properties, to predict the properties of all 118,000 sequences in the recombination libraries, and to engineer ChR sequences with desirable properties.
Materials and Methods
Design and Construction of Parental ChRs and Recombination Library.
The three ChR parent genes were built using a consistent vector backbone (pFCK) (37) with the same promoter (CMV), trafficking signal (TS) sequence (38), and fluorescent protein (mKate2.5) (39). For the SpyTag/SpyCatcher membrane localization assay, it was necessary to add the SpyTag sequence close to the N terminus of each of the parental proteins but C-terminal to the signal peptide sequence cleavage site. Assembly-based methods and traditional cloning were used for vector construction and parental gene insertion. Annotated vector sequences of the three SpyTagged parental constructs are included as Datasets S3–S5.
SCHEMA was used to design 10-block contiguous and noncontiguous recombination libraries of the three parent ChRs that minimize the library-average disruption of the ChR structure (10, 23, 24). Both recombination library designs were made using software packages for calculating SCHEMA energies openly available at cheme.che.caltech.edu/groups/fha/Software.htm. The SCHEMA software outputs the amino acid sequences of all chimeras in a library. The amino acid sequence for each chimera chosen for experimental testing was converted into a nucleotide sequence such that all chimeras had consistent codon use. Gene sequences for the 223-chimera set were synthesized by Twist Bioscience, cloned in the pFCK vector by a homology-based cloning strategy, and transformed into Stbl3 cells (Invitrogen) or Endura cells (Lucigen). Individual clones were picked and sequence verified by next-generation sequencing (NGS). Purified plasmid DNA of each chimera was prepared for HEK cell transfection.
Measuring ChR Expression, Localization, and Photocurrents.
HEK 293T cells were transfected with purified, ChR variant DNA using Fugene6 reagent according to the manufacturer’s recommendations. Cells were given 48 h to express before being assayed for expression, localization, or photocurrents. To assay localization level, transfected cells were subjected to the SpyCatcher-GFP labeling assay, as described by Bedbrook et al. (44). Transfected HEK cells were then imaged for mKate and GFP fluorescence using a Leica DMI 6000 microscope. We used conventional whole-cell patch-clamp recordings in transfected HEK cells to measured light-activated inward currents using methods and equipment described in ref. 51.
Parental ChR Constructs
Each of the three ChR library parent genes was built using a consistent vector backbone (pFCK) with the same promoter (CMV), trafficking signal (TS) sequence, and fluorescent protein (mKate). We used the pFCK vector from the construct FCK-CheRiff-eGFP [Addgene plasmid #51693 (41)]. A TS sequence (42) was inserted between the opsin and the fluorescent protein. The TS sequence has been shown to enhance opsin membrane trafficking (42). The GFP was replaced with mKate2.5 (43). Use of a red fluorescent protein as the marker for the opsin expression enabled use of SpyCatcher-GFP labeling for membrane-localized proteins. mKate2.5 is a monomeric far-red fluorescent protein that shows no aggregation. The mKate2.5 sequence was synthesized by IDT with overhangs for cloning into the desired vector system.
For the SpyTag/SpyCatcher membrane localization assay, it was necessary to add the SpyTag sequence close to the N terminus of each of the parental proteins and C-terminal to the signal peptide sequence cleavage site. For C1C2, an optimal position of the SpyTag had already been published. The SpyTag-C1C2 gene was amplified from the construct pLenti-CaMKIIa-SpyTag-C1C2-TS-mCherry (44) and inserted into the pFCK backbone. For CheRiff and CsChrimR, it was necessary to test various N-terminal SpyTag locations. The CheRiff gene was first amplified from FCK-CheRiff-eGFP [Addgene plasmid #51693 (41)], and the SpyTag sequence was added at different N-terminal positions by assembly PCR methods. The CsChrimR gene was built by assembly of the Cs N-terminal sequence (synthesized by IDT) with the C-terminal end of ChrimsonR amplified from the FCK-ChrimsonR-GFP construct [Addgene plasmid #59049 (39)]. The sequence of CsChrimR was designed to be identical to the previously published sequence (39). The SpyTag sequence was then inserted at different positions in the N-terminal region of the protein using assembly PCR methods. We tested three different pFCK-SpyTag-CheRiff-TS-mKate designs and three different pFCK-SpyTag-CsChrimR-TS-mKate designs and selected the design that showed expression and localization levels most similar to the nontagged parent.
Assembly-based methods and traditional cloning were used for vector construction and parental gene insertion. Annotated vector sequences of the three SpyTagged parental constructs are included as Datasets S3–S5.
Library Design
SCHEMA was used to design recombination libraries of the three parental ChRs to minimize the library-average disruption of the ChR structure (10, 25, 28). For the contiguous library, the SCHEMA-predicted block definitions were not modified. This 10-block library had roughly even-length blocks (14–43 residues), a relatively low average E value (E = 25), and whose sequences have an average of 73 mutations from the nearest parent. For the noncontiguous library, the SCHEMA-predicted block definitions were modified to group the N- or C-terminal domains into single blocks, maintain the presumptive dimer interface, and minimize the number of small blocks (less than five mutations). Specifically, a 13-block noncontiguous recombination library was generated for which two N-terminal blocks were combined, two C-terminal blocks were combined, two of four blocks in TM 5 were combined, and two residues of TM 3 were switched to the same block as TM 4 (where TM 3 and 4 make up the dimer interface observed for C1C2). The two loops that were not modeled in the C1C2 structure, between TM 1 and TM 2 and in the β-turn of the C-terminal motif, were added to the block containing TM2 and the C-terminal block, respectively. The unmodeled residues of the N and C termini were added to the N- and C-terminal blocks. The resulting noncontiguous library had 10 blocks, an average E value of 23, an average of 71 mutations, and block size similar to the contiguous library (Fig. 2 C and D).
Among the three ChR parents, five unique N-linked glycosylation sites have been predicted by the NetNGlyc 1.0 (www.cbs.dtu.dk/services/NetNGlyc/) and GlycoEP servers (52). C1C2 harbors four of these sites with by far the highest confidence at each site. With one exception, the putative N-linked glycosylation sites do not overlap with recombination block borders. The exception site (SpyTag-C1C2 N95) is located in between the N-terminal domain and the first TM helix.
Contiguous recombination design was done using a software package for calculating SCHEMA energies and running the RASPP algorithm (23) openly available at cheme.che.caltech.edu/groups/fha/Software.htm (53). Noncontiguous recombination design was done using a software package for performing noncontiguous protein recombination (24) openly available at cheme.che.caltech.edu/groups/fha/Software.htm (54). Both software packages are written in the Python programming language.
Construction of Chimeras
The SCHEMA software outputs the amino acid sequences of all chimeras in a library. The amino acid sequence for each chimera chosen for experimental testing was converted into a nucleotide sequence using the following method to define codon use:
-
1.
Align the amino acid sequence to the C1C2 parent.
-
2.
Assign conserved amino acids in the alignment to the C1C2 parental codon.
-
3.
Assign nonconserved amino acids to the parental codon from which the amino acid is derived.
This method was used for all chimeras to ensure that codon use was consistent. Once amino acid sequences were converted into nucleotide sequences, additional 3′ and 5′ sequences containing a BamHI and a NotI restriction enzyme cut site, respectively, were appended to the gene sequence. These sequences were necessary for cloning in the pFCK vector using either restriction ligation or homology-based cloning strategies. Gene sequences for the 223-chimera set were synthesized by Twist Bioscience, using its proprietary silicon-based DNA writing technology. After assembly, each fragment was cloned in the pFCK vector by homology-based cloning strategy and transformed into Stbl3 cells (Invitrogen) or Endura cells (Lucigen). Individual clones were picked and sequenced by NGS. Perfect clones were stored as individual glycerol stocks. Eight of the single-block swap sequences failed either the synthesis or cloning steps; these were not included in the chimera set.
Purified plasmid DNA of each chimera was prepared for HEK cell transfection. Each construct was streaked onto LB-amp plates from a glycerol stock, and an individual colony from each construct was picked and used to inoculate a 5-mL LB-ampicillin liquid media. Cultures were then grown overnight to reach saturation. Plasmid DNA for each construct was then purified using the QIAprep Spin Miniprep Kit. DNA concentrations for all constructs were measured and normalized before HEK cell transfection.
HEK Cell Maintenance and Transfection
HEK 293T cells were cultured at 37 °C and 5% CO2 in D10 [DMEM supplemented with 10% (vol/vol) FBS, 1% sodium bicarbonate, and 1% sodium pyruvate]. For 96-well transfections, HEK cells were plated on poly-d-lysine–coated glass-bottom 96-well plates at 20–30% confluency. Cells were left to divide until they reached 70–80% confluency. HEK cells were then transfected with one library variant per well at a prenormalized DNA concentration using Fugene6 reagent according to the manufacturer’s recommendations. Cells were given 48 h to express and then subjected to the SpyCatcher-GFP labeling assay and imaged.
Recombinant SpyCatcher-GFP Expression and Purification
The SpyCatcher-GFP was produced from a previously published construct—pQE80l-T5::6xhis-SpyCatcher-Elp-GFP [for details, see Bedbrook et al. (44)]. E. coli expression strain BL21(DE3) harboring the pQE80l-T5::6xhis-SpyCatcher-Elp-GFP plasmid was grown at 37 °C in TB medium to an optical density of 0.6–0.8 at 600 nm, and protein expression was induced using 1 mM isopropyl β-d-1-thiogalactopyranoside at 30 °C. After 4 h of induction, cells were harvested and frozen at −80 °C before protein purification. Protein purification was carried out using HisTrap columns (GE Healthcare) following the column manufacturer’s recommendations. Protein was buffer exchanged into sterile PBS at 4 °C. Protein was stable through multiple freeze/thaws and over many months.
SpyCatcher Labeling of HEK Cells
HEK cells were subjected to SpyCatcher labeling 48 h posttransfection. Labeling was done in a 96-well format using multichannel pipettes. SpyCatcher-GFP was added directly into the D10 media of wells containing HEK cells at a final concentration of 30 μM, and the cells were then incubated for 45 min at 25 °C. To avoid variability in labeling in the 96-well format screen, we used a saturating concentration of the SpyCatcher (30 μM) for labeling experiments. After labeling, HEK cells were washed with D10 three times, and then cells were incubated at 37 °C for 1 h to allow any remaining SpyCatcher to diffuse off of the well surface. For cell imaging, D10 medium was replaced with extracellular buffer (in mM: 140 NaCl, 5 KCl, 10 Hepes, 2 MgCl2, 2 CaCl2, 10 glucose; pH 7.35) to avoid the high autofluorescence of the D10. Cells were washed two times with extracellular buffer to fully remove any residual D10 before imaging.
Imaging and Image Processing of ChR Expression and Localization
Imaging of ChR expression and localization was done using a Leica DMI 6000 microscope. Four positions in each well were imaged in all 96-well plates using a fully automated system with motorized stage and automated z focus. Three channels were imaged at each position (mKate, GFP, and bright field). Cell segmentation was done using CellProfiler (55), an open-source image-processing software, and whole population intensity measurements were done using custom image-processing scripts written using open-source packages in the SciPy ecosystem (56–58). Both processing methods require a series of filtering steps and background subtraction. Whole population intensity measurements required a thresholding step when defining a pixel mask for image processing. We used wells containing nontransfected HEK cell that went through the labeling experiment as a background for establishing a threshold. A threshold was set to 2 SDs above the mean intensity values calculated in these background wells for each channel (mKate and GFP). For each image, a mask was defined for each channel (mKate and GFP) as the pixels above a set threshold. The masks for the two channels were then combined so that the mask included any pixel that was above threshold in the GFP channel or the mKate channel. This combined pixel mask was used to calculate the mean mKate fluorescence intensity (expression) and mean GFP fluorescence intensity (localization) across the pixels in the mask. The ratio mean mKate intensity/mean GFP intensity is the localization efficiency.
Electrophysiology for ChR Photocurrents
Conventional whole-cell patch-clamp recordings were done in cultured HEK cells at 2 d posttransfection. Cells were continuously perfused with extracellular solution at room temperature (in mM: 140 NaCl, 5 KCl, 10 Hepes, 2 MgCl2, 2 CaCl2, 10 glucose; pH 7.35) while mounted on the microscope stage. Patch pipettes were fabricated from borosilicate capillary glass tubing (1B150-4; World Precision Instruments) using a model P-2000 laser puller (Sutter Instruments) to resistances of 2–5 MΩ. Pipettes were filled with intracellular solution containing the following (in mM): 134 K gluconate, 5 EGTA, 10 Hepes, 2 MgCl2, 0.5 CaCl2, 3 ATP, and 0.2 GTP. Whole-cell patch-clamp recordings were made using a Multiclamp 700B amplifier (Molecular Devices), a Digidata 1440 digitizer (Molecular Devices), and a PC running pClamp (version 10.4) software (Molecular Devices) to generate current injection waveforms and to record voltage and current traces.
Patch-clamp recordings were done with short light pulses to measure photocurrents. Photocurrents for each chimera were induced by three different wavelengths of light (473 ± 10, 560 ± 25, and 650 ± 13 nm) at 2 mW (∼0.1 mW⋅mm−2). Photocurrents were recorded from cells in voltage clamp held at −50 mV with one light pulse for 1 s with each wavelength of light tested sequentially with 2 min between light exposures. Because ChRs show some level of desensitization to light after continued light exposure, we ran all colors in one direction (red → green → blue) and then again in the other direction (blue → green → red). The means of peak and steady-state currents were calculated for each color between the two trials for a given cell. Light wavelengths were produced using LED illumination using a Lumencor SPECTRAX light engine with quad band 387/485/559/649-nm excitation filter, quad band 410/504/582/669-nm dichroic mirror, and quad band 440/521/607/700-nm emission filter (all SEMROCK).
Electrophysiology data were analyzed using custom data-processing scripts written using open-source packages in the Python programming language to do baseline adjustments, find the peak inward currents, and find the steady-state currents.
Supplementary Material
Acknowledgments
We thank Dr. John Bedbrook for critical reading of the manuscript. Imaging was performed in the Biological Imaging Facility, with the support of the Caltech Beckman Institute and the Arnold and Mabel Beckman Foundation. This work is funded by the National Institute for Mental Health Grant R21MH103824 (to V.G. and F.H.A.); the Beckman Institute for CLARITY, Optogenetics and Vector Engineering Research for technology development and broad dissemination: www.beckmaninstitute.caltech.edu/clover.shtml (V.G.); and the Institute for Collaborative Biotechnologies through Grant W911F-09-0001 from the US Army Research Office (to F.H.A.). C.N.B. and A.J.R. are funded by Ruth L. Kirschstein National Research Service Awards F31MH102913 and F32GM116319. K.K.Y. is a trainee in the Caltech Biotechnology Leadership Program and has received financial support from the Donna and Benjamin M. Rosen Bioengineering Center. The content is solely the responsibility of the authors and does not necessarily reflect the position or policy of the National Center for Research Resources, the National Institutes of Health, or the Government, and no official endorsement should be inferred.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700269114/-/DCSupplemental.
References
- 1.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5(12):993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Urban DJ, Roth BL. DREADDs (designer receptors exclusively activated by designer drugs): Chemogenetic tools with therapeutic utility. Annu Rev Pharmacol Toxicol. 2015;55:399–417. doi: 10.1146/annurev-pharmtox-010814-124803. [DOI] [PubMed] [Google Scholar]
- 3.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71(1):9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Zhang F, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147(7):1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andréll J, Tate CG. Overexpression of membrane proteins in mammalian cells for structural studies. Mol Membr Biol. 2013;30(1):52–63. doi: 10.3109/09687688.2012.703703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lluis MW, Godfroy JI, 3rd, Yin H. Protein engineering methods applied to membrane protein targets. Protein Eng Des Sel. 2013;26(2):91–100. doi: 10.1093/protein/gzs079. [DOI] [PubMed] [Google Scholar]
- 7.Cymer F, von Heijne G, White SH. Mechanisms of integral membrane protein insertion and folding. J Mol Biol. 2015;427(5):999–1022. doi: 10.1016/j.jmb.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapple JP, Cheetham ME. The chaperone environment at the cytoplasmic face of the endoplasmic reticulum can modulate rhodopsin processing and inclusion formation. J Biol Chem. 2003;278(21):19087–19094. doi: 10.1074/jbc.M212349200. [DOI] [PubMed] [Google Scholar]
- 9.Conn PM, Ulloa-Aguirre A. Trafficking of G-protein-coupled receptors to the plasma membrane: Insights for pharmacoperone drugs. Trends Endocrinol Metab. 2010;21(3):190–197. doi: 10.1016/j.tem.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voigt CA, Martinez C, Wang ZG, Mayo SL, Arnold FH. Protein building blocks preserved by recombination. Nat Struct Biol. 2002;9(7):553–558. doi: 10.1038/nsb805. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki T, et al. Archaeal-type rhodopsins in Chlamydomonas: Model structure and intracellular localization. Biochem Biophys Res Commun. 2003;301(3):711–717. doi: 10.1016/s0006-291x(02)03079-6. [DOI] [PubMed] [Google Scholar]
- 12.Sineshchekov OA, Jung KH, Spudich JL. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2002;99(13):8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spudich JL, Yang CS, Jung KH, Spudich EN. Retinylidene proteins: Structures and functions from archaea to humans. Annu Rev Cell Dev Biol. 2000;16:365–392. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 14.Schneider F, Grimm C, Hegemann P. Biophysics of channelrhodopsin. Annu Rev Biophys. 2015;44:167–186. doi: 10.1146/annurev-biophys-060414-034014. [DOI] [PubMed] [Google Scholar]
- 15.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8(9):1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 16.Ishizuka T, Kakuda M, Araki R, Yawo H. Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci Res. 2006;54(2):85–94. doi: 10.1016/j.neures.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Scott DJ, Kummer L, Tremmel D, Plückthun A. Stabilizing membrane proteins through protein engineering. Curr Opin Chem Biol. 2013;17(3):427–435. doi: 10.1016/j.cbpa.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar CA, et al. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc Natl Acad Sci USA. 2008;105(39):14808–14813. doi: 10.1073/pnas.0803103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newstead S, Kim H, von Heijne G, Iwata S, Drew D. High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104(35):13936–13941. doi: 10.1073/pnas.0704546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trudeau DL, Smith MA, Arnold FH. Innovation by homologous recombination. Curr Opin Chem Biol. 2013;17(6):902–909. doi: 10.1016/j.cbpa.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 2009;10(12):866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drummond DA, Silberg JJ, Meyer MM, Wilke CO, Arnold FH. On the conservative nature of intragenic recombination. Proc Natl Acad Sci USA. 2005;102(15):5380–5385. doi: 10.1073/pnas.0500729102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Endelman JB, Silberg JJ, Wang ZG, Arnold FH. Site-directed protein recombination as a shortest-path problem. Protein Eng Des Sel. 2004;17(7):589–594. doi: 10.1093/protein/gzh067. [DOI] [PubMed] [Google Scholar]
- 24.Smith MA, Romero PA, Wu T, Brustad EM, Arnold FH. Chimeragenesis of distantly-related proteins by noncontiguous recombination. Protein Sci. 2013;22(2):231–238. doi: 10.1002/pro.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer MM, Hochrein L, Arnold FH. Structure-guided SCHEMA recombination of distantly related beta-lactamases. Protein Eng Des Sel. 2006;19(12):563–570. doi: 10.1093/protein/gzl045. [DOI] [PubMed] [Google Scholar]
- 26.Crameri A, Raillard SA, Bermudez E, Stemmer WP. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998;391(6664):288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- 27.Otey CR, et al. Structure-guided recombination creates an artificial family of cytochromes P450. PLoS Biol. 2006;4(5):e112. doi: 10.1371/journal.pbio.0040112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, et al. A diverse family of thermostable cytochrome P450s created by recombination of stabilizing fragments. Nat Biotechnol. 2007;25(9):1051–1056. doi: 10.1038/nbt1333. [DOI] [PubMed] [Google Scholar]
- 29.Romero PA, et al. SCHEMA-designed variants of human arginase I and II reveal sequence elements important to stability and catalysis. ACS Synth Biol. 2012;1(6):221–228. doi: 10.1021/sb300014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinzelman P, et al. A family of thermostable fungal cellulases created by structure-guided recombination. Proc Natl Acad Sci USA. 2009;106(14):5610–5615. doi: 10.1073/pnas.0901417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagel G, et al. Channelrhodopsin-1: A light-gated proton channel in green algae. Science. 2002;296(5577):2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 32.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA. 2003;100(24):13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang F, et al. Red-shifted optogenetic excitation: A tool for fast neural control derived from Volvox carteri. Nat Neurosci. 2008;11(6):631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kianianmomeni A, Stehfest K, Nematollahi G, Hegemann P, Hallmann A. Channelrhodopsins of Volvox carteri are photochromic proteins that are specifically expressed in somatic cells under control of light, temperature, and the sex inducer. Plant Physiol. 2009;151(1):347–366. doi: 10.1104/pp.109.143297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ernst OP, et al. Photoactivation of channelrhodopsin. J Biol Chem. 2008;283(3):1637–1643. doi: 10.1074/jbc.M708039200. [DOI] [PubMed] [Google Scholar]
- 36.Govorunova EG, Spudich EN, Lane CE, Sineshchekov OA, Spudich JL. New channelrhodopsin with a red-shifted spectrum and rapid kinetics from Mesostigma viride. MBio. 2011;2(3):e00115-11. doi: 10.1128/mBio.00115-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou SY, et al. Diversity of Chlamydomonas channelrhodopsins. Photochem Photobiol. 2012;88(1):119–128. doi: 10.1111/j.1751-1097.2011.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Govorunova EG, Sineshchekov OA, Li H, Janz R, Spudich JL. Characterization of a highly efficient blue-shifted channelrhodopsin from the marine alga Platymonas subcordiformis. J Biol Chem. 2013;288(41):29911–29922. doi: 10.1074/jbc.M113.505495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11(3):338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato HE, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482(7385):369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hochbaum DR, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11(8):825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141(1):154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shemiakina II, et al. A monomeric red fluorescent protein with low cytotoxicity. Nat Commun. 2012;3:1204. doi: 10.1038/ncomms2208. [DOI] [PubMed] [Google Scholar]
- 44.Bedbrook CN, et al. Genetically encoded spy peptide fusion system to detect plasma membrane-localized proteins in vivo. Chem Biol. 2015;22(8):1108–1121. doi: 10.1016/j.chembiol.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zakeri B, et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci USA. 2012;109(12):E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krause A, Golovin D. Tractability: Practical Approaches to Hard Problems. Cambridge Univ Press; Cambridge, UK: 2014. Submodular function maximization; pp. 71–104. [Google Scholar]
- 47.Wagner S, Bader ML, Drew D, de Gier JW. Rationalizing membrane protein overexpression. Trends Biotechnol. 2006;24(8):364–371. doi: 10.1016/j.tibtech.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci. 2009;12(2):229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 49.Smith MA, Bedbrook CN, Wu T, Arnold FH. Hypocrea jecorina cellobiohydrolase I stabilizing mutations identified using noncontiguous recombination. ACS Synth Biol. 2013;2(12):690–696. doi: 10.1021/sb400010m. [DOI] [PubMed] [Google Scholar]
- 50.Heinzelman P, et al. SCHEMA recombination of a fungal cellulase uncovers a single mutation that contributes markedly to stability. J Biol Chem. 2009;284(39):26229–26233. doi: 10.1074/jbc.C109.034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flytzanis NC, et al. Archaerhodopsin variants with enhanced voltage-sensitive fluorescence in mammalian and Caenorhabditis elegans neurons. Nat Commun. 2014;5:4894. doi: 10.1038/ncomms5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chauhan JS, Rao A, Raghava GP. In silico platform for prediction of N-, O- and C-glycosites in eukaryotic protein sequences. PLoS One. 2013;8(6):e67008. doi: 10.1371/journal.pone.0067008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith MA, Arnold FH. Designing libraries of chimeric proteins using SCHEMA recombination and RASPP. Methods Mol Biol. 2014;1179:335–343. doi: 10.1007/978-1-4939-1053-3_22. [DOI] [PubMed] [Google Scholar]
- 54.Smith MA, Arnold FH. Noncontiguous SCHEMA protein recombination. Methods Mol Biol. 2014;1179:345–352. doi: 10.1007/978-1-4939-1053-3_23. [DOI] [PubMed] [Google Scholar]
- 55.Carpenter AE, et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walt SVD, Colbert SC, Varoquaux G. The NumPy array: A structure for efficient numerical computation. Comput Sci Eng. 2011;13(2):22–30. [Google Scholar]
- 57.Oliphant TE. Python for scientific computing. Comput Sci Eng. 2007;9(3):10–20. [Google Scholar]
- 58.Hunter JD. Matplotlib: A 2D graphics environment. Comput Sci Eng. 2007;9(3):90–95. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.